Abstract
Short hydrogen bonds (H-bonds) have been proposed to play key functional roles in several proteins. The location of the proton in short H-bonds is of central importance, as proton delocalization is a defining feature of low barrier hydrogen bonds (LBHBs). Experimentally determining proton location in H-bonds is challenging. Here, bond length analysis of atomic (1.15–0.98 Å) resolution X-ray crystal structures of the human protein DJ-1 and its bacterial homolog YajL was used to determine the protonation states of H-bonded carboxylic acids. DJ-1 contains a buried, dimer-spanning 2.49 Å H-bond between Glu15 and Asp23 that satisfies standard donor-acceptor distance criteria for a LBHB. Bond length analysis indicates that the proton is localized on Asp24, excluding a LBHB at this location. However, similar analysis of the E. coli homolog YajL shows both residues may be protonated at the H-bonded oxygen atoms, potentially consistent with an LBHB. A PDB-wide screen identifies candidate carboxylic acid H-bonds in approximately 14% of proteins, which are typically short (<dO-O>=2.542(2) Å). Chemically similar H-bonds between hydroxylated residues (Ser/Thr/Tyr) and carboxylates show a trend of lengthening O-O distance with increasing H-bond donor pKa. This trend suggests that conventional electronic effects provide an adequate explanation for short, charge-assisted carboxylic acid-carboxylate H-bonds in proteins, without the need to invoke LBHBs in general. This study demonstrates that bond length analysis of atomic resolution X-ray crystal structures provides a useful experimental test of certain candidate LBHBs.
There is long-standing interest in the role of short H-bonds (donor-acceptor distances ≤2.5 Å) in protein stability and enzyme catalysis. These short H-bonds often form between ionizable groups and have been postulated to be particularly strong in certain circumstances 1. Oppositely charged groups can form short charge-assisted H-bonds (sometimes called short ionic H-bonds or salt bridges), where the favorable electrostatic component drives the donor-acceptor pKa values even further apart 2. An H-bond with either an ionized donor or acceptor is also a charge-assisted, and is typically stronger than one between neutral groups. Carboxylic acids, which are the focus of this work, often form short H-bonds and can serve as either H-bond donors or acceptors, depending on their ionization state and interacting group.
A H-bond between two carboxylic acids has the potential to be a low barrier H-bond (LBHB). LBHBs are named in reference to their low activation barrier to proton transfer from the donor to the acceptor atoms 1, 3, 4. Because the proton can exchange between two atoms in a LBHB, a close matching of pKa values for donor and acceptor has been proposed to be a prerequisite for LBHB formation 1. In many cases, a symmetrical double-well potential surface is also cited as an important feature of LBHBs, although this is not strictly required. In the most rigorous LBHB definition, the activation barrier for proton transfer is smaller than the zero point energy of the H-bond, thus leading to a complete delocalization of the proton nuclear wavepacket over the bond. This type of interaction is energetically similar to a single well H-bond, where the proton is located midway between the two interacting atoms. However, any H-bond where the proton transfer barrier height is comparable to or lower than either the zero point energy or kT would permit some degree of proton delocalization, even in an asymmetric potential. Therefore, LBHBs exist on a continuum of H-bond energetics and are distinguished from conventional H-bonds by exhibiting proton delocalization permitted by a transversable proton transfer barrier. The extensive literature on various classes of H-bonds in small molecules, including LBHBs, is reviewed in 5. Because the definition of a LBHB is somewhat variable in the expansive literature on this topic, we choose a LBHB definition that requires proton transfer between the donor to acceptor but does not require strict symmetry of the underlying potential.
The biological significance of LBHBs has been highly controversial. Seminal proposals that LBHBs may dramatically stabilize transition states and thus enhance enzymatic catalysis are supported by some experiments 4, 6–11, however others have argued that a major catalytic role for LBHBs is unlikely given both experimental and computational evidence 12–16. Despite the two decade-long history of this debate, the existence and potential importance of protein LBHBs remains unresolved and is of considerable interest 17. As carboxylic acids have similar solution pKa values and their H-bonds involve an acid/base couple of the same functional group as both donor and acceptor, they are more likely to meet the matching pKa and symmetry conditions of an LBHB than are most other pairs of biological H-bond donors and acceptors.
One difficulty in studying LBHBs is that their definition is rooted in features of the potential energy function of the proton along the H-bond coordinate, which is difficult to experimentally characterize. Direct localization of the proton by neutron diffraction is the most reliable evidence for a LBHB but is a challenging experiment to perform, requiring large, well-diffracting crystals and access to high flux neutron sources. Consequently, a number of more accessible observable proxies for LBHBs have been used, including donor-acceptor distances less than or equal to approximately 2.5 Å, deshielded proton resonances in NMR, perturbed IR spectra, and anomalously low H/D substitution factors in hydrogen-deuterium exchange experiments 3. While each has advantages and reports on some aspect of LBHB energetics, these empirical criteria suffer from limitations that have complicated the reliable identification of LBHBs in proteins.
The donor-acceptor distance of a candidate H-bond is one of the easiest LBHB criteria to observe, and short H-bonds between carboxylic acids are common 18, 19. Small molecule carboxylic acids form tightly associated H-bonded cyclic dimers in the gas phase, and crystal structures of various carboxylic acids have typically have short H-bonds 20. In proteins, the active sites of aspartyl proteases are well-known for the short H-bonds formed between catalytic residues 21. Additionally, ultra-high resolution (0.79–0.89 Å) crystal structures of CTM-X β-lactamase identified a 2.54 Å H-bond between Asp233 and Asp246 that is conserved in other β-lactamases and is proposed to serve a structural role 22. Salmonella typhimurium glucose/galactose-binding protein possesses a solvent-exposed Asp121-Glu165 interaction of 2.54 Å, 18, and rhamnogalacturonan acetylesterase has a 2.48 Å interaction between Asp75 and Asp 87 19. Furthermore, Langkilde et al., 19 surveyed the Cambridge Structural Database (CSD) 19 and both Flocco and Mowbray 18 and Langkilde et al. surveyed the Protein Data Bank (PDB), finding many examples of short carboxylic acid H-bonds. The ~2.5 Å O-O distance of these H-bonds suggests that this class of interaction may be unusually strong and merits closer study.
In this work, we use atomic resolution X-ray crystallography, site-directed mutagenesis, and solution stability analysis to study a conserved 2.49 Å carboxylic acid H-bond at the dimer interface of the parkinsonism-associated human protein DJ-1. Bond length analysis of a 1.15 Å resolution crystal structure shows that Glu15 and Asp24 have different protonation states, inconsistent with a LBHB. However, this H-bond plays an important role in stabilizing DJ-1, as the D24N mutation mildly destabilizes the protein and the D24L mutation results in a population of poorly folded protein. The Glu-Asp H-bond is conserved in a prokaryotic homolog of DJ-1, where bond lengths suggest that both residues are protonated, potentially consistent with LBHB. A computational survey of the PDB indicates that approximately 14% of proteins contain candidate carboxylic acid H-bonds, making this type of interaction fairly common. As these H-bonds have a short ~2.5 Å average O-O distance and are unlikely to all be LBHBs, a donor-acceptor distance criterion should not be used for LBHB identification. We suggest that bond length analysis could be a routine tool for characterizing putative LBHBs involving carboxylic acids.
Experimental procedures
Protein Expression and Purification
Hexahistidine-tagged human DJ-1 and E. coli YajL were expressed from pET15b constructs in BL21(DE3) E. col, purified using Ni2+-metal affinity chromatography, and thrombin-cleaved to remove the histidine tag as described previously 23, 24. The hexahistidine tag was removed by thrombin cleavage and the final protein contains the vector-derived sequence GSH- before the first methionine in the native protein sequence. The D24N, D24L, E15Q and E15Q/D24N DJ-1 mutations were generated by site-directed mutagenesis and were purified in the same manner as wild-type DJ-1. The purified proteins were dialyzed into storage buffer (25 mM HEPES pH 7.5, 100 mM KCl, 2 mM DTT) and concentrated to 20 mg/ml using a centrifugal concentrator (Millipore) with a 10-kDa cutoff. Protein concentration was determined by absorbance at 280 nm using calculated extinction coefficients of 4470 M−1 cm−1 for DJ-1 and 17,200 M−1 cm−1 for YajL. Proteins were divided into aliquots, flash cooled in liquid nitrogen, and stored at −80 °C until needed.
Crystallization and Data Collection
All proteins were crystallized using the sitting drop vapor diffusion method by mixing 2 μL of protein at 20 mg/ml with 2 μL of the reservoir solution. Crystals grew in 1–4 days at room temperature. Crystals of wild-type DJ-1 in space group P3121 grew from a reservoir of 20–25% PEG 3000, 100 mM HEPES pH 7.5, 200 mM NaCl, 10 mM dithiothreitol (DTT) and similar D24N crystals grew from 23–27% polyethylene glycol (PEG) 4000, 0.2 M Sodium Citrate, and 0.1 M Tris-HCl (pH 8.5). Plate-like crystals of E15Q DJ-1 in space group C2221 grew from a reservoir of 25–28% PEG 4000, 0.1 M Tris-HCl (pH 8.5), 0.1 M sodium citrate. E. coli YajL crystals grew from a reservoir of 30% polyethylene glycol 4000, 0.1 M Tris–HCl (pH 8.5), 0.225 M MgCl2 in space group P212121 24, 25. YajL crystals were obtained by microseeding in order to improve their growth habit and reduce local depletion zone hollowing at the ends of the prismatic crystals. All crystals were cryoprotected by serial transfer through their respective reservoir solutions supplemented with ethylene glycol increasing in 5% concentration increments to a final concentration of 25% (v/v). Cryoprotected crystals were cooled by submersion in liquid nitrogen whose cool vapor layer was blown away immediately before the crystal was cooled.
Diffraction data for wild-type DJ-1 were collected at the Advanced Photon Source (APS), BioCARS beamline 14-BMC, data for D24N and E15Q DJ-1 were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) beamline 9-2, and data for E. coli YajL were collected at SSRL beamline 12-2. In all cases, data were collected from a single crystal maintained at 100 K. The data for wild-type DJ-1 were collected in multiple passes with differing exposures on an ADSC Q315 CCD detector, while E15Q, D24N, and YajL diffraction was measured using a Pilatus 6M PAD with shutterless data collection. All crystals were subjected to in situ annealing by interrupting the cold nitrogen stream flow for 5–7 seconds, which improved diffraction and reduced mosaicity 25. Data for wild-type DJ-1 were integrated and scaled using HKL2000 26; data for E15Q, D24N, and YajL were integrated and scaled using XDS 27, and Pointless, and Aimless 28 in the CCP4 suite 29.
Structure Refinement and Validation
All models were refined in Refmac5 30, part of the CCP4 suite 29, using geometric restraints and anisotropic atomic displacement parameters (ADPs) against a maximum likelihood target based on structure factor amplitudes. Wild-type DJ-1 (PDB code 1P5F 31) and E. coli YajL (PDB code 2AB0 24) were used as starting models. A test set of 2–5% of reflections were chosen at random and sequestered in the test set for calculation of the Rfree value 32. Distinct test sets were chosen for each DJ-1 mutant, both because E15Q DJ-1 crystallized in a different space group and thus could not have the same test reflections as the wild-type and D24N structures and because the same starting model 1P5F was used for all three structures. Therefore, test set bias could not propagate from the wild-type to the mutant structures, since they were refined in parallel starting from a common, solvent-free, isotropic model derived from 1P5F. The models were inspected and manually improved in reference to 2mFo-DFc and mFo-DFc electron density maps in COOT 33. Where electron density provided evidence of multimodal disorder, residues were built in two alternative conformations whose occupancies were refined and constrained to sum to unity. Weights for the B-factor restraints and geometric term were optimized using automated refinement through the PDB_REDO webserver 34. Final models were validated using COOT 33, MolProbity 35, and PDB_REDO 34. As is common for proteins in the DJ-1 superfamily, the reactive cysteine residue (Cys106) in DJ-1 and YajL is a Ramachandran outlier but well-supported by electron density. The model statistics are provided in Supplementary Table 1. Structural figures were made with POVScript+36.
Bond length analysis
Bond lengths were determined by subjecting the final models from Refmac5 (see above) to an additional 20 cycles of conjugate gradient least squares refinement in SHELXL 37 without geometric restraints on Asp or Glu residues. The observation-to-parameter ratio for fully anisotropic refinement of these structures was ~5.5:1, justifying this treatment. A separate cycle of least squares (SHELX instruction L.S.) refinement without geometric or ADP restraints was then performed to calculate the inverted Hessian matrix, which was restricted to coordinates using the BLOC 1 instruction. The eigenvalues of this inverted matrix were used to calculate the estimated standard uncertainties (ESUs) on bond lengths in SHELX 37. Differences in C-O bond lengths were considered significant if they equaled or exceeded three ESU 38–40. Only fully occupied and well-ordered residues were used for analysis. 40
Thermofluor assay
The thermal stabilities of wild type, D24N, E15Q, E15Q/D24N, and D24L DJ-1 were measured in 25 mM HEPES (pH 7.5), 100 mM KCl, and 1 mM DTT using the Thermofluor assay 41. Dilutions of a 5000× stock of Sypro Orange (Sigma-Aldrich) in DMSO were added to multiple concentrations of protein (1.0, 2.5, and 5.0 mg/mL) in optically clear PCR tube strips (Bio-Rad). Measurements were made at multiple protein and dye concentrations for each protein. The samples were heated from 10 to 95 °C at a rate of 2 °C/min, with fluorescence excitation at 490 nm and emission at 575 nm using an iCycler iQ real-time thermal cycler (Bio-Rad). The region between ± 20°C of the melting transition (inflection point) in the raw fluorescence data was fitted using:
| Eqn 1 |
Where f(T) is the measured fluorescence, T is temperature, fN=aN+bNT is the baseline fluorescence of the native protein, fU=aU+bUT is the baseline fluorescence of the denatured protein, Tm is the melting temperature, and ΔT is a fit parameter quantifying the width of the melting transition that relates enthalpy to Tm as below:
| Eqn 2 |
Reported errors are calculated for fitted Tm values obtained from the multiple dye and protein concentration curves for each sample. A program that performs this analysis is available at BitBucket (https://bitbucket.org/bullseye_maurice/epydsf)
Secondary structure content determination using far-UV circular dichroism
Proteins were dialyzed overnight at 4°C against 10 mM sodium phosphate pH 7.2, and the final protein concentration was adjusted to 0.5 mg/mL. CD spectra were measured using a Jasco J-815 CD spectrophotometer (Jasco Inc., Easton, MD, USA) in a 1 mm pathlength quartz cuvette. Spectra were measured in continuous scanning mode with a grating of 3400 lines cm-1, 1 nm bandwidth, and a rate of 20 nm min−1. Four scans were accumulated per spectrum with a data pitch of 0.1 nm and a data integration time of 1 s. The mean molar residue ellipticity (deg cm2 dmol−1 residue−1) was calculated using Scopes’ method 42 for protein concentration determination as previously described 43.
Search of the PDB for candidate COOH-−OOC and COH-−OOC H-bonds
The set of all X-ray crystal structures determined to a resolution of 2.0 Å or better in the August 2016 release of the PDB was used for analysis. Initially, potential hydrogen bonds were identified by applying a distance cutoff of 3.5 Å between oxygen atoms of all Asp/Glu sidechains. For each potential hydrogen bond, the distance between oxygen atoms, the C-O-O angle and C-C-O-O torsion angle were recorded. One- and two-dimensional frequency distributions were calculated with each putative hydrogen bond weighted according to the number of times it was encountered in the dataset (to account for redundancies due to oligomerization and multiple instances of the protein and its close homologues in the PDB). Analysis of the resulting distributions revealed the presence of a peak in the two-dimensional distribution centered around specific values of interatomic distance and C-O-O angle. This peak was fitted to a two-dimensional normal distribution to determine mean and variance. In addition, the proton was required to be located close to the plane formed by carboxylate groups in the C-C-O-O torsion angle. Hydrogen bonds formed at protein-protein interfaces were identified by querying against all oligomeric assemblies predicted by the PISA 44 algorithm. Predicted monomeric structure were downloaded directly from the PDB, while putative dimers, trimers, tetramers and pentamers were downloaded from PISA. COH-−OOC H-bonds, where the donor could be the hydroxyl sidechains of Ser, Thr, or Tyr, were identified in the PDB as described above, and the corresponding distributions were studied separately for the Ser, Thr, and Tyr donors
Results
Human DJ-1 contains a short but conventional dimer-spanning hydrogen bond between Asp24 and Glu15
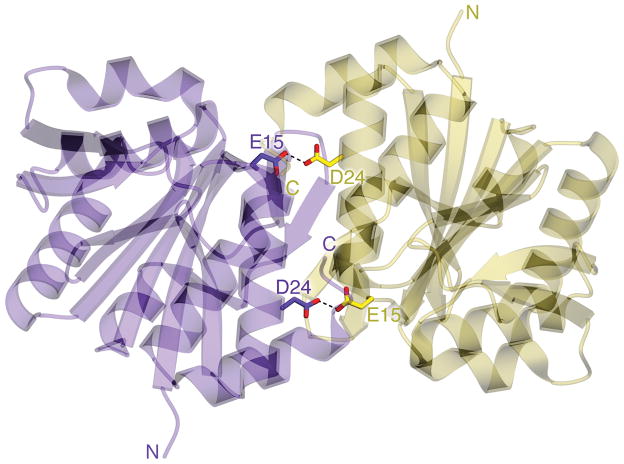
Human DJ-1 dimerization is important for its cytoprotective function 45. The dimer interface contains a buried pair of highly conserved residues, Glu15 and Asp24, whose sidechain oxygen atoms are separated by 2.49 Å, indicating an H-bond (Fig 1). For H-bonding to occur, at least one of these residues must be protonated. Although hydrogen atoms can be difficult to visualize in X-ray crystallographic electron density maps even at high resolution, carboxylic acid geometry is sensitive to protonation, providing an indirect but reliable means to identify the donor and acceptor in this interaction. Fully protonated carboxylic acids have unequal carboxylic acid bond lengths of 1.21 Å for a C=O bond and 1.30 Å for a C-OH bond, while deprotonated carboxylates have identical C-O bond lengths of 1.26 Å due to resonance 46. The difference in C-O bond lengths for protonated carboxylic acids (~0.1 Å) is approximately 5–10 times larger than the estimated standard uncertainties (ESUs) on bond lengths for atomic resolution (dmin≤1.2 Å) X-ray crystal structures. Therefore, carboxylic acid protonation states can be confidently determined from X-ray diffraction data when the data extend to atomic resolution and the residues of interest are well-ordered 40, 47–49. We note, however, that other factors may influence carboxylic acid bond lengths (such as accepting H-bonds) and are not well-studied.
Figure 1.
A carboxylic acid H-bond spans the core of the DJ-1 dimer. A ribbon diagram of the DJ-1 dimer is shown with protomers colored purple and gold. The two dimer-spanning H-bonds at Glu15-Asp24 are shown in dotted lines.
To perform this analysis on human DJ-1, a new 1.15 Å resolution crystal structure was determined in space group P3121. Although nearly identical to prior DJ-1 structures refined at similar resolutions (PDB codes 1P5F 31, 1SOA 50, 2OR3 49), this structure has a predominantly reduced Cys106 residue, which is prone to spontaneous oxidation and is often present as the cysteine-sulfinate in other structures 50, 51. While there is difference (mFo-DFc) electron density supporting partial oxidation to Cys106-SO2−, it is less pronounced than is observed in many other comparable DJ-1 structures. In addition, DTT is bound to DJ-1 in this crystal structure, likely the result of the high concentration (10 mM) of DTT used as a reductant in the buffer. The occupancy of the DTT is correlated with occupancies of alternative conformations of Asn76 but not expected to impact the protonation state of Glu15 or Asp24.
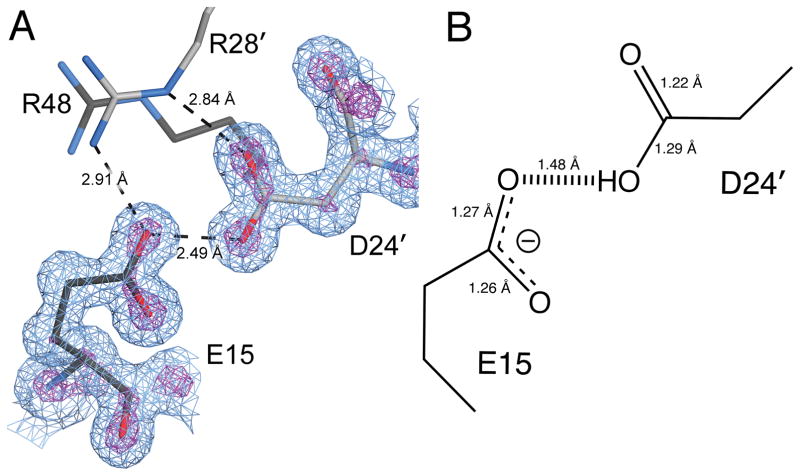
The DJ-1 model was refined in REFMAC5 using standard protocols followed by unrestrained refinement and bond length analysis in SHELX (see Methods). Carboxylic acid bond length analysis shows that Glu15 is deprotonated and accepts an H-bond from a protonated Asp24 (Table 2 and Fig. 2). The pronounced differences in the bond lengths of these two residues (Table 2) indicates that their pKas are markedly different and that the proton is located predominantly on Asp24 Oδ2. The program PROPKA 52 supports this conclusion, with predicted carboxylic pKa values of 2.6 for Glu15 and 11.3 for Asp24. These anomalous carboxylic acid pKa values are not surprising, as both residues are completely buried in the core of the DJ-1 dimer. The depressed pKa for Glu15 is likely due to its proximity to and interaction with the Arg28/Arg48 dyad (Fig. 2A), whose presumed cationic character would stabilize the Glu15 carboxylate anion and thus facilitate ionization. The importance of the microenvironment of an ionizable residue for both its charge state and H-bonding properties is well-known in the small molecule community, as exemplified by the influence that different cations have on the proton location in the intramolecular H-bond of the hydrogen maleate monoanion 53. In DJ-1, both the calculated pKa values and experimentally determined bond lengths indicate that the Glu15-Asp24 interaction is a conventional charge assisted COOH-−OOC H-bond with a short donor-acceptor distance rather than a LBHB.
Table 2.
Protonation status of Asp/Glu residues of interest
| Protein | Residue | C-O1 (Å)a | C-O2 (Å)a | Δ(C-O)/<ESU>b | Status |
|---|---|---|---|---|---|
| wtDJ-1 | Glu15 | 1.256(13) | 1.273(13) | 1.31 | Deprotonated |
| wtDJ-1 | Asp24 | 1.220(13) | 1.290(12) | 5.39 | Protonated |
| E15Q DJ-1 | Asp24 | 1.270(14) | 1.289(13) | 1.46 | Deprotonated |
| D24N DJ-1 | Glu15 | 1.236(13) | 1.262(13) | 2.00 | Deprotonated |
| YajL | Glu14 (mol. A; mol. B) | 1.229(13); 1.232(11) | 1.292(14); 1.333(12) | 4.50; 8.42 | Protonated |
| YajL | Asp23 (mol. A; mol. B) | 1.239(12); 1.226(12) | 1.278(12); 1.289(13) | 3.25; 4.85 | Protonated |
ESUs are given in parenthetical error notation for each bond length
The ratio of the absolute value of the difference between C-O1 and C-O2 to the averaged ESUs on these bonds determined by inversion of the full least squares matrix. This value is analogous to an unsigned Z score, except the average ESU is used instead of a single population σ.
Figure 2.
The proton is located on Asp24 in the short H-bond with Glu15. In panel A, the immediate environment of the Glu15-Asp24 H-bond is shown. Glu15 makes stabilizing contacts with Arg28 and Arg48 at the dimer interface in addition to its 2.49 Å H-bond with Asp24. Primes indicate residues from the other protomer. 2mFo-DFc electron density at 1.15 Å resolution contoured at 1.0σ (blue) and 5.3σ (purple) are shown. Panel B shows the refined bond lengths for Glu15 and Asp24 with estimated standard uncertainties in parenthesis. These bond lengths indicate that Glu15 is deprotonated and Asp24 is protonated.
The hydrogen bond between Glu15 and Asp24 is important for human DJ-1 stability
To determine the importance of the Glu15-Asp24 COOH-−OOC H-bond for DJ-1 structure and stability, this interaction was modulated by creating E15Q, D24N, E15Q/D24N, and D24L mutations. The D24L mutation eliminates the H-bond, while E15Q, D24N, and E15Q/D24N are expected to reduce the H-bond strength by varying degrees. E15Q and D24N behave similarly to wild-type protein throughout protein purification. In contrast, the D24L and E15Q/D24N mutants precipitated extensively during purification, were obtained in lower yields, and could not be crystallized, indicative of diminished stabilities.
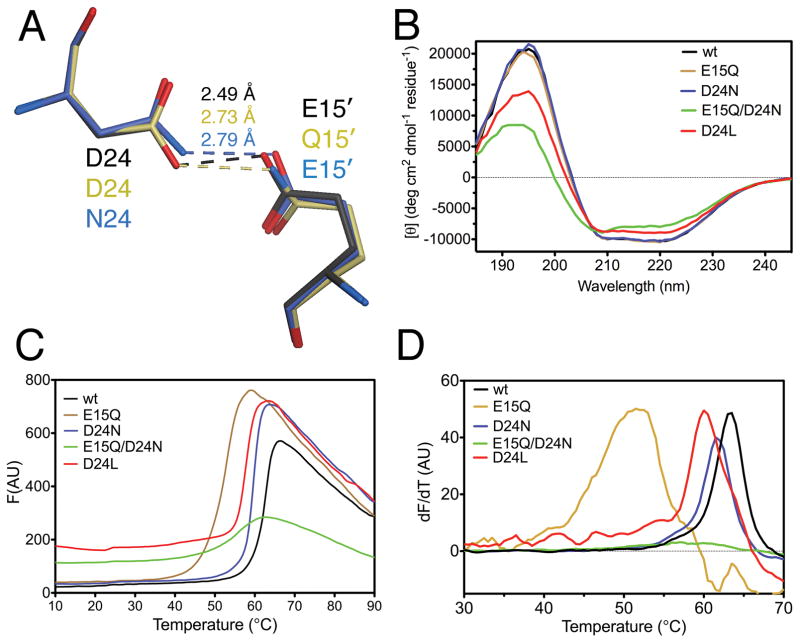
E15Q and D24N DJ-1 were crystalized and their structures solved by X-ray crystallography at 1.1 Å resolution. Both mutant structures are nearly identical to wild-type DJ-1, with Cα RMSD 0.21 Å for E15Q and 0.12 Å for D24N (Fig. 3A). As expected, bond length analysis confirms that Glu15 remains deprotonated in the D24N mutation, where it accepts the longer 2.79 Å H-bond donated by Asn24 (Table 2; Fig. 3A). In contrast, Asp24 becomes deprotonated in the converse E15Q mutation (Table 2 and Fig 3A), although both C-O bonds are slightly longer than anticipated for a carboxylate. Deprotonation of Asp24 is expected because the structurally conservative E15Q mutation forces Gln15 to be the H-bond donor at the Nε2 atom, rather than an acceptor at the Oε2 atom of Glu, as in the wild-type protein. Presumably, the H-bond stabilization energy is greater than any energetic cost incurred by Asp24 ionization in the E15Q mutant.
Figure 3.
The Glu15-Asp24 COOH--OOC H-bond is important for DJ-1 stability. Panel A shows a superposition of the structures of wild-type (black), E15Q (beige), and D24N (blue) DJ-1 at the location of the short H-bond. Substitution of an amide for a carboxylic acid at either location lengthens the H-bond. Panel B shows CD spectra for each mutant, with loss of structure in D24L and E15Q/D24N DJ-1 evident by decreased signal at 190–200 and 210–220 nm. Panel C shows Thermofluor analysis each mutant as raw fluorescence traces, where the inflection point in the Tm. Panel D is the first derivative of the data in Panel C to emphasize location of the melting transitions as maxima.
Far UV circular dichroism spectroscopy was used to characterize the overall secondary structural changes introduced by the H-bond mutations. Wild-type DJ-1, D24N, and E15Q had comparable CD spectra, indicating that these mutations do not globally effect DJ-1 structure (Fig. 3B). In contrast, D24L and E15Q/D24N DJ-1 both show a modest decrease in overall secondary structural content, going from approximately 22% α-helix, 30% β-sheet in the wild-type protein to 18% α-helix, 31% β-sheet in D24L and 14% α-helix, 30% β-sheet in E15Q/D24N DJ-1 as determined using K2D3 54. Thus, both E15Q/D24N and D24L DJ-1 exhibit some loss of α-helical content relative to the wild-type protein (Fig. 3B).
The contribution of the Glu15-Asp24 H-bond on DJ-1 thermal stability was determined using the Thermofluor assay with the E15Q, D24N, D24L, and E15Q/D24N mutants (Fig. 3C). The melting temperature (Tm) was determined by fitting of Eqn. 2 to the F vs. T curve in Fig. 3C. The Tm of D24N is 60.1(1.1)°C, 2.4°C lower than the 62.5(0.4) Tm of the wild type protein. The D24N mutation preserves the H-bond between residues 15 and 24 but lengthens it by 0.3 Å in going from a stronger −OH…−O- to a weaker −NH2…−O- interaction. The modest decrease in Tm indicates that the short Glu15-Asp24 H-bond is not much stronger than a conventional H-bond, contrary to what has been hypothesized for LBHBs. In contrast, E15Q DJ-1 is markedly destabilized, with a Tm of 51.9(2.0)°C (ΔTm=−10.6 °C), even though it also preserves but weakens the H-bond (Fig. 3C,D). This lowered Tm indicates that other stabilizing contacts (likely involving Arg28 and Arg48; see above) are disrupted by the E15Q mutation. The double E15Q/D24N mutant shows no obvious melting transition in the dF/dT plot (Fig. 3D), although the small transition in the raw fluorescence curve can be fitted (see Methods) with a Tm of 58.9(1.1)°C (ΔTm=−3.6°C). The Tm of E15Q/D24N DJ-1 is unexpectedly higher than that of E15Q, however the smaller amplitude of the signal and its higher baseline indicates that a fraction of E15Q/D24N DJ-1 is poorly structured. The substantial loss of stability for both E15Q and E15Q/D24N is consistent with a major contribution of stabilizing contacts between Glu15 and Arg28/48 to DJ-1 stability. E15Q/D24N DJ-1 could not be crystallized, so the detailed structural impact of this double mutation on the H-bond could not be determined. The D24L mutation, which eliminates the hydrogen bond by removing the donor, has a Tm of 59.4(1.4)°C (ΔTm=−3.1°C) (Fig. 3D), which is surprisingly high considering its diminished CD signal. However, like E15Q/D24N DJ-1, the D24L total fluorescence signal from Sypro orange is higher at baseline (Fig. 3C), indicating a population of poorly structured DJ-1 that can bind to the fluorophore at low temperatures. Therefore, the loss of the Glu15-Asp24 H-bond destabilizes DJ-1 but does not cause complete loss of dimerization or structural integrity.
The bacterial DJ-1 homolog YajL contains an unusual H-bond between carboxylic acids
DJ-1 has homologs in many organisms, including a close homolog in E. coli (YajL). A new 0.98 Å resolution crystal structure of YajL was determined and subjected to bond length analysis as described above. The H-bonded residues of interest in YajL are Asp23 and Glu14, which make a 2.43–2.45 Å H-bond. This structure contains two molecules in the asymmetric unit (ASU), thus providing two of crystallographically independent Glu14-Asp23 interactions for analysis. Unexpectedly, both Glu14 and Asp23 have bond lengths consistent with protonation in both molecules in the ASU (Table 2). Because the H-bond exists between the Asp23 Oδ2 and Glu14 Oε2 atoms (Fig. 4), both residues being protonated means either that there are hydrogen atoms on both residues, only one of which is involved in the Glu14-Asp23 H-bond, or that a proton is being shared between Glu14 and Asp23 in a LBHB.
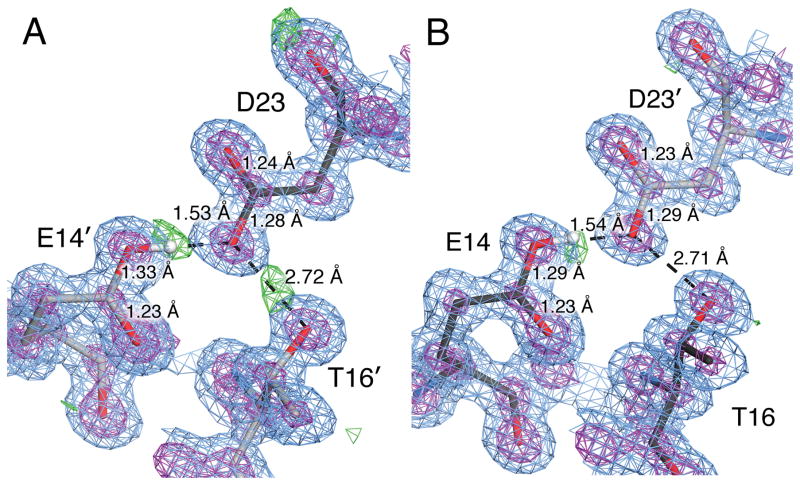
Figure 4.
E. coli YajL has a COOH--OOC H-bond with ambiguous protonation state. Panels A and B show the two independent Glu14-Asp23 H-bonds in the ASU. 2mFo-DFc electron density at 0.98 Å resolution contoured at 1.0σ (blue) and 5.3σ (purple) and mFo-DFc electron density contoured at 2.5σ (green). Primes indicate residues contributed by the other protomer, whose carbon atoms are colored grey. The modeled hydrogen atom on Glu14 is shown in white in each panel and agrees closely with the location of a mFo-DFc electron density peak. However, bond length analysis indicates both Glu14 and Asp23 are protonated, which is supported by the continuous 2mFo-DFc electron density for C=O bonds and broken density for C-O bonds in both instances.
Experimental evidence in favor of conventional H-bonding between Glu14 and Asp23 is provided by inspection of the mFo-DFc electron density around these residues (Fig. 4). A 3σ positive difference electron density peak is observed near to the Oε2 atom of Glu14 in both molecules in the asymmetric unit. Interestingly, this is contrary to DJ-1, where the corresponding residue (Glu15) is the H-bond acceptor, even though both YajL and DJ-1 have the same Arg28/Arg48 (Arg27/Arg49 in YajL) dyad that we postulate stabilizes the anionic form of Glu15 in DJ-1 (see above). PROPKA predicts an Asp23 pKa value of 2.4 and a Glu14 pKa value of 10.7 in YajL, consistent with a localized proton on Glu14. The mFo-DFc electron density peak is close to the ideal calculated position of the hydrogen in a conventionally protonated carboxylic acid as modeled using PDB2PQR (Fig. 4). Therefore, the location of this feature is more consistent with a fully localized hydrogen on the donor Glu14 than a delocalized proton in a LBHB, supporting the PROPKA analysis.
Importantly, because hydrogen has only one electron, the electron density for a hydrogen atom is polarized toward the bond rather than spherically distributed around the nuclear position. Therefore, it is likely that the true nuclear position of the hydrogen is further away from the donor than is the mFo-DFc peak, possibly consistent with lengthening of the O-H distance expected for a LBHB. However, it is not clear that a significant peak in electron density would be expected for a LBHB, as the shared proton can not scatter X-rays and its bonding electron density would be distributed between the donor and acceptor. This would decrease the electron density for the hydrogen atom in an LBHB and make a corresponding mFo-DFc electron density peak difficult to observe.
The protonation state of Asp23 is ambiguous, as bond length analysis indicates that it is also protonated (~4–8σ, Table 2) but it should be the H-bond acceptor and is predicted to be deprotonated by PROPKA. Therefore, although the mFo-DFc electron density map supports a conventional H-bond, we cannot rule out a potential LBHB here, as proton delocalization would result in the observed protonated carboxylic acid geometries for both residues. However, because protonation of a carboxylic acid can occur with a “syn” or an “anti” geometry at the oxygen atom, it is possible, though unlikely, that a hydrogen atom on the Oδ2 of Asp23 could be “backward-facing” (anti configuration) and thus not in conflict with the syn hydrogen donated by Glu14. Inspection of the local H-bond network does not resolve this issue, as there is a H-bond involving Asp23 and Thr16′ (with prime indicating a residue from the other protomer), and where Asp23 could either donate or accept a H-bond with Thr16′. One of the two molecules in the ASU has a mFo-DFc electron density map peak consistent H-bond donation by Thr16′ (Fig. 4). However, this mFo-DFc peak is absent in the other molecule in the ASU. Therefore, these data do not rule out the possibility of a LBHB at this location in YajL, although on balance they suggest a localized hydrogen on Glu14.
A screen for COOH-−OOC H-bonds in the PDB
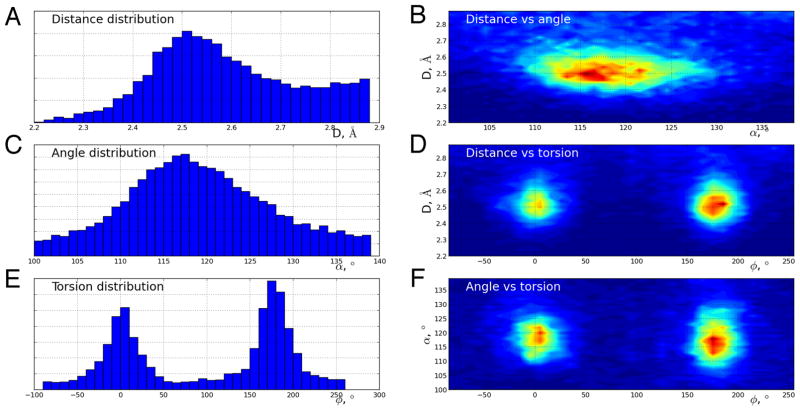
The total of 41,214 structural models were searched for candidate COOH-−OOC H-bonds (see Methods). Of these, the PISA server 44 classified these as follows: 20,255 monomers, 13,619 dimers, 2,335 trimers, 4,902 tetramers, 103 pentamers. The one- and two-dimensional distributions are shown in Fig. 5 and are calculated using the resulting dataset of 70,857 candidate bonds. We estimate that <10% of these are unique and contribute 95% to the major peak seen in the distance-vs-angle distribution. When fitted to a 2D Gaussian distribution, the average bond length is found to be 2.542(2) Å, with the standard deviation of 0.12 Å. This indicates that most COOH-−OOC H-bonds are short and would qualify as candidate LBHB when using the standard 2.5 Å distance criterion. The torsional preferences are as expected with peaks at 0° and 180°, corresponding to “syn” and “anti” orientations of the two carboxylic acids. The average length of the hydrogen bond formed by aspartate/aspartic acid (2.531(8) Å) is slightly shorter than that of glutamate/glutamic acid (2.561(7) Å). Interestingly, the average length of a bond formed by different amino acids (i.e. Asp and Glu) is 2.526(6) Å, which is closer to the value for an Asp-Asp bond.
Figure 5.
Distributions of geometric parameters for carboxylic acid H-bonds identified in the PDB. In panels A,C, and E, histograms of O-O distances (A), C-O-O angles (C), and C-C-O-O torsion angles (E) for Asp-Glu interactions that satisfied H-bonding geometric criteria (see Methods) are shown. In panels B, D, and F, two-dimensional distributions are shown for the indicated parameters.
COOH-−OOC H-bonds are markedly shorter than COH-−OOC H-bonds
The unusually short length of the COOH-−OOC H-bond has been noted previously 19 and is one reason that these interactions have been suggested to be potential LBHBs. In light of our results with DJ-1, it is clear that at least some, and possibly most, of these interactions are conventional H-bonds with a localized proton. To determine how COOH-−OOC H-bond lengths compare with chemically similar COH-−OOC H-bonds in proteins, the PDB was screened for examples of Ser/Thr/Tyr-Asp/Glu interactions that satisfy H-bonding criteria. These interactions are much more abundant than COOH-−OOC H-bonds and have longer mean O-O distances (Table 3). The small differences between the mean dO-O values are statistically significant because the relevant error parameter is the standard error of the mean (in parenthesis, Table 3), which is quite small for these large populations. All of the COH-−OOC H-bonds are markedly longer than the 2.542(2) Å O-O distance average for Asp/Glu-Asp/Glu interactions (Supplemental Data, Fig S1–3). In addition, they agree very closely with a prior comprehensive analysis of similar classes of H-bonds found in small molecule structures deposited in the Cambridge Structure Database (CSD) 19. These results indicate that COOH-−OOC H-bonds are uncommonly short among chemically similar interactions and thus have high potential to be erroneously characterized has LBHBs based on the commonly cited dO-O≤2.5 Å distance criterion.
Table 3.
Population mean H-bond geometric parameters from the PDB
| Bond type | Mean O-O distance (Å) | Mean C-O-O angle (°) | Donor model compound pKa |
|---|---|---|---|
|
| |||
| Asp/Glu to Asp/Glu | 2.542(2) | 117.7(1) | propanoic acid; 4.9 |
| Tyr to Asp/Glu | 2.609(1) | 114.7(1) | 4-ethyl phenol; 10.0 |
| Ser to Asp/Glu | 2.651(1) | 107.8(1) | ethanol; 16.0 |
| Thr to Asp/Glu | 2.664(1) | 111.1(1) | isopropanol; 17.1 |
Discussion
In this work, we used bond length analysis of atomic (dmin<1.2 Å) resolution X-ray crystal structures to determine the protonation state of Glu15 and Asp23 in human DJ-1, which compose a short dimer-spanning H-bond. COOH-−OOC H-bonds have attracted interest due to their 2.4–2.5 Å O-O distances and potential for pKa matching, both features that increase the likelihood of a LBHB. We show that in human DJ-1, the Glu15-Asp23 H-bond is not a LBHB and that the proton is localized on Asp23. The corresponding H-bond in the prokaryotic homolog YajL is ambiguous, however, and we can not rule out a LBHB. Bond length analysis of atomic resolution X-ray crystal structures is well-established as a means to determine the protonation states of carboxylic acids and has been used for this purpose in several macromolecular systems 38–40, 47–49, 55. However, to the best of our knowledge, this is the first time that it has been used to test for the existence of a protein LBHB, despite its clear value for this application. Advances in synchrotron X-ray sources and detector technology have made collecting atomic resolution diffraction data from macromolecular crystals more common, with 2856 examples with a resolution of ≤1.2 Å in the August 2016 release of the PDB. In principle, bond length analysis could be applied to all atomic resolution crystal structures in the PDB using automated refinement methods to globally assess proton location in carboxylic acids, although that was beyond the scope of this work.
We propose some basic criteria for using bond length analysis to determine if H-bonded carboxylic acids are candidate LBHBs. Based on guidelines from prior work, C-O bond length differences that exceed three ESUs for a fully occupied and well-ordered (Beq≤ ~10Å2) carboxylic acid indicate that it is likely protonated 38–40. Conversely, C-O bond length differences below three ESUs indicate a deprotonated (ionized) carboxylate. The bond length ESUs should be determined from inversion of the unrestrained full least squares Hessian matrix (see Methods), which requires atomic resolution X-ray diffraction data in order to provide sufficient observations. If statistically rigorous bond length analysis supports a protonated-deprotonated COOH-−OOC H-bonding pair, then this should be interpreted as evidence against an LBHB, as the proton is fully localized to one of the carboxylic acids and not shared. Therefore, the pKa values of these two residues must be different and the proton delocalization that is a defining feature of LBHBs is unlikely. However, as described in the Introduction, a proton does not need to be equally shared between donor and acceptor to participate in a LBHB. Therefore, it is possible that bond length analysis may produce values that are intermediate between protonated and ionized carboxylic acid bond lengths in certain cases. More study is needed to determine how well carboxylic acid bond lengths correlate with proton occupancy and thus how precisely this method can discriminate between fully and partially localized protons. Because this correlation is presently unknown, bond length analysis is used as a binary test in this work, although more nuanced applications may be possible. Additionally, accepting an H-bond may also modulate carboxylic acid bond lengths, although this remains untested. In cases where bond length analysis suggests both residues are fully protonated (such as we observed in YajL, see above) a LBHB is possible and is a candidate for additional study, preferably using neutron diffraction where possible.
As potential confounding factors, we note that pKa values are temperature-dependent and thus it is possible that the protonation states of H-bonding moieties could be influenced by the cryocooling that is commonly performed to collect X-ray diffraction data at ~100 K. However, different Asp/Glu residues in proteins are expected to have very similar pKa temperature dependencies and thus cryocooling may not alter their relative protonation states in COOH-−OOC H-bonds. In addition, a proton transfer barrier whose height is comparable to kT at physiological temperature (thus permitting proton delocalization) may become insurmountable at 100 K, thereby converting an LBHB into a conventional H-bond at cryogenic temperature, although proton tunneling through the barrier may counteract this effect. Furthermore, if the proton transfer barrier is comparable to the H-bond zero point energy, the interaction should be a LBHB at any temperature. Ambient temperature crystallography is an increasingly popular way of addressing cryocooling-related artifacts 56, however the vulnerability of carboxylic acids to radiation-induced decarboxylation 57, 58 may complicate bond length analysis in ambient temperature structures and thus would require careful data collection and radiation damage-mitigation strategies. Neutron diffraction is typically performed at room temperature and is thus less susceptible to cryogenic artifacts. Despite these considerations, X-ray bond length analysis is comparatively straightforward to conduct if atomic resolution diffraction data are available and is thus an economical way of testing for LBHBs when carboxylic acids are involved. We propose that it should be used routinely to address these questions.
Although apparently not an LBHB, the short COOH-−OOC H-bond between Glu15 and Asp24 is important for DJ-1 dimer stability. The D24N mutation preserves but lengthens this H-bond by 0.3 Å and destabilizes the protein by 2.4°C. Therefore, protein COOH-−OOC H-bonds appear to be only slightly stronger than similar NH2-−OOC H-bonds. Despite the ability of this interaction to stabilize the DJ-1 dimer, it is not essential, as some homologs lack this H-bond. In Schizosaccharaomyces pombe DJ-1, the corresponding residues are hydrophobic although the dimer interface is very similar to human DJ-1 59. The potential for COOH-−OOC H-bonds to stabilize protein structure has been previously noted 22, 48, 60, and DJ-1 is now one of the few systems for which quantitative experimental data about the contribution of a COOH-−OOC H-bond to protein stability is available. Our data indicate that COOH-−OOC H-bonds appear to be comparable to (or slightly stronger than) conventional NH2-−OOC H-bonds and thus are not unusually strongly stabilizing interactions, even when buried in the protein interior.
Our PDB search shows that COOH-−OOC H-bonds are present in ~14% of protein structures and are typically short (~2.5 Å). Our updated result is in good agreement with prior estimates of the prevalence of protein COOH-−OOC H-bonds 18, 19. If they are not all LBHBs, why are COOH-−OOC H-bonds characterized by such short O-O distances? H-bonds remain surprisingly poorly understood at a theoretical level, however one explanation is that COOH-−OOC H-bond is short due to a conventional electronic effect. The carbonyl of the COOH moiety is electron-withdrawing due to resonance. This increases the partial positive charge density on the protonated oxygen atom of carboxylic acids, thus making them better H-bond donors by enhancing the dipolar contribution to the H-bond interaction. This would strengthen an H-bond, especially when the acceptor has a formal negative charge, as in COOH-−OOC charge-assisted H-bonds. A similar though less pronounced effect is expected for Tyr, whose aromatic system is slightly electron-withdrawing for the -OH substituent and has the next-shortest average OH-−OOC H-bond length in our search. As expected based on this reasoning, the O-O distances for H-bonds with carboxylate acceptors increase as the pKa values of model compounds for the H-bond donors increase (Table 3). Although the dependence of H-bonding strength on donor/acceptor pKa values is well-known 61, it does not seem to have been fully integrated into the discussion of short H-bonds in proteins. Therefore, the average COOH--OOC H-bond lies at the short end of a spectrum of conventional COH-−OOC charge-assisted H-bonds. These results also emphasize that H-bond donor-acceptor distance is a poor proxy for a LBHB and should not be used for this purpose.
Despite the interest they have generated, bona fide LBHBs have been difficult to experimentally observe in macromolecules for many reasons. The strongest experimental evidence to date for a putative protein LBHB is in the neutron diffraction crystal structure of photoactive yellow protein (PYP) 62, although a recent study has indicated that may also be a conventional short H-bond 63. Other systems, such as GFP, also have short H-bonds that have been suspected of being LBHBs. However, recent work by Oltrogge et al. using non-natural amino acid substitution to tune the pKa of the chromophore H-bond donor in GFP did not find evidence of the proton delocalization expected for a LBHB, even when donor and acceptor pKa values were closely matched 64. Moreover, the symmetric potential often cited as an important feature of a LBHB is possible in the highly symmetric lattice environments of certain small molecule crystals or in gas phase complexes 65, but is less likely in the complex structures of proteins or in solution. The ease with which environmental effects can disturb the symmetry of the associated free energy surface (and thus the pKa values of donors and acceptors) has been appreciated in the small molecule community for some time 53. Combining these considerations, buried carboxylic acids near dimer interfaces, as in DJ-1, should be among the more likely donor-acceptor pairs to display features consistent with LBHBs in protein environments. Therefore, the failure to observe an LBHB in this system as well as in others where conditions for their formation have been optimized may indicate that protein LBHBs are generally rare.
Supplementary Material
Table 1.
Crystallographic data and model statistics
| Protein | Wild-type DJ-1 | E15Q DJ-1 | D24N DJ-1 | E. coli YajL |
|---|---|---|---|---|
| PDB code | 5SY6 | 5SY9 | 5SYA | 5SY4 |
| Diffraction source | APS BioCars 14BM-C | SSRL beamline 9-2 | SSRL beamline 9-2 | SSRL beamline 12-2 |
| Wavelength (Å) | 0.90 | 0.88 | 0.88 | 0.73 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Detector | ADSC Q315 CCD | Pilatus 6M PAD | Pilatus 6M PAD | Pilatus 6M PAD |
| Space group | P3121 | C2221 | P3121 | P212121 |
| a, b, c (Å) | 75.23, 75.23, 75.23 | 86.28, 121.96, 43.51 | 74.97, 74.97, 75.34 | 43.88, 78.31, 99.43 |
| Mosaicity (°) | 0.20 | 0.14 | 0.10 | 0.08 |
| Resolution range (Å) | 49-1.15 | 37-1.10 | 38-1.10 | 38-0.98 |
| No. of unique reflections | 87636 (4315) | 91327 (3854) | 98874 (4755) | 191363 (9275) |
| Completeness (%) | 100.0 (99.7) | 98.1 (85.4) | 99.8 (97.1) | 97.6 (96.0) |
| Multiplicity | 11.1 (8.4) | 5.1 (3.8) | 5.5 (5.4) | 10.2 (9.5) |
| 〈I/σ(I)〉 | 16.3 (1.7) | 14.9 (1.4) | 20.4 (1.8) | 15.9 (1.1) |
| CC1/2a | 1.000 (0.614) | 0.999 (0.569) | 1.000 (0.738) | 1.000 (0.479) |
| Rmeas | 0.088 (1.667) | 0.070 (1.066) | 0.042 (0.919) | 0.085 (2.393) |
|
| ||||
| Refinement program | Refmac5 | Refmac5 | Refmac5 | Refmac5 |
|
| ||||
| Resolution range (Å) | 49-1.15 | 37-1.10 | 38-1.10 | 38-0.98 |
| Completeness (%) | 100.0 | 97.9 | 99.7 | 97.3 |
| No. of reflections, working set | 83306 | 88556 | 93885 | 187313 |
| No. of reflections, test set | 4296 | 2751 | 4940 | 3842 |
| Rwork | 0.1097 | 0.1178 | 0.1132 | 0.1140 |
| Rfree b | 0.1295 | 0.1374 | 0.1277 | 0.1266 |
| Rall c | 0.1107 | 0.1184 | 0.1139 | 0.1143 |
| No. of non-H atoms d | ||||
| Protein | 1517 | 1611 | 1588 | 3539 |
| Water | 258 | 308 | 230 | 429 |
| Heteroatoms | 8 (DTT) | 40 (EDO, CSD) | 20 (EDO, CSD) | 2 (MG) |
| R.m.s. deviations | ||||
| Bonds (Å) | 0.013 | 0.014 | 0.015 | 0.018 |
| Angles (°) | 1.675 | 1.821 | 1.699 | 1.937 |
| Average B factors (Å2) | ||||
| Protein | 16.8 | 11.1 | 15.5 | 11.4 |
| Water | 38.0 | 33.4 | 33.6 | 29.7 |
| Heteroatoms | 21.7 | 19.1 | 16.3 | 18.4 |
| ADP anisotropye | ||||
| Protein | 0.51 | 0.44 | 0.55 | 0.44 |
| Water | 0.36 | 0.32 | 0.40 | 0.33 |
| Heteroatoms | 0.58 | 0.54 | 0.59 | 0.62 |
CC1/2 was used to determine the resolution limit of the data
Rfree was calculated as Rwork using a test set of randomly chosen sequestered reflections
Rall includes both test and working reflections in the statistic
Includes partially occupied atoms
Anisotropy is the ratio of the smallest to largest eigenvalue of the anisotropic ADP tensor
Acknowledgments
Funding Information: This work was supported in part by NIH R01GM092999 to M.A.W. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of BioCARS was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number R24GM111072. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
We thank Dr. Donald Becker and Dr. Joseph Barycki (University of Nebraska-Lincoln) for discussions about H-bonding. We thank Dr. Javier Seravalli (University of Nebraska-Lincoln Biophysical Core Facility) for assistance with the CD measurements.
Abbreviations
- ASU
asymmetric unit
- DTT
dithiothreitol
- ESU
estimated standard uncertainty
- H-bond
hydrogen bond
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LBHB
low barrier hydrogen bond
- PEG
polyethyleneglycol
- Tris
Tris(hydroxymethyl)aminomethane
Footnotes
Supporting Information Available
Figures S1–S3 show the one- and two-dimensional distributions of geometric parameters for Ser- −OOC, Thr-−OOC, and Tyr-−OOC H-bonds.
References
- 1.Cleland WW, Kreevoy MM. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 2.Fuhrmann CN, Daugherty MD, Agard DA. Subangstrom crystallography reveals that short ionic hydrogen bonds, and not a His-Asp low-barrier hydrogen bond, stabilize the transition state in serine protease catalysis. J Am Chem Soc. 2006;128:9086–9102. doi: 10.1021/ja057721o. [DOI] [PubMed] [Google Scholar]
- 3.Cleland WW. Low-barrier hydrogen bonds and low fractionation factor bases in enzymatic reactions. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]
- 4.Frey PA, Whitt SA, Tobin JB. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 5.Hibbert F, Emsley J. Hydrogen Bonding and Chemical Reactivity. Advances in Physical Organic Chemistry. 1990;26:255–379. [Google Scholar]
- 6.Cassidy CS, Lin J, Frey PA. A new concept for the mechanism of action of chymotrypsin: the role of the low-barrier hydrogen bond. Biochemistry. 1997;36:4576–4584. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- 7.Lin J, Cassidy CS, Frey PA. Correlations of the basicity of His 57 with transition state analogue binding, substrate reactivity, and the strength of the low-barrier hydrogen bond in chymotrypsin. Biochemistry. 1998;37:11940–11948. doi: 10.1021/bi980278s. [DOI] [PubMed] [Google Scholar]
- 8.Das A, Prashar V, Mahale S, Serre L, Ferrer JL, Hosur MV. Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates. Proc Natl Acad Sci USA. 2006;103:18464–18469. doi: 10.1073/pnas.0605809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Northrop DB. Follow the protons: a low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Acc Chem Res. 2001;34:790–797. doi: 10.1021/ar000184m. [DOI] [PubMed] [Google Scholar]
- 10.Zheng YJ, Bruice TC. Is strong hydrogen bonding in the transition state enough to account for the observed rate acceleration in a mutant of papain? Proc Natl Acad Sci USA. 1997;94:4285–4288. doi: 10.1073/pnas.94.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hur O, Leja C, Dunn MF. Evidence of a low-barrier hydrogen bond in the tryptophan synthase catalytic mechanism. Biochemistry. 1996;35:7378–7386. doi: 10.1021/bi960240k. [DOI] [PubMed] [Google Scholar]
- 12.Schutz CN, Warshel A. The low barrier hydrogen bond (LBHB) proposal revisited: the case of the Asp... His pair in serine proteases. Proteins. 2004;55:711–723. doi: 10.1002/prot.20096. [DOI] [PubMed] [Google Scholar]
- 13.Warshel A, Papazyan A. Energy considerations show that low-barrier hydrogen bonds do not offer a catalytic advantage over ordinary hydrogen bonds. Proc Natl Acad Sci USA. 1996;93:13665–13670. doi: 10.1073/pnas.93.24.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warshel A, Papazyan A, Kollman PA. On low-barrier hydrogen bonds and enzyme catalysis. Science. 1995;269:102–106. doi: 10.1126/science.7661987. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz B, Drueckhammer DG. A Simple Method for Determining the Relative Strengths of Normal and Low-Barrier Hydrogen-Bonds in Solution - Implications to Enzyme Catalysis. J Am Chem Soc. 1995;117:11902–11905. [Google Scholar]
- 16.Perrin CL. Are short, low-barrier hydrogen bonds unusually strong? Acc Chem Res. 2010;43:1550–1557. doi: 10.1021/ar100097j. [DOI] [PubMed] [Google Scholar]
- 17.Graham JD, Buytendyk AM, Wang D, Bowen KH, Collins KD. Strong, Low-Barrier Hydrogen Bonds May Be Available to Enzymes. Biochemistry. 2014;53:344–349. doi: 10.1021/bi4014566. [DOI] [PubMed] [Google Scholar]
- 18.Flocco MM, Mowbray SL. Strange bedfellows: interactions between acidic side-chains in proteins. J Mol Biol. 1995;254:96–105. doi: 10.1006/jmbi.1995.0602. [DOI] [PubMed] [Google Scholar]
- 19.Langkilde A, Kristensen SM, Lo Leggio L, Molgaard A, Jensen JH, Houk AR, Navarro Poulsen JC, Kauppinen S, Larsen S. Short strong hydrogen bonds in proteins: a case study of rhamnogalacturonan acetylesterase. Acta Crystallogr. 2008;D64:851–863. doi: 10.1107/S0907444908017083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilli P, Bertolasi V, Ferretti V, Gilli G. Covalent Nature of the Strong Homonuclear Hydrogen-Bond - Study of the O-H---O System by Crystal-Structure Correlation Methods. J Am Chem Soc. 1994;116:909–915. [Google Scholar]
- 21.Das A, Prashar V, Mahale S, Serre L, Ferrer JL, Hosur MV. Crystal structure of HIV-1 protease in situ product complex and observation of a low-barrier hydrogen bond between catalytic aspartates. Proc Natl Acad Sci USA. 2006;103:18464–18469. doi: 10.1073/pnas.0605809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols DA, Hargis JC, Sanishvili R, Jaishankar P, Defrees K, Smith EW, Wang KK, Prati F, Renslo AR, Woodcock HL, Chen Y. Ligand-Induced Proton Transfer and Low-Barrier Hydrogen Bond Revealed by X-ray Crystallography. J Am Chem Soc. 2015;137:8086–8095. doi: 10.1021/jacs.5b00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakshminarasimhan M, Madzelan P, Nan R, Milkovic NM, Wilson MA. Evolution of new enzymatic function by structural modulation of cysteine reactivity in Pseudomonas fluorescens isocyanide hydratase. J Biol Chem. 2010;285:29651–29661. doi: 10.1074/jbc.M110.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson MA, Ringe D, Petsko GA. The atomic resolution crystal structure of the YajL (ThiJ) protein from Escherichia coli: a close prokaryotic homologue of the Parkinsonism-associated protein DJ-1. J Mol Biol. 2005;353:678–691. doi: 10.1016/j.jmb.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 25.Yeh JI, Hol WG. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Crystallogr. 1998;54:479–480. doi: 10.1107/s0907444998004697. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol. 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. Xds. Acta Crystallogr. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. 2013;D69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci USA. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
- 37.Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 38.Erskine PT, Coates L, Mall S, Gill RS, Wood SP, Myles DA, Cooper JB. Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides. Prot Sci. 2003;12:1741–1749. doi: 10.1110/ps.0305203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wlodawer A, Li M, Gustchina A, Dauter Z, Uchida K, Oyama H, Goldfarb NE, Dunn BM, Oda K. Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase. Biochemistry. 2001;40:15602–15611. doi: 10.1021/bi011817n. [DOI] [PubMed] [Google Scholar]
- 40.Singh RN, Kumar A, Tiwari RK, Rawat P. Synthesis, molecular structure, hydrogen-bonding, NBO and chemical reactivity analysis of a novel 1,9-bis(2-cyano-2-ethoxycarbonylvinyl)-5-(4-hydroxyphenyl)-dipyrromethane: a combined experimental and theoretical (DFT and QTAIM) approach. Spectrochimica acta Part A, Molecular and biomolecular spectroscopy. 2013;113:378–385. doi: 10.1016/j.saa.2013.04.121. [DOI] [PubMed] [Google Scholar]
- 41.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 42.Scopes RK. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 43.Prahlad J, Hauser DN, Milkovic NM, Cookson MR, Wilson MA. Use of cysteine-reactive cross-linkers to probe conformational flexibility of human DJ-1 demonstrates that Glu18 mutations are dimers. J Neurochem. 2014 doi: 10.1111/jnc.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Miller DW, Ahmad R, Hague S, Baptista MJ, Canet-Aviles R, McLendon C, Carter DM, Zhu PP, Stadler J, Chandran J, Klinefelter GR, Blackstone C, Cookson MR. L166P mutant DJ-1, causative for recessive Parkinson’s disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 46.Engh RA, Huber R. Accurate Bond and Angle Parameters for X-Ray Protein-Structure Refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- 47.Coates L, Erskine PT, Crump MP, Wood SP, Cooper JB. Five atomic resolution structures of endothiapepsin inhibitor complexes: implications for the aspartic proteinase mechanism. J Mol Biol. 2002;318:1405–1415. doi: 10.1016/s0022-2836(02)00197-3. [DOI] [PubMed] [Google Scholar]
- 48.Coates L, Tuan HF, Tomanicek S, Kovalevsky A, Mustyakimov M, Erskine P, Cooper J. The catalytic mechanism of an aspartic proteinase explored with neutron and X-ray diffraction. J Am Chem Soc. 2008;130:7235–7237. doi: 10.1021/ja801269x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witt AC, Lakshminarasimhan M, Remington BC, Hasim S, Pozharski E, Wilson MA. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47:7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeffrey GA. An Introduction to Hydrogen Bonding. Oxford University Press; New York, NY USA: 1997. [Google Scholar]
- 54.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 55.Coates L, Erskine PT, Mall S, Gill R, Wood SP, Myles DA, Cooper JB. X-ray, neutron and NMR studies of the catalytic mechanism of aspartic proteinases. Eur Biophys J. 2006;35:559–566. doi: 10.1007/s00249-006-0065-7. [DOI] [PubMed] [Google Scholar]
- 56.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci USA. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. 2000;D56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 58.Weik M, Ravelli RBG, Kryger G, McSweeney S, Raves ML, Harel M, Gros P, Silman I, Kroon J, Sussman JL. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci USA. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madzelan P, Labunska T, Wilson MA. Influence of peptide dipoles and hydrogen bonds on reactive cysteine pKa values in fission yeast DJ-1. FEBS J. 2012;279:4111–4120. doi: 10.1111/febs.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pozharski E, Wilson MA, Hewagama A, Shanafelt AB, Petsko G, Ringe D. Anchoring a cationic ligand: The structure of the fab fragment of the anti-morphine antibody 9B1 and its complex with morphine. J Mol Biol. 2004;337:691–697. doi: 10.1016/j.jmb.2003.12.084. [DOI] [PubMed] [Google Scholar]
- 61.Gilli P, Pretto L, Bertolasi V, Gilli G. Predicting Hydrogen-Bond Strengths from Acid-Base Molecular Properties. The pK(a) Slide Rule: Toward the Solution of a Long-Lasting Problem. Acc Chem Res. 2009;42:33–44. doi: 10.1021/ar800001k. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi S, Kamikubo H, Kurihara K, Kuroki R, Niimura N, Shimizu N, Yamazaki Y, Kataoka M. Low-barrier hydrogen bond in photoactive yellow protein. Proc Natl Acad Sci USA. 2009;106:440–444. doi: 10.1073/pnas.0811882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graen T, Inhester L, Clemens M, Grubmuller H, Groenhof G. The Low Barrier Hydrogen Bond in the Photoactive Yellow Protein: A Vacuum Artifact Absent in the Crystal and Solution. J Am Chem Soc. 2016 doi: 10.1021/jacs.6b05609. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 64.Oltrogge LM, Boxer SG. Short Hydrogen Bonds and Proton Delocalization in Green Fluorescent Protein (GFP) ACS Cent Sci. 2015;1:148–156. doi: 10.1021/acscentsci.5b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley J. Very Strong Hydrogen-Bonding. Chem Soc Rev. 1980;9:91–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.