Abstract
The early life microbiome plays important roles in host immunological and metabolic development. Because the incidence of type 1 diabetes (T1D) has been increasing substantially in recent decades, we hypothesized that early-life antibiotic use alters gut microbiota, which predisposes to disease. Using non-obese diabetic mice that are genetically susceptible to T1D, we examined the effects of exposure to either continuous low-dose antibiotics or pulsed therapeutic antibiotics (PAT) early in life, mimicking childhood exposures. We found that in mice receiving PAT, T1D incidence was significantly higher, and microbial community composition and structure differed compared with controls. In pre-diabetic male PAT mice, the intestinal lamina propria had lower Th17 and Treg proportions and intestinal SAA expression than in controls, suggesting key roles in transducing the altered microbiota signals. PAT affected microbial lipid metabolism and host cholesterol biosynthetic gene expression. These findings show that early-life antibiotic treatments alter the gut microbiota and its metabolic capacities, intestinal gene expression and T-cell populations, accelerating T1D onset in non-obese diabetic mice.
Type 1 diabetes (T1D), an autoimmune disease characterized by pancreatic β-cell destruction, is increasing in incidence worldwide1, with decreasing age of onset2, suggesting that early-life environmental exposures are critical. Early-life interactions between hosts and their microbiota, key for immunological development, may have lasting effects3. Epidemiological studies support a ‘window of opportunity’: microbial exposures during pregnancy/early childhood may protect against developing allergic disorders4, whereas caesarean section and early-life antibiotic use, perturbing the microbiome, may have opposite effects5,6, consistent with decreased gut microbial biodiversity with human socioeconomic development7 and in children at high risk for T1D who develop diabetes-specific autoantibodies8. Epidemiological studies have associated prior antibiotic exposure with increased risk for T1D and type 2 diabetes (T2D)9.
In non-obese diabetic (NOD) mice that spontaneously develop autoimmune (T1D-like) diabetes, microbial exposures affect diabetes incidence, with increased exposures (‘dirty’ conditions) being protective10,11. Conversely, T1D incidence is higher in NOD mice reared in specific-pathogen-free facilities than in those raised conventionally12. Although Myd88−/− NOD mice have reduced T1D incidence, germ-free (GF) status or broad-spectrum antibiotic treatment from birth restores diabetes development11, indicating the interplay between microbiota and immunity. In C57BL/6 mice, antibiotic-induced early-life microbial perturbations alter metabolic phenotypes13–15, and recent NOD mouse studies show strong antibiotic effects on T1D development16–18. We now examine the effects of sub-therapeutic continuous (STAT) or therapeutic-dose pulsed (PAT) antibiotics on immune development and T1D in NOD mice. We propose that PAT alters the developing microbiome, affecting early-life immune responses, tipping the balance toward T1D.
Results
PAT-accelerated T1D and insulitis development
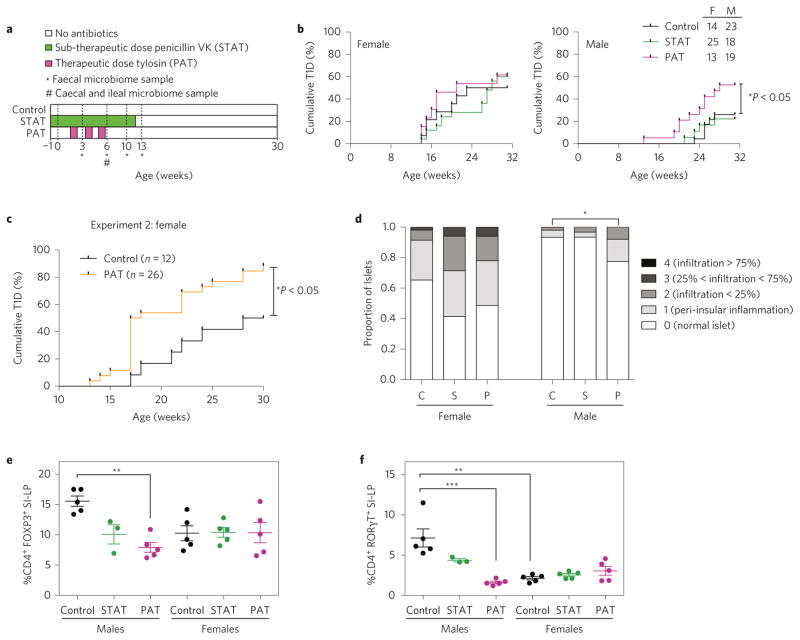
To determine whether antibiotic use affects T1D incidence, NOD mice were exposed to STAT, PAT or no antibiotics (control) (Fig. 1a). As expected12, control females developed higher T1D incidence than males, but among males, PAT mice had higher T1D incidence (53%) than controls (26%) (P < 0.05) (Fig. 1b). No significant effects were seen in PAT females or in the STAT groups. In a later cohort of females only (males were not studied) in a separate facility (experiment 2; Fig. 1c), T1D incidence was also significantly higher in PAT than in controls. With experiments providing evidence that PAT promotes T1D development, we focused subsequent analyses on the first experiment with the male-specific phenotype; we did not formally compare STAT and PAT.
Figure 1. Effect of PAT/STAT on the development of T1D and insulitis severity in NOD mice.
a, Schematic of NOD PAT/STAT study design. Pregnant NOD/ShiLTj mice were randomized into three groups: control, STAT and PAT, and maintained and treated as described in the Methods. At 6 weeks of age, mice were killed for immune-phenotyping by flow cytometry, and ileal gene expression analysis by microarray and qPCR. From 10 to 31 weeks of age, mice were tested weekly for diabetes by blood glucose measurement. *Weeks at which faecal samples were sequenced. #Timepoint at which a subset of mice were killed for collection of ileal and caecal samples for sequencing. b, T1D incidence in female and male NOD mice. Diabetes incidence curves were compared using the log-rank test (*P < 0.05). c, T1D incidence in control and PAT female NOD mice housed at a different animal facility (experiment 2). NOD dams and their litters were randomized into either control or PAT groups and treated and maintained as described in the Methods. Statistical significance was determined by the log-rank test. d, Pancreatic insulitis in female and male NOD mice. Upon necropsy, pancreata were preserved, stained, and insulitis was scored and compared in control (C), STAT (S) and PAT (P) mice, as described in the Methods. *P < 0.05. e,f, Proportions of CD4+ FOXP3+ T-regulatory (Treg) cells and CD4+ RORγT+ Th17 cells in the small intestinal lamina propria (SI-LP) in 6-week-old mice. CD3 and CD4 surface stains and a viability stain were used to gate on live Thelper cells. Nuclear staining for FOXP3 and RORγT was performed to identify Treg and Th17 cells, respectively. Cells were gated sequentially on singlets, total lymphocytes, live cells, CD3+, CD4+ cells and finally on the nuclear stains. For all groups, n = 5, except for STAT males, n = 3. Quantitation of the proportion of Treg cells (e) or Th17 cells (f) in the SI-LP. Statistical analysis was performed using the Kruskal–Wallis test followed by Dunn’s multiple comparison test. Error bars represent the mean + standard error of the mean (s.e.m.) for the biological replicates. **P < 0.01, ***P < 0.001.
Assessing insulitis in 6-week-old mice, 35% of pancreatic islets in control females had inflammation, versus 7% in males (Fig. 1d). Among females, there were no significant insulitis differences across treatment groups, but male PAT mice had significantly higher scores than controls. In total, the 6-week-old insulitis scores were consistent with the treatment and sex-specific differential T1D development rates.
Antibiotic treatment alters T-cell subtypes in the small intestinal lamina propria (SI-LP)
Examining the effects on immunological development, we focused on Treg and Th17 cells19,20 in the spleen and SI-LP of 6-week-old mice. Splenic Treg and Th17 cell proportions did not significantly differ between treatment groups for either sex. In the control SI-LP, male mice had higher Th17 proportions (P < 0.01) than females, and trended towards higher numbers of Treg cells. PAT male, but not female, mice had reduced SI-LP Treg and Th17 cell proportions compared to controls (Fig. 1e,f and Supplementary Fig. 1a,b). These T-helper population changes are consistent with the PAT-enhanced T1D incidence in males. In STAT males, Treg and Th17 cell proportions were intermediate between controls and PAT.
PAT alters ileal gene expression
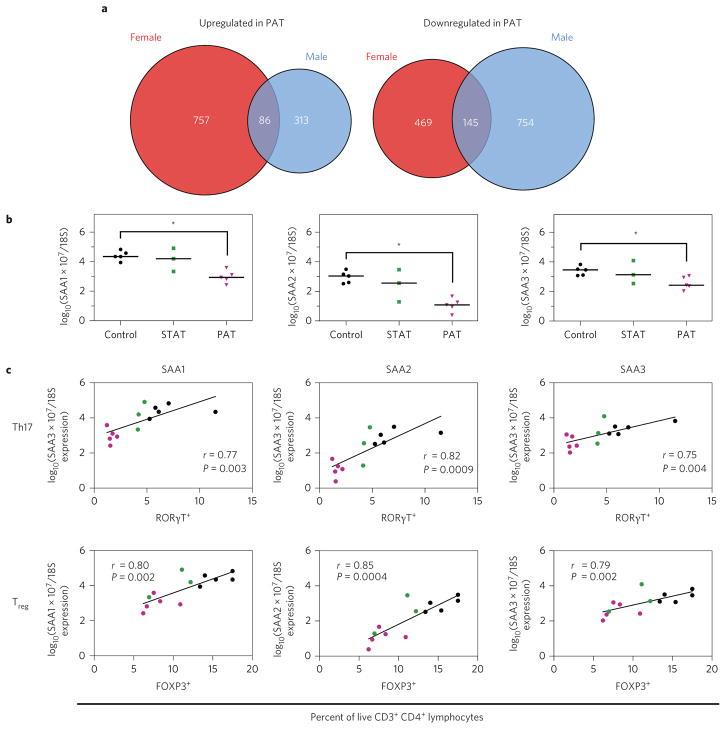
Next, we hypothesized that the antibiotic-altered gut microbiota mediated systemic effects via altered ileal gene expression. After false discovery rate (FDR) correction, no genes were significantly altered in STAT compared with controls, versus >1,000 in PAT (Supplementary Table 1). Despite the identical antibiotic treatment, the proportions of up- and downregulated genes in male and female PAT mice compared to their controls differed significantly (χ2 < 0.0001). Most differentially regulated genes were sex-specific (Fig. 2a), consistent with recent findings21. Of the male-specific gene functionality, sterol, lipid, steroid and cholesterol metabolic processes were the most significantly upregulated (Supplementary Tables 2 and 3).
Figure 2. PAT alters ileal gene expression.
The microarray experiment was performed on RNA extracted from the terminal ileum of biological replicate control, STAT and PAT mice (female, n = 3 per group; male, n = 5 per group; except STAT, n = 3). a, Comparison of PAT-mediated ileal gene expression changes in male and female mice. Venn diagram of number of genes upregulated (left) and downregulated (right) in PAT samples compared with controls. b, Expression of SAA1, SAA2 and SAA3 in male control, STAT and PAT mice. RT-qPCR was used to verify the expression of serum amyloid A1 (SAA1), 2 (SAA2) and 3 (SAA3). Expression values were determined using known standards and normalized to 18S rRNA expression. Statistical significance was determined using the Kruskal–Wallis test followed by Dunn’s multiple comparison test. The horizontal bar represents the median. *P < 0.05. c, Correlation of SAA1, SAA2 and SAA3 expression with the proportion of Th17 and Treg cells determined by flow cytometry. r and P values were determined using non-parametric Spearman correlations.
Focusing on the differentially expressed genes in the PAT males compared with their controls (Supplementary Table 1), the serum amyloid A (SAA) genes were among the most downregulated. quantitative reverse transcription PCR (RT-qPCR) analyses confirmed reduced SAA 1, 2 and 3 expression (Fig. 2b); all were significantly correlated with SI-LP Th17 and Treg proportions (Fig. 2c), consistent with the role of SAAs as immune mediators20. Segmented filamentous bacteria (SFB) induce SAA expression20; that no control and PAT males were SFB-colonized (Supplementary Fig. 1c) indicates that the observed differences are SFB-independent.
The microarray identified other genes significantly impacted by the PAT exposure, including those involved in innate immunity (for example, amphiregulin (upregulated)) and in adaptive (CD8) immunity (granzyme A and B (downregulated)). RAB30, the most highly induced (19-fold) gene in PAT males participates in early Th1 and Th2 cell differentiation22. PAT exposure significantly affected T-cell function pathways including those unique to males or females, or shared (FDR-corrected P < 0.05) (Supplementary Table 4).
PAT upregulates cholesterol biosynthesis and altered metabolomic profiles
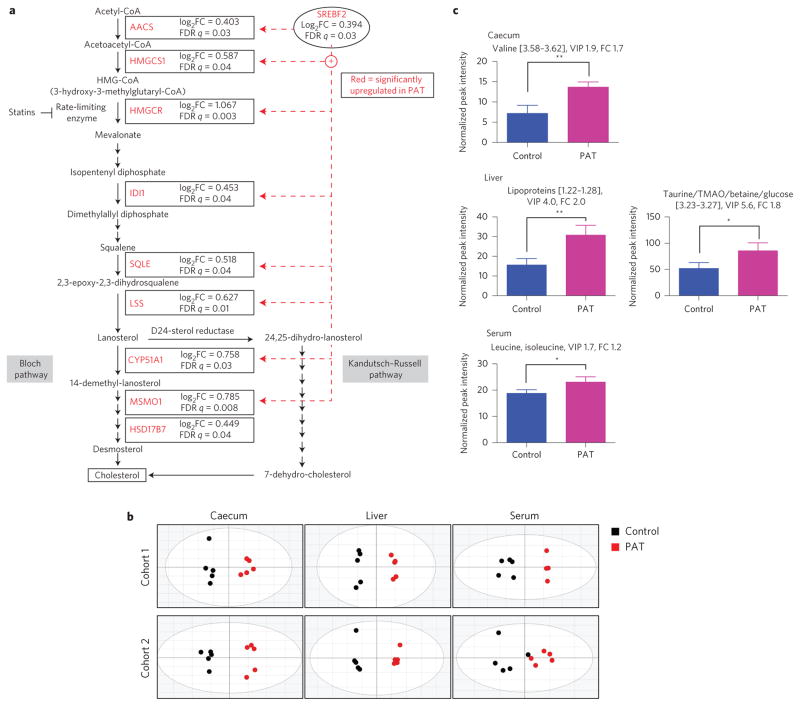
Gene expression in the cholesterol biosynthetic superpathway was significantly increased in PAT male ilea compared with controls (P < 0.0001) (Fig. 3a). The major transcription factor Srebf2, which controls cholesterol biosynthetic gene expression and pathway genes including the rate-limiting Hmgcr, was upregulated in PAT. Metabolomic analysis of caecal, hepatic and serum samples from two 6-week-old cohorts consistently separated PAT from control in all three matrices (Fig. 3b). The differences reflected numerous small changes; however, across experiments, caecal valine, hepatic lipoproteins, taurine, trimethylamine N-oxide (TMAO) and betaine, and gut-microbiota-regulated23 serum leucine and isoleucine (Supplementary Table 5), were consistently increased (Fig. 3c).
Figure 3. PAT alters metabolic gene expression pathways as well as metabolite composition in caecum, liver and serum.
Analyses were performed on 6-week-old male control and PAT NOD mice (biological replicates). a, PAT increases the expression of ileal genes involved in cholesterol biosynthesis. Microarray analysis as described in Fig. 2. Ingenuity pathway analysis was performed to identify differentially regulated gene pathways. Acetoacetyl-CoA synthetase, AACS; HMG-CoA synthase 1, HMGCS1; HMG-CoA reductase, HMGCR; isopentenyl diphosphate isomerase 1, IDI1; squalene epoxidase, SQLE; lanosterol synthase, LSS; lanosterol 14 α-demethylase, CYP51A1; methylsterol monooxygenase 1, MSMO1; 17-β-hydroxysteroid dehydrogenase, HSD17B7. b, Metabolomic composition in caecal, liver and serum samples of control and PAT mice. Score plots of OPLS-DA analysis of caecal, liver and serum samples show separation of PAT (black circles, left) from control mice (red circles, right). The statistics for the model fits of the OPLS-DA: caecal samples: [Cohort 1, R2X = 0.912, R2Y = 0.968, Q2 = 0.892; Cohort 2, R2X = 0.952, R2Y = 0.972, Q2 = 0.906]; liver samples: [Cohort 1, R2X = 0.859, R2Y = 0.992, Q2 = 0.956; Cohort 2, R2X = 0.886, R2Y = 0.991, Q2 = 0.891]; serum samples: [Cohort 1, R2X = 0.734, R2Y = 0.953, Q2 = 0.776; Cohort 2, R2X = 0.732, R2Y = 0.758, Q2 = 0.120]. c, Select differential metabolites (using NMR bin intensities) in caecum, liver and serum of control and PAT mice. Error bars represent mean + s.d.
Antibiotics alter intestinal microbial communities
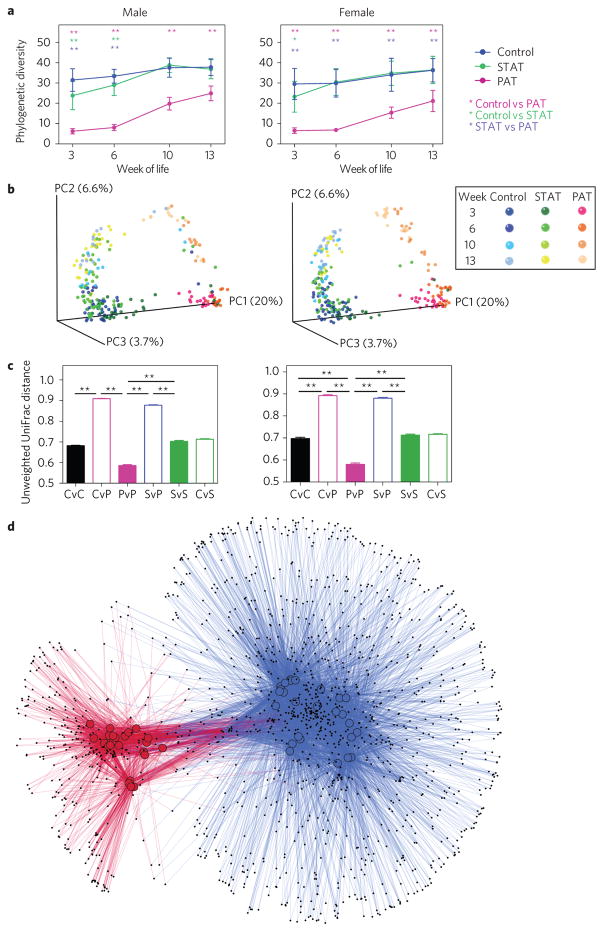
To determine the antibiotic effects on the intestinal microbiota, we examined 16S rRNA genes. At all timepoints, all sample types and in both males and females, the phylogenetic diversity in PAT was lower than in controls (FDR-corrected P values <0.05) (Fig. 4a and Supplementary Fig. 2a). By 13 weeks of age, seven weeks after antibiotic cessation, richness partially recovered. For the STAT mice, richness was intermediate early, but recovered after cessation.
Figure 4. Microbiota characteristics of control, STAT and PAT NOD mice.
a, Phylogenic diversity (PD) over time. Samples (n = 555) were rarefied to a depth of 4,150, which only excluded five (<1%) samples with lower coverage from the analysis. PD from biological replicate female and male mice for four faecal time points is presented. Statistical significance was determined using the non-parametric t-test with 999 Monte Carlo permutations, followed by Bonferroni’s correction for multiple comparisons. Error bars represent mean + s.e.m., *P < 0.05, **P < 0.01. b, Principal coordinates analysis (PCoA) of unweighted UniFrac distances. PCoA of unweighted UniFrac distances was performed to visualize differences in community structure at sequence depth >4,150. Each point represents the microbiota in a single sample: controls (blue), STAT (green) and PAT (pink). All faecal sample timepoints are displayed together and colour-coded in shades of blue, green or pink as indicated in the key. c, β-diversity, as measured by unweighted UniFrac analysis of samples from control (C), STAT- (S) and PAT- (P) treated mice. Statistical significance of the inter- and intra-group β-diversity was determined by permutation testing. The P value was calculated using the fraction of the random permutations of 10,000 that yielded a greater difference than in the one observed (t-statistic), followed by Bonferroni’s correction for multiple comparisons. Error bars represent mean + s.e.m., **P < 0.01. d, OTU co-occurrence network. Cytoscape was used to visualize sample-OTU co-occurrence patterns for male PAT and control mice at 6 weeks. In the network, samples were coloured by treatment group (control samples, blue; PAT samples, red). Individual OTUs are represented by small black nodes. Lines connecting a sample to an OTU, coloured by treatment group, indicate that that particular OTU was observed in that specific sample, forming the network.
At all timepoints in both sexes, control and STAT faecal samples showed substantial overlap in community structure (β-diversity), but the PAT were distinct. Communities in all samples progressively matured, with PAT converging towards controls (Fig. 4b and Supplementary Table 6). In both control and STAT mice, community structure intragroup diversities were stable up to week 13, but were reduced early in life in PAT mice, recovering later (Fig. 4c and Supplementary Fig. 2b). At all timepoints and for both sexes, pairwise intergroup distances between control and PAT were significantly higher than the intragroup variation (Fig. 4c and Supplementary Fig. 2b). In total, these results indicate that PAT selected for distinctive microbial community structure, persisting weeks after antibiotic cessation, before T1D onset.
Using operational taxonomic unit (OTU) co-occurrence network analysis of faecal samples from 6-week-old males, PAT deviated substantially from controls, with fewer constituent OTUs and relatively little overlap (Fig. 4d), and lower diversity scores (P < 0.0001; Supplementary Fig. 2c,d). Hierarchical clustering of samples from 6-week-old males showed deep branching separating PAT from the intermixed control and STAT samples (Supplementary Fig. 3a). Although minor branches correspond to litters (Supplementary Fig. 3b), each treatment group had ≥5 litters, reducing individual cage effects.
PAT effects on taxa abundances
Male and female control (and STAT) samples had similar, but not identical, compositions over time (Supplementary Fig. 3c). Notably, the 3-week-old PAT males had nearly complete caecal and ileal loss of Bacteroidetes and Actinobacteria, and increased Proteobacteria and Akkermansia mucinophila (Supplementary Figs 3c and 4a). Akkermansia relative abundance progressively decreased in PAT samples but remained significantly greater than in controls. The S24-7 family (Bacteroidales) was decreased in early PAT samples, recovering after antibiotic cessation (Supplementary Fig. 4a).
Using LDA effect size (LefSe) to determine differentially enriched taxa, at week 3, males and females were similar, as expected, because pups were co-housed and pre-pubertal. At weeks 6–13, particular taxa (including genus Bifidobacterium) were over-represented in control male faeces compared to PAT, and in control caecal and ileal samples at 6 weeks (Supplementary Figs 4b, 5 and 6). In contrast, Akkermansia was enriched in PAT males at all times (Supplementary Figs 4b, 5 and 6).
Three defined Bifidobacterium species were detected: adolescentis, animalis and pseudolongum, with abundances paralleling the composite genus Bifidobacterium (Supplementary Figs 4c and 7a). Ileal Bifidobacterium spp. including unnamed Bifidobacterium, which were present at higher relative abundances in control males than the named species, were much more abundant in males than females (Supplementary Fig. 7a,b). Bifidobacterium genus-specific qPCR correlated highly with 16S sequencing abundances (Supplementary Figs 4d and 7c). In mice that developed diabetes, Bifidobacterium levels determined by qPCR directly correlated with age of disease onset (Supplementary Fig. 4e).
Predictive microbial models for T1D
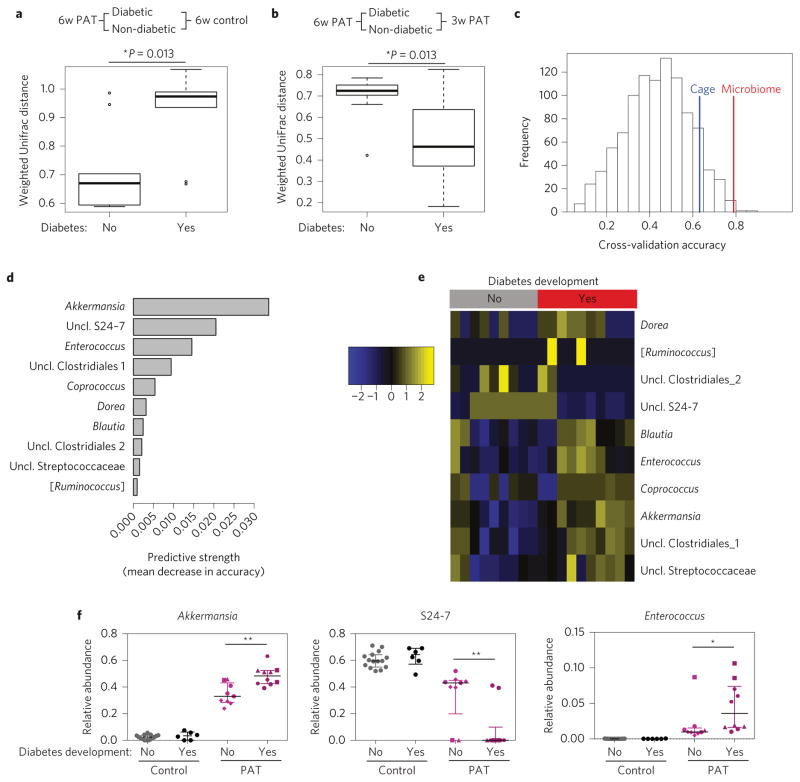
The extent of microbiota perturbation at 3-week-old PAT mice relative to controls was similar in those ultimately becoming diabetic or not (P = 0.11). By 6 weeks, mice that ultimately developed diabetes were more compositionally different from same-age controls than those not developing diabetes (P = 0.013, Fig. 5a). Faecal samples from 6-week-old PAT mice that later developed diabetes were more similar to their 3-week populations than mice that did not (P = 0.013, Fig. 5b). A predictive model for T1D development in the 6-week-old PAT males (Fig. 5c) was significantly better than random, or than a model based on cage membership. The ten most predictive genera include Akkermansia, S24-7, Enterococcus and Dorea (Fig. 5d,e). In PAT males that developed diabetes, S24-7 was lower and Akkermansia and Enterococcus were higher than in those that did not develop diabetes (Fig. 5f). In experiment 2, the relative abundances of Akkermansia and Enterococcus were increased and S24-7 reduced in PAT females (Supplementary Fig. 7d), paralleling experiment 1 diabetes development trends (Supplementary Fig. 7e).
Figure 5. Microbiota perturbation and composition in relation to the development of T1D.
a, Pairwise weighted UniFrac distances of faecal samples from 6 week control versus PAT mice that developed diabetes or not. Boxes represent median and interquartile range (IQR) and whiskers represent 1.5 IQR intervals. b, Pairwise intragroup weighted UniFrac distances of faecal samples from PAT mice at 3 (3w PAT) and 6 weeks (6w PAT) that developed diabetes or not. Boxes and whiskers are as in a. c, Histogram of null distribution of classifier accuracy levels. Use of random forests analysis for classification of taxa in faecal samples from 6-week-old males, associated with protection against diabetes development. The histogram shows the expected distribution of accuracy levels from classifiers predicting diabetes onset, if the microbiome contained no useful information. To generate this null distribution, we used bacterial genera as the predictors and randomized class labels as the outcome to be predicted. Vertical lines indicate the observed accuracy of the classifier compared with true outcomes, when trained on bacterial genera (red line) or on cage membership alone (blue line). The area on the tail to the right of each vertical line represents the reported P value corresponding to the test that that classifier is better than random. In this analysis, the P value for microbiome (P = 0.017) is smaller than for cage (P = 0.149), indicating that the microbiome provides significant predictive information. d, Feature importance scores for the ten most predictive genera in the random forests classifier. Feature importance was measured as the mean decrease in model accuracy when that feature’s values were permuted randomly. e, Heatmap of predictive taxa in the PAT samples. The heat map is coloured by the relative abundance (standardized to have mean 0 and variance 1 for each identified taxon) of the ten bacterial genera most highly predictive of diabetes onset. Samples were first stratified (by diabetes development, columns) and hierarchical clustering was performed within each outcome group. f, Relative abundances for three discriminative taxa: Akkermansia, S24-7 and Enterococcus. Different symbols for the PAT samples represent different cages. The Mann–Whitney U test was used to determine significance, comparing those that develop diabetes and those that do not. Error bars represent median + IQR. *P < 0.05, **P < 0.01.
PAT alters predicted intestinal metagenomic pathways
Predicting metagenomic content in faecal specimens using PICRUSt, we identified KEGG level 3 pathways enriched in 6-week-old PAT males (Supplementary Fig. 8a). Biosynthesis of total and unsaturated fatty acids and branched chain amino acid (BCAA) metabolism was significantly increased in PAT samples at all timepoints (Supplementary Fig. 8b–d). PAT significantly decreased the relative abundance of genes involved in metabolizing butanoate, with reciprocal changes for propionate (Supplementary Fig. 8e,f). By metabolomic analysis, caecal butyrate was found to be lower in PAT males, consistent with the PICRUSt findings (Supplementary Fig. 8g). In summary, PAT significantly affected intestinal metagenomic content, with both transient and long-lasting changes after antibiotic cessation.
Microbiota transfer to germ-free mice
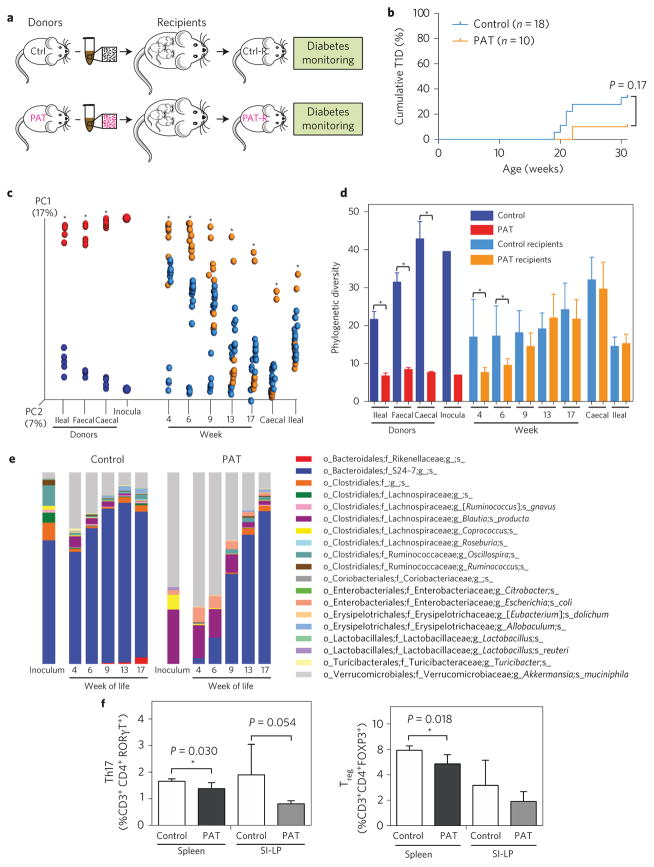
We transferred caecal contents from 6-week-old male control or PAT mice (donor pools) to pregnant germ-free NOD recipients via oral gavage and studied their male pups (Fig. 6a). For pups receiving the control inoculum via their mothers, T1D development was as expected23, but for pups receiving the PAT inoculum, T1D development was not increased (Fig. 6b). As expected, the microbiota of recipient pups resembled that of their respective donors; however, the recipient communities overlapped (Fig. 6c). For the PAT recipients, early α-diversity was lower than for control recipients; however, the groups converged by week 9 (Fig. 6d). At early timepoints, the PAT recipient pups had higher Akkermansia, Blautia and Escherichia coli and lower S24-7 relative abundances compared with controls (Fig. 6e); thus, transfer of caecal contents from 6-week-old PAT mice to pregnant germ-free mice conventionalized pups with microbiota enriched in opportunistic organisms.
Figure 6. Inoculation of germ-free NOD mice with control or PAT microbiota.
a, Microbiota transfer experimental design. Donor mice: controls (Ctrl) were maintained on non-acidified sterilized water; PAT mice were given three courses of tylosin, as described. At 6 weeks of age, control and PAT donor male mice (n = 5 each) were euthanized, and caecal contents were collected and separately pooled (control and PAT inoculum, respectively), then transferred into NOD germ-free pregnant dams by oral gavage. The male pups of these dams were designated the control and PAT recipient mice (Ctrl-R, n = 18; PAT-R, n = 10) and monitored for diabetes development. For microbiome analysis, faecal samples were collected from the biological replicate recipient mice between 4 and 17 weeks of life, and caecal and ileal samples at sacrifice (30 weeks). b, Diabetes incidence in germ-free NOD recipients of control or PAT caecal microbiota. Mice were tested for diabetes weekly as above: 33% (6/18) of the control recipients developed diabetes versus 10% (1/10) of the PAT recipients (P = 0.17, log-rank test). c, Intestinal microbial composition of control and PAT donors and recipients. PCoA was performed on unweighted UniFrac distances to visualize community structure differences. Each point represents the microbiota in a single sample, coloured by group: donors, control samples (dark blue), PAT samples (red); recipients, control samples (light blue), PAT samples (orange). Statistical significance was determined using the Adonis test, *P < 0.05. d, Phylogenetic diversity of control and PAT donors and recipients. Statistical significance was determined using the non-parametric t-test with 999 Monte Carlo permutations, *P < 0.05. e, Taxa relative abundances in donor inocula and control and PAT recipients over time. f, Transfer of caecal microbiota from NOD mice at 17 days of life (two days after a single PAT or control exposure) to germ-free C57BL/6 recipients. Graphs show proportions of CD4+ FOXP3+ Treg cells and CD4+ RORγT+ Th17 cells in the spleens or SI-LP in biological replicate recipient mice 6 weeks after transfer, using flow cytometry, as indicated in Fig. 1. For both PAT and control groups, n = 4. Statistical analysis of the a priori hypothesis was performed using the one-tailed t-test. Error bars represent mean + s.d.
We performed a second transfer experiment to examine immune phenotypes in recipient germ-free animals, using C57BL/6 mice because they develop immunological abnormalities with early-life antibiotic exposure14 and because prior studies have shown that conventionalization across mouse strain types yields the expected phenotypes14. In this experiment, caecal contents from four (three female and one male) NOD mice at day of life (dol) 17 that had received only a single course of PAT from dol 10–15 (or were untreated controls) were transferred to 3-week-old germ-free C57BL/6 mice. Recipient mice were killed 6 weeks after conventionalization, and splenic and SI-LP lymphocytes were studied (Fig. 6f). In each of the four comparisons, the PAT microbiota recipients showed lower Treg and Th17 cell proportions than the recipients of the control microbiota; although the numbers were very small, the results in male and female recipients were similar (Supplementary Fig. 9). This experiment provides preliminary evidence that early-in-life antibiotic-altered microbiota are sufficient to confer the altered immune phenotypes.
Discussion
We have shown that when initiated early in life in NOD mice, therapeutic antibiotic pulses (PAT), based on the doses (and pharmacokinetics) used in young children, can increase autoimmune diabetes (T1D) development and worsen its histopathological hallmark, insulitis. The macrolide-based PAT15 model had consistently greater effects than the β-lactam-based STAT model13,14 on intestinal microbiota, ileal gene expression, T-helper subtypes and T1D development, but we are not comparing them directly because they differ in both dose and antibiotic class. The PAT model is clinically relevant, because US children receive approximately one antibiotic course annually, frequently macrolides24. Previous T1D studies involving antibiotic exposures, differing in design, yielded inconsistent results11,25,26. Although multiple T1D-protective factors are known12, fewer disease-accelerating factors have been identified in NOD mice. However, three recent studies16–18 have shown that antibiotic regimens severely disrupting the microbiota enhance T1D in NOD mice. Exposure to the STAT regimen13,14 produced few immunological effects, consistent with the reduced intensity of antibiotic exposure and microbiome effects; therapeutic antibiotic doses have more profound and long-lasting effects15.
In our first experiment, PAT increased T1D development in males but not females. Because T1D rates are lower in NOD males than in females10,21, our findings suggest that males may have protective microbiota that are eliminated by PAT; this is consistent with the finding that transfer of adult male microbiota to female NOD mice conferred T1D protection21. Future experiments could test this directly, by including PAT exposure before the transfer. However, among female mice bred elsewhere, PAT also promoted T1D, illustrating facility-dependent differences10 and showing that the antibiotic effects were not male-restricted. Despite environmental differences, this work provides evidence that early-in-life PAT exposures accelerate T1D development in NOD mice.
By studying tissues of 6-week-old mice, we examined events in the intestine and pancreas before T1D development. We focused on the ileum because microbial colonization is more variable than in the colon, with multiple genes being inducible27. The PAT-mediated decreased proportions of intestinal FOXP3+ Treg and RORγt+ Th17 cells in male, but not female, mice were consistent with the experiment 1 sex-specific T1D incidence differences. Treg cell imbalances may promote autoimmune diseases, including T1D28, with defective small intestinal FOXP3+ T-cell differentiation29. Although Th17 cells contribute to other autoimmune diseases30, both protection and pathogenicity have been reported for T1D30,31. In a recent study, intensive antibiotic exposure reduced ileal and colonic lamina propria Th17 populations and increased diabetes development16. SI-LP Th17 cell differentiation induced by SFB and Lactobacillus johnsonii has been associated with T1D protection32,33, consistent with our inverse finding of reduced Th17 cells with T1D acceleration. Because Th17 cells promote intestinal barrier function34, their diminution may lead to defective barrier function in T1D patients35.
Although partially recovered by 13 weeks, the PAT-exposed microbiota remained reduced in α-diversity and compositionally distinct from control mice, paralleling observations in human children with T1D36 and autoantibodies before onset8. Our analyses identified taxa that may modulate T1D development. Bifidobacterium was enriched in the relatively protected control males, but essentially absent in all three relatively at-risk groups. In humans and other mammals, more than 30 Bifidobacterium species have been identified37, and broad conservation suggests functional importance. Bifidobacterium species dominate the gut microbiota of healthy infants38, especially when breast-fed38, consistent with Bifidobacterium digesting breast milk oligosaccharides. Both breast-feeding39 and vaginal birth40 protect against T1D development, and B. adolescentis levels inversely correlate with diabetes-associated autoantibody prevalence41. Bifidobacterium induction of Treg cells and REGIIIγ secretion correlates with their reduced levels in PAT males. Thus, both epidemiological and mechanistic studies suggest potential Bifidobacterium protection against T1D. The less well-known S24-7 family42 was associated with decreased T1D development in both experiments, warranting further investigation. S24-7 was the most highly over-represented taxon in uncontacted Amerindians compared to US subjects7, suggesting its continuing disappearance with modernization. Conversely, informatic analyses identified Akkermansia and Enterococcus as significantly T1D-accelerating in PAT, consistent with the concept that ‘dirty’ protects in NOD mice12. These studies also indicate both microbial population-specific and immune-specific differences between male and female mice that might explain the sex differences in T1D incidence.
We found significant differences between control and PAT males in microbiome-predicted functions, affecting carbohydrate, lipid and amino-acid metabolism. Increased representation of BCAA metabolism genes in the PAT metagenome as well as increased serum levels of leucine and isoleucine is consistent with higher BCAA serum levels in mice43 and in humans who progress T1D44. The choline metabolites, TMAO and betaine, derived from gut microbiota, have associations with atherosclerosis45. Previous work has suggested that the microbiome can alter lipid metabolism46. Here, PAT significantly increased the representation of pro-inflammatory free fatty acid lipids and increased the expression of cholesterol biosynthesis in the ileum, a major inducible site of cholesterol biosynthesis. Cholesterol enhances inflammation47 and inhibition of their synthesis by statins, HMG-CoA reductase inhibitors, is anti-inflammatory and counters autoimmunity47. In total, the increased ileal cholesterol synthesis and metabolites in PAT may promote inflammation.
In addition to global PAT effects, we observed decreased caecal 48 and consistent butyrate, known to induce Treg cell differentiation with the reduced intestinal Treg cells in PAT. The parallel reduction in predicted butanoate metabolism we observed is consistent with reduced butyrate-producing bacteria in T1D children49. NOD progressors to T1D had elevated circulating lipids43, consistent with our observation of increased liver-specific and serum-detectable lipid species in PAT mice. In total, the metabolic characteristics we observed in the PAT mice are consistent with findings in both humans and in NOD models for accelerated T1D. Nevertheless, these changes are only associations, and causal relationships must be established.
In our agnostic approach to identify ileal genes affected by PAT, the SAAs, acute phase reactants produced by intestinal epithelial cells that also promote Th17 cell differentiation20 were strongly differentiating. The consistent decreases in PAT of SAA1, SAA2 and SAA3 ileal expression suggest reductions in stimulatory bacterial species, consortia and/or metabolites50. Because our mice lack the stimulatory SFB20, other bacteria must be relevant to this pathway. We propose that intestinal SAA proteins are important innate effectors connecting the microbiota to adaptive immunity. We speculate that PAT reduces antigenic stimulation during a critical window in immunological development, leading to reduced intestinal immune effectors, including innate molecules such as SAAs, antimicrobial peptides (REGIIIγ) and altering particular T-helper populations (for example, via RAB30)22, with important effector functions. Such changes could reduce intestinal barrier function, allowing translocation of luminal bacteria or products promoting systemic inflammation. Barrier function loss, suspected in human T1D and in animal models35, has been related to microbiota composition and presumed functions49.
To isolate the effects of the microbiota from those directly mediated through antibiotics, we transferred caecal contents from either PAT or control mice to germ-free recipient pregnant NOD dams, so that pups would be exposed from birth. We did not observe accelerated T1D in the PAT caecal-content recipients, but three non-exclusive hypotheses might be operant. First, conventionalizing pregnant dams may have direct immunological effects. Second, the initial microbial selection occurred in the mothers, affecting the inocula the pups actually received. Such phenomena could explain the observed intermediate microbial state in the pups, regardless of donor source, and are consistent withT1D protection in offspring of NOD mothers treated during gestation17. A third explanation is that >5 weeks after PAT initiation, opportunistic bacteria had already bloomed in the PAT mice, providing a compromised inoculum derived after the critical immunological window. The trend to protection in the recipient mice is consistent with earlier observations that ‘dirty protects’, specifically Akkermansia25, and with our informatic findings in PAT mice without transfer (Fig. 5f). Performing caecal transfers from younger mice (dol 17), immediately after antibiotic exposure may be more germane to the pathogenesis of PAT-induced accelerated T1D. Our experiment, although across mouse strains and of small size (Fig. 6f), provides preliminary evidence supporting this point. Previously, antibiotic-altered microbiota transferred diminished Th17 phenotypes across strain lines (C57BL/6 to Swiss Webster)14. Together, these findings suggest that despite the genetic differences between the tested mouse strains, the antibiotic-altered microbiota might differentially signal immunological effectors, providing preliminary evidence that early-life microbiome changes due to PAT may be sufficient for the T1D effect.
In conclusion, this work shows that early-in-life antibiotic exposure in NOD mice can accelerate T1D development, modifying the disease. This study, along with other recent investigations16–18, raises questions regarding widespread antibiotic use in early childhood24. However, in other studies, antibiotics were found to have the opposite effects24,25; thus, antibiotic type, dose and timing are all relevant variables that might influence effects. Environmental changes, independent of antibiotic-induced effects, could also contribute to increasing T1D incidence in human children1,2. Roles for altered environmental exposures and microbiota need not be exclusive. We identify potentially important intestinal bacterial taxa affected by antibiotic exposures and downstream immune and metabolic pathways. Additional targeted metabolomic and metagenomic analyses are warranted to extend these findings. Further studies will help understand generalizability, pathway independence or synergy, and whether such pathways can be modified to prevent or delay T1D development.
Methods
Mice
For experiment 1, male and female 8- to 10-week-old NOD/Shiltj mice (no. 001976, Jackson Labs) were bred in an SPF vivarium at New York University Langone Medical Center (NYULMC) to obtain sufficient litters. The pups used for experiments were born in the first litter of each dam and were randomly assigned to each of the three experimental groups. The litters were not intermixed following weaning, and this determined the cages in which post-weaning mice were housed. Mice were housed in autoclaved Thoren polycarbonate cages (Hazelton PA) with autoclaved bedding, and maintained on a 12 h light/dark cycle at 70 ± 2 °F, according to the guidelines of the Division of Laboratory Animal Resources (DLAR) at NYULMC. Mice were fed ad libitum using an irradiated laboratory chow (LabDiet JL Rat and Mouse/Irr 6F 5LG4). All animal procedures were approved by the NYULMC Institutional Animal Care and Use Committee (IACUC protocol no. 110711).
In experiment 2, mice were bred and housed at Memorial Sloan Kettering Cancer Center (MSKCC) animal facility. As above, NOD dams and their litters were randomly assigned to either control or PAT groups. The female pups were monitored for diabetes development and the other methods are as described for experiment 1.
Antibiotic treatments
Dams and their litters had been randomly divided into control, STAT (sub-therapeutic antibiotic treatment) and PAT (pulsed antibiotic treatment) groups (Fig. 1a). The control mice received non-acidified sterilized water (NASW) until the end of the antibiotic treatments, and for the two antibiotic groups (STAT and PAT) the antibiotics were dissolved in NASW. For STAT13,14, penicillin V potassium salt (Sigma Aldrich) was used at a concentration of 6.67 mg l–1 to a dose of ~1 mg per kg bodyweight per day (based on a mouse average consumption of 150 ml water per kg body weight51), which is a sub-therapeutic dose approved by the FDA for agriculture52. STAT was begun in late pregnancy, and then continued until day 84 of the pups’ lives. For PAT15, three pulses of therapeutic doses of the macrolide tylosin tartrate (Sigma Aldrich) were given to the mice in their drinking water, dissolved at a concentration of 333 mg l–1 to achieve a dose of about 50 mg per kg body weight per day, as routinely given as a therapeutic to rodents53. The first pulse was given on dol 10–15, so pups received the antibiotic via their mothers’ milk, and the following two pulses were given post-weaning, on dol 28–31 and 37–40.
Diabetes monitoring
By measuring blood glucose (FreeStyle Lite meter and test strips; Abbott) obtained via a tail vein nick starting at week 10 of life, all mice were monitored for diabetes onset, defined as two consecutive values >250 mg dl–1 (refs 11,54).
Histology
Pancreata from 6-week-old control, STAT and PAT mice were fixed in modified Bouin’s fixative, paraffin-embedded, cut, and stained with aldehyde fuchsin for insulitis scoring12. Each of five sections of each pancreas was 5 μm thick, with a 70 μm gap between sections. Insulitis was evaluated on a 0–4 scale (0, normal islet; 1, peri-insular leukocyte aggregates; 2, leukocyte infiltration <25%; 3, leukocyte infiltration >25%, but <75%; 4, leukocyte infiltration >75% and β-cell destruction)55. Thirty islets per mouse were scored by a board-certified veterinary pathologist (ABR) blinded to treatment groups. The distributions of insulitis severity scores in each treatment group were compared with the same-sex control group using the generalized Wald-type test56 in the HMP R package57.
Tissue collection
The distal ileum (1 cm) with Peyer’s patches removed was collected from each mouse at sacrifice for RNA extraction. The tissue was placed in RNAlater (Life Technologies) overnight at 4 °C and then after RNAlater removal, stored at −80 °C, until RNA extraction was performed.
Microarray gene expression profiling
Ileal gene expression profiling was performed using the Affymetrix Genechip system. Total RNA was extracted from mouse tissues using the RNeasy Mini Kit with on-column DNase treatment to remove contaminating gDNA (Qiagen). Total RNA quality and quantity were determined using the Nanodrop ND-1000 (Wilmington DE), agarose gel and Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA (100 ng) was used to prepare cDNA using the Affymetrix 3′IVT Express Kit labelling protocol. Standardized array processing procedures recommended by Affymetrix were performed, including hybridization, fluidics processing and scanning of the Affymetrix Mouse Genome 430 2.0 Array chips. Raw probe intensities were normalized by Robust Multi-array Average (RMA) algorithm R58 using the affy package in R and the limma package used to identify regulated genes using linear models and empirical Bayes methods with FDR cutoff of P < 0.05 (ref. 59). Gene expression pathway analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, NIH)60 Ingenuity Pathway Analysis (IPA, Qiagen)61 and clusterProfiler62.
Gene expression analysis by qPCR
From the total extracted RNA described above, 1 μg was reverse transcribed to cDNA using Super Script II (Life Technologies). To generate standards for expression analysis of each target gene, the cDNA region of interest was PCR-amplified and the product cloned into pGEM-T easy (Promega). qPCR was performed (primer sequences are provided in Supplementary Table 7) with Power SYBR Green PCR Master mix (Roche) and run in a LightCycler 480 system (Roche). Target mRNA was normalized to 18S rRNA as an internal control in each sample. For group mean comparisons, the Kruskal–Wallis test followed by Dunn’s multiple comparison test was performed, and for correlations, non-parametric Spearman methods were used.
Metabolomics
Serum and liver samples were prepared as described in ref. 63, with minor adjustments. Briefly, serum samples were thawed on ice and vortexed for 30 s. An aliquot (50 μl) of each study sample was transferred into labelled tubes. Three aliquots (50 μl each) of study-independent mouse sera (Sigma, no. S7273) were also transferred into labelled tubes for use as analytical quality control (QC) samples. Freshly prepared 0.9% saline (wt/vol) solution in deuterium oxide (D2O) was added to each tube (150 μl), with vortexing for 30 s. Freshly prepared 10 mM formate solution containing 0.2% (wt/vol) NaN3 was added to each tube (50 μl) to serve as an internal standard. Tubes were vortexed for 30 s, centrifuged at 12,000 r.c.f. for 5 min at 4 °C and a 200 μl aliquot of each supernatant was transferred into 3 mm NMR tubes (Bruker-Biospin).
Frozen liver samples were weighed (50–100 mg) into labelled homogenizer bead tubes (stainless-steel beads). Cold acetonitrile:water (1:1) was added to tissue based on weight to make 100 mg ml–1 homogenates, then samples were extracted by homogenization (Spex Geno/Grinder) for two 45 s pulses at 1,750 r.p.m., then centrifuged at 12,000 r.c.f. for 5 min, and total liver supernatants were transferred into new labelled tubes. A 500 μl aliquot (50 mg tissue equivalent) of supernatant/sample was transferred into a second set of labelled tubes for further processing. Study-independent homogenized liver supernatants were pooled, and three aliquots were transferred into labelled tubes for use as analytical QC samples. All samples were dried on a SpeedVac (Eppendorf, V-AL setting) at 30 °C and stored at −80 °C overnight. The dried liver extracts were reconstituted in 630 μl D2O. Chenomx Internal Standard solution (Chenomx ISTD) containing 5 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS, chemical shift indicator), 100 mM Imidazole (pH indicator) and 0.2% NaN3 was added, and samples were vortex-mixed on a multi-tube vortexer for 10 min at speed 5. Tubes were centrifuged at 12,000 r.c.f. for 5 min, and a 600 μl aliquot of each sample supernatant was transferred into 5 mm NMR tubes (Bruker-BioSpin).
Caecal content samples were prepared as described in ref. 64, with minor adjustments. Briefly, frozen samples were weighed into labelled homogenizer bead tubes. D2O was added (500 μl) and the samples were homogenized for two 30 s pulses at 1,750 r.p.m., centrifuged at 12,000 r.c.f. for 5 min and 450 μl supernatants transferred into 2.0 ml tubes containing 0.2 μm nylon filters and centrifuged at 16,000 r.c.f. until the homogenate was completely filtered. Study-independent homogenized caecal supernatants were pooled and filtered, and three aliquots were transferred into labelled tubes for use as analytical QC samples. A volume of filtered supernatant equal to 50 mg per sample and a calculated volume of D2O to bring the total sample volume to 630 μl was transferred into new labelled tubes. Chenomx ISTD was added (70 μl) and the tubes were vortexed for 30 s, centrifuged at 12,000 r.c.f. for 5 min and a 600 μl aliquot of each supernatant was transferred into 5 mm NMR tubes.
1H NMR spectra were acquired on a Bruker Avance III 700 MHz NMR spectrometer (Bruker-Biospin) located at the David H. Murdock Research Institute at Kannapolis (NC) using a cryogenically cooled 5 mm ATMA probe and an ambient temperature of 25 °C. For liver and caecal samples, a standard 1D pulse sequence (noesypr1d; ref. 63) of a NOESY pulse program was used with water pre-saturation during a relaxation delay of 2 s and mixing time of 100 ms. For serum samples, a CPMG pulse sequence with water pre-saturation (cpmgpr1d; ref. 63) was used. For each serum sample, 256 transients were collected into 64,000 data points using a spectral width of 19.5 kHz (20.5 ppm), 2 s relaxation delay, 400 μs fixed echo time, loop for T2 filter (l4) = 80 and an acquisition time of 2.32 s per free induction decay (FID). For each liver or caecal sample, 64 transients were collected into 64,000 data points using a spectral width of 19.5 kHz (20.5 ppm), 2 s relaxation delay and an acquisition time of 2.32 s per FID. FIDs were zero-filled by a factor of 2, and a line broadening factor of 0.5 Hz was applied before Fourier transformation. Spectra were manually phased, baseline corrected, and calibrated to DSS (δ 0) for liver and caecal samples and to formate (δ 8.43) for serum samples.
Metabolomics analysis was performed as described in refs 63 and 65–70. Briefly, following data acquisition, spectra were binned (0.50–9.0 ppm) using intelligent bucketing integration with a 0.04 ppm bucket width and a 50% looseness factor in ACD NMR Processor 12.0 (ACD Labs) software. Chemical shift regions for water (caecal: 4.70–4.90 ppm; liver: 4.70–4.85 ppm; serum: 4.7–6.35 ppm), imidazole (caecal: 7.22–7.34 ppm; liver: 7.25–7.39 ppm) and formate (8.40–8.5 in serum) were excluded from binning. Each of the bin integrals was normalized to the total integral of each spectrum. Principal component analysis (PCA) and orthogonal partial least-squares projection (OPLS-DA) of binned data were performed using pareto-scaling and mean centring, using SIMCA 13.0 (Umetrics). Variable importance for projection plots (VIP ≥1.0) were used to identify the important bins responsible for the separation of the study groups. These bins were matched to the metabolites using the metabolite library in Chenomx NMR Suite 7.7 Professional (Edmonton) software.
Isolation of SI-LP leukocytes
After euthanasia of the mice by CO2 narcosis followed by cervical dislocation, the small intestines were dissected and immediately placed in cold calcium and magnesium-free Dulbecco’s phosphate-buffered saline (dPBS), followed by removal of mesenteric fat and Peyer’s patches. The intestines were opened longitudinally and the contents were removed by washing thoroughly with cold dPBS, then cut into 2 cm pieces and treated with 1 mM dithiothreitol (Sigma Aldrich) in dPBS, for 10 min at room temperature, to remove contaminating mucus and intraepithelial lymphocytes. The intestinal tissue was then treated twice with 30 mM EDTA and 10 mM HEPES in dPBS to remove epithelial cells. To isolate lamina propria lymphocytes, intestinal tissue was incubated for 1.5 h at 37 °C in digestion mix (0.5 mg ml−1 collagenase/dispase (Roche), 150 μg ml−1 DNase 1 (Sigma Aldrich) in complete RPMI (RPMI 1640 containing 10% fetal calf serum, 2 mM L-glutamine, penicillin (100 IU ml−1), streptomycin (100 μg ml−1) and 0.05 mM 2-mercaptoethanol), then cells passed through a 100 μm diameter nylon mesh filter (BD Bioscience). For further purification of leukocytes, intestinal cell suspensions were resuspended in 40% Percoll, under-layered with 80% Percoll (GE Healthcare Life Sciences) and centrifuged at 974g for 25 min at room temperature. The leukocytes, collected at the interphase of the 40/80% discontinuous Percoll gradient, were washed and resuspended in dPBS for staining, as described, with minor modifications, in ref. 71.
Isolation of splenic leukocytes
Spleens were homogenized between frosted slides into RPMI and then passed through a 40 μm nylon mesh filter (BD Biosciences). Cells were pelleted at 300g for 5 min, supernatants were removed, followed by red blood cell lysis with ACK (ammonium-chloride-potassium) lysis buffer (Life Technologies) for 7 min at room temperature. Cells were washed with RPMI and then resuspended in dPBS for staining.
Lymphocyte staining and subset analysis
Cells were incubated with LIVE/DEAD Fixable Blue dead cell stain (Life Technologies) for 10 min at 4 °C to identify live cells. Intestinal leukocytes were phenotyped using the following antibodies: CD3-APC-Cy 7 (BD Biosciences), CD4-PE-Alexa Fluor 610 (Invitrogen), CD8-V500 (BD Biosciences), RORγt-PE (eBioscience) and FOXP3-PE-Cy7 (eBioscience). Cells were first incubated with surface antibodies (each at 1:50 in FACS buffer) along with FC block (anti-mouse CD16/CD32, eBioscience) at 1:200 for 30 min at 4 °C, then fixed and permeabilized with fixation/permeabilization buffer (eBioscience) and subsequently incubated with the nuclear antibodies in permeabilization buffer (eBioscience) for 30 min at 4 °C. All cells were acquired on an LSRII cell analyser (Becton-Dickinson) and analysed using FlowJo software (Tree Star). To determine statistical significance between groups, we used the Kruskal–Wallis test followed by Dunn’s multiple comparison test.
Microbiome assessment
Freshly voided faecal pellets were collected at weaning (day 22 of life) and then weekly for the remainder of the experiments, by placing each mouse in a plastic container in a fume hood. Between each mouse, the container was cleaned with MB-10. The pellets were placed into sterilized microcentrifuge tubes with forceps cleaned in 70% ethanol and frozen at −80 °C. Caecal and ileal samples were collected following necropsy then frozen at −80 °C; contents and tissue were collected together to allow sampling of both mucosal-associated and luminal bacteria. DNA was extracted from faecal, caecal and ileal samples using the PowerLyzer PowerSoil DNA isolation kit and the PowerSoil-htp 96 Well Soil DNA isolation kit (MoBio), and DNA concentration was quantified using the Nanodrop ND-1000. Sequencing analysis was conducted for experiment 1. The microbiota associated with the enhanced phenotype in experiment 2 have not been determined. The V4 region of the bacterial 16S rRNA gene was amplified by triplicate PCR (F515/R806) using barcoded fusion primers, as described in ref. 72. The DNA concentration of the V4 amplicons for each sample was measured using the Quant-iT PicoGreen dsDNA assay kit and the Tecan infinite plate reader or the PerkinElmer plate reader. Samples were pooled in sets with a maximum of 96 samples in equal quantities. These set pools were then purified using the Qiaquick PCR purification kit (Qiagen) to remove primers, quantified using the high-sensitivity dsDNA assay kit and the Qubit 2.0 Fluorometer (Life Technologies) and then combined at equal concentrations to form the sequencing library. Paired-end sequencing of the amplicon library was performed on the Illumina MiSeq plateform, with a 15% PhiX spike to improve the signal from a low-diversity library. The sequencing generated 151-base-pair forward and reverse reads and a 12-base-pair barcode read; reads passing the Illumina quality filter were used for downstream analysis.
Bioinformatic analysis of microbiome sequences
The forward and reverse paired-end reads were trimmed using fastq-mcf, then joined using the fastq-join function from EA-utils73. Only reads with a minimum overlap of 30 bases and with perfect matching of bases between reads were retained, then demultiplexed, filtered and analysed using QIIME74. Reads with more than three consecutive low-quality bases (Phred score <20) were truncated and retained only if the read was >75% of the original length. Open reference OTU picking was performed using the uclust method and taxonomy was assigned using the RDP classifier with a confidence interval of 50%, using the GreenGenes May 2013 database release as reference75.
Unweighted UniFrac distances and α-diversity metrics (richness and phylogenetic diversity) were calculated and principal coordinate (PCoA) and relative abundance analyses were generated using QIIME. The R vegan package Adonis test was used to determine β-diversity differences76,77. Statistical significance of the inter-and intra-group β-diversity was determined by permutation testing. The P value was calculated using the fraction of the random permutations out of 10,000 that yielded a greater difference than in the one observed (t-statistic), followed by Bonferroni’s correction for multiple comparisons. For α-diversity, statistical significance was determined using the non-parametric t-test with 999 Monte Carlo permutations at a depth of 4,000 followed by Bonferroni’s correction.
OTU co-occurrence and diversity in male control and PAT samples
To identify co-association of mouse intestinal microbiome samples with sets of taxonomic groups (OTUs), we visualized the sample–OTU co-occurrence network in Cytoscape (version 3.1.0) with edge-weighted spring-embedded layout. The input files were from the QIIME script make_otu_network.py performed on male control and PAT 6-week-old faecal samples.
Determination of differential taxa
To determine significantly different taxa between sexes and treatment groups, the linear discriminate analysis (LDA) effect size (LEfSe) algorithm on the Galaxy browser was used78. LEfSe first performs the non-parametric Kruskal–Wallis test to determine taxa that were significantly different in abundances and then uses LDA to determine the effect size. Taxa were considered significantly different if the P value for the factorial Kruskal–Wallis sum-rank test was <0.05 and the logarithmic LDA score was >2.0.
Bifidobacterium quantitation by qPCR
Bifidobacterium species were quantified in faecal samples using genus-specific primers as described previously (F 5′ CTCCTGGAAACGGGTGG 3′; R 5′ GGTGTTCTTCCCGATATCTACA 3′)79. Bifidobacterium breve genomic DNA (AATC 15700-D5) was used as the standard to absolutely quantitate Bifidobacterium abundance. qPCR was performed with LightCycler 480 SYBR Green I Master mix (Roche) and run in a LightCycler 480 system (Roche) with an annealing temperature of 55 °C. Under these conditions, Bifidobacterium was quantifiable in a dynamic range from 101 to 106 copies.
SFB quantitation
SFB were quantified by qPCR in faecal samples using the described 736F/844R primer set80 (Supplementary Table 7). To obtain absolute quantities of SFB, the PCR product was cloned using the pGEM-T easy Vector System I (Promega). Colonies were confirmed to have the correct insert by PCR followed by sequencing. The plasmid was then purified using the Qiagen Plasmid Midi kit and DNA concentration was determined by Nanodrop. qPCR was performed with LightCycler 480 SYBR Green I Master mix (Roche) and run in a LightCycler 480 system (Roche) with an annealing temperature of 62 °C. Under these conditions, SFB were quantifiable in a dynamic range from 102 to 106 copies.
Measurement of PAT-induced microbial perturbation
To test the hypothesis that non-species-specific perturbation of the microbiota was related to the increase in subsequent diabetes risk, we used the Mann–Whitney U-test to compare the weighted UniFrac β-diversity in mice that ultimately developed diabetes or not. We measured perturbation in terms of weighted UniFrac distances, comparing faecal samples from control mice at 6 weeks to PAT mice at 6 weeks that develop diabetes or not. Similarly, we compared those distances from paired samples of PAT mice obtained at 3 and 6 weeks by whether the mice ultimately developed diabetes or not.
Development of a predictive model for T1D in PAT male mice
We built a classifier for predicting the development of T1D using 6-week-old male faecal samples, determined using the randomForest package in R with 2,000 trees and default settings81,82. In contrast to traditional classification models, supervised learning methods are inherently designed to generalize to unseen input data rather than optimal model fit. When benchmarked against other supervised learning methods, random forest was the strongest performer in classifying human microbiome data83 and has been used robustly to classify western and non-western human gut microbiomes38. The mean and standard deviation of the expected classifier accuracy were calculated using tenfold cross-validation error estimates nested within a tenfold jackknifing procedure. Microbiome compositions, represented by the relative abundances of species-level taxon tables, were used as predictors of T1D development. We assessed accuracy by training the classifier on 90% of the samples (‘training set’), then measuring the accuracy in predicting the correct label for the other 10% of the samples (‘test set’), repeating this process ten times, so that every sample was included in the test set exactly once. This cross-validation procedure produced a single estimation of the expected predictive accuracy of the model. To determine both the final estimate of the expected accuracy and the standard deviation of this estimate, we nested this tenfold cross-validation procedure within tenfold jackknifing. As such, we performed the entire train/test tenfold cross-validation procedure ten times while withholding a different 10% of the data each time, to produce ten accuracy estimates. The mean and standard error of the accuracy (76.5 ± 6.2%) were calculated from these ten jackknifed estimates. We measured feature importance as the mean decrease in model accuracy when that feature’s values were permuted randomly. The feature importance scores were reported by the randomForest package. Genera with mean 1% accuracy decrease (corresponding to an importance score of 0.01) were considered highly discriminative.
Metagenomic predictions
The PICRUSt bioinformatics tool was used to predict the metagenomic content from the 16S rRNA sequencing data of the gut microbial samples, with slight modifications, including closed-reference OTU picking, as suggested by the developers84. The predicted KEGG orthologues were summarized at hierarchy level 3, then differential abundances by group were determined and displayed using the STAMP algorithm85. For pairwise comparisons, White’s non-parametric t-test was used, and for multiple group comparisons, the Kruskal–Wallis H-test was used followed by the Tukey–Kramer post-hoc test. All P values were corrected for multiple comparisons using the Benjamini–Hochberg FDR correction.
Microbiota transfer
At age 6 weeks, control and PAT male NOD mice (n = 5 each) raised at NYULMC were killed, and caecal samples were collected and frozen at −80 °C. From each mouse, one-third of the caecal contents with tissue was thawed in an anaerobic chamber and diluted in pre-reduced anaerobically sterilized liquid dental transport media (Anaerobe Systems), the tissue was removed, and the supernatants were pooled to yield control or PAT caecal inoculum (7 ml). These suspensions were transferred into NOD germ-free pregnant dams at random by oral gavage (150 μl per breeder), and these now-conventionalized mice and their pups were raised in sterilized ventilated cages with autoclaved food and drinking water at the University of Gothenburg (Sweden) until weaning, when the male pups were transferred to new sterilized cages. The male pups were designated the control or PAT recipient mice (Ctrl-R, n = 18; PAT-R, n = 10) and were monitored for diabetes development weekly by blood glucose measurement, as defined above. From faecal samples collected at weeks of life 4, 6, 9, 13 and 17, and caecal and ileal samples at sacrifice at 31 weeks of life, DNA was extracted and analysed as described above. In a second experiment, caecal contents were collected from dol 17 NOD male or female mice that had received the PAT tylosin regime from dol 10–15 or untreated controls, creating two pools (PAT and control), and each was transferred to recipient 3-week-old germ-free C57BL/6 mice (n = 4 recipients per pool). These now-conventionalized mice were followed for the next six weeks, then killed, and flow cytometry was performed on isolated splenocytes and SI-LP cells, as already described. Because our a priori hypothesis was that the PAT-altered microbiota would reduce Th17 and Treg populations, we used a one-tailed t-test to determine statistical significance.
Accession codes
16S rRNA sequencing data has been deposited at EBI with accession number ERP016357 or using Qiita with accession number 10508. Microarray data has been deposited in GEO with accession number GSE81648.
Supplementary Material
Acknowledgments
Research support funding was provided by the Juvenile Diabetes Research Foundation, the Diane Belfer Program for Human Microbial Ecology, the Knapp Family, the Ziff Family and C&D Funds (to M.J.B.), the Howard Hughes Medical Institute and the Defendi Fellowship (to A.E.L.). Sequencing was performed at the NYUMC Genome Technology Center, partially supported by a Cancer Center Support Grant (P30CA016087) at the Laura and Isaac Perlmutter Cancer Center. The authors thank T. Battaglia and P. Meyn for informatic and technical assistance.
Footnotes
Author contributions
A.E.L. and M.J.B. conceived and designed the study. A.E.L., T.U.G., P.V., W.P., D.S., J.C., J.S., S.K., Z.G., C.B., J.K., S.N., A.R. and X.-S.Z. acquired the data. A.E.L., S.M., H.L., A.B.R., S.S., X.-S.Z., K.C., D.K., A.A., F.B. and M.J.B. analysed and interpreted the data. A.E.L., T.U.G., S.M., H.L., A.B.R., S.S., X.-S.Z., K.C., D.K., A.A., F.B. and M.J.B. drafted or revised the article. A.E.L., T.U.G., P.V., W.P., D.S., S.M., H.L., J.C., J.S., S.K., Z.G., C.B., J.K., S.N., A.B.R., S.S., X.-S.Z., K.C., D.K., A.A., F.B. and M.J.B. approved the final manuscript.
Additional information
Supplementary information is available online.
Competing interests
The authors declare no competing financial interests.
References
- 1.Diamond Project Group. Incidence and trends of childhood type 1 diabetes worldwide 1990–1999. Diabetic Med. 2006;23:857–866. doi: 10.1111/j.1464-5491.2006.01925.x. [DOI] [PubMed] [Google Scholar]
- 2.Patterson CC, et al. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373:2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 3.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azad MB, Kozyrskyj AL. Perinatal programming of asthma: the role of gut microbiota. Clin Dev Immunol. 2012;2012:932072. doi: 10.1155/2012/932072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozyrskyj AL, Ernst P, Becker AB. Increased risk of childhood asthma from antibiotic use in early life. Chest. 2007;131:1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 6.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between caesarean section and childhood asthma. Clin Exp Allergy. 2008;38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 7.Clemente JC, et al. The microbiome of the uncontacted Amerindians. Sci Adv. 2015;1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic AD, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boursi B, Mamtani R, Haynes K, Yang YX. The effect of past antibiotic exposure on diabetes risk. Eur J Endocrinol. 2015;172:639–648. doi: 10.1530/EJE-14-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world—recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 11.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiter EH. The NOD mouse: a model for insulin-dependent diabetes mellitus. Curr Protoc Immunol. 2001;Ch. 15(Unit 15.9) doi: 10.1002/0471142735.im1509s24. [DOI] [PubMed] [Google Scholar]
- 13.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobel YR, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candon S, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE. 2015;10:e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, et al. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol. 2015;195:4176–4184. doi: 10.4049/jimmunol.1500884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown K, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016;10:321–332. doi: 10.1038/ismej.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 22.Lund RJ, et al. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J Immunol. 2007;178:3648–3660. doi: 10.4049/jimmunol.178.6.3648. [DOI] [PubMed] [Google Scholar]
- 23.Greiner TU, Hyotylainen T, Knip M, Backhed F, Oresic M. The gut microbiota modulates glycaemic control and serum metabolite profiles in non-obese diabetic mice. PLoS ONE. 2014;9:e110359. doi: 10.1371/journal.pone.0110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai G, et al. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics. 2012;130:23–31. doi: 10.1542/peds.2011-2879. [DOI] [PubMed] [Google Scholar]
- 25.Hansen CH, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 26.Tormo-Badia N, et al. Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol. 2014;80:250–260. doi: 10.1111/sji.12205. [DOI] [PubMed] [Google Scholar]
- 27.Larsson E, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckner JH. Mechanisms of impaired regulation by CD4+CD25+FOXP3+ regulatory T cells in human autoimmune diseases. Nat Rev Immunol. 2010;10:849–859. doi: 10.1038/nri2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badami E, et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60:2120–2124. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedoya SK, Lam B, Lau K, Larkin J. III Th17 cells in immunity and autoimmunity. Clin Dev Immunol. 2013;2013:986789. doi: 10.1155/2013/986789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emamaullee JA, et al. Inhibition of Th17 cells regulates autoimmune diabetes in NOD mice. Diabetes. 2009;58:1302–1311. doi: 10.2337/db08-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriegel MA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau K, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 34.Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosi E, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 36.Giongo A, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardwell CR, et al. Breast-feeding and childhood-onset type 1 diabetes: a pooled analysis of individual participant data from 43 observational studies. Diabetes Care. 2012;35:2215–2225. doi: 10.2337/dc12-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardwell CR, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 41.De Goffau MC, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzman NH, et al. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology. 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- 43.Grapov D, et al. Diabetes associated metabolomic perturbations in NOD mice. Metabolomics. 2015;11:425–437. doi: 10.1007/s11306-014-0706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oresic M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin FP, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Youssef S, et al. The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature. 2002;420:78–84. doi: 10.1038/nature01158. [DOI] [PubMed] [Google Scholar]
- 48.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 49.Brown CT, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowell CA, Stearman RS, Morrow JF. Transcriptional regulation of serum amyloid A gene expression. J Biol Chem. 1986;261:8453–8461. [PubMed] [Google Scholar]
- 51.Harkness JE, Wagner JE. The Biology and Medicine of Rabbits and Rodents. Lea & Febiger; 1989. [Google Scholar]
- 52.Jukes TH. The present status and background of antibiotics in the feeding of domestic animals. Ann NY Acad Sci. 1971;182:362–379. doi: 10.1111/j.1749-6632.1971.tb30672.x. [DOI] [PubMed] [Google Scholar]
- 53.Lewicki J. Tylosin: a review of pharmacokinetics, residues in food animals and analytical methods. United Nations Food and Agriculture Organization; 2006. ftp://ftp.fao.org/ag/agn/food/tylosin_2006.pdf. [Google Scholar]
- 54.Ize-Ludlow D, et al. Progressive erosion of β-cell function precedes the onset of hyperglycemia in the NOD mouse model of type 1 diabetes. Diabetes. 2011;60:2086–2091. doi: 10.2337/db11-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forestier C, et al. Improved outcomes in NOD mice treated with a novel Th2 cytokine-biasing NKT cell activator. J Immunol. 2007;178:1415–1425. doi: 10.4049/jimmunol.178.3.1415. [DOI] [PubMed] [Google Scholar]
- 56.Wilson JR, Koehler KJ. Testing of equality of vectors of proportions for several cluster samples. Proc Joint Statist Assoc Meet Surv Res Meth. 1984;39:201–206. [Google Scholar]
- 57.La Rosa PS, et al. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS ONE. 2012;7:e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 59.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statist Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 61.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, Wang LG, Han Y, He Q. Y clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 64.Le Gall G, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 65.Banerjee R, Pathmasiri W, Snyder R, McRitchie S, Sumner S. Metabolomics of brain and reproductive organs: characterizing the impact of gestational exposure to butylbenzyl phthalate on dams and resultant offspring. Metabolomics. 2012;8:1012–1025. [Google Scholar]
- 66.Church RJ, et al. A systems biology approach utilizing a mouse diversity panel identifies genetic differences influencing isoniazid-induced microvesicular steatosis. Toxicol Sci. 2014;140:481–492. doi: 10.1093/toxsci/kfu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pathmasiri W, et al. Integrating metabolomic signatures and psychosocial parameters in responsivity to an immersion treatment model for adolescent obesity. Metabolomics. 2012;8:1037–1051. [Google Scholar]
- 68.Sumner S, et al. Metabolomics in the assessment of chemical-induced reproductive and developmental outcomes using non-invasive biological fluids application to the study of butylbenzyl phthalate. J Appl Toxicol. 2009;29:703–714. doi: 10.1002/jat.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sumner SC, Fennell TR, Snyder RW, Taylor GF, Lewin A. H 14 Distribution of carbon-14 labeled C60 ([ C]C60) in the pregnant and in the lactating dam and the effect of C60 exposure on the biochemical profile of urine. J Appl Toxicol. 2010;30:354–360. doi: 10.1002/jat.1503. [DOI] [PubMed] [Google Scholar]
- 70.Sumner SJ, Burgess JP, Snyder RW, Popp JA, Fennell TR. Metabolomics of urine for the assessment of microvesicular lipid accumulation in the liver following isoniazid exposure. Metabolomics. 2010;6:238–249. doi: 10.1007/s11306-010-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanov II, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 72.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aronesty E. Comparison of sequence utility programs. Open Bioinformatics. 2013;7:1–8. [Google Scholar]
- 74.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 77.McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- 78.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuki T, et al. Quantitative PCR with 16S rRNA-gene-targeted species specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barman M, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 82.Breiman L. Manual on setting up, using, and understanding random forests v3.1. 2002 https://www.stat.berkeley.edu/~breiman/Using_random_forests_V3.1.pdf.
- 83.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 84.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.