 We present a novel strategy for the highly selective detection of single-nucleotide variation at room temperature, based on an extremely specific interaction between Lambda exonuclease (λ exo) and a chemically modified DNA structure.
We present a novel strategy for the highly selective detection of single-nucleotide variation at room temperature, based on an extremely specific interaction between Lambda exonuclease (λ exo) and a chemically modified DNA structure.
Abstract
We demonstrate a new powerful tool to detect single-nucleotide variation in DNA at room temperature with high selectivity, based on predetermined specific interactions between Lambda exonuclease and a chemically modified DNA substrate structure which comprises two purposefully introduced mismatches and a covalently attached fluorophore. The fluorophore not only acts as a signal reporter in the detection system, but also plays a notable role in the specific molecular recognition between the enzyme and the probe/target hybrid substrate. The method is single-step, rapid, and can be easily adapted to different high-throughput micro-devices without the need for temperature control.
Selective detection of rare base substitutions or low-abundance point mutations in a large background of DNA sequences with single-base differences is regarded as one of the major issues to be addressed in the screening of single nucleotide polymorphisms (SNPs) in genomic studies or the tracking of DNA damage in clinical diagnosis.1–5 A number of elegantly designed DNA probes, such as molecular beacons,6,7 binary probes,8 triple-stem probes9 and other hybridization probes10,11 have been reported in recent years for the discrimination of single-base substitutions in DNA sequences.12–14 However, most of these hybridization-based DNA assays have limited power for the specific identification of single-base variants because of the small difference between the stability of a perfectly matched duplex and a duplex containing only one mismatched base. By inserting an extra mismatch at certain positions of the probes, a significant increase in the specificity of DNA hybridization has been demonstrated by Smith and co-workers.15 A novel type of discontinuous double-stranded DNA (dsDNA) probes have also been developed by Seelig et al. for specific DNA detection via a double-stranded toehold exchange mechanism.16 These probes showed promising results for a broad range of applications, though the sensitivity and the assay speed were not so competitive. Utilizing an isocysteine-mediated peptide nucleic acid (PNA) ligation reaction, Seitz et al. achieved a single-nucleotide selectivity as high as 3450 : 1.17 However, the assay has some practical limitations, such as the high cost of the PNA probes and the requirement for careful manipulation of the reactions to avoid exposure to oxygen. Many efforts have also been made to improve the selectivity and reduce the time and sample consumption by using various signal amplification reactions18–21 or PCR-based discrimination techniques such as allele-specific amplification PCR,22 coamplification at lower denaturation temperature PCR,23 wild-type blocking PCR24 and digital PCR.25 However, these approaches rely on precise temperature control and primer design.
In recent years, enzyme-mediated oligonucleotide-based fluorescent sensing methods have shown promising results in both DNA detection and nuclease measurement.21,26–31 Recently, we have investigated the effects of a purposefully incorporated mismatch in the DNA duplexes on the discrimination ability of nucleases to the presence of a second mismatch,27,29 which afforded remarkably enhanced selectivity in comparison to the hybridization-based probes. However, these established assays could not work efficiently at room temperature.
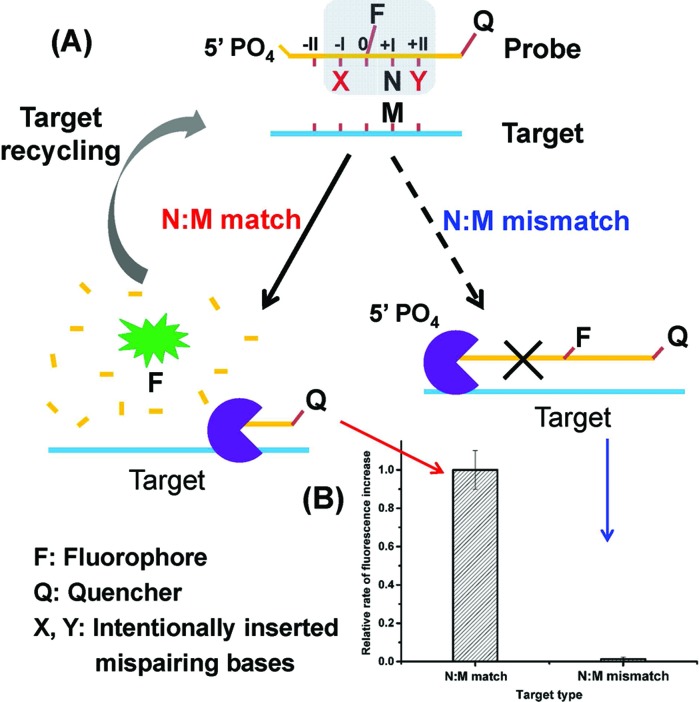
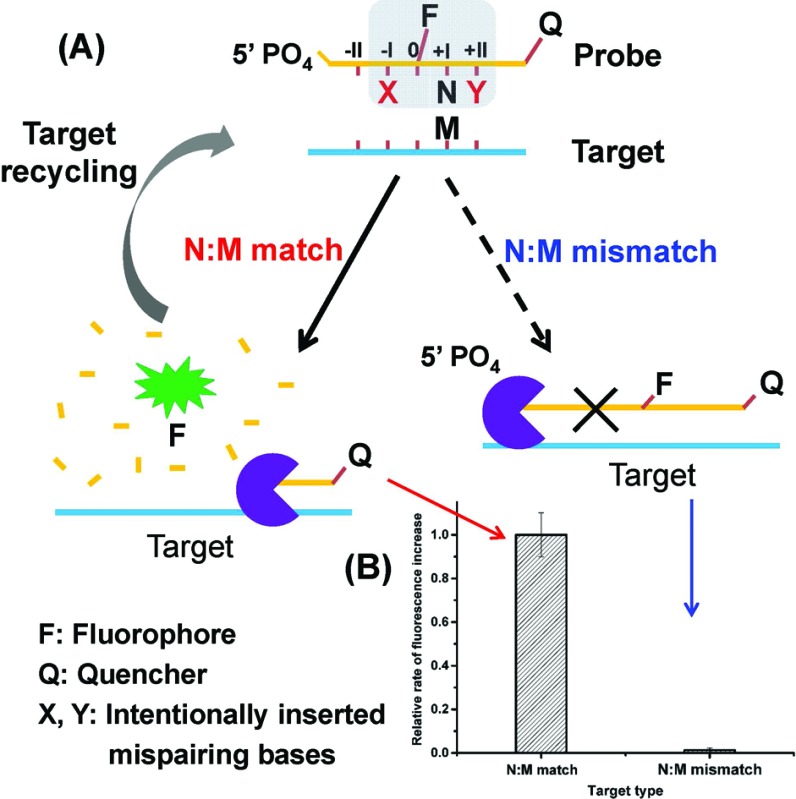
Herein, we present a novel strategy for the detection of single-nucleotide variation at room temperature with high selectivity. It is based on an extremely specific interaction between Lambda exonuclease (λ exo) and a chemically modified DNA structure. As depicted in Fig. 1A, a 5′-phosphorylated single-stranded DNA (ssDNA) probe is hybridized to a target strand that contains a polymorphic base (represented by M). A fluorophore (FAM) is covalently attached to the nucleoside at the 5′ side of N (the base site opposite M) and a quencher is attached at the 3′ end of the strand. With the fluorophore-tagged nucleoside designated as position 0, two mismatched bases are intentionally inserted at position –I (represented by X) and position +II (represented by Y), respectively. λ exo is an exonuclease that prefers double-stranded DNA (dsDNA) with a 5′-phosphorylated end as a substrate, and catalyzes stepwise hydrolysis of the 5′-phosphorylated strand in the 5′ to 3′ direction by forming a symmetrical toroid homotrimer.32 Despite the existence of the two purposefully inserted mismatched bases, we found that the probe/target hybrid containing a matched N:M base pair at position +I can still be efficiently hydrolyzed by λ exo and emits strong fluorescent signals. By contrast, when M is substituted by a mismatched base, the hydrolytic reaction by λ exo was almost completely inhibited. More importantly, since λ exo has little activity on ssDNA or non-5′-phosphorylated dsDNA substrates,33 excess amounts of the probe can be added to the reaction system. Thus, after the hybridized probe was digested, the remaining intact target strand could hybridize with another probe and trigger the next round of enzymatic digestion, resulting in significantly amplified differential effects between the two single-base different target DNA strands (see Fig. 1B).
Fig. 1. (A) Schematic depiction of the principle of the enzyme-mediated DNA detection at single-base difference level by using a double-mismatch/fluorophore modified DNA probe and lambda exonuclease. M represents the polymorphic base in the target strand. N is the base opposite M in the probe. (B) Comparison of the fluorescence response signals of two single-base different DNA strands. For the probe/target hybrid containing a matched N:M basepair, the probe was efficiently hydrolyzed by λ exo and emitted strong fluorescent signals. By contrast, when M was a mismatched base, the hydrolytic reaction carried out by λ exo was almost completely prohibited.
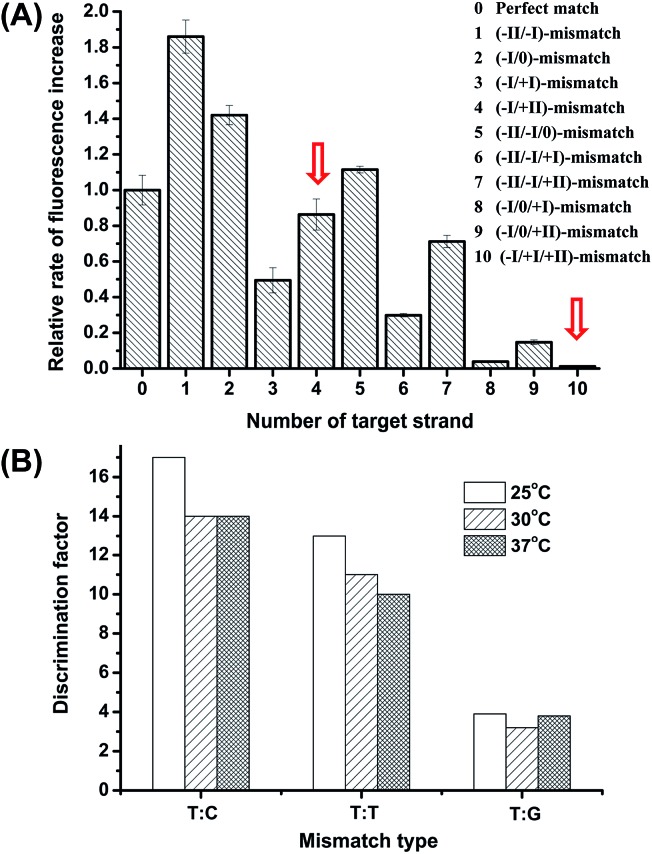
The high discrimination power of λ exo between a double mismatch containing DNA duplex and a triple mismatch containing DNA duplex in the presence of a covalently labelled fluorophore were unexpectedly observed in our lab recently during a systematic study of the fidelity of λ exo.34 By using a 21-nt probe (P-3FAM-4C, see Table 1) in which the fluorophore (FAM) is attached to the third base from the 5′ end, the presence of double or triple mismatched bases adjacent to the fluorophore-tagged nucleoside resulted in different effects on the digestion rate of the probe/target hybrid by λ exo (Fig. 2A). In comparison with the perfectly matched probe/target hybrid, the simultaneous presence of double or triple mismatched bases at the 5′ side of the fluorophore-tagged nucleoside all led to an increase of the digestion rates. By contrast, a dramatic decline of the digestion rate was observed for probe/target duplexes containing mismatches at both the 5′ and 3′ sides of the fluorophore-tagged nucleoside. By comparing the signal observed with the (–I/0/+I)-triple-mismatch strand with that observed with the (–I/0)-double-mismatch strand, substitution of the base at position +I with a mismatch in the presence of two existing mismatched bases induced an abrupt decrease of the digestion rate. Though a similar effect could be seen by comparing the signal of the (–I/+I/+II)-triple-mismatch strand with that of the (–I/+II)-mismatch strand, the single-mismatch discrimination factor (DF, defined as the ratio of the rate of increase in fluorescence intensity observed with the target containing a matched base at the polymorphic site to that observed with the target mismatches with the probe at the polymorphic site) between the (–I/+II)-double-mismatch and the (–I/+I/+II)-triple-mismatch was about two-fold that between the (–I/0)-double-mismatch and the (–I/0/+I)-triple-mismatch. These results imply great potential of the (–I/+II)-double-mismatch probe for ultra selective discrimination of single-nucleotide variations.
Table 1. The sequences and discrimination factors (DF) of duplexes containing different types of 5′,3′ mismatches.
| Probe a | Type of mismatched base pair (N:M) | Discrimination factor (DF) b |
| P-3FAM-4C | C:C | 320 |
| C:A | 165 | |
| C:T | 140 | |
| P-3FAM-4G | G:T | 52 |
| G:A | 35 | |
| G:G | 17 | |
| P-3FAM-4T | T:C | 38 |
| T:T | 25 | |
| T:G | 4.0 | |
| P-3FAM-4A | A:C | 7.2 |
| A:A | 5.6 | |
| A:G | 4.2 |
aThe probe sequence is 5′PO4-TCT(-FAM)NCACAGACACATACTCCA-BHQ1; the target sequences are 5′GTTTTAAA TTATGGAGTATGTGTCTGTTMAAACGAGAGTAAG; the two purposefully inserted mismatched bases are underlined. M represents the SNP site. N is the base opposite M in the probe, which is C, G, T and A for P-3FAM-4C, P-3FAM-4G, P-3FAM-4T and P-3FAM-4A, respectively.
b1× ThermoPol Reaction Buffer with additional (NH4)2SO4 and KCl was used as the reaction buffer. For reactions with P-3FAM-4G, the concentration of (NH4)2SO4 and KCl was 30 mM. For the other three probes, the concentration of (NH4)2SO4 and KCl was 20 mM.
Fig. 2. (A) Comparison of the rates of the fluorescence increase for the probe/target hybrid containing double or triple mismatches at different positions in comparison with the perfectly matched duplex. The Roman numerals in the brackets indicate the positions of the mismatch bases in the target strands when hybridized to probe P-3FAM-4C (the sequences is 5′PO4-TCT(-FAM)CCACAGACACATACTCCA-BHQ1). The two red arrows denote the data observed for the probe/target hybrids containing the (–I/+II)-double-mismatch and the (–I/+I/+II)-triple-mismatch, respectively. (B) Comparison of the DFs between the (–I/+II)-double-mismatch and the (–I/+I/+II)-triple-mismatch at different temperatures. P-3FAM-4T(5′PO4-TCT(-FAM)TCACAGACACATACTCCA-BHQ1) was used for the test. The two purposefully inserted mismatched bases are underlined and the base opposite the SNP site is indicated in bold italics.
To confirm the generality of the phenomena, similar experiments were conducted using two new probes in which the fluorophore was attached to the fourth nucleoside from the 5′ end (referred to as P-4FAM, see Table S1†) and the tenth nucleoside from the 5′ end (referred to as P-10FAM, see Table S1†), respectively. As shown in Fig. S1 in the ESI,† remarkable discrimination effects were also observed for the two reference probes.
To elucidate the main reasons for the above discrimination effects, we performed several comparison studies. First, we synthesized a new 29-nt probe (P-15FAM-4C, see Table S1†) comprising the same sequence as P-3FAM-4C (Table 1) near the 5′-end, while the fluorophore was attached to the fifteenth nucleoside from the 5′ end of the probe. Then we measured the fluorescence signals of the probe in the presence of three different target strands. As shown in Fig. S2A in the ESI,† in comparison with the perfectly matched probe/target hybrid, the (–I/+II)-double-mismatched probe/target duplex showed a generally similar decreased signal compared with that observed with P-3FAM-4C. However, the (–I/+I/+II)-triple-mismatched probe/target duplex did not show a significantly reduced signal compared with that observed with P-3FAM-4C (Fig. 1B), suggesting a considerable contribution of the fluorophore to the inhibition of the enzymatic digestion. To confirm this, we further synthesized a probe P-3digoxin-15FAM-4C, which had the same sequence as P-15FAM-4C but with an additional label of digoxin at position 3 from the 5′ end. Fig. S2B† shows the considerable discrimination ability of this probe between the (–I/+II)-double-mismatched probe/target duplex and the (–I/+I/+II)-triple-mismatched probe/target duplex. We also compared the reaction rates of the perfectly-matched P-3digoxin-15FAM-4C/target duplex and the perfectly-matched P-15FAM-4C/target duplex, which were found to be at a ratio of 0.9 : 1.0, indicating that, without the two inserted mismatches, modification of the nucleotide with digoxin only slightly influenced the reaction rate. Taken together, a synergistic effect between the covalently modified fluorophore and two adjacent mismatched bases seems to play a key role in the dramatic change of the interactions between the probe/target hybrid and λ exo. Further investigation into the exact mechanism of these effects is underway and will be reported later.
The commonly used buffer for λ exo reaction is 1× Lambda Exonuclease Buffer (67 mM glycine–KOH, 2.5 mM MgCl2, 50 μg mL–1 BSA, pH 9.4@25 °C). However, the above detection system showed better performance in 1× ThermoPol Reaction Buffer (20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% TritonX-100, pH 8.8@25 °C). We further tested different concentrations of ThermoPol Reaction Buffer (from 0.5× to 5×). As shown in Table S2,† the largest DF was obtained in 3× ThermoPol Reaction Buffer. Then we tried to add (NH4)2SO4–KCl, MgSO4, and Triton X-100 respectively to the 1× ThermoPol Reaction Buffer to elucidate the major factor in the enhancement effect. It was found that additional MgSO4 in the buffer increased the enzyme activity but decreased the DF, while additional Triton X-100 slightly decreased both the enzyme activity and the DF. Mg2+ is the essential ion in the active center of λ exo. With more Mg2+, the digestion rate of the (–I/+I/+II)-mismatched DNA duplex by λ exo likely increased more than that of the (–I/+II)-mismatched duplex, resulting in a decrease of the DF. By contrast, additional (NH4)2SO4 and KCl slightly decreased the enzyme activity but obviously improved the DF (see Table S2†). This may be due to the increase of the ionic strength of the solution, which decreased the digestion rate of the (–I/+I/+II)-mismatched substrate more than that of the (–I/+II)-mismatched substrate, thus leading to an increase of the DF. Accordingly, we chose 1× ThermoPol Reaction Buffer with 20–30 mM (NH4)2SO4 and 20–30 mM KCl as the reaction buffer.
The influence of temperature on the discrimination effects was examined in the range of 25 °C to 48 °C. From Fig. 2B, the assay shows high DFs in the temperature range of 25 °C to 37 °C, suggesting the working temperature of the method is widely adaptable. At temperatures higher than 42 °C, the enzyme activity significantly decreased. Considering the potential use of the method in high-throughput microfluidic chips or other micro-devices without a temperature-control system, we chose 25 °C (room temperature conditions) to perform the detection.
Under the optimized reaction conditions, we tested twelve types of target with match or mismatched bases at the +I position. As shown in Table 1, the DFs obtained by our new probes are remarkably higher than those obtained by the simple hybridization approaches. As an example, the highest DF (320) observed with the probe/target hybrid containing a C:C mismatch is more than ten times higher than that obtained by the triple-stem probe for the same type of mismatch (28.4).9
The specificity of hybridization-based probes was usually believed to be determined by their thermodynamic stability, and insertion of an additional mismatch has been found to cause a significant decrease in the melting temperature (T m) of the hybrid.15 But as aforementioned, without the covalently attached fluorophore, the presence of double mismatches in the probe–target hybrid did not offer significant discrimination power. To clarify whether the exceptionally large DFs of the new detection system was mainly due to the difference in melting temperatures (T m) or the specific interaction between the enzyme and the modified structure of the probe, we measured the T ms of the probe/target duplexes containing different mismatches. The difference in melting temperature for the (–I/+II)-double-mismatch vs. the (–I/+I/+II)-triple-mismatch (57.5 °C vs. 56.8 °C) was observed to be much smaller than that for the (–I/0)-double-mismatch vs. the (–I/0/+I)-triple-mismatch (62.5 °C vs. 59.5 °C), suggesting that the stability of the hybrid is not the major factor in the high discrimination between the (–I/+II)-double-mismatch and the (–I/+I/+II)-triple-mismatch. We also measured the T ms of the perfectly-matched probe/target duplexes with and without the fluorophore label, which were found to be 65.0 °C and 63.5 °C, respectively. Clearly, the difference in T m was not the major cause of the large DFs of the new method. We conferred that the modification of the chemical structure of the probe by the fluorophore and the two mispaired bases might have altered the interactions between the enzyme and the probe/target hybrid, and led to a sensitive discrimination effect against a further single-base substitution at the 3′ side of the fluorophore labeled nucleoside.
It is worth mentioning that the fluorophore in our probe not only acts as a signal reporter in the detection system, but also plays a notable role in the specific molecular recognition between the enzyme and the probe/target hybrid. Moreover, the amplification process of our assay is not a simple enzyme-driven amplification process. In fact, each time the probe strand is digested by λ exo, the enzyme will precisely differentiate the special structure of the probe/target hybrid. Thus the amplification process resulted in an accumulated effect of multiple differentiation processes. So the high discrimination power actually originates from a more complicated protein/DNA interaction rather than a simple hybridization between two single strands.
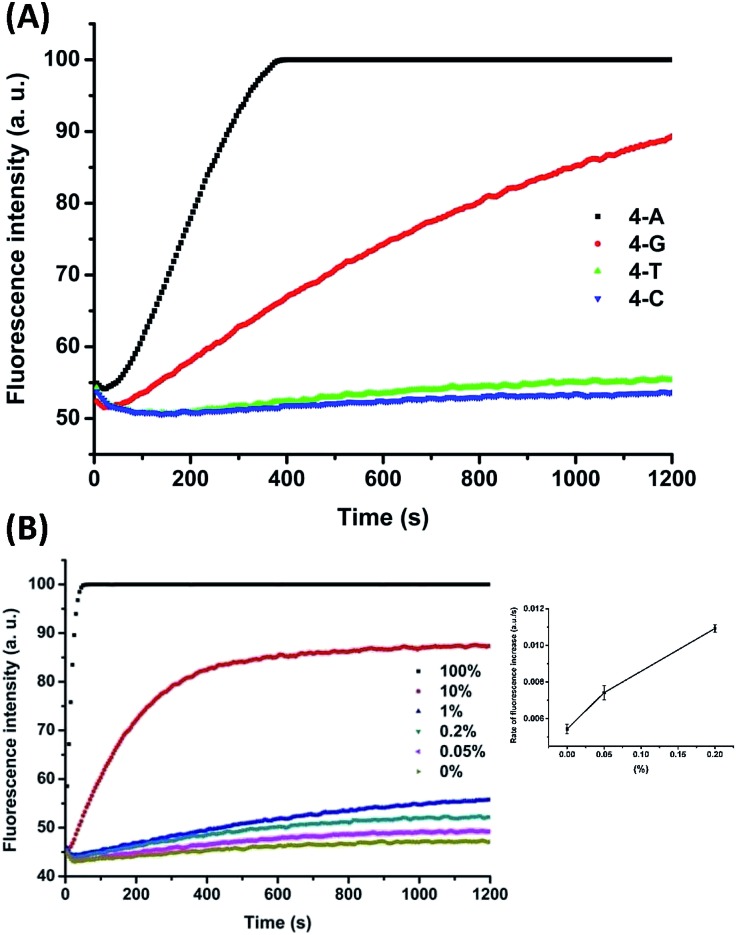
To demonstrate the applicability of the system in SNP detection, we used P-3FAM-4T to detect different types of base at position +I. From the fluorescence intensity responses over time, shown in Fig. 3A, the four hybrids formed with normal and single-nucleotide-variant DNA targets are clearly differentiated. Then we further applied the method to detect low-abundance mutations in the presence of different amounts of single-base-different wild-type strands. The probe was designed to be matched with the mutant type at the +I position, thus the wild-type will mismatch with the probe at position +I. From Fig. 3B, the C:C mismatched target (G → C & C → G mutant types) can be clearly differentiated from the matched target at an abundance as low as 0.05% (S/N = 4.4 > 3 as shown in the inset of Fig. 3B) at room temperature within 20 min.35 The assay sensitivity was determined by using P-3FAM-4C as the probe and 5′-GTTTTAAATTATGGAGTATGTGTCTGTTGAAACGAG-AGTAAG as the target sequence. From the results shown in Fig. S3,† the limit of detection of the assay was 30 pM (1.5 fmol in 50 μL) (S/N = 4.9 > 3). Other mutant types were also measured and the detailed results were summarized in Fig. S4 in the ESI.†
Fig. 3. (A) Fluorescence intensity responses of the double-mismatch/fluorophore modified DNA probe (P-3FAM-4T) in the detection of different SNPs at position +I; (B) fluorescence intensity responses of P-3FAM-4C in the detection of DNA point mutations at different abundances. The inset shows the mean values and standard deviations of the rate of fluorescence increase over the time period from 100 s to 600 s for the background solution and the two tested solutions with the lowest abundances (i.e. 0.05% and 0.2%). The probe sequence is 5′PO4-TCT(-FAM)NCACAGACACATACTCCA-BHQ1. The target sequence is 5′GTTTTAAATTATGGAGTATGTGTCTGTTMAAACGA-GAGTAAG. 100% means the tested strands are all mutant type (N:M = C:G). 0% means the tested strands are all wild-type (N:M = C:C).
For an arbitrary analyte, the double-mismatch/fluorophore modified DNA probe containing different types of intentionally inserted mismatch bases at the –I/+II positions may result in different DFs. Based on our experimental results, introduction of C:C, C:T or C:A mismatches in the probe/target hybrids usually leads to high DFs. In the case that the base in the target strand opposite –I/+II is G, introduction of T:G mismatches may generate relatively high DFs. To provide a simpler design rule for the probe, we then tested the use of a universal mismatch base in the probe. 3-Nitropyrrole and 5-nitroindole are two commonly used artificial mismatch bases.15,16,36 By substituting the artificial base for the natural base, previous studies found that 5-nitroindole offered better stabilization properties in the DNA duplex than 3-nitropyrrole. So we further employed 5-nitroindole as a universal mismatch base and investigated its suitability for the detection.
V617F point mutation (1849G → T) in the Janus kinase 2 (JAK2) gene has been shown to be associated with several myeloproliferative disorders.37–39 The sequences of the JAK2 wild-type and V617F mutant were listed in Table S1.† Employing the strand complementary to the sequence of V617F mutant type as the target, we designed P-JAK2-DD, in which 5-nitroindole (represented by D) was inserted at the –I/+II positions as the universal mismatch base (see Table S1†). Thus, in addition to the two purposely inserted mismatches, the third T:C mismatch at position +I between P-JAK2-DD and the wild-type would significantly prohibit the reactions between the probe and the wild-type DNA. For comparison, we also synthesized P-JAK2-CC, in which we used the native base C to form mismatches at positions –I/+II (see Table S1†). The reaction buffer was chosen to be 1× ThermoPol Reaction Buffer with 30 mM (NH4)2SO4 and 30 mM KCl after optimization.
From the results shown in Table S3,† P-JAK2-DD achieved a DF of 26, which was higher than that of P-JAK2-CC (DF = 9.3), indicating better performance of the artificial base than native bases in the enzyme-mediated single-base difference discrimination test. To confirm this, we further tested two other target strands in which the two bases opposite the two artificial bases at positions –I/+II in the probe were changed from C/A to T/A and T/T, respectively (see Table S3†). Interestingly, the DFs with P-JAK2-DD were both higher than those with P-JAK2-CC.
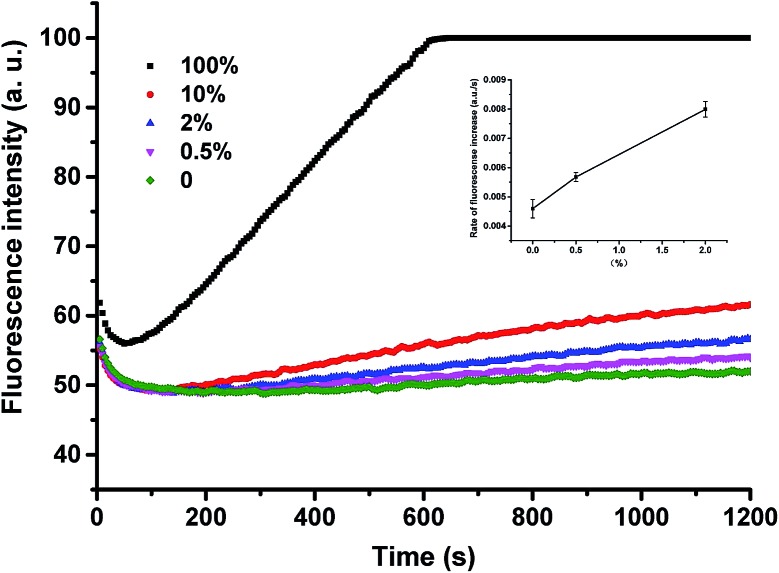
Next, we used P-JAK2-DD to detect the JAK2V617F mutant-type at different abundances. From the data shown in Fig. S5,† V617F mutant-type targets could be successfully identified in the presence of wild-type strands at an abundance as low as 0.5%. Since the ThermoPol Reaction Buffer used for the above detection is suitable for many polymerases such as Taq polymerase, our method can also be readily coupled to various PCR procedures. As shown in Fig. 4, the assay could also be applied for low-abundance mutation detection in PCR amplicons with high selectivity.
Fig. 4. Fluorescence intensity responses of P-JAK2-DD to the detection of JAK2V617F mutant-type at different abundances in PCR amplicons. 100% means the tested strands are all JAK2V617F mutant-type. 0% means the tested strands are all JAK2 wild-type. The inset shows the mean values and standard deviations of the rate of fluorescence increase over 300–1200 s for the two tested solutions with the lowest abundances (i.e. 0.5% and 2%).
The above results clearly demonstrate the high single-base substitution selectivity of the new system. It offers a very simple and rapid tool for highly selective single-base mutant detection at room temperature. The probe sequence can be flexibly adjusted to various target sequences without any strict length constraints, as long as the fluorophore and the two adjacent auxiliary mismatched bases are located at suitable positions. Moreover, we found that 5-nitroindole could be utilized as a universal mismatch base for different target sequences, which further simplified the probe design work. As no temperature control is required, the assay can be easily adapted to different high-throughput micro-devices without the need for complicated instrument design.
Conclusions
In this work, we have demonstrated that the combination of two purposefully introduced mismatches and a fluorophore tag in the probe sequence offers a special chemical structure that shows specific interactions with Lambda exonuclease (λ exo). By taking advantage of this new property, a simple and powerful system for the detection of single-nucleotide variation has been developed with extraordinarily high DFs (320–4.0). The method is single-step, rapid, and applicable at room temperature with minimal sample consumption. It can be used individually or flexibly coupled to microfluidic instruments for the sensitive detection of SNPs and low-abundance point mutations without the need for temperature control.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (91132717, 21175007 and 21375004).
Footnotes
References
- Dong L. M., Potter J. D., White E., Ulrich C. M., Cardon L. R., Peters U. JAMA, J. Am. Med. Assoc. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A. Nat. Rev. Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan R. S. Circulation. 2006;113:2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Cancer Res. 1988;48:1697–1701. [PubMed] [Google Scholar]
- Luo J. D., Chan E. C., Shih C. L., Chen T. L., Liang Y., Hwang T. L., Chiou C. C. Nucleic Acids Res. 2006;34:e12. doi: 10.1093/nar/gnj008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Kramer F. R. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- He Y. Q., Zeng K., Gurung A. S., Baloda M., Xu H., Zhang X. B., Liu G. D. Anal. Chem. 2010;82:7169–7177. doi: 10.1021/ac101275s. [DOI] [PubMed] [Google Scholar]
- Kolpashchikov D. M. Chem. Rev. 2010;110:4709–4723. doi: 10.1021/cr900323b. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Plakos K. J. I., Lou X. H., White R. J., Qian J. R., Plaxco K. W., Soh H. T. Angew. Chem., Int. Ed. 2009;48:4354–4358. doi: 10.1002/anie.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J., Gerasimova Y. V., Kolpashchikov D. M. Angew. Chem., Int. Ed. 2010;49:8950–8953. doi: 10.1002/anie.201004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Y., Chen S. X., Yin P. Nat. Chem. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbury C. A., Li J., Makrigiorgos G. M. Clin. Chem. 2009;55:632–640. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesova L., Belsanova B., Suchanek S., Kopeckova M., Minarikova P., Lipska L., Levy M., Visokai V., Zavoral M., Minarik M. Anal. Biochem. 2013;433:227–234. doi: 10.1016/j.ab.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Knez K., Spasic D., Janssen K. P. F., Lammertyn J. Analyst. 2014;139:353–370. doi: 10.1039/c3an01436c. [DOI] [PubMed] [Google Scholar]
- Guo Z., Liu Q. H., Smith L. M. Nat. Biotechnol. 1997;15:331–335. doi: 10.1038/nbt0497-331. [DOI] [PubMed] [Google Scholar]
- Chen S. X., Zhang D. Y., Seelig G. Nat. Chem. 2013;5:782–789. doi: 10.1038/nchem.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose C., Ficht S., Seitz O. Angew. Chem. 2006;45:5369–5373. doi: 10.1002/anie.200600464. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski R. W., Lyamichev V., de Arruda M., Neri B. Mol. Diagn. 1999;4:353–364. doi: 10.1016/s1084-8592(99)80012-5. [DOI] [PubMed] [Google Scholar]
- Lyamichev V., Mast A. L., Hall J. G., Prudent J. R., Kaiser M. W., Takova T., Kwiatkowski R. W., Sander T. J., de Arruda M., Arco D. A., Neri B. P., Brow M. A. D. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- Hall J. G., Eis P. S., Law S. M., Reynaldo L. P., Prudent J. R., Marshall D. J., Allawi H. T., Mast A. L., Dahlberg J. E., Kwiatkowski R. W., de Arruda M., Neri B. P., Lyamichev V. I. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11673–11673. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Xiao X., Zhang C., Zhao M. Appl. Spectrosc. 2012;66:1249–1262. doi: 10.1366/12-06803. [DOI] [PubMed] [Google Scholar]
- Wu D. Y., Ugozzoli L., Pal B. K., Wallace R. B. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang L. L., Mamon H., Kulke M. H., Berbeco R., Makrigiorgos G. M. Nat. Med. 2008;14:579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- Dominguez P. L., Kolodney M. S. Oncogene. 2005;24:6830–6834. doi: 10.1038/sj.onc.1208832. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K. W. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. Z., Chan D. S., Leung C. H., Ma D. L. Chem. Commun. 2012;48:9462–9464. doi: 10.1039/c2cc32253f. [DOI] [PubMed] [Google Scholar]
- Xiao X. J., Zhang C., Su X., Song C., Zhao M. P. Chem. Sci. 2012;3:2257–2261. [Google Scholar]
- Su X., Zhang C., Zhu X., Fang S., Weng R., Xiao X., Zhao M. Anal. Chem. 2013;85:9939–9946. doi: 10.1021/ac402615c. [DOI] [PubMed] [Google Scholar]
- Xiao X. J., Liu Y., Zhao M. P. Chem. Commun. 2013;49:2819–2821. doi: 10.1039/c3cc40902c. [DOI] [PubMed] [Google Scholar]
- He H. Z., Leung K. H., Wang W., Chan D. S., Leung C. H., Ma D. L. Chem. Commun. 2014;50:5313–5315. doi: 10.1039/c3cc47444e. [DOI] [PubMed] [Google Scholar]
- Gerasimova Y. V., Kolpashchikov D. M. Chem. Soc. Rev. 2014;43:6405–6438. doi: 10.1039/c4cs00083h. [DOI] [PubMed] [Google Scholar]
- Kovall R., Matthews B. W. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- Little J. W. J. Biol. Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- Subramanian K., Rutvisuttinunt W., Scott W., Myers R. S. Nucleic Acids Res. 2003;31:1585–1596. doi: 10.1093/nar/gkg266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDouglas D., Crummett W. B. Anal. Chem. 1980;52:2242–2249. [Google Scholar]
- Loakes D., Brown D. M., Linde S., Hill F. Nucleic Acids Res. 1995;23:2361–2366. doi: 10.1093/nar/23.13.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C., Ugo V., Le Couedic J. P., Staerk J., Delhommeau F., Lacout C., Garcon L., Raslova H., Berger R., Bennaceur-Griscelli A., Villeval J. L., Constantinescu S. N., Casadevall N., Vainchenker W. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- James C., Ugo V., Casadevall N., Constantinescu S. N., Vainchenker W. Trends Mol. Med. 2005;11:546–554. doi: 10.1016/j.molmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Levine R. L., Pardanani A., Tefferi A., Gilliland D. G. Nat. Rev. Cancer. 2007;7:673–683. doi: 10.1038/nrc2210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.