Abstract
How do Ras isoforms attain oncogenic specificity at the membrane? Oncogenic KRas, HRas and NRas (K-Ras, H-Ras, N-Ras) differentially populate distinct cancers. How they selectively activate effectors and why is KRas4B the most prevalent are highly significant questions. Here we consider determinants that may bias isoform-specific effector activation and signaling at the membrane. We merge functional data with a conformational view to provide mechanistic insight. Cell-specific expression levels, pathway cross-talk and distinct interactions are the key; but conformational trends can modulate selectivity. There are two major pathways in oncogenic Ras-driven proliferation: MAPK (Raf/MEK/ERK) and PI3Kα/Akt/mTOR. All membrane-anchored, proximally-located, oncogenic Ras isoforms can promote Raf dimerization and fully activate MAPK signaling. So why the differential statistics of oncogenic isoforms in distinct cancers and what makes KRas so highly oncogenic? Many cell-specific factors may be at play, including higher KRAS mRNA levels. As a key factor, we suggest that because only KRas4B binds calmodulin, only KRas can fully activate PI3Kα/Akt signaling. We propose that full activation of both MAPK and PI3Kα/Akt proliferative pathways by oncogenic KRas4B – but not by HRas or NRas – may help explain why the KRas4B isoform is especially highly populated in certain cancers. We further discuss pharmacological implications.
Keywords: K-RAS, H-RAS, N-RAS, K-RAS4A, K-RAS4B, KRas4A, small molecule drug
Introduction
A question which has been at the center stage of cancer biology is why the differential statistics of Ras oncogenic isoforms in distinct cancers. Here we argue that even though conformational heterogeneity at the membrane will influence signaling preferences – it will not fully explain them. The more likely reasons are certain determinants of cell specificity. Below, we review possible cell-specific factors differentiating KRas4B signaling from HRas and NRas, and suggest that these may help unravel Ras isoforms oncogenic specificity.
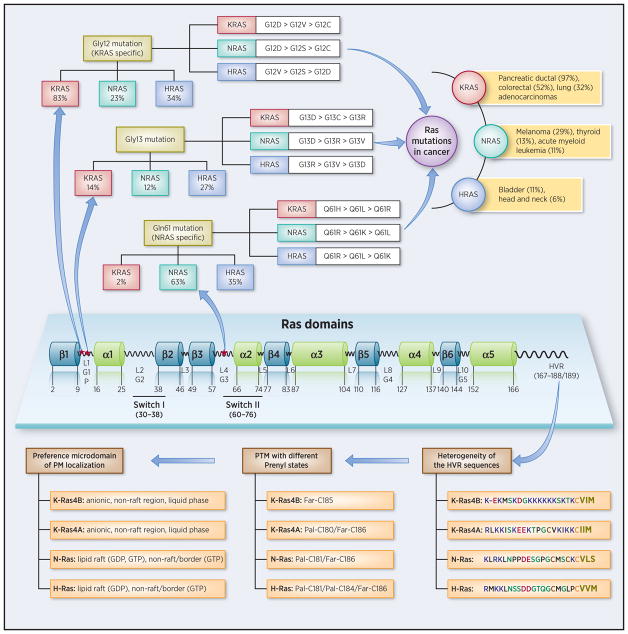
HRas, NRas, and alternatively spliced KRas4A and KRas4B control key cell proliferation pathways (1–3). They bind a common set of activators and effectors (4, 5), but are not functionally redundant and certain oncogenic isoforms are more commonly observed in distinct cancer types (4, 6–8). Interest has largely focused on oncogenic mutants of KRas4B because of its relatively high frequency in cancers, particularly adenocarcinomas in pancreas, colorectal and lung cancers (9), although early (10–12) and more recent work (13–15) affirmed that KRas4A can also be widely expressed in human cancer cell lines, and that the relationship between KRas4A/KRas4B may be the reason for the higher frequency of mutations observed in KRas4A (10).
Ras isoforms signal effectively when anchored and enriched at the plasma membrane (PM) (16–21). Signaling pathways include mitogen-activated protein kinase (MAPK), phosphoinositide-3-kinase (PI3K), Ras-Ral guanine nucleotide dissociation stimulator (RalGDS), phospholipase C ε (PLCε), Cdc42/Rac, Ras association domain family 5 (RASSF5, also known as NORE1A) mediating mammalian sterile 20-like kinase 1/2 (MST1/2) Hippo pathway (albeit with limited direct experimental data supporting RASSF5 repressing Yes-associated protein 1 (YAP1) activity (22)), as well as the emerging pathway cross-talk (23).
Insight into how differential isoform signaling at the membrane is achieved is among the most coveted aims of Ras cancer biology (1, 6, 15). Ras proteins consist of the soluble catalytic G-domain and the hypervariable region (HVR). The sequences and structures of the catalytic domains of the isoforms are highly similar, in contrast to their disordered C-terminal HVRs that attach them to the PM (24–26). Nature stapled the HVRs with distinct combinations of lipid post-translational modifications (PTMs) – farnesyl (or geranylgeranyl) and palmitoyl. Membrane attachment can also be regulated by cell-specific mechanisms, such as monoubiquitination (27). PM lipids segregate laterally into microdomains with distinct composition and organization (28–30) which appear to be differentially preferred by Ras isoforms, resulting in homogeneous dimerization and nanoclustering. The microdomain environments can influence Ras structure and orientation at the membrane, which influences Ras isoforms accessibility, thus association states with its effectors (31). Recent saturation mutagenesis of the catalytic domain unraveled the selection pressures to retain (or prevent loss of) Ras function (32). The similarity of the catalytic domains and heterogeneity of the HVRs along with their combinations of lipid PTMs speak of encoding unique functional roles (33). A key question is thus how the HVRs assist in biasing isoform signaling selectivity, and how they partner with the conserved catalytic domains to execute function. To date, attention has largely focused on revealing the interplay between Ras isoform-specific lipidation motifs and the membrane (34). Here we discuss how they can also help to accomplish Ras isoform-specific signaling. Insight into these mechanisms may help tumor-specific drug discovery.
An added level of selectivity is among oncogenic mutants of the same isoform (24). As documented for KRAS, specific mutations can be more frequent in distinct adenocarcinomas (9), leading to questions like how mutations in the catalytic domain can bias the conformational ensemble, and whether distinct mutations may promote oncogenic signaling specificity via the farnesylated HVR behavior. Allosteric effects elicited by mutations can affect the conformation of the effector binding site (35, 36) as well as its exposure (14, 37); they can affect regulatory sites (38), enhance nanoclustering (39), and be mediated by the HVR at the PM (33, 40). Mutant Ras molecules are also greatly affected by cell-specific environments (14) and genetics (41, 42).
Here we address the long-standing question of how Ras isoforms (and their distinct mutants) acquire oncogenic specificity despite their high structural homology. Our model integrates functional cell biology data with conformational theory. Our key underlying hypotheses are that (i) the conformational behavior of Ras at the membrane is important in modulating specificity but is unable to explain isoform and oncogenic mutant signaling preferences in specific cancers which we believe is determined by the cell-specific environment; and (ii) even though all adjacent membrane-anchored oncogenic Ras isoforms can stimulate Raf’s dimerization and fully activate MAPK, only oncogenic KRas4B can fully activate PI3Kα/Akt signaling. This may explain why KRas4B (and KRas4A) – rather than HRas or NRas – is particularly oncogenic. Perceiving differential isoform signaling at the membrane requires a grasp of Ras conformational landscape. Below we provide the basis, and then discuss isoform selectivity from this standpoint.
The Importance of a Structural View
Numerous pathway databases exist for Ras and its signaling pathways (43). The mammoth task of assembling cellular, biochemical, clinical and high resolution structural data results in an extremely useful overall picture; however, it still falls short. An organized and informative cartoon still cannot decipher the relationship between distinct isoforms and their preferred signaling pathways; nor are compilations able to explain the root causes of signaling trends which can be correlated with specific cancers. Compilations are critical; but insufficient. A conformational view of the data can build a structural map of Ras pathways, and consider concepts that underlie such isoform signaling enigmas.
Among these concepts, is the idea of the energy landscape (44); that is, that biomolecules are not static sculptures. Molecules continuously interconvert between different states, and the statistics of these states reflect their relative populations. These depend on mutational events, membrane composition, post-translational modifications, ion concentrations, and more. Thus, the most populated state of KRasG12D differs from that of the KRasG12V mutant (45, 46); and the most populated state of KRas4B at the membrane differs from that of NRas because they favor altered interactions with membrane phospholipids (14, 24, 37). Ras molecules constantly fluctuate, and the fluctuations depend on their specific sequence and the molecules that they bind, e.g. lipids, proteins, ions and water. The fluctuations are critical for the preferred orientational states with respect to the membrane as well as the time spent in these states, and the consequent exposure of the effector binding site. Forecasting them calls for deciphering atomic-scale interactions and conformational distributions, which in principle may be achieved by molecular dynamics simulations. Current simulations are handicapped by their affordable timescales, by the idealized membrane systems whose kinetics may differ from those of complex lipid mixtures found in nature, inaccurate forcefields, and simulations of monomers rather than the functional states.
The Chemistry of the Farnesyl and Palmitoyl Biases Signaling
Ras signaling initiates at the membrane. Thus, the heterogeneity of HVR sequences across the isoforms and the distinct combinations of the lipid PTMs (Figure 1) should be considered within this framework (15). The palmitoyl is a 16-carbon saturated fatty acid attached to a cysteine residue via thioester linkage (S-palmitoylation) (47). Saturation coupled with chain length makes it a major driving force for insertion into lipid microdomains, including lipid rafts or liquid ordered phase (48–52). The thioester linkage can be hydrolyzed by palmitoyl protein thioesterases (PTEs) (53). By contrast, the thioether-linked 15-carbon farnesyl (and 20-carbon geranylgeranyl) isoprenoid group prefers to insert into liquid ordered phase membranes rich in unsaturated fatty acids (54, 55). Its hydrocarbon chain contains cis double bonds, making farnesyl (and geranylgeranyl) insertion reversible. In the absence of palmitoyl in KRas4B (or in depalmitoylated KRas4A), the HVR’s strong positive charge stabilizes membrane anchorage (56). The PTMs’ favored membrane environments coupled with their combinatorial codes and HVR properties point to distinct favored orientations of the Ras isoforms. The HVRs of HRas and NRas are electrostatically similar (13, 15). However, HRas has a farnesyl and two palmitoyls and NRas one. Like NRas, KRas4A has a farnesyl and a single palmitoyl; however, its positive charge may result in altered favored orientations (14, 15). With only a farnesyl and charged HVR, KRas4B may prefer different orientations on the anionic membranes. Why are the distinct HVR-PM states important for isoform-specific signaling at the membrane? Binding affinity reflects the strength of the interaction between the molecules. Because all isoforms bind the same set of effectors, even slight differences in the surfaces that they expose to the cytoplasm – largely due to the differential occlusion by the membrane - may alter their affinities to their cognate receptors, thus bias their effector selectivity (56–58). Hydrolysis of the palmitoyl linkage will disengage HRas and NRas whose HVR-membrane interaction is weak, and result in shuttling to the Golgi for re-palmitoylation (16). KRas4A also anchors during its palmitoylation/depalmitoylation cycle (13). Calmodulin (CaM) binding (59) or Ser181 phosphorylation by protein kinase C (PKC) or cyclic GMP (cGMP)-dependent protein kinase (PKG) (60) disengage, or reduce the interaction of KRas4B with the membrane; nonetheless, phosphorylated molecules are still able to form nanoclusters and their lateral membrane organization is unaffected (61).
Figure 1.
Domain structure of Ras. The catalytic domain (effector lobe: residues 1–86, allosteric lobe: residues 87–166) sequence is highly homologous and the structure is identical among Ras isoforms with five helices and six β-strands connected by ten loops, and the G1-loop (or P-loop) to G5-loop enclosing the nucleotide. Ras isoforms differ in their flexible C-terminal HVRs (residues 167–188/189), which undergo distinct posttranslational modification (PTM) events including prenylation, methylation, and acylation (hydrophobic, polar/glycine, positively charged, and negatively charged residues are colored black, green, blue, and red, respectively; farnesylated cysteine residues are denoted by yellow, and palmitoylated cysteine residues are denoted by orange. The hollowed letters denote “AAX” motif). Ras mutations data were collected from the literature (4, 9). The preferred membrane lodging is also indicated, with ‘lipid rafts’ described as membrane domains whose their lipid components are in a liquid-ordered (lo) state as distinct from their coexisting liquid-disordered (ld) ‘non-raft’ regions of the bilayer (109).
The functional importance (62, 63) and different chemical properties and linkages of the palmitoyl and the farnesyl evolved varied means for their transportation (33, 64); vesicular for HRas, NRas and palmitoylated KRas4A (65), and phosphodiesterase-δ (PDEδ) for KRas4B (13, 66). Farnesyl contributes dominantly to the PDEδ–Ras interaction (66). Experimental data indicated that PDEδ can extract from the PM NRas – but not KRas4A; modeling suggested that the stronger interaction with the PM of KRas4A versus NRas can explain these extraction data (67).
Accessibility of the Effector Binding Site on the Membrane
Experiments and simulations suggest that the same catalytic domain surface can be engaged in binding protein effectors and in facing – albeit not securely interacting with – the membrane (37, 40, 58, 68). The interaction of the catalytic domain with the PM is weak. Instead, evolution has endowed proteins such as Ras and G protein subunit αi1 (69) with hydrophobic PTM-decorated termini (C- or N-), which are disordered when in the free, uncomplexed state (26). This elegant solution also permits multiple states, creates space for large complexes and facilitates lateral mobility (70) and nanocluster formation. Accessibility can be defined in terms of the residence time of the ensemble in an ‘open state’; occlusion denotes conformations with the binding site facing the PM and fluctuating against it, or against the HVR. Even if GTP-bound, occlusion of this site may hamper signaling of weak-binding effectors (14). Thus, whereas the classical definition of an active Ras state is based on its nucleotide (GTP/GDP) binding status, conformational reasoning argues that on its own, GTP-loading may not be sufficient for downstream effector activation from the PM (58).
In vitro and in silico, albeit not tested in vivo when competing with the membrane or PDEδ, the HVR can directly interact with the effector binding site of inactive KRas4B, and this interaction is almost 100-fold tighter than that of the active state (68). Oncogenic mutations away from this site can allosterically amplify this exposed state to facilitate effector binding (71). In vivo, KRas has a high degree of specificity for phosphatidylserine (PS), particularly with certain amino acid side-chain/lipid interactions (17). In vitro, the HVRs of GDP-bound KRas4BG12V/G12D mutants interact selectively with phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 4-phosphate (PI4P), phosphaditylinositol 5-phosphate (PI5P) and phosphatidic acid (PA), suggesting that mutations in the catalytic domain can allosterically modulate the HVR-phospholipid binding specificity, tuning KRas4B selectivity (33, 40).
Organization, Dynamics, Mobility and Signaling at the Membrane
Ras molecules favor distinct organizations in the PM, diffuse, and can dimerize. Unlike HRasG12V in 2-dimyristoylglycero-3-phosphocholine (DMPC), the catalytic domains in GTP- and GDP-bound KRas4B adopt similar membrane orientations (57). Whereas HRas-GTP (but not HRas-GDP) orientations are stabilized by helix α4, KRas4BG12V appears stabilized by the HVR. KRas4A (in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1,2-palmitoyl-oleoyl-sn-glycero-3-phosphoserine (POPC/POPS)) resembles KRas4B (14, 37, 58). This different behavior of HRas (and presumably NRas as well), is in line with the larger sequence differences and preferred orientations observed in dimer interfaces at the membrane for KRas4B versus HRas (72). Catalytic domain orientations are also influenced by interactions with the PM (33, 73), suggesting that they partner with HVR-phospholipid binding to accomplish signaling (74). KRas4B-GTP dimers interacting through the allosteric lobe favor interfaces involving helices α3 and α4; those of HRas-GTP may populate α4 and α5. Effector lobe dimer interfaces are mostly β-sheet extension with additional species through β-sandwich (72). Raf’s and RASSF5 (NORE1A) activations involve the helical dimer interface. Oncogenic mutations may alter this picture.
The lateral diffusion of Ras is as fast as lipid probes and significantly faster than a typical membrane protein (75–77). However, Ras dimerization/nanoclustering, and its assemblies including high-affinity (but not low-affinity) effectors/regulators and scaffolding protein platforms, is likely to constrain its lateral dynamics and mobility in the PM. Two main factors are at play. The first is the sheer increase in size and molecular weight. The second involves additional membrane anchorage by some of those other molecules. A KRas4B dimer will have two anchorages. If a membrane-anchored Raf is bound to each Ras molecule, anchoring of the two Raf molecules will also be involved. RASSF5 tumor suppressor presents a similar scenario (78). In the complex of KRas4B–PI3Kα both species will be anchored; binding to CaM, which does not interact with the PM, will increase the assembly size (Figure 2). Signaling complexes involving e.g. galectin-3, apoptosis-stimulating of p53 protein 2 (ASPP2) (79–81), or isoleucine-glutamine GTP-activating protein 1 (IQGAP1) (82), and its associated kinases such as Akt, and even actin (83), will slow lateral mobility and increase residence times of certain states.
Figure 2.
Ras signaling pathways. Under physiological conditions, Ras forms nanoclusters and activates the Raf/MEK/ERK (MAPK) pathway, leading to cell proliferation. In the PI3K/Akt pathway, both Ras and RTK activate PI3K, recruiting it to the plasma membrane, leading to cell growth. Under oncogenic conditions, a constitutively active Ras (KRas, HRas or NRas) cluster boosts Raf/MEK/ERK signaling. In adenocarcinoma of KRAS-driven cancer, CaM specifically recruits oncogenic KRas4B (and possibly KRas4A, but not HRas or NRas) from the membrane to PI3Kα and substitutes for the missing RTK signal, consequently fully activating the PI3Kα/Akt/mTOR pathway.
Most effectors function as single entities. For these, Ras dimerization or proximity status appears irrelevant. For those whose function involves dimerization, such as Raf (84), or for mediating auto-cross-phosphorylation, such as RASSF5 (mediating MST) kinase activation (78, 85, 86)), Ras dimerization (or productive nanocluster organization which leads to similar outcomes (20, 21, 39, 87)), is vital (Figure 2). Multiple mechanisms modulate C-Raf kinase activity including phosphorylation (by Src family kinases and PAK) of Ser338, dephosphorylation of Ser259 (by serine phosphatases), interactions with 14-3-3, with lipids (such as PS and PA), and more. Single molecule fluorescence resonance energy transfer (FRET) data suggested that Raf is recruited to immobile sites on the PM (88, 89). All data point to Raf’s activation requiring membrane anchorage which is assisted by Raf’s cysteine-rich domain (CRD). Ras membrane-anchored nanoclusters (or dimers) increase the chance of Raf’s dimerization, and from the structural standpoint, activation has been fully explained by kinase domain dimerization (84, 90). Ras nanoclusters are homogeneous, segregated by isoform and nucleotide type, likely reflecting distinct preferred local membrane composition. Blocking Ras dimerization blocks nanocluster formation resulting in loss Ras nanoclusters and MAPK signaling (91). The rate of diffusion, or mobility, of Ras monomers is a key factor in Ras dimerization (or nanoclustering). Higher mobility of Ras monomers leads to faster dimerization, promoting Raf’s dimerization, thus activation and MAPK signaling. Slower mobility delays dimerization, slowing down MAPK response times. Notably, if Ras is overexpressed, MAPK signaling response times will be fast irrespective of the diffusion rate. By contrast, Ras dimerization is not required for PI3K activation. This is because its activation does not require Ras dimers. PI3K effectively acts a monomeric unit. Under physiological conditions it is activated by a single Ras molecule binding to the p110 subunit, and a phosphorylated RTK C-terminal motif which binds the SH2 domains of the p85 subunit (92). In line with this, microdomains were observed to regulate HRas-driven MAPK signaling - but not PI3K signaling (93).
The linkage between the lipid content and KRas binding has also been investigated. The inner PM leaflet is often enriched with anionic phospholipids such as PS, and cell signaling lipid molecules like phosphatidylinositols (PIs). KRas activation level in cancer cells has been linked to PS content (94) and the membrane potential influences the organization of the phospholipids in the PM, which in turn regulates the localization and activity of KRas4B (20, 95). HVR amino acid side-chains may favor specific lipid head groups; one example is PA and KRas4B (33); another is the non-equivalency of lysine and arginine despite both being positively charged (17). Thus, taken together, mobility, and PM composition and localization may modulate Ras action (33, 96–98), substantiating the role of PM microdomains as signaling platforms (99) (Figure 1). Further substantiating this notion, relationships between inhibitory specificities of Ras isoforms and microlocalization (100) and between NRas clustering and activation patterns (101) were also observed.
Additional factors that govern membrane localization and Ras signaling include e.g., HVR motifs (102, 103), the influence of domain structure (104), PM scaffolding proteins such as galectins (105–108), partitioning of membrane molecules between raft and non-raft domains (109), and nanoclusters segregation within these (110).
Mechanisms Extracting KRas4B from the Membrane and Their Distinct Functions
Complex mechanisms coevolved to accomplish isoform-specific signaling at the membrane. At their core is the distinct chemical nature of the HVR sequences and PTMs. It is thus not surprising that the three main mechanisms extracting KRas4B, but not HRas or NRas, from the PM to block Raf’s activation and MAPK signaling, also involve the distinct properties of the KRas4B HVR (16, 92, 111–113).
Phosphorylation
Modeling suggests that the strong electrostatic interaction of the phosphoryl with the HVR’s lysine side-chains bends the HVR and collapses the C-terminal residues around it (56). This results in the phosphoryl releasing the farnesyl-PM interaction and increasing the farnesyl-water and farnesyl-peptide interactions. The phosphoryl also generates electrostatic repulsion between the HVR and the phospholipid headgroups. Phosphorylation reduces but does not inhibit membrane binding and clustering of KRas4B (60, 61). Electrostatic repulsion may however operate when KRas attempts to rebind the membrane after removal (56).
Ca2+-CaM
Ca2+-CaM has numerous roles in oncogenic signaling (114, 115). KRas4B’s positively charged HVR – but not HRas or NRas – binds tightly to Ca2+-CaM, and its farnesyl docks into a CaM pocket (116). CaM’s binding extracts KRas4B from the PM (59); CaM also interacts with PI3Kα and activates it (117, 118). Under normal physiological conditions, a receptor tyrosine kinase (RTK) phosphorylated C-terminal peptide motif couples with KRas4B (as well as other Ras isoforms, such as HRas (119)) to fully activate PI3Kα. Because the affinity of Ras to the Ras binding domain (RBD) of PI3Kα is low (in the high micromolar range), PI3Kα’s activation requires that Ras is also attached to the membrane, which increases the proximity and stabilizes the Ras–PI3Kα interaction. In cancer, KRas4B–CaM is the driving force in Akt activation and PI3K/Akt pathway (120, 121). Oncogenic KRas4B–CaM–PI3Kα ternary complex (121), where CaM replaces the RTK motif, can explain how PI3Kα gets fully activated by oncogenic KRas4B in the absence of a signaling cue (112). CaM’s binding to the SH2 domains of the p85 subunit of PI3Kα (117) releases its autoinhibition and together with KRas4B promotes its full activation, KRas4B/PI3Kα/Akt/mTOR signaling and cell proliferation (59, 112) (Figure 2). Even though non-phosphorylated CaM molecules can bind, CaM can be phosphorylated including at Tyr99 (114, 115), and the affinity of phosphorylated CaM is higher (92, 118, 122). We expect that other, still-to-be discovered, proteins help PI3Kα activation by oncogenic HRas and NRas.
PDEδ
Cytosolic PDEδ promotes effective KRas4B signaling by securing its continuous sequestration from endomembranes (16). It shuttles KRas4B to the PM to enrich local KRas4B concentration and enhance signaling of MAPK and Hippo (though only limited direct experimental data for Ras effector RASSF5 (NORE1A) repressing YAP1 activity (22)). PDEδ binding to KRas4B is tight, with high affinity (66, 67). Upon contact with a heterogeneous membrane, KRas4B–PDEδ releases the KRas4B (113). A spatial organizing cycle, facilitated by PDEδ, maintains KRas4B on the PM, countering entropic redistribution to the endomembrane (16). HRas and NRas are extracted through depalmitoylation, followed by trafficking to the Golgi and a new signaling cycle.
Factors Differentiating KRas4B from HRas and NRas Signaling
Why are the differential statistics of oncogenic isoforms in distinct cancers? That is, why is oncogenic KRas the most prevalent isoform in pancreatic cancer (over 95%) whereas NRas is the one in melanoma (15%) and HRas in bladder cancer (5%)? Further, why is oncogenic KRAS the most abundant overall in Ras-driven cancers? What makes it so highly oncogenic? To date, these challenging and profound questions – which are the focus of hundreds of laboratories world-wide – are still unresolved. KRas4B is more highly expressed than other isoforms (7); thus, it is not surprising that its oncogenic form is more abundant. Nonetheless, this would not explain these statistics. Cell-specific functions are largely controlled by the cellular environment and genetics (42). Thus, addressing these questions only by assessing the expression levels of the isoforms may not solve them; however, neither would consideration of only cell types, as analyses of patients’ variability – even with the same type of cancer – indicate. To decipher these questions, here we ask which factors appear favorable for KRas4B signaling – but not HRas or NRas – viewing some recent observations from a conformational standpoint (Figure 1).
Oncogenic Ras isoforms may not signal equally effectively via the PI3Kα/Akt pathway
The two major pathways in oncogenic Ras-driven proliferation are the MAPK (Raf/MEK/ERK) and the PI3Kα/Akt/mTOR (123) (Figure 2). PI3Kα has two subunits, p85 and p110; their heterodimer acts as a monomeric unit (124–126). By contrast, Raf’s activation occurs via kinase domains auto-cross-phosphorylation (84, 90), which argues for dimerization or proximity (nanoclustering) of the respective Ras molecules (127–130). Oncogenic PM-anchored KRas4A, KRas4B, HRas and NRas cluster (and can dimerize) (21, 131); thus, MAPK signaling output is likely to be similar for all. This may not be the case for PI3Kα/Akt/mTOR signaling (92, 112). In the absence of the RTK signal, oncogenic Ras may be unable to fully activate PI3Kα (124, 132). If available, Ca2+-CaM can replace the RTK signal (117) – but only KRas4B (and presumably depalmitoylated KRas4A (15)) can bind CaM (114, 121, 133–135). Thus, only KRas can fully activate PI3Kα/Akt/mTOR signaling (Figure 2).
PDEδ availability
To signal, Ras isoforms must be translocated to the PM, and the translocation mechanisms vary. PDEδ plays a key role in KRas4B shuttling to the PM. HRas, NRas and palmitoylated KRas4A are shuttled by vesicles. In those cells where PDEδ availability is limited, KRas4B maybe mislocalized (16).
Appropriate membrane composition to accommodate KRas4B
Because KRas, but not other isoforms, favors acidic disordered membranes (involving PS, PIs, and other microdomain components, some of which are detailed above), membrane composition is also an important signaling factor (136).
Oncogenic KRas4B may be less populated in HRas and NRas cancers
Cell-specific factors, including processing enzymes of lipidation motifs (47), isoform-specific scaffolding proteins, localization factors (137) and other cell-and isoform-specific factors (138) may play a role. Comparisons across cell types, e.g. acute myeloid leukemia, melanoma and bladder cancers versus pancreatic cancer, may help in factor identification.
Differential behavior of isoforms of wild-type versus their respective mutants
In an apparent twist, studies of mouse model carcinogenesis as well as patient tumors seemed to indicate that wild-type Ras can act as tumor suppressor of its mutant form, particularly in KRAS cancers as compared to NRAS (1, 139, 140). KRas is the most abundantly-expressed isoform, and Ras nanoclusters are homogeneous. Thus, we reason that the inhibition of the oncogenic isoform by its respective wild-type (which in the absence of a signaling cue is inactive), may simply reflect the reduction in the number of the oncogenic (active) Ras molecules in the cluster.
Notably, there is also evidence that wild-type KRas (and HRas and NRas) may also collaborate with oncogenic isoforms to promote cancer (141–143). In these cases, wild-type Ras can be both tumor-suppressing and -promoting, and these effects, as well as clonal fitness, appear context- and tissue-dependent.
Concluding Remarks and Pharmacological Implications
How then to explain Ras isoform and oncogenic mutant signaling preferences in specific cancers and what pharmacology can it suggest? The conformational ensemble of the catalytic domains in solution is broad (24, 35, 45, 144). It is further broadened in full-length Ras on membranes with different local compositions (14, 72, 145). These may reflect distinct orientation preferences of their signaling assemblies. Thus, significant questions are to what extent these determine selectivity and druggability. The mobile nature of Ras molecules at the membrane, the fleeting residence times, and the absence of clear dominant states – at least in the time scales and PM composition that current molecular dynamics simulations of these huge systems can afford – question the efficacy of targeting preferred membrane interaction states. By contrast, productive cell-specific signaling pathways and cross-talk (23, 146, 147) appear more promising candidates; however, ferreting the complexes and poses to drug presents an even more daunting challenge. Lower drug toxicity calls for specificity; yet, working out the key factors that differentiate among isoforms is demanding. Along these lines, we proposed a ternary complex of KRas4B–CaM–PI3Kα (148). Among the recent promising isoform-specific efforts are the palmitoylation/depalmitoylation cycle (149), specific Gal-1 – an HRas scaffolding protein inhibitors (87, 150, 151), monobody targeting the α4-β6-α5 dimeric interface, recently shown to reduce the interaction of oncogenic KRas, but not HRas, with Raf, as well as KRas PM localization (152) and PDEδ (153). Past efforts have shown that the paths are treacherous; here we foraged for a merged conformational and cellular view which we believe may better reflect Ras behavior, and thus help in addressing the pharmacology challenge. Still, the ultimate challenge resides in the inevitable emergence of drug resistance, which we believe calls for preorganized combinatorial drug regimens aiming at parallel pathways (123, 154–156).
Acknowledgments
Grant Support
This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of NIH, Frederick National Laboratory, Center for Cancer Research.
This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol. 2016;58:60–9. doi: 10.1016/j.semcdb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Cheong JK. The renewed battle against RAS-mutant cancers. Cell Mol Life Sci. 2016;73:1845–58. doi: 10.1007/s00018-016-2155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussinov R, Tsai CJ, Jang H, Korcsmaros T, Csermely P. Oncogenic KRAS signaling and YAP1/beta-catenin: Similar cell cycle control in tumor initiation. Semin Cell Dev Biol. 2016;58:79–85. doi: 10.1016/j.semcdb.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–92. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakhaeizadeh H, Amin E, Nakhaei-Rad S, Dvorsky R, Ahmadian MR. The RAS-Effector Interface: Isoform-Specific Differences in the Effector Binding Regions. PLoS One. 2016;11:e0167145. doi: 10.1371/journal.pone.0167145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–31. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newlaczyl AU, Coulson JM, Prior IA. Quantification of spatiotemporal patterns of Ras isoform expression during development. Sci Rep. 2017;7:41297. doi: 10.1038/srep41297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker JA, Mattos C. The Ras-Membrane Interface: Isoform-specific Differences in The Catalytic Domain. Mol Cancer Res. 2015;13:595–603. doi: 10.1158/1541-7786.MCR-14-0535. [DOI] [PubMed] [Google Scholar]
- 9.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–67. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voice JK, Klemke RL, Le A, Jackson JH. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem. 1999;274:17164–70. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 11.Patek CE, Arends MJ, Wallace WA, Luo F, Hagan S, Brownstein DG, et al. Mutationally activated K-ras 4A and 4B both mediate lung carcinogenesis. Exp Cell Res. 2008;314:1105–14. doi: 10.1016/j.yexcr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–45. doi: 10.1002/path.2625. [DOI] [PubMed] [Google Scholar]
- 13.Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A. 2015;112:779–84. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li ZL, Buck M. Computational Modeling Reveals that Signaling Lipids Modulate the Orientation of K-Ras4A at the Membrane Reflecting Protein Topology. Structure. 2017;25:679–89. e2. doi: 10.1016/j.str.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nussinov R, Tsai CJ, Chakrabarti M, Jang H. A New View of Ras Isoforms in Cancers. Cancer Res. 2016;76:18–23. doi: 10.1158/0008-5472.CAN-15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, et al. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157:459–71. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Prakash P, Liang H, Cho KJ, Gorfe AA, Hancock JF. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell. 2017;168:239–51. e16. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blazevits O, Mideksa YG, Solman M, Ligabue A, Ariotti N, Nakhaeizadeh H, et al. Galectin-1 dimers can scaffold Raf-effectors to increase H-ras nanoclustering. Sci Rep. 2016;6:24165. doi: 10.1038/srep24165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najumudeen AK, Posada IM, Lectez B, Zhou Y, Landor SK, Fallarero A, et al. Phenotypic Screening Identifies Protein Synthesis Inhibitors as H-Ras-Nanocluster-Increasing Tumor Growth Inducers. Biochemistry. 2015;54:7212–21. doi: 10.1021/acs.biochem.5b00724. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Wong CO, Cho KJ, van der Hoeven D, Liang H, Thakur DP, et al. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–6. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta. 2015;1853:841–9. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhou XH, Yang CQ, Zhang CL, Gao Y, Yuan HB, Wang C. RASSF5 inhibits growth and invasion and induces apoptosis in osteosarcoma cells through activation of MST1/LATS1 signaling. Oncol Rep. 2014;32:1505–12. doi: 10.3892/or.2014.3387. [DOI] [PubMed] [Google Scholar]
- 23.Rauch J, Kolch W. Spatial regulation of ARAF controls the MST2-Hippo pathway. Small GTPases. 2017 doi: 10.1080/21541248.2017.1288686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, et al. Ras Conformational Ensembles, Allostery, and Signaling. Chem Rev. 2016;116:6607–65. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 25.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nussinov R, Jang H, Tsai CJ, Liao TJ, Li S, Fushman D, et al. Intrinsic protein disorder in oncogenic KRAS signaling. Cell Mol Life Sci. 2017;74:3245–61. doi: 10.1007/s00018-017-2564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baietti MF, Simicek M, Abbasi Asbagh L, Radaelli E, Lievens S, Crowther J, et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 2016;8:288–303. doi: 10.15252/emmm.201505972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–95. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157:1071–81. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino R, Yamashita H, et al. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys J. 2004;86:4075–93. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bandaru P, Shah NH, Bhattacharyya M, Barton JP, Kondo Y, Cofsky JC, et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. Elife. 2017;6 doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Jang H, Nussinov R, Gaponenko V. The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr Opin Struct Biol. 2016;36:10–7. doi: 10.1016/j.sbi.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erwin N, Patra S, Dwivedi M, Weise K, Winter R. Influence of isoform-specific Ras lipidation motifs on protein partitioning and dynamics in model membrane systems of various complexity. Biol Chem. 2017;398:547–63. doi: 10.1515/hsz-2016-0289. [DOI] [PubMed] [Google Scholar]
- 35.Fetics SK, Guterres H, Kearney BM, Buhrman G, Ma B, Nussinov R, et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23:505–16. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ting PY, Johnson CW, Fang C, Cao X, Graeber TG, Mattos C, et al. Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding. FASEB J. 2015;29:3750–61. doi: 10.1096/fj.15-271510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash P, Zhou Y, Liang H, Hancock JF, Gorfe AA. Oncogenic K-Ras Binds to an Anionic Membrane in Two Distinct Orientations: A Molecular Dynamics Analysis. Biophys J. 2016;110:1125–38. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin G, Kistler S, George SD, Kuhlmann N, Garvey L, Huynh M, et al. A KRAS GTPase K104Q Mutant Retains Downstream Signaling by Offsetting Defects in Regulation. J Biol Chem. 2017;292:4446–56. doi: 10.1074/jbc.M116.762435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solman M, Ligabue A, Blazevits O, Jaiswal A, Zhou Y, Liang H, et al. Specific cancer-associated mutations in the switch III region of Ras increase tumorigenicity by nanocluster augmentation. Elife. 2015;4:e08905. doi: 10.7554/eLife.08905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci U S A. 2015;112:6625–30. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–92. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smithson LJ, Anastasaki C, Chen R, Toonen JA, Williams SB, Gutmann DH. Contextual signaling in cancer. Semin Cell Dev Biol. 2016;58:118–26. doi: 10.1016/j.semcdb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 45.Lu S, Jang H, Nussinov R, Zhang J. The Structural Basis of Oncogenic Mutations G12, G13 and Q61 in Small GTPase K-Ras4B. Sci Rep. 2016;6:21949. doi: 10.1038/srep21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buhrman G, Holzapfel G, Fetics S, Mattos C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc Natl Acad Sci U S A. 2010;107:4931–6. doi: 10.1073/pnas.0912226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Resh MD. Covalent lipid modifications of proteins. Curr Biol. 2013;23:R431–5. doi: 10.1016/j.cub.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–90. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 50.Kapoor S, Weise K, Erlkamp M, Triola G, Waldmann H, Winter R. The role of G-domain orientation and nucleotide state on the Ras isoform-specific membrane interaction. Eur Biophys J. 2012;41:801–13. doi: 10.1007/s00249-012-0841-5. [DOI] [PubMed] [Google Scholar]
- 51.Lingwood D, Kaiser HJ, Levental I, Simons K. Lipid rafts as functional heterogeneity in cell membranes. Biochem Soc Trans. 2009;37:955–60. doi: 10.1042/BST0370955. [DOI] [PubMed] [Google Scholar]
- 52.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–9. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 53.Marlier JF, Fogle EJ, Redman RL, Stillman AD, Denison MA, Robins LI. A mechanistic study of thioester hydrolysis with heavy atom kinetic isotope effects. J Org Chem. 2015;80:1905–8. doi: 10.1021/jo502472m. [DOI] [PubMed] [Google Scholar]
- 54.Lane KT, Beese LS. Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res. 2006;47:681–99. doi: 10.1194/jlr.R600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 55.Khmelinskaia A, Ibarguren M, de Almeida RF, Lopez DJ, Paixao VA, Ahyayauch H, et al. Changes in membrane organization upon spontaneous insertion of 2-hydroxylated unsaturated fatty acids in the lipid bilayer. Langmuir. 2014;30:2117–28. doi: 10.1021/la403977f. [DOI] [PubMed] [Google Scholar]
- 56.Jang H, Abraham SJ, Chavan TS, Hitchinson B, Khavrutskii L, Tarasova NI, et al. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J Biol Chem. 2015;290:9465–77. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci U S A. 2010;107:1130–5. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang H, Banerjee A, Chavan TS, Lu S, Zhang J, Gaponenko V, et al. The higher level of complexity of K-Ras4B activation at the membrane. FASEB J. 2016;30:1643–55. doi: 10.1096/fj.15-279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sperlich B, Kapoor S, Waldmann H, Winter R, Weise K. Regulation of K-Ras4B Membrane Binding by Calmodulin. Biophys J. 2016;111:113–22. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho KJ, Casteel DE, Prakash P, Tan L, van der Hoeven D, Salim AA, et al. AMPK and Endothelial Nitric Oxide Synthase Signaling Regulates K-Ras Plasma Membrane Interactions via Cyclic GMP-Dependent Protein Kinase 2. Mol Cell Biol. 2016;36:3086–99. doi: 10.1128/MCB.00365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang SY, Sperlich B, Li FY, Al-Ayoubi S, Chen HX, Zhao YF, et al. Phosphorylation Weakens but Does Not Inhibit Membrane Binding and Clustering of K-Ras4B. ACS Chem Biol. 2017;12:1703–10. doi: 10.1021/acschembio.7b00165. [DOI] [PubMed] [Google Scholar]
- 62.Eisenberg S, Laude AJ, Beckett AJ, Mageean CJ, Aran V, Hernandez-Valladares M, et al. The role of palmitoylation in regulating Ras localization and function. Biochem Soc Trans. 2013;41:79–83. doi: 10.1042/BST20120268. [DOI] [PubMed] [Google Scholar]
- 63.Wang M, Casey PJ. Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol. 2016;17:110–22. doi: 10.1038/nrm.2015.11. [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Otto JC, Bergo MO, Young SG, Casey PJ. The C-terminal polylysine region and methylation of K-Ras are critical for the interaction between K-Ras and microtubules. J Biol Chem. 2000;275:41251–7. doi: 10.1074/jbc.M006687200. [DOI] [PubMed] [Google Scholar]
- 65.Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res. 2015;21:1819–27. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dharmaiah S, Bindu L, Tran TH, Gillette WK, Frank PH, Ghirlando R, et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEdelta. Proc Natl Acad Sci U S A. 2016;113:E6766–E75. doi: 10.1073/pnas.1615316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muratcioglu S, Jang H, Gursoy A, Keskin O, Nussinov R. PDEδ Binding to Ras Isoforms Provides a Route to Proper Membrane Localization. J Phys Chem B. 2017;121:5917–27. doi: 10.1021/acs.jpcb.7b03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chavan TS, Jang H, Khavrutskii L, Abraham SJ, Banerjee A, Freed BC, et al. High-Affinity Interaction of the K-Ras4B Hypervariable Region with the Ras Active Site. Biophys J. 2015;109:2602–13. doi: 10.1016/j.bpj.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Keulen SC, Rothlisberger U. Effect of N-Terminal Myristoylation on the Active Conformation of Galphai1-GTP. Biochemistry. 2017;56:271–80. doi: 10.1021/acs.biochem.6b00388. [DOI] [PubMed] [Google Scholar]
- 70.Patra S, Erwin N, Winter R. Translational Dynamics of Lipidated Ras Proteins in the Presence of Crowding Agents and Compatible Osmolytes. Chemphyschem. 2016;17:2164–9. doi: 10.1002/cphc.201600179. [DOI] [PubMed] [Google Scholar]
- 71.Lu S, Banerjee A, Jang H, Zhang J, Gaponenko V, Nussinov R. GTP Binding and Oncogenic Mutations May Attenuate Hypervariable Region (HVR)-Catalytic Domain Interactions in Small GTPase K-Ras4B, Exposing the Effector Binding Site. J Biol Chem. 2015;290:28887–900. doi: 10.1074/jbc.M115.664755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R. Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem J. 2016;473:1719–32. doi: 10.1042/BCJ20160031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, et al. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–35. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sunshine H, Iruela-Arispe ML. Membrane lipids and cell signaling. Curr Opin Lipidol. 2017;28:408–13. doi: 10.1097/MOL.0000000000000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodwin JS, Drake KR, Remmert CL, Kenworthy AK. Ras diffusion is sensitive to plasma membrane viscosity. Biophys J. 2005;89:1398–410. doi: 10.1529/biophysj.104.055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, et al. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–46. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol. 2002;157:865–72. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao TJ, Tsai CJ, Jang H, Fushman D, Nussinov R. RASSF5: An MST activator and tumor suppressor in vivo but opposite in vitro. Curr Opin Struct Biol. 2016;41:217–24. doi: 10.1016/j.sbi.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–70. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plowman SJ, Hancock JF. Ras signaling from plasma membrane and endomembrane microdomains. Biochim Biophys Acta. 2005;1746:274–83. doi: 10.1016/j.bbamcr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Posada IM, Serulla M, Zhou Y, Oetken-Lindholm C, Abankwa D, Lectez B. ASPP2 Is a Novel Pan-Ras Nanocluster Scaffold. PLoS One. 2016;11:e0159677. doi: 10.1371/journal.pone.0159677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol. 2016;18:1324–35. doi: 10.1038/ncb3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LeCour L, Jr, Boyapati VK, Liu J, Li Z, Sacks DB, Worthylake DK. The Structural Basis for Cdc42-Induced Dimerization of IQGAPs. Structure. 2016;24:1499–508. doi: 10.1016/j.str.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jambrina PG, Rauch N, Pilkington R, Rybakova K, Nguyen LK, Kholodenko BN, et al. Phosphorylation of RAF Kinase Dimers Drives Conformational Changes that Facilitate Transactivation. Angew Chem Int Ed Engl. 2016;55:983–6. doi: 10.1002/anie.201509272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donninger H, Schmidt ML, Mezzanotte J, Barnoud T, Clark GJ. Ras signaling through RASSF proteins. Semin Cell Dev Biol. 2016;58:86–95. doi: 10.1016/j.semcdb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanchez-Sanz G, Tywoniuk B, Matallanas D, Romano D, Nguyen LK, Kholodenko BN, et al. SARAH Domain-Mediated MST2-RASSF Dimeric Interactions. PLoS Comput Biol. 2016;12:e1005051. doi: 10.1371/journal.pcbi.1005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Posada IMD, Lectez B, Sharma M, Oetken-Lindholm C, Yetukuri L, Zhou Y, et al. Rapalogs can promote cancer cell stemness in vitro in a Galectin-1 and H-ras-dependent manner. Oncotarget. 2017;8:44550–66. doi: 10.18632/oncotarget.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, et al. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci U S A. 2004;101:7317–22. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hibino K, Watanabe TM, Kozuka J, Iwane AH, Okada T, Kataoka T, et al. Single- and multiple-molecule dynamics of the signaling from H-Ras to cRaf-1 visualized on the plasma membrane of living cells. Chemphyschem. 2003;4:748–53. doi: 10.1002/cphc.200300731. [DOI] [PubMed] [Google Scholar]
- 90.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–5. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 91.Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol. 2017;13:62–8. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nussinov R, Wang G, Tsai CJ, Jang H, Lu S, Banerjee A, et al. Calmodulin and PI3K Signaling in KRAS. Cancers Trends Cancer. 2017;3:214–24. doi: 10.1016/j.trecan.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Michael JV, Wurtzel JG, Goldfinger LE. Regulation of H-Ras-driven MAPK signaling, transformation and tumorigenesis, but not PI3K signaling and tumor progression, by plasma membrane microdomains. Oncogenesis. 2016;5:e228. doi: 10.1038/oncsis.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cho KJ, van der Hoeven D, Zhou Y, Maekawa M, Ma X, Chen W, et al. Inhibition of Acid Sphingomyelinase Depletes Cellular Phosphatidylserine and Mislocalizes K-Ras from the Plasma Membrane. Mol Cell Biol. 2015;36:363–74. doi: 10.1128/MCB.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gregory MC, McLean MA, Sligar SG. Interaction of KRas4b with anionic membranes: A special role for PIP2. Biochem Biophys Res Commun. 2017;487:351–5. doi: 10.1016/j.bbrc.2017.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolfman A. Ras isoform-specific signaling: location, location, location. Sci STKE. 2001;2001:pe2. doi: 10.1126/stke.2001.96.pe2. [DOI] [PubMed] [Google Scholar]
- 97.Goldfinger LE, Michael JV. Regulation of Ras signaling and function by plasma membrane microdomains. Biosci Trends. 2017;11:23–40. doi: 10.5582/bst.2016.01220. [DOI] [PubMed] [Google Scholar]
- 98.Novotny CJ, Hamilton GL, McCormick F, Shokat KM. Farnesyltransferase-Mediated Delivery of a Covalent Inhibitor Overcomes Alternative Prenylation to Mislocalize K-Ras. ACS Chem Biol. 2017;12:1956–62. doi: 10.1021/acschembio.7b00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matallanas D, Arozarena I, Berciano MT, Aaronson DS, Pellicer A, Lafarga M, et al. Differences on the inhibitory specificities of H-Ras, K-Ras, and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J Biol Chem. 2003;278:4572–81. doi: 10.1074/jbc.M209807200. [DOI] [PubMed] [Google Scholar]
- 101.Eisenberg S, Beckett AJ, Prior IA, Dekker FJ, Hedberg C, Waldmann H, et al. Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol Cell Biol. 2011;31:3938–52. doi: 10.1128/MCB.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gentry LR, Nishimura A, Cox AD, Martin TD, Tsygankov D, Nishida M, et al. Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of Ral GTPases. J Biol Chem. 2015;290:22851–61. doi: 10.1074/jbc.M115.656710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakhaei-Rad S, Nakhaeizadeh H, Kordes C, Cirstea IC, Schmick M, Dvorsky R, et al. The Function of Embryonic Stem Cell-expressed RAS (E-RAS), a Unique RAS Family Member, Correlates with Its Additional Motifs and Its Structural Properties. J Biol Chem. 2015;290:15892–903. doi: 10.1074/jbc.M115.640607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, et al. Visualizing association of N-ras in lipid microdomains: influence of domain structure and interfacial adsorption. J Am Chem Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- 105.Astorgues-Xerri L, Riveiro ME, Tijeras-Raballand A, Serova M, Neuzillet C, Albert S, et al. Unraveling galectin-1 as a novel therapeutic target for cancer. Cancer Treat Rev. 2014;40:307–19. doi: 10.1016/j.ctrv.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 106.Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem. 2004;279:34922–30. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]
- 107.Shalom-Feuerstein R, Levy R, Makovski V, Raz A, Kloog Y. Galectin-3 regulates RasGRP4-mediated activation of N-Ras and H-Ras. Biochim Biophys Acta. 2008;1783:985–93. doi: 10.1016/j.bbamcr.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol Biol Cell. 2008;19:1404–14. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Silvius JR. Partitioning of membrane molecules between raft and non-raft domains: insights from model-membrane studies. Biochim Biophys Acta. 2005;1746:193–202. doi: 10.1016/j.bbamcr.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 110.Janosi L, Li Z, Hancock JF, Gorfe AA. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc Natl Acad Sci U S A. 2012;109:8097–102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, et al. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell. 2015;163:1237–51. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 112.Nussinov R, Muratcioglu S, Tsai CJ, Jang H, Gursoy A, Keskin O. The Key Role of Calmodulin in KRAS-Driven Adenocarcinomas. Mol Cancer Res. 2015;13:1265–73. doi: 10.1158/1541-7786.MCR-15-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weise K, Kapoor S, Werkmuller A, Mobitz S, Zimmermann G, Triola G, et al. Dissociation of the K-Ras4B/PDEdelta complex upon contact with lipid membranes: membrane delivery instead of extraction. J Am Chem Soc. 2012;134:11503–10. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 114.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014;1843:398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 115.Anguita E, Villalobo A. Src-family tyrosine kinases and the Ca2+ signal. Biochim Biophys Acta. 2017;1864:915–32. doi: 10.1016/j.bbamcr.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 116.Jang H, Banerjee A, Chavan T, Gaponenko V, Nussinov R. Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling K-Ras4B membrane extraction. J Biol Chem. 2017;292:12544–59. doi: 10.1074/jbc.M117.785063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joyal JL, Burks DJ, Pons S, Matter WF, Vlahos CJ, White MF, et al. Calmodulin activates phosphatidylinositol 3-kinase. J Biol Chem. 1997;272:28183–6. doi: 10.1074/jbc.272.45.28183. [DOI] [PubMed] [Google Scholar]
- 118.Chaudhuri P, Rosenbaum MA, Sinharoy P, Damron DS, Birnbaumer L, Graham LM. Membrane translocation of TRPC6 channels and endothelial migration are regulated by calmodulin and PI3 kinase activation. Proc Natl Acad Sci U S A. 2016;113:2110–5. doi: 10.1073/pnas.1600371113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Siempelkamp BD, Rathinaswamy MK, Jenkins ML, Burke JE. Molecular mechanism of activation of class IA phosphoinositide 3-kinases (PI3Ks) by membrane-localized HRas. J Biol Chem. 2017;292:12256–66. doi: 10.1074/jbc.M117.789263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liao J, Wolfman JC, Wolfman A. K-ras regulates the steady-state expression of matrix metalloproteinase 2 in fibroblasts. J Biol Chem. 2003;278:31871–8. doi: 10.1074/jbc.M301931200. [DOI] [PubMed] [Google Scholar]
- 121.Liao J, Planchon SM, Wolfman JC, Wolfman A. Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular ras isoforms. J Biol Chem. 2006;281:29730–8. doi: 10.1074/jbc.M600668200. [DOI] [PubMed] [Google Scholar]
- 122.Zhang M, Jang H, Gaponenko V, Nussinov R. Phosphorylated Calmodulin Promotes PI3K Activation by Binding to the SH2 Domains. Biophys J. 2017 doi: 10.1016/j.bpj.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nussinov R, Tsai CJ, Jang H. A New View of Pathway-Driven Drug Resistance in Tumor Proliferation. Trends Pharmacol Sci. 2017;38:427–37. doi: 10.1016/j.tips.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci Signal. 2011;4:re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 125.Echeverria I, Liu Y, Gabelli SB, Amzel LM. Oncogenic mutations weaken the interactions that stabilize the p110alpha-p85alpha heterodimer in phosphatidylinositol 3-kinase alpha. FEBS J. 2015;282:3528–42. doi: 10.1111/febs.13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci U S A. 2007;104:7809–14. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, et al. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci U S A. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, et al. GTP-Dependent K-Ras Dimerization. Structure. 2015;23:1325–35. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cho KJ, Kasai RS, Park JH, Chigurupati S, Heidorn SJ, van der Hoeven D, et al. Raf inhibitors target ras spatiotemporal dynamics. Curr Biol. 2012;22:945–55. doi: 10.1016/j.cub.2012.03.067. [DOI] [PubMed] [Google Scholar]
- 130.Sarkar-Banerjee S, Sayyed-Ahmad A, Prakash P, Cho KJ, Waxham MN, Hancock JF, et al. Spatiotemporal Analysis of K-Ras Plasma Membrane Interactions Reveals Multiple High Order Homo-oligomeric Complexes. J Am Chem Soc. 2017;139:13466–75. doi: 10.1021/jacs.7b06292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guzman C, Solman M, Ligabue A, Blazevits O, Andrade DM, Reymond L, et al. The efficacy of Raf kinase recruitment to the GTPase H-ras depends on H-ras membrane conformer-specific nanoclustering. J Biol Chem. 2014;289:9519–33. doi: 10.1074/jbc.M113.537001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller MS, Schmidt-Kittler O, Bolduc DM, Brower ET, Chaves-Moreira D, Allaire M, et al. Structural basis of nSH2 regulation and lipid binding in PI3Kalpha. Oncotarget. 2014;5:5198–208. doi: 10.18632/oncotarget.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Villalonga P, Lopez-Alcala C, Bosch M, Chiloeches A, Rocamora N, Gil J, et al. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol Cell Biol. 2001;21:7345–54. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lopez-Alcala C, Alvarez-Moya B, Villalonga P, Calvo M, Bachs O, Agell N. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J Biol Chem. 2008;283:10621–31. doi: 10.1074/jbc.M706238200. [DOI] [PubMed] [Google Scholar]
- 135.Fivaz M, Meyer T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J Cell Biol. 2005;170:429–41. doi: 10.1083/jcb.200409157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chavan TS, Muratcioglu S, Marszalek R, Jang H, Keskin O, Gursoy A, et al. Plasma membrane regulates Ras signaling networks. Cell Logist. 2015;5:e1136374. doi: 10.1080/21592799.2015.1136374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Siljamaki E, Abankwa D. SPRED1 Interferes with K-ras but Not H-ras Membrane Anchorage and Signaling. Mol Cell Biol. 2016;36:2612–25. doi: 10.1128/MCB.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bender RH, Haigis KM, Gutmann DH. Activated k-ras, but not h-ras or N-ras, regulates brain neural stem cell proliferation in a raf/rb-dependent manner. Stem Cells. 2015;33:1998–2010. doi: 10.1002/stem.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Macrae M, Neve RM, Rodriguez-Viciana P, Haqq C, Yeh J, Chen C, et al. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–8. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 140.Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–23. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 141.Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, et al. KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer. Cell. 2017;168:817–29. e15. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 143.Doherty GJ, Kerr EM, Martins CP. KRAS Allelic Imbalance: Strengths and Weaknesses in Numbers. Trends Mol Med. 2017;23:377–8. doi: 10.1016/j.molmed.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 144.Mao Y, Yao H, Wang H, Cheng P, Long D. Microsecond Timescale Dynamics of GDP-Bound Ras Underlies the Formation of Novel Inhibitor-Binding Pockets. Angew Chem Int Ed Engl. 2016;55:15629–32. doi: 10.1002/anie.201608653. [DOI] [PubMed] [Google Scholar]
- 145.Lu S, Jang H, Gu S, Zhang J, Nussinov R. Drugging Ras GTPase: a comprehensive mechanistic and signaling structural view. Chem Soc Rev. 2016;45:4929–52. doi: 10.1039/c5cs00911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rauch J, Vandamme D, Mack B, McCann B, Volinsky N, Blanco A, et al. Differential localization of A-Raf regulates MST2-mediated apoptosis during epithelial differentiation. Cell Death Differ. 2016;23:1283–95. doi: 10.1038/cdd.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Choi S, Anderson RA. And Akt-ion! IQGAP1 in control of signaling pathways. EMBO J. 2017;36:967–9. doi: 10.15252/embj.201796827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nussinov R, Muratcioglu S, Tsai CJ, Jang H, Gursoy A, Keskin O. K-Ras4B/calmodulin/PI3Kα: A promising new adenocarcinoma-specific drug target? Expert Opin Ther Targets. 2016;20:831–42. doi: 10.1517/14728222.2016.1135131. [DOI] [PubMed] [Google Scholar]
- 149.Lin DTS, Davis NG, Conibear E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem Soc Trans. 2017;45:913–21. doi: 10.1042/BST20160303. [DOI] [PubMed] [Google Scholar]
- 150.Shih TC, Liu R, Fung G, Bhardwaj G, Ghosh PM, Lam KS. A Novel Galectin-1 Inhibitor Discovered through One-Bead Two-Compound Library Potentiates the Antitumor Effects of Paclitaxel in vivo. Mol Cancer Ther. 2017;16:1212–23. doi: 10.1158/1535-7163.MCT-16-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michael JV, Wurtzel JG, Goldfinger LE. Inhibition of Galectin-1 Sensitizes HRAS-driven Tumor Growth to Rapamycin Treatment. Anticancer Res. 2016;36:5053–61. doi: 10.21873/anticanres.11074. [DOI] [PubMed] [Google Scholar]
- 152.Spencer-Smith R, Li L, Prasad S, Koide A, Koide S, O’Bryan JP. Targeting the alpha4-alpha5 interface of RAS results in multiple levels of inhibition. Small GTPases. 2017 doi: 10.1080/21541248.2017.1333188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Papke B, Murarka S, Vogel HA, Martin-Gago P, Kovacevic M, Truxius DC, et al. Identification of pyrazolopyridazinones as PDEdelta inhibitors. Nat Commun. 2016;7:11360. doi: 10.1038/ncomms11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nussinov R, Tsai CJ, Mattos C. ‘Pathway drug cocktail’: targeting Ras signaling based on structural pathways. Trends Mol Med. 2013;19:695–704. doi: 10.1016/j.molmed.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 155.Tong CWS, Wu WKK, Loong HHF, Cho WCS, To KKW. Drug combination approach to overcome resistance to EGFR tyrosine kinase inhibitors in lung cancer. Cancer Lett. 2017;405:100–10. doi: 10.1016/j.canlet.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 156.Guri Y, Hall MN. mTOR Signaling Confers Resistance to Targeted Cancer Drugs. Trends Cancer. 2016;2:688–97. doi: 10.1016/j.trecan.2016.10.006. [DOI] [PubMed] [Google Scholar]