Abstract
O-Linked β-N-acetylglucosamine (O-GlcNAc) is a critical post-translational modification (PTM) of thousands of intracellular proteins. Reversible O-GlcNAcylation governs many aspects of cell physiology and is dysregulated in numerous human diseases. Despite this broad pathophysiological significance, major aspects of O-GlcNAc signaling remain poorly understood, including the biochemical mechanisms through which O-GlcNAc transduces information. Recent work from many laboratories, including our own, has revealed that O-GlcNAc, like other intracellular PTMs, can control its substrates’ functions by inhibiting or inducing protein–protein interactions. This dynamic regulation of multiprotein complexes exerts diverse downstream signaling effects in a range of processes, cell types, and organisms. Here, we review the literature about O-GlcNAc-regulated protein–protein interactions and suggest important questions for future studies in the field.
Graphical abstract
O-Linked β-N-acetylglucosamine (O-GlcNAc) is an abundant intracellular post-translational modification (PTM), reversibly decorating serine and threonine side chains of thousands of nuclear, cytoplasmic, and mitochondrial proteins in animals, plants, and perhaps some groups of fungi (Figure 1).1–5 In many ways, O-GlcNAcylation is analogous to phosphorylation. In both cases, dedicated enzymes respond to physiological cues by adding or removing a small covalent moiety to control target proteins’ functions. O-GlcNAc is added by O-GlcNAc transferase (OGT) and removed by O-GlcNAcase (OGA) (Figure 1).1–3 O-GlcNAc cycling controls myriad processes, including cell metabolism, cell cycle progression, and cell death,1,2 and is essential, as genetic ablation of OGT or OGA is lethal in mice.6–8 UDP-GlcNAc, the nucleotide-sugar cofactor used by OGT (Figure 1), is created through the hexosamine biosynthetic pathway (HBP) from other essential metabolites, including glucose, glutamine, acetyl-coenzyme A, uridine, and ATP.3,9,10 Because of this, O-GlcNAc serves in part as a sentinel for cell metabolism, linking nutrient status to signaling.1–3,9,11,12 In addition, aberrant O-GlcNAc cycling is implicated in numerous human diseases, including cancer,2,13–15 diabetes,16–18 cardiac dysfunction,19–22 and neurodegeneration.23–26
Figure 1.
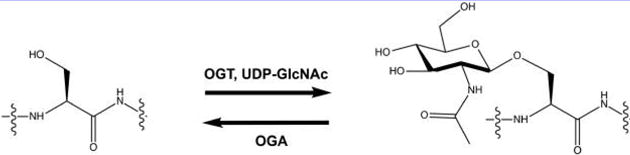
O-Linked β-N-acetylglucosamine (O-GlcNAc) is added to serine and threonine side chains of intracellular proteins by the O-GlcNAc transferase (OGT) using the nucleotide-sugar donor UDP-GlcNAc and is removed by O-GlcNAcase (OGA).
Despite its importance, key aspects of O-GlcNAc signaling remain poorly understood, such as the functional effects it exerts on most substrates.1,2,9 Like phosphorylation and other intracellular PTMs, O-GlcNAc can cause a wide range of biochemical changes to its targets, including activation, inhibition, conformational changes, relocalization, or destruction.1,2,6,7,9,12 However, the biochemical consequences of the vast majority of O-GlcNAcylation events remain unknown. More work is needed to understand the complex mechanistic impacts that O-GlcNAcylation has on its thousands of substrates. Recently, we and others have approached this larger problem by studying the specific question of how O-GlcNAc governs protein–protein interactions. As described below, O-GlcNAc can inhibit or induce functionally important protein–protein interactions on a range of substrates, and it is likely that many more instances of this mode of regulation remain to be discovered.
PROTEIN–PROTEIN INTERACTIONS INHIBITED BY O-GLCNAC
Gene expression is controlled in part by the regulated assembly of multiprotein complexes on DNA to modulate its epigenetic state and to control transcription initiation. Because many chromatin and transcriptional regulatory proteins are reversibly O-GlcNAcylated,1–3 it is perhaps not surprising that the disruption of protein–protein interactions by glycosylation can influence gene expression (Figure 2). (Because O-GlcNAc is the sole known form of intracellular glycosylation in most eukaryotes, we will use “O-GlcNAcylation” and “glycosylation” interchangeably in this Perspective). An early example of this mode of regulation was reported by the Kudlow lab, who demonstrated that glycosylation of transcription factor Sp1 blocks its interaction with the TATA-binding protein-associated factor TAF110 in both Drosophila and human cell systems, thereby inhibiting Sp1-mediated transcription.27,28 Extending this observation, Lim and Chang29 subsequently showed that Sp1 O-GlcNAcylation also prevents its interaction with transcription factor NF-Y, inhibiting the expression of their cooperatively regulated target genes, including cytochrome P450 family members.
Figure 2.
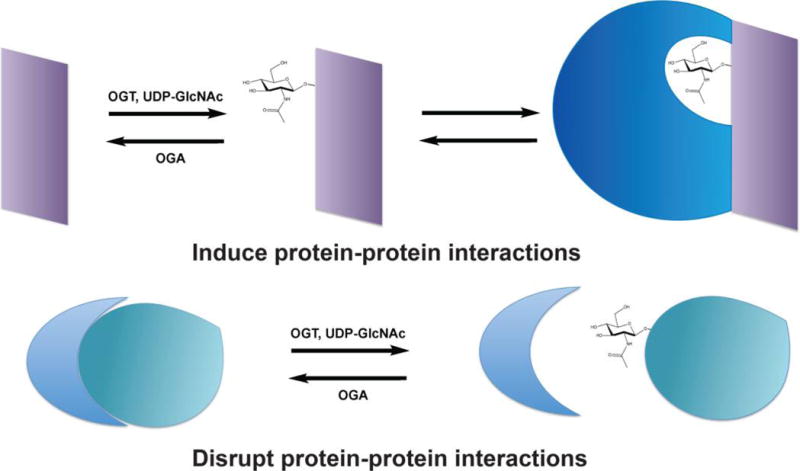
O-GlcNAc modifications can induce or inhibit protein–protein interactions on a wide variety of substrates, in myriad biological processes, and in a diverse range of organisms.
Remarkably, dysregulated glycosylation can also disrupt normal protein–protein interactions in transcriptional signaling. For example, Latorre et al. recently showed that aberrant O-GlcNAcylation abrogates the interactions of transcription factor PGC1α, a key regulator of metabolic function and mitochondrial biogenesis.30 A common polymorphism in pigs results in a Cys → Ser mutation in PGC1α, creating a new glycosylation site at residue 430.30 Ser430-O-GlcNAc stabilizes PGC1α but prevents its interaction with its binding partner, PPARγ.30 This disruption hampers the expression of gluconeogenic genes that are normally upregulated by the PGC1α–PPARγ complex, potentially having an impact on downstream intramuscular fat content and muscle pH.30 Whether analogous polymorphisms or mutations cause harmful de novo glycosylation of human PGC1α or other OGT substrates is not yet known.
O-GlcNAc also governs important protein–protein interactions among transcription factors in embryonic stem cell (ESC) differentiation. Myers et al.31 recently reported that SOX2, a transcription factor known to regulate pluripotency in ESCs, is O-GlcNAcylated on Ser248 in its transactivation domain and is deglycosylated in response to differentiation stimuli. The authors showed that an unglycosylatable Ser248Ala SOX2 mutant can replace wild type SOX2 in mouse ESCs but reduces their reprogramming efficiency in response to differentiation signals, indicating that SOX2 O-GlcNAcylation promotes pluripotency.31 Ser248 O-GlcNAcylation influences the genome occupancy and gene expression signatures of SOX2, an effect that may be due in part to disrupted protein–protein interactions.31 In particular, the authors show the SOX2 glycosylation reduces its interaction with poly-ADP ribose polymerase 1 (PARP1) both in vitro and in vivo, providing a possible mechanistic explanation for at least some of these transcriptional effects.31 However, proteomics experiments demonstrated that numerous proteins co-purified differently with wild type versus Ser248Ala SOX2, with the mutation reducing some associations and strengthening others, beyond PARP1.31 More work will be required to fully dissect the functional implications of O-GlcNAcylation on SOX2 and other pluripotency factors in ESCs.
O-GlcNAcylation can also inhibit interactions among transcription factors and their upstream regulators. The Montminy lab discovered one such example during their investigation of hyperglycemia-induced hepatic gluconeogenesis, a hallmark of uncontrolled diabetes.32 They showed that hyperglycemic conditions cause the accumulation and nuclear localization of CREB-regulated transcription coactivator 2 (CRTC2, also called TORC2), leading to the induction of gluconeogenic genes.32 In its inactive state, CRTC2 is phosphorylated by AMP-activated protein kinases (AMPK) and is sequestered in the cytoplasm by subsequent binding to 14-3-3 proteins.32 The authors found that hyperglycemia triggered the O-GlcNAcylation of two key AMPK sites, Ser70 and Ser171.32 Because glycosylation and phosphorylation are mutually exclusive on the same residue, increasing the level of O-GlcNAcylation at Ser70 and Ser171 prevents their phosphorylation, breaking the interaction with 14-3-3 and allowing CRTC2 to translocate to the nucleus and activate transcription.32 Importantly, the level of CRTC2 O-GlcNAcylation was increased in two different mouse models of diabetes, as compared to control animals, suggesting that hyperglycemia-induced glycosylation might be a pathological feature of human diabetes, as well.32
In another similar example, the Cho lab demonstrated that O-GlcNAcylation of NFκB family transcription factor p65 reduces its interaction with the inhibitory protein IκBα, which masks p65’s nuclear localization signal, keeping it sequestered in the cytoplasm and therefore inactive.33 Interestingly, hyperglycemic conditions induce p65 glycosylation at multiple sites, with Thr352 being the most important for relieving IκBα inhibition and boosting NFκB transcriptional activation.33 These results demonstrate that glucose availability can influence NFκB signaling through direct p65 O-GlcNAcylation.33
The Cho group has also reported a similar mode of regulation of tumor suppressor protein p53.34 In response to genotoxic stress, such as DNA-damaging drugs, p53 is O-GlcNAcylated on Ser149, stabilizing it and allowing it to perform its cell cycle arrest and apoptotic functions.34 The authors demonstrated that Ser149-O-GlcNAc disrupts the interaction of p53 with MDM2, which mediates p53 ubiquitination and destruction.34 Indeed, the authors showed that pharmacologically potentiating global O-GlcNAc inhibited the p53–MDM2 interaction and lowered the level of p53 ubiquitination for the wild type protein, but not for an unglycosylatable Ser149Ala mutant p53.34 In future studies, it will be important to determine the extent of crosstalk, if any, between O-GlcNAcylation and the numerous other PTMs of p53 in DNA damage signaling.
O-GlcNAc-inhibited protein–protein interactions control transcriptional signaling beyond the animal kingdom, as well. For example, the Sun lab recently demonstrated that DELLA family protein REPRESSOR OF ga1-3 (RGA) is O-GlcNAcylated in Arabidopsis by one of its two OGT homologues, SECRET AGENT (SEC).35 RGA and other DELLA proteins are transcriptional regulators, inhibiting signaling by the hormone gibberellin, among other pathways, by binding and antagonizing transcription factors. The authors showed that RGA glycosylation blocks its binding to at least four different transcription factor-binding partners that function in light sensing, jasmonate, and brassinosteroid signaling pathways.35 These biochemical results were bolstered by genetic evidence showing that a hypomorphic sec mutant is less responsive to gibberellin but could be partially rescued by null mutations in two DELLA-encoding genes.35 Therefore, SEC-mediated O-GlcNAcylation of DELLA proteins relieves repression in several important developmental signaling pathways in plants.
Taken together, the examples mentioned above illustrate how dynamic O-GlcNAcylation can disrupt functionally important interactions among chromatin proteins, transcription factors, and their upstream regulators in a wide variety of organisms. Often, this mode of regulation serves in part as a nutrient-sensitive form of cell signaling, connecting metabolic and gene expression pathways. In mammalian cells, O-GlcNAc is most abundant in the nucleus, perhaps accounting for the large proportion of glycosylation-inhibited interactions described among nuclear proteins. In future studies, it will be interesting to determine whether similar modes of regulation are as widespread among OGT substrates in the cytoplasm, mitochondria, and other organelles.
PROTEIN–PROTEIN INTERACTIONS INDUCED BY O-GLCNAC
Like other intracellular PTMs, O-GlcNAc glycosylation can induce, as well as inhibit, protein–protein interactions (Figure 2). Again, this form of regulation has been best described in the context of transcriptional signaling. One early example focused on STAT5, a transcription factor and OGT substrate that responds to receptor tyrosine kinase signaling to influence gene expression programs regulating cell proliferation, apoptosis, and inflammation. Gewinner et al.36 demonstrated that the transcriptional coactivator cyclic AMP response element-binding protein (CREB)-binding protein (CBP) binds to O-GlcNAcylated, but not unmodified, STAT5 after cytokine stimulation of epithelial cells. STAT5 glycosylation does not alter its signal-induced nuclear import but instead potentiates the transactivation of some (but not all) STAT5 target genes, because of O-GlcNAc-induced CBP binding.36 Importantly, the authors also showed that the STAT5 homologues STAT1, STAT3, and STAT6 are similarly glycosylated.36 Because more than 40 mitogens and cytokines signal through the STAT family,37 O-GlcNAcylation, like phosphorylation, may be a broad and conserved mode of regulating STAT-driven gene expression in a wide variety of biological contexts.
Several other groups have characterized additional functionally important O-GlcNAc-induced protein–protein interactions during transcriptional control. For example, the Yang lab reported that OGT is recruited to and glycosylates wild type PGC1α, a transcription factor described above.38 O-GlcNAcylation of PGC1α induces the binding of the deubiquitinating enzyme BRCA1-associated protein 1 (BAP1), which removes ubiquitin marks from PGC1α, stabilizing it and promoting downstream transcriptional activation of gluconeogenic genes.38 Furthermore, the authors found that high-glucose conditions potentiated PGC1α glycosylation, suggesting that this O-GlcNAc-induced PGC1α–BAP1 complex may provide a functional explanation for the aberrant induction of hepatic gluconeogenesis in hyperglycemic patients.38 Indeed, the authors showed that knockdown of either OGT or host cell factor 1 (HCF1, which promotes recruitment of OGT to PGC1α) in the liver ameliorated glucose homeostasis in a diabetic mouse model, suggesting that the HCF1/OGT/PGC1α/BAP1 axis could be a target for therapeutic intervention in diabetes.38
In another example of transcriptional control through O-GlcNAc-mediated protein–protein interactions, the Hart lab discovered that the retinoblastoma proteins (pRB), a family of tumor suppressive transcription factors, are glycosylated in a cell cycle phase-dependent manner.39 The authors showed that the level of pRB O-GlcNAcylation is high in G1, coinciding with both its activation and its hypophosphorylation.39 Moreover, glycosylation of pRB was correlated with its binding to and inhibition of E2F-1, a transcription factor that controls S phase entry.39 While the inhibitory relationship between pRB and E2F-1 during G1 was already well-known, the authors’ work revealed a potential new layer to this regulation, as O-GlcNAcylation of pRB proteins themselves may directly promote E2F-1 binding and inhibition.39
Transcriptional control through O-GlcNAc-induced protein–protein interactions is not limited to mammals, or even animals. For example, Xiao et al.40 discovered that such an interaction in wheat regulates transcription-dependent vernalization, the response to prolonged cold that occurs in many plants during winter months. It had been reported previously that vernalization induces transcriptional activator TaVRN1 in monocots, but the mechanistic explanation for this observation remains unclear.41–43 Xiao et al. found that vernalization induces the O-GlcNAcylation of TaGRP2, an RNA-binding protein that associates directly with TaVRN1 pre-mRNA, thereby inhibiting TaVRN1 expression.40 This glycosylation event recruits jacalin-like lectin VER2 to bind TaGRP2, reducing its levels in the nucleus and prompting its dissociation from the TaVRN1 pre-mRNA, promoting TaVRN1 translation.40 Therefore, the O-GlcNAc-mediated VER2–TaGRP2 interaction de-represses TaVRN1 expression, allowing it to accumulate and drive the vernalization response.40 It will be interesting to see whether future studies reveal the upstream mechanism through which seasonal environmental and/or metabolic cues induce TaGRP2 O-GlcNAcylation to initiate vernalization.
O-GlcNAc-induced interactions have also emerged as an important mode of regulating chromatin. For example, Fujiki et al.44 reported that the O-GlcNAcylation of histone H2B at Ser112 recruits ubiquitin ligase Bre1A, which in turn induces the monoubiquitination of H2B Lys120, an important gene-activating PTM. The authors used chromatin immunoprecipitation and genomewide profiling approaches to show that H2B glycosylation peaks near the transcription start site of many genes, especially those encoding metabolic proteins.44 Histone glycosylation fluctuates in response to extracellular glucose availability and HBP flux.1–3 On this basis, the authors proposed that the O-GlcNAc-dependent recruitment of Bre1A to histone H2B may be an important, nutrient-sensitive chromatin modification for adjusting the global transcription of metabolic genes.44
Though this hypothesis remains to be thoroughly tested, subsequent studies by the authors and other groups have further elucidated the mechanism and functional impact of histone H2B O-GlcNAcylation. For example, Chen et al.45 showed that the enzyme Tet methylcytosine dioxygenase 2 (TET2) binds to OGT and recruits it to specific chromatin sites, increasing the level of local histone H2B Ser112 glycosylation and triggering transcriptional changes. These results implicate the TET family of dioxygenases, which are well-known OGT interactors, as potentially important upstream regulators of O-GlcNAc-mediated histone–protein interactions. In complementary work, Wang et al.46 recently discovered that histone H2B Ser112 glycosylation is triggered by double-strand DNA breaks and is required for homologous recombination and nonhomologous end joining, both major DNA repair pathways. Moreover, the authors showed that H2B Ser112-O-GlcNAc recruits the Nijmegen breakage syndrome 1 (NBS1) DNA repair protein, providing a potential mechanistic link between OGT signaling and the repair process.46 Future work will likely determine exactly how Bre1A and NBS1 bind H2B Ser112-O-GlcNAc (directly vs indirectly, through similar vs distinct biophysical contacts, etc.) and characterize the relationship, if any, between glucose availability and OGT/NBS1-dependent DNA repair processes. However, despite these results, it is important to note that the glycosylation of mammalian histones has been disputed,47 perhaps because of subtle discrepancies across experimental systems or analytical techniques. Clearly, more work is needed to resolve the apparent contradiction among these studies.
Because O-GlcNAc is most abundant in the nucleus, it likely governs many interactions among nuclear proteins beyond chromatin and transcription factors. For example, it has long been known that the nuclear pore complex is extensively decorated with O-GlcNAc moieties on multiple nucleoporins (Nups) and that glycosylation is required for the nuclear import and permeability barrier functions of the pore.48–53 The mechanistic underpinnings of these observations are not completely understood, but O-GlcNAc may contribute to both the bulk material properties of the pore and specific biochemical interactions among Nups. In support of the former, the Görlich group showed that nuclear pore complexes reconstituted from Xenopus egg extracts, containing native O-GlcNAcylation, recapitulate key permeability barrier features of the intact pore.54 O-GlcNAc is required for this property, because biochemical depletion of glycosylated Nups from the extracts compromised the active transport and passive barrier properties of the resulting pore complexes.54 Interestingly, the authors found that the heavily glycosylated FG domain of Nup98 forms a hydrogel with nuclear pore-like permeability properties in vitro, suggesting that O-GlcNAc may contribute to these effects in part through its bulk biophysical properties.54,55
Mizuguchi-Hata et al.56 subsequently provided evidence of a role for specific O-GlcNAc-mediated interactions in pore architecture, reporting that the Nup62–Nup88 subcomplex in particular is regulated by O-GlcNAcylation. The authors showed that, in mammalian cells, Nup88 preferentially binds to glycosylated (vs unmodified) Nup62, and Nup88 protein levels are reduced by siRNAs directed against either Nup62 or OGT.56 These results indicate that Nup62 glycosylation is required to bind and stabilize Nup88. Indeed, more recent work from the Vocadlo group suggests that O-GlcNAc-mediated interactions among Nups may indeed be required for overall nuclear pore architecture.57 The authors showed that pharmacological inhibition or genetic deletion of OGT eliminated Nup glycosylation, enhanced Nup ubiquitination, and shortened Nup protein half-lives.57 As a result, the nuclear pore selective permeability barrier, which gates nuclear–cytoplasmic trafficking, collapsed in both mitotic and non-dividing cells.57 Taken together, these results suggest that O-GlcNAc-mediated protein–protein interactions among Nups are required for their stability, and for pore structure and function, at least in vertebrate cells.
O-GlcNAc-mediated protein–protein interactions also have signaling roles outside the nucleus. For example, Ha et al.58 demonstrated that glycosylation of Ser23 on β-catenin, a bifunctional cell adhesion and transcriptional regulatory protein, promotes its recruitment to the plasma membrane and its binding to E-cadherin. Membrane sequestration is a well-known mechanism of inhibiting the transcriptional activity of β-catenin and activating its cell adhesion function, and the authors showed that pharmacological potentiation of O-GlcNAc levels reduced the level of expression of a β-catenin reporter construct and inhibited the anchorage-independent growth of a human prostate cancer cell line.58 Moreover, Ser23 glycosylation is likely required for the transcription and cell proliferation effects of increased global O-GlcNAc levels in this system, because a Ser23Gly β-catenin mutant was unable to effect these responses.58 Interestingly, O-GlcNAcylation may be a more general mode of regulating β-catenin interactions, because the Lefebvre group demonstrated that glycosylation of β-catenin stabilizes it in human colon cell lines and promotes its interaction with α-catenin, forming an adherens junction subcomplex required for the integrity of mucosa.59 Whether β-catenin glycosylation directly promotes E-cadherin and/or α-catenin binding and how this is accomplished remains to be determined.58,59
Our lab has also characterized O-GlcNAc-mediated protein–protein interactions in a variety of cell biological contexts. For example, we recently discovered an unexpected connection between redox stress signaling and glycosylation-dependent protein complexes.60 In a genomewide expression experiment, we found that pharmacological inhibition of OGT triggered the concerted induction of many targets of NRF2, a transcription factor and master regulator of oxidative stress signaling in mammals.60 Guided by this observation, we determined that OGT inhibition stabilized NRF2 by reducing its level of ubiquitination, a well-known mode of NRF2 pathway regulation.60 O-GlcNAcylation of particular proteasome substrates has long been known to inhibit their degradation.61–65 Surprisingly, however, we discovered through a variety of chemical biology, biochemical, and cellular approaches that O-GlcNAcylation is required for the optimal activity of the ubiquitin E3 ligase complex that targets NRF2 for destruction under unstressed conditions.60 We showed that OGT directly modifies Ser104 of KEAP1, the adaptor protein in this complex, and that Ser104-O-GlcNAc is required for both optimal binding of KEAP1 to E3 ligase CUL3 and for NRF2 ubiquitination by the KEAP1–CUL3 complex.60 In addition, KEAP1 glycosylation co-varies with extracellular glucose levels, revealing an unanticipated connection between nutrient sensing by OGT and redox stress signaling.60 Under nutrient-replete conditions, KEAP1 is glycosylated and the O-GlcNAc-mediated KEAP1–CUL3 complex effects NRF2 ubiquitination and degradation.60 Conversely, during glucose starvation, KEAP1 is deglycosylated, hindering NRF2 ubiquitination and allowing its accumulation and transcriptional activity.60 Therefore, KEAP1 glycosylation may provide a general mechanism for activating the NRF2 stress response pathway in the face of limiting nutrients. These observations may also have implications beyond the KEAP1–NRF2 pathway. KEAP1 belongs to the large family of Kelch-like proteins (KLHL), many of which serve as regulators of the ubiquitin–proteasome pathway. Interestingly, the Ser104 glycosylation site is conserved both across KLHL orthologs in other organisms and among 37 of the 42 human KLHL proteins.60 This observation suggests that O-GlcNAc-mediated protein–protein interactions may be a broadly conserved mode of regulating the KLHL family, connecting nutrient sensing by OGT with cellular proteostasis, an important question for future studies.60
In each of the examples mentioned above, O-GlcNAc-induced protein–protein interactions were discovered through directed biochemical experiments on a known glycoprotein of interest. These studies illustrate the potentially broad physiological significance of O-GlcNAc-mediated protein–protein interactions but also suggest that unbiased methods of detecting and characterizing such interactions would be a powerful tool for discovering unanticipated interactions and new cell biology. However, physiological O-GlcNAc-mediated interactions are often low-affinity, substoichiometric, and transient, presenting a technical barrier to their study.1,2,6,7,9,12 To overcome these difficulties, the Kohler lab recently described a chemical biology strategy for covalently capturing O-GlcNAc-mediated protein–protein interactions (Figure 3). In this method, cells are metabolically labeled with a GlcNAc analogue bearing a diazirine photo-cross-linking moiety, abbreviated “GlcNDAz”.66 A protected, 1-phosphorylated precursor form of GlcNDAz is accepted by the cellular GlcNAc salvage pathway, converted to UDP-GlcNDAz, and used by OGT to decorate its native substrates.66 Briefly, ultraviolet (UV) treatment of GlcNDAz-labeled live cells triggers the elimination of molecular nitrogen from the diazirine moiety and the formation of a highly reactive carbene, resulting in the covalent cross-linking of O-GlcNDAz to any binding partner proteins within ~2–4 Å of the sugar.66 Because of this short radius, GlcNDAz cross-linking occurs exclusively at sites where the glycan contributes to the interaction interface, without cross-linking to distant or nonspecific proteins.66 The authors’ work established GlcNDAz as a powerful tool for identifying direct, glycosylation-mediated interactions between endogenous proteins in live cells.66
Figure 3.
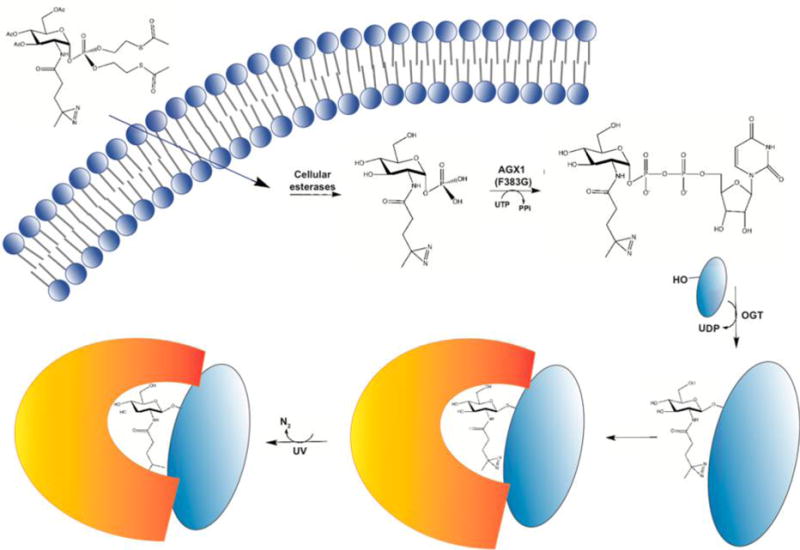
GlcNDAz permits the covalent capture of O-GlcNAc-mediated protein–protein interactions in live cells. The GlcNDAz reagent is a GlcNAc analogue with a diazirine moiety appended to position 2. An S-acetyl-2-thioethyl-protected, 1-phosphorylated, acetylated derivative of GlcNDAz [Ac3GlcNDAz-1-P(Ac-SATE)2] serves as a cell-permeable precursor to GlcNDAz. Upon diffusion across the cell membrane, cytosolic esterases remove the acetyl protecting groups, revealing GlcNDAz-1P. An expressed, designed mutant pyrophosphorylase, AGX1(F383G), then uses GlcNDAz-1P to create the nucleotide-sugar UDP-GlcNDAz. OGT accepts UDP-GlcNDAz as a glycosyl donor, adding O-GlcNDAz moieties to its native substrates. Brief treatment of GlcNDAz-labeled cells with UV light affords the carbene-mediated cross-linking of O-GlcNDAz-modified proteins and any binding partner proteins within ~2–4 Å of the glycan. For additional details, see the text and ref 66.
Our lab has adapted the GlcNDAz system to interrogate O-GlcNAc-mediated protein–protein interactions in several new experimental contexts. For instance, we report in this issue of Biochemistry that O-GlcNAc mediates interactions of proteins in the COPII trafficking pathway, which transports protein and lipid cargoes from the endoplasmic reticulum (ER) toward the Golgi. Several groups, including our own, had observed previously that multiple core COPII proteins are reversibly O-GlcNAcylated, but the biochemical effects of these glycosylation events remained unknown.67–71 Using GlcNDAz, we demonstrated that O-GlcNAc mediates multiple protein–protein interactions of COPII components. We mapped O-GlcNAc sites on several COPII proteins via mass spectrometry and used GlcNDAz and site-directed mutagenesis to pinpoint specific residues on the COPII protein Sec23A that are required for O-GlcNAc-mediated protein–protein interactions. Sec23A is required in both humans and model organisms for the COPII-dependent trafficking of collagen, a major component of the extracellular matrix in the skeleton and other tissues.72–74 Indeed, mutations in the SEC23A gene cause collagen mistrafficking and skeletogenesis defects in the congenital human disease cranio-lenticulo-sutural dysplasia (CLSD).72–74 Interestingly, we found that mutations in particular Sec23A O-GlcNAc sites required for its protein–protein interactions were also defective in collagen trafficking and skeletal development in a human cell culture system and a zebrafish model of CLSD, respectively. These results indicate that O-GlcNAc-mediated protein–protein interactions of Sec23A and other COPII proteins may be a key mode of regulation in the early secretory pathway.
In recent work, our lab has also combined GlcNDAz with biophysical, biochemical, proteomic, and cellular approaches to characterize O-GlcNAc-mediated protein–protein interactions in new contexts. For instance, we recently found that site-specific O-GlcNAcylation of the intermediate filament (IF) protein vimentin governs its assembly state and function in processes such as cell migration and bacterial infection (H. J. Tarbet and M. Boyce, manuscript under review). The human genome encodes more than 70 IF proteins, and many of these are known OGT substrates.75–87 Our results suggest that O-GlcNAcylation may be a broadly conserved mode of governing the IF cytoskeleton form and function. In another example, we addressed the long-standing question of whether O-GlcNAc “reader” proteins exist, recognizing and binding to O-GlcNAc moieties to transduce functional signals. It is well-established that other intracellular PTMs, such as phosphorylation or acetylation, are recognized by such reader proteins,88,89 but no analogous proteins for O-GlcNAc have been reported. We designed a biochemical assay to discover candidate O-GlcNAc readers, discovered several human proteins that bind O-GlcNAc directly and specifically, and determined the crystal structures of multiple candidate readers with model glycopeptides (C. A. Toleman, M. Schumacher, and M. Boyce, manuscript under review). We anticipate that these results will open new avenues of research by identifying the proteins that mediate O-GlcNAc signaling and by providing the first insight into the structural basis of O-GlcNAc recognition.
Together, the examples mentioned above demonstrate how reversible O-GlcNAcylation can mediate the assembly of multiprotein complexes in a wide variety of signaling contexts. We anticipate that some of these binding interactions absolutely require O-GlcNAc, whereas O-GlcNAc likely plays a facilitating or regulatory, but nonessential, role in other instances. The biophysical aspects of these interactions remain to be investigated thoroughly and may reveal a wealth of new information about the regulation of macromolecular recognition. It will also be important to determine through future work how the kinetics and stoichiometry of O-GlcNAcylation impact multiprotein complex formation and downstream signaling in vivo. Finally, as the example of the nuclear pore complex illustrates, extensive glycosylation may impart biologically important bulk material properties to its substrates, especially in the context of low-complexity or intrinsically disordered protein domains. It will be very interesting to learn whether the extensive O-GlcNAcylation of nucleoporins or other cellular structures governs hydrogel formation, phase transitions, or similar biophysical separations in living cells.
CONCLUSION AND OUTLOOK
O-GlcNAcylation is a major mode of cell signaling with broad pathophysiological significance. Nevertheless, the biochemical effects that O-GlcNAc has on most substrates are still largely obscure. The regulation of protein–protein interactions is only one of several mechanisms by which O-GlcNAc influences protein function, but the research discussed above demonstrates that this mode of regulation is widespread within the cell, common among diverse signaling pathways, and conserved across evolution. Despite this recent progress, major questions remain. For example, the stoichiometry and kinetics of O-GlcNAcylation are poorly characterized for nearly all endogenous substrates, precluding a comprehensive understanding of how O-GlcNAc cycling dynamically regulates protein function. In addition, the biophysical basis of O-GlcNAc binding and recognition is poorly understood. Structural studies are needed to elucidate the atomic details of O-GlcNAc-mediated protein–protein complexes and to determine what (if any) common principles underlie these diverse glycosylation-induced interactions. Importantly, new technological advances in areas like mass spectrometry-based proteomics90 and the semisynthesis of homogeneous glycoproteins91,92 will likely empower new studies and spur fresh discoveries. In the very long term, understanding dynamic O-GlcNAc-mediated protein–protein interactions will be an essential part of our system-level understanding of intracellular signaling in both health and disease.
Acknowledgments
Funding
Research in the Boyce lab has been supported by the Rita Allen Foundation, the Mizutani Foundation, and National Institute of General Medical Sciences Grants 1R01GM118847 and 1R01GM117473.
Footnotes
ORCID
Michael Boyce: 0000-0002-2729-4876
Author Contributions
H.J.T. and C.A.T. contributed equally to this work.
Notes
The authors declare no competing financial interest.
References
- 1.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta, Gen Subj. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol. 2014;5:183. doi: 10.3389/fendo.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta, Gen Subj. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh HJ, Moon HY, Cheon SA, Hahn Y, Kang HA. Functional analysis of recombinant human and Yarrowia lipolytica O-GlcNAc transferases expressed in Saccharomyces cerevisiae. J Microbiol. 2016;54:667–674. doi: 10.1007/s12275-016-6401-4. [DOI] [PubMed] [Google Scholar]
- 6.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keembiyehetty C, Love DC, Harwood KR, Gavrilova O, Comly ME, Hanover JA. Conditional knockout reveals a requirement for O-GlcNAcase in metabolic homeostasis. J Biol Chem. 2015;290:7097–7113. doi: 10.1074/jbc.M114.617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ, Moon HY, Byun HY, Kim EK, Kim DH, Lee MN, Koh A, Ghim J, Choi JH, Lee-Kwon W, Kim KT, Ryu SH, Suh PG. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012;11:439–448. doi: 10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- 9.Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan HB, Singh JP, Li MD, Wu J, Yang X. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondoux MA, Love DC, Ghosh SK, Fukushige T, Bond M, Weerasinghe GR, Hanover JA, Krause MW. O-linked-N-acetylglucosamine cycling and insulin signaling are required for the glucose stress response in Caenorhabditis elegans. Genetics. 2011;188:369–382. doi: 10.1534/genetics.111.126490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Z, Vosseller K. O-GlcNAc in cancer biology. Amino Acids. 2013;45:719–733. doi: 10.1007/s00726-013-1543-8. [DOI] [PubMed] [Google Scholar]
- 14.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh JP, Zhang K, Wu J, Yang X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015;356:244–250. doi: 10.1016/j.canlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaidyanathan K, Wells L. Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. J Biol Chem. 2014;289:34466–34471. doi: 10.1074/jbc.R114.591560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20:208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, Copeland RJ, Despa F, Hart GW, Ripplinger CM, Bers DM. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erickson JR. Mechanisms of CaMKII Activation in the Heart. Front Pharmacol. 2014;5:59. doi: 10.3389/fphar.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dassanayaka S, Jones SP. O-GlcNAc and the cardiovascular system. Pharmacol Ther. 2014;142:62–71. doi: 10.1016/j.pharmthera.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darley-Usmar VM, Ball LE, Chatham JC. Protein O-linked beta-N-acetylglucosamine: a novel effector of cardiomyocyte metabolism and function. J Mol Cell Cardiol. 2012;52:538–549. doi: 10.1016/j.yjmcc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 24.Vaidyanathan K, Durning S, Wells L. Functional O-GlcNAc modifications: implications in molecular regulation and pathophysiology. Crit Rev Biochem Mol Biol. 2014;49:140–163. doi: 10.3109/10409238.2014.884535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuzwa SA, Vocadlo DJ. O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem Soc Rev. 2014;43:6839–6858. doi: 10.1039/c4cs00038b. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-GlcNAc and Alzheimer disease. J Biol Chem. 2014;289:34472–34481. doi: 10.1074/jbc.R114.601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roos MD, Su K, Baker JR, Kudlow JE. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol Cell Biol. 1997;17:6472–6480. doi: 10.1128/mcb.17.11.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci U S A. 2001;98:6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim K, Chang HI. O-GlcNAcylation of Sp1 interrupts Sp1 interaction with NF-Y. Biochem Biophys Res Commun. 2009;382:593–597. doi: 10.1016/j.bbrc.2009.03.075. [DOI] [PubMed] [Google Scholar]
- 30.Latorre P, Varona L, Burgos C, Carrodeguas JA, Lopez-Buesa P. O-GlcNAcylation mediates the control of cytosolic phosphoenolpyruvate carboxykinase activity via Pgc1alpha. PLoS One. 2017;12:e0179988. doi: 10.1371/journal.pone.0179988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers SA, Peddada S, Chatterjee N, Friedrich T, Tomoda K, Krings G, Thomas S, Maynard J, Broeker M, Thomson M, Pollard K, Yamanaka S, Burlingame AL, Panning B. SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. eLife. 2016;5:e10647. doi: 10.7554/eLife.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 33.Yang WH, Park SY, Nam HW, Kim DH, Kang JG, Kang ES, Kim YS, Lee HC, Kim KS, Cho JW. NFkappaB activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci U S A. 2008;105:17345–17350. doi: 10.1073/pnas.0806198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 35.Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, Cockrell S, Boyce M, Shabanowitz J, Hunt DF, Olszewski NE, Sun TP. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016;30:164–176. doi: 10.1101/gad.270587.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 37.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan HB, Han X, Li MD, Singh JP, Qian K, Azarhoush S, Zhao L, Bennett AM, Samuel VT, Wu J, Yates JR, 3rd, Yang X. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 2012;16:226–237. doi: 10.1016/j.cmet.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells L, Slawson C, Hart GW. The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids. 2011;40:877–883. doi: 10.1007/s00726-010-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun. 2014;5:4572. doi: 10.1038/ncomms5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci U S A. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P, Peng C, Liu X, Liu H, Chen Y, Zheng L, Han B, Pei H. OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response. J Genet Genomics. 2015;42:467–475. doi: 10.1016/j.jgg.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Gagnon J, Daou S, Zamorano N, Iannantuono NV, Hammond-Martel I, Mashtalir N, Bonneil E, Wurtele H, Thibault P, Affar EB. Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics. 2015;10:677–691. doi: 10.1080/15592294.2015.1060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 50.Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 1986;261:8049–8057. [PubMed] [Google Scholar]
- 51.Kearse KP, Hart GW. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc Natl Acad Sci U S A. 1991;88:1701–1705. doi: 10.1073/pnas.88.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sterne-Marr R, Blevitt JM, Gerace L. O-linked glycoproteins of the nuclear pore complex interact with a cytosolic factor required for nuclear protein import. J Cell Biol. 1992;116:271–280. doi: 10.1083/jcb.116.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 54.Hulsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 55.Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizuguchi-Hata C, Ogawa Y, Oka M, Yoneda Y. Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochim Biophys Acta, Mol Cell Res. 2013;1833:2682–2689. doi: 10.1016/j.bbamcr.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Y, Liu TW, Madden Z, Yuzwa SA, Murray K, Cecioni S, Zachara N, Vocadlo DJ. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J Mol Cell Biol. 2016;8:2–16. doi: 10.1093/jmcb/mjv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ha JR, Hao L, Venkateswaran G, Huang YH, Garcia E, Persad S. beta-catenin is O-GlcNAc glycosylated at Serine 23: implications for beta-catenin’s subcellular localization and transactivator function. Exp Cell Res. 2014;321:153–166. doi: 10.1016/j.yexcr.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 59.Olivier-Van Stichelen S, Dehennaut V, Buzy A, Zachayus JL, Guinez C, Mir AM, El Yazidi-Belkoura I, Copin MC, Boureme D, Loyaux D, Ferrara P, Lefebvre T. O-GlcNAcylation stabilizes beta-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 2014;28:3325–3338. doi: 10.1096/fj.13-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen PH, Smith TJ, Wu J, Siesser PF, Bisnett BJ, Khan F, Hogue M, Soderblom E, Tang F, Marks JR, Major MB, Swarts BM, Boyce M, Chi JT. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–2250. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17:303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, Kong C, Li B, Peng C, Wong CC, Hou F, Zhang Y. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7) PLoS One. 2015;10:e0145023. doi: 10.1371/journal.pone.0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baldini SF, Wavelet C, Hainault I, Guinez C, Lefebvre T. The Nutrient-Dependent O-GlcNAc Modification Controls the Expression of Liver Fatty Acid Synthase. J Mol Biol. 2016;428:3295–3304. doi: 10.1016/j.jmb.2016.04.035. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Wang L, Liu J, Zhang P, An M, Han C, Li Y, Guan X, Zhang K. O-GlcNAcylation modulates Bmi-1 protein stability and potential oncogenic function in prostate cancer. Oncogene. 2017:6293–6305. doi: 10.1038/onc.2017.223. [DOI] [PubMed] [Google Scholar]
- 66.Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zachara NE, Molina H, Wong KY, Pandey A, Hart GW. The dynamic stress-induced ″O-GlcNAc-ome″ highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids. 2011;40:793–808. doi: 10.1007/s00726-010-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dudognon P, Maeder-Garavaglia C, Carpentier JL, Paccaud JP. Regulation of a COPII component by cytosolic O-glycosylation during mitosis. FEBS Lett. 2004;561:44–50. doi: 10.1016/S0014-5793(04)00109-7. [DOI] [PubMed] [Google Scholar]
- 70.Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee A, Miller D, Henry R, Paruchuri VD, O’Meally RN, Boronina T, Cole RN, Zachara NE. Combined Antibody/Lectin Enrichment Identifies Extensive Changes in the O-GlcNAc Sub-proteome upon Oxidative Stress. J Proteome Res. 2016;15:4318–4336. doi: 10.1021/acs.jproteome.6b00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyadjiev SA, Kim SD, Hata A, Haldeman-Englert C, Zackai EH, Naydenov C, Hamamoto S, Schekman RW, Kim J. Cranio-lenticulo-sutural dysplasia associated with defects in collagen secretion. Clin Genet. 2011;80:169–176. doi: 10.1111/j.1399-0004.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, Orci L, Eyaid W. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 75.King IA, Hounsell EF. Cytokeratin 13 contains O-glycosidically linked N-acetylglucosamine residues. J Biol Chem. 1989;264:14022–14028. [PubMed] [Google Scholar]
- 76.Chou CF, Smith AJ, Omary MB. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J Biol Chem. 1992;267:3901–3906. [PubMed] [Google Scholar]
- 77.Ku NO, Toivola DM, Strnad P, Omary MB. Cytoskeletal keratin glycosylation protects epithelial tissue from injury. Nat Cell Biol. 2010;12:876–885. doi: 10.1038/ncb2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong DL, Xu ZS, Hart GW, Cleveland DW. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J Biol Chem. 1996;271:20845–20852. doi: 10.1074/jbc.271.34.20845. [DOI] [PubMed] [Google Scholar]
- 79.Dong DL, Xu ZS, Chevrier MR, Cotter RJ, Cleveland DW, Hart GW. Glycosylation of mammalian neurofilaments. Localization of multiple O-linked N-acetylglucosamine moieties on neurofilament polypeptides L and M. J Biol Chem. 1993;268:16679–16687. [PubMed] [Google Scholar]
- 80.Ludemann N, Clement A, Hans VH, Leschik J, Behl C, Brandt R. O-glycosylation of the tail domain of neurofilament protein M in human neurons and in spinal cord tissue of a rat model of amyotrophic lateral sclerosis (ALS) J Biol Chem. 2005;280:31648–31658. doi: 10.1074/jbc.M504395200. [DOI] [PubMed] [Google Scholar]
- 81.Deng Y, Li B, Liu F, Iqbal K, Grundke-Iqbal I, Brandt R, Gong CX. Regulation between O-GlcNAcylation and phosphorylation of neurofilament-M and their dysregulation in Alzheimer disease. FASEB J. 2008;22:138–145. doi: 10.1096/fj.07-8309com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J Biol Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol Biol Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, Pandey A, Hart GW. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell Proteomics. 2007;6:1365–1379. doi: 10.1074/mcp.M600453-MCP200. [DOI] [PubMed] [Google Scholar]
- 85.Srikanth B, Vaidya MM, Kalraiya RD. O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J Biol Chem. 2010;285:34062–34071. doi: 10.1074/jbc.M109.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kakade PS, Budnar S, Kalraiya RD, Vaidya MM. Functional Implications of O-GlcNAcylation-dependent Phosphorylation at a Proximal Site on Keratin 18. J Biol Chem. 2016;291:12003–12013. doi: 10.1074/jbc.M116.728717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tao GZ, Kirby C, Whelan SA, Rossi F, Bi X, MacLaren M, Gentalen E, O’Neill RA, Hart GW, Omary MB. Reciprocal keratin 18 Ser48 O-GlcNAcylation and Ser52 phosphorylation using peptide analysis. Biochem Biophys Res Commun. 2006;351:708–712. doi: 10.1016/j.bbrc.2006.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol. 2013;14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 89.Wozniak GG, Strahl BD. Hitting the ’mark’: interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta, Gene Regul Mech. 2014;1839:1353–1361. doi: 10.1016/j.bbagrm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 90.Myers SA, Daou S, Affar EB, Burlingame A. Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein host cell factor C1. Proteomics. 2013;13:982–991. doi: 10.1002/pmic.201200332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raj R, Lercher L, Mohammed S, Davis BG. Synthetic Nucleosomes Reveal that GlcNAcylation Modulates Direct Interaction with the FACT Complex. Angew Chem, Int Ed. 2016;55:8918–8922. doi: 10.1002/anie.201603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, Zachara N, Etzkorn FA, Hart GW, Jeong JS, Blackshaw S, Zhu H, Cole PA. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]


