Abstract
Hepatocellular carcinoma is the third leading cause of cancer-related death worldwide and late diagnosis is the main cause of death in HCC patients. In this study expression patterns of HSP70, GPC3 and GS and their relationships with pathogenesis of HCC in Iranian patients were investigated. The expression of HSP70, GPC3 and GS were determined by immunohistochemistry and quantitative real-time PCR (q-PCR) methods, using 121 cases from patients with HBV alone, HCC without HBV, HBV+HCC and 30 normal tissues as control group. HSP70, GPC3 and GS were expressed in higher levels in HBV-related HCC samples compared to HBV alone group. The results showed that the labeling index of HSP70, GPC3 and GS are correlated with immunohistochemical and molecular expressions of HSP70, GPC3 and GS. The sensitivity and specificity for HCC diagnosis were 43.4% and 89.7% for HSP70, 64.3% and 90.4% for GPC3, and 60.7% and 94.3% for GS, respectively. The sensitivity and specificity of the panels with 3, 2 and 1 positive markers, regardless of which one, were 21.6% and 100%, 51.3% and 100% and 93.4% and 80.5% respectively. The current study demonstrated an association between HSP70, GPC3 and GS expressions and HBV-related HCC in our population. It was concluded that HSP70, GPC3 and GS expressions could be useful biomarkers for increasing the specificity and sensitivity of HCC diagnosis to acceptable level. Also, proper combinations of these 3 markers could improve diagnostic accuracy.
Keywords: HSP70, GPC3, GS, hepatocellular carcinoma, immunohistochemistry, quantitative real-time PCR
Introduction
Hepatocellular carcinoma (HCC) is an important disease of the liver in the world. It is the 5th common cancer and the third leading cause of cancer-related death.1 Chronic hepatitis B infection (HBV), hepatitis C (HCV), cirrhosis and alcohol consumption are common etiologies leading to HCC occurrence and also cancer development. 2,3 In regard to the environmental risk factor of HCC development Zhang et al.4 identified specific genetic and mutation features of HCCs associated with aflatoxin (AFB1) exposure, including mutations in ADGRB1, compared to HCCs from general populations. They associated these mutations with increased vascularization and expression of PD-L1 in HCC tissues. Aflatoxin B1 invokes induces apoptosis via death receptor pathway in hepatocytes.4-6 These findings might be used to identify patients with HCC due to aflatoxin exposure, and select therapies. However, more than half of the HCC patients have an evidence of chronic HBV infection. HBV induces liver inflammation followed by fibrosis and cirrhosis and subsequently increases the risk of HCC.7,8 Currently, Alavian et al.9 in a review article revealed that HBV infection was the main cause of HCC in Iran, Lebanon, Turkey and Yemen, while in North African nations, Saudi Arabia and Pakistan, HCV was strongly related to HCC. Late diagnosis is the main cause of death in HCC patients; therefore, early detection is crucial to preserve the patients.10 Liver biopsy and surgical procedures are routine methods for HCC diagnosis but these methods only report the morphological changes such as wide cell plates, loss of the reticular framework, small cell change, mitotic activity, stromal invasion, and vascular invasion. These features may be absent in some conditions and therefore are not accurate and sensitive enough, especially when limited tissue is available for evaluation.11 HCC development has a long process: it initiates by chronic liver diseases that spreads to the cirrhosis and finally HCC. The “trilogy” of HCC emphasizes that the risk of HCC is high in HBV infected patients and it is necessary to monitor the disease development from HBV to malignant conditions.12 Tracking the HCC-related biomarkers by immunohistochemistry is a reliable way to know pathogenesis of HCC13-16 and should be considered more than ever. Heat-shock proteins (HSP) are cell protectors against stress and chaperones for proteins. Hepatitis B infection acts as a stressful condition and induces HSP synthesis. HSP70 is a housekeeping gene which can block apoptosis and induces a carcinogenic role.17 Antiapoptotic effect of HSP70 with HSP27 ensures the survival of cells and promotes tumor cell proliferation.18 HSP70 sensitizes cancer cells to apoptosis and destroy tumors such as breast and colon carcinomas.19 Increased expression of HSP70 gene has been reported in patients suffering from HCC20 and also it was upregulated in early HCC.21 Overexpression of HSP70 protein has been studied in HCC by immunohistochemistry22 and it was related to the vascular invasion, high stage, and high Ki-67 index in HCC.22,23 HSP70 expression increases with the evolution of hepatocarcinogenesis and its immunoreactivity is nucleocytoplasmic and mostly focal.21 Due to the role of HSP70 in pathology of HCC, study of its molecular expression and immunohistochemical expression pattern can be useful for monitoring of HCC in an early stage. β-catenin is a dual function protein regulating cell adhesion and proliferation via the Wnt-signaling pathway. Genetic changes in exon 3 of the β-catenin gene are associated with HCC. More than 20% of HCC patients have mutations in β-catenin gene.24 These mutations affect the nuclear translocation of β-catenin protein followed by upregulation of glutamine synthetase (GS). In the mammalian liver, GS catalyzes synthesis of glutamine from glutamate and ammonia. Glutamine is a source of energy for tumor cells. In normal liver of human and murine, GS is localized in the hepatocytes surrounding the terminal hepatic venules,25 but in hepatocellular tumors GS has a diffuse expression pattern that indicates mutations in β-catenin gene.26 The gradual increased expression of GS was found by immunohistochemistry in precancerous lesions, early and advanced HCCs.27 It seems that, due to carcinogens native hepatocytes change to the GS positive cells which can produce GS positive tumor cells. Therefore, GS might be an important marker in HCC diagnosis.28 In this regard Christa et al.29 reported an increased expression pattern of GS mRNA, protein, and activity in human HCC.
Glypican-3 (GPC3) is a member of the glypican family of glycosyl-phosphatidylinositol- anchored cell surface heparan sulfate proteoglycans. It has cytoplasmic/ membranous staining pattern and is not expressed in normal liver or hepatocellular adenoma.30 GPC3 can reduce cell proliferation and induce apoptosis in specific tissues; 31 it has a tumor-suppressive role through gene methylation in tumorigenesis.32 Also, an overexpression of GPC3 has been seen in HCC; Sung et al.33and Capurro et al.27 showed increased expression of GPC3 messenger and protein in HCC patients. Moreover, immunohistochemical methods revealed a weak and focal staining pattern of GPC3 in HCC precursor lesions and diffuse pattern in HCC.34
Di Tommaso et al.35,36 and Tremosini et al.37 have reported usefulness of the triple panel include of HSP70, GPC3, and GS to distinguish HCC from high-grade dysplastic nodule. In another study, Lagana et al.38 implemented a plan to determine whether the HSP70, GPC3, and GS were useful in the distinction of HCC and hepatocellular adenoma. They concluded that GPC-3 and HSP-70 were helpful in separating carcinomas from adenomas but GS was not useful.
In this study, we evaluated diagnostic importance of this panel (HSP70, GPC3 and GS) in liver biopsies of HBV-related HCC patients. The expressions of biomarkers were determined by immunohistochemistry and quantitative real-time PCR (q-PCR) methods. The aim was to evaluate whether the application of this panel can increase its diagnostic power for the detection of HCC in an early stage in HBV infected patients.
Materials and Methods
Patients and tissue samples
Patients pathologically diagnosed with only chronic HBV infection (HBV), patients with only early hepatocellular carcinoma (single tumor < 5 cm without vascular invasion) (HCC) and patients with early HBV-related HCC (HBV+HCC), in accordance with WHO criteria. Healthy subject consisted of 30 subjects, 6 females and 24 males with a mean age of 52.33±6.216. A total of 121 cases were included in this study; 40 patients of the cases had HBV infection (28 males and 12 females, 53.85±9.582 mean age years), 41 patients with HCC (32 males and 9 females, 55.44±10.305 mean age years), and 40 patients with HBV+HCC (29 males and 11 females, 57.13±9.819 mean age years).
Samples were obtained from Namazi Hospital in Shiraz, Iran, and Shaheed Labbafinezhad Hospital, Tehran, Iran, during the period from September 2015 to May 2016. Also thirty healthy subjects who were voluntary donor for liver transplant with negative test for HBV, HCV and HCC serology and had normal values for alanine aminotransferase (ALT) were enrolled. Clinicopathological data were gathered from the medical records and pathologic data (Table 1). The study was approved by the ethics committee of the Zahedan University of Medical Sciences (IR.ZAUMS.REC.1394.211), and carried out in Infectious Diseases and Tropical Medicine Research Center, Zahedan, Iran and Histology Department, Zahedan University of Medical Sciences, Zahedan, Iran. The informed consent was obtained from all participants. The liver samples were divided into two pieces; one of them was immediately snap-frozen and stored at -80°C for subsequent RNA extraction. The other one was fixed in formalin (10%) and embedded in paraffin and processed for histological examination and immunohistochemistry. Histological diagnosis of HCC was done according to the international criteria. 39 All HBV patients were positive for HBsAg with ELISA and HBV-DNA with RT-PCR.
Table 1.
Demographic and clinical data of control (C), HBV infected (HBV), hepatocellular carcinoma (HCC) and HBV-related HCC (HBV+HCC) groups.
| Parameters | C, N (%) | HBV, N (%) | HCC, N (%) | HBV+HCC, N (%) | P |
|---|---|---|---|---|---|
| Age (years) | Mean age | Mean age | Mean age | Mean age | P=0.161 |
| 33±6.216 | 53.85±9.582 | 55.44±10.305 | 57.13±9.819 | F=1.740 | |
| Age range | Age range | Age range | Age range | ||
| 37-61 | 31-71 | 30-72 | 37-72 | ||
| Median | Median | Median | Median | ||
| 51.50 years | 58 years | 56 years | 59 years | ||
| Sex | |||||
| Male | 24(80.0) | 28(70.0) | 32(78.0) | 29(72.5) | |
| Female | 6(20.0) | 12(30.0) | 9(22.0) | 11(27.5) | P=0.738 |
| Hepatocellular carcinoma: | - | - | |||
| Well or moderately differentiated | 37(90.2) | 35(87.5) | |||
| Poorly differentiated | 4(9.8) | 5(12.5) | |||
| HCC grading: | - | ||||
| Early | 39(95.1) | 38(95.0) | |||
| G1 | - | 1(2.4) | 2(5.0) | ||
| G2-G3 | 1(2.4) | 0 | |||
| Total bilirubin (μ mol/l) | 15.43±5.65 | 18.76±6.75 | 28.45±12.24 | 33.10±11.77 | |
| ALT (U/I) | 26.23±10.90 | 45.76±32.03 | 88.25±95.32 | 117.76±102.54 | |
| AFP (ng/mL) | 2.12±1.14 | 3.12±2.79 | 421.21±104.33 | 534.54±420.76 | |
| Serum HBV DNA level | - | 7.6±0.8 | - | 7.8±0.1 | |
| Mean, log IU/mL (1SD) | |||||
| HBs-Ag positive | - | 40(100.0) | - | 40(100.0) | |
| HBe-Ab positive | - | 12(30.0) | - | 15(37.5) |
Total RNA isolation and q-PCR Analysis
Total RNA was extracted from fresh frozen liver tissue samples, using the CinnaPure RNA Kit (SinaClon BioScience, Tehran, Iran), according to the manufacturer’s instructions. In summary, 10 mg fresh tissues were grinded by mortar and pestle in liquid nitrogen then added 400 μL lysis solution immediately. After it was completely homogenized, 300 μL precipitation solution was added and centrifuged (at 12,000g, 13,000RPM) for 1 min. The upper aqueous phase was then removed and 400 μL wash buffer I was added to the precipitate and centrifuged (at 12,000g, 13,000RPM) for 1 min. The precipitate was washed with wash buffer II and then centrifuged (at 12,000g, 13,000RPM) for 1 min. Finally, 100 μL 55°C pre heated RNase-free water was placed to the precipitate and incubated for 3-5 min at 55°C. Thereafter, the solution was centrifuged (at 12,000g, 13,000RPM) for 1 min to elute the RNA and digested with DNase I to prevent DNA contamination. The RNA was stored at -80°C for experimental use. The quantity and quality of the total RNA samples were determined using an ultraviolet spectrophotometer and electrophoresis before reverse transcription. Complementary DNA (cDNA) was synthesized, using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Inc., Foster City, CA, USA) with the 2-step RT-PCR kit (vivantis), based on the manufacturer’s instructions. Briefly, 1 μg of total RNA was reverse-transcribed in 20 μL reaction solution using oligonucleotide (dT)-tailed primer and M-MuLV Reverse Transcriptase. The expression of HSP70, GPC3 and GS were detected with the LightCycler ABI 7500 system (Applied Biosystems, Inc.). GAPDH, as a reference gene was used for normalization. The sequences of primers used were showed in Table 2. The qRT-PCR cycles were as follows: initial denaturation at 94°C for 2 min; 94°C for 30 s and annealing at 60°C for 30 s through 40 cycles and 72°C for 30s. The 2-ΔΔCT method40 was used to determine the relative copy number and to count the gene expression as compared to the control. Each qPCR run was performed in duplicate and the ΔΔCT value was calculated for each sample.
Table 2.
The primer sequences used for the HSP70, GPC3 and GS expression (qRTPCR) analyses.
| PCR product size (bp) | Primers and probe (5’-3’) | Gene |
|---|---|---|
| HSP70 | F: GGTGGTGGGCATAGACCTG | 147 |
| R: GCTGCTCCAATTGAACGATTC | ||
| P: AGAGCTGCTACGTCGCTGTGG | ||
| GPC3 | F: AAGGGCCCTGAGCCAGTG | 138 |
| R: GCAGTCTCCACTTTCAAACC | ||
| P: AATTATTGACAAACTGAAGCACATTA | ||
| GS | F: ATGATGCTGTGCATATACCTAG | 109 |
| R: TAAAAACAATCACTATTGCCCAC | ||
| P: CAATTATTGGACACATTGGAGTGC | ||
| GAPDH (internal control) | F: CATGAGAAGTATGACAACAGCC | 70 |
| R: GGGGTGCTAAGCAGTTGGTG | ||
| P: CATCAGCAATGCCTCCTGCACC |
Immunohistochemistry
For deparaffinization, tissue sections were processed with xylene (10 min, 3 times). Then, sections were hydrated in graded alcohol. Endogenous peroxidase was blocked with aqueous 0.3% H2O2 for 30 min. Sections were heated in 10 mmol/L sodium citrate buffer solution, pH 6.0 at 120°C for 20 min by autoclave for antigen retrieval and allowed to cool for 20 min at RT. Monoclonal primary antibodies to HSP70 (1:100, mouse monoclonal antibody, Santa Cruz Biotechnologies, Inc., Santa Cruz, CA, USA), GPC3 (1:100, mouse monoclonal antibody, Santa Cruz Biotechnologies, Inc.) and GS (1:200, mouse monoclonal antibody, Santa Cruz Biotechnologies, Inc.) were used for immunostaining. Sections were incubated overnight at 4°C. Then the samples were exposed to the secondary antibodies (1:200, ABC mouse IgG, Santa Cruz Biotechnologies, Inc.) for 30 min at room temperature. Finally, sections were stained with DAB chromogen and counterstained with hematoxylin. The slides were passed through graded ethanol, sealed and evaluated microscopically for the intensity of biomarkers expressions afterwards. Biomarkers labeling indices were estimated based on the staining intensity and the percentage of positive cells on coded slides by two expert histologists. Randomly, 10 fields in each section were selected (meanly 500 hepatocytes). Staining pattern for HSP70, GPC3 and GS were nucleocytoplasmic, cytoplasmic, and diffuse and unrelated to vascular areas staining, respectively. External positive controls were always included in the batch of slides. HSP70 immunoreactive ductules were also used as an internal control. GPC3 is not usually immunoreactive in non-lesional hepatocytes or ductal cells. Only cases showing lesional areas of strong and diffuse immunoreactivity unrelated to vascularization were considered positive for GS. Nonlesional immunoreactive pericentral hepatocytes were used as an internal control.
Positive cells were counted in sequential high-powered fields (x400) and the results were expressed as the mean number of positive cells per surface area. Staining density was graded as follow: no staining or less than 5%, mild staining (<5-25%), moderate (25-75%), and severe (>75%)(41). The staining intensity was assigned a score of 0-3 (0-absent, 1-weak, 2-moderate, and 3- strong).
Statistical analysis
Statistical analysis was done using SPSS program ver. 2.0 statistical software package. To compare the intensity of biomarkers expressions between the groups, non-parametric Mann-Whitney test and Kruskal-Wallis test were used. Chi-square analysis was used for contingency table analysis and Fisher’s exact testing proportion independence. One-way ANOVA test was used to compare the mean expression levels among the groups with different expressions profiles. Finally, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of HSP70, GPC3 and GS were calculated. Bonferroni correction was used for multiple testing. P values less than 0.05 were considered as statistically significant. All P values were two-sided.
Results
The characteristics of the study cases are summarized in Table 1. Demographic data of HBV, HCC and HBV+HCC groups were compared with the control group. All groups were matched in terms of age and gender therefore there were no significant difference between the four groups (P>0.05). No correlation was shown between the staining patterns of the lesions with demographic data.
Analysis of relative HSP70, GPC3 and GS expression
The expressions levels of HSP70, GPC3 and GS genes were determined in all participants to identify whether HBV-related hepatocellular carcinoma was correlated with the expressions of these proteins. We founded that the levels of mRNA expressions of HSP70, GPC3 and GS were significantly different between cases (HBV, HCC, HBV+HCC) and healthy controls.
As shown in Figure 1, HSP70 gene expression was increased in cases, HBV, 1.39±0.22; HCC, 1.71±0.34; HBV+HCC, 2.72±0.42 compared to the controls, 0.66±0.26. These differences were statistically significant (P<0.001) compared with the controls. Moreover, HSP70 gene expression level was significantly different between HBV+HCC and HBV groups (P<0.001).
Figure 1.
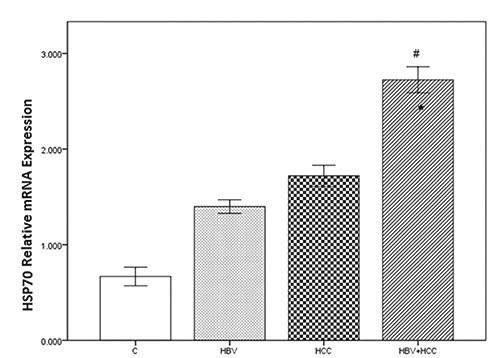
The HSP70 mRNA expression levels significantly increased in HBV+HCC liver tissue samples, compared to controls (#P Value <0.001) and HBV infected cases (*P Value <0.001). Bonferroni correction PBC<0.001.
As shown in Figure 2, GPC3 gene expression was increased in cases, HBV, 0.19±0.24; HCC, 0.90±0.25; HBV+HCC, 1.22±0.35 compared to the controls, 0.00. These differences were statistically significant (P<0.001). Moreover, GPC3 gene expression level was significantly different between HBV+HCC and HBV groups (P<0.001).
Figure 2.
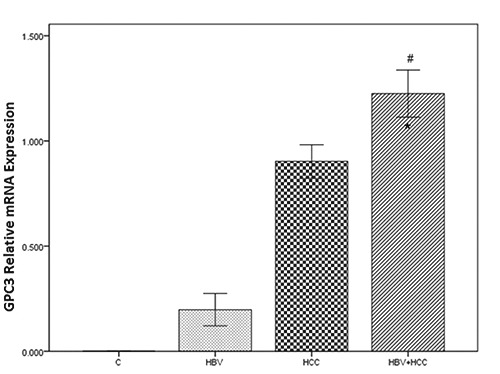
The GPC3 mRNA expression levels significantly increased in HBV+HCC liver tissue samples, compared to controls (#P Value <0.001) and HBV infected cases (*P Value <0.001). Bonferroni correction PBC<0.001.
The results indicated that the level of mRNA expression of GS was significantly different between cases and the matched normal tissues. As shown in Figure 3, GS gene expression was increased in HBV, 9.37±2.39; HCC, 10.90±2.69; HBV+HCC, 15.70±3.00, compared to the matching normal tissues 6.85±2.17, and the difference was statistically significant (P < 0.001).
Figure 3.

The GS mRNA expression levels significantly increased in HBV+HCC liver tissue samples, compared to controls (#P Value <0.001) and HBV infected cases (*P Value <0.001). Bonferroni correction PBC<0.001.
Immunohistochemical expression of the biomarkers
The expressions of the HSP70, GPC3 and GS genes were compared between the four groups; liver tissue samples from patients with HBV, HCC, HBV+HCC and healthy controls (Table 3). HSP70 immunoreactivity was localized in the nuclei/cytoplasm, and GPC3 and GS were localized in cytoplasm. The results showed that in patients with HBV+HCC, average HSP70 expression was higher than HBV (P<0.001) and HCC groups (P<0.001) (Table 3). GS expression was significantly higher in the specimens with HBV+HCC compared to those with only HBV infection (P<0.001) or HCC (P<0.001), (Table 3). Healthy controls were not immunostained against GPC3 antibody while cases groups (HBV, HCC, HBV+HCC) were immunoreactive against GPC3. GPC3 expression was significantly higher in the specimens with HBV+HCC compared to those with only HBV infection (P<0.001) or HCC (P<0.001, (Table 3).
Table 3.
Comparing the expression levels of HSP70, GPC3 and GS in liver tissue samples of HBV-related HCC, HBV infected, HCC and healthy control groups.
| Group | N | HSP70 positive, (mean±SEM) | P value | GPC3 positive, (mean±SEM) | P value | GS positive, (mean±SEM) | P value |
|---|---|---|---|---|---|---|---|
| C | 30 | 1.90±0.62 | <0.001 | - | 0 | 6.26±2.06 | <0.001 |
| HBV | 40 | 4.17±1.20 | <0.001 | 1.42±0.63 | =0.000 | 7.85±1.90 | <0.001 |
| HCC | 41 | 5.57±1.48 | <0.001 | 3.73±1.22 | =0.000 | 9.26±1.84 | <0.001 |
| HBV+HCC | 40 | 8.70±2.54* | <0.001 | 5.07±1.50** | =0.000 | 13.02±1.87*** | <0.001 |
*P<0.001, Compared with C, HBV and HCC groups. Bonferroni correction PBC<0.001
**P<0.001, Compared with C, HBV and HCC groups. Bonferroni correction PBC<0.001
***P<0.001, Compared with C, HBV and HCC groups. Bonferroni correction PBC <0.001.
In healthy liver sections there were a limited number of both HSP70 and GS positive cells as shown in Table 3, with mean expression level 1.90±0.62 and 6.26±2.06, respectively. The number of HSP70 and GS positive cells were increased significantly in HBV+HCC group compared to the HBV and HCC groups (P<0.05).
In Figure 4 A-D, Figure 5A-D and Figure 6 A-D the immunohistochemical stainings of HSP70, GPC3 and GS positive cells in the four groups (HBV+HCC, HCC, HBV and C) are shown; in HBV+HCC group HSP70, GPC3 and GS expressions were significantly higher compared to HBV and HCC groups (P<0.001).
Figure 4.
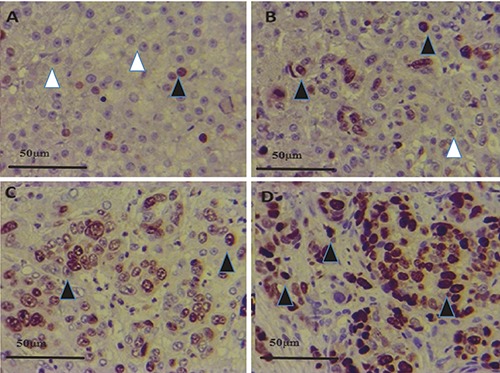
HSP70 expression in control (A), HBV (B), HCC (C) and HBV+HCC (D) liver tissue (immunperoxidase x400). HSP70 positive expression in hepatocytes (black arrowheads) and healthy cells (white arrowhead) are shown.
Figure 6.
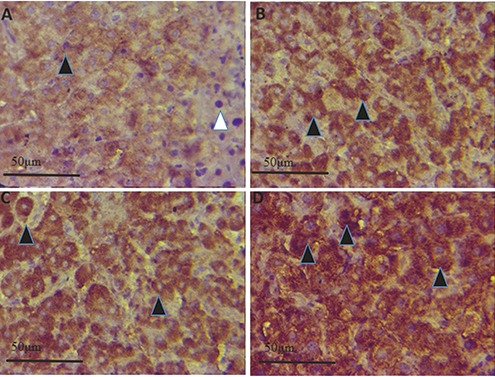
GS expression in control (A), HBV (B), HCC (C) and HBV+HCC (D) liver tissue (immunperoxidase x400). GS positive expression in hepatocytes (black arrowheads) and healthy cells (white arrowhead) are shown.
Overall, the sensitivity, specificity, positive and negative predictive values and accuracy of the three biomarkers for HCC detection were listed in Figure 7. HBV or control cases were not immunostained while immunoreactive cases for at least 1 marker steadily increased from HCC to HBV+HCC groups. However, at least 2 immunoreactive markers (++-), regardless of which two, were seen in HBV, HCC and HBV+HCC cases. Also, the diagnostic accuracy of the panel was considered. In a panel with at least 2 positive markers (++-), regardless of which two, the sensitivity for the detection of malignancy was 51.3% with 100% specificity. In other hand, in a panel with single marker immunoreactivity (+ -) the sensitivity increased (94.3%) but the specificity reduced markedly (80.5%). At least 2 positive markers regardless of which two, were able to identify HCC in more than 50% cases with 100% specificity.
Figure 7.
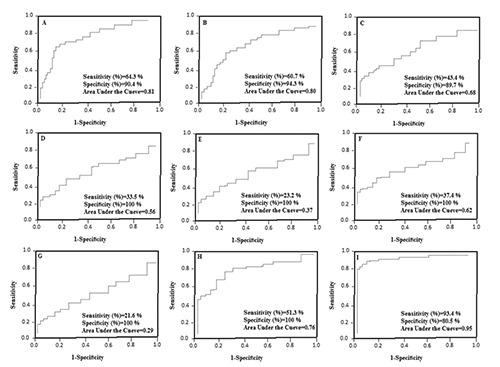
Diagnostic accuracy for detection of hepatocellular carcinoma using one, two or three markers. A) GPC3, (PPV=87.5%, NPV=65.8%, accuracy=78.4). B) GS, (PPV=92.2%, NPV=64.3%, accuracy =77.8). C) HSP70, (PPV=80.7%, NPV=54.8%, accuracy= 65.1). D) Hsp70+/GS+ (PPV=100%, NPV=58.6%, accuracy=65.3). E) Hsp70+/GPC3+ (PPV=100%, NPV=54.8%, accuracy=59.5). F) GPC3+/GS+ (PPV=100%, NPV=59.1%, accuracy=67.8). G) all 3 positive markers (PPV=100%, NPV=54.6%, accuracy=59.4). H) At least 2 positive markers (PPV=100%, NPV=65.4%, accuracy=74.2). I) At least 1 positive markers (PPV=84.3%, NPV=91.7%, accuracy= 88.6). GPC3, glypican 3; GS, glutamine synthetase; HSP70, heat shock protein 70; NPV, negative predictive value; PPV, positive predictive value.
Discussion
In some HCC cases, radiologic information are ambiguous and liver biopsy is the best way to establish the HBV-related HCC. In this regard, there are some difficulties in distinguishing the nature of a lesion especially in nodules with well-differentiated histology. For example, a dysplastic nodule can be introduced as malignant but a well-differentiated lesion may be suggested as normal liver; both conclusions have clinical implications for patients.35 In order to resolve these problems, molecular and immunohistochemical features have been used to increase the accuracy of diagnosis. Although, a good strategy has not been reported still for the use of biomarkers, but these compounds can improve the pathological diagnosis and real positive results.
In this study, we designed an approach to evaluate the diagnostic value of three markers of malignancy (HSP70, GPC3, and GS), in HBV-related HCC patients, either as individual marker or as a panel as a whole. We used qRT-PCR and immunohistochemistry because these two methods can confirm and complement the results of each other and also they are available techniques and reliable to be applied on fresh, paraffin embedded and formalin-fixed tissue sections. Our findings showed that the expressions of HSP70, GPC3, and GS significantly increased in HBV+HCC, HCC patients compared to the HBV infection, which can be used to detect early-stage HCC. The HSP70, GPC3, and GS could obviously predict HCC incidence in chronic hepatitis B patients. Expressions of these biomarkers were correlated with HCC risk in chronic HBV infection and were highly expressed in HBV infected patients. Furthermore, proper combination of HSP70, GPC3, and GS could increase the accuracy and precision of HCC diagnosis, which would help choose the best cure for patients and disease management, rather than use of only one marker.
Studies have shown more than 10% decrease in diagnosis sensitivity of the panel contains HSP70, GPC3, and GS in hepatocellular nodules in cirrhosis and it seems that heterogeneous pattern of HSP70 and GPC3 may be responsible for this issue.35 In our study, when comparing diagnostic accuracy of panels using one, two or three markers for detection of HCC, the sensitivity, specificity and accuracy of at least two immunoreactive markers (++-) was 51.3%, 100% and 74.2% respectively. On the other hand, the sensitivity of the panel with 3 markers (+++) lowered (21.6%), while specificity remained intact (100%). These results show that the maximum accuracy for HCC detection is related to the panel with at least 2 markers; regardless of which two are positive (++-). Since our samples were early stage hepatocellular lesions and did not have an aggressive essence, we propose that this panel may be appropriate for the understanding the natural history of challenging differentiated lesions in HBV patients.
The diagnosis of HBV-related HCC patients mainly depends on histological examinations using very tiny samples. Moreover, the stromal invasion as the main sign of HCC, is not found in some biopsies. 42 To address this challenge, use of analogous of p53 and/or p16 in Barrett’s esophagus and cervical cancer might be beneficial.43,44 In our results, at least two markers (++-); regardless of which two, overexpressed in about more than half of the HBV and HBV-related HCC cases with 74.2% accuracy. Also, there were samples with only one marker in both HBV and HBV-related HCC groups with strong and diffuse staining pattern. These results revealed that in HBV liver tissues at least 2 positive markers (++-) support the susceptibility to the HBV-related HCC but the expression of one positive marker was not enough significant in a way that true reported conclusion depends on which biomarker is positive and the staining pattern.
In regard to the useful biomarkers, Abdul-Al et al. revealed GPC3 overexpression in non-neoplastic HCV,45 but Di Tommaso36 did not see this issue in their cases which was consistent with our results. This inconsistency in results could be due to the different nature of various hepatitis infections. In addition, our findings revealed optimal results in fine needle biopsies for GS staining compared to resection in which GS has lower sensitivity against GPC3 and HSP70 markers. Malfunctions in the Wnt mechanism, and mutations in β-catenin genes, are related to the HCC. It seems that these phenomena are associated to the GS overexpression46 and also HCC may blurt GS overexpression as well as β-catenin mutations.47 However, HCC could not be ruled out by lack of immunoreactivity, but, the immunoreactivity of at least two markers can support the malignancy. In facing with illusive morphological features, the use of immunonegative profile is suggested. In other words, the use of two markers is recommended for the diagnosis of suspected cases even in minimal samples. Even in cases where hematoxylin and eosin staining could not rule out an obvious conclusion, this panel can probably highlight the nature of nodules.
As our results showed, the accuracy of the panel in regard to the combination of two markers out of three, was enough reliable and might be helpful in facing with challenging lesions. Some other immunohistochemical studies have reported diagnostic accuracy and specificity close to 100% in discrimination between well differentiated HCC and dysplastic nodules using the surgically resected specimens.48,49 Paradis et al.49 validated a molecular index for the diagnosis of HCC based on RT-PCR and 13 genes to report adequate diagnostic accuracies. Liovet et al.48 identified a molecular signature to diagnose early HCC. They were assessed transcriptional profiles of 55 candidate genes by RT-PCR and introduced a useful panel consisted of three genes, Glypican 3, BIRC5 and XLKD1. This panel which was based on gene transcriptional profiles of a 3-gene set allows a reliable diagnosis of early HCC with an overall accuracy of 94.60% and 94% specificity.
In conclusion, our observations clearly demonstrate the association between the HSP70, GPC3, and GS expression and the HCC risk in HBV infected patients. Our findings also emphasized that HSP70, GPC3, and GS proteins could be used to distinguish HCC at an early stage in peoples who suffered from hepatitis B. Finally, the proper combination of some biomarkers is more useful for the management of the liver malignancy in different stages. In other words, our panel can be improved by addition of some other markers to increase the accuracy for the HCC diagnosis. Given the high prevalence of HBV infection and malignant chronic diseases of liver worldwide, further studies are required in this field. These biomarkers could be used with some other specific proteins in HCC diagnosis in early stages.
Figure 5.
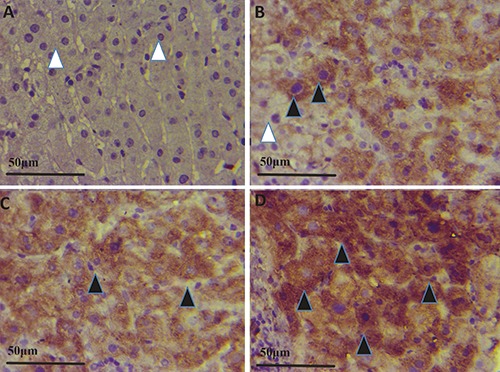
GPC3 expression in control (A), HBV (B), HCC (C) and HBV+HCC (D) liver tissue (immunperoxidase x400). GPC3 positive expression in hepatocytes (black arrowheads) and healthy cells (white arrowhead) are shown.
Acknowledgements
This study was supported by a dissertation grant (PhD thesis of BM #7262, IR.ZAUMS.REC.1394.211) from the deputy for Research, Zahedan University of Medical Sciences. The authors would like to thank all the subjects who willingly participated in the study.
References
- 1.Saber MA, MM AbdelHafiz S, Khorshed FE, Aboushousha TS, Hamdy HE, Seleem MI, Soliman AH. Differential expression of glypican-3 and insulin-like growth factor-II mRNAs and alpha-fetoprotein and Ki-67 markers in HCV related hepatocellular carcinomas in Egyptian patients. Asian Pac J Cancer Prev 2017;18:121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji X, Zhang Q, Du Y, Liu W, Li Z, Hou X, et al. Somatic mutations, viral integration and epigenetic modification in the evolution of hepatitis B virusinduced hepatocellular carcinoma. Current genomics. 2014;15(6):469-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M. Association between IL-10 gene promoter polymorphisms (-592 A/C, -819 T/C, -1082 A/G) and susceptibility to HBV infection in an Iranian population. Hepat Mon. 2016;16. Doi: 10.5812/.hepatmon. 32427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, He H, Zang M, Wu Q, Zhao H, Lu LL, et al. Genetic features of aflatoxin- associated hepatocellular carcinoma. Gastroenterology 2017;153:249-62.e2. [DOI] [PubMed] [Google Scholar]
- 5.Mughal MJ, Xi P, Yi Z, Jing F. Aflatoxin B1 invokes apoptosis via death receptor pathway in hepatocytes. Oncotarget 2017;8:8239-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Wang L, Yang X, Zeng H, Zhang R, Pu C, et al. Environmental microcystin exposure increases liver injury risk induced by hepatitis B virus combined with aflatoxin: A Cross-sectional study in southwest China. Environ Sci Technol 2017;51:6367-78. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76. [DOI] [PubMed] [Google Scholar]
- 8.Moudi B, Heidari Z, Mahmoudzadeh- Sagheb H. Impact of host gene polymorphisms on susceptibility to chronic hepatitis B virus infection. Infect Genet Evol 2016;44:94-105. [DOI] [PubMed] [Google Scholar]
- 9.Alavian SM, Haghbin H. Relative importance of hepatitis B and C Viruses in hepatocellular carcinoma in EMRO countries and the Middle East: A systematic review. Hepat Mon 2016;16: e35106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tawada A, Kanda T, Yokosuka O. Current and future directions for treating hepatitis B virus infection. World J Hepatol 2015;7:1541-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrell LD, Crawford JM, Dhillon AP, Scheuer PJ, Nakanuma Y. Proposal for standardized criteria for the diagnosis of benign, borderline, and malignant hepatocellular lesions arising in chronic advanced liver disease. Am J Surg Pathol 1993;17:1113-23. [DOI] [PubMed] [Google Scholar]
- 12.Lun-Gen L. Antiviral Therapy of liver cirrhosis related to hepatitis B virus infection. J Clin Translat Hepatol 2014;2:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelfattah MR, Abaalkhail F, Al- Manea H. Misdiagnosed or Incidentally detected hepatocellular carcinoma in explanted livers: Lessons learned. Ann Transplant 2015;20:366-72. [DOI] [PubMed] [Google Scholar]
- 14.Cioca A, Ceausu AR, Marin I, Raica M, Cimpean AM. The multifaceted role of podoplanin expression in hepatocellular carcinoma. Eur J Histochem 2017;61: 2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanni D, Manchia M, Lai F, Gerosa C, Ambu R, Faa G. Immunohistochemical markers of CYP3A4 and CYP3A7: A new tool towards personalized pharmacotherapy of hepatocellular carcinoma. Eur J Histochem 2016;60:2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerriero E, Capone F, Accardo M, Sorice A, Costantini M, Colonna G, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J Histochem 2015;59:2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle (Georgetown, Tex) 2006; 5:2592-601. [DOI] [PubMed] [Google Scholar]
- 18.Gehrmann M, Cervello M, Montalto G, Cappello F, Gulino A, Knape C, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immun 2014;5:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett 2012;325:117-24. [DOI] [PubMed] [Google Scholar]
- 20.Luk JM, Lam CT, Siu AF, Lam BY, Ng IO, Hu MY, et al. Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics 2006;6:1049-57. [DOI] [PubMed] [Google Scholar]
- 21.Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology 2003;37:198-207. [DOI] [PubMed] [Google Scholar]
- 22.Tan GS, Lim KH, Tan HT, Khoo ML, Tan SH, Toh HC, et al. Novel proteomic biomarker panel for prediction of aggressive metastatic hepatocellular carcinoma relapse in surgically resectable patients. J Proteome Res 2014;13:4833-46. [DOI] [PubMed] [Google Scholar]
- 23.Shin E, Ryu HS, Kim SH, Jung H, Jang JJ, Lee K. The clinicopathological significance of heat shock protein 70 and glutamine synthetase expression in hepatocellular carcinoma. J Hepatobiliary Pancreat Sci 2011;18: 544-50. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Fujii H, Sankila A, Mahler- Araujo BM, Matsuda M, Cathomas G, et al. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am J Pathol 1999;155:1795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorman AF, Vermeulen JL, Charles R, Lamers WH. Localization of ammoniametabolizing enzymes in human liver: ontogenesis of heterogeneity. Hepatology 1989;9:367-72. [DOI] [PubMed] [Google Scholar]
- 26.Evason KJ, Grenert JP, Ferrell LD, Kakar S. Atypical hepatocellular adenoma- like neoplasms with beta-catenin activation show cytogenetic alterations similar to well-differentiated hepatocellular carcinomas. Human Pathol 2013;44:750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology 2003;125:89-97. [DOI] [PubMed] [Google Scholar]
- 28.Gebhardt R, Tanaka T, Williams GM. Glutamine synthetase heterogeneous expression as a marker for the cellular lineage of preneoplastic and neoplastic liver populations. Carcinogenesis 1989;10:1917-23. [DOI] [PubMed] [Google Scholar]
- 29.Christa L, Simon MT, Flinois JP, Gebhardt R, Brechot C, Lasserre C. Overexpression of glutamine synthetase in human primary liver cancer. Gastroenterology 1994;106:1312-20. [DOI] [PubMed] [Google Scholar]
- 30.Wang XY, Degos F, Dubois S, Tessiore S, Allegretta M, Guttmann RD, et al. Glypican-3 expression in hepatocellular tumors: diagnostic value for preneoplastic lesions and hepatocellular carcinomas. Human Pathol 2006;37:1435-41. [DOI] [PubMed] [Google Scholar]
- 31.Filmus J. Glypicans in growth control and cancer. Glycobiology 2001;11:19r- 23r. [DOI] [PubMed] [Google Scholar]
- 32.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene 2001;20:7408-12. [DOI] [PubMed] [Google Scholar]
- 33.Sung YK, Hwang SY, Park MK, Farooq M, Han IS, Bae HI, et al. Glypican-3 is overexpressed in human hepatocellular carcinoma. Cancer Sci 2003;94:259-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamauchi N, Watanabe A, Hishinuma M, Ohashi K, Midorikawa Y, Morishita Y, et al. The glypican 3 oncofetal protein is a promising diagnostic marker for hepatocellular carcinoma. Mod Pathol 2005;18:1591-8. [DOI] [PubMed] [Google Scholar]
- 35.Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 2007;45:725-34. [DOI] [PubMed] [Google Scholar]
- 36.Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, et al. The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J Hepatol 2009; 50:746-54. [DOI] [PubMed] [Google Scholar]
- 37.Tremosini S, Forner A, Boix L, Vilana R, Bianchi L, Reig M, et al. Prospective validation of an immunohistochemical panel (glypican 3, heat shock protein 70 and glutamine synthetase) in liver biopsies for diagnosis of very early hepatocellular carcinoma. Gut 2012;61:1481-7. [DOI] [PubMed] [Google Scholar]
- 38.Lagana SM, Salomao M, Bao F, Moreira RK, Lefkowitch JH, Remotti HE. Utility of an immunohistochemical panel consisting of glypican-3, heatshock protein-70, and glutamine synthetase in the distinction of low-grade hepatocellular carcinoma from hepatocellular adenoma. Appl Immuno - histochem Mol Morphol 2013; 21:170-6. [DOI] [PubMed] [Google Scholar]
- 39.Gibson JB, Sobin LH. Histological typing of tumors of the liver, biliary tract and pancreas. International histological classification of tumors, Number 20. WHO; Geneva; 1978; p. 12-30. [Google Scholar]
- 40.Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics 2006;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohamed WS, Omar MM, Khayri TM, Fakhr IM. Assessment of the proliferative marker Ki-67 and p53 protein expression in HBV- and HCV-related hepatocellular carcinoma cases in Egypt. Int J Health Sci 2008;2:27-34. [PMC free article] [PubMed] [Google Scholar]
- 42.Park YN, Kojiro M, Di Tommaso L, Dhillon AP, Kondo F, Nakano M, et al. Ductular reaction is helpful in defining early stromal invasion, small hepatocellular carcinomas, and dysplastic nodules. Cancer 2007;109:915-23. [DOI] [PubMed] [Google Scholar]
- 43.Lorinc E, Jakobsson B, Landberg G, Veress B. Ki67 and p53 immunohistochemistry reduces interobserver variation in assessment of Barrett’s oesophagus. Histopathology 2005;46:642-8. [DOI] [PubMed] [Google Scholar]
- 44.Dray M, Russell P, Dalrymple C, Wallman N, Angus G, Leong A, et al. p16(INK4a) as a complementary marker of high-grade intraepithelial lesions of the uterine cervix. I: Experience with squamous lesions in 189 consecutive cervical biopsies. Pathology 2005;37: 112-24. [DOI] [PubMed] [Google Scholar]
- 45.Abdul-Al HM, Makhlouf HR, Wang G, Goodman ZD. Glypican-3 expression in benign liver tissue with active hepatitis C: implications for the diagnosis of hepatocellular carcinoma. Human Pathol 2008;39:209-12. [DOI] [PubMed] [Google Scholar]
- 46.Audard V, Grimber G, Elie C, Radenen B, Audebourg A, Letourneur F, et al. Cholestasis is a marker for hepatocellular carcinomas displaying beta-catenin mutations. J Pathol 2007;212:345-52. [DOI] [PubMed] [Google Scholar]
- 47.Bioulac-Sage P, Rebouissou S, Thomas C, Blanc JF, Saric J, Sa Cunha A, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology 2007;46:740-8. [DOI] [PubMed] [Google Scholar]
- 48.Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology 2006;131:1758-67. [DOI] [PubMed] [Google Scholar]
- 49.Paradis V, Bieche I, Dargere D, Laurendeau I, Laurent C, Bioulac Sage P, et al. Molecular profiling of hepatocellular carcinomas (HCC) using a large-scale real-time RT-PCR approach: determination of a molecular diagnostic index. Am J Pathol 2003;163:733-41. [DOI] [PMC free article] [PubMed] [Google Scholar]


