Abstract
In Escherichia coli, proteins found in the periplasm or the outer membrane are exported from the cytoplasm by the general secretory, Sec, system before they acquire stably folded structure. This dynamic process involves intricate interactions among cytoplasmic and membrane proteins, both peripheral and integral, as well as lipids. In vivo, both ATP hydrolysis and protonmotive force are required. Here, we review the Sec system from the inception of the field through the present day, including biochemical, genetic and structural data.
Keywords: Sec system, protein export, translocation, precursors
Introduction
E. coli has eight different systems for export of protein. This chapter focuses on the general secretory system, the Sec system, which handles the great majority of proteins that are exported from the site of synthesis of all bacterial proteins, the cytoplasm, into the cytoplasmic membrane, the outer membrane or the aqueous space between, the periplasm. The periplasm is the final destination for some proteins but for others it is only the initial step. Proteins destined for the outer membrane engage several periplasmic chaperones. A multiprotein complex, the β barrel assembly machine, BAM, facilitates transfer and insertion of the β-barrel proteins into the outer membrane. Lipoproteins arrive in the periplasm and are sorted either to the cytoplasmic membrane or to the outer membrane by the Lol transport system.
Membrane proteins integral to the cytoplasmic membrane are inserted co-translationally while undergoing elongation on the ribosome and are recruited to the Sec system by the signal recognition particle (SRP). YidC functions in concert with the SRP-Sec pathway in insertion of cytoplasmic membrane proteins and assembly of multimeric membrane proteins. YidC can also function independently as an insertase for proteins with short transmembrane sequences.
The Sec system transports proteins before they acquire stable tertiary structure. Proteins that are folded are transferred to the periplasm via the twin arginine translocation, TAT, system. Some proteins secreted to the periplasm by either the Sec system or TAT system are further exported across the outer membrane into the extracellular environment by specialized apparatuses of the type II or type V systems. Other proteins are exported across both the cytoplasmic and outer membranes in one step by the type I, type III, type IV or type VI pathways.
The interested reader is referred to Table 1 for reviews of systems involved in export of proteins. We begin our discussion of the Sec system from a historical perspective, describing basic principles that were established before any of the components of the system had been identified.
Table 1.
Systems of Protein Export
|
Transfer across the Cytoplasmic Membrane:
| ||
| System | Features | Reviews |
|
| ||
| General Secretory System (Sec) | Export of polypeptides during or after synthesis, but before acquisition of stable structure | (368–370) |
| Twin Arginine Transport (TAT) | Export of fully folded proteins | (371, 372) |
|
Insertion of Membrane Proteins:
| ||
| System | Features | Reviews |
|
| ||
| Signal Recognition | Co-translational insertion of polypeptides into inner membrane | (373, 374) |
| Particle (SRP) Pathway | ||
| YidC | Insertion of polypeptides into inner membrane | (375, 376) |
|
Transfer Proteins from Periplasm:
| ||
| System | Features | Reviews |
|
| ||
| Type II | Export of folded proteins from the periplasm through the outer membrane | (377) |
| Type V | Autotransporter; two partner secretion | (378) |
| BAM | Insertion of β-barrel proteins into the outer membrane | (379) |
| Lol | Localization of lipoproteins from periplasm to membranes | (380, 381) |
|
Transfer through Both Membranes:
| ||
| System | Features | Reviews |
|
| ||
| Type I | One step protein translocation to the extracellular space | (382) |
| Type IV | Translocate DNA and proteins into prokaryotic and eukaryotic cells by mechanism dependent on cell-to-cell contact | (383) |
| Type VI | Delivers protein effectors into prokaryotic and eukaryotic cells using a contractile mechanism | (384) |
| Type III | Contiguous channel through the bacterial and host membranes | (385) |
Early Studies
By the mid-1970s it had been well established that in eukaryotic cells secreted proteins are synthesized on ribosomes bound to the membrane of the endoplasmic reticulum and the energy for transfer through the membrane has its origin in the energy of protein synthesis (1–3). However, it was controversial as to whether or not bacteria contained physiologically significant membrane-bound ribosomes. Early studies in the 1960s used a high concentration of lysozyme to break the cells (4–8), which was later shown to cause non-specific association between ribosomes and membranes (9). Evidence for physiological function of membrane-bound ribosomes came in 1977 from two different approaches. Membrane-bound polysomes were isolated from cells without use of lysozyme and were shown to synthesize both outer membrane and periplasmic proteins whereas the major product of free polysomes was a cytoplasmic protein, EF-Tu (10). In a different approach, nascent polypeptide chains associated with ribosomes were labelled from outside of the cell, demonstrating that they spanned the membrane (11).
Studies in mammalian systems that led to the signal hypothesis (3) demonstrated that secreted proteins were synthesized as precursors carrying amino-terminal extensions (signals) and subsequently were processed to the mature form in the endoplasmic reticulum. In E. coli, precursor forms of exported proteins were detected by several techniques: by labelling of toluene treated cells (12, 13), by examining the translational products of genes for exported proteins using in vitro systems (14) and by pulse-labelling of cell cultures in vivo (15). An early review by Michaelis and Beckwith (16) catalogues the numerous precursor proteins that had been demonstrated by 1982. Isolation of mutants having alterations in the amino-terminal sequences of precursor species confirmed that these signal sequences were essential for export (17–19).
Transport into the endoplasmic reticulum in mammalian cells occurs during synthesis of the polypeptide chain and thus the energy of synthesis can provide the energy for the vectorial transfer. In prokaryotes (20, 21), as well as some eukaryotes (22, 23), transfer can occur post-translationally, after synthesis is complete, necessitating a different source of energy. In E. coli, even when transfer of protein into the periplasm occurs co-translationally, while the polypeptides are still undergoing elongation on the ribosome, the export is only temporally coupled to synthesis, it is not mechanistically coupled (20, 24). Approximately 80% of the polypeptide chain is elongated before transfer occurs. Thus it is post-translational from the point of view of mechanism. There is no evidence that the details of transfer differ between the two temporal modes of transfer in E. coli. In recent years, the Sec pathway that uses SecA and SecB to export proteins to the periplasm has been referred to as occurring entirely post-translationally to distinguish it from the SRP-mediated pathway that inserts proteins into the cytoplasmic membrane and occurs during synthesis of polypeptides. However, it must be emphasized that the fundamental difference between SRP-mediated export and that which uses the SecA-SecB pathway is not a difference in the timing relative to protein synthesis but rather the mechanistic tight coupling of synthesis to translocation.
Protein export in E. coli requires both ATP and the electrochemical potential of protons, , or protonmotive force. The first indication of the nature of the energy for export came from studies in vivo using uncouplers or ionophores that indicated a role for protonmotive force (25–28). The electrical potential (ΔΨ) and the chemical potential (ΔpH) appear to function interchangeably (29). Demonstration of the requirement for ATP came after the establishment of an in vitro assay for post-translational translocation so that the ATP requirement for protein synthesis could be separated from effects on translocation (30–33). Reconstitution of the export system with purified components led to identification of SecA as an ATPase with a crucial role in export (34–36). The precise role of each source of energy remains a subject of investigation (Section Models of Translocation).
The discovery that fully elongated proteins can be translocated led to studies of the relationship between the structure of polypeptides and their translocation. The competence of a polypeptide for export is correlated to a lack of stable tertiary structure (37). A kinetic partitioning (38) exists between productive export and non-productive folding of precursors in the cytoplasm. The signal sequence, also called the leader peptide, retards folding of precursors (39–41) allowing time for the chaperone SecB to bind and maintain the precursor in an unfolded state (Section SecB, a Chaperone).
By the mid-1980s, Jon Beckwith (for a review of the approach see (42)) and his colleagues had developed elegant genetic techniques that led to the identification of the genes that encode the proteinaceous components of the export apparatus (Table 2). Subsequent genetic, biochemical and structural inquiry resulted in detailed understanding of much of the phenomena of protein secretion through the Sec apparatus.
Table 2.
Proteins of the Sec system
| Identification | Protein | Purification | Molar Mass | pI | Location | Binding Partnersd |
|---|---|---|---|---|---|---|
| 1980 (318) | Lep | 1980 (318) | 35960 | 6.85 | membrane | |
| 1981 (386) | SecA | 1989 (387) | 102022 | 5.43 | cytoplasm, membrane | precursor, SecB, SecY, lipids |
| 1983 (48) | SecB | 1989 (309) | 17277 | 4.26 | cytoplasm | precursor, SecA |
| 1984 (388)a; 2001 (297) | SecM | 2006 (389) | 18880 | 9.98 | Function in cytoplasm; degraded in periplasm | Ribosome exit tunnel, SRP |
| 1984 (390); (391) | SecY | 1990 (212) | 48512 | 9.89 | membrane | SecE, SecG, SecF, SecA, YajC, Syd |
| 1987 (226) | SecD | 1992 (231) | 66632 | 8.62 | membrane | SecF, YidC |
| 1988 (392) | SecE | 1990 (212) | 13643 | 10.55 | membrane | SecY, SecG |
| 1990 (227); (240)b | SecF | 1992 (231) | 35382 | 5.57 | membrane | SecY, SecD, YajC, YidC |
| 1990 (212); 1993 (213); 1994 (393)c | SecG | 1993 (213) | 11365 | 6.09 | membrane | SecY, SecE |
| 1994 (234) | YajC | 2011 (394) | 11887 | 9.57 | membrane | SecF, SecY |
| 2000 (285); (395) | YidC | 2008 (396) | 61526 | 7.70 | membrane | SecF |
Identification of geneX (1984) and of secM (2001)
Contains a replacement figure with corrected sequence
Identification of Prot1 (1990), P12 (1993) and secG (1994)
See relevant section of text for references
Overview
The proteins involved in the Sec system that have known structures are shown in Figure 1. Table 3 presents the Protein Data Bank codes for all structures solved to date. The number of copies of each protein per cell can be found in Table 4. Before presenting a detailed survey of the proteins and the current understanding of their roles we give a brief overview of export through the Sec system.
Figure 1. Composite of the structures of the proteins of the Sec system.
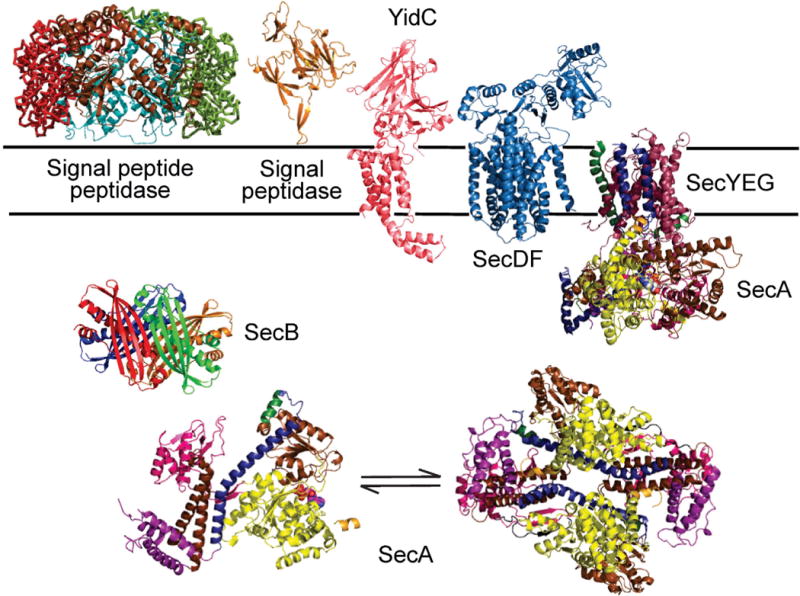
The structures are shown in ribbon representation of SecYEG in complex with SecA from T. maritima PDB 3DIN; SecDF from T. thermophilus PDB 3AQO; YidC from E. coli PDB 1B12; signal peptide peptidase soluble domain PDB 3BF0; SecA dimer from B. subtilis PDB 1M6N; SecA monomer from B. subtilis 1TF5; SecB from E. coli PDB 1QYN.
Table 3.
Structures of the Proteins in the Sec System
| Protein | PDB | Method | Resolution | Species | Reference |
|---|---|---|---|---|---|
| SecA | 1M6N | X-ray | 2.70 Å | B. subtilis | (159) Hunt et al, 2002 |
| 1M74 | X-ray | 3.00 Å | B. subtilis | (159) Hunt et al, 2002 | |
| 1TF2 | X-ray | 2.9 Å | B. subtilis | (164) Osborne et al, 2004 | |
| 1TF5 | X-ray | 2.18 Å | B. Subtilis | (164) Osborne et al, 2004 | |
| 2IBM | X-ray | 3.20 Å | B. subtilis | (161) Zimmer et al, 2006 | |
| 3JV2 | X-ray | 2.50 Å | B. Subtilis | (167) Zimmer et al, 2009 | |
| 3JV2 | X-ray | 2.50 Å | B. Subtilis | (167) Zimmer et al, 2009 | |
| 3IQM | X-ray | 3.4 Å | B. subtilis | (397) Kim et al, 2013 | |
| 3IQY | X-ray | 3.30 Å | B. subtilis | (397) Kim et al,, 2013 | |
| 1TM6 | NMR | E. coli | (136) Matsousek et al, 2004 | ||
| 2VDA | NMR | E. coli | (111) Gelis et al, 2007 | ||
| 2FSI | X-ray | 2.11 Å | E. coli | (163) Papanikolau et al, 2007 | |
| 2FSF | X-ray | 2.0 Å | E. coli | (163) Papanikolau et al, 2007 | |
| 2FSG | X-ray | 2.2 Å | E. coli | (163) Papanikolau et al, 2007 | |
| 2FSH | X-ray | 2.0 Å | E. coli | (163) Papanikolau et al, 2007 | |
| 3BXZ | X-ray | 3.00 Å | E. coli | (398) Nithianantham et al, 2008 | |
| 1NL3 | X-ray | 2.80 Å | M. tuberculosis | (160) Sharma et al, 2003 | |
| 1NKT | X-ray | 2.60 Å | M. tuberculosis | (160) Sharma et al, 2003 | |
| 4UAQ* | X-ray | 2.80 Å | M. tuberculosis | (399) Swanson et al, 2015 | |
| 4YS0 | X-ray | 1.90 Å | T. maritima | (168) Chen et al, 2015 | |
| 2IPC | X-ray | 2.80 Å | T. thermophilus | (162) Vassylyev et al, 2006 | |
| 3JUX | X-ray | 3.1 Å | T. maritima | (167) Zimmer et al, 2009 | |
| SecB | 1QYN | X-ray | 2.35 Å | E. coli | (68) Dekker et al, 2003 |
| 1FX3 | X-ray | 2.50 Å | H. influenzae | (67) Xu et al, 2000 | |
| 1OZB | X-ray | 2.80 Å | H. influenzae | (135) Zhou & Xu, 2003 | |
| SecYEG | 2AKH | EM | 14.9 Å | E. coli | (400) Mitra et al, 2005 |
| 2AKI | EM | 14.9 Å | E. coli | (400) Mitra et al, 2005 | |
| 1RH5 | X-ray | 3.20 Å | M. jannaschii | (189) Van den Berg et al, 2004 | |
| 1RHZ | X-ray | 3.50 Å | M. jannaschii | (189) Van den Berg et al, 2004 | |
| 2YXQ | X-ray | 3.50 Å | M. jannaschii | (192) Li et al, 2007 | |
| 2YXR | X-ray | 3.60 Å | M. jannaschii | (192) Li et al, 2007 | |
| 3MP7 | X-ray | 2.90 Å | P. furiosus | (190) Egea et al, 2010 | |
| 2ZJS | X-ray | 3.20 Å | T. thermophilus | (191) Tsukazaki et al, 2008 | |
| 2ZQP | X-ray | 6.00 Å | T. thermophilus | (191) Tsukazaki et al, 2008 | |
| 5AWW | X-ray | 2.72 Å | T. thermophilus | (206) Tanaka et al, 2015 | |
| 5CH4 | X-ray | 3.64 Å | T. thermophilus | (206) Tanaka et al, 2015 | |
| Complexes between Components | |||||
| 3DL8 | X-ray | 7.5 Å | B. subtilis SecA: A. aeolicus SecYEG | (166) Zimmer et al, 2008 | |
| 3DIN | X-ray | 4.5 Å | T. maritima SecA:SecYEG | (166) Zimmer et al, 2008 | |
| 3BO0 | EM | 9.60 Å | E. coli SecYEG: 23S ribosome protein fragments | (401) Menetret et al, 2007 | |
| 3BO1 | EM | 9.60 Å | E. coli SecYEG: 23S ribosome protein fragments | (401) Menetret et al, 2007 | |
| 4V6M | EM | 7.10 Å | E. coli SecYEG: 30S and 50S ribosome protein fragments | (401) Menetret et al, 2007 | |
| 4V7I | EM | 9.60 Å | E. coli SecYEG: 30S and 50S ribosome protein fragments | (402) Gumbart et al, 2009 | |
| 3J45 | EM | 9.5 A | E. coli SecYEG: 23S and 50S ribosome protein fragments | (193) Park et al, 2014 | |
| 3J46 | EM | 10.10 Å | E. coli SecYEG: 23S and 50S ribosome protein fragments | (193) Park et al, 2014 | |
| 5EUL | X-ray | 3.7 Å | Geobacillus thermodenitrificans SecYEG: B. subtilis SecA | (210) Li et al, 2016 | |
| 4V4N | EM | 9.0 Å | M. jannaschii SecYEG: 23S and 50S ribosome protein fragments | (193)Park et al, 2014 | |
| 4UTQ | EM | 8.00 Å | E. coli YidC membrane domain:ribosome fragment | (403) Wickles et al, 2014 | |
| SecDF | 2RRN | NMR | T. thermophilus | (233) Tsukazaki et al, 2011 | |
| 3AQO | X-ray | 2.60 Å | T. thermophilus | (233) Tsukazaki et al, 2011 | |
| 3AQP | X-ray | 3.30 Å | T. thermophilus | (233) Tsukazaki et al, 2011 | |
| YajC | 2RDD | X-ray | 3.50 Å | E. coli | (252) Tornroth-Horsefield et al, 2007 |
| YidC | 3WO6 | X-ray | 2.4 Å | Bacillus halodurans | (404) Kumazaki et al, 2014 |
| 3WO7 | X-ray | 3.20 Å | Bacillus halodurans | (404) Kumazaki et al, 2014 | |
| 3BLC | X-ray | 2.50 Å | E. coli periplasmic domain | (405) Oliver et al, 2008 | |
| 3BS6 | X-ray | 1.80 Å | E. coli periplasmic domain | (406) Ravaud et al, 2008 | |
| 3WVF | X-ray | 3.20 Å | E. coli | (404) Kumazaki et al, 2014 | |
| Signal Peptidase soluble domain | 1B12 | X-ray | 1.95 Å | E. coli | (407) Paetzel et al, 1998 |
| 1KN9 | X-ray | 2.4 Å | E. coli | (408) Paetzel et al, 2002 | |
| 1T7D | X-ray | 2.47 Å | E. coli, Streptomyces sp | (409) Paetzel et al, 2004 | |
| 3IIQ | X-ray | 2 Å | E. coli, Streptomyces sp | (410) Luo et al, 2009 | |
| 3S04 | X-ray | 2.44 Å | E. coli, Streptomyces sp | (411) Liu et al, 2011 | |
| Signal Peptide | 3BF0 | X-ray | 2.55 Å | E. coli | (336) Kim et al, 2008 |
| Peptidase soluble domain | 3BEZ | X-ray | 2.76 Å | E. coli | (336) Kim et al, 2008 |
Structure is SecA2, which is involved in pathogenesis
Table 4.
Level of the Sec proteins in E. coli
| Protein | Copies/cell | Molarity* | Reference |
|---|---|---|---|
| 200 – 400 | (231) | ||
| SecY | 0.3 μM | ||
| 300 | (310) | ||
|
| |||
| 300 – 600 | (231) | ||
| SecE | 0.4 μM | ||
| 250 – 500 | (197) | ||
|
| |||
| SecG | 650 –1300 | 1.0 μM | (412) |
|
| |||
| SecD | 7 – 30 | 20 nM | (234) |
|
| |||
| 30 – 60 | (231) | ||
| SecF | 30 nM | ||
| 7 – 30 | (234) | ||
|
| |||
| 150 – 300 | |||
| YajC | 0.2 μM | (234) | |
| (5 – 10X SecDF) | |||
|
| |||
| YidC | 2500 | 2.5 μM | (413) |
|
| |||
| 500 | (323) | ||
| Lep | 0.75 μM | ||
| 1000 | (414) | ||
|
| |||
| SecA | 2500 – 5000 | 4 μM | (231) |
|
| |||
| SecB | |||
using an approximation of 1 molecule/cell is 1 nM (415)
Passage through the cytosolic membrane is mediated by the SecYEG translocon. Proteins exported to the periplasm or to the outer membrane are delivered to the translocon in a nonnative state by interaction with the cytosolic chaperone SecB and the peripheral membrane component of the translocation apparatus, SecA, an ATPase. Transfer can occur either after the protein is completely synthesized, i.e., post-translationally, or during translation, but only after the majority of the polypeptide has been finished. The activity of SecA as an ATPase is enhanced by its binding to its partners, SecB, precursors, lipids and SecYEG. When bound to the translocon SecA undergoes cycles of ATP binding, hydrolysis and release of ADP. During this cycle SecA inserts into the membrane and deinserts coupled to an inversion of SecG. Both ATP hydrolysis and protonmotive force are required for in vivo export. The signal peptide is cleaved on the periplasmic side of the membrane by signal peptidase. The cleaved signal peptides are degraded by signal peptide peptidase. Following release, periplasmic proteins fold whereas outer membrane proteins enter the Bam pathway for assembly into the outer membrane.
The discussion of the individual proteins begins with SecB since this chaperone participates in the early steps of export. During the export cycle all the protein components of the system participate in many interactions so that it is impossible to discuss one independently of the others. For this reason, when discussing one protein we frequently refer the reader to other sections.
SecB, a Chaperone
SecB is one of a wide family of proteins, termed molecular chaperones, whose general function is to ensure correct interactions within and between other polypeptides (43). Chaperones interact with a diverse group of polypeptides and are involved in many processes including proper folding, assembly of oligomeric proteins, prevention of aggregation during stress and facilitating proper localization. SecB plays a crucial role as a chaperone during protein secretion by binding precursors and delivering them to the membrane for translocation. In the course of its function SecB displays two modes of binding. When SecB acts as a chaperone to capture its precursor ligands it binds promiscuously, capable of interacting tightly with many polypeptides. To deliver the bound ligand to the translocon, SecB demonstrates specific binding to a unique partner, SecA. In E. coli, SecB facilitates export only through the Sec system; however, in Serratia marcescens secretion of HasA, a hemoprotein, is strictly dependent on the chaperone function of SecB for secretion through the type 1 secretion system (44–47).
The secB gene was the second gene of the Sec system isolated by selection for mutants with pleiotropic defects in protein export, secA was the first (48). Whereas secA is an essential gene, secB is not. Null mutants are not lethal and export of some proteins is unaffected (49). In the publication describing the isolation of the secB null it was reported that the strain could grow on minimal media, but not on rich media. Subsequently, it was shown that the growth defect was not related to export, but arose from a polar effect on the adjacent gene encoding sn-glycerol-3-phosphate dehydrogenase, gpsA (50).
The secB gene encodes 155 aminoacyl residues (51) which form the monomeric unit of the functional homotetramer (total mass 68.6 kDa) (51–54). The oligomer is arranged as a dimer of dimers (54–57). Although the dimers exchange (56), the equilibrium constant for the wild-type is well below 20 nM (55). Given that the concentration of SecB in vivo is in the micromolar range, dimers are not populated.
Role of a Soluble SecB:SecA Complex
The targeting role of SecB requires specific binding to SecA. In solution SecB binds SecA with affinity in the micromolar range as determined by fluorescence anisotropy spectroscopy (58) and by isothermal titration calorimetry (59, 60). The binding affinity of SecB for SecA is greatly increased if the SecA is bound to SecYEG (Kd 30 nM (61) to 200 nM (62)). If SecB has a precursor bound there is a further increase in affinity of two- to three-fold (Kd 10 nM (61) to 60 nM (62)). This enhancement of the affinity is due to the presence of the signal which increases affinity even if it is bound as a separate peptide (61).
The 100-fold higher affinity of SecB:precursor for SecA bound to the translocon relative to cytoplasmic SecA has brought into question the existence of physiologically significant soluble complexes between SecB:precursor and SecA. There is no question that SecB is present in the cytoplasm complexed with precursors since SecB binds not only to fully synthesized precursors but also to polypeptides in the process of elongation on the ribosome (63, 64). Even given the much higher affinity of SecB:precursor for membrane-bound SecA, cytoplasmic complexes of SecA:SecB will exist since the amount of SecYEG in the cell is limiting. SecA and SecB are present in a 10-fold molar excess over SecYEG (Table 4). If SecYEG were saturated SecB:precursor:SecA, complexes in the cytoplasm could wait until a free translocon became available. The ternary complex of SecA:SecB:precursor has an affinity for SecYEG (Kd ~60 nM) similar to that of SecA alone for SecYEG (Kd ~40 nM) (62). A complex isolated from the cytoplasm containing SecB and a factor later shown to be SecA was active in translocation of a precursor into inverted membrane vesicles (53, 65, 66).
Structure of SecB
The X-ray structures of SecB from both Haemophilus influenza (169 residues) (67) and E. coli (155 residues) (68), which have 59% sequence identity over a stretch of 132 residues, confirm the conclusion from biochemical studies (55, 56) that the homotetramer is organized as a dimer of dimers (Fig. 2). The protomer, from N to C terminus, comprises a four-stranded, Greek key, antiparallel β-sheet followed by a long α-helix (α−1) and a long antiparallel linking strand ending in a short α-helix (α−2). The dimer is a flat molecule formed by rotation of one protomer relative to the other so that the β1 strands of each are antiparallel and come together to result in an eight-stranded β-sheet on one surface with the α-helices packed against the opposite hydrophobic surface. The only differences between the structures are found in the loops. The dimers associate through the α-helices to form a tetramer with flat β-sheets on the opposite faces and α-helices at the interface. The interface of the tetramer is stabilized by hydrogen bonds between β1 of one dimer and α1 of the opposite dimer and hydrogen bonds between the two α1 helices. Stabilization by hydrogen bonding via water molecules at the interface may explain the free exchange of dimers in solution. The extreme N- and C-terminal residues are not resolved in either structure. Both proton nuclear magnetic resonance (NMR) spectroscopy (69) and electron paramagnetic resonance (EPR) spectroscopy (70) show that the C-terminal region is highly mobile in solution.
Figure 2. Structure of SecB, a dimer of dimers.
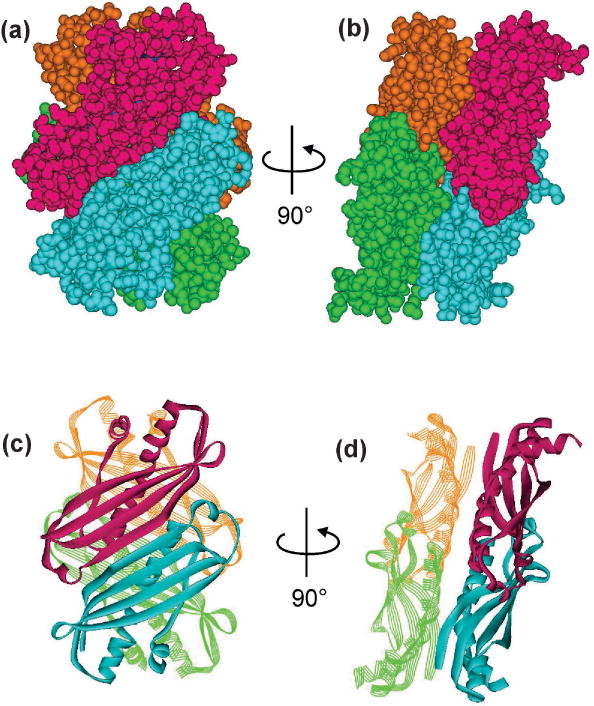
SecB is a tetramer organized as a dimer of dimers. (a) and (c) show the two protomers that make eight-stranded β sheets on the flat sides of SecB. (b) and (d) are related to (a) and (c) by 90° rotation to show the interface of the dimer of dimers. Each protomer is shown as a different color. PDB 1QYN.
SecB Binding to Ligands
To be exported via the Sec pathway proteins must be devoid of their final stable tertiary structure (37, 71). In vivo, SecB is the cytosolic factor that captures the majority of precursors before they fold (for exceptions see section SecB: Dependence on SecB) (72). Influenced by the fact that secretion into the eukaryotic endoplasmic reticulum involves the signal recognition particle, SRP, which specifically binds the signal sequence to bring its ligands to the translocon, it was thought that SecB would also recognize and bind the signal. This notion was supported by demonstration that SecB could form a stable complex with precursor maltose-binding protein (pMBP), but not with the mature form (73). The only apparent difference between the two forms was the presence or absence of the signal, thus, it was mistakenly concluded that SecB binds the signal sequence. The missing element was time, which is not readily visible. The signal sequence is crucial to binding, but it is not directly recognized (74). The signal retards folding (39, 40, 52) exposing the mature region for binding (41, 72, 74–79). In vivo studies show that the retardation of folding is a physiologically significant role of the signal sequence (41). The signal does not contribute significantly to the binding energy since proteins that can be maintained in an unfolded state show equally tight binding to SecB with or without the signal (80). The Kd for both species of unfolded MBP is approximately 30 nM (80, 81).
These observations lead to the idea that SecB modulates a partitioning among the pathways open to a newly synthesized precursor: 1) aggregation, 2) intracellular folding which may result in the correct fold, but in the wrong compartment, and 3) properly engaging the translocon while still in a nonnative state for productive export followed by folding. The role of SecB to capture nonnative proteins during this partitioning is dominated by the kinetics (rates) of the competing events (38). Thermodynamics plays an important role in respect to the affinities (Kds) of the protein:protein interactions such as the binding of SecB carrying the ligand to SecA and binding of the ternary complex to SecYEG (62).
SecB, like other members of the chaperone family, has the remarkable ability to recognize ligands because they are in a nonnative state (82). The binding is rapid, near diffusion limited (83–85), and promiscuous. SecB will bind to proteins that are not its natural ligands if they are unfolded (38, 41). Binding occurs at multiple subsites on the surface of SecB (81). There is no obvious consensus in sequence of the ligands tested. Peptide studies show a binding preference for sequences of eight or nine aminoacyl residues that are enriched in aromatic and basic amino acids (82, 86, 87). If the nonnative proteins are small, such as RBPTI (reduced bovine pancreatic trypsin inhibitor, 58 residues) (88) or a 131 residue fragment of staphylococcus nuclease (81), multiple copies can bind one tetrameric SecB. Most of the natural ligands of SecB are larger than these model ligands, having masses of approximately 20 kDa to 50 kDa. For polypeptides tested with masses ranging from 36 kDa to 61 kDa, the stoichiometry is one polypeptide per tetramer (76, 80, 81, 89).
The length of the natural ligands combined with the multiple subsites for binding on SecB account for high affinity binding (60, 80) with low specificity (84). The long polypeptide chain can wrap around SecB to simultaneously occupy several low specificity sites resulting in high affinity from the sum of binding energies. The residues on SecB that are in contact with ligands were mapped by site-directed spin-labeling and EPR spectroscopy (70, 90). Contact residues on SecB were found not only in the deep cleft identified as a candidate for binding from visual inspection of the X-ray structure (67, 68), but also at the ends of the tetramer and on the flat β-sheets located at the sides. The pattern of contact sites suggests that there are many possible pathways around SecB allowing one ligand to bind on both sides (Fig. 3). The EPR-based technique is an ensemble approach that averages the results from all complexes in the sample. It is not likely that each polypeptide ligand would follow precisely the same path around the chaperone or occupy all the potential sites. A cryo-electron microscopy study concluded that the binding of proOmpA was confined to one side of SecB (91).
Figure 3. Possible pathways of ligand binding.
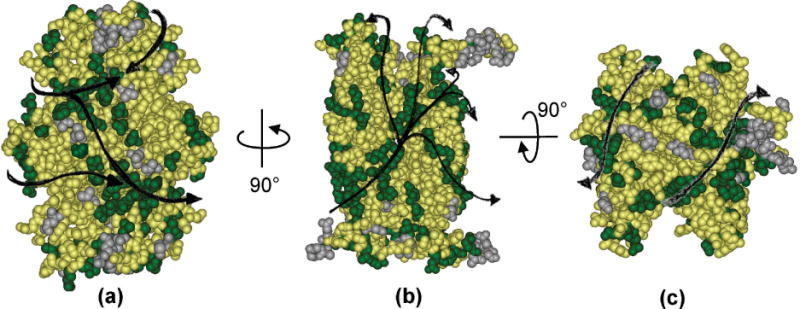
Site-directed spin labeling and electron paramagnetic resonance spectroscopy were used to map the contact sites between SecB and polypeptide ligands. The sites of contact are shown in green. Residues that showed no contact are shown in gray and resides not tested in yellow. (a) Flat eight-stranded β sheet on the side of the tetramer. (b) is related to (a) by a 90° rotation around the vertical axis to show the channel at the interface between the dimers. (c) the end view of the tetramer shows the depth of the channel. The structure was generated by threading the E. coli sequence through the H. influenza structure (PDB 1FX3) which has more C-terminal residues resolved than does the E. coli structure.
The residues on SecB that make contact with ligands are the same for all ligands tested: galactose-binding protein (GBP), both the precursor and mature forms, and several forms of the outer membrane protein OmpA, full-length precursor, mature and a truncated form that includes the transmembrane domain, but lacks both the signal peptide and the periplasmic domain (70, 90). SecB makes contact with approximately 50% of the length of its ligands centered around the middle of the mature region as determined for pMBP (92), pGBP (93) and precursor oligopeptide binding protein (proOppA) (89). The large interactive surfaces of both SecB and the ligands accounts for the near diffusion limited rate of association that allows SecB to bind proteins before they fold (83–85).
Thermodynamic and Kinetic Aspects of Protein Export
The central premise that SecB modulates a kinetic partitioning of proteins among several pathways does not mean that thermodynamics are unimportant. Consider the effects of thermodynamic (affinity) and kinetic (rate) parameters during protein export. The strength of binding between two proteins is given by the dissociation constant, Kd, a thermodynamic parameter that can be expressed as the ratio of…mb the kinetic parameters koff, the rate constant for dissociation, and kon, the rate constant for association, Kd = koff/kon. The Kd dictates the probability that at equilibrium a complex exists. During the initial stage of export it is not the Kd of binding that is crucial; there are overriding kinetic aspects. In the subsequent targeting of the precursor to the translocon, thermodynamics play a dominant role. The stability of binding between SecB carrying a precursor and SecA, which will shuttle the precursor into the export pathway, is determined by the affinity (Kd).
SecB cannot bind if the rate of folding of a polypeptide to its native state exceeds the rate of association with SecB. When the folding reaction is at equilibrium, the polypeptide is no longer a ligand. Many studies in vitro demonstrate that SecB binds only the nonnative state (41, 74, 93, 94). Additionally, the rate of folding has been shown to be crucial to binding SecB in vivo (41, 95–97). The importance in vivo of kinetics of folding is strongly supported by the successful isolation of mutations that slow folding by selection for intragenic suppressors of signal sequence mutations both in precursor ribose-binding protein (pRBP) (96) and in pMBP (98).
The central idea of kinetic partitioning, that the interaction of SecB with its ligands is modulated by the rate of folding of the polypeptide ligands relative to the rate of association with SecB (38, 84) was quantitatively tested and validated using rate constants determined empirically for the interaction of pMBP and SecB (99). However, the idea that SecB distinguishes proteins destined for export based on the partitioning has been questioned (99). It was proposed that since binding is near diffusion limited (kon for MBP is 1 × 107 M−1s−1) SecB could capture cytoplasmic proteins unless they folded with a kf 10-fold higher than the binding rate, i.e., kf greater than 40s−1 (for assumptions made see (85, 99)). Even so, the majority of proteins would escape capture by SecB based on the folding rates given in the comprehensive data base of protein folding, ACPro (100) (https://www.ats.amherst.edu/protein), which currently lists 126 proteins, 57% of which have a kf greater than 40s−1. Although such calculations are useful to determine general feasibility, it must be remembered that rates determined in vitro in dilute solution are different from what occurs in the cytoplasm of cells. The extreme macromolecular crowding that exists in cells (protein concentration in E. coli is between 200 and 300 mg/mL(101)) generally enhances the rates of reactions such as protein folding that lead to a reduction of volume whereas diffusion is slower than in dilute solution (102, 103). Therefore the number of proteins that fold faster than they can bind SecB is likely to be much higher than estimated. If cytosolic proteins were to bind SecB they would not enter the export pathway because they would lack a signal peptide. SecB would not become saturated with such proteins. The ligands, if unfolded, could be degraded by proteases as observed for a SecB-bound precursor that lacks a leader (95) or they might be capable of refolding on the surface of SecB (99).
Dependence on SecB
When a strain completely lacking SecB was first examined for protein export, the proteins studied fell into two classes: 1) proteins that were exported normally, RBP and alkaline phosphatase (PhoA), and 2) those that accumulated as precursors, MBP, outer membrane protein F (OmpF) and maltoporin (LamB). It was concluded that SecB is required for efficient export of a subset of proteins (49). Since that time exported proteins often have been classified as SecB-dependent and SecB-independent. This is misleading since the involvement of SecB is not so clearly demarcated. The physiological conditions of growth greatly influence the need for SecB. PhoA is a good example. It has been reported to be SecB independent (49, 72), slightly SecB dependent (75) and SecB dependent (104). In the case of PhoA it is likely that the observed difference in SecB dependence resulted from differences in growth temperature. As discussed below (Interplay of Chaperones), PhoA uses the heat shock proteins DnaK/DnaJ as its primary chaperones. RBP does not require SecB in normal growth conditions, but when export is compromised either by mutation in the signal sequence, which blocks export, paired with a suppressor mutation in the mature region, which slows folding to restore export (96), or in the presence of an uncoupler which dissipates protonmotive force, SecB facilitates translocation (105). Most proteins classified as SecB dependent are not absolutely dependent on SecB for export but rather SecB enhances the export. The degree of stimulation by SecB varies with the protein species. The need for the chaperone to maintain a nonnative state is decreased if the precursor folds sufficiently slowly (106). The need for the targeting function of SecB depends on the affinity of the precursor to bind SecA directly relative to the affinity to bind SecA via SecB. For example, precursor PhoA directly binds the SecA:SecYEG complex with a Kd of 230 nM (107). The affinity of other precursors delivered via a complex of precursor with SecB is only approximately four-fold higher (Kd 60 nM) (62). SecA itself acts as a chaperone through binding that involves the mature portion of ligands (108) as well as the signal peptide (109–112). There may be other as yet undefined functions of SecB during the actual translocation step. When SecB mediates export of RBP it does not function by increasing the lifetime of the competent structural state, but might stabilize binding of RBP at the translocon through interaction with SecA (113). In addition, SecB might act directly on SecA to enhance precursor-activated ATPase (114).
The number of proteins that are known to utilize SecB was increased through a comparative proteomic approach combined with Western blotting and pulse-chase experiments (115) (Table 5). The list of secreted periplasmic and outer membrane proteins using other chaperones to enter the Sec system has not increased, but that does not mean that there are no others.
Table 5.
Proteins that Utilize SecB
| Protein | Localization | Reference | |
|---|---|---|---|
| 1985 | MBP | periplasmic | (49) |
| LamB (maltoporin) | outer membrane | (49) | |
| OmpF | outer membrane | (49) | |
| 1988 | OppA | periplasmic | (72) |
| OmpA | outer membrane | (72, 104) | |
| 1989 | PhoA | periplasmic | (49)a(75)b(104)c |
| PhoE | outer membrane | (416) | |
| 1995 | GBP | periplasmic | (417) |
| 2006 | HtrA | periplasmic | (115) |
| FhuA | outer membrane | (115) | |
| FkpA | periplasmic | (115) | |
| OmpT | outer membrane | (115) | |
| OmpX | outer membrane | (115) | |
| TolB | periplasmic | (115) | |
| TolC | outer membrane PSORT predicts periplasmic or outer |
(115) | |
| YbgFd | membrane PSORT predicts periplasmic or outer |
(115) | |
| YcgKd | membrane PSORT predicts periplasmic or outer |
(115) | |
| YgiWd | membrane PSORT predicts |
(115) | |
| YncEd | periplasmic or outer membrane | (115) | |
| BtuBe | Outer membrane | (115) | |
| FhuEe | Outer membrane | (115) | |
| FadLe | Outer membrane | (115) | |
SecB independent
Minimally dependent on SecB
Dependent on SecB at 30°C
Hypothetical proteins
Effect on the rate of translocation
Interplay of Chaperones
In a living cell there is a great deal of overlap and interplay among chaperones. The cell does not maintain neatly compartmentalized roles for each, but takes advantage of the entire available pool to adapt to changes and maintain viability. The first evidence that heat shock proteins play a role in export through the Sec system was the demonstration that overproduction of DnaK facilitated export of a fusion protein between precursor LamB and β-galactosidase, which normally jams the apparatus because it cannot be translocated (116). Additionally, synthesis of heat shock proteins is induced when precursors that fail to be exported accumulate in the cytoplasm (117, 118). The involvement of heat shock proteins is also implicated by the finding that overproduction of σ32, the transcriptional factor that regulates the production of heat shock proteins can substitute for SecB in a secB null strain and restore export of proteins which normally would use SecB (119).
Several proteins that were originally identified in a secB null strain as independent of SecB for export through the Sec system (49) use other chaperones. DnaK and DnaJ are the primary chaperones in the secretion of periplasmic PhoA in vivo (120). However, when cells are grown at 30°C, SecB plays a role. Perhaps at the low temperature there is insufficient DnaK and DnaJ (104). Export of the lipoprotein (Lpp) which was not affected when SecB was depleted uses SRP (121). Even so, in a secB null strain a low level of aggregated Lpp was detected within the cytoplasm (115) indicating that SecB is involved to some degree. The chaperone pair GroEL and GroES mediate export of TEM β-lactamase (104, 122, 123). No other exported proteins have been identified that use the GroEL/GroES chaperones.
The singular ability of chaperones to recognize and bind nonnative structure results in promiscuous binding which can apply in both directions. Ligands for SecB will bind other chaperones in the absence of SecB and SecB will interact with unstructured proteins in the absence of their normal binding partner. In a previous review (124) we attributed early insight into this phenomenon to Jagger and Richards who pointed out in 1969, “You can’t always get what you want, but if you try sometimes you just might find you get what you need.” (125)
Evidence that SecB serves as a general buffer of nonnative proteins when the need arises comes from several observations. The loss of heat shock chaperones GroEL, GroES, DnaK or DnaJ results in increased production of SecB (126). SecB can aid in dissociation of aggregates of misfolded polypeptides. Through a thermodynamic coupling, in vitro SecB uses the energy of binding to monomers to pull the equilibrium and disrupt aggregates of insulin B chains (127). In vivo, SecB might use this mechanism to disrupt aggregates and upon dissociation from SecB, the monomer might either fold properly or enter a pathway for degradation. Strains lacking both DnaK/DnaJ and TF (trigger factor, the product of the tig gene), display a temperature-sensitive phenotype and the proteins produced are prone to aggregate. Both problems are suppressed by overproduction of SecB, even if the SecB is a variant that is defective in secretion. Thus the general chaperone function of SecB is independent of its role in translocation (128).
SecB:SecA Complex
Two areas of contact on SecB stabilize the specific binding between SecA and SecB, either with or without a precursor bound: 1) the flat sides of the tetramer and 2) the 13 C-terminal, highly mobile residues of each protomer. The binding site on the β-sheet (termed the side site) was first identified by isolation of mutants defective in export in vivo that had alterations at residues D20, E24, L75 and E77 (129, 130). The site on SecA that interacts with the flat side of SecB is the carboxyl-terminal 21 aminoacyl residues that contain an essential bound zinc (61, 131–134). A structure of H. influenzae SecB in complex with a peptide representing the C-terminal residues of H. influenzae SecA shows that the role of the zinc is to impart structure to the peptide so that the side chains are in position to contact residues in the β-sheet (135). The structure of the C-terminal zinc domain of SecA does not change upon binding SecB (136, 137).
The C terminus of SecA was incorrectly considered to be the sole binding site for SecB based on binding studies using a fusion between the C-terminal 22 residues with zinc bound and glutathione-S-transferase immobilized on a column (61). Wild-type SecB bound to the column whereas SecB variants with substitutions in the side site did not. The fundamental problem with this reasoning is that the remainder of the large SecA protein (molar mass 101 kDa) was not present and so of course was not tested for interaction. In the complete absence of the interaction between the side of SecB and the C terminus of SecA there are additional sites of contact that stabilize a SecA:SecB complex (138). These contacts are between the C-terminal 13 mobile residues of SecB (termed the tail sites) and both the amino-terminal 10 residues of SecA and the aminoacyl residues in a stretch that connects the nucleotide binding fold 2 and the helix scaffold domain of SecA (residues 600 – 610, see section SecA for structure) (59, 139, 140). The energy of stabilization between the wild-type species (Kd ~ 2 μM) and substituted variants lacking contact at the side sites is small. The weakest binding was seen with SecBL75Q binding wild-type SecA and with wild-type SecB binding SecA that has a deletion of the C-terminal zinc-containing region (Kds ~ 4 μM) (60, 139). It should be noted that because energy is a function of the log of the Ka (association constant), the difference in binding energy between 2 μM and 4 μM is only 5% of the total energy of stabilization. The small difference results from enthalpic/entropic compensation, i.e., the loss of enthalpy when binding sites are missing is compensated by an increase in entropy (60, 139). Heterotetramers, i.e., hybrids, formed by mixing two species of SecB carrying different binding sites, allowed determination of the relative contribution to binding energy from interaction at the side and tail sites on SecB (57).
Even though the complexes stabilized by all contacts and those stabilized by a subset of interactions show very similar stabilities, they differ in stoichiometry of SecA:SecB. With all contacts present the complex has two protomers of SecA bound to a tetramer of SecB, whereas species that are missing one of the two contact sites have only one SecA protomer bound (59). When complexes were first identified that contain only one protomer of SecA they were considered to be inactive (59, 133, 138). However, the complex having only one SecA protomer is active in translocation but it is much less efficient in the coupling of ATP hydrolysis to translocation as compared to the complex with two SecA protomers bound (141). When SecB binds SecA, the protomers do not maintain the SecA dimer interface, but bind separately (114, 140).
The contacts between SecA and SecB are distributed asymmetrically even though the complex of two protomers of SecA bound to one tetramer of SecB is itself symmetrical. Breaking the contact at the side site on SecB within a wild-type complex results in dissociation of only one protomer (59). The site of asymmetry has been defined: only one SecA protomer contacts the side site of SecB, whereas both protomers interact with SecB tails (57). A model was proposed in which the contacts between SecA and the tails of SecB involve SecB protomers that lie directly across the dimer interface (57).
After a precursor is bound to SecB it must be passed to SecA and on to the translocon. In free solution SecB has a higher affinity for precursor than does SecA. For transfer to occur the relative affinities must be reversed. Binding of the model ligand, BPTI, causes an opening of the tetrameric structure of SecB across the dimer interface (142, 143). The proposed interactions of the SecA amino termini on opposite dimers of SecB would allow binding of SecA to exert a force across the interface to reverse the change exerted by ligand binding and thereby weaken the precursor interaction (57). Additional energy to effect a conformational change might come from binding and hydrolysis of ATP by SecA or by interaction of the complex with SecYEG.
SecA, an ATPase
SecA is a large (901 aminoacyl residues), multidomain protein that is essential for export of protein (35). In the cell 50% of SecA is free in the cytoplasm and 50% is associated with the cytoplasmic membrane (144). Of the membrane-associated SecA, approximately 30% is integral to the bilayer as demonstrated by many approaches (144–148). Association of SecA with SecYEG at the membrane increases the affinity of SecA for precursors 20 to 30 fold (62, 110). Nonetheless, the soluble fraction of SecA has a crucial role. During translocation through a single SecYEG translocon multiple molecules of SecA function in translocation through exchange with SecA from the cytosol (149).
The SecA in the cytoplasm is in equilibrium between monomers and dimers. The dissociation constant (Kd) is sensitive to temperature and to salt concentration (150, 151). In solutions chosen to mimic intracellular conditions (buffer at pH 7.5, 5 mM Mg2+acetate, 300 mM K+ acetate), the Kd was approximately 1 μM at 8°C. Potassium acetate was used at 300 mM because the thermodynamic activity of potassium has been determined to vary between 0.14 M and 0.76 M depending on the osmolarity of the growth media (101, 152). Cells maintain intracellular sodium at low concentration, usually 10 – 20 mM (153, 154). As the salt concentration decreases the association of the protomers to dimers is strengthened (Kd ~ 0.1 μM). The weakest binding is observed at 8°C, the lowest temperature tested (150), whereas at 20°C in low salt, Kds between 0.6 μM and 14 nM have been determined (151, 155). The monomer-dimer equilibrium is shifted toward monomer when SecA binds phospholipids (156, 157) or SecY (158).
The Structures
SecA has been crystallized in five dimeric forms (159–163). Whereas SecA from different organisms reveal different contacts at the dimer interface, the protomer structures are closely related (Fig. 4) (159, 164). SecA is organized in multiple domains, which undergo rearrangements during translocation. (The N-terminal segment (residues 1 – 619) comprises three folded domains. There are two nucleotide-binding folds, Nucleotide Binding Domains 1 and 2 (NBD1, yellow and NBD2, red/brown). The Precursor Binding Domain (PBD, pink), originally identified as a region that crosslinked overlapping fragments of SecA to a precursor (109), emerges from the NBD1 between β-strands 5 and 6 of that domain. The carboxyl-terminal segment of ~30 kDa (residues 620 – 901) forms α-helices. The α-helical domain termed the Helix Scaffold Domain (HSD, blue) is connected to NBD2 via a short Linker Helix (green). The HSD comprises three antiparallel α-helices. A long helix at the amino terminal end and two shorter helices toward the C terminus. The two short helices have been termed the Intramolecular Regulator of ATPase, (IRA1, brown) (165) or the two-helix finger (161). The amino-terminal region of the long helix has one face packed onto NBD1 and NBD2. The C-terminal helices of the HSD make contact with the PBD in the closed conformation seen in all the dimeric structures except E. coli, in which the PBD was modeled based on the Bacillus open monomer (Fig. 5 for a comparison of open and closed) (163). Structures of SecA bound to ADP in solution and bound to SecYEG indicate that the PBD undergoes rigid body movements (Section SecYEG for further discussion) (166–168). A second helical domain, the Helical Wing Domain (HWD, purple) is inserted between helices 1 and 2 in the HSD. Twenty-five residues, the C-terminal linker (CTL, green), form a β-strand bound to the two antiparallel β-strands that connect NBD1 to the PBD. The remaining 40 residues of SecA are flexible and not observed in the structure. In E. coli SecA the last 22 residues function to bind phospholipids (132) and to also bind SecB due to the coordination of zinc through three cysteines and one histidine (132, 134).
Figure 4. Structures of SecA monomers.
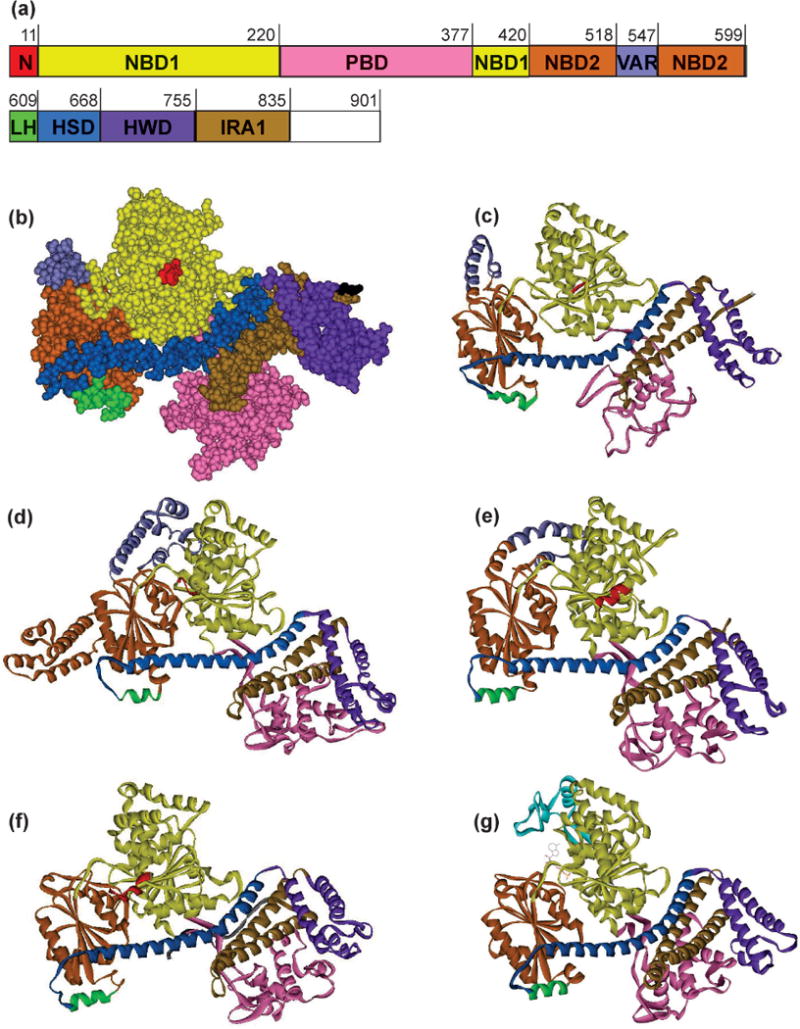
(a) The sequence is E. coli SecA with the domains colored as in the structures. See text for domain abbreviations. (b) SecA from E. coli in CPK representation, PDB 2FSF with the PBD modeled in based on B. subtilis SecA, PDB ITF5, by A. Economou. (c) Ribbon representation of E. coli SecA shown in (b). (d) – (g) Ribbon representation of SecA from the following species: (d) T. thermophilus, PDB 2IPC, (e) M. tuberculosis, PDB 1NL3, (f) B. subtilis, PDB 1M6N, and (g) T. maritima, PDB 3JUX.
Figure 5. Comparison of the open and closed structures of SecA.
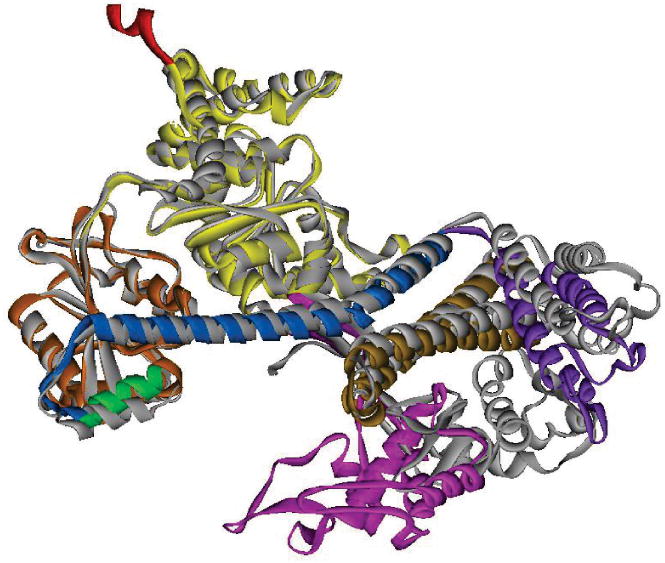
The closed conformation of B. subtilis SecA (PDB 1M6N) is shown as the gray ribbon. The open conformation of B. subtilis SecA (PDB 1TF5) is shown in ribbon representation with the domains colored as in Figure 4. The Protein Binding Domain (pink) is the only domain that has moved.
The structures of the dimers show distinct interfaces. The extreme N-terminal residues 2 – 11 and one face of the HSD are involved at the interface of the antiparallel B. subtilis dimer (Fig. 6a, PDB 1M6N) and the parallel dimer from T. thermophilus (Fig. 6b, PDB 2IPC). The arrangement of the protomers in the dimer of M. tuberculosis (Fig. 6c, PBD 1NL3) is antiparallel and the interface involves contacts between loops at the end of the β-sheet of the PBD of one protomer with the HSD of the opposite protomer. The final two arrangements differ drastically from the others. The interface of the E. coli dimer (Fig. 6d PDB 2FSF) (163) is the variable domain (VAR, lilac), which lies within the NBD2 (residues 519 – 547) (169). This domain is present in E. coli, T. thermophilus and in M. tuberculosis, but is lacking in B. subtilis and in T. maritima (169). SecA from B. subtilis has been crystallized in a dimer form(Fig. 6e, PDB 2IBM) that is stabilized by contacts between the two-stranded β-sheet that forms the stem connecting the PBD to NBD1 and a short segment from the opposite protomer that interacts to form the third strand of the β-sheet. The equivalent region is α-helical in the other protomer and corresponds to the Linker Helix.
Figure 6. Dimeric forms of SecA.
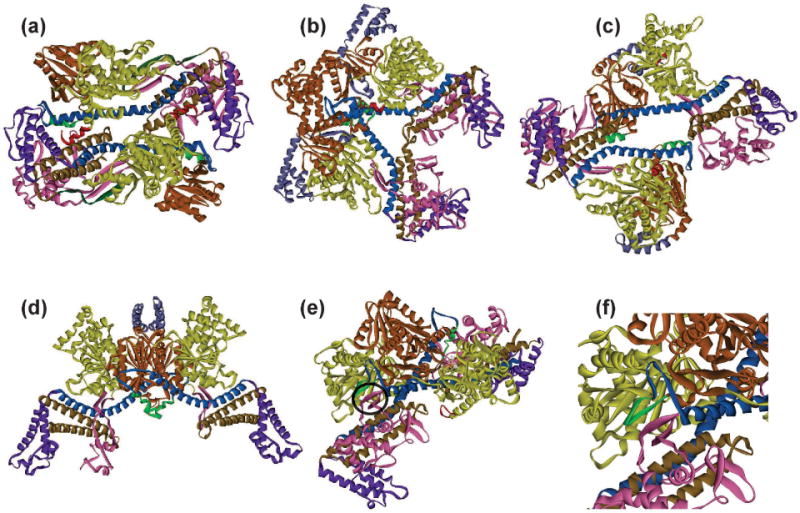
Dimeric structures of SecA with the domains colored as in Figure 4. The SecA species are from: (a) B. subtilis PDB 1M6N, (b) T. thermophilus PDB 2IPC, (c) M. tuberculosis PDB 1NL3, (d) E. coli PDB 2FSF, (e) B. subtilis PDB 2IB; the three-stranded β sheet that forms the interface is circled and enlarged in (f).
Many attempts have been made to determine which of the five dimer interfaces represents the physiological dimer in E. coli. As pointed out in the excellent work of Auclair, Oliver and Mukerj (170) many of the studies used non-equilibrium techniques such as crosslinking that distort normal equilibria and could trap dimeric forms that may exist as minor populations. The authors present a well-controlled study of SecA from E. coli using Förster resonance energy transfer (FRET) which does not perturb the equilibrium. They determined 15 distances which were sufficient to distinguish among the five dimeric forms. The only reasonable fit for the data obtained using E. coli SecA was the antiparallel dimer seen in the structure of B. subtilis SecA (Fig. 6). A separate, extensive in vivo photocrosslinking study identified crosslinks that were not consistent with any one dimer (171). This may mean that in vivo minor populations of several dimeric forms exist and were trapped.
Role of SecA during Translocation
During protein translocation SecA binds precursors and delivers them to the membrane-embedded translocon through its binding affinity for SecYEG. Precursors that rapidly aggregate or acquire folded structure are first captured by SecB. SecB with precursor bound interacts with SecA and the ternary complex engages the translocon (Section SecB: SecB Binding to Ligands and SecB:SecA Complex). SecA binds precursor polypeptides directly to the HSD and the PBD through interaction with both the signal and mature domains (107, 108, 172, 173). The binding site of the signal peptide has been mapped by two techniques, NMR (111) and Förster Resonance Energy Transfer (FRET) (174, 175). The two binding sites are in the same general area, but differ in the precise location and the orientation. The NMR study used a model signal peptide, KRR – LamB, which is the signal of LamB extended at the N terminus by a lysine and two arginine residues. This peptide bound within a groove at the interface of the HWD and the PBD. The peptide was oriented perpendicular to the IRA subdomain (two helix finger) of NBD1. The FRET studies located the signal peptide of PhoA in a region bounded by the PBD, the HSD and NBD1. This peptide was oriented parallel to the two helix finger (IRA) subdomain. These differences may reflect the species of signal peptide studied or the conditions used: NMR requires high concentrations of the proteins while FRET allows studies of proteins at much lower concentrations.
SecA hydrolyzes ATP during translocation of precursors (36). In solution, SecA exhibits a low level of ATPase activity defined as the basal level. The basal level is stimulated by binding to lipids (172), referred to as lipid ATPase and reaches a maximum when SecA is bound to both a precursor and SecYEG, which is denoted translocation ATPase (36).
Conformational Changes in Activation of ATPase
Hydrolysis of ATP by SecA is coupled to productive translocation. There are several ideas as to precisely the work that is done (Section Models of Translocation). The following discussion is limited to the mechanism of activation of ATP hydrolysis without speculation about the work done. We begin with the conformational changes in the active site and the propagation of those changes between domains.
The two nucleotide binding domains NBD1 and NBD2 are structurally similar to the DEAD-box helicases named for the crucial residues Asp, Glu, Ala, Asp (176). As in the related helicases, the high-affinity binding site for ATP is located at the interface of these two domains. The residues comprising the high affinity ATP binding site had been identified by mutagenesis prior to the determination of the structure (177). The X-ray structures of Bacillus SecA with and without ADP bound show no difference (159). Conformational changes do occur but they are too small to be resolved in the structures.
NMR identified two disordered regions in intact SecA that are involved in conformational changes during function. One region is the extreme C-terminal segment (residues 864 – 901) and the other lies within NBD2 (residues 564 – 579) (178). An extensive study (179) of the N-terminal fragment of SecA, which includes the mobile residues in NBD2, combined thermodynamic analyses with NMR spectra to show that the surfaces at the interface of NBD1 and NBD2 are dynamic, undergoing order – disorder transitions. The binding of ATP favors the ordered state whereas after hydrolysis the binding cleft remains closed with ADP bound but disorder increases making this conformation less stable. The high ATPase activity characteristic of SecA poised for translocation is suppressed by the association of the HSD with the NBDs. The interface between the HSD and the NBDs is stabilized by two salt bridges, one between Asp649 and Arg220 (179) and the other between Arg642 and E400 (180). If the association loosens, the equilibrium is shifted toward disordered and the ATPase activity is increased. In the extreme, removal of the entire region spanning the start of the HSD to the C terminus, termed the C domain, results in high, unregulated ATPase activity (181, 182).
A structural element comprising two antiparallel helices (residues 756 – 835), originally considered to be part of the HSD, was renamed IRA1, intramolecular regulator of ATPase, when it was discovered to be primarily responsible for the suppression of ATPase activity by mediating the association of the HSD with the N domain (165). The same region on SecA is a site of interaction with SecY (183). Since the binding site on IRA1 for the HSD and for SecY overlap, binding of SecA to SecY might loosen the IRA1 – HSD contact thereby activating SecA ATPase (For an alternative model of the role of IRA1, also called the two helix finger, see section Models of Translocation).
ATP binds at the interface between NBD1 and NBD2 at the top of the cleft. At the bottom of the cleft, a salt bridge between two aspartate residues (D212, D217) and an arginine (R566), referred to as Gate 1, connects the two NBDs and stabilizes the interface (184). This stabilization imposes an energy barrier to activation of ATP hydrolysis in addition to that imposed by association of the HSD and the NBDs. The barrier provided by the HSD is dominant but when it is loosened, then the barrier of Gate 1 regulates allosteric communication between the PDB and the NBDs. When the salt bridge is broken, the NBDs go from a high affinity, closed state, to a low affinity, open state. Thus Gate 1 controls the rate-limiting step in the ATP hydrolysis cycle of SecA, the release of ADP (185). Binding of precursor to the PBD causes conformational changes that are propagated allosterically through Gate 1 to activate ATPase. The propagation occurs through the two antiparallel β-strands that form the stem of the PBD. The stem emerges from NBD1 very near Gate 1. The binding of precursor to HSD may also be involved in the loosening of interactions between the HSD and NBDs. Site-directed spin labelling coupled with EPR spectroscopy shows that precursor binds the HSD in addition to the PBD. Additionally, interactions of SecA with SecYEG, with lipids and with SecB result in mobilization of residues in the HSD, consistent with loosening of the structure (108). Thus, when SecA engages the translocon the barrier provided by the C-terminal domain may be overcome, poising the system for full activation by binding precursors. Further evidence for the conformational relay between the NBD and PBD comes from an NMR study (179) demonstrating that nucleotide binding induces slow dynamics at residues that have been shown to be crucial to binding precursors such as Tyr326 (186) and Trp349 (110).
The ATPase is also regulated in E. coli SecA by the variable subdomain (VAR) (169). Deletion of VAR results in azide resistance and in accelerated release of ADP. Both effects can be explained if deletion of the region loosens the contact of NBD1 and NBD2 to enhance release of ADP. In ATPases that are structurally related to SecA, azide blocks hydrolysis in the presence of ADP by occupying the site that normally binds the γ phosphate of ATP, thereby tightening the site and stabilizing the binding of ADP (187).
SecYEG
The Structures
After 25 years of accumulation of data from biochemical, genetic and biophysical studies, which provided insights into the mechanism of export, the first structure of the membrane translocon was reported in 2002 with a cryoelectron microscopic study of 2-D crystals of E. coli SecYEG in a lipid bilayer. The 8Å structure allowed identification of 15 transmembrane helices in a monomer (188). The first high resolution structure was the X-ray structure of the archaeal SecYEβ from Methanocaldococcus jannaschii (189). The SecY and SecE subunits are evolutionarily conserved among eukaryotes, eubacteria and archaea. The third subunit, Sec61β in mammals, Sbh1p in S. cerevisiae and Secβ in archaea, shows no homology to SecG, the third subunit in eubacteria. To date, X-ray structures with resolution of 3.6 Å or better have been solved for the translocon from two archaea (M. jannaschii and Pyrococcus furiosus) (190) and two thermophilic eubacteria Thermotoga maritima (166) and Thermus thermophilus (191) (Table 3 gives a complete list of structures, including solution NMR and electron microscopy, and references).
All translocons display a common structure of the SecYE core (Fig. 7). A central channel is formed by 10 α-helical transmembrane (TM) segments arranged in pseudosymmetrical halves, TM1 to 5 and TM6 to 10. The halves are hinged by a flexible loop between TM5 and TM6. A lateral gate lies opposite the hinge between helices TM2b and TM7. The central channel has an hourglass shape. On the cytoplasmic face of the membrane a large funnel-like cavity leads into a constriction at the middle formed by a ring of six hydrophobic residues, which in E. coli are all isoleucine. The ring closes the channel to block the passage of precursors (192, 193).
Figure 7. Structure of SecYE.
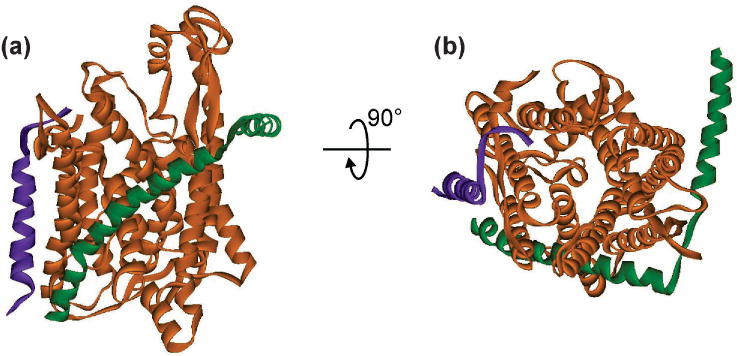
The structure of SecYEβ (PDB 1RHZ) is shown as an example of the common structure of the SecYE core. SecY is shown as the orange ribbon, SecE as the green ribbon and Secβ (SecG in E coli) as purple. The view in (a) is in the plane of the membrane with the cytoplasmic face at the top and the periplasmic face at the bottom. The view in (b) results from a 90° rotation toward the viewer to show the channel in the translocon from the cytoplasmic face. The plug can be seen in the middle of the channel at the periplasmic side.
The fully open channel with the lateral gate widened can translocate proOmpA conjugated to rigid spherical organic molecules of sizes up to diameters 22 to 24 Å. When the spheres have a diameter of 29 Å, translocation is blocked (194). On the periplasmic face of the membrane a short α-helical plug extends into the channel and blocks the exit. When the channel opens and is occupied by a translocating polypeptide, ions can pass (195). If this were to happen in vivo the dissipation of protonmotive force by passage of protons would lead to cell death. However, the presence of a physiological level of membrane potential (ΔΨ) drives the channel to close thereby maintaining protonmotive force (196). It is not clear what senses the voltage and closes the channel.
In E. coli the two small subunits of the translocon, SecE and SecG differ from those in M. jannaschii. The SecE subunit in M. jannaschii comprises two helices, an N-terminal amphipathic helix that lies on the cytoplasmic surface of the membrane and a second long transmembrane helix that crosses the membrane at an approximately 35° angle making contacts with TM1, TM5, TM6 and TM10 of SecY, thereby clamping the two halves together. The SecE subunit from E. coli has 127 amino acyl residues whereas the M. jannaschii SecE has 74 amino acyl residues. The last 62 amino acyl residues of E. coli SecE (amino acid 65 to 127) correspond to the M. jannaschii SecE subunit. In E. coli the additional amino acids at the N terminus form two transmembrane helices that are connected to the third transmembrane helix through the surface, amphipathic helix. Only the third transmembrane helix is essential for viability and function (197, 198). In E.coli the third subunit, SecG, contains three transmembrane helices whereas M. jannaschii Secβ has only two.
The extensive similarity of the two translocons allowed the X-ray structure of SecYEβ to be visually docked into the electron density map of the 2-D crystals from E. coli SecYEG. The overall fit was excellent indicating that the general architecture of the two translocon complexes is the same (189). Subsequently an atomic model of E. coli SecYEG was built using the X-ray structure of M. jannaschii taking into account insertions and deletions in the two sequences. The homology model was then docked into the 8Å map of the E. coli dimer of SecYEG in the lipid bilayer. Interesting differences were observed between the visual fit (189) and the refined homology model docking (199). Interactions between monomers within the lipid bilayer cause a 9Å displacement of TM6. The change propagates through the complex so that the plug moves closer to the membrane surface on the periplasmic side and movements of TM2b and TM7 result in a slight widening of the channel. The hydrophobic ring and the plug maintain a seal in the channel so that translocation of polypeptides would require additional factors to open the channel fully.
Conformational Changes in SecYEG and SecA
The binding of SecA to SecYEG further opens the translocon as indicated by the structure of SecA in complex with SecYEG. (T. maritima) (166) Relative to the closed channel seen in M. jannaschii, the SecY has a gap of 5Å at the lateral gate between TM7 and TM2b. The plug moves away from its position at the center of the channel. However, the channel remains sealed toward the periplasmic surface.
A second example of a structure showing a “pre-open” state is that of SecYE from T. thermophilus in complex with an anti-SecY Fab fragment (191). The Fab fragment binds to the same site as does SecA in the highly conserved C5 loop and can be considered to mimic SecA binding. In comparison to the M. jannaschii structure the helices in the C-terminal half of SecY occupy different locations, opening a hydrophobic crack toward the cytoplasm with dimensions of ~6Å by 15Å between TM2, TM7 and TM8.
The binding of SecA to SecYEG not only widens the channel of the translocon but also induces changes in SecA. Comparison of three structures of SecA from T. maritima: 1) SecA free in solution, 2) SecA bound to SecYEG and 3) SecA in solution with ADP bound shows the PBD in different positions relative to the remainder of SecA (Fig. 8). These structures suggest that binding of SecA to SecY results in movement of PBD that would trap a polypeptide in a clamp between the PBD, NBD2 and the HSD to position it directly over the mouth of the channel in SecY ready for translocation. In the structure of SecA in solution (PDB 3JUX) (167), the clamp is completely open with the PBD closely associated with the HWD and the HSD at a distance of 36Å from the NBD2. When SecA, occupied by ADP and BeFx, is bound to SecY the PBD moves to contact NBD2 and close the clamp (PDB 3DIN) (166). In the structure of SecA with ADP bound, the PBD is in a position intermediate between fully open and closed (PDB 4YSO) (168). The two β strands that connect NBD1 and the PBD act as a hinge to allow the movements. When SecY interacts with SecA, the loop between TM6 and TM7 of SecY inserts at the interface of PBD and the HSD/HWD, breaking contacts between domains of SecA. The intramolecular contacts are replaced by interactions between SecA and SecY. The IRA1 (two-helix finger) moves from its association with the HSD to insert into the entrance of the channel in SecY (200). The PBD in SecA from B. subtilis has also been crystallized in two positions (Fig. 5). It is associated with the HWD/HSD in a “closed” state in the dimeric form (PDB 1M6N) (159) and in an “open” state in the monomer (PDB 1TF2) (164).
Figure 8. Movement of the Protein Binding Domain.
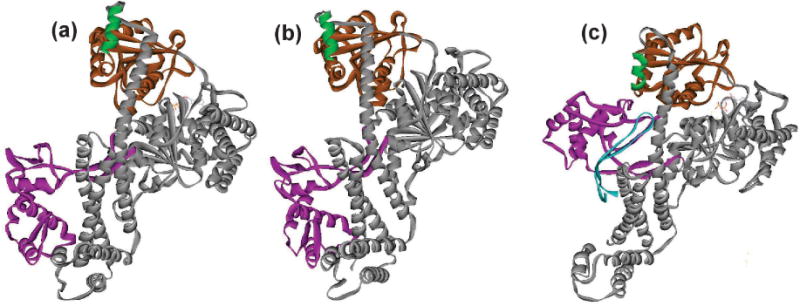
Ribbon representation of SecA from T. maritima with the PBD in different positions. The Linker Helix is shown in green and the NBD2 in brown to serve as references for movement of the PBD, shown in magenta. The reminder of the SecA is represented in gray. The SecY loop between TM6 and TM7 which inserts into SecA is shown in cyan in (c). (a) SecA in solution, PDB 3JUX, (b) SecA in solution with ADP bound, PDB 4YS0, and (c) SecA with ADP and BeFx bound in complex with SecY, PDB 3DIN.
Biochemical Studies
Movement of the PBD in solution has been demonstrated by fluorescence energy transfer studies of SecA (201). The movement of the PBD from the open position seen in idle SecA to closed position upon binding to SecYEG is crucial to initiation of the translocation process (202). When the clamp was held in the open position by intramolecular crosslinks, the basal ATPase of SecA was unchanged but activation by SecYEG was reduced. In contrast, when the clamp was fixed in the closed position, the kcat and Km of ATP hydrolysis were increased to the same degree as that seen when activated by SecYEG (203, 204). Both species of SecA with intramolecular crosslinks were inactive in mediating translocation. It is likely that the crosslinks interfered with transfer of the polypeptide into the channel. Stabilization of the closed position by a crosslink at a different site between SecY and SecA activated the ATPase and supported translocation. Therefore the PBD does not need to make large movements during translocation, but must initially move from open to closed to initiate the process (202).
The interactions of polypeptides in the process of moving from SecA through the SecY channel were mapped using disulfide crosslinking. Two cysteines were introduced into the polypeptide, one that would crosslink to the pore ring near the center of the SecY channel and a second at a distance appropriate to crosslink to SecA. The results defined the pathway taken through the clamp of SecA, past the two-helix finger and into the channel (205). Sites of interaction between SecA free in solution and polypeptide ligands, including both precursor and mature forms, were mapped using site-directed spin-labelling and EPR spectroscopy. Contacts were identified on the surface of the PBD facing the NBD2, which forms the wall of the proposed clamp, and were also seen along the HSD (108).
Insertion of Signal Peptide to open the Lateral Gate
It has been proposed that the signal peptide inserts into the lateral gate to effect a widening of the channel. Support for this idea is found in two structures that show insertion of polypeptides accompanied by opening of the lateral gate. In the structure of SecYEβ from P. furiosus (190) the C-terminal α-helical region of an adjacent SecY in the crystal was inserted to open the gate laterally, but the plug still occluded the pore. This structure suggests a route for lateral exit of cytoplasmic membrane proteins that follow the SRP pathway, but may also represent early stages of insertion of polypeptides destined for the periplasm. A high resolution structure of T. thermophilus SecYEG obtained from crystals in a lipidic cubic phase (206) also shows a peptide from a symmetrically related molecule inserted into SecY. In this case it is the amino-terminal hydrophobic residues of SecE that insert into the cytoplasmic hydrophobic crack seen in the structure of T. thermophilus SecYEG in complex with the Fab fragment that mimics SecA binding. The result is an expansion of the crack between TM2 and TM8. The gate remains unchanged on the periplasmic side suggesting that the insertion represents an early stage of interaction.
Two structures have been solved that have authentic signal peptides bound outside of the lateral gate. A structure of dimeric SecYEG in membranes solved by cryo-electron microscopy of two-dimensional crystals (207), shows one copy of SecY in the dimer associated with a truncated preprotein comprising the signal sequence of the outer membrane protein LamB and 15 aminoacyl residues of the mature protein. The signal is not intercalated but binds outside the lateral gate in contact with TM2b. The helix TM7 is relocated to bring the periplasmic end toward TM5 and TM10 resulting in displacement of the plug. The unoccupied copy of SecYEG is more tightly closed than are the unoccupied SecYEG dimers observed in the 2-D crystals grown without peptides (188). This result supports the idea that whereas two units of SecYEG may bind to one SecA, only one SecYEG functions as a channel (208, 209).
A crystal structure has been solved that has a fragment of the N-terminal region of a precursor within the channel of SecY. (PDB 5EUL, (210)) This was accomplished by fusing the signal sequence of OmpA with a short polypeptide following it into the tip of the two helix finger of B. subtilis SecA and forming a complex with SecYE from Geobacillus thermodenitrificans. The hydrophobic core of the signal forms a helix and docks into a groove outside of the lateral gate in contact with lipids. The following stretch of polypeptide is within the channel in extended form. The strategy involving the fusion of SecA to a signal sequence was based on the previous observation that in a SecA:SecY complex the two helix finger was positioned over the mouth of the channel (166).
Implications for Mechanism of Translocation
The multiple structures of SecYEG are static representations of unique moments in the dynamic interactions of the protein complex. These structures, when considered in combination with the vast body of data accumulated from studies of the translocon in action, can provide insight into the mechanism.
Only SecY and SecE are absolutely required to observe translocation in vitro when reconstituted into proteoliposomes (211), however, the level is very low compared to inner membrane vesicles (IMV). Addition of SecG (originally called band1 or p12) stimulated activity twenty-fold to reach the level seen in membrane vesicles (212, 213). The original demonstration of stimulation by SecG was carried out in proteoliposomes in the absence of protonmotive force (213). If protonmotive force is present then the requirement for SecG in membrane vesicles is less strong indicating an overlap in function of SecG and protonmotive force (214).
SecA undergoes a cycle of membrane insertion and deinsertion driven by ATP binding and hydrolysis (215, 216) that is associated with movement of precursor into the translocon channel (217). SecA inserts into the membrane when ATP binds, and the hydrolysis of ATP causes deinsertion. The cycle of insertion – deinsertion is coupled to inversion of the topology of SecG in the membrane (218). When the system is idle, the C terminus of SecG is exposed on the periplasmic side of the membrane, whereas when SecA is in the process of translocating precursors SecG inverts to move the C terminus to the cytoplasmic side of the membrane (Fig. 9). To recover the original topology of SecG, SecA must hydrolyze ATP and deinsert. The evidence for this inversion is robust. In idle inverted membrane vesicles that are not actively translocating, the C terminus of SecG is inside the vesicle on the periplasmic side and inaccessible to antibodies specific for the C-terminal region. However, if the vesicles are in the presence of precursor, SecA and ATP and are undergoing translocation, addition of the antibody inhibits the transfer indicating that the C terminus of SecG becomes accessible to the antibody, and therefore is on the cytoplasmic side. Translocation-dependent changes are also documented for proteinase K sensitivity. The inversion exposes a protease-sensitive site found between the two transmembrane segments. The site is sequestered as ATP is hydrolyzed and SecG recovers the original topology (218). A translocation-dependent cycle of accessibility and inaccessibility of single cysteine variants of SecG to a membrane-impermeant reagent was seen in studies using spheroplasts as well as inverted membrane vesicles (219).
Figure 9. Inversion of SecG in the cytoplasmic membrane.
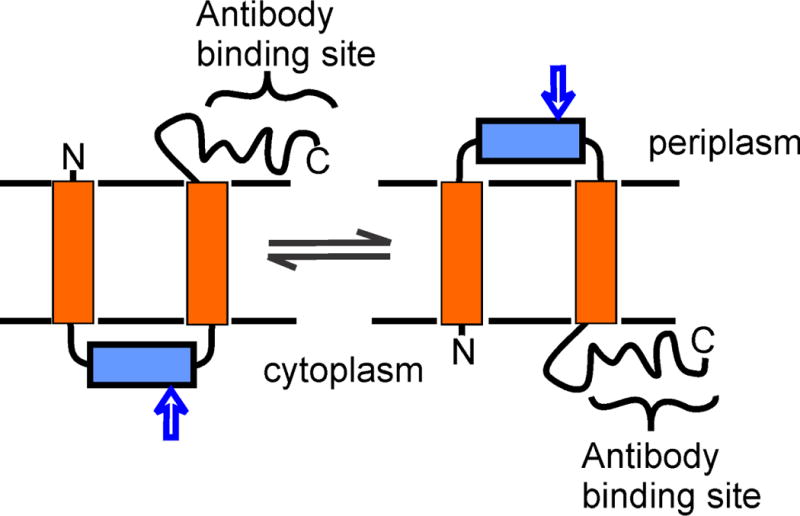
SecG has two transmembrane domains represented by orange and a segment connecting the two membrane domains represented by blue. The N and C termini lie on the same side of the membrane. The left hand image represents SecG in the idle state. During protein translocation SecG inverts as shown on the right hand side. The open arrows indicate sites of protease cleavage. The region at the C terminus recognized by anti-SecG antibody is indicated.
Membrane vesicles derived from cells that overexpress both SecY and SecE without accompanying overexpression of SecG show a high level of SecA translocation ATPase that is not coupled to translocation (220). Not only is the presence of SecG necessary for ATP hydrolysis to be productive, but the SecG must also be able to invert. If SecG is present, but inversion is blocked by fusion of alkaline phosphatase to the C terminus of SecG, ATP hydrolysis is not accompanied by translocation (221). The simplest interpretation is that inversion of SecG is essential for the coupling.
The inversion of topology was questioned when SecG was fixed by crosslinking and no effect on translocation was seen (222). However, the vesicles used were isolated from cells overexpressing SecYEG. It was later shown that the SecG inversion does not occur if SecYEG is overproduced or reconstituted into proteoliposomes (223). This suggests a need for a factor that is limiting. The factor was determined to be a glycolipid that is involved in membrane protein integration and also stimulates protein export. The lipid was named membrane protein integrase or MPIase, a glycolipozme (224). Co-reconstitution of purified MPIase with SecYEG restored inversion of SecG.
The extreme amino-terminal region of SecA is involved in the function of SecG. Deletion of the first eight aminoacyl residues of SecA resulted in defective translocation of three precursors assayed in the absence of protonmotive force. The defect was strongest for the precursor that was most dependent on protonmotive force (225). SecA with the amino-terminal deletion is also less efficient in coupling its ATP cycle to the topology inversion of SecG indicating that SecA may directly interact with SecG. Interaction of the two proteins has been directly demonstrated by examining crosslinks between SecA and SecG as a function of the cycle of inversion. Crosslinking increased when the topology was inverted (219).
SecDFYajC
Discovery of the secD Locus
The secD locus was identified in 1987 by isolation of mutants that were cold sensitive for growth and expressed defects in protein export in vivo (226). The locus includes an operon encoding three genes secD, secF and yajC (227, 228). The three proteins form a heterotrimeric complex, SecDFYajC (229) that associates with SecYEG via interactions of SecY with SecF. YajC is in direct contact with both SecY and SecF (229, 230). When SecDF was purified and added to SecYEG in proteoliposomes it showed no stimulation (231). This was unexpected because of the growth and export defects observed in vivo (226). The problems with demonstration of a stimulation of export in vitro will be discussed later. The growth defect observed in vivo is not a direct effect on secretion into the periplasm, but rather is a secondary effect on insertion of Sec-dependent proteins into the cytoplasmic membrane, which requires YidC. In the absence of SecDF, YidC does not efficiently associate with SecYEG (230, 232). This idea is confirmed by the finding that overproduction of YidC suppresses the growth defect in strains depleted of SecDF (230).
Insights from the Structure of Thermus thermophilus SecDF
We shall begin with the X-ray structure of SecDF from Thermus thermophilus solved in 2011 (233) and proceed to discuss the accumulated knowledge from past contributions within the framework of that structure. SecDF from T. thermophilus (TtSecDF) is encoded as a single polypeptide chain of mass 80.5 kDa whereas in E. coli SecD (66.6 kDa) and SecF (35.4 kDa) are produced as separate polypeptides from one operon. The topology of the SecD and SecF proteins, which have sequence similarity, was mapped by determining the alkaline phosphatase activity of constructs with phoA introduced within the secD gene. Alkaline phosphatase is only active if it is secreted to the periplasm so that the native disulfide bonds form. Thus, transmembrane and periplasmic domains can be distinguished. The activity of the fusions in combination with hydropathy plots of both proteins show that each contains six transmembrane segments (234). The X-ray structure of the TtSecDF polypeptide reveals a transmembrane domain that is a pseudo-symmetrical arrangement of 12 transmembrane α-helices with periplasmic domains (Fig. 10) (233). The large periplasmic P1 domain in TtSecDF corresponds to the predicted periplasmic domain of the homologous E. coli SecD. The P1 domain is between TM1 and TM2 and the periplasmic P4 domain in TtSecDF is between TM7 and TM8 (in E. coli, SecF). The P1 domain has two subdomains, a head and a base. The relative positions of the subdomains are different in the isolated P1 domain as compared to the positions in the full length TtSecDF. Docking of the base subdomain of the isolated P1 onto the base subdomain in the full length protein revealed two possible conformations differing in configuration of the head. The full length has the head bent toward the membrane surface and is designated the F form. The structure revealed by the docking of P1 has the head rotated 120° to be directly above the transmembrane domain and is referred to as the I form (Fig. 11). Direct evidence that SecDF undergoes a conformational transition is provided by electron tomography and single particle reconstruction (235) showing the presence of both the I and F forms in solution.
Figure 10. Structure of SecDF.
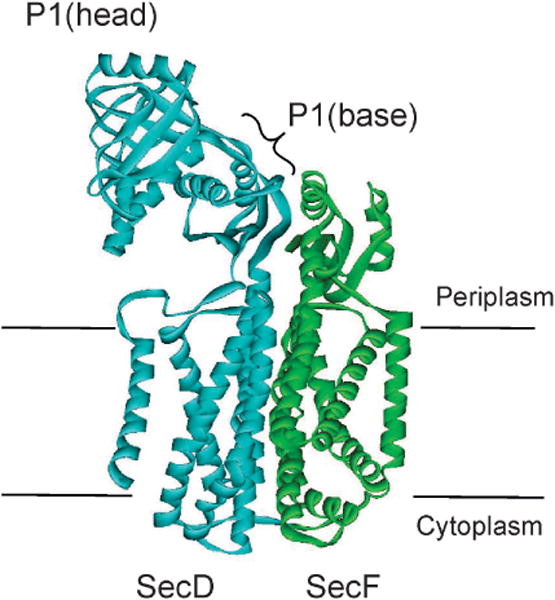
The structure of T. thermophilus SecDF, which is encoded as a single polypeptide chain, is colored to represent the individual SecD and SecF polypeptides found in E. coli. Transmembrane helices 1 – 6 (blue) represent E. coli SecD. The periplasmic P1 domain between TM1 and TM2 is shown at the top of the figure. The head and the base subdomains are indicated. Transmembrane helices 7 – 8 represent E. coli SecF.
Figure 11. Two conformations of SecDF.
The P1 domain of SecDF, shown extending into the periplasm comprises two subdomains: a P1 head (orange) and P1 base (blue). The protein was crystallized in the F form (left hand side) with the P1 domain positioned so that the head is bent toward the membrane. The I form shows the head directly above the base. This structure is a model built from superimposing the base subdomain of the isolated P1 structure onto that of the full-length SecDF. (used with permission from Tsukazaki et al. (233)
SecDF belongs to the RND (resistance nodulation and cell division) superfamily of proteins. Another member of the RND superfamily is AcrB, a multi-drug efflux transporter, which couples the energy of the electrochemical potential of protons (protonmotive force) to export of its substrates (236). The proton flux in AcrB can be abolished by changes in conserved charged residues thought to be directly involved in proton transport (237). The structural counterparts of these residues in TtSecDF and in E. coli SecDF lie on the interface of SecD and SecF and in the base under the periplasmic head. Changes in three of the conserved residues from aspartate to asparagine or from arginine to methionine eliminated the stimulation of protein export by SecDF both in T. thermophilus and in E. coli, consistent with the idea that SecDF conducts protons (233). Further evidence for coupling of ion flux to protein export comes from studies on a marine halophilic bacterium.
A study in 1990 with Vibrio alginolyticus, long before SecDF had been crystallized, showed that in vitro protein translocation into inverted membrane vesicles from V. alginolyticus to which E.coli SecA was added could be stimulated by the electrochemical potential of Na+ (238). This halophile has two sets of SecDF genes, V.SecDF1 and V.SecDF2, which couple different species of cations to transport. V.SecDF2, like TtSecDF, uses a H+ gradient whereas V.SecDF1 is coupled to a Na+ gradient (239). Expression of the Vibrio, Na+-driven SecDF1 in E. coli that is depleted of endogenous SecDF results in Na+-driven enhancement of export. Additionally, changes of the conserved aspartate and arginine residues in V.SecDF, which are discussed above, eliminate the Na+ stimulation of protein translocation by V.SecDF1 expressed in E. coli (233). These observations provide strong evidence that SecDF couples cation movement to protein export, although the mechanism is undefined.
Difficulties in Demonstration of a Role for SecDF in vitro
When the SecDF complex was originally purified and added to SecYEG in proteoliposomes it showed no stimulation (231), even though the strain with a deletion of SecDF was severely defective in export in vivo (240). There are several reasons that account for the difficulties in the demonstration that SecDF couples protonmotive force to translocation. Although in vivo ATP hydrolysis is likely to be involved only in the early steps, in vitro SecA and ATP can drive transfer of precursors to completion in the absence of protonmotive force (172, 217, 241, 242). Therefore, the role of SecDF and/or protonmmotive force in later steps can be masked in experiments by the use of saturating amounts of ATP and SecA. When chemical quantities of precursors were used, as opposed to the high specific activity, radiochemical quantities used in many early assays, the presence of SecDF stimulated the rate of translocation two-fold (243). Assessing the rate of translocation, instead of the final level as was done in most earlier experiments, and use of a high concentration of precursor to render the SecYEG limiting allowed the stimulation to be observed.
More Confusion
Progress in sorting out the relationship between protonmotive force and SecDF was hindered for several years when it was reported in 1994 that SecDF was required to maintain protonmotive force (244). This finding was frequently cited until it was shown to be an artefact in 2001 (245). In the earlier work the cells had been grown on glucose to deplete the level of SecDFYajC. Glucose represses the synthesis of the succinate dehydrogenase complex. Thus the lack of protonmotive force in the vesicles was not the result of depletion of SecDF, but rather was because succinate had been used as the energy source. Generation of protonmotive force by NADH in vesicles depleted of SecDFYajC showed normal levels of protonmotive force. Nonetheless, the relationship between SecDF and protonmotive force remained confused because stimulation by protonmotive force derived from NADH was observed with or without SecDF (245).
Functional Roles of SecDF
In the two decades following the discovery of SecD and SecF much research was directed toward defining the role of these proteins and several proposals have been set forth. One role suggested for SecDF is the release of the proteins from the membrane after export is complete. Treatment of spheroplasts with anti-SecD IgG specifically inhibited the release of the matured form of maltose-binding protein (246). The release of exported protein into the periplasm also has been shown to be dependent on protonmotive force (247, 248). A proposal for a mechanism of release of proteins mediated by SecDF comes from: 1) demonstration by patch clamp studies that TtSecDF conducts protons, 2) observed enhancement of the conductance by inclusion in the assay of an unfolded protein that binds the P1 head domain, 3) observation in solution of both the F form and I form of SecDF and 4) demonstration that introduction of a disulfide bond to constrain movement between the two forms causes a defect in export. Tsukazaki and his colleagues (233) have proposed that the transition of SecDF from the F form to the I form is coupled to protonmotive force. The translocating polypeptide might emerge from the translocon and bind to the head domain of SecD on the periplasmic side of the membrane. The transition from F to I could include a conformational change that would release the bound polypeptide. A slight modification of this model could explain how SecDF could couple protonmotive force directly to pull the polypeptide through the channel as discussed below (Section Models of Translocation: Energy Source of Translocation).
A third possible role of SecDF overlaps with the function of SecG. Both are involved in the cycle of SecA insertion and deinsertion. SecDF acts to stabilize the inserted state by slowing the deinsertion (249, 250). Since stabilization of the inserted state of SecA requires that a precursor be bound, it is possible that these results reflect interaction of the translocating polypeptide with the head domain of SecD as suggested in the role of polypeptide release. Such an interaction would stabilize insertion as well as prevent backsliding.
YajC, a protein of unknown function
SecYEG is ubiquitous, found in all bacteria, archaea and eukaryotes, whereas SecDFYajC is limited to prokaryotes (251). Not all prokaryotic translocons contain YajC. For example, SecDF from T. thermophilus is not associated with YajC. In E. coli, SecDF copurifies as a complex with YajC, but no function has been assigned to YajC. YajC is produced at five to ten fold the amount of SecDF (Table 4) (234), which may mean it has roles outside of protein export. The structurally related protein AcrB, which functions in drug efflux, was co-crystallized with YajC (252). However, deletion of YajC had only a modest effect on sensitivity to antibiotics. Thus YajC remains a protein of unknown function.
Models of Translocation
Overlap of Function
There is a high degree of interaction among the proteins which mediate export and an apparent overlap of function; thus, the roles are not easily disentangled. Much research indicates that not all components of the system are absolutely required in all conditions. We focus our discussion on what the roles of each might be. The presence of one component may compensate for the lack of another. Perhaps SecDF, SecG and protonmotive force have overlapping roles. Without protonmotive force, the hydrolysis of ATP, which involves SecG inversion, can drive translocation. Since there is evidence that when SecG is present it must invert to stimulate translocation (221), could it be that protonmotive force can also mediate inversion of SecG either through SecDF or directly? In the absence of SecG, SecDF stimulates translocation four-fold, whereas when SecG and SecYE are present in stoichiometric amounts no stimulation can be seen (229). It appears that SecG masks the role of SecDF.
Insights from Studies of Suppressor Mutations
Clues of overlap and a common underlying principle come from studies of suppressors of signal sequence mutations. The mutations, designated prl, have been isolated in secY (prlA) (253), in secA (prlD) (254), in secE (prlG) (255) and in secG (prlH) (256). As the suppressors were identified, different ideas were put forth. Early work proposed that in prlA mutants (changes in SecY) precursors carrying defective signal sequences were translocated because of loss of a proofreading function of SecY (257, 258). Later it was proposed that the prlA mutants increased the affinity of SecYEG for SecA (259). Mutations in secG (prlH) restore export of precursors with defective leaders and were thought to function by recognition of the altered signals (256). The strongest suppressors are alleles of prlA. These suppressors restore export of proteins completely lacking signal sequences (260) and also enhance the rate of translocation of normal precursors (261, 262). Moreover, defects in export that are not derived from altered signals but from folded elements in the mature portion of precursors are suppressed (262).
Demonstration that prl mutations in SecY and SecE destabilize the association of SecYEG led to the simple unifying explanation that all effects of suppressors result from changes in rigidity of the SecYEG complex (262, 263). What seem to be disparate functions may in fact be directed to the same end, opening the channel of the translocon to allow efficient transfer. While reading the literature of the field in preparation for writing this review we began to wonder if it might be that many of the diverse components that stimulate protein translocation work through the same fundamental mechanism: loosening the SecYEG structure to open the channel.
First, consider SecA and SecG. Hydrolysis of ATP by SecA results in the inversion of SecG. The change in the contacts between SecG and SecYE as it inverts might destabilize the channel and allow it to open. Thus, one role of ATP hydrolysis might be to open the channel indirectly. Since, as discussed earlier (Section SecDFYajC, Functional Roles of SecDF) SecDF modulates SecA insertion and deinsertion, it could also be involved in cycles of relaxing the tranlocation channel. It is likely that protonmotive is involved since it is clear that SecDF couples proton movement to translocation. Protonmotive force many not only work through SecDF, but it might also act directly on SecYEG (264). If so, we could reconcile the confusing result that protonmotive force stimulates export in the absence of SecDF both in vesicles (245) and in proteoliposomes (212). Such an overlap would allow the system to function lacking some components, but at lower efficiency.
Energy Source of Translocation
Citations of the work that support the cycle described here are given in the previous sections. Precursors to be exported enter the Sec pathway through interactions with SecB, a cytosolic chaperone, and SecA, an ATPase that is a peripheral membrane component of the translocation apparatus. The channel which provides the pathway through the membrane is formed by SecY within one trimeric unit of the translocon, SecYEG (158, 265). In the idle state the channel is closed and provides a permeability barrier to polypeptides as well as to ions and small compounds. In the initial stage of translocation the channel is widened by interactions of SecYEG with SecA and precursor polypeptides. The binding of SecA to SecYEG also releases the suppression of the active site on SecA for ATP hydrolysis. The precursor is transferred from SecB and binds to SecA in a clamp between the wall of NBD2 and PBD to be positioned over the mouth of the translocation channel. Up to this stage there is general agreement. Nonetheless, even in these initial steps, there is ongoing discussion concerning the details. Much less defined is the exact mechanism and source of the energy for the movement of the precursor polypeptide through the channel.
In order to discuss the diverse and very distinct models we will divide them into two groups. The models in the first group postulate that the chemical energy, whether it be hydrolysis of ATP or protonmotive force (an electrochemical potential energy), is converted to mechanical work. An early model, proposed long before any structural data were available, proposed that the cyclic membrane insertion – deinsertion of SecA is accompanied by insertion of the precursor, in steps of 20 – 30 aminoacyl residues for each round of ATP hydrolysis (215–217, 266). This model implicates SecA as providing the pushing force, but does not speculate as to the region of SecA involved. A recent, closely related model introduced a refinement based on a structure of SecA bound to SecY. In the structure the two helix finger (also referred to as IRA1) of SecA is inserted into the wide opening at the top of the channel in SecY. It is posited that a translocating polypeptide is bound at the tip and hydrolysis of ATP results in movement of the finger further into the channel to push the precursor through. The finger would release the precursor and withdraw to rebind in another cycle (200). Both models propose that when released from SecA, the polypeptide chain is free to diffuse either forward or backward (referred to as sliding). Whereas the earlier model proposed a defined step size based on the observation of discrete intermediates during translocation, the two helix finger model suggests that the length of the polypeptide chain that is moved in a single step depends on the distance between stretches of aminoacyl residues that make sufficiently strong contact with the two helix finger to be pushed into the channel. Indirect support for this idea is that a precursor undergoing translocation can be crosslinked to the tip of the finger (200). Further support is found in the model of signal peptide binding determined by FRET studies that shows a parallel alignment of the peptide with the two helix finger positioning the two elements for interaction (174, 175). There is no evidence to date for the purported movement of the two helix finger into and out of the channel. Inconsistent with the model is an independent study in which very little effect on translocation was observed when the two helix finger was immobilized by crosslinking it to the edge of the channel (267), indicating that a high degree of flexibility cannot be required.
A model has also been proposed that couples protonmotive force to mechanical work through SecDF. As the polypeptide emerges from the channel it binds SecDF on the periplasmic side of the membrane (233). If protonmotive force acts to induce the conformational changes observed in the head of the P1 domain, then SecDF might pull the precursor through the translocon (233). In the extreme, ATP would be required only to push a length of residues sufficient to reach the SecD head. Both proposed mechanisms, pushing by SecA and pulling by SecDF, might function together. It has long been known, but frequently forgotten, that in vivo the energy of ATP hydrolysis alone is not sufficient to translocate precursors through the membrane; proton motive force is also required.
A model different from those discussed above proposes that no exogenous energy is required for the actual movement through the channel. Kinetic studies of translocation in an in vitro fluorescence-based real time assay (268, 269) led to proposal of a model in which the source of energy for movement of polypeptides is Brownian diffusion after an initial opening of the channel. The diffusion would be directional if the channel were blocked at the cytoplasmic side, either by association of SecA or closure by a different mechanism. Since diffusion over the short distance from SecA through the channel in SecY would be complete in a few seconds (~5 seconds), it was proposed that passage through the channel is not the rate-limiting step but rather the rate observed (~25 seconds) is slower because it is limited by the probability of abortive transport, which might be due to the polypeptide becoming incompetent within the channel. The kinetics show no evidence of step-wise transfer predicted by the models that propose a mechanical coupling of either ATP or protonmotive force to provide the power to push or pull the precursor. However, if, as it is speculated in the diffusion model, the rate were limited by abortive translocation, which would require clearing of incompetent precursors from the channel, then ATP would be necessary through the entire process for reinitiation (269). Although this aspect of the diffusion model appears to be inconsistent with observations that translocation can be completed in vitro by protonmotive force in the absence of ATP, it is not. In this case, the intermediates in transfer are not incompetent necessitating removal from the translocon, but remain in the channel and can bypass the initiation step. If diffusion were to drive translocation, then the energy of ATP hydrolysis and/or protonmotive force might function to open the channel. Speculation on how this might happen was given in the section which discusses insights from studies of suppressor mutations.
Although it is not known what step limits the rate, evidence that the rate-limiting step in vitro involves proton-transfer reactions is found in the demonstration that the rate of translocation in vitro is approximately three-fold slower in deuterium oxide relative to water (270).
The Translocon: a Modular Assembly
The essential core unit of the translocon that is capable of secretion of protein into the periplasm is the heterotrimer, SecYEG. In vivo the SecY and SecE subunits are stably associated showing no exchange. The SecG subunit exchanges under some conditions (271). There is an equilibrium between monomers and dimers of SecYEG (272, 273) that can be modified by translocation-binding partners (274). There are two dimeric forms of SecYEG. One form has SecG at the interface (referred to as back-to-back) and the other has the lateral gate of SecY at the interface (front-to-front). Both forms exist in vivo (275, 276). While it is clear that a monomeric unit of SecYEG contains the translocon channel, there is continuing discussion as to whether for activity SecYEG must be dimeric (208, 209, 277) or can act alone (275, 278). Two studies carried out in vivo using very similar approaches came to conflicting conclusions. One using disulfide crosslinking to detect SecYEG dimers showed that when translocating polypeptides jammed the translocon the level of dimeric SecYEG dropped (275). The other study utilized both in vivo photo-crosslinking and disulfide crosslinking to determine the oligomeric state of active translocons (276). The results show that the level of SecYEG dimers was constant whether the SecYEG was actively engaged in translocation or the translocons were cleared of precursor substrates. In addition, a complex of SecA bound to a dimer of SecY in the process of translocating proOmpA was observed. Clearly a dimer is functional, but it cannot be concluded that to be functional SecY must be a dimer. The answer may not be one or the other. It is likely that the functional oligomeric state varies depending on whether SecYEG is functioning in the SecB:SecA pathway for secretion or works in conjunction with the SRP mediated co-translational pathway for insertion of membrane proteins (272). To insert membrane proteins YidC must associate with SecYEG through association with SecDF. This “holotranlocon” contains a monomeric unit of SecYEG (279). Additionally, within either pathway of precursor undergoing translocation may dictate the stoichiometry of the SecYEG (280).
Two chaperones that function in the periplasm, PpiD and YfgM, interact with SecYEG. Each is tethered to the membrane via a single transmembrane helix. PpiD is positioned near the lateral gate of SecY and interacts with secreted proteins as they emerge from the channel on the periplasmic side (281, 282). When the SecYEG functions to insert membrane proteins, the PpiD dissociates allowing YidC to approach the lateral gate. YfgM interacts with both PpiD and SecYEG (283, 284). These chaperones aid the entrance of newly translocated proteins into the periplasmic chaperone network.
The SecYEG in complex with SecDFYajC and YidC, which has been termed the “holotranslocon,” (279) must represent a minor fraction of the assemblies containing SecYEG since in vivo there are approximately ten copies of SecYEG for every one SecDF (Table 4). The “holo” complex allows coupling of protonmotive force through the SecDF unit and insertion of membrane proteins via YidC. It is not known whether in vivo the holotranslocon exists as a static stable subpopulation of the translocation apparatuses or if a dynamic equilibrium exists, allowing modules to associate as needed. YidC, which is in large excess (~ 2500 copies per cell, Table 4), can function either with SecYEG or it can act alone to insert cytoplasmic membrane proteins (285).
Perhaps the term holotranslocon is a misnomer in that there are many possible modular assemblies. So, in fact, the complete or “holo” translocon will depend on the function.
Level of Proteins in the Cell
Regulation of Expression
Regulation of the synthesis of several components of the Sec system in E. coli is dependent on growth rate. The production of SecA, SecY and SecE has been shown to increase during rapid growth in rich media relative to slower growth in minimal media. The greatest difference observed was a three-fold increase in SecY. SecA showed a 60% increase and SecE an increase of 30% (286). Such growth-rate dependent regulation is characteristic of “housekeeping” genes such as the genes encoding ribosomal proteins. Thus the increased expression of SecY and SecE can be explained since the SecY gene is located at the promoter-distal part of the spc operon and is co-transcribed with ribosomal proteins (287) and SecE is transcribed with nusG, which encodes a protein involved in transcription anti-termination (288). The production of SecY, SecF and SecA show differential expression dependent on growth phase. The levels of SecY and SecF are lower in stationary phase compared to the levels in exponential phase; whereas the level of SecA is higher in exponential phase (289).
Production of SecA is very tightly regulated to produce the optimal amount for the cellular conditions. This is achieved by an upstream gene, secM, which exerts regulation at the level of translation (290). Years before the mechanism was worked out it was known that the synthesis of SecA is derepressed 10 to 20 fold when secretion is blocked either genetically or physiologically (291, 292). Sequencing of the secA gene (293) revealed a gene of unknown function geneX immediately upstream. A mutation in that gene had a polar effect on secA expression indicating that geneX and secA are in the same operon. The protein encoded by geneX is a precursor polypeptide containing 170 aminoacyl residues. Regulation of secA expression was shown to depend on the secretion status of the geneX preprotein (294). Therefore, geneX was renamed secM for “secretion monitor.”
SecM is exported to the periplasm where it is rapidly degraded by Prc, a periplasmic protease (295, 296). SecM, which has no function in the periplasm, performs its function in the cytoplasm as a monitor of secretion. When the sequence 150FX4WIX4GIRAGP166 (where X can be any amino acid) interacts with the exit tunnel of the ribosome, translation is transiently arrested (297). The interaction of the nascent peptide stalled at Pro166, the fifth position from the C terminus, with both the 23S rRNA and with L22, a ribosomal protein (298), propagates changes through the 50S ribosomal subunit. Rearrangements have been observed by cryo-electron microscopy of both the ribosomal RNA (299) and of the ribosomal proteins (300). These changes reach the site of mRNA and tRNA binding at the interface of the 30S and 50S subunits. A structure with 5.6Å resolution (300) indicates changes in the geometry of the peptidyl transferase center that can explain the arrest.
The translational arrest occurs in the cytoplasm requiring no exogenous factors. Under normal growth conditions the arrest is transient. The signal sequence is recognized by SRP and the nascent SecM is targeted to SecYEG. When the nascent SecM preprotein undergoes transfer through the SecYEG channel the arrest is released (301). This provides a mechanism for translational regulation of secA, the next gene in the operon. The region of mRNA between the two genes forms a stem-loop structure, which masks the Shine-Dalgarno sequence of the secA gene. When the ribosome is stalled it occupies sequences that comprise the 5′ end of the structure so that the stem-loop is opened and SecA is translated (298, 302). In this way, if secretion is blocked, SecA is expressed at a high level. If the arrest is short-lived, as occurs when the secretion system is functioning normally, SecA is produced at a basal level. To achieve an appropriate basal level of SecA the timing between the onset and the release of the arrest is critical. Delivery to the membrane translocon too soon after arrest does not allow sufficient time to synthesize the basal quantity of SecA; whereas an arrest that persists too long results in overproduction of SecA. The precise timing between arrest of translation and delivery to the translocon for release is controlled by the signal sequence of the SecM preprotein. The property of the signal that achieves the timing is the unusually long amino-terminal region (303). This signal sequence targets the nascent SecM-ribosome complex to the translocon at a rate intermediate between that of signals for co-translational and signals for post-translational export (304).
In addition to regulating the level of SecA synthesized, SecM appears to play a role in ensuring that SecA has maximal activity. The co-translational targeting of SecM to the SecYEG translocon would tether the entire polysome so that SecA would also be produced in close proximity to the membrane (305). It has been suggested that biosynthesis at the membrane helps SecA adapt a functional conformation. In this context, it is important to note that SecA has been shown to exist in the cell free in the cytoplasm and associated with membrane, both peripherally and integrally (144, 148, 306). Additionally, when SecY and SecA are simultaneously co-assembled into lipids from detergent the resulting proteoliposomes have a six-fold increase in the number of SecYEG units that are active compared to reconstituted proteoliposomes containing SecYEG only with SecA added after assembly (280). The activation of secretion by membrane localization of SecA via cotranslation with SecM might reflect an in vivo assembly in the membrane of SecA with SecY.
SecB shows no evidence of regulation dependent on growth rate or growth phase, but the transcription of the secB gene is subject to catabolite repression (307). Cells grown with glycerol as the carbon source contain more SecB than do cells grown with glucose. The secB gene is transcribed from two promoters. The distal promoter, P1, transcribes an operon that contains an open reading frame (ORF) designated yibN followed by grxC, secB and gpsA. The gene grxC encodes a minor glutaredoxin and gpsA is the gene for sn-glycerol-3-phosphate dehydrogenase, an enzyme involved in lipid synthesis. The proximal promoter, P2, transcribes only the secB and gpsA genes. Whereas the P1 promoter expresses secB constitutively, the P2 promoter is responsive to the carbon source via the regulatory protein CRP, the cyclic AMP (cAMP) receptor protein. Growth on glucose decreases the cellular level of cAMP, which in turn results in a decreased level of the CRP-cAMP complex. The transcription is decreased because CRP-cAMP is not bound near the promoter. When transcribed from the P2 promoter in the absence of P1, addition of cAMP increases expression three to four fold.
Expression of SecB is increased two to four fold in response to depletion of the chaperones DnaK, DnaJ, GrpE, GroEL and GroES (126). Unlike SecA, SecB is not regulated in response to defects in the translocation machinery unless the defect results in an accumulation of precursors of exported proteins that interact with SecB. Overproduction of an exported precursor with a defective leader or of a cytoplasmic protein does not result in elevated levels of SecB (308).
Stability of Proteins
The amount of protein in a cell is determined not only by regulation of transcription and/or translation, but also by the stability of the polypeptide. SecA, SecB, SecE and SecF are stable in the absence of other proteins involved in export and can be overproduced individually (34, 309–311). The levels of the other subunits of the translocation complex in the membrane, SecY, SecD and SecG, are dependent on their protein binding partners for stabilization.
SecY is the most dependent on assembly into a complex for stability. It cannot be overproduced in the absence of overproduction of SecE. It is degraded with a half-life of 2 minutes (310, 312) by the FtsH protease (313). When both SecY and SecE are overproduced, SecY is completely stabilized. YajC overproduction has a slight stabilizing effect on SecY, lengthening the half-life from two minutes to 10 minutes. SecF also has a stabilizing effect. In the absence of SecE, overproduction of SecF 5,400 fold allowed accumulation of SecY at 54-fold the chromosomal level. This extent of stabilization is much less than that produced by SecE, which when overproduced 176 fold results in a 265-fold increase in SecY (311).
As discussed in an earlier section (Models of Translocation), SecDF and SecG appear to overlap in function and the stability of SecG is dependent on SecDF. The level of SecG in membrane decreases to about half when SecDFYajC is depleted. Physical interaction between SecG and SecDFYajC is suggested by the inhibition of crosslinking between two molecules of SecG by the presence of SecDFYajC (314).
Removal and Degradation of Signal Peptides
Proteins exported by the Sec system are synthesized as precursors, which are proteolytically processed to remove the amino-terminal extensions of 20 to 25 residues termed signal sequences or leader peptides. These sequences begin with a short (five residues), basic domain (N region), followed by a central hydrophobic core (H region) and end with a region (C region), which serves as the recognition site for the protease that removes the signal (315). The enzyme cleaves at sites that have small residues at the −3 and −1 positions relative to the scissile bond (316). Recognition involves the five to nine amino acyl residues preceding the cleavage site, including a helix-disrupting glycyl or prolyl residue (317). The specific protease, signal (leader) peptidase I was first purified in 1980 (318) and the gene lepB (leader peptidase) was identified in 1981 (319). Signal peptidase I functions at the periplasmic face of the cytoplasmic membrane (320, 321) and is not necessary for transfer of the precursor across the cytoplasmic membrane, but rather for the release of the matured protein from the surface of the membrane into the periplasm (322). Construction of a strain which overproduces signal peptidase I approximately 75-fold (323) was crucial in obtaining sufficient quantities of highly pure protein for studies of the enzymic activity.
Signal peptidase I from E. coli belongs to a family of type 1 signal peptidases that use a serine/lysine catalytic mechanism (324). The crucial catalytic residues are Ser91 and Lys146. The numbering of these residues does not correspond to the numbering in many early publications (Ser90 and Lys145) because there was an error in the original DNA sequence published in 1983 (321), which caused the numbers to be off by one starting with Asp44. Crystal structures of signal peptidase I, with and without inhibitors, (Table 3, ((325)) in combination with mutational and biochemical studies (324, 326–329) have resulted in the proposal of a mechanism in which Ser91Oγ serves as the nucleophile and Lys146Nζ as the general base. Signal peptidase I processes not only precursors exported by the Sec system, but also those transported by the twin-arginine translocation (TAT) pathway (330).
Lipid-modified proteins (lipoproteins) in E. coli are processed by a specialized protease, signal peptidase II, the product of the lspA gene (331). Before precursors can be cleaved by signal peptidase II, the cysteine at the cleavage site must be modified by diacylglyceryl transferase, which adds a thioether-linked diacylglycerol (332, 333). Cleavage generates an amino-terminal cysteine which is modified by an N-linked fatty acid. The major lipoprotein in E. coli, lpp, Braun’s lipoprotein, is anchored to the outer membrane and associates with the peptidoglycan. This protein is considered the prototype for the numerous lipoproteins in bacteria. Ninety different lipoproteins of diverse function have been identified; some associated with the periplasmic face of the cytoplasmic membrane and others the outer membrane (334).
After cleavage by either signal peptidase I or signal peptidase II to generate the mature proteins, the signal peptides remain in the membrane. Several signal peptide hydrolases have been implicated in the degradation of the signals. In 1988, signal peptide peptidase, SppA, (previously called protease IV) was identified as an endopeptidase that cleaves within the H region of the peptide (335). SppA is anchored to the cytoplasmic membrane by an N-terminal transmembrane segment. Crystallization of the soluble catalytic domain revealed that SppA is a tetrameric assembly with a bowl-shaped structure (336). The interior of the inverted bowl is predominantly non-polar and contains four active sites. Each of the protomers mediates proteolysis using a catalytic dyad with Ser409 serving as the nucleophile and Lys209 as the general base. The wide opening of the inverted bowl is oriented over the surface of the membrane by transmembrane N-terminal segments (residues 39–45), which are missing in the crystallized protein. This would position the active site approximately 45 Å away from the surface of the membrane. The SppA assembly exhibits architecture similar to that of ClpP, an ATP-dependent protease that interacts with chaperones ClpA and ClpX to unfold and degrade misfolded proteins. Based on this observation it has been proposed (336) that SppA may interact with protein partners to extract the signal peptide from the membrane. The induction of HexII phase by leader peptides (Section Role of Lipid) might play a role in rendering the peptides more accessible to proteins at the surface. Another signal peptide hydrolase involved in degradation of signal peptides is RseP (originally YaeL) (337). RseP, a metalloprotease, is a member of the S2P family of proteins which cleave within membrane spanning regions of proteins and has its active site embedded in the membrane. RseP requires a C-terminal hydrophobic residue to activate the protease (338) and therefore can cleave only after cleavage by signal peptidase I or signal peptidase II.
Role of Lipid
Not only are the proteinaceous components of the translocation apparatus important, but the proper lipid environment is crucial for activity. The cytoplasmic membrane of E. coli comprises two main groups of phospholipids. Phosphatidylethanolamine, PE, a zwitterionic, neutral lipid, makes up 75–80 % of the total phospholipids. The remainder consists of negatively charged lipids, approximately 20% is phosphatidylglycerol (PG) and approximately 5% is cardiolipin (diphophatidylglycerol, CL). Both classes of lipids are necessary for efficient translocation of proteins across the cytoplasmic membrane.
The importance of anionic lipids in protein translocation was first demonstrated in vivo by using strains of E. coli with severe defects in pgsA, the gene which encodes phosphatidylglycerophosphate synthase, resulting in extremely low levels of anionic lipids (339). Under these conditions the processing of the precursors of two outer membrane proteins, OmpA and PhoE was retarded. A more severe effect was demonstrated by an in vitro assay of translocation using prePhoE and inner membrane vesicles from the lipid depleted strain. Translocation was restored to vesicles that had a low content of PG and CL by introduction of anionic lipids. The efficiency of translocation is directly proportional to the amount of anionic lipids present (340). Several anionic lipids in addition to PG restore function indicating that the crucial feature of the lipid is a negative charge on the head group. Cardiolipin is not specifically required since deletion of the cardiolipin synthase gene (clsA) showed no effect on translocation (339). For a lucid discussion of the complications surrounding the establishment of a pgsA null mutant and the origin of apparently conflicting conclusions concerning the viability of strains lacking a functional pgsA gene see Dowhan (341). Strains constructed in 1989 (342) and in 2000 (343) that have the pgsA gene under control of inducible promoters were used in later studies in place of a null mutant, allowing regulation of the level of the anionic lipids.
The requirement of the translocation system for anionic lipids has several origins. The best documented interactions are those with SecA. As discussed previously the full ATPase activity of SecA, the translocation ATPase, occurs in the presence of a precursor protein and inner membrane vesicles containing SecYEG (172). SecA can be stimulated to a level lower than that of full translocation ATPase by liposomes alone if they contain PG and CL. Liposomes comprising zwitterionic lipids, phosphatidylcholine (PC) and phosphatidyl ethanolamine (PE) are inactive (172) in binding SecA and in activating the ATPase. SecA not only binds the surface of the membrane but penetrates into the hydrocarbon region (131, 145, 148, 344). This penetration requires anionic lipids and is inhibited by ATP (344). SecA exists in both a freely soluble form in the cytoplasm but also in a tightly bound membrane form (144). Association with the membrane induces changes in conformation (148, 306). Numerous studies by P.C. Tai and his collaborators have suggested an unconventional role for membrane-inserted SecA as a low efficiency, low specificity channel (306, 345, 346) that evolved by the recruitment of SecYEG and SecDF into the present day highly specific and efficient translocon.
It is difficult to separate the effects of anionic lipids on SecA from possible requirements for direct interaction of anionic lipids with SecYEG since SecA is itself required for translocation. However, it is clear the cardiolipin specifically associates with SecYEG to stabilize the dimeric form (347). Extraction of SecYEG with its surrounding annular lipids from native membrane using styrene- malic acid lipid particles showed enrichment of PG and CL, whereas extraction of SecDF or YidC by the same method showed no specific enrichment of the anionic lipids (348).
Phosphatidylethanolamine, the major lipid constituent of the cytoplasmic membrane, has a preference for non-lamellar structures, such as the hexagonal phase (HexII), which has negative curvature. Nonetheless, the biological membrane functions as a lamellar bilayer. Although it is clear that the activity of many membrane proteins is influenced by nonbilayer lipids, the mechanism of action of these lipids is not clearly defined. The effects of nonbilayer lipids on peripheral and integral proteins are discussed in a review by de Kruijff and colleagues (349). These lipids may exert their influence via aspects of curvature stress, which arises because nonbilayer lipids tend to form structures with a negative curvature. Thus, each monolayer has a tendency for negative curvature, but in a bilayer they are forced to be planar, resulting in curvature stress. The stress has effects on properties of the bilayer such as packing defects at the interface and changes in lateral pressure across the bilayer. In vivo PE is not essential for protein translocation; however, in vitro translocation is very low in membrane vesicles having low levels of PE (349). Translocation can be stimulated in these membranes by introduction of PE or other nonbilayer lipids. Derivations of PE that adopt the lamellar, bilayer structures do not stimulate translocation (350), indicating that it is the nonbilayer property of PE that is necessary.
During passage of protein through the channel, SecYEG undergoes conformation changes (166, 189–191, 351–353). The nonbilayer properties of PE might be involved in allowing function through changes in lateral pressure in the bilayer. The stimulation of translocation by PE cannot be entirely attributed to interaction with SecYEG since PE has effects on SecA, stimulating the ATPase activity (354), and on leader peptidase. The insertion of the soluble catalytic domain of leader peptidase into the membrane is facilitated by PE (355–357). This penetration may be functionally significant since signal sequences would not reach the periplasmic surface of the membrane if they were helical.
Investigations have been carried out that compare the properties of isolated signal peptides having sequences that function to mediate efficient export of precursors with signal peptides having altered sequences that are not functional. Active signal peptides are distinguished from those of low activity by two features: a higher tendency to form α-helices in the presence of membrane-mimetic systems and a higher affinity for bilayers that contain anionic lipids (358–363).
After initial interaction with the surface of a lipid bilayer or monolayer, signal peptides penetrate into the hydrocarbon region where the α-helical conformation forms. The penetration of the peptide into the hydrocarbon lipid tails induces lateral segregation resulting in clustering of PG (364). The hydrophobic region of signals ideally comprises 10 leucine or isoleucine residues (365), which is too short to span the bilayer as an α-helix. This hydrophobic mismatch induces nonlamellar Hex II lipid structures (366). Although there is no evidence for a function in vivo, it has been suggested that this destabilization of the bilayer may be involved in some phase of protein export.
Closing Remarks
We have endeavored to present an accurate picture of the current understanding of the Sec system of protein export. However, during the time that we have paused to write this review, research has continued. Keep in mind that the models presented here involve speculation and were based on knowledge available at the time. It is difficult to incorporate all accumulated data into one comprehensive model. Results from different laboratories may appear to be contradictory; however, as progress is made new ideas arise and what seemed to be contradictions may be resolved. We hope we have been successful in bringing the robust data from the past forward to be incorporated into current ideas about the Sec system. There are many approaches used in research and we see only fragments of the entire picture at a time: we view export in broken images. We are optimistic that as more is learned we shall, to paraphrase what Robert Graves wrote in the poem “In Broken Images,” come to a new understanding of our confusion (367).
Note to Readers.
Our goal in this chapter is to cover the field of export via the Sec system from its inception to the time of writing. We have read every paper cited; therefore, we are as certain as possible that if the reader goes to the paper, data will be found to support the statements made. The reader will find references to other relevant publications within the papers we have cited. We apologize to the authors of the papers we did not cite.
Our hope is that this review will be useful to investigators new to the field as well as to those who are currently deeply involved in studies of protein export. My laboratory has been investigating export in Escherichia coli since 1977 and in writing this chapter, I learned much that I never knew, or have forgotten.
Sometimes the early literature is overlooked because powerful, new techniques have been developed and structures have become available. However, the early data are robust and hold an enormous amount of valuable information. Perhaps early interpretations were wrong, but well-conceived and carefully carried out experiments produce valid data that should not be forgotten. One may discover important insight into interpretation of early data when seen in retrospect and armed with knowledge gained from subsequent work, including determination of protein structures. If this chapter functions to encourage investigators to look back at previous work to facilitate movement forward, we will have succeeded in our goal.
Acknowledgments
We thank Chunfeng Mao for her help surveying the scientific literature and Wing-Cheung Lai for his assistance in the preparation of the figures. This work was supported by an endowment from the Hugo Wurdack Trust at the University of Missouri and National Institutes of Health Grant GM29798 (to L.L.R.).
References
- 1.Redman CM, Siekevitz P, Palade GE. Synthesis and transfer of amylase in pigeon pancreatic micromosomes. J Biol Chem. 1966;241:1150–1158. [PubMed] [Google Scholar]
- 2.Andrews TM, Tata JR. Protein synthesis by membrane-bound and free ribosomes of secretory and non-secretory tissues. Biochem J. 1971;121:683–694. doi: 10.1042/bj1210683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlessinger D. Protein Synthesis by Polyribosomes on Protoplast Membranes of B. Megaterium. J Mol Biol. 1963;7:569–582. doi: 10.1016/s0022-2836(63)80103-5. [DOI] [PubMed] [Google Scholar]
- 5.Hendler RW, Tani J. On the Cytological Unit for Protein Synthesis in Vivo in E. Coli. Ii. Studies with Intact Cells of Type B. Biochim Biophys Acta. 1964;80:294–306. doi: 10.1016/0926-6550(64)90101-x. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger D, Marchesti VT, Kwan BC. Binding of Ribosomes to Cytoplasmic Reticulum of Bacillus Megaterium. J Bacteriol. 1965;90:456–466. doi: 10.1128/jb.90.2.456-466.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron EZ, Kohler RE, Davis BD. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966;153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 8.Moore LD, Kocun FJ, Umbreit WW. Cell-free protein synthesis: effects of age and state of ribosomal aggregation. Science. 1966;154:1350–1353. doi: 10.1126/science.154.3754.1350. [DOI] [PubMed] [Google Scholar]
- 9.Cancedda R, Schlesinger MJ. Localization of polyribosomes containing alkaline phosphatase nascent polypeptides on membranes of Escherichia coli. J Bacteriol. 1974;117:290–301. doi: 10.1128/jb.117.1.290-301.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randall LL, Hardy SJ. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur J Biochem. 1977;75:43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith WP, Tai PC, Thompson RC, Davis BD. Extracellular labeling of nascent polypeptides traversing the membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1977;74:2830–2834. doi: 10.1073/pnas.74.7.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halegoua S, Sekizawa J, Inouye M. A new form of structural lipoprotein of outer membrane of Escherichia coli. J Biol Chem. 1977;252:2324–2330. [PubMed] [Google Scholar]
- 13.Sekizawa J, Inouye S, Halegoua S, Inouye M. Precursors of major outer membrane proteins of Escherichia coli. Biochem Biophys Res Commun. 1977;77:1126–1133. doi: 10.1016/s0006-291x(77)80095-8. [DOI] [PubMed] [Google Scholar]
- 14.Inouye H, Beckwith J. Synthesis and processing of an Escherichia coli alkaline phosphatase precursor in vitro. Proc Natl Acad Sci U S A. 1977;74:1440–1444. doi: 10.1073/pnas.74.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall LL, Hardy SJ, Josefsson LG. Precursors of three exported proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:1209–1212. doi: 10.1073/pnas.75.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaelis S, Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- 17.Emr SD, Schwartz M, Silhavy TJ. Mutations altering the cellular localization of the phage lambda receptor, an Escherichia coli outer membrane protein. Proc Natl Acad Sci U S A. 1978;75:5802–5806. doi: 10.1073/pnas.75.12.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassford P, Beckwith J. Escherichia coli mutants accumulating the precursor of a secreted protein in the cytoplasm. Nature. 1979;277:538–541. doi: 10.1038/277538a0. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Bassford PJ, Jr, Beckwith J. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell. 1981;24:707–717. doi: 10.1016/0092-8674(81)90097-0. [DOI] [PubMed] [Google Scholar]
- 20.Josefsson LG, Randall LL. Different exported proteins in E. coli show differences in the temporal mode of processing in vivo. Cell. 1981;25:151–157. doi: 10.1016/0092-8674(81)90239-7. [DOI] [PubMed] [Google Scholar]
- 21.Koshland D, Botstein D. Evidence for posttranslational translocation of beta-lactamase across the bacterial inner membrane. Cell. 1982;30:893–902. doi: 10.1016/0092-8674(82)90294-x. [DOI] [PubMed] [Google Scholar]
- 22.Hansen W, Garcia PD, Walter P. In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent posttranslational translocation of the prepro-alpha-factor. Cell. 1986;45:397–406. doi: 10.1016/0092-8674(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 23.Waters MG, Blobel G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J Cell Biol. 1986;102:1543–1550. doi: 10.1083/jcb.102.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randall LL. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell. 1983;33:231–240. doi: 10.1016/0092-8674(83)90352-5. [DOI] [PubMed] [Google Scholar]
- 25.Daniels CJ, Bole DG, Quay SC, Oxender DL. Role for membrane potential in the secretion of protein into the periplasm of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:5396–5400. doi: 10.1073/pnas.78.9.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enequist HG, Hirst TR, Harayama S, Hardy SJ, Randall LL. Energy is required for maturation of exported proteins in Escherichia coli. Eur J Biochem. 1981;116:227–233. doi: 10.1111/j.1432-1033.1981.tb05323.x. [DOI] [PubMed] [Google Scholar]
- 27.Pages JM, Lazdunski C. Maturation of exported proteins in Escherichia coli. Requirement for energy, site and kinetics of processing. Eur J Biochem. 1982;124:561–566. [PubMed] [Google Scholar]
- 28.Zimmermann R, Wickner W. Energetics and intermediates of the assembly of Protein OmpA into the outer membrane of Escherichia coli. J Biol Chem. 1983;258:3920–3925. [PubMed] [Google Scholar]
- 29.Bakker EP, Randall LL. The requirement for energy during export of beta-lactamase in Escherichia coli is fulfilled by the total protonmotive force. EMBO J. 1984;3:895–900. doi: 10.1002/j.1460-2075.1984.tb01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Tai PC. ATP is essential for protein translocation into Escherichia coli membrane vesicles. Proc Natl Acad Sci U S A. 1985;82:4384–4388. doi: 10.1073/pnas.82.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geller BL, Movva NR, Wickner W. Both ATP and the electrochemical potential are required for optimal assembly of pro-OmpA into Escherichia coli inner membrane vesicles. Proc Natl Acad Sci U S A. 1986;83:4219–4222. doi: 10.1073/pnas.83.12.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller M, Fisher RP, Rienhofer-Schweer A, Hoffschulte HK. DCCD inhibits protein translocation into plasma membrane vesicles from Escherichia coli at two different steps. EMBO J. 1987;6:3855–3861. doi: 10.1002/j.1460-2075.1987.tb02723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamane K, Ichihara S, Mizushima S. In vitro translocation of protein across Escherichia coli membrane vesicles requires both the proton motive force and ATP. Journal of Biological Chemistry. 1987;262:2358–2362. [PubMed] [Google Scholar]
- 34.Cabelli RJ, Chen L, Tai PC, Oliver DB. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver D. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lill R, Cunningham K, Brundage LA, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall LL, Hardy SJ. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 38.Hardy SJ, Randall LL. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science. 1991;251:439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- 39.Park S, Liu G, Topping TB, Cover WH, Randall LL. Modulation of folding pathways of exported proteins by the leader sequence. Science. 1988;239:1033–1035. doi: 10.1126/science.3278378. [DOI] [PubMed] [Google Scholar]
- 40.Liu GP, Topping TB, Cover WH, Randall LL. Retardation of folding as a possible means of suppression of a mutation in the leader sequence of an exported protein. J Biol Chem. 1988;263:14790–14793. [PubMed] [Google Scholar]
- 41.Liu G, Topping TB, Randall LL. Physiological role during export for the retardation of folding by the leader peptide of maltose-binding protein. Proc Natl Acad Sci U S A. 1989;86:9213–9217. doi: 10.1073/pnas.86.23.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckwith J, Ferro-Novick S. Genetic studies on protein export in bacteria. Curr Top Microbiol Immunol. 1986;125:5–27. doi: 10.1007/978-3-642-71251-7_2. [DOI] [PubMed] [Google Scholar]
- 43.Ellis RJ, Hemmingsen SM. Molecular chaperones: proteins essential for the biogenesis of some macromolecular structures. Trends Biochem Sci. 1989;14:339–342. doi: 10.1016/0968-0004(89)90168-0. [DOI] [PubMed] [Google Scholar]
- 44.Delepelaire P, Wandersman C. The SecB chaperone is involved in the secretion of the Serratia marcescens HasA protein through an ABC transporter. EMBO J. 1998;17:936–944. doi: 10.1093/emboj/17.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debarbieux L, Wandersman C. Folded HasA inhibits its own secretion through its ABC exporter. EMBO J. 2001;20:4657–4663. doi: 10.1093/emboj/20.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolff N, Sapriel G, Bodenreider C, Chaffotte A, Delepelaire P. Antifolding activity of the SecB chaperone is essential for secretion of HasA, a quickly folding ABC pathway substrate. Journal of Biological Chemistry. 2003;278:38247–38253. doi: 10.1074/jbc.M302322200. [DOI] [PubMed] [Google Scholar]
- 47.Sapriel G, Wandersman C, Delepelaire P. The SecB chaperone is bifunctional in Serratia marcescens: SecB is involved in the Sec pathway and required for HasA secretion by the ABC transporter. J Bacteriol. 2003;185:80–88. doi: 10.1128/JB.185.1.80-88.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumamoto CA, Beckwith J. Mutations in a new gene, secB, cause defective protein localization in Escherichia coli. J Bacteriol. 1983;154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumamoto CA, Beckwith J. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu H, Nishiyama K, Tokuda H. Expression of gpsA encoding biosynthetic sn-glycerol 3-phosphate dehydrogenase suppresses both the LB- phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol Microbiol. 1997;26:1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x. [DOI] [PubMed] [Google Scholar]
- 51.Kumamoto CA, Nault AK. Characterization of the Escherichia coli protein-export gene secB. Gene. 1989;75:167–175. doi: 10.1016/0378-1119(89)90393-4. [DOI] [PubMed] [Google Scholar]
- 52.Weiss JB, Ray PH, Bassford PJ., Jr Purified secB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc Natl Acad Sci U S A. 1988;85:8978–8982. doi: 10.1073/pnas.85.23.8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe M, Blobel G. Cytosolic factor purified from Escherichia coli is necessary and sufficient for the export of a preprotein and is a homotetramer of SecB. Proc Natl Acad Sci U S A. 1989;86:2728–2732. doi: 10.1073/pnas.86.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith VF, Schwartz BL, Randall LL, Smith RD. Electrospray mass spectrometric investigation of the chaperone SecB. Protein Sci. 1996;5:488–494. doi: 10.1002/pro.5560050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murén EM, Suciu D, Topping TB, Kumamoto CA, Randall LL. Mutational alterations in the homotetrameric chaperone SecB that implicate the structure as dimer of dimers. J Biol Chem. 1999;274:19397–19402. doi: 10.1074/jbc.274.27.19397. [DOI] [PubMed] [Google Scholar]
- 56.Topping TB, Woodbury RL, Diamond DL, Hardy SJ, Randall LL. Direct demonstration that homotetrameric chaperone SecB undergoes a dynamic dimer-tetramer equilibrium. J Biol Chem. 2001;276:7437–7441. doi: 10.1074/jbc.M009584200. [DOI] [PubMed] [Google Scholar]
- 57.Suo Y, Hardy SJS, Randall LL. The Basis of Asymmetry in the SecA:SecB Complex. Journal of Molecular Biology. 2015;427:887–900. doi: 10.1016/j.jmb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.den Blaauwen T, Terpetschnig E, Lakowicz JR, Driessen AJ. Interaction of SecB with soluble SecA. FEBS Lett. 1997;416:35–38. doi: 10.1016/s0014-5793(97)01142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJ. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol. 2005;348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 60.Patel CN, Smith VF, Randall LL. Characterization of three areas of interactions stabilizing complexes between SecA and SecB, two proteins involved in protein export. Protein Sci. 2006;15:1379–1386. doi: 10.1110/ps.062141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fekkes P, van der Does C, Driessen AJ. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 63.Kumamoto CA, Francetic O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Randall LL, Topping TB, Hardy SJ, Pavlov MY, Freistroffer DV, Ehrenberg M. Binding of SecB to ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc Natl Acad Sci U S A. 1997;94:802–807. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Müller M, Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoffschulte HK, Drees B, Muller M. Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J Biol Chem. 1994;269:12833–12839. [PubMed] [Google Scholar]
- 67.Xu Z, Knafels JD, Yoshino K. Crystal structure of the bacterial protein export chaperone secB. Nat Struct Biol. 2000;7:1172–1177. doi: 10.1038/82040. [DOI] [PubMed] [Google Scholar]
- 68.Dekker C, de Kruijff B, Gros P. Crystal structure of SecB from Escherichia coli. J Struct Biol. 2003;144:313–319. doi: 10.1016/j.jsb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 69.Volkert TL, Baleja JD, Kumamoto CA. A highly mobile C-terminal tail of the Escherichia coli protein export chaperone SecB. Biochem Biophys Res Commun. 1999;264:949–954. doi: 10.1006/bbrc.1999.1590. [DOI] [PubMed] [Google Scholar]
- 70.Crane JM, Suo Y, Lilly AA, Mao C, Hubbell WL, Randall LL. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J Mol Biol. 2006;363:63–74. doi: 10.1016/j.jmb.2006.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diamond DL, Strobel S, Chun SY, Randall LL. Interaction of SecB with intermediates along the folding pathway of maltose-binding protein. Protein Sci. 1995;4:1118–1123. doi: 10.1002/pro.5560040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collier DN, Bankaitis VA, Weiss JB, Bassford PJ., Jr The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe M, Blobel G. High-affinity binding of Escherichia coli SecB to the signal sequence region of a presecretory protein. Proc Natl Acad Sci U S A. 1995;92:10133–10136. doi: 10.1073/pnas.92.22.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Randall LL, Topping TB, Hardy SJ. No specific recognition of leader peptide by SecB, a chaperone involved in protein export. Science. 1990;248:860–863. doi: 10.1126/science.2188362. [DOI] [PubMed] [Google Scholar]
- 75.Gannon PM, Li P, Kumamoto CA. The mature portion of Escherichia coli maltose-binding protein (MBP) determines the dependence of MBP on SecB for export. J Bacteriol. 1989;171:813–818. doi: 10.1128/jb.171.2.813-818.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford PJ, Jr, Kumamoto CA, Wickner W. Three pure chaperone proteins of Escherichia coli–SecB, trigger factor and GroEL–form soluble complexes with precursor proteins in vitro. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altman E, Bankaitis VA, Emr SD. Characterization of a region in mature LamB protein that interacts with a component of the export machinery of Escherichia coli. J Biol Chem. 1990;265:18148–18153. [PubMed] [Google Scholar]
- 78.Weiss JB, Bassford PJ., Jr The folding properties of the Escherichia coli maltose-binding protein influence its interaction with SecB in vitro. J Bacteriol. 1990;172:3023–3029. doi: 10.1128/jb.172.6.3023-3029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Cock H, Overeem W, Tommassen J. Biogenesis of outer membrane protein PhoE of Escherichia coli. Evidence for multiple SecB-binding sites in the mature portion of the PhoE protein. J Mol Biol. 1992;224:369–379. doi: 10.1016/0022-2836(92)91001-6. [DOI] [PubMed] [Google Scholar]
- 80.Randall LL, Topping TB, Suciu D, Hardy SJ. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci. 1998;7:1195–1200. doi: 10.1002/pro.5560070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Randall LL, Hardy SJ, Topping TB, Smith VF, Bruce JE, Smith RD. The interaction between the chaperone SecB and its ligands: evidence for multiple subsites for binding. Protein Sci. 1998;7:2384–2390. doi: 10.1002/pro.5560071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Randall LL. Peptide binding by chaperone SecB: implications for recognition of nonnative structure. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]
- 83.Fekkes P, den Blaauwen T, Driessen AJ. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry. 1995;34:10078–10085. doi: 10.1021/bi00031a032. [DOI] [PubMed] [Google Scholar]
- 84.Randall LL, Hardy SJ. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem Sci. 1995;20:65–69. doi: 10.1016/s0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 85.Kulothungan SR, Das M, Johnson M, Ganesh C, Varadarajan R. Effect of crowding agents, signal peptide, and chaperone SecB on the folding and aggregation of E. coli maltose binding protein. Langmuir. 2009;25:6637–6648. doi: 10.1021/la900198h. [DOI] [PubMed] [Google Scholar]
- 86.Knoblauch NT, Rudiger S, Schonfeld HJ, Driessen AJ, Schneider-Mergener J, Bukau B. Substrate specificity of the SecB chaperone. J Biol Chem. 1999;274:34219–34225. doi: 10.1074/jbc.274.48.34219. [DOI] [PubMed] [Google Scholar]
- 87.Kim J, Kendall DA. Identification of a sequence motif that confers SecB dependence on a SecB-independent secretory protein in vivo. J Bacteriol. 1998;180:1396–1401. doi: 10.1128/jb.180.6.1396-1401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vekshin NL. Protein sizes and stoichiometry in the chaperone SecB--RBPTI complex estimated by ANS fluorescence. Biochemistry (Mosc) 1998;63:485–488. [PubMed] [Google Scholar]
- 89.Smith VF, Hardy SJ, Randall LL. Determination of the binding frame of the chaperone SecB within the physiological ligand oligopeptide-binding protein. Protein Sci. 1997;6:1746–1755. doi: 10.1002/pro.5560060815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lilly AA, Crane JM, Randall LL. Export chaperone SecB uses one surface of interaction for diverse unfolded polypeptide ligands. Protein Sci. 2009;18:1860–1868. doi: 10.1002/pro.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Q, Sun S, Tai P, Sui S-F. Structural Characterization of the Complex of SecB and Metallothionein-Labeled proOmpA by Cryo-Electron Microscopy. PLoS ONE. 2012;7:e47015. doi: 10.1371/journal.pone.0047015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Topping TB, Randall LL. Determination of the binding frame within a physiological ligand for the chaperone SecB. Protein Sci. 1994;3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khisty VJ, Munske GR, Randall LL. Mapping of the binding frame for the chaperone SecB within a natural ligand, galactose-binding protein. J Biol Chem. 1995;270:25920–25927. doi: 10.1074/jbc.270.43.25920. [DOI] [PubMed] [Google Scholar]
- 94.Topping TB, Randall LL. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligands. J Biol Chem. 1997;272:19314–19318. doi: 10.1074/jbc.272.31.19314. [DOI] [PubMed] [Google Scholar]
- 95.Cover WH, Ryan JP, Bassford PJ, Jr, Walsh KA, Bollinger J, Randall LL. Suppression of a signal sequence mutation by an amino acid substitution in the mature portion of the maltose-binding protein. J Bacteriol. 1987;169:1794–1800. doi: 10.1128/jb.169.5.1794-1800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teschke CM, Kim J, Song T, Park S, Park C, Randall LL. Mutations that affect the folding of ribose-binding protein selected as suppressors of a defect in export in Escherichia coli. J Biol Chem. 1991;266:11789–11796. [PubMed] [Google Scholar]
- 97.Khisty VJ, Randall LL. Demonstration in vivo that interaction of maltose-binding protein with SecB is determined by a kinetic partitioning. J Bacteriol. 1995;177:3277–3282. doi: 10.1128/jb.177.11.3277-3282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chun SY, Strobel S, Bassford P, Jr, Randall LL. Folding of maltose-binding protein. Evidence for the identity of the rate-determining step in vivo and in vitro. J Biol Chem. 1993;268:20855–20862. [PubMed] [Google Scholar]
- 99.Krishnan B, Kulothungan SR, Patra AK, Udgaonkar JB, Varadarajan R. SecB-mediated protein export need not occur via kinetic partitioning. J Mol Biol. 2009;385:1243–1256. doi: 10.1016/j.jmb.2008.10.094. [DOI] [PubMed] [Google Scholar]
- 100.Wagaman AS, Coburn A, Brand-Thomas I, Dash B, Jaswal SS. A comprehensive database of verified experimental data on protein folding kinetics. Protein Sci. 2014;23:1808–1812. doi: 10.1002/pro.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cayley S, Lewis BA, Guttman HJ, Record MT., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 102.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar M, Mommer MS, Sourjik V. Mobility of cytoplasmic, membrane, and DNA-binding proteins in Escherichia coli. Biophys J. 2010;98:552–559. doi: 10.1016/j.bpj.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J, Lee Y, Kim C, Park C. Involvement of SecB, a chaperone, in the export of ribose-binding protein. J Bacteriol. 1992;174:5219–5227. doi: 10.1128/jb.174.16.5219-5227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Strobel SM, Cannon JG, Bassford PJ., Jr Regions of maltose-binding protein that influence SecB-dependent and SecA-dependent export in Escherichia coli. J Bacteriol. 1993;175:6988–6995. doi: 10.1128/jb.175.21.6988-6995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. Signal peptides are allosteric activators of the protein translocase. Nature. 2009;462:363–367. doi: 10.1038/nature08559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli. J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- 110.Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280:43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 111.Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural Basis for Signal-Sequence Recognition by the Translocase Motor SecA as Determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Musial-Siwek M, Rusch SL, Kendall DA. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J Mol Biol. 2007;365:637–648. doi: 10.1016/j.jmb.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Francetić O, Kumamoto CA. Escherichia coli SecB stimulates export without maintaining export competence of ribose-binding protein signal sequence mutants. J Bacteriol. 1996;178:5954–5959. doi: 10.1128/jb.178.20.5954-5959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim J, Miller A, Wang L, Muller JP, Kendall DA. Evidence that SecB enhances the activity of SecA. Biochemistry. 2001;40:3674–3680. doi: 10.1021/bi002617z. [DOI] [PubMed] [Google Scholar]
- 115.Baars L, Ytterberg AJ, Drew D, Wagner S, Thilo C, van Wijk KJ, de Gier J-W. Defining the Role of the Escherichia coli Chaperone SecB Using Comparative Proteomics. Journal of Biological Chemistry. 2006;281:10024–10034. doi: 10.1074/jbc.M509929200. [DOI] [PubMed] [Google Scholar]
- 116.Phillips GJ, Silhavy TJ. Heat-shock proteins DnaK and GroEL facilitate export of LacZ hybrid proteins in E. coli. Nature. 1990;344:882–884. doi: 10.1038/344882a0. [DOI] [PubMed] [Google Scholar]
- 117.Ito K, Akiyama Y, Yura T, Shiba K. Diverse effects of the MalE-LacZ hybrid protein on Escherichia coli cell physiology. J Bacteriol. 1986;167:201–204. doi: 10.1128/jb.167.1.201-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wild J, Walter WA, Gross CA, Altman E. Accumulation of secretory protein precursors in Escherichia coli induces the heat shock response. J Bacteriol. 1993;175:3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Altman E, Kumamoto CA, Emr SD. Heat-shock proteins can substitute for SecB function during protein export in Escherichia coli. EMBO J. 1991;10:239–245. doi: 10.1002/j.1460-2075.1991.tb07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wild J, Altman E, Yura T, Gross CA. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 121.Froderberg L, Houben EN, Baars L, Luirink J, de Gier JW. Targeting and translocation of two lipoproteins in Escherichia coli via the SRP/Sec/YidC pathway. J Biol Chem. 2004;279:31026–31032. doi: 10.1074/jbc.M403229200. [DOI] [PubMed] [Google Scholar]
- 122.Bochkareva ES, Lissin NM, Girshovich AS. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature. 1988;336:254–257. doi: 10.1038/336254a0. [DOI] [PubMed] [Google Scholar]
- 123.Laminet AA, Ziegelhoffer T, Georgopoulos C, Pluckthun A. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the beta-lactamase precursor. EMBO J. 1990;9:2315–2319. doi: 10.1002/j.1460-2075.1990.tb07403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Randall LL, Hardy SJ. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stones R. You Can’t Always Get What You Want. Decca Records London Records 1969 [Google Scholar]
- 126.Müller JP. Influence of impaired chaperone or secretion function on SecB production in Escherichia coli. J Bacteriol. 1996;178:6097–6104. doi: 10.1128/jb.178.21.6097-6104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Panse VG, Vogel P, Trommer WE, Varadarajan R. A thermodynamic coupling mechanism for the disaggregation of a model peptide substrate by chaperone secB. J Biol Chem. 2000;275:18698–18703. doi: 10.1074/jbc.275.25.18698. [DOI] [PubMed] [Google Scholar]
- 128.Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:7583–7588. doi: 10.1073/pnas.0402398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gannon PM, Kumamoto CA. Mutations of the molecular chaperone protein SecB which alter the interaction between SecB and maltose-binding protein. J Biol Chem. 1993;268:1590–1595. [PubMed] [Google Scholar]
- 130.Kimsey HH, Dagarag MD, Kumamoto CA. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- 131.Rajapandi T, Oliver D. Carboxy-terminal region of Escherichia coli SecA ATPase is important to promote its protein translocation activity in vivo. Biochem Biophys Res Commun. 1994;200:1477–1483. doi: 10.1006/bbrc.1994.1617. [DOI] [PubMed] [Google Scholar]
- 132.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 133.Fekkes P, de Wit JG, van der Wolk JP, Kimsey HH, Kumamoto CA, Driessen AJ. Preprotein transfer to the Escherichia coli translocase requires the co- operative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 134.Fekkes P, de Wit JG, Boorsma A, Friesen RH, Driessen AJ. Zinc stabilizes the SecB binding site of SecA. Biochemistry. 1999;38:5111–5116. doi: 10.1021/bi982818r. [DOI] [PubMed] [Google Scholar]
- 135.Zhou J, Xu Z. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nat Struct Biol. 2003;10:942–947. doi: 10.1038/nsb980. [DOI] [PubMed] [Google Scholar]
- 136.Matousek WM, Alexandrescu AT. NMR structure of the C-terminal domain of SecA in the free state. Biochim Biophys Acta. 2004;1702:163–171. doi: 10.1016/j.bbapap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 137.Dempsey BR, Wrona M, Moulin JM, Gloor GB, Jalilehvand F, Lajoie G, Shaw GS, Shilton BH. Solution NMR structure and X-ray absorption analysis of the C-terminal zinc-binding domain of the SecA ATPase. Biochemistry. 2004;43:9361–9371. doi: 10.1021/bi0493057. [DOI] [PubMed] [Google Scholar]
- 138.Woodbury RL, Topping TB, Diamond DL, Suciu D, Kumamoto CA, Hardy SJ, Randall LL. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions. J Biol Chem. 2000;275:24191–24198. doi: 10.1074/jbc.M002885200. [DOI] [PubMed] [Google Scholar]
- 139.Randall LL, Crane JM, Liu G, Hardy SJ. Sites of interaction between SecA and the chaperone SecB, two proteins involved in export. Protein Sci. 2004;13:1124–1133. doi: 10.1110/ps.03410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suo Y, Hardy SJS, Randall LL. Orientation of SecA and SecB in complex, derived from disulfide cross-linking. Journal of Bacteriology. 2011;193:190–196. doi: 10.1128/JB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mao C, Hardy SJS, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. Journal of Bacteriology. 2009;191:978–984. doi: 10.1128/JB.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Panse VG, Beena K, Philipp R, Trommer WE, Vogel PD, Varadarajan R. Electron spin resonance and fluorescence studies of the bound-state conformation of a model protein substrate to the chaperone SecB. J Biol Chem. 2001;276:33681–33688. doi: 10.1074/jbc.M104466200. [DOI] [PubMed] [Google Scholar]
- 143.Haimann Michaela M, Akdogan Y, Philipp R, Varadarajan R, Hinderberger D, Trommer Wolfgang E. Conformational changes of the chaperone SecB upon binding to a model substrate – bovine pancreatic trypsin inhibitor (BPTI) 2011;392:849. doi: 10.1515/BC.2011.151. [DOI] [PubMed] [Google Scholar]
- 144.Cabelli RJ, Dolan KM, Qian LP, Oliver DB. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J Biol Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 145.Ulbrandt ND, London E, Oliver DB. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 146.Chen X, Xu H, Tai PC. A significant fraction of functional SecA is permanently embedded in the membrane. SecA cycling on and off the membrane is not essential during protein translocation. J Biol Chem. 1996;271:29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 147.Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 148.Chen X, Brown T, Tai PC. Identification and characterization of protease-resistant SecA fragments: secA has two membrane-integral forms. J Bacteriol. 1998;180:527–537. doi: 10.1128/jb.180.3.527-537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morita K, Tokuda H, Nishiyama K. Multiple SecA molecules drive protein translocation across a single translocon with SecG inversion. Journal of Biological Chemistry. 2012;287:455–464. doi: 10.1074/jbc.M111.301754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wowor AJ, Yu D, Kendall DA, Cole JL. Energetics of SecA dimerization. J Mol Biol. 2011;408:87–98. doi: 10.1016/j.jmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rhoads DB, Waters FB, Epstein W. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J Gen Physiol. 1976;67:325–341. doi: 10.1085/jgp.67.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Castle AM, Macnab RM, Shulman RG. Measurement of intracellular sodium concentration and sodium transport in Escherichia coli by 23Na nuclear magnetic resonance. J Biol Chem. 1986;261:3288–3294. [PubMed] [Google Scholar]
- 154.Castle AM, Macnab RM, Shulman RG. Coupling between the sodium and proton gradients in respiring Escherichia coli cells measured by 23Na and 31P nuclear magnetic resonance. J Biol Chem. 1986;261:7797–7806. [PubMed] [Google Scholar]
- 155.Doyle SM, Braswell EH, Teschke CM. SecA folds via a dimeric intermediate. Biochemistry. 2000;39:11667–11676. doi: 10.1021/bi000299y. [DOI] [PubMed] [Google Scholar]
- 156.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem. 2003;278:3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 157.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 160.Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci U S A. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zimmer J, Li W, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J Mol Biol. 2006;364:259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 162.Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol. 2006;364:248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 163.Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366:1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 164.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci U S A. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 166.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zimmer J, Rapoport TA. Conformational Flexibility and Peptide Interaction of the Translocation ATPase SecA. Journal of Molecular Biology. 2009;394:606–612. doi: 10.1016/j.jmb.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen Y, Bauer BW, Rapoport TA, Gumbart JC. Conformational Changes of the Clamp of the Protein Translocation ATPase SecA. Journal of Molecular Biology. 2015;427:2348–2359. doi: 10.1016/j.jmb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Das S, Grady LM, Michtavy J, Zhou Y, Cohan FM, Hingorani MM, Oliver DB. The variable subdomain of Escherichia coli SecA functions to regulate SecA ATPase activity and ADP release. J Bacteriol. 2012;194:2205–2213. doi: 10.1128/JB.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Auclair SM, Oliver DB, Mukerji I. Defining the Solution State Dimer Structure of Escherichia coli SecA Using Förster Resonance Energy Transfer. Biochemistry. 2013;52:2388–2401. doi: 10.1021/bi301217t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu D, Wowor AJ, Cole JL, Kendall DA. Defining the Escherichia coli SecA dimer interface residues through in vivo site-specific photo-cross-linking. J Bacteriol. 2013;195:2817–2825. doi: 10.1128/JB.02269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 173.van Voorst F, Vereyken IJ, de Kruijff B. The high affinity ATP binding site modulates the SecA-precursor interaction. FEBS Lett. 2000;486:57–62. doi: 10.1016/s0014-5793(00)02209-2. [DOI] [PubMed] [Google Scholar]
- 174.Auclair SM, Moses JP, Musial-Siwek M, Kendall DA, Oliver DB, Mukerji I. Mapping of the signal peptide-binding domain of Escherichia coli SecA using Forster resonance energy transfer. Biochemistry. 2010;49:782–792. doi: 10.1021/bi901446r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang Q, Li Y, Olson R, Mukerji I, Oliver D. Conserved SecA Signal Peptide-Binding Site Revealed by Engineered Protein Chimeras and Forster Resonance Energy Transfer. Biochemistry. 2016 doi: 10.1021/acs.biochem.5b01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci U S A. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 178.Chou YT, Swain JF, Gierasch LM. Functionally significant mobile regions of Escherichia coli SecA ATPase identified by NMR. J Biol Chem. 2002;277:50985–50990. doi: 10.1074/jbc.M209237200. [DOI] [PubMed] [Google Scholar]
- 179.Keramisanou D, Biris N, Gelis I, Sianidis G, Karamanou S, Economou A, Kalodimos CG. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat Struct Mol Biol. 2006;13:594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- 180.Mori H, Ito K. The long alpha-helix of SecA is important for the ATPase coupling of translocation. J Biol Chem. 2006;281:36249–36256. doi: 10.1074/jbc.M606906200. [DOI] [PubMed] [Google Scholar]
- 181.Price A, Economou A, Duong F, Wickner W. Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J Biol Chem. 1996;271:31580–31584. doi: 10.1074/jbc.271.49.31580. [DOI] [PubMed] [Google Scholar]
- 182.Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20:961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 184.Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song XJ, Chou YT, Gierasch LM, Hunt JF. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 186.Kourtz L, Oliver D. Tyr-326 plays a critical role in controlling SecA-preprotein interaction. Mol Microbiol. 2000;37:1342–1356. doi: 10.1046/j.1365-2958.2000.02078.x. [DOI] [PubMed] [Google Scholar]
- 187.Bowler MW, Montgomery MG, Leslie AG, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci U S A. 2006;103:8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 189.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 190.Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proceedings of the National Academy of Sciences. 2010;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 193.Park E, Menetret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. Structure of the SecY channel during initiation of protein translocation. Nature. 2014;506:102–106. doi: 10.1038/nature12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bonardi F, Halza E, Walko M, Du Plessis F, Nouwen N, Feringa BL, Driessen AJM. Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7775–7780. doi: 10.1073/pnas.1101705108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Saparov SM, Erlandson K, Cannon K, Schaletzky J, Schulman S, Rapoport TA, Pohl P. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell. 2007;26:501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 196.Knyazev DG, Winter L, Bauer BW, Siligan C, Pohl P. Ion Conductivity of the Bacterial Translocation Channel SecYEG Engaged in Translocation. The Journal of Biological Chemistry. 2014;289:24611–24616. doi: 10.1074/jbc.M114.588491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nishiyama K, Mizushima S, Tokuda H. The carboxyl-terminal region of SecE interacts with SecY and is functional in the reconstitution of protein translocation activity in Escherichia coli. J Biol Chem. 1992;267:7170–7176. [PubMed] [Google Scholar]
- 199.Bostina M, Mohsin B, Kuhlbrandt W, Collinson I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J Mol Biol. 2005;352:1035–1043. doi: 10.1016/j.jmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 200.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ding H, Mukerji I, Oliver D. Nucleotide and phospholipid-dependent control of PPXD and C-domain association for SecA ATPase. Biochemistry. 2003;42:13468–13475. doi: 10.1021/bi035099b. [DOI] [PubMed] [Google Scholar]
- 202.Gold Vicki AM, Whitehouse S, Robson A, Collinson I. The dynamic action of SecA during the initiation of protein translocation. Biochemical Journal. 2013;449:695–705. doi: 10.1042/BJ20121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gold VA, Robson A, Clarke AR, Collinson I. Allosteric regulation of SecA: magnesium-mediated control of conformation and activity. J Biol Chem. 2007;282:17424–17432. doi: 10.1074/jbc.M702066200. [DOI] [PubMed] [Google Scholar]
- 204.Robson A, Gold VA, Hodson S, Clarke AR, Collinson I. Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc Natl Acad Sci U S A. 2009;106:5111–5116. doi: 10.1073/pnas.0809592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bauer BW, Rapoport TA. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci U S A. 2009;106:20800–20805. doi: 10.1073/pnas.0910550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tanaka Y, Sugano Y, Takemoto M, Mori T, Furukawa A, Kusakizako T, Kumazaki K, Kashima A, Ishitani R, Sugita Y, Nureki O, Tsukazaki T. Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State. Cell Rep. 2015;13:1561–1568. doi: 10.1016/j.celrep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 207.Hizlan D, Robson A, Whitehouse S, Gold Vicki A, Vonck J, Mills D, Kühlbrandt W, Collinson I. Structure of the SecY complex unlocked by a preprotein mimic. Cell Reports. 2012;1:21–28. doi: 10.1016/j.celrep.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 209.Deville K, Gold VAM, Robson A, Whitehouse S, Sessions RB, Baldwin SA, Radford SE, Collinson I. The oligomeric state and arrangement of the active bacterial translocon. Journal of Biological Chemistry. 2011;286:4659–4669. doi: 10.1074/jbc.M110.175638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Li L, Park E, Ling J, Ingram J, Ploegh H, Rapoport TA. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature. 2016 doi: 10.1038/nature17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 213.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hanada M, Nishiyama K-i, Tokuda H. SecG plays a critical role in protein translocation in the absence of the proton motive force as well as at low temperature. FEBS Letters. 1996;381:25–28. doi: 10.1016/0014-5793(96)00066-x. [DOI] [PubMed] [Google Scholar]
- 215.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 216.Eichler J, Wickner W. The SecA subunit of Escherichia coli preprotein translocase is exposed to the periplasm. Journal of Bacteriology. 1998;180:5776–5779. doi: 10.1128/jb.180.21.5776-5779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 218.Nishiyama K, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 219.Nagamori S, Nishiyama K, Tokuda H. Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J Biochem. 2002;132:629–634. doi: 10.1093/oxfordjournals.jbchem.a003266. [DOI] [PubMed] [Google Scholar]
- 220.Kawasaki S, Mizushima S, Tokuda H. Membrane vesicles containing overproduced SecY and SecE exhibit high translocation ATPase activity and countermovement of protons in a SecA- and presecretory protein-dependent manner. J Biol Chem. 1993;268:8193–8198. [PubMed] [Google Scholar]
- 221.Sugai R, Takemae K, Tokuda H, Nishiyama K. Topology inversion of SecG is essential for cytosolic SecA-dependent stimulation of protein translocation. J Biol Chem. 2007;282:29540–29548. doi: 10.1074/jbc.M704716200. [DOI] [PubMed] [Google Scholar]
- 222.van der Sluis EO, van der Vries E, Berrelkamp G, Nouwen N, Driessen AJ. Topologically fixed SecG is fully functional. J Bacteriol. 2006;188:1188–1190. doi: 10.1128/JB.188.3.1188-1190.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Moser M, Nagamori S, Huber M, Tokuda H, Nishiyama K-i. Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9734–9739. doi: 10.1073/pnas.1303160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nishiyama K-i, Maeda M, Yanagisawa K, Nagase R, Komura H, Iwashita T, Yamagaki T, Kusumoto S, Tokuda H, Shimamoto K. MPIase is a glycolipozyme essential for membrane protein integration. Nat Commun. 2012;3:1260. doi: 10.1038/ncomms2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mori H, Sugiyama H, Yamanaka M, Sato K, Tagaya M, Mizushima S. Amino-terminal region of SecA is involved in the function of SecG for protein translocation into Escherichia coli membrane vesicles. J Biochem. 1998;124:122–129. doi: 10.1093/oxfordjournals.jbchem.a022070. [DOI] [PubMed] [Google Scholar]
- 226.Gardel C, Benson S, Hunt J, Michaelis S, Beckwith J. secD, a new gene involved in protein export in Escherichia coli. J Bacteriol. 1987;169:1286–1290. doi: 10.1128/jb.169.3.1286-1290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gardel C, Johnson K, Jacq A, Beckwith J. The secD locus of E.coli codes for two membrane proteins required for protein export. EMBO J. 1990;9:3209–3216. doi: 10.1002/j.1460-2075.1990.tb07519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nouwen N, Driessen AJM. SecDFyajC forms a heterotetrameric complex with YidC. Molecular Microbiology. 2002;44:1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 231.Matsuyama S, Fujita Y, Sagara K, Mizushima S. Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochimica et Biophysica Acta (BBA) 1992;1122:77–84. doi: 10.1016/0167-4838(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 232.Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry. 2006;45:13401–13408. doi: 10.1021/bi060826z. [DOI] [PubMed] [Google Scholar]
- 233.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pogliano KJ, Beckwith J. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol. 1994;176:804–814. doi: 10.1128/jb.176.3.804-814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mio K, Tsukazaki T, Mori H, Kawata M, Moriya T, Sasaki Y, Ishitani R, Ito K, Nureki O, Sato C. Conformational variation of the translocon enhancing chaperone SecDF. Journal of Structural and Functional Genomics. 2014;15:107–115. doi: 10.1007/s10969-013-9168-4. [DOI] [PubMed] [Google Scholar]
- 236.Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48:5801–5812. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 238.Tokuda H, Kim YJ, Mizushima S. In vitro protein translocation into inverted membrane vesicles prepared from Vibrio alginolyticus is stimulated by the electrochemical potential of Na+ in the presence of Escherichia coli SecA. FEBS Lett. 1990;264:10–12. doi: 10.1016/0014-5793(90)80751-4. [DOI] [PubMed] [Google Scholar]
- 239.Ishii E, Chiba S, Hashimoto N, Kojima S, Homma M, Ito K, Akiyama Y, Mori H. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci U S A. 2015;112:E5513–5522. doi: 10.1073/pnas.1513001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Gardel C, Johnson K, Jacq A, Beckwith J. The secD locus of E. coli codes for two membrane proteins required for protein export. EMBO J. 1990;9:4205–4206. doi: 10.1002/j.1460-2075.1990.tb07645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tani K, Shiozuka K, Tokuda H, Mizushima S. In vitro analysis of the process of translocation of OmpA across the Escherichia coli cytoplasmic membrane. A translocation intermediate accumulates transiently in the absence of the proton motive force. Journal of Biological Chemistry. 1989;264:18582–18588. [PubMed] [Google Scholar]
- 242.Tani K, Tokuda H, Mizushima S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J Biol Chem. 1990;265:17341–17347. [PubMed] [Google Scholar]
- 243.Nouwen N, Driessen AJM. Inactivation of Protein Translocation by Cold-Sensitive Mutations in the yajC-secDF Operon. Journal of Bacteriology. 2005;187:6852–6855. doi: 10.1128/JB.187.19.6852-6855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Arkowitz RA, Wickner W. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. Embo J. 1994;13:954–963. doi: 10.1002/j.1460-2075.1994.tb06340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nouwen N, van der Laan M, Driessen AJM. SecDFyajC is not required for the maintenance of the proton motive force. FEBS Letters. 2001;508:103–106. doi: 10.1016/s0014-5793(01)03033-2. [DOI] [PubMed] [Google Scholar]
- 246.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Geller BL. Electrochemical potential releases a membrane-bound secretion intermediate of maltose-binding protein in Escherichia coli. J Bacteriol. 1990;172:4870–4876. doi: 10.1128/jb.172.9.4870-4876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ueguchi C, Ito K. Escherichia coli sec mutants accumulate a processed immature form of maltose-binding protein (MBP), a late-phase intermediate in MBP export. Journal of Bacteriology. 1990;172:5643–5649. doi: 10.1128/jb.172.10.5643-5649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 250.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Saier MH., Jr Protein secretion and membrane insertion systems in gram-negative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 252.Tornroth-Horsefield S, Gourdon P, Horsefield R, Brive L, Yamamoto N, Mori H, Snijder A, Neutze R. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 2007;15:1663–1673. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 253.Emr SD, Hanley-Way S, Silhavy TJ. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 254.Bankaitis VA, Bassford PJ., Jr Proper interaction between at least two components is required for efficient export of proteins to the Escherichia coli cell envelope. J Bacteriol. 1985;161:169–178. doi: 10.1128/jb.161.1.169-178.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Stader J, Gansheroff LJ, Silhavy TJ. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev. 1989;3:1045–1052. doi: 10.1101/gad.3.7.1045. [DOI] [PubMed] [Google Scholar]
- 256.Bost S, Belin D. prl mutations in the Escherichia coli secG gene. J Biol Chem. 1997;272:4087–4093. doi: 10.1074/jbc.272.7.4087. [DOI] [PubMed] [Google Scholar]
- 257.Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Flower AM, Doebele RC, Silhavy TJ. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol. 1994;176:5607–5614. doi: 10.1128/jb.176.18.5607-5614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.van der Wolk JP, Fekkes P, Boorsma A, Huie JL, Silhavy TJ, Driessen AJ. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Derman AI, Puziss JW, Bassford PJ, Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Perez-Perez J, Barbero JL, Marquez G, Gutierrez J. Different PrlA proteins increase the efficiency of periplasmic production of human interleukin-6 in Escherichia coli. J Biotechnol. 1996;49:245–247. doi: 10.1016/0168-1656(96)83990-3. [DOI] [PubMed] [Google Scholar]
- 262.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci U S A. 1996;93:5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Nishiyama K, Fukuda A, Morita K, Tokuda H. Membrane deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. Embo J. 1999;18:1049–1058. doi: 10.1093/emboj/18.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:30862–30868. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 267.Whitehouse S, Gold VA, Robson A, Allen WJ, Sessions RB, Collinson I. Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex. J Cell Biol. 2012;199:919–929. doi: 10.1083/jcb.201205191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Liang F-C, Bageshwar UK, Musser SM. Bacterial Sec Protein Transport Is Rate-limited by Precursor Length: A Single Turnover Study. Molecular Biology of the Cell. 2009;20:4256–4266. doi: 10.1091/mbc.E09-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Liang F-C, Bageshwar UK, Musser SM. Position-dependent Effects of Polylysine on Sec Protein Transport. The Journal of Biological Chemistry. 2012;287:12703–12714. doi: 10.1074/jbc.M111.240903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Driessen AJ, Wickner W. Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc Natl Acad Sci U S A. 1991;88:2471–2475. doi: 10.1073/pnas.88.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Joly JC, Leonard MR, Wickner WT. Subunit Dynamics in Escherichia coli Preprotein Translocase. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4703–4707. doi: 10.1073/pnas.91.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Boy D, Koch H-G. Visualization of Distinct Entities of the SecYEG Translocon during Translocation and Integration of Bacterial Proteins. Molecular Biology of the Cell. 2009;20:1804–1815. doi: 10.1091/mbc.E08-08-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Sanganna Gari RR, Frey NC, Mao C, Randall LL, King GM. Dynamic Structure of the Translocon SecYEG in Membrane: DIRECT SINGLE MOLECULE OBSERVATIONS. The Journal of Biological Chemistry. 2013;288:16848–16854. doi: 10.1074/jbc.M113.471870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Scheuring J, Braun N, Nothdurft L, Stumpf M, Veenendaal AK, Kol S, van der Does C, Driessen AJ, Weinkauf S. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 275.Park E, Rapoport TA. Bacterial protein translocation requires only one copy of the SecY complex in vivo. J Cell Biol. 2012;198:881–893. doi: 10.1083/jcb.201205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zheng Z, Blum A, Banerjee T, Wang Q, Dantis V, Oliver D. Determination of the Oligomeric State of SecYEG Protein Secretion Channel Complex using in vivo Photo-and Disulfide-crosslinking. J Biol Chem. 2016 doi: 10.1074/jbc.M115.694844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Dalal K, Chan CS, Sligar SG, Duong F. Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc Natl Acad Sci U S A. 2012;109:4104–4109. doi: 10.1073/pnas.1117783109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kedrov A, Kusters I, Krasnikov VV, Driessen AJ. A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 2011;30:4387–4397. doi: 10.1038/emboj.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Schulze RJ, Komar J, Botte M, Allen WJ, Whitehouse S, Gold VAM, Lycklama a Nijeholt JA, Huard K, Berger I, Schaffitzel C, Collinson I. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holotranslocon SecYEG–SecDF–YajC–YidC. Proceedings of the National Academy of Sciences. 2014;111:4844–4849. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Mao C, Cheadle CE, Hardy SJS, Lilly AA, Suo Y, Gari RRS, King GM, Randall LL. Stoichiometry of SecYEG in the active translocase of escherichia coli varies with precursor species. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11815–11820. doi: 10.1073/pnas.1303289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Antonoaea R, Fürst M, Nishiyama K-i, Müller M. The Periplasmic Chaperone PpiD Interacts with Secretory Proteins Exiting from the SecYEG Translocon†. Biochemistry. 2008;47:5649–5656. doi: 10.1021/bi800233w. [DOI] [PubMed] [Google Scholar]
- 282.Sachelaru I, Petriman N-A, Kudva R, Koch H-G. Dynamic Interaction of the Sec Translocon with the Chaperone PpiD. Journal of Biological Chemistry. 2014;289:21706–21715. doi: 10.1074/jbc.M114.577916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Maddalo G, Stenberg-Bruzell F, Gotzke H, Toddo S, Bjorkholm P, Eriksson H, Chovanec P, Genevaux P, Lehtio J, Ilag LL, Daley DO. Systematic analysis of native membrane protein complexes in Escherichia coli. J Proteome Res. 2011;10:1848–1859. doi: 10.1021/pr101105c. [DOI] [PubMed] [Google Scholar]
- 284.Gotzke H, Palombo I, Muheim C, Perrody E, Genevaux P, Kudva R, Muller M, Daley DO. YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J Biol Chem. 2014;289:19089–19097. doi: 10.1074/jbc.M113.541672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Seoh HK, Tai PC. Carbon source-dependent synthesis of SecB, a cytosolic chaperone involved in protein translocation across Escherichia coli membranes. J Bacteriol. 1997;179:1077–1081. doi: 10.1128/jb.179.4.1077-1081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ito K, Wittekind M, Nomura M, Shiba K, Yura T, Miura A, Nashimoto H. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell. 1983;32:789–797. doi: 10.1016/0092-8674(83)90065-x. [DOI] [PubMed] [Google Scholar]
- 288.Downing WL, Sullivan SL, Gottesman ME, Dennis PP. Sequence and transcriptional pattern of the essential Escherichia coli secE-nusG operon. J Bacteriol. 1990;172:1621–1627. doi: 10.1128/jb.172.3.1621-1627.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yang C-K, Lu C-D, Tai P. Differential Expression of Secretion Machinery During Bacterial Growth: SecY and SecF Decrease While SecA Increases During Transition from Exponential Phase to Stationary Phase. Current Microbiology. 2013;67:682–687. doi: 10.1007/s00284-013-0421-7. [DOI] [PubMed] [Google Scholar]
- 290.Nakatogawa H, Murakami A, Ito K. Control of SecA and SecM translation by protein secretion. Curr Opin Microbiol. 2004;7:145–150. doi: 10.1016/j.mib.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 291.Oliver DB, Beckwith J. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell. 1982;30:311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- 292.Rollo EE, Oliver DB. Regulation of the Escherichia coli secA gene by protein secretion defects: analysis of secA, secB, secD, and secY mutants. J Bacteriol. 1988;170:3281–3282. doi: 10.1128/jb.170.7.3281-3282.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Schmidt MG, Rollo EE, Grodberg J, Oliver DB. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Oliver D, Norman J, Sarker S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J Bacteriol. 1998;180:5240–5242. doi: 10.1128/jb.180.19.5240-5242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Silber KR, Keiler KC, Sauer RT. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A. 1992;89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Nakatogawa H, Ito K. Secretion Monitor, SecM, Undergoes Self-Translation Arrest in the Cytosol. Molecular Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 298.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 299.Mitra K, Schaffitzel C, Fabiola F, Chapman MS, Ban N, Frank J. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 300.Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Butkus ME, Prundeanu LB, Oliver DB. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sarker S, Oliver D. Critical Regions of secM That Control Its Translation and Secretion and Promote Secretion-Specific secA Regulation. Journal of Bacteriology. 2002;184:2360–2369. doi: 10.1128/JB.184.9.2360-2369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sarker S, Rudd KE, Oliver D. Revised Translation Start Site for secM Defines an Atypical Signal Peptide That Regulates Escherichia coli secA Expression. Journal of Bacteriology. 2000;182:5592–5595. doi: 10.1128/jb.182.19.5592-5595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yap MN, Bernstein HD. The translational regulatory function of SecM requires the precise timing of membrane targeting. Mol Microbiol. 2011;81:540–553. doi: 10.1111/j.1365-2958.2011.07713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Nakatogawa H, Murakami A, Mori H, Ito K. SecM facilitates translocase function of SecA by localizing its biosynthesis. Genes Dev. 2005;19:436–444. doi: 10.1101/gad.1259505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.You Z, Liao M, Zhang H, Yang H, Pan X, Houghton JE, Sui SF, Tai PC. Phospholipids Induce Conformational Changes of SecA to Form Membrane-Specific Domains: AFM Structures and Implication on Protein-Conducting Channels. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0072560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Seoh HK, Tai PC. Catabolic repression of secB expression is positively controlled by cyclic AMP (cAMP) receptor protein-cAMP complexes at the transcriptional level. J Bacteriol. 1999;181:1892–1899. doi: 10.1128/jb.181.6.1892-1899.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Müller JP. Effects of pre-protein overexpression on SecB synthesis in Escherichia coli. FEMS Microbiol Lett. 1999;176:219–227. doi: 10.1111/j.1574-6968.1999.tb13665.x. [DOI] [PubMed] [Google Scholar]
- 309.Kumamoto CA, Chen L, Fandl J, Tai PC. Purification of the Escherichia coli secB gene product and demonstration of its activity in an in vitro protein translocation system. J Biol Chem. 1989;264:2242–2249. [PubMed] [Google Scholar]
- 310.Matsuyama S, Akimaru J, Mizushima S. SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett. 1990;269:96–100. doi: 10.1016/0014-5793(90)81128-b. [DOI] [PubMed] [Google Scholar]
- 311.Sagara K, Matsuyama S, Mizushima S. SecF stabilizes SecD and SecY, components of the protein translocation machinery of the Escherichia coli cytoplasmic membrane. J Bacteriol. 1994;176:4111–4116. doi: 10.1128/jb.176.13.4111-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Taura T, Baba T, Akiyama Y, Ito K. Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J Bacteriol. 1993;175:7771–7775. doi: 10.1128/jb.175.24.7771-7775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci U S A. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kato Y, Nishiyama K-i, Tokuda H. Depletion of SecDF-YajC causes a decrease in the level of SecG: implication for their functional interaction. FEBS Letters. 2003;550:114–118. doi: 10.1016/s0014-5793(03)00847-0. [DOI] [PubMed] [Google Scholar]
- 315.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 316.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 317.Dierstein R, Wickner W. Requirements for substrate recognition by bacterial leader peptidase. EMBO J. 1986;5:427–431. doi: 10.1002/j.1460-2075.1986.tb04228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zwizinski C, Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980;255:7973–7977. [PubMed] [Google Scholar]
- 319.Date T, Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981;78:6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Zimmermann R, Watts C, Wickner W. The biosynthesis of membrane-bound M13 coat protein. Energetics and assembly intermediates. J Biol Chem. 1982;257:6529–6536. [PubMed] [Google Scholar]
- 321.Wolfe PB, Wickner W, Goodman JM. Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope. J Biol Chem. 1983;258:12073–12080. [PubMed] [Google Scholar]
- 322.Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- 323.Wolfe PB, Silver P, Wickner W. The isolation of homogeneous leader peptidase from a strain of Escherichia coli which overproduces the enzyme. J Biol Chem. 1982;257:7898–7902. [PubMed] [Google Scholar]
- 324.Tschantz WR, Sung M, Delgado-Partin VM, Dalbey RE. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 325.Paetzel M. Structure and mechanism of Escherichia coli type I signal peptidase. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843:1497–1508. doi: 10.1016/j.bbamcr.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 326.Sung M, Dalbey RE. Identification of potential active-site residues in the Escherichia coli leader peptidase. J Biol Chem. 1992;267:13154–13159. [PubMed] [Google Scholar]
- 327.Black MT. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J Bacteriol. 1993;175:4957–4961. doi: 10.1128/jb.175.16.4957-4961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Paetzel M, Strynadka NC, Tschantz WR, Casareno R, Bullinger PR, Dalbey RE. Use of site-directed chemical modification to study an essential lysine in Escherichia coli leader peptidase. J Biol Chem. 1997;272:9994–10003. doi: 10.1074/jbc.272.15.9994. [DOI] [PubMed] [Google Scholar]
- 329.Klenotic PA, Carlos JL, Samuelson JC, Schuenemann TA, Tschantz WR, Paetzel M, Strynadka NC, Dalbey RE. The role of the conserved box E residues in the active site of the Escherichia coli type I signal peptidase. J Biol Chem. 2000;275:6490–6498. doi: 10.1074/jbc.275.9.6490. [DOI] [PubMed] [Google Scholar]
- 330.Luke I, Handford JI, Palmer T, Sargent F. Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch Microbiol. 2009;191:919–925. doi: 10.1007/s00203-009-0516-5. [DOI] [PubMed] [Google Scholar]
- 331.Yamagata H, Taguchi N, Daishima K, Mizushima S. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli. Mol Gen Genet. 1983;192:10–14. doi: 10.1007/BF00327640. [DOI] [PubMed] [Google Scholar]
- 332.Tokunaga M, Tokunaga H, Wu HC. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci U S A. 1982;79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 334.Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem. 2004;279:39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- 335.Novak P, Dev IK. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988;170:5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kim AC, Oliver DC, Paetzel M. Crystal structure of a bacterial signal Peptide peptidase. J Mol Biol. 2008;376:352–366. doi: 10.1016/j.jmb.2007.11.080. [DOI] [PubMed] [Google Scholar]
- 337.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc Natl Acad Sci U S A. 2011;108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Inaba K, Suzuki M, Maegawa K, Akiyama S, Ito K, Akiyama Y. A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli. J Biol Chem. 2008;283:35042–35052. doi: 10.1074/jbc.M806603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- 340.Kusters R, Dowhan W, de Kruijff B. Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol-depleted Escherichia coli inner membranes. J Biol Chem. 1991;266:8659–8662. [PubMed] [Google Scholar]
- 341.Dowhan W. A retrospective: use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim Biophys Acta. 2013;1831:471–494. doi: 10.1016/j.bbalip.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Heacock PN, Dowhan W. Alteration of the phospholipid composition of Escherichia coli through genetic manipulation. J Biol Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- 343.Kikuchi S, Shibuya I, Matsumoto K. Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J Bacteriol. 2000;182:371–376. doi: 10.1128/jb.182.2.371-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Breukink E, Demel RA, de Korte-Kool G, de Kruijff B. SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study. Biochemistry. 1992;31:1119–1124. doi: 10.1021/bi00119a021. [DOI] [PubMed] [Google Scholar]
- 345.Hsieh Y-h, Zhang H, Lin B-r, Cui N, Na B, Yang H, Jiang C, Sui S-f, Tai PC. SecA Alone Can Promote Protein Translocation and Ion Channel Activity: SecYEG INCREASES EFFICIENCY AND SIGNAL PEPTIDE SPECIFICITY. Journal of Biological Chemistry. 2011;286:44702–44709. doi: 10.1074/jbc.M111.300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Hsieh Y-h, Zhang H, Wang H, Yang H, Jiang C, Sui S-f, Tai PC. Reconstitution of functionally efficient SecA-dependent protein-conducting channels: Transformation of low-affinity SecA-liposome channels to high-affinity SecA-SecYEG-SecDF·YajC channels. Biochemical and Biophysical Research Communications. 2013;431:388–392. doi: 10.1016/j.bbrc.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 347.Gold VA, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Prabudiansyah I, Kusters I, Caforio A, Driessen AJM. Characterization of the annular lipid shell of the Sec translocon. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2015;1848:2050–2056. doi: 10.1016/j.bbamem.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 349.van den Brink-van der Laan E, Antoinette Killian J, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 350.van der Does C, Swaving J, van Klompenburg W, Driessen AJM. Non-bilayer Lipids Stimulate the Activity of the Reconstituted Bacterial Protein Translocase. Journal of Biological Chemistry. 2000;275:2472–2478. doi: 10.1074/jbc.275.4.2472. [DOI] [PubMed] [Google Scholar]
- 351.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Frauenfeld J, Gumbart J, Sluis EOvd, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome–SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Lycklama a Nijeholt JA, Wu ZC, Driessen AJ. Conformational dynamics of the plug domain of the SecYEG protein-conducting channel. J Biol Chem. 2011;286:43881–43890. doi: 10.1074/jbc.M111.297507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Ahn T, Kim H. Effects of nonlamellar-prone lipids on the ATPase activity of SecA bound to model membranes. J Biol Chem. 1998;273:21692–21698. doi: 10.1074/jbc.273.34.21692. [DOI] [PubMed] [Google Scholar]
- 355.van Klompenburg W, Paetzel M, de Jong JM, Dalbey RE, Demel RA, von Heijne G, de Kruijff B. Phosphatidylethanolamine mediates insertion of the catalytic domain of leader peptidase in membranes. FEBS Lett. 1998;431:75–79. doi: 10.1016/s0014-5793(98)00733-9. [DOI] [PubMed] [Google Scholar]
- 356.van den Brink-van der Laan E, Dalbey RE, Demel RA, Killian JA, de Kruijff B. Effect of nonbilayer lipids on membrane binding and insertion of the catalytic domain of leader peptidase. Biochemistry. 2001;40:9677–9684. doi: 10.1021/bi002903a. [DOI] [PubMed] [Google Scholar]
- 357.Wang Y, Bruckner R, Stein RL. Regulation of signal peptidase by phospholipids in membrane: characterization of phospholipid bilayer incorporated Escherichia coli signal peptidase. Biochemistry. 2004;43:265–270. doi: 10.1021/bi034535r. [DOI] [PubMed] [Google Scholar]
- 358.McKnight CJ, Briggs MS, Gierasch LM. Functional and nonfunctional LamB signal sequences can be distinguished by their biophysical properties. J Biol Chem. 1989;264:17293–17297. [PubMed] [Google Scholar]
- 359.Jones JD, McKnight CJ, Gierasch LM. Biophysical studies of signal peptides: implications for signal sequence functions and the involvement of lipid in protein export. J Bioenerg Biomembr. 1990;22:213–232. doi: 10.1007/BF00763166. [DOI] [PubMed] [Google Scholar]
- 360.Killian JA, Keller RC, Struyve M, de Kroon AI, Tommassen J, de Kruijff B. Tryptophan fluorescence study on the interaction of the signal peptide of the Escherichia coli outer membrane protein PhoE with model membranes. Biochemistry. 1990;29:8131–8137. doi: 10.1021/bi00487a021. [DOI] [PubMed] [Google Scholar]
- 361.Hoyt DW, Gierasch LM. A peptide corresponding to an export-defective mutant OmpA signal sequence with asparagine in the hydrophobic core is unable to insert into model membranes. J Biol Chem. 1991;266:14406–14412. [PubMed] [Google Scholar]
- 362.Hoyt DW, Gierasch LM. Hydrophobic content and lipid interactions of wild-type and mutant OmpA signal peptides correlate with their in vivo function. Biochemistry. 1991;30:10155–10163. doi: 10.1021/bi00106a012. [DOI] [PubMed] [Google Scholar]
- 363.Keller RC, Killian JA, de Kruijff B. Anionic phospholipids are essential for alpha-helix formation of the signal peptide of prePhoE upon interaction with phospholipid vesicles. Biochemistry. 1992;31:1672–1677. doi: 10.1021/bi00121a014. [DOI] [PubMed] [Google Scholar]
- 364.Ahn T, Oh DB, Kim H, Park C. The phase property of membrane phospholipids is affected by the functionality of signal peptides from the Escherichia coli ribose-binding protein. J Biol Chem. 2002;277:26157–26162. doi: 10.1074/jbc.M203445200. [DOI] [PubMed] [Google Scholar]
- 365.Kendall DA, Kaiser ET. A functional decaisoleucine-containing signal sequence. Construction by cassette mutagenesis. J Biol Chem. 1988;263:7261–7265. [PubMed] [Google Scholar]
- 366.Killian JA, de Jong AM, Bijvelt J, Verkleij AJ, de Kruijff B. Induction of non-bilayer lipid structures by functional signal peptides. EMBO J. 1990;9:815–819. doi: 10.1002/j.1460-2075.1990.tb08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Graves R. Collected poems, 1975. Cassell; London: 1975. [Google Scholar]
- 368.Denks K, Vogt A, Sachelaru I, Petriman N-A, Kudva R, Koch H-G. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Molecular Membrane Biology. 2014;31:58–84. doi: 10.3109/09687688.2014.907455. [DOI] [PubMed] [Google Scholar]
- 369.Lycklama A, Nijeholt JA, Driessen AJM. The bacterial Sec-translocase: structure and mechanism. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Chatzi KE, Sardis MF, Economou A, Karamanou S. SecA-mediated targeting and translocation of secretory proteins. Biochim Biophys Acta. 2014;1843:1466–1474. doi: 10.1016/j.bbamcr.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 371.Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol. 2012;10:483–496. doi: 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 372.Patel R, Smith SM, Robinson C. Protein transport by the bacterial Tat pathway. Biochim Biophys Acta. 2014;1843:1620–1628. doi: 10.1016/j.bbamcr.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 373.Saraogi I, Shan SO. Co-translational protein targeting to the bacterial membrane. Biochim Biophys Acta. 2014;1843:1433–1441. doi: 10.1016/j.bbamcr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Zhang X, Shan SO. Fidelity of cotranslational protein targeting by the signal recognition particle. Annu Rev Biophys. 2014;43:381–408. doi: 10.1146/annurev-biophys-051013-022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Dalbey RE, Kuhn A, Zhu L, Kiefer D. The membrane insertase YidC. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843:1489–1496. doi: 10.1016/j.bbamcr.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 376.Hennon SW, Soman R, Zhu L, Dalbey RE. YidC/Alb3/Oxa1 Family of Insertases. J Biol Chem. 2015;290:14866–14874. doi: 10.1074/jbc.R115.638171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Nivaskumar M, Francetic O. Type II secretion system: a magic beanstalk or a protein escalator. Biochim Biophys Acta. 2014;1843:1568–1577. doi: 10.1016/j.bbamcr.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 378.van Ulsen P, Rahman S, Jong WS, Daleke-Schermerhorn MH, Luirink J. Type V secretion: from biogenesis to biotechnology. Biochim Biophys Acta. 2014;1843:1592–1611. doi: 10.1016/j.bbamcr.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 379.Ricci DP, Silhavy TJ. The Bam machine: A molecular cooper. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 381.Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Thomas S, Holland IB, Schmitt L. The Type 1 secretion pathway - the hemolysin system and beyond. Biochim Biophys Acta. 2014;1843:1629–1641. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 383.Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta. 2014;1843:1664–1673. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 385.Burkinshaw BJ, Strynadka NC. Assembly and structure of the T3SS. Biochim Biophys Acta. 2014;1843:1649–1663. doi: 10.1016/j.bbamcr.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 386.Oliver DB, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 387.Kawasaki H, Matsuyama S, Sasaki S, Akita M, Mizushima S. SecA protein is directly involved in protein secretion in Escherichia coli. FEBS Lett. 1989;242:431–434. doi: 10.1016/0014-5793(89)80516-2. [DOI] [PubMed] [Google Scholar]
- 388.Sullivan NF, Donachie WD. Transcriptional organization within an Escherichia coli cell division gene cluster: direction of transcription of the cell separation gene envA. J Bacteriol. 1984;160:724–732. doi: 10.1128/jb.160.2.724-732.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Garza-Sanchez F, Janssen BD, Hayes CS. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ito K. Identification of the secY (prlA) gene product involved in protein export in Escherichia coli. Mol Gen Genet. 1984;197:204–208. doi: 10.1007/BF00330964. [DOI] [PubMed] [Google Scholar]
- 391.Shiba K, Ito K, Yura T, Cerretti DP. A defined mutation in the protein export gene within the spc ribosomal protein operon of Escherichia coli: isolation and characterization of a new temperature-sensitive secY mutant. EMBO J. 1984;3:631–635. doi: 10.1002/j.1460-2075.1984.tb01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Riggs PD, Derman AI, Beckwith J. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Hanada M, Nishiyama KI, Mizushima S, Tokuda H. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12) J Biol Chem. 1994;269:23625–23631. [PubMed] [Google Scholar]
- 394.Fang J, Wei Y. Expression, Purification and Characterization of the Escherichia coli Integral Membrane Protein YajC. Protein and Peptide Letters. 2011;18:601–608. doi: 10.2174/092986611795222713. [DOI] [PubMed] [Google Scholar]
- 395.Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 396.Martinez Molina D, Lundback AK, Niegowski D, Eshaghi S. Expression and purification of the recombinant membrane protein YidC: a case study for increased stability and solubility. Protein Expr Purif. 2008;62:49–52. doi: 10.1016/j.pep.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 397.Kim DM, Zheng H, Huang YJ, Montelione GT, Hunt JF. ATPase active-site electrostatic interactions control the global conformation of the 100 kDa SecA translocase. J Am Chem Soc. 2013;135:2999–3010. doi: 10.1021/ja306361q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Nithianantham S, Shilton BH. Analysis of the isolated SecA DEAD motor suggests a mechanism for chemical-mechanical coupling. J Mol Biol. 2008;383:380–389. doi: 10.1016/j.jmb.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 399.Swanson S, Ioerger TR, Rigel NW, Miller BK, Braunstein M, Sacchettini JC. Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2. J Bacteriol. 2015;198:720–730. doi: 10.1128/JB.00696-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Menetret JF, Schaletzky J, Clemons WM, Jr, Osborne AR, Skanland SS, Denison C, Gygi SP, Kirkpatrick DS, Park E, Ludtke SJ, Rapoport TA, Akey CW. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 402.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Wickles S, Singharoy A, Andreani J, Seemayer S, Bischoff L, Berninghausen O, Soeding J, Schulten K, van der Sluis EO, Beckmann R. A structural model of the active ribosome-bound membrane protein insertase YidC. Elife. 2014;3:e03035. doi: 10.7554/eLife.03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Kumazaki K, Kishimoto T, Furukawa A, Mori H, Tanaka Y, Dohmae N, Ishitani R, Tsukazaki T, Nureki O. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep. 2014;4:7299. doi: 10.1038/srep07299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Oliver DC, Paetzel M. Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J Biol Chem. 2008;283:5208–5216. doi: 10.1074/jbc.M708936200. [DOI] [PubMed] [Google Scholar]
- 406.Ravaud S, Stjepanovic G, Wild K, Sinning I. The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J Biol Chem. 2008;283:9350–9358. doi: 10.1074/jbc.M710493200. [DOI] [PubMed] [Google Scholar]
- 407.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 408.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser-Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 409.Paetzel M, Goodall JJ, Kania M, Dalbey RE, Page MG. Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based inhibitor. J Biol Chem. 2004;279:30781–30790. doi: 10.1074/jbc.M401686200. [DOI] [PubMed] [Google Scholar]
- 410.Luo C, Roussel P, Dreier J, Page MG, Paetzel M. Crystallographic analysis of bacterial signal peptidase in ternary complex with arylomycin A2 and a beta-sultam inhibitor. Biochemistry. 2009;48:8976–8984. doi: 10.1021/bi9009538. [DOI] [PubMed] [Google Scholar]
- 411.Liu J, Luo C, Smith PA, Chin JK, Page MG, Paetzel M, Romesberg FE. Synthesis and characterization of the arylomycin lipoglycopeptide antibiotics and the crystallographic analysis of their complex with signal peptidase. J Am Chem Soc. 2011;133:17869–17877. doi: 10.1021/ja207318n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Matsuyama S, Mizushima S. Biochemical Analyses of Components Comprising the Protein Translocation Machinery of Escherichia coli. In: Tartakoff AM, Dalbey RE, editors. Advances in Cell and Molecular Biology of Membranes and Organelles: Protein Export and Membrane Biogenesis. Vol. 4. JAI Press Ltd.; Hampton Hill, Middlesex, England: 1995. pp. 61–84. [Google Scholar]
- 413.Urbanus ML, Fröderberg L, Drew D, Björk P, de Gier J-WL, Brunner J, Oudega B, Luirink J. Targeting, Insertion, and Localization of Escherichia coli YidC. Journal of Biological Chemistry. 2002;277:12718–12723. doi: 10.1074/jbc.M200311200. [DOI] [PubMed] [Google Scholar]
- 414.van Klompenburg W, Whitley P, Diemel R, von Heijne G, de Kruijff B. A quantitative assay to determine the amount of signal peptidase I in E. coli and the orientation of membrane vesicles. Mol Membr Biol. 1995;12:349–353. doi: 10.3109/09687689509072437. [DOI] [PubMed] [Google Scholar]
- 415.Milo R, Phillips R. Cell biology by the numbers. Garland Science; New York, NY: 2016. [Google Scholar]
- 416.Kusters R, de Vrije T, Breukink E, de Kruijff B. SecB protein stabilizes a translocation-competent state of purified prePhoE protein. J Biol Chem. 1989;264:20827–20830. [PubMed] [Google Scholar]
- 417.Powers EL, Randall LL. Export of periplasmic galactose-binding protein in Escherichia coli depends on the chaperone SecB. J Bacteriol. 1995;177:1906–1907. doi: 10.1128/jb.177.7.1906-1907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


