Genes controlled by imprinting were evolutionarily conserved during wheat polyploidization.
Abstract
Genomic imprinting is an epigenetic phenomenon that causes genes to be differentially expressed depending on their parent of origin. To evaluate the evolutionary conservation of genomic imprinting and the effects of ploidy on this process, we investigated parent-of-origin-specific gene expression patterns in the endosperm of diploid (Aegilops spp), tetraploid, and hexaploid wheat (Triticum spp) at various stages of development via high-throughput transcriptome sequencing. We identified 91, 135, and 146 maternally or paternally expressed genes (MEGs or PEGs, respectively) in diploid, tetraploid, and hexaploid wheat, respectively, 52.7% of which exhibited dynamic expression patterns at different developmental stages. Gene Ontology enrichment analysis suggested that MEGs and PEGs were involved in metabolic processes and DNA-dependent transcription, respectively. Nearly half of the imprinted genes exhibited conserved expression patterns during wheat hexaploidization. In addition, 40% of the homoeolog pairs originating from whole-genome duplication were consistently maternally or paternally biased in the different subgenomes of hexaploid wheat. Furthermore, imprinted expression was found for 41.2% and 50.0% of homolog pairs that evolved by tandem duplication after genome duplication in tetraploid and hexaploid wheat, respectively. These results suggest that genomic imprinting was evolutionarily conserved between closely related Triticum and Aegilops species and in the face of polyploid hybridization between species in these genera.
INTRODUCTION
During double fertilization, a phenomenon unique to flowering plants, the egg cell (1n) and central cell (2n) fuse with two sperm cells (1n) to generate the diploid embryo (2n) and the triploid endosperm (3n), respectively. The resulting endosperm, a functional analog of the placenta in mammals, facilitates embryogenesis and supports seedling growth by providing the embryo with nutrients. The endosperm tissue also interacts dynamically with the embryo over the course of development by activating important signaling pathways that are required for embryo development (Yang et al., 2008; Fouquet et al., 2011; Costa et al., 2014; Xu et al., 2014). Therefore, proper endosperm development is essential for coordinating embryo and seed growth.
Genomic imprinting, which refers to monoallelic gene expression in a parent-of-origin-dependent manner, generally involves epigenetic regulation. In plants, imprinting primarily occurs in the endosperm; however, recent studies have shown that a portion of genes are also imprinted in the embryo of Arabidopsis thaliana (Raissig et al., 2013), rice (Oryza sativa) (Luo et al., 2011), and maize (Zea mays) (Meng et al., 2017). This uniparental transcription pattern indicates that, to some extent, parental genomes might not contribute equally to the filial genome, at least for some specific loci, if not at the genome-wide level (Vielle-Calzada et al., 2000; Grimanelli et al., 2005; Autran et al., 2011).
Two major hypotheses have been proposed to explain the extensive occurrence and convergent evolution of genomic imprinting across flowering plants and mammals. One hypothesis, the parental conflict theory, argues that paternally derived alleles promote the transport of resources from maternal tissue to the offspring to improve their fitness, whereas maternally derived alleles tend to share resources equally to balance nutrient allocation among embryos (Haig and Westoby, 1989; Wilkins and Haig, 2003). The other hypothesis, the maternal-offspring coadaptation model, proposes adaptive integration rather than a struggle for resources between the maternal tissue and the offspring (Curley et al., 2004; Wolf and Hager, 2006; Swaney et al., 2007; Keverne and Curley, 2008). However, the biological relevance of the parent-of-origin expression pattern of a gene remains a matter of considerable debate, and the potential effects of allele-specific expression on the embryo and seedling remain ambiguous, since genomic imprinting mainly occurs in the terminal tissue of the endosperm, which does not genetically contribute to the next generation.
The biological implications of genomic imprinting can be inferred from the results of reciprocal crosses of plants with different ploidy levels, thus providing different dosages of the parental genomes. Specifically, paternal-excess crosses strongly promote seed development, resulting in the production of big seeds, whereas maternal-excess crosses dramatically inhibit endosperm growth, resulting in the production of small seeds (Lin, 1984; Scott et al., 1998). These findings indicate that the correct balance between maternally and paternally derived genomes is responsible for proper embryo and endosperm development. Furthermore, A. thaliana plants with loss of function of the maternally inherited alleles of the imprinted genes MEDEA (MEA) and FERTILIZATION INDEPENDENT SEED2 (FIS2), which encode components of the Polycomb Repressive Complex 2 (PRC2), exhibit overproliferation of endosperm after fertilization. This finding suggests that MEA and FIS2 restrain seed growth (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999). However, other studies have argued that the expression level rather than the imprinting pattern of a gene is likely indispensable for phenotypic variation, as plants with loss of function of MULTICOPY SUPPRESSOR OF IRA1, which encodes a nonimprinted subunit of PRC2, exhibit the same phenotypes as those with mutations in MEA and FIS2 (Köhler et al., 2003; Guitton and Berger, 2005; Leroy et al., 2007). In addition, emerging evidence suggests that paternally expressed genes are involved in establishing postzygotic hybridization barriers in A. thaliana, as downregulating the expression of the paternally imprinted genes ADMETOS, SU(VAR)3-9, HOMOLOG7, PATERNALLY EXPRESSED IMPRINTED GENE2 (PEG2), and PEG9 can partially rescue triploid seed development (Kradolfer et al., 2013; Wolff et al., 2015). In maize, the maternal expression of Meg1 in basal endosperm transfer cells was directly shown to be functionally relevant for seed development and growth. This study not only verified the necessity and sufficiency of Meg1 in regulating transfer cell differentiation, but also demonstrated the importance of imprinted gene expression in controlling seed size, as revealed through the development of transgenic lines with reduced expression, ectopic expression, and nonimprinted expression of Meg1 (Costa et al., 2014).
RNA-seq analyses have identified hundreds of imprinted genes in A. thaliana, maize, rice, castor bean (Ricinus communis), and sorghum (Sorghum bicolor; Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Waters et al., 2011; Wolff et al., 2011; Zhang et al., 2011, 2016; Waters et al., 2013; Xin et al., 2013; Pignatta et al., 2014; Xu et al., 2014). However, the amount of overlap among imprinted genes of various plant species is limited (Waters et al., 2013; Pires and Grossniklaus, 2014; Hatorangan et al., 2016). For example, only 14% of MEGs and 29% of PEGs in Capsella rubella were commonly imprinted in A. thaliana (Hatorangan et al., 2016). Subsequent study indicated that genes controlled by imprinting are highly conserved between Arabidopsis lyrata and A. thaliana (Klosinska et al., 2016). In addition, the consistently imprinted expression of two paralogous maize genes, Fertilization-independent endosperm1 (Fie1) and Fie2, suggests that parent-of-origin-dependent allelic expression can be maintained during tetraploidization or gene duplication events (Danilevskaya et al., 2003).
Hexaploid wheat (AABBDD; Triticum aestivum) is a typical allopolyploid species with three distinct subgenomes that has undergone two separate allopolyploidization events. The first event involved a cross between Triticum urartu (AA genome) and an unidentified species (BB genome) 0.36 to 0.50 million years ago (MYA) (Dvořák, 1976; Huang et al., 2002; Dvorak and Akhunov, 2005; Pont and Salse, 2017). The resulting tetraploid wheat species, Triticum turgidum (AABB), then hybridized with Aegilops tauschii (DD genome) to generate hexaploid wheat (AABBDD) ∼10,000 years ago (Kihara, 1944; McFadden and Sears, 1946; Dvorak et al., 1998; Huang et al., 2002). These wheat species provide an excellent model system for studying the genomics of polyploid plants.
In this study, we performed genome-wide identification of imprinted genes in wheat species with different ploidy levels using reciprocal endosperm at different developmental stages. We then analyzed the conservation of genomic imprinting among diploid, tetraploid, and hexaploid wheat species, as well as their homoeologous genes. Our findings demonstrate that parent-of-origin-dependent allelic expression was evolutionarily conserved during wheat polyploidization.
RESULTS
Transcriptome Sequencing, Data Processing, and SNP Calling
To assess the allelic expression patterns of genes in diploid (DD), tetraploid (AABB), and hexaploid wheat (AABBDD) endosperm, we performed deep transcriptome sequencing of reciprocally crossed developing endosperm from wheat species with different ploidy levels, including diploid wheat (DD; Y177 [Y]×RM220 [R] and R×Y at 15 and 20 d after pollination [DAP]), tetraploid wheat (AABB; Jinying8 [J]×SCAUP [S] and S×J at 15 and 20 DAP), and hexaploid wheat (AABBDD; Doumai [D] and Keyi5214 [K] and K×D at 15, 20, and 25 DAP), together with their respective parental lines. In total, we obtained 3539.3 million paired-end reads, with an average of ∼111.1, ∼149.2, and ∼242.2 million reads per sample which, on average, covered 14,550, 27,983, and 40,489 endosperm-expressed genes (fragments per kilobase of transcript per million mapped reads > 1) in diploid, tetraploid, and hexaploid wheat, respectively. To identify high-quality single-nucleotide polymorphisms (SNPs) between the parental lines, we mapped the corresponding RNA-seq reads of the parental lines to the wheat reference genome (TGACv1 [Chinese Spring, CS]; Clavijo et al., 2017) using Bowtie 2 (v2.2.9; Langmead and Salzberg, 2012); ∼33.7% (diploid wheat, 31.5–35.8% mapped to the CS_D genome), ∼50.0% (tetraploid wheat, 46.3–53.5% mapped to the CS_A and B genomes), and ∼49.9% (hexaploid wheat, 42.0–59.5% mapped to the CS_A, B, and D genomes) of uniquely mapped reads were retained for subsequent analysis (Supplemental Table 1). We then performed SNP calling using Samtools (v1.4; Li et al., 2009) and BCFtools (v1.4; Li, 2011).
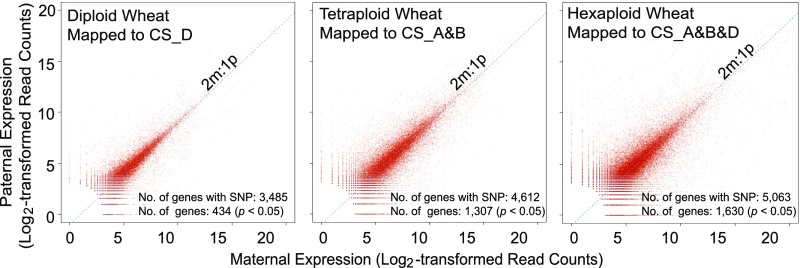
The accuracy of SNP identification will be reduced in polyploid wheat due to the widespread presence of homoeologs, which might cause ambiguous mapping. Therefore, to improve the reliability of the SNPs in polyploid wheat, we only considered RNA-seq reads that were uniquely mapped to the A, B, or D subgenome under the condition that reference sequence information was available for all three homoeologous loci (see Methods). Ultimately, we identified 7109 (mapped to the CS_D genome), 14,995 (mapped to the CS_A and B genome), and 13,085 (mapped to the CS_A, B, and D genome) high-confidence SNPs located in 3485, 4612, and 5063 genes in diploid, tetraploid, and hexaploid wheat species, respectively.
Next, we aligned the uniquely mapped RNA-seq reads from reciprocal crosses to the reference sequence with SNP information to distinguish their parental origin. To determine whether the ratio of allele-specific reads properly reflected the ratio of maternal-to-paternal transcripts, we calculated the correlation coefficient between each pair (∼0.91 on average) after excluding genes with fewer than 10 reads. This analysis indicated that the allelic expression patterns inferred from parent-specific reads were proportional to total gene expression. By plotting paternal versus maternal expression for all retained genes, we found that the majority of SNP-containing genes exhibited the expected ratio of 2m:1p, whereas ∼9.5 to 31.1% exhibited allele-biased expression patterns, i.e., the ratio of maternally to paternally derived reads significantly deviated from 2:1 in both reciprocal crosses (χ2 test, false discovery rate [FDR]-adjusted P value ≤ 0.05).
Genome-Wide Survey of Imprinted Genes in Hybrid Endosperm from Diploid, Tetraploid, and Hexaploid Wheat Species
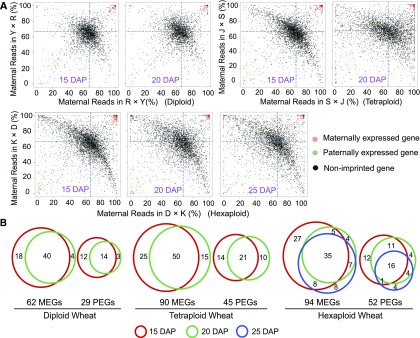
To identify high-confidence imprinted genes in wheat species with different ploidy levels, we performed a χ2 test to determine whether these selected genes possessed parent-specific expression patterns in both reciprocal crosses. At a significance level of P value = 0.05, 434 genes (mapped to the CS_D genome) were determined to have maternally or paternally preferred expression patterns in reciprocally crossed diploid endosperm during at least one stage of development. Correspondingly, 1307 (mapped to the CS_A and B genome) and 1630 (mapped to the CS_A, B, and D genome) genes had either maternally or paternally preferred expression patterns in tetraploid and hexaploid wheat, respectively (Figure 1; Supplemental Figure 1). To determine the parent-of-origin expression status of allele-specific genes, we further filtered the candidate imprinted genes, with maternally and paternally expressed genes (MEGs and PEGs, respectively) defined as genes that had 90% maternal reads or 70% paternal reads among all SNP-associated reads in both reciprocal crosses (with a minimum of 10 SNP-associated reads per cross and a FDR-adjusted P value of 0.05), respectively. We calculated the proportion of maternal/paternal reads separately for each reciprocal cross based on the criterion that the values for both crosses had to be above the threshold (see Methods). Using this more stringent, ratio-based criterion, we finally identified 372 imprinted genes (Figure 2), with 91 (62 MEGs and 29 PEGs), 135 (90 MEGs and 45 PEGs), and 146 (94 MEGs and 52 PEGs) in diploid, tetraploid, and hexaploid wheat species, respectively (Figures 2A and 2B; Supplemental Data Sets 1 and 2). Of these 372 imprinted genes, 176 genes (47.3%) exhibited consistent imprinted expression patterns across all developmental stages (40 MEGs and 14 PEGs for diploid wheat, 50 MEGs and 21 PEGs for tetraploid wheat, and 35 MEGs and 16 PEGs for hexaploid wheat). The remaining genes were considered to be imprinted in a stage-specific manner (30 MEGs and 7 PEGs for diploid wheat, 39 MEGs and 25 PEGs for tetraploid wheat, and 59 MEGs and 36 PEGs for hexaploid wheat) (Figure 2B; Supplemental Data Set 1), indicating that a portion of imprinted wheat genes exhibited dynamic expression patterns during endosperm development. Further investigation revealed that all of these stage-specific imprinted genes were also expressed during other developmental stages, but their parental alleles exhibited biallelic expression patterns. Thus, the major cause of stage-specific imprinting is not a lack of expression, but can instead be attributed to biallelic expression patterns.
Figure 1.
Most Genes Exhibited the Expected Expression Ratios in Developing Wheat Endosperm.
Parental expression ratios plot for each reciprocal cross in diploid, tetraploid, and hexaploid wheat species. The expression levels of paternal (y axis) and maternal (x axis) alleles are represented by the log2-transformed read counts of the paternally and maternally derived reads in the reciprocal crosses, respectively. The expression patterns of 3485, 4612, and 5063 genes with SNPs were analyzed in reciprocally crossed endosperm for diploid, tetraploid, and hexaploid wheat, respectively. Of these, 434, 1,307, and 1630 genes were identified as parental biased expressed genes in diploid, tetraploid, and hexaploid wheat endosperm, respectively, according to a χ2 goodness-of-fit test (FDR-adjusted P < 0.05). The dashed diagonal line represents the expected 2m:1p ratio.
Figure 2.
Computational Identification of Imprinted Genes in Wheat Endosperm.
(A) Ratio-based cutoff to identify MEGs and PEGs. Spots clustered in the upper-right corners have more than 90% maternal reads (red, MEGs), whereas spots clustered in the lower-left corners have more than 70% paternal reads (green, PEGs). Black dots represent nonimprinted genes. The intersection of the dashed lines indicates a 2m:1p ratio. Dots representing MEGs and PEGs are semitransparent. Y, Y177; R, RM220; J, Jinying 8; S, SCAUP; D, Doumai; K, Keyi5214.
(B) Venn diagram analysis of imprinted genes. The number of imprinted genes identified at 15, 20, and 25 DAP are shown in the red, green, and blue circles, respectively.
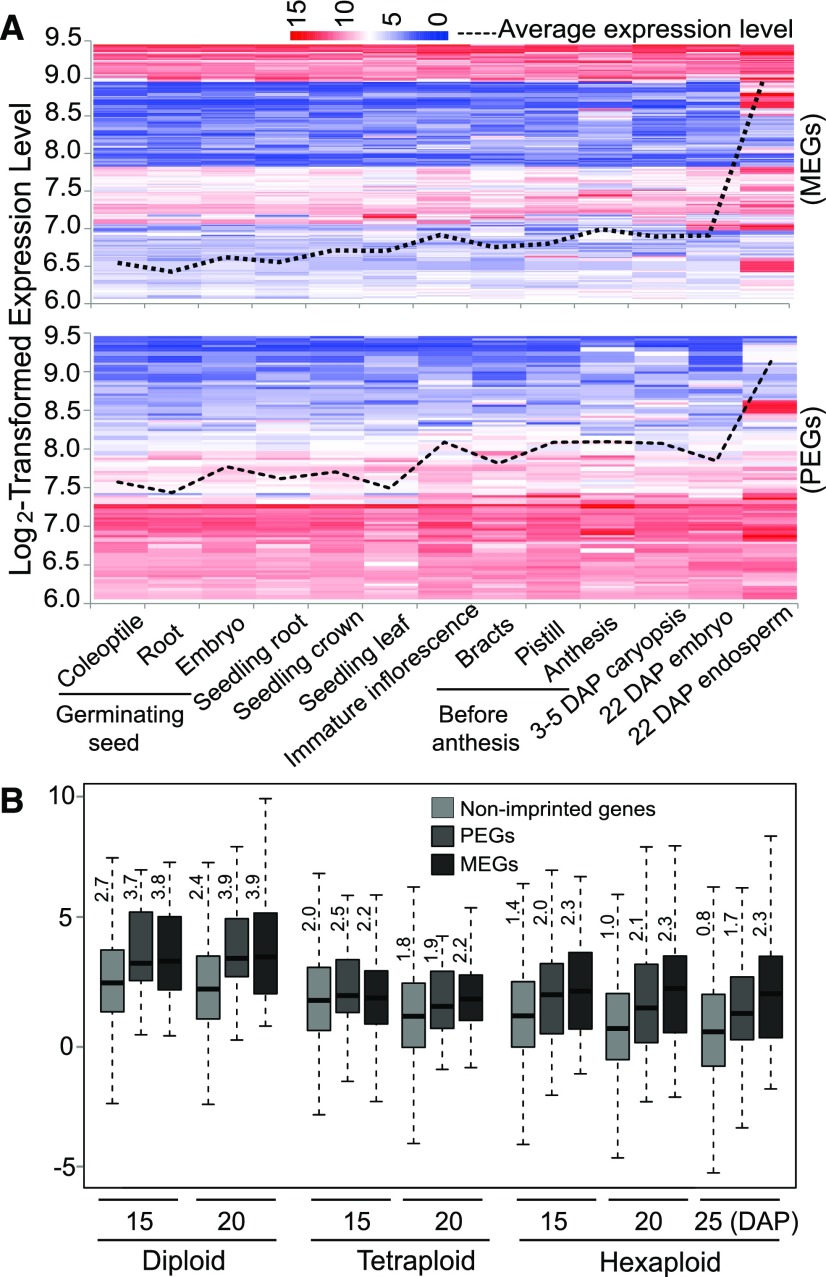
Next, we investigated whether the imprinted genes exhibit tissue-specific expression patterns in wheat (Figure 3). By examining previously published data sets (http://www.plexdb.org/plex.php?database=Wheat) (Schreiber et al., 2009), we found that both PEGs and MEGs were more abundantly expressed in endosperm than in other tissues, and MEGs appeared to exhibit more endosperm-specific expression compared with PEGs (the fold change of gene expression [endosperm versus other tissues] was 4.14 and 2.58 for MEGs and PEGs, respectively) (Figure 3A). In addition, MEGs and PEGs were expressed at higher levels in developing endosperm than nonimprinted genes (Figure 3B). An analysis of previously published laser capture microdissection data revealed that MEGs were more highly expressed in all endosperm compartments compared with PEGs (Pfeifer et al., 2014) (Supplemental Figure 2). Gene Ontology (GO) analysis showed that MEGs were enriched in the categories “regulation of nitrogen compound metabolic process” (GO:0051171), “regulation of macromolecule biosynthetic process” (GO:0010556), and “regulation of primary metabolic process” (GO:0080090). By contrast, the significantly enriched GO categories for PEGs were “RNA biosynthetic process” (GO:0032774) and “transcription, DNA-dependent” (GO:0006351). The high expression levels of MEGs, together with their roles in regulating nutrient biosynthesis and metabolism based on GO analysis, suggest that they play important roles in regulating nutrient accumulation in wheat endosperm. This notion is consistent with the finding that MEGs are rapidly upregulated in maize endosperm at the filling stage (Xin et al., 2013), which partially supports the parental conflict theory, i.e., that MEGs tend to control the growth of their offspring by limiting nutrient allocation to their offspring (Haig and Westoby, 1989; Wilkins and Haig, 2003).
Figure 3.
Expression of Imprinted Genes in Different Wheat Tissues.
(A) The expression levels of MEGs and PEGs were examined in 13 wheat tissues. The average expression level is higher in endosperm than in other tissues. MEGs appeared to be more endosperm-specific than PEGs. Dashed line indicates the average expression level of imprinted genes in different tissues. The color scale from blue (low) to red (high) indicates relative gene expression level.
(B) The expression levels of MEGs and PEGs are higher than those of nonimprinted genes in diploid, tetraploid, and hexaploid wheat species at all stages examined. The number indicates the average expression level (log2-transformed fragments per kilobase of transcript per million mapped reads).
Experimental Validation of Imprinted Genes in Diploid, Tetraploid, and Hexaploid Wheat Species
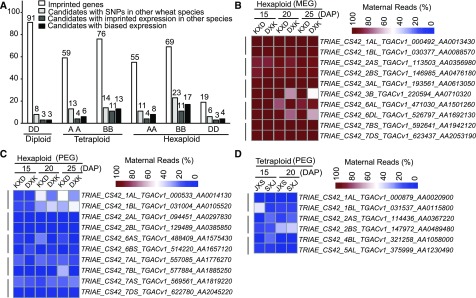
To validate the bioinformatically identified imprinted genes, we performed RT-PCR followed by cleaved amplified polymorphic sequence (CAPS) assays or sequencing. We examined 10 diploid imprinted candidate genes (six MEGs and four PEGs), among which six were sequenced, seven were cleaved with SNP-sensitive restriction enzymes, and three (TRIAE_CS42_5DS_TGACv1_457221_AA1483900, TRIAE_CS42_1DL_TGACv1_062392_AA0213710, and TRIAE_CS42_3DS_TGACv1_272480_AA0921380) were confirmed by both methods (Supplemental Figures 3 and 4). Consistent with the RNA-seq data, all 10 putative imprinted genes showed the expected parent-of-origin expression patterns, which exactly coincided with the imprinting predictions at different developmental stages. For example, TRIAE_CS42_7DL_TGACv1_602576_AA1961380 was predicted to be a PEG at 15 DAP and 20 DAP in diploid wheat, although with a few maternal reads detected in its 20-DAP endosperm of the Y×R cross, and CAPS analysis confirmed these expression patterns at both developmental stages (Supplemental Figure 3). In addition, although TRIAE_CS42_3DS_TGACv1_272480_AA0921380 was identified as a PEG according to our ratio-based criteria, a few maternal reads appeared in the Y×R cross at 15 and 20 DAP but not in the reciprocal cross; both the CAPS assay and sequencing confirmed our observation (Supplemental Figures 3 and 4 and Supplemental Data Set 1).
Although it is more difficult to experimentally confirm imprinted genes in tetraploid and hexaploid wheat than in diploid wheat due to the presence of homoeologous genes, but we overcame this challenge by using homoeolog-specific primer pairs. In tetraploid reciprocal crosses, six putative imprinted genes (five MEGs and one PEG) showing parent-of-origin expression status were confirmed via CAPS or sequencing (Supplemental Figures 3 and 4). Of these six genes, TRIAE_CS42_4BS_TGACv1_329466_AA1101720 was predicted to be maternally expressed only in 15-DAP endosperm based on a 90% ratio cutoff, and consistently, paternally derived reads were abundant at 20 DAP according to sequencing validation (Supplemental Figure 4 and Supplemental Data Set 1). For hexaploid wheat, five candidate imprinted genes were verified in 15-, 20-, and 25-DAP reciprocally crossed endosperm, including four MEGs and one PEG. These five candidates were considered to be consistently imprinted at 15, 20, and 25 DAP based on the RNA-seq data, and the CAPS experiment revealed a clear maternally or paternally imprinted pattern at all three developmental stages in both reciprocal crosses. Together, the experimental validation of imprinted genes in diploid, tetraploid, and hexaploid wheat confirmed the efficiency of our strategy for identifying genomic imprinting in polyploid plants. We also performed sequencing analysis of two other genes (TRIAE_CS42_1DL_TGACv1_061444_AA0195450 and TRIAE_CS42_5AS_TGACv1_393185_AA1269500) that showed imprinted expression patterns only in one biological replicate due to limited read counts in the other replicate. The maternal or paternal expression patterns of these two genes were clearly confirmed in developing wheat endosperm (Supplemental Figure 5), indicating that a subset of imprinted genes were not identified in wheat endosperm at the current sequencing depth.
Imprinted Wheat Genes Were Evolutionarily Conserved during Polyploidization
Genes controlled by genomic imprinting are poorly conserved between A. thaliana and monocots as well as between rice and maize, possibly because this biased expression pattern is partially dependent on the presence of transposable elements (Luo et al., 2011; Waters et al., 2011, 2013; Rodrigues and Zilberman, 2015). The paternally expressed auxin biosynthesis-related genes YUCCAs and Tryptophan aminotransferase related1 (TAR1) are rare examples of conserved imprinted genes present in rice, maize, and A. thaliana endosperm (Hsieh et al., 2011; Luo et al., 2011; Zhang et al., 2011; Chen et al., 2017). In addition, only 14% of MEGs and 29% of PEGs in C. rubella were commonly imprinted in A. thaliana (Hatorangan et al., 2016). However, 50% of imprinted genes were subsequently found to be conserved between A. lyrata and A. thaliana, which diverged ∼13 MYA (Klosinska et al., 2016). The paralogs of 10 imprinted genes (resulting from the recent whole-genome duplication) also exhibit parent-of-origin-dependent allelic expression patterns in maize endosperm (Waters et al., 2013), which prompted us to investigate the conservation of imprinted genes during the polyploidization of wheat.
Hexaploid wheat has undergone two separate allopolyploidization events and has arisen from the convergence of three diploid ancestors (AA, BB, and DD). To investigate the conservation of imprinted genes during wheat evolutionary history, we only considered genes with high sequence identity (>90%, E-value < 1e-10) and syntenic chromosome regions among different wheat species. For example, if gene X in position Y of the B subgenome in hexaploid wheat was imprinted, we only examined the expression patterns of its homolog in position Y of the B subgenome in tetraploid wheat and not its homolog in position Y of the A subgenome of tetraploid wheat or the D subgenome of diploid wheat, and vice versa. We examined the imprinted expression patterns of individual MEGs and PEGs in wheat species with different ploidy levels (Figure 4). Of the 135 imprinted genes (59 in the A subgenome and 76 in the B subgenome) in tetraploid reciprocal crosses (AABB), 27 genes (13 in the A subgenome and 14 in the B subgenome) were found to possess SNPs between parental lines of hexaploid wheat. Among these, 15 genes (55.6%; 4 in the A subgenome and 11 in the B subgenome) showed conserved parent-of-origin expression patterns based on our criteria, including 10 MEGs and 5 PEGs (Figure 4A; Supplemental Data Set 3). Similarly, of the 91 imprinted genes in diploid reciprocal crosses, eight candidates happened to possess SNPs between D and K, and three (37.5%) genes were also imprinted in hexaploid wheat (Supplemental Data Set 3). No overlap of imprinted genes between diploid wheat (DD) and tetraploid wheat (AABB) was observed, since these species have different subgenomes. Thus, in total, 18 genes (18/35, 51.4%) exhibited conserved parent-of-origin-dependent expression patterns between diploid and hexaploid wheat or between tetraploid and hexaploid wheat when considering both sequence identity and syntenic chromosome regions. Interestingly, at a significance level of FDR-adjusted P = 0.05, 62.9% (22/35) of genes showed conserved maternally or paternally preferred expression patterns during wheat hexaploidization (Figure 4A). For example, TRIAE_CS42_2BS_TGACv1_147693_AA0486590 met the requirement of 70% paternal reads for PEGs in both reciprocal crosses of hexaploid wheat, but it was excluded from the imprinted gene sets due to the limited number of SNP-associated reads, whereas TRIAE_CS42_6AS_TGACv1_487181_AA1568730 exhibited significant maternally biased expression at 15, 20, and 25 DAP in the reciprocal crosses of hexaploid wheat based on the χ2 test (P value < 0.05), but it did not pass the cutoff criterion of 90% maternal reads in one cross.
Figure 4.
Imprinted Genes That Were Evolutionarily Conserved during Hexaploidization.
(A) Parent-of-origin expression patterns of imprinted genes are highly conserved among wheat species. The white bar indicates the number of imprinted genes in different subgenomes of diploid, tetraploid, and hexaploid wheat; light-gray bar indicates the number of imprinted genes with SNPs in other wheat species; dark-gray bar indicates the number of conserved imprinted genes in different wheat species; black bar indicates the number of conserved candidate imprinted genes with biased expression patterns in different wheat species only considering the criterion of FDR-adjusted P value.
(B) to (D) Thirteen pairs of homoeologs show similar imprinted expression patterns in tetraploid and hexaploid wheat. Vertical lines indicate the 13 groups of homoeologous wheat genes. Blue (low), white (medium), and red (high) represent the relative expression levels of maternal or paternal alleles. S, SCAUP; J, Jinying 8; D, Doumai; K, Keyi5214.
Polyploid wheat has experienced one or two rounds of allopolyploidization events, resulting in thousands of homoeologs due to whole-genome duplication. Thus, we next investigated whether the parent-of-origin-dependent expression pattern is conserved among these homoeologous genes. Interestingly, we successfully distinguished the parental origins of reads for 25 pairs of homoeologous genes in reciprocally crossed hexaploid wheat, as supported by SNP information. Among these gene pairs, 10 pairs simultaneously exhibited imprinted expression patterns, accounting for 40% of the total, whereas the proportion for tetraploid wheat was 23.1% (3/13) (Figures 4B to 4D; Supplemental Data Set 4). In addition, although SNP information might have been unavailable for the homoeologs of imprinted genes in one wheat species, it might have been available for other wheat species for a subset of homoeologs. In total, 37 pairs of such homoeologs simultaneously exhibited imprinted expression patterns in two wheat species (Supplemental Data Set 5).
In addition to homoeologs resulting from whole-genome duplication, many homologs evolved via tandem duplication after whole-genome duplication, such as Fie1 and Fie2 in maize, which both exhibit imprinted expression patterns in developing endosperm, although in a stage-specific manner for Fie2 (Dickinson et al., 2012). As expected, we identified 34 and 20 homologous pairs (identity > 90%, E-value < 1e-10) of imprinted genes containing SNP information in tetraploid and hexaploid wheat, respectively, 14 and 10 of which also showed conserved imprinted expression patterns in the respective hybrid endosperm (Supplemental Data Set 6). For example, the homologous genes TRIAE_CS42_6AS_TGACv1_487181_AA1568730 and TRIAE_CS42_6AS_TGACv1_485362_AA1544140, which are located on the short arm of chromosome 6A in tetraploid wheat, encode an F-box domain-containing protein, and both exhibited paternally biased expression patterns in J and S reciprocal endosperm. Furthermore, TRIAE_CS42_3B_TGACv1_220627_AA0712550, TRIAE_CS42_3B_TGACv1_220627_AA0712570, TRIAE_CS42_3B_TGACv1_220627_AA0712610, and TRIAE_CS42_3B_TGACv1_220627_AA0712620 are located close to each other on chromosome 3B in hexaploid wheat and encode a putative E3 ubiquitin-protein ligase. Supported by SNP information, we found that all of these genes showed maternally preferred expression patterns in K and D reciprocal crosses. In conclusion, our results suggest that the expression patterns of imprinted genes were largely conserved throughout the evolutionary history of wheat.
DISCUSSION
Genomic Imprinting Occurs Extensively in Wheat Species of Various Ploidy Levels
Hexaploid wheat (AABBDD; T. aestivum) has undergone two allopolyploidization episodes during its evolutionary history, including tetraploidization and hexaploidization, which involved the hybridization of T. urartu (AA), an unidentified species (BB), and A. tauschii (DD) (Kihara, 1944; McFadden and Sears, 1946; Dvorak et al., 1998; Huang et al., 2002). Thus, it is difficult to identify imprinted genes in polyploid wheat due to the widespread presence of homoeologs resulting from genome duplication. In this study, we performed genome-wide identification of imprinted genes in reciprocally crossed endosperm from diploid, tetraploid, and hexaploid wheat, and detected 91, 135, and 146 imprinted genes in reciprocal endosperm, respectively, including 246 MEGs and 126 PEGs. We validated 21 out of 23 imprinted genes by RT-PCR followed by CAPS or sequencing, suggesting that our strategy was highly effective for identifying imprinted genes in polyploid plants. In addition, we experimentally confirmed the parent-of-origin expression patterns of two genes (TRIAE_CS42_1DL_TGACv1_061444_AA0195450 and TRIAE_CS42_5AS_TGACv1_393185_AA1269500), which were identified as imprinted genes in only one biological replicate (Supplemental Figure 5 and Supplemental Data Set 2), indicating that our criteria for identifying imprinted genes might have been too stringent for a subset of genes expressed in the endosperm.
The imprinted genes identified in this study are unevenly distributed on the chromosomes, with the D subgenome containing the smallest number of imprinted genes in hexaploid wheat (55, 69, and 19 imprinted genes for the A, B, and D subgenome, respectively). This is likely due to the reduced genetic diversity of the D subgenome compared with the A and B subgenome that arose during hexaploid wheat evolution and domestication, since we found less SNP information in the D subgenome (1533) than in the A (4863) and B subgenome (6197). This observation is consistent with the previous finding that the D subgenome has the lowest nucleotide diversity among the three subgenomes in hexaploid wheat (Akhunov et al., 2010). Accordingly, the proportion of genes that could be evaluated for imprinting was 15.7, 16.8, and 5.1%, respectively. Therefore, the reduced SNP information in the D subgenome among wheat lines used in this study probably led us to underestimate the number of imprinted genes in the D subgenome. In addition, only 125 MEGs and 51 PEGs exhibited a persistent imprinted expression pattern during endosperm development (Figure 2B), and a large proportion (52.7%) of genes showed stage-specific imprinting patterns due to their biallelic expression during other developmental stages. These results are consistent with findings for rice, maize, A. thaliana, castor bean, and sorghum (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Waters et al., 2011, 2013; Wolff et al., 2011; Zhang et al., 2011, 2016; Xin et al., 2013; Pignatta et al., 2014; Xu et al., 2014). Since we only considered reads that mapped to specific subgenomes and due to the dynamic nature of genomic imprinting as well as the limited availability of SNP information, it is reasonable to assume that we underestimated the number of imprinted genes in wheat endosperm. Nevertheless, this study, which provides a genome-wide survey of imprinted genes in various wheat species involving the use of high-throughput RNA-seq analysis, indicates that genomic imprinting is widespread among wheat species.
Imprinted Genes Were Evolutionarily Conserved during Wheat Hexapolyploidization
Triticum and Aegilops species provide an ideal system for studying polyploid genome evolution because hexaploid wheat has extant diploid and tetraploid progenitors with well-established phylogenetic relationships (Levy and Feldman, 2004; Feldman and Levy, 2005). In a comprehensive study of genomic imprinting in maize, 10 pairs of homologous genes exhibited conserved maternally biased expression patterns in endosperm, suggesting that genomic imprinting might have been maintained during the various genome duplication events (Waters et al., 2013). This prompted us to investigate the conservation of genomic imprinting among wheat species with various ploidy levels. We found that the parent-of-origin expression patterns are evolutionarily conserved among wheat species with different ploidy levels, as 51.4% (18/35) of imprinted genes in diploid or tetraploid wheat were also imprinted in hexaploid wheat when considering the criteria of both sequence identity and syntenic chromosome region (Figure 4A; Supplemental Data Set 3). These imprinted genes, which are conserved among wheat species, might play crucial roles in regulating seed development, among which, TRIAE_CS42_2BS_TGACv1_147972_AA0489480, a homolog of A. thaliana DA (LARGE IN CHINESE) 1, was identified as a PEG in both hexaploid and tetraploid wheat endosperm at 15 DAP. Interestingly, one amino acid change in DA1 (arginine to lysine at position 358) dramatically increases seed size by extending the duration of proliferative growth, suggesting that the imprinted DA1 gene might help regulate seed development (Li et al., 2008). Notably, TRIAE_CS42_5BL_TGACv1_410079_AA1366740 encodes a component of PRC2 in wheat (homolog to EMBRYONIC FLOWER 2 in A. thaliana and rice) and exhibits maternally biased expression patterns in both tetraploid and hexaploid wheat (Supplemental Data Set 3), indicating parent-of-origin effects of PRC2 are conserved during seed development among wheat, rice, maize, and A. thaliana, although the imprinted PRC2 genes might be various between each other. In addition, 50 pairs of homoeologs (13 in one wheat species and 37 in another wheat species) exhibited conserved genomic imprinting in reciprocally crossed endosperm (Supplemental Data Sets 4 and 5).
There is relatively little overlap between imprinted genes in A. thaliana versus monocots and rice versus maize, indicating that parent-of-origin expression patterns tend to vary during evolutionary history (Waters et al., 2013; Pires and Grossniklaus, 2014), with the exception of paternally expressed YUCCA genes and TAR1 in rice, maize, and A. thaliana endosperm (Hsieh et al., 2011; Luo et al., 2011; Zhang et al., 2011; Chen et al., 2017). In addition, the proportion of commonly imprinted genes is also limited between C. rubella and A. thaliana (Hatorangan et al., 2016). However, extensive conservation of imprinted expression patterns was revealed between A. lyrata and A. thaliana (Klosinska et al., 2016). We compared the conservation of imprinted genes among diploid wheat, tetraploid wheat, hexaploid wheat, maize, rice, sorghum, castor bean, and A. thaliana (sequence identity > 50%, E-value < 1e-10). As shown in Supplemental Data Set 7, we found 52 homologous pairs (20 between hexaploid and tetraploid wheat, 16 between hexaploid and diploid wheat, and 16 between diploid and tetraploid wheat) with conserved imprinted expression patterns between two wheat species. Furthermore, the overlap of genomic imprinting among diploid, tetraploid, and hexaploid wheat was the most significant among the plant species examined according to Fisher’s exact test. We also detected statistically significant overlaps in imprinted genes between wheat species and maize, as well as between wheat and rice, but not between wheat and A. thaliana (Supplemental Figure 6, Supplemental Table 2, and Supplemental Data Set 7). This finding is not unexpected, since these three wheat species are closely related, and hexaploid wheat appeared only ∼10,000 years ago due to the hybridization of diploid and tetraploid wheat (Feldman, 1995). Furthermore, monocots branched off from dicots 140 to 150 MYA, whereas wheat, rice, and maize diverged from a common ancestor ∼40 MYA (Gill et al., 2004). In summary, our analyses indicated that the degree of between-species overlap of genes exhibiting parent-of-origin expression biases is correlated with their phylogenetic relationship.
Interplay among Subgenomes Might Influence Genomic Imprinting in Hexaploid Wheat
It is thought that homoeologs make unequal contributions to total gene expression levels in polyploid wheat and that gene expression is regulated in a complex manner during grain development, possibly due to crosstalk between genomes during polyploidization (Akhunova et al., 2010; Chagué et al., 2010; Leach et al., 2014; Liu et al., 2015; Han et al., 2016). In this study, a major proportion of homoeologous gene pairs (65.8%) indeed exhibited divergent expression patterns in terms of genomic imprinting in polyploid wheat. Genomic imprinting is a contributing factor to the divergence in expression patterns of duplicated genes due to the silencing of one allele in a parent-of-origin-specific manner (Qiu et al., 2014). We also found that the silencing of the parental allele varied among homologs after polyploidization. For example, the maternal allele of TRIAE_CS42_3DL_TGACv1_249702_AA0854790 was preferentially expressed in diploid wheat endosperm, whereas its homolog, TRIAE_CS42_1AL_TGACv1_000533_AA0014130, exhibited paternally biased expression after polyploidization in hexaploid wheat. In addition, many MEGs and PEGs arose after the hexaploidization event, e.g., 22 imprinted genes in hexaploid wheat exhibited biallelic expression patterns in diploid or tetraploid wheat (Figure 4). These findings indicate that parent-of-origin gene expression is more prevalent in hexaploid wheat than in diploid and tetraploid wheat and that interplay among subgenomes might play a role in regulating genomic imprinting, a topic that merits further investigation.
METHODS
Plant Materials
Hexaploid wheat (Triticum aestivum; AABBDD) cultivars Keyi5214 (K) and Doumai (D) and tetraploid wheat (Triticum turgidum; AABB) cultivars SCAUP (S) and Jinying8 (J), as well as diploid goatgrass (Aegilops tauschii, DD) lines Y177 (Y) and RM220 (R), were sown in a field at China Agricultural University, Beijing, China. Reciprocal crosses and self-pollination were performed as follows: Spikelets at the base and very top of the spike, as well as florets from the central part of the spike, were removed before anthesis, and the top of the florets was cut off and bagged. Pollination was performed 1 to 2 d later using the appropriate pollen. Endosperm tissues were collected from at least three different ears to create three biological replicates at 15, 20, and 25 DAP; the endosperm tissues were isolated by hand dissection and immediately frozen in liquid nitrogen.
RNA Extraction
Total RNA was extracted from 60 (20 samples × 3 replicates) plant samples using the SDS-phenol method (Shirzadegan et al., 1991) with some modifications. Endosperm tissue (∼0.5 g) was ground to a fine powder in liquid nitrogen and mixed with 6 mL of buffer containing 1% SDS, 50 mM Tris-HCl (pH 8.0), 150 mM LiCl, 5 mM EDTA, and 10 mM DTT. The sample was combined with 6 mL phenol:chloroform (5:1, pH 4.5; Ambion AM9720) and incubated on ice for 5 min. The mixture was centrifuged at 5000 rpm for 10 min at 4°C and the aqueous phase was transferred to a new tube. These steps were repeated using phenol:chloroform (1:1), followed by chloroform alone. The RNA was then precipitated with 2.5 M LiCl at 4°C overnight, washed with ice-cold 2 M LiCl, dissolved in TE, mixed with a 1/9 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethanol, and incubated at −80°C for at least 4 h, after which the RNA was pelleted by centrifugation at 14,000 rpm for 15 min at 4°C. After rinsing with 75% ethanol and air-drying, the RNA was dissolved in diethylpyrocarbonate-treated water. DNA was removed with TURBO DNase I (Ambion), and the RNA was purified using an RNeasy column (Qiagen).
Illumina Sequencing
The RNA samples were sent to Berry Genomics for mRNA library construction and deep sequencing using the Illumina HiSeq 2000 platform. Before library construction, the quality of the RNA samples was examined using an Agilent 2100 Bioanalyzer. High-quality mRNA from three biological replicates per sample was sequenced. FastQC software (v0.11.5; http://www.bioinformatics.babraham.ac.uk) was used to examine the sequencing quality of the reads in each sample (Andrews, 2010). Then, raw data were processed using Trimmomatic (v0.36; http://www.usadellab.org/cms/index.php?page=trimmomatic) to trim adaptor sequence and low-quality end (Bolger et al., 2014), and only high-quality reads were retained for further analysis. In total, 125.5, 400.4, and 430.1 Gb of high-quality RNA-seq data were generated from parental and reciprocally crossed endosperm from diploid, tetraploid, and hexaploid wheat, respectively. Because the sequencing depth of the first replicate was not as high as that of the other two, data from the first two replicates were combined, and three sets of sequenced transcriptomes were considered to represent two biological replicates. The correlation coefficient of two biological replicates was 0.977 to 0.998. The biological replicates were treated independently and imprinted genes were identified separately and then compared with each other; only overlapping candidates were considered to represent imprinted genes. The RNA-seq reads used in this study were deposited in the National Center for Biotechnology Information Short Read Archive under accession number SRP075528.
SNP Calling and Identification of Imprinted Genes
RNA-seq data from each parent (K and D for hexaploid wheat, J and S for tetraploid wheat, and Y and R for diploid goatgrass) were used for SNP identification. High-quality reads were mapped to the reference gene sequence (TGACv1; Clavijo et al., 2017) using Bowtie2 (v2.2.9; Langmead and Salzberg, 2012) with the parameters “–end-to-end–reorder–score-min L, -0.6,-0.3 -L 15.” To improve credibility, only reads that uniquely mapped to one subgenome with no more than two mismatches were considered. SNP calling was then performed using the mpileup function of Samtools (v1.4; Li et al., 2009) and the call function of BCFtools (v1.4; Li, 2011). SNPs supported by ≥10 reads, ≥95% of the total SNP site-mapped reads, and a genotype-likelihood of ≥95% in each parent were identified as SNPs between parents and used for subsequent allele-specific expression analysis in hybrids.
RNA-seq reads from reciprocal crosses of both biological replicates were mapped to the reference genes separately using Bowtie2 (v2.2.9; Langmead and Salzberg, 2012), and only uniquely mapped reads with no more than two mismatches were retained. SNP-containing reads originating from different parents were then distinguished based on maternal and paternal SNPs identified in the previous step and counted using customized Perl scripts. Genes with a ratio deviating from 2m:1p (χ2 goodness-of-fit test, FDR-adjusted P value < 0.05) and ≥90% of total SNP-containing reads that were maternally derived or ≥70% that were paternally derived in two reciprocal crosses of both biological replicates (with a minimum of 10 SNP-associated reads per cross) were identified as imprinted genes.
CAPS Assay
RNA samples from reciprocal crosses of wheat species with different ploidy levels were independently prepared to validate imprinted gene expression patterns using CAPS assays, as previously described (Konieczny and Ausubel, 1993), or by sequencing. RT-PCR was performed using the gene- or homoeolog-specific primers listed in Supplemental Table 3. The amplification products were digested with the restriction enzymes listed in Supplemental Table 3.
Accession Numbers
The RNA-seq reads used in this study were deposited in the National Center for Biotechnology Information Short Read Archive under accession number SRP075528.
Supplemental Data
Supplemental Figure 1. Parental expression ratio plot for endosperm-expressed genes at different developmental stages for each reciprocal cross in diploid, tetraploid, and hexaploid wheat species.
Supplemental Figure 2. The expression levels of MEGs are higher than those of PEGs in the aleurone layer and transfer cells of 20- and 30-DAP wheat endosperm.
Supplemental Figure 3. Experimental validation of the 13 imprinted genes in diploid, tetraploid, and hexaploid wheat by CAPS assays.
Supplemental Figure 4. Experimental validation of 11 imprinted genes in diploid, tetraploid, and hexaploid wheat by sequencing.
Supplemental Figure 5. Experimental validation of candidate imprinted genes identified in only one biological replicate of diploid and hexaploid wheat by sequencing.
Supplemental Figure 6. Conservation of imprinted genes among different species.
Supplemental Table 1. Summary of RNA-seq data and reads mapping results.
Supplemental Table 2. Overlaps between wheat imprinted genes and those of maize, rice, A. thaliana, sorghum, and castor bean.
Supplemental Table 3. Primers and enzymes used for sequencing and CAPS assays.
Supplemental Data Set 1. Allele-specific expression of the 91, 135, and 146 imprinted genes in diploid, tetraploid, and hexaploid wheat, respectively.
Supplemental Data Set 2. Allele-specific expression of imprinted candidate genes from each replicate in diploid, tetraploid, and hexaploid wheat, respectively.
Supplemental Data Set 3. Parent-of-origin expression of imprinted genes is highly conserved between diploid/tetraploid and hexaploid wheat.
Supplemental Data Set 4. Thirteen pairs of homoeologs show similar imprinted expression patterns in tetraploid and hexaploid wheat.
Supplemental Data Set 5. Expression patterns of homoeologs of imprinted genes in different wheat species.
Supplemental Data Set 6. Imprinted homologous pairs resulting from tandem duplication after polyploidization in tetraploid and hexaploid wheat.
Supplemental Data Set 7. Conserved imprinted genes in various plant species.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFD0101004), the Major Program of the National Natural Science Foundation of China (31290210), the National Natural Science Foundation of China (31471479), and Chinese Universities Scientific Fund (2017TC035).
AUTHOR CONTRIBUTIONS
M.X., Z.N., and Q.S. conceived the project. G.Y., Y.Y., and H.P. collected the plant materials. G.Y. and M.F. performed the research. Z.L. and K.Y. analyzed the data. M.X., Q.S., and Z.N. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Akhunov E.D., et al. (2010). Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11: 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunova A.R., Matniyazov R.T., Liang H., Akhunov E.D. (2010). Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Autran D., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagué V., Just J., Mestiri I., Balzergue S., Tanguy A.M., Huneau C., Huteau V., Belcram H., Coriton O., Jahier J., Chalhoub B. (2010). Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 187: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Chaudhury A.M., Ming L., Miller C., Craig S., Dennis E.S., Peacock W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Li T., Zhu S., Liu Z., Shi Z., Zheng X., Chen R., Huang J., Shen Y., Luo S., Wang L., Liu Q.Q., E Z. (2017). Characterization of imprinted genes in rice reveals post-fertilization regulation and conservation at some loci of imprinting in plant species. bioRxiv doi/10.1101/143214. [Google Scholar]
- Clavijo B.J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L.M., et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Curley J.P., Barton S., Surani A., Keverne E.B. (2004). Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc. Biol. Sci. 271: 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Hermon P., Hantke S., Muszynski M.G., Kollipara K., Ananiev E.V. (2003). Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson H., Costa L., Gutierrez-Marcos J. (2012). Epigenetic neofunctionalisation and regulatory gene evolution in grasses. Trends Plant Sci. 17: 389–394. [DOI] [PubMed] [Google Scholar]
- Dvořák J. (1976). The relationship between the genome of Triticum urartu and the A and B genomes of Triticum aestivum. Can. J. Genet. Cytol. 18: 371–377. [Google Scholar]
- Dvorak J., Akhunov E.D. (2005). Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Luo M.C., Yang Z.L., Zhang H.B. (1998). The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97: 657–670. [Google Scholar]
- Feldman H.A. (1995). On the allometric mass exponent, when it exists. J. Theor. Biol. 172: 187–197. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2005). Allopolyploidy--a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109: 250–258. [DOI] [PubMed] [Google Scholar]
- Fouquet R., Martin F., Fajardo D.S., Gault C.M., Gómez E., Tseung C.W., Policht T., Hueros G., Settles A.M. (2011). Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell 23: 4280–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill B.S., et al. (2004). A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450. [DOI] [PubMed] [Google Scholar]
- Guitton A.E., Berger F. (2005). Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15: 750–754. [DOI] [PubMed] [Google Scholar]
- Haig D., Westoby M. (1989). Parent-specific gene-expression and the triploid endosperm. Am. Nat. 134: 147–155. [Google Scholar]
- Han Y., Xin M., Huang K., Xu Y., Liu Z., Hu Z., Yao Y., Peng H., Ni Z., Sun Q. (2016). Altered expression of TaRSL4 gene by genome interplay shapes root hair length in allopolyploid wheat. New Phytol. 209: 721–732. [DOI] [PubMed] [Google Scholar]
- Hatorangan M.R., Laenen B., Steige K.A., Slotte T., Köhler C. (2016). Rapid evolution of genomic imprinting in two species of the Brassicaceae. Plant Cell 28: 1815–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M.J., Hashimoto M., Kirkbride R.C., Harada J.J., Zilberman D., Fischer R.L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., Gornicki P. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keverne E.B., Curley J.P. (2008). Epigenetics, brain evolution and behaviour. Front. Neuroendocrinol. 29: 398–412. [DOI] [PubMed] [Google Scholar]
- Kihara H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hortic. 19: 889–890. [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska M., Picard C.L., Gehring M. (2016). Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat. Plants 2: 16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Kradolfer D., Wolff P., Jiang H., Siretskiy A., Köhler C. (2013). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26: 525–535. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach L.J., Belfield E.J., Jiang C., Brown C., Mithani A., Harberd N.P. (2014). Patterns of homoeologous gene expression shown by RNA sequencing in hexaploid bread wheat. BMC Genomics 15: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy O., Hennig L., Breuninger H., Laux T., Köhler C. (2007). Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134: 3639–3648. [DOI] [PubMed] [Google Scholar]
- Levy A.A., Feldman M. (2004). Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. Lond. 82: 607–613. [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B.Y. (1984). Ploidy barrier to endosperm development in maize. Genetics 107: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xin M., Qin J., Peng H., Ni Z., Yao Y., Sun Q. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Taylor J.M., Spriggs A., Zhang H., Wu X., Russell S., Singh M., Koltunow A. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden E.S., Sears E.R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–, 107.. [DOI] [PubMed] [Google Scholar]
- Meng D., Zhao J., Zhao C., Luo H., Xie M., Liu R., Lai J., Zhang X., Jin W. (2017). Sequential gene activation and gene imprinting during early embryo development in maize. Plant J. 10.1111/tpj.13786 [DOI] [PubMed] [Google Scholar]
- Pfeifer M., Kugler K.G., Sandve S.R., Zhan B., Rudi H., Hvidsten T.R., Mayer K.F., Olsen O.A.; International Wheat Genome Sequencing Consortium (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Pignatta D., Erdmann R.M., Scheer E., Picard C.L., Bell G.W., Gehring M. (2014). Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N.D., Grossniklaus U. (2014). Different yet similar: evolution of imprinting in flowering plants and mammals. F1000Prime Rep. 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C., Salse J. (2017). Wheat paleohistory created asymmetrical genomic evolution. Curr. Opin. Plant Biol. 36: 29–37. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Liu S.L., Adams K.L. (2014). Frequent changes in expression profile and accelerated sequence evolution of duplicated imprinted genes in Arabidopsis. Genome Biol. Evol. 6: 1830–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Bemer M., Baroux C., Grossniklaus U. (2013). Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 9: e1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J.A., Zilberman D. (2015). Evolution and function of genomic imprinting in plants. Genes Dev. 29: 2517–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber A.W., Sutton T., Caldo R.A., Kalashyan E., Lovell B., Mayo G., Muehlbauer G.J., Druka A., Waugh R., Wise R.P., Langridge P., Baumann U. (2009). Comparative transcriptomics in the Triticeae. BMC Genomics 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.J., Spielman M., Bailey J., Dickinson H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Shirzadegan M., Christie P., Seemann J.R. (1991). An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res. 19: 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney W.T., Curley J.P., Champagne F.A., Keverne E.B. (2007). Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc. Natl. Acad. Sci. USA 104: 6084–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Baskar R., Grossniklaus U. (2000). Delayed activation of the paternal genome during seed development. Nature 404: 91–94. [DOI] [PubMed] [Google Scholar]
- Waters A.J., Bilinski P., Eichten S.R., Vaughn M.W., Ross-Ibarra J., Gehring M., Springer N.M. (2013). Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl. Acad. Sci. USA 110: 19639–19644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.J., Makarevitch I., Eichten S.R., Swanson-Wagner R.A., Yeh C.T., Xu W., Schnable P.S., Vaughn M.W., Gehring M., Springer N.M. (2011). Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins J.F., Haig D. (2003). What good is genomic imprinting: the function of parent-specific gene expression. Nat. Rev. Genet. 4: 359–368. [DOI] [PubMed] [Google Scholar]
- Wolf J.B., Hager R. (2006). A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Jiang H., Wang G., Santos-González J., Köhler C. (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. eLife 4: 10.7554/eLife.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M.T., Spillane C., Nordborg M., Rehmsmeier M., Köhler C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., et al. (2013). Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25: 3212–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dai M., Li F., Liu A. (2014). Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res. 42: 6987–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Johnston N., Talideh E., Mitchell S., Jeffree C., Goodrich J., Ingram G. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509. [DOI] [PubMed] [Google Scholar]
- Zhang M., Li N., He W., Zhang H., Yang W., Liu B. (2016). Genome-wide screen of genes imprinted in sorghum endosperm, and the roles of allelic differential cytosine methylation. Plant J. 85: 424–436. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhao H., Xie S., Chen J., Xu Y., Wang K., Zhao H., Guan H., Hu X., Jiao Y., Song W., Lai J. (2011). Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. USA 108: 20042–20047. [DOI] [PMC free article] [PubMed] [Google Scholar]