Abstract
Genomic variation in the untranslated region (UTR) has been shown to influence human leukocyte antigen (HLA) class I expression level, and associate with disease outcomes. Sequencing of the 3’UTR of common HLA-A alleles indicated the presence of two polyadenylation signals (PAS). The proximal PAS is conserved, whereas the distal PAS is disrupted within certain alleles by sequence variants. Using 3’ rapid amplification of cDNA ends (3’RACE), we confirmed expression of two distinct forms of the HLA-A 3’UTR based on use of either the proximal or the distal PAS, which differ in length by 100 base pairs. Specific HLA-A alleles varied in the usage of the proximal vs. distal PAS, with some alleles using only the proximal, and others using both the proximal and distal PAS to differing degrees. We show that the short and the long 3’UTR produced similar mRNA expression levels. However, the long 3’UTR conferred lower luciferase activity as compared to the short form, indicating translation inhibition of the long 3’UTR. RNA affinity pull down followed by mass spectrometry (MS) analysis as well as RNA co-immunoprecipitation indicated differential binding of Syncrip to the long vs. short 3’UTR. Depletion of Syncrip by siRNA increased surface expression of an HLA-A allotype that uses primarily the long 3’UTR, whereas an allotype expressing only the short form was unaffected. Further, specific blocking of the proximal 3’UTR reduced surface expression without decreasing mRNA expression. These data demonstrate HLA-A allele-specific variation in PAS usage, which modulates their cell surface expression post-transcriptionally.
Introduction
HLA class I molecules are expressed on virtually all nucleated cells and present peptides to cytotoxic T lymphocytes (CTL), initiating an adaptive immune response. They also serve as ligands for the killer cell immunoglobulin-like receptors expressed on natural killer (NK) cells, thereby regulating NK cell responses. HLA class I genes are the most polymorphic loci in the human genome, and along with the HLA class II genes, they associate with more human diseases than any other locus genome wide (GWAS catalogue; http://www.ebi.ac.uk/gwas). Disease associations with HLA class I variation have largely been attributed to variants encoding polymorphic amino acid positions in the peptide binding groove of the class I molecules. We have shown previously that a polymorphic microRNA (miRNA) binding site in the 3’UTR of HLA-C contributes to allele-specific variation in expression levels and associates with both human immunodeficiency virus (HIV) viral control and risk of Crohn’s disease independently of individual HLA allelic effects(1). More recently, a variant in an Oct1 binding site 800bp upstream of the HLA-C coding region was also shown to regulate HLA-C expression levels(2), and this variant along with the polymorphic miRNA binding site account for 40% of the differential cell surface expression levels of HLA-C. Thus, variation in regulatory regions of the HLA class I loci that affect their expression levels may influence the immune response and disease susceptibility.
The mRNA expression levels of HLA-A alleles were recently shown to vary in an allele-dependent manner as a function of the degree of methylation in the promoter region of each allele (3). Thus, epigenetic mechanisms account for a portion of the differential mRNA expression patterns across HLA-A alleles. Here we describe a novel mechanism, the use of alternative polyadenylation (APA) signals in the 3’UTR of HLA-A, which modulates expression levels of HLA-A protein through regulation of translation. These data underscore the complex regulatory mechanisms that dictate HLA class I expression levels.
Materials and Methods
Samples
Healthy European American (EA) donors recruited at the National Cancer Institute, Frederick, MD were used for determination of HLA-A genotypes and the length of the HLA-A 3’UTR. The respective institutional review boards approved the study, and all subjects gave written informed consent.
HLA genotyping
DNA samples were genotyped for HLA-A, -B, and -C genes by sequence based typing of exons 2 and 3 and/or the polymerase chain reaction (PCR)-sequence-specific oligonucleotide probe typing protocol as recommended by the 13th International Histocompatibility Workshop(4). The entire HLA-A 3’UTR was amplified from genomic DNA by PCR specific primers (Supplemental Table I). The amplicons were sequenced in both directions using the same primers by capillary electrophoresis using an ABI-31730XL DNA analyzer (Applied Biosystems).
3’Rapid amplification of cDNA ends (3’RACE) and quantitative PCR (qPCR)
Peripheral blood was obtained from healthy donors and lymphocytes were separated using lymphocyte separation medium as per manufacturer’s instructions (Lonza). Total RNA was extracted from peripheral blood lymphocytes (PBLs; RNeasy Universal kit, Qiagen). Each sample was treated with gDNA eliminator to remove genomic DNA. The RNAs were quantitated using HT RNA Lab Chip (Caliper, Life Sciences), and all samples had an RNA Quality score of >8. In order to determine the length of the 3’UTR, 3’RACE was performed using a kit (Invitrogen) according to the manufacturer’s instructions. One microgram of RNA was used to initiate the first strand cDNA synthesis at the poly(A) tail of the mRNA. The HLA-A 3’UTR was amplified using a gene specific forward primer (HLA-A 3’UTR 3’RACE Fw) and a universal primer (UAP) that targets the 3’end as the reverse primer (Supplemental Table I). Amplicons were electrophoresed on 2% agarose gels to determine their length, and then they were cloned and sequenced to confirm the specificity of amplification.
The mRNA expression level was measured by quantitative (qPCR). Briefly, reverse transcription was performed with 900ng of total RNA using the high capacity RNA to cDNA kit (Applied Bioscience) in a volume of 10µl. HLA-A coding region [amplified using previously published primers (3)], HLA-A 3’UTR and GAPDH transcripts were amplified (Primer sequences in Table S1) by SYBR green qPCR using the threshold cycle (CT) method(5) in a Viia7 machine (Applied Bioscience). Each qPCR reaction included 6µl of power SYBR green PCR mastermix (Applied Biosystems), 200nM primers that specifically amplified the gene of interest (HLA-A) or the housekeeping gene (GAPDH), and 2 µl of cDNA (1:20 dilution) in a total volume of 12 µl. The genes were amplified using the following conditions: 50°C for 2 minutes, 95°C for 2 minutes followed by 40 cycles of 95°C 15 seconds and 60°C for 30 seconds. The specificity of primers was verified by melt curve analysis using a dissociation step following the qPCR protocol. Specificity of the primers was confirmed by sequencing the HLA-A amplicons. The primers were found to amplify HLA-A specifically and did not cross react with any other locus. The average expression levels of the genes were normalized to that of GAPDH RNA using the 2−ΔΔCt method (5). Oligonucleotide sequences of the primers are listed in Supplemental Table I.
Construction of HLA-A 3’UTR luciferase reporters
The synthetic polyA signal downstream of Renilla luciferase gene in a psicheck2 vector (Promega) was deleted. The complete 3’UTR fragments from two HLA-A alleles (A*01 and A*03) were amplified from genomic DNA and inserted downstream of the Renilla luciferase gene in the modified psicheck2 vector. In order to create a functional distal promoter only (PASD), mutations were introduced in the proximal PAS of HLA-A*03 3’UTR using a site directed mutagenesis kit (Stratagene). Sequential deletions in the PASD sequence were carried out and a strong PAS (AAUAAA) was added at the end of each fragment: 375bp (PASD375), 350bp (PASD350), 325 bp (PASD325) and 300bp (PASD300), respectively. A 25bp region between the proximal and the distal PAS spanning from position 300 to 325 bp (i.e. immediately downstream of the proximal PAS [PASP]) was deleted to construct the PASDΔ300–325 mutant. All mutant amplicons were cloned downstream of Renilla luciferase in the modified psicheck2 vector. Oligonucleotide sequences for the cloning primers used are listed in Supplemental Table I.
Construction of the HLA-A ORF-c-Flag vector
HLA-A ORF with a c-terminal DYKDDK (Flag) tag was amplified using PCR (Genscript clone OHu21196; NM_002116). The Renilla luciferase sequence was replaced by Flag tagged HLA-A ORF in the PASP, PASPD and PASD vectors described above. Oligonucleotide sequences for the cloning primers used are listed in the Supplemental Table I.
Cell lines, cell transfection, luciferase reporter assays and western blots
Transformed B cell lines from three individuals homozygous for HLA-A*11, A*03 or HLA-A*26, and the human T cell line Jurkat were grown in RPMI 1640 (Gibco) medium with 10% heat inactivated fetal bovine serum (FBS; Atlanta Biologicals). PBLs were plated at a density of 1×106 cells/well in a 96 well plate and transfected with 1µM final concentration of either the Apro1 morpholino-oligo or the control morpholino-oligo (Supplemental Table I) using AMAXA 4Dnucleofector (Lonza). The cells were incubated for 36 hours in a 37°C CO2 incubator before determining cell-surface expression of HLA-A. Jurkat cells were plated at a density of 0.5×106 cells/well in a 12 well plate. 1µg/well of the 3’UTR reporter constructs in the modified psichek2 vectors were transfected using the optimized TransIT-X2® (Mirus Bio LLC) protocol. The transfected Jurkat cells were incubated for 18 hours at 37°C in a CO2 incubator. The cells were lysed and the Firefly and Renilla luciferase activity were measured using the Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activity was normalized relative to the Firefly luciferase activity for each transfection. Renilla luciferase activity of each reporter construct is calculated as fold change relative to the normalized activity of plasmid containing the 3’UTR of A*01. All experiments were performed with six replicates in three independent experiments. Jurkat cells transfected with HLA-AORF-Flag plasmids were lysed 48 hours after transfection. Western blot was carried out with DYKDDDK (Flag) tag antibody (Genscript) and anti-firefly luciferase (GenTex) antibody.
Antibodies and flow cytometry
HLA-A expression on the surface of PBLs from an HLA-A*26 homozygous donor was analyzed by staining with a biotinylated monoclonal antibody that specifically binds the HLA-A*26 molecule (BIH0048, One Lambda) used in conjunction with Steptavidin-APC (Biolegend), and expression was measured using an LSRII flow cytometer (BD Biosciences). Transformed B cell lines expressing HLA-A*03 and HLA-A*11 were stained using respective antibodies (BIH0269, One Lambda in conjunction with Streptavidin-APC, Biolegend for A*03; 0544HA, One lambda with PE labelled secondary antibody, Biolegend for A*11) and expression was measured using an AccuriC6 flow cytometer. The histograms were plotted using the FlowJo software version 10.
RNA affinity pull down, polyacrylamide gel electrophoresis PAGE and Western blot analyses
An RNA affinity pull down protocol was adapted from a previously published method (6). The 3’UTR fragments of canonical A*03 (PASPD) and PASDΔ300–325 were in vitro transcribed, labelled with biotin (Biotin labeling mix, Sigma Aldrich) and bound to the Avidin agarose beads (Sigma Aldrich). The RNA coated beads were incubated with Jurkat cell lysate and washed to remove unbound proteins. The proteins captured on the beads were separated with Tris-glycine 4–12% gradient protein gels (Invitrogen) electrophoresis and detected by coomassie-blue stain. Western blot was carried out using anti-Syncrip antibody (Invitrogen).
Identification of proteins by nano LC-tandem Mass Spectrometry (nLC-MS/MS)
The gel bands were excised with gel cutting racked tips and digested with trypsin as described previously (7). Tryptic digests of proteins extracted from the gels (“in-gel” digests), were separated with a reversed-phase column using a linear gradient. Eluted peptides were subjected to reverse-phase micro-capillary nLC-MS/MS analysis using an Eksigent HPLC system (Eksigent) directly interfaced with an Orbitrap LTQ XL mass spectrometer (Thermo Fisher). The eluted ions were analyzed by full precursor MS scans acquired with the FT Orbitrap analyzer operated at a resolving power of 30,000 (400–2,000 m/z). MS spectrum was followed by eight MS/MS spectra, where the eight most abundant multiply charged ions were selected for MS/MS sequencing. Raw data were analyzed with Proteome Discoverer 1.4 (Thermo Fisher; https://www.thermofisher.com/order/catalog/product/IQLAAEGABSFAKJMAUH) software and searched against the SwissProt database restricted to human entries by using the MASCOT http://www.matrixscience.com/help/seq_db_setup_sprot.html) search engine. The precursor-ion tolerance was 10 ppm and the fragment-ion tolerance was 0.8 Da. Enzymatic digestion was specified as trypsin, with up to 2 missed cleavages allowed.
RNA immuno-precipitation (RIP)
EBV transformed B lymphoblastoid cell lines derived from individuals with known HLA-A genotypes (HLA-A*11 or A*03 homozygotes) were transfected with a plasmid encoding Syncrip cDNA with a Myc-tag (RC217902, Origene) or a control plasmid (pcDNA3.1-eGFP, Addgene). For RNA immunoprecipitation of ribonucleoprotein (RNP) complexes from whole-cell extracts, the transfected cells were lysed in 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5mM MgCl2 and 0.5% NP-40 for 10 min on ice and centrifuged at 10,000 RPM for 15 min at 4°C. The supernatants were incubated with magnetic beads coated with anti-Myc antibodies (Invitrogen) overnight at 4°C. After the beads were washed with TBS-T buffer, the complexes were incubated with 20 units of RNase-free DNase I (15 min at 37°C) and further incubated with 0.1% SDS/0.5 mg/ml Proteinase K (15min at 55 °C) to remove DNA and proteins, respectively. The RNPs isolated from the RIP were further assessed by Western blot using an anti-Myc antibody (Invitrogen) for detection of the Myc tagged proteins and qPCR for detection of HLA-A 3’UTR.
Results
Genomic DNA from Caucasian individuals homozygous for common HLA-A alleles were used to amplify and sequence the HLA-A 3’UTR using sequence specific primers. HLA-A 3’UTRs encode two PAS, a canonical polyadenylation signal motif (AATAAA) as well as its common variant (ATTAAA; Supplemental Fig.1). The distal PAS (AATAAA; 395–400bp downstream of the stop codon) was found to be polymorphic and disrupted in some alleles, including HLA-A*01 and A*11 (Supplemental Fig.1), but the proximal PAS (AATAAA; 294–299bp) was conserved across alleles. 3’RACE was carried out using RNA from PBLs of healthy donors, confirming the presence of two distinct forms of the 3’UTR that differ in length by 100bp (Fig. 1), the sequence of which indicated usage of either the proximal PAS (294–299bp; short 3’UTR) or the distal PAS (395–400bp; long 3’UTR). Interestingly, HLA-A alleles vary in expression ratios of the short vs. long forms, with some alleles expressing only the short form due to a polymorphism in the distal PAS (HLA-A*01 & A*11), some expressing predominantly the long form (HLA-A*03), and others expressing both forms to differing degrees. HLA-B and -C alleles, on the other hand, encode only the distal PAS (Supplemental Fig. 2A) and express the long form of 3’UTR (Supplemental Fig. 2B, 2C). The patterns of alternative polyA usage by distinct HLA-A alleles was consistent across CD4+ T cells, CD8+ T cells, and monocytes from peripheral blood, as well as multiple cells lines (Jurkat, HEK293T, transformed B cell lines) (Supplemental Fig. 2B, 2C).
Figure 1. Variation in the relative abundance of the two forms of the HLA-A 3’UTR.
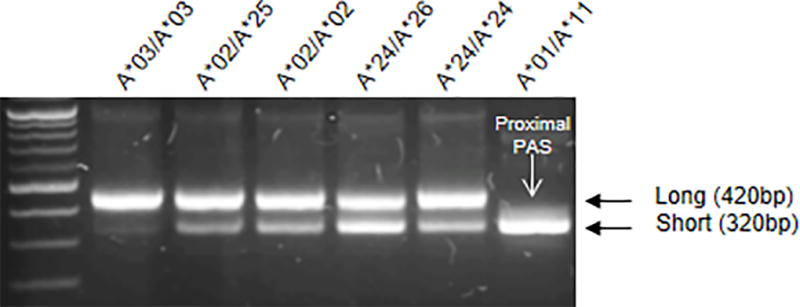
RNA was obtained from peripheral blood lymphocytes of healthy donors. Rapid amplification of cDNA ends (3’ RACE) of 3’UTRs of HLA-A alleles from 6 individuals with distinct HLA-A genotypes showed preferential use of either the proximal or distal PAS. HLA-A*01 and A*11 use only the proximal PAS, resulting in the short form of the 3’UTR, whereas others use various fractions of each. HLA-A genotypes of the individuals are indicated below each amplicon.
To determine if the length of the 3’UTR affects HLA-A mRNA expression levels, constructs were cloned that contained HLA-A 3’UTR sequences downstream of Renilla luciferase in a modified psicheck2 vector where the synthetic PAS downstream of Renilla luciferase was deleted (Supplemental Fig. 3A). Three clones were constructed using the following 3’UTR sequences: HLA-A*01 3’UTR, which has an intact proximal PAS and mutated distal PAS (PASP); A*03 3’UTR, which carries intact proximal and distal PAS (PASPD); and HLA-A*03 3’UTR with an experimentally mutated proximal PAS, which uses only the distal PAS (PASD; Fig. 2A). The three constructs did not differ significantly in mRNA expression levels of luciferase (Fig. 2B). However, expression level of Renilla luciferase protein was a function of the length of the 3’UTR, with the long form (PASD) showing the least luciferase activity, presence of both intact forms (PASPD) showing intermediate activity, and the short form (PASP) producing the greatest luciferase activity (Fig. 2C). We further confirmed the effect of length of the 3’UTR on HLA-A protein expression by replacing the Firefly luciferase sequence with an HLA-A ORF sequence and C-terminal Flag tag (Supplementary Fig. 3B) in the PASP, PASPD and PASD vectors (A-PASD, A-PASPD and A-PASD; Fig. 2D). The HLA-A mRNA expression levels of the three plasmids were similar (Fig. 2E). Concordant with the luciferase assay, however, A-PASD showed the least HLA-A protein expression as determined by Western blot analysis (Fig. 2F). The effect of the length of the 3’UTR on HLA-A protein expression in Jurkat (Fig. 2D–F) was confirmed in HEK293 cells (Supplemental Fig. 3C).
Figure 2. The long HLA-A 3’UTR results in less luciferase activity and protein expression as compared to the short form.
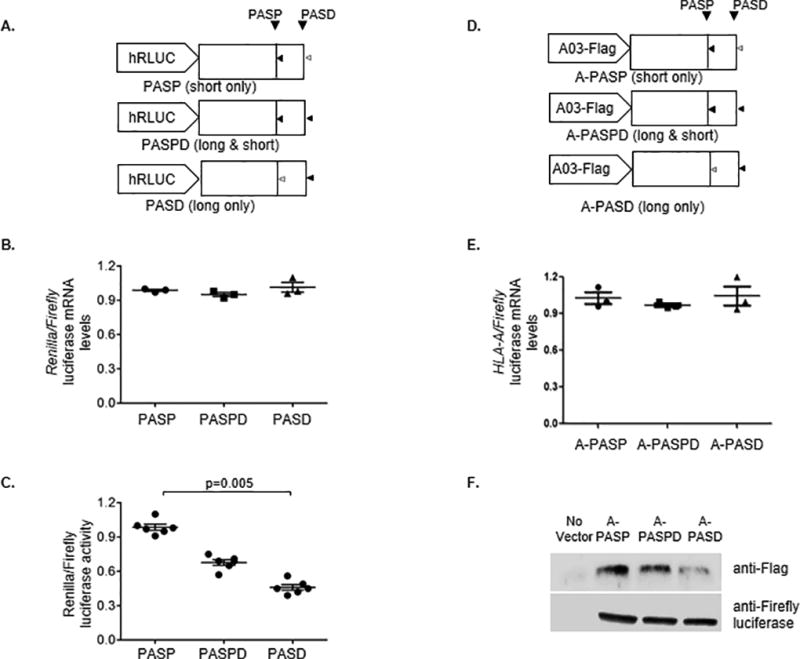
(A) Design of the luciferase reporter constructs used in this study is shown. The active PAS are indicated by black triangles, whereas disrupted PAS are indicated by open grey triangles. (B) The HLA-A 3’UTR luciferase constructs were transfected into Jurkat cell lines. Total RNA was extracted from the transfected cells after 18 hours. Specific primers were used to measure the abundance of Renilla and Firefly luciferase transcripts by qPCR. The normalized mRNA abundance is presented as a relative ratio Renilla/Firefly mRNA. The data represent triplicates in each experimental group. (C) Renilla and firefly luciferase activity was estimated by dual luciferase assays and presented as the normalized ratio of Renilla vs Firefly luciferase activity. The data represent six replicates in each group. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments that were performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated. (D) Design of the HLA-AORF-Flag constructs used in this study is shown. The active PAS are indicated by black triangles, whereas disrupted PAS are indicated by open grey triangles. (E) Jurkat cells were transfected with HLA-AORF-Flag plasmids. Total mRNA was extracted from the transfected cells, and cell lysates were extracted from the transfected cells after 24 hours. Specific primers were used to measure the abundance of HLA-A and Firefly luciferase transcripts by qPCR. The normalized mRNA abundance is presented as a relative ratio of HLA-A/Firefly mRNA. The data represent triplicates in each experimental group. (F) Total cell lysates of Jurkat cells transfected with HLA-AORF-Flag plasmids were used to carry out Western blot analysis with anti-Flag and anti-firefly luciferase antibodies. One of two independent experiments is shown.
In order to determine whether the length of the HLA-A 3’UTR has an effect on endogenous HLA-A surface expression levels, PBLs from an HLA-A*26 homozygous donor were transfected with either an anti-sense morpholino-oligonucleotide that blocks the proximal PAS of HLA-A (APro1) or with a control morpholino (control), and HLA-A mRNA as well as surface expression levels were measured 36 hours later (Fig. 3A). While there was no effect of APro1 on expression levels of HLA-A*26 mRNA (Fig. 3B), an allele that uses both the proximal and distal PAS, cell surface expression of A*26 was significantly decreased upon blockage of the proximal PAS (Fig. 3C). Taken together with the data showing an effect of 3’UTR length on luciferase activity and total protein expression of the constructs without a change in mRNA abundance (Fig. 2), these data indicate that the length of the 3’UTR affects protein expression through a mechanism involving translation of HLA-A, and not through differential stability of HLA-A transcripts.
Figure 3. Blocking the proximal PAS inhibited surface expression of HLA-A on peripheral blood lymphocytes.
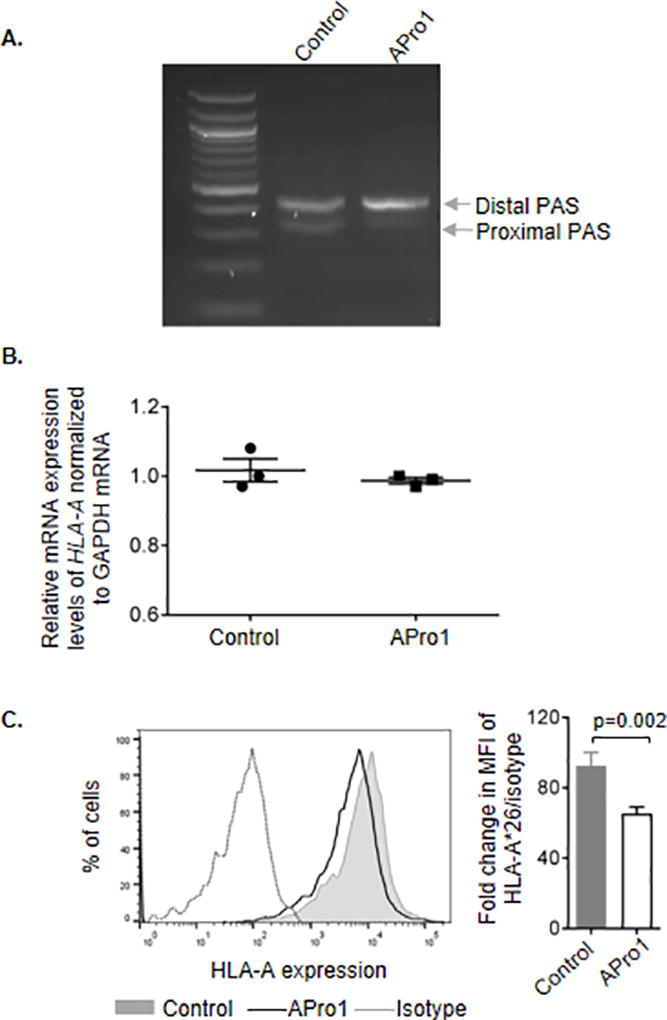
(A) PBLs from an individual expressing A*26 were transfected with either a morpholino-oligonucleotide blocking the proximal PAS of HLA-A (APro1) or with a control morpholino and cultured for 36 hours. Apro1 inhibited the production of the short (proximal PAS) form of the HLA-A 3’UTR, but the control morpholino does not. (B) HLA-A*26 mRNA expression is not affected by blockade of the proximal PAS. (C) Cell surface expression of HLA-A*26 was reduced on cells transfected with APro1 relative to the expression level on cells transfected with the control morpholino. One of three comparable experiments performed is depicted in the histogram. Fold change in expression levels shown in the bar graph was calculated as ratio of mean fluorescence intensity (MFI) of HLA-A*26 vs isotype control. The data represent three replicates in each experimental group. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively, Student t test was used for statistical comparisons and a two tailed p value is indicated.
The lower level of luciferase expression associating with the usage of the distal PAS suggested the presence of a translation inhibitory sequence located between the proximal and distal PAS sequences, which are separated by 100bp (nucleotide positions 300–400 of the 3’UTR). We therefore sequentially deleted 25bp segments of the region between the proximal and distal PAS in the PASD construct, leaving the distal PAS intact, but disrupting the proximal PAS (Fig. 4A). Deletion of the 25bp (PASD375), 50bp (PASD350), and 75bp (PASD325) immediately upstream of the distal PAS resulted in low luciferase expression similar to that of PASD, but deletion of the remaining 25bp (PASD300) resulted in high levels of luciferase expression similar to that observed for PASP, in which only the proximal PAS is intact (Fig. 4B). Deletion of only the 25bp immediately downstream of the proximal PAS and retaining the remaining downstream 75bp (PASDΔ300–325) resulted in high luciferase expression. Together these data point to the presence of an inhibitory sequence located between 300–325bp.
Figure 4. A translation inhibitor sequence is encoded within the 300–325bp segment immediately downstream of the proximal PAS in the HLA-A 3’UTR.
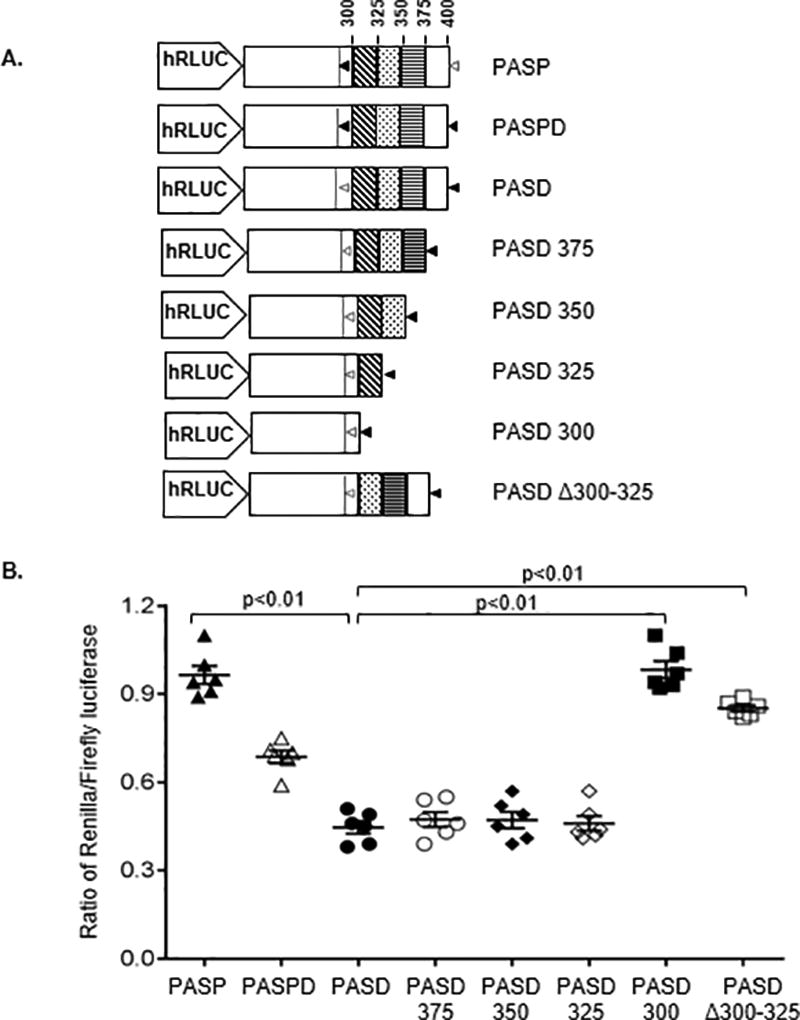
(A) Schematic representations of the luciferase reporter constructs used to identify the region causing inhibition of translation. Portions of PASD, the construct in which the proximal PAS of HLA-A*03 is mutated, were sequentially deleted by 25bp increments with an intact PAS added at the end of each deletion. An intact PAS is shown as black triangles, and a disrupted PAS is shown as open grey triangles. (B) The various 3’UTRs were cloned into the luciferase vector and transfected into Jurkat cells, and luciferase activity of each was compared to the full length, canonical 3’UTRs of HLA-A*01 (PASP) and A*03 (PASPD). Deletion of the 25bp immediately downstream of the disrupted proximal PAS resulted in significantly higher luciferase activity than any other PAS construct in which this 25bp segment was present, indicating the presence of an inhibitory sequence between 300–325bp. The normalized luciferase activity is presented as the relative ratio of Renilla/Firefly luciferase. The data represent six replicates in each experimental group. The mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated.
RNA binding proteins (RBPs)(8, 9) and miRNAs (10, 11) are known to bind the 3’UTR of many mRNA species and regulate their translation. Bioinformatic analyses did not predict a strong miRNA-binding site within the 300–325bp inhibitory sequence, raising the possibility that an RBP may be involved in translation inhibition of the long form. In order to identify the RBP(s) specific for the inhibitory sequence, we performed RNA pull-down experiments. The canonical A*03 3’UTR (PASPD) and the Δ300–325 mutant (PASDΔ300–325) sequences were in vitro transcribed and biotinylated. Jurkat cell lysates were mixed with the biotinylated RNAs, precipitated with streptavidin beads, and separated by PAGE. To identify the RBPs that were precipitated specifically with the canonical A*03 3’UTR (PASPD), we excised a protein band (~70Kd) that was selectively precipitated with the PASPD (Supplemental Fig. 3D). We also excised the corresponding gel regions in the PASDΔ300–325 and the bead only lane. The proteins in the excised gel bands were analyzed by MS, identifying Syncrip as the protein that bound to the canonical long form (Supplemental Fig. 3E; Fig. 5A). Further, the siRNA-mediated knockdown of Syncrip (Fig. 5B) enhanced luciferase activity of the constructs expressing the long form (PASPD and PASD), but had no significant effect on luciferase activity of the construct that is missing the distal PAS (PASP) or is missing the Syncrip binding region (PASDΔ300–325; Fig. 5C).
Figure 5. Syncrip binds the long form of HLA-A 3’UTR and inhibits luciferase activity of the constructs.
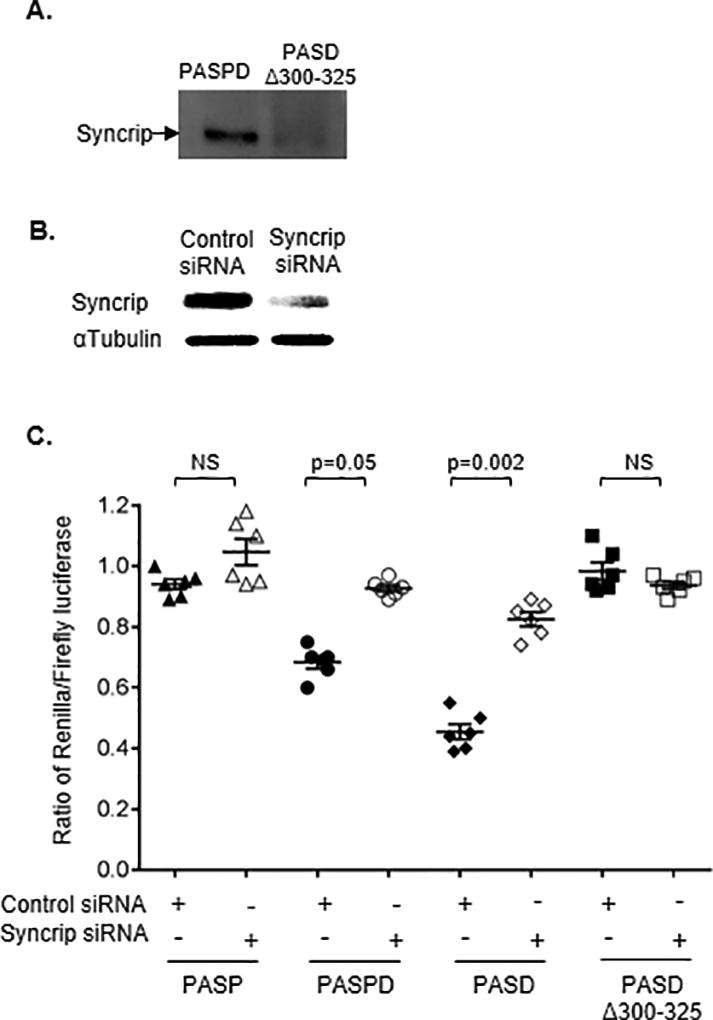
(A) Western blot indicated the presence of Syncrip bound to the long form of HLA-A 3’UTR. RNA affinity pulldown was carried out with the in vitro transcribed 3’UTR fragments of canonical A*03 (PASPD) and PASDΔ300–325, labelled with biotin, and incubated with Jurkat cell lysate. RNA bound protein was separated on PAGE and western blot was carried out using anti-Syncrip antibody.
(B) Jurkat cells were transfected with either an siRNA targeting Syncrip (Syncrip siRNA) or a control siRNA. Knockdown of Syncrip protein was confirmed after 72 hours post-transfection using western blot. (C) Seventy two hours post siRNA transfection, Jurkat cells were transfected with various HLA-A 3’UTR constructs and luciferase activity was measured after 18 hours. Inhibition of Syncrip increased luciferase activity of the constructs encoding the distal PAS (PASPD; PASD). Luciferase activity of the constructs containing only the proximal PAS (PASP) or lacking the inhibitory sequence (PASDΔ300–325) were not affected. The mean ±SE (n=6) are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated.
In order to confirm the interaction between Syncrip and cellular HLA-A 3’UTR, a vector encoding Syncrip tagged with cMyc or a GFP control was transfected into transformed B cell lines (Fig. 6A) from individuals homozygous for HLA-A alleles that use either the long 3’UTR (HLA-A*03) or the short 3’UTR (HLA-A*11), and RIP was performed. The cMyc-tagged Syncrip was immunoprecipitated using anti-Myc antibody coated magnetic beads and specific pull-down was confirmed using Western blot (Fig. 6B). Quantitative amplification (qPCR) of the co-precipitated, protein-bound RNA following RIP with anti-cMyc antibody coated magnetic beads showed that the 3’UTR of the allele expressing the long form (A*03), but not the short form (A*11), co-precipitated with Syncrip (Fig. 6C), confirming the specific association of the long form of 3’UTR with Syncrip. Finally, we transfected B cell lines expressing HLA-A*03 or A*11 with siRNA targeting Syncrip or a control siRNA. Syncrip knockdown was confirmed using Western blot (Fig. 6D). Reduction in Syncrip increased surface expression of HLA-A*03, which uses the long form of the 3’UTR, whereas expression of A*11, which uses only the short form of 3’UTR, was unaffected (Fig. 6E). These data strongly point to Syncrip as the RBP involved in decreased translation of HLA-A alleles that use the distal PAS (i.e. long form).
Figure 6. Differential binding of Syncrip to the long form of HLA-A 3’UTR regulates expression.
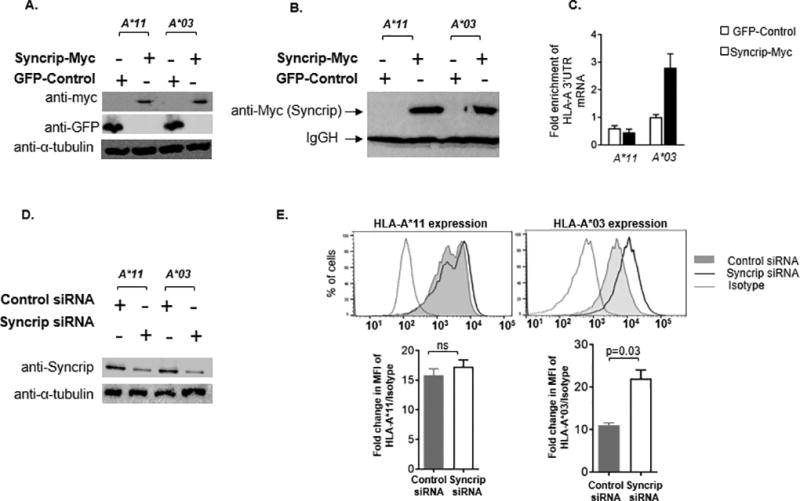
RNA immunoprecipitation was carried out to confirm differential binding of Syncrip to the long 3’UTR of cellular HLA-A mRNA. (A) Transformed B cell lines from individuals with HLA-A*11 or HLA-A*03 homozygous genotypes were transfected with a c-Myc tagged Syncrip encoding vector (Syncrip-Myc) or with pcDNA3-GFP (Control). Cells were lysed after 48 hours and expression of c-Myc tagged Syncrip as well as GFP-control was confirmed using Western blot. (B) RNA immunoprecipitation was performed with the anti-c-Myc antibody coated magnetic beads. Specific pull down of myc-tagged Syncrip was confirmed by Western blot. (C) Fold enrichment of HLA-A 3’UTR in the immunoprecipitated RNA was analyzed by qPCR. (D) EBV transformed B cell lines from individuals with HLA-A*11 or HLA*03 homozygous genotypes were transfected with a Syncrip targeting siRNA and cultured for 72 hours. Syncrip knockdown was confirmed by Western blot. (E) Cell surface expression levels of the HLA-A allotypes treated with the Syncrip siRNA (open black curve) or a control siRNA (gray curve) were measured using allele specific antibodies. Cell surface expression of HLA-A*03 (long form of 3’UTR) was enhanced on the cells transfected with Syncrip targeting siRNA, whereas expression of HLA-A*11 (short form of 3’UTR) remained unaffected. The dotted curves indicate isotype control. Fold change in expression levels was calculated as the ratio of mean fluorescence intensity (MFI) of HLA-A vs isotype control. A histogram of one of three comparable experiments performed is shown for each allele. The mean ±SE (n=3) are depicted as horizontal and vertical bars for each group, respectively. Student t tests were used for statistical comparisons and two tailed p value is indicated.
Discussion
Polyadenylation is essential for the stability, appropriate cellular localization, and efficient translation of mRNA transcripts (12). Transcriptome-wide analyses have revealed that approximately 70% of human genes utilize alternative polyadenylation to generate transcript isoforms with varying lengths of 3’UTRs (13). Here we describe the identification of two distinct forms of the HLA-A 3’UTR that differ in length by 100bp. Multiple cell subsets and cell lines of distinct tissue origins showed consistent alternative polyA usage by the given HLA-A alleles. Using a luciferase reporter assay, we showed that both forms produced similar mRNA expression levels, but the long 3’UTR resulted in lower luciferase activity compared to the short form, suggesting the presence of a motif within the intervening sequence that results in translation inhibition of the long 3’UTR. Sequential deletion of segments within the 100bp intervening sequence mapped the inhibitory sequence immediately downstream of the proximal PAS. 3’UTRs encode docking sites for miRNAs (10, 11) as well as RBPs(8, 9), which are the major determinants of post-transcriptional gene regulation. Several reports indicate a role of 3’UTR associated RBPs in the post-transcriptional regulation of HLA genes (14–17), including another member of the heterogeneous nuclear ribonucleoprotein family, HNRNP-R (17). We identified the RBP Syncrip as the translational inhibitor of the long form of the HLA-A 3’UTR. Depletion of Syncrip increased surface expression of HLA-A*03, an allotype that normally uses the distal PAS (i.e. long 3’UTR), but had no effect on expression level of HLA-A*11, an allotype associated with only the short form. Syncrip (alias HNRNPQ1, Nsap1) is a ubiquitously expressed, cytoplasmic isoform of the RBP HNRNPQ (18–20), and is known to regulate splicing (21–24), editing (25–27), transport (28–32), translation (33–42), and stability (43–48) of mRNA. Syncrip binding sites are enriched in two core consensus sequences (AYAAYY and UAUYRR; Y=C/U and R=A/G) (29) as well as AU rich elements (ARE)(25). HLA-A 3’UTR encodes one ARE (AUUUA; 318–322bp) located within the translation inhibitory sequence that we identified herein. Inherent limitations of the RNA pulldown technique, such as incorrect folding of synthetic biotinylated RNAs that does not allow optimal binding of RBPs and insufficiency of the pulled down protein for detection by MS, may limit identification of all the endogenous RNA bound proteins. In addition, any quantitative differences in total RBPs pulled down with the long or mutant 3’UTR were not considered for this analysis. Thus, additional RBPs acting in association with or independently of Syncrip may have a role in determining which PAS is used, and in translation efficiency of HLA-A mRNA.
The regulation of differential APA is yet unclear and several possible mechanisms have been proposed. Based on bioinformatic analysis or limited functional evidence in cell lines, several factors have been shown to influence the choice of the polyA site, including the strength of the polyA signals (49), expression levels of polyA binding factors(50), sequence motifs surrounding the polyA signals(51), and epigenetic alterations(52–54). Several studies indicate coupling of APA regulation with transcription(55–57), which may be related to the relative increase in short 3’UTRs across the transcriptome observed upon immune cell activation(58). Recently, vesicular stomatitis virus infection was shown to induce shortening of 3’UTRs globally and especially in immune genes (59). The global shortening of 3’UTRs is believed to facilitate rapid change in protein expression as well as increase in proliferation. Thus, this mechanism may enhance the strength of immune responses under certain conditions, such as exposure to pathogens and autoimmune antigens. It would be interesting to determine whether the distinct patterns of PAS usage seen for HLA-A alleles are stable across individuals and tissues, and whether they vary with cell differentiation or activation. For example, HLA-A*03, which predominantly uses the distal PAS, may potentially switch to using the proximal PAS upon exposure to certain pathogens. HLA-A*01 and HLA-A*11 carry variants in the distal PAS that prevent its use, so these alleles express only the short 3’UTR and have no flexibility in their surface expression levels through differential use of PAS. This may pose a disadvantage to individuals carrying these alleles if alternating expression levels of a given allele is beneficial depending on the immune environment at any given point.
Differential HLA expression levels have been associated with infectious (60–63), autoimmune and inflammatory (61, 64–67) disease outcome. Our data indicate that alternative polyadenylation is one mechanism by which HLA-A cell surface expression levels are regulated, potentially affecting HLA-A-mediated immune responses. Blocking of the proximal or distal PAS has the potential to be used as a tool for experimental or therapeutic modulation of HLA-A surface expression in order to regulate HLA-A mediated immune responses.
Supplementary Material
Acknowledgments
We would like to thank the participants from the Research Donor Program at the Frederick National Laboratory for Cancer Research.
The project was supported by Institutional funds from the Texas Biomedical Research Institute and the Ragon Institute of MGH, MIT and Harvard. Veron Ramsuran is supported by the South African Medical Research Council (grant number MRC-RFA-UFSP-01-2013/UKZN-HIVEPI). Sukhvinder Singh is supported by the Cowles post-doctoral fellowship. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, and Center for Cancer Research.
References
- 1.Kulkarni S, Qi Y, O'HUigin C, Pereyra F, Ramsuran V, McLaren P, Fellay J, Nelson G, Chen H, Liao W, Bass S, Apps R, Gao X, Yuki Y, Lied A, Ganesan A, Hunt PW, Deeks SG, Wolinsky S, Walker BD, Carrington M. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20705–20710. doi: 10.1073/pnas.1312237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vince N, Li H, Ramsuran V, Naranbhai V, Duh FM, Fairfax BP, Saleh B, Knight JC, Anderson SK, Carrington M. HLA-C Level Is Regulated by a Polymorphic Oct1 Binding Site in the HLA-C Promoter Region. American journal of human genetics. 2016;99:1353–1358. doi: 10.1016/j.ajhg.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsuran V, Kulkarni S, O'Huigin C, Yuki Y, Augusto DG, Gao X, Carrington M. Epigenetic regulation of differential HLA-A allelic expression levels. Human molecular genetics. 2015;24:4268–4275. doi: 10.1093/hmg/ddv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gokhale A, Kunder R, Goel A, Sarin R, Moiyadi A, Shenoy A, Mamidipally C, Noronha S, Kannan S, Shirsat NV. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. Journal of cancer research and therapeutics. 2010;6:521–529. doi: 10.4103/0973-1482.77072. [DOI] [PubMed] [Google Scholar]
- 5.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 6.Thakor N, Holcik M. RNA Affinity Chromatography. In: Magdeldin DS, editor. Affinity Chromatography. InTech; 2012. [Google Scholar]
- 7.Josic D, Brown MK, Huang F, Lim YP, Rucevic M, Clifton JG, Hixson DC. Proteomic characterization of inter-alpha inhibitor proteins from human plasma. Proteomics. 2006;6:2874–2885. doi: 10.1002/pmic.200500563. [DOI] [PubMed] [Google Scholar]
- 8.Szostak E, Gebauer F. Translational control by 3'-UTR-binding proteins. Brief Funct Genomics. 2013;12:58–65. doi: 10.1093/bfgp/els056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends in cell biology. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends in cell biology. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 12.Sachs A. The role of poly(A) in the translation and stability of mRNA. Current opinion in cell biology. 1990;2:1092–1098. doi: 10.1016/0955-0674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browne SK, Roesser JR, Zhu SZ, Ginder GD. Differential IFN-gamma stimulation of HLA-A gene expression through CRM-1-dependent nuclear RNA export. J Immunol. 2006;177:8612–8619. doi: 10.4049/jimmunol.177.12.8612. [DOI] [PubMed] [Google Scholar]
- 15.Cano F, Bye H, Duncan LM, Buchet-Poyau K, Billaud M, Wills MR, Lehner PJ. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. The EMBO journal. 2012;31:3596–3606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cano F, Rapiteanu R, Sebastiaan Winkler G, Lehner PJ. A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat Commun. 2015;6:8670. doi: 10.1038/ncomms9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, Seliger B, Mandelboim O. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J Immunol. 2016;196:4967–4976. doi: 10.4049/jimmunol.1501550. [DOI] [PubMed] [Google Scholar]
- 18.Harris CE, Boden RA, Astell CR. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J Virol. 1999;73:72–80. doi: 10.1128/jvi.73.1.72-80.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizutani A, Fukuda M, Ibata K, Shiraishi Y, Mikoshiba K. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J Biol Chem. 2000;275:9823–9831. doi: 10.1074/jbc.275.13.9823. [DOI] [PubMed] [Google Scholar]
- 20.Svitkin YV, Yanagiya A, Karetnikov AE, Alain T, Fabian MR, Khoutorsky A, Perreault S, Topisirovic I, Sonenberg N. Control of translation and miRNA-dependent repression by a novel poly(A) binding protein, hnRNP-Q. PLoS Biol. 2013;11:e1001564. doi: 10.1371/journal.pbio.1001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabat JL, Barberan-Soler S, Zahler AM. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J Biol Chem. 2009;284:28490–28497. doi: 10.1074/jbc.M109.023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. The EMBO journal. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 25.Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J Biol Chem. 2001;276:10272–10283. doi: 10.1074/jbc.M006435200. [DOI] [PubMed] [Google Scholar]
- 26.Lau PP, Chang BH, Chan L. Two-hybrid cloning identifies an RNA-binding protein, GRY-RBP, as a component of apobec-1 editosome. Biochem Biophys Res Commun. 2001;282:977–983. doi: 10.1006/bbrc.2001.4679. [DOI] [PubMed] [Google Scholar]
- 27.Quaresma AJ, Oyama S, Jr, Barbosa JA, Kobarg J. The acidic domain of hnRNPQ (NSAP1) has structural similarity to Barstar and binds to Apobec1. Biochem Biophys Res Commun. 2006;350:288–297. doi: 10.1016/j.bbrc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 28.Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, Inoue T, Mikoshiba K. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J Biol Chem. 2004;279:53427–53434. doi: 10.1074/jbc.M409732200. [DOI] [PubMed] [Google Scholar]
- 29.Chen HH, Yu HI, Chiang WC, Lin YD, Shia BC, Tarn WY. hnRNP Q regulates Cdc42-mediated neuronal morphogenesis. Mol Cell Biol. 2012;32:2224–2238. doi: 10.1128/MCB.06550-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 32.McDermott SM, Meignin C, Rappsilber J, Davis I. Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol Open. 2012;1:488–497. doi: 10.1242/bio.2012885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol Cell Biol. 2007;27:368–383. doi: 10.1128/MCB.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duning K, Buck F, Barnekow A, Kremerskothen J. SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J Neurochem. 2008;105:351–359. doi: 10.1111/j.1471-4159.2007.05138.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim DY, Kim W, Lee KH, Kim SH, Lee HR, Kim HJ, Jung Y, Choi JH, Kim KT. hnRNP Q regulates translation of p53 in normal and stress conditions. Cell Death Differ. 2013;20:226–234. doi: 10.1038/cdd.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DY, Kwak E, Kim SH, Lee KH, Woo KC, Kim KT. hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic acids research. 2011;39:8901–8914. doi: 10.1093/nar/gkr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim DY, Woo KC, Lee KH, Kim TD, Kim KT. hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic acids research. 2010;38:7068–7078. doi: 10.1093/nar/gkq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol. 2004;24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol. 2012;32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyabin DN, Nigmatullina LF, Doronin AN, Eliseeva IA, Ovchinnikov LP. Identification of proteins specifically interacting with YB-1 mRNA 3' UTR and the effect of hnRNP Q on YB-1 mRNA translation. Biochemistry (Mosc) 2013;78:651–659. doi: 10.1134/S0006297913060102. [DOI] [PubMed] [Google Scholar]
- 41.Vincendeau M, Nagel D, Brenke JK, Brack-Werner R, Hadian K. Heterogenous nuclear ribonucleoprotein Q increases protein expression from HIV-1 Rev-dependent transcripts. Virol J. 2013;10:151. doi: 10.1186/1743-422X-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xing L, Yao X, Williams KR, Bassell GJ. Negative regulation of RhoA translation and signaling by hnRNP-Q1 affects cellular morphogenesis. Mol Biol Cell. 2012;23:1500–1509. doi: 10.1091/mbc.E11-10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 46.Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moser JJ, Eystathioy T, Chan EK, Fritzler MJ. Markers of mRNA stabilization and degradation, and RNAi within astrocytoma GW bodies. J Neurosci Res. 2007;85:3619–3631. doi: 10.1002/jnr.21439. [DOI] [PubMed] [Google Scholar]
- 48.Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Molecular cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 50.Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 51.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3' end RNA polyadenylation. Genome Res. 2008;18:1224–1237. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Molecular cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreira A. Integrating transcription kinetics with alternative polyadenylation and cell cycle control. Nucleus. 2011;2:556–561. doi: 10.4161/nucl.2.6.18064. [DOI] [PubMed] [Google Scholar]
- 57.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Molecular cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P, Chen S, Xu A. The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun. 2017;8:14605. doi: 10.1038/ncomms14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ, Kirk GD, Hoots WK, Wolinsky S, Carrington M. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M, Mallal S, Nelson G, Bosch R, Heckerman D, Stein JL, Soderberg KA, Moody MA, Denny TN, Zeng X, Fang J, Moffett A, Lifson JD, Goedert JJ, Buchbinder S, Kirk GD, Fellay J, McLaren P, Deeks SG, Pereyra F, Walker B, Michael NL, Weintrob A, Wolinsky S, Liao W, Carrington M. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blais ME, Zhang Y, Rostron T, Griffin H, Taylor S, Xu K, Yan H, Wu H, James I, John M, Dong T, Rowland-Jones SL. High frequency of HIV mutations associated with HLA-C suggests enhanced HLA-C-restricted CTL selective pressure associated with an AIDS-protective polymorphism. J Immunol. 2012;188:4663–4670. doi: 10.4049/jimmunol.1103472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, Pereyra F, Goldstein D, Wolinsky S, Walker B, Young HA, Carrington M. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raj P, Rai E, Song R, Khan S, Wakeland BE, Viswanathan K, Arana C, Liang C, Zhang B, Dozmorov I, Carr-Johnson F, Mitrovic M, Wiley GB, Kelly JA, Lauwerys BR, Olsen NJ, Cotsapas C, Garcia CK, Wise CA, Harley JB, Nath SK, James JA, Jacob CO, Tsao BP, Pasare C, Karp DR, Li QZ, Gaffney PM, Wakeland EK. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife. 2016:5. doi: 10.7554/eLife.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersdorf EW, Gooley TA, Malkki M, Bacigalupo AP, Cesbron A, Du Toit E, Ehninger G, Egeland T, Fischer GF, Gervais T, Haagenson MD, Horowitz MM, Hsu K, Jindra P, Madrigal A, Oudshoorn M, Ringden O, Schroeder ML, Spellman SR, Tiercy JM, Velardi A, Witt CS, O'Huigin C, Apps R, Carrington M T. International Histocompatibility Working Group in Hematopoietic Cell. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood. 2014;124:3996–4003. doi: 10.1182/blood-2014-09-599969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petersdorf EW, Malkki M, O'HUigin C, Carrington M, Gooley T, Haagenson MD, Horowitz MM, Spellman SR, Wang T, Stevenson P. High HLA-DP Expression and Graft-versus-Host Disease. N Engl J Med. 2015;373:599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wissemann WT, Hill-Burns EM, Zabetian CP, Factor SA, Patsopoulos N, Hoglund B, Holcomb C, Donahue RJ, Thomson G, Erlich H, Payami H. Association of Parkinson disease with structural and regulatory variants in the HLA region. American journal of human genetics. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


