Abstract
Chronic lymphocytic leukaemia (CLL) consists of two biologically and clinically distinct subtypes defined by the abundance of somatic hypermutation (SHM) affecting the Ig variable heavy-chain locus (IgHV). The molecular mechanisms underlying these subtypes are incompletely understood. Here, we present a comprehensive whole-genome sequencing analysis of somatically acquired genetic events from 46 CLL patients, including a systematic comparison of coding and non-coding single-nucleotide variants, copy number variants and structural variants, regions of kataegis and mutation signatures between IgHVmut and IgHVunmut subtypes. We demonstrate that one-quarter of non-coding mutations in regions of kataegis outside the Ig loci are located in genes relevant to CLL. We show that non-coding mutations in ATM may negatively impact on ATM expression and find non-coding and regulatory region mutations in TCL1A, and in IgHVunmut CLL in IKZF3, SAMHD1,PAX5 and BIRC3. Finally, we show that IgHVunmut CLL is dominated by coding mutations in driver genes and an aging signature, whereas IgHVmut CLL has a high incidence of promoter and enhancer mutations caused by aberrant activation-induced cytidine deaminase activity. Taken together, our data support the hypothesis that differences in clinical outcome and biological characteristics between the two subgroups might reflect differences in mutation distribution, incidence and distinct underlying mutagenic mechanisms.
Introduction
Chronic lymphocytic leukaemia (CLL) is the most common form of adult leukaemia in the Western world,1 accounting for one-third of new cases of leukaemia each year.2 CLL is characterised by significant clinical heterogeneity. While one-third of patients never require any treatment, others invariably progress and ultimately develop chemorefractoriness. This clinical heterogeneity is reflected at least in part by characteristic biological features. Chromosomal deletions and amplifications are among the most common genomic aberrations described in CLL3 and have been associated with diverse phenotypes and contrasting clinical behaviour.4
Genome5, 6 and exome-wide7 sequencing efforts have identified a number of recurrent acquired mutations in the coding regions of genes. Mutations in the coding regions of TP53,8, 9, 10 SF3B1,7, 11, 12 NOTCH1,13, 14, 15 RPS1516 and SAMHD117 have been associated with chemorefractoriness, advanced disease, poor prognosis and/or transformation to high-grade lymphoma.
A recent study, using a combination of whole-genome sequencing (WGS) and whole-exome sequencing, addressed for the first time the potential significance of non-coding mutations in CLL18 and, in doing so, characterised a region of kataegis affecting an enhancer site 300 kb upstream of the PAX5 gene. Kataegis is characterised by hotspots of somatically acquired mutations that are scattered throughout the genome. The study also described splice-site mutations in NOTCH1, which have since been correlated with clinical outcome.19 The study illustrates the potential of WGS to reveal important driver mutations outside the protein-coding regions.
During B-cell development, somatic point mutations are introduced at the Ig variable gene loci via the action of activation-induced cytidine deaminase (AID). CLL is divided into two distinct subgroups: those with little or no somatic hypermutation (SHM) and unmutated Ig variable heavy-chain locus (IgHVunmut), defined by ⩾98% homology to the germline IgHV sequence, and those with IgHV hypermutated CLL (IgHVmut) (<98% germline homology). IgHVmut patients have significantly better treatment-free and overall survival than IgHVunmut patients.20 Importantly, there is accumulating evidence that at least a proportion of IgHVmut patients might achieve cure following chemoimmunotherapy.21, 22 Using targeted deep sequencing of the IgHV locus, our group recently demonstrated that 25% of CLL carry additional small IgHV subclones that further refine the prognostic value of the IgHV status.23 IgHVunmut and IgHVmut CLL also differ in their gene expression24, 25 and genome-wide methylation profiles.26 These differences in transcriptional output and clinical behaviour might be due to differences in B-cell receptor signalling response to antigens.27 Alternatively, cell-autonomous and antigen-independent signalling could be a crucial pathogenic mechanism in both types of CLL, independent of IgHV status.28
Recent WGS studies have identified various mutation signatures in CLL,29, 30 including AID- and aging-related signatures, and there is evidence for the full range of AID activity in both IgHVunmut and IgHVmut CLL.31 We therefore hypothesised that the clinical and biological differences seen might be due to differences in the activity and target loci of AID and/or other mutagenic mechanisms between these two groups. One previous study examined mutation signatures in good-risk CLL with isolated deletion of 13q and IgHVmut.32 Here, we therefore included patients with IgHVunmut poor-risk CLL and compared the coding and non-coding mutation spectrum between the two groups. To perform a more rigorous analysis of the non-coding space, we used experimentally determined CLL-specific chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) data in combination with existing data from the ENCODE project33, 34 to produce bespoke non-coding annotations. We then developed an in silico annotation pipeline for these mutations that included the identification of regions of kataegis, and revealed distinct patterns in the occurrence and distribution of mutations between IgHVunmut and IgHVmut subgroups. From our experimental data, we also identify non-coding and mutations in regulatory elements of known cancer-related genes that we link to IgHVunmut CLL. Finally, we show the potential association of these non-coding mutations with gene expression using paired RNA-seq data from the same patients. Taken together, our data support the hypothesis that the difference in clinical outcomes observed between IgHVunmut and IgHVmut is due, at least in part, to differences in the genomic footprint between these two subgroups. Further, it reveals known cancer-linked genes frequently mutated in the IgHVunmut subtype that may be candidates for a deeper understanding of these differences.
Patients and methods
Patient and sample characteristics
We isolated tumour and germline DNA from peripheral blood and saliva samples, respectively, of 46 CLL patients diagnosed at the Oxford University Hospitals, Oxford, or the Karolinska Institute, Stockholm. Informed consent was obtained in line with the Declaration of Helsinki and local research ethics committees. The median age was 69 (range 49–94) years. Fifty-nine per cent (27/46) were male. Sixteen cases were IgHVmut and 27 were IgHVunmut. Patients with the IgHV 3–21 rearrangement have been shown to display the same poor prognostic indications as those with unmutated Ig genes.35 Thus, three cases with <98% homology to the Ig germline sequence, harbouring the V3–21 rearrangement, were also classified as IgHVunmut. Of the 16 IgHVmut patients, 5 were refractory and 11 naïve, and of the 30 IgHVunmut samples, 17 were refractory and 13 naïve. We also subjected 31 samples to targeted deep sequencing of the IgHV gene as described previously.23 Eight patients had been treated with at least one round of chemotherapy before WGS (Supplementary Figures 5 and 6). Twenty-two cases developed chemorefractoriness subsequent to sequencing, defined by either relapse ⩽24 months following initial treatment or the presence of TP53 disruption. Twenty-two were chemosensitive, while the status of the remaining two was unknown (Supplementary Table 1).
WGS and annotation
Matched tumour and germline WGS libraries were prepared from 2 μg DNA using the TruSeq DNA PCR Free Library Preparation Kit (Illumina, San Diego, CA, USA). Libraries were subjected to 2 × 35 bp paired-end sequencing on a HiSeq 2500 instrument (Illumina), to a mean sequencing depth of 39x for tumour (range 35–54) and 36x for germline (range 28–54) samples. Alignment of sequencing reads to the hg19 human genome build was performed using iSAAC v.01.13.04.04.36 SNVs and indels were called using Starling v.2.0.3 and Strelka v.2.0.10.37 Germline mutations were subtracted from the matched tumour and filtered and annotated using our in-house pipeline (Supplementary Methods).
Experimentally determined CLL-specific epigenetics data was generated in the context of the Blueprint Project by ATAC-seq and ChIP-seq on six histone modification marks with non-overlapping functions (H3K4me3, H3k27ac, H3K4me1, H3K27me3, H3K36me3 and H3K9me3). Both methods were performed according to standard protocols available through the Blueprint Portal (http://www.blueprint-epigenome.eu/index.cfm?p=7BF8A4B6-F4FE-861A-2AD57A08D63D0B58). Chromatin states were identified using the chromHMM Software,38 and include Active Promoter, Weak Promoter, Poised Promoter, Strong Enhancer, Weak Enhancer, Transcription Transition, Transcription Elongation, Weak Transcription and Heterochromatin (Supplementary Methods).
Whole transcriptome sequencing
Whole transcriptome sequencing libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation Kit (Illumina). Libraries underwent 2 × 76 bp paired-end sequencing on a HiSeq 2500 instrument (Illumina). Sequence data was aligned to the hg19 build of the human genome using TopHat.39
Data analysis
Copy number alterations (CNAs) and structural rearrangements were identified using Nexus (Biodiscovery Inc., El Segundo, CA, USA) and DELLY,40 respectively. Regions of kataegis within individual patients were identified using methods described previously.41 A further in-house pipeline was used to identify regions of higher mutational load across the cohort (Supplementary Methods). The presence of mutational signatures were determined using methods described previously.42, 43 (Supplementary Methods).
Statistical analysis
Statistical analyses were performed using SPSS version 22 (IBM, Armonk, NY, USA) and Prism version 5 (GraphPad, La Jolla, CA, USA). Categorical variables were compared using either the χ2 test or Fisher's exact test as appropriate. Survival analysis was performed using the Kaplan–Meier method. P-values were considered significant at the 0.05 level.
Results
Somatic mutation distribution and density
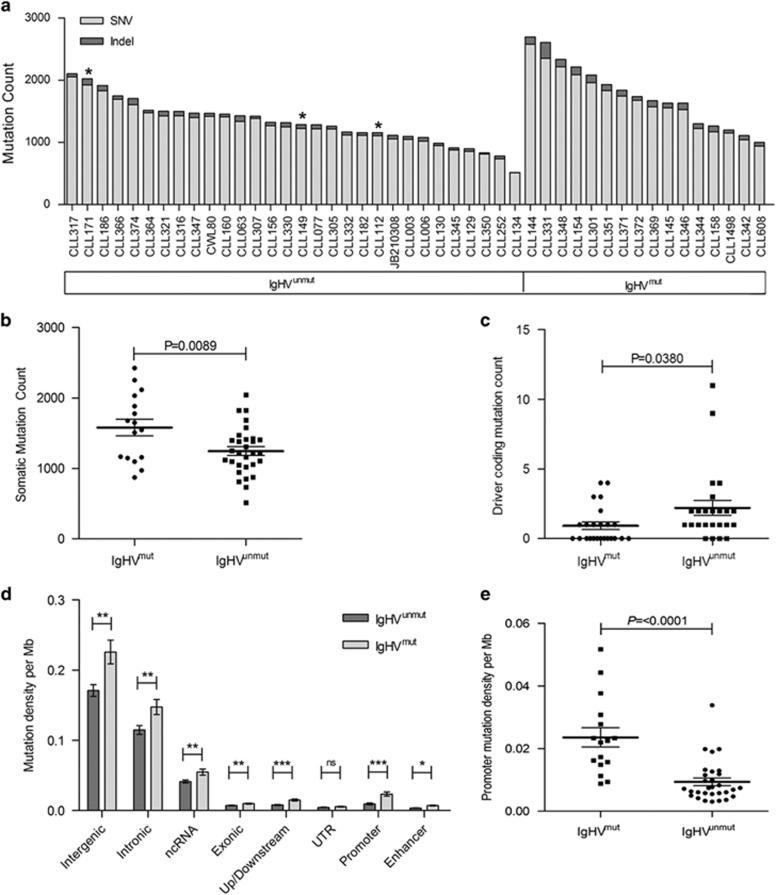
After imposing a rigorous QC step, the number of bona fide mutations per tumour ranged from 516 to 2697 per sample (median, 1426) (Figure 1a and Supplementary Table 2), corresponding to a mean mutation rate of 0.49±0.16 per megabase (range 0.17–0.9). Of these, 823 missense, 62 nonsense and 85 splice-site mutations occurred in protein-coding regions. Of the 3260 insertion/deletions detected across the genomes, 62 affected protein-coding regions. While the overall mutation rate outside the Ig loci was higher in IgHVmut tumours compared with that in IgHVunmut samples (P=0 0089, unpaired t-test; Figure 1b), there was no difference across all coding regions (P=0.1191, unpaired t-test). IgHVunmut cases did, however, show an enrichment for coding mutations in the known CLL driver genes16, 44 (P=0.0380, unpaired t-test; Figure 1c), consistent with a more aggressive phenotype. Furthermore, upon examining the genomic context of the mutations, we discovered significant enrichment for variants within gene promoter regions in IgHVmut patients (P<0.0001, unpaired t-test; Figures 1d and e).
Figure 1.
Overview of the mutational landscape in IgHVmut and IgHVunmut CLL. (a) Total number of SNVs and indels per sample and by IgHV mutation status. Asterisks denote cases with IgHV 3-21 rearrangement. (b) Dot plot of total somatic mutation count between IgHV subgroups. Patients with IgHVmut CLL harbour higher total mutational load, but reduced driver coding (c) and higher promoter variant counts (d and e) than IgHVunmut CLL. Error bars show±s.e.m.
Copy number alterations
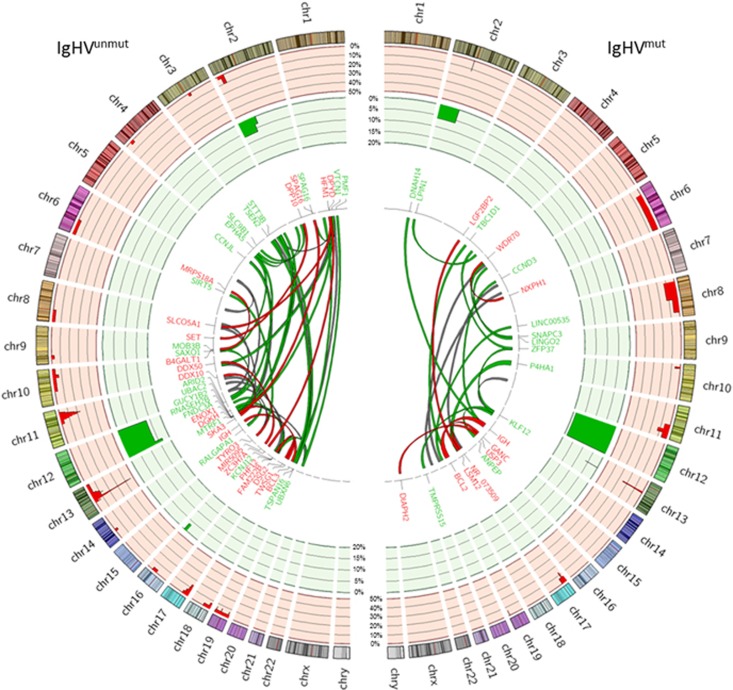
Thirty-five of the 46 CLL cases (76%) harboured at least one CNA (median, 2) (Figure 2 and Supplementary Table 3), with IgHVunmut tumours tending to feature more CNAs than IgHVmut cases (1.3 vs 0.8 per sample, P=0.0651). The most frequent recurrent CNAs were del(13q) (41%, 19/46), del(11q) (20%, 9/46) and del(17p) (20%, 9/46). Trisomy 12 was seen in 20% of tumours (9/46). Seventy-eight per cent of del(17p) cases were IgHVunmut. The minimally deleted region at del(11q) encompassed ATM, with four tumours also carrying ATM mutations (two IgHVmut, two IgHVunmut). Partial or full gain of 2p45 (70–90.5 Mb) was seen in three tumours, but was not associated with differential expression of REL, ALK or MYCN, which mapped to the minimally deleted region. Del(8p) was also seen in three patients (CLL364, CLL154 and CLL372). It was noteworthy that del(8q) was associated with other CNAs—del(11q), gain(12) and del(17p) (CLL364), del(11q) and del(17p) (CLL372) and del(13q) (CLL154). This concurs with other studies, which have reported that del8p deletions often coexist with other CNAs.46, 47 Tumour CLL364 was also the only one to display chromothripsis.
Figure 2.
Structural rearrangements in IgHVunmut and IgHVmut CLL. Circos plot depicting the frequency and locations of CNAs and translocations detected in 46 CLL genomes. The two outermost tracks represent CNAs: CN loss (red) and CN gain (green). Copy number data is displayed as the percentage of each IgHV subgroup affected. The centre plot shows translocations identified in the cohort. Red links indicate tier 1 events (gene:gene), green links indicate tier 2 events (gene:intergenic) and grey indicate tier 3 (intergenic:intergenic). The widths of the bands are indicative of the number of patients affected by the translocation.
Structural variants
We detected 79 interchromosomal translocations in the tumours from 30 of the 46 patients (average of 1.7 per case, range 1–9; Figure 2 and Supplementary Table 4). Seventeen of the 79 events (22%) were predicted to generate gene fusion proteins (tier 1). Two IgHVmut cases (CLL301 and CLL348) had IGH/BCL2 t(14;18) fusions, and a single IgHVunmut case had IGH/BCL3 t(14;19). RNA-seq data was available in 22 out of the 30 patients (73%, 22/30). The other interchromosomal translocations involved either the coding region of a gene (tier 2, 44%, 35/79) or intergenic regions (tier 3, 34%, 27/79). RNA expression supported the functional consequences of t(14;18) (IGH/BCL2), t(2;9) (DPP10;SET) and both t(1;2) and t(3;19), linking SPAG16 and UBXN6 to intergenic regions (Supplementary Figure 1).
Somatic mutation profiles
Coding mutations
As expected, somatic mutations in the coding regions of SF3B1, TP53, ATM and NOTCH1 were the most frequent, occurring in 30.4% (14/46), 19.6% (9/46), 15.2% (7/46) and 13% (6/46) of patients, respectively. The missense Lys700Glu substitution in SF3B1 (28%, 4/14), between the third and fifth HEAT domains, affected mainly IgHVunmut patients, although this did not reach significance (P=0.316). Missense mutations were also the most frequent type found in TP53 (82%, 9/11), occurring in the DNA-binding domain and in all previously described in the COSMIC database. Two cases harboured indels within TP53, one 4 bp deletion in exon 4 and one 19 bp deletion in exon 5. Again, TP53 mutations were more common in IgHVunmut patients (9/11, 82%), but not to a significant level (P=0.463). Seven cases contained eight mutations within the coding region of ATM, comprising six missense mutations, a 5 bp deletion in exon 53 and one stop gain mutation in exon 25, resulting in the loss of 60% of the coding sequence. Mutations in NOTCH1 were confined to exon 34, with 71% (5/7) comprising the c.7541_7542delCT variant resulting in early termination of the PEST domain. In addition, we identified a number of low-frequency recurrent mutations. The tumour suppressor gene FAT1 harboured protein-coding mutations in 11% of samples (5/46), encompassing both chemorefractory and untreated cases.48 Most mutations in FAT1 occurred within exon 2 (60%, 3/5), including a nonsense mutation at residue Lys125, resulting in the loss of >97% of the coding region. We detected five missense and two synonymous mutations in KLHL6 across two patients. In one case, three mutations (Arg66Gly, Met67Thr and Val88Glu) were clustered close together in exon 1, and both cases were IgHVmut as described previously.5 Other previously described candidate driver genes harboured low-frequency variants, including two mutations each in MYD88, SAMHD1,DDX3X and BIRC3, with one each in FBXW7, XPO1, CHD2 and POT1.5, 6, 12, 14, 17, 44
Mutations in non-coding and regulatory regions
Localised regions of increased mutation density, also referred to as kataegis, have been described previously in other types of cancer. Here, we used both previously described methods41 and our own approaches to identify these regions in our cohort (Supplementary Methods).
To this end, we first examined each CLL genome individually, where we detected 74 kataegic regions across 31 samples affecting 14 chromosomes. Some chromosomal regions were recurrently affected, most notably, and as expected, those encoding the Ig light and heavy chains on chromosomes 2 (11/74, 15%), 14 (33/74, 45%) and 22 (13/74, 18%). These regions accounted for 88% (1397/1591) of all kataegic mutations. IgHVmut cases harboured significantly more kataegic regions than IgHVunmut cases (P=0.0004), a relationship that continued even after removing regions found at the Ig loci (P=0.0308). The 19 non-Ig regions occurred in 11 patients and ranged in length from 219 bp to 159 kbp. Fourteen were located within or across gene boundaries. Importantly, almost half of these (6/14, 43%) affected non-coding regions of genes were known to also carry coding mutations in CLL including KLHL6,5 MEGF9,49 CDH12,44 CADPS2,5, 44 LRP1B44, 49 and ATM50, 51 (Table 1 and Supplementary Table 5).
Table 1. Regions of kataegis affecting gene regulatory and exonic regions in individual CLL cases.
| Patient ID | IgHV status | Chr. | Start (bp) | End (bp) | No. of mutations | Affected genes |
|---|---|---|---|---|---|---|
| CLL156 | Unmutated | 2 | 140 885 911 | 141 045 820 | 8 | LRP1B |
| CLL154 | Mutated | 3 | 157 290 339 | 157 295 704 | 6 | PQLC2L |
| CLL348 | Mutated | 3 | 183 273 058 | 183 273 364 | 6 | KLHL6 |
| CLL063 | Unmutated | 5 | 21 810 369 | 21 843 504 | 9 | CDH12 |
| CLL301 | Mutated | 7 | 122 433 699 | 122 517 296 | 12 | CADPS2 |
| CLL301 | Mutated | 7 | 122 622 586 | 122 638 447 | 6 | TAS2R16 |
| CLL351 | Mutated | 9 | 123 416 699 | 123 479 606 | 43 | MEGF9 |
| CLL144 | Mutated | 11 | 108 121 624 | 108 129 499 | 9 | ATM |
| CLL307 | Unmutated | 13 | 51 664 141 | 51 665 954 | 6 | LINC00371 |
| CLL156 | Unmutated | 14 | 21 835 327 | 21 835 546 | 6 | SUPT16H |
| CLL252 | Unmutated | 18 | 9 284 149 | 9 374 417 | 6 | ANKRD12 |
| CLL301 | Mutated | 18 | 60 873 525 | 60 988 029 | 12 | BCL2 |
| CLL348 | Mutated | 18 | 60 906 440 | 60 988 117 | 10 | BCL2 |
| CLL301 | Mutated | X | 98 765 633 | 98 769 284 | 10 | XRCC6P5 |
Abbreviations: CLL, chronic lymphocytic leukaemia; IgHV, Ig variable heavy-chain locus.
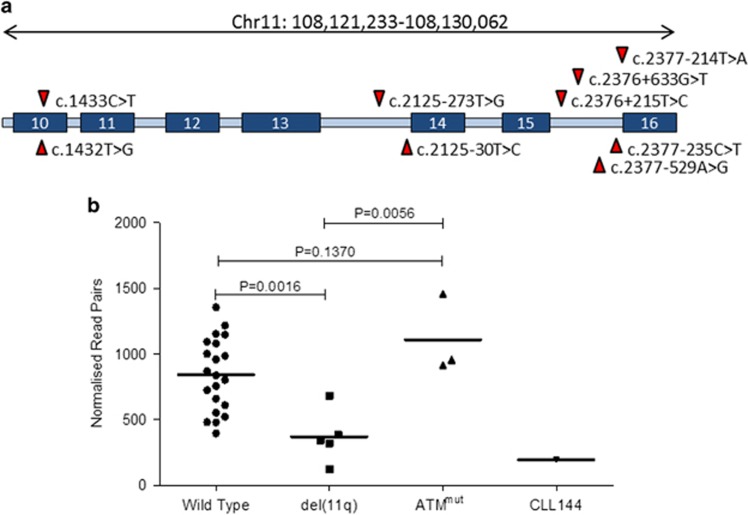
In one IgHVmut case, CLL144, a kataegic region was identified within ATM, comprising 10 mutations in a 9 kb span between exon 10 and intron 16. Eighty per cent (8/10) of the mutations occurred in intronic regions, with two more in exon 10. The two coding mutations affected adjacent bases, but resulted in a single serine to valine residue substitution (Ser478Val), predicted as tolerated by the sorting tolerant from intolerant algorithm. Experimentally determined ChIP-seq data revealed that all 10 mutations occurred in regions of strong H3K36 trimethylation, which is an indication of transcription elongation.38 Analysis of paired RNA-seq data from CLL144 revealed a similarly reduced level of ATM expression as seen in cases harbouring del(11q). Furthermore, cases with del(11q) showed significantly lower ATM expression levels than ATM wild-type cases (P=0.0016), as expected50 (Figure 3).
Figure 3.
Kataegic ATM mutations in CLL144. (a) Graphical representation of ATM showing the distribution of kataegic mutations in CLL144. Blue boxes represent exonic regions of the ATM transcript. (b) Dot plot comparing the ATM transcript expression levels of CLL cases with no 11q disruption (wild type), those with del(11q), mutations in ATM (ATMmut) and CLL144. Expression levels are shown as reads per million aligned reads. Error bars show±s.e.m.
Sites of kataegis have been shown to colocalise with structural rearrangements.42, 52 As such, the 19 kataegic regions that were not at sites of SHM were correlated with CNAs and translocations. Five were found to be in close proximity to structural alteration events (Supplementary Table 6). In line with previous observations,42, 52 this suggests that kataegis can indicate the presence of structural rearrangements.
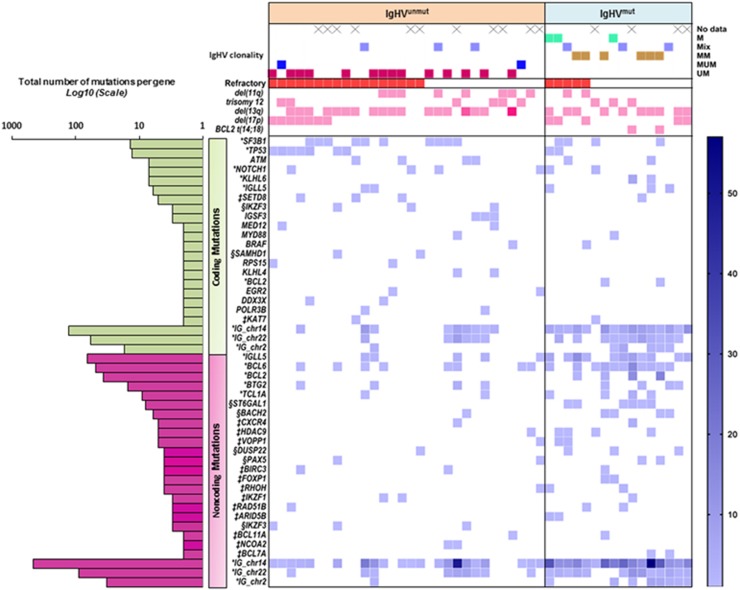
Second, we combined the WGS data from all 46 CLL samples, and used a combination of methods to highlight strict regions of kataegis, alongside wider mutational hotspots (Supplementary Methods). Through this approach, we identified 159 additional kataegic regions, which were further annotated for the presence of coding and non-coding regulatory regions using a combination of functional data obtained from normal B cells, CLL cell lines and primary CLL. Sites were defined as active regulatory regions if they fell within chromatin states containing H3K27 acetylation (that is, active promoters, strong enhancers or transcription transition), as determined using the CLL-specific ChIP-seq data (see Supplementary Methods). With the exception of the 9q34.3 locus of NOTCH1, 51 regions were excluded from further analysis because of their centromeric or telomeric location. In addition to the expected clustering of coding mutations in SF3B1, TP53, NOTCH1 and KLHL6, occurring predominantly in IgHVunmut patients, the remaining 108 regions included 21 clusters of non-coding mutations lying in active regulatory promoter or enhancer regions of genes expressed in CLL cells. The majority of these regions occurred in either the Ig loci (52%, 11/21) or within regulatory regions of known targets of SHM, including BCL6, TCL1A, BTG2 and BACH2,53, 54 and were more common in IgHVmut patients (Figure 4). We were unable to find previously described mutations in the splice-site region of NOTCH1.18 This is likely due to their low incidence of ~2%.18
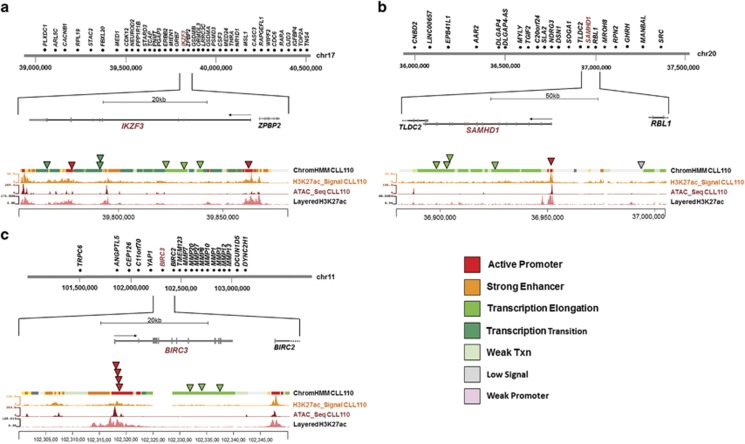
Figure 4.
Non-coding mutations confirmed experimentally with ChIP-seq and ATAC-seq. Diagrams of non-coding mutations in three genes and the corresponding annotation data. All lower panels; correlation of mutation loci with H3K27Ac and ATAC-seq signal of CLL110 and ChromHMM annotation (all three layers obtained from experiments with primary CLL cells) and layered H3K27Ac data from the ENCODE database (GM12878). (a) IKZF3 locus and surrounding region on chromosome 17, with the position of all mutations detected in our cohort, (b) SAMHD1 locus and surrounding region on chromosome 20, with the position of all mutations detected in our cohort and (c) BIRC3 locus and surrounding region on chromosome 11, with the position of all mutations detected in our cohort.
Third, we performed a further hotspot analysis to identify recurrently mutated regions and genes with fewer variants than are required to be classified as kataegis. This revealed IKZF3 mutations in three IgHVunmut patients and a further five with mutations in regulatory regions, only one of which was IgHVmut. We also detected SAMHD1 coding mutations in two IgHVunmut cases and three mutations in active regulatory elements in two IgHVunmut and one IgHVmut case (Figures 4a and b and Supplementary Figures 2 and 3). Both IKZF3 and SAMHD1 have been identified as driver genes in CLL.16, 17, 44 Furthermore, we identified clusters of mutations within active enhancer and promoter regions of ST6GAL1 in nine IgHVmut patients and three IgHVunmut patients. Overexpression of ST6GAL1 has been linked to chemoresistance in leukaemia55 and indeed two of the affected cases were confirmed chemorefractory with otherwise good-risk features. Mutations affecting the PAX5 enhancer region have been recently described in CLL,18, 56 and, indeed, in our cohort a single IgHVmut patient, CLL351, harboured a mutation in this region. Additionally, we identified eight mutations within the boundaries of the PAX5 gene, and which correlated with non-coding elements, in our cohort, comprising seven mutations across six IgHVunmut patients and one mutation in one IgHVmut patient. Six of these mutations were located in sites of transcription elongation, and the remaining two in promoter and enhancer regions (Supplementary Figure 4).
Finally, we focussed on the regulatory regions of genes with strong a priori relevance to CLL biology. This analysis was restricted to sites at active regulatory regions (Supplementary Methods) in genes expressed in CLL cells. Here, only instances that harboured mutations in at least two patients in a given region were counted. This analysis revealed a number of recurrently mutated active regulatory regions in genes involved in key pathways including B-cell development (BCL6, IKZF1, PAX5), nuclear factor-κB signalling (TCL1A, BACH2, BIRC3, VOPP1), NOTCH signalling (HDAC9) and DNA damage response (BTG2, BCL2) (Figure 5). With the exception of a subset of BCL-2 promoter mutations that were linked to t(14;18) in two of three patients, paired RNA-seq data from the same patients did not reveal any differences in gene expression compared with patients without mutations in these regions (data not shown). As it can be difficult to uncouple treatment effect with biological effect, an association plot was constructed dividing data by chemoresistance and specifying previously treated patients (Supplementary Figures 5 and 6); however, no clustering was found within these groups.
Figure 5.
Overview of the mutational landscape in IgHVmut and IgHVunmut CLL. Top panel—Type and number of IgHV subclones present in each patient. Where several clones are present, IgHV status is determined by the identity of the dominant clone. Samples where only Sanger sequencing data were available are labelled as ‘not known’. Middle panels—Presence of copy number aberrations within the cohort. Lower panel—Incidence of both coding and non-coding mutations within the cohort. Colour spectrum (light to dark blue or light to dark pink) corresponds with mutational or CNA load per patient, respectively. *Genes containing kataegic mutations. ‡Genes with multiple mutations in regulatory regions and are involved in important B-cell pathways. §Genes with smaller mutational hotspots. Panel left: Number of mutations per gene across the entire cohort.
Mutation signatures
Finally, we asked whether the difference in mutation distribution could arise as a consequence of the action of distinct mutagenic mechanisms. By using complex mathematical modelling, it has become possible to isolate a number of mutational patterns, or signatures, within the molecular landscape of cancer.29, 30, 42, 57
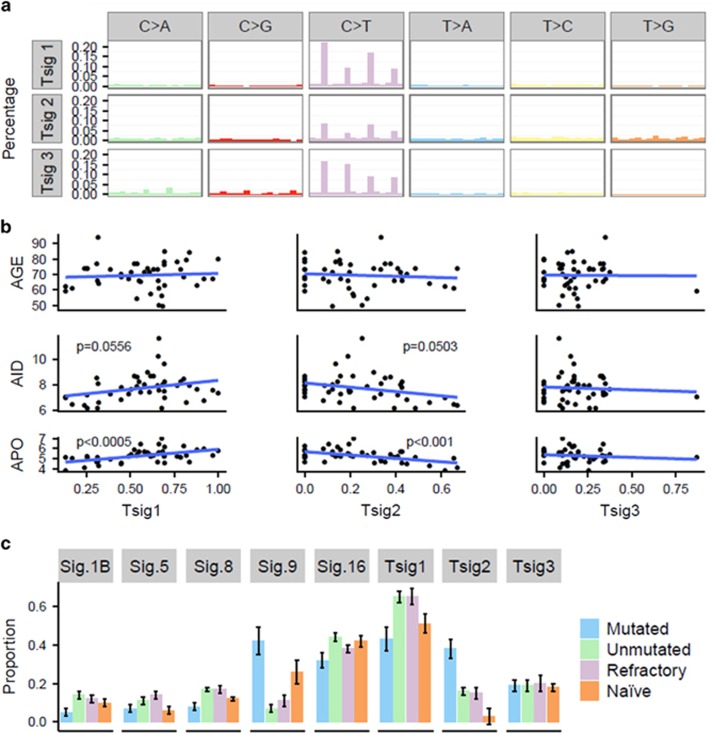
Although previously derived mutation signatures are extensive, they focus solely on exome data, and not whole genomes. Here, our aim was to compare the age, AID content and apolipoprotein B mRNA editing enzyme catalytic polypeptide (APOBEC) content of our samples with the mutational signatures found in our complete data. Thus, for our initial analysis we derived novel mutation signatures. We used our WGS data to extract three distinct mutational signatures from our cohort. Tumour signature 1 (Tsig1) is dominated by C>T transitions in NpCpG and NpCpC contexts accounting for 56.8 and 5.5% of the signature, respectively (Figure 6a). Tumour signature 2 (Tsig2) is again defined primarily by C>T transitions in NpCpG trinucleotides; however, the occurrence of T>G transversions and T>C transitions at NpTpN sites differentiates it from Tsig1 (Figure 6a). Tsig3 is characterised by substitutions involving cytosine residues, with C>T, C>A and C>G variants at NpCpG trinucleotides accounting for 63% of the signature.
Figure 6.
Mutation signature analysis. (a) Three mutation signatures identified across 46 patients using non-negative matrix factorisation (NMF). (b) Correlations between signatures identified in 46 patients and the corresponding age, or proportion of mutated canonical AID or APOBEC sites per subtracted somatic sample. Blue lines=regression lines, and P-values from the glm. (c) Proportions of mutation signatures from Alexandrov (2013),29,43 where Sig.1B corresponds to Alexandrov Signature 1B, and so on, and signatures found across 46 patients.
Previously, three mutational signatures were described in CLL,29 attributed to the ageing process, and the ongoing presence of AID and APOBEC activity. To examine the relationship between these factors and the signatures in our data, we used a generalised linear model to correlate the proportion of each signature present in each sample with the levels of APOBEC and AID mutations, and the age of the patient (Figure 6b). We identified a strong positive correlation between Tsig1 and APOBEC mutations and a correlation between Tsig2 and the noncanonical AID signature.
We also examined our data for the presence of any of the 21 Alexandrov signatures, of which we identified 5 (1b, 5, 8, 9 and 16) (Figure 6c). Signature 1b is generally attributed to ageing, and showed a strong correlation with our Tsig1. However, in our cohort, neither signature 1b nor Tsig1 correlated with the age of the patient. Instead, we saw a significant increase in the proportion of Tsig1 in the refractory and IgHVunmut patient groups. Owing to the overlap between these groups in our cohort, it was impossible to ascertain whether this correlation was independent. However, it strongly suggests that the underlying mechanism for the increase in Tsig1 in these patients might be exposure to previous chemotherapy and/or genomic instability. Tsig2-matched signature 9 is attributed to noncanonical AID activity and Ig hypermutation. As expected, both signature 9 and Tsig2 in our cohort are present at significantly higher levels in the IgHV-mutated cohort (Figure 6c). In summary, we were able to identify statistically significant differences in the proportion of mutation signatures between IgHVunmut and IgHVmut for Sig1b and Tsig1, Sig8, Sig9 and Tsig2, and Sig16.
Discussion
Here we have used WGS to provide a comprehensive genome-wide description of the landscape of non-coding mutations of both IgHVmut and IgHVunmut CLL. We reveal significant differences in the distribution of both somatic mutations and mutation signatures between these two subgroups. Previous WGS studies of CLL have focussed on good-risk treatment-naïve patient groups, such as those with monoclonal B-cell lymphocytosis or early stage CLL18 or IgHVmut CLL with concurrent del(13q).56 Here we have extended these published data sets and were able to compare it with the WGS data from poor-risk patients, many of whom were either IgHVunmut and/or refractory to chemoimmunotherapy.
First, this genome-wide comparison of the mutation distribution between IgHVmut and IgHVunmut patients revealed a distinctive pattern for each subtype, with exonic CLL driver gene mutations being more common in IgHVunmut patients, whereas mutations in regulatory elements of B-cell master regulators were more frequently observed in IgHVmut patients. We developed and implemented a highly stringent filtering pipeline that excluded a large proportion of the genome, including repetitive, centromeric and telomeric domains to reduce the number of false-positive variants. Importantly, we only included mutations in DNAse hypersensitivity sites or in regions of H3K4me3, H3K9ac, H3K4me1 or H3K27ac marks and complemented these with RNA-seq, and experimental CLL-specific data derived from ChIP-seq and ATAC-seq (see Supplementary Methods). Annotations were assigned to the ‘active non-coding element’ classes of active promoter, strong enhancer or transcription transition sites from these primary CLL cells using chromHMM. This analysis revealed that at least 50% of somatically acquired mutation hotspots in regulatory regions of individual patients are located in genes found to also carry mutations in coding regions in CLL. In the case of non-coding mutations in ATM, these appeared to associate with altered RNA expression levels of these genes.
By performing a cohort-wide kataegis analysis, we were able to corroborate the findings of other groups with regard to the presence of mutations affecting the enhancer region of PAX5 in IgHVmut patients.18, 56
Transcription elongation (H3K36me3) and active promoter mutations were seen in PAX5 in 6/27 (22%) of IgHVunmut patients (Supplementary Figure 4). H3K36 trimethylation has been noted in yeast as a mechanism for the deacetylation of coding regions, thus stopping overactive transcription,58 and there is some evidence to suggest that it might have a similar role in humans.59 However, no effect on gene expression was found in our data (data not shown). We also found that 6 of 46 (13%) patients carried at least one mutation in the promoter region of TCL1A (Supplementary Figure 7). TCL1A is a direct activator of nuclear factor-κB. Translocations leading to TCL1A overexpression (t(14;14)(q11;q32)) by putting TCL1A expression under the control of the IgHV promoter have been described as rare events in lymphomas. Besides, the TCL1A promoter has previously been shown to display hypomethylation in CLL cells, providing an alternative mechanism for TCL1A upregulation in CLL.60, 61 Moreover, microRNA dysregulation has also been proposed as a cause for TCL1A overexpression.62 In our series of patients, there was no difference in the TCL1A RNA expression in patients with or without mutations in the TCL1A promoter.
Importantly, we describe for the first time a number of somatically acquired genomic events that occurred predominantly or exclusively in patients in IgHVunmut patients:
(1) We identified for the first time coding and non-coding mutations in IKZF3 in almost 30% of IgHVunmut patients (8 mutations in 8/27 patients), making IKZF3 one of the most frequently mutated genes in high-risk CLL (Figure 4a). IKZF3 is known to be overexpressed in CLL, and epigenetic modification of its promoter region has previously been described as one of the underlying mechanisms.63 In the same study, overexpression or downregulation of IKZF3 was shown to affect the expression of some Bcl-2 family members including BCL-2, thereby regulating apoptosis and cell survival. We can speculate that direct mutations in promoter and/or enhancer regions of IKZF3 might represent either an alternative or complementary mechanism driving IKZF3 deregulation in CLL. Further functional work will be required to understand their precise impact on CLL biology. In addition, mutations in the IKZF3 paralogue IKZF1 were found in active promoter regions in two IgHVunmut patients (Supplementary Figure 8). Similar to IKZF3, IKZF1 is a regulator of lymphocyte differentiation and has been linked to childhood B-cell precursor acute lymphoblastic leukaemia.64
(2) Coding mutations in SAMHD1 occurred in two IgHVunmut patients, with three mutations in active regulatory regions in another three patients (Figure 4b). Somatically acquired germline mutations in the SAMHD1 coding region have been shown to exist as founder events, regulating cell proliferation and survival, in relapsed and refractory CLL patients.17, 44
(3) BIRC3 mutations in an active promoter or transcription elongation site occurred in 4/27 IgHVunmut patients and one IgHVmut patient (Figure 4c). BIRC3 is a known cancer driver gene involved in nuclear factor-κB signalling. Exonic SNV mutations and deletions have been described previously and are associated with poor-risk disease.65
(4) In all cases of regulatory mutations, further experimental analyses are required to fully link mutations to altered gene expression.
(5) Finally, we identify differences in the distribution of a number of mutation signatures, in particular the AID and aging signatures, pinpointing different pathogenic mechanisms of leukemogenesis. First, although the AID signature is more abundant in IgHVmut CLL, it is also prevalent in IgHVunmut CLL. These data are consistent with recent studies, which found that AID expression and full functional activity are intact in both IgHVmut and IgHVunmut CLL,31 and that canonical AID activity is an ongoing process in IgHVmut patients.32 Conversely, IgHVunmut and chemoimmunotherapy refractory CLL are dominated by the aging signature. This was independent of the age of the patient and implies that the increased incidence of driver mutations in this subgroup might be caused by repeated cell divisions, genomic instability and prior exposure to DNA-damaging agents rather than aberrant AID activity.
In conclusion, we provide evidence that in addition to recurrent mutations in the coding elements of the genome, non-coding mutations in known regulatory regions of critical B-cell transcription factors such as PAX5 or IKZF3 occur in significant subsets of CLL. On the basis of their mutation frequency in regulatory regions in several patients with IgHVunmut CLL, we propose a number of genes including IKZF3, BIRC3 and SAMHD1 as warranting further functional investigation into the differences between IgHVmut and IgHVunmut CLL. Finally, we identify for the first time different dominant mutagenic mechanisms between these different types of CLL. The identification of the differences in the distribution of non-coding mutations between IgHVmut and IgHVunmut CLL is the first step towards an in-depth functional analysis. However, the fact that certain non-coding regions are recurrently targeted by mutations in a significant number of patients and that the majority of these occur in genes also targeted by mutations in the exome is in itself evidence for their functional relevance in CLL pathogenesis. Our data therefore supports the hypothesis that the biological and clinical differences observed between IgHVunmut and IgHVmut CLL are due, at least in part, to differences in the genomic footprint between these two subgroups and that regulatory mutations present a crucial avenue for further investigation.
Acknowledgments
RA receives funding from the Saudi Arabian Government. This work was supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme. AS, JT and SJLK are supported also by the Health Innovation Challenge Fund (HICF-1009-026), a parallel funding partnership between the Wellcome Trust and the Department of Health. SJLK is also supported by the Wellcome Trust Core Award Grant (090532/Z/09/Z). The views expressed in this publication are those of the authors and not necessarily those of the Department of Health or the Wellcome Trust. EC and JIM-S are supported by the European Union's Seventh Framework Program through the Blueprint Consortium (grant agreement 282510) and by the Spanish Instituto de Salud Carlos III (grant ID PMP15/00007). JIM-S is also supported by Worldwide Cancer Research (grant number 16-1285). Additional annotation data were provided with permission from the ENCODE Consortium.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
JB and DB are employees of Illumina Inc., a public company that develops and markets systems for genetic analysis. The other authors declare no conflict of interest.
11/21/2017
This article has been corrected since Advance Online Publication and a corrigendum is also printed in this issue
Supplementary Material
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et alWHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. World Health Organization: Geneva, Switzerland, 2008.
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62: 10–29. [DOI] [PubMed] [Google Scholar]
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Seiler T, Döhner H, Stilgenbauer S. Risk stratification in chronic lymphocytic leukemia. Semin Oncol 2006; 33: 186–194. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 2012; 120: 4191–4196. [DOI] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52. [DOI] [PubMed] [Google Scholar]
- Zenz T, Vollmer D, Trbusek M, Smardova J, Benner A, Soussi T et al. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia 2010; 24: 2072–2079. [DOI] [PubMed] [Google Scholar]
- Zenz T, Mohr J, Edelmann J, Sarno A, Hoth P, Heuberger M et al. Treatment resistance in chronic lymphocytic leukemia: the role of the p53 pathway. Leuk Lymphoma 2009; 50: 510–513. [DOI] [PubMed] [Google Scholar]
- Zenz T, Kröber A, Scherer K, Häbe S, Bühler A, Benner A et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 2008; 112: 3322–3329. [DOI] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 2011; 118: 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2011; 119: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011; 208: 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Rasi S, Spina V, Fangazio M, Monti S, Greco M et al. Different impact of NOTCH1 and SF3B1 mutations on the risk of chronic lymphocytic leukemia transformation to Richter syndrome. Br J Haematol 2012; 158: 426–429. [DOI] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015; 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 2014; 123: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015; 526: 519–524. [DOI] [PubMed] [Google Scholar]
- Larrayoz M, Rose-Zerilli MJJ, Kadalayil L, Parker H, Blakemore S, Forster J et al. Non-coding NOTCH1 mutations in chronic lymphocytic leukemia; their clinical impact in the UK CLL4 trial. Leukemia 2016; 31: 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–1854. [PubMed] [Google Scholar]
- Tam CS, O’Brien S, Plunkett W, Wierda W, Ferrajoli A, Wang X et al. Long-term results of first salvage treatment in CLL patients treated initially with FCR (fludarabine, cyclophosphamide, rituximab). Blood 2014; 124: 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Bahlo J, Fink AM, Goede V, Herling CD, Cramer P et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood 2016; 127: 208–215. [DOI] [PubMed] [Google Scholar]
- Stamatopoulos B, Timbs A, Bruce D, Smith T, Clifford R, Robbe P et al. Targeted deep sequencing reveals clinically relevant subclonal IgHV rearrangements in chronic lymphocytic leukemia. Leukemia 2016; 31: 837–845. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med 2001; 194: 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med 2001; 194: 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet 2016; 48: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood 2011; 118: 4313–4320. [DOI] [PubMed] [Google Scholar]
- Minden MD, Übelhart R, Schneider D, Wossning T, Bach MP, Buchner M et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012; 489: 309–312. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, Staaf J et al. The topography of mutational processes in breast cancer genomes. Nat Commun 2016; 7: 11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten PEM, Chu CC, Albesiano E, Damle RN, Yan X-J, Kim D et al. IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions. Blood 2012; 120: 4802–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat Commun 2015; 6: 8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004; 306: 636–640. [DOI] [PubMed] [Google Scholar]
- Hong EL, Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS et al. Principles of metadata organization at the ENCODE data coordination center. Database 2016; 2016: baw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsélius M, Kröber A, Murray F, Thunberg U, Tobin G, Bühler A et al. Strikingly homologous immunoglobulin gene rearrangements and poor outcome in VH3-21-using chronic lymphocytic leukemia patients independent of geographic origin and mutational status. Blood 2006; 107: 2889–2894. [DOI] [PubMed] [Google Scholar]
- Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013; 29: 2041–2043. [DOI] [PubMed] [Google Scholar]
- Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012; 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012; 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 2009; 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012; 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012; 149: 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013; 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapiro E, Leporrier N, Radford-Weiss I, Bastard C, Mossafa H, Leroux D et al. Gain of the short arm of chromosome 2 (2p) is a frequent recurring chromosome aberration in untreated chronic lymphocytic leukemia (CLL) at advanced stages. Leuk Res 2010; 34: 63–68. [DOI] [PubMed] [Google Scholar]
- Forconi F, Rinaldi A, Kwee I, Sozzi E, Raspadori D, Rancoita PMV et al. Genome-wide DNA analysis identifies recurrent imbalances predicting outcome in chronic lymphocytic leukaemia with 17p deletion. Br J Haematol 2008; 143: 532–536. [DOI] [PubMed] [Google Scholar]
- Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res 2012; 18: 3791–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina M, Del Giudice I, Khiabanian H, Rossi D, Chiaretti S, Rasi S et al. Genetic lesions associated with chronic lymphocytic leukemia chemo-refractoriness. Blood 2014; 123: 2378–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ramsay AJ, Rodríguez D, Puente XS, Campo E, López-Otín C. The genomic landscape of chronic lymphocytic leukemia: clinical implications. BMC Med 2013; 11: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd PJ et al. Inactivation of ataxia telangiectasia mutated gene in B-cell chronic lymphocytic leukaemia. Lancet (London, England) 1999; 353: 26–29. [DOI] [PubMed] [Google Scholar]
- Schaffner C, Stilgenbauer S, Rappold GA, Döhner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood 1999; 94: 748–753. [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Murison A, Boyle EM, Begum DB, Dahir NM et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun 2015; 6: 6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabakhshi AH, Morin RD, Fejes AP, Mungall AJ, Mungall KL, Bolger-Munro M et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget 2012; 3: 1308–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Küppers R et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001; 412: 341–346. [DOI] [PubMed] [Google Scholar]
- Ma H, Cheng L, Hao K, Li Y, Song X, Zhou H et al. Reversal effect of ST6GAL 1 on multidrug resistance in human leukemia by regulating the PI3K/Akt pathway and the expression of P-gp and MRP1. PLoS One 2014; 9: e85113. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rose-Zerilli MJJ, Gibson J, Wang J, Tapper W, Davis Z, Parker H et al. Longitudinal copy number, whole exome and targeted deep sequencing of ‘good risk’ IGHV-mutated CLL patients with progressive disease. Leukemia 2016; 30: 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016; 534: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005; 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010; 143: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garding A, Bhattacharya N, Haebe S, Müller F, Weichenhan D, Idler I et al. TCL1A and ATM are co-expressed in chronic lymphocytic leukemia cells without deletion of 11q. Haematologica 2013; 98: 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuille MR, Condie A, Stone EM, Wilsher J, Bradshaw PS, Brooks L et al. TCL1 is activated by chromosomal rearrangement or by hypomethylation. Genes Chromosomes Cancer 2001; 30: 336–341. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 2006; 66: 11590–11593. [DOI] [PubMed] [Google Scholar]
- Billot K, Soeur J, Chereau F, Arrouss I, Merle-Beral H, Huang M-E et al. Deregulation of Aiolos expression in chronic lymphocytic leukemia is associated with epigenetic modifications. Blood 2011; 117: 1917–1927. [DOI] [PubMed] [Google Scholar]
- Boer JM, van der Veer A, Rizopoulos D, Fiocco M, Sonneveld E, de Groot-Kruseman HA et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia 2016; 30: 32–38. [DOI] [PubMed] [Google Scholar]
- Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, Cresta S et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood 2012; 119: 2854–2862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.