Abstract
Follicular lymphoma (FL) is an indolent malignancy of germinal center B cells. Although the overall survival of FL patients has recently improved with the introduction of novel therapies, there is significant heterogeneity in patient outcome and a need for rationally designed therapeutic strategies that target disease biology. Next-generation sequencing studies have identified chromatin modifying gene (CMG) mutations as a hallmark of FL, highlighting epigenetic modifiers as an attractive therapeutic target in this disease. Understanding the complex roles of these mutations will be central to identifying and adaptively targeting associated vulnerabilities. Recent studies have provided insight into the functional consequences of the most frequently mutated CMGs (KMT2D, CREBBP, and EZH2) and point to a role for these events in modifying normal B-cell differentiation programs and impeding germinal center exit. However, the majority of FL tumors serially acquire multiple CMG mutations, suggesting that there is a level of cross talk or cooperation between these events that has not yet been defined. Here, I review the current state of knowledge on CMG mutations in FL, discuss their potential as therapeutic targets, and offer my perspective on unexplored areas that should be considered in the future.
Introduction
Follicular lymphoma (FL) is an indolent malignancy of germinal center B (GCB) cells, and the second most common form of non-Hodgkin lymphoma (NHL). It is characterized by a unique histology, in which tumor B cells form follicle-like structures with large numbers of nonmalignant immune cells infiltrating within the follicular and interfollicular regions.1 As such, the tumor microenvironment of FL is central to disease etiology and affects response to therapy and patient outcome.2-5 There is considerable heterogeneity in patient outcome; ∼20% of patients have rapid disease progression following treatment and a poor prognosis.6 Advancing our understanding of FL disease biology and harnessing this information to develop rationally targeted therapies is therefore a priority.
The most frequent genetic event in FL is the t(14;18)(q32;q21) translocation that places BCL2 under control of the immunoglobulin heavy-chain enhancer. This occurs in ∼90% of FL patients7 but is also observed within a subset of cells in healthy individuals. The presence of BCL2 translocation is associated with a significantly increased risk, particularly in those individuals with a high frequency of translocation-positive cells in the blood, but the majority of these individuals never develop FL.8,9 Furthermore, BCL2 translocations are likely acquired in pro- or pre-B cells,10 and in healthy individuals these cells can terminally differentiate via the GC to become memory B cells.8 Together, this suggests that secondary alterations are required for FL disease genesis, and these may play a role in stalling B-cell development at the GCB stage. In 2011, Morin et al described the frequent mutation of KMT2D (aka MLL2) and other chromatin modifying genes (CMGs) in FL.11 Since then, CMG mutations have emerged as a hallmark of FL and have been the subject of recent studies that have provided insight into their function. Here I will discuss the role of CMG mutations in FL, with a focus on EZH2, KMT2D, and CREBBP.
The unique genetic landscape of FL
Epigenetic deregulation is a salient feature of multiple hematologic malignancies, but those derived from GCB cells are unique. Myeloid malignancies are in part characterized by somatic mutations that alter DNA methylation status.12 Among others, these include IDH1/2, TET2, and DNMT3A mutation.13-15 Interestingly, mutations in these genes have also been observed in some T-cell malignancies,16,17 despite these cells aligning to a different hematopoietic lineage. The homeostatic control of DNA methylation is therefore important in both myeloid and T-lymphoid malignancies, but mutations affecting these processes are largely absent from GCB-derived malignancies such as FL. In contrast, FL preferentially acquires mutations in genes with a role in catalyzing the posttranslational modification of histones, such as the histone H3 lysine 4 (H3K4) methyltransferases KMT2D and KTM2C, the histone acetyltransferases CREBBP and EP300, and the histone H3 lysine 27 (H3K27) methyltransferase EZH2. Other CMGs that are mutated at a lower frequency in FL include those that control higher-order chromatin structure, such as the Switch/sucrose nonfermentable (SWI/SNF) complex components and multiple genes in the HIST1H1/2 linker histone family.18-20 Taken together, the somatic genome of FL shows a highly significant enrichment for mutations in CMGs.19 This makes understanding the functional role of these genes and mutations critical to the understanding of FL etiology and highlights this axis as a potentially attractive therapeutic target in this disease.
FL is not the only NHL subtype to align with a GCB differentiation state, nor is it the only subtype to acquire CMG mutations. Burkitt lymphoma (BL) and the GCB-like subtype of diffuse large B-cell lymphoma (DLBCL) also align with the GCB cell differentiation state and acquire mutations in CMGs.21,22 Table 1 shows the frequencies of CMG mutations from whole genome sequencing (WGS) and whole-exome sequencing (WES) studies of 71 FL,19,23-26 82 BL,27-29 and 1046 DLBCL,30-32 including a subset of 331 GCB-like DLBCL.32 Notably, there are key contrasts between FL and other GCB-derived malignancies. For example, BL more commonly acquires mutations in components of the SWI/SNF chromatin remodeling complex (SMARCA4 [∼21%] and ARID1A [∼7%]) than it does in genes that posttranslationally modify histones22,27,29 (Table 1). A recent targeted sequencing study suggests that SWI/SNF complex components may be recurrently mutated in FL,20 including frequent mutation of BCL7A. However, many of the other genes encoding the complex were mutated only within a single tumor and analogous approaches have not been applied to BL and DLBCL that would allow direct comparison of mutation frequencies across diseases. Furthermore, WES/WGS studies have only identified a low frequency of mutations in genes such as SMARCA4 (∼1% of tumors) in FL. The CMGs that are recurrently mutated in DLBCL show a similar rank order to those mutated in FL.11,33,34 However, even when restricting the analysis to the GCB-like subtype of DLBCL, the frequency of CMG mutations is markedly lower in this disease. The final characteristic that distinguishes FL from other GCB-derived malignancies is the high rate at which FLs acquire multiple CMG mutations in the same tumor, at variant allele frequencies which indicate that they co-occur within the same cell.19 This could be ascribed to a greater degree of genetic homogeneity in FL compared with other GCB-derived malignancies, but my alternative hypotheses are that (1) CMG mutations have a unique role in the context of FL compared with other GCB-derived malignancies, and/or (2) the microenvironment that is so distinctive of FL may provide selective pressure for the acquisition of a unique spectrum of mutations. Each of these scenarios may be influenced by the interplay of multiple CMG mutations within the same tumor cell, which is yet to be investigated in murine models or other experimental systems. Recent studies have, however, cast light onto the role of individual alterations of EZH2, KMT2D, and CREBBP in B-cell lymphomagenesis and are briefly discussed subsequently.
Table 1.
Characteristics of select CMG mutations
| CMG | Function | Mutation frequency | Percent missense: disruptive* | Percent occurring early in evolution† (no. early: no. late) | Murine models | ||
|---|---|---|---|---|---|---|---|
| FL, % | DLBCL, % (GCB-like, %) | BL, % | |||||
| KMT2D | H3K4 methyltransferase | 72 | 24 (28) | 2 | 17:83 | 62 (43:26) | Knockout,49 knockdown50 |
| CREBBP | Lysine acetyltransferase | 65 | 11 (16) | 6 | 81:19 | 89 (42:5) | Knockout,56-58,60 knockdown61 |
| EZH2 | H3K27 methyltransferase | 25 | 6 (12) | 2 | 100:0 | 41 (9:13) | Point-mutation knock-in38,42 |
| EP300 | Lysine acetyltransferase | 15 | 6 (6) | 0 | 100:0 | 57 (8:6) | — |
| KMT2C | H3K4 methyltransferase | 13 | 5 (5) | 1 | 75:25 | 60 (3:2) | — |
| HIST1H1E | Linker histone | 14 | 17 (17) | 0 | 100:0 | 21 (3:11) | — |
| ARID1A | SWI/SNF component | 11 | 9 (10) | 7 | 0:100 | 31 (4:9) | — |
| SMARCA4 | SWI/SNF component | 1 | 8 (10) | 21 | ‡ | ‡ | — |
Missense mutations include single amino acid substitutions and coding indels. Disruptive mutations include premature stop codon and frameshift mutations. Data are from FL WES/WGS studies with available information19,24,25 and are represented as relative percentage.
The percent of mutations occurring early in evolution is calculated from studies of clonal ancestry across 65 patients with paired diagnosis and relapse biopsies.19,24-26 Early events are defined as those shared between all biopsies from the same patient. Late events are defined as those not present in all paired biopsies from the same patient.
Only 1 evaluable mutation (disruptive and clonal).
What we know (and do not know) about the function of CMG mutations in lymphomagenesis
EZH2
The EZH2 gene encodes a lysine methyltransferase enzyme that catalyzes trimethylation of H3K27 (H3K27me3) as part of the polycomb repressor 2 (PRC2) complex. The H3K27me3 mark is transcriptionally repressive and has an important role in the control of developmentally regulated genes through the formation of bivalent promoters. These promoters possess both the activating H3K4 trimethylation (H3K4me3) mark and the repressive H3K27me3 mark and can be rapidly activated through loss of H3K27me3 or stably repressed through loss of H3K4me3. EZH2 and other components of the PRC2 complex are highly expressed in GCB cells,35-37 and conditional deletion of Ezh2 within this compartment significantly attenuates GC development.38 The normal function of EZH2 in GCB cells is to repress the expression of a set of genes that are highly transcribed in naïve B cells, either through addition of H3K27me3 to H3K4me3-marked promoters to create bivalency or through addition of H3K27me3 to promoters that are not marked with H3K4me3. Genes with bivalent promoters in GCB cells include those involved in terminal differentiation such as PRDM1, IRF4, and XBP1,38,39 and negative regulators of the cell cycle such as CDKN1A and CDKN1B.38 Silencing of these genes by EZH2 occurs in cooperation with BCL6 and BCOR40 and temporarily suspends the B-cell differentiation program during the GC reaction. Notably, silencing of the BCL6 target gene, CDKN1A, by EZH2 is particularly important for promoting B-cell proliferation and GC formation, and deletion of Cdkn1a is sufficient to rescue GC formation in Ezh2 conditional knockout mice.41
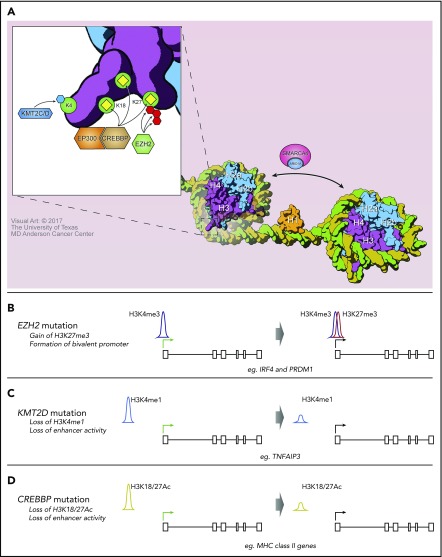
Mutations of EZH2 were the first recurrent CMG mutation to be reported in FL42 and were the first to be investigated using murine models.38-40 These are most often missense mutations of tyrosine 641 (Y641) within the Su(var)3-9, Enhancer-of-zeste and Trithorax (SET) domain of EZH2, resulting in a neomorphic change with increased activity for catalyzing the addition of the third methyl group to H3K27me2.43,44 However, the mutant protein has reduced activity for addition of the first and second methyl groups, potentially explaining why it is always found as a heterozygous mutation in patient tumors. This is believed to be because wild-type and mutant EZH2 act cooperatively to promote H3K27me3, but there is contrasting evidence about whether maintenance of the wild-type EZH2 allele is necessary for increased H3K27me3 and tumor development.39,40,45 However, it is clear that B-cell–specific expression of Ezh2 Y641 point mutants can cooperate with Bcl2 overexpression to promote B-cell lymphoma.38,39 B-cell–specific expression of mutant Ezh2 resulted in increased H3K27me3 at promoter and promoter-proximal regions of genes (Figure 1) that are normally repressed in GCB cells, such as PRDM1 and IRF4. As many of these genes are involved in GC exit and terminal differentiation, their stable repression likely contributes to lymphomagenesis at least in part by stalling B-cell differentiation at the GCB stage.38,39
Figure 1.
The consequences of CMG mutations on histone posttranslational modifications. (A) A depiction of the normal role of CMGs that are recurrently mutated in FL. KMT2C/D methylates H3K4, CREBBP and EP300 acetylate multiple residues including H3K18 and H3K27, and EZH2 catalyzes trimethylation of H3K27. (B) EZH2 Y641 mutation was predominantly associated with the gain of H3K27me3 at promoters already marked with H3K4me3, to form bivalent promoters. (C) KMT2D loss was associated with reduced H3K4 methylation at enhancer elements, including that for the TNFAIP3 gene. (D) CREBBP loss was associated with reduced H3K18Ac/H3K27Ac at enhancer elements, including those of major histocompatibility complex (MHC) class II genes.
Murine models of Ezh2 Y641 mutations have characterized a relatively strong role for these events in promoting B-cell lymphoma, especially in comparison with models of Kmt2d or Crebbp loss (discussed subsequently). However, the majority of EZH2 mutations are subclonal events in FL at diagnosis19,23,24,26 (Table 1) and remain subclonal at relapse.19 In contrast, other subclonal drivers such as TP53 mutations in chronic lymphocytic leukemia are subject to strong selection and expand to become clonal at disease progression.46 This difference may be related to a stronger role for TP53 mutations in promoting therapeutic resistance. However, an alternative hypothesis is that there is a yet unexplored contextual advantage of EZH2 mutations within the background of KMT2D and/or CREBBP mutations. This may result in a preferential acquisition of EZH2 mutations at a later stage of genomic evolution after the early driver mutations have been acquired. Alternatively, there may be an advantage to EZH2 mutation heteroclonality. In this scenario, only a minor number of EZH2 mutant cells would be required to benefit the broader tumor population, and overpopulation by EZH2 mutant cells may be deleterious. Understanding the context-specific roles of EZH2 mutation and heteroclonality will be particularly important because of the emerging potential for EZH2 inhibitors as a novel therapy for FL.
KMT2D
The KMT2D gene (aka MLL2) encodes an SET domain–containing lysine methyltransferase that is part of the KMT2 family. KMT2D and KMT2C are conserved members of this family that are recruited to genomic loci by nuclear coreceptors or transcription factors and are thought to be responsible for the majority of H3K4 monomethylation at enhancer elements.47 In addition to their ability to catalyze H3K4 methylation independently, KMT2D and KMT2C can also perform this function as part of multiprotein complex of proteins associated with Set1 (COMPASS)–like complexes that are predominantly associated with developmentally regulated genes.48 The monomethylation of enhancer elements leads to the recruitment of other coactivators that culminate in enhancer-promoter looping, activation of RNA pol II, and gene transcription. Knockout studies have pointed to a partial genetic redundancy between Kmt2d and Kmt2c, although many of their functions are nonredundant.49
The KMT2D gene is the most recurrently mutated CMG in FL (∼72% of cases) and is also recurrently mutated in a lower frequency of DLBCL (∼24% of cases). The majority of these mutations are nonsense or frameshift events that result in a loss of KMT2D protein50 (Table 1). The remaining coding mutations predominantly affect the C-terminal portion of the gene containing the SET domain and reduce methyltransferase activity.50 Two recent studies have interrogated the functional consequence of Kmt2d loss in murine models.50,51 Knockout or knockdown of Kmt2d in murine B cells resulted in no obvious changes in normal B-cell development but led to a marked increase in the frequency of GCB cells and reduced numbers of class-switched B cells following immunization. This is indicative of a defect in B-cell maturation and/or class-switch recombination in the absence of Kmt2d. These studies also showed a loss of H3K4 methylation at GCB-specific enhancers that were bound by Kmt2d (Figure 1) and confirmed a role for Kmt2d loss in promoting lymphomagenesis in the context of Bcl2 overexpression.
Notably, KMT2C is also mutated in ∼13% of FL. These mutations do not mutually exclude KMT2D mutations, suggesting that these genes may function nonredundantly in FL. However, the question of redundancy between KMT2D and KMT2C activity remains controversial because of contrasting results with respect to the effect of Kmt2d loss on the global abundance of H3K4 methylation.50,51 In addition, it is not clear whether C-terminal missense mutations that reduce KMT2D methyltransferase activity, but maintain the protein expression, would have the same functional consequence as nonsense or frameshift mutations that result in a loss of protein. This is because KMT2D methyltransferase activity has been found to be dispensable for the transcription of enhancer RNAs, which indicate enhancer activity.52 The scaffolding function of KMT2D, in addition to its methyltransferase activity, is therefore very important for enhancer activation. This may explain why FLs select for a higher rate of nonsense/frameshift mutations in KMT2D than in other CMGs and also points to a possible functional discrepancy between loss-of-protein and missense mutations. The role of KMT2C mutation, and the function of KMT2D SET domain missense mutations in FL, will be important areas to address in future studies.
CREBBP
The CREBBP gene (aka CBP) encodes a lysine acetyltransferase (KAT) that acetylates histone 3 at lysines 18 (H3K18Ac) and 27 (H3K27Ac), as well as acetylates nonhistone proteins. It functions as a transcriptional coactivator by acetylating histones at regulatory elements following its recruitment by DNA-binding transcription factors and other coactivator complexes.53 Histone acetylation alters their charge and loosens their association with DNA to make it more accessible to transcription factors. In addition, the acetyl mark is also recognized by bromodomain-containing proteins such as adenosine triphosphate–dependent chromatin remodelers and factors that promote transcriptional elongation.53,54
Mutations of CREBBP occur in ∼65% of FLs and ∼11% of DLBCLs.22 Approximately 80% of the mutations observed in FL create missense changes in the KAT domain, with 26% of all mutations altering a single KAT domain amino acid (R1446).19,55-57 These lead to reduced acetyltransferase activity.55,56 Multiple studies including our own have shown that CREBBP mutations arise as early events during FL disease genesis and likely reside within tumor cell progenitors that propagate disease relapse.19,23-26 A recent study also showed in a single patient that a CREBBP mutation could be detected within the hematopoietic stem and progenitor cell pool,58 although this requires further validation. In primary human FL tumors, these mutations are associated with a marked reduction in MHC class II expression and reduced tumor-infiltrating T-cell number and function.19 Importantly, reducing tumor cell MHC class II expression was found to enhance lymphomagenesis,59 suggesting CREBBP mutation–associated reductions in antigen presentation may be important for disease pathogenesis.
Recent murine studies found that loss of Crebbp promotes B-cell lymphoma,58,60 particularly in cooperation with Bcl2 overexpression,57,61,62 and that regions of reduced histone acetylation associated with Crebbp loss were primarily located at distal enhancer elements57,58,61,62 (Figure 1). This included regulatory elements of MHC class II genes and was also associated with reduced MHC class II expression.59,61 However, the magnitude of this change was less than that observed in primary human FLs.19 In addition, regions of reduced H3K27Ac associated with Crebbp loss were found to be enriched for loci that are also bound by BCL661,62 and repressed by the recruitment of HDAC3.61 In this context, loss of CREBBP may promote B-cell lymphoma by interfering with the derepression of BCL6 target genes and thereby preventing GC exit and terminal differentiation.
Reduced acetylation of nonhistone proteins such as p53 and BCL6 were also previously described as important features associated with CREBBP mutation.56 A recent study expanded on this observation by showing that Crebbp knockout led to a significant attenuation of p53 acetylation in response to DNA damage, and reduced DNA damage response.58 Although it is likely that the reduced acetylation of nonhistone proteins plays a contributing role in disease pathogenesis, the relative importance of these changes and their possible interplay with patterns of reduced histone acetylation remains to be explored. In addition, our recent study suggests that CREBBP mutations show a different spectrum in FL compared with DLBCL.57 Loss of CREBBP was associated with induction of MYC expression,57,58 which is more characteristic of DLBCL than FL.57 It is likely that the expression of a catalytically inactive CREBBP protein resulting from KAT domain missense mutations has different functional consequences than mutations resulting in the loss of CREBBP protein expression. It will therefore be important to characterize the functional consequence of CREBBP KAT domain mutations that are more prevalent in FL and compare this with loss-of-protein events that were modeled in recent studies57,61,62 and are more frequent in DLBCL.57
Coassociation of CMG mutations
Our recent study using deep-sequencing of purified tumor cells from FL revealed that ∼70% of tumors possess ≥2 different CMG mutations.19 This same pattern has been noted in other studies utilizing whole tumor specimens, although at a lower frequency.24-26 We and others have expanded on this observation by analyzing paired biopsies from diagnosis and relapse and showing that CMG mutations are serially acquired during the genomic evolution of FL.19,23-26 It is therefore clear that the majority of FLs undergo genomic evolution that culminates in multiple CMG mutations. Some might suggest that coassociation of CMG mutations within individual FL tumors is a random consequence of their high individual gene mutation rates. In contrast, my perspective is that the individual gene mutation rates in this disease are high because it is of evolutionary benefit for FLs to acquire multiple CMG mutations.
There is considerable cross talk between epigenetic marks and complex interactions among different CMGs. Although the purpose of this article is not to review these interactions, some are particularly pertinent to our consideration of the possible function of cosegregating CMG mutations. For example, there is a well-defined antagonistic relationship between H3K27Ac and H3K27me3.63,64 This suggests that CREBBP mutations that result in a loss of H3K27Ac may also alter the landscape of H3K27me3, because of normal PRC1/2 activity with wild-type EZH2 and/or enhanced PRC2 activity resulting from EZH2 mutation. KMT2D and KMT2C can also indirectly influence both H3K27Ac and H3K27me3. The COMPASS-like complexes formed by KMT2D and KMT2C recruit the UTX histone demethylating enzyme to enhancers, thereby facilitating the removal of H3K27me3.48 This indicates that loss of KMT2C/D may lead to the accumulation of H3K27me3 at enhancers that are normally bound by these proteins. Although many of the changes in H3K27me3 catalyzed by mutant EZH2 were observed at promoter-proximal elements, a loss of KMT2D and COMPASS-like complex occupancy may allow PRC1/2 to additionally silence enhancer elements and may open a new dimension of epigenetic changes catalyzed by mutant EZH2 in the presence of KMT2C/D loss-of-protein mutations.38,39 Enhancer occupancy by KMT2C/D, but not their methyltransferase activity, is also required for the recruitment of cofactors such as CREBBP and EP300 that catalyze H3K27Ac and promote enhancer activation.65 This suggests that the distribution of CREBBP and EP300 at enhancers, and therefore the functional consequence of their mutation or loss, may be different in the context of KMT2C/D nonsense/frameshift mutations than it would be in the presence of KMT2C/D protein expression. Importantly, ∼50% of FLs possess mutations in both KMT2D and CREBBP.19 These observations again raise the questions of whether KMT2C and KMT2D can act redundantly to recruit UTX or CREBBP/EP300, and whether inactivating missense mutations of KMT2D would be functionally equivalent to nonsense/frameshift mutations.
Mutations in HIST1H1/2 linker histone family genes are also likely to have significant cross talk with mutations in genes that control histone posttranslational modification. Linker histones bind to DNA at the entry and exit sites of the nucleosome and are required for the stability of higher-order chromatin structure. Recent data suggest that the histone tails of oligonucleosomes that are assembled with linker histones are poorer substrates for a range of posttranslational modifiers including CREBBP and EZH2,66 although alternative findings suggest that they are better substrates for EZH2.67 In addition, the most frequently mutated linker histone, HIST1H1E, can be methylated at lysine 26 by EZH2 to create a docking site for HP1 and facilitate heterochromatin formation. Linker histones can also recruit DNA methyltransferases, and recruitment DNMT3B is impaired by somatic mutation.18 In many cases, linker histones therefore facilitate heterochromatin formation, possibly in tandem with EZH2, and their inactivation by somatic mutation is inconsistent with the promotion of heterochromatin formation by activating EZH2 mutations and loss-of-function KMT2D and CREBBP mutations. However, acetylation of HIST1H1E at lysine 34 by GCN5 can also lead to recruitment of TAF1 and promote transcriptional activation, meaning that loss-of-function mutations may have a transcriptionally repressive effect. As the biology of linker histones is not as well defined as core histones, and each family member plays a unique role in gene-specific activation and inactivation,68 detailed functional studies will be required to understand the precise role of these mutations in FL and their cross talk with other CMG mutations.
Despite these interactions only representing the “tip of the iceberg” with respect to epigenetic cross talk, they clearly indicate a potential for CMG mutations in the same tumor cell to interact and promote unique epigenetic phenotypes. This would suggest a level of codependency between CMG mutations wherein the product of the mutations is greater than (or different from) the sum of the parts, and their serial acquisition progressively “locks in a phenotype.” That is, the critical genes that are altered by each CMG mutation may be largely overlapping, and serial acquisition of multiple CMG mutations may therefore lead to progressively deeper repression of these common genes that is more difficult to reverse. An alternative viewpoint may be that, despite epigenetic cross talk, each CMG mutation has a unique and mutually exclusive functional consequence and their serial acquisition acts to “build a phenotype.” That is, the critical genes that are altered by each CMG mutation may be nonoverlapping, and serial acquisition of multiple CMG mutations may therefore lead to the repression of progressively larger sets of genes. I would suggest that the truth is probably somewhere between these 2 scenarios.
Targeting FL with epigenetic modifying agents
Therapeutic targeting of epigenetic deregulation in FL is a theoretically attractive concept. However, the most common CMG mutations (ie, those in KMT2D and CREBBP) are loss-of-function/loss-of-protein events, which are difficult to drug. This has made targeting activating EZH2 mutations the lowest hanging fruit. Multiple companies have developed inhibitors for EZH2,69-71 and these have entered early phase clinical trials in NHL and other settings (www.clinicaltrials.gov; #NCT02082977, #NCT02395601, #NCT01897571). Cell lines bearing EZH2 mutations are more sensitive to EZH2 inhibitors than wild-type cell lines,69,70 providing some credence to precision targeting of EZH2 mutations. However, genetic studies of FL have shown that EZH2 mutations are late events in disease evolution and are present only in a subclonal population.19,23,24,26 In this respect, EZH2 mutations may not be a good therapeutic target, because they only reside in a subset of tumor cells. Despite this, a recent update of a phase 2 study of an EZH2 inhibitor, tazemetostat, reported a 92% objective response rate in FL patients bearing EZH2 mutations,72 which was higher than EZH2 wild-type patients (26%). These trends are based on early observations in a small cohort of patients and require further follow-up and validation, but we should also be careful to discriminate between targeting EZH2 vs targeting EZH2 mutations in FL. Although there may be flaws in the rationale for targeting EZH2 mutations, multiple studies have demonstrated that wild-type EZH2 is a critical regulator of GCB development35,38,40,73 and therefore also represents an attractive therapeutic target. The acquisition of an EZH2 mutation may be restricted to cases in which the EZH2-driven epigenetic program has a strong oncogenic role. This would mean that even tumor cells with wild-type EZH2 may be sensitive to its inhibition, and responses will be observed regardless of the clonal representation of the EZH2 mutation. In other words, EZH2 mutational status may be a biomarker for the likelihood and/or depth of response to EZH2 inhibition, rather than a strict criterion for whether EZH2 inhibition should be used. Alternatively, the association between EZH2 mutation and better clinical outcome74,75 may mean that EZH2 mutation is associated with superior responses to a range of therapies, and the use of EZH2 mutation as a predictive biomarker for response may not be restricted to EZH2 inhibitors.
As previously discussed, there may also be a role for KMT2D and/or CREBBP mutations in altering the landscape of EZH2-catalyzed H3K27me3 in FL tumors through epigenetic cross talk. It is currently unclear how this may modify sensitivity to EZH2 inhibition. In addition, EZH2 plays an important role in the development of other immune cells, including T cells76-79 and natural killer cells.80 Inhibiting EZH2 may therefore have additional tumor cell–independent therapeutic activity through the alteration of T-cell polarization or natural killer cell development/activation, similar to that described for ibrutinib.81,82 The notion that EZH2 may be a good therapeutic target in FL, independent of its mutational status, is supported by some of the most pronounced responses to tazemetostat being observed in EZH2 wild-type patients.72 The complexity of epigenetic cross talk and the potential role of nonmalignant immune cells may make it difficult to fully predict the efficacy of EZH2 inhibition in FL using genotypic information alone.
Histone deacetylase (HDAC) inhibitors are also a logical avenue for continued exploration in FL, because of their potential to counteract the loss of histone acetylation that results from CREBBP or EP300 mutations.55,56,62 Pan-HDAC inhibitors have shown some promise in FL, with objective responses rates of 47% to 49% and 64% for vorinostat and abexinostat, respectively.60,83,84 These response rates are similar to the reported frequency of CREBBP mutations,19,24,56 but this may be purely coincidental as the mutational status of CREBBP was not reported in these studies. It will be interesting to evaluate whether histone acetyltransferase gene mutations are predictive of response to pan-HDAC inhibitors. Isoform-specific inhibitors, such as those that are HDAC3-specific,61 may also provide a similar therapeutic benefit while limiting the toxicity associated with pan-HDAC inhibition.85 HDAC3 participates in nuclear receptor corepressor 1 (NCOR) and 2 (SMRT) complexes that are recruited by BCL6 to repress target genes in GCB cells via deacetylation of H3K27,86 and it was hypothesized that mutation of CREBBP/EP300 impedes the reversal of this HDAC3-mediated target-gene repression in GCB cells. As a proof of concept, knockdown or chemical inhibition of HDAC3 was capable of inducing the expression of genes that were silenced by Crebbp loss, including MHC class II genes.61 Specific inhibition of HDAC3 may therefore be efficient in targeting CREBBP or EP300 mutations and may have efficacy in FL.
Although epigenetic modifying agents represent a logical and exciting avenue for treatment of FL, the complete response rates for these therapeutics as single agents have been low.60,72,83,84 This may be a product of the relapsed/refractory population used in the trials but might also indicate that their most effective use may be as part of a combination regimen. An area of importance is understanding the functional implications of these therapeutics, including both sensitizing pathways and mechanisms of resistance, so as to identify rational combination strategies.
Conclusion
There is strong evidence that KMT2D, CREBBP, and EZH2 mutations share a common function in controlling GCB-cell differentiation programs. This likely contributes to lymphomagenesis by stalling FL tumor cells within the GC, where they can persist as a result of BCL2 overexpression and are subject to continuous activation-induced cytidine deaminase activity that promotes further genomic evolution. However, I find it unlikely that CMG mutations act redundantly in FL, because of the large degree of cosegregation of these mutations within the same tumor and the huge potential for epigenetic cross talk. Serial mutation of unique CMGs likely “builds” a complex epigenetic program by altering discrete sets of genes that would normally be deleterious to the prolonged residence of a B cell within the GC, with a subset of their effects complementarily “locking in” 1 or more critical epigenetic programs. This includes programs that (1) drive germinal center exit, (2) are required for terminal differentiation, or (3) control interactions with cells in the microenvironment. The latter may modulate B-cell deletion from the GC resulting from neglect by T follicular helper (TFH) cells87 or promote antigen-driven antitumor immunity.88 Box 1 highlights some suggested avenues of exploration to provide greater understanding of the role of CMG mutations in FL and how we may target them therapeutically. Although it is difficult to model, it will be imperative to somehow account for the complex role of the tumor microenvironment in FL etiology and its interplay with the tumor cell–intrinsic phenotypes that are governed by CMG mutations. This is an incredibly challenging task ahead of us, but one that may unravel a targetable biology of FL and improve the lives of countless patients.
Box 1.
Potential future avenues of exploration:
Investigate the role of combinations of CMG mutations using patient samples, cell lines, and animal models.
Related to the first avenue, perform single-cell analysis of genotype/phenotype relationships for combinations of CMG mutations and single low variant allele frequency mutations.
Discriminate between the functional consequences of missense and disruptive mutations in frequently mutated CMGs.
Interrogate the relationship between the tumor microenvironment and somatic mutations using comparisons of both inter- and intrapatient heterogeneity.
Identify mechanisms of sensitivity and resistance to epigenetic modifying agents and rational therapeutic combination strategies.
Acknowledgments
The figure was created with assistance from The University of Texas MD Anderson Cancer Center Medical Graphics Department.
M.R.G. receives funding from the National Cancer Institute, National Institutes of Health (grant R01CA201380).
Authorship
Contribution: M.R.G. wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Michael R. Green, Department of Lymphoma/Myeloma, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 0903, Houston, TX 77030; e-mail: mgreen5@mdanderson.org.
References
- 1.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122(10):3424-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159-2169. [DOI] [PubMed] [Google Scholar]
- 3.Blaker YN, Spetalen S, Brodtkorb M, et al. The tumour microenvironment influences survival and time to transformation in follicular lymphoma in the rituximab era. Br J Haematol. 2016;175(1):102-114. [DOI] [PubMed] [Google Scholar]
- 4.Wahlin BE, Aggarwal M, Montes-Moreno S, et al. A unifying microenvironment model in follicular lymphoma: outcome is predicted by programmed death-1--positive, regulatory, cytotoxic, and helper T cells and macrophages. Clin Cancer Res. 2010;16(2):637-650. [DOI] [PubMed] [Google Scholar]
- 5.Kiaii S, Clear AJ, Ramsay AG, et al. Follicular lymphoma cells induce changes in T-cell gene expression and function: potential impact on survival and risk of transformation. J Clin Oncol. 2013;31(21):2654-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casulo C, Byrtek M, Dawson KL, et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the National LymphoCare Study. J Clin Oncol. 2015;33(23):2516-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zelenetz AD, Chu G, Galili N, et al. Enhanced detection of the t(14;18) translocation in malignant lymphoma using pulsed-field gel electrophoresis. Blood. 1991;78(6):1552-1560. [PubMed] [Google Scholar]
- 8.Roulland S, Kelly RS, Morgado E, et al. t(14;18) Translocation: a predictive blood biomarker for follicular lymphoma. J Clin Oncol. 2014;32(13):1347-1355. [DOI] [PubMed] [Google Scholar]
- 9.Roulland S, Navarro JM, Grenot P, et al. Follicular lymphoma-like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis [published correction appears in J Exp Med. 2006;203(11):2563]. J Exp Med. 2006;203(11):2425-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce CM. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229(4720):1390-1393. [DOI] [PubMed] [Google Scholar]
- 11.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12(9):599-612. [DOI] [PubMed] [Google Scholar]
- 13.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95-96. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Kaminski MS, Li Y, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123(10):1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci USA. 2015;112(10):E1116-E1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krysiak K, Gomez F, White BS, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017;129(4):473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5(4):251-262. [DOI] [PubMed] [Google Scholar]
- 22.Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Reports. 2014;6(1):130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kridel R, Chan FC, Mottok A, et al. Histological transformation and progression in follicular lymphoma: a clonal evolution study. PLoS Med. 2016;13(12):e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter J, Schlesner M, Hoffmann S, et al. ; ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316-1320. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin RD, Mungall K, Pleasance E, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasqualucci L, Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol. 2015;52(2):67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velichutina I, Shaknovich R, Geng H, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raaphorst FM, van Kemenade FJ, Fieret E, et al. Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol. 2000;164(1):1-4. [DOI] [PubMed] [Google Scholar]
- 37.van Galen JC, Dukers DF, Giroth C, et al. Distinct expression patterns of polycomb oncoproteins and their binding partners during the germinal center reaction. Eur J Immunol. 2004;34(7):1870-1881. [DOI] [PubMed] [Google Scholar]
- 38.Béguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23(5):677-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souroullas GP, Jeck WR, Parker JS, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med. 2016;22(6):632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Béguelin W, Teater M, Gearhart MD, et al. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell. 2016;30(2):197-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Béguelin W, Rivas MA, Calvo Fernández MT, et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun. 2017;8:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107(49):20980-20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooun A, Gajiwala KS, Deng YL, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun. 2016;7:11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ford DJ, Dingwall AK. The cancer COMPASS: navigating the functions of MLL complexes in cancer. Cancer Genet. 2015;208(5):178-191. [DOI] [PubMed] [Google Scholar]
- 49.Lee JE, Wang C, Xu S, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorighi KM, Swigut T, Henriques T, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell. 2017;66(4):568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586(17):2692-2704. [DOI] [PubMed] [Google Scholar]
- 55.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.García-Ramírez I, Tadros S, González-Herrero I, et al. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood. 2017;129(19):2645-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horton SJ, Giotopoulos G, Yun H, et al. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat Cell Biol. 2017;19(9):1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashwah H, Schmid CA, Kasser S, et al. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc Natl Acad Sci USA. 2017;114(36):9701-9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evens AM, Balasubramanian S, Vose JM, et al. A phase I/II multicenter, open-label study of the oral histone deacetylase inhibitor abexinostat in relapsed/refractory lymphoma. Clin Cancer Res. 2016;22(5):1059-1066. [DOI] [PubMed] [Google Scholar]
- 61.Jiang Y, Ortega-Molina A, Geng H, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discov. 2017;7(1):38-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Vlasevska S, Wells VA, et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov. 2017;7(3):322-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tie F, Banerjee R, Stratton CA, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasini D, Malatesta M, Jung HR, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38(15):4958-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Lee JE, Lai B, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci USA. 2016;113(42):11871-11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stützer A, Liokatis S, Kiesel A, et al. Modulations of DNA contacts by linker histones and post-translational modifications determine the mobility and modifiability of nucleosomal H3 tails. Mol Cell. 2016;61(2):247-259. [DOI] [PubMed] [Google Scholar]
- 67.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem. 2006;281(13):8365-8370. [DOI] [PubMed] [Google Scholar]
- 68.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16(11):1439-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890-896. [DOI] [PubMed] [Google Scholar]
- 70.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108-112. [DOI] [PubMed] [Google Scholar]
- 71.Bradley WD, Arora S, Busby J, et al. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol. 2014;21(11):1463-1475. [DOI] [PubMed] [Google Scholar]
- 72.Morschhauser F, Salles G, McKay P, et al. Interim report from a phase 2 multicenter study of Tazemetostat, an EZH2 inhibitor, in patents with relapsed or refractory B-cell non-Hodgkin lymphomas. Hematol Oncol. 2017;35(suppl 2):24-25. [Google Scholar]
- 73.Su IH, Basavaraj A, Krutchinsky AN, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4(2):124-131. [DOI] [PubMed] [Google Scholar]
- 74.Pastore A, Jurinovic V, Kridel R, et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015;16(9):1111-1122. [DOI] [PubMed] [Google Scholar]
- 75.Huet S, Xerri L, Tesson B, et al. EZH2 alterations in follicular lymphoma: biological and clinical correlations. Blood Cancer J. 2017;7(4):e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tumes DJ, Onodera A, Suzuki A, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39(5):819-832. [DOI] [PubMed] [Google Scholar]
- 77.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280(36):31470-31477. [DOI] [PubMed] [Google Scholar]
- 78.Christofides A, Karantanos T, Bardhan K, Boussiotis VA. Epigenetic regulation of cancer biology and anti-tumor immunity by EZH2. Oncotarget. 2016;7(51):85624-85640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang XP, Jiang K, Hirahara K, et al. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci Rep. 2015;5:10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin J, Leavenworth JW, Li Y, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci USA. 2015;112(52):15988-15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sagiv-Barfi I, Kohrt HE, Burckhardt L, Czerwinski DK, Levy R. Ibrutinib enhances the antitumor immune response induced by intratumoral injection of a TLR9 ligand in syngeneic mouse lymphoma model. Blood. 2015;125(13):2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sagiv-Barfi I, Kohrt HE, Czerwinski DK, Ng PP, Chang BY, Levy R. Therapeutic antitumor immunity by checkpoint blockade is enhanced by ibrutinib, an inhibitor of both BTK and ITK. Proc Natl Acad Sci USA. 2015;112(9):E966-E972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kirschbaum M, Frankel P, Popplewell L, et al. Phase II study of vorinostat for treatment of relapsed or refractory indolent non-Hodgkin’s lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29(9):1198-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogura M, Ando K, Suzuki T, et al. A multicentre phase II study of vorinostat in patients with relapsed or refractory indolent B-cell non-Hodgkin lymphoma and mantle cell lymphoma. Br J Haematol. 2014;165(6):768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gryder BE, Sodji QH, Oyelere AK. Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med Chem. 2012;4(4):505-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hatzi K, Jiang Y, Huang C, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Reports. 2013;4(3):578-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayer CT, Gazumyan A, Kara EE, et al. The microanatomic segregation of selection by apoptosis in the germinal center. Science. 2017;358(6360):eaao2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khodadoust MS, Olsson N, Wagar LE, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543(7647):723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]