Abstract
PARP inhibitors (PARPi) benefit only a fraction of breast cancer patients. Several of those patients exhibit intrinsic/acquired resistance mechanisms that limit efficacy of PARPi monotherapy. Here we show how the efficacy of PARPi in triple-negative breast cancers (TNBC) can be expanded by targeting MYC-induced oncogenic addiction. In BRCA-mutant/sporadic TNBC patients, amplification of the MYC gene is correlated with increased expression of the homologous DNA recombination enzyme RAD51 and tumors overexpressing both genes are associated with worse overall survival. Combining MYC blockade with PARPi yielded synthetic lethality in MYC-driven TNBC cells. Using the cyclin-dependent kinase inhibitor dinaciclib, which downregulates MYC expression, we found that combination with the PARPi niraparib increased DNA damage and downregulated homologous recombination, leading to subsequent downregulation of the epithelial-mesenchymal transition (EMT) and cancer stem-like cell phenotypes. Notably, dinaciclib re-sensitized TBNC cells, which had acquired resistance to niraparib. We found that the synthetic lethal strategy employing dinaciclib and niraparib was also highly efficacious in ovarian, prostate, pancreatic, colon and lung cancer cells. Taken together, our results show how blunting MYC oncogene addiction can leverage cancer cell sensitivity to PARPi, facilitating the clinical use of c-myc as a predictive biomarker for this treatment.
Keywords: MYC, PARP, Synthetic Lethal, RAD51, TNBC
Introduction
The proto-oncogene MYC is amplified in several different cancer types, including triple negative breast cancer (TNBC), pancreas, ovarian and prostate cancers (1). The variety of cellular processes that MYC regulates (e.g. metabolism, cell cycle, metastasis & DNA repair) suggests a crucial role in the oncogenic transformation of MYC driven cancers, resulting in an addiction to MYC expression (2). One of the mechanisms by which MYC mediates oncogenic potential is induction of replication stress, causing increased γH2AX expression and subsequent upregulation of homologous recombination (HR) via increased RAD51 activity (3). However, MYC’s specific role in regulating DNA repair mechanisms as a function of response to therapy has often been overlooked. Treatment with poly (ADP-ribose) polymerase (PARP) inhibitors (PARPis) is associated with a favorable response in tumors lacking BRCA1/2 function. Several studies have recently demonstrated that BRCA mutant (BRCAmut) cancer cells frequently retain an intrinsic RAD51 dependent HR repair mechanism that imparts de novo resistance to PARPis and platinum therapeutic agents (4–6). In addition, upregulation of the DNA repair pathway is often overlooked as a sign of decreased response to chemotherapy. Moreover, because RAD51 expression is involved in several non-DNA repair pathways (e.g. increased metastasis of TNBC) (7), we hypothesized that MYC positive tumors upregulate the HR DNA repair pathway causing resistance to DNA damaging agents including PARPis. Therefore, using RAD51 as a marker of de novo resistance to PARPis we classified TNBC breast cancer cell lines as either PARPi sensitive or resistant independent of BRCA status. Furthermore, we showed that MYC directly regulates HR via several DNA repair proteins including RAD51, whereas inhibition (or downregulation) of MYC expression induces PARPi sensitivity independent of BRCA status. These findings suggest that TNBC patients with high c-myc and RAD51 expression, which have poor prognoses and are unresponsive to neoadjuvant chemotherapy, are likely to be sensitive to agents that downregulate c-myc (e.g. dinaciclib) and PARPis independent of BRCA mutational status.
Materials and Methods
Cell lines and culture conditions
All parental cancer cell lines used in this study were purchased from the ATCC. The TNBC cell lines MDA-MB-231, MDA-MB-468, HCC1937, HCC1806, SUM149, SUM1315, MDA-MB-436, and MDA-MB-157 and human mammary epithelial cell lines MCF-10A were cultured as described previously (8, 9), The non-small cell lung cancer cell lines PC3, DU145, A549, Calu-1, H1299, and H1993 were cultured in RPMI medium in the presence of 10% fetal bovine serum. The head and neck squamous cell carcinoma cell lines OVCAR3, 59M, FUOV1, BxPC3, PANC-1, HCT116, and SW620 were cultured in Dulbecco’s modified Eagle’s medium in the presence of 10% fetal bovine serum and growth factors. All cells were free of mycoplasma contamination. Cell lines were identified and authenticated according to karyotype and using short tandem repeat analysis in the MD Anderson Characterized Cell Line Core facility every 6 months.
Acquired treatment resistance
Cells were cultured in normal growth media supplemented with the PARPi niraparib at increasing concentrations (MDA-MB-436, 0.1 nM–2.0 μM; HCC1806, 0.5–15.0 μM) for 6 months. At the final concentrations, cells were maintained in media supplemented with niraparib. All experiments were conducted in the absence of niraparib-supplemented media unless otherwise noted.
siRNA
In vitro cell transfections were carried out in six-well plates seeded (5 x 104) and then transfected with 5 μM MYC siRNA( 4609), (SMART pool; Dharmacon, Lafayette, CO, USA; 5′-ACGGAACUUGUGCGUAA-3′, 5′-GAACACACAACGUCUUGGA-3′, 5′-AACGUUAGCUUCACCAACA-3′, and 5′-CGAUGUUGUUUCUGUGGAA-3′), 5 μM RAD51 (5888), 5 μM RAD51 siRNA (SMART pool; 5′-UAUCAUCGCCCAUGCAUCA-3′, 5′-CUAAUCAGGUGGUAGCUCA-3′, 5′-GCAGUGAUGUCCUGGAUAA-3′, and 5′-CCAACGAUGUGAAGAAAAUU-3′), or a non targeting pool 5 μM siRNA Cells were incubated at 36°C in 5% CO2 for 48 h, and the media were removed. Briefly, in vitro siRNA transfections were performed using the jetPRIME transfection reagent (Polyplus, New York, NY, USA) following the manufacturer’s protocol.
Short hairpin and open reading frame constructs and viral infection
The pGIPZ-shRNA and MYC overexpression plasmids were purchased from Dharmacon and used to produce lentiviruses (shBRCA1 and sh53BP1) by transfecting 293T cells shRNA plasmids. TNBC cells were infected with viral particles in complete media in the presence of hexadimethrine bromide (Polybrene, 8 mg/ml; EMD Millipore, Billerica, MA, USA) overnight. The next day, media containing the viruses were washed and replaced with fresh media. Puromycin (Invitrogen, Carlsbad, CA, USA) selection for infected cells was performed for 7 d.
High Throughput Survival Assay (HTSA)
Cells were seeded at a density of 1000 cells/well in a 96-well plate and treated with single drugs or drug combinations at the indicated concentrations for 72 h. Cells were then released into a complete drug-free medium for 9 d, and the medium was changed every 48 h. On the day of cell harvest, 100 μl/well 2.5 mg/ml MTT (Sigma, St. Louis, MO, USA) was added to the serum-free medium and incubated at 37°C for 3–4 h. After incubation, the medium was removed, and 100 μl of a solubilization solution (0.04 mmol/l HCl and 1% sodium dodecyl sulfate in isopropyl alcohol) was added to each well. Plates were lightly rocked at room temperature for 1 h and read using a plate reader (Gen5 Epoch Microplate Spectrophotometer and software program; BioTek, Winooski, VT, USA) at a wavelength of 590 nm. Combination indices were calculated using the Calcusyn software program.
Quantitative RT-PCR
Total RNA was isolated from cell cultures using an RNeasy Kit with DNase treatment according to the manufacturer’s protocol (QIAGEN, Germantown, MD, USA). Two micrograms of the RNA samples was reverse-transcribed using a cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA). RT-PCR was done with aliquots of cDNA samples mixed with SYBR Green Master Mix (Sigma). Reactions were carried out in triplicate. The fold difference in transcripts was calculated using the ΔΔCT method with GAPDH as a control. The following primers were used: MYC forward, 5′-GGCTCCTGGCAAAAGGTCA; MYC reverse, 5′-CTGCGTAGTTGTGCTGATGT; RAD51 forward, 5′-CAACCCATTTCACGGTTAGAGC; RAD51 reverse, 5′-TTCTTTGGCGATAGGCAACA; BRCA2 forward, 5′-CACCCACCCTTAGTTCTACTGT; BRCA2 reverse, 5′-CCAATGTGGTCTTTGCAGCTAT; RAD54L forward, 5′-TTGAGTCAGCTAACCAATCAACC; RAD54L reverse, 5′-GGAGGCTCATACAGAACCAAGG; E2F1 forward, 5′-CATCCCAGGAGGTCACTTCTG; E2F1 reverse, 5′-GACAACAGCGGTTCTTGCTC; BRCA1 forward, 5′ ACCTTGGAACTGTGAGAACTCT; BRCA1 reverse, 5′-TCTTGATCTCCCACACTGCAATA; RAD21 forward, 5′-GGATAAGAAGCTAACCAAAGCCC; and RAD21 reverse, 5′-CTCCCAGTAAGAGATGTCCTGAT. FEN1 forward, 5′-ATGACATCAAGAGCTACTTTGGC; and FEN1 reverse, 5′-GGCGAACAGCAATCAGGAACT. APEX1 forward, 5′-CAATACTGGTCAGCTCCTTCG; and APEX1 reverse, 5′-TGCCGTAAGAAACTTTGAGTGG. The following conditions were used for quantitative RT-PCR: denaturation: 95°C for 10 min; 40 cycles: 95°C for 30 s, 58°C for 10 s, and 72°C for 30 s; extension: 72°C for 10 min as described previously to be optimum for data analysis of quantitative real-time PCR using GAPDH as a internal control (10, 11).
Western blot analysis
Western blot analyses were performed as described previously (12) with the following modifications. The cell pellet was lysed in RIPA buffer with a cocktail of protease/phosphatase inhibitors (250 μg/ml leupeptin, 250 μg/ml aprotinin, 100 μg/ml pepstatin, 1 mM benzamidine, 100 μg/ml soybean trypsin inhibitor, 5 mM phenylmethylsulfonyl fluoride, 50 mM sodium fluoride, and 0.5 mM sodium orthovanadate). The primary antibodies used were 53BP1 (ab21083; Abcam, Cambridge, UK), Axl (#8661; Cell Signaling Technology, Danvers, MA, USA), β-actin (MAB1501R; EMD Millipore), BRCA1 (07-434; EMD Millipore), c-myc (Y69, ab32072), E-cadherin (#3195; Cell Signaling Technology), E2F1 (KH-92, sc-251; Santa Cruz Biotechnology, Santa Cruz, CA, USA), γH2AX (05-36; EMD Millipore), N-cadherin (#4061; Cell Signaling Technology), phospho c-myc (Ser62, 13748; Cell Signaling Technology), phospho c-myc (Thr58) (ab28842; Abcam), phospho S6 (Ser235/6, #2211; Cell Signaling Technology), RAD21 (ab992; Abcam), RAD51 (H-92, sc-8349), vimentin (#3932; Cell Signaling Technology), Zeb1 (#3396; Cell Signaling Technology), PARP (polyclonal, #9542; Cell Signaling Technology), caspase 3 (polyclonal, #9662; Cell Signaling Technology), caspase 7 (polyclonal, #9492; Cell Signaling Technology), Bcl2 (M-0887; Dako, Carpinteria, CA, USA), Mcl1 (polyclonal, sc-819; Santa Cruz Biotechnology), p53 (OP43; Oncogene), and phospho p53 (S15, #9284; Cell Signaling Technology).
Immunofluorescence
To measure DNA damage repair using γH2AX staining, cells were plated at 10,000 cells/well in an eight-well chamber slide. After 24 h, cells were treated with dinaciclib and/or niraparib, incubated for 72 h, and fixed in 4% paraformaldehyde for 20 min before staining. For quantification of RAD51 foci, cells were plated at 10,000 cells/well in an eight-well chamber slide. After 72 h, cells were treated with DMSO, dinaciclib, or niraparib and incubated for 48 h. For immunofluorescence of MYC siRNA, 10,000 cells/well were plated in an eight-well chamber and treated with doxorubicin as described above. Following treatment, cells were fixed in 4% formaldehyde for 20 min before staining. For immunofluorescent staining, fixed cells were rinsed in phosphate-buffered saline (PBS) and permeabilized for 5 min in 0.1% Triton X-100. The cells were then blocked in 1% bovine serum albumin for 1 h at room temperature. Primary antibodies prepared in 1% bovine serum albumin were added to the cells according to the antibody manufacturer’s instructions, followed by a 2-h incubation at room temperature in a moist chamber. Primary antibodies used were those against γH2AX (mouse monoclonal; EMD Millipore) and RAD51 (rabbit polyclonal; a gift from Dr. Junjie Chen, MD Anderson). A secondary antibody was added after the cells were washed thoroughly in PBS. Cells were incubated with secondary antibodies tagged with Alexa Fluor dyes (goat-anti-mouse-Alexafluor-488 and goat-anti-rabbit-Alexa fluor-594; Invitrogen) for 1 h at room temperature. After being rinsed and washed thoroughly with PBS, slides were mounted using VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA, USA) containing DAPI and sealed. Cells were visualized using a 1 × 81 DSU confocal microscope (Olympus, Waltham, MA, USA), and images were analyzed using the SlideBook software program (Intelligent Imaging Innovations, Denver, CO, USA). At least 150 cells were counted in each experiment. Each experiment was performed in triplicate, and the average count was taken. Statistical analysis was performed using the Student t-test (significance, P = 0.05).
Immunohistochemistry
5 μm-thick sections deparaffinized in xylene and hydrated in a series of graded alcohol dilutions. Slides were boiled in a microwave at 650 V for 20 minutes in 10 mm sodium citrate (pH 6.5) for antigen retrieval and subsequently cooled at room temperature for 30 minutes. 3% hydrogen peroxide used to block endogenous peroxidase activity. Sections were incubated overnight with primary antibodies against Ki67 (mouse monoclonal, clone MIB-1; DAKO, 1:100 dilution), Rad51 (rabbit polyclonal, clone H92; Santa Cruz, 1:500 dilution), c-myc (rabbit polyclonal, clone N-262; Santa Cruz, 1:500 dilution) and γ-H2AX (Ser139) (mouse monoclonal, clone 05-636; Millipore, 1:100 dilution). After the sections were rinsed, antibodies were detected with a secondary antibody from the Vectastain Elite ABC Kit (PK6101 and PK6102; Vector Laboratories, Burlingame, CA) Color development was performed using 3,3-diaminobenzidine. Counterstaining was provided by staining with hematoxylin. All washing steps were performed in PBS alone and PBS with 0.1% Tween.
Homologous Recombination (DR-GFP) assay
To measure HR efficiency in cells, a two-plasmid HR assay was used as described previously (8). Briefly, cells were plated at about 0.4 × 106 cells/well in a six-well plate. After overnight incubation, the plasmids pSce-I and pDR-GFP were transfected into the cells using jetPRIME transfection reagent. 24 h after transfection, cells were treated with dinaciclib, 10058-F4, or MYC siRNA. Cells were harvested 72 h later via trypsinization, resuspended in PBS, and analyzed using a Gallios flow cytometer. The percentage of GFP-positive cells was used as a measure of HR efficiency.
Statistical analyses
Statistical analyses were performed using the Prism software program (version 5.04). Tumor volume, FACS, and annexin V data were analyzed using one-way ANOVA and Tukey’s multiple comparison test. Survival curves were analyzed using a log-rank (Mantel-Cox) test. P values below 0.05 were considered statistically significant.
Results
PARPi sensitivity is independent of BRCA status
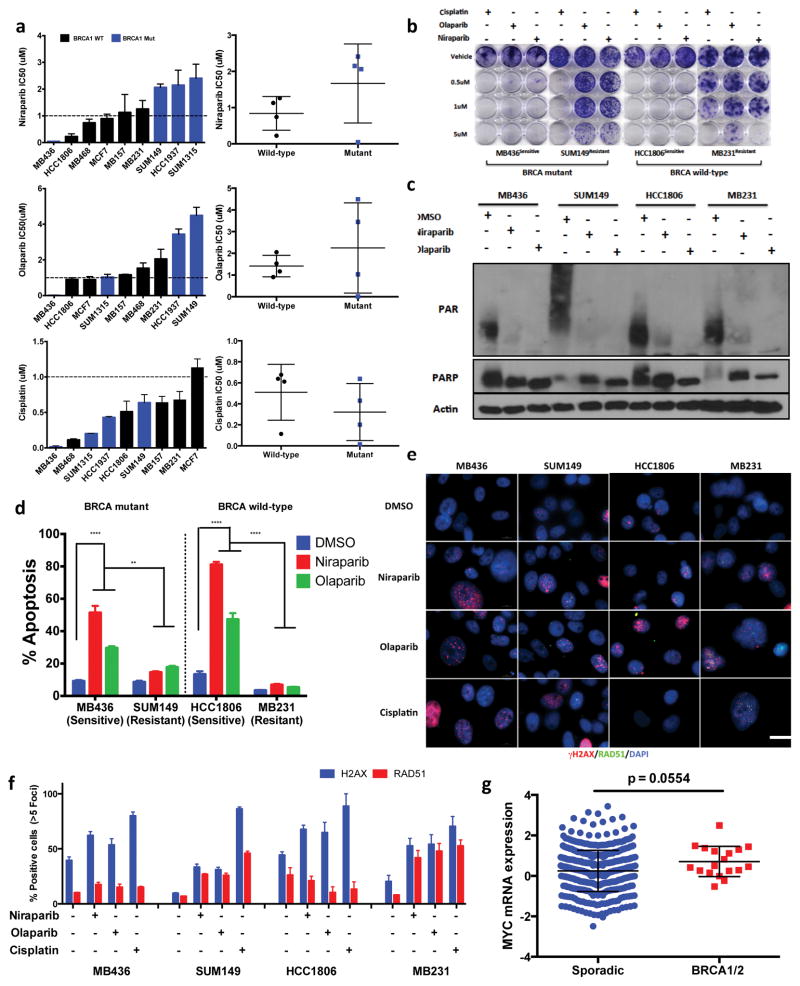
We used an in vitro High Throughput Survival Assay (HTSA) to examine the long-term (i.e. 12 days) response of PARPi (Supplementary Fig 1a), in PARPi-sensitive BRCAmut MDA-MB-436 (MB436), and resistant BRCA wild-type (BRCAwt) MDA-MB-MB231 (MB231) cell lines. In this assay, we treated the cells with a PARPi for 24 h with 11-d release (24-h HTSA), 72 h with a 9-d release (72-h HTSA) or continuously until day 12 (Continuous HTSA), we then assessed survival. For both cell lines the response to PARPi-based treatment differed markedly in the 24- and 72-h HTSAs, but the response did not differ significantly in 72-h and continuous HTSAs (Supplementary Fig 1b–g). Furthermore, the response was enhanced in the 72-h HTSA, suggesting that the effect of PARPis is maintained during assay’s recovery phase (Supplementary Fig 2a–c). Hence, the therapeutic effects of PARPis are sustained following drug removal and at least 72-h of treatment is required to sustain response. Thus PARPi sensitivity is independent of BRCA status while PARPi sensitive cells have impaired RAD51 foci formation.
Next, we examined a panel of breast cancer cell lines treated with two PARPis (niraparib and olaparib) and cisplatin via 72-h HTSA (72-h of treatment followed by 9-d of recovery) and clonogenic assay (Fig. 1a,b). With the exception of MB436 cells, BRCA status did not dictate sensitivity to PARPis (<1 μM). All BRCAmut cell lines demonstrated increased sensitivity to cisplatin (Fig. 1a,b and Supplementary Fig. 2b), supporting divergence in the response of TNBC to PARPis versus platinum agents as a function of BRCAmut status. Poly (ADP-ribose), levels in sensitive and resistant cells did not differ significantly under different treatment conditions (Fig 1c). Apoptotic analysis also demonstrated a difference in sensitivity and resistance irrespective of BRCA status in cells treated with PARPis for 72-h with a 72-h recovery (Fig 1d). We next used RAD51 foci formation as an indicator of homologous recombination (HR) to interrogate if HR defects are associated with BRCA1 mutation status. Our results revealed that the PARPi sensitive, MB436 and HCC1806, cells had impaired RAD51 foci, whereas PARPi de novo resistant SUM149 and MB231 cells exhibited increased RAD51 foci in response to PARPi exposure independent of BRCA1 mutational status (Fig 1e–1f and Supplementary Fig 3a–f). Collectively these experiments validate the sustained effect of PARPis following drug removal (Supplementary Fig 3g, 4a–h) and suggest that PARPi sensitive cells have impaired RAD51 foci formation. Consistently, BRCA1 knockdown revealed PARPi resistance to be an independent function of BRCA1 loss (Supplementary Fig 5a–m).
Figure 1.
RAD51 predicts PARP inhibitor sensitivity in TNBC. (a) HTSA IC50 values of breast cancer cell lines screened for Niraparib, Olaparib, Cisplatin & DMSO (ctrl) Error bars represent mean ± s.d. (n ≥ 3 independent experiments) (b–f) Analysis of PARP inhibitor Sensitive (MB436, HCC1806) & Resistant (SUM149, MB231) TNBC cell lines (b) Clonogenic assay of cells treated with 0.5, 1 & 5μM Niraparib, Olaparib, Cisplatin and DMSO (ctrl) 72hr followed by 9-day release (c) Immunoblot analysis of breast cancer cells treated 72hr with 1μM PARP inhibitor (Niraparib, Olaparib) for PAR and PARP. Actin was used as a loading control (d) Average cell viability using a FACS-based assay with Annexin V and propidium iodide staining of cells treated with 10μM Niraparib, Olaparib, DMSO 72hrs followed by 72hr release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments) two-way ANOVA with Sidak post-test correcting for multiple comparisons (e–f) Immunofluorescence analysis of cells treated with 1μM Niraparib, Olaparib, Cisplatin & DMSO (ctrl) 72hr (e) Immunofluorescence images of γH2AX (red), RAD51 (green), nuclear DAPI (blue) in breast cancer cells. Scale bar, 10 μm. Representative images from three independent experiments. (f) Quantification of γH2AX, RAD51. Data are mean ± s.d. of biological replicates and analyzed by unpaired two-sided t-tests (g) TCGA analysis of MYC mRNA expression of sporadic versus BRCA 1/2 mutant TNBC breast cancer patient cohort, statistical analysis unpaired two-sided t-test, *P < 0.05; **P < 0.01 and ***P < 0.001
MYC regulates DNA repair in TNBC
The oncogene MYC transcriptionally regulates several DNA repair genes and correlates with decreased overall survival rates in several rates for types of cancer, therefore we next sought to determine the utility of MYC as a mediator of DNA repair in PARPi resistant models of TNBC (13). MYC amplification was higher in the BRCAmut patients than in sporadic TNBC (p=0.0554) (p, not significant) (Fig 1g).
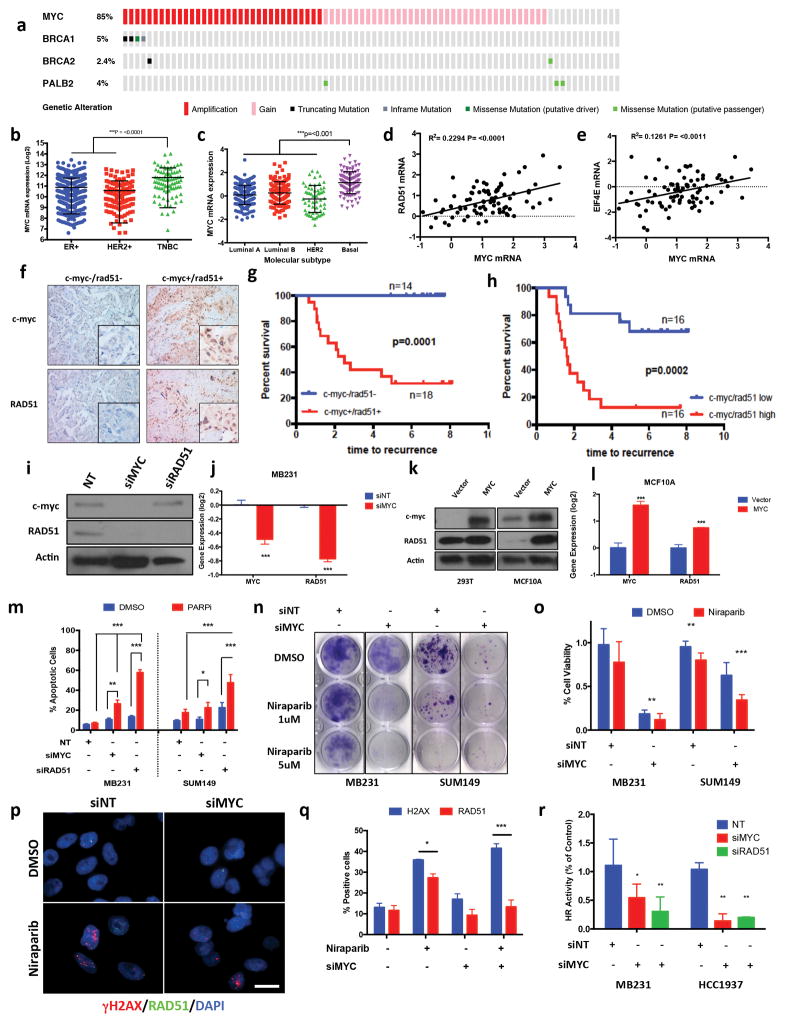
The prevalence of BRCA mutational status has been estimated at approximately 10–20% of patients who present with TNBC (14–16). Furthermore, patients with BRCA1 mutational status have a 60–80% chance of having high grade TNBC and/or a basal subtype (17–20). Similarly, TCGA patient cohort analysis also revealed that patients with BRCAmut disease are typically presented with TNBC and/or a basal molecular subtype, with high MYC amplification and gene expression (Fig 2a–c, and Supplementary Fig 6a). Specifically, MYC was amplified/gained in >85% of TNBC patient samples, which correlated with (69%) of BRCA1/2 and PALB2 (directly interacts with RAD51 in HR) (21) mutation carriers (Fig 2a).
Figure 2.
MYC regulates DNA repair in TNBC (a) Oncoprint analysis of MYC copy number gain, BRCA 1/2, PALB2 mutations in TCGA, TNBC breast cancer cohort (b–c) MYC mRNA expression in breast cancer subtypes as determined by (b) ER/PR/HER-2 expression via IHC, or (c) PAM50 signature (d–e) MYC mRNA correlation with (d) RAD51 and (e) EIF4E mRNA expression in TCGA breast cancer (f) IHC of TNBC patient cohort stained for c-myc/RAD51 from either pre or post neoadjuvant chemotherapy treatment samples (g, h) Kaplan-Meyer curve of c-myc+/RAD51+ vs. c-myc-/RAD51- of Pre (g) and Post (h) neoadjuvant chemotherapy treated TNBC breast cancer patients (i–j) MB231 cells treated with siMYC, siNT (ctrl) for 48hrs (i) Immunoblot for c-myc, RAD51, Actin (ctrl) (j) RT-PCR analysis of c-myc, RAD51 (n = 3) (k–l) HEK293T & MCF10A cells transfected with MYC ORF cDNA (k) Immunoblot analysis for c-myc, RAD51, Actin (ctrl) (l) RT-PCR analysis of c-myc, RAD51. (m) Annexin V and propidium iodide staining of MB231 and SUM149 cells treated with Non Targeting, MYC & RAD51 siRNA 48hrs followed by Niraparib/DMSO treatment 72hrs followed by 72hr release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments) (n) Clonogenic assay MB231 & SUM149 cells treated with siNT (ctrl), siMYC & siRAD51 for 48hrs followed by 48hr treatment with DMSO (ctrl), Niraparib (1, 5μM) treatment 48hrs followed by release for 6 days (o) Cell viability assay, MB231 & SUM149 cells transfected with siNT, siMYC 48hr, followed by 72hr Niraparib (2.5μM) then drug release for 7 days. Error bars represent mean ± s.d. (n ≥ 3 independent experiments) (p–q) Immunofluorescence of MB231 cells treated with siNT (ctrl), siMYC 48hr, followed by 72hr Niraparib (5μM), DMSO (ctrl) treatment (p) Immunofluorescence of γH2AX (red), RAD51 (green), nuclear DAPI (blue). Scale bar, 10μm. Representative images of three independent experiments. (q) Quantification of γH2AX, RAD51 staining. Data are mean ± s.d. of two biological replicates (r) DR-GFP homologous recombination repair assay. MB231 & HCC1937 were treated with siNT, siMYC 48hr, followed by transfection of DR-GFP reporter assay then analyzed by FACS for GFP+ cells. Values are normalized by control group. Error bars represent mean ± s.d. (n ≥ 3 independent experiments). b, c, two-tailed unpaired t-test was performed; in b, c, the multiple t-test was performed. n.s, (non significant), *P < 0.05, **P < 0.01, ***P < 0.001
MYC amplification also trended to an increased expression of several DNA repair genes, including RAD21, RAD54L and RAD51, in both breast and ovarian cancers (Supplementary Fig 6b). As an alternative objective to identify a secondary target in patients with BRCAmut breast cancer that can be used as a biomarker for PARPi response, we mined The Cancer Genome Atlas (TCGA) databases for BRCAmut breast (n=27) and ovarian (n=69) cancer patients, revealing that MYC was the most frequently amplified gene in these aggressive cancers (Supplementary Fig 6c, 6d).
Pearson correlation analysis demonstrated that RAD51 was the third most significant DNA repair gene associated with MYC expression in TNBC breast tumor samples, (Supplementary Fig 6e, Pearson correlation =1.022). Analysis of both TNBC and Ovarian cancer tumors supported an upregulation of DNA repair genes including RAD21, RAD54L and RAD51, in both breast and ovarian cancers (Supplementary Fig 6f–i). Moreover, gene expression analysis of 817 human breast tumor samples demonstrated a significant correlation between MYC and RAD51 expression (R2=0.2294, p=<0.001) (Fig 2d, R2=0.2294, p=<0.001), more so than MYC target gene EIF4E (Fig 2e, R2=0.124, p=<0.011). Collectively, these analyses point to MYC as a mediator of DNA repair via HR in TNBC.
To directly test this hypothesis in patient samples, we interrogated whether the correlation between c-myc and RAD51 persists at the protein level in human TNBC samples obtained before and after neoadjuvant chemotherapy using immunohistochemistry (IHC) (Fig 2f). Fifty-three TNBC patients with stage II–III TNBC enrolled in a prospective study at MD Anderson Cancer Center received neoadjuvant chemotherapy with anthracyclins, taxanes or a combination. Pretreatment core biopsy samples from 40 patients and post-chemotherapy surgical samples from 37 of those patients were available for immunohistochemical staining for c-myc and RAD51 (Supplementary Fig 7a). We recorded their clinical and pathological treatment responses then evaluated and compared the patient, tumor and treatment characteristics in the two groups as a function of c-myc and RAD51 staining (Supplemental Table 1). Pre and post-treatment tumors expressing both c-myc and RAD51 (Fig 2f) exhibited poor treatment response whereas absence of both markers indicated favorable response (Fig 2g,h, P=0.0001 and 0.0002, respectively). Although, c-myc and RAD51 expression individually predicted poor overall response, dual expression predicted the strongest correlation in both the pre- and post-treatment cohorts (Supplementary Fig 7b–d, p = 0.0001). Of the 53 patients, 24 had residual disease following chemotherapy allowing for comparison of RAD51 and c-myc expression in matched pre/post chemotherapy samples. Whereas c-myc expression was rarely altered in response to treatment (Supplementary Fig 7e), RAD51 expression was altered (in both directions) (Supplementary Fig 7f). However all patients with c-myc-ve/RAD51-ve tumors retained the phenotype in response to chemotherapy, whereas only 35% of c-myc+ve/RAD51+ve tumors reverted to a c-myc-ve/RAD51-ve phenotype (Supplementary Fig 7g). Furthermore, assessment of pathological complete response (pCR) in pre-treatment tumor samples supported previous findings that the c-myc-ve/RAD51-ve phenotype predicted the highest chemotherapy response rate (71%), whereas the c-myc+ve/RAD51+ve phenotype predicted a lower pCR response (Supplementary Fig 7h). These observations suggested that low c-myc/RAD51 expression predicts response of TNBC patients to standard-of-care therapy; whereas high expression can be used to identify tumors likely responsive to c-myc targeted therapy.
We next investigated whether MYC regulates RAD51 expression and the DNA repair pathway in breast cancer. MYC small interfering RNA (siRNA) downregulated RAD51 mRNA and protein expression in MB231 cells (Fig 2i, j) consistent with previous observations of transcriptional regulation of RAD51 via MYC (2). Consistently, MYC overexpression in normal breast cells (MCF10A) increased RAD51 protein and mRNA expression (Fig 2k, l). To examine if downregulation of MYC alters the sensitivity of cells to PARPis, we treated BRCAwt MDA231, and BRCAmut SUM149 cells with siRNAs for MYC and RAD51 for 48hrs and then cultured them with the PARPi niraparib for 72-h followed by 72-h of recovery. MYC and RAD51 siRNA in combination with PARPi increased apoptosis (Fig 2m), decreased clonogenicity (Fig 2n) and decreased cell proliferation (Fig 2o) of MB231 and SUM149 cells. Additionally, treatment of TNBC BRCAmut (SUM149, HCC1937) and BRCAwt (MDA231) with MYC siRNA in combination with either platinum DNA damaging agent, cisplatin or a taxane, docetaxel, revealed only increased efficacy of cisplatin in combination with MYC siRNA, but not with docetaxel. Furthermore, the immortalized MCF-10A cells were resistant to the combination treatments with MYC siRNA as revealed in minimal increased cell death (Supplementary Fig 8a). Cell cycle changes as a result of MYC siRNA treatments had no effect on PARPi response (Supplementary Fig 8b). However, MYC siRNA markedly impaired RAD51 foci formation in PARPi treated cells as compared to control (Fig 2p, q and Supplementary Fig 8c–d). Finally, MYC downregulation impaired HR activity in MB231 and HCC1937 cells as measured using a DR-GFP reporter assay (Fig 2r). Collectively these results suggested that MYC inhibition of HR is comparable with RAD51 loss in both MB231 and HCC1937 cells, providing further evidence of the HR-regulatory role of MYC in TNBCs.
Pharmacological inhibition of MYC expression induces PARPi sensitivity
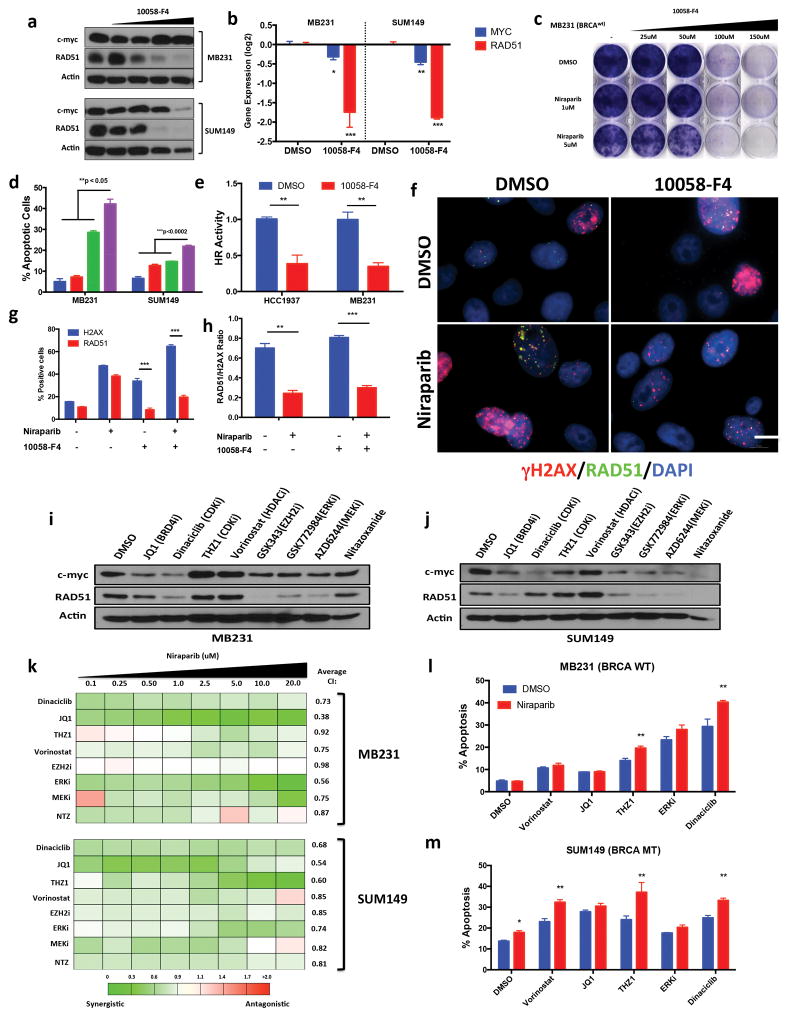
The downstream oncogenic function of MYC is primarily dependent on its heterodimerization with basic helix-loop-helix protein MAX, resulting in the activation of transcriptional targets (22). The small molecule 10058-F4 inhibited MYC-MAX binding (23), which resulted in the downregulation of RAD51 protein (Fig 3a) and RNA (Fig 3b) expression in MB231 and SUM149 cells. Treatment with 10058-F4 also resulted in transcriptional downregulation of MYC in both MB231 and SUM149 cells (Fig 3b). Treatment of these cells with combination of 10058-F4 and niraparib induced synergistic growth inhibition (Fig 3c, and Supplementary Fig 9a, b), increased apoptosis (Fig 3d) and decreased DR-GFP HR activity (Fig 3e) resulting in a decreased RAD51-γH2AX ratio (Fig 3f–h, and Supplementary Fig 9c). Collectively, these results suggest the following criteria for identifying agents that when combined with PARPi could lead to synergistic activity in TNBC cells: 1) downregulates MYC/RAD51 expression 2) induces synergistic growth inhibition in HTSAs and 3) increases apoptosis in combination with PARPi (Fig 3i–m, and Supplementary Fig 10). We tested several compounds including JQ1 a BRD4 inhibitor, and known MYC expression inhibitor (24), dinaciclib a pan cyclin-dependent kinase (CDK) 1,2,5,9 inhibitor with potent anti proliferative activity (25), THZ1, a CDK7 inhibitor that transcriptionally regulates MYC expression (26) and nitazoxanide, an anti-parasitic agent that inhibits c-myc expression (27). Of all these agents, the only one that met all three criteria- reducing the expression of both MYC and RAD51 in both MB231 and SUM149 cells (Fig 3i–j), exhibiting synergism with niraparib HTSAs (Fig 3k), and significantly inducing apoptosis (Fig 3l–m) in combination therapy was dinaciclib. Dinaciclib has an extensive clinical profile in several types of cancer including breast cancer (28).
Figure 3.
Inhibition of MYC in TNBC induces PARP inhibitor sensitivity (a) Immunoblot analysis for c-myc, RAD51, Actin (ctrl) of MB231 & SUM149 cells treated with DMSO, MYC inhibitor 10058-F4 25–150μM 72hr (b) RT-PCR analysis of c-myc, RAD51 in cells treated with 10058-F4, MB231 (100μM) SUM149 (50μM). Values are normalized to the levels of actin RNA expression in control (DMSO) cells (n = 3). Statistical analysis via unpaired two-sided t-test (c) Clonogenic assay: MB231 cells treated 72hr with DMSO (ctrl) 10058-F4 (25–150μM) and Niraparib (1, 5mM) with 9 day release (d) Annexin V and propidium iodide staining of cells treated 72hr with 10058-F4 (MB231: 100μM, SUM149: 50uM), Niraparib 1μM followed by drug release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments). (e) DR-GFP homologous recombination repair assay. MB231 & HCC1937 cells treated with 10058-F4 72hr, followed by transfection of DR-GFP & analyzed by FACS. Values are normalized with those from control group. Error bars represent mean ± s.d. (n ≥ 3 independent experiments). (f–h) Immunofluorescence analysis of MB231 cells treated 72hr with DMSO (ctrl), 10058-F4 (100μM), Niraparib (5mM) (f) Immunofluorescence images of γH2AX (red), RAD51 (green), nuclear DAPI (blue). Scale bar, 10μm. Representative images of three independent experiments. (g–h) Quantification of γH2AX, RAD51 staining. Representative images of three different experiments. Data are mean ± s.d. of biological replicates. (i–j) Immunoblot analysis for c-myc, RAD51 and Actin of (i) MB231 & (j) SUM149 cells treated 72hr with DMSO, JQ1 (1μm), Dinaciclib (25nM), THZ1 (50nM), Vorinostat (2.5μM ), GSK343 (10μM), GSK772983 (1μM), AZD6244 (10μM) Nitazoxanide (10μM) (k) Synergistic analysis of MB231 & SUM149 cells treated with Niraparib (0.1–20μM) in combination with the panel of drugs from i–j. 72hr HTSA was performed and synergism was determined using Calcusyn (<0.9 = synergism, 0.9–1.1 = Additive, >1.1 + Antagonistic) (l–m) Annexin V and propidium iodide staining of cells treated 72hr with Niraparib 1uM, in combination with panel of drugs followed by 72 drug release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments). *P < 0.05, **P < 0.01 two-way ANOVA with Sidak post-test correcting for multiple comparisons.
Dinaciclib Induces TNBC sensitivity to PARPis
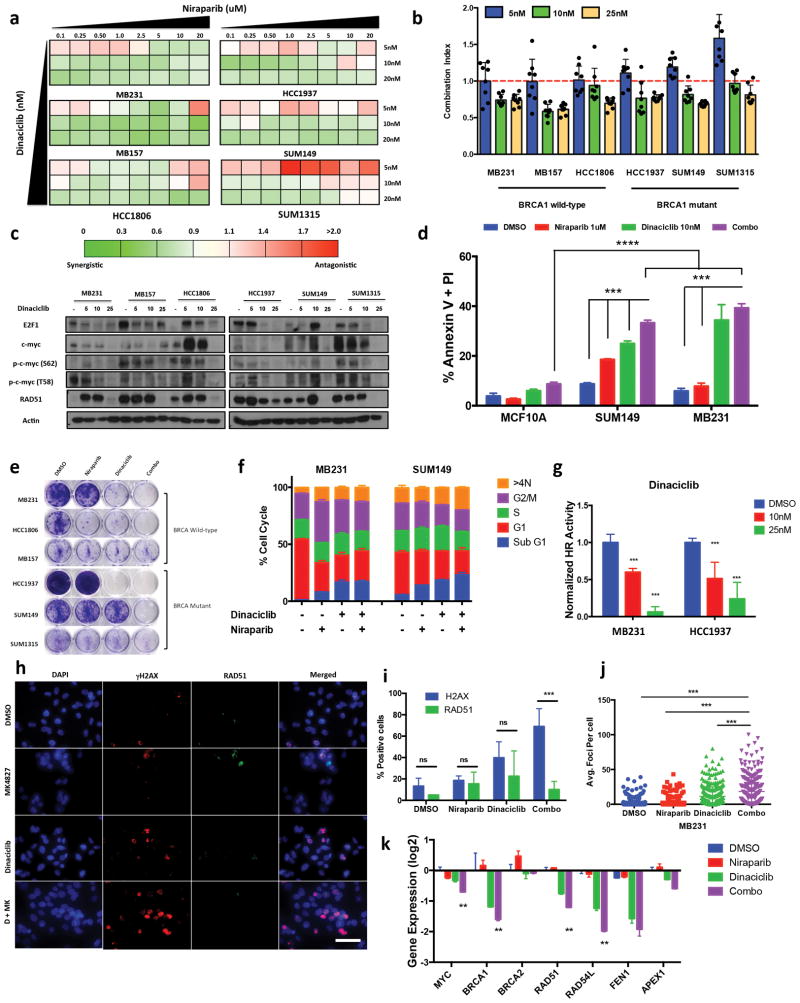
Treatment of a panel of TNBC BRCAmut and BRCAwt TNBC cell lines with dinaciclib and niraparib, revealed that the synergism of this combination therapy (assessed via HTSA) increased proportionally with the dinaciclib concentration (Fig 4a–b) that resulted in c-myc downregulation (Fig 4c), increased apoptosis (Fig 4d), a decrease in proliferation (Fig 4e) and an increase sub G1 cell cycle population (Fig 4f). Other PARPis such as olaparib and velaparib exhibited similar synergism in TNBC cell lines when combined with dinaciclib (Supplementary Fig 11a–c). The combination therapy was not synthetically lethal in the immortalized mammary epithelial cell line MCF10A, suggesting that induction of apoptosis by this combination strategy is tumor specific and dependent upon MYC expression (Fig 4d). Dinaciclib + Niraparib combination therapy consistently inhibited growth in both BRCA wild-type (MB231, MB157) and BRCA mutant (SUM149, HCC1937) (Supplementary Fig 11d). Induced MYC expression in MB231 cells partially reversed the effect of dinaciclib and niraparib combination therapy (Supplementary Fig 11e–f). Protein expression for several proapoptotic and anti-apoptotic markers including cleaved PARP, caspase 3, caspase 7, Mcl-1 and Bcl-2 did not delineate activation of a conventional apoptotic pathway in the cells treated with monotherapy versus combination therapy (Supplementary Fig 11g). On the other hand, the DR-GFP assay demonstrated impaired HR activity in the dinaciclib treated cells (Fig 4g). Dinaciclib inhibited RAD51 foci formation in MB231, HCC1937 and SUM149 cells (Fig 4g–j, and Supplementary Fig 12a–d). Combination therapy with dinaciclib and niraparib further increased the number of γH2AX positive cells (Fig4i, and Supplementary Fig 12a–g). Therefore, the mechanism of synthetic lethality of dinaciclib and niraparib is manifested through increased γH2AX DNA damage concomitant with downregulation of RAD51 foci formation. BRCA1 foci formation was also impaired by dinaciclib and niraparib in MB231 cells combination (Supplementary Fig 12h). Therefore, dinaciclib can impair the HR pathway by inhibiting BRCA1 and RAD51 activity via c-myc in BRCAwt cells. However, in cells with retained RAD51 functionality in the absence of BRCA1, dinaciclib and niraparib also induces synthetic lethality via RAD51 inhibition. Downregulation of MYC target gene expression in response to treatment with dinaciclib alone or combined with niraparib supports the effect of dinaciclib on c-myc and transcriptional regulation of the DNA repair pathway (Fig 4k).
Figure 4.
CDK inhibitor Dinaciclib induces PARP inhibitor sensitivity in TNBC (a–b) 72hr HTSA was performed on a panel of TNBC cell lines were treated with Dinaciclib in combination with Niraparib and synergism was determined using Calcusyn software (<0.9 = synergism, 0.9–1.1 = Additive, >1.1 + Antagonistic) (a) Synergistic analysis of TNBC cells treated with Niraparib (0.1–20μM) in combination with a panel of drugs. (b) Calcusyn combination index averages of Dinaciclib (5, 10, 25nM) treatment in with Niraparib (0.1–20μM) (c) Immunoblot analysis for E2F1, c-myc, p-c-myc (S62), p-c-myc (T58), RAD51 and Actin of TNBC cells treated 72hr with DMSO, Dinaciclib (5, 10, 25nM) (d) Annexin V and propidium iodide staining of MCF10A, MB231 & SUM149 treated 72hr with Dinaciclib (10nM) Niraparib (1μM). Error bars represent mean ± s.d. (n ≥ 3 independent experiments). (e) Clonogenic assay, TNBC cells treated 72hr with DMSO (ctrl) Niraparib (1uM), Dinaciclib (10nM) or combo followed by with 9-day release. (f) FACS cell cycle analysis of TNBC cells treated 72hr with Dinaciclib 10nM, Niraparib 1μM followed by 72 drug release (g) DR-GFP homologous recombination repair assay, MB231 & HCC1937 cells were transfected 24hr with DR-GFP reporter assay, then treated 72hr with Dinaciclib (10, 25nM) followed by FACS analysis for GFP+ cells. Values are normalized by control group. Error bars represent mean ± s.d. (n ≥ 3 independent experiments) (h–j) Immunofluorescence analysis of MB231 cells treated 72hr with DMSO (ctrl), Niraparib (5μM), Dinaciclib (10nM) or Combo (h) Immunofluorescence images of γH2AX (red), RAD51 (green), nuclear DAPI (blue). Scale bar, 2.5 μm. Representative images of three independent experiments. (i) Quantification of γH2AX, RAD51 staining. Representative images of three different experiments. Data are mean ± s.d. of biological replicates. unpaired two-sided t-tests (j) Quantification of average γH2AX foci per cell. A minimum of 250 cells were counted. (k) RT-PCR analysis of DNA repair genes in MB231 cells treated with DMSO, Niraparib (1μM), Dinaciclib (10nM), Combo. ***P < 0.001, n.s. (non significant) *P < 0.05, **P < 0.01, ***P < 0.001 two-way ANOVA with Sidak post-test correcting for multiple comparisons
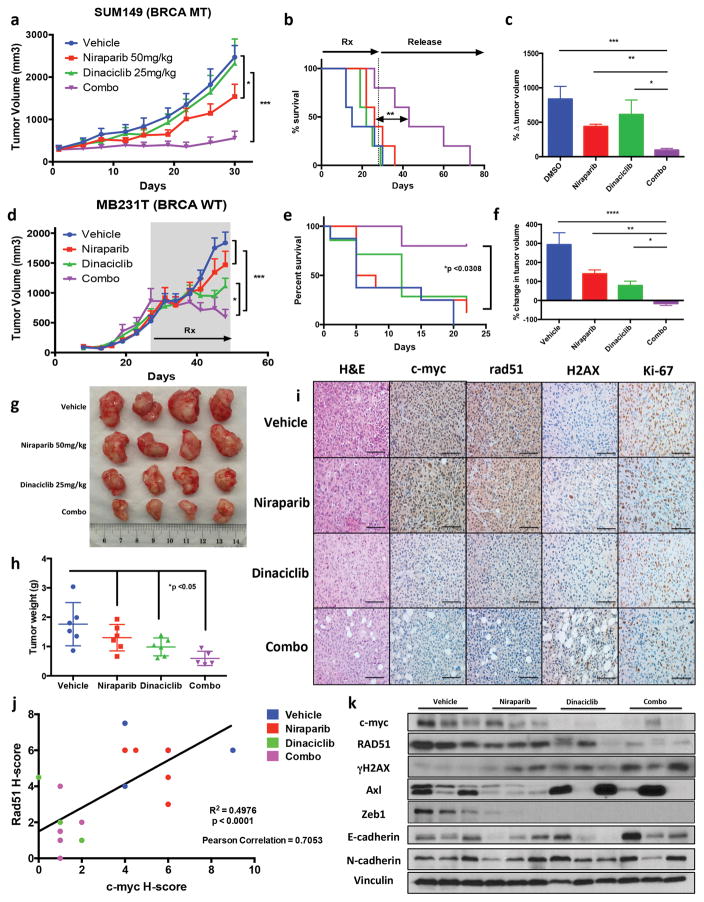
We next examined the in vivo efficacy of dinaciclib and niraparib against de novo treatment resistant BRCAmut SUM149 (Fig 5a–c, and Supplementary Fig 13a–c), and BRCAwt MB231T cells (Fig 5d–f, and Supplementary Fig 13d–e). In both cell types, combination therapy produced superior growth inhibition versus either agent alone (Fig 5a–f). We also determined whether growth inhibition during this combination therapy (four cycles) was sustained in BRCAmut SUM149 xenografts after treatment (Fig 5b, and Supplementary Fig 13a). Only the dinaciclib + niraparib combination therapy arm showed significant increases in median survival duration of 43-d as compared to vehicle, niraparib and dinaciclib arms with 15-d, 26-d and 23-d median survival times, respectively (P = 0.0115) (Fig 5b).
Figure 5.
Dinaciclib + Niraparib treatment in vivo (a–c) SUM149 cells were injected into immunocompromised mice and allowed to grow to ~200mm3, xenografts were treated with either Vehicle, Dinaciclib 25mg/kg 3 times weekly, Niraparib 5mg/kg 5 times weekly or Dinaciclib + Niraparib (Combo) therapy for 4 weeks (a) Tumor volume measurements and (b) Kaplan Meyer survival analysis of mice treated with indicated drug treatment arms for 4 weeks (28 days), followed by release without treatment until mice reached maximum allowed tumor size (c) Percent change in tumor volume normalized by day 1 of treatment. *P < 0.05, **P < 0.01, ***P < 0.001 two-way ANOVA with Sidak post-test correcting for multiple comparison (d–j) MB231T cells were injected into nude mice and cells were allowed to grow to >500mm3 before treatment was initiated. Mice were treated with Vehicle, Dinaciclib 25mg/kg 3 times weekly, Niraparib 5mg/kg 5 times weekly or Dinaciclib + Niraparib (Combo) therapy for 3 weeks. Upon completion tumors were extracted and analyzed by IHC and RPPA (d) Tumor volume measurements and (e) Kaplan Meyer survival analysis of mice treated with indicated drug arms for 3 weeks (f) Percent change in Tumor volume normalized by Day 1 of treatment. *P < 0.05, **P < 0.01, ***P < 0.001 two-way ANOVA with Sidak post-test correcting for multiple comparison (g–h) Tumors from mice treated with each of the treatment arms were extracted and weighed and subjected to (i) Immunohistochemical analysis of treated tumors for H&E, Ki67, c-myc, RAD51, and γHAX resulting in a (j) comparison the H-score of c-myc with RAD51 expression in MB231T treated xenografts (k) immunoblot analysis for c-myc, RAD51, γH2AX, Zeb1, Axl, E-cadherin, N-cadherin and Actin.
We injected BRCAwt MB231T cells into nude mice and allowed the resulting tumors to grow >500mm3 before initiating treatment with dinaciclib and niraparib to determine whether the initial tumor size had any bearing on therapeutic efficacy (Fig 5d). Treatment with dinaciclib and niraparib resulted in significant tumor growth inhibition (Fig 5d, and Supplementary Fig 13d), increased overall survival (Fig 5e), decreased percent change in tumor volume (Fig 5f, and Supplementary Fig 13f) and a reduced tumor weight (Fig 5g, h). Immunohistochemical analysis demonstrated lower c-myc and RAD51 in dinaciclib and combination treated tumors than in the niraparib and vehicle treatment arms (Fig 5i, and Supplementary 13g–i). Tumors from the combination treatment arm also demonstrated the greatest upregulation of γH2AX expression (Fig 5i and Supplementary Fig 13j). Downregulation of c-myc expression correlated with RAD51 and Ki67 downregulation in treated xenografts (Fig 5j, Supplementary Fig 13g, k). Western blot analysis revealed downregulation of c-myc and RAD51 in both dinaciclib and combination treatment arms (Fig 5k). Additionally, we observed loss of programmed cell death ligand 1 (PD-L1) expression in treated tumors (Supplementary Fig 13l). Consistent with down regulation of EMT markers, dinaciclib given either alone or in combination therapy inhibited the CD44hi/CD24lo cancer stem cell (CSC) population in vitro, whereas niraparib alone had no effect on it (Supplementary Fig 13m). Additionally, we observed a reduction of mammosphere formation of MB231 cells treated with siMYC and Dinaciclib compared to control, supporting a role of MYC on the CSC phenotype in TNBC (Supplementary Fig 13n). Although MYC itself is a marker of the CSC phenotype, several others (e.g. Vimentin, ALDH) were downregulated at the protein level. Reverse Phase Protein Array (RPPA) analysis demonstrated upregulation of the proapoptotic pathway but downregulation of the DNA Repair, anti-apoptotic, and epithelial-mesenchymal transition proteins (Supplementary Fig 13o–q). Downregulation of the EMT pathway was confirmed by protein downregulation of Axl, Vimentin, Zeb1 and N-cadherin protein expression and upregulation of E-cadherin expression (Fig 5k). These alterations correlated with downregulation of c-myc and RAD51 expression (Fig 5j,i). Collectively, our in vivo analysis suggested that c-myc is a useful predictive biomarker for treatment with the combination of dinaciclib and niraparib having synthetic lethality via HR repair and dinaciclib inhibits the EMT and CSC pathways in TNBC xenografts.
Dinaciclib resensitizes PARPi-resistant cells to PARP inhibition
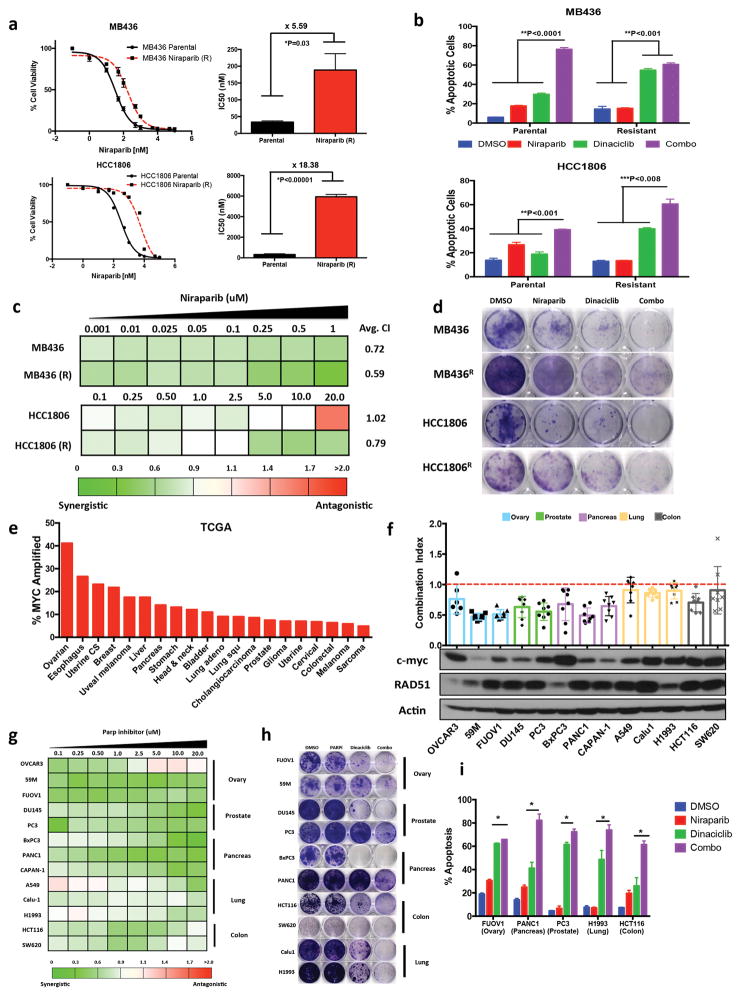
Next, we sought to determine whether therapy with dinaciclib and niraparib is also effective against TNBC cells with acquired resistance to PARP inhibition. We generated PARPi-sensitive BRCAmut, MB436 and BRCAwt, HCC1806 TNBC cell lines (Fig 1a), resistant to increasing concentrations of the PARPi niraparib in a step-wise fashion resulting in 5- to18-fold increased resistance according to half maximal inhibitory concentration (IC50) values (Fig 6a). Dinaciclib alone reversed the resistance of these cells to PARP inhibition, while inducing a synthetically lethal increase in apoptosis incidence (Fig 6b). Also, the combination therapy induced synergistic growth inhibition (Fig 6c–d). To further analyze the efficacy of combination therapy of dinaciclib and niraparib we induced PARPi resistance via 53BP1 loss. Knockdown of 53BP1 expression in parental HCC1806 cells induced PARPi resistance by two fold (Supplementary Fig 14 a, b). The combination treatment also induced synergistic growth inhibition response in 53BP1 knockdown cells (Supplementary Fig 14c, d). Conversely, 53BP1 knockdown by short hairpin RNA did not induce PARP inhibitor resistance in MB436 cells (data not shown). Thus, treatment with dinaciclib and niraparib can overcome both acquired and induced resistance to PARPi.
Figure 6.
Dinaciclib induces synthetic lethality in Niraparib acquired resistant & high MYC cancers (a) 72hr HTSA for Niraparib was performed on acquired resistance and parental MB436 & HCC8106 cells (b) Annexin V and propidium iodide staining of MB436 (parental), MB436 (Resistant), HCC1806 & HCC1806 (Resistant) treated 72hr with DMSO (ctrl), Dinaciclib 10nM, Niraparib 1μM & Combo followed by 72 drug release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments) (c) 72hr HTSA was performed on MB436, MB436 (R), HCC1806 & HCC1806 (R) treated with Dinaciclib 10nM in combination with Niraparib and synergism was determined using Calcusyn software (<0.9 = synergism, 0.9–1.1 = Additive, >1.1 Antagonistic) (d) Clonogenic assay of MB436, MB436 (R), HCC1806 & HCC1806 (R) treated 72hr with DMSO (ctrl), Dinaciclib 10nM, Niraparib 1μM & Combo followed by 72 drug release (e) TCGA analysis of MYC amplification across several cancer types (f–g) 72hr HTSA was performed on c-myc high cancers with Dinaciclib 10nM in combination with Niraparib and synergism was determined using Calcusyn software (h) Clonogenic assay of MYC high cancer cell lines treated 72hr with DMSO (ctrl), Dinaciclib 10nM, Niraparib 1μM & Combo followed by 9 drug release (i) Annexin V and propidium iodide staining of MYC high cancer cell lines treated 72hr with DMSO (ctrl), Dinaciclib 10nM, Niraparib 1mM & Combo followed by 72 drug release. Error bars represent mean ± s.d. (n ≥ 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001 two-way ANOVA with Sidak post-test correcting for multiple comparisons
Dinaciclib and PARP inhibition are effective against MYC driven cancers
MYC is a potent oncogene that is amplified in several cancer types. Analysis of several TCGA patient cohorts revealed high MYC gene expression in uveal, colorectal, head and neck, lung and ovarian tumors (Fig 6e, and Supplementary Fig 15). Often, patients with these high MYC expressing cancers have very poor response rates to standard of care therapies indicating a need for novel therapeutic strategies (13). Therefore, we sought to determine the applicability of c-myc and PARP inhibition for other aggressive cancer types besides TNBC. We selected a panel of high MYC expressing ovarian, prostate, pancreatic, lung and colon cancer cell lines (Fig 6f). In vitro analysis of dinaciclib and niraparib revealed synergistic growth inhibition (i.e. combination index < 1) in all cell lines examined (Fig 6f–h). Additionally, the combination therapy significantly increased apoptosis incidence, as compared to either single agent, in several cell lines tested irrespective of cancer type (Fig 6i). These results suggested that the combination of dinaciclib and niraparib is an effective treatment strategy extending to many tumors with high MYC expression.
Discussion
Clinical PARPi use is steadily advancing with 3 drugs receiving FDA approval thus far and a high probability of more approvals to come. As the first therapeutics approved for BRCAmut tumors, their safe clinical profiles render them a paradigm-changing modality versus conventional chemotherapy. However, most cancer patients do not benefit from PARPi use, as the BRCAmut population represents only a small percentage of cancer patients overall. De novo and acquired resistance mechanisms further limit PARPi efficacy (29), demonstrating the need for increased clinical PARPi use in combination therapy. We developed and tested a promising treatment strategy based on MYC oncogene addiction and DNA repair. Oncogene MYC induction induces replication stress causing double strand DNA breaks which require the activation of HR pathway to repair lesions (3). Our study demonstrates that MYC expression directly invokes HR activation in response to DNA damage and can potentially be used as a predictor of response of TNBC to therapy with PARPi combination.
The connection between MYC and RAD51 (as an indicator of HR activity) is based upon (i) correlation between TNBC and ovarian cancer tumor tissue gene expression of MYC and several DNA repair genes including RAD51 in the TCGA cohort (Supplementray fig. 6e–i). (ii) IHC analysis of MYC and RAD51 in TNBC patient cohorts accrued at MD Anderson Cancer Center (Fig 2g–h, Supplementary Fig a–d). (iii) Downregulation of RAD51 in response to MYC knockdown in MDA231 cells (Fig 2I–j). (iv) MYC regulation of RAD51 in knock-in experiments (Fig 2k–l). Collectively these results, which are consistent with published reports showing MYC is a transcriptional regulator of RAD51 (30–33), provide experimental evidence for the functional relationship between MYC and RAD51.
Although, oncogenic MYC transformation embodies several aspects of the “Hallmarks of Cancer”, it is designated as an undruggable target (1). Therefore, targeting of MYC driven cancers through a synthetically lethal approach has therapeutic potential. MYC expression has long been associated with BRCA mutation frequency in breast cancer cases (34, 35). Studies of transgenic mouse models have demonstrated that MYC facilitates BRCAmut progression of ovarian cancer (36). The frequency of MYC upregulation in BRCAmut tumors and correlation with HR repair genes suggests that MYC compensates for BRCA loss via upregulation of a compensatory HR pathway, via increased RAD51 expression (37). The correlation of MYC and RAD51 gene expression in sporadic TNBCs (Fig 2d) supports MYC dependence on HR repair as a function of MYC-driven tumors. RAD51 also regulates TNBC resistance to PARPis via the CSC resistance phenotype in TNBC (38). Inhibition of RAD51 expression in combination with PARPi-based treatment abrogates this CSC resistance phenotype, supporting our use of RAD51 as a marker of response to PARPis. Although RAD51 is classified as a tumor suppressor, high RAD51 expression compensates for BRCA loss in tumors, causing decreased sensitivity to irradiation and chemotherapy (37). Independent of MYC, increased RAD51 expression promotes TNBC metastasis, supporting high MYC, and RAD51 individual expression as negative predictors of poor outcome of TNBC (7).
The clinical use of CDK inhibitors to treat cancer has yielded minimal success until recently with the approval of the CDK4/6 inhibitors palbociclib and riibociclib (39). Dinaciclib, a pan CDK 1/2/5/9 inhibitor, has undergone several rounds of clinical investigation for several types of cancer including breast cancer (28, 40). However, the lack of a suitable biomarker for patient selection has plagued the advancement of dinaciclib beyond Phase II clinical trials. Additionally, dinaciclib combined with conventional chemotherapy has compounded patient toxicity, leading to treatment discontinuation, which further supports the use of dinaciclib in combination with targeted therapy to minimize patient toxicity profiles (28). Although dinaciclib was not developed as an agent targeted against MYC, several studies have corroborated our observation of dinaciclib’s regulation of MYC expression (41, 42). Ideally, MYC downregulation would induce synthetic lethality in MYC driven tumors, however studies have demonstrated that MYC loss alone is insufficiently lethal (31–33) further strengthening the need for combination therapy.
The precise mechanism of dinaciclib-induced MYC downregulation remains unclear. CDK1 and CDK9 inhibition are both synthetic lethal in high MYC-expressing cancer cells (43). Several CDKs (CDK2, CDK1, CDK5, CDK12) have been linked to DNA repair regulation, with several CDKs phosphorylating DNA repair genes directly (BRCA1, BRCA2) (44–48). CDK5 has been directly linked to regulation of DNA repair in cancer (49). Transcriptional downregulation of MYC in response to dinaciclib exposure points to a transcriptional CDK regulating MYC expression. Recently, Johnson et al have suggested that CDK12 mediates dinaciclib-induced sensitivity to PARP inhibition in TNBC (50). In support of these findings, loss of CDK12 expression is synthetic lethal in c-myc overexpressing cells, thus characterizing CDK12 as an upstream effector of MYC expression via RNA polymerase phosphorylation (50). Loss of CDK12 expression also predicts sensitivity of ovarian cancer to PARPis (51). Traditionally, the degree of PARP trapping by PARPis plays a significant role in the efficacy of PARPis against HR deficient cells (52). The present study demonstrated a synergistic growth inhibitory effect of dinaciclib on TNBC cells when given with several different PARPis, suggesting that PARP trapping does not dictate sensitivity to this combination therapy.
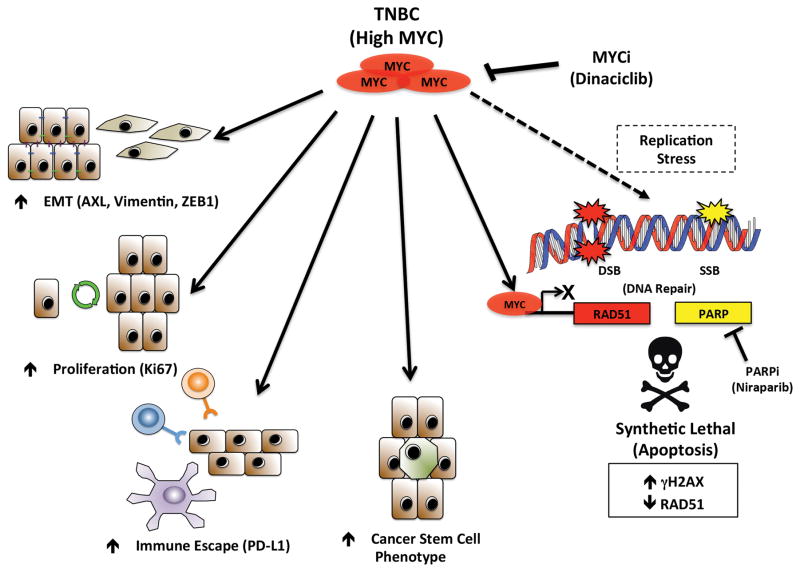
We have presented a novel mechanism of stratifying TNBC patients for therapy with PARPis based upon MYC expression (Supplementary Fig 16). This model corrects a number of deficiencies in treatment options for TNBC patients by making MYC druggable, enhancing the therapeutic effect of PARPis in non-BRCAmut TNBC patients while targeting several cancer hallmarks including DNA repair, EMT & the CSC phenotype, which lie at the crux of conventional chemotherapeutic efficacy (Figure 7).
Figure 7.
A graphical representation of targeting both MYC and PARP in MYC high TNBC could elicit anti tumor effects
Predictive immunohistochemical analysis of c-myc/RAD51 expression points to a less favorable response to chemotherapy of tumors that express both c-myc and RAD51 (Fig 2f–h). By harnessing this knowledge we seek to improve patient outcomes in both c-myc-/RAD51− (via chemotherapy) and c-myc+/RAD51+ (via treatment with dinaciclib and niraparib) patient cohorts. Additionally, implementation of dinaciclib and niraparib as a neoadjuvant therapeutic treatment that downregulates both c-myc and RAD51 expression infers subsequent response to chemotherapy and/or radiation therapy, if necessary. As a proto-oncogene, upregulation of MYC can drive therapeutic resistance to other targeted therapeutic agents (53, 54). Therefore the combination strategy with dinaciclib and niraparib may be a secondary treatment option for patients who have developed MYC dependent resistance to therapy.
Supplementary Material
Acknowledgments
Financial Support
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award (P30CA016672) to The University of Texas MD Anderson Cancer Center, R01 grants CA87548 and CA1522218 and Cancer Prevention and Research Institute of Texas (CPRIT) RP170079 grants to K. Keyomarsi, the Susan G. Komen for the Cure grant KG100521 to K.K. Hunt, the Susan G. Komen post-doctoral fellowship grant PDF14302675 to J.P.W. Carey and a CPRIT training award RP170067 to S. Vijayaraghavan.
We thank Dr. Debu Tripathy, Dr. Banu Arun and Dr. Gabriel Hortobagyi for their insightful comments during the course of this study.
Footnotes
Conflicts of Interest: No conflicts to disclose
References
- 1.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–39. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–7. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cottini F, Hideshima T, Suzuki R, Tai YT, Bianchini G, Richardson PG, et al. Synthetic Lethal Approaches Exploiting DNA Damage in Aggressive Myeloma. Cancer Discov. 2015;5:972–87. doi: 10.1158/2159-8290.CD-14-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AlHilli MM, Becker MA, Weroha SJ, Flatten KS, Hurley RM, Harrell MI, et al. In vivo anti-tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol. 2016;143:379–88. doi: 10.1016/j.ygyno.2016.08.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.RING-less BRCA1 Induces PARP Inhibitor and Platinum Resistance. Cancer Discov. 2016;6:OF6. [Google Scholar]
- 6.Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, Nicolas E, et al. The BRCA1-Delta11q Alternative Splice Isoform Bypasses Germline Mutations and Promotes Therapeutic Resistance to PARP Inhibition and Cisplatin. Cancer Res. 2016;76:2778–90. doi: 10.1158/0008-5472.CAN-16-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiegmans AP, Al-Ejeh F, Chee N, Yap PY, Gorski JJ, Da Silva L, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5:3261–72. doi: 10.18632/oncotarget.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaji K, Vijayaraghavan S, Diao L, Tong P, Fan Y, Carey JP, et al. AXL Inhibition Suppresses the DNA Damage Response and Sensitizes Cells to PARP Inhibition in Multiple Cancers. Mol Cancer Res. 2017;15:45–58. doi: 10.1158/1541-7786.MCR-16-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanos-Webb A, Jabbour NA, Multani AS, Wingate H, Oumata N, Galons H, et al. Targeting low molecular weight cyclin E (LMW-E) in breast cancer. Breast Cancer Res Treat. 2012;132:575–88. doi: 10.1007/s10549-011-1638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravalli RN, Talbot NC, Steer CJ. Gene expression profiling of MYC-driven tumor signatures in porcine liver stem cells by transcriptome sequencing. World J Gastroenterol. 2015;21:2011–29. doi: 10.3748/wjg.v21.i7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nanos-Webb A, Bui T, Karakas C, Zhang D, Carey JP, Mills GB, et al. PKCiota promotes ovarian tumor progression through deregulation of cyclin E. Oncogene. 2016;35:2428–40. doi: 10.1038/onc.2015.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung M, Russell AJ, Liu B, George J, Liu PY, Liu T, et al. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res. 2017;77:971–81. doi: 10.1158/0008-5472.CAN-15-2906. [DOI] [PubMed] [Google Scholar]
- 14.Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118:2787–95. doi: 10.1002/cncr.26576. [DOI] [PubMed] [Google Scholar]
- 15.Kwon JS, Gutierrez-Barrera AM, Young D, Sun CC, Daniels MS, Lu KH, et al. Expanding the criteria for BRCA mutation testing in breast cancer survivors. J Clin Oncol. 2010;28:4214–20. doi: 10.1200/JCO.2010.28.0719. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–9. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–8. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayraktar S, Gluck S. Systemic therapy options in BRCA mutation-associated breast cancer. Breast Cancer Res Treat. 2012;135:355–66. doi: 10.1007/s10549-012-2158-6. [DOI] [PubMed] [Google Scholar]
- 19.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peshkin BN, Alabek ML, Isaacs C. BRCA1/2 mutations and triple negative breast cancers. Breast Dis. 2010;32:25–33. doi: 10.3233/BD-2010-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–54. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurlin PJ, Dezfouli S. Functions of myc:max in the control of cell proliferation and tumorigenesis. Int Rev Cytol. 2004;238:183–226. doi: 10.1016/S0074-7696(04)38004-6. [DOI] [PubMed] [Google Scholar]
- 23.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg Med Chem Lett. 2009;19:807–10. doi: 10.1016/j.bmcl.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Shao Q, Kannan A, Lin Z, Stack BC, Jr, Suen JY, Gao L. BET protein inhibitor JQ1 attenuates Myc-amplified MCC tumor growth in vivo. Cancer Res. 2014;74:7090–102. doi: 10.1158/0008-5472.CAN-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Molecular cancer therapeutics. 2010;9:2344–53. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 26.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–20. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitri Z, Karakas C, Wei C, Briones B, Simmons H, Ibrahim N, et al. A phase 1 study with dose expansion of the CDK inhibitor dinaciclib (SCH 727965) in combination with epirubicin in patients with metastatic triple negative breast cancer. Invest New Drugs. 2015;33:890–4. doi: 10.1007/s10637-015-0244-4. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–50. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton FK, Patterson MJ, Elstob CJ, Fordham S, Herriott A, Wade MA, et al. Common cancer-associated imbalances in the DNA damage response confer sensitivity to single agent ATR inhibition. Oncotarget. 2015;6:32396–409. doi: 10.18632/oncotarget.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Kim DK, Ko JJ, Kim KP, Park KS. Rad51 Regulates Reprogramming Efficiency through DNA Repair Pathway. Dev Reprod. 2016;20:163–9. doi: 10.12717/DR.2016.20.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gravina GL, Festuccia C, Popov VM, Di Rocco A, Colapietro A, Sanita P, et al. c-Myc Sustains Transformed Phenotype and Promotes Radioresistance of Embryonal Rhabdomyosarcoma Cell Lines. Radiat Res. 2016;185:411–22. doi: 10.1667/RR14237.1. [DOI] [PubMed] [Google Scholar]
- 33.Ambrosio S, Amente S, Napolitano G, Di Palo G, Lania L, Majello B. MYC impairs resolution of site-specific DNA double-strand breaks repair. Mutat Res. 2015;774:6–13. doi: 10.1016/j.mrfmmm.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Grushko TA, Dignam JJ, Das S, Blackwood AM, Perou CM, Ridderstrale KK, et al. MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res. 2004;10:499–507. doi: 10.1158/1078-0432.ccr-0976-03. [DOI] [PubMed] [Google Scholar]
- 35.Rhei E, Bogomolniy F, Federici MG, Maresco DL, Offit K, Robson ME, et al. Molecular genetic characterization of BRCA1- and BRCA2-linked hereditary ovarian cancers. Cancer Res. 1998;58:3193–6. [PubMed] [Google Scholar]
- 36.Xing D, Orsulic S. A mouse model for the molecular characterization of brca1-associated ovarian carcinoma. Cancer Res. 2006;66:8949–53. doi: 10.1158/0008-5472.CAN-06-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67:9658–65. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Burness ML, Martin-Trevino R, Guy J, Bai S, Harouaka R, et al. RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res. 2017;23:514–22. doi: 10.1158/1078-0432.CCR-15-1348. [DOI] [PubMed] [Google Scholar]
- 39.Hortobagyi GN. Ribociclib for HR-Positive, Advanced Breast Cancer. N Engl J Med. 2017;376:289. doi: 10.1056/NEJMc1615255. [DOI] [PubMed] [Google Scholar]
- 40.Chen XX, Xie FF, Zhu XJ, Lin F, Pan SS, Gong LH, et al. Cyclin-dependent kinase inhibitor dinaciclib potently synergizes with cisplatin in preclinical models of ovarian cancer. Oncotarget. 2015;6:14926–39. doi: 10.18632/oncotarget.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory GP, Hogg SJ, Kats LM, Vidacs E, Baker AJ, Gilan O, et al. CDK9 inhibition by dinaciclib potently suppresses Mcl-1 to induce durable apoptotic responses in aggressive MYC-driven B-cell lymphoma in vivo. Leukemia. 2015;29:1437–41. doi: 10.1038/leu.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209:679–96. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800–14. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Li J, Song YS, Li Y, Jia YH, Zhao HD. Cdk5 links with DNA damage response and cancer. Mol Cancer. 2017;16:60. doi: 10.1186/s12943-017-0611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Kim NH. CDK2 Is Required for the DNA Damage Response During Porcine Early Embryonic Development. Biol Reprod. 2016;95:31. doi: 10.1095/biolreprod.116.140244. [DOI] [PubMed] [Google Scholar]
- 46.Trovesi C, Manfrini N, Falcettoni M, Longhese MP. Regulation of the DNA damage response by cyclin-dependent kinases. J Mol Biol. 2013;425:4756–66. doi: 10.1016/j.jmb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 47.Cerqueira A, Santamaria D, Martinez-Pastor B, Cuadrado M, Fernandez-Capetillo O, Barbacid M. Overall Cdk activity modulates the DNA damage response in mammalian cells. J Cell Biol. 2009;187:773–80. doi: 10.1083/jcb.200903033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson N, Shapiro GI. Cyclin-dependent kinases (cdks) and the DNA damage response: rationale for cdk inhibitor-chemotherapy combinations as an anticancer strategy for solid tumors. Expert Opin Ther Targets. 2010;14:1199–212. doi: 10.1517/14728222.2010.525221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shupp A, Casimiro MC, Pestell RG. Biological functions of CDK5 and potential CDK5 targeted clinical treatments. Oncotarget. 2017;8:17373–82. doi: 10.18632/oncotarget.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep. 2016;17:2367–81. doi: 10.1016/j.celrep.2016.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74:287–97. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao D, Yue M, Su H, Ren P, Jiang J, Li F, et al. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol Cell. 2016;64:493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 54.Liu H, Ai J, Shen A, Chen Y, Wang X, Peng X, et al. c-Myc Alteration Determines the Therapeutic Response to FGFR Inhibitors. Clin Cancer Res. 2017;23:974–84. doi: 10.1158/1078-0432.CCR-15-2448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.