Abstract
For over 50 years, the International HLA and Immunogenetics Workshops (IHIW) have advanced the fields of histocompatibility and immunogenetics (H&I) via community sharing of technology, experience and reagents, and the establishment of ongoing collaborative projects. Held in the fall of 2017, the 17th IHIW focused on the application of next generation sequencing (NGS) technologies for clinical and research goals in the H&I fields. NGS technologies have the potential to allow dramatic insights and advances in these fields, but the scope and sheer quantity of data associated with NGS raise challenges for their analysis, collection, exchange and storage. The 17th IHIW adopted a centralized approach to these issues, and we developed the tools, services and systems to create an effective system for capturing and managing these NGS data. We worked with NGS platform and software developers to define a set of distinct but equivalent NGS typing reports that record NGS data in a uniform fashion. The 17th IHIW database applied our standards, tools and services to collect, validate and store those structured, multi-platform data in an automated fashion. We have created community resources to enable exploration of the vast store of curated sequence and allele-name data in the IPD-IMGT/HLA Database, with the goal of creating a long-term community resource that integrates these curated data with new NGS sequence and polymorphism data, for advanced analyses and applications.
Keywords: International Workshop, 17th IHIW, Next Generation Sequencing, HLA, Database, Data Management, XML, HML
1. Introduction
1.1. The Histocompatibility Workshops
Since their introduction in 1964, the Histocompatibility Workshops have been forums for the exchange of community knowledge and experience, allowing histocompatibility and immunogenetics (H&I) researchers, clinicians and technologists to evaluate new methods and technologies, establish standards and advance ongoing collaborative projects. Sixteen International HLA and Immunogenetics Workshop (IHIW) meetings have been held on five continents over the last half-century[1–16], and the 17th IHIW was held in northern California in the fall of 2017, continuing many long-standing workshop projects.
The advent of next-generation sequencing (NGS) based genotyping technologies has allowed new insights and innovations for the fields of histocompatibility, immunogenetics and immunogenomics. The 17th IHIW’s ultimate goals were to advance H&I basic research and clinical efforts through the application and evaluation of NGS HLA and KIR genotyping technologies, and to foster the development of NGS technologies tailored to meet the H&I community’s needs, building on the technological and scientific momentum of the previous sixteen workshops.
Toward those ends, we developed systems, standards and tools for the collection, storage and management of NGS HLA genotyping data (the HLA genotype and associated consensus sequences) generated for 17th IHIW projects. The goals of this effort were to build on the data-collection and -storage experiences of previous workshops, and produce NGS data-managing tools that will support IHIW efforts and persist as public resources after the 17th IHIW. Here, we provide a brief overview of the challenges faced in organizing coordinated data-generation and -collection efforts, the strategies we applied, and the tools, standards and services we developed to address these challenges.
1.2. The Challenges of Coordinated Data Collection
The collection, storage and analysis of data have been key issues of all workshops. Many workshops have used centralized databases[17–21], while in several of the more recent workshops, individual components and projects were responsible for collecting, managing and analyzing data[22–32]. Centralized data-management requires close communication between workshop participants and leaders, instrument and software vendors, and database developers to achieve consensus regarding required data content, data formats, reporting guidelines and quality standards. Sufficient time is also required for all parties involved to develop both the systems and tools to manage data, and the preliminary data on which to test the tools.
1.2.1. Reference Data Management
The specifics of the H&I field bring additional challenges that any data-management and analysis approach, centralized or decentralized, must address[33]. The body of HLA sequence data and associated allele names curated by the IPD-IMGT/HLA Database[34] (Reference Database) increases every four months; because workshop data-generation efforts often span multiple years, the details of the pertinent Reference Database version under which each HLA genotype was generated must be collected along with the genotyping data. The collection and management of such genotyping meta-data (Table 1) can be just as important for the workshop effort as the genotyping data themselves; without them it may not be possible to determine the extent to which datasets generated years apart or using different methods are equivalent. When workshop efforts span time periods that include major changes to the nomenclature[35, 36], these problems are only compounded.
Table 1.
Data-Elements in 17th IHIW Typing Reports
| Data-Elements | Data Type | Description | Typing Report Formata |
|---|---|---|---|
| 17th IHIW Lab-code | Identifier | A 6-character code provided by the 17th IHIW to identify each participating laboratory. | ABCDE |
| Report ID | Identifier | A code provided by the originating lab to identify each report. | ABCDE |
| Specimen ID | Identifier | A 17th IHIW code that uniquely identifies the specimen that was genotyped. | ABCDE |
| Instrument | Meta-data | Parameters that document the name, manufacturer, model, and on-board software of each instrument used to generate the typing. | AB |
| Reagent Protocol | Meta-data | Parameters that document the name, manufacturer, and reference source for any reagents or kits used to generate the typing, along with protocol deviations. | AB |
| Software | Meta-data | Parameters that document the name, manufacturer and version of each program used to generate the typing, along with the use to which that program was applied, and any non-default parameters applied. | ABC |
| Reference Database Version | Meta-data | Documentation of the IPD-IMGT/HLA Database release version(s) used for the sequence alignment and base-calling that generated the consensus sequence and genotype. | ABCDE |
| Reference Sequence | Meta-data | The identifiers for the reference sequences used for the sequence alignment and base calling that generated the consensus sequence and genotype | CDE1 |
| Locus | Genotyping data | The locus associated with each genotype and consensus sequence. | ABCDE |
| Genotype | Genotyping data | A genotype written in GL-String format[40] for each locus typed. | ABDE2 |
| Consensus Sequence | Genotyping data | A nucleotide sequence representing a contiguous phased region of DNA. | ABCDE |
| Sequence Coordinate | Meta-data | The start and end positions of the consensus sequence(s) with respect to the reference sequence. | ABCDE |
| Phasing | Meta-data | Parameters that describe the phase relationships between the consensus sequences at each locus. | ABCDE |
| Sequence Feature | Meta-data | The gene feature or features (exons, introns or untranslated regions) represented by the consensus sequence | AB3 |
| Sequence Quality | Meta-data | The mean depth of reads used to generate a given consensus sequence | AC |
| Typing Annotation | Meta-data | A structured notation for identifying instances when allele names included in the genotype are the closest matches to the consensus sequence, but do not correspond exactly to the reported consensus sequence. | A4 |
| Novel Polymorphism | Genotyping data | A description of any novel polymorphism detected. | ABCDE |
| FASTQ Location | Meta-data | The name and location (in the WS Database, or online) of the primary (“raw”) FASTQ data for each genotype | ACDE |
IHIW: International HLA and Immunogenetics Workshop
GL: Genotype List
IPD-IMGT: ImmunoPolymorphism Database-ImMunoGeneTics
For each data-element, the typing report format in which it is found it is listed. As referenced in Figure 1, A: Manual IHIW XML; B: Illumina and laboratory-generated IHIW XML; C: GenDx XML; D: HLA Twin, HistoGenetics, MIA FORA and TypeStream Visual HML; E: HLA Twin and MIA FORA HML.
The A and B formats use the reference sequences in Table 3.
The WS Database conversion daemon generates GL Strings for format C.
The C, D, and E formats use the “Genomic - Unknown Location” sequence feature.
The hlaPoly tool identifies this information for all typing report formats.
1.2.2. Primary Data Management
The nature of the primary or “raw” data, from which all experimental data and meta-data are ultimately derived, can vary widely from method to method and from project to project. This was particularly pronounced for the molecular genotyping methods applied in the 11th through the 16th workshops, where multiple reference strand conformation analysis (RSCA), sequence-specific (SS) oligo (SSO), reverse SSO (rSSO), SS priming (SSP) and sequence-based typing (SBT) methods were in use, each with its own distinct type of primary data.
1.2.3. Allele Name Data Management
Allele name data must be recorded and managed in a standard manner to facilitate meaningful data-analysis. For many previous workshops, the management of HLA allele names was performed by humans, and involved data recorded in paper documents or spreadsheets in a variety of different ways. Humans are adept at “figuring out” the true meaning of unusual notations and spreadsheet-initiated errors that may occur, but machines are not. For example, “HLA-A*02:99” and “HLA-A*03:01:02” are often recorded as “02:99” or “03:01:02” in spreadsheet columns labeled “HLA-A”, “A”, etc.; however, common spreadsheet applications may change “02:99” to “0.152083333333333” or “3:39”, and “03:01:02” to “3:01:02”, all of which erroneously represent times instead of alleles. The range of potential human-generated transcription errors is large. Previous workshops devoted considerable manual effort to review, identify and correct errors, and standardize allele-name notations prior to analysis. However, the analysis, collection, exchange and storage of NGS genotyping data requires machines (computers) that are able to process allele name data, and the accompanying nucleotide sequence data, without the human ability to identify and correct errors.
1.2.4. Describing Novel Polymorphism
The description of previously unknown (novel) HLA sequence variants has been a long-standing challenge for the H&I community. Until a novel sequence is named by the World Health Organization Nomenclature Committee for Factors of the HLA System (Nomenclature Committee)[37], it is very difficult to discuss that sequence in the context of the HLA nomenclature. The common practice, associated with pre-NGS genotyping, has been to append a “novel-allele” identifier to a truncated version of a related allele name (e.g. “HLA-A*02V”, “HLA-A*02:NEW”, “HLA-A*02:01new”, etc.). The World Marrow Donor Association guidelines for the use of HLA nomenclature (WMDA guidelines) indicate that “NEW” should be reported for alleles that have not been named by the Nomenclature Committee[38]. However, the absence of a standard for describing novel HLA alleles and associated nucleotide sequences represents a considerable challenge for the collection of NGS HLA genotyping data.
2. Meeting the Challenge
The 17th IHIW adopted a centralized data-storage approach, in which all specimen-related data, reference data, genotyping data and associated meta-data were stored in a single database system. The goal of this effort was to facilitate data and analysis access for workshop participants, with these workshop products and the database itself made available to the H&I community after the workshop. The 17th IHIW focus on NGS provided an advantage for centralized data collection in that there are currently only a small number NGS platforms, which generate primary data in FASTQ[39] format, and associated genotyping software. A key 17th IHIW goal was to collect machine-generated HLA data for consumption by IHIW informatics services, with minimal human intervention. We worked with NGS software developers to define a small number of equivalent and interchangeable data reporting formats that allowed genotyping data and meta-data to be collected using a “uniform NGS data-collection” approach. This approach built on the genotype list (GL) string format[40] and the GL Service[41], the Minimum Information for Reporting Immunogenomic NGS Genotyping (MIRING) reporting guidelines and messaging standard[42], and the MIRING-compliant Histoimmunogenetics Markup Language (HML) version 1.0 messaging format[43].
2.1. Uniform NGS Data Collection
The 17th IHIW did not require all workshop projects or participating laboratories to use the same NGS platform, typing kit or protocol. NGS instruments manufactured by Illumina (e.g., MiSeq), One Lambda (e.g., S5XL), Pacific Biosciences (e.g., PacBio RSII) and Roche 454 (e.g., GS FLX) were used in 17th IHIW NGS genotyping efforts. The goal in uniform NGS data collection was that all NGS HLA genotyping data and associated meta-data (which together constitute a “typing report”) be compatible and comparable, so that all collected data were equally interpretable, regardless of the format in which those data were exchanged. This allowed data generated by different laboratories, in different countries, using different platforms and software, to be stored in one database and made available for multiple projects.
Toward this end, the 17th IHIW accepted NGS genotyping data and meta-data in three MIRING-compliant eXtensible Markup Language (XML)[44] based typing report document formats – HML (version 1.0.1); GenDx XML, exported by GenDx NGS Engine version 2.4.0; and IHIW XMLA, a format developed specifically for the 17th IHIW (Supplements A and B). HML was generated by HistoGenetics, Omixon HLA Twin (version 1.1.4.2), Immucor MIA FORA (version 3.1) and One Lambda TypeStream Visual (version 1.1) software. IHIW XML typing reports were generated using the 17th IHIW Database (WS Database) system (described in section 2.2), by individual laboratories (using the Supplementary Materials), and by Illumina, using a “.cgp” file exported by TruSight HLA Assign version 2.1 RUO. We were unable to define a specific typing report document format for data generated on Pacific Biosciences instruments, but presumably such data could have been reported in HML or IHIW XML format. In addition, the WS Database accepted HLA genotypes in a comma-separated values (CSV) file generated by Scisco Genetics.
GenDx XML, HML and IHIW XML typing reports include subsets of the NGS genotyping data and meta-data elements described in Table 1. These data-elements are equivalent to MIRING elements 1–8[42]. HML or GenDx XML typing reports may include additional data, but because these document formats include equivalent 17th IHIW data-elements, all submitted HML and GenDx XML HLA typing reports were converted into IHIW XML typing reports (described in section 2.2.1), which were subsequently stored in the WS Database. In addition to these typing reports, the primary FASTQ data, too large to include in a report, were stored on a secure File Transfer Protocol (sFTP) server linked to the WS Database.
2.2. 17th IHIW Database
The WS Database included an Oracle SQL database (12c Standard Edition) and a web application built with APEX 5.0, running on a multi-core Linux CentOS 6 platform with 960GB of storage, expandable up to 3TB. The 17th IHIW sFTP server was an IBM high-performance computing cluster running Linux RedHat 6, with a 1Gbps Ethernet connection. The server comprised a management node, three compute nodes, two storage nodes and 15 TB of storage. The database and archived redo logs were backed up to the sFTP server with Oracle Recovery Manager (RMAN) on a daily basis, with each backup maintained for five days. The WS Database schema is illustrated in Supplementary Figure S1. WS Database tools and services were scripted in the Perl, R or Python programming languages. Both the database and the sFTP server were housed in the high-performance computing Stanford Data Center facility on the Stanford Linear Accelerator Center campus, and were managed by the Stanford Research Computing staff.
The WS Database’s structure reflected the workshop’s organization and the defined roles of workshop participants. Each of the six 17th IHIW components – NGS of HLA, NGS of KIR, Hematopoietic Cell Transplantation, Mapping of Serologic Epitopes, Informatics of Genomic Data, and Quality Control & Quality Assurance – was led by a Component Chair (or Chairs). Projects were associated with each component, with a Principal Investigator (PI) for each project. PIs enrolled Lab Members, and could enroll in Components and Projects. Lab Members uploaded and managed data, and could enroll as Project and Component Affiliates. Further details of these participant roles can be found onlineB.
The WS Database systemC stored data from typing reports and Scisco CSV tables, FASTQ files, subject and specimen data, and pedigrees (PED format[45]), and managed the accounts and data-access privileges for 17th IHIW principal investigators and lab members, project leaders, and component affiliates and chairs. When laboratory-initiated subject IDs were submitted to the WS Database, those IDs were anonymized and linked to unique 17th IHIW IDs, which were used to identify those subjects in genotyping and analysis efforts, to avoid the distribution of protected health information. The WS Database also stored project-specific data, using custom document formats, and analytic results.
2.2.1. Participant Initiated Management of Typing Reports
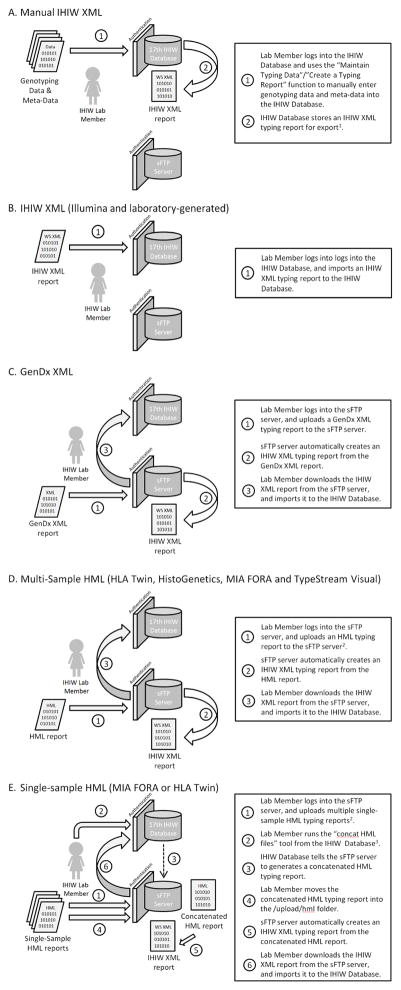
The submission and management of typing reports is illustrated in Figure 1. Genotyping data and meta-data could be manually entered into the WS Database, and laboratory-generated and Illumina IHIW XML typing reports were submitted directly to the WS Database. HML and GenDx XML typing reports were converted to IHIW XML reports by uploading them to the sFTP server, and using the WS Database tools to generate IHIW XML reports from them. These converted IHIW XML typing reports were stored in the WS Database, where they were available for download by participants. Regardless of their source, all IHIW XML typing reports were submitted to and stored in the WS Database. Detailed instructions on the 17th IHIW data-submission process are available onlineD.
Figure 1. Typing Report Submission Procedures.
Cartoon descriptions describing the key steps of the five 17th IHIW typing report submission processes are shown, with the steps defined in each inset box.
HML: Histoimmunogenetics Markup Language
IHIW: International HLA and Immunogenetics Workshop
sFTP: secure File Transfer Protocol
XML: eXtensible Markup Language
1: Though this final step is only shown in panel A, an IHIW XML typing report remains available for download from the WS Database after an IHIW XML report of any source is loaded into the WS Database.
2: WS Database watcher daemons monitor the sFTP server’s /upload/hml directory for the arrival of HML typing reports, and trigger the automatic conversion HML reports into IHIW XML reports. Multi-sample (project-level) HML reports should be loaded directly into the /upload/hml directory. Single-sample HML reports should be loaded into a user-created subdirectory of the /upload directory.
3: For the “concat HML files” tool to run, the Lab Member must supply the user-created subdirectory of the /upload directory in which the single-sample HML reports have been loaded.
A. Manual entry of genotyping data and meta-data to the WS Database
B. Illumina-generated or laboratory-generated IHIW XML
C. GenDx XML
D. Multi-sample (project-level) HML generated by HistoGenetics, HLA Twin, MIA FORA and TypeStream Visual
E. Single-sample HML generated by MIA FORA or HLA Twin
2.3. 17th IHIW Standards and Tools
To facilitate uniform NGS data collection for the 17th IHIW, we have adopted specific data-standards and conventions for the validation of typing reports, and the analysis of workshop data. The tools described in sections 2.3.1 to 2.3.4 are available on GitHubE,F.
2.3.1. Typing Report Validation
Given the number of typing report formats accepted by the WS Database, we developed a number of tools and services for validating the format and content of each. Several of these tools were built into the WS Database, and ran when typing reports were uploaded or created in the system. The semantic validations and WS Database functions applied to each typing report format are listed in Table 2. Because HML and GenDx XML typing reports were converted into IHIW XML reports, the validation and database functions listed for IHIW XML format were applied to all typing reports. In addition, software developers generating HML typing reports were encouraged to use the public MIRING validator for HML service (miring-validatorE)) as part of their development efforts. This validator determines if a potential HML typing report follows basic HML and MIRING rules of syntax, and if it contains MIRING dataelements. Because this validator operates as a web-service, it can be built into an HML typing report generation pipeline.
Table 2.
Validation and Database Functions Applied to each typing report format
| HML | Validation |
|
| Functions |
|
|
| GenDx | Functions |
|
| IHIW XML/Manual Entry | Validation |
|
| Functions |
|
HML: Histoimmunogenetics Markup Language
IHIW: International HLA and Immunogenetics Workshop
IUPAC: International Union of Pure and Applied Chemistry
XML: eXtensible Markup Language
Because HML and GenDx XML typing reports are converted to IHIW XML reports upon submission, the validation and functions listed for IHIW WS format are applied to all typing reports.
Validation results and details are provided in the WS Database system under “Lab Member”/”Tools”/”Job Log”.
2.3.2. IPD-IMGT/HLA Database Versions
As noted in section 1.2.1, the Reference Database is updated quarterly; the number of alleles increases with each release, and the extent of sequence known for a given allele, as well as the number of fields in a given allele name, can increase between database releases. We addressed this by “freezing” all WS Database functions at Reference Database version 3.25.0 (released July 2016). While HLA allele names described in other Reference Database versions could have been submitted using HML or GenDx XML, the WS Database translated those names to their 3.25.0 counterparts upon submission (described in section 2.3.4), and all HLA-related data were analyzed using Reference Database version 3.25.0, which was also the source of all reference allele sequences. Restricting the WS Database to a single Reference Database version in this way streamlined database functions, facilitated uniform data management and analysis, and will allow the final WS Database product to be updated to later Reference Database versions in future workshops. The Reference Database resources described below are available from the Reference Database FTP siteG.
To facilitate the use of Reference Database 3.25.0 for the 17th IHIW, we defined a set of full-length (“genomic”) version 3.25.0 reference alleles (Table 3) for use in generating and aligning consensus sequences, and identifying novel polymorphism. Though some alleles in this set have names with fewer than four fields, indicating that no synonymous or non-coding polymorphism has been identified for those alleles as of Reference Database version 3.25.0, genomic sequence was available for all of them. When possible, a reference allele was been identified for each allele family at a locus, but for some loci only a single full-length reference allele was identified.
Table 3.
Full-length HLA Reference Alleles in IPD-IMGT/HLA Database Version 3.25.0
| Locus | Accession Number | Allele Name | Descriptiona |
|---|---|---|---|
| HLA-A | HLA00001 | HLA-A*01:01:01:01 | HLA-A Reference A*01 Reference A*36 Reference |
| HLA00005 | HLA-A*02:01:01:01 | A*02 Reference | |
| HLA00037 | HLA-A*03:01:01:01 | A*03 Reference | |
| HLA00043 | HLA-A*11:01:01:01 | A*11 Reference | |
| HLA00048 | HLA-A*23:01:01 | A*23 Reference | |
| HLA00050 | HLA-A*24:02:01:01 | A*24 Reference | |
| HLA00071 | HLA-A*25:01:01 | A*25 Reference A*26 Reference |
|
| HLA00085 | HLA-A*29:01:01:01 | A*29 Reference | |
| HLA00089 | HLA-A*30:01:01 | A*30 Reference | |
| HLA00097 | HLA-A*31:01:02:01 | A*31 Reference | |
| HLA00101 | HLA-A*32:01:01 | A*32 Reference | |
| HLA00104 | HLA-A*33:01:01 | A*33 Reference | |
| HLA00108 | HLA-A*34:01:01 | A*34 Reference | |
| HLA00112 | HLA-A*66:01:01 | A*43 Reference A*66 Reference |
|
| HLA05918 | HLA-A*68:01:01:02 | A*68 Reference A*69 Reference |
|
| HLA05527 | HLA-A*74:02:01:02 | A*74 Reference | |
| HLA00130 | HLA-A*80:01:01:01 | A*80 Reference | |
| HLA-B | HLA00132 | HLA-B*07:02:01 | HLA-B Reference B*07 Reference B*82 Reference B*83 Reference |
| HLA00146 | HLA-B*08:01:01:01 | B*08 Reference | |
| HLA00152 | HLA-B*13:01:01 | B*13 Reference | |
| HLA00157 | HLA-B*14:01:01 | B*14 Reference | |
| HLA00162 | HLA-B*15:01:01:01 | B*15 Reference | |
| HLA00213 | HLA-B*18:01:01:01 | B*18 Reference | |
| HLA00221 | HLA-B*27:02:01 | B*27 Reference | |
| HLA00237 | HLA-B*35:01:01:01 | B*35 Reference | |
| HLA00265 | HLA-B*37:01:01 | B*37 Reference | |
| HLA00267 | HLA-B*38:01:01 | B*38 Reference B*39 Reference |
|
| HLA00292 | HLA-B*40:01:02 | B*40 Reference | |
| HLA13397 | HLA-B*40:305 | B*41 Reference | |
| HLA00315 | HLA-B*42:01:01 | B*42 Reference | |
| HLA00318 | HLA-B*44:02:01:01 | B*44 Reference | |
| HLA00329 | HLA-B*45:01:01 | B*45 Reference | |
| HLA00331 | HLA-B*46:01:01 | B*46 Reference | |
| HLA14088 | HLA-B*47:01:01:03 | B*47 Reference | |
| HLA00335 | HLA-B*48:01:01 | B*48 Reference B*81 Reference |
|
| HLA00340 | HLA-B*49:01:01 | B*49 Reference B*50 Reference |
|
| HLA00344 | HLA-B*51:01:01:01 | B*51 Reference B*52 Reference B*78 Reference |
|
| HLA00364 | HLA-B*53:01:01 | B*53 Reference B*58 Reference |
|
| HLA00367 | HLA-B*54:01:01 | B*54 Reference B*55 Reference B*56 Reference B*59 Reference |
|
| HLA00381 | HLA-B*57:01:01 | B*57 Reference | |
| HLA00390 | HLA-B*67:01:01 | B*67 Reference | |
| HLA00392 | HLA-B*73:01 | B*73 Reference | |
| HLA-C | HLA00401 | HLA-C*01:02:01 | HLA-C Reference C*01 Reference |
| HLA00405 | HLA-C*02:02:02:01 | C*02 Reference | |
| HLA01543 | HLA-C*03:02:02:01 | C*03 Reference | |
| HLA00420 | HLA-C*04:01:01:01 | C*04 Reference | |
| HLA00427 | HLA-C*05:01:01:01 | C*05 Reference | |
| HLA00430 | HLA-C*06:02:01:01 | C*06 Reference | |
| HLA00433 | HLA-C*07:01:01:01 | C*07 Reference | |
| HLA00445 | HLA-C*08:01:01 | C*08 Reference | |
| HLA00454 | HLA-C*12:02:02 | C*12 Reference | |
| HLA00462 | HLA-C*14:02:01:01 | C*14 Reference | |
| HLA00467 | HLA-C*15:02:01:01 | C*15 Reference | |
| HLA00475 | HLA-C*16:01:01:01 | C*16 Reference | |
| HLA00481 | HLA-C*17:01:01:01 | C*17 Reference | |
| HLA00483 | HLA-C*18:01 | C*18 Reference | |
| HLA-DPA1 | HLA06604 | HLA-DPA1*01:03:01:02 | HLA-DPA1 Reference DPA1*01 Reference DPA1*03 Reference DPA1*04 Reference |
| HLA00505 | HLA-DPA1*02:01:02 | DPA1*02 Reference | |
| HLA-DPB1 | HLA00517 | HLA-DPB1*02:01:02 | HLA-DPB1 Reference |
| HLA-DQA1 | HLA00601 | HLA-DQA1*01:01:01:01 | HLA-DQA1 Reference |
| HLA-DQB1 | HLA00622 | HLA-DQB1*02:01:01 | HLA-DQB1 Reference |
| HLA-DRB1 | HLA00664 | HLA-DRB1*01:01:01 | HLA-DRB1 Reference DRB1*01 Reference DRB1*10 Reference |
| HLA00671 | HLA-DRB1*03:01:01:01 | DRB1*03 Reference | |
| HLA00685 | HLA-DRB1*04:01:01:01 | DRB1*04 Reference | |
| HLA00719 | HLA-DRB1*07:01:01:01 | DRB1*07 Reference | |
| HLA00727 | HLA-DRB1*08:03:02 | DRB1*08 Reference | |
| HLA09928 | HLA-DRB1*09:21 | DRB1*09 Reference | |
| HLA00751 | HLA-DRB1*11:01:01:01 | DRB1*11 Reference | |
| HLA14829 | HLA-DRB1*12:01:01:02 | DRB1*12 Reference | |
| HLA00797 | HLA-DRB1*13:01:01:01 | DRB1*13 Reference | |
| HLA00837 | HLA-DRB1*14:05:01 | DRB1*14 Reference | |
| HLA-DRB1 | HLA03453 | HLA-DRB1*15:01:01:02 | DRB1*15
Reference DRB1*16 Reference |
| HLA-DRB3 | HLA00887 | HLA-DRB3*01:01:02:01 | HLA-DRB3 Reference |
| HLA-DRB4 | HLA00905 | HLA-DRB4*01:01:01:01 | HLA-DRB4 Reference |
| HLA-DRB5 | HLA00915 | HLA-DRB5*01:01:01 | HLA-DRB5 Reference |
While each locus has at least one reference allele (e.g., HLA-A Reference), reference alleles for some allele families at a given locus are also identified (e.g., A*01 Reference). In some cases, the same allele may serve as a reference for multiple allele families.
Full gene sequence (i.e., for all exons, introns and UTRs) is available for all alleles on this table. Allele names for those alleles that include only two or three fields (e.g., HLA-B*73:01 or HLA-B*07:02:01) indicate that no synonymous or non-coding polymorphism, respectively, has been identified for those alleles as of Reference Database release version 3.25.0. Each allele family reference was selected on the basis of close sequence-identity between that reference allele and the alleles in that family.
2.3.3. Genotype Format and Validation
The large variety of formats used in the H&I community to record HLA genotypes and describe typing ambiguity made it difficult to collect genotyping data in a uniform manner. We addressed this by collecting all HLA genotypes in GL string format[40], using the strict-mode GL Service[41] to validate the allele content of the GL string, and applying python scripts (pyglstringE) to validate the structure of the GL string. Data submitters were notified when GL strings failed validation (see section 2.3.5.2.1), and were requested to modify them accordingly.
These structural validation scripts we applied include an exception for the DRB3, DRB4 and DRB5 loci (the secondary DRB loci), permitting combinations of alleles at these loci to be connected by the GL string “+” operator (e.g., “HLA-DRB3*01:26N+HLA-DRB5*01:01:01”), whereas for other loci, the “+” operator connects only alleles of a single locus. When GL string format was introduced[40], the “+” operator denoted the number of copies of a given gene present in an individual. Ideally, given the structural haplotype variation known for the DRB loci[46], when the absence of a DRB3, DRB4 or DRB5 locus can be determined, the absence of that locus should be noted in a GL string. The WMDA guidelines indicate that the absence of any allele at a secondary DRB locus be reported using “NNNN” (e.g., “HLADRB1* NNNN”)[38], but this is not a widely used approach. Without a standard nomenclature for describing the confirmed absence of a secondary DRB gene, we treat these loci as alleles of a single locus. The development of a nomenclature for describing the confirmed absence of a locus (e.g. “HLA-DRB3* NNNN”, “HLA-DRB3*00:00” or “HLA-DRB3*ABSENT”) should be considered by the Nomenclature Committee.
2.3.4. LiftOver Tool
As typing reports were accepted into the WS Database, HLA genotypes identified under Reference Database versions other than 3.25.0 were translated to their 3.25.0 counterparts via a LiftOver tool (IHIW17LiftOver.pmF). Non-3.25.0 alleles are translated on the basis of their Reference Database accession numbers, as related in the Allelelist_history.txt fileG. In cases of alleles named after version 3.25.0 (e.g., HLA-A*01:01:01:05, identified in Reference Database version 3.27.0), the submitted allele name is translated to either the lowest-numbered 3.25.0 allele name with the greatest number of matching lower-order fields to the submitted allele (e.g., HLA-A*01:01:01:01 is chosen to replace HLA-A*01:01:01:05), or the reference allele for that locus (Table 3) when there are no matching lower-order fields, and the submitted allele is noted in the “Novelpolymorphism” field for that genotype (e.g. as “IPD-IMGT/HLA-3270-HLA-A*01:01:01:05”). We note that it remains unclear if a typing system that e.g., identified HLA-A*01:01:01:05 under Reference Database version 3.27.0 would return a result of HLA-A*01:01:01:01 for the given specimen under version 3.25.0. However, we felt that storage of the submitted allele name in the Novelpolymorphism field would be sufficient to allow these instances to be investigated. In cases where allele name changes that occurred in Reference Database versions prior to 3.25.0 resulted in accession number changes (e.g., HLA-DRB1*08:01:03, with accession number HLA02257, was changed to HLA-DRB1*08:01:01, with accession number HLA00723, as part of Reference Database version 3.24.0, as detailed in Table 4), the version 3.25.0 allele name was used.
Table 4.
HLA Allele Remapping for WS Database LiftOver and Consensus Linking Functions
| Reference Database Release in which the change occurred |
Rationale | Original Allele Name | Original Accession Number |
Current Allele Name | Current Accession Number |
Current Allele Present in Version 3.25.0? |
Accession Number Change? |
WS Database Action When Original Allele Name Has Been Submitted |
|---|---|---|---|---|---|---|---|---|
| 3.17.0 | Sequence identical | B*49:15 | HLA05834 | B*49:01:01 | HLA00340 | YES | YES | Use 3.25.0 version of Current Allele Name |
| 3.20.0 | Sequence renamed | A*26:03:02 | HLA04741 | A*26:111 | HLA04741 | YES | NO | |
| 3.21.0 | Sequence error | DRB1*11:11:02 | HLA02157 | DRB1*11:11:01 | HLA00765 | YES | YES | |
| 3.22.0 | Sequence error | A*03:194 | HLA11939 | A*03:213 | HLA12966 | YES | YES | |
| 3.24.0 | Sequence error | A*23:69 | HLA12676 | A*23:01:01:01 | HLA00048 | YES1 | YES | |
| Sequence identical | DRB1*08:01:03 | HLA02257 | DRB1*08:01:01 | HLA00723 | YES | YES | ||
| 3.25.0 | Sequence renamed | DRB1*04:94:02N | HLA14178 | DRB1*04:212N | HLA14178 | YES | NO | |
| 3.26.0 | Sequence renamed | A*30:02:12 | HLA09547 | A*30:100 | HLA14873 | YES | YES | Use 3.25.0 version of Original Allele Name |
| Sequence error | C*17:01:01:01 | HLA00481 | C*17:01:01:02 | HLA04311 | YES | YES | ||
| 3.27.0 | Sequence renamed | DPB1*35:01:02 | HLA04110 | DPB1*621:01 | HLA04110 | NO2 | NO | |
| Sequence identical | DQB1*06:220 | NA3 | DQB1*06:217 | HLA16016 | NO2 | NA3 | Change to DQB1*06:01:014 | |
| 3.28.0 | Sequence identical | DPA1*02:02:01 | HLA00508 | DPA1*02:07:01:01 | HLA15619 | NO5 | YES | Use 3.25.0 version of Original Allele Name |
| 3.29.0 | Sequence identical | DQB1*03:01:01:13 | HLA07476 | DQB1*03:01:01:07 | HLA17167 | NO6 | YES | Change to DQB1*03:01:01:017 |
Changes to HLA allele names and their associated accession numbers that occurred in Reference Database versions 3.15.0 – 3.29.0, and the action taken by the WS Database when the original allele name is encountered are shown. The data in all but the last column are derived from the hla_nom.txt and allelelist_history.txt files in Reference Database version 3.28.0.
NA: Not applicable.
The allele name in Reference Database version 3.25.0 is A*23:01:01.
This current allele name was assigned in Reference Database version 3.27.0.
No accession number was released for the DQB1*06:220 allele; this allele name has not appeared in any release version.
DQB1*06:01:01 is the lowest numbered allele sharing a common prefix (DQB1*06) with DQB1*06:220 (or DQB1*06:217).
This current allele name was assigned in Reference Database version 3.28.0.
Both the original and current allele names were assigned in Reference Database version 3.29.0.
DQB1*03:01:01:01 is the lowest numbered allele sharing a common prefix (DQB1*03:01:01) with DQB1*03:01:01:13 (or DQB1*03:01:01:07)
When ambiguous HLA genotypes were submitted, the LiftOver tool evaluated ambiguous alleles (delimited with the GL string slash [/] operator) and ambiguous genotypes (delimited with the GL string pipe [|] operator), and identified alleles and genotypes that could be translated to their 3.25.0 counterparts (Figure 2). These alleles were translated, and the GL string consolidated to eliminate duplications. If an ambiguous HLA genotype consisted entirely of alleles named after Reference Database version 3.25.0, the LiftOver tool translated those alleles to the corresponding lowest-numbered 3.25.0 alleles with the greatest number of matching lower-order fields, as described above, and consolidated the GL string. In all cases, the submitted non-3.25.0 GL strings were stored in the “Original_GL” field for that genotype. These allelic and GL string LiftOver functions were accomplished using a modified version of the Allelelist_history.txt file that includes data from the hla_nom.txtG files and Table 3 (IHIW17_AllelelistGgroups_history.txtF). This LiftOver process occurred when HML and GenDx XML typing reports were converted to IHIW XML reports. All collected IHIW XML typing reports corresponded to version 3.25.0. However, the LiftOver tool can be modified to allow all collected typing reports to be updated to a future Reference Database version.
Figure 2. Example of LiftOver of Ambiguous HLA-A Genotypes.
Examples of the LiftOver process for three ambiguous HLA-A genotypes consistent with Reference Database version 3.28.0 are shown. Allele names that are included in Reference Database 3.25.0 are shown in boldface.
A. LiftOver process for slash-delimited ambiguous allele strings that include 3.25.0 alleles.
B. LiftOver process for pipe-delimited ambiguous genotype strings that include 3.25.0 alleles.
C. LiftOver process for slash-delimited allele strings that do not include 3.25.0 alleles.
2.3.5. 17th IHIW Database Tools and Functions
Reference Database version 3.25.0 included 12.9 million bases of sequence for 14,957 HLA alleles at 19 HLA loci. Of this, more than 40,000 exons comprised 9 million bases of sequence, making this a rich, but very complex, data resource. We developed user-facing front end tools to assist 17th IHIW participants in working with these data, and data-facing back end tools to facilitate the integration of the large quantities of new sequence that were be generated via NGS.
2.3.5.1. Front End Tools
2.3.5.1.1. hlaPoly
The absence of a standard method for describing novel nucleotide polymorphism in consensus sequences posed challenges for our uniform data collection approach. Typing reports generated for the same specimen using different genotyping software could include identical consensus sequences and genotypes, but when a consensus sequence includes novel polymorphism, the Reference Database version, reference allele sequence, and sequence coordinate system used to describe that polymorphism could vary between software applications, and typing reports generated by different software may have identified different novel polymorphism for identical consensus sequences. For example, the nucleotide sequence of the HLA-A*01:01:01:05 allele differs from the HLA-A*01:01:01:01 allele in Reference Database 3.25.0 at three intron 2 nucleotide positions, and differs from the HLA-A*01:01:01:03 allele in Reference Database 3.25.0 at those same three positions as well as at an intron 1 position; the reference allele used to align the HLA-A*01:01:01:05 consensus sequence informed the description of novel polymorphism.
To standardize novel polymorphism description for the 17th IHIW, we developed the hlaPoly R packageF, which identifies novel polymorphism for a given consensus sequence, when provided with the closest matching allele name (which was usually included in the genotype) and the Reference Database version (version 3.25.0). The hlaPoly tool was deployed online as a Shiny applicationH. As illustrated in Supplementary Figure S2, hlaPoly uses the DECIPHER R package[47] to generate a multiple sequence alignment for the full-length HLA reference allele sequence (Table 3), the sequence of the pertinent allele in the genotype (closest allele) and the consensus sequence, and then retrieves the mismatches and indels between the consensus sequence and the called allele as novel polymorphism. If no sequence is known for the called allele in an aligned region, the mismatches and indels between the consensus sequence and the full-length HLA reference allele are retrieved. For each novel polymorphism, the feature number and start/end position relative to that feature are also calculated. The WS Database stored these novel polymorphism data in both a tabular form (see the bottom of Figure S1) and a string format (described in Supplement A).
2.3.5.1.2. Quick Calculation of Feature Position
To assist in manual entry of genotyping data and meta-data into the WS Database, we developed a tool for the calculation of gene-feature information. Given an allele name and the nucleotide position relative to the start of known nucleotide sequence for that allele, this tool returned the feature name (e.g. Exon 2), feature ID and the relative nucleotide position in that feature. This tool was available in the WS Database under “Lab Member”/”Tools”/”IMGT/HLA Feature List”.
2.3.5.1.3. Concatenate HML files
Each HML file uploaded to the sFTP server was treated as single typing report, and as suggested in Figure 1, some HML typing reports were generated for individual samples. Rather than requiring that hundreds or thousands of individual-sample HML files be converted to IHIW XML files, each of which would need to be manually loaded into the WS Database, we provided a tool (concathml.plF) that concatenates multiple HML files into a single HML file, which can be converted into a single IHIW XML file for loading. This “Concat HML files” tool was available in the WS Database under “Lab Member”/”Tools”.
2.3.5.1.4 “Convert HML to IHIW XML” and “Convert GenDX XML to IHIW XML”
As noted in Figure 1, the sFTP server automatically generated an IHIW XML typing report when an HML report was loaded into the /hml directory. The server also generated an IHIW XML report when an GenDX XML report was loaded into the /gendx directory. The “Convert HML to IHIW XML” and “Convert GenDx XML to IHIW XML” tools could be used to force these automatic functions to run immediately, or to manually convert HML or GenDx XML typing reports that had been loaded into other directories. Both tools were available in the WS Database under “Lab/Member”/”Tools”.
2.3.5.2. Back End Tools
2.3.5.2.1. Watcher Daemons
To monitor activity on the sFTP server, we developed daemons that detected new HML and GenDx XML files as they were uploaded to the sFTP server, automatically converted them to IHIW XML files, and validated them during the conversion. Any validation errors were logged and made available under “Lab Member”/”Tools”/”Job Log” in the WS Database. A second set of daemons performed daily checks for new typing reports in the WS Database. These daemons ran hlaPoly for newly added or edited consensus sequences, and stored the novel polymorphism results in the WS Database.
2.3.5.2.2. Consensus Linking
Genotypes and consensus sequences are recorded separately in HML typing reports. Each consensus sequence is associated with the reference allele used align it, which is usually a full-length allele, but is not directly linked to specific alleles in the associated genotype. For cases when these reference alleles are not included in the genotype, we developed a consensus linking tool that identified the allele in the genotype that most closely matched the reference allele, using the same approach applied for the LiftOver process (described in section 2.3.4). For example, if HLA-A*11:01:01:01 and *31:01:02:01 are the respective reference alleles for consensus sequences A and B, which are associated with the HLA-A*11:01:28+HLA-A*31:01:07, the consensus linking tool would associate HLA-A*11:01:28 with consensus sequence A and *31:01:07 with consensus sequence B.
2.4. Support for 17th IHIW Projects
In addition to its collection, validation and storage functions, the WS Database supported 17th IHIW projects by integrating tools for HLA data analysis and exchange. An updated version of PyPop[48]I, supporting colon-delimited allele names, with increased multi-locus analysis capacity, was accessible through the WS Database system. Similarly, integration of Gene Feature Enumeration[49] (GFE) functions (e.g., the feature-serviceE,J, GFE serviceE,K and Allele-Calling ToolE,L) allows full-gene HLA sequences to be exchanged and analyzed in the absence of an HLA allele name.
2.5. Validation of Tools and Functions
The data collections tools and functions described in section 2.3 were validated iteratively, as part of the process of working with NGS vendors to develop each typing report format. As new submitters provided data (e.g, a new vendor generating HML, or a new laboratory generating IHIW XML) the initial data imported to the WS Database were compared manually via comparison with the submitted typing reports; in instances of discrepancies, either feedback was provided to the submitter, or the tools and functions were updated to accommodate the “new” typing report format.
2.5.1 Genotyping Expectations
In 2014, a panel of 50 IHWG reference cell lines that had been well-characterized using pre-NGS HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1, -DRB3, -DRB4 and -DRB5 typing methods was assembled for a 17th IHIW NGS genotyping pilot project. HLA genotyping for this pilot project cell panel was independently performed by 15 laboratories using multiple NGS methods. This set of 15 NGS genotyping results was used to establish concordance expectations for the initial quality control (QC) and ongoing proficiency testing (PT) required for all 17th IHIW participants performing NGS genotyping. Random subsets of 24 pilot project cell lines were sent to each genotyping laboratory for QC/PT, and we used these QC/PT results to validate the genotype-data manipulations and transformations performed by the tools, functions and services described in section 2.3.
2.5.2 Manual Validation of Performance
Parsers and converters that did not depend on algorithmic transformations were validated by manual inspection of the results. For example, during hlaPoly development, output tables were validated manually, by comparison with novelPolymorphism results included in typing reports. The LiftOver process was validated using the typing reports generated by each laboratory as part of the QC/PT process.
3. Conclusions
We addressed several of the long-standing challenges to uniform NGS HLA data-collection and -storage for the 17th IHIW by developing new tools and formats, and adopting existing standards and services. NGS vendors worked with us to develop equivalent NGS HLA typing report formats that ensured data-portability across 17th IHIW projects. We ensured data-quality by validating all typing reports before they were loaded to the WS Database. All HLA genotyping data were recorded using the same Reference Database version, and novel HLA polymorphism was described using the same reference alleles. This approach facilitated the basic and clinical research aims of 17th IHIW HLA Projects, and can be applied by the larger H&I community. The 17th IHIW was held in September of 2017. We hope to work with the organizers of the 18th IHIW so that the WS Database and its associated tools will serve as a persistent central H&I community resource that will ensure research and data continuity with future IHIW efforts.
Supplementary Materials
Acknowledgments
This work was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) grant R01AI128775 (BM, MM, SM), NIH National Institute of General Medical Sciences (NIGMS) grant R01GM109030 (BM, MM, SM), Office of Naval Research (ONR) grant N00014-08-1-1207 (BM and MM), and an overseas project funded by the Taiwanese Ministry of Science and Technology (MST) (CC). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the MST, NIAID, NIGMS, NIH, ONR, Taiwanese government or United States government. We thank the Stanford Blood Center for the support and promotion of the 17th IHIWS endeavor, Ken Yamaguchi for helpful discussions and manuscript review, Tamara Vayntrub for her tremendous administrative support of 17th IHIW efforts, and the histocompatibility and immunogenetics community and the International HLA and Immunogenetics Workshop Council for their continued dedication to and support of the International Workshops. We also thank President Barack H. Obama for his support and appreciation of American science and basic research; several of the authors owe their continuing scientific careers to the passage of the American Recovery and Reinvestment Act (ARRA), and while ARRA funds did not support this work, it would not have been possible without ARRA.
Abbreviations
- CSV
Comma-Separated Values
- GFE
Gene Feature Enumeration
- GL
Genotype List
- HLA
Human Leukocyte Antigen
- HML
Histoimmunogenetics Markup Language
- H&I
Histocompatibility and Immunogenetics
- IHIW
International HLA and Immunogenetics Workshop
- IMGT
ImMunoGeneTics
- IPD
ImmunoPolymorphism Database
- IUPAC
International Union of Pure and Applied Chemistry
- KIR
Killer-cell Immunoglobulin-like Receptor
- MIRING
Minimum Information for Reporting Immunogenomic NGS Genotyping
- NGS
Next Generation Sequencing
- PI
Principal Investigator
- RMAN
Recovery Manager
- RSCA
Reference Strand Conformation Analysis
- rSSO
Reverse Sequence-Specific Oligo
- SBT
Sequence-Based Typing
- sFTP
secure File Transfer Protocol
- SS
Sequence-Specific
- SSO
Sequence-Specific Oligo
- SSP
Sequence-Specific Priming
- WMDA
World Marrow Donor Association
- WS
Workshop
- XML
eXtensible Markup Language
Footnotes
ONLINE RESOURCES
A. BioSharing.org record for International HLA and Immunogenetics Workshop XML; https://biosharing.org/bsg-s000700l Accessed April 24, 2017.
B. Instructions for using the 17th IHIWS Database; https://ihiws17.stanford.edu/ihiw_docs/17WS_Database_Manual_Registration_v1.pdf; Accessed April 25, 2017.
C. Login portal for the 17th IHIW Database; http://workshop.ihiws.org/; Accessed April 25, 2017.
D. Instructions for entering & uploading IHIW Typing reports; http://ihiws.org/wpcontent/uploads/2017/02/Instructions-for-entering_uploading-IHIWS-Typing-report_Version7.pdf; Accessed April 25, 2017.
E. NMDP/Be The Match Bioinformatics Research GitHub repository; https://github.com/nmdp-bioinformatics; Accessed April 25, 2017.
F. 17th IHIW GitHub repository; https://github.com/IHIW/bioinformatics/tree/master/db_related; Accessed December 10, 2017; referenced files are in the “/data”, “/hlaPoly”, and “/scripts” directories.
G. IPD-IMGT/HLA Database FTP site; ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/; Accessed April 25, 2017; some referenced files are in the “/wmda” directory.
H. hlaPoly: Identifying Novel Polymorphisms in HLA Sequences; http://hlapoly.immunogenetics.org; Accessed April 25, 2017.
I. PyPop GitHub repository; https://github.com/alexlancaster/pypop; Accessed May 28, 2017.
J. Feature service web-interface; http://feature.b12x.org; Accessed May 15, 2017; the Feature service allows individual gene feature sequences to be registered, returning an accession number for that sequence, and dereferences accession numbers to identify specific gene feature sequences; code is available on the NMDP GitHub repository under “service-feature”.
K. GFE service; http://gfe.b12x.org; Accessed May 15, 2017; the GFE service accepts full-gene or multi-feature consensus sequence, splits it into individual features, which are registered with the Feature service, and returns a GFE notation; code is available on the NMDP GitHub repository under “service-gfe- submission”.
L. GFE Allele-Calling Tool; http://act.b12x.org/; Accessed May 18, 2017; the Allele-Calling Tool accepts full-gene consensus sequence, and identifies the closest matching HLA allele and corresponding GFE notation; code is available on the NMDP GitHub repository under “service-act”.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Scandinavian Journal of Haematology; Histocompatibility Testing 1965: Report of a Conference and Workshop Sponsored by the Boerhaave Courses for Postgraduate Medical Education, Unviersity of Leiden; Aug 15–21, 1965; Copenhagen: Munksgaard; 1965. [Google Scholar]
- 2.Histocompatibility Testing; Report of a Conference and Workshop Sponsored by the Division of Medical Sciences, National Academy of Sciences, National Research Council; 7–12 June, 1964; Washington: National Academy of Sciences; 1965. [Google Scholar]
- 3.Curtoni ES, Mattiuz PL, Tosi RM. Histocompatibility Testing 1967. Copenhagen: Munksgaard; 1967. [Google Scholar]
- 4.Terasaki PI. Histocompatibility Testing 1970. Copenhagen: Munksgaard; 1970. [Google Scholar]
- 5.Dausset J, Colombani J. Histocompatibility Testing 1972. Copenhagen: Munksgaard; 1973. [Google Scholar]
- 6.Kissmeyer-Nielsen F. Histocompatibility Testing 1975. Copenhagen: Munksgaard; 1975. [Google Scholar]
- 7.Bodmer WF, Batchelor JR, Bodmer JG, Festenstein H, Morris PJ. Histocompatibility Testing 1977. Copenhagen: Munksgaard; 1978. [Google Scholar]
- 8.Terasaki PI. Histocompatibility Testing 1980. Los Angeles: UCLA Tissue Typing Laboratory; 1980. [Google Scholar]
- 9.Albert ED, Baur MP, Mayer WR. Introductory Remarks. In: Albert ED, Baur MP, Mayer WR, editors. Histocompatibility Testing 1984. Heidelberg: Springer-Verlag; 1984. [Google Scholar]
- 10.Dupont B. Overview of Experimental Desgin for the Tenth International Histocompatibility Workshop. In: Dupont B, editor. Immunobiology of HLA. Volume I. Histocompatibility Testing. Vol. 1. Springer Verlag; 1987. p. 3. [Google Scholar]
- 11.Tsuji K. Overview of the Eleventh International Histocompatibility Workshop and Conference. In: Kimiyoshi Tsujki MA, Sasazuki Takehiko, editors. HLA 1991: Proceedings of the Eleventh International Histocompatibility Workshop and Conference. Vol. 1. Oxford Science Publications; 1991. p. 3. [Google Scholar]
- 12.Charron D, Fauchet R. The 12th International Histocompatibility Workshop. In: Charron D, editor. HLA Volume 1. Genetic Diversity of HLA: Functional and Medical Implication. Vol. 1. EDK; 1997. p. XXV. [Google Scholar]
- 13.Hansen JA. Foreword: Immunobiology of the Human MHC. Proceedings of the 13th International Histocompatibility Workshop and Conference. In: Hansen JA, editor. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. Volume 1. Vol. 1. IHWG Press; 1997. p. xxv. [Google Scholar]
- 14.McCluskey J, Tait CB, Christiansen FT, Holdsworth R. 14th International HLA and Immunogenetics Workshop Reports: Introduction. Tissue Antigens. 2007;69:1. doi: 10.1111/j.1399-0039.2006.762_1.x. [DOI] [PubMed] [Google Scholar]
- 15.Tissue Antigens; Abstracts for the 15th International Histocompatibility and Immunogenetics Workshop and Conference; Rio de Janeiro, Brazil. September 13–20, 2008; 2008. p. 231. [Google Scholar]
- 16.Middleton D, Marsh SGE. 16th International HLA and Immunogenetics Workshop (IHIW) Introduction. Int J Immunogenet. 2013;40:1. doi: 10.1111/iji.12034. [DOI] [PubMed] [Google Scholar]
- 17.Baur MP, Albert ED, Mayr WR. The Central Data Analysis of the Ninth Workshop. In: Albert ED, Baur MP, Mayr WR, editors. Histocompatibility Testin 1984. Heidelberg: Springer-Verlag; 1984. p. 37. [Google Scholar]
- 18.Lalouel J-M, Ferguson M, Wheeler R, King N. Tenth International Histocompatibility Workshop: Overview of Data Processing and Analysis. In: Dupont B, editor. Immunobiology of HLA. Volume I. Histocompatibility Testing 1987. Vol. 1. Springer-Verlag; 1987. p. 83. [Google Scholar]
- 19.Inoko H, Sato K, Takata H, Imanishi T, Ina Y, Saitou N, et al. Data Flow from the Collection of Raw Data to the Construction of the MHC Database. In: Kimiyoshi Tsujki MA, Sasazuki Takehiko, editors. HLA 1991. Proceedings of the Eleventh International Histocompatibility Workshop and Conference. Vol. 1. Oxford Science Publications; 1991. p. 65. [Google Scholar]
- 20.Cambon-Thomsen A, Albert E, Bodmer JG, Piazza A, Thouzellier Y, Clayton JF. Database, Communication, Analysis (DCA) of the 12th International Histocompatibility Workshop: General Overivew. In: Charron D, editor. HLA Volume 1. Genetic Diversity of HLA: Functional and Medical Implications. Vol. 1. EDK; 1997. p. 469. [Google Scholar]
- 21.Schoch G, Kesten M, McKallor C, Mickelson E, Hansen JA. 13th IHWS Shared Resources Joint Report. In: Hansen JA, editor. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. Volume I. Vol. 1. IHWG Press; 2007. p. 482. [Google Scholar]
- 22.Nunes JM. Tools for analysing ambiguous HLA data. Tissue Antigens. 2007;69:203. doi: 10.1111/j.1399-0039.2006.00808.x. [DOI] [PubMed] [Google Scholar]
- 23.Mack SJ, Sanchez-Mazas A, Single RM, Meyer D, Hill J, Dron HA, et al. Population samples and genotyping technology. Tissue Antigens. 2007;69:188. doi: 10.1111/j.1399-0039.2006.00768.x. [DOI] [PubMed] [Google Scholar]
- 24.Single RM, Meyer D, Mack SJ, Lancaster A, Erlich HA, Thomson G. 14th International HLA and Immunogenetics Workshop: Report of progress in methodology, data collection, and analyses. Tissue Antigens. 2007;69:185. doi: 10.1111/j.1399-0039.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 25.Steenkiste A, Valdes AM, Feolo M, Hoffman D, Concannon P, Noble J, et al. 14th International HLA and Immunogenetics Workshop: Report on the HLA component of type 1 diabetes. Tissue Antigens. 2007;69:214. doi: 10.1111/j.1399-0039.2006.00772.x. [DOI] [PubMed] [Google Scholar]
- 26.Leffell MS, Cao K, Coppage M, Hansen JA, Hart JM, Pereira N, et al. Incidence of humoral sensitization in HLA partially mismatched hematopoietic stem cell transplantation. Tissue Antigens. 2009;74:494. doi: 10.1111/j.1399-0039.2009.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton D, Gonzalez F, Fernandez-Vina M, Tiercy JM, Marsh SGE, Aubrey M, et al. A bioinformatics approach to ascertaining the rarity of HLA alleles. Tissue Antigens. 2009;74:480. doi: 10.1111/j.1399-0039.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 28.Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A, et al. Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the co-evolution of KIR and HLA. Tissue Antigens. 2010;76:9. doi: 10.1111/j.1399-0039.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- 29.Nunes JM, Riccio ME, Buhler S, Di D, Currat M, Ries F, et al. Analysis of the HLA population data (AHPD) submitted to the 15th International Histocompatibility/Immunogenetics Workshop by using the Gene[rate] computer tools accommodating ambiguous data (AHPD project report) Tissue Antigens. 2010;76:18. doi: 10.1111/j.1399-0039.2010.01469.x. [DOI] [PubMed] [Google Scholar]
- 30.Naumova E, Ivanova M, Pawelec G, Constantinescu I, Bogunia-Kubik K, Lange A, et al. Tissue Antigens; ‘Immunogenetics of Aging’: report on the activities of the 15th International HLA and Immunogenetics Working Group and 15th International HLA and Immunogenetics Workshop; 2011. p. 187. [DOI] [PubMed] [Google Scholar]
- 31.Riccio ME, Buhler S, Nunes JM, Vangenot C, Cuénod M, Currat M, et al. 16th IHIW: Analysis of HLA Population Data, with updated results for 1996 to 2012 workshop data (AHPD project report) Int J Immunogenet. 2013;40:21. doi: 10.1111/iji.12033. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez-Galarza FF, Mack SJ, Hollenbach J, Fernandez-Vina M, Setterholm M, Kempenich J, et al. 16th IHIW: Extending the number of resources and bioinformatics analysis for the investigation of HLA rare alleles. Int J Immunogenet. 2013;40:60. doi: 10.1111/iji.12030. [DOI] [PubMed] [Google Scholar]
- 33.Hollenbach JA, Mack SJ, Gourraud PA, Single RM, Maiers M, Middleton D, et al. A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens. 2011;78:333. doi: 10.1111/j.1399-0039.2011.01777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J, Soormally AR, Hayhurst JD, Marsh SG. The IPD-IMGT/HLA Database - New developments in reporting HLA variation. Hum Immunol. 2016;77:233. doi: 10.1016/j.humimm.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2002. Tissue Antigens. 2002;60:407. doi: 10.1034/j.1399-0039.2002.600509.x. [DOI] [PubMed] [Google Scholar]
- 36.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Committee WN Nomenclature for factors of the HL-a system. Bull World Health Organ. 1968;39:483. [PMC free article] [PubMed] [Google Scholar]
- 38.Bochtler W, Maiers M, Oudshoorn M, Marsh SG, Raffoux C, Mueller C, et al. World Marrow Donor Association guidelines for use of HLA nomenclature and its validation in the data exchange among hematopoietic stem cell donor registries and cord blood banks. Bone Marrow Transplant. 2007;39:737. doi: 10.1038/sj.bmt.1705672. [DOI] [PubMed] [Google Scholar]
- 39.Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milius RP, Mack SJ, Hollenbach JA, Pollack J, Heuer ML, Gragert L, et al. Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens. 2013;82:106. doi: 10.1111/tan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milius RP, Heuer M, George M, Pollack J, Hollenbach JA, Mack SJ, et al. The GL service: Web service to exchange GL string encoded HLA & KIR genotypes with complete and accurate allele and genotype ambiguity. Hum Immunol. 2016;77:249. doi: 10.1016/j.humimm.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mack SJ, Milius RP, Gifford BD, Sauter J, Hofmann J, Osoegawa K, et al. Minimum information for reporting next generation sequence genotyping (MIRING): Guidelines for reporting HLA and KIR genotyping via next generation sequencing. Hum Immunol. 2015;76:954. doi: 10.1016/j.humimm.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milius RP, Heuer M, Valiga D, Doroschak KJ, Kennedy CJ, Bolon YT, et al. Histoimmunogenetics Markup Language 1. 0: Reporting next generation sequencing-based HLA and KIR genotyping. Hum Immunol. 2015;76:963. doi: 10.1016/j.humimm.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray T, Paoli J, Sperberg-McQueen CM, Maler E, Yergeau F. Extensible markup language (XML) World Wide Web Consortium Recommendation REC-xml-19980210. 1998;16:16. http://www.w3.org/TR/1998/REC-xml-19980210. [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersson G. Evolution of the human HLA-DR region. Front Biosci. 1998;27:d739. doi: 10.2741/a317. [DOI] [PubMed] [Google Scholar]
- 47.Wright ES. Using DECIPHER v2. 0 to analyze big biological sequence data in R. The R Journal. 2016;8:352. [Google Scholar]
- 48.Lancaster AK, Single RM, Nelson MP, Solberg O, Thomson G. PyPop update - a software pipeline for large-scale multi-locus population genomics. Tissue Antigens. 2007;69:192. doi: 10.1111/j.1399-0039.2006.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mack SJ. A gene feature enumeration approach for describing HLA allele polymorphism. Hum Immunol. 2015;76:975. doi: 10.1016/j.humimm.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.