Abstract
Regions with complex geological histories often have diverse and highly endemic biotas, yet inferring the ecological and historical processes shaping this relationship remains challenging. Here, in the context of the taxon cycle model of insular community assembly, we investigate patterns of lineage diversity and habitat usage in a newly characterized vertebrate radiation centred upon the world's most geologically complex insular region: island arcs spanning from the Philippines to Fiji. On island arcs taxa are ecologically widespread, and provide evidence to support one key prediction of the taxon cycle, specifically that interior habitats (lowland rainforests, montane habitats) are home to a greater number of older or relictual lineages than are peripheral habitats (coastal and open forests). On continental fringes, however, the clade shows a disjunct distribution away from lowland rainforest, occurring in coastal, open or montane habitats. These results are consistent with a role for biotic interactions in shaping disjunct distributions (a central tenant of the taxon cycle), but we find this pattern most strongly on continental fringes not islands. Our results also suggest that peripheral habitats on islands, and especially island arcs, may be important for persistence and diversification, not just dispersal and colonization. Finally, new phylogenetic evidence for subaerial island archipelagos (with an associated biota) east of present-day Wallace's Line since the Oligocene has important implications for understanding long-term biotic interchange and assembly across Asia and Australia.
Keywords: ecological evolution, island arcs, island biogeography, Melanesia, montane relict, taxon cycle
1. Introduction
Regions with complex geological histories often have diverse and highly endemic biotas [1,2]. The historical and ecological processes that have shaped this relationship are complex and may vary across space and time. For instance, in rapidly uplifting mountains, new high-elevation habitats have inflated regional diversity pools by providing opportunities for ecological diversification and long-distance colonization [3,4]. In contrast, older mountains have a higher proportion of relictual endemic diversity [2]. Island arcs fringing continental plates are also characterized by highly dynamic geological histories [5,6], and often famously diverse and endemic biotas [7,8]. However, resolving the historical processes that have shaped island-arc biotas is particularly challenging as the geographical signature of evolutionary processes in these comparatively small and unstable landmasses may be obliterated by a combination of geological and biotic processes driving dispersal and extinction [9].
One process hypothesized to shape biotic turnover on islands is the taxon cycle [10], a dynamic community-assembly model for faunal ‘development’ that has recently received renewed support [11,12]. The taxon cycle proposes that taxa with certain phenotypes or ecological strategies, often termed ‘generalist’, are predisposed to disperse and colonize new habitats (early stage). These taxa are often associated with peripheral habitats such as small islands, or disturbed and coastal forests. Subsequent to colonization, these lineages tend to specialize ecologically, resulting in range fracture and contraction into more interior habitats (late stage), with many lineages ultimately going extinct, leaving a pattern of geographically disjunct, old lineages [10]. The key driver of this process is hypothesized to be biotic interactions with an ongoing influx of new immigrants into coastal peripheral habitats that push earlier colonists inwards and continually reset the cycle [10]. Some recent studies have supported the taxon cycle in more isolated island systems [11,12]. However, others have identified surprisingly deep divergences in widespread insular lineages from disturbed habitats [13,14]. The latter observations suggest that peripheral habitats have the potential to be important zones for not just colonization, but also persistence and diversification, however this idea remains little tested.
In the tropical West Pacific, the islands spanning Wallacea, the Philippines, and Melanesia have the most complex biogeographic history of any insular region in the world [15]. Bordered to the west by Wallace's Line (or the eastern edge of the Sunda Shelf) and to the south by the northern edge of the Australian Plate, this region has played a prominent and ongoing role in the generation and testing of evolutionary theory, including the taxon cycle [7,8,10]. Historical geological reconstructions indicate a near-continuous chain of island arcs in the Oligocene [15], which subsequently fractured and was incorporated into present-day landforms extending from the Philippines to Fiji (sometimes termed the Vitiaz Arc) [16,17]. These arcs have been linked to increased rates of floral and faunal dispersal between Australia and Asia [18–20] and biotic distributions spanning from the Philippines to Melanesia [17,19,20]. However, while there are exceptions [17], in many recent molecular phylogenetic analyses a role for island arcs in generating endemic terrestrial biological diversity has been overlooked [21], not supported [22] or contested [18].
Here we provide new, robust statistical evidence that a radiation of small, often secretive and rarely observed lizards [23,24] of the genus Lepidodactylus and allied genera originally colonized early island arcs of the West Pacific in the Oligocene before going through a period of extensive diversification and localized persistence. A synthesis of distributional and phylogenetic data for this newly characterized radiation shows no prevailing pattern that interior habitats are home to more phylogenetically divergent lineages, which contrasts with predictions of the taxon cycle. However, on or near continental margins there is strong evidence of a non-random distribution, with taxa concentrated away from lowland rainforest into open, coastal or montane habitats (here grouped together into the catchall ‘marginal’) that are likely to be less species rich for lizards. Taken together, these data suggest that relatively species-poor ‘marginal’ habitats and island arcs in the geologically dynamic western Pacific may have played an important, often overlooked role in the persistence and generation of regional diversity since the mid-Cenozoic.
2. Material and methods
(a). Sampling and sequence data
Through extensive fieldwork across the Philippines and Melanesia over two decades we were able to obtain tissue samples (mainly liver) of 21 of 33 recognized species of Lepidodactylus (including two subspecies of L. herrei), plus an additional 22 candidate species (determined by mitochondrial sequence divergences ≥10% and/or morphological differentiation). Total ingroup sampling includes 206 specimens (electronic supplementary material, figure S1 and table S1), including closely related taxa in the genera Luperosaurus (n = five taxa) and Pseudogekko (n = seven taxa) [23,25]. In combined mitochondrial- and nuclear-data alignments, outgroup samples from the most closely related genera Gekko and Ptychozoon (electronic supplementary material, table S1) were included from recent studies [23,25,26]. The full alignment consisted of 2,481 base pairs (bp) of sequence data comprised of the mitochondrial NADH dehydrogenase subunit 2 (ND2:1041 bp) gene and the nuclear loci phosducin (PDC: 395 bp) and recombination-activating gene 1 (RAG-1:1035 bp) generated using primers and protocols outlined elsewhere [26,27]. Sequence data were aligned using the MUSCLE algorithm [28] and subsequently checked by eye.
(b). Phylogenetic analyses
We estimated phylogenetic relationships using Bayesian and maximum likelihood (ML) approaches as implemented in BEAST v. 1.8.2 [29] and RAxML-VI-HPC v. 8.2.10 [30], respectively. Topologies and support values from three different subsets of the data (mitochondrial, nuclear, and mitochondrial + nuclear) were compared across analyses. We used partitioning schemes as suggested by Partitionfinder2 [31]: nuclear first and second positions; nuclear thirds; mitochondrial firsts, mitochondrial seconds and mitochondrial thirds. We employed the GTR-CAT model in RAxML [30], and used the GTR+Γ and HKY+Γ models for mitochondrial and nuclear partitions, respectively, in Bayesian analyses. Bayesian analyses were run for 50 million generations, with confirmation of run stationarity and effective samples sizes above 200 (using Tracer v. 1.6 [32]). The first 20% of each chain was discarded as burn-in. All phylogenetic analyses were run in the CIPRES gateway (http://www.phylo.org/).
(c). Bayesian dating
Bayesian dating analyses were performed in BEAST v. 1.8.2 [33]. For initial estimation of the crown-radiation age of Lepidodactylus and two genera nested within this clade (hereafter Lepidodactylus sensu lato [s.l.]; see results) we used alignments for nuclear genes only and excluded mitochondrial data to reduce the probability of this rapidly saturating gene inflating date estimates [34]. First, we estimated basal-divergence ages in Lepidodactylus s.l. using a published dataset comprising five nuclear genes and including almost all recognized gecko genera and key examplars spanning the diversity of Lepidodactylus s.l. [35]. Partitioning strategies, model choices, four fossil calibrations and one root age prior were taken from the source study [35] (electronic supplementary material, tables S2 and S3). Other younger fossil and biogeographic calibrations used in the source study have been shown to be of questionable reliability inasmuch as dating constraints were not used [36,37]. Subsequently, to better understand basal-age estimates within Lepidodactylus s.l., we focused on a nuclear-gene alignment that included fewer genes (PDC + RAG1), the same calibrations (electronic supplementary material, table S3), but more taxa from within the Lepidodactylus clade.
To generate a lineage-complete phylogeny for downstream biogeographic analyses we used a concatenated dataset (ND2 + PDC + RAG1) that included a single exemplar of all species/candidate species. We ran analyses with, and without, third positions from the mitochondrial data alignment to assess age inflation from saturated sites [34]. Basal ages for crown Lepidodactylus and its sister lineage Gekko were constrained secondarily using the most conservative (youngest) crown-age priors (mean and 95% HPD) derived from analyses of the dataset comprising five nuclear genes (electronic supplementary material, table S3). We ran all four possible combinations of the strict and uncorrelated lognormal clock models and Yule and Birth–Death models. Results were identical across combinations (electronic supplementary material, table S3), and we only report results with the highest marginal likelihoods.
(d). Geographical-range evolution
To estimate whether ancestral biogeographic ranges for taxa in both Lepidodactylus s.l. and its sister clade (Gekko + Ptychozoon) were on island arcs or continents we used BioGeoBEARS v. 0.2.1 [38,39] and BEAST v. 1.8.2 [33]. We evaluated the fit of three alternative models implemented in BioGeoBEARS (DEC, DIVALIKE and BAYAREALIKE) with and without the jump (J) parameter, and we selected best models using maximum-likelihood model comparisons (AIC; electronic supplementary material, table S4). These analyses were implemented on a lineage-complete maximum-credibility tree obtained from BEAST. To assess whether topological uncertainty at the base of the tree confounded geographical-range estimation we also ran additional analyses in BEAST, including areas as unordered states with a simple stochastic model of equal probability of all transitions and otherwise using parameters and settings identical to the dating analyses above.
We coded regions according to their underlying geology (three states): (i) continental, including the Sunda Shelf and the central and southern portions of New Guinea; (ii) island arcs, including the Philippines, northern New Guinea, eastern Melanesia; and finally (iii) oceanic islands, meaning small islands (less than 1000 km2) with no history of connection to any larger landmasses (Christmas Island, Micronesia and French Polynesia). Several lineages were of potentially ambiguous placement under this scheme, and we dealt with them as follows: portions of the western Philippines (Palawan, Zamboanga Peninsula of Mindanao) are of continental origins and defined as crustal fragments that have rifted to their current locations [40], so we ran separate analyses coding taxa endemic to these areas as either continental or island arcs; New Guinea likewise is a conglomerate of continental and island-arc fragments, so we assigned geological codes based on where the majority of taxa in each major lineage occur [41]; finally, the East Papuan Composite Terrane (EPCT) of eastern New Guinea may not be derived from island arcs but is also not continental [41], so we again ran separate analyses to explore the impact that alternative coding schemes might have on ancestral-state estimation.
(e). Evolutionary shifts in habitat types
The taxon cycle predicts an ongoing transition of lineages from marginal to interior habitats. To understand patterns and evolutionary shifts of Lepidodactylus habitat usage in the context of the taxon cycle we undertook three sets of analyses. First, we simultaneously estimated the evolution of habitat type (at collection localities) and the phylogeny using BEAST. Second, we tested the prediction that rainforest and montane habitats should contain more deeply divergent lineages than do coastal and open habitats (here equating deeply divergent lineages with relictual/specialized under the taxon cycle). Third, our newly synthesized distributional data suggested that continental Lepidodactylus were non-randomly distributed away from lowland rainforests and their rich biotic communities (electronic supplementary material, table S5), so we tested if the number of taxa occurring in different habitat types in regions with different geological underpinnings (island arcs versus continental) departed from predictions of a null model.
For initial ancestral-state estimation, we used four ecological states: (i) lowland rainforest, (ii) coastal forest, including entire islands less than 200 km2, (iii) montane forest, and (iv) open habitats, such as beaches, disturbed anthropogenic landscapes, savannahs and swamp forests (electronic supplementary material, table S5). Taxa occurring across multiple habitats were coded as occupying each. Second, because our ancestral-state analyses suggested continental Lepidodactylus were non-randomly distributed away from lowland rainforests, we also undertook a subsequent two-state estimation contrasting lowland rainforest versus a combination of the other three habitat states. We did not code outgroups for ecology, and we estimated ancestral states as per geological regions (see above) using BEAST.
To test the critical taxon cycle assumption of shifts between habitat types, we focused on the prediction that interior habitats would be dominated by older lineages than would peripheral (coastal and open) habitats. We coded primary lowland rainforest and montane habitats as interior and all other habitats―such as coastal habitats, strand forests, disturbed or open habitats including towns, gardens, savannahs, swamp forests―as peripheral. We extracted tip branch lengths for all taxa (using functions in the R package ape [42]) and then tested if interior taxa were statistically associated with longer terminal branches, both across the entire phylogeny and within three relatively diverse and geographically cohesive regions (Philippines, New Guinea, Pacific Islands + eastern Melanesia).
To test if continental taxa are non-randomly distributed away from lowland rainforest, we coded lowland rainforest as core and all other habitats as marginal. We grouped taxa according to Geology (see above: continental, island arcs and oceanic), with the EPCT alternately coded as ‘continental’ or ‘island arc’ in separate runs. We excluded oceanic island taxa, as they all occur on small islands where, under our definition, core lowland rainforest habitats are unavailable. We then generated null distributions for expected numbers of marginal taxa occurring in the two focal geological categories. First, we estimated the rate of transitions between habitats (q) on a trimmed topology (no oceanic taxa) with Geiger [43], using both equal-rate (ER) and all-rates-different (ARD) functions in separate analyses. The equal-rate function had the best AICc value (67.256 versus 69.417) and was used to conduct 1000 simulations (function simchar in [41]) of evolutionary shifts in habitat type, with the root set to marginal, on the assumption that the earliest colonists of an island arc must have dispersed through coastal marginal habitats. From these simulations we obtained distributions for the expected number of marginal taxa in each of the two geological categories.
3. Results
(a). Phylogenetic analyses
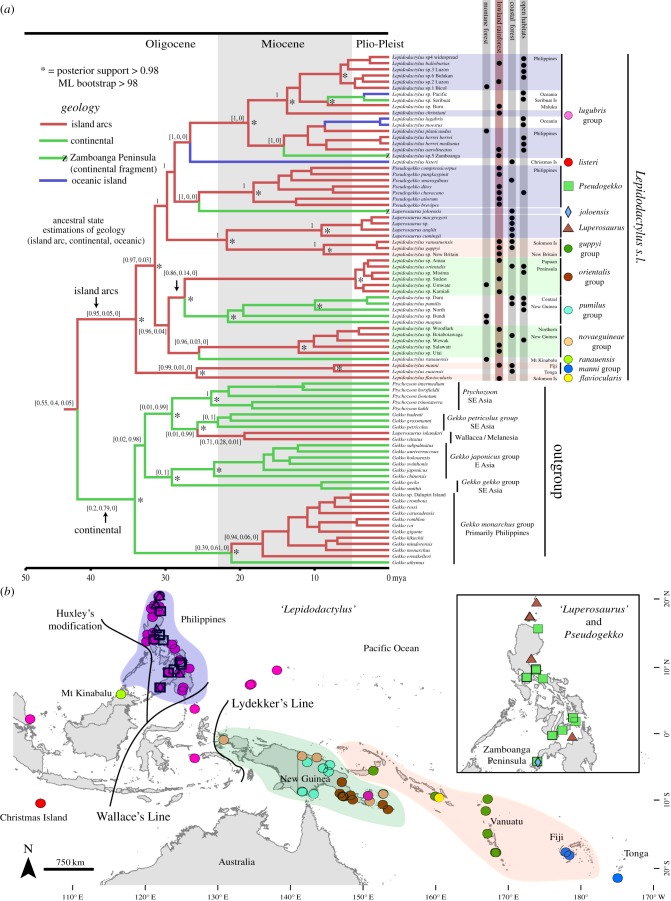
All phylogenetic analyses (Bayesian and ML) identify a well-supported clade (hereafter Lepidodactylus s.l.) composed of 12 major lineages, including all sampled Lepidodactylus (nine deeply divergent and geographically cohesive lineages), Luperosaurus (in part, two lineages) and Pseudogekko (one lineage) (figure 1; electronic supplementary material, figure S1). A sister-taxon relationship between Lepidodactylus s.l. and a clade centred to the west of Wallace's Line, comprising the genera Gekko, Ptychozoon and one lineage of ‘Luperosaurus’ from Sulawesi (generally referred to as the Gekko group), is strongly supported. Deeper relationships among the 12 major lineages of Lepidodactylus s.l. are generally not well supported (figure 1; electronic supplementary material, figure S1). Only three lineage pairs receive high support (posterior probability (pp) > 0.95); the L. manni group (Fiji/Tonga) + L. flaviocularis (Solomons), true Luperosaurus (Philippines) + L. guppyi group (East Melanesia), and the L. orientalis group (New Guinea, EPCT) + the L. pumilus group (central New Guinea) (figure 1). A further pairing of Lepidodactylus listeri (Christmas Island) and the L. lugubris group (widespread) receives support in analyses of nuclear data.
Figure 1.
(a) Detailed chronogram for Lepidodactylus s.l. and outgroups, * = Bayesian support >0.95, branches coloured according to ancestral state estimation of geological origins of landmass and ancestral states probabilities shown in brackets at major nodes (island arc, continental, oceanic). Clusters of taxa concentrated on key regions with arc origins are highlighted: Philippines (blue), New Guinea (green) and East Melanesia (red). Taxa currently placed in Lepidodactylus s.s. are indicated by circular symbols, other ‘genera’ by squares, diamonds or triangles. Summary of habitat types based on field observations and published literature. (b) Localities for the 12 major lineages of Lepidodactylus s.l., excluding samples of the globally distributed, anthropogenically dispersed parthenogen Lepidodactylus lugubris. Major regions with arc origins are highlighted as per tips of phylogenetic tree: Philippines (blue), New Guinea (green) and East Melanesia (red).
(b). Bayesian dating
Fossil-calibrated divergence-date estimation using nuclear genes recovers an initial mid-Cenozoic (mean 34.7 Ma, 95% HPD 23.3–44.4 Ma) radiation of Lepidodactylus s.l., preceded by divergence from a primarily Asian sister lineage (51.9, 41.1–62.6) (see table 1 and electronic supplementary material, table S3 for full details of age ranges). Analyses of the five-nuclear-gene dataset estimate the age of radiation for Lepidodactylus s.l. as older than co-occurring gekkonid clades, including Cyrtodactylus (22.3, 14.9–29.7), Gehyra (30.6, 21.3–40.6) and Nactus (15.6, 7.8–24.4) (electronic supplementary material, figure S2).
Table 1.
Summary of posterior age estimates for major lineages from dating analyses. All ages are in millions of years ago. Mean values for posteriors are given first followed by 95% higher posterior density intervals in brackets. Ages for Lepidodactylus s.l. are highlighted in bold. Only results for model combinations with highest marginal likelihoods are shown. Full summary of all analyses and details of fossil and secondary calibration strategies are provided in electronic supplementary material, tables S2 and S3.
| nucleotide data | calibrations |
crown age estimates |
|||
|---|---|---|---|---|---|
| Gekkota | Gekko + Lepidodactylus | Gekko | Lepidodactylus s.l. | ||
| nuclear (5 genes) | fossil | 118.8 (112.7–127.3) | 51.9 (41.1–62.6) | 36.2 (27.4–45.9) | 34.7 (23.3–44.4) |
| nuclear (2 genes) | fossil | 115 (112.4–121.3) | 46.5 (43.8–60.4) | 36.7 (28.0–45.0) | 35.2 (28.3–42.3) |
| combined | secondary | n.a. | n.a. | 34.0 (28.7–40.4) | 32.4 (26.9–37.6) |
| combined (no mtDNA third codons) | secondary | n.a. | n.a. | 33.6 (27.9–39.9) | 33.0 (26.9–38.3) |
Analyses of both nuclear-only and nuclear + mitochondrial datasets infer all major lineages of Lepidodactylus s.l. to have diverged before the start of the Miocene (23 Ma; figures 1 and 2; electronic supplementary material, figure S3). Mean estimates of crown divergences within two Philippines-centred lineages (Pseudogekko, the L. lugubris group) and one New Guinean lineage (L. pumilis group) both cluster around the early Miocene (16–28 Ma), further indicating long histories of diversification within offshore island arcs (figure 1).
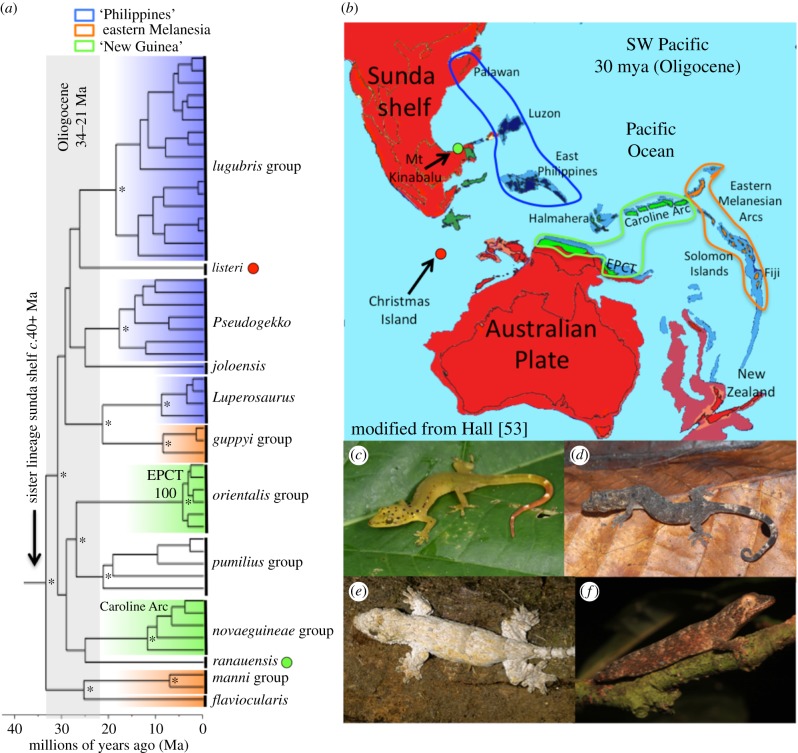
Figure 2.
Historical biogeography of Lepidodactylus s.l. (a) Simplified chronogram (*Bayesian support <0.95) with major lineage-specific distributions centred on island arcs highlighted. (b) Simplified geological reconstructions of Australasia c.30 Ma with key island arcs indicated (after reference [53]). Representative taxa: (c) Pseudogekko smaragdinus (Philippines); (d) Lepidodactylus sp. New Britain; (e) Luperosaurus cumingii (Philippines); and (f) Lepidodactylus flaviocularis (Solomon Islands). EPCT, East Papuan Composite Terrane. Photographs: (c) CDS, (d) Stephen Richards, (e) RMB, (f) SLT.
(c). Geographical-range evolution
The distribution of major lineages in Lepidodactylus s.l. is conspicuously centred on island arcs or former island arcs that have accreted to continents: the Philippines (four lineages total), northern New Guinea (two lineages) and eastern Melanesia (three lineages; figure 1). One additional lineage occurs across a geological composite of arc and continental geology in central New Guinea. The two remaining major lineages have restricted distributions: Christmas Island in the Indian Ocean and lower-montane areas of Mt. Kinabalu, Borneo. The latter is thus far the only lineage with mid-Cenozoic origins in this clade that is also endemic to the Sunda Shelf.
Geographical analyses using model-choice-based ML (figures 1 and 2; electronic supplementary material, figure S4) and Bayesian methods strongly infer island arcs as the ancestral geographical range for Lepidodactylus s.l. Three shifts from island arcs (the Philippines, northern New Guinea, and East Melanesia) to more isolated oceanic islands are inferred, as are three shifts from island arcs to continental areas, including central New Guinea (figure 1). Within these island arcs, an early Miocene (see below) dispersal event between the Philippines and eastern Melanesia receives strong support. In contrast, among the sister lineage of Lepidodactylus s.l. (Gekko, Ptychozoon), most initial diversification is continental, with more recent and limited colonization of island arcs in the West Pacific (electronic supplementary material, figure S4).
(d). Evolutionary shifts in habitat types
Lowland rainforest is the most commonly observed of the four habitat states (figure 1). However, few continental taxa occur in lowland rainforest: Depending on whether the EPCT was or was not coded as island arc the number was respectively 2 out of 15 or 0 out of 7. Instead, lowland rainforest taxa are concentrated in island arcs, primarily in the Philippines and, to a lesser extent, along northern New Guinea: (EPCT coded as island arc, 21 out of 42 taxa; EPCT coded as continental, 17 out 36 taxa). Ancestral habitat estimation using four habitat types infers Lepidodactylus s.l. as having a lowland rainforest ancestral state (figure 1). Additional ancestral-habitat estimation based on two ecological states (lowland rainforest versus all other habitats) supports habitats other than lowland rainforest as ancestral for the entire radiation (electronic supplementary material, figure S5).
There was no significant difference in lineage length between interior (lowland rainforest and montane forest) and non-interior taxa (all other habitat types) in New Guinea and Philippine taxa (electronic supplementary material, table S6); however, interior lineages were significantly older in combined analyses of taxa from the eastern Melanesian islands and the Pacific (although note small number of taxa n = 9).
The observed dearth of continental taxa from lowland rainforest is outside the 95% confidence limits predicted from our simulations of ecological state evolution, regardless of whether the EPCT is considered island arc or continental (p-values 0.03 and 0.01, respectively). Conversely, the distribution of species across lowland rainforest and other habitats on island arcs does not significantly differ from the null model (electronic supplementary material, figure S6 and table S6).
4. Discussion
(a). Can species-poor ‘marginal’ habitats facilitate persistence and diversification?
In the original formulation of the taxon cycle, coastal and disturbed peripheral habitats were hypothesized to play a key role in facilitating ongoing colonization, followed by evolutionary shifts into more interior habitats such as lowland and montane forests [10]. Recent work has provided supporting evidence of ecological displacement of older lineages into interior habitats in small island biotas, as predicted by the taxon cycle [11,12]. Here, however, our strongest result is an absence of Lepidodactylus s.l. species inhabiting lowland rainforest on continental plates. Instead, continental taxa have disjunct distributions across montane forest and open or disturbed habitats (beaches, swamp forests or mangroves). Strikingly, some montane lineages are older than current estimates for the uplift of the mountains on which they occur, especially in New Guinea [41]. Ecological displacement away from lowland-forest environments having diverse squamate communities [44] is one possible explanation for the outwardly disjunct ecological distributions of these divergent continental taxa. Thus, this pattern is potentially consistent with the underlying biotic displacement processes, but not the geographical context, of the traditional taxon cycle model [10,11]; to wit, it is occurring on the peripheries of continents instead of on islands.
We also found little evidence that older lineages were concentrated in lowland rainforest and montane habitats on insular landmasses. Across Lepidodactylus s.l. nearly half of the species, including phylogenetically ancient taxa, occur in habitats that can be regarded as peripheral under Taxon Cycle models (especially savannahs, swamps and coastal forest, which are predicted to hold fewer deeply divergent lineages). Several other unrelated lizard groups of the West Pacific are also rare or marginalized in continental systems yet are abundant in disturbed or peripheral habitats on islands and show substantial phylogenetic divergences despite clearly having been capable of overseas dispersal [13,45]. One important caveat here is that in Lepidodactylus from islands of eastern Melanesia and the Pacific (the region from which the taxon cycle was originally formulated), there is some evidence that interior lineages are more deeply divergent. More sampling is required to investigate patterns in this area more carefully. Broadly, however it would appear that while peripheral habitats certainly play an important role in community assembly and colonization, they may also allow colonizing lineages to persist and even diversify without becoming restricted to interior habitats. One potential explanation of this may be that in geologically complex insular systems newly forming and often species-poor habitats may be the only continuously and reliably accessible habitats. Alternatively, in groups that disperse rarely, new colonists may simply be too infrequent to drive a taxon cycle.
(b). Life of the lost arcs
Geological models indicate that fragments of present-day landmasses extending from the Philippines, across much of Melanesia, and as far east as Fiji formed a near continuous chain in the late Eocene (approximately 35 Ma) (figure 2) [15], but they do not provide clear evidence as to whether the key landforms were subaerial or submarine. Our late Eocene estimates of initial diversification ages in Lepidodactylus s.l. provide the first dated phylogenetic corroboration of these geological models and support earlier, often overlooked inferences that West Pacific's island arcs have been generating biodiversity since at least the Oligocene [6,20]. Indeed, many of the lineages we identified are old enough that persistence and transfer across the West Pacific on geologic fragments of a formerly more continuous arc [15] seems a likely explanation for extant distributional patterns spanning from the Philippines to New Guinea. This is not to say that overseas dispersal, with subsequent speciation on isolated islands, has not occurred, as it clearly has in both Lepidodactylus (oceanic islands such Christmas Island, French Polynesia, Micronesia) and other overlapping lizard lineages [46]. However, in the Philippines, four ancient lineages of Lepidodactylus s.l. with disparate phenotypes and ecological attributes co-occur [7]. Likewise, in New Guinea and Eastern Melanesia endemic lineages widely pre-date estimates for the current configuration (or even existence) of key landmasses such as New Guinea [15,41]. These patterns suggest palaeotransport and accretion of formerly isolated arc fragments with associated biotas. They also support phylogenetic studies suggesting that elements of the highly diverse and endemic New Guinean biota have Miocene (or earlier) island arc origins [47], while standing in stark contrast with recent phylogenetic studies that have either not supported [22] or have contested this idea [18].
The antiquity of Lepidodactylus s.l. further underlines the potential long-term role island arcs may have played in shaping dispersal and diversification across Asia and Australia. Indeed, the evolutionary diversity of Lepidodactylus s.l. is of comparable (or older) age to most of Australia's diverse continental vertebrate radiations [17,18,48,49]. In contrast, lineages from the more intensively studied Wallacean region [21] appear to be relatively recently derived. The importance of island arcs in Asian-Australian biogeography may have been frequently overlooked in part because recent dated biogeographic studies have often focused on vertebrates that may not preserve signatures of early island-arc diversification (i.e. they may disperse either too readily [18], or too poorly [50]). However, while island arcs will certainly not explain diversity patterns in all groups, further work on historical dispersal and diversification across the West Pacific should more explicitly incorporate island arcs as both a potential zone of long-term persistence and a source of diversity for nearby areas such as the Australian continental plate [51].
5. Conclusion
Our results are consistent with a role for biotic interactions (as predicted by the taxon cycle) in shaping distributions, but they suggest resultant patterns may be highly contextual across taxa of varying dispersal ability, and island systems of varying size, complexity and proximity to continents. Furthermore, in geologically complex and dynamic settings such as island arcs the ongoing formation of marginal, ephemeral and relatively species-poor habitats may not only mediate dispersal, but also play a role in long-term persistence and evolutionary diversification. Finally, our results also indicate that diversification, persistence and accretion on formerly isolated Oligo-Miocene islands with their associated endemic biotas may be an important mechanism underpinning regional diversity not just in Wallacea [52], but across the West Pacific and Australasia [20,51].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
Bee Gunn undertook laboratory work, Nicolas Friedman, Dan Rosauer and Emma Sherratt assisted with statistical analyses, and Allen Allison, Angel Alcala, Arvin Diesmos, Chris Austin, Indraneil Das, Steve Donnellan, Lee Grismer and Steve Richards provided key tissues. We thank Craig Moritz and Leo Tedeschi for improving earlier versions of this manuscript.
Data accessibility
All alignments, supplementary methods, tables and figures are provided in the electronic supplementary material.
Authors' contributions
P.M.O., R.M.B. and F.K. conceived the paper; P.M.O., C.D.S., R.M.B., F.K., S.L.T. and E.R. contributed samples and generated data; P.M.O. led writing and analyses, with major contributions from all other authors.
Competing interests
We declare we have no competing interests.
Funding
The Australian Research Council, a CBA Ignition Grant, and the Australian Pacific Science Foundation (P.M.O.), the National Science Foundation (NSF) (DEB 0073199, 1701952 and 0743491 to R.M.B.; DEB 0103794, 0743890 and 1145453 to F.K.; DBI 1402285 to E.R.; DEB 0804115 and IOS 1353683 to C.D.S.), and Fulbright and Fulbright-Hayes Fellowships (C.D.S.).
References
- 1.Badgley C, et al. 2017. Biodiversity and topographic complexity: modern and geohistorical perspectives. Trends Ecol. Evol. 32, 211–226. ( 10.1016/j.tree.2016.12.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fjeldså J, Bowie RCK, Rahbek C. 2012. The role of mountain ranges in the diversification of birds. Annu. Rev. Ecol. Evol. Syst. 43, 249–265. ( 10.1146/annurev-ecolsys-102710-145113) [DOI] [Google Scholar]
- 3.Price TD, et al. 2014. Niche filling slows the diversification of Himalayan songbirds. Nature 509, 222–225. ( 10.1038/nature13272) [DOI] [PubMed] [Google Scholar]
- 4.Merckx VSFT, et al. 2015. Evolution of endemism on a young tropical mountain. Nature 524, 347–350. ( 10.1038/nature14949) [DOI] [PubMed] [Google Scholar]
- 5.Hamilton WB. 1988. Plate tectonics and island arcs. Geol. Soc. Am. Bull. 100, 1503–1527. ( 10.1130/0016-7606(1988)100%3C1503:PTAIA%3E2.3.CO;2) [DOI] [Google Scholar]
- 6.Polhemus DA. 1996. Island arcs, and their influence on Indo-Pacific biogeography. In The origin and evolution of pacific island biotas, New Guinea to eastern Polynesia: patterns and processes (eds Keast A, Miller SE), pp. 51–66. Amsterdam, The Netherlands: Academic Publishing. [Google Scholar]
- 7.Brown RM, et al. 2013. Evolutionary processes of diversification in a model island archipelago. Annu. Rev. Ecol. Evol. Syst. 44, 411–435. ( 10.1146/annurev-ecolsys-110411-160323) [DOI] [Google Scholar]
- 8.Wallace AR. 1860. On the zoological geography of the Malay Archipelago. Zool. J. Linn. Soc. 4, 172–184. ( 10.1111/j.1096-3642.1860.tb00090.x) [DOI] [Google Scholar]
- 9.MacArthur RM, Wilson E. 1967. The theory of island biogeography. Monographs in population biology, vol. 1 Princeton, NJ: Princeton University Press. [Google Scholar]
- 10.Wilson E. 1961. The nature of the taxon cycle in Melanesian Ants. Am. Nat. 95, 169–193. ( 10.1086/282174) [DOI] [Google Scholar]
- 11.Economo EP, Sarnat EM. 2012. Revisiting the ants of Melanesia and the taxon cycle: historical and human-mediated invasions of a tropical archipelago. Am. Nat. 180, E1–E16. ( 10.1086/665996) [DOI] [PubMed] [Google Scholar]
- 12.Jønsson KA, Irestedt M, Christidis L, Clegg SM, Holt BG, Fjeldså J. 2014. Evidence of taxon cycles in an Indo-Pacific passerine bird radiation (Aves: Pachycephala). Proc. R. Soc. Lond. B 281, 20131727 ( 10.1098/rspb.2013.1727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linkem CW, et al. 2013. Stochastic faunal exchanges drive diversification in widespread Wallacean and Pacific island lizards (Squamata: Scincidae: Lamprolepis smaragdina). J. Biogeogr. 40, 507–520. ( 10.1111/jbi.12022) [DOI] [Google Scholar]
- 14.Gillespie RG, Brewer MS, Roderick GK. 2017. Ancient biogeography of generalist predators on remote oceanic islands. J. Biogeogr. 44, 1098–1109. ( 10.1111/jbi.12967) [DOI] [Google Scholar]
- 15.Hall R. 2002. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer based reconstructions, model and animations. J. Asian Earth Sci. 20, 353–431. ( 10.1016/S1367-9120(01)00069-4) [DOI] [Google Scholar]
- 16.Buerki S, Forest F, Stadler T, Alvarez N. 2013. The abrupt climate change at the Eocene—Oligocene boundary and the emergence of South-East Asia triggered the spread of sapindaceous lineages. Ann. Bot. 112, 151–160. ( 10.1093/aob/mct106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jønsson KA, Fabre P, Ricklefs RE, Fjeldså J. 2010. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc. Natl Acad. Sci. USA 108, 2328–2333. ( 10.1073/pnas.1018956108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyle RG, Oliveros CH, Andersen MJ, Hosner PA, Benz BW, Manthey JD, Travers SL, Brown RM, Faircloth BC. 2016. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat. Commun. 7, 1–7. ( 10.1038/ncomms12709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown WC. 1997. Biogeography of amphibians in the Islands of the southwest Pacific. Proc. Calif. Acad. Sci. 50, 21–38. [Google Scholar]
- 20.Boer AJ De, Duffels JP. 1996. Historical biogeography of the cicadas of Wallacea, New Guinea and the West Pacific: a geotectonic explanation. Palaeogeogr. Palaeoclimatol. Palaeoecol. 124, 153–177. ( 10.1016/0031-0182(96)00007-7) [DOI] [Google Scholar]
- 21.Lohman DJ, de Bruyn M, Page T, von Rintelen K, Hall R, Ng PKL, Shih H-T, Carvalho GR, von Rintelen T. 2011. Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226. ( 10.1146/annurev-ecolsys-102710-145001) [DOI] [Google Scholar]
- 22.Toussaint EFA, Hall R, Monaghan MT, Sagata K, Ibalim S, Shaverdo HV, Vogler AP, Pons J, Balke M. 2014. The towering orogeny of New Guinea as a trigger for arthropod megadiversity. Nat. Commun. 5, 4001 ( 10.1038/ncomms5001) [DOI] [PubMed] [Google Scholar]
- 23.Brown RM, Siler CD, Das I, Min Y. 2012. Testing the phylogenetic affinities of Southeast Asia's rarest geckos: flap-legged geckos (Luperosaurus), flying geckos (Ptychozoon) and their relationship to the pan-Asian genus Gekko. Mol. Phylogenet. Evol. 63, 915–921. ( 10.1016/j.ympev.2012.02.019) [DOI] [PubMed] [Google Scholar]
- 24.Oliver PM, Parker F, Tallowin O. 2015. Further records of reptiles and amphibians utilising ant plant (Rubiaceae) domatia in New Guinea. Herpetol. Notes 8, 239–241. [Google Scholar]
- 25.Siler CD, Dececchi TA, Merkord CL, Davis DR, Christiani TJ, Brown RM. 2014. Cryptic diversity and population genetic structure in the rare, endemic, forest-obligate, slender geckos of the Philippines. Mol. Phylogenet. Evol. 70, 204–209. ( 10.1016/j.ympev.2013.09.014) [DOI] [PubMed] [Google Scholar]
- 26.Heinicke MP, Greenbaum E, Jackman TR, Bauer AM. 2012. Evolution of gliding in Southeast Asian geckos and other vertebrates is temporally congruent with dipterocarp forest development. Biol. Lett. 8, 994–997. ( 10.1098/rsbl.2012.0648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver PM, Skipwith P, Lee MSY. 2014. Crossing the line: increasing body size in a trans-Wallacean lizard radiation. Biol. Lett. 10, 20140479 ( 10.1098/rsbl.2014.0479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. 2004. MUSCLE User Guide. Nucleic Acids Res. 32, 1–15. ( 10.1093/chemse/bjs128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 31.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 32.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. See http://tree.bio.ed.ac.uk/software/tracer.
- 33.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukoschek V, Scott Keogh J, Avise JC. 2012. Evaluating fossil calibrations for dating phylogenies in light of rates of molecular evolution: a comparison of three approaches. Syst. Biol. 61, 22–43. ( 10.1093/sysbio/syr075) [DOI] [PubMed] [Google Scholar]
- 35.Gamble T, Greenbaum E, Jackman TR, Bauer AM. 2015. Into the light: diurnality has evolved multiple times in geckos. Biol. J. Linn. Soc. 115, 896–910. ( 10.1111/bij.12536) [DOI] [Google Scholar]
- 36.Renner SS. 2016. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. J. Biogeogr. 43, 1479–1487. ( 10.1111/jbi.12755) [DOI] [Google Scholar]
- 37.Lee MSY, Oliver PM, Hutchinson MN. 2009. Phylogenetic uncertainty and molecular clock calibrations: a case study of legless lizards (Pygopodidae, Gekkota). Mol. Phylogenet. Evol. 50, 661–666. ( 10.1016/j.ympev.2008.11.024) [DOI] [PubMed] [Google Scholar]
- 38.Matzke NJ. 2013. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 5.4, 242–248. [Google Scholar]
- 39.Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst. Biol. 63, 951–970. ( 10.1093/sysbio/syu056) [DOI] [PubMed] [Google Scholar]
- 40.Yumul GP, Dimalanta CB, Maglambayarw VB, Marquez EJ. 2008. Tectonic setting of a composite terrane: a review of the Philippine island arc system. Geosci. J. 12, 7–17. ( 10.1007/s12303-008-0002-0) [DOI] [Google Scholar]
- 41.van Ufford AQ, Cloos M. 2005. Cenozoic tectonics of New Guinea. Am. Assoc. Pet. Geol. Bull. 89, 119–140. ( 10.1306/08300403073) [DOI] [Google Scholar]
- 42.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 43.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131. ( 10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 44.Tallowin O, Allison A, Algar AC, Kraus F, Meiri S. 2017. Papua New Guinea terrestrial-vertebrate richness: elevation matters most for all except reptiles. J. Biogeogr. 44, 1734–1744. ( 10.1111/jbi.12949) [DOI] [Google Scholar]
- 45.Rocha S, Ineich I, Harris J. 2009. Cryptic variation and recent bipolar range expansion within the Stumped-Toed Gecko Gehyra mutilata across Indian and Pacific Ocean Islands. Contrib. Zool. 78, 1–8. [Google Scholar]
- 46.Oliver PM, Travers SL, Richmond JQ, Pikacha P, Fisher R. 2017. At the end of the line: independent overwater colonisations of the Solomon Islands by a hyperdiverse trans-Wallacean lizard lineage (Cyrtodactylus: Gekkota: Squamata). Zool. J. Linn. Soc. zlx047. [Google Scholar]
- 47.Unmack PJ, Allen GR, Johnson JB. 2013. Phylogeny and biogeography of rainbowfishes (Melanotaeniidae) from Australia and New Guinea. Mol. Phylogenet. Evol. 67, 15–27. ( 10.1016/j.ympev.2012.12.019) [DOI] [PubMed] [Google Scholar]
- 48.Brennan IG, Oliver PM. 2017. Mass turnover and recovery dynamics of a diverse Australian continental radiation. Evolution 71, 1352–1365. ( 10.1111/evo.13207) [DOI] [PubMed] [Google Scholar]
- 49.Oliver PM, Hugall AF. 2017. Phylogenetic evidence for mid-Cenozoic turnover of a diverse continental biota. Nat. Ecol. Evol. 12, 1896 ( 10.1038/s41559-017-0355-8) [DOI] [PubMed] [Google Scholar]
- 50.Mitchell KJ, et al. 2014. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol. Biol. Evol. 31, 2322–2330. ( 10.1093/molbev/msu176) [DOI] [PubMed] [Google Scholar]
- 51.Strickland JL, Carter S, Kraus F, Christopher L. 2016. Snake evolution in Melanesia: origin of the Hydrophiinae (Serpentes, Elapidae), and the evolutionary history of the enigmatic New Guinean elapid Toxicocalamus. Zool. J. Linn. Soc. 178, 663–678. ( 10.1111/zoj.12423) [DOI] [Google Scholar]
- 52.Stelbrink B, Albrecht C, Hall R, Rintelen TV. 2012. The biogeography of Sulawesi revisited: is there evidence for a vicarient origin of taxa on Wallace's ‘anomalous island’? Evolution 66, 2252–2271. ( 10.5061/dryad.7nk1nc63) [DOI] [PubMed] [Google Scholar]
- 53.Hall R. 1998. The plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In Biogeography and geological evolution in South-East Asia (eds Holloway JD, Hall R), pp. 99–131. Kerkwerve, The Netherlands: Backhuys Publishers. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All alignments, supplementary methods, tables and figures are provided in the electronic supplementary material.