Abstract
Trauma is the leading cause of death worldwide for individuals under the age of 55. Interpatient genomic differences, in the form of candidate single nucleotide polymorphisms (SNPs), have been associated previously with adverse outcomes after trauma. However, the utility of these SNPs to predict outcomes based on a meaningful endpoint such as survival is as yet undefined. We hypothesized that specific SNP haplotypes could segregate trauma survivors from non-survivors. Genomic DNA samples were obtained from 453 blunt trauma patients, for whom complete daily clinical and biomarker data were available for 397. Of these, 13 patients were non-survivors and the remaining 384 were survivors. All 397 DNA samples were amplified, fragmented, and examined for 551,839 SNPs using the Illumina Infinium CoreExome-24 v1.1 BeadChip (Illumina). To enrich for likely important SNPs, we initially compared SNPs of the 13 non-survivors vs. 13 matched survivors, who were matched algorithmically for injury severity score (ISS), age, and gender ratio. This initial enrichment yielded 126 SNPs; a further comparison to the haplotypes of the remaining 371 survivors yielded a final total of 7 SNPs that distinguished survivors from non-survivors. Furthermore, severely injured survivors with the same seven SNPs as non-survivor exhibited distinct inflammatory responses from similarly injured survivors without those SNPs, and specifically had evidence of altered Th17 cell phenotypes based on computational modeling. These studies suggest an interaction between genetic polymorphism, injury severity, and initial inflammatory responses in driving trauma outcomes.
INTRODUCTION
Traumatic injury induces an inflammatory response which is thought to impair organ function. If it is appropriate in magnitude, character, and duration, injury-induced inflammation is not in and of itself detrimental; rather, it is in most cases a homoeostatic, well-coordinated communication network. However, the feed-forward loop of inflammation → tissue damage/dysfunction → inflammation can lead to organ dysfunction, nosocomial infection, and death (1). To date, there are no definitive molecular diagnostics for trauma-induced inflammatory derangement and organ dysfunction, nor is there a therapeutic option besides supportive care. Thus, there is a major need for both accurate diagnosis and novel therapeutic options.
The need for novel diagnostic and therapeutic modalities for trauma-induced critical illness, combined with the extensive advances in understanding of, and data on, the human genome has led to the hypothesis that genetic variability can drive outcomes following injury (2). Genetic variability is typically observed via single nucleotide polymorphisms (SNPs). SNPs are defined as single variants of one base in the genome, present in more than 1% of the population. Depending on their locus in the DNA, SNPs can have different effects. They can alter the mRNA sequence, potentially leading to a varied amino acid sequence, which might end in a functionally different protein. Furthermore, SNPs have the potential to affect the regulation of gene expression, alter the alternative splicing of the transcribed mRNA, or change mRNA half-life, thereby influencing the amount or the function of the encoded protein (3).
The Inflammation and the Host Response to Injury large-scale collaborative research program was the largest-scale effort in the shock community dedicated to defining the genomic underpinnings of the response to injury. Seminal work from this effort has shown that the magnitude of the changes in differentially expressed genes in circulating leukocytes assessed by a novel scoring system correlated with adverse outcomes in blunt trauma patients (4). Numerous other efforts have suggested candidate SNPs, typically in inflammatory pathways, to be associated with adverse outcomes following trauma (2, 5).
Despite these advances at the systems biology level, the complex interaction between genotype, the host’s inflammatory response, and ultimate clinical outcomes remains elusive. Our group has been focused on deciphering the complexity of the inflammatory response to traumatic injury, in part by elucidating principal drivers and dynamic networks of inflammation and organ dysfunction in both experimental animals and trauma patients (6, 7). We have leveraged a 493-patient observational study of human blunt trauma to help define dynamic inflammatory responses in highly matched patients that differed in etiology or outcome, including survival vs. death (7), to glean insights into how these SNPs might interplay with host-specific differences in post-injury inflammation.
MATERIALS AND METHODS
Patients
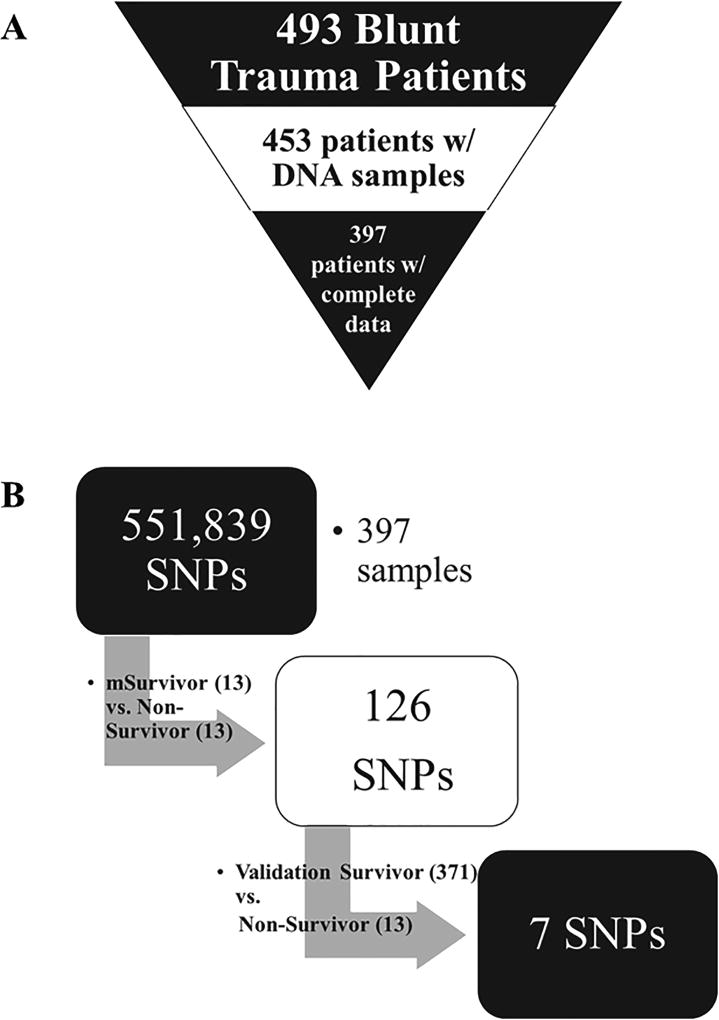
493 blunt trauma patients were enrolled in this study following admission to the emergency department of the Presbyterian University hospital (a Level 1 trauma center), following Institutional Review Board approval and obtaining informed consent. DNA samples were obtained from 453 of the 493 patients. 397 of these 453 patients had complete daily clinical and biomarker records (Fig. 1A). Of these 397 patients, 145 were female and 252 were male. The mean age was 48.8 ± 0.9 years (min: 18 years, max: 90 years). The mean ISS in this patient cohort was 19.8 ± 0.5 (min: 1, max: 54). Out of the 397 patients, there were 13 non-survivors (age: 64.3 ± 5.2 years [min: 19 years, max: 86 years]; gender: 10 males, 3 females; ISS: 18.4 ± 1.7 [min: 9, max: 33]), which were matched algorithmically (see below) with 13 survivors (age: 64.3 ± 5.2 years [min: 21 years, max: 88 years]; gender: 10 males, 3 females; ISS: 18.3 ± 1.8 [min: 9, max: 35]). Ten survivors (referred to as “S-7 SNP”; age: 44.4 ± 4.5 years [min: 28 years, max: 73 years]; gender: 6 males, 4 females; ISS: 30.1 ± 4.6 [min: 10, max: 54]) carried the same 7 SNP genotypes as the non-survivors. To avoid demographic differences, these 10 survivors were matched with 10 survivors (referred to as “matched S-0 SNP”; age: 43 ± 5.8 years [min: 20 years, max: 78 years]; gender: 7 males, 3 females; ISS: 25.5 ± 2.1 [min: 18, max: 35]) carrying none of the same SNP genotypes as the non-survivors (Table 2). Due to the small n-number of both patient groups (S-7 SNP: n=10; S-0 SNP: n=19), the matching was performed manually and confirmed by using Fisher’s exact test, Student t-test and Mann-Whitney U test (see below).
Figure 1. Study schematic and enrichment strategy.
A) Patient cohort derivation: 493 blunt trauma patients were enrolled in this study, 453 from whom DNA samples were obtained. 397 of these patients had complete daily clinical and biomarker records. B) Enrichment strategy: Allele frequencies for each genotype of all 551,839 SNPs were compared between the 13 non-survivors vs the 13 matched survivors. Based on this initial step, 126 SNPs were identified. In a second step, the frequencies of the discriminative 126 SNPs were compared between the non-survivors and the remaining 371 survivors, resulting in a total of 7 SNPs that were present at 100% in the 13 non-survivors and less than 50% in the 384 survivors.
Table 2.
Demographics and clinical outcomes of S-7SNP vs matched S-0 SNP patients. There were no statistically significant differences in total hospital length of stay, ICU length of stay, requirement for mechanical ventilation, and days on ventilation.
| S-7 SNP (n=10) | matched S-0 SNP (n=10) | |
|---|---|---|
| Age (years) | 44.9 ± 5.1 | 44.5 ± 5.6 |
| ISS | 31.6 ± 4.9 | 22.1 ± 2.1 |
| Gender | Male: 5 | Male: 7 |
| Female: 4 | Female: 4 | |
|
| ||
| ICU LOS (days) | 12.7 ± 4.1 | 6.8 ± 1.7 |
| total LOS (days) | 22.4 ± 6.0 | 13.3 ± 2.5 |
| Days of Ventilation (days) | 8.6 ± 3.6 | 3.3 ± 1.5 |
| Requirement for Ventilation | 0.56 | 0.55 |
Patient sub-group matching
To compare trauma non-survivors with a highly matched group of survivors, we performed a pairwise, retrospective case-control study with 1:1 matching using clinical data from the 397 blunt trauma patients detailed above. Of these patients, 13 (3.2%) were non-survivors (mean age: 64.3 ± 5.2 [min: 19 years, max: 86 years]; ISS: 18.4 ± 1.7 [min: 9, max: 33]) with a male to female ratio of 10:3. The other 384 patients (96.8%) were survivors who were discharged following hospitalization in the ICU. We next sought to select a control sub-cohort of stringently matched survivors according to the following matching criteria: age (±2 years), sex ratio (exact), and ISS score (±2 points) calculated upon hospital arrival. To do so, the 13 non-survivors were matched to 13 survivors using IBM SPSS Statistics® case-control matching (controlling for age, gender ratio, and injury severity). This resulted in the following 13 highly matched survivors (mean age: 64.3 ± 5.2 [min: 21 years, max: 88 years]; p= 0.93 vs. non-survivors), with an equivalent male to female ratio of 10:3, and a highly matched injury severity (ISS: 18.3 ± 1.8 [min: 9, max: 35]; p= 0.87 vs. non-survivors).
DNA sampling and single-nucleotide polymorphism genotyping
Whole blood samples were collected into heparinized tubes. DNA was extracted using the QIAamp® DNA Blood Midi Kit (QIAGEN, Valencia, CA) as per manufacturer’s specifications. Single nucleotide polymorphism genotyping was performed with 200 ng of genomic DNA input using the Human Core Exome-24 v1.1 BeadChip (Illumina, San Diego, CA) following the manufacturer’s Infinium® HTS Assay protocol. Briefly, DNA was denatured in 0.1N NaOH and neutralized prior to isothermal amplification. Amplified DNA was fragmented and then hybridized to locus-specific 50mers that make up the array for 16–24 h with rocking at 48°C. After removal of unbound or non-specifically annealed DNA, single base extension of the 50mer oligonucleotides was performed with labeled nucleotides, which were scanned using an Illumina iScan with autoloader 2.x. Data analysis was performed using Illumina Genome Studio 2.0.
Enrichment strategy
In order to enrich our dataset for SNPs likely to be associated with death following trauma, allele frequencies (homozygous: AA or BB; heterozygous: AB) for each genotype of all 551,839 SNPs were compared between the 13 non-survivors vs the 13 matched survivors. In the first step, those SNPs that had an allele frequency of 100% of one type and a difference greater than 50% between non-survivors and matched survivors were discriminated. Based on this initial step, 126 SNPs were identified: 44 SNPs had an allele frequency of 100% of one genotype in the non-survivors but <50% in the matched survivors, and 82 SNPs had the reciprocal allele frequency. In a second step, the frequencies of the discriminative 126 SNPs were compared between the non-survivors and the remaining 371 survivors, resulting in a total of 7 SNPs that were present at 100% in the 13 non-survivors and less than 50% in the 384 survivors (Fig. 1B).
Serial analysis of inflammatory mediators
Whole blood samples were withdrawn in heparinized tubes 3 times in the first 24 h after admission, and then daily for 7 days. The samples were kept on ice and centrifuged to obtain plasma, and then stored at −80 °C until assayed for inflammatory mediators. The Luminex™ 100 IS analyzer (Luminex, Austin, TX) and Human Cytokine/Chemokine MILLIPLEX™ Panel kit (Millipore Corporation, Billerica, MA) were used to measure plasma levels of Eotaxin (CCL11), interleukin (IL)-1β, IL-1 receptor antagonist (IL-1RA), IL-2, soluble IL-2 receptor-α (sIL-2Rα), IL-4, IL-5, IL-6, IL-7, IL-8 (CCL8), IL-10, IL-13, IL-15, IL-17, interferon (IFN)-α, IFN-γ, IFN-γ inducible protein (IP)-10 (CXCL10), monokine induced by gamma interferon (MIG; CXCL9), macrophage inflammatory protein (MIP)-1α (CCL3), MIP-1β (CCL4), monocyte chemotactic protein (MCP)-1 (CCL2), granulocyte-macrophage colony stimulating factor (GM-CSF) and tumor necrosis factor alpha (TNF-α). Human Th17 MILLIPLEX™ Panel kit (Millipore Corporation, Billerica, MA) was used to measure IL-9, IL-21, IL-22, IL-23, IL-17E/25, and IL-33. NO2−/NO3 levels were measured by a Greiss Reagent colorimetric assay (Cayman Chemical, Ann Arbor, MI). IL-1 receptor-like 1 (ST2) was measured by a sandwich ELISA assay (R&D Systems, Minneapolis, MN). All cytokine/chemokine mediator concentrations are given in pg/ml; NO2−/NO3− concentrations are in µM. Experimental data are shown as mean ± SEM.
Statistical and computational analyses
To define if patient sub-groups differed with regard to demographics, clinical outcomes, or dynamic inflammatory responses, our analytic strategy was to apply a stepwise series of statistical and data-driven modeling techniques aimed at discovering significant differences, principal drivers, interconnected networks, and potential key regulatory nodes. We detail these analyses below:
D'Agostino & Pearson normality test was used to identify if the patient demographics and outcomes were distributed normally, using GraphPad Prism 7 (Graphpad Software, Inc., San Diego, CA). A p-value of less than 0.05 was considered significant.
Student’s-t test was used to compare differences between groups of patients with regard to normally distributed demographics and outcomes, using GraphPad Prism 7. A p-value of less than 0.05 was considered significant.
The Mann-Whitney U test was used to compare differences between groups of patients with regard to non-normally distributed patient demographics and outcomes, using Graph Pad Prism 7. A p-value of less than 0.05 was considered significant.
Fisher’s Exact test was used to compare patient demographics and outcomes organized in contingency tables, using GraphPad Prism 7. A p-value of less than 0.05 was considered significant.
Two-Way ANOVA was used to determine time-dependent changes of circulating inflammatory mediators as a function of patient sub-group, using GraphPad Prism 7. A p-value of less than 0.05 was considered significant.
Principal Component Analysis (PCA) (6) was carried out to identify those inflammatory mediators that were the most characteristic of the overall dynamic, multivariate response of a given patient sub-group using Matlab® software (The MathWorks, Inc., Natick, MA). To perform this analysis, the data were first normalized for each inflammatory mediator (i.e. a given value divided by the maximum value for a given inflammatory mediator), so that all mediator levels were converted into the same scale (from 0 to 1). In this way, any artifactual effects on variance due to the different ranges of concentration observed for different cytokines were eliminated. Only sufficient components to capture at least 70% of the variance in the data were considered. From these leading principal components, the coefficient (weight) associated with each inflammatory mediator was multiplied by the eigenvalue associated with that principal component. This product represented the contribution of a given mediator to the variance accounted for in that principal component. The overall score given to each mediator is the sum of its scores in each component, depicted as a stacked bar graph. This gives a measure of a given inflammatory mediator’s contribution to the overall variance of the system. The mediators with the largest scores are the ones which contributed most to the variance of the process being studied (6).
Dynamic Network Analysis (DyNA) (6) was used to define the central inflammatory network mediators as a function of both time and patient sub-group. Using inflammatory mediator measurements of at least three time-points for experimental group, networks were created over seven consecutive time periods (Admission-D1, D1–D2, D2–D4, D4–D5 and D5–D6) using Matlab® software. Connections ([network edges] represent trajectories of inflammatory mediators [network nodes] that move in parallel; positive: same direction; negative: opposite direction) were created if the Pearson correlation coefficient between any two nodes (inflammatory mediators) at the same time-interval was greater or equal to a threshold of 0.7, as indicated. The network complexity for each time-interval was calculated using the following formula: Sum (N1 + N2 +…+ Nn)/n−1, where N represents the number of connections for each mediator and n is the total number of mediators analyzed. The total number of connections represents the sum of the number of connections across all time intervals for all patients in a given sub-group. In previous studies, we showed, that rising network complexity is associated with rising MODScores when comparing trauma survivors vs non-survivors (7).
Spearman’s correlation was performed to measure the strength of the association between the Luminex™ data for two different mediators using a modified version of a Matlab®-based toolbox described recently (7). A p-value of less than 0.05 was considered significant.
RESULTS
Differential dynamic inflammatory responses in matched blunt trauma survivors and non-survivors
Our primary goal in the present study was to determine if SNP differences could be observed between trauma survivors and non-survivors. DNA samples of 453 were obtained from a parent cohort of 493 blunt trauma patients, of whom 397 patients had complete daily clinical and biomarker data (Fig. 1A). Out of the 397, there were 13 non-survivors (age: 64.3 ± 5.2 years [min: 19 years, max: 86 years; gender: 10 males, 3 females; ISS: 18.4 ± 1.7 [min: 9, max: 35]), which were matched algorithmically (see Materials and Methods) with 13 survivors (age: 64.3 ± 5.2 years [min: 21 years, max: 88 years]; gender: 10 males, 3 females; ISS: 18.3 ± 1.8 [min: 9, max: 35]).
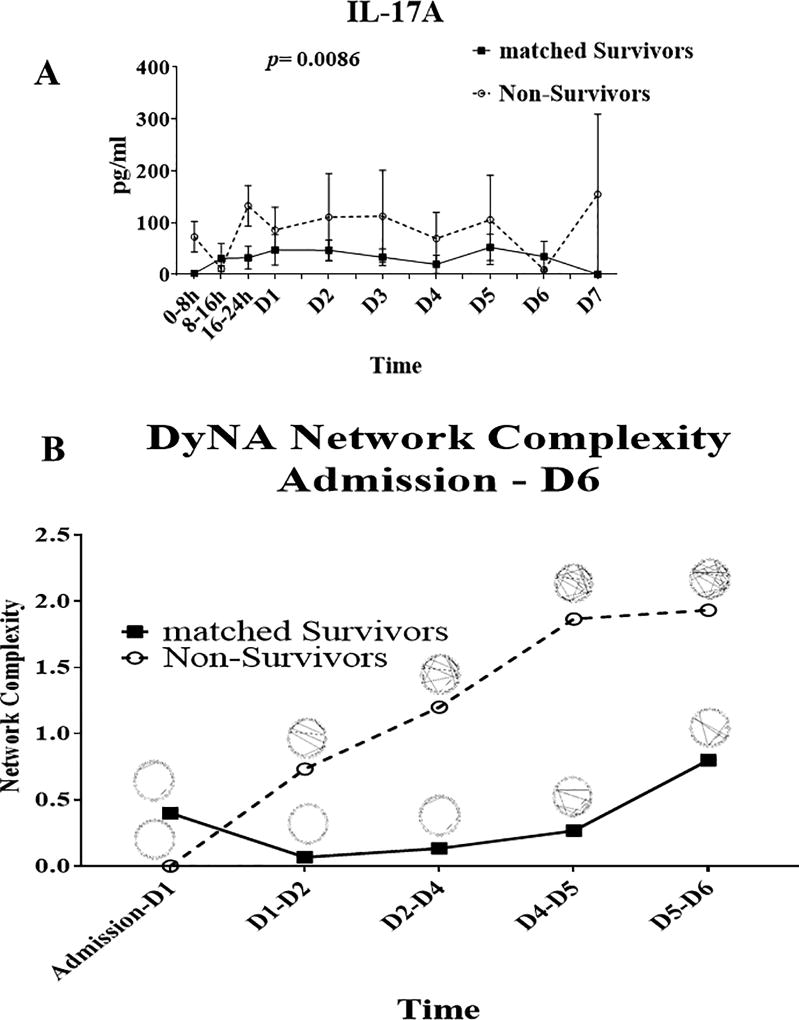
We sought initially to confirm key differences in systemic inflammatory responses between trauma survivors and non-survivors before proceeding with genomic analyses. In a recent study, we have demonstrated that, when compared to highly matched survivors, trauma non-survivors exhibit elevated IL-17A responses and rising dynamic inflammatory network complexity (7). In the present study, our data confirmed those results in the 13 non-survivors vs. 13 matched survivors in our study cohort (Figs. 2 A and B, respectively).
Figure 2. Inflammatory properties of trauma non-survivors and matched survivors.
A) IL-17A response of non-survivors and matched survivors: trauma non-survivors (n=13) exhibit significantly higher IL-17A responses compared to algorithmically matched survivors (n=13; see Materials and Methods) over a time course of seven days after admission; p= 0.0086 by two-way ANOVA. B) Trauma non-survivors (n=13) showed rising dynamic inflammatory network complexity as compared to matched survivors (n=13) over a time course of six days.
An enrichment strategy yields seven novel single-nucleotide polymorphisms that distinguish blunt trauma survivors from non-survivors
Genomic DNA samples were obtained from 453 patients, for whom complete daily clinical and biomarker data were available for 397, including the 13 non-survivors and 13 matched survivors described above (Fig. 1A). All 397 DNA samples were amplified, fragmented, and examined for 551,839 SNPs using the Infinium CoreExome-24 v1.1 BeadChip (Illumina). Allele frequencies (homozygous: AA or BB; heterozygous: AB) for each genotype of each SNP were compared between the matched groups. This initial step resulted in a total of 126 SNPs that discriminated between non-survivors and matched survivors (Fig. 1B). The discriminative SNPs were compared between the 13 non-survivors and further 371 survivors, resulting in a total of 7 SNPs that were present at 100% in the 13 non-survivors (Fig. 1 B, Table 1).
Table 1.
List of the seven SNPs that discriminate trauma survivors from non-survivors.
| Reference SNP | Position | Base Variation | Gene Name | Chromosome | Sequence |
|---|---|---|---|---|---|
| AA | |||||
| rs10741668 | 15277383 | [T/C] | ? | Chr 11, p14.1 | CAGCGTTTTACAGATGAAGAATCCA[A/G]GGTACAGAGATGTCAAAGGGCTTGG |
| rs10790334 | 98895933 | [A/G] | ? | Chr 11, q14 | GTTAAAATTCAAACTTTTGTCTGTA[C/T]GTGTATGATTTCCAAGCTATTTCTA |
| rs2065418 | 30400521 | [A/C] | MPPED2 | Chr 11, p14.1 | GGCTAATATTAACACTGACATCTGC[A/C]AAGTAATATTGGAATGGACATCCAA |
|
| |||||
| AB | |||||
| rs2241777 | 103400160 | [T/G] | SLC25A32 | Chr 8, q22.3 | AACGTAGAAATCTGTGAAACTCTAT[A/C] CTTCGTGTCAGTTTTAACATTGTGT |
| rs3134287 | 103411258 | [T/C] | SLC25A32 | Chr 8, q22.3 | CCCACCTTAGTTAGATACGTTACTC[C/T] TTATCCTCCTGCCTCCATTTCCCAA |
| rs3098223 | 103434877 | [A/G] | DCAF13 | Chr 8, q22.3 | TACTGGTGATATGTAAGAGTGAACA[C/T]GGCCTTTCAAAGGGTGAATCAAAAT |
| rs906790 | 76161264 | [A/G] | ? | Chr 13, q21 | CTTCACTCAGTCAAAAAATTTCATG[C/T] TAAGCCAGCCAGGTTTACACACATT |
Of these, three SNPs are located on chromosome 11 and have an AA genotype in the non-survivors: Reference (ref) SNP rs10741668 (Chromosome [Chr] 11; telomere p14.1; position [pos] 15,277,383; base T/C), ref SNP rs10790334 (Chr 11; q14; pos 98,895,933; A/G) and ref SNP rs2065418 (Chr 11; p14.1; pos 30,400,521; A/C). Ref SNP rs2065418 is located inside an intron region of the MPPED2 gene, and encodes metallophosphoesterase domain-containing protein 2. The other two SNPs on chromosome 11 are unknown in both name and function (Table 1). Three SNPs are located on chromosome 8 and have an AB genotype in the non-survivors: Ref SNP rs2241777 (Chr 8; q22.3; pos 103400160; T/G), Ref SNP rs3098223 (Chr 8; q22.3; pos 103,434,877; A/G) and ref SNP rs3134287 (Chr 8; q22.3; pos 103,411,258; T/C). Ref SNP rs3098223 is located inside an intron region of the DCAF13 gene, coding for DDB1- and CUL4-associated factor 13. Ref SNP rs2241777 and Ref SNP rs3134287 are both located in the SLC25A32 gene, coding for Solute carrier family 25 member 32. While Ref SNP rs2241777 is located inside an untranslated 3' end of an exon region, Ref SNP rs3134287 is located inside an intron region of the SLC25A32 gene. Ref SNP rs906790 is located on chromosome 13 (Chr 13; q21; pos 76,161,264; A/G) and is unknown in both name and function (Table 1).
Blunt trauma survivors with non-survivor SNPs showed no differences in clinical outcomes compared to survivors without those SNPs
We next hypothesized that survivors with the same seven SNPs as the non-survivors would have worse clinical outcomes and a different dynamic inflammatory response as compared to survivors with none of the 7 SNPs characteristic of the 13 non-survivors in our cohort. A total of 10 survivors were found with the same 7 SNPs as non-survivors (referred to as “S-7 SNP”). Accordingly, we matched the S-7 SNP patients manually with 10 survivors, none of whom carried any of the 7 SNP genotypes as the non-survivors (referred to as “matched S-0 SNP”). Contrary to our hypothesis, there were no statistically significant differences between S-7 SNP and matched S-0 SNP patients with regard to total hospital length of stay, ICU length of stay, requirement for mechanical ventilation, and days on ventilation (Table 2). In addition, there were no significant differences in nosocomial infections (Suppl. Fig. 1A), degree of hypotension (Suppl. Fig. 1B), shock index (Suppl. Fig. 1C), MODScore trajectories over 7 days (Suppl. Fig. 1D), or comorbidities (Suppl. Fig. 1E) between S-7 SNP and matched S-0 SNP patients.
S-7 SNP patients exhibit distinct inflammatory responses from matched S-0 SNP patients
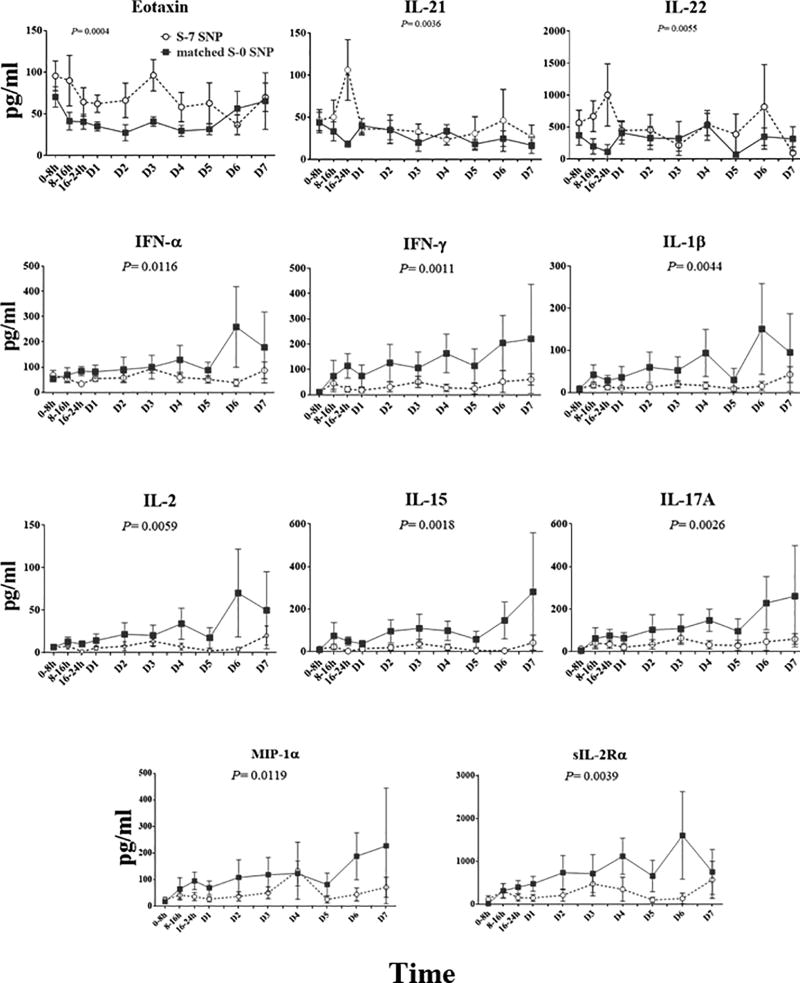
We next compared the dynamic inflammatory responses of these two patient sub-groups. We measured plasma levels of 31 inflammatory mediators over a time course of 7 days following injury. Eleven of these mediators showed statistically significant differences between S-7 SNP and matched S-0 SNP patients: S-7 SNP patients expressed higher levels of Eotaxin (p= 0.0036), IL-21 (p= 0.0255) and IL-22 (p= 0.0351) vs. matched S-0 SNP patients, but lower plasma levels of IFN-α (p= 0.0116), IFN-γ (p= 0.0011), IL-1β (p= 0.044), IL-2 (p= 0.0059), IL-15 (p= 0.0018) and IL-17A (p= 0.0026), MIP-1a (p= 0.0119) and sIL-2Ra (0.0039) (Fig. 3).
Figure 3. Circulating levels of inflammatory mediators over a time course of seven days after admission of S-7 SNP vs matched S-0 SNP patients.
Survivors with the same 7 SNPs as non-survivors (S-7 SNP patients; n = 10) expressed significantly different levels of multiple circulating inflammatory mediators as compared to survivors with none of the same SNPs as non-survivors (matched S-0 SNP; n = 10).
Computational modeling suggests a role for altered Th17 responses in S-7 SNP patients vs. matched S-0 SNP patients, which in turn differ from those of non-survivors
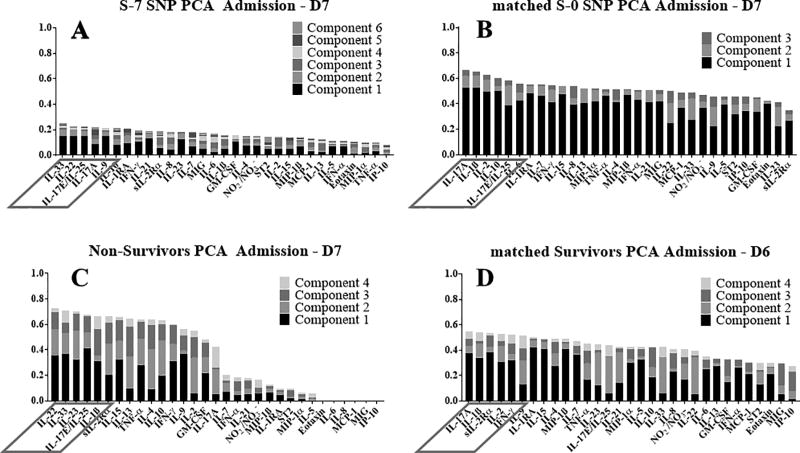
Our next goal was to identify the principal characteristics, and thus possibly identify main drivers, of the inflammatory responds of both S-7 SNP and matched S-0 SNP patients as well as of matched survivors and non-survivors in an unbiased way over the first 7 days following admission. Therefore, we utilized Principal Component Analysis (PCA) of the dynamic, multivariate data of all 4 cohorts. In this analysis, the overall score given to each mediator is the sum of its scores in each principal component, depicted as a stacked bar graph (Fig. 4). This gives a measure of a given inflammatory mediator’s contribution to the overall variance of the system. The mediators with the largest scores are the ones which contributed most to the variance of the process being studied
Figure 4. Principal component analysis suggests a role for type 17 immunity in the circulating inflammatory response to traumatic injury.
Principal component analysis was carried out using the data on all four patient sub-groups (non-survivors, matched survivors, S-7 SNP, and matched S-0 SNP) as described in the Materials and Methods. In S-7 SNP patients, IL-33, IL-22, IL-17E/25, IL-17A, and IL-9 were the most relevant inflammatory mediators (A). In the matched S-0 SNP patients, IL-17A. IL-1β, IL-2, IL-10, and IL-17E/25 were the most relevant inflammatory mediators (B). In the non-survivos, IL-22, IL-33, IL-23, IL-17E/25, and IL-1β appeared as the principal characteristics of the inflammatory response (C). In the matched survivors, sIL-2Rα, IL-4, IFN-γ, IL-7 and IL-17 appeared as the principal characteristics (D).
This analysis pointed to type 17-related immune mediators (8) in all patient groups (Fig. 4). In S-7 SNP patients, IL-33, IL-22, IL-17E/25, IL-17A, and IL-9 were the most relevant inflammatory mediators (Fig. 4A). In the matched S-0 SNP patients, IL-17A, IL-1β, IL-2, IL-10, and IL-17E/25 were the most relevant inflammatory mediators (Fig. 4B). In non-survivors, IL-22, IL-33, IL-23, IL-17E/25, and IL-1β appeared as the principal characteristics of the inflammatory response (Fig. 4C). In the matched survivors, IL-17A, IL-1β, sIL-2Rα, IL-2 and IFN-γ appeared as the principal characteristics (Fig. 4D).
While the mediators of S-7 SNP patients had a low weighting of less than 0.25 in the PCA (Fig. 4A), the other three cohorts (matched S-0 SNP, matched survivors, non-survivors) had higher weighting levels of greater than 0.5 (Fig. 4B–D). Six components contributed to the profile of S-7 SNP patients (Fig. 4A), while there were four in both non-survivors and matched survivors (Fig. 4C+D) and three in matched S-0 SNP patients (Fig. 4B). Non-survivors showed a condensed PCA pattern of inflammatory mediators with a drop in PCA weighting after 16 mediators (Fig. 4C), while the pattern of the other three cohorts (matched survivors, S-7 SNP, matched S-0 SNP) had a more spread profile with only small drops in PCA weightings (Fig. 4A, B+D).
Taken together, these results suggest that non-survivors exhibit a systemic inflammatory response that is comprised of a more restricted number of pathways recruited over time as compared to the other three groups. Furthermore, this analysis suggests that S-7 SNP patients undergo a dampened systemic inflammatory response as compared to the other three groups.
The PCA and two-way ANOVA results suggested that type 17 responses play a central role in the post-trauma systemic inflammatory response of all patient sub-groups in the present study. Kuchroo and co-workers have described a sub-population of type 17 cells known as pathogenic Th17 cells that, in addition to expressing IL-17A, upregulate GM-CSF and down-regulate IL-10 in a manner that is potentiated by IL-23; a reciprocal cell population, non-pathogenic Th17 cells, co-express IL-17A and IL-10 (9). As mentioned above, we demonstrated recently that trauma non-survivors exhibit a positive correlation between IL-17A and GM-CSF – while exhibiting a negative correlation between IL-17A and IL-10 – thus suggesting a shift toward pathogenic Th17 cells that characterize, and may be involved in, the systemic inflammation associated with mortality in trauma patients (7). In that study, matched survivors showed no correlation between IL-17A and either GM-CSF or IL-10 (7).
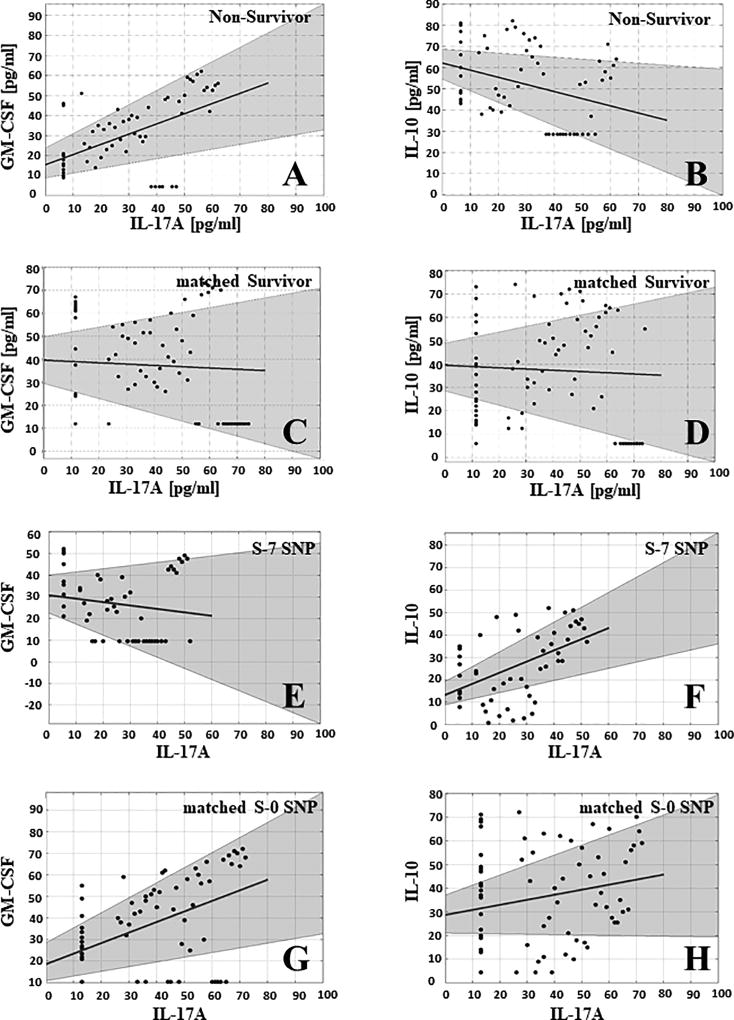
We therefore hypothesized that one potential mechanism by which S-7 SNP patients avoid the runaway, self-sustaining systemic inflammation following injury that is characteristic of non-survivors is due to an altered Th17 balance as compared to non-survivors. As in our prior study (7), non-survivors showed a significant, positive correlation between IL-17A and GM-CSF (r= 0.51, p<0.0001; Fig. 5A), while matched survivors showed no correlation between those two mediators (r= −0.06; p= 0.63; Fig. 5C). In contrast, IL-17A and IL-10 exhibited a significant, negative correlation in non-survivors (r= −0.34; p=0.007; Fig. 5B), but not in matched survivors (r= −0.05; p= 0.66; Fig. 5D). Thus, non-survivors appear to exhibit a bias towards pathogenic Th17 cells.
Figure 5. Spearman correlations of IL-17A vs. GM-CSF or IL-10 in non-survivors, matched survivors, S-7 SNP and matched S-0 SNP patients suggest differential Th17 responses.
Non-survivors showed a significant, positive correlation between IL-17A and GM-CSF (r= 0.51, p<0.0001) (A), and a significant, negative correlation between IL-17A and IL-10 (r= −0.34; p=0.007) (B). In contrast, matched survivors showed no correlations between either IL-17A and GM-CSF (r= −0.06; p= 0.63) (C) or between IL-17A and IL-10 (r= −0.05; p= 0.66) (D). S-7 SNP patients exhibited no significant correlation between IL-17A and GM-CSF (r= −0.17; p= 0.21; Fig. 4E), but showed a significant, positive correlation between IL-17A and IL-10 (r= 0.51; p<0.0001) (F). In contrast, matched S-0 SNP patients showed a significant, positive correlation between IL-17A and GM-CSF (r= 0.52; p< 0.0001) (G), but no significant correlation between IL-17A and IL-10 (r= 0.21; p= 0.08) (H).
In contrast to non-survivors, S-7 SNP patients exhibited no significant correlation between IL-17A and GM-CSF (r= −0.17; p= 0.21; Fig. 5E), but showed a significant, positive correlation (r= 0.51; p <0.0001; Fig. 5F) between IL-17A and IL-10. Thus, S-7 SNP patients appear to exhibit a bias toward non-pathogenic Th17 cells. In contrast to S-7 SNP, matched S-0 SNP patients showed a significant, positive correlation between IL-17A and GM-CSF (r= 0.52; p< 0.0001; Fig. 5G), but no significant correlation between IL-17A and IL-10 (r= 0.21; p= 0.08; Fig. 5H).
These results suggest that the hallmark of non-survival involves, at minimum, the presence of seven particular SNPs along with the upregulation of pathogenic Th17 cells and down-regulation of non-pathogenic Th17 cells relative to the general non-survivor cohort. These results further suggest that survival post-trauma in the presence of a non-survivor SNP haplotype may require a commensurate shift in the Th17 response to both upregulate non-pathogenic Th17 cells and down-regulate pathogenic Th17 cells, associated with elevation in cytokines that downregulate the type 17 response and potentially a shift toward type 2 inflammation. Interestingly, the data in matched S-0 SNP patients suggests that upregulation of pathogenic Th17 cells, while a hallmark of non-survivors, may be compensated by both the absence of the non-survivor SNPs as well as the lack of downregulation of non-pathogenic Th17 cells, and thus allow a more balanced inflammatory response to take place.
DISCUSSION
Over the past three decades, the search for early biomarkers that are prognostic of trauma outcomes has led to an extensive and growing list of cell populations and their inflammatory mediators (1). More recently, this search has been extended to examine the potential role of individual genetic variability, in the form of SNPs, on trauma outcomes (2, 5). These genomic studies have defined the roles of candidate SNPs, typically in genes encoding inflammatory cytokines (e.g. TNF-α (10), IL-6 (11), and IL-10 (12)); pattern recognition receptors (e.g. Toll-like receptors (13)), and downstream signaling intermediates (e.g. kinases downstream of Toll-like receptors (14)).
The main limitation of these studies is their focus on investigator-defined candidate genes, and, thus, these studies represent tests of specific hypotheses regarding the role of previously established mediators or signaling pathways. The alternative approach is to carry out a genome-wide association study to identify SNPs associated with specific outcomes. The advantage of this approach is that it is substantially less biased. The main disadvantage, however, is that this unbiased approach requires a very large number of patients to deal with inter-patient variability as well as the fact that the number of variables (SNPs) is much larger than the number of available subjects (15).
In the present study, we hypothesized that an enrichment strategy based on identifying SNPs that discriminate trauma non-survivors from highly matched survivors would allow for a relatively unbiased screen for SNPs that characterize these two key trauma outcomes. At its core, our strategy depends on the identification of highly matched subjects whose outcomes are drastically different. We therefore suggest that this novel enrichment approach could be useful in multiple other disease settings, both in the critical care arena (e.g. sepsis) and other diseases in which dramatically different outcomes are observed along with large interpatient variability.
This enrichment strategy led to the identification of 126 SNPs initially, a list which was reduced to seven SNPs upon comparison of non-survivors to a broader population of trauma survivors. For three of these seven SNPs, neither the genes nor their products are known.
For the remaining four SNPs, more information is available. Ref SNP rs2065418 is located inside an intron region of the MPPED2 gene, and encodes metallophosphoesterase domain-containing protein 2. Currently, there are only 13 PubMed entries for MPPED2. A recent genome-wide study suggested that this gene influences the estimated glomerular filtration rate (eGFR), and appears to be associated with renal function in patients with chronic kidney disease (16). Another study associated MPPED2 with the pathobiology of experimental ventilator-induced lung injury (17). Yet other studies suggested a loss of the anti-proliferative functions of MPPED2 in tumor neogenesis (18), and found that increased expression of MPPED2 is associated with better outcomes in neuroblastoma patients (19). We therefore speculate that a certain MPPED2 genotype, likely AA, possibly impacts the renal function of trauma patients and/or leads to lung damage in patients receiving prolonged mechanical ventilation. Due to its putative anti-proliferative function, it is also possible that MPPED2 AA impairs the tissue repair mechanisms of trauma patients.
Ref SNP rs2241777 is located inside a not translated 3' end of an exon and Ref SNP rs3134287 is located inside an intron region of the SLC25A32 gene, encoding for the protein solute carrier family 25 member 32, known alternatively as mitochondrial folate transporter/carrier. There are 13 entries on PubMed for SLC25A32. The SLC25A32 protein transports flavin adenine dinucleotide (FAD) over the mitochondrial membrane, ultimately helping drive the production of adenosine triphosphate (ATP). SLC25A32 is thought to play a role in multiple acyl-CoA dehydrogenase deficiency (MADD)-like diseases (20). Since recovery from severe injuries is a highly energetic challenge for the body (21), an altered activity of this FAD transporter due to a distinct genotype (according to our findings, the AB genotype) could lead to worse outcomes after trauma caused by an impaired supply of ATP.
Ref SNP rs3098223 is located inside an intron region of the DCAF13 gene, and encodes for DDB1- and CUL4-associated factor 13. To date, there are no entries for DCAF13 on PubMed. However, DDB1 and CUL4-associated factors (DCAFs) are substrates of the CUL4-DDB1 ubiquitin ligase, which is a regulator of cell proliferation and survival (22). Alterations in the gene could lead to impaired tissue repair after trauma.
A key aspect of the present studies was the use of computational modeling. Our group has previously combined transcriptomic and genomic analyses with computational modeling to gain a greater understanding of the role of inflammation following traumatic injury (23). We have described the potential utility of PCA to gain insights into experimental trauma/hemorrhage in mice (6). In the present study, we carried out PCA to define the principal characteristics (and potential drivers) of the dynamic, multivariate inflammatory responses of non-survivors, matched survivors, S-7 SNP, and matched S-0 SNP patients. In a broad sense, this analysis suggested that non-survivors exhibit a systemic inflammatory response that is comprised of a more restricted number of pathways following injury as compared survivors in general.
This analysis also led us to the hypothesis that the type 17 immune response, and more specifically the Th17 response, is a central characteristic of these patient groups. Notably, none of the SNPs we identified are those previously identified in the Th17 pathway (24–29), though these SNPs were included in our array (data not shown). Combined with subsequent correlation analyses, our results lead us to posit that there is an interaction between genetic predisposition for inadequate tissue repair or dysfunctional energetics, along with early (or pre-existing) immune/inflammatory state, in driving survival or death following severe traumatic injury. Indeed, it is tempting to speculate that having the seven SNPs we identified as distinguishing non-survivors from survivors may lead to a degree of tissue repair. This deficit may act in concert with a robust, self-sustaining inflammatory response (an “inflammation accelerator”) driven by pathogenic Th17 cells, together leading to death. We would further speculate that patients who survive despite having the same seven SNPs as non-survivors do so because their immune responses are dominated by non-pathogenic Th17 cells, which act to limit inflammation induced by the initial injury as well as secondary organ dysfunction (an “inflammation brake pedal”). Indeed, the systemic inflammatory responses of S-7 SNP patients exhibit elevations in the types of cytokines that limit Th17 responses (e.g. IL-21 and IL-22) combined with lower levels of IL-17A. Furthermore, PCA implicated the anti-inflammatory IL-17E/IL-25 as a central driver of this process.
In the matched S-0 SNP patients, it is tempting to speculate that upregulation of pathogenic Th17 cells is compensated for by the absence of all 7 SNPs that are characteristic of non-survivors, what might act in a tissue-protective manner. This may explain why matched S-0 SNP and S-7 SNP patients do not exhibit significant differences in clinical outcomes such as ICU length of stay or duration of mechanical ventilation. Indeed, none of those patients have both a pathogenic Th17 response and a higher tissue vulnerability simultaneously. These hypotheses are also reflected by the MODScores of each group over a time course of seven days (Suppl. Fig. 2), but given the overall low number of trauma non-survivors as well as survivors exhibiting the seven non-survivor SNPs within our inclusion/exclusion criteria, the must be tested in a larger patient cohort.
Another key cytokine that was implicated in the responses of S-7 SNP patients by PCA, and which is known to synergize with IL-17E/IL-25 in driving type 2 innate lymphoid cell responses, is IL-33 (30). Tissue damage can lead to the release of damage-associated molecular pattern molecules (DAMPs) that alert the immune system to potential breeches in barrier integrity (1). Interleukin-33, a member of the IL-1 cytokine super-family, is expressed constitutively in the epithelium and acts as a DAMP after tissue injury by activating immune cells through its cognate receptor, ST2 (31). Recent studies from our group in trauma patients demonstrated elevated circulating levels of IL-33 early post-injury, which were correlated with increases in the type 2 cytokines IL-4, IL-5, and IL-13 (32). Based on studies in a mouse model of trauma/hemorrhagic shock, IL-33 drives IL-5 expression by Group 2 innate lymphoid cells (ILC2) and CXCR2+ neutrophils in the lung (32). Thus, it is possible that IL-33 also helps limit inflammation in S-7 SNP patients by downregulating Th17 responses (33) and promoting type 2 inflammation (34); our finding of elevated IL-5 in S-7 SNP patients would support this hypothesis.
This study must be interpreted in the context of several limitations. A key limitation is the size of the patient sub-groups we have identified; clearly, this study must be validated in a larger and more diverse trauma patient population, ideally sampled at multiple institutions. Another limitation involves the number of SNPs studied. Although quite large, and involving both known disease-associated SNPs and SNPs spanning the human genome, it is entirely possible that additional SNPs might be identified if full genome sequencing were to be carried out on each patient. At present, that endeavor is a daunting one due to considerations of cost and patient recruitment. Finally, additional circulating inflammatory mediators could be assayed, as could the full transcriptome, metabolome, etc. to gain further insights into factors that differentiate trauma survivors from non-survivors.
In conclusion, our study suggests a novel enrichment strategy by which to identify SNPs associated with trauma outcomes. We demonstrate a SNP profile, whose absence in any given blunt trauma patient we construe to imply patients survive an episode of moderate to severe trauma. This study reinforces conclusions from prior work which suggests a key, early role for the type 17 immune pathway in driving survival or death after trauma (7). Notably, our study raises potentially thorny ethical questions regarding how this type of genomic information and inflammatory cell information could be used, especially in targeting care or triaging severely injured trauma patients. As such, further work is needed before the full significance of the genetic polymorphisms we have identified is understood.
Supplementary Material
Non-survivors (n=13) showed a significant elevated MODScore over a time course of 7 days compared to the other cohorts (matched survivors [n=13], matched S-0 SNP [n=10], S-7 SNP [n=10]) (p< 0.0001)
There were no significant differences in nosocomial infections (A), degree of hypotension (B), shock index (C), MODScore trajectories over 7 days (D), or comorbidities (E) between S-7 SNP (n=10) and matched S-0 SNP patients (n=10).
Acknowledgments
This work was supported by NIH grant P50-GM-53789. This project used the University of Pittsburgh HSCRF Genomics Research Core Illumina Infinium genotyping service.
ABBREVIATIONS
- ANOVA
Analysis of Variance
- ATP
adenosine triphosphate
- DAMP
damage-associated molecular pattern molecule
- DCAF
DDB1- and CUL4-associated factor
- DyNA
Dynamic Network Analysis
- eGFR
estimated glomerular filtration rate
- FAD
Flavin adenine dinucleotide
- GM-CSF
Granulocyte-Macrophage Colony-Stimulating Factor
- ICU
Intensive Care Unit
- IFN
Interferon
- IL
Interleukin
- IL-1RA
IL-1 receptor antagonist
- IP-10
Interferon-γ-inducible Protein of 10 kDa
- ISS
Injury Severity Score
- MADD
Multiple Acyl-CoA Dehydrogenase Deficiency
- AMCP-1
Monocyte Chemoattractant Protein-1
- MIG
Monokine induced by Interferon-γ
- MIP
Macrophage Inflammatory Protein
- MPPED2
metallophosphoesterase domain containing protein 2
- NO2−
nitrite
- NO3−
nitrate
- SEM
standard error of the mean
- sIL-2Rα
Soluble Interleukin-2 receptor α-chain
- SLC25A32
solute carrier family 25 member 32
- SNP
Single Nucleotide Polymorphism
- ST2
IL-1 receptor-like 1
- Th17
T helper 17 cells
- PCA
Principal Component Analysis
- TNF-α
Tumor Necrosis Factor-α
References
- 1.Namas RA, Mi Q, Namas R, Almahmoud K, Zaaqoq AM, Abdul-Malak O, Azhar N, Day J, Abboud A, Zamora R, et al. Insights into the role of chemokines, damage-associated molecular patterns, and lymphocyte-derived mediators from computational models of trauma-induced inflammation. Antiox. Redox Signaling. 2015;10:1370–1387. doi: 10.1089/ars.2015.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrand F, Mommsen P, Frink M, van Griensven M, Krettek C. Genetic predisposition for development of complications in multiple trauma patients. Shock. 2011;35(5):440–8. doi: 10.1097/SHK.0b013e31820e2152. [DOI] [PubMed] [Google Scholar]
- 3.Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001;2(12):930–42. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 4.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. A genomic storm in critically injured humans. J. Exp. Med. 2011;208(13):2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronkhorst MW, P Patka P, van Lieshout EM. Effects of sequence variations in innate immune response genes on infectious outcome in trauma patients: A comprehensive review. Shock. 2015;44(5):390–6. doi: 10.1097/SHK.0000000000000450. [DOI] [PubMed] [Google Scholar]
- 6.Mi Q, Constantine G, Ziraldo C, Solovyev A, Torres A, Namas R, Bentley T, Billiar TR, Zamora R, Puyana JC, et al. A dynamic view of trauma/hemorrhage-induced inflammation in mice: Principal drivers and networks. PLoS ONE. 2011;6:e19424. doi: 10.1371/journal.pone.0019424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abboud A, Namas RA, Ramadan M, Mi Q, Almahmoud K, Abdul-Malak O, Azhar N, Zaaqoq A, Namas R, Barclay DA, et al. Computational analysis supports an early, type 17 cell-associated divergence of blunt trauma survival and mortality. Crit Care Med. 2016;44:e1074–e1081. doi: 10.1097/CCM.0000000000001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: Lessons from genetics and therapeutic interventions. Immunity. 2015;43(6):1040–51. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–9. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majetschak M, Obertacke U, Schade FU, Bardenheuer M, Voggenreiter G, Bloemeke B, Heesen M. Tumor necrosis factor gene polymorphisms, leukocyte function, and sepsis susceptibility in blunt trauma patients. Clin Diagn Lab Immunol. 2002;9(6):1205–11. doi: 10.1128/CDLI.9.6.1205-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heesen M, Obertacke U, Schade FU, Bloemeke B, Majetschak M. The interleukin-6 G(−174)C polymorphism and the ex vivo interleukin-6 response to endotoxin in severely injured blunt trauma patients. Eur Cytokine Netw. 2002;13(1):72–7. [PubMed] [Google Scholar]
- 12.Upperman JS, Pillage G, Siddiqi MQ, Zeevi A, Kelly N, Ford HR, Kammerer C, Spolarics Z. Dominance of high-producing interleukin 6 and low-producing interleukin 10 and interferon gamma alleles in glucose-6-phosphate dehydrogenase-deficient trauma patients. Shock. 2005;23(3):197–201. [PubMed] [Google Scholar]
- 13.Barber RC, Aragaki CC, Rivera-Chavez FA, Purdue GF, Hunt JL, Horton JW. TLR4 and TNF-alpha polymorphisms are associated with an increased risk for severe sepsis following burn injury. J Med Genet. 2004;41(11):808–813. doi: 10.1136/jmg.2004.021600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperry JL, Zolin S, Zuckerbraun BS, Vodovotz Y, Namas R, Neal MD, Ferrell RE, Rosengart MR, Peitzman AB, Billiar TR. X-chromosome linked IRAK1 polymorphism is a strong predictor of multiple organ failure and mortality post-injury. Ann Surg. 2014;260:698–703. doi: 10.1097/SLA.0000000000000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraft P, Cox DG. Study designs for genome-wide association studies. Adv Genet. 2008;60:465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]
- 16.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012;8(3):e1002584. doi: 10.1371/journal.pgen.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kompass KS, Deslee G, Moore C, McCurnin D, Pierce RA. Highly conserved transcriptional responses to mechanical ventilation of the lung. Physiol Genomics. 2010;42(3):384–96. doi: 10.1152/physiolgenomics.00117.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen L, Liu L, Ge L, Xie L, Liu S, Sang L, Zhan T, Li H. miR-448 downregulates MPPED2 to promote cancer proliferation and inhibit apoptosis in oral squamous cell carcinoma. Exp Ther Med. 2016;12(4):2747–2752. doi: 10.3892/etm.2016.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liguori L, Andolfo I, de Antonellis P, Aglio V, di Dato V, Marino N, Orlotti NI, De Martino D, Capasso M, Petrosino G, et al. The metallophosphodiesterase Mpped2 impairs tumorigenesis in neuroblastoma. Cell Cycle. 2012;11(3):569–81. doi: 10.4161/cc.11.3.19063. [DOI] [PubMed] [Google Scholar]
- 20.Spaan AN, Ijlst L, van Roermund CW, Wijburg FA, Wanders RJ, Waterham HR. Identification of the human mitochondrial FAD transporter and its potential role in multiple acyl-CoA dehydrogenase deficiency. Mol Genet Metab. 2005;86(4):441–7. doi: 10.1016/j.ymgme.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Poulose N, Raju R. Aging and injury: alterations in cellular energetics and organ function. Aging Dis. 2014;5(2):101–8. doi: 10.14336/AD.2014.0500101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26(6):775–80. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Brown D, Namas RA, Almahmoud K, Zaaqoq A, Sarkar J, Barclay DA, Yin J, Ghuma A, Abboud A, Constantine G, et al. Trauma in silico: individual-specific mathematical models and virtual clinical populations. Sci Transl Med. 2015;7:285ra61. doi: 10.1126/scitranslmed.aaa3636. [DOI] [PubMed] [Google Scholar]
- 24.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Nakamura M, Yoshioka D, Arima Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28(1):44–9. doi: 10.1007/s10875-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 25.Marwa OS, Kalthoum T, Wajih K, Kamel H. Association of IL17A and IL17F genes with rheumatoid arthritis disease and the impact of genetic polymorphisms on response to treatment. Immunol Lett. 2017;183:24–36. doi: 10.1016/j.imlet.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Abdollahi E, Tavasolian F, Momtazi-Borojeni AA, Samadi M, Rafatpanah H. Protective role of R381Q (rs11209026) polymorphism in IL-23R gene in immune-mediated diseases: A comprehensive review. J Immunotoxicol. 2016;13(3):286–300. doi: 10.3109/1547691X.2015.1115448. [DOI] [PubMed] [Google Scholar]
- 27.Lill CM, Schilling M, Ansaloni S, Schröder J, Jaedicke M, Luessi F, Schjeide BM, Mashychev A, Graetz C, Akkad DA, et al. Assessment of microRNA-related SNP effects in the 3' untranslated region of the IL22RA2 risk locus in multiple sclerosis. Neurogenetics. 2014;15(2):129–34. doi: 10.1007/s10048-014-0396-y. [DOI] [PubMed] [Google Scholar]
- 28.Shi ZF, Fang QB, Limu S, Jiareke T, Ge XH. Association Between Three SNPs and Thromboangiitis Obliterans in Xinjiang Uyghur Population. Genet Test Mol Biomarkers. 2016;20(2):55–62. doi: 10.1089/gtmb.2015.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YG, Ihm CG, Lee TW, Lee SH, Jeong KH, Moon JY, Chung JH, Kim SK, Kim YH. Association of genetic polymorphisms of interleukins with new-onset diabetes after transplantation in renal transplantation. Transplantation. 2012;93(9):900–7. doi: 10.1097/TP.0b013e3182497534. [DOI] [PubMed] [Google Scholar]
- 30.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, Huang LC, Johnson D, Scanlon ST, McKenzie AN, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210(13):2939–50. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peine M, Marek RM, Lohning M. IL-33 in T cell differentiation, function, and immune homeostasis. Trends Immunol. 2016;37(5):321–33. doi: 10.1016/j.it.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Guardado J, Hoffman R, Xu H, Namas R, Vodovotz Y, Xu L, Ramadan M, Brown J, Turnquist HR, et al. IL-33 drives lung injury via ILC2 promotion of neutrophil IL-5 production after polytrauma. PLoS Med. 2017 doi: 10.1371/journal.pmed.1002365. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vocca L, Di Sano C, Uasuf CG, Sala A, Riccobono L, Gangemi S, Albano GD, Bonanno A, Gagliardo R, Profita M. IL-33/ST2 axis controls Th2/IL-31 and Th17 immune response in allergic airway diseases. Immunobiology. 2015;220(8):954–63. doi: 10.1016/j.imbio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–90. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Non-survivors (n=13) showed a significant elevated MODScore over a time course of 7 days compared to the other cohorts (matched survivors [n=13], matched S-0 SNP [n=10], S-7 SNP [n=10]) (p< 0.0001)
There were no significant differences in nosocomial infections (A), degree of hypotension (B), shock index (C), MODScore trajectories over 7 days (D), or comorbidities (E) between S-7 SNP (n=10) and matched S-0 SNP patients (n=10).