Abstract
Inflammation is known to play a key role in preterm and term parturition. Cell-free fetal DNA (cff-DNA) is present in the maternal circulation and increases with gestational age and some pregnancy complications (e.g. preterm birth, preeclampsia). Microbial DNA and adult cell-free DNA can be pro-inflammatory through DNA-sensing mechanisms such as Toll-like receptor 9 and the Stimulator of Interferon Genes (STING) pathway. However, the pro-inflammatory properties of cff-DNA, and the possible effects of this on pregnancy and parturition are unknown. Clinical studies have quantified cff-DNA levels in the maternal circulation in women who deliver preterm and women who deliver at term and show an association between preterm labor and higher cff-DNA levels in the 2nd, 3rd trimester and at onset of preterm birth symptoms. Together with potential pro-inflammatory properties of cff-DNA, this rise suggests a potential mechanistic role in the pathogenesis of spontaneous preterm birth. In this review, we discuss the evidence linking cff-DNA to adverse pregnancy outcomes, including preterm birth, obtained from preclinical and clinical studies.
Introduction
Preterm birth (PTB), defined as delivery before 37 weeks of gestation, is the leading cause of neonatal morbidity and mortality (Goldenberg et al. 2008, Blencowe et al. 2012). Spontaneous preterm birth (spPTB) contributes to 70% of preterm births (Goldenberg et al. 2008). The pathogenesis of spPTB is largely unknown and worryingly, the incidence of PTB is rising globally (Blencowe et al. 2012). There are no effective therapies or markers to predict PTB. Indeed, only three preventative treatments have proposed potential benefit, and evidence of clinical effectiveness is conflicting (Stock & Ismail 2016). A better understanding of the pathogenesis of spPTB is urgently required to develop more effective therapies.
It is recognized that parturition is an inflammatory event. Inflammatory cells and pro-inflammatory cytokines are found in maternal and fetal tissues during labor (Christiaens et al. 2008, Goldenberg et al. 2008, Bollapragada et al. 2009, Cappelletti et al. 2016). Pro-inflammatory cytokines initiate a cascade of inflammatory mediator production, including matrix metalloproteinases and prostaglandins, which in turn lead to cervical dilation, rupture of membranes and uterine contractions (Christiaens et al. 2008, Goldenberg et al. 2008, Bollapragada et al. 2009). Recently, interest has grown in the potential of Cell-free fetal DNA (cff-DNA) to elicit inflammation and the parturition cascade (Phillippe 2014, Nadeau-Vallée et al. 2016, Herrera et al. 2017).
In 1997, fetal DNA was found in the maternal circulation by quantifying the male SRY gene in 43 different pregnant women (Lo et al. 1997). This new finding led to the development of the non-invasive prenatal test (NIPT), a test used to detect chromosomal abnormalities in the fetus by sampling of maternal blood. More recently, it has been shown that cff-DNA is increased in maternal blood in association with pregnancy complications including early pregnancy loss (Lim et al. 2013), preeclampsia (Contro et al. 2016), fetal growth restriction (Hahn et al. 2005, Taglauer et al. 2014) and preterm labor (Leung et al. 1998, Farina et al. 2005, Illanes et al. 2011, Jakobsen et al. 2012, Sifakis et al. 2015, Quezada et al. 2015, Dugoff et al. 2016). This has led to research into the potential for cff-DNA concentrations to be used as a biomarker for pregnancy complications (Bauer et al. 2006, Sifakis et al. 2015, Contro et al. 2016, Dugoff et al. 2016, Nadeau-Vallée et al. 2016). The putative mechanisms linking cff-DNA to the pathogenesis of pregnancy complications are also a new area of investigation (Scharfe-Nugent et al. 2012, Goulopoulou et al. 2016, Nadeau-Vallée et al. 2016, Conka et al. 2017).
The aim of this review is to examine the role of cff-DNA in the pathogenesis of spPTB. We will present evidence relating to (a) the biological and pro-inflammatory activities of cell-free DNA; (b) cff-DNA concentrations in the circulation of women who deliver preterm and (c) preclinical studies which examine the causative link between cff-DNA and preterm birth.
Cell-free DNA in physiology and pathology
The human body releases cell-free DNA (cfDNA) into the circulation through cell death. In healthy people cfDNA, principally of hematopoietic origin, can be found circulating in both the plasma and serum (Jung et al. 2010). Indeed, circulating cfDNA may have biological functions, such as messaging functions after transcription to RNA (Jung et al. 2010). Although cfDNA is found free in the circulation, the majority is adherent to the surface of blood cells (Jung et al. 2010, Hahn et al. 2014). cfDNA has been shown to have a short half-life in the body and can be cleared as quickly as half an hour (Lo et al. 1999). Detection of cfDNA in healthy individuals is therefore normal, and a cutoff to distinguish ‘pathogenic’ from ‘normal’ values has not yet been established, as values between 6 and 650 ng/mL have been measured in healthy men (Jung et al. 2010). This is in part due to high intra-patient variance, but also due to different methods of cfDNA quantification that are used (Fernando et al. 2010, Jung et al. 2010, Manokhina et al. 2014).
High levels of cfDNA have been reported in infectious, ischemic, malignant and autoimmune diseases, obesity and during pregnancy (Jung et al. 2010, Dwivedi et al. 2012, Nishimoto et al. 2016). This likely relates to high levels of tissue necrosis (e.g. in sepsis) or abnormal high cell turnover in tissues (e.g. from tumors, or from the placenta in pregnancy) (Enninga et al. 2015).
A protective host response to ‘non-self’ or pathogenic DNA (viral or bacterial DNA) is essential for an adequate host defence response and underpins the aspects of our immune systems. However, inappropriate inflammatory responses against ‘self-DNA’ can be detrimental, and break immunological tolerance, leading to autoimmune diseases (Bauer 2006, Ori et al. 2017). Humans therefore have critical mechanisms to discriminate between ‘self’ and ‘non-self’-DNA. Nevertheless, in certain in vivo models, cfDNA originating from tumor cells and from adipocytes has been shown to elicit an inflammatory response on epithelial cells and by attracting macrophages to adipocytes, respectively (Fűri et al. 2015, Nishimoto et al. 2016). These data suggest that under certain conditions cfDNA can bypass protective mechanisms and be pro-inflammatory.
DNA-sensing mechanisms – TLR9 and STING pathways
There are two primary DNA-sensing pathways in cells that have been linked to cfDNA sensing: the Toll-like receptor (TLR) 9 pathway and the Stimulator of Inteferon Genes (STING) pathway (Fűri et al. 2015).
TLR9
TLRs are pattern recognition receptors (PRRs), which activate the innate immune response when they sense damage-associated molecular patterns (DAMPs) (O’Neill et al. 2013). TLR9 is an intracellular receptor that senses DNA, specifically hypomethlated CpG DNA sequences found in high frequency in viral and bacterial DNA. Hypomethylated microbial DNA can be identified by membrane receptors, such as Fc Receptors, and transported to the intracellular compartment where it can be sensed by TLR9 (Bauer 2006). Under normal conditions, adult vertebrate DNA is a poor TLR9 ligand (Marsman et al. 2016). However, placental and fetal DNA is more hypomethylated than adult DNA and may therefore have the potential to be a TLR9 ligand (Scharfe-Nugent et al. 2012, Phillippe 2015). Other changes in DNA morphology can also influence TLR9 affinity, such as bending of the DNA backbone of vertebrate DNA (e.g. by nucleosomes found in cell-free DNA or binding to antibodies or antimicrobial peptides). In addition, when ‘self-DNA’ is artificially transfected into dendritic cells (DC), it can induce a TLR9 response (Yasuda et al. 2006). A number of mechanisms may introduce DNA into the cell cytoplasm (Fig. 1), including binding to antimicrobial peptides (human-beta defensin-3 or human cathelicidin LL-37), anti-DNA antibodies, DNA-like receptors (RAGE) or histones (Nakagawa & Gallo 2015, Marsman et al. 2016, McGlasson et al. 2017).
Figure 1.
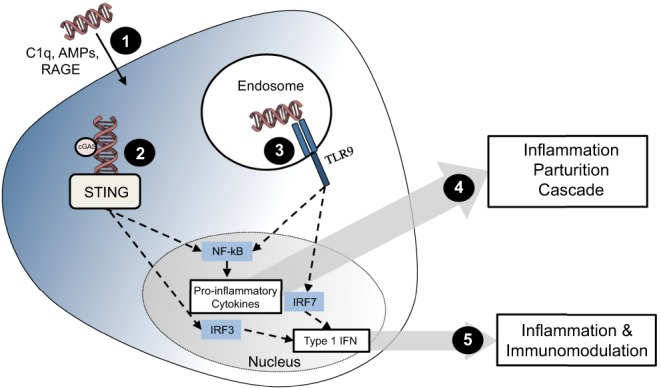
DNA sensing through STING and TLR9. (1) DNA enters the cells through a variety of mechanisms, including interactions with C1q, antimicrobial peptides (AMPs) and receptor for advanced glycation end-products (RAGE). (2) This DNA can then be sensed by binding to STING directly or by firstly biding to cyclic GMP-AMP synthase (cGAS). STING activation produces type 1 interferons through transcription factor interferon receptor factor 3 (IRF3) and, to a lesser extent, pro-inflammatory cytokines via activation of NF-κB. (3) TLR9 is found in the endosome and unmethylated DNA (CpG) or DNA with a modified back bone are typical ligands. TLR9 activation produces type 1 interferons and pro-inflammatory cytokines through IRF7 and NF-κB. (4) Pro-inflammatory cytokines are hypothesized to elicit a potent inflammatory response that can lead to the parturition cascade. (5) Type 1 interferons are known to play a role in inflammation and immunomodulation.
Successful activation of TLR9 by DNA will recruit the myeloid differentiation primary response gene 88 (MyD88). This associates with interleukin-1 receptor-associated kinase (IRAK) 4 and IRAK2 and subsequently recruits tumor-necrosis factor-α receptor factor 6 (TRAF6). This leads to the activation of pro-inflammatory signaling cascades including NF-ĸB activation, by transforming growth factor-β-associated kinase 1 (TAK1), mediated phosphorylates of IĸB (Krieg 2006, Vollmer 2006). These pathways result in changes in gene transcription and the production of pro-inflammatory cytokines (Krieg 2006, Vollmer 2006). The specificity of these responses is cell dependent, with activation of TLR9 in plasmatocytoid DC, for example, resulting primarily in interferon-α production, through recruitment of interferon regulatory factor-7 (IRF-7) (Vollmer 2006, Nakagawa & Gallo 2015).
STING
Activation of the STING pathway has been implicated in viral, bacterial and ‘self’-DNA-mediated stimulation and is independent of TLR9 activation (Barber 2015). STING is an intracellular protein that is expressed in hematopoietic cells (including T cells, macrophages and DC) as well as endothelial and epithelial cells. DNA activates the STING pathway by binding to cyclic GMP-AMP synthase (cGAS) and the subsequent production of cyclic dinucleotides. In addition, double-stranded DNA can directly bind to STING (Fűri et al. 2015). Once activated, STING changes in conformation and complexes with TANK-binding kinase (TBK1). TBK1 in turn phosphorylates transcription factors IRF3 and NF-ĸB. This results in the release of IFN-β and IFN-α, and other pro-inflammatory mediators, dependent upon cell type and stimulation (Barber 2015). Similar to TLR9, DNA must be transported into the cytosol in order to activate STING and these downstream cascades (Fig. 1).
Both TLR9 and STING therefore present mechanisms by which the detection of cfDNA might be pro-inflammatory. However, there is little direct evidence that cff-DNA actually does stimulate TLR9 and whether cff-DNA can elicit inflammation through other pathways, including STING, remains unknown. As such, the effect of these pathways on spPTB remains to be determined.
Cell-free fetal DNA
Cff-DNA originates from the placenta via cell death, likely by apoptosis in normal pregnancies (Masuzaki et al. 2004, Reddy et al. 2008, Manokhina et al. 2015, Nadeau-Vallée et al. 2016) or necrosis during infection (Hahn et al. 2005) in the syntiotrophoblast and cytotrophoblast layers. In addition, an association between cff-DNA and placental microparticles (such as exosomes) is clear, with various studies having found that these microparticles can release cff-DNA and demonstrating fetal DNA in, or bound to, placental microparticles (Orozco et al. 2008, Tong & Chamley 2015). In contrast, cfDNA (from non-fetal origin) is known to be present unbound and free in the plasma (Gold et al. 2015). It is however unknown how much the contribution of placental microparticles (bound or released by) contributes to the total cff-DNA found in the maternal plasma. Recently, placental extracellular vesicles have been shown to activate endothelial cells in a TLR9-dependent manner (Tong et al. 2017). Further clarity on the state of cff-DNA in the maternal circulation is required, as this may influence the manner in which cff-DNA can interact with immune cells and receptors (Tong & Chamley 2015).
In normal pregnancies, cff-DNA steadily increases throughout gestation (Birch et al. 2005) and is cleared as quickly as two hours after delivery of the placenta (Lo et al. 1999). An increase in non-fetal cfDNA was associated with labor, where 75.3% of the cell-free DNA is found to be maternal (quantified by whole-genome bisulfite sequencing) compared to 73.8% in term non-labor (Herrera et al. 2017). The percentage of cff-DNA in total cfDNA is estimated to be about 19–26.2% in normal, non-labouring pregnancies between 10 and 36 weeks (Hui & Bianchi 2011, Herrera et al. 2017). Pregnancies complicated with placental pathologies such as IUGR and preeclampsia have increased amounts of cff-DNA in the maternal circulation (Hahn et al. 2005, Reddy et al. 2008, Taglauer et al. 2014, Contro et al. 2016).
Cell-free fetal DNA concentrations and preterm birth: evidence and study limitations
Within a year of the discovery of cff-DNA in the maternal circulation, high cff-DNA concentrations in maternal circulation were correlated with spPTB (Leung et al. 1998). In this first report, cff-DNA levels were calculated using the SRY gene quantification (whereby a quantification could be done of cff-DNA from women pregnant with a male fetus) method in 32 women with spPTB symptoms. Women who delivered preterm had significantly higher cff-DNA levels than those who did not (Leung et al. 1998). In a similar study, Farina and colleagues quantified cff-DNA levels of 50 women at onset of spPTB symptoms and found an association of increased cff-DNA levels and spPTB deliveries (Farina et al. 2005).
Subsequently, eight studies have investigated the association between cff-DNA and adverse pregnancy outcomes in asymptomatic women, with quantification from first trimester (concurrent with prenatal testing for fetal anueploides) up to 25 weeks of pregnancy (Table 1).
Table 1.
Publications investigating the association between cell-free fetal DNA (cff-DNA) and adverse pregnancy outcomes including spontaneous preterm birth (spPTB).
| Publication | Total N and (spPTB cases) | Study setup | Methods | Main findings |
|---|---|---|---|---|
| Leung et al. (1998) | 32 (20) | Prospective cohort study to assess association with cff-DNA and spPTB | Quantification of SRY gene in maternal plasma at onset of PTB symptoms | Significantly higher SRY detection in women who deliver preterm (P = 0.042). Lower concentration of cff-DNA associated with successful tocolytic therapy (P = 0.017) |
| Farina et al. (2005) | 71 (50) | Cross-sectional study of women at high risk for spPTB | Quantification of DYS1 gene in maternal serum at onset of PTB symptoms | Higher DYS1 detection in women who deliver preterm, by regression analysis of cff-DNA and gestational age at delivery (P = 0.003), significant maker when using cutoff of 1.82 MoM DYS1 gene |
| Bauer et al. (2006) | 84 (7) | Prospective analysis for Cff-DNA as an indicator for adverse pregnancy outcomes | Quantifying the SRY gene and short tandem sequence from maternal plasma at amniocentesis (average 15 weeks) with blood sample | No significant increase in women who later delivered preterm (gestational age of 15.7 ± 0.5 at time of Cff-DNA quantification) |
| Illanes et al. (2011) | 56 (14) | Case-control study to assess cff-DNA and risk of spPTB | DYS gene quantification from maternal plasma at 22–24 weeks in combination with a cervical length measurement | No correlation between cff-DNA levels and gestational age at delivery (r = −0.23; P = 0.07) |
| Stein et al. (2013) | 611 (76) | Prospective cohort study to assess cff-DNA and adverse pregnancy outcome in low risk pregnancies | RhD gene quantification at 25 weeks (mean gestational age at quantification) | No significant increase in women who delivered preterm |
| Jakobsen et al. (2012) | 876 (19) | Prospective cohort study to assess association with cff-DNA and spPTB | RhD gene quantification at 25 weeks of gestation | Strong association between cff-DNA levels above the 95th centile and subsequent spPTB (odds ratio of 6.3; 95% confidence interval: 1.9–20.9) |
| Poon et al. (2013) | 1949 (20) | Prospective cohort study to assess cff-DNA and adverse pregnancy outcome | Chromosome selective assay at 11–13 weeks of gestation | No significant increase in regression analysis (20 deliveries <34 weeks of gestation P = 0.46) |
| Quezada et al. (2015) | 3169 (103) | Cross-sectional study to assess cff-DNA and prediction of spPTB | Fetal Fraction quantified at 10–14 weeks with chromosome selective assay | No significant increase in women who deliver preterm |
| Dugoff et al. (2016) | 1653 (119) | Retrospective cohort study at increased risk for aneuploidy | Methylation method and regional read depth counts from autosomes generated by whole-genome low coverage massively parallel single-end sequencing at 10–20 weeks | Elevated fetal fraction levels at 14.1–20 weeks were significantly associated with incidence of preterm birth (adjusted odds ratio, 4.59; 95% confidence interval, 1.39–15.2) |
| Thurik et al. (2016) | 527 (49) | Nested case-control study to assess cff-DNA and adverse pregnancy outcome | Quantification of DYS14 gene at 8–14 weeks of gestation | No association with spPTB (49, P = 0.19) |
Four studies have quantified cff-DNA in early pregnancy (<16 weeks average gestational age at cff-DNA quantification) (Bauer et al. 2006, Poon et al. 2013, Quezada et al. 2015, Thurik et al. 2016). These included a total of 5729 women with 159 spontaneous preterm deliveries (defined as either <34 (Poon et al. 2013) or <37 weeks (Bauer et al. 2006) of gestation at delivery (Quezada et al. 2015, Thurik et al. 2016)). No association between cff-DNA levels and subsequent spPTB was seen in any of these studies; however, three of the four studies only looked for an association between cff-DNA and overall adverse pregnancy outcomes and were not sufficiently powered to specifically detect an effect of cff-DNA on spPTB. Two of these studies found associations between maternal parameters and cff-DNA. Thurik et al. found an inverse association between cff-DNA and maternal obesity and smoking (Thurik et al. 2016). Bauer et al found an association between increased cff-DNA and fetal abnormalities, HELLP syndrome, IUGR, gestational diabetes (Bauer et al. 2006, Thurik et al. 2016). An important potential confounder in these associations was gestational age at sampling – as neither of these studies adjusted for gestational age in their calculated associations with pregnancy outcomes, despite the well-recognized relationship between gestational age and amount of cff-DNA (Birch et al. 2005). In the largest study of early gestational cff-DNA, Quezada and colleagues were limited to the gestational age of 11–13 weeks and also found no association between cff-DNA and spPTB <34, 34–37 and <37 weeks, although the quantification used combined cff-DNA and cfDNA levels to quantify the fetal fraction (Quezada et al. 2015).
Quantification of cff-DNA levels at later points in gestation have provided conflicting evidence regarding the relationship between cff-DNA and spPTB. Illanes and colleagues investigated the predictive value of cff-DNA for spPTB in 56 women by quantifying the DYS gene (a gene found on the Y chromosome) at 22–24 weeks of gestation in combination with cervical length screening. They found no correlation between cff-DNA levels and gestational age at delivery (P = 0.07) and showed that this was not a predictive marker in combination with cervical length measurement (Illanes et al. 2011), although the numbers of spPTBs were small. Stein and colleagues reached a similar conclusion after quantifying cff-DNA using RhD fetal-specific PCR in 611 low-risk pregnancies between 19 and 32 weeks of gestation. No statistically significant difference in cff-DNA levels was seen in women who developed preeclampsia (n = 44), IUGR (n = 22) or PTB (n = 76) compared to those who had uncomplicated pregnancies (Stein et al. 2013). However, no adjustment for gestational age at sampling was made. A study by Jakobsen and colleagues investigated a larger group of women, but was limited to the gestational age of 25 weeks, found a lower prevalence of spPTB (19 cases) and observed a strong association between cff-DNA levels above the 95th centile and subsequent spPTB (P = 0.002). This raises the possibility that gestational age at sampling might be critical. To assess this, another study directly compared the fetal fraction levels between 11–14 weeks and 14–20 weeks. This quantification calculates the ratio of cff-DNA to total cell-free DNA. In this study, only the latter group showed a significant association between high levels of cff-DNA and the likelihood of spPTB (Dugoff et al. 2016).
It is difficult to conduct a formal comparative analysis and make accurate conclusions of the studies described above due to the substantial heterogeneity, not only in participants, but also in the methodologies used to extract and quantify cff-DNA. Furthermore, given that here is currently no consensus on the normal range of cff-DNA at any point in gestation, adjustments for gestational age are difficult to make.
Participants
Studies had different inclusion and exclusion criteria. Many studies were small, and not able to adjust for variables known to influence the levels of cff-DNA, which include gestational age at sample collection, intra-uterine growth restriction, preeclampsia, smoking status and obesity (Urato et al. 2008, Wang et al. 2013, Taglauer et al. 2014). It will be important that future studies are sufficiently well powered to enable detailed interrogation, and ideally, allow longitudinal sample collection across multiple gestational ages.
Extraction method
The method of cff-DNA extraction is not standardized, and different methods were used in these studies, including commercial DNA extraction kits (Qiagen), quantification directly from the maternal serum or plasma and chemagic magnetic separation. Different blood tubes were used for blood collection and storage (EDTA, sodium citrate, lithium heparin), and cff-DNA was analyzed from either fresh or stored samples, and cff-DNA was quantified from either serum or plasma. Recently, these factors have shown to have an influence on the total cff-DNA yield in different fields (Fernando et al. 2010, Manokhina et al. 2014). Direct comparisons of different methodologies on the same samples and subsequent standardization of approach will be necessary to enable more effective conclusions in the future.
Quantification
A further complication to meta-analysis is the use of different measures of DNA quantity. For example, some studies use the fetal fraction (ratio of fetal DNA to total DNA) while others measure the total amount of cff-DNA in maternal plasma (Manokhina et al. 2015, Dugoff et al. 2016). The method of quantification also varied between studies. Initial studies limited their approach to researching only pregnancies with male fetuses in order to differentiate cff-DNA (DNA from the Y chromosome) from maternal DNA. However, more recent methodologies include using placenta-specific methylation characteristics (Manokhina et al. 2014, Dugoff et al. 2016) or cff-DNA-specific short tandem repeats (Bauer et al. 2006). Both these methods have capacity to quantify cff-DNA irrespective of fetal sex or blood group status. Furthermore, molecular techniques used in quantification have also varied; including PCR, massively parallel single-end sequencing and chromosome selective assays. These differences have the potential to impact the yield of cff-DNA and consequently alter findings.
In summary, cff-DNA has been measured throughout pregnancy. Four out of ten studies concluded a significant increase in cff-DNA levels in women who delivered preterm when compared to women who delivered at term, when measurements were made in the 2nd and 3rd trimester or at the onset of spPTB symptoms. However, these studies have generally looked at small populations with numbers that were insufficient to confirm the prognostic ability of cff-DNA. Nevertheless, these higher levels of circulating cff-DNA detected in later gestation and in labor (including preterm labor in four studies described above), do raise the possibility of role in the onset of parturition (Birch et al. 2005, Phillippe 2014, 2015, Herrera et al. 2017). In this context, in additional to further, more definitive studies, it is important to consider potential mechanisms of action, if indeed this is indeed a causative, not simply associated, phenomenon. It has been hypothesized that cff-DNA is pro-inflammatory and can stimulate, or contribute to the stimulation of, the inflammation-parturition cascade, following detection by intracellular DNA receptors, including TLR9 (Scharfe-Nugent et al. 2012). Studies aiming to evaluate this mechanistically in vivo in the context of pregnancy and spPTB have been conducted in animals (Thaxton et al. 2009, Scharfe-Nugent et al. 2012, Sun et al. 2013, Lin et al. 2014, Conka et al. 2017).
Cell-free fetal DNA as a pro-inflammatory stimulus
We found no studies that have investigated the role of the STING pathway in adverse pregnancy outcomes in vitro or in vivo. However, six studies have investigated the effects of TLR9 activation in pregnancy and parturition in mice, with varied results.
Our group has investigated the effect of time to delivery by intra-uterine injected mouse placental DNA (van Boeckel et al. 2017). Ultrasound-guided intra-uterine injections of 3–300 μg/dam of mouse placental DNA was administered on day 17 to wild-type C57Bl/6 mice. We found no significant decrease in time to delivery in the mouse placental DNA administered groups, while mice treated with 1 μg of LPS all delivered preterm (P < 0.0001), similar to previously published findings (Rinaldi et al. 2015). We examined the effects of cell-free placental DNA in-vitro using DNA extracted from a placental explant method (van Boeckel et al. 2017) and found that up to 500 ng/mL elicited no inflammatory response in peripheral blood mononuclear cells (PBMCs) from pregnant women. Furthermore, we compared the total amount of unmethylated CpG motifs in cff-DNA extracted from the supernatant of human placental explant culture, E-coli DNA and adult human DNA extracted from human blood. We demonstrated the E-coli had 9.1% of unmethylated CpGs compared to 0.06% unmethylated CpGs in cff-DNA and 0% of adult DNA (van Boeckel et al. 2017). This demonstrates that E coli DNA is a better TLR9 ligand compared to cff-DNA and may be a reason for the lack of pro-inflammatory properties seen in our in vitro and in vivo experiments (van Boeckel et al. 2017).
These findings differ from the findings in a 2012 study by Scharfe and colleagues. Scharfe and coworkers used human fetal genomic DNA, which is larger in size than cff-DNA, but their findings suggest fetal DNA is less methylated than human adult DNA (however, no comparison with E coli DNA was made). Using PBMCs from non-pregnant women, they observed that fetal DNA stimulation resulted in the production of the inflammatory cytokine interleukin (IL) 6 with a similar magnitude to that induced by synthetic unmethylated CpG oligonucleotides (CpG). Further activation of TLR9 was demonstrated by IΚBα degradation in both CpG and fetal DNA-stimulated PBMCs. PBMCs from pregnant women showed a significant increase in IL-6 after fetal DNA stimulation, but no comparison to CpG was made. Subsequently, pregnant C57Bl/6 mice were given a single intra-peritoneal injection of 300 μg/dam human fetal DNA between gestational days 10 and 14. There were higher rates of fetal resorption when compared to control groups or mice injected with human adult DNA. This effect was significantly reduced in TLR9-knockout mice. However, despite these findings, this study found no systemic inflammatory response, no true PTB and no in vitro inflammatory response of murine macrophages to fetal DNA (Scharfe-Nugent et al. 2012). Nevertheless, these data implicated a TLR9-mediated effect on fetal viability specific to stimulation with fetal DNA in vivo. TLR9 is strongly expressed in the decidua of spontaneous abortion compared to normal human pregnancies (Kang et al. 2015), making it possible that targeted inflammatory stimuli to the reproductive tract might elicit an inflammatory response leading to PTB. However, the finding that CpG/TLR9 simulation can cause fetal resorption and/or PTB in healthy wild-type mice has not been replicated in subsequent studies in specific genetically modified mouse models.
Thaxton and coworkers found that hypomethylated CpG DNA (CpG) caused adverse pregnancy outcomes in IL-10-deficient mice, in the context of a study investigating the safety profile of CpG as a vaccine adjuvant in pregnancy. Intraperitoneal injections of dams with 25 μg CpG on days 6 or 14 of gestation, induced significant levels of fetal resorption and/or spPTB compared to controls. However, these adverse responses were not seen in wild-type C57Bl/6 mice, indicating that IL-10 had a protective function against harmful responses to, or recognition of this hypomethylated CpG DNA. The CpG-treated IL-10-deficient mice had higher numbers of uterine neutrophils and macrophages than the control IL-10-deficient mice and higher levels of tumor necrosis factor (TNF) in the maternal circulation. Interestingly, the adverse pregnancy outcomes in the CpG-treated IL-10-deficient mice could be reversed by depleting macrophages or by neutralizing maternal TNF, suggesting a macrophage-driven mechanism, occurring in the absence of IL-10-mediated inhibition (Thaxton et al. 2009).
Sun and colleagues used CpG to mimic bacterial DNA and the effects this might have on spontaneous abortion and PTB, in the context of infection. They administered intra-peritoneal injections of CpG (25–400 µg/dam) on gestational days 6 or 14 to wild-type BALB/c and natural killer (NK) cell-deficient non-obese diabetic (NOD) BALB/c mice. NOD mice showed 80–90% rates to CpG-mediated PTB and fetal resorption, while the CpG-treated wild-type mice showed no adverse effects. The adverse effects could be significantly diminished using TLR9 competitive antagonist ODN 2088 or by repletion of NK cells. These data highlight the importance of NK cells and the essential role of TLR9 in the CpG-abortion and PTB pathway (Sun et al. 2013).
In 2014, Lin and colleagues further investigated the ‘CpG-mediated pregnancy failure’ pathway in NOD mice. They use the same model as the Sun and colleagues, and showed that administration of interleukin 10 (IL-10) or T-regulatory cells could prevent the CpG-induced PTB/fetal resorption. These data reinforce the protective importance of IL-10 in these models. In line with the previous findings, wild-type mice did not have any adverse response to the injected CpG (Lin et al. 2014).
The studies described above do suggest that in the context of specific impairments to the normal immune system, CpG can induce adverse pregnancy outcomes. However, the studies were not using fetal DNA (as used by Scharfe and colleagues), nor placental DNA, which is the origin of cff-DNA (Bianchi 2004). In contrast, Čonka and colleagues did include use of DNA from placental origin in a preeclampsia mouse model. They administered daily intra-peritoneal injections of different types of DNA (human fetal, mouse placental and mouse adult DNA) or lipopolysaccharide (LPS) as a positive control once a day during gestation days 14–18 in wild-type C57Bl/6 mice. Whereas LPS induced fetal resorption in all cases (P < 0.001), none of the DNA-injected mice showed significant decreases in litter size. In this model, they hypothesized that TLR9 activation could cause preeclampsia; however, the different types of DNA were not able to induce preeclampsia-like symptoms (Conka et al. 2017). This study therefore contributes to the weight of evidence that systemic exposure of a healthy mouse to mammalian DNA (irrespective of methylation status) cannot induce adverse pregnancy outcomes without additional impairments of the immune system.
In conclusion, DNA has the potential to be pro-inflammatory, through activation of TLR9 and STING pathways. However, whether it can act as a stimulus that directly induces the parturition cascade is unknown, with conflicting in vitro and in vivo data. However, given that cff-DNA is found in the maternal circulation, DNA-sensing mechanisms exist and that these may function incorrectly in pregnancy in the context of additional immune system compromise, further investigation into the pro-inflammatory properties of cff-DNA and the consequences of cff-DNA-induced inflammation in pregnancy is merited.
Summary
In this review, we have summarized the studies that have quantified cff-DNA in the maternal blood and examined those associated with PTB. There is evidence that PTB is associated with increased cff-DNA levels after 20 weeks or with the onset of spPTB symptoms. Further investigation is needed to standardize cff-DNA and fetal fraction quantification and be able to establish a normal range according to gestational age. The increase in cff-DNA suggests potential for a causative link between cff-DNA and spPTB. There are mechanisms by which DNA, and possibly cff-DNA, can be pro-inflammatory. It is however unclear how cff-DNA can reach the intracellular compartment where cell-sensing mechanisms (TLR9 and STING pathway) exist, and whether any consequent TLR9 and STING activation would elicit a response that can lead to spPTB. In vivo studies have given conflicting results and further investigation should be performed using cff-DNA from placental origin and in a manner that the potential pathway of inflammation can be identified. The effects of this pathway on PTB must then be established to be able to prove causative link between cff-DNA and PTB. As DNA can be pro-inflammatory and pregnancy yields in a unique type of cfDNA in the maternal circulation, more research is merited to further unravel the possible inflammatory role of cff-DNA in pregnancy and parturition.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research was made possible by funding from Tommy’s Baby Charity and the Medical Research Council Centre for Reproductive Health. Donald J Davidson was supported by a Medical Research Council Senior Non-clinical Fellowship (G1002046).
References
- Barber GN. 2015. STING: infection, inflammation and cancer. Nature Reviews Immunology 15 760–770. ( 10.1038/nri3921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S. 2006. Toll-erating self DNA. Nature Immunology 7 13–15. ( 10.1038/nbib106-13) [DOI] [PubMed] [Google Scholar]
- Bauer M, Hutterer G, Eder M, Majer S, Leshane E, Johnson KL, Peter I, Bianchi DW, Pertl B. 2006. A prospective analysis of cell-free fetal DNA concentration in maternal plasma as an indicator for adverse pregnancy outcome. Prenatal Diagnosis 26 831–836. ( 10.1002/pd.1513) [DOI] [PubMed] [Google Scholar]
- Bianchi DW. 2004. Circulating fetal DNA: its origin and diagnostic potential-a review. Placenta 25 (Supplement A) S93–S101. ( 10.1016/j.placenta.2004.01.005) [DOI] [PubMed] [Google Scholar]
- Birch L, English CA, O’Donoghue K, Barigye O, Fisk NM, Keer JT. 2005. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clinical Chemistry 51 312–320. ( 10.1373/clinchem.2004.042713) [DOI] [PubMed] [Google Scholar]
- Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L. et al 2012. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet 379 2162–2172. ( 10.1016/S0140-6736(12)60820-4) [DOI] [PubMed] [Google Scholar]
- Bollapragada S, Youssef R, Jordan F, Greer I, Norman J, Nelson S. 2009. Term labor is associated with a core inflammatory response in human fetal membranes, myometrium, and cervix. American Journal of Obstetrics and Gynecology 200 104.e101–111.e101. ( 10.1016/j.ajog.2008.08.032) [DOI] [PubMed] [Google Scholar]
- Cappelletti M, Della Bella S, Ferrazzi E, Mavilio D, Divanovic S. 2016. Inflammation and preterm birth. Journal of Leukocyte Biology 99 67–78. ( 10.1189/jlb.3MR0615-272RR) [DOI] [PubMed] [Google Scholar]
- Christiaens I, Zaragoza DB, Guilbert L, Robertson SA, Mitchell BF, Olson DM. 2008. Inflammatory processes in preterm and term parturition. Journal of Reproductive Immunology 79 50–57. ( 10.1016/j.jri.2008.04.002) [DOI] [PubMed] [Google Scholar]
- Conka J, Konečná B, Lauková L, Vlková B, Celec P. 2017. Fetal DNA does not induce preeclampsia-like symptoms when delivered in late pregnancy in the mouse. Placenta 52 100–105. ( 10.1016/j.placenta.2017.02.008) [DOI] [PubMed] [Google Scholar]
- Contro E, Bernabini D, Farina A. 2017. Cell-free fetal DNA for the prediction of pre-eclampsia at the first and second trimesters: a systematic review and meta-analysis. Molecular Diagnosis and Therapy 21 125 ( 10.1007/s40291-016-0245-9) [DOI] [PubMed] [Google Scholar]
- Dugoff L, Barberio A, Whittaker PG, Schwartz N, Sehdev H, Bastek JA. 2016. Cell-free DNA fetal fraction and preterm birth. American Journal of Obstetrics and Gynecology 215 231.e1–231.e7. ( 10.1016/j.ajog.2016.02.009) [DOI] [PubMed] [Google Scholar]
- Dwivedi DJ, Toltl LJ, Swystun LL, Pogue J, Liaw KL, Weitz JI, Cook DJ, Fox-Robichaud AE, Liaw PC. & Canadian Critical Care Translational Biology Group 2012. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Critical Care 16 R151 ( 10.1186/cc11466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga EA, Nevala WK, Holtan SG, Markovic SN. 2015. Immune reactivation by cell-free fetal DNA in healthy pregnancies re-purposed to target tumors: novel checkpoint inhibition in cancer therapeutics. Frontiers in Immunology 6 424 ( 10.3389/fimmu.2015.00424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, LeShane ES, Romero R, Gomez R, Chaiworapongsa T, Rizzo N, Bianchi DW. 2005. High levels of fetal cell-free DNA in maternal serum: a risk factor for spontaneous preterm delivery. American Journal of Obstetrics and Gynecology 193 421–425. ( 10.1016/j.ajog.2004.12.023) [DOI] [PubMed] [Google Scholar]
- Fernando MR, Chen K, Norton S, Krzyzanowski G, Bourne D, Hunsley B, Ryan WL, Bassett C. 2010. A new methodology to preserve the original proportion and integrity of cell-free fetal DNA in maternal plasma during sample processing and storage. Prenatal Diagnosis 30 418–424. ( 10.1002/pd.2484) [DOI] [PubMed] [Google Scholar]
- Fűri I, Kalmár A, Wichmann B, Spisák S, Schöller A, Barták B, Tulassay Z, Molnár B. 2015. Cell free DNA of tumor origin induces a 'metastatic' expression profile in HT-29 cancer cell line. PLoS ONE 10 e0131699 ( 10.1371/journal.pone.0131699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. 2015. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. Journal of Molecular Diagnostics 17 209–224. ( 10.1016/j.jmoldx.2015.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg RL, Culhane JF, Iams JD, Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371 75–84. ( 10.1016/S0140-6736(08)60074-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulopoulou S, Wenceslau CF, McCarthy CG, Matsumoto T, Webb RC. 2016. Exposure to stimulatory CpG oligonucleotides during gestation induces maternal hypertension and excess vasoconstriction in pregnant rats. American Journal of Physiology-Heart and Circulatory Physiology 310 H1015–H1025. [DOI] [PubMed] [Google Scholar]
- Hahn S, Huppertz B, Holzgreve W. 2005. Fetal cells and cell free fetal nucleic acids in maternal blood: new tools to study abnormal placentation? Placenta 26 515–526. ( 10.1016/j.placenta.2004.10.017) [DOI] [PubMed] [Google Scholar]
- Hahn S, Giaglis S, Buser A, Hoesli I, Lapaire O, Hasler P. 2014. Cell-free nucleic acids in (maternal) blood: any relevance to (reproductive) immunologists? Journal of Reproductive Immunology 104–105 26–31. ( 10.1016/j.jri.2014.03.007) [DOI] [PubMed] [Google Scholar]
- Herrera CA, Stoerker J, Carlquist J, Stoddard GJ, Jackson M, Esplin S, Rose NC. 2017. Cell-free DNA, inflammation, and the initiation of spontaneous term labor. American Journal of Obstetrics and Gynecology 217 583.e1–583.e8. ( 10.1016/j.ajog.2017.05.027) [DOI] [PubMed] [Google Scholar]
- Hui L, Bianchi DW. 2011. Cell-free fetal nucleic acids in amniotic fluid. Human Reproduction Update 17 362–371. ( 10.1093/humupd/dmq049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illanes S, Gomez R, Fornes R, Figueroa-Diesel H, Schepeler M, Searovic P, Serra R, Perez A, Nien JK. 2011. Free fetal DNA levels in patients at risk of preterm labour. Prenatal Diagnosis 31 1082–1085. ( 10.1002/pd.2838) [DOI] [PubMed] [Google Scholar]
- Jakobsen TR, Clausen FB, Rode L, Dziegiel MH, Tabor A. 2012. High levels of fetal DNA are associated with increased risk of spontaneous preterm delivery. Prenatal Diagnosis 32 840–845. ( 10.1002/pd.3917) [DOI] [PubMed] [Google Scholar]
- Jung K, Fleischhacker M, Rabien A. 2010. Cell-free DNA in the blood as a solid tumor biomarker – a critical appraisal of the literature. Clinica Chimica Acta 411 1611–1624. ( 10.1016/j.cca.2010.07.032) [DOI] [PubMed] [Google Scholar]
- Kang X, Zhang X, Liu Z, Xu H, Wang T, He L, Zhao A. 2015. Excessive TLR9 signaling contributes to the pathogenesis of spontaneous abortion through impairment of Treg cell survival by activation of Caspase 8/3. International Immunopharmacology 29 285–292. ( 10.1016/j.intimp.2015.11.004) [DOI] [PubMed] [Google Scholar]
- Krieg AM. 2006. Therapeutic potential of toll-like receptor 9 activation. Nature Reviews Drug Discovery 5 471–484. ( 10.1038/nrd2059) [DOI] [PubMed] [Google Scholar]
- Leung TN, Zhang J, Lau TK, Hjelm NM, Lo YM. 1998. Maternal plasma fetal DNA as a marker for preterm labour. Lancet 352 1904–1905. ( 10.1016/S0140-6736(05)60395-9) [DOI] [PubMed] [Google Scholar]
- Lim JH, Kim MH, Han YJ, Lee DE, Park SY, Han JY, Kim MY, Ryu HM. 2013. Cell-free fetal DNA and cell-free total DNA levels in spontaneous abortion with fetal chromosomal aneuploidy. PLoS ONE 8 e56787 ( 10.1371/journal.pone.0056787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Liu X, Shan B, Wu J, Sharma S, Sun Y. 2014. Prevention of CpG-induced pregnancy disruption by adoptive transfer of in vitro-induced regulatory T cells. PLoS ONE 9 e94702 ( 10.1371/journal.pone.0094702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. 1997. Presence of fetal DNA in maternal plasma and serum. Lancet 350 485–487. ( 10.1016/S0140-6736(97)02174-0) [DOI] [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. 1999. Rapid clearance of fetal DNA from maternal plasma. American Journal of Human Genetics 64 218–224. ( 10.1086/302205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokhina I, Singh TK, Peñaherrera MS, Robinson WP. 2014. Quantification of cell-free DNA in normal and complicated pregnancies: overcoming biological and technical issues. PLoS ONE 9 e101500 ( 10.1371/journal.pone.0101500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokhina I, Wilson SL, Robinson WP. 2015. Noninvasive nucleic acid-based approaches to monitor placental health and predict pregnancy-related complications. American Journal of Obstetrics and Gynecology 213 S197–S206. ( 10.1016/j.ajog.2015.07.010) [DOI] [PubMed] [Google Scholar]
- Marsman G, Zeerleder S, Luken BM. 2016. Extracellular histones, cell-free DNA, or nucleosomes: differences in immunostimulation. Cell Death and Disease 7 e2518 ( 10.1038/cddis.2016.410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzaki H, Miura K, Yoshiura K, Yoshimura S, Niikawa N, Ishimaru T. 2004. Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. Journal of Medical Genetics 41 289–292. ( 10.1136/jmg.2003.015784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlasson SL, Semple F, MacPherson H, Gray M, Davidson DJ, Dorin JR. 2017. Human β-defensin 3 increases the TLR9-dependent response to bacterial DNA. European Journal of Immunology 47 658–664. ( 10.1002/eji.201646799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau-Vallée M, Obari D, Palacios J, Brien MÈ, Duval C, Chemtob S, Girard S. 2016. Sterile inflammation and pregnancy complications: a review. Reproduction 152 R277–R292. ( 10.1530/REP-16-0453) [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Gallo RL. 2015. Endogenous intracellular cathelicidin enhances TLR9 activation in dendritic cells and macrophages. Journal of Immunology 194 1274–1284. ( 10.4049/jimmunol.1402388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Fukuda D, Higashikuni Y, Tanaka K, Hirata Y, Murata C, Kim J-R. 2016. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. Science Advances 2 e1501332 ( 10.1126/sciadv.1501332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Golenbock D, Bowie AG. 2013. The history of Toll-like receptors – redefining innate immunity. Nature Reviews Immunology 13 453–460. [DOI] [PubMed] [Google Scholar]
- Ori D, Murase M, Kawai T. 2017. Cytosolic nucleic acid sensors and innate immune regulation. International Reviews of Immunology 36 74–88. ( 10.1080/08830185.2017.1298749) [DOI] [PubMed] [Google Scholar]
- Orozco AF, Jorgez CJ, Horne C, Marquez-Do DA, Chapman MR, Rodgers JR, Bischoff FZ, Lewis DE. 2008. Membrane protected apoptotic trophoblast microparticles contain nucleic acids: relevance to preeclampsia. American Journal of Pathology 173 1595–1608. ( 10.2353/ajpath.2008.080414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippe M. 2014. Cell-free fetal DNA – a trigger for parturition. New England Journal of Medicine 370 2534–2536. ( 10.1056/NEJMcibr1404324) [DOI] [PubMed] [Google Scholar]
- Phillippe M. 2015. Cell-free fetal DNA, telomeres, and the spontaneous onset of parturition. Reproductive Sciences 22 1186–1201. ( 10.1177/1933719115592714) [DOI] [PubMed] [Google Scholar]
- Poon LC, Musci T, Song K, Syngelaki A, Nicolaides KH. 2013. Maternal plasma cell-free fetal and maternal DNA at 11–13 weeks’ gestation: relation to fetal and maternal characteristics and pregnancy outcomes. Fetal Diagnosis and Therapy 33 215–223. ( 10.1159/000346806) [DOI] [PubMed] [Google Scholar]
- Quezada MS, Francisco C, Dumitrascu-Biris D, Nicolaides KH, Poon LC. 2015. Fetal fraction of cell-free DNA in maternal plasma in the prediction of spontaneous preterm delivery. Ultrasound in Obstetrics and Gynecology 45 101–105. ( 10.1002/uog.14666) [DOI] [PubMed] [Google Scholar]
- Reddy A, Zhong XY, Rusterholz C, Hahn S, Holzgreve W, Redman CW, Sargent IL. 2008. The effect of labour and placental separation on the shedding of syncytiotrophoblast microparticles, cell-free DNA and mRNA in normal pregnancy and pre-eclampsia. Placenta 29 942–949. ( 10.1016/j.placenta.2008.08.018) [DOI] [PubMed] [Google Scholar]
- Rinaldi SF, Makieva S, Frew L, Wade J, Thomson AJW, Moran CM, Norman JE, Stock SJ. 2015. Ultrasound-guided intrauterine injection of lipopolysaccharide as a novel model of preterm birth in the mouse. American Journal of Pathology 185 1201–1206. ( 10.1016/j.ajpath.2015.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfe-Nugent A, Corr SC, Carpenter SB, Keogh L, Doyle B, Martin C, Fitzgerald KA, Daly S, O’Leary JJ, O’Neill LA. 2012. TLR9 provokes inflammation in response to fetal DNA: mechanism for fetal loss in preterm birth and preeclampsia. Journal of Immunology 188 5706–5712. ( 10.4049/jimmunol.1103454) [DOI] [PubMed] [Google Scholar]
- Sifakis S, Koukou Z & Spandidos DA 2015. Cell-free fetal DNA and pregnancy-related complications (review). Molecular Medicine Reports 11 2367–2372. ( 10.3892/mmr.2014.3118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein W, Müller S, Gutensohn K, Emons G, Legler T. 2013. Cell-free fetal DNA and adverse outcome in low risk pregnancies. European Journal of Obstetrics and Gynecology and Reproductive Biology 166 10–13. ( 10.1016/j.ejogrb.2012.09.006) [DOI] [PubMed] [Google Scholar]
- Stock SJ, Ismail KM. 2016. Which intervention reduces the risk of preterm birth in women with risk factors? BMJ 355 i5206 ( 10.1136/bmj.i5206) [DOI] [PubMed] [Google Scholar]
- Sun Y, Qin X, Shan B, Wang W, Zhu Q, Sharma S, Wu J, Lin Y. 2013. Differential effects of the CpG-Toll-like receptor 9 axis on pregnancy outcome in nonobese diabetic mice and wild-type controls. Fertility and Sterility 99 1759–1767. ( 10.1016/j.fertnstert.2013.01.121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglauer ES, Wilkins-Haug L, Bianchi DW. 2014. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 35 S64–S68. ( 10.1016/j.placenta.2013.11.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaxton JE, Romero R, Sharma S. 2009. TLR9 activation coupled to IL-10 deficiency induces adverse pregnancy outcomes. Journal of Immunology 183 1144–1154. ( 10.4049/jimmunol.0900788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurik FF, Lamain-de Ruiter M, Javadi A, Kwee A, Woortmeijer H, Page-Christiaens GC, Franx A, van der Schoot CE, Koster MP. 2016. Absolute first trimester cell-free DNA levels and their associations with adverse pregnancy outcomes. Prenatal Diagnosis 36 1104–1111. ( 10.1002/pd.4940) [DOI] [PubMed] [Google Scholar]
- Tong M, Chamley LW. 2015. Placental extracellular vesicles and feto-maternal communication. Cold Spring Harbor Perspectives in Medicine 5 a023028 ( 10.1101/cshperspect.a023028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M, Chen Q, Cree LM, Chamley LW. 2017. Antiphospholipid antibodies increase the levels of mitochondrial DNA in placental extracellular vesicles: alarming for preeclampsia. Scientific Reports 7 16556 ( 10.1038/s41598-017-16448-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urato AC, Peter I, Canick J, Lambert-Messerlian G, Pulkkinen A, Knight G, Jeong YJ, Johnson KL, Bianchi DW. 2008. Smoking in pregnancy is associated with increased total maternal serum cell-free DNA levels. Prenatal Diagnosis 28 186–190. ( 10.1002/pd.1950) [DOI] [PubMed] [Google Scholar]
- van Boeckel SR, MacPherson H, Davidson DJ, Norman JE, Stock SJ. 2017. Evaluating the Role of Cell-free Fetal DNA in Inflammation and Spontaneous Preterm Birth. bioRxiv . ( 10.1101/191528) [DOI] [Google Scholar]
- Vollmer J. 2006. TLR9 in health and disease. International Reviews of Immunology 25 155–181. ( 10.1080/08830180600743107) [DOI] [PubMed] [Google Scholar]
- Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. 2013. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenatal Diagnosis 33 662–666. ( 10.1002/pd.4119) [DOI] [PubMed] [Google Scholar]
- Yasuda K, Rutz M, Schlatter B, Metzger J, Luppa PB, Schmitz F, Haas T, Heit A, Bauer S, Wagner H. 2006. CpG motif-independent activation of TLR9 upon endosomal translocation of "natural" phosphodiester DNA. European Journal of Immunology 36 431–436. ( 10.1002/eji.200535210) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a