Abstract
Increasing evidence indicates that many small secretory preproteins can undergo post-translational translocation across the membrane of the endoplasmic reticulum. Although the cellular machinery involved in post-translational translocation of small secretory preproteins has begun to be elucidated, the intrinsic signals contained within these small secretory preproteins that contribute to their efficient post-translational translocation remain unknown. Here, we analyzed the eukaryotic secretory proteome and discovered the small secretory preproteins tend to have a higher probability to harbor the positive charge in the n-region of the signal peptide (SP). Eliminating the positive charge of the n-region blocked post-translational translocation of newly synthesized preproteins and selectively impaired translocation efficiency of small secretory preproteins. The pathophysiological significance of the positive charge in the n-region of SP was underscored by recently identified preproinsulin SP mutations that impair translocation of preproinsulin and cause maturity onset diabetes of youth (MODY). Remarkably, we have found that slowing the polypeptide elongation rate of small secretory preproteins could alleviate the translocation defect caused by loss of the n-region positive charge of the signal peptide. Together, these data reveal not only a previously unrecognized role of the n-region's positive charge in ensuring efficient post-translational translocation of small secretory preproteins, but they also highlight the molecular contribution of defects in this process to the pathogenesis of genetic disorders such as MODY.
Keywords: positive charge, signal peptide, n-region, secretory preprotein, translocation, signal peptide, n-region, protein translocation, protein translocation, protein synthesis, protein translocation, protein domain, protein translocation, protein export, protein inport, protein processing, protein targeting
Introduction
As established by the signal hypothesis, many newly-synthesized secretory proteins possess signal peptides (SP)4 that direct translocation across the membrane of the endoplasmic reticulum (ER) (1–3). Despite the lack of a precise consensus sequence, all SPs share three common structural features: 1) a positively charged region positioned at the N terminus of the SP (the so-called “n-region”); 2) a hydrophobic region composed of 10–15 nonpolar residues (“h-region”) that can enter the lipid bilayer of the ER membrane and initiate interaction with the Sec61 protein translocon; and 3) residues with small side chains located near the cleavage site of the SP (“c-region”) (3–6). Each of these SP regions interact with cellular translocation and processing machinery contained within the ER membrane.
A secretory preprotein may undergo translocation across the ER membrane via co-translational translocation or via post-translational translocation. In mammalian cells, most secretory preproteins are co-translationally translocated (i.e. translocation is coupled to protein synthesis) using a process in which the ribonucleoprotein signal recognition particle (7–9) can interact with the SP (10, 11) and initiate delivery of the elongating polypeptide chain to the Sec61 translocon. As the nascent polypeptides enter the ER lumen, the SPs are efficiently removed by signal peptidase on the lumenal side of the ER membrane (12–14).
Compared with this well-characterized process, post-translational translocation is less well understood. It was initially thought that post-translational translocation occurred mostly in prokaryotes or in simple eukaryotes such as yeast (8). However, increasing evidence indicates that in higher eukaryotic cells there are many secretory preproteins that can undergo a post-translational translocation (15–18). Over the past 2 decades, several cellular components have been identified in mediating post-translational translocation (19–22), but the intrinsic signals of small secretory preproteins that contribute to their selection and efficiency for post-translational translocation have not been elucidated.
In this report, we show that eliminating the positive charge in the SP n-region blocks post-translational translocation and selectively impairs the translocation of small secretory preproteins across the ER membrane. The orientation of SPs of many secretory preproteins appears to follow a “positive-inside rule,” by which the positively charged residues in the n-region of the SPs are preferably oriented toward the cytosolic side of the ER (11, 23–28). Eliminating the positive charge of the n-region could result in mis-orientation of preproteins in the Sec61 translocon, thereby leading to abnormal nascent polypeptide translocation (with consequent failure of SP cleavage by signal peptidase). This appears to be the case for a recently identified diabetogenic mutation preproinsulin-R6C, in which a positively charged residue in the SP n-region is mutated, leading to inefficient post-translational translocation of preproinsulin and maturity onset diabetes of youth (29–31). Thus, the data in this report reveal not only the importance of the positive charge in the n-region of the SP in maintaining efficient translocation of smaller secretory proteins (including preproinsulin), but they also help to better understand the molecular basis of genetic disorders caused by inefficient ER translocation of newly synthesized preproteins.
Results
Eliminating positive charge in the n-region of the signal peptide selectively impairs translocation for small secretory preproteins
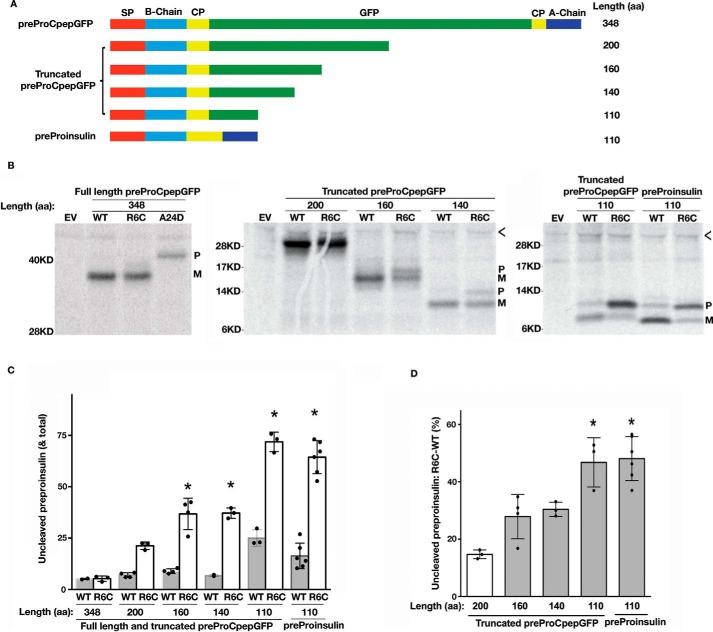
Recent studies have suggested that larger secretory preproteins may be insensitive to the loss of positive charge in the n-region of the signal peptide (30). To directly test this hypothesis, we constructed GFP-tagged preproinsulin (preproCpepGFP) wildtype (WT) and diabetogenic preproinsulin-R6C mutants that were truncated to different lengths in the mature polypeptide region (Fig. 1A). We were not looking for functionality of the expressed constructs, only whether the nascent translation product underwent SP cleavage, which is tightly coupled to preproinsulin translocation into the ER lumen and thus serves as a convenient assay of translocation (30). Importantly, we found that the full-length preproCpepGFP-R6C was successfully processed to proCpepGFP, similar to that seen with the precursor bearing the WT signal peptide (Fig. 1B, left panel, an SP cleavage site mutant preproCpepGFP-A24D (33) was used as a control to show the position of uncleaved preprotein). Similarly, no detectable translocation defect was found in truncated preproCpepGFP with 200 residues (Fig. 1B, middle panel). Remarkably, however, as the polypeptides were truncated to ≤160 residues, the translocation defect caused by the R6C mutation became increasingly apparent (Fig. 1B, middle panel). At a length of 110 residues, ∼70% of the preprotein failed to be translocated to allow conversion to the mature signal-cleaved form (Fig. 1, B–D). The translocation defect caused by R6C was not strictly dependent on the sequence of the mature protein because the R6C mutation caused a similar defect in two distinct preproteins with 110 residues (Fig. 1B, right panel). These results suggest that eliminating the positive charge in the n-region of the SP selectively impairs the translocation of small secretory proteins.
Figure 1.
Translocation defect caused by R6C mutation occurs only in the GFP-tagged preproinsulin shorter than 160 amino acids. A, full-length and truncated GFP-tagged preproinsulins with different lengths ranging from 348 to 110 amino acids (aa). The signal peptide (SP, red), insulin B-chain (light blue), C-peptide (yellow), insulin A-chain (blue), and the insertion site of GFP (green) are indicated. Authentic preproinsulin without tag is shown in the lower panel. B, 293T cells were transfected with plasmids encoding different lengths of preproteins shown in A with wildtype (WT) or R6C signal sequence. At 40 h post-transfection, the cells were labeled with [35S]Met/Cys for 10 min without chase. The newly synthesized preproteins were immunoprecipitated with anti-insulin antibody. Uncleaved preproteins (P) and cleaved mature proteins (M) are indicated. The SP cleavage site mutation preproCpepGFP-A24D was used as a control of full-length signal-uncleaved preproCpepGFP. < indicates nonspecific bands. C, newly synthesized preproteins shown in B from as least three independent experiments were quantified using ImageJ. The percentages of signal-uncleaved preproteins in the corresponding total newly synthesized wildtype (WT) and R6C mutant preproteins were calculated and are shown as mean ± S.D. *, p < 0.05 compared with WT. D, increases of percentages of signal-uncleaved preproteins of the mutants minus that of the WT were calculated and shown as mean ± S.D. *, p < 0.05 comparing to mutant preprotein with a length of 200 residues.
R6C mutation does not affect preproinsulin signal peptide cleavage or ER targeting but affects preproinsulin translocation across the ER membrane
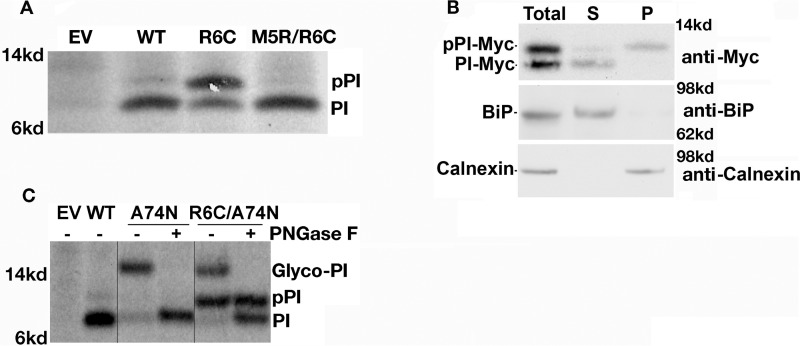
To further explore the fate of preproinsulin-R6C, we first generated the double mutant preproinsulin-M5R/R6C and found this was completely processed to proinsulin (PI, Fig. 2A), indicating full rescue of the defect upon re-introduction of a positive charge in the n-region despite persistence of the R6C mutation. Next, HEK293T cells expressing preproinsulin-R6C were extracted with sodium carbonate, followed by centrifugation that can separate content proteins of the ER lumen from ER membrane-associated proteins (30, 33). Although the signal-cleaved proinsulin was recovered in the ER lumen fraction (supernatant), along with the ER luminal protein BiP, the signal-uncleaved preproinsulin-R6C was recovered with ER membrane protein fraction (pellet) along with the ER membrane protein calnexin (Fig. 2B). These data indicate that preproinsulin-R6C is targeted and associated with the ER membrane. To establish whether preproinsulin-R6C could be translocated across the ER membrane, we introduced a glycosylation site at the 74th position of preproinsulin. As shown in Fig. 2C, whereas signal-cleaved proinsulin acquired N-linked glycosylation (Glyco-PI with a glycan that is sensitive to PNGase F, Fig. 2C), signal-uncleaved preproinsulin-R6C was not glycosylated, indicating that the signal-uncleaved fraction of preproinsulin-R6C molecules was not delivered into the ER lumen. Altogether, the data in Fig. 2, along with our previously published findings (30), suggest that the positive charge in the n-region of the SP is not required for targeting to the ER membrane but is required for efficient preproinsulin translocation across the ER membrane.
Figure 2.
R6C mutation impairs preproinsulin translocation across the ER membrane. A, 293T cells were transfected with plasmids encoding preproinsulin-WT, R6C, or M5R/R6C. At 40 h post-transfection, the cells were labeled with [35S]Met/Cys for 10 min without chase. Newly synthesized preproinsulin-WT and mutants were immunoprecipitated (IP) and resolved using SDS-4–12% NuPAGE and phosphorimaging. B, 293T cells were transfected to express preproinsulin-R6C. The cells were lysed at 40 h post-transfection; membrane association of preproinsulin-R6C was examined using sodium carbonate extraction as described under “Experimental procedures.” The supernatants (S) and pellets (P) were analyzed by Western blotting using anti-Myc for detecting Myc-tagged preproinsulin-R6C, anti-BiP for detecting an ER luminal protein BiP, and anti-calnexin for detecting an ER membrane protein calnexin. C, 293T cells were transfected with plasmids encoding preproinsulin-WT, a glycosylated mutant A74N, and double mutant R6C/A74N. Newly synthesized preproinsulin-WT and mutants were labeled and immunoprecipitated as in A. The immunoprecipitations were digested with or without peptide:N-glycosidase (PNGase-F) before analysis as in A. EV, empty vector; PI, proinsulin.
Natural design of many small secretory preproteins requires the presence of positive charge in the n-region of the SP for efficient ER translocation
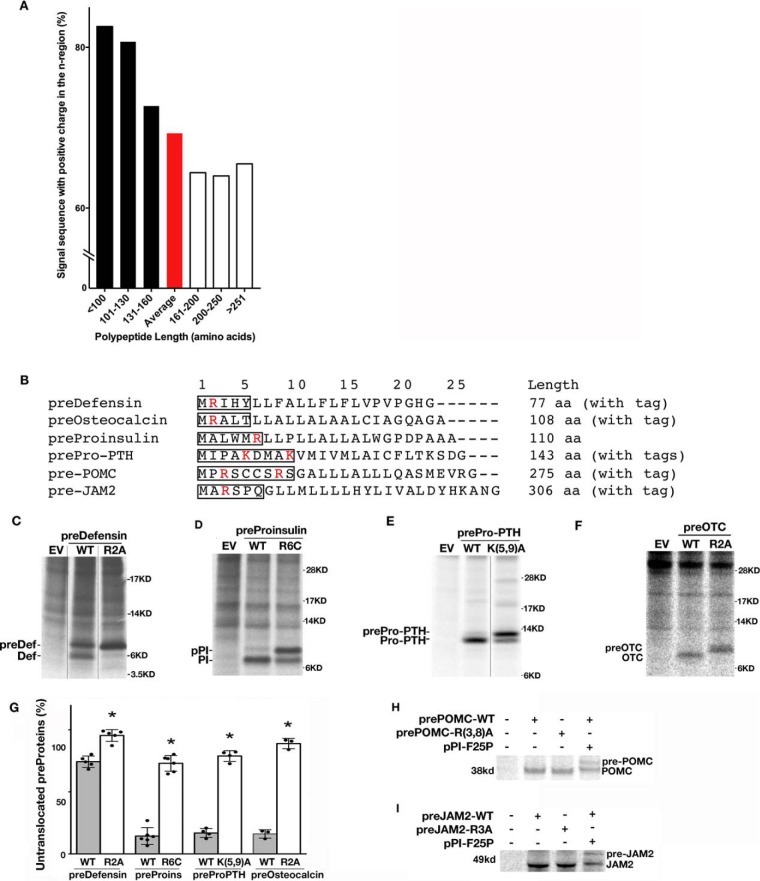
To broaden our understanding of the general significance of the positive charge in the n-region of the SP, we examined the eukaryotic secretome (34) and found that among 1877 secretory preproteins, 69.3% overall had positive charge in the n-region of SP (Table S1). However, when grouped by length, the fraction of such proteins with the positive charge in the n-region of the SP was found to increase as the polypeptide becomes shorter, reaching ≥80% for proteins smaller than 130 residues (Fig. 3A and Table S1). To test whether the translocation efficiency of small secretory preproteins is more dependent on the positive charge of the n-region, we mutated positively charged residues in the n-region of six secretory preproteins with different lengths, including FLAG-tagged junction adhesion molecule 2 (pre-Jam2, 306 amino acids); FLAG-tagged preproopiomelanocotin (pre-POMC, 275 amino acids); Myc- and FLAG-tagged preproparathyroid hormone (prepro-PTH, 143 amino acids); untagged preproinsulin (pPI, 110 amino acids); FLAG-tagged preosteocalcin (pre-OTC, 108 amino acids); and Myc-tagged predefensin (pre-Def, 77 amino acids) (Fig. 3B). Indeed, loss of the positive charge in the n-region of the SP exerted no detectable impact on the translocation of the two large preproteins, pre-Jam2 and pre-POMC (Fig. 3, H and I). However, loss of the positive charge in the n-region of the SP caused more than 60% of all four tested small secretory preproteins: predefensin, pPI, prepro-PTH, and pre-OTC failed to be translocated across the ER membrane (Fig. 3, C–F, quantified in G). Interestingly, even wildtype (WT) predefensin was not rapidly translocated into the ER during a 10-min translation period labeled with 35S-labeled amino acids (Fig. 3C), indicating that such a short secretory preprotein has low co-translational translocation efficiency. Taken together, these results suggest that small secretory preproteins are intrinsically predisposed to inefficient co-translational translocation and are susceptible to further translocational impairment as a consequence of loss of the positive charge in the n-region of the SP.
Figure 3.
Small secretory proteins have more positive charge in the n-region's SP and are more susceptible to the loss of n-region's positive charge. A, analysis of the percentage of the secretory proteins with the positive charge in the n-region of the SP in 1877 eukaryotic secretory proteins. Among these proteins, on average, 69.3% (red bar) had positive charge in the n-region of their SP. A trend indicated that the percentage of proteins with the positive charge in the n-region of the SP as the polypeptide became shorter. B, signal sequences and lengths of predefensin, preosteocalcin, preproinsulin, prepro-PTH, pre-POMC, and pre-JAM2. The n-regions of signal sequences are boxed. The positively charged residues in the n-region are highlighted in red. C–G, 293T cells transfected with plasmids encoding small secretory preproteins, including wildtype (WT) or mutant predefensin (preDef), preproinsulin (pPI), prepro-PTH, and preosteocalcin (preOTC), were pulse-labeled with 35S-labeled amino acids (aa) for 10 min without chase. The newly synthesized preproteins were immunoprecipitated, resolved in SDS-4–12% NuPAGE gels, phosphorimaged (C–F), and quantified using ImageJ. The percentages of uncleaved preproteins from at least three independent experiments were calculated and shown as mean ± S.D. in G. *, p < 0.05 compared with WT preproteins. H and I, 293T cells transfected with plasmids encoding large secretory preproteins pre-POMC and pre-JAM2 were pulse-labeled with 35S-labeled amino acids for 10 min without chase. The newly synthesized WT and mutant preproteins were immunoprecipitated, resolved using 10% SDS-PAGE, and phosphorimaged. EV, empty vector.
Positive charge in the SP n-region plays an important role in post-translational translocation of small secretory preproteins
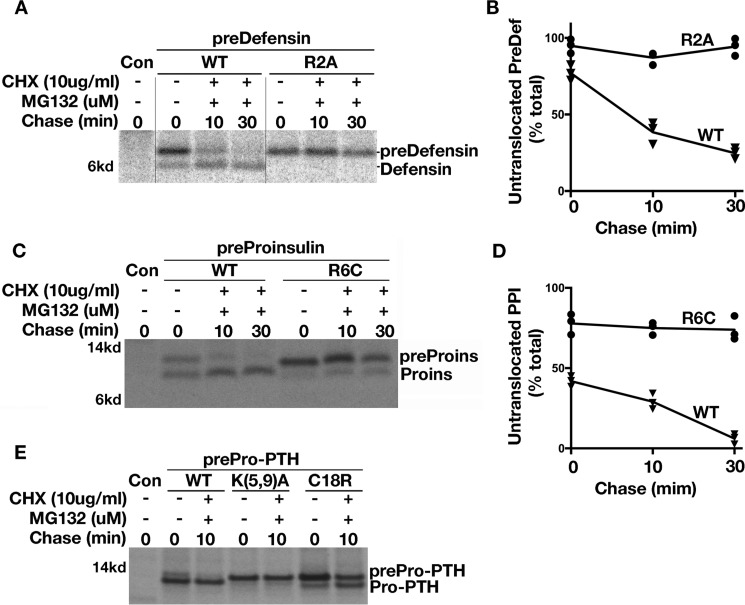
Because preproteins shorter than 160 residues can undergo post-translational translocation (19), we asked whether the positive charge of the n-region exerts a unique influence on such preproteins. To increase detection of preproteins that are kinetically impaired in translocation, we used metabolic incorporation of radioactive amino acids with a pulse-labeling time limited to 5 min. At the zero chase time, up to 75% of fully synthesized predefensin molecules had not yet been translocated. During 10 and 30 min after the translation period (in the presence of MG132 to prevent proteasomal degradation of untranslocated preproteins and cycloheximide to prevent further protein synthesis after labeling), most of the predefensin molecules underwent subsequent translocation, forming defensin. By contrast, mutant predefensin-R2A molecules, eliminating the positive charge in the n-region of the SP, were blocked from further translocation into the ER (Fig. 4, A and B). This phenomenon was not unique to predefensin, as similar results were obtained for two other small secretory preproteins, preproinsulin-R6C and prepro-PTH-K5A/K9A (Fig. 4, C–E). We also noted that the prepro-PTH n-region K5A/K9A mutant exhibited a more severe post-translational translocation defect than that of another disease-causing mutation that is located in the h-region of prepro-PTH SP (35, 36), prepro-PTH-C18R (Fig. 4E). Together, these data strongly suggest that the positive charge in the n-region of the SP plays a general and important role in post-translational translocation of small secretory preproteins.
Figure 4.
Loss of positive charge in the n-region of SP impairs post-translational translocation of small secretory preproteins. A, 293T cells transfected with plasmids encoding predefensin-WT or n-region mutant R2A were labeled for 5 min followed by 0, 10, or 30 min of chase in the presence of 10 μm MG132 and 10 μg/ml CHX. Post-translational translocation of predefensin-WT and mutant were analyzed in 4–12% NuPAGE gel. B, newly synthesized signal-uncleaved and -cleaved predefensin-WT and R2A from three experiments were quantified using ImageJ. The percentages of signal-uncleaved predefensin-WT and R2A at 0-, 10-, or 30-min chase time points were calculated and shown. C, 293T cells transfected with plasmids encoding preproinsulin-WT or R6C were labeled for 5 min followed by a 0-, 10-, or 30-min chase in the presence of 10 μm MG132 and 10 μg/ml CHX. Post-translational translocation of preproinsulin-WT and R6C were analyzed in 4–12% NuPAGE gel. D, newly synthesized signal-uncleaved and -cleaved preproinsulin-WT and R2A from three experiments were quantified using ImageJ. The percentages of signal-uncleaved preproinsulin-WT and R6C at a 0-, 10-, or 30-min chase time points were calculated and shown. E, 293T cells transfected with plasmids encoding prepro-PTH wildtype (WT), n-region mutant K5A/K9A, or h-region mutant C18R were labeled for 5 min followed by a 0- and 10-min chase in the presence of 10 μm MG132 and 10 μg/ml CHX. Untranslocated but fully synthesized WT predefensin, preproinsulin, and prepro-PTH could clearly undergo post-translational translocation and were further processed during the 10–30-min chase. However, post-translational translocation of all three SP n-region mutants were essentially blocked. By contrast, the mutation in the h-region of prepro-PTH C18R could undergo post-translational translocation during a 10-min chase.
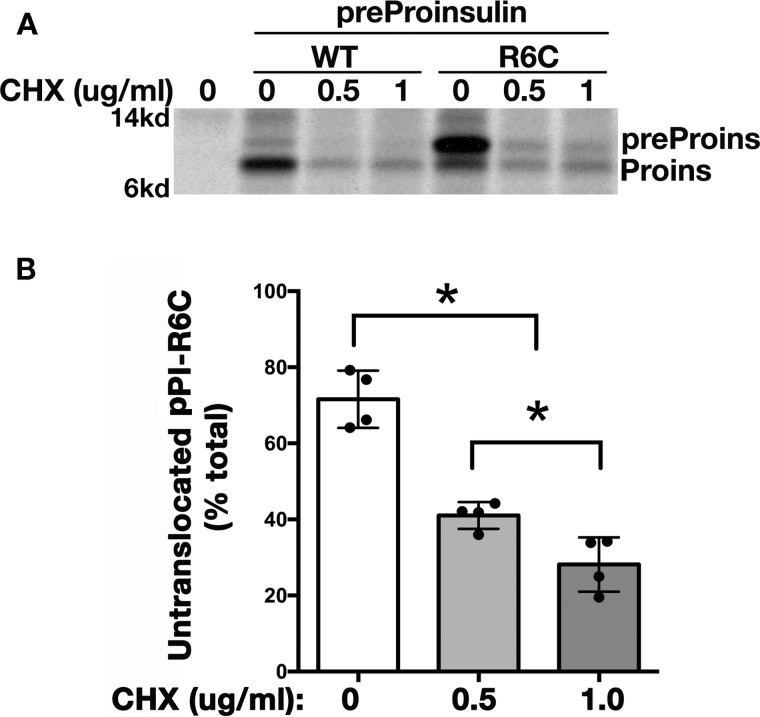
Reducing the nascent polypeptide elongation rate partially rescues the translocation defect caused by loss of positive charge in the n-region of the SP
We reasoned that a small secretory preprotein may rapidly complete translation of the entire polypeptide before being properly targeted to the ER membrane translocation machinery. In such a case, the fully synthesized preprotein would be released into the cytosol, accounting for the need to enter the ER via a post-translational mechanism (22). For this reason, we wished to test whether slowing the rate of polypeptide chain elongation might provide more time for small secretory preproteins to be recognized and targeted to the ER translocon, thereby enhancing co-translational translocation (37). With this in mind, we treated cells with differing low doses of cycloheximide that have been shown to decrease the rate of polypeptide chain elongation (37, 38). Cells expressing preproinsulin-R6C (or WT) and treated with cycloheximide were pulse-labeled without chase to monitor the efficiency of co-translational translocation. Although total protein synthesis was of course decreased upon cycloheximide treatment, the percentage of untranslocated preproinsulin-R6C declined from 71% in the cells without treatment to 26% in cells treated with 1 μg/ml cycloheximide (Fig. 5A and quantified in B). These data suggest that one reason secretory preproteins utilize the post-translational translocation route is because they complete their synthesis too quickly for efficient targeting to the ER translocation machinery. Co-translational translocation of small secretory preproteins can partially rescue the inefficient translocation, but such small preproteins are especially susceptible to translocation defects caused by a loss of positive charge in the n-region of SP.
Figure 5.
Reducing translation elongation rate partially rescues the translocation defect caused by loss of positive charge in the n-region of the SP. A, 293T cells were transfected with preproinsulin-WT or R6C. At 40 h post-transfection, the cells were labeled with [35S]Met/Cys in the presence of 0, 0.5, or 1 μg/ml CHX for 10 min without chase. The newly synthesized preproinsulin molecules were immunoprecipitated with anti-insulin and analyzed using SDS-4–12% NuPAGE. B, untranslocated preproinsulin-R6C and processed proinsulin from three independent experiments were quantified using ImageJ. The percentages of the untranslocated pPI-R6C in total newly synthesized preproinsulin are shown as mean ± S.D., * indicates p < 0.05.
Discussion
Protein targeting into the ER is essential for the normal function of many secretory proteins. Secretory preproteins can enter the ER through co-translational and/or post-translational translocation (8, 15). In this study, we demonstrate a loss of co-translational translocation efficiency for short secretory preproteins as well as an increased translocational dependence on the positive charge in the n-region of the SP (Fig. 1). Whereas co-translational translocation efficiency appears excellent for secretory preproteins of more than 160 residues, the fraction of newly-synthesized untranslocated molecules increases for smaller secretory preproteins. Indeed, as much as 75% of secretory preproteins of ∼77 residues are not translocated during the translation period (Fig. 3, C and G) but are translocated into the ER lumen during the post-translational period (Fig. 4, A–D). The most parsimonious explanation of the present data is that the positive charge in the n-region of the SP is important for post-translational translocation of small secretory preproteins.
A recently identified diabetogenic mutation, preproinsulin-R6C that causes SP misorientation during translocation (30, 31), highlights a critical role of the positive charge in the n-region of the SP for efficient translocation (Fig. 2A). Interestingly, while eliminating the n-region's positive charge caused up to 70% of the newly synthesized 110-residue preproinsulin-R6C to fail to translocate across the ER membrane, it did not appear critical for translocation of the 348-residue GFP-tagged preproinsulin (Fig. 1, B and C). When the GFP-tagged preproinsulin was truncated to different lengths, the translocation defect caused by the R6C substitution only became apparent when the secretory preprotein was ≤160 residues, and this was not critically dependent on the sequence of the downstream domain (Fig. 1). We conclude that the positive charge in the n-region of the SP is important for the efficient translocation of small secretory preproteins.
One of the potential differences between large and small secretory preproteins is their mode of translocation. Because of the short time window to complete translation, small secretory preproteins may be released from the ribosome before they have been recognized and targeted to the translocon. These preproteins entering the cytosol then need to translocate into the ER post-translationally (22). Growing evidence suggests that post-translational translocation is more prevalent than previously thought (19, 20, 22, 39), via a process that can engage multiple helper proteins, including SecA/B, Kar2P (BiP), Sec62/63, calmodulin, TRC40, Bag6 and HSP70 (19–22, 40–44). Over 200 human secretory preproteins have the potential to undergo post-translational translocation (15, 45), and most of these are small secretory preproteins.
Along with this, smaller secretory preproteins have a higher probability to harbor the positive charge in the n-region of the SP than do large secretory preproteins (Fig. 3A). In this study examining six distinct polypeptides, including two large preproteins (pre-JAM2 and pre-POMC) and four small preproteins (preproinsulin, prepro-PTH, preosteocalcin, and predefensin), we provide experimental evidence confirming that the translocation defect caused by loss of the positive charge in the n-region of the SP is specific for small secretory preproteins (Fig. 3). We propose that the positive charge in the n-region of the SP is likely to play a significant role for all small secretory preproteins whose net ER translocation efficiency is increasingly dependent upon a post-translational translocation mechanism.
Consistent with this hypothesis, we observed that post-translational translocation of small secretory preproteins across the ER membrane was virtually completely blocked when the positive charge in the n-region of the SP was eliminated (Fig. 4). Slowing the polypeptide chain elongation rate (with low-dose cycloheximide treatment) provides additional time for the nascent chain to emerge from the ribosome and be directed to the ER translocon; remarkably, while decreasing the overall protein synthesis rate, this maneuver significantly improved the fraction of molecules that were successfully translocated despite loss of the positive charge in the n-region of the SP (Fig. 5). Altogether, we posit that because of a limited time for efficient targeting to the ER translocon prior to the rapid completion of secretory protein translation, small secretory preproteins are intrinsically predisposed to lower efficiency in co-translational translocation, which is readily demonstrated by measuring translocation during the synthesis period marked by pulse-labeling. In such cases, post-translational translocation becomes an important backup mechanism to enhance delivery of small secretory preproteins into the ER lumen. Correlating precisely with this backup mechanism, small secretory preproteins are susceptible to further translocational impairment as a consequence of the loss of the positive charge in the n-region of the SP. Such considerations are important, as there are serious pathophysiological consequences to defective ER translocation that has been linked mechanistically to multiple human diseases, including diabetes (29–31), hypoparathyroidism (36), chondrodysplasia (46), and Ehlers-Danlos syndrome (47).
Experimental procedures
Materials
Rabbit anti-Myc and anti-GFP were from Immunology Consultants Laboratories; guinea pig anti-human insulin was made by Novus; zysorbin was from Zymed Laboratories Inc.; 35S-amino acid mixture (Met + Cys) was from ICN; dithiothreitol (DTT), protein A-agarose, MG132, and anti-FLAG M2 were from Sigma; Met/Cys-deficient Dulbecco's modified Eagle's medium (DMEM) and all other tissue culture reagents were from Invitrogen. The plasmid encoding human pre-POMC was from Drs. Mencarelli and Di Blasio at Instituto Auxologico Italiano, Italy.
Mutagenesis, cell culture and transfection, and metabolic labeling and immunoprecipitation
The plasmids encoding human wildtype and mutant preproCpepGFP, in which GFP was inserted into the C-peptide of human preproinsulin, were constructed as described previously (30, 32). To make truncated preproCpepGFP, stop codons at designed sites were introduced by a site-directed mutagenesis kit (Stratagene). FLAG-tagged human predefensin, prepro-PTH, preosteocalcin, and junction adhesion molecule 2 (pre-Jam2) were synthesized by Integrated DNA Technologies (IDT) and were subcloned into pcmsGFP using EcoRI and XbaII. Predefensin, preproinsulin, prepro-PTH, preosteocalcin, pre-JAM2, and pre-POMC mutants were generated by the site-directed mutagenesis kit (Stratagene). HEK293T cells were plated onto 12-well plates 1 day before transfection with Lipofectamine (Invitrogen) using 1–2 mg of plasmid DNA. At 48 h after transfection, cells were pulse-labeled with [35S]Met/Cys for 5 or 10 min as indicated. In some experiments, the labeled cells were chased in the presence or absence of MG132 or cycloheximide (CHX) for different times as indicated.
Sodium carbonate extraction and Western blotting
For sodium carbonate extraction, the transfected 293T cells were suspended in 0.1 m sodium carbonate, pH 12, homogenized, and incubated on ice for 1 h followed by sedimentation at 50,000 rpm at 4 °C for 1 h. The supernatants and pellets were collected. For Western blotting, the collected supernatants and pellets were boiled in SDS sample buffer containing 100 mmol/liter DTT, resolved by 4–12% NuPAGE, electrotransferred to nitrocellulose, and blotted with anti-Myc, anti-GRP78 (BiP), and anti-calnexin followed by appropriate secondary antibodies conjugated with HRP, with development by enhanced chemiluminescence.
Statistical analyses
Statistical analyses were carried out by analysis of variance followed by using GraphPad Prism 5. A p value of < 0.05 was taken as statistically significant.
Author contributions
H. G., J. S., X. L., and Y. X. data curation; H. G., J. S., X. L., Y. X., H. W., H. S., Q. L., Y. H., R. M., Y. W., and J. C. methodology; J. S., X. L., R. Z., Y. H., R. M., and M. L. formal analysis; P. A. and M. L. funding acquisition; P. A. and M. L. writing-review and editing; M. L. conceptualization; M. L. supervision; M. L. investigation; M. L. writing-original draft.
Supplementary Material
Acknowledgments
We thank Drs. Shu-ou Shan (Caltech) and Peter Walter (University of California at San Francisco) for helpful discussions.
This work was supported by National Natural Science Foundation of China Research Grants 81070629, 81570699, 81620108004, and 81370895, National Institutes of Health Grants RO1-DK088856 (to M. L.), RO1-DK-48280 (to P. A.), and R01-DK111174, and the Protein Folding Disease Initiative of the University of Michigan. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Table S1.
- SP
- signal peptide
- ER
- endoplasmic reticulum
- CHX
- cycloheximide
- pPI
- preproinsulin.
References
- 1. Nobel Prize Committee (1999) Physiology or Medicine for 1999–Press Release, October 11, 1999, https://www.nobelprize.org/nobel_prizes/medicine/laureates/1999/press.html
- 2. Blobel G., and Sabatini D. (1971) Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 68, 390–394 10.1073/pnas.68.2.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gennity J., Goldstein J., and Inouye M. (1990) Signal peptide mutants of Escherichia coli. J. Bioenerg. Biomembr. 22, 233–269 10.1007/BF00763167 [DOI] [PubMed] [Google Scholar]
- 4. Nothwehr S. F., and Gordon J. I. (1989) Eukaryotic signal peptide structure/function relationships. Identification of conformational features which influence the site and efficiency of co-translational proteolytic processing by site-directed mutagenesis of human pre(delta pro)apolipoprotein A-II. J. Biol. Chem. 264, 3979–3987 [PubMed] [Google Scholar]
- 5. von Heijne G. (1990) The signal peptide. J. Membr. Biol. 115, 195–201 10.1007/BF01868635 [DOI] [PubMed] [Google Scholar]
- 6. Clérico E. M., Maki J. L., and Gierasch L. M. (2008) Use of synthetic signal sequences to explore the protein export machinery. Biopolymers 90, 307–319 10.1002/bip.20856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rapoport T. A. (2008) Protein transport across the endoplasmic reticulum membrane. FEBS J. 275, 4471–4478 10.1111/j.1742-4658.2008.06588.x [DOI] [PubMed] [Google Scholar]
- 8. Rapoport T. A. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 10.1038/nature06384 [DOI] [PubMed] [Google Scholar]
- 9. Elvekrog M. M., and Walter P. (2015) Dynamics of co-translational protein targeting. Curr. Opin. Chem. Biol. 29, 79–86 10.1016/j.cbpa.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Batey R. T., Rambo R. P., Lucast L., Rha B., and Doudna J. A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287, 1232–1239 10.1126/science.287.5456.1232 [DOI] [PubMed] [Google Scholar]
- 11. Peterson J. H., Woolhead C. A., Bernstein H. D. (2003) Basic amino acids in a distinct subset of signal peptides promote interaction with the signal recognition particle. J. Biol. Chem. 278, 46155–46162 10.1074/jbc.M309082200 [DOI] [PubMed] [Google Scholar]
- 12. Liu M., Wright J., Guo H., Xiong Y., and Arvan P. (2014) in Vitamins & Hormones (Gerald L., ed) pp. 35–62, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 13. Nyathi Y., Wilkinson B. M., and Pool M. R. (2013) Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2392–2402 10.1016/j.bbamcr.2013.02.021 [DOI] [PubMed] [Google Scholar]
- 14. Cui J., Chen W., Sun J., Guo H., Madley R., Xiong Y., Pan X., Wang H., Tai A. W., Weiss M. A., Arvan P., and Liu M. (2015) Competitive inhibition of the endoplasmic reticulum signal peptidase by non-cleavable mutant preprotein cargos. J. Biol. Chem. 290, 28131–28140 10.1074/jbc.M115.692350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson N., Powis K., and High S. (2013) Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2403–2409 10.1016/j.bbamcr.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 16. Johnson N., Hassdenteufel S., Theis M., Paton A. W., Paton J. C., Zimmermann R., and High S. (2013) The signal sequence influences post-translational ER translocation at distinct stages. PLoS ONE 8, e75394 10.1371/journal.pone.0075394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ast T., Cohen G., and Schuldiner M. (2013) A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 152, 1134–1145 10.1016/j.cell.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 18. Zimmermann R., Eyrisch S., Ahmad M., and Helms V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924 10.1016/j.bbamem.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 19. Lakkaraju A. K., Thankappan R., Mary C., Garrison J. L., Taunton J., and Strub K. (2012) Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 23, 2712–2722 10.1091/mbc.E12-03-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson N., Vilardi F., Lang S., Leznicki P., Zimmermann R., and High S. (2012) TRC40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 125, 3612–3620 10.1242/jcs.102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deltour L., Montagutelli X., Guenet J.-L., Jami J., and Páldi A. (1995) Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev. Biol. 168, 686–688 10.1006/dbio.1995.1114 [DOI] [PubMed] [Google Scholar]
- 22. Shao S., and Hegde R. S. (2011) A calmodulin-dependent translocation pathway for small secretory proteins. Cell 147, 1576–1588 10.1016/j.cell.2011.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. von Heijne G. (1986) Net N-C charge imbalance may be important for signal sequence function in bacteria. J. Mol. Biol. 192, 287–290 10.1016/0022-2836(86)90365-7 [DOI] [PubMed] [Google Scholar]
- 24. Fujita H., Kida Y., Hagiwara M., Morimoto F., and Sakaguchi M. (2010) Positive charges of translocating polypeptide chain retrieve an upstream marginal hydrophobic segment from the endoplasmic reticulum lumen to the translocon. Mol. Biol. Cell 21, 2045–2056 10.1091/mbc.E09-12-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Higy M., Junne T., and Spiess M. (2004) Topogenesis of membrane proteins at the endoplasmic reticulum, Biochemistry 43, 12716–12722 10.1021/bi048368m [DOI] [PubMed] [Google Scholar]
- 26. Goder V., Junne T., and Spiess M. (2004) Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell 15, 1470–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parks G. D., and Lamb R. A. (1991) Topology of eukaryotic type II membrane proteins: Importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64, 777–787 10.1016/0092-8674(91)90507-U [DOI] [PubMed] [Google Scholar]
- 28. von Heijne G. (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341, 456–458 10.1038/341456a0 [DOI] [PubMed] [Google Scholar]
- 29. Liu M., Sun J., Cui J., Chen W., Guo H., Barbetti F., and Arvan P. (2015) INS-gene mutations: From genetics and beta cell biology to clinical disease. Mol. Aspects Med. 42, 3–18 10.1016/j.mam.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo H., Xiong Y., Witkowski P., Cui J., Wang L. J., Sun J., Lara-Lemus R., Haataja L., Hutchison K., Shan S. O., Arvan P., and Liu M. (2014) Inefficient translocation of preproinsulin contributes to pancreatic beta cell failure and late-onset diabetes. J. Biol. Chem. 289, 16290–16302 10.1074/jbc.M114.562355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meur G., Simon A., Harun N., Virally M., Dechaume A., Bonnefond A., Fetita S., Tarasov A. I., Guillausseau P.-J., Boesgaard T. W., Pedersen O., Hansen T., Polak M., Gautier J.-F., Froguel P., et al. (2010) Insulin gene mutations resulting in early-onset diabetes: marked differences in clinical presentation, metabolic status, and pathogenic effect through endoplasmic reticulum retention. Diabetes 59, 653–661 10.2337/db09-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu M., Hodish I., Rhodes C. J., and Arvan P. (2007) Proinsulin maturation, misfolding, and proteotoxicity. Proc. Natl. Acad. Sci. U.S.A. 104, 15841–15846 10.1073/pnas.0702697104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu M., Lara-Lemus R., Shan S. O., Wright J., Haataja L., Barbetti F., Guo H., Larkin D., and Arvan P. (2012) Impaired cleavage of preproinsulin signal peptide linked to autosomal-dominant diabetes. Diabetes 61, 828–837 10.2337/db11-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choo K., and Ranganathan S. (2008) Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinformatics 9, S15 10.1186/1471-2105-9-S12-S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karaplis A. C., Lim S.-K., Baba H., Arnold A., and Kronenberg H. M. (1995) Inefficient membrane targeting, translocation, and proteolytic processing by signal peptidase of a mutant preproparathyroid hormone protein. J. Biol. Chem. 270, 1629–1635 10.1074/jbc.270.4.1629 [DOI] [PubMed] [Google Scholar]
- 36. Datta R., Waheed A., Shah G. N., and Sly W. S. (2007) Signal sequence mutation in autosomal dominant form of hypoparathyroidism induces apoptosis that is corrected by a chemical chaperone. Proc Natl. Acad. Sci. U.S.A. 104, 19989–19994 10.1073/pnas.0708725104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang D., and Shan S.-O. (2012) Translation elongation regulates substrate selection by the signal recognition particle. J. Biol. Chem. 287, 7652–7660 10.1074/jbc.M111.325001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., Merrick W. C., Green R., Shen B., and Liu J. O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., and Schleiff E. (2010) Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402, 510–523 10.1016/j.jmb.2010.07.052 [DOI] [PubMed] [Google Scholar]
- 40. Economou A., and Wickner W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843 10.1016/S0092-8674(94)90582-7 [DOI] [PubMed] [Google Scholar]
- 41. Panzner S., Dreier L., Hartmann E., Kostka S., and Rapoport T. A. (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 10.1016/0092-8674(95)90077-2 [DOI] [PubMed] [Google Scholar]
- 42. Rapoport T. A., Matlack K. E., Plath K., Misselwitz B., and Staeck O. (1999) Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol. Chem. 380, 1143–1150 [DOI] [PubMed] [Google Scholar]
- 43. Hessa T., Sharma A., Mariappan M., Eshleman H. D., Gutierrez E., and Hegde R. S. (2011) Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475, 394–397 10.1038/nature10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Randall L. L., and Hardy S. J. (1995) High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem. Sci. 20, 65–69 10.1016/S0968-0004(00)88959-8 [DOI] [PubMed] [Google Scholar]
- 45. Frith M. C., Forrest A. R., Nourbakhsh E., Pang K. C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T. L., and Grimmond S. M. (2006) The abundance of short proteins in the mammalian proteome. PLoS Genet. 2, e52 10.1371/journal.pgen.0020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikegawa S., Nakamura K., Nagano A., Haga N., and Nakamura Y. (1997) Mutations in the N-terminal globular domain of the type X collagen gene (COL10A1) in patients with Schmid metaphyseal chondrodysplasia. Hum. Mutat. 9, 131–135 10.1002/(SICI)1098-1004(1997)9:2%3C131::AID-HUMU5%3E3.0.CO%3B2-C [DOI] [PubMed] [Google Scholar]
- 47. Symoens S., Malfait F., Renard M., André J., Hausser I., Loeys B., Coucke P., and De Paepe A. (2009) COL5A1 signal peptide mutations interfere with protein secretion and cause classic Ehlers-Danlos syndrome. Hum. Mutat. 30, E395–E403 10.1002/humu.20887 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.