Abstract
Introduction
Although a growing list of essential genomic/immune-based biomarkers are linked to approved non-small-cell lung cancer (NSCLC) therapies worldwide, few reports have detailed the evolution of NSCLC predictive biomarker assessment in routine clinical practice.
Methods
We retrospectively reviewed the first one thousand plus NSCLC patient specimens from our institution analyzed for predictive biomarkers from 2004 to 2017 and evaluated patterns of testing as well as correlation with clinical-pathologic characteristics.
Results
The majority of 1009 NSCLC patients had advanced stages of adenocarcinoma with most tissues obtained from the lung, mediastinal/hilar nodes, or pleura. The majority of testing was performed on cytology or small biopsy specimens. All were tested for EGFR mutations, 895 for ALK rearrangement, 841 for KRAS mutation, 537 for ROS1 rearrangement, and 179 using comprehensive genomic profiling. Implementation of near-universal genomic biomarker testing at our institution for EGFR, ALK, ROS1 and PD-L1 all occurred within the first year following evidence of clinical activity or regulatory body approval of an associated inhibitor. The overall testing failure rate after use of the best specimen for the most common tests was ≤5.5%. A quarter of tumors had a driver oncogene identified (EGFR/ALK/ROS1/BRAF V600E) with an approved oral targeted therapy, with the highest prevalence in those patients with no or light (≤15 pack-years) history of tobacco use.
Conclusions
Tumor biomarker testing using clinical NSCLC specimens in routine oncologic care evolves rapidly following approval of targeted therapies linked to diagnostic assays. Our practice's decade plus experience highlights the rapid evolution of biomarker testing and confirms the therapeutic relevance of such testing in all patients—particularly those patients with light/no history of tobacco use.
Keywords: Keywords: lung cancer, adenocarcinoma, biomarker testing, smoking history, EGFR, ALK
1. Introduction
As recently as a decade ago, the management of advanced non-small-cell lung cancer (NSCLC) was relatively uniform, with limited/absent ability to optimally match patients with best selected systemic therapies using tumor-based predictive biomarkers. Much has changed since then, with tumor genomic and/or immunologic biomarker testing now imperative in the initial assessment and management of advanced NSCLC, particularly adenocarcinomas. The growing list of essential biomarkers that are linked to approved therapies worldwide include: epidermal growth factor receptor (EGFR) gene mutations, anaplastic lymphoma receptor tyrosine kinase (ALK) gene rearrangements, ROS proto-oncogene 1 receptor tyrosine kinase (ROS1) gene rearrangements, B-Raf proto-oncogene serine/threonine kinase (BRAF) V600E gene mutations, and programmed death-ligand 1 (PD-L1) expression using immunohistochemistry (IHC). In addition, the rapid evolution of tumor genotyping platforms with the advent of commercially-available comprehensive genomic profiling sequencing technologies has allowed for the identification of other potentially actionable driver oncogenes such as: MET proto-oncogene receptor tyrosine kinase (MET) gene mutations or amplification, Erb-B2 receptor tyrosine kinase 2 (ERBB2) gene mutations and amplifications, Ret proto-oncogene (RET) gene rearrangements, and neurotrophic receptor tyrosine kinase (NTRK) gene rearrangements among others. However, the most common genomic events in lung cancer-tumor protein P53 (TP53) and KRAS proto-oncogene GTPase (KRAS) gene mutations—remain elusive drug targets.
The rapid pace of drug approvals with matched companion diagnostic assays has been documented in intermittent snapshots focusing on a particular year, technology, or therapeutic agent. Few, if any, reports have described the evolution of biomarker assessment in routine clinical practice. The Cancer Genome Atlas (TCGA) dataset represents the most comprehensive genomic profiling efforts in NSCLC to date; however, specimens analyzed were from surgically-resected tumors and thus may not fully capture the process and outcomes of tumor genomic profiling in de novo advanced/recurrent metastatic disease, where genomic profiling and therapeutic stratification is often accomplished using much smaller pathologic specimens from metastatic sites of disease. Therefore, we sought to compile our medical center's decade plus evolving experience with diagnostic tumor-based predictive biomarker testing in routine clinical care in order to provide a historical perspective on and highlight future opportunities for the implementation of precision oncology into thoracic oncology clinics.
2. Methods
2.1 Tumor and data collection
Patient and tumor specimen pairs diagnosed and/or followed at Beth Israel Deaconess Medical Center (BIDMC) with a diagnosis of NSCLC were recorded through an ongoing Institutional Review Board-approved study. The genomic cohort of this report was designed to match evolving evidence-based genomic biomarker testing in advanced NSCLC. EGFR genotyping, either through single gene assay or next generation sequencing (NGS), as the initial predictive biomarker receiving evidence-base status was a pre-requisite for initial inclusion of our tumor-patient pairs [1,2]. As such, this design results in a skewing of the data towards testing in non- squamous tumors [1,2]. When multiple tumors were tested, only the best available diagnostic specimen for testing was entered. Clinical-pathologic data, tumor genotype, and other tumor biomarkers were obtained from retrospective electronic chart extraction. Data was managed using the REDCap electronic data capture held at BIDMC. The dates for data assessment for this study spanned from January 1st, 2004 through April 19th, 2017.
2.2 Tumor genomic analyses and other biomarker tests
Tumor genotype was performed by analyzing EGFR (at least Sanger sequencing of exons 18-21 until 2016 or multiplex PCR for common exon 18-21 mutations since 2016), ALK (fluorescence in situ hybridization [FISH] break-apart probe, IHC, or NGS), ROS1 (FISH break-apart probe, IHC, or NGS), KRAS (sequencing of codons 12-13 or NGS), BRAF (sequencing of exon 15 or NGS) in tumor samples using a commercial vendor, as described previously [1]. NGS-based comprehensive genomic profiling and other FISH-based assays were evaluated using different commercially-available assays as described previously [2]. PD-L1 IHC testing was performed and interpreted by a commercial vendor using the PD-L1 clone 22C3 pharmDx kit as described previously [3].
2.3 Statistical methods
Fisher's exact test was used to compare categorical variables. All p-values reported are two-sided, and tests were conducted at the 0.05 significance level.
3. Results
3.1 Patient and tumor characteristics
The majority of patients in our cohort with NSCLC were older than 65 years, more frequently women, of White self-reported race, and former smokers (median 30 pack-years). Their tumors were most often of advanced stages with adenocarcinoma histology, obtained from thoracic sites (lung, mediastinal nodes, or pleura), and collected by minimally invasive techniques (small biopsies or needle aspirates/fluids made into formalin fixed paraffin embedded cell blocks). However, the studied population broadly includes a diverse and representative patient population (Table 1). The most frequent biomarker tested for was EGFR mutation at 100% testing rate, as this was an initial prerequisite for inclusion at the inception of this cohort in 2004 (Table 1). ALK rearrangement testing was ordered in 88.7% of cases, KRAS mutation testing in 83.3%, ROS1 rearrangement testing in 53.2%, and NGS-based testing and/or additional genotyping in 17.7% of tumors (Table 1).
Table 1. Baseline Clinical, Pathologic, and Biomarker Testing Characteristics of Lung Adenocarcinomas from 2004-2017.
| Age at time of tissue acquisition Median years-old (range) | 66 (27-92) |
|---|---|
| Gender | |
| Women n (%) | 594 (58.9%) |
| Men n (%) | 415 (41.1%) |
| Race n (%) | |
| White | 789 (78.2%) |
| Asian | 108 (10.7%) |
| Black | 74 (7.3%) |
| Other/Multiple | 38 (3.7%) |
| Ethnicity n (%) | |
| Non-Hispanic | 980 (97.1%) |
| Hispanic | 29 (2.9%) |
| Smoking status n (%) | |
| Current smoker | 240 (23.8%) |
| Former smoker | 529 (52.5%) |
| Never smoker | 238 (23.6%) |
| Pack-years smoking | |
| Median (range) | 30 (0-240) |
| Stage at tumor analyses n (%) | |
| I | 47 (4.7%) |
| II | 67 (6.7%) |
| III | 145 (14.4%) |
| recurrent | 61 (6.1%) |
| IV | 687 (68.2%) |
| Histology n (%) | |
| Adenocarcinoma | 899 (89.1%) |
| NSCLC (NOS) | 55 (5.5%) |
| Squamous cell carcinoma | 38 (3.8%) |
| Other | 17 (1.7%) |
| Type of tissue n (%) | |
| Surgical specimen | 359 (35.6%) |
| Small biopsy | 264 (26.2%) |
| Cytology block from aspirate/fluid | 385 (38.2%) |
| Anatomic site of tissue acquisition n (%) | |
| Lung | 445 (44.1%) |
| Mediastinal/hilar lymph node | 224 (22.2%) |
| Pleura | 132 (13.1%) |
| Soft tissue/bone | 57 (5.6%) |
| Brain | 53 (5.3%) |
| Liver | 31 (3.1%) |
| Extra-thoracic lymph node | 29 (2.9%) |
| Adrenal | 8 (0.8%) |
| Other | 30 (3.0%) |
| Tumor biomarker testing n (%, from total cases) | |
| EGFR mutation (exons 18-21) | 1009 (100%) |
| ALK rearrangement (FISH, IHC or NGS) | 895 (88.7%) |
| ROS1 rearrangement (FISH, IHC or NGS) | 537 (53.2%) |
| KRAS mutation | 841 (83.3%) |
| BRAF mutation | 143 (14.2%) |
| ERBB2 mutation | 144 (14.3%) |
| NGS-based testing/other technology | 179 (17.7%) |
3.2 Temporal trends in tumor genotyping and PD-L1IHC testing
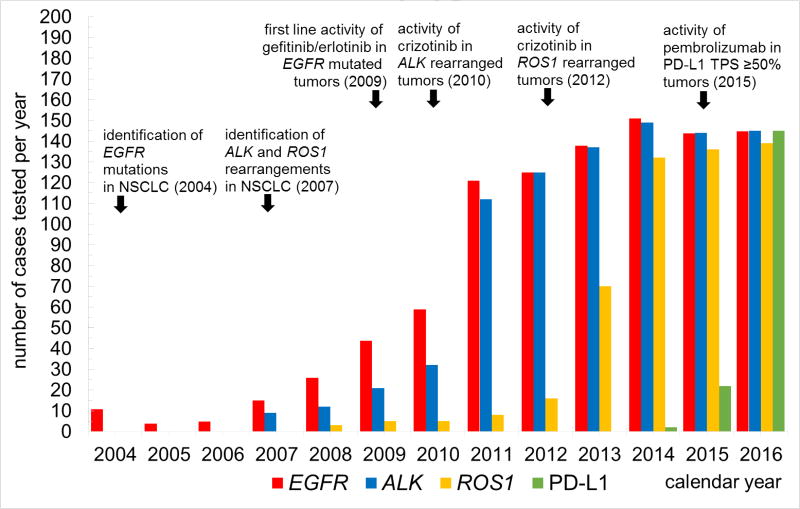
In order to understand the temporal trends within our institution of biomarker testing over time, annual testing volumes were plotted in Figure 1. Testing for EGFR mutations commenced in 2004 at an extremely low rate, increasing steadily as data from advanced phase trials accumulated showing that these mutations are strongly predictive of responses to EGFR- directed tyrosine kinase inhibitors (TKIs). Near-universal testing of all advanced stage adenocarcinomas for EGFR mutations was only achieved in 2011, following several key events: publication of the seminal IPASS and EURTAC trials in 2009 and 2011, respectively [4,5], and European Union approval of gefitinib and erlotinib for EGFR exon 19 deletion or L858R-bearing tumors in the first line setting. An even more striking uptake testing pattern occurred for ALK rearrangement testing, where occasional testing started in 2007 after the initial description of this genomic change in lung cancer. ALK testing quickly escalated to near-universal status in 2011 following the publication of the seminal PROFILE 1001 study in 2010 demonstrating brisk clinical responses to crizotinib in tumors with ALK rearrangements as identified by FISH [6] as well as approval of the drug for this NSCLC subset in 2011 by the United States Food and Drug Administration (FDA). Trends for ROS1 rearrangement testing shared similar characteristics, with near universal testing of advanced adenocarcinomas by 2014 following preliminary reports of the activity of crizotinib and commensurate with the publication of the activity of crizotinib in this subgroup in 2014 [7] leading to expanded FDA approval of the drug by 2016.
Figure 1. Annual biomarker testing volumes reflect evidence-based milestones in targeted therapies for non-small-cell lung cancer.
To compare genomic to immune-based biomarker uptake patterns, we have also included in Figure 1 data from our cohort of PD-L1 IHC testing. Although PD-L1 IHC testing increased in 2015 following the October 2015 FDA approval of pembrolizumab for previously treated NSCLC with any detectable PD-L1 by IHC, it had increased to levels that match currently recommended testing guidelines by 2016, even before the FDA expanded its approval of pembrolizumab to the first line setting for highly-expressing PD-L1 tumors (tumor proportion score [TPS] by IHC of ≥50%) in October 2016. Although data is preliminary for 2017 (data not shown), near universal testing of all advanced/recurrent NSCLCs for PD-L1 IHC was performed in initial diagnostic specimens as was EGFR/ALK/ROS1/BRAF genomic testing in all advanced or recurrent lung adenocarcinomas.
3.3 Evaluation of commonly ordered genomic biomarkers
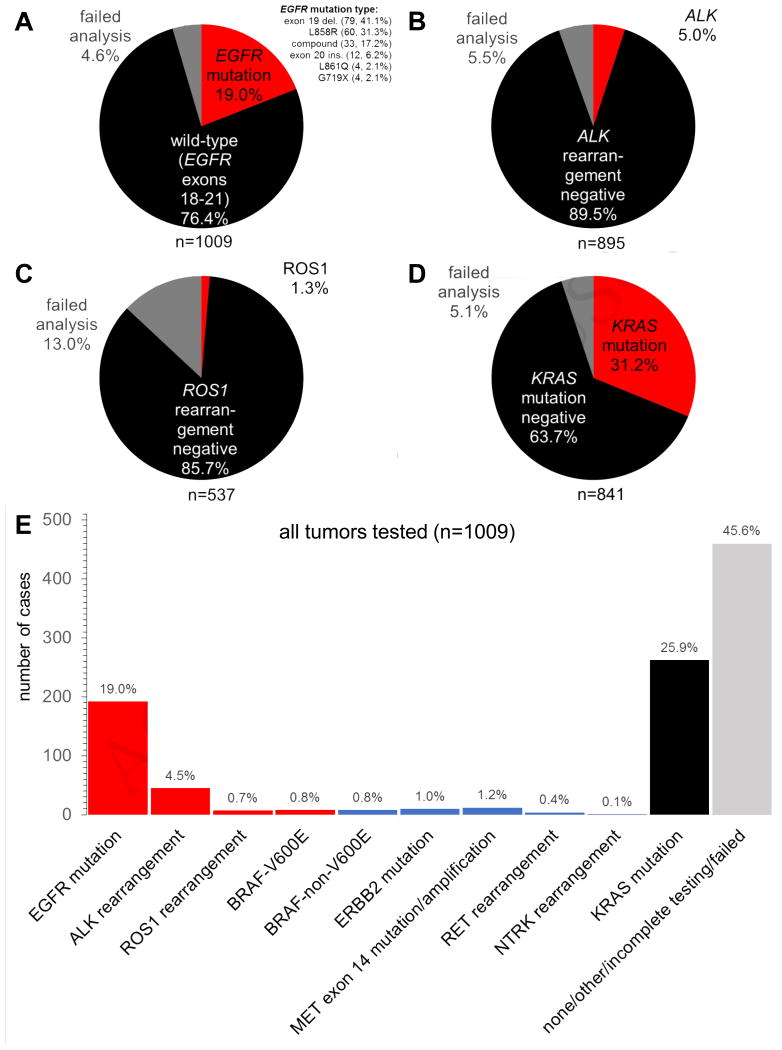
As with most groups, our experience is most robust with EGFR mutation testing, with an institutional genotyping success rate for EGFR exceeding 95% (Figure 2A) when the best available clinical specimen was selected. Overall, rates of positive genomic findings were consistent with other large series in the published literature to date. 19% of all 1009 tumors had an EGFR mutation, with EGFR exon 19 deletions (79, 41.1%) and L858R mutations (60, 31.1%) more frequently encountered than compound (33, 17.2%), exon 20 insertion (12, 6.2%), L861Q (4, 2.1%), or G719X (4, 2.1%) mutations (Figure 2A). The failed analysis frequency for ALK FISH was similarly low in our clinical specimens at 5.5%. A total of 5% of the 895 tumors harbored an ALK rearrangement (Figure 2B). The overall failure rate for ROS1 FISH was higher at 13% of the 537 tumors sent; 1.3% of tumors had a ROS1 rearrangement identified (Figure 2C). The failed analysis frequency for KRAS mutation was low at 5.1%, and 31.2% of the 841 tested tumors harbored a KRAS mutation (Figure 2D). Overlap of EGFR, ALK, ROS1, and KRAS genomic aberrations was not seen in tumors where any initial discrepancies in single gene assays were re-analyzed by NGS (data not shown). Therefore, our data support the practice of clinically obtained diagnostic NSCLC specimens for use in evidence-based biomarker assessment and are generally suitable for therapeutic decision making with low failure rates.
Figure 2.
Genomic testing results and failure rates for non-small-cell lung cancer. A. EGFR mutation results. B. ALK FISH results. C. ROS1 FISH results. D. KRAS mutation results. E. Distribution of testing results for the entire testing cohort; red bars indicate genomic alterations with approved matched therapies, blue bars with emerging biomarker targets, and black/grey bars with no approved matched therapies.
When we evaluate all tumors tested in our real-world setting and take into consideration all results obtained (i.e. success, failure, or incomplete/not tested), more than half of tumors had a recognized driver mutation (Figure 2E). Further, a quarter of all tumors had an actionable driver oncogene (EGFR/ALK/ROS1/BRAF-V600E) aberration associated with an approved oral targeted therapy (Figure 2E). However, biomarker testing as per current consensus guidelines in 45.6% of our 1009 patient-tumors pairs did not yield a driver mutation (Figure 2E), highlighting that a significant portion of patients with advanced NSCLC continue to receive standard oncologic care that is not biomarker-driven.
3.4 Genomic biomarkers in enriched cohorts
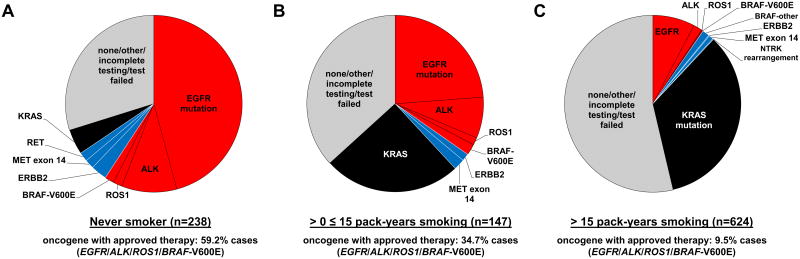
A prior study from our group had confirmed that cohorts with no history of tobacco use (0 pack- years) or light (1 to 15 pack-years) tobacco use are enriched for actionable oncogenes [8]. As such, we further evaluated these groups to evaluate the frequency of actionable oncogenes. Never smokers (n=238) had a driver oncogene (EGFR, ALK, ROS1, or BRAF-V600E) with an approved oral targeted therapy identified almost 60% of the time (Figure 3A). This was reduced to approximately 35% in the subset with a history of light tobacco use (≤15 pack-years, n=147), and down to 9.5% in patients with >15 pack-years tobacco history (n=624) (Figures 3B and 3C, respectively). When considering driver oncogenes (ERBB2, MET, RET, NTRK) where precision therapies hold developmental therapeutic promise, these alterations were found in 6.3% of never smokers, 3.3% of light smokers, and 1.0% of patients with >15 pack-years tobacco history (Figure 3A-C).
Figure 3. Genomic testing results for non-small-cell lung cancer stratified by tobacco use. A. Never smokers. B. 1-15 pack years. C. > 15 pack years.
4. Discussion
In this brief report, we provide a retrospective snapshot of the evolution of biomarker testing in advanced NSCLC over more than a decade at our institution. Given the dire prognosis associated with this disease, rapidly evolving, patient-/tumor-specific treatment options are a welcome reality in the day-to-day care of patients in the thoracic oncology clinic. Identifying those patients most likely to benefit from targeted therapies has led to increasing reliance on now mandatory biomarker testing in routine clinical lung cancer specimens.
Over the past decade, this testing has largely followed an add-on paradigm, with additional single gene assays performed sequentially on targets once efficacy has been shown in the rigorous, advanced phase trials and regulatory approval is granted for new drugs in specified populations. However, as we rely more heavily on minimally invasive techniques for obtaining diagnostic small biopsy or cytology specimens that are shared as biomarker testing substrates, this one-biomarker one-test model may be reaching practical limits. Overall, the testing failure rate in our experience has been low, ranging from <5% to 13% depending on the target, and this considering that nearly 2/3 of tested specimens over the past decade have been performed on cytology cell block or small biopsy specimens. Testing success on these limited specimens can be maximized by developing institutional measures to optimize specimen acquisition, specimen processing in the pathology laboratory, specimen selection for molecular testing, and processing of the specimen in the molecular laboratory [9]. Systems for multi-disciplinary collaboration with colleagues in procedural disciplines obtaining biopsies, evaluating pathologists, and treating oncologists are imperative to ensure the success of this effort. Moving forward, the testing paradigm will inevitably move towards comprehensive molecular profiling through large NGS-based panels, given the pragmatic need for multiplex biomarker assessment on small diagnostic specimens. Indeed, the clinical utility of a more comprehensive NGS testing approach has already proven effective in a large cohort of patients with metastatic lung cancer, resulting in a significant proportion of patients treated with a matched therapy that was guided by their tumor molecular profile [10].
An additional important highlight from this large cohort of patients with NSCLC is the importance of smoking status with respect to the frequency and type of genomic alterations. Lung cancers in never smokers and those with less than 15 pack-years tobacco history as a group are biologically different from those with more extensive tobacco history. Our cohort confirms the observation of many other studies that EGFR and ALK alterations are much more frequently observed in never/light smokers, whereas KRAS mutations are more frequently found in heavy smokers. However, the frequency of potentially targetable genomic alterations in never/light smokers extends beyond just these common biomarkers, as similar trends were also seen for less common genomic alterations (i.e. ROS1, BRAF V600E, and ERBB2). For patients with less than 15 pack-years tobacco use, comprehensive genomic profiling appears to be particularly beneficial, with actionable genomic alterations identified in up to 65% of tumors when no targetable mutations were identified using targeted/single gene molecular and FISH testing [11].
Ultimately, the goal of tumor molecular testing is to identify the appropriate targeted therapeutic approaches that improve patients' survival. Prolonged median overall survivals have been reported for patients with NSCLC that receive precision oncology when a genomic biomarker can be identified [12,13]. Although the data collected in this overall patient testing cohort was not empowered to follow median survival trends for genomically-defined groups of NSCLC, prior small subgroup analyses of patients with EGFR mutated and ALK rearranged tumors have demonstrated impressive median overall survival times that exceed 3 years [14,15].
As we are now more than a decade into the era of targeted therapies for the management of advanced NSCLC, tumor genomic profiling to optimally pair patients with best therapies is the standard of care and an ever evolving arena with regards to molecular diagnostics and therapeutic applications. New molecular targets will continue to be identified, and testing modalities along with institutional testing processes must adapt accordingly. Minimally invasive procedures for tissue procurement will remain a cornerstone for cytologic and pathologic diagnosis of lung cancer, and these small specimens must be judiciously used to extract the growing amount of clinically necessary data to guide best practice treatment decisions.
Highlights.
Review of biomarker testing on over 1000 patients with non-small cell lung cancer.
Biomarker testing patterns evolve with approval of new targeted therapies.
Smoking status affects the frequency of targetable oncogenic drivers.
Acknowledgments
Funding/Grant Support: This work was funded in part through an American Cancer Society grant RSG 11-186 (DBC), National Cancer Institute grants P50CA090578 (DBC), R01CA169259 (SSK) and R21CA17830 (SSK), and internal donations to Beth Israel Deaconess Medical Center.
Footnotes
Conflict of interest: DBC has received consulting fees and honoraria from Pfizer, Boehringer Ingelheim and Ariad pharmaceuticals; outside the submitted work. PAV has received consulting fees from Gala Therapeutics; outside the submitted work. No other conflict of interest is stated.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.VanderLaan PA, Yamaguchi N, Folch E, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer. 2014;84:39–44. doi: 10.1016/j.lungcan.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rangachari D, VanderLaan PA, Le X, et al. Experience with targeted next generation sequencing for the care of lung cancer: insights into promises and limitations of genomic oncology in day-to-day practice. Cancer Treat Commun. 2015;4:174–181. doi: 10.1016/j.ctrc.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangachari D, VanderLaan PA, Shea M, et al. Correlation between Classic Driver Oncogene Mutations in EGFR, ALK, or ROS1 and 22C3-PD-L1 >/=50% Expression in Lung Adenocarcinoma. J Thorac Oncol. 2017;12:878–883. doi: 10.1016/j.jtho.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 6.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small- cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROSI-Rearranged Non-Small-Cell Lung Cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi N, VanderLaan PA, Folch E, et al. Smoking status and self-reported race affect the frequency of clinically relevant oncogenic alterations in non-small-cell lung cancers at a United States-based academic medical practice. Lung Cancer. 2013;82:31–37. doi: 10.1016/j.lungcan.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker Testing in Lung Carcinoma Cytology Specimens: A Perspective From Members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2016 Apr;:15. doi: 10.5858/arpa.2016-0091-SA. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rangachari D, Yamaguchi N, VanderLaan PA, et al. Brain metastases in patients with EGFR-mutated or ALK-rearranged non-small-cell lung cancers. Lung Cancer. 2015;88:108–11. doi: 10.1016/j.lungcan.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangachari D, Le X, Shea M, et al. Cases of ALK-Rearranged Lung Cancer with 5-Year Progression-Free Survival with Crizotinib as Initial Precision Therapy. J Thorac Oncol. 2017;12:e175–e177. doi: 10.1016/j.jtho.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]