Abstract
Many studies have discussed clinical practice guidelines for the treatment of cystitis and pyelonephritis. Treatment of uncomplicated urinary tract infections (UTIs) can be based on empiric antibiotic therapy. For complicated or recurrent UTIs, therapy can be based on laboratory-controlled culture and sensitivity (C&S) reports. The diagnosis of UTI by clinical criteria alone has an error rate of up to 33%. In addition, positive laboratory culture results do not always indicate a diagnosis of UTI. Comparison of urine in a conventional culture model versus DNA next-generation sequencing (NGS) to accurately identify and provide information on resistance factors (mobile genetic elements) is warranted. Our study was a head-to-head comparative phase II study of standard urine C&S versus DNA NGS testing for the diagnosis and treatment efficacy in patients with symptoms of acute cystitis based on short-term outcomes.
Keywords: Cystitis, Pyelonephritis, Urinary tract infection, DNA next-generation sequencing
Many studies have discussed clinical practice guidelines for the treatment of cystitis and pyelonephritis.1-5 Treatment in many instances of uncomplicated urinary tract infections (UTIs) can be based on empiric antibiotic therapy. For complicated or recurrent UTIs, therapy can be based on laboratory-controlled culture and sensitivity (C&S) reports.1,2 The diagnosis of UTI by clinical criteria alone has an error rate of up to 33%.6,7 In addition, positive laboratory culture results do not always indicate a diagnosis of UTI.8-10
In many patients, it is important to properly identify the causal agents in the urine of those who have infections. Inaccurate and insufficient therapeutic coverage can lead to increased healthcare costs, hours of work lost, and decreased patient satisfaction. For these reasons, comparison of urine in a conventional culture model versus DNA next-generation sequencing (NGS) to accurately identify and provide information on resistance factors (mobile genetic elements) is warranted.10,11 The improved diagnosis of UTIs may also lead to a decrease in resistant strains of bacteria, which reached a level up to 34% in certain areas of the United States.2,12,13
Any new diagnostic test must meet a threshold that surpasses the gold standard of urine C&S testing.7,10 The aim of our study was to conduct a head-to-head comparative phase II study of standard urine C&S versus DNA NGS testing for the diagnosis and treatment efficacy in patients with symptoms of acute cystitis based on shortterm outcomes.
Material and Methods
Study Design and Participants
After receiving institutional review board (IRB) approval, we performed a standard prospective, randomized, open-label, controlled repeated-measures design study. Randomization was performed using the Research Randomizer software (http://www.randomizer.org). The population under investigation had symptoms of acute cystitis including urinary frequency, urgency, dysuria, and abdominal pain, as well as possible signs of hematuria. Patients were divided into uncomplicated and complicated acute cystitis.
Complicated cases included men and patients with an indwelling Foley catheter. Exclusion criteria included patients with fever and clinical symptoms of acute pyelonephritis, acute or chronic prostatitis, urethritis, and epididymitis. Other exclusion criteria included patients who were treated for a UTI in the past month.
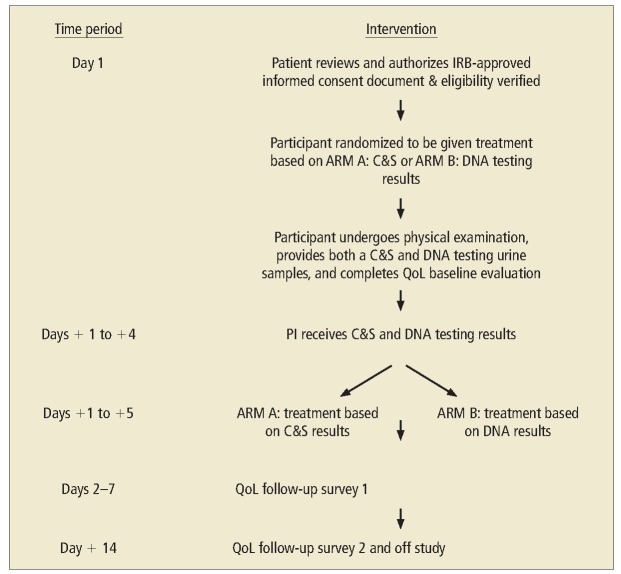
Between January 2016 and December 2016, 56 patients entered the study with symptoms of acute cystitis; 12 patients were removed from the study based on IRB-approved protocol criteria and 44 patients completed the study. Twenty-two asymptomatic volunteers entered the control group. Patients were instructed on how to perform standardized midstream urine collection samples. Nursing staff collected urine samples from catheterized patients. All specimens underwent C&S and DNA NGS testing. The study schema is presented in Figure 1.
Figure 1.
Study schema. C&S, culture and sensitivity; IRB, institutional review board; PI, physician investigator; QoL, quality of life.
Patients were randomized into Arm A (treatment based on C&S) and Arm B (treatment based on DNA NGS) results. If patients had a negative C&S result in Arm A, treatment could be initiated on day 8 depending on DNA sequencing results, and vice versa. Treatment was based on sensitivity reports as determined on culture results or antibiograms and resistance results in the case of DNA sequencing. All treatment was with a single antibiotic only for 7 days in duration.
The UTI Symptom Assessment questionnaire (Table 1) is a selfadministered, validated instrument used to ascertain the effect of UTIs on the individual’s quality of life.14 This survey was administered to participants on days 1 through 7, and on day 14, after completion of antibiotic therapy. Follow-up surveys were done through telephone or email contact. Two types of molecular microbial diagnostic testing “levels” by MicroGen DX (Orlando, FL) are performed as noted below.
Table 1.
A Portion of the Urinary Tract Infection Symptom Assessment Questionnaire
| About Your Symptoms and Their Impact on Your Life (For Use After Visit 1) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Since you last completed this questionnaire, please indicate whether you have had the following symptoms/problems and how severe they were (please circle one number for each symptom): | Since you last completed this questionnaire, if you have experienced these symptoms/problems, please indicate how bothersome they were (please circle one number for each symptom): | |||||||
| Did not have | Mild | Moderate | Severe | Not at all | A little | Moderately | A lot | |
| 0 | 1 | 2 | 3 | Frequency of urination (going to the toilet very often) | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Urgency of urination (a strong and uncontrollable urge to pass urine) | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Pain or burning when passing urine | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Not being able to empty your bladder completely/passing only small amounts of urine | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Pain or uncomfortable pressure in the lower abdomen/pelvic area caused by your urinary tract infection | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Low back pain caused by your urinary tract infection | 0 | 1 | 2 | 3 |
| 0 | 1 | 2 | 3 | Blood in your urine | 0 | 1 | 2 | 3 |
| Please give an overall rating of the severity of your urinary tract infection symptoms as they are (Please circle the number of your answer) | ||||||||
| 0 No symptoms at all | 1 Mild | 2 Moderate | 3 Severe | |||||
Level 1: Rapid Screen Using Quantitative Polymerase Chain Reaction Methods
The Level 1 panel is a quantitative real-time polymerase chain reaction (PCR) test for bacteria and fungi. The panel also includes a qualitative real-time PCR test, which can assess for genetic factors conferring resistance to bacteria. Presently, resistance factors for vancomycin, methicillin, β-lactam, carbapenem, macrolide, aminoglycoside, fluoroquinolones, and tetracycline are determined by these methods. The panel utilizes unique genes present in each organism to identify how much of that organism is present in each patient sample. This concentration is achieved in a multistep process. The urine specimen is tested in Level 1 for Escherichia coli, Enterococcus faecalis, E faecium, Klebsiella pneumoniae, group B streptococcus (S agalactiae), group A streptococcus (S pyogenes), Candida albicans, Pseudomonas aeruginosa, Staphylococcus aureus, Serratia marcescens, Proteus species, Citrobacter species, Enterobacter species, and Morganella species.
The resistance factors are run on a qualitative assay, which reports them as present or absent. The 16S universal bacteria is a semiquantitative assay, which reports the total bacteria load as low, medium, or high. These ranges for general bacteria copy number per mL or mg are <105, 105 to 107, and >107 for low, medium, and high, respectively.
Level 2: DecodEXTM DNA Pyrosequencing Method
The Level 2 test detects virtually all microbial organisms and fungal pathogens that may be present in patient specimens. The use of 16S ribosomal RNA is pivotal in this process. In order to decide which pathogens need to be treated, we analyzed results of rapid screening and comprehensive identification defining the amount of bacterial/fungal load with resistance genes detected and presence of predominant bacteria with higher quantitative level (%). Full sequence is based on the national database of 25,000 species. An antibiotics report providing research-based recommendations for antibiotics and antifungal drugs determined by each patient’s specific diagnostic report is presented for each patient.
Culture and Sensitivity Testing
Urine collection for C&S follows the protocol of the Florida Hospital pathology laboratory. Antibiotic susceptibility testing is provided when the following uropathogens are present in significant amounts (>105): gram-negative rods (eg, E coli, K pneumoniae, P aeruginosa) Staphylococcus species, yeasts, β-hemolytic Streptococcus species, Enterococcus species, Aerococcus urinae, Corynebacterium urealyticum, Proteus species, Citrobacter species, Enterobacter species, and Morganella species.
Results
A total of 44 patients and 22 control subjects completed the study, over a 14-day commitment. Twenty-two patients were randomly allocated to each treatment arm (A and B; Table 2). The constituency is as follows: Group A = 14 women and 8 men (treatment based on C&S), and Group B = 15 women and 7 men (treatment based on DNA).
Table 2.
Designations Based on Treatment and by Result
| Group A | Subset 1 | Subset 2 |
| 22 Patients Treatment based on C&S results | (1C&S, 1DNA 7/22) | (2C&S, 1DNA 15/22) patients were treated day 8 based on DNA NGS results |
| Group B | Subset 3 | Subset 4 |
| 22 Patients Treatment based on DNA NGS results | (1DNA, 1C&S 6/22) | (1DNA, 2C&S 16/22) |
C&S, culture and sensitivity; NGS, next-generation sequencing.
In total, 13 of 44 patients had positive urine culture results whereas 44 of 44 patients had positive DNA NGS results. All 15 men with positive laboratory findings were designated as having a complicated UTI, whereas 4 women were given that designation. In total, 19 of 44 patients were determined to have complicated UTIs.
In total, 13 of 44 patients had positive urine culture results whereas 44 of 44 patients had positive DNA NGS results.
The patient symptom severity score was measured at entry for a viable comparison of C&S to DNA-based treatment. Symptom scores ranged from 0 to 21, with 21 being the worst score possible. The mean score for Arm A on entry is 9.00 and Arm B, 10.22. The distribution of scores is similar, with Arm B having a mean score of 1.22 larger than Arm A, but this difference is not significant as discerned from a t-test and Wilcoxon signed-rank test.
For purposes of comparing C&S versus DNA NGS, we compared the change in symptom severity from entry to 14 days completion.
For treatment in Arm A, Subset 1, 7 of the 22 patients exhibited a positive culture result; 15 of 22 patients in Arm A, Subset 2 had negative culture results and positive DNA NGS results. This subset began treatment on day 8 based on DNA NGS results. In contrast, all 22 patients in Arm B had a positive DNA NGS test result. In Arm B, Subset 3, a urine culture test result was positive in 6 of the 22 patients, whereas the remaining 16 patients (Arm B, subset 4) revealed no growth in their urine cultures.
For purposes of comparing C&S versus DNA NGS, we compared the change in symptom severity from entry to 14 days completion. Figure 2 provides the corresponding results, with larger values indicative of greater improvement. The difference in average improvement of 8.5 in Arm B is highly significant, as indicated in Figure 2. On average, scores decreased in Arm A from 9 to 5.3, in Arm B, Subset 3, from 11.2 to 4.3 and in Arm B, Subset 4, from 11.6 to 3.7.
Figure 2.
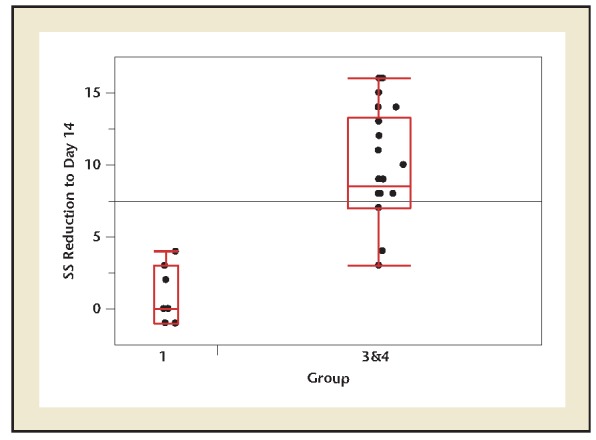
Symptom severity reduction at day 14 in treatment arms. Two-sample t-test P value is <.001. SS, severity score.
Additional information on the DNA NGS performance as a diagnostic test can be gleaned by considering the patients in Arm A, Subset 2, whose culture results were negative. Arm A, Subset 2 had the benefit of a DNA determination that indicated a treatment protocol that was initiated on day 8 of the study. The 15 patients from Arm A, Subset 2 exhibited an average improvement score of 7.4 that was intermediate to the score improvements for Arm B, Subsets 3 and 4 (6.67 and 10.56, respectively) and substantially better than Arm A, Subset 1.
The DNA NGS found on average three microbes in each sample (each registering >2%). DNA testing results revealed 34 of 44 patients with polyorganisms (ie, 2 or more bacteria or fungal organisms). In contrast, 2 of 13 culture reports revealed 2 or more organisms; 20 DNA NGS results revealed anaerobic bacteria either primarily or as part of a polymicrobial sequence.
Of the 22 control subjects, 5 had positive urine culture results and 21 of 22 had positive DNA NGS results. The five subjects with positive C&S and DNA NGS findings all had similar organisms. However, in three of these subjects, DNA NGS results reported two or more organisms in addition to the common one. In patients with symptoms of acute cystitis and positive DNA NGS findings, the average number of organisms present in each sample was 105.89 compared with control subjects who had an average of 105.09 (P <.001; Figure 3).
Figure 3.
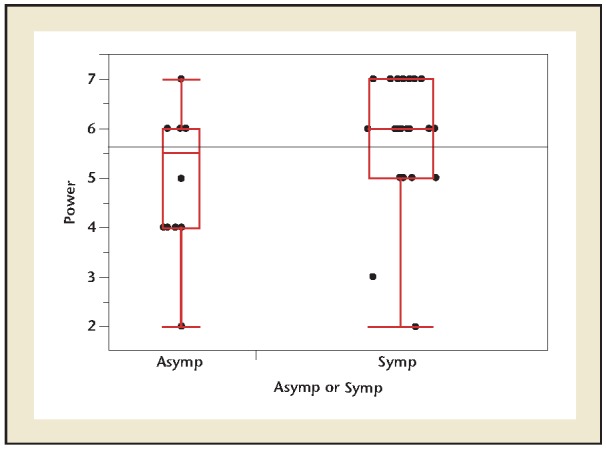
One-way symptomatic versus asymptomatic comparison of power of DNA at entry. Two-sample t-test P value = .0216. Asymp, asymptomatic; Symp, symptomatic.
Of the 22 control subjects, 5 had positive urine culture results and 21 of 22 had positive DNA NGS results.
Discussion
Koch and Petri developed plating and culture models in the 1880s. Considered the gold standard in the diagnosis of UTIs, there are several reasons for re-evaluation of this established technology.15,16 One disadvantage of urine evaluated by culture techniques is contamination.11,17 Urine cultures are considered spoiled if the test yields more than two isolates in quantities ≥10,000 colony forming units (CFU)/mL.18 Contamination of urine ranges from 0.8% to 41.7% based on review of current laboratory practices, whereas the median institution in the United States has a contamination rate of 15.0%.18 DNA NGS revealed a vast majority of specimens in this study (34/44) as multimicrobial (average 2.8), whereas only 2 of 13 culture specimens showed more than one organism.
For typical urine cultures, many laboratories define 105 CFU/mL urine as the threshold. However, this threshold misses many relevant infections.19 There are, therefore, other recommendations that suggest the diagnosis of UTIs from a count of 103 CFU/mL, depending on the types of bacteria detected.9 This will also not identify sexually transmitted diseases, which are prevalent in 10% to 50% of women who present with symptoms of acute cystitis.20,21 No fixed bacterial count can be considered conclusive for significant bacteriuria in all types of UTIs and under all circumstances.1,7,11
DNA NGS reports on bacterial loads as low (102–104), medium, (105–107), and high quantities (>107).7,10 The culture reports in our study were quantified as 105 to 107. Several urine specimens diagnosed with DNA NGS techniques will include many aerobic and anaerobic bacteria with as little as 102 to 104 organisms. Fungal organisms are displayed only as present or absent.
The term antibiotic stewardship was introduced 2 decades ago to search for the best methods necessary to prevent and control the problem of antimicrobial resistance.7,22 Despite an implementation of new antibiotics in clinical practice, the problem of increased resistance reduces the chance of efficient prophylaxis and treatment. It can be achieved by better identification of true uropathogens through better understanding of the urinary tract microbiome. The clinical application of DNA NGS may allow us to improve the targeting of pathogenic bacterial strains while avoiding eradication of beneficial commensal flora.
This study reveals several new concepts in the diagnosis and treatment of acute cystitis. All 44 patients showed positive results in DNA NGS, whereas only 13 of 44 patients had positive urine C&S tests. On a head-to-head comparison, symptom scores were significantly better for those patients whose treatment was based on DNA NGS versus traditional C&S. Patients treated in Arm A, Subset 2, (culture-negative, DNA NGS-positive) improved with respect to symptom scores when they started treatment on day 8.
The clinical application of DNA NGS may allow us to improve the targeting of pathogenic bacterial strains while avoiding eradication of beneficial commensal flora.
We note that 20 of 44 DNA NGS results had anaerobic bacteria; 10 of the 20 patients had an anaerobic bacteria accounting for the primary infectious component of the urine. Another 10 were secondary, associated with aerobic or facultative anaerobic bacteria. Historically, anaerobic UTIs were often overlooked due to the inconsistent use of adequate methods to isolate and identify them. More recently, several studies have confirmed the presence of anaerobic bacteria with the use of 16S ribosomal RNA sequencing in both symptomatic and asymptomatic patients.23-25 This has led to the suggestion of a microbiota (ie, microorganisms in a particular environment), in this case, urine that is either normal or potentially pathologic26,27
The economics of a new diagnostic test are extremely important in establishing an effective tool in treating UTIs.
Participants who were asymptomatic had an average of 105.09 organisms with DNA NGS whereas symptomatic patients averaged 105.89 bacteria—a significant finding. Asymptomatic bacteriuria is common but varies widely in the general population based on age, sex, and the presence of genitourinary abnormalities; its prevalence can range from 1% to 5% in healthy premenopausal women, to 27% in women with diabetes, based on traditional C&S studies.16
The significant limitation of the study is the small sample size, with low statistical power of the results; a larger study needs to be done with a bigger sample size to achieve more robust conclusions of this promising study.
The economics of a new diagnostic test are extremely important in establishing an effective tool in treating UTIs. The cost of the DNA NGS including PCR and 16S ribosomal RNA testing totals US$199. The cost of an aerobic urine culture in the Florida Hospital system is US$60. When including fungal and anaerobic cultures the total cost is over US$500.
Certain clinical scenarios may present an opportunity to utilize DNA NGS in the future. Patients with simple uncomplicated UTIs are normally treated empirically. However, patients who have continued symptoms of acute cystitis despite treatment and with negative urine culture results may benefit from DNA NGS. In those patients with suspected atypical or anaerobic bacteria, DNA NGS may also prove more beneficial in diagnosing a UTI. DNA NGS can help to evaluate for low bacterial loads or resistant infections not otherwise diagnosed with traditional methods.11
Conclusions
Based on a head-to-head comparison, symptom scores were statistically significantly better for those patients whose treatment was based on DNA NGS results versus traditional C&S studies. All 44 patients showed positive results in DNA NGS, whereas only 13 of 44 patients had positive urine C&S results. In the cohort of patients in whom treatment was based on culture results with culture-negative and DNA-positive findings, treatment outcomes were improved with respect to symptom scores when they started treatment on day 8. Ultimately, in this study DNA NGS allowed for better treatment outcomes in patients treated for primary anaerobic, aerobic, or a combination of bacteria. DNA NGS may help when diagnosing and treating symptoms of acute cystitis, especially when urine culture results are negative.
Main Points.
Treatment of uncomplicated urinary tract infections (UTIs) can be based on empiric antibiotic therapy. For complicated or recurrent UTIs, therapy can be based on laboratory-controlled culture and sensitivity (C&S) reports. However, the diagnosis of UTI by clinical criteria alone has an error rate of up to 33%. In addition, positive laboratory culture results do not always indicate a diagnosis of UTI.
Our study was a head-to-head comparative phase II study of standard urine C&S versus DNA next-generation sequencing (NGS) testing for the diagnosis and treatment efficacy in patients with symptoms of acute cystitis.
In our study, 13 of 44 patients had positive urine culture results whereas 44 of 44 patients had positive DNA NGS results.
Despite an implementation of new antibiotics in clinical practice, the problem of increased resistance reduces the chance of efficient prophylaxis and treatment. The clinical application of DNA NGS may allow us to improve the targeting of pathogenic bacterial strains while avoiding eradication of beneficial commensal flora.
As an initial study, results are promising, but much more research is needed to determine if DNA NGS is a diagnostic tool worthy of pursuing. Further research needs to include multi-institutional trials, evidence of improved treatment based on resistant bacterial genes, and the overall economics of the test.
The results of this study were presented at the plenary session of the American Urological Association annual meeting in Boston, Massachusetts, on May 13, 2017.
References
- 1. Badalato G, Kaufmann M. Medical student curriculum: Adult UTI. American Urological Association website. http://www.auanet.org/education/educational-programs/medical-student-education/medical-student-curriculum/adult-uti. Update July 2016. Accessed November 19, 2017.
- 2.Jancel T, Dudas V. Management of uncomplicated urinary tract infections. West J Med. 2002;176:51–55. doi: 10.1136/ewjm.176.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nickel JC. Practical management of recurrent urinary tract infections in premenopausal women. Rev Urol. 2005;7:11–17. [PMC free article] [PubMed] [Google Scholar]
- 4. Grabe M, Bartoletti R, Bjerklund Johansen TE. EAU guidelines on urological infections. European Association of Urology website. http://uroweb.org/guideline/urological-infections/. Accessed November 19, 2017.
- 5. Gupta K, Hooton TM, Naber KG, et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. American Urological Association website. http://www.auanet.org/guidelines/urinary-tract-infection. Accessed November 19, 2017.
- 6.Schmiemann G, Kniehl E, Gebhardt K, et al. The diagnosis of urinary tract infection: a systematic review. Dtsch Arztebl Int. 2010;107:361–367. doi: 10.3238/arztebl.2010.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjerklund Johansen TE, Bonicat G, Cai T, et al. Grey scale in the field of urinary tract infections. Eur UrolFocus. 2016;2:460–462. doi: 10.1016/j.euf.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Kunin CM, White LV, Hua TH. A reassessment of the importance of “low-count” bacteriuria in young women with acute urinary symptoms. Ann Intern Med. 1993;119:454–460. doi: 10.7326/0003-4819-119-6-199309150-00002. [DOI] [PubMed] [Google Scholar]
- 9.Arav-Boger R, Leibovici L, Danon YL. Urinary tract infections with low and high colony counts in young women. Spontaneous remission and singledose vs multiple-day treatment. Arch Intern Med. 1994;154:300–304. [PubMed] [Google Scholar]
- 10.Smelov V, Naber K, Bjerklund Johansen TE. Letter to the Editor: diagnostic criteria in urological diseases do not always match with findings by extended culture techniques and metagenomic sequencing of 16S rDNA. Open Microbiol J. 2016;10:23–26. doi: 10.2174/1874285801610010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smelov V, Naber K, Bjerklund J, ohansen TE. Improved classification of urinary trcat infection: future consideration. Eur Urol Suppl. 2016;15:71–80. [Google Scholar]
- 12.Wagenlehner F, Tandogdu Z, Bartoletti R, et al. The Global Prevalence of Infections in Urology Study: a long-term, worldwide surveillance study on urological infections. Pathogens. 2016;5:E10. doi: 10.3390/pathogens5010010. doi: 10.3390/pathogens5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagenlehner FM, Naber KG. Treatment of bacterial urinary tract infections: presence and future. Eur Urol. 2006;49:235–244. doi: 10.1016/j.eururo.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Clayson D, Wild D, Doll H, et al. Validation of a patient-administered questionnaire to measure the severity and bothersomeness of lower urinary tract symptoms in uncomplicated urinary tract infection (UTI): the UTI Symptom Assessment questionnaire. BJU Int. 2005;96:350–359. doi: 10.1111/j.1464-410X.2005.05630.x. [DOI] [PubMed] [Google Scholar]
- 15.Hooton TM, Roberts PL, Cox ME, Stapleton AE. Voided midstream urine culture and acute cystitis in premenopausal women. N Engl J Med. 2013;369:1883–1891. doi: 10.1056/NEJMoa1302186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolle LE, Bradley S, Colgan R, et al. Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin Infect Dis. 2005;40:643–654. doi: 10.1086/427507. [DOI] [PubMed] [Google Scholar]
- 17.Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis. 2004;38:1150–1158. doi: 10.1086/383029. [DOI] [PubMed] [Google Scholar]
- 18.Bekeris LG, Jones BA, Walsh MK, Wagar EA. Urine culture contamination: a College of American Pathologists Q-Probes study of 127 laboratories. Arch Pathol Lab Med. 2008;132:913–917. doi: 10.5858/2008-132-913-UCCACO. [DOI] [PubMed] [Google Scholar]
- 19.Price TK, Dune T, Hilt EE, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54:1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai T, Gallelli L, Cocci A, et al. Antimicrobial prophylaxis for transrectal ultrasound-guided prostate biopsy: fosfomycin trometamol, an attractive alternative. World J Urol. 2017;35:221–228. doi: 10.1007/s00345-016-1867-6. [DOI] [PubMed] [Google Scholar]
- 21.Tomas ME, Getman D, Donskey CJ, Hecker MT. Overdiagnosis of urinary tract infection and underdiagnosis of sexually transmitted infection in adult women presenting to an emergency department. J Clin Microbiol. 2015;53:2686–2692. doi: 10.1128/JCM.00670-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Høiby N, Bjarnsholt T, Moser C, et al. ESCMID Study Group for Biofilms and Consulting External Expert Werner Zimmerli ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21(suppl 1):S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5:e01283–14. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willner D, Low S, Steen JA, et al. Single clinical isolates from acute uncomplicated urinary tract infections are representative of dominant in situ populations. MBio. 2014;5:e01064–13. doi: 10.1128/mBio.01064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70(suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]