Abstract
Microbial single cell analysis has led to discoveries that are beyond what can be resolved with population-based studies. It provides a pristine view of the mechanisms that organize cellular physiology, unbiased by population heterogeneity or uncontrollable environmental impacts. A holistic description of cellular functions at the single cell level requires analytical concepts beyond the miniaturization of existing technologies, defined but uncontrolled by the biological system itself. This review provides an overview of the latest advances in single cell technologies and demonstrates their potential. Opportunities and limitations of single cell microbiology are discussed using selected application-related examples.
Keywords: single cell analysis, heterogeneity, single cell dynamics, microfluidics, technical bias, lab-on-a-chip
Analyzing single microbial cells reveals functionalities inaccessible via bulk-population studies. Studying individual cells requires special tools, but it also opens new horizons. Our current mechanistic understanding of individual cell behavior is still limited by available technologies.
INTRODUCTION
What distinguishes a single and isolated microbial cell from a cell as a member of a microbial population or even a microbiome? The answer to this question is hidden in the cellular functionality of an isolated microbial cell. It is governed by cell internal parameters and the interaction of the cell with its immediate environment, without the influences of cell-to-cell interactions. Scaled down analytical technology and new lab-on-a-chip platforms are starting to reveal previously hidden phenomena in single cell physiology and metabolism. This provides new empirical and holistic access to microbiology and complements synthetic biology—which in turn is building up our understanding from the genome. Single cell studies provide new concepts in regulation and functioning of the genome, proteome and metabolome, inaccessible in bulk population studies. Concepts and discoveries from single cell studies can thus largely complement those from more classical approaches.
The single cell represents the basic functional unit in biology. The cumulative activity of cells comprises the measurable macroscopic output of microbial populations. What remains inaccessible with population analyses is the wide range of individual behavior, which can only be revealed by studying living single cells (Elowitz et al.2002; Lidstrom and Konopka 2010). Individuality is a fundamental feature of any cellular biological system and manifests in cell-to-cell heterogeneity (Lidstrom and Konopka 2010). Even isogenic microbial populations show extensive phenotypic heterogeneity among cells, evident from gene expression patterns, protein levels and metabolic activity (Mueller 2008; Heine et al.2009; Mueller, Harms and Bley 2010; Koch, Harms and Mueller 2014). The underlying mechanisms of cellular individuality are manifold (Ackermann 2015). Besides spontaneous genetic mutations, cell-to-cell differences arise from external perturbations or fluctuations in the cellular surroundings, which result in concerted physiological responses of the cell (Kussell and Leibler 2005; Acar, Mettetal and Van Oudenaarden 2008). Hence, cells will react heterogeneously when they experience individual differences in extracellular conditions due to, for example, spatial gradients (Wang et al. 2010b). However, a wide range of intracellular regulatory processes manifest in phenotypic heterogeneity despite homogeneous environmental conditions (Huh and Paulsson 2011). These processes comprise stochastic effects, multi-stability, regulatory oscillations, or partitioning of central control molecules with low abundance upon cell division (Jahn, Guenther and Mueller 2015). The biological importance of phenotypic heterogeneity has been attributed to increasing population fitness or survival chances upon environmental changes. The extent of phenotypic heterogeneity can also be relevant for efficient technical application of microbes (Arnoldini et al.2012; Arnoldini et al.2014; Delvigne and Goffin 2014).
The identification and differentiation of biological mechanisms underlying phenotypic heterogeneity are very challenging. One example is the classification of phenotypic heterogeneity within populations in a quantitative manner. Due to the lack of a common mathematical formalism for describing heterogeneity, it proves difficult to compare results from different studies or experimental series (Delvigne et al.2017). This fact has to be addressed, for example by using simple biological key figures. The Gini coefficient, a parameter describing general heterogeneity, was recently proposed as a conceivable solution for describing the degree of phenotypic heterogeneity within microbial populations (Westerwalbesloh et al.2017). Our microbiological toolbox needs to be extended to address interdependent parameters and internal vs environmental control mechanisms of cellular functionality. This includes gradients, mass and energy transfer processes and their influence on physiology. New technologies are required to enable the quantitative analysis of single cells in controlled environments (Dusny and Schmid 2015a). Such emerging tools make use of tailored microfluidic environments, where individual cells can be cultivated with reduced bias due to chemical gradients or influences of neighboring cells. Microfluidic systems can be integrated in lab-on-a-chip platforms, enabling integral analysis of single cell physiology (Fritzsch et al.2012; Fig. 1). The application of technologies for analyzing single cells opens up fascinating opportunities. At the same time, as always, these technologies introduce bias due to the peculiarities of the created microhabitats. One has to exercise care to not overinterpret physiological data from single cells, as these might be artifacts from the artificial microhabitat. Carefully designed control experiments can remedy this and reduce the risk of false interpretations. Hence, we also outline current challenges, pitfalls and limitations of single cell technologies.
Figure 1.
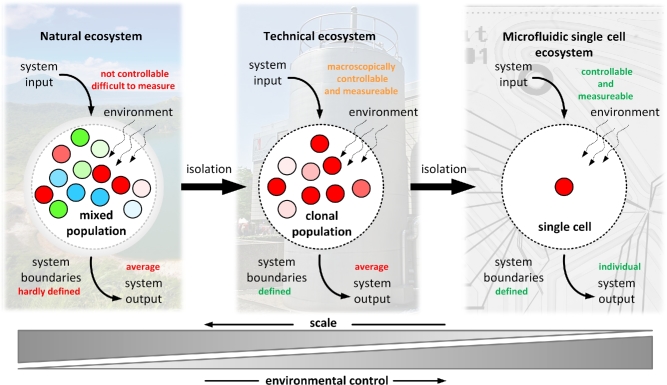
Single cell microbiology represents the final reductionist stage of microbiology and biotechnology. In natural ecosystems, system input and output cannot be quantitatively assigned to catalytic activity of individual community members or classes due to the open nature of the system. In population-based artificial ecosystems, system boundaries are defined and the catalytic activity can be linked to the genetic identity of the population, but again not to individual cells. In single cell ecosystems, environmental conditions can be stringently controlled and linked to single cell activity.
Individual cells can be studied in ‘single cell ecosystems’. Within the single cell ecosystem, the cell is uncoupled from the activity of surrounding cells by means of spatial isolation (Probst et al.2013b). The boundaries of the single cell ecosystem are technically defined by microstructures or microfluidic networks around individual microbes. The cells are put in place to facilitate the determination of the single cell system's input, output and dynamics (Rusconi, Garren and Stocker 2014). The exploitation of physics at the microscale allows control of the physicochemical properties of the extracellular environment to the level of bacterial cells (Weibel, Di Luzio and Whitesides 2007; Westerwalbesloh et al.2015). This control is of importance since it allows differentiating intrinsic from extrinsic factors, or determining origins of individual dynamics (Dusny et al.2012; Nikel et al.2014). Microfluidic concepts have mostly been developed to control individual cell microhabitats. An example is the prediction of mass transfer rates and substrate concentrations within microbial microcolonies with different microfluidic cultivation concepts via simulations (Westerwalbesloh et al.2017).
Microfabrication is now within reach of standard microbiological laboratories and requires only moderate financial and temporal investments. Mature and commercialized microfluidic technologies can be used and several template microbioreactor structures for soft lithography are available (Whitesides et al.2001; Qin, Xia and Whitesides 2010). (For further details on microbioreactor concepts for single cell microbiology, see the excellent reviews of Zare and Kim (2010) and Rusconi, Garren and Stocker (2014).) Despite useful microfluidic structures, the analytical and conceptual challenges for quantitatively analyzing cellular parameters of individual microbial cells are still immense (Schmid et al.2010; Fritzsch et al.2012; Dittrich and Jakubowski 2014). The handling of minute analyte volumes and amounts from single cells necessitates comprehensive adaptions of existing analytical tools. Recent analytics have become available that allow single cell genome, transcriptome or metabolome analysis (Kortmann, Blank and Schmid 2011; Fritzsch et al.2012; Vasdekis and Stephanopoulos 2015). Quantitative time-lapse microscopy, e.g. by applying fluorescent markers, is the most important and widely used analytical method for single cell analysis (Locke and Elowitz 2009). Optical approaches are simple to use and powerful, when the right markers or readouts are given. Physiological dynamics of single cells can be deduced from time-lapse microscopy imaging (Locke and Elowitz 2009). Cellular parameters can be measured that remain hidden in the bulk of a population, such as cell morphology. Many classical microbial physiological parameters like specific growth rates or production and uptake rates are not easily deduced from single cell studies (Gruenberger, Wiechert and Kohlheyer 2014; Dusny and Schmid 2015a,b). Yet, this is necessary for a holistic description of microbial parameters at a single cell level to complement population level studies.
The goal of this review is to provide the reader with an overview of the recent advances in single cell analyses, tools and concepts. We will focus first on studies linking cellular physiology with environmental physicochemical conditions, second on studies addressing biophysical properties of single cells, and third on those characterizing biochemical, metabolic and genomic aspects. Finally, we will describe specific environmental and technological aspects that are needed for single cell studies. Although we use many examples from bacterial single cell studies, the methods and concepts presented are not restricted to those. We do not cover technologies that analyze single cells in bulk populations. What emerges is a picture of single cells in isolation that gives us fascinating new insights into the properties and mechanisms of microbial physiology.
Shaping the environment of single microbes
Single cell microfluidics can stringently control the cellular environment during cultivation and analysis with regard to nutrient and metabolite concentrations, osmolality, pH, shear force, temperature, or other physicochemical parameters (Ong et al.2008; Marques and Fernandes 2011).
For this the cells have to be kept in place during cultivation and analysis. A variety of methods for cell retention have been tested, comprising mechanical, hydrodynamic, electric, optical, acoustic, or magnetic cell manipulation (Fritzsch et al.2012; Lo and Yao 2015). Depending on the design of the microhabitat and the method of trapping, the experimenter can focus on one cell to obtain mechanistic insight or perform parallel experiments with hundreds of single cells. The underlying concepts have been described in detail elsewhere (Mustafi et al.2012; Benavente-Babace et al.2014; Mustafi et al.2014; Riordon et al.2014). Here we will focus on studies that retain single microbes in microstructured habitats for controlled perturbation experiments.
Perfusion enables the complete exchange of medium around isolated cells within seconds. This is a feature that cannot be realized with suspended microbial cultures and constitutes one of the most important aspects of microfluidic single cell analysis for analyzing cellular responses to environmental changes. Single cells can be trapped hydrodynamically in a microfluidic system and perfused with a fluid of defined chemical composition to trigger cell growth or division, perturbation, or metabolic production (Benavente-Babace et al.2014; Mustafi et al.2014; Riordon et al.2014). The principle of hydrodynamic trapping is based on the exploitation of altered fluidic resistances, which can be achieved by the creation of bypass channels with lower flow velocities or by sieve-like physical structures in microchannels (Benavente-Babace et al.2014; Riordon et al.2014; Khalili and Ahmad 2015). Many trapping structures restrict cell growth to a monolayer in one focal plane for optical analysis by matching channel heights and dimensions of microbial cells (Gruenberger et al.2012; Probst et al.2013a; Benavente-Babace et al.2014; Stratz et al.2014; Dusny et al.2015). Such microfluidic devices are particularly useful for massive parallelization of a large number of cell cultivations that can be observed optically. They have been used for measuring and tracking single cell responses to variations in medium composition in a time-resolved manner (Mustafi et al.2012; Gruenberger et al.2013; Probst et al.2013b; Unthan et al.2014). Differences in growth rate, gene expression or size change in single cells can be identified with high throughput. Single cell experiments with hydrodynamic trapping technologies provide statistically safe information on the heterogeneity of biological function among single cells and its interconnection to nutritional status or cell lineage. Further detailed examples will be described in the following sections.
The so called ‘mother machine’ is a prominent example of a hydrodynamic cell trapping structure. It consists of dead-end, cell-sized growth channels that are connected to a main feeding trench for medium supply and removal of surplus cells. Rod-shaped microbes are entrapped in the growth channels, allowing for division and cell elongation (Wang et al.2010a). Dynamics and fates of single mother and daughter cells can be followed before the daughter cells are displaced into the main trench. The mother machine has been used to study the robustness of single cell growth and cell size homeostasis (Wang et al.2010a; Jun and Taheri-Araghi 2015), physical and biochemical properties of chromosomes (Pelletier et al.2012; Youngren et al.2014), stochastic switching of motile cells into a chained, sessile state (Norman et al.2013) and dependency of cell wall growth on mechanical stress (Amir 2014) in Escherichia coli and Bacillus subtilis. Transient oscillations in constitutive gene expression and cell sizing could be analyzed in different E. coli strains during more than 20 000 individual cell cycles (Tanouchi et al.2015, 2017). In fact, the mother machine concept enables unique analyses of cell behavior, molecular partitioning, aging and cell fate in rod-shaped bacteria and fission yeasts. The continuous nature of the device, with supply of fresh medium and constant cell removal, allows long-term single cell growth experiments from days up to weeks for the first time. For example, growth regulation was shown to be robust in a single mother cell pole over hundreds of generations and decoupled from its replicative age.
One disadvantage of physical cell retention structures is that individual cells are influenced by surface contact or metabolic activity of neighboring cells, which can change their behavior (Geng et al.2014). Contactless cell trapping concepts have been developed to avoid this. One of the gentlest concepts for contactless trapping is cell manipulation by negative dielectrophoresis, which was originally developed for trapping larger mammalian cells, but was adapted for smaller microbial cells (yeast and bacteria). The weak electric field enables specific isolation and manipulation (alignment, isolation, trapping) of individual cells (Voldman 2006; Qian et al.2014). One particular embodiment (the so-called ‘Envirostat’ system, for environment–static) provides constant extracellular conditions for isolated cells via perfusion (Kortmann et al.2009a). The extracellular environment is shaped by laminar medium flow (Fritzsch et al.2013; Rosenthal et al.2015). The Envirostat concept enabled the determination of direct connections between the phenotypes of individual cells with their environmental conditions (Dusny et al.2015). It was found that isolated microbes respond to the constant microfluidic environment with higher specific growth rates than observed in populations (Dusny et al.2012). Using the Envirostat, a yeast-specific promoter system was proven to be ultrasensitive to carbon-catabolite repression. Repressing-sugar concentrations for the MOX promoter had hitherto been underestimated by almost four orders of magnitude within populations (Dusny and Schmid 2016). This accurate and quantitative description of promoter regulation could be achieved by decoupling cell and population activity with microfluidics.
Cells can also be manipulated contactlessly and isolated with optical tweezers using a focused laser beam (Zhang and Liu 2008). In contrast to negative dielectrophoresis, optical tweezers cannot be used for retaining and culturing single cells in isolation for longer time periods, because the high laser intensity induces heat and photodamage (Svoboda and Block 1994). Nevertheless, the combined application of optical tweezers and microfluidic cultivation is interesting, because a cell can be relocated to desired zones in the microfluidic system for further cultivation, analysis or enrichment (Wang et al.2011b; Probst et al.2013b). Umehara and coworkers followed growth of E. coli cells in microchambers and relocated daughter cells after cell division into spatially separated microchambers by using optical tweezers (Umehara et al.2003). Growth of the mother cell could be maintained for more than 90 h. It was observed that cells stopped elongation within 20 min independent of their cell cycle when changing from nutrient-rich to nutrient-free medium. The cells started to elongate again upon restoration of nutrient-rich medium within 30 min. This cycle was repeated three times and resulted in consistent adaptation dynamics of dividing cells, granting insight into the connection of growth and nutrient conditions.
The simplest method for single cell cultivation uses semi-solid growth supports such as agarose pads (Reinhard and van der Meer 2010). During cultivation on semi-solid agarose pads, cells are confined between the agarose surface and a glass coverslide. This entails a spatial restriction during cultivation, while nutrients diffuse from the agar to the cells (Young et al.2012). Agarose pads enable the simultaneous culturing of many cells, yet at the expense of limited cell isolation and lack of control of the cellular microenvironment (Dusny et al.2015). This cultivation concept was successfully applied for single cell-based toxicity assays and for cellular differentiation studies (e.g. Reinhard et al. 2013), and constitutes a convenient alternative in terms of throughput and accuracy to conventional but laborious and time-consuming 96-well-plate assays (Li et al.2014b). The spatial distances at which bacteria are typically spread on agarose pads can also be exploited for studying transfer processes between cells. This concept was used to investigate lateral gene transfer associated to integrating and conjugative elements in Pseudomonas (Reinhard et al.2013). Integrating and conjugative elements were found to induce host cell differentiation towards transfer competence in only a small proportion of cells. Limiting transfer competence to a few cells of the population was interpreted as being beneficial for the population fitness status, as integrating and conjugative element horizontal transmission is thus associated with little cost in terms of vertical transmission. This was a previously unknown mechanism for controlling host properties via transferable DNA elements to facilitate horizontal gene transmission.
Dynamics in inhibition of single cells were studied with agarose pads capped with polydimethylsiloxane covers with imprinted microfluidic channels (Li et al.2014a,b). The channels contained different solutions that diffused through the agarose pad and created a concentration profile. Escherichia coli cells were positioned on the agarose surface between source and sink channels and were monitored via microscopy to measure time- and concentration-dependent inhibitory effects of antibiotics on growth (Li et al.2014b). Additional data such as concentration-dependent morphological changes such as filamentation and bulge formation were collected. Furthermore an opportunistic persistence was observed, meaning that inhibited E. coli cells benefited from lysed cells in close proximity, recovered and started to re-grow (Li et al.2014a,b). Given the simplicity of basal cultivation systems such as agarose pads, labor can be focused on revealing biological mechanisms instead of on the sometimes tedious design and implementation of complex microfluidics. However, this only applies to cases where the envisaged studies do not require the stringent control of the cellular surrounding.
Every microsystem has its specific and distinct degrees of analytical freedom. Agarose pads and some of the mentioned hydrodynamic systems allow parallel investigation of many individual microbial cells. This is achieved at the expense of tight environmental control. Single cell perfusion methods, such as the Envirostat technology, have limited throughput, but can be used to more specifically manipulate and control individual cells, enabling mechanistic studies uncoupled from the activity of other cells (Kortmann et al.2009b; Rosenthal et al.2015). However, single cell ecosystems are artificial, and possible biases caused by the specific retention method or by interference of the materials used for constructing the microfluidic device have to be considered carefully.
Environmental impacts on cellular physiology
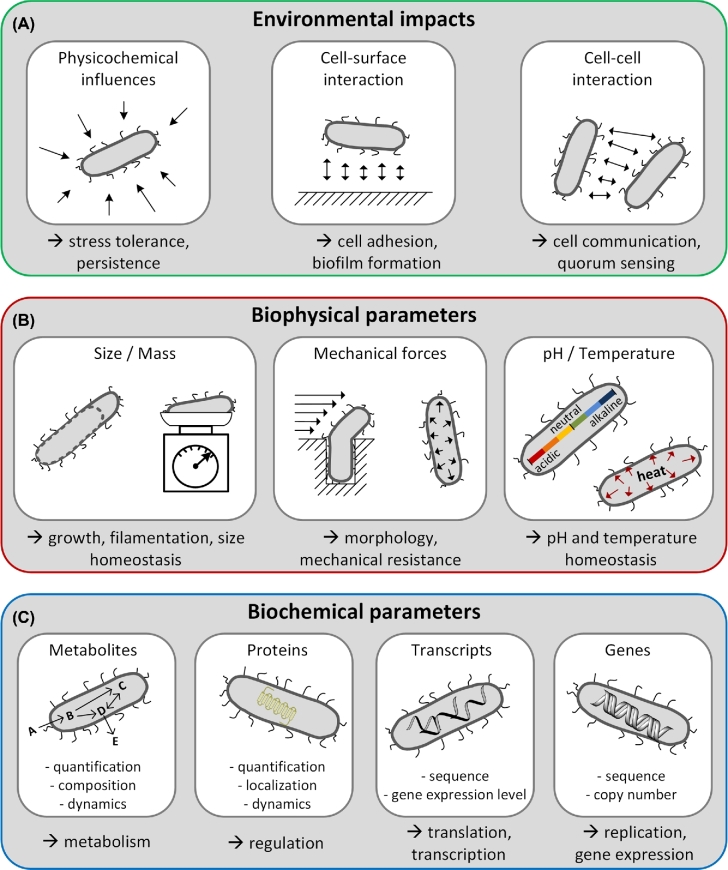
Single cell cultivation technologies enable linking cellular physiology with controlled extracellular physicochemical conditions. Extracellular factors influencing the cell's behavior include, for instance, cell-surface interactions and cell-to-cell interactions. Technologies and corresponding application examples are reviewed next (Fig. 2A).
Figure 2.
Current broad aspects of single cell research. (A) Studies focusing on environmental impacts, such as adhesion or signaling. (B) Studies focusing on biophysical parameters of single cells, such as size, mass and mechanical properties. (C) Studies of biochemical aspects, such as single cell metabolites, proteins or genetics.
Cell-to-cell interactions
In both natural and artificial ecosystems, microbial cells are in close contact and continuously interact with each other (Fig. 2A; Li and Tian 2012; Neu and Lawrence 2015; Schlafer et al.2015). Cell-to-cell interactions comprise sharing metabolites, excreting defense molecules and synchronizing physiological activities (Caro et al.2007; Stepanauskas 2012). Such cell-to-cell interactions were studied with agarose pads with imprinted parallel sub-micrometer growth tracks (Moffitt, Lee and Cluzel 2012). Inserted cells grew in the tracks and outgrowing cells were flushed into a gutter trench. As an example of a symbiotic relationship, growth of two E. coli mutants, each auxotrophic for different amino acids, was followed in parallel tracks. Secreted amino acids diffused through the porous agarose sidewalls of the channels, which allowed mutual exchange of essential metabolites (Moffitt, Lee and Cluzel 2012). The elongation rate of single E. coli cells was dependent on the culture composition and on the spatial distances between both auxotrophic mutants. Auxotrophs separated by distances of less than ∼20 μm grew 3- to 5-fold faster than cells separated by longer distances (Moffitt, Lee and Cluzel 2012). This example has implications for cell-to-cell metabolic interactions and mass transfer for establishing symbiotic lifestyles.
Cell–cell communication by quorum-sensing (QS) and its physiological consequences can be excellently studied at the single cell level (Waters and Bassler 2005; Keller and Surette 2006). QS enables a collective, multicellular organism-like behavior of the population (Bassler and Losick 2006). It is regulated by extracellular signaling molecules called autoinducers. Their levels correlate with cell densities in populations and cells alter gene expression when the autoinducer concentration exceeds or falls below a certain threshold (Waters and Bassler 2005). Examples of some QS-regulated processes are the production of virulence factors or antibiotics, exoproteolytic activity, biofilm formation, bioluminescence production and swarming motility (Hammer and Bassler 2003; Waters and Bassler 2005; Anetzberger, Pirch and Jung 2009; Long et al.2009; Perez and Hagen 2010; Anetzberger, Schell and Jung 2012; Castillo-Juarez et al.2015).
Single cell technologies are useful for understanding the mechanistic principles of QS. Pseudomonas aeruginosa cells have been captured in aqueous droplets for analysis of the variability of QS (Boedicker, Vincent and Ismagilov 2009). The droplets were generated by pumping a suspension with low cell density through a microfluidic channel with tiny wells. Subsequently, an air bubble was introduced that removed excess liquid and formed individual aqueous droplets with a volume of merely 100 fL per well. Each droplet contained one cell or a small number of cells (max. 14) and QS sensing was monitored by a genetically encoded fluorescence reporter (Hentzer et al.2002). The initiation of QS was found to be highly variable among P. aeruginosa cells and even single cells were able to initiate QS on their own when the droplet volume was small enough (Boedicker, Vincent and Ismagilov 2009). QS communication between two cells was monitored with cells trapped in double droplets (Bai et al.2013). A monodisperse emulsion of droplets was created with a microfluidic droplet generator (Bai et al.2010). Droplet pairs were confined in traps to form a droplet interface bilayer, which enabled the diffusion of molecules between the droplets (Bai et al.2013). Two recombinant E. coli strains were investigated, which either secreted or sensed the autoinducer N-(3-oxododecanoyl)-L-homoserine lactone (OdDHL) (Andersen et al.2001; Bai et al.2013). OdDHL sensing was detected with a genetically encoded fluorescence reporter (Andersen et al.2001). The pair of droplets with an OdDHL-producing cell in one and an OdDHL-sensing cell in the other was trapped and successful induction of QS was detected upon diffusion of OdDHL across the droplets. With this approach, intra-species QS was proven at the single cell level for the first time. Unfortunately, these droplet technologies for studying cell–cell interactions are currently not applicable for high throughput as they are limited to the simultaneous analysis of maximally two droplets.
Microscopy studies of individual cells in growing populations of bioluminescent Vibrio harveyi and Vibrio fischeri revealed QS heterogeneity (Anetzberger, Pirch and Jung 2009; Perez and Hagen 2010; Plener et al.2015). QS in V. harveyi and V. fischeri is regulated by the lux operon (Fig. 3; Anetzberger, Schell and Jung 2012) leading to bioluminescence as a direct output of the lux regulatory cascade (Plener et al.2015). Light intensities varied between individual cells and variability increased with increasing cell densities. Individual cells of cultures with a high cell density exhibited increased bioluminescence compared with those from cultures with low cell densities. Furthermore, bioluminescence was more heterogeneous in the case of V. harveyi, which also produced more biofilm (Anetzberger, Pirch and Jung 2009). Thus, QS is related to individual bioluminescence and biofilm formation. Autoinducers were also found to be involved in the formation of heterogeneous gene expression in clonal populations of V. harveyi (Fig. 3; Anetzberger, Schell and Jung 2012). Expression of luxC, the first gene of the fatty acid reductase complex of the lux operon, was quantified by fluorescence microscopy targeting several bioluminescence-related genes fused to the green fluorescent protein gene. The number of cells expressing luxC increased over the cultivation period. Furthermore, induction of luxC expression of individual population members was heterogeneous (Anetzberger, Schell and Jung 2012). The knowledge obtained about QS mechanisms can be utilized to manipulate microbial communication. This is particularly important to avoid one species taking over control in a microbial multispecies community, e.g. in a number of bacterial pathogenic processes (Vikram et al.2011) or controlling population compositions in waste water treatment (Lade, Paul and Kweon.2014a,b).
Figure 3.
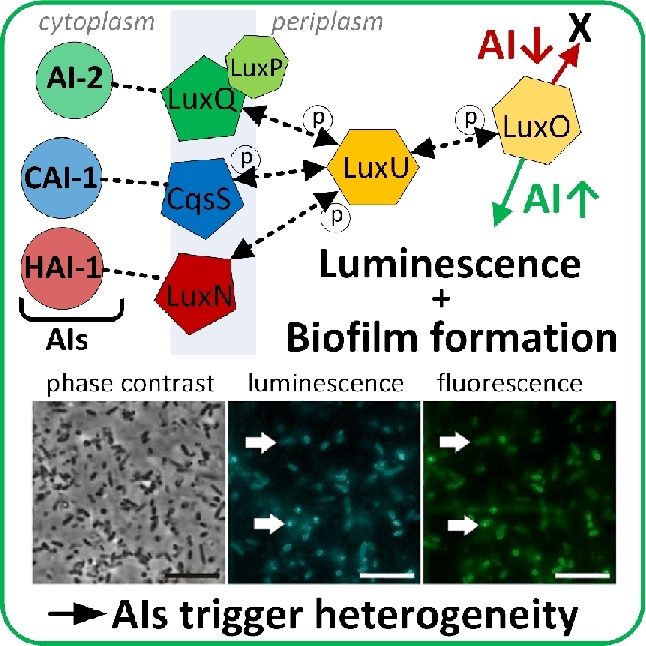
Single cell heterogeneity as a response to auto inducer molecules (AIs). The images show bioluminescent cells of V. harveyi that respond to the presence of AIs with bioluminescence and biofilm formation. The length of the scale bar is 2.5 μm. (Adapted from Anetzberger, Schell and Jung 2012 with permission from BioMed Central.)
Tolerance and adaptation
Mechanisms for adaptation during environmental stress exposure ensure the survival of microbial populations. Bet-hedging is a mechanism that involves stochastic switching of phenotypes, and the transition between phenotypes is actively induced upon recognition of a stress signal (Beaumont et al.2009). A bimodal on/off-switching of gene expression supports the formation of two subpopulations, and the switching rate is variable among individual population members (Balaban et al.2004; Acar, Mettetal and Van Oudenaarden 2008; Nikel et al.2014). The resistant subpopulation has a higher survival probability when environmental perturbations occur. A more complex counter-mechanism to stress was reported for eukaryotic cells based on switching among several phenotypes (Levy, Ziv and Siegal 2012). Various subpopulations are formed prior to the appearance of a stress signal due to stochastic gene expression and deterministic factors, which results in at least a small fraction of stressed cells that are able to survive (Balaban et al.2004; Nikel et al.2014; Martins and Locke 2015).
As an example for bacterial microorganisms, Streptococcus pneumoniae has evolved several co-existing phenotypes that enable it to resist the effect of antibiotics (Sorg and Veening 2015). Exposure of S. pneumoniae cultures to eight different bacteriostatic or bactericidal antibiotics resulted in distinct inhibition profiles: the cells grew unimpaired for certain time periods after exposure to the bactericides, but the periods of unimpaired growth became shorter as the bactericide concentrations increased. Some of the cells responded with a complete shutdown of gene expression activity, and a large cell fraction was irreparably damaged or died during growth arrest. The addition of bacteriostatic compounds led to reduced growth velocities. Growth-arrested cells exhibited higher metabolic activities after exposure to bacteriostatic antibiotics than untreated cells. Bacteriostatics and bactericides provoked different types of adaptation mechanisms, indistinguishable at a population scale. The recovery times of cells exposed to bacteriostatics (indicated by longer lag phases) scaled with exposure time. The extended lag phases of the cultures treated with bacteriostatics were associated with increased phenotypic heterogeneity. Cells whose parental cell metabolism had adapted to the bactericide cephalexin displayed a lower susceptibility to this antibiotic. These cells survived and resumed growth although most cells died after cephalexin treatment (Sorg and Veening 2015). It thus appears that pre-adaptation of the culture to a certain antibiotic might facilitate stochastic switching between phenotypes. We learn from these examples that cellular responses to stress cannot be fully resolved with suspended cultures.
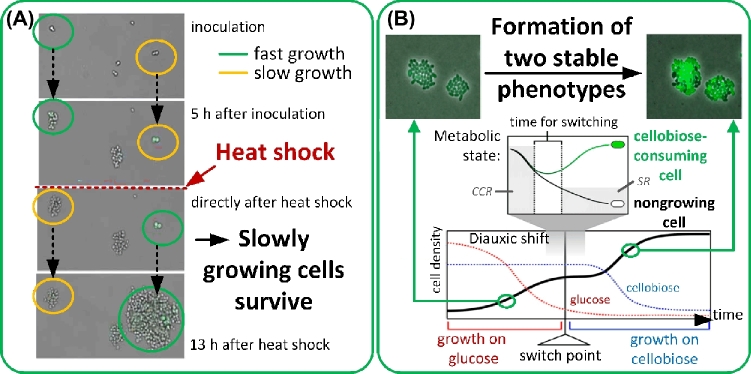
Phenotypic heterogeneity in Saccharomyces cerevisiae was beneficial for the survival of heat stress (Levy, Ziv and Siegal 2012). Gene expression levels of the regulator protein Tsl1were variable population-wide and responsible for counteracting high temperatures. Tsl1p is a trehalose-synthesis regulator that is part of the general stress response of S. cerevisiae (Winderickx et al.1996; Singer and Lindquist 1998). Slowly growing cells produced higher amounts of the regulator protein and survived heat shock (Fig. 4A). The population-wide variance in the regulator protein level during stress thus confirmed the benefits of phenotypic intrapopulation heterogeneities for survival during stress (Levy, Ziv and Siegal 2012).
Figure 4.
Growth arrest in single cells as a survival strategy under stress. (A) Cells of S. cerevisiae with different growth phenotypes. Slow growing cells survive a heat shock and take over the population after termination of the heat treatment (70 min at 60°C) (adapted from Levy, Ziv and Siegal 2012). (B) Heterogeneity in growth of individual L. lactis cells as a bet-hedging mechanism. Upon change of the carbon source, the rearrangement of the metabolism to the new substrate is dependent on the available energy (metabolic state). Cells with high levels of available energy overcome the regulatory burden of carbon catabolite repression (CCR) much faster than cells with a low available energy. (Adapted from Solopova et al. 2014.)
The investigation of individual cellular responses to environmental stress has enabled redefinition of the origin of lag phases during diauxic shifts from one substrate to another (Boulineau et al.2013; Solopova et al.2014; Stratford et al.2014). Classical population-based theory of diauxic growth teaches that all cells in the population rearrange their metabolism to adapt their enzymatic machinery to a new carbon source upon depletion of the preferred carbon source (Monod 1949). Populations continue to grow exponentially after metabolic rearrangement (Monod 1949). Bet-hedging mechanisms were identified as a further possible reason for prolonged lag phases of populations during diauxic growth. Kotte et al. (2014) reported the formation of two subpopulations of an isogenic E. coli population with either a growing or a non-growing/dormant phenotype after switching from glucose to gluconeogenetic carbon sources. In addition, experiments with Zygosaccharomyces bailii (Stratford et al.2014), as well as Lactococus lactis (Fig. 4B) (Solopova et al.2014) and E. coli (Boulineau et al.2013) demonstrated that growing cells were pre-equipped to metabolize the new carbon source. These subpopulations continued to grow with specific growth rates similar to those of the population prior to the shift of growth substrates, while the bulk of the population members stopped growth or died owing to the shift. The lag phase observed at a population scale was therefore an artifact due to the delay in monitoring biomass and not a biological response itself.
A recent example demonstrated the protective role of loosely controlled, so-called noisy response of cells to stress and its role for survival during a subsequent stress (Mitosch, Rieckh and Bollenbach 2017). It was found that antibiotic treatment triggered the noisy expression of the gadBC acid resistance operon in single E. coli cells. With microfluidic shift experiments, a clear link between enhanced survival during acid stress, gadBC expression and a previous antibiotic treatment could be identified. The cross-protection between antibiotics and other stressors could hence be uncovered.
Single cell cultivation systems can also be used as powerful screening tools for identifying mutants with beneficial phenotypes at high throughput and accuracy. Individual mutants of the cyanobacterium Synechococcus elongatus PCC 7942 with beneficial phenotypes, especially a high tolerance against alcohols, were recently identified by Arai et al. (2017) using a single cell-based screening concept. Synechococcus elongatus PCC 7942 is known to have a generally low tolerance towards alcohols and has consequently not been applied in the production of photobioalcohol from sunlight and CO2. Via UV-C-induced random mutagenesis, a mutant library was established and enriched in medium containing 10 g L−1 isopropanol. A subsequent single cell-based microarray cultivation step enabled identification of the fastest growing mutants (Arai et al.2016). The strain finally isolated was able to grow in the presence of up to 30 g L−1 isopropanol and showed increased tolerance towards other alcohols such as ethanol, 1-butanol, isobutanol and 1-pentanol. This strain is currently under investigation as a potential next-generation biocatalyst for the high level production of alcohols from CO2.
Cell surface interactions
Microorganisms interact with surfaces in their surroundings. In nature, an immense number of microorganisms thrive on solid surfaces or interfaces as biofilms (Vigeant et al.2002). Biofilm formation allows the physiological interaction of several species in spatially close proximity, which can be either competitive or cooperative. (For more information about biofilm formation and social interactions within biofilms see the excellent reviews of Wimpenny, Manz and Szewzyk (2000) and Li and Tian (2012).) Biofilms protect cells from extracellular influences, which can be critical in biofilm-related bacterial infections (Wu et al.2015b). Biotechnological production processes with solvents benefit from biofilms due to their increased resistance compared with suspended cells (Halan, Buehler and Schmid 2012).
The initial cell attachment mechanism and the formation of a stable biofilm are central research topics in single cell microbiology. To evaluate the involvement of electrostatic, van der Waals and hydrodynamic forces in the cell attachment mechanism, distances between cells and solid surfaces before cell attachment have been quantified (Berke et al.2008; Lauga et al.2006; Vigeant et al.2002). A cell suspension was introduced between two glass coverslips and the attraction of motile cells to surfaces was determined via total internal reflection fluorescence microscopy at high optical resolution (Lauga et al.2006; Berke et al.2008). The probability of cell adhesion and biofilm formation increased when cells stayed near surfaces (Vigeant et al.2002). Electrostatic and van der Waals forces were found to be responsible for the initial, irreversible cell attachment, as these forces are known to have an effect in short ranges of ∼50 nm. In contrast, hydrodynamic forces range over several micrometers and were identified as keeping the cells in proximity to the surfaces (Vigeant et al.2002). With this knowledge, microbial colonization of solid surfaces can now be understood from its initial stages on. Studying physical interactions of isolated living microbes with matter might lead to new perspectives on the initiation of surface-attached lifestyles such as microbial biofilms in natural and technical environments.
Lin, Crosson and Scherer (2010) analyzed the swarming motility and surface adhesion mechanisms of single Caulobacter crescentus cells within a microfluidic device by microscopy. The device consisted of a rectangular polydimethylsiloxane flow channel attached to a glass coverslip. Adhesion frequencies were found to be dependent on expression levels of the multiple actuator divJ. The actuator codes for a histidine kinase (EC 2.7.13.3) that regulates cell division and differentiation (Wheeler and Shapiro 1999). High DivJ levels inhibited surface adhesion and decreased swarming in semi-solid media. Swimming motility, analyzed by measuring the cell swimming speed in micrometers per second, was not impeded by high DivJ concentrations and was thus excluded as a cause for the observed swarming limitation (Lin, Crosson and Scherer 2010).
The assembly and composition of cell surfaces and their biophysical properties have been investigated in order to disclose cell adhesion mechanisms (Dufrêne 2014). Cell surface properties were studied using atomic force microscopy, in which samples are scanned with a nanoscale tip attached to a flexible cantilever. The movement of the cantilever was mapped with a reflected laser beam and translated into a three-dimensional topological image with nanometer resolution (Dufrêne 2002). Several surface structures of cells were described at a high resolution using atomic force microscopy. By localizing cell wall components, conclusions on their functionality and physiological role could be drawn (Andre et al.2010, 2011). It was found that the heterogeneous distribution of teichoic acids in the outer cell wall of Lactococcus plantarum controls cell division (Dufrêne 2014). Furthermore the number of flagella of Bacillus thuringiensis directly correlated with their swarming motility (Gillis et al.2012). Atomic force microscopy was further developed to single cell force spectroscopy by replacing the atomic force microscopy cantilever tip by a cell (Benoit and Gaub 2002). The immobilization of microbes to the atomic force microscopy cantilever can be applied with, for example, adhesive (Zeng, Mueller and Meyer 2014). This is a quick and easy method; the simultaneous immobilization of several cells cannot, however, be excluded and the precise positioning of cells on the cantilever is challenging. Beaussart et al. (2013) improved the attachment of cells by bonding polydopamine-coated colloids to the cantilever and immobilizing single cells to the colloids. This prevented multiple cell attachment, cell surface denaturation and cell loss due to weak cell-cantilever bonding. Furthermore, the use of coated colloids for cell bonding reduced cell damage due to heat transfer caused by the laser beam. Individual Lactobacillus plantarum cells were immobilized with this technique to measure the adhesion forces between cells and surfaces. Interestingly, the strength of bonding of L. plantarum to a hydrophobic (abiotic) surface was time-independent, while bonding to lectin (biotic) surfaces got stronger over time. This observation could be attributed to glucose-based polysaccharides on the cell surface that caused slower formation of lectin bonds (Beaussart et al.2013). Hence, adhesive properties of individual cells on defined surfaces can be easily quantified in order to identify adhesion influencing parameters.
Single cell biophysics
Technologies for the quantification of biophysical cellular parameters are required for describing global physiological functions and mechanisms such as growth, morphology, regulation and homeostasis (Fig. 2B). In the following section, we review recent technologies for measuring biophysical cellular parameters such as size, mass, morphology, mechanical forces, pH and temperature of individual microbes.
Cell growth
Microbial growth directly depends on to the conditions prevailing in the cells’ microenvironmental surroundings, as well as the intracellular constitution (Schaechter 2015). It unrestrictedly reflects physiological changes, e.g. in gene expression patterns, medium compositions, cell sizes and ribosome concentrations per cell, with minimal delay (Scott et al.2010; Klumpp and Hwa 2014; Schaechter 2015). Growth is thus particularly interesting as a global physiological readout. Microbial growth on the population scale is typically analyzed by measuring the increase of the optical density of a cell suspension over time or by automatic high-throughput cell counting in a coulter counter (Bryan et al.2012). This enables determination of specific growth rates of whole populations, whereby dynamics in individual growth rates are pooled. Individual cell growth can be analyzed and quantified by applying distinct single cell analysis methods. These methods are based on the determination of the cell number, cell size (volume, area, elongation) and cell mass.
Growth of small micropopulations can be described by cell numbers determined by cell counting. Computer-assisted processing of microscopy images even enables automatic quantification of cell numbers (Probst et al.2013b). Growth from initially one bacterium up to microcolonies consisting of more than 500 cells was followed by applying this method. Cell counting can also be used for analyses on a population level. Cell counting is thus a suitable method for comparing population- and single cell-based results (Gruenberger et al.2013; Unthan et al.2014). Unthan et al. used cell counting at single cell and population levels and revealed an iron-chelating medium compound as a factor for biphasic growth during bioreactor cultivation of Corynebacterium glutamicum in defined medium.
However, the analysis of cell growth via cell counting is based on the assumption that all considered cells are similar in size and length. This is probably only true for cells in constant cultivation conditions (Probst et al.2013b). Additionally, cell counting is limited to growth analyses of microcolonies or populations, because growth of individual cells does not affect cell numbers.
Quantitative measurements of cell geometry, such as cell volume, enable the following of growth of a cell over time, even if it is not dividing. The growth behavior of individual cells can thus be quantitatively compared. A straightforward approach to measure total cell volume is the manual determination of cell geometry from microscopy images. This method was shown to be universally applicable for the calculation of single cell growth rates of microbes that were cultivated in different microfluidic structures (Dusny et al.2012, 2015). Semi-solid agarose pads, microfluidic monolayer growth chambers and non-contact cell traps driven by negative electrophoresis (Envirostat) were applied for single cell cultivation (Fig. 5), which provided distinct environmental conditions. Individual C. glutamicum cells were isolated from an exponentially growing population. The investigated cells exhibited similar specific volumetric growth rates independent of the cultivation technology used. Specific volumetric growth rates of isolated cells were equal to or even higher than in population cultivations (Dusny et al.2015). This demonstrates that maximal specific growth rates obtained in population-based ecosystems do not reflect the maximal possible growth rates of cells that experience optimal growth conditions. Morphological aspects such as division rates, division angles and division symmetry of cells were inconsistent in the three devices. The morphology of cells grown on semi-solid agarose pads significantly differed from cell morphologies cultivated in the monolayer growth chambers and the Envirostat (Dusny et al.2015). The authors attributed those differences to spatial constriction, local substrate/nutrient depletion and accumulation of inhibiting products in semi-solid agarose pads. Hence, non-optimal growth conditions were better compensated by the regulation of the specific growth rate than of the cellular morphology (Dusny et al.2015). This study demonstrates that growth and morphology are directly dependent on the environmental conditions around the individual cell. This suggests a deliberated choice of cultivation technology, as every technique imposes technology-specific conditions on the cell investigated.
Figure 5.
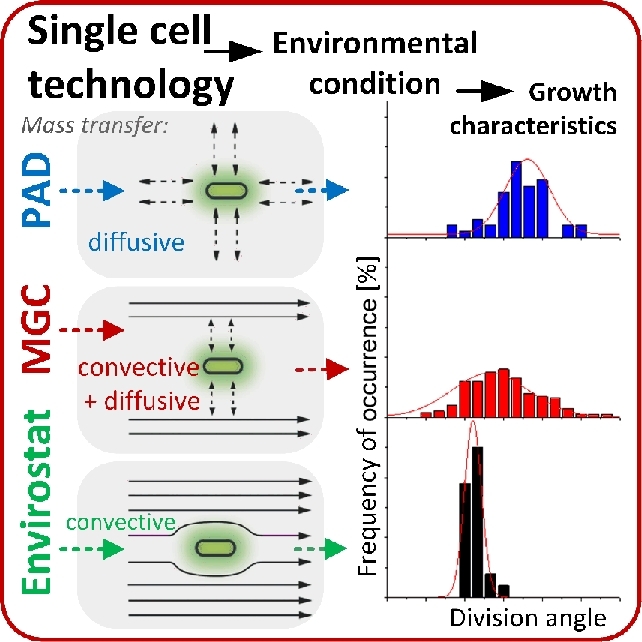
Technological concepts for producing controlled single cell environmental conditions. The examples illustrate three distinct technologies for single cell microbiology and their respective properties in terms of mass transfer: semi-solid agarose pad (PAD), microfluidic monolayer growth chamber (MGC) and non-contact traps driven by negative electrophoresis (Envirostat). Growth studies with C. glutamicum revealed that the different microenvironmental conditions with these technologies determine growth characteristics such as division angles and division times. (Reproduced from Dusny et al. 2015 with permission from the Royal Society of Chemistry.)
The laborious and time-consuming manual determination of cell volumes limits the applicability of this method in terms of time. Automated segmentation is faster, but requires computational solutions for image processing, which are prone to error. Most of the available analysis software solutions, such as Schnitzcells or MicrobeTracker, are limited to certain cell types as they are based on cell segmentation with defined morphological boundary conditions (Sliusarenko et al.2011; Young et al.2012; Chowdhury et al.2013). The detection of cell areas at high precision is especially reliable for rod-shaped bacteria, because most software algorithms are adjusted to the specific morphology of common laboratory strains (Sliusarenko et al.2011; Young et al.2012). Many industrially relevant bacteria are rod-shaped, such as E. coli, C. glutamicum, B. subtilis and Pseudomonas sp., but many other morphological manifestations of microorganisms exist that are not quantifiable with common software packages. In this sense, the recently developed image analysis software Oufti is particularly worth mentioning (Paintdakhi et al.2016). Oufti allows the quantification of various cell morphologies, irregular shapes and even the identification of individual cells that form confluent monolayers by using powerful and flexible segmentation algorithms. Furthermore, Oufti offers post-processing features that enable the identification of differential growth behavior among single cells, e.g. abnormal exponential growth, slow or fast growing cells. The assignment of these observations to cell physiology is, however, often difficult as the molecular causes can be difficult to assess. Next to Outfi, the recently released software MicrobeJ provides a framework for intensity, size and morphology measurements, as well as septa, foci, pole and organelle analyses from microscopy images (Ducret, Quardokus and Brun 2016). The obtained data can be processed and also visualized, with a strong focus on integrated tools for data integrity verification. Besides these generalized software solutions, highly specialized tools have been developed as well. For instance, the software toolbox Molyso has been specifically designed to process time-lapse images obtained from mother machine experiments for growth studies (Sachs et al.2016).
In general, the above described software tools allow automated high-throughput analyses of single cell traits from images and have become invaluable for processing the massive data amounts from time-lapse experiments. However, automated image analysis algorithms are still error-prone and careful inspection of segmentation results remains inevitable to date.
Besides image-based approaches, growth of single microbes can also be quantified via physical biomass measurements. Cell mass quantification with high resolution is especially useful for uncovering mechanisms involved in, for example, cell death or responses to physicochemical perturbations (Weng et al.2011; Zangle and Teitell 2014). Cell mass is either quantified as dry mass or as buoyant mass. The dry mass of living single cells can be determined with quantitative phase imaging (Popescu et al.2014). This technique is based on optical interferometry and enables the differentiation of the refractive index of the cell and the non-aqueous cell content (Popescu et al.2008). Spatial light interference microscopy is a prominent quantitative phase imaging technology that provides highly sensitive data on a spatial scales from micrometers to millimeters and temporal scaling from seconds to days (Mir et al.2011). Mir et al. applied this technology to profile biomass and to quantify specific growth rates of individual E. coli cells that grew on agarose pads. Specific growth rates varied between cells, which demonstrated the individual contributions to the macroscopic biomass increase of a population (Mir et al.2011). An advantage of quantitative phase imaging is the simultaneous analysis of several cells, regardless of whether the cells are adherent or form biofilms. Suspended microbes are not analyzable with this technology due to artifacts arising from movement. Hence, different technologies are required for quantifying single cell masses in suspension. Godin et al. used a suspended microchannel resonator for measuring buoyant masses of individual suspended microbes and human blood cells. The suspended microchannel resonator consisted of a cantilever with an integrated microfluidic channel in an on-chip vacuum (Godin et al.2007; Weng et al.2011). Cells were rinsed through the microfluidic channel and detected by the small frequency change of the cantilever's resonance frequency due to the cell. The frequency change is induced by the density difference between the cell and the medium, which directly corresponds to the cell's buoyant mass (Godin et al.2007; Weng et al.2011). This particular technology provided subfemtogram-level mass resolution, which makes it applicable to smallest microbial cell types. The technology was applied for studying individual yeast cells in flow-through configuration and for time-related mass analyses of trapped yeast and bacterial cells retained with mechanical barriers (Godin et al.2007; Bryan et al.2010; Weng et al.2011). As an important result, Bryan et al. revealed a cell density increase before bud formation of yeast. Such observations can significantly support the understanding of how cells coordinate growth, division and cell cycle progression (Bryan et al.2010). Suspended microchannel resonators were also applied for measuring the biomass of single marine bacteria (Cermak et al.2017). The knowledge of biomass composition and contribution by various taxonomic groups provided an estimate of the total marine biomass. With the aid of nutrient flux models, the analyses of single cells might be used to estimate biomass and carbon fluxes in the world's oceans.
The investigation of individual cell growth uncovered the characteristics of distinct physiological growth states of populations. The stationary growth phase of microbial populations is indicated by a constant optical density after nutrient depletion (Monod 1950). This can be for two reasons: cells arrest growth or an equilibrium state between growing and lysing cells is reached (Gefen et al.2014). Gefen et al. monitored single E. coli cells that reached starvation conditions in order to distinguish between the two proposed mechanisms. The majority of investigated cells showed arrested growth, while the remaining cells lysed (max. 7%) or grew extremely slowly (max. 5%) (Gefen et al.2014). The absence of growth was thus verified as the main cause for the stationary growth phase during starvation. Furthermore, the individual metabolic activity of cells during starvation was characterized over several hours by time-resolved investigations of protein synthesis (Gefen et al.2014). Interestingly, non-growing bacteria maintained a constant metabolic activity over several days under starvation conditions. These investigations proved that the metabolic activity of E. coli in the stationary growth phase is not restricted to a small subpopulation of slowly growing cells, but is homogeneously distributed among individuals during extended periods of starvation (Gefen et al.2014).
Biological membrane organization
Microbial membranes are asymmetric and heterogeneous, dependent on their lipid as well as protein composition (Lingwood and Simons 2010; Elani et al.2015). Biological membranes are among the most important features of cellular life, as they allow the active differentiation of the cell from its surrounding. Microbial growth can only be accomplished when the biological membranes within the cell are intact. In this context, the organization of biological membranes is important as it allows the correct positioning and thus functioning of transport proteins. Membrane-associated proteins are organized in microdomains that are enriched by lipid assemblies called lipid rafts. Those lipid rafts represent a kind of compartmentalization (Schneider et al.2015). Specific proteins exhibit higher activities when they are arranged in lipid rafts and are thus more efficient (Lopez and Kolter 2010; Schneider et al.2015). Currently, the existence of lipid rafts in living cells is accepted, but their occurrence in the microbial cytoplasmic membrane was controversially discussed in the past (Munro 2003; Shaw 2006). One reason is the small size of lipid rafts and the difficulty of visualizing them by microscopy technologies. The existence of lipid rafts in eukaryotic microbes was first discovered by in vivo single cell studies (Wachtler, Rajagopalan and Balasubramanian 2003). Lipid rafts were visualized by using a fluorescent probe that forms specific complexes with membrane proteins, which was detected with fluorescence microscopy. Later, the existence of microscale domains with equal structures and functions to eukaryotic lipid rafts was proven in bacteria (Lopez and Kolter 2010). Schneider et al. investigated the diversity of lipid rafts in individual B. subtilis cells (Fig. 6; Schneider et al.2015). Distinct lipid rafts were responsible for regulatory tasks in cellular membranes. This diversity of functionalized microdomains facilitates the strategic organization of membrane connected signaling networks in a cell. Schneider et al. (2015) concluded that bacteria are organized in a more complex way than expected, as bacterial membranes were originally thought to be homogeneous, compartment-free structures. The complexity of membrane-related processes can now be investigated from a completely new perspective.
Figure 6.
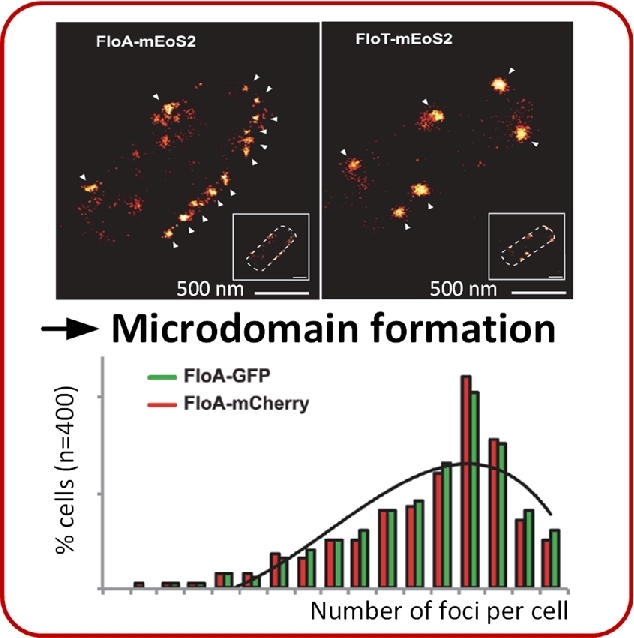
Microdomain formation in growing cells of Bacillus subtilis. The two proteins FloA and FloT arrange locally in distinct microdomains in B. subtilis cells. Protein localization was revealed by photoactivated localization microscopy of mEoS2-labelled proteins (top). Variability in foci formation of FloA is indicated by fluorescence labeling with green fluorescent protein (GFP) or the red fluorescent protein mCherry (bottom). (Adapted from Schneider et al. 2015.)
Cell shape adaptation
Cells are able to adapt their shape to spatial restrictions. The adaptability of cell shape to confined cultivation spaces was investigated with a microfluidic device containing microchannels with different heights (Maennik et al.2009). The device had several adjacent growth chambers, which were connected by microchannels with decreasing diameters. Escherichia coli cells were able to actively swim through channels up to widths equal to their own diameter. Cells passed through channels with diameters smaller than their own by growing into the channel entry. The newborn cell in the channel exhibited a smaller diameter than the mother and was able to traverse the small channel and propagate into the next growth chamber (Maennik et al.2009). Escherichia coli cells are obviously able to adapt their size to promote the colonization of their environment. Escherichia coli cells that grew in the channels with smaller heights than their own diameter exhibited anomalous broadening shapes compared with the typical rod-shaped morphology (Maennik et al.2012). Interestingly, these irregularly shaped cells divided into rod-shaped daughter cells of equal size. This demonstrates that cell volume partitioning is robust and accurate during division, even when the cell morphology is temporarily impaired.
A specific definition of cell geometry was achieved by using agarose pads with polydimethylsiloxane ceilings that contained imprinted microchambers. Single E. coli cells grew in these chambers with prescribed shapes such as crescents, zigzags, sinusoids and spirals (Takeuchi et al.2005). The unusually shaped cells were motile and retained their shape after release (Takeuchi et al.2005). A similar microfluidic device was used to study the adaptation of Min protein dynamics in E. coli cells exhibiting anomalous shapes (squares, rectangles, triangles and circles) (Wu et al.2015a). Min proteins oscillate from pole to pole over the length of the cells and are responsible for the accurate localization of the cell septum before division in many bacteria (Wu et al.2015a). Wu et al. visualized Min oscillation using fluorescent fusion proteins in differently shaped cells in order to determine the influence of cell size and geometry. Min protein oscillations aligned to symmetry axes in the cells regardless of the cell shape. The symmetry axes were oriented in such a way that the total length of the axis was between 3 and 6 μm. The Min proteins predominantly rotated in the cell when the symmetry axes were shorter. Disordered oscillations were observed for longer symmetry axes. Overall, the formation of highly artificial cell shapes enabled discovery of the dependency of Min protein oscillation on geometrical parameters and allowed the study of molecular interactions that are dependent on cell morphology.
The filamentous cell shape is a naturally occurring anomalous morphology of bacteria arising without spatial restriction during growth. Bacterial filamentation is a growth phenomenon where cells exclusively grow by elongation without division (Jaimes-Lizcano, Hunn and Papadopoulos 2014). Filamentation is advantageous in waste water treatment, because it is required for flocculation (Aonofriesei and Petrosanu 2007). In biofilms, filamentous bacteria increase the film thickness and roughness. This is disadvantageous in industry because it entails, for example, energy loss in heat exchangers due to fouling (McCoy et al.1981). Understanding mechanisms provoking filamentous bacterial growth is therefore important. Population-based studies showed bacterial filamentation to be a result of the SOS response (Justice et al.2008). Individual recombinant E. coli cells with a constitutively suppressed SOS response were investigated to validate its role in filamentation (Wang et al.2010a). Filamentation rates were reduced in cells with a suppressed SOS response. The suppression of the SOS response also inhibited filamentous cell elongation. Furthermore, experiments with isolated E. coli cells that grew in the mother machine revealed a stable, filamentation-free growth for more than 50 generations under steady-state growth conditions. Filamentation was only observed for cells exhibiting an elevated replicative age (Fig. 7; Wang et al.2010a). The filamentous phenotype was shown to be reversible in E. coli (Probst et al.2013b). This was investigated by following growth and morphology over time in a monolayer growth chamber. A filamentous E. coli cell of a microcolony was picked with optical tweezers and relocated into the center of a monolayer growth chamber. Astonishingly, the filamentous cells resumed normal growth after a few generations with specific growth rates similar to the original microcolony (Probst et al.2013b). The formation mechanism of bacterial filamentation is far from being understood, but the basis has been established. The origin of filamentous bacterial growth will be pursued in future single cell research.
Figure 7.
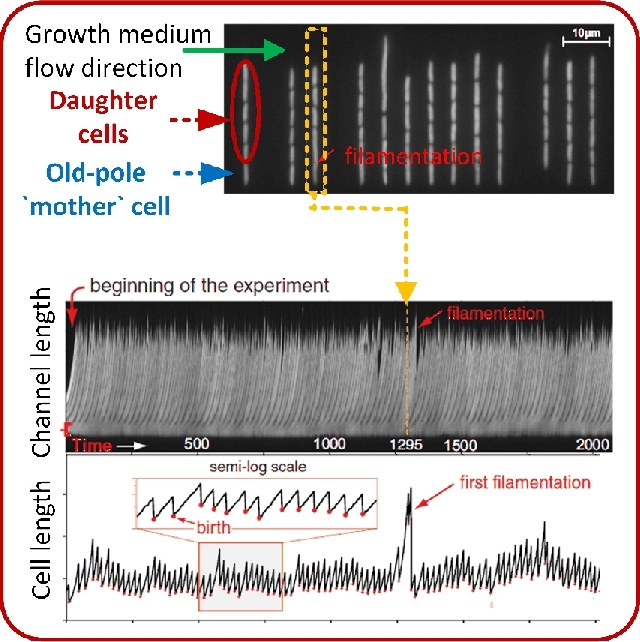
Cell shape maintenance and filamentous growth of E. coli is coupled to the replicative age of cells. Filamentation of individual mother cells only occurs at elevated replicative age. (Adapted and republished from Wang et al.2010a with permission of Elsevier, Copyright © 2010.)
Cell wall properties
Cell walls are involved in cell shape maintenance. The cell wall is subject to internal pressure and extracellular restrictions caused by shear forces and spatial boundaries (Takeuchi et al.2005; Maennik et al.2009; Pelletier et al.2012; Amir 2014). The cytosol is crowded due to the large number of macromolecules such as proteins, nucleic acids and carbohydrates (Amir 2014; Nakano, Miyoshi and Sugimoto 2014). Storage of the bacterial chromosome in the restricted space of the cytosol causes entropic forces, which were investigated by Pelletier et al. (2012). Individual plasmolyzed E. coli cells were loaded into microchannels of the mother machine, digested with lysozyme and lysed by an osmotic shock (Pelletier et al.2012; Wegner et al.2012). The rapid cell lysis caused the direct chromosome expansion into the microchannels. Chromosomes were visualized via functional fusion of a fluorescent protein to the nucleoid-associated protein HupA (Marceau et al.2011). The mechanical properties of the chromosome were quantified by compressing and releasing the chromosome in a closed microchannel using a polystyrene microbead as a piston that was moved by optical tweezers (Pelletier et al.2012). The forces were approximated with entropic spring models, which depend on compressed chromosome size, the equilibrium length after expansion and characteristic constants. For the first time, the mechanical energy stored in the chromosome was quantified. The mechanical energy amounted to ∼105kBT and repeated chromosome compression required a force of ∼100 pN (Pelletier et al.2012).
The cell wall has hence to withstand high internal pressures. Simultaneously, the cell wall has to be flexible for adaptation to external forces such as mechanical stresses caused by shear forces (Amir 2014). The resistance of single E. coli and B. subtilis cells against shear was monitored with cells that were grown in the microchannels of the mother machine. Cells were exposed to filamentation-inducing conditions, which suppressed cell division (Amir 2014). Filamentous cells protruded out of the microchannels into the main trench and were subjected to shear forces, which led to cell bending. Resulting deformations were measured to describe the shape adaptation and recovery capacity of cells. Cells deformed elastically when they were subjected to temporary forces. In contrast, cells deformed plastically when bending forces were constantly applied during growth (Amir 2014). The cell wall composition also affected the bending stiffness of cells (Wang et al.2010b). Cell wall mutants of Escherichia coli were immobilized on a polyethylenimine-coated coverslip and poly-lysine-coated beads were immobilized on the cell tips. Bending forces were then applied using optical tweezers that manipulated the beads at the cell tips. In this way structural proteins could be identified that contributed to the rigidity of the cell wall (Wang et al.2010b).
Cell size homeostasis
Cell sizes vary in a narrow range within the same species (Amir 2014; Campos et al.2014; Jun and Taheri-Araghi 2015). The size of bacteria rarely exceeds the two-fold difference between cell division events when they are cultivated under constant environmental conditions (Jun and Taheri-Araghi 2015). How microbes maintain their size is a central question in microbiology since cellular physiological growth states are still under investigation (Kjeldgaard, Maaloe and Schaechter 1958). Distinct theoretical models were postulated for the underlying intrinsic regulatory mechanisms. It was assumed that cells divide at a critical threshold, dependent on an increase in size, time or volume (Schaechter et al.1962; Cooper and Helmstetter 1968; Donachie 1968). Validation of these models was impossible with population-based analyses, because they merely deliver averaged data and neglect individual physiological effects (Jun and Taheri-Araghi 2015). Additionally, monitoring a huge number of cell divisions under non-fluctuating environmental conditions was not realizable (Campos et al.2014).
Time-lapse investigations of cell division in individual E. coli and C. crescentus cells in a microfluidic device (Ullman et al.2013) has uncovered the intrinsic principles of bacterial cell size homeostasis (Fig. 8; Campos et al.2014). Both species exhibited distinct cell division characteristics. C. crescentus cells that were shorter than the population average produced daughter cells that were longer than the mother cell. Inversely, cells that were longer than the population average divided into shorter daughter cells. In contrast, E. coli cells grew on average to a common length, independent of their original size (Campos et al.2014). Both species did not divide at a critical size threshold, which refuted the theoretical model of a critical cell size-based bacterial cell size homeostasis. Interestingly, cells of both species constantly elongated before division. This verified the theoretical model of a constant extension mechanism that was hypothesized in the same study (Campos et al.2014). The constant extension mechanism is based on a constant elongation rate during steady-state growth conditions, in which cells divide when a target length increase is reached (Campos et al.2014). Experiments with individual E. coli, B. subtilis and C. crescentus cells supported a very similar model (Iyer-Biswas et al.2014; Jun and Taheri-Araghi 2015). Individual cell divisions of E. coli and B. subtilis were quantitatively analyzed under seven controlled environmental conditions. Cells added a constant volume before they divided, independent on their original cell size (Jun and Taheri-Araghi 2015). This so-called ‘adder’ behavior was further investigated and linked to the molecular mechanisms of chromosome replication (Wallden et al.2016). It was found that chromosome replication is activated after the addition of a fixed volume per chromosome. These observations could be translated into a growth and division model for E. coli that links cell-to-cell differences in division timing and cell size to variations in specific growth rates. The size of C. crescentus cells was found to increase exponentially during cell cycle progression and division occurred when cells reached a multiple of 1.8 of their initial size (Iyer-Biswas et al.2014). In contrast to mother machine experiments, Iyer-Biswas et al. achieved single cell fixation over several generations via inducible cell adhesion. Cells and medium that contained the adhesion inducer were constantly flushed through a microfluidic device. Inducer free medium was supported after cell adhesion, which removed the daughter cells formed that did not adhere (Iyer-Biswas et al.2014). The authors transferred the experimental findings obtained to a generally valid single cell scaling law for bacteria. Large datasets were implemented into a mathematical model that perceived fluctuations in cell sizes. The established single-cell scaling laws comprise a proportional correlation between the mean division time and the inverse of the mean growth rate, the temperature independency of the mean division-time distribution, and scaling of the coefficient of variation of cell size with the square root of time for a given initial cell size (Iyer-Biswas et al.2014). With this, a relatively simple, but generally valid mathematical framework was developed for the description of the cell sizing during stochastic growth and division.
Figure 8.
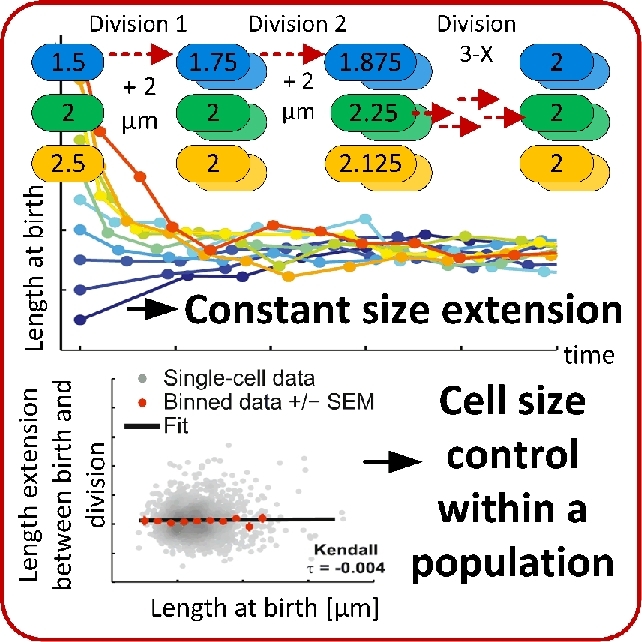
Mechanisms of cell size control. Cell size homeostasis during growth of E. coli is achieved by a constant size extension prior to cell division. (Adapted and republished from Campos et al. 2014 with permission of Elsevier, Copyright © 2014.)
To conclude, the cell size threshold model was refuted. A model that is based on a constantly added volume difference was confirmed to describe the mechanisms of cell size homeostasis correctly.
Intracellular pH regulation
The intracellular pH (pHi) is tightly associated with the cytosolic buffer capacity and linked to the integrity of cell membranes and the membrane potential (Aabo et al.2011; David et al.2012; Orij et al.2012; Bencina 2013). The pHi level affects essential biological processes and enzyme functionalities, regulates metabolic processes such as cellular redox metabolism and molecular transport across membranes, and affects cell vitality and viability (Brett, Donowitz and Rao 2006; Weigert et al.2009; Aabo et al.2011; Orij et al.2012). Earlier population-based attempts to discover the relationship between oscillations in the microbial cell cycle and pH homeostasis were misleading. Karagiannis and Young (2001) revealed that the reported dynamic pHi changes were caused by synchronization steps of the population, which perturbed the cells. Such complex synchronization procedures are obsolete when individual cells are investigated. The pHi of single cells and their cellular compartments are accessible via genetically encoded biosensors such as pHluorin. pHluorin is a green fluorescent protein-derived biosensor that reacts with conformer switching to the surrounding pH (Miesenboeck et al.1998; Orij et al.2009; Valkonen et al.2013). The conformer switch manifests in a changed spatial arrangement of the protein caused by the protonation and deprotonation of its amino acids. The switch induces a detectable shift in fluorescence intensitiy at two distinct excitation wavelengths and can be used as a ratiometric pH probe. The linear response of pHluorin is between pH 5.5 and 8.0, which covers most of the physiological pHi values that have been reported for living microbial cells (Brett, Donowitz and Rao 2006).
pHluorin was used as a probe to measure the pHi of individual Schizosaccharomyces pombe cells. Contrary to the results obtained with synchronized populations, the pHi turned out to be constant during the cell cycle of exponentially growing, unperturbed single cells. These results clearly demonstrate how experimental data can be biased by population effects and averaged values.
Maintaining a constant pHi is an indicator for cellular fitness, which in turn affects the quality of biotechnological manufacturing processes (Valli et al.2006; Valkonen et al.2013; Valkonen, Penttila and Bencina 2014). Individual S. cerevisiae cells were investigated to perform a rational and effective strain improvement strategy regarding lactic acid production (Valli et al.2006). Accordingly, a correlation between the lactic acid production capability and the pHi maintenance capacity of single cells was established. The pHi was determined by staining with the fluorescent probe carboxy-SNARF-4F AM (SNARF-4F 5-(and-6)-carboxylic acid, acetoxymethyl ester, acetate; Valli et al.2005, 2006). Cells with improved pHi maintenance capacities were more robust against cytosolic acidification (Valli et al.2006). These cells exhibited higher pHi values and simultaneously higher lactic acid productivities. Cells with improved lactic acid productivities were isolated and applied for further process optimizations (Valli et al.2006).
Heat production
Temperature is an important parameter in industry as it can determine the (economic) success of a bioprocess, since microbial metabolic heat entails a temperature increase in bioreactors that has to be counteracted. Elevated temperatures might cause a deactivation of the cellular biocatalyst, which can be prevented by the installation of cooling technologies (Maskow et al.2010). Cells emit heat to dissipate excess energy. The magnitude of heat formation depends on the stoichiometry of growth and product formation. Heat emission thus reflects the metabolic activity of the cell (Von Stockar et al.2008; Maskow et al.2010). Furthermore, various velocities of biochemical reactions in microbial metabolism are known to be temperature-dependent (Heijnen 1994; Cornish-Bowden 2014). The intracellular temperature thus correlates with biological reactions and functions such as transcription factor binding, mRNA structure, protein folding and membrane permeability (Nobel 1969; John and Weeks 2000; McCabe et al.2011; Tsuji et al.2013).
Macroscopic analyses of microbial heat production are commonly based on calorimetric methods combined with theoretical thermodynamic energy balancing (Von Stockar and Briou 1989; Von Stockar et al.1993; Lechner, Maskow and Wolf 2008). However, the heat power resolution of calorimeters (about 10 mW L−1 in a chip calorimeter; Lechner, Maskow and Wolf 2008) is far below the resolution required to detect heat production of single cells (e.g. 7.8 pW for an E. coli cell; Lechner, Maskow and Wolf 2008). To our knowledge, only two studies are currently published that report the intracellular temperature quantification in single living microbial cells (McCabe et al.2011; Tsuji et al.2013). In the first study, a temperature-sensitive vector was developed. The vector included a promoter that regulated the temperature-dependent lacZ expression. Expression of lacZ, and thus the production of LacZ (β-galactosidase, EC 3.1.26.12) increased with increasing temperature. LacZ converted fluorogenic substrates into chromophores and its quantity was used to approximate the intracellular temperature (McCabe et al.2011). The detection range of this method was between 35 and 45°C with an average sensitivity of 0.7°C in E. coli cells (McCabe et al.2011). However, gene expression fluctuations in individual cells were not taken into account with this method.
Next to the genetic thermometer, a cationic fluorescence polymeric thermometer was developed for measuring temperatures inside cells. The fluorescent polymer is spontaneously taken up by S. cerevisiae cells and retained within the cytoplasm (Tsuji et al.2013). The fluorescent polymer exhibited a temperature-dependent fluorescence lifetime. A valid correlation was reported for a temperature range between 15 and 35°C, with a resolution of 0.09–0.78°C (Tsuji et al.2013).
The development of intracellular thermometers is still at a proof-of-concept stage. Novel insight into biological mechanisms has not yet been obtained by the application of the described techniques. Nevertheless, the presented studies demonstrate fascinating opportunities for the future. One example could be the contribution of cells to global warming, because microbial heat production effects are known to be involved in thawing rates of the arctic permafrost (Schaefer et al.2014; Hollesen et al.2015).
Single cell biochemistry
In this section we review single cell tools and their applications for analyzing biochemical parameters at the subcellular level. Biochemical parameters are involved in functions and mechanisms on all hierarchical cell organization levels, from genome to metabolome (Fig. 2C). Single cell analyses are important for advancing the omics fields, but considering every element of this field would go beyond the scope of this review. Hence, we focus on current technologies and their applications.
Genome level
The genome comprises the heritable information of every living organism. Population-based genome studies prevalently provide metagenome data of microbial communities obtained from environmental samples without culturing (Clingenpeel et al.2015; Oulas et al.2015). Metagenomes contain data about the entire community without individual differentiation between cells. In contrast, single cell genomics (SCG) makes genome sequences of individual cells accessible, which reveals the metabolic potential of the community (Clingenpeel et al.2015). SCG therefore links function to phylogeny and assigns gene organization structures to individual genomes within complex environments (Siegl et al.2011; Stepanauskas 2012; Clingenpeel et al.2015).
SCG studies have been frequently performed to discover hitherto unknown species of symbiotically living organisms (Siegl et al.2011; Embree et al.2014; Kashtan et al.2014), for establishing genomic blueprints of microbial communities (Probst et al.2015), and for deciphering genomic heterogeneity in monocultures (De Bourcy et al.2014). The high resolution of SCG allows comparison of the genetic repertoire of individual population members in monocultures or closely related strains that occupy the same habitat. In contrast, population-based metagenomic analyses often fail to assign analytical information to specific cells due to the inability to cultivate or study them in isolation. In addition, the origin of pooled information is still not precisely traceable with metagenome analyses (Embree et al.2014). SCG technologies can also be used for analyzing unculturable microbes and especially for deciphering their individual traits (Grindberg et al.2011; Swan et al.2011; Martinez-Garcia et al.2012), which expands the range of applications of population-based genomic methods. As an example, new pathways for microbial CO2 fixation were discovered by SCG studies of prokaryotes that occupy subtropical gyres (Swan et al.2011). Microbial CO2 fixation constitutes an alternative way to store the atmospheric carbon surplus (Jiao et al.2014). Atmospheric CO2 affects processes like global warming and climate change, which in turn affect the environmental conditions of natural microbial habitats (Soon et al.1999). Finding a way to decrease the accumulation rate of CO2 in the atmosphere has far-reaching consequences for the ecosphere. Atmospheric carbon storage with maritime chemoautotrophic cyanobacteria is an additional alternative for microbial CO2 fixation (Li et al.2012b). The fixation mechanisms are not yet completely understood, because methods for the identification of hitherto unculturable microorganisms were lacking for a long time (Li et al.2012b). The mechanism of microbial CO2 fixation in the dark ocean may also be analyzed with population-based metagenomics. However, single cell technologies enable pure cultures to be provided for further analyses (Swan et al.2011).
The discovery of new metabolic pathways or related genes of unculturable microbes also supports the exploitation of catalytic potential in biotechnology. The microbes currently used in biotechnological applications represent only a minority of microorganisms present in nature. Only 1% of all bacteria are culturable at present (Lasken and McLean 2014). Hence, the catalytic potential of uncultured microorganisms is still far from being exhausted for industrial biotechnological production processes and will serve as a target for further SCG research in future.
The identification of microbes in microbial communities or hitherto unculturable microorganisms and the amplification of their isolated genomes provide an important basis for SCG (Li et al.2012a; Clingenpeel et al.2015). Technologies for identifying microorganisms in their natural environment include fluorescence in situ hybridization (FISH) combined with microautoradiography (MAR) (Kong, Nielsen and Nielsen 2005; Wagner et al.2006), Raman microspectroscopy (Haider et al.2010; Li et al.2012b; Berry et al.2015; Zhang et al.2015) and nanometer-scale secondary ion mass spectrometry (nanoSIMS) (Milucka et al.2012; Berry et al.2013; McGlynn et al.2015). New microbial species and their functional properties in waste water treatment were discovered using FISH-MAR (Kong, Nielsen and Nielsen 2005). In FISH cells are exposed to fluorescently labeled oligonucleotides that specifically bind to rRNA. The cells are then analyzed by fluorescence microscopy and identified according to their staining pattern. MAR technology can be used simultaneously to quantify the uptake of specific molecules. For this purpose, microbes are incubated with radiolabeled substrates prior to analysis and are then exposed to an autoradiographic emulsion. Silver grains are formed on active cells and the amount of silver grains can be correlated with the amount of radiolabeled substrates (Musat et al.2012). Kong, Nielsen and Nielsen (2005) used FISH-MAR to screen for polyphosphate-accumulating cells in wastewater. New Actinobacteria species were thereby identified, which were able to take up and store polyphosphate (Kong, Nielsen and Nielsen 2005). The use of such polyphosphate-accumulating organisms helps to protect wastewater-receiving waters against eutrophication.
An alternative technology for cell identification is nanoSIMS. This is based on mass spectrometry of secondary ions, which are formed by sample bombardment with primary ions. The well-focused beam has a high lateral resolution for sample scanning (Musat et al.2012). Even subcellular structures like the pigment formed within archaea and proteobacteria from marine sediments can be studied with nanoSIMS (Milucka et al.2012). However, nanoSIMS is an invasive technology that excludes further analyses of the same cell.
In contrast, Raman microspectroscopy is a non-destructive cell identification method. Molecules inside cells are excited with monochromatic light by a laser. This causes a wavelength shift of the scattered light, which is recorded as a Raman spectrum. The spectrum peaks are characteristic for the composition and conformation of the sample analyzed. Hence, this technology can be used for cell identification or analyses of cell components such as storage compounds (Musat et al.2012; Majed et al.2012; Milucka et al.2012). The non-destructive character of Raman microscopy allows further analyses of the same cell after identification. Berry et al. (2015) combined Raman microspectroscopy with FISH to detect heavy water (D2O) that was incorporated in distinct active bacterial and archaeal cells. D2O-labeled metabolically active cells were isolated with optical tweezers. The genomes of the isolated cells were amplified and subsequently sequenced. The method was applied for identifying individual microbes in the mouse cecum as a proof-of-principle. It is important to understand the compound utilization and composition of the microbial gut community, because it is directly involved in the pathogen defense system. Single cell Raman spectroscopy is also a powerful tool that enables assigning metabolic cell states to genomics (Song et al.2017). Individual carotenoid-containing cells from Red Sea water samples were isolated with a Raman-activated cell sorting system. The subsequent gene sequencing revealed putative genes responsible for carotenoid biosynthesis. The analysis of metabolism and genome hence allows understanding of the physiological interrelations of biological information and its utilization in cellular catalytic activity.
The amplification of whole genomes or low amounts of nucleic acids contained in a single cell is mostly performed with multiple displacement amplification (MDA). MDA is based on random primer annealing to denatured DNA strands and strand displacement synthesis at a constant temperature catalyzed by a ϕ29 DNA polymerase (Liang, Cai and Sun 2014). MDA provides minor amplification bias, generates long amplicons and has a low error rate of 1 per 106–107 bases (Blainey 2013; Liang, Cai and Sun 2014). Multiple annealing and looping-based amplification cycles (MALBAC) is an alternative single cell gene amplification method (Liang, Cai and Sun 2014). MALBAC is based on multiple cycles of strand displacement pre-amplification with random primers and subsequent amplification by polymerase chain reaction. The amplicons form loops ensuring quasilinear pre-amplification, because the amplification product cannot be used as a new template by the polymerase (Liang, Cai and Sun 2014). Both methods, MDA and MALBAC, were compared for whole genome amplification of single E. coli cells with regard to single-nucleotide and copy number variations (De Bourcy et al.2014). MALBAC was more robust for copy number variant measurements. MDA analysis was more suitable for single-nucleotide variant analyses than MALBAC, since it has a lower amplification-based error rate.
Amplified single cell genomes are mostly sequenced with next generation sequencing technologies, such as the commercially available 454 pyrosequencing (Rothberg and Leamon 2008; Shendure and Ji 2008; Liu et al.2012) and with Solexa/Illumina (Shendure and Ji 2008) and SOLiD sequencers (Shendure and Ji 2008; Liu et al.2012). (For a comprehensive description of sequencing technologies and bioinformatics data analysis tools, see Oulas et al.2015.) In general, next generation sequencing is a reliable, relatively cost-effective and readily available state-of-the-art technology that enables high-throughput sequencing. The capabilities of single cell genome analyses present scientist with new tasks, namely to provide vital single cells suspended in adequate liquid volumes that are free of exogenous DNA. The application of microfluidic lab-on-a-chip devices is one option to separate cells, encapsulate them into droplets, sort them and export them into adequate compartments for subsequent genome analysis (Zhang et al.2017). However, single cell sequencing technologies require further development in terms of accuracy to unfold their full potential for systems-level analyses of genomes in correlation with physiological data.
Transcriptome level
The genome of cells is fixed, meaning that the entire DNA that a cell contains is already determined during cell division. In contrast, the transcriptome is highly dynamic, because the transcript composition varies from cell to cell and from time to time. Transcripts are all kinds of RNA molecules that are synthesized by transcription of DNA, such as tRNA, rRNA, mRNA, as well as the non-coding RNA in the form of introns in eukaryotic cells. Variations in transcript abundance thus affect protein stability, localization, translation and functionality (Pelechano, Wei and Steinmetz 2013).
Transcript variations were detected by investigations of individual yeast cells originating from an isogenic population. The variations were induced by stochastic processes in gene expression and environmental fluctuations (Becskei, Kaufmann and Van Oudenaarden 2005; Colman-Lerner et al.2005). The biological significance of such cell heterogeneities can be described in more detail by analyzing entire transcriptomes of individual cells (Lidstrom and Meldrum 2003; Wang et al.2015). The first transcriptome analysis of a single bacterium was performed with Burkholderia thailandensis (Kang et al.2011). Individual cells were isolated with optical tweezers and displaced onto a membrane. Membrane pieces with attached cells were cut and transferred into lysis buffer for further processing. Released mRNA was enriched with a terminator 5΄-phosphate-dependent exonuclease, which selectively degraded rRNA and tRNA. Enriched mRNA was transcribed into circularized cDNA, which was amplified using MDA (Kang et al.2011). This novel strategy for transcript amplification was chosen because earlier linear and exponential amplification methods required immense laboratory work, had a low reproducibility and had extensive gene expression bias (Kang et al.2011). The long processing times often resulted in RNA loss due to extremely low transcript contents contained in a single cell and the short half-life of RNA (Gao, Zhang and Meldrum 2011). The circularized cDNA was analyzed by DNA microarray technology; 94–96% of transcripts from single B. thailandensis cells were reliably sequenced with this approach and the individual induction and repression of gene product syntheses by the inhibitor glyphosate were detected (Kang et al.2011).
Burkholderia thailandensis cells contain extremely high transcript levels of about 2 pg, which is not representative for the majority of bacterial cells (Kang et al.2011). Lower amounts of RNA, about 5 fg, are sufficient for sequencing with the RNA sequencing method BaSiC (Bacterial Single Cell) (Wang et al.2015). This method is simple to use and requires no specialized equipment, because all sequencing steps can be performed with commercially available kits with minor modifications. Individual cells are separated with a micromanipulator, RNA is isolated with a Zymo RNA Isolation Kit and RNA is translated to cDNA with the One-Direct RNA Amplification System. Resulting cDNA is amplified with the NuGen WT-Ovation One-Direct RNA Amplification System and sequenced by using Illumina's Solexa sequencer. The applicability of the method was validated by transcriptome analyses of individual Synechocystis sp. PCC 6803 cells; 82–98% of single cell transcriptomes were sequenced within a processing time of 8 h per cell. The duration of the sequencing process for a single cell transcriptome has to be significantly reduced, because high throughput is required to distinguish between stochastic and significant differences in gene expression. Alternatively, the process could be optimized by parallelization to achieve high throughput and to yield statistically relevant data (Wang et al.2015).
Proteome level
Proteins, in their role as enzymes, are responsible for catalyzing metabolic reactions, replicating DNA, the cellular response to stimuli, the regulation of intracellular biochemical processes and the transport of molecules (Schmid et al.2010). The entire protein content of a cell at a specific time point represents the proteome. The proteome decisively affects the physiological state and the ability to respond to changing environmental conditions. Interestingly, a correlation between the protein and transcript copy number of any gene was not detected within single E. coli cells, although transcriptome and proteome are dependent on the same gene expression mechanisms (Taniguchi et al.2010). This contradiction was ascribed to different lifetimes of mRNA and proteins. Transcripts in the form of mRNA are mostly degraded within minutes, whereas most proteins have longer lifetimes than the cell cycle. The transcriptome thus reflects the recent transcriptional activity, while the proteome represents accumulated gene expression over the lifetime of the cells (Taniguchi et al.2010). A single microbial cell contains more than 1000 different protein species depending on environmental conditions (Schmid et al.2010). A lack of methods for the simultaneous recording of the entire protein content of a single cell makes whole proteome analyses extremely challenging and currently impossible. Nevertheless, parts of the proteome of single cells have already been successfully analyzed.
Enzymes of individual E. coli cells were quantified with an enzyme-linked immunosorbent assay and monitored via fluorescence microscopy (Stratz et al.2014). Individual cells were mechanically trapped between two pillar barriers of a sealable fluidic microchamber. The trapped cells were lysed and enzymes, in this case a β-galactosidase, were bound to antibodies that were previously immobilized by avidin linkers and poly(L-lysine)-g-poly(ethylene glycol) biotin (Stratz et al.2014). The enzyme quantity was detected by a fluorescence signal, which was emitted after the addition of fluorescein di-β-D-galactopyranoside. The β-galactosidase hydrolyzed fluorescein di-β-D-galactopyranoside into fluorescent fluorescein and galactose. The limit of detection was as low as 200 enzymes. The β-galactosidase concentration depended on the composition of the growth medium and the β-galactosidase abundance in individual E. coli cells was variable (Stratz et al.2014). This demonstrates the phenotypic heterogeneity in the proteome of isogenic populations.
Intracellular proteins can also be localized and quantified with single molecule localization microscopy (SMLM). Stochastic optical reconstruction microscopy and photoactivated localization microscopy are the most important SMLM techniques for protein analyses in single cells. Both technologies are based on the sequential activation of photoswitchable fluorophores (Lukyanov et al.2005; Bates, Jones and Zhuang 2013). The high spatial resolution of such technologies allows single molecule detection and hence cellular functions can be assigned to specific proteins.
SMLM technologies were used, for example, to describe how genetic material is organized in bacteria (Dame, Wyman and Goosen 2000; Jensen and Shapiro 2003; Strunnikov 2006; Sullivan, Marquis and Rudner 2009; Schwartz and Shapiro 2011; Wang et al.2011a; Scolari, Sclavi and Cosentino Lagomarsino 2015; Song and Loparo 2015). Similar to DNA condensation in chromosomes of eukaryotes (Misteli 2007), a global chromosome organization mechanism enables plugging of long DNA in small bacterial cells. The plugging mechanism is related to protein–DNA interactions (Nolivos and Sherratt 2014; Song and Loparo 2015). The contribution of nucleoid-associated proteins (NAPs) and structural maintenance of chromosomes proteins (SMCs) to the bacterial chromosome organization was identified with population-based analyses (Song and Loparo 2015). However, these analyses were not sufficient to decrypt the underlying mechanisms, due to the limited resolution for studying protein localization.
Experiments with single C. crescentus cells proved that SMCs interact with DNA in an ordered pattern. SMCs aggregated as condensing complex foci in the bacterial chromosome (Jensen and Shapiro 2003). SMC dynamics during the cell cycle and cell division have been successfully discovered in different bacteria (Mascarenhas et al.2002; Jensen and Shapiro 2003; Strunnikov 2006). SMC foci localized at the cell poles prior to cell division of individual C. crescentus cells and immediately disappeared after division (Fig. 9; Jensen and Shapiro 2003). Hence, the polar SMC aggregates dissociated during cell division. In contrast, NAPs were found to form DNA clusters by bridging and wrapping DNA strands. Variable DNA-condensation degrees, which are regulated by NAPs, suggested a smart polymer function for the bacterial nucleoid. This was revealed by analyzing single cells with SMLM (Scolari, Sclavi and Cosentino Lagomarsino 2015). Smart polymers react to external stimuli with large changes in their conformational transition. External stimuli can be small changes in the environmental temperature, pH values or salt concentrations. NAPs thus cause either the extension or the condensation of the chromosome in response to small changes in environmental conditions (Scolari, Sclavi and Cosentino Lagomarsino 2015). Wang et al. reported that the NAP H-NS forms compact clusters in the bacterial nucleoid that are separated from each other, which was analyzed with fluorescent fusion proteins (Wang et al.2011a). In a further study, the fusion of distinct fluorescence marker proteins to the same NAP was identified to originate from methodological artifacts. The large, discrete clusters were caused by dimerization of fusion proteins (Wang et al.2014; Song and Loparo 2015). In truth, the abundance of H-NS is much more dispersed than expected. This demonstrates the importance of a deliberated choice of analysis method and the design of control experiments.
Figure 9.
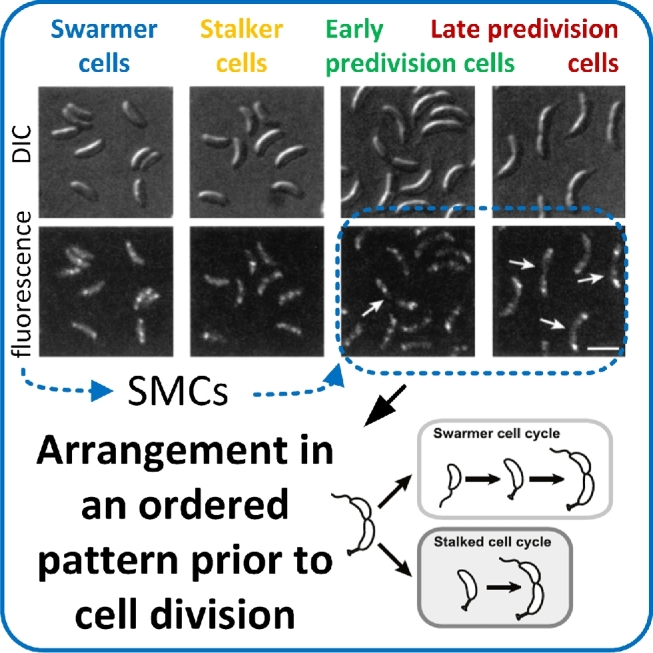
Bacterial chromosome organization in C. crescentus. The example shows the structural maintenance of chromosomes proteins (SMCs) that are involved in bacterial DNA condensation. The SMCs arrange in an ordered manner prior to cell division. (Adapted and republished with permission of American Society for Microbiology — Journals, from Jensen and Shapiro (2003); Adapted and republished from Campos et al. 2014 with permission of Elsevier, Copyright © 2014.)
The knowledge obtained about the bacterial chromosome organization formed the basis for pursuing single cell studies in other research areas, e.g. studies of cell division, DNA replication and DNA repair (Britton et al.2007; Simmons et al.2008; Gupta et al.2014; Uphoff and Kapanidis 2014; Stracy et al.2015). Escherichia coli cells divide in a morphologically symmetric manner under optimal growth conditions, while the intracellular material is stochastically distributed between the dividing cells (Yu and Margolin 1999; Gupta et al.2014). The division of individual E. coli cells was followed for nine generations by automated time-lapse microscopy of monolayer colonies in order to investigate cell aging (Stewart et al.2005). Cells with inherited old poles (the end of the cell pre-existing from a previous division) grew more slowly, produced fewer daughter cells and had an increased probability of dying. Interestingly, morphological characteristics were identical between cells with old and new poles (Stewart et al.2005). This means that the seemingly identical cells resulting from the same division exhibit functional asymmetries and proved that E. coli is prone to aging. Escherichia coli is not, however, capable of compensating those functional asymmetries under external stress exposure (Gupta et al.2014). Gupta et al. visualized morphologically asymmetric cell divisions and an increased uneven inheritance of matter by analyzing fluorescently tagged NAPs (Fig. 10). A temperature increase stimulated the smart polymer function of the nucleoid, which resulted in nucleoid expansion in dividing cells. In consequence, the mean distances between the nucleoids increased. The uncertainty of the point of cell division consequently increased, because more space was available to set the division point. The division points varied from cell to cell, caused morphological asymmetry and hence resulted in phenotypic heterogeneity within isogenic populations (Gupta et al.2014).
Figure 10.
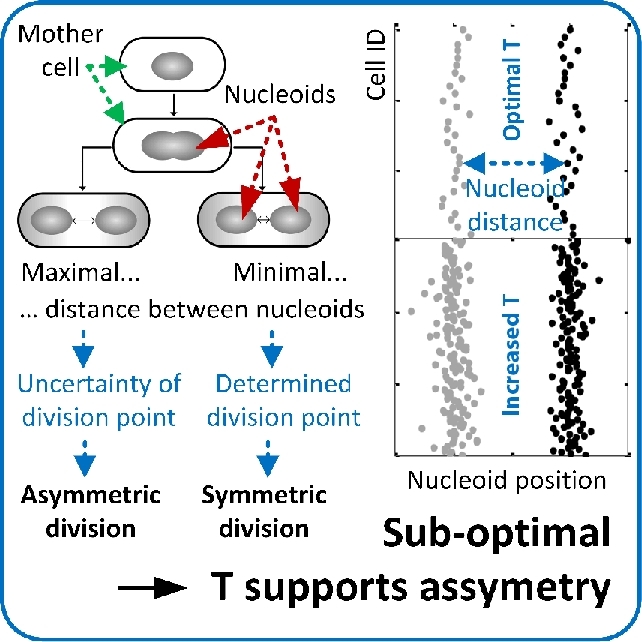
Consequence of nucleoid distances for the symmetry of cell division. Increased mean distances between the nucleoids support asymmetric cell division in E. coli. Non-optimal temperature favors increased nucleoid mean distances as well as symmetry breaking during cell division. (Adapted from Gupta et al. 2014 © IOP Publishing. Reproduced with permision. All rights reserved.)
The decryption of unequal cell division mechanisms might enable a specific manipulation of phenotypic intrapopulation heterogeneities in future. The ability to control phenotypic heterogeneity has significant consequences for biotechnological processes, because it can influence the robustness to process conditions (Delvigne and Goffin 2014), as well as the survival of microbial populations in fluctuating natural environments (Bishop et al.2007; Chakraborty and Li 2011).
Metabolome level
The metabolome encompasses substrates, intermediates and products of the entire metabolism with molecular masses of typically less than 2 kDa (Zenobi 2013). This includes amino acids, organic acids, fatty acids, carbohydrates, nucleotides, adenosine triphosphate (ATP) as energy carrier and redox equivalents like NAD(P)(H) and FAD(H) as cofactors (Brett, Donowitz and Rao 2006). The intracellular metabolite composition changes at time scales between milliseconds and seconds (Zenobi 2013). Monitoring of the metabolome condition is challenging because of its dynamic character. The various chemical molecule classes within the metabolome even complicate accessing the entirety of metabolites. Metabolite quantification of a single cell is especially challenging, because the minute sample volume corresponds to the dimensions of the cell (size 1–10 μm, volume 1 fL to 1 pL) and very low total molecule concentrations prevail inside a cell (from a few hundred up to 1010 molecules per cell) (Schmid et al.2010; Zenobi 2013).
Fluorescently labeled metabolites, biosensors or mass spectrometry (MS)-based methods have been developed for metabolite quantification at the single cell level. The quantification of substrate uptake can be simply realized in real-time by tagging substrates with fluorescent labels. Glucose and toluene were, for example, tagged with N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino (NBD) resulting in the fluorescent molecules 2-NBD-2-D-glucose and 3-NBD-3-toluene. Those molecules were used for analyzing individual glucose and toluene uptake of E. coli cells (Natarajan and Srienc 1999; Straeuber et al.2010; Nikolic, Barner and Ackermann 2013). The attachment of fluorescent labels to metabolites might, however, affect their biochemical function (Zenobi 2013). Biosensors, which provide correlations between metabolite concentrations and readouts such as fluorescence intensity levels, do not affect the metabolite function. Biosensors are even suitable for real-time detection of intracellular metabolite concentrations (Van Engelenburg and Palmer 2008). Various types of biosensors have been developed for quantifying different metabolites. Such sensors can be based on the production of fluorescent proteins that is under the control of a promoter, which is induced by the presence of specific metabolites. Nicotinamide adenine dinucleotide (NADH), hydrogen peroxide or several amino acids were, for example, quantified with these kinds of sensors (Belousov et al.2006; Binder et al.2012; Mustafi et al.2012; Knudsen, Carlquist and Gorwa-Grauslund 2014; Mustafi et al.2014). Other biosensors, such as circularly permuted fluorescent protein-based and Förster resonance energy transfer-based sensors, exploit conformational changes or interactions of fluorescent proteins caused by binding of specific metabolites (Berg, Hung and Yellen 2009; Yaginuma et al.2014; Boersma, Zuhorn and Poolman 2015). The conformational changes cause switches in the fluorescence spectrum or intensity. Although the application of biosensors is limited when the simultaneous reporting of different metabolites in one single cell is intended, the analyses of metabolites with biosensors are in general highly sensitive and enable a quantitative, time-resolved monitoring of metabolite levels in living single cells (Zenobi 2013).
This was realized in a biotechnological context, where biosensors were applied for process development. Valuable metabolites in biotechnological processes include, for example, amino acids, which are mainly produced by C. glutamicum (Hermann 2003; Eggeling and Bott 2015). Interestingly, the production of the amino acid L-valine by C. glutamicum cells varies with distinct cell viability states (Fig. 11; Mustafi et al.2014).L-Valine production, which was initiated by a medium change, was decoupled from growth in the L-valine producer strain. The interplay between growth, physiology and metabolite production was investigated via single cell cultivations in monolayer growth chambers (Mustafi et al.2014). Growth and viability were detected by time-resolved live cell imaging and intracellular L-valine production was monitored with a fluorescent biosensor. L-Valine production was heterogeneous during the production phase and the heterogeneity distribution width increased during cultivation (Mustafi et al.2014). Some cells were viable and productive in the beginning of the production phase and then suddenly lysed. Other cells did not start to produce, because they were either dead or dormant. Another type of non-producing cell continued to grow without L-valine production, although the preferred growth substrate was not delivered after medium transition (Mustafi et al.2014). The occurrence of these distinct cell types illustrates a complex regulation during the absence of growth substrates, which underlies the formation of non-producing cells. The non-producing cells might negatively influence the productivity of industrial biotechnological production processes. The reduction of such phenotypes might prospectively lead to the enhancement of industrial process efficiency.
Figure 11.
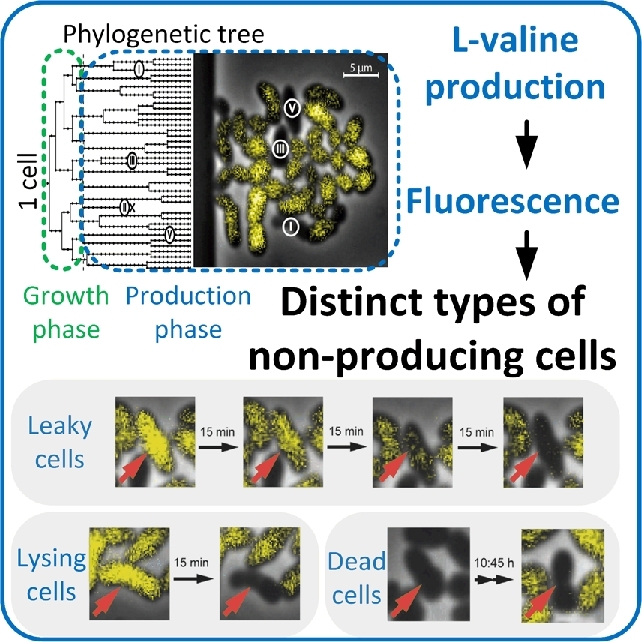
Metabolic activity of single cells. The example shows the connection between metabolic activities and amino acid production capacities in C. glutamicum cells. The occurrence of distinct non-producing phenotypes in a heterogeneous culture was revealed by time-resolved productivity studies employing a genetically encoded fluorescent metabolite sensor. (Adapted from Mustafi et al. 2014.)
Beyond the exploitation of the natural metabolite production of microbes, new synthetic metabolic pathways can be created that do not exist in nature. This adds novel functions to well-known gene networks or improves the activity of the target pathways (Fritz et al.2010). Single cell microbiology allows a precise characterization of novel pathway properties. The non-native D-xylonate production pathway from D-xylose in S. cerevisiae is an example of a synthetic gene network (Toivari et al.2012; Nygard et al.2014). D-Xylonate is an organic acid that serves as an important platform chemical for the production of biomass-derived plastics or as a concrete additive (Chun et al.2006; Toivari et al.2010). Cells exhibiting the synthetic pathway accumulate D-xylonate intracellularly (Toivari et al.2012). Nygard et al. hypothesized that this product accumulation causes a vitality/viability loss during D-xylonate production, which is highly undesirable for an industrial production process. Mechanistic characteristics of the new metabolic pathways were analyzed in order to test this hypothesis (Fig. 12). The lactone ring opening reaction prior to D-xylonate production was identified as the rate-limiting step, revealed with time-resolved analyses of individual cells containing a biosensor (Nygard et al.2014). These restrictions of the heterologous production pathways can now be used for specific process optimization by debottlenecking metabolic conversions via targeted genetic engineering.
Figure 12.
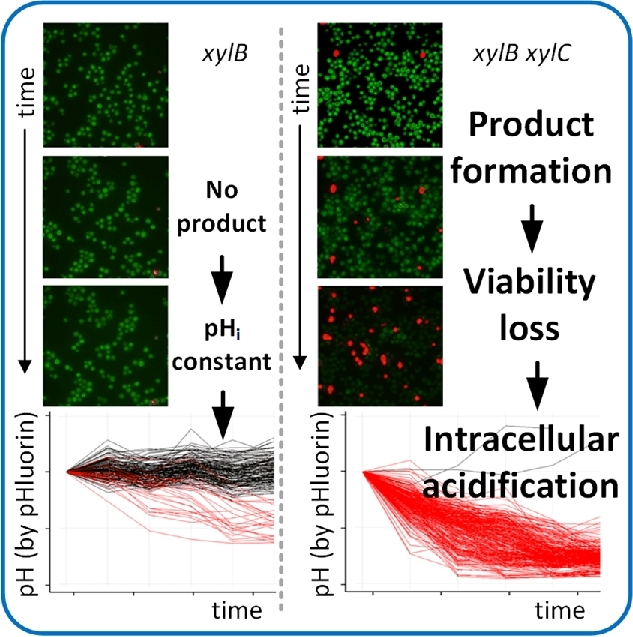
Monitoring intracellular properties of single microbes. The example shows D-xylonate-producing S. cerevisae cells that were equipped with pHluorin (green) for monitoring the intracellular pH. The cells were additionally stained with propidium iodide (red) for visualizing non-viable cells. It was found that cells lose their viability as a result of intracellular acidification caused by D-xylonate accumulation. (Adapted and republished from Nygard et al. 2014 with permission of Elsevier, Copyright © 2014.)
MS-based technologies constitute a good alternative for the simultaneous analysis of several metabolites, because they provide quantitative information and deliver structural descriptions of molecules at the same time (Lottspeich and Engels 2006). The most commonly used ionization technologies for analyzing small amounts of metabolites are matrix assisted laser desorption/ionization (MALDI) (Urban et al.2010; Passarelli and Ewing 2013) and electrospray ionization (Mizuno et al.2008; Heinemann and Zenobi 2011). MALDI-MS technologies enable the simultaneous sensitive, label-free detection of numerous metabolites. Nevertheless, the application of MS for single cell analysis is currently restricted to one measurement time point as the analysis compulsorily consumes the cell. Also the full ionization of minute amounts of target molecules, which are usually present in chemically complex solutions, is challenging. Low-abundant molecules could be concentrated with repeated ion accumulation in ion trap mass spectrometers to overcome this challenge (Si et al.2017). This method was successfully validated for concentrating ATP up to 22-fold. High-density microarrays for MS (MAMS) were developed to aliquot small volumes of solutions or suspensions. This technique enabled high-throughput metabolite analyses of single cells with a detection limit of 100 amol to 10 fmol (Urban et al.2010; Ibanez et al.2013). A total of 26 intracellular metabolites of individual S. cerevisiae cells were simultaneously measured with this approach (Ibanez et al.2013; Zenobi 2013). Ibanez et al. further validated the biological information that can be obtained with MAMS. Intrinsic metabolic cell-to-cell differences that emerge from cell size, cell age or cell cycle stage were described by the metabolite analysis of individual cells. Two different phenotypes were thus identified in an isogenic S. cerevisiae population by MS-based measurements of the intracellular levels of the glycolytic metabolite fructose-1,6-bisphosphate. In addition, differences in metabolic network regulations were disclosed in single S. cerevisiae cells upon perturbation with 2-deoxy-D-glucose, which blocks glycolysis. The shift of metabolic fluxes was determined by correlating metabolites pairwise. Perturbed cells exhibited a more active pentose phosphate pathway. Metabolites that were too far away from the metabolic entry point of 2-deoxy-D-glucose were not affected by the perturbation (Ibanez et al.2013). Such metabolite correlations of individual cells enabled description of metabolic regulatory mechanisms at a high resolution and provide a glimpse of future applications of single cell metabolomics.
To conclude, the combination of single cell analyses with standard population-based analyses enables a mechanistic understanding of cellular biology (Table 1). Several fundamental biological questions have already been addressed with this approach and some of the complex underlying biological mechanisms are currently under investigation. The answer to one biological question mostly gives rise to several new ones, often in relation to other physiological parameters. Thus, a network of knowledge about the idealized cell in its environment is established step by step and non-idealities are minimized or describable for the first time. This is in accordance with the demand of synthetic biology that claims abstractable and quantifiable biological elements that can be used to synthesize novel functional cell systems.
Table 1.
Examples for new biological concepts obtained with single cell microbiology which would not have been possible from population-based microbiology.
| Technology for cultivation/analysis | Analysis/data | New concepts | References |
|---|---|---|---|
| High-throughput microscopy | Growth rate and gene expression | Intrapopulation heterogeneity is beneficial for surviving e.g. heat stress | Levy, Ziv and Siegal (2012) |
| Microfluidic growth chamber, agarose pads, microscopy | Growth and gene expression | Lag phases during diauxic shifts result from persistent cells that continue to grow slowly resulting in a delay in biomass monitoring | Boulineau et al. (2013), Solopova et al. (2014) |
| Microfluidic cultivation device, microscopy | Intracellular pH and gene expression | A cross-protection between antibiotics and other stressors enhances cell survival | Mitosch, Rieckh and Bollenbach (2017) |
| Microscopy | Viability, bioluminescence and population cell density | Variabilities in quorum sensing mechanisms are related to individual bioluminescence and biofilm formation | Anetzberger, Pirch and Jung (2009) |
| Microscopy | Bioluminescence, cell size and gene expression | Autoinducer concentration correlates with population cell densities. Autoinducers are involved in the formation of intrapopulation heterogeneity | Anetzberger, Schell and Jung (2012) |
| Agarose pad, microfluidic monolayer growth chamber, Envirostat, time-resolved microscopy | Specific growth rates | Individual cells that experience optimal growth conditions have higher specific growth rates compared with population scale analyses | Dusny et al. (2015) |
| Microscopic cell monitoring in a microfluidic device | Elongation rate | During stationary growth phase conditions, the majority of cells do not grow; a minority of cells lyse or grows slowly | Gefen et al. (2014) |
| Agarose pad, microfluidic monolayer growth chamber, Envirostat, time-resolved microscopy | Division rates, division angles and division symmetry | Non-optimal growth conditions are better compensated by specific growth rates than by morphological characteristics | Dusny et al. (2015) |
| Microscopy, photo-activated localization microscopy | Gene expression | Bacteria are organized in a more complex way than expected as membrane proteins are arranged in microdomains as a kind of compartmentalization | Schneider et al. (2015) |
| Mothermachine, microscopy | Cell shape | Filamentous growth is dependent on the replicative cell age | Wang et al. (2010a) |
| Monolayer growth chamber, optical tweezers, microscopy | Growth and morphology | The filamentous growth phenotype is not stable but reversible | Probst et al. (2013b) |
| Agarose pads, time-lapse microscopy | Cell length, elongation rate and relative growth rates | Bacterial cells elongate in a constant manner before cell division | Campos et al. (2014) |
| Mother machine, time-lapse microscopy | Growth rate, elongation rate, cell size, generation time | Bacterial cells add a constant volume before they divide | Taheri-Araghi et al. (2015b) |
| Microfluidic cultivation device, microscopy | Growth rate, cell volume, generation time | Chromosome replication is initiated after the addition of a fixed volume per chromosome | Wallden et al. (2016) |
| Microfluidic flow chamber, fluorescence-activated cell sorting, time-lapse microscopy, biosensor | Cell cycle, intracellular pH level, cell length | Exponentially growing cells maintain a constant intracellular pH level during the cell cycle | Karagiannis and Young (2001) |
| Flow cytometry, microscopy, pH staining | Metabolite secretion, intracellular pH level | Cells with improved intracellular pH maintenance capacities exhibit higher intracellular pH levels during lactic acid production resulting in production rates | Valli et al. (2006) |
| Microscopy | Intracellular temperature | Proof of concept of temperature measurement technologies for measuring heat production in single cells | McCabe et al. (2011), Tsuji et al. (2013) |
| Microfluidic device, microscopy | Growth, growth arrest and gene expression | The metabolic activity in the stationary growth phase is not restricted to a small subpopulation of slowly growing cells but is homogeneously distributed among individuals | Gefen et al. (2014) |
| Monolayer growth chamber, microscopy, biosensor | Intracellular metabolite concentration, gene expression, viability, growth | Non-producing cells exhibit distinct variability states | Mustafi et al. (2014) |
| Envirostat, microscopy | Gene expression | A promoter system was proven to be extremely sensitive to carbon-catabolite repression, which was previously severely underestimated with population-based analyses | Dusny and Schmid (2016) |
| Single cell genomics | Genome information | Discovery of hitherto unknown species of symbiotic living organisms and pathways of non-cultivable organisms | Siegl et al. (2011), Grindberg et al. (2011), Swan et al. (2011), Martinez-Garcia et al. (2012), Embree et al. (2014), Kashtan et al. (2014) |
| Single molecule localization microscopy | Molecule localization | Bacterial chromosome organization mechanisms exist, similar to DNA condensation in chromosomes of eukaryotes | Strunnikov (2006), Wang et al. (2011a), Song & Loparo (2015) |
| Immunofluorescence microscopy | Molecule/protein localization | Proteins interact with DNA in an ordered pattern and arrange at the cell poles prior to cell division | Jensen and Shapiro (2003) |
| Microfluidic flow chamber, time-lapse microscopy, nucleoid staining | Molecule localization, morphological (a)symmetry, cell segmentation | Symmetry breaking during cell division depends on the mean distance between bacterial nucleoids. Division symmetry is temperature dependent | Gupta et al. (2014) |
TECHNICAL BIAS IN SINGLE CELL MICROBIOLOGY
The examples discussed above demonstrate the importance of single cell microbiology and its contribution to obtaining new biological insights beyond populations. However, single cell technologies also have technology-specific restraints. There is little knowledge about how technical bias affects microbial physiology. Very few experimental studies shed light on these aspects despite obvious physiological effects imposed by the technologies of single cell cultivation and analysis. From our experience, bias from single cell technologies is commonly discussed amongst researches, but often omitted in scientific publications. We therefore think it is important to discuss some major aspects of this topic within the frame of this article.
Microhabitats
The most commonly occurring complications in single cell microbiology are growth arrest, loss of viability, changing cell morphologies or altered gene expression patterns upon the introduction of cells into the artificial cultivation microhabitat. The reasons for such phenomena can be manifold and their identification often requires tedious trial-and-error experiments. Attention should be given to the choice of the device material in the first place to identify the origin of these complications.
Biocompatible polymers and adhesives, primarily polydimethylsiloxane, are typically used for manufacturing microfluidic devices. Secreted molecules or medium compounds can be sequestered into the polymer, as polymeric substances such as polydimethylsiloxane adsorb small hydrophobic molecules and proteins (Halldorsson et al.2015). Polymers might be prone to monomer, oligomer or additive leaching as well. The interaction of such leached molecules with cells potentially entails adverse effects on cellular physiology (Regehr et al.2009). The magnitude of leaching is much more pronounced in microdevices compared with macroscopic cultivations, because microstructured devices have high surface-to-liquid volume ratios. Material compatibility tests should be performed during the device design stage or whenever leaching is suspected to affect microbes. Molecule leaching or sequestration in the device material can be avoided by altering surface characteristics. Inherently hydrophobic polymers such as polydimethylsiloxane are often hydrophilized by plasma treatment during manufacture of the microdevice (Bodas and Khan-Malek 2007; Zhou et al.2012). Priming of the microfluidic network with sacrificial molecules, e.g. with albumin-spiked growth medium or Tween, can effectively passivate surfaces and restore surface biocompatibility after activation (Turner et al.2010; Taheri-Araghi et al.2015a). Alternatively, biocompatible aqueous materials such as agarose gels can be considered for microdevice manufacture (Moffitt, Lee and Cluzel 2012). Direct cell-surface contact further enhances the exposure of cells to leaching substances and might promote adverse cellular behavior (Petrova and Sauer 2012). Surface contact triggers regulatory cascades associated with cell adhesion, which usually remain inactive in liquid cultures (Geng et al.2014). Non-contactless trapping of single microbes with, for example, negative dielectrophoresis serves here as an alternative (Fritzsch et al.2012).
Microdroplets used for cultivation occupy an exceptional position among the single cell microhabitats. The aqueous cultivation volume is typically confined by a second immiscible continuous phase instead of solid channel walls (Joensson and Andersson Svahn 2012). Technical bias in microdroplets primarily arises from adsorption effects at the liquid–liquid interface (Roach, Song and Ismagilov 2005). Proteins and hydrophobic molecules accumulate at the interface and are depleted in the aqueous cultivation volume owing to the hydrophobic nature of the continuous phase. This effect can be significant due to the high surface-to-volume ratio of the microdroplets. In addition, the physiology of the confined cells is influenced by traces of the continuous phase that can dissolve in the aqueous droplet. Surfactants for droplet stabilization also play a critical role, as they were shown to induce detrimental processes such as cell lysis in eukaryotes (Clausell-Tormos et al.2008).
Cell trapping and manipulation technologies
Single cell retention, relocation or isolation technologies potentially impose technical bias on microbial physiology. Directed cell manipulation technologies such as optical manipulation, acoustic trapping and electrokinetic trapping involve the exposure of single microbes to a focused light beam, ultrasound or an electric field, respectively (Eriksson et al.2007, 2010; Evander et al.2007; Kortmann et al.2009a). The impact on the cellular physiology depends on the magnitude of field intensity, exposure time and the properties of the biological system. Exposure times and field intensities should be as low as possible to avoid cell damage. Optical tweezers, for example, are only suitable for quickly relocating living cells over short distances within microhabitats, but not for longer trapping of single microbes. Extended optical manipulations of E. coli resulted in a rapid viability loss (Probst et al.2013b). Growth immediately ceases after the shortest exposure times to the light beam for some microorganisms that are particularly sensitive to light irradiation, such as C. glutamicum (C. Probst, personal communication). Single cell manipulation technologies based on electric fields, such as dielectrophoresis, entail inherent technical bias as well. Local resistive heating, called Joule heating, in dielectrophoresis trapping structures arises from the passaging current through the growth medium. In consequence, temperatures inside the trapping structures can be several degrees above the actual chip temperature, which entails physiological responses of trapped cells (Jaeger, Mueller and Schnelle 2007). Quantitative knowledge on the magnitude of heating is necessary for the effective compensation of joule heating with temperature control systems (Rosenthal et al.2015). Joule heating compensation has already allowed long-term single cell cultivations of several yeast and bacteria species in negative dielectrophoresis traps without technology-borne effects on microbial physiology (Dusny et al.2015).
It is of course always advisable to perform control experiments in order to identify and circumvent biological artifacts that might arise from technical bias during cell trapping and manipulation. Reliable comparative experiments can involve population-based controls, but also single cell experiments with complementary cultivation technologies. A simple and cheap variant for control experiments can, for instance, be performed with agarose pads as single cell growth supports (Young et al.2012).
Mode of cultivation
Unexpected cellular behavior can be imposed by the mode of cultivation. Many microfluidic devices enable the continuous perfusion of single cells with growth medium, which allows controlling and changing of the chemical composition in the single cell microenvironment (Di Carlo, Wu and Lee 2006; Kortmann et al.2009a; Gruenberger et al.2012). Perfusion, however, creates a highly artificial situation for the trapped cells, where secreted substances are immediately removed due to the continuous medium flow. In contrast, secreted molecules accumulate to various extents during population-based cultivations. Low quantities of molecules accumulate in the microenvironmental cell surroundings even in continuous chemostat fermentations on a population scale. In consequence, microbial strains show impaired growth behavior when they rely on the extracellular presence of specific secreted substances. Such an impaired growth behavior was described for B. subtilis cells, which were cultivated with minimal medium in a microfluidic perfusion device (Taheri-Araghi et al.2015b). Growth was restored upon introduction of sterile-filtered, cell-free minimal medium obtained from the late exponential or stationary phase of previous cultivations. Growth did not arrest when complex medium was applied in the single cell perfusion system. The authors attributed this observation to secreted ion carrier molecules that are essential for an effective uptake of extracellular metal salts and hence for cell growth (Langdahl and Ingvorsen 1997). Similar observations were made by our group when phototrophic microorganisms were cultivated in single cell perfusion systems (unpublished). Growth was immediately interrupted upon perfusion of cells with fresh minimal medium, but could be restored by the addition of minor amounts of cell-free medium from a former batch culture. These observations in microfluidic systems demonstrate how microbes shape their environment. The application of conditioned minimal medium or the addition of essential nutrients can provide information about the necessity of secreted molecules for growth (Taheri-Araghi et al.2015a).
Analytics
Technical bias from the analysis technique itself might also result in biological artifacts, which leads to wrong conclusions on the cellular mechanism of interest. The validation of single cell experiments with carefully designed control experiments is especially necessary when novel analysis technologies are used. Single cell experiments are presumably more strongly affected by technical bias than experiments with whole populations, because disturbances from analysis technologies do not equally affect all cells in populations. Biological artifacts can hence be compensated by measuring averages from a large number of individuals.
Optical methods are presumably the most prominent technologies for analyzing physiological dynamics in single microbial cells. Fundamental biological mechanisms have been unraveled in single microorganisms with time-lapse brightfield or phase contrast microscopy complemented with fluorescence imaging of fluorescent dyes or proteins (Lee et al.2012; Young et al.2013; Vasdekis and Stephanopoulos 2015). Although optical analysis technologies are defined as being non-invasive, this is not always the case. Light-induced phototoxicity can affect all types of microorganisms. Phototoxicity comprises a number of potential mechanisms, in which the formation of highly reactive oxygen species or radicals is probably the most damaging (Zipfel, Williams and Webb 2003; Davies 2004). Even white illumination used for standard brightfield/phase contrast imaging can entail photo-induced effects on microbial metabolism (Woodward, Cirillo and Edmunds 1978; Merbt et al.2012). Photo-induced physiological effects can be especially critical when high energy UV light from filament-based white light lamps is applied. Appropriate filters for filament-based lamps or LED illumination with a defined light spectrum prevents such UV-induced effects.
Things become more critical when fluorescence imaging is used. The physiology of many microbial cell types is susceptible to changes or damage by fluorescence excitation light (Tinevez et al.2012). A non-linear negative correlation between the dose of green fluorescent protein excitation light and single cell division times was demonstrated for E. coli (Jun and Taheri-Araghi 2015). The minimization of phototoxic effects can be accomplished by reducing light exposure time or excitation light intensity or by increasing detector sensitivity via, for example, detector pixel binning. Excellent guidelines on avoiding pitfalls during fluorescence imaging of live cells are already available (Dixit and Cyr 2003; Frigault et al.2009). The basic principles for avoiding technical bias of fluorescence imaging can also be applied for microbes, although most work is based on cell cultures. Another aspect of fluorescence imaging is the application of fluorescent proteins or dyes. Genetically encoded fluorescent proteins can impose a metabolic burden for microbes (Wendland and Bumann 2002). Fusion of fluorescent proteins to cellular proteins allows monitoring of the abundance, localization and dynamics of specific proteins, but might influence the native physiological function of the tagged protein (Shaner, Steinbach and Tsien 2005). The delay in signal occurrence due to synthesis and maturation times of fluorescent proteins in biological system has to be considered as well when dynamic cellular processes are analyzed (Hebisch et al.2013). Chemical dyes can also be applied, for example to selectively stain cell organelles or cellular components or to determine cell viability. However, chemical dyes often intercalate DNA or impair the function of the stained molecules (Terai and Nagano 2013). These considerations suggest that controls are mandatory for identifying and eliminating analysis-borne technical bias on microbial physiology.
We are convinced that extensive control experiments help to identify and circumvent biological artifacts that might arise from the technical bias described, regardless of whether bias is caused by the microhabitat, cell manipulation, mode of cultivation, or the analysis technology used.
FUTURE CHALLENGES OF MICROBIAL SINGLE CELL ANALYSIS
Single cell analysis has changed the way we look at microbial physiology. However, the capability of current single cell technologies is limited for the investigation of some specific research questions. An extensive discussion of these limits would go beyond the scope of this review. As such, we focus on a few representative examples.
Culturability of diverse microorganisms present in the natural and technical ecosystems is in general an important issue in single cell microbiology. The majority of published single cell studies focus on the analysis of domesticated model bacteria and yeast. The cultivation and analysis technologies are often specifically tailored for their analysis. Cultivation and analysis of their natural relatives raise new issues in terms of technological compatibility.
Very few single cell studies with obligate anaerobic microorganisms have been published yet, although these microbes are highly abundant in nature and important for medical and technical processes (Steinhaus et al.2007; Fievet et al.2015). The main challenge for cultivating obligate anaerobic microorganisms is posed by the necessity for absolute oxygen absence from the microhabitat. The complete desorption of oxygen from growth medium and from polymers used for microfabrication is difficult to realize. However, anaerobic conditions could be established by placing the complete analysis system and its periphery in an anaerobic enclosure.
The maximal possible cultivation time of microbes is another important aspect. Long-term cultivations are necessary to follow dynamic processes in cells that have a very slow metabolism. This can be problematic when technical restraints impede long-term single cell cultivations, such as evaporation or irreversible and adverse changes of the microhabitat material. Nevertheless, technical solutions for rarely investigated microbes such as obligate anaerobes or slow-growing microbes will emerge with growing interest in single cell microbiology.
Technological development constitutes the main driver for progress in single cell analysis. Novel and sophisticated analytical technologies provide ever deeper insights into cellular mechanisms of microbes. In addition usable technology is the key, not the existence of proof-of-concept technology alone (Andersson and van den Berg 2006). Many essential cellular parameters have already been successfully determined and others will hopefully follow in the upcoming decade (Love et al.2012; Hammar et al.2015; Krone et al.2016). This knowledge opens the door to new approaches for improving the efficiency of microbes in a technical context.
CONCLUSIONS
Single cell microbiology has added a new conceptual dimension to microbiology. In this article, we discussed the advances and exciting possibilities of single cell technologies and demonstrate how they enable formulating and testing new hypotheses. Single cell technologies enable the analysis of cellular mechanisms in individual microbes with spatiotemporal resolution unbiased by population effects. Causal links between environmental parameters and cellular physiology can be established because of the precisely controlled microenvironment within microfluidics. Microbiologists, biotechnologists and biochemical engineers working with microbes can benefit from applying these technologies for answering research questions hidden in the bulk of microbial populations. Numerous studies demonstrate how single cell technologies can be used for approaching biological questions concerning mechanisms in microbial metabolism, its regulatory circuits and interconnections, gene expression, protein synthesis, homeostasis in size, shape and intracellular properties, cell-to-cell communication, as well as mass and energy transfer. We can learn from these analyses how natural microbial concepts are set up in order to efficiently perform biological functions, like growth, product formation, survival or adaptation to environments in natural and artificial ecosystems. This information can be used to identify, understand and abstract the constraints that control microbial functional modules in order to maximize the efficiency of their biological functioning. These considerations clearly show that technological developments are the main driver for obtaining novel biological insights with single cell microbiology. However, some studies demonstrate the importance of considering technical bias from novel single cell cultivation and analysis technologies. This reminds us to critically question the results obtained with carefully designed control experiments. Single cell analysis has matured to be an integral part of microbiology and future technological advances will give us new and exciting insights into the properties and mechanisms of the smallest functional unit of life, the microbial single cell.
Acknowledgements
Funding via the Graduate Cluster Industrial Biotechnology (CLIB 2021) and the BeBasic research program is acknowledged. The authors are also grateful for financial support from the European Union (EFRE), the German Federal Ministry for Economic Affairs and Energy, as well as the Ministry of Innovation, Science, Research and Technology of North-Rhine Westphalia.
Conflict of interest. None declare.
REFERENCES
- Aabo T, Glueckstad J, Siegumfeldt H et al. Intracellular pH distribution as a cell health indicator in Saccharomyces cerevisiae. J R Soc Interface 2011;8:1635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acar M, Mettetal JT, Van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 2008;40:471–5. [DOI] [PubMed] [Google Scholar]
- Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 2015;13:497–508. [DOI] [PubMed] [Google Scholar]
- Amir A. Cell size regulation in bacteria. Phys Rev Lett 2014;112:208102. [Google Scholar]
- Andersen JB, Heydorn A, Hentzer M et al. GFP-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol 2001;67:575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H, van den Berg A. Where are the biologists? Lab Chip 2006;6:467–70. [DOI] [PubMed] [Google Scholar]
- Andre G, Deghorain M, Bron PA et al. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem Biol 2011;6:366–76. [DOI] [PubMed] [Google Scholar]
- Andre G, Kulakauskas S, Chapot-Chartier MP et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat Commun 2010;1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anetzberger C, Pirch T, Jung K. Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol Microbiol 2009;73:267–77. [DOI] [PubMed] [Google Scholar]
- Anetzberger C, Schell U, Jung K. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiol 2012;12:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aonofriesei F, Petrosanu M. Activated sludge bulking episodes and dominant filamentous bacteria at waste water treatment plant Constanţa Sud. Proc Rom Acad 2007;B:83–7. [Google Scholar]
- Arai S, Hayashihara K, Kanamoto Y et al. Alcohol-tolerant mutants of cyanobacterium Synechococcus elongatus PCC 7942 obtained by single-cell mutant screening system. Biotechnol Bioeng 2017;114:1771–8. [DOI] [PubMed] [Google Scholar]
- Arai S, Okochi M, Shimizu K et al. A single cell culture system using lectin-conjugated magnetite nanoparticles and magnetic force to screen mutant cyanobacteria. Biotechnol Bioeng 2016;113:112–9 [DOI] [PubMed] [Google Scholar]
- Arnoldini M, Mostowy R, Bonhoeffer S et al. Evolution of stress response in the face of unreliable environmental signals. PLoS Comput Biol 2012;8:e1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldini M, Vizcarra IA, Pena-Miller R et al. Bistable expression of virulence genes in salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol 2014;12:e1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, He X, Liu D et al. A double droplet trap system for studying mass transport across a droplet-droplet interface. Lab Chip 2010;10:1281–5. [DOI] [PubMed] [Google Scholar]
- Bai Y, Patil SN, Bowden SD et al. Intra-species bacterial quorum sensing studied at single cell level in a double droplet trapping system. Int J Mol Sci 2013;14:10570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R et al. Bacterial persistence as a phenotypic switch. Science 2004;305:1622–5. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell 2006;125:237–46. [DOI] [PubMed] [Google Scholar]
- Bates M, Jones SA, Zhuang X. Stochastic optical reconstruction microscopy (STORM): A method for superresolution fluorescence imaging. Cold Spring Harb Protoc 2013;2013:498–520. [DOI] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C et al. Experimental evolution of bet hedging. Nature 2009;462:90–3. [DOI] [PubMed] [Google Scholar]
- Beaussart A, El-Kirat-Chatel S, Herman P et al. Single-cell force spectroscopy of probiotic bacteria. Biophys J 2013;104:1886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Kaufmann BB, Van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet 2005;37:937–44. [DOI] [PubMed] [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 2006;3:281–6. [DOI] [PubMed] [Google Scholar]
- Benavente-Babace A, Gallego-Perez D, Hansford DJ et al. Single-cell trapping and selective treatment via co-flow within a microfluidic platform. Biosens Bioelectron 2014;61:298–305. [DOI] [PubMed] [Google Scholar]
- Bencina M. Illumination of the spatial order of intracellular pH by genetically encoded pH-sensitive sensors. Sensors 2013;13:16736–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods 2009;6:161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke AP, Turner L, Berg HC et al. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys Rev Lett 2008;101:038102. [DOI] [PubMed] [Google Scholar]
- Berry D, Mader E, Lee TK et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci U S A 2015;112:194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Stecher B, Schintlmeister A et al. Host-compound for aging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A 2013;110:4720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Schendzielorz G, Stabler N et al. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 2012;13:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Rab FA, Sumner ER et al. Phenotypic heterogeneity can enhance rare-cell survival in ‘stress-sensitive’ yeast populations. Mol Microbiol 2007;63:507–20. [DOI] [PubMed] [Google Scholar]
- Blainey PC. The future is now: Single-cell genomics of bacteria and archaea. FEMS Microbiol Rev 2013;37:407–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit M, Gaub HE. Measuring cell adhesion forces with the atomic force microscope at the molecular level. Cells Tissues Organs 2002;172:174–89. [DOI] [PubMed] [Google Scholar]
- Bodas D, Khan-Malek C. Hydrophilization and hydrophobic recovery of PDMS by oxygen plasma and chemical treatment—An SEM investigation. Sensor Actuat B Chem 2007;123:368–73. [Google Scholar]
- Boedicker JQ, Vincent ME, Ismagilov RF. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Ed Engl 2009;48:5908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma AJ, Zuhorn IS, Poolman B. A sensor for quantification of macromolecular crowding in living cells. Nat Methods 2015;12:227–9. [DOI] [PubMed] [Google Scholar]
- Boulineau S, Tostevin F, Kiviet DJ et al. Single-cell dynamics reveals sustained growth during diauxic shifts. PLoS One 2013;8:e61686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Donowitz M, Rao R. Does the proteome encode organellar pH? FEBS Lett 2006;580:717–9. [DOI] [PubMed] [Google Scholar]
- Britton RA, Kuster-Schock E, Auchtung TA et al. SOS induction in a subpopulation of structural maintenance of chromosome (SMC) mutant cells in Bacillus subtilis. J Bacteriol 2007;189:4359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AK, Engler A, Gulati A et al. Continuous and long-term volume measurements with a commercial coulter counter. PLoS One 2012;7: e29866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AK, Goranov A, Amon A et al. Measurement of mass, density, and volume during the cell cycle of yeast. Proc Natl Acad Sci U S A 2010;107:999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Surovtsev IV, Kato S et al. A constant size extension drives bacterial cell size homeostasis. Cell 2014;159:1433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro A, Gros O, Got P et al. Characterization of the population of the sulfur-oxidizing symbiont of Codakia orbicularis (bivalvia, lucinidae) by single-cell analyses. Appl Environ Microbiol 2007;73:2101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Juarez I, Maeda T, Mandujano-Tinoco EA et al. Role of quorum sensing in bacterial infections. World J Clin Cases 2015;3:575–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak N, Becker JW, Knudsen SM et al. Direct single-cell biomass estimates for marine bacteria via Archimedes' principle. ISME J 2017;11:825–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Li BL. Contribution of biodiversity to ecosystem functioning: A non-equilibrium thermodynamic perspective. J Arid Land 2011;3:71–4. [Google Scholar]
- Chowdhury S, Kandhavelu M, Yli-Harja O et al. Cell segmentation by multi-resolution analysis and maximum likelihood estimation (MAMLE). BMC Bioinformatics 2013;14(Suppl 10):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun BW, Dair B, Macuch PJ et al. The development of cement and concrete additive: Based on xylonic acid derived via bioconversion of xylose. Appl Biochem Biotechnol 2006;129–132:645–58. [PubMed] [Google Scholar]
- Clausell-Tormos J, Lieber D, Baret JC et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem Biol 2008;15:427–37. [DOI] [PubMed] [Google Scholar]
- Clingenpeel S, Clum A, Schwientek P et al. Reconstructing each cell's genome within complex microbial communities—Dream or reality? Front Microbiol 2015;5:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature 2005;437:699–706. [DOI] [PubMed] [Google Scholar]
- Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 1968;31:519–40. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Fundamentals of Enzyme Kinetics. 5th ed., London: Elsevier, 2014. [Google Scholar]
- Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res 2000;28:3504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David F, Hebeisen M, Schade G et al. Viability and membrane potential analysis of Bacillus megaterium cells by impedance flow cytometry. Biotechnol Bioeng 2012;109:483–92. [DOI] [PubMed] [Google Scholar]
- Davies MJ. Reactive species formed on proteins exposed to singlet oxygen. Photochem Photobiol Sci 2004;3:17–25. [DOI] [PubMed] [Google Scholar]
- De Bourcy CF, De Vlaminck I, Kanbar JN et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One 2014;9:e105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvigne F, Baert J, Sassi H et al. Taking control over microbial populations: Current approaches for exploiting biological noise in bioprocesses. Biotechnol J 2017;12:1600549. [DOI] [PubMed] [Google Scholar]
- Delvigne F, Goffin P. Microbial heterogeneity affects bioprocess robustness: Dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol J 2014;9:61–72. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip 2006;6:1445–9. [DOI] [PubMed] [Google Scholar]
- Dittrich P, Jakubowski N. Current trends in single cell analysis. Anal Bioanal Chem 2014;406:6957–61. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: Effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J 2003;36:280–90. [DOI] [PubMed] [Google Scholar]
- Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature 1968;219:1077–9. [DOI] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 2016;1:16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF. Atomic force microscopy, a powerful tool in microbiology. J Bacteriol 2002;184:5205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF. Atomic force microscopy in microbiology: New structural and functional insights into the microbial cell surface. MBio 2014;5:e01363–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusny C, Schmid A. Microfluidic single-cell analysis links boundary environments and individual microbial phenotypes. Environ Microbiol 2015a;17:1839–56. [DOI] [PubMed] [Google Scholar]
- Dusny C, Schmid A. Challenging biological limits with microfluidic single cell analysis. Microb Biotechnol 2015b;8:23–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusny C, Fritzsch FSO, Frick O et al. Isolated microbial single cells and resulting micropopulations grow faster in controlled environments. Appl Environ Microbiol 2012;78:7132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusny C, Gruenberger A, Probst C et al. Technical bias of microcultivation environments on single-cell physiology. Lab Chip 2015;15:1822–34. [DOI] [PubMed] [Google Scholar]
- Dusny C, Schmid A. The MOX promoter in Hansenula polymorpha is ultrasensitive to glucose-mediated carbon catabolite repression. FEMS Yeast Res 2016;16, DOI: 10.1093/femsyr/fow067. [DOI] [PubMed] [Google Scholar]
- Eggeling L, Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol 2015;99:3387–94. [DOI] [PubMed] [Google Scholar]
- Elani Y, Purushothaman S, Booth PJ et al. Measurements of the effect of membrane asymmetry on the mechanical properties of lipid bilayers. Chem Commun 2015;51:6976–9. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED et al. Stochastic gene expression in a single cell. Science 2002;297:1183–6. [DOI] [PubMed] [Google Scholar]
- Embree M, Nagarajan H, Movahedi N et al. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J 2014;8:757–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson E, Enger J, Nordlander B et al. A microfluidic system in combination with optical tweezers for analyzing rapid and reversible cytological alterations in single cells upon environmental changes. Lab Chip 2007;7:71–6. [DOI] [PubMed] [Google Scholar]
- Eriksson E, Sott K, Lundqvist F et al. A microfluidic device for reversible environmental changes around single cells using optical tweezers for cell selection and positioning. Lab Chip 2010;10:617–25. [DOI] [PubMed] [Google Scholar]
- Evander M, Johansson L, Lilliehorn T et al. Noninvasive acoustic cell trapping in a microfluidic perfusion system for online bioassays. Anal Chem 2007;79:2984–91. [DOI] [PubMed] [Google Scholar]
- Fievet A, Ducret A, Mignot T et al. Single-cell analysis of growth and cell division of the anaerobe Desulfovibrio vulgaris Hildenborough. Front Microbiol 2015;6:1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault MM, Lacoste J, Swift JL et al. Live-cell microscopy—Tips and tools. J Cell Sci 2009;122:753–67. [DOI] [PubMed] [Google Scholar]
- Fritz BR, Timmerman LE, Daringer N et al. Biology by design: From top to bottom and back. J Biomed Biotechnol 2010;2010:232016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch FSO, Dusny C, Frick O et al. Single-cell analysis in biotechnology, systems biology, and biocatalysis. Annu Rev Chem Biomol Eng 2012;3:129–55. [DOI] [PubMed] [Google Scholar]
- Fritzsch FS, Rosenthal K, Kampert A et al. Picoliter nDEP traps enable time-resolved contactless single bacterial cell analysis in controlled microenvironments. Lab Chip 2013;13:397–408. [DOI] [PubMed] [Google Scholar]
- Gao WM, Zhang WW, Meldrum DR. RT-qPCR based quantitative analysis of gene expression in single bacterial cells. J Microbiol Methods 2011;85:221–7. [DOI] [PubMed] [Google Scholar]
- Gefen O, Fridman O, Ronin I et al. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci U S A 2014;111:556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Beloin C, Ghigo JM et al. Bacteria hold their breath upon surface contact as shown in a strain of Escherichia coli, using dispersed surfaces and flow cytometry analysis. PLoS One 2014;9:e102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A, Dupres V, Delestrait G et al. Nanoscale imaging of Bacillus thuringiensis flagella using atomic force microscopy. Nanoscale 2012;4:1585–91. [DOI] [PubMed] [Google Scholar]
- Godin M, Bryan AK, Burg TP et al. Measuring the mass, density, and size of particles and cells using a suspended microchannel resonator. Appl Phys Lett 2007;91:123121. [Google Scholar]
- Grindberg RV, Ishoey T, Brinza D et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS One 2011;6:e18565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberger A, Paczia N, Probst C et al. A disposable picolitre bioreactor for cultivation and investigation of industrially relevant bacteria on the single cell level. Lab Chip 2012;12:2060–8. [DOI] [PubMed] [Google Scholar]
- Gruenberger A, Van Ooyen J, Paczia N et al. Beyond growth rate 0.6: Corynebacterium glutamicum cultivated in highly diluted environments. Biotechnol Bioeng 2013;110:220–8. [DOI] [PubMed] [Google Scholar]
- Gruenberger A, Wiechert W, Kohlheyer D. Single-cell microfluidics: Opportunity for bioprocess development. Curr Opin Biotechnol 2014;29:15–23. [DOI] [PubMed] [Google Scholar]
- Gupta A, Lloyd-Price J, Oliveira SMD et al. Robustness of the division symmetry in Escherichia coli and functional consequences of symmetry breaking. Phys Biol 2014;11:066005. [DOI] [PubMed] [Google Scholar]
- Haider S, Wagner M, Schmid MC et al. Raman microspectroscopy reveals long-term extracellular activity of chlamydiae. Mol Microbiol 2010;77:687–700. [DOI] [PubMed] [Google Scholar]
- Halan B, Buehler K, Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol 2012;30:453–65. [DOI] [PubMed] [Google Scholar]
- Halldorsson S, Lucumi E, Gomez-Sjoberg R et al. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron 2015;63:218–31. [DOI] [PubMed] [Google Scholar]
- Hammar P, Angermayr SA, Sjostrom SL et al. Single-cell screening of photosynthetic growth and lactate production by cyanobacteria. Biotechnol Biofuels 2015;8:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 2003;50:101–14. [DOI] [PubMed] [Google Scholar]
- Hebisch E, Knebel J, Landsberg J et al. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PLoS One 2013;8:e75991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen SJ. Thermodynamics of microbial-growth and its implications for process design. Trends Biotechnol 1994;12:483–92. [Google Scholar]
- Heine F, Stahl F, Sträuber H et al. Prediction of flocculation ability of brewing yeast inoculates by flow cytometry, proteome analysis, and mRNA profiling. Cytometry A 2009;75A:140–7. [DOI] [PubMed] [Google Scholar]
- Heinemann M, Zenobi R. Single cell metabolomics. Curr Opin Biotechnol 2011;22:26–31. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Riedel K, Rasmussen TB et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002;148:87–102. [DOI] [PubMed] [Google Scholar]
- Hermann T. Industrial production of amino acids by coryneform bacteria. J Biotechnol 2003;104:155–72. [DOI] [PubMed] [Google Scholar]
- Hollesen J, Henning M, Moller AB et al. Permafrost thawing in organic arctic soils accelerated by ground heat production. Nat Clim Change 2015;5:574–8. [Google Scholar]
- Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet 2011;43:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez AJ, Fagerer SR, Schmidt AM et al. Mass spectrometry-based metabolomics of single yeast cells. Proc Natl Acad Sci U S A 2013;110:8790–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Biswas S, Wright CS, Henry JT et al. Scaling laws governing stochastic growth and division of single bacterial cells. Proc Natl Acad Sci U S A 2014;111:15912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger MS, Mueller T, Schnelle T. Thermometry in dielectrophoresis chips for contact-free cell handling. J Phys D Appl Phys 2007;40:95–105. [Google Scholar]
- Jahn M, Guenther S, Mueller S. Non-random distribution of macromolecules as driving forces for phenotypic variation. Curr Opin Microbiol 2015;25:49–55. [DOI] [PubMed] [Google Scholar]
- Jaimes-Lizcano YA, Hunn DD, Papadopoulos KD. Filamentous Escherichia coli cells swimming in tapered microcapillaries. Res Microbiol 2014;165:166–74. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J Bacteriol 2003;185:3068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao N, Robinson C, Azam F et al. Mechanisms of microbial carbon sequestration in the ocean—Future research directions. Biogeosciences 2014;11:5285–306. [Google Scholar]
- Joensson HN, Andersson Svahn H. Droplet microfluidics—a tool for single-cell analysis. Angew Chem Int Ed Engl 2012;51:12176–92. [DOI] [PubMed] [Google Scholar]
- John DM, Weeks KM. van’t Hoff enthalpies without baselines. Protein Sci 2000;9:1416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Taheri-Araghi S. Cell-size maintenance: Universal strategy revealed. Trends Microbiol 2015;23:4–6. [DOI] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Cegelski L et al. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol 2008;6:162–8. [DOI] [PubMed] [Google Scholar]
- Kang Y, Norris MH, Zarzycki-Siek J et al. Transcript amplification from single bacterium for transcriptome analysis. Genome Res 2011;21:925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis J, Young PG. Intracellular pH homeostasis during cell-cycle progression and growth state transition in Schizosaccharomyces pombe. J Cell Sci 2001;114:2929–41. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 2014;344:416–20. [DOI] [PubMed] [Google Scholar]
- Keller L, Surette MG. Communication in bacteria: An ecological and evolutionary perspective. Nat Rev Microbiol 2006;4:249–58. [DOI] [PubMed] [Google Scholar]
- Khalili AA, Ahmad MR. Numerical analysis of hydrodynamic flow in microfluidic biochip for single-cell trapping application. Int J Mol Sci 2015;16:26770–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldgaard NO, Maaloe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol 1958;19:607–16. [DOI] [PubMed] [Google Scholar]
- Klumpp S, Hwa T. Bacterial growth: Global effects on gene expression, growth feedback and proteome partition. Curr Opin Biotechnol 2014;28:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen JD, Carlquist M, Gorwa-Grauslund M. NADH-dependent biosensor in Saccharomyces cerevisiae: Principle and validation at the single cell level. AMB Express 2014;4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Harms H, Mueller S. Dynamics in the microbial cytome-single cell analytics in natural systems. Curr Opin Biotechnol 2014;27C:134–41. [DOI] [PubMed] [Google Scholar]
- Kong Y, Nielsen JL, Nielsen PH. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl Environ Microbiol 2005;7:4076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann H, Blank LM, Schmid A. Single cell analytics: An overview. Adv Biochem Eng Biotechnol 2011;124:99–122. [DOI] [PubMed] [Google Scholar]
- Kortmann H, Chasanis P, Blank LM et al. The Envirostat—A new bioreactor concept. Lab Chip 2009a;9:576–85. [DOI] [PubMed] [Google Scholar]
- Kortmann H, Kurth F, Blank LM et al. Towards real time analysis of protein secretion from single cells. Lab Chip 2009b;9:3047–9. [DOI] [PubMed] [Google Scholar]
- Kotte O, Volkmer B, Radzikowski JL et al. Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol Syst Biol 2014;10:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone KM, Warias R, Ritter C et al. Analysis of enantioselective biotransformations using a few hundred cells on an integrated microfluidic chip. J Am Chem Soc 2016;138:2102–5. [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005;309:2075–8. [DOI] [PubMed] [Google Scholar]
- Lade H, Paul D, Kweon JH. N-Acyl homoserine lactone-mediated quorum sensing with special reference to use of quorum quenching bacteria in membrane biofouling control. Biomed Res Int 2014a;2014:162584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lade H, Paul D, Kweon JH. Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci 2014b;10:547–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdahl BR, Ingvorsen K. Temperature characteristics of bacterial iron solubilisation and C-14 assimilation in naturally exposed sulfide ore material at Citronen Fjord, North Greenland (83°N). FEMS Microbiol Ecol 1997;23:275–83. [Google Scholar]
- Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet 2014;15:577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauga E, Di Luzio WR, Whitesides GM et al. Swimming in circles: Motion of bacteria near solid boundaries. Biophys J 2006;90:400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Maskow T, Wolf G. Chip calorimetry and its use for biochemical and cell biological investigations. Chem Eng Process 2008;47:991–9. [Google Scholar]
- Lee SS, Avalos Vizcarra I, Huberts DH et al. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc Natl Acad Sci U S A 2012;109:4916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol 2012;10:e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qiu Y, Glidle A et al. Single cell growth rate and morphological dynamics revealing an “opportunistic” persistence. Analyst 2014a;139:3305–13. [DOI] [PubMed] [Google Scholar]
- Li B, Qiu Y, Glidle A et al. Gradient microfluidics enables rapid bacterial growth inhibition testing. Anal Chem 2014b;86:3131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MQ, Canniffe DP, Jackson PJ et al. Rapid resonance Raman microspectroscopy to probe carbon dioxide fixation by single cells in microbial communities. ISME J 2012b;6:875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MQ, Xu J, Romero-Gonzalez M et al. Single cell Raman spectroscopy for cell sorting and imaging. Curr Opin Biotechnol 2012a;23:56–63. [DOI] [PubMed] [Google Scholar]
- Li YH, Tian XL. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012;12:2519–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Cai W, Sun Z. Single-cell sequencing technologies: Current and future. J Genet Genomics 2014;41:513–28. [DOI] [PubMed] [Google Scholar]
- Lidstrom ME, Meldrum DR. Life-on-a-chip. Nat Rev Microbiol 2003;1:158–64. [DOI] [PubMed] [Google Scholar]
- Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol 2010;6:705–12. [DOI] [PubMed] [Google Scholar]
- Lin Y, Crosson S, Scherer NF. Single-gene tuning of Caulobacter cell cycle period and noise, swarming motility, and surface adhesion. Mol Syst Biol 2010;6:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010;327:46–50. [DOI] [PubMed] [Google Scholar]
- Liu L, Li Y, Li S et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012;2012:251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SJ, Yao DJ. Get to understand more from single cells: Current studies of microfluidic-based techniques for single-cell analysis. Int J Mol Sci 2015;16:16763–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol 2009;7:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang YF et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol 2009;7:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev 2010;24:1893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottspeich F, Engels JW. Bioanalytik. Munich, Heidelberg: Elsevier, Spektrum Akademischer Verlag, 2006. [Google Scholar]
- Love KR, Politano TJ, Panagiotou V et al. Systematic single-cell analysis of Pichia pastoris reveals secretory capacity limits productivity. PLoS One 2012;7:e37915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukyanov KA, Chudakov DM, Lukyanov S et al. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol 2005;6:885–91. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia M, Brazel DM, Swan BK et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 2012;7:e35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV et al. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J 2002;21:3108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KM, Lacherndo EJ, Albino-Flores I et al. LacI(Ts)-regulated expression as an in situ intracellular biomolecular thermometer. Appl Environ Microbiol 2011;77:2863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy WF, Bryers JD, Robbins J et al. Observations of fouling biofilm formation. Can J Microbiol 1981;27:910–7. [DOI] [PubMed] [Google Scholar]
- McGlynn SE, Chadwick GL, Kempes CP et al. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 2015;526:531–5. [DOI] [PubMed] [Google Scholar]
- Maennik J, Driessen R, Galajda P et al. Bacterial growth and motility in sub-micron constrictions. Proc Natl Acad Sci U S A 2009;106:14861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maennik J, Wu FB, Hol FJH et al. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc Natl Acad Sci U S A 2012;109:6957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majed N, Chernenko T, Diem M et al. Identification of functionally relevant populations in enhanced biological phosphorus removal processes based on intracellular polymers profiles and insights into the metabolic diversity and heterogeneity. Environ Sci Technol 2012;46:5010–7. [DOI] [PubMed] [Google Scholar]
- Marceau AH, Bahng S, Massoni SC et al. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J 2011;30:4236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques MP, Fernandes P. Microfluidic devices: Useful tools for bioprocess intensification. Molecules 2011;16:8368–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins BMC, Locke JOW. Microbial individuality: How single-cell heterogeneity enables population level strategies. Curr Opin Microbiol 2015;24:104–12. [DOI] [PubMed] [Google Scholar]
- Maskow T, Kemp R, Buchholz F et al. What heat is telling us about microbial conversions in nature and technology: From chip- to megacalorimetry. Microb Biotechnol 2010;3:269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbt SN, Stahl DA, Casamayor EO et al. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 2012;327:41–6. [DOI] [PubMed] [Google Scholar]
- Miesenboeck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998;394:192–5. [DOI] [PubMed] [Google Scholar]
- Milucka J, Ferdelman TG, Polerecky L et al. Zero-valent sulphur is a key intermediate in marine methane oxidation. Nature 2012;491:541–6. [DOI] [PubMed] [Google Scholar]
- Mir M, Wang Z, Shen Z et al. Optical measurement of cycle-dependent cell growth. Proc Natl Acad Sci U S A 2011;108:13124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: Cellular organization of genome function. Cell 2007;128:787–800. [DOI] [PubMed] [Google Scholar]
- Mitosch K, Rieckh G, Bollenbach T. Noisy response to antibiotic stress predicts subsequent single-cell survival in an acidic environment. Cell Syst 2017;4:393–403. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Tsuyama N, Date S et al. Live single-cell metabolomics of tryptophan and histidine metabolites in a rat basophil leukemia cell. Anal Sci 2008;24:1525–7. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Lee JB, Cluzel P. The single-cell chemostat: An agarose-based, microfluidic device for high-throughput, single-cell studies of bacteria and bacterial communities. Lab Chip 2012;12:1487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu Rev Microbiol 1949;3:371–94. [Google Scholar]
- Monod J. La technique de culture continue theorie et applications. Ann I Pasteur Paris 1950;79:390–410. [Google Scholar]
- Mueller S. Cytomics reaches microbiology—population heterogeneity on the protein level caused by chemical stress. Cytometry A 2008;73:3–4. [DOI] [PubMed] [Google Scholar]
- Mueller S, Harms H, Bley T. Origin and analysis of microbial population heterogeneity in bioprocesses. Curr Opin Biotechnol 2010;21:100–13. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: Elusive or illusive? Cell 2003;115:377–88. [DOI] [PubMed] [Google Scholar]
- Musat N, Foster R, Vagner T et al. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev 2012;36:486–511. [DOI] [PubMed] [Google Scholar]
- Mustafi N, Gruenberger A, Kohlheyer D et al. The development and application of a single-cell biosensor for the detection of l-methionine and branched-chain amino acids. Metab Eng 2012;14:449–57. [DOI] [PubMed] [Google Scholar]
- Mustafi N, Gruenberger A, Mahr R et al. Application of a genetically encoded biosensor for live cell imaging of l-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS One 2014;9:e85731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Miyoshi D, Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev 2014;114:2733–58. [DOI] [PubMed] [Google Scholar]
- Natarajan A, Srienc F. Dynamics of glucose uptake by single Escherichia coli cells. Metab Eng 1999;1:320–33. [DOI] [PubMed] [Google Scholar]
- Neu TR, Lawrence JR. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol 2015;23:233–42. [DOI] [PubMed] [Google Scholar]
- Nikel PI, Silva-Rocha R, Benedetti I et al. The private life of environmental bacteria: Pollutant biodegradation at the single cell level. Environ Microbiol 2014;16:628–42. [DOI] [PubMed] [Google Scholar]
- Nikolic N, Barner T, Ackermann M. Analysis of fluorescent reporters indicates heterogeneity in glucose uptake and utilization in clonal bacterial populations. BMC Microbiol 2013;13:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel PS. The Boyle-Van’t Hoff relation. J Theor Biol 1969;23:375–9. [DOI] [PubMed] [Google Scholar]
- Nolivos S, Sherratt D. The bacterial chromosome: Architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev 2014;38:380–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Lord ND, Paulsson J et al. Memory and modularity in cell-fate decision making. Nature 2013;503:481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygard Y, Maaheimo H, Mojzita D et al. Single cell and in vivo analyses elucidate the effect of xylC lactonase during production of d-xylonate in Saccharomyces cerevisiae. Metab Eng 2014;25:238–47. [DOI] [PubMed] [Google Scholar]
- Ong SE, Zhang S, Du HJ et al. Fundamental principles and applications of microfluidic systems. Front Biosci 2008;13:2757–73. [DOI] [PubMed] [Google Scholar]
- Orij R, Postmus J, Ter Beek A et al. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009;155:268–78. [DOI] [PubMed] [Google Scholar]
- Orij R, Urbanus ML, Vizeacoumar FJ et al. Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHC in Saccharomyces cerevisiae. Genome Biol 2012;13:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulas A, Pavloudi C, Polymenakou P et al. Metagenomics: Tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol Insights 2015;9:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M et al. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 2016;99:767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli MK, Ewing AG. Single-cell imaging mass spectrometry. Curr Opin Chem Biol 2013;17:854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013;497:127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Halvorsen K, Ha BY et al. Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc Natl Acad Sci U S A 2012;109:2649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez PD, Hagen SJ. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One 2010;5:e15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Sticky situations: Key components that control bacterial surface attachment. J Bacteriol 2012;194:2413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plener L, Lorenz N, Reiger M et al. The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J Bacteriol 2015;197:1747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G, Park Y, Lue N et al. Optical imaging of cell mass and growth dynamics. Am J Physiol Cell Physiol 2008;295:C538–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu G, Park K, Mir M et al. New technologies for measuring single cell mass. Lab Chip 2014;14:646–52. [DOI] [PubMed] [Google Scholar]
- Probst AJ, Weinmaier T, DeSantis TZ et al. New perspectives on microbial community distortion after whole-genome amplification. PLoS One 2015;10:e0124158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst C, Gruenberger A, Wiechert W et al. Polydimethylsiloxane (PDMS) sub-micron traps for single-cell analysis of bacteria. Micromachines 2013a;4:357–69. [Google Scholar]
- Probst C, Gruenberger A, Wiechert W et al. Microfluidic growth chambers with optical tweezers for full spatial single-cell control and analysis of evolving microbes. J Microbiol Methods 2013b;95:470–6. [DOI] [PubMed] [Google Scholar]
- Qian C, Huang HB, Chen LG et al. Dielectrophoresis for bioparticle manipulation. Int J Mol Sci 2014;15:18281–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc 2010;5:491–502. [DOI] [PubMed] [Google Scholar]
- Regehr KJ, Domenech M, Koepsel JT et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009;9:2132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard F, van der Meer JR. Microcolony growth assays. In: Timmis KN, de Lorenzo V, McGenity T. et al. (eds). Handbook of Hydrocarbon and Lipid Microbiology. Springer, 2010, 3562–70. [Google Scholar]
- Reinhard F, Miyazaki R, Pradervand N et al. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr Biol 2013;23:255–9. [DOI] [PubMed] [Google Scholar]
- Riordon J, Nash M, Jing W et al. Quantifying the volume of single cells continuously using a microfluidic pressure-driven trap with media exchange. Biomicrofluidics 2014;8:011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach LS, Song H, Ismagilov RF. Controlling nonspecific protein adsorption in a plug-based microfluidic system by controlling interfacial chemistry using fluorous-phase surfactants. Anal Chem 2005;77:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal K, Falke F, Frick O et al. An inert continuous microreactor for the isolation and analysis of a single microbial cell. Micromachines 2015;6:1836–55. [Google Scholar]
- Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat Biotechnol 2008;26:1117–24. [DOI] [PubMed] [Google Scholar]
- Rusconi R, Garren M, Stocker R. Microfluidics expanding the frontiers of microbial ecology. Annu Rev Biophys 2014;43:65–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs CC, Grunberger A, Helfrich S et al. Image-based single cell profiling: High-throughput processing of mother machine experiments. PLoS One 2016;11:e0163453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si XY, Xiong XC, Zhang SC et al. Detecting lowabundance molecules at single-cell level by repeated ion accumulation in ion trap mass spectrometer. Anal Chem 2017;89:2275–81. [DOI] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell 1998;5:639–48. [DOI] [PubMed] [Google Scholar]
- Schaechter M. A brief history of bacterial growth physiology. Front Microbiol 2015;6:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Williams JP, Koch AL et al. Growth, cell and nuclear divisions in some bacteria. J Gen Microbiol 1962;29:421–34. [DOI] [PubMed] [Google Scholar]
- Schaefer K, Lantuit H, Romanovsky VE et al. The impact of the permafrost carbon feedback on global climate. Environ Res Lett 2014;9:085003. [Google Scholar]
- Schlafer S, Garcia JE, Greve M et al. Ratiometric imaging of extracellular pH in bacterial biofilms with C-SNARF-4. Appl Environ Microbiol 2015;81:1267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Kortmann H, Dittrich PS et al. Chemical and biological single cell analysis. Curr Opin Biotechnol 2010;21:12–20. [DOI] [PubMed] [Google Scholar]
- Schneider J, Klein T, Mielich-Suss B et al. Spatio-temporal remodeling of functional membrane microdomains organizes the signaling networks of a bacterium. PLoS Genet 2015;11:e1005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, Shapiro L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol Microbiol 2011;82:1359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolari VF, Sclavi B, Cosentino Lagomarsino M. The nucleoid as a smart polymer. Front Microbiol 2015;6:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM et al. Interdependence of cell growth and gene expression: Origins and consequences. Science 2010;330:1099–102. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods 2005;2:905–9. [DOI] [PubMed] [Google Scholar]
- Shaw AS. Lipid rafts: Now you see them, now you don’t. Nat Immunol 2006;7:1139–42. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol 2008;26:1135–45. [DOI] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 2011;5:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Foti JJ, Cohen SE et al. The SOS regulatory network. Ecosal Plus 2008;2008: 10.1128/ecosalplus.5.4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T et al. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 2011;80:612–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solopova A, Van Gestel J Weissing FJ et al. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A 2014;111:7427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Loparo JJ. Building bridges within the bacterial chromosome. Trends Genet 2015;31:164–73. [DOI] [PubMed] [Google Scholar]
- Song Y, Kaster AK, Vollmers J et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb Biotechnol 2017;10:125–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon W, Baliunas SL, Robinson AB et al. Environmental effects of increased atmospheric carbon dioxide. Climate Res 1999;13:149–64. [Google Scholar]
- Sorg RA, Veening JW. Microscale insights into pneumococcal antibiotic mutant selection windows. Nat Commun 2015;6:8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhaus B, Garcia ML, Shen AQ et al. A portable anaerobic microbioreactor reveals optimum growth conditions for the methanogen Methanosaeta concilii. Appl Environ Microbiol 2007;73:1653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R. Single cell genomics: An individual look at microbes. Curr Opin Microbiol 2012;15:613–20. [DOI] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul G et al. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol 2005;3:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracy M, Uphoff S, De Leon FG et al. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett 2015;589:787. [DOI] [PubMed] [Google Scholar]
- Straeuber H, Huebschmann T, Jehmlich N et al. NBDT (3-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-3-toluene)—A novel fluorescent dye for studying mechanisms of toluene uptake into vital bacteria. Cytometry A 2010;77:113–20. [DOI] [PubMed] [Google Scholar]
- Stratford M, Steels H, Nebe-von-Caron G et al. Population heterogeneity and dynamics in starter culture and lag phase adaptation of the spoilage yeast Zygosaccharomyces bailii to weak acid preservatives. Int J Food Microbiol 2014;181:40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratz S, Eyer K, Kurth F et al. On-chip enzyme quantification of single Escherichia coli bacteria by immunoassay-based analysis. Anal Chem 2014;86:12375–81. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV. SMC complexes in bacterial chromosome condensation and segregation. Plasmid 2006;55:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 2009;137:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Biological applications of optical forces. Annu Rev Biophys Biomol Struct 1994;23:247–85. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 2011;333:1296–300. [DOI] [PubMed] [Google Scholar]
- Taheri-Araghi S, Brown SD, Sauls JT et al. Single-cell physiology. Annu Rev Biophys 2015a;44:123–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT et al. Cell-size control and homeostasis in bacteria. Curr Biol 2015b;25:385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, Di Luzio WR, Weibel DB et al. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett 2005;5:1819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010;329:533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanouchi Y, Pai A, Park H. et al, A noisy linear map underlies oscillations in cell size and gene expression in bacteria. Nature 2015;523:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanouchi Y, Pai A, Park H. et al, Long-term growth data of Escherichia coli at a single-cell level. Sci Data 2017;4:170036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai T, Nagano T. Small-molecule fluorophores and fluorescent probes for bioimaging. Pflugers Arch 2013;465:347–59. [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Dragavon J, Baba-Aissa L et al. A quantitative method for measuring phototoxicity of a live cell imaging microscope. Methods Enzymol 2012;506:291–309. [DOI] [PubMed] [Google Scholar]
- Toivari M, Nygard Y, Kumpula EP et al. Metabolic engineering of Saccharomyces cerevisiae for bioconversion of d-xylose to d-xylonate. Metab Eng 2012;14:427–36. [DOI] [PubMed] [Google Scholar]
- Toivari MH, Ruohonen L, Richard P et al. Saccharomyces cerevisiae engineered to produce d-xylonate. Appl Microbiol Biotechnol 2010;88:751–60. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Yoshida S, Yoshida A et al. Cationic fluorescent polymeric thermometers with the ability to enter yeast and mammalian cells for practical intracellular temperature measurements. Anal Chem 2013;85:9815–23. [DOI] [PubMed] [Google Scholar]
- Turner L, Zhang R, Darnton NC et al. Visualization of flagella during bacterial swarming. J Bacteriol 2010;192:3259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman G, Wallden M, Marklund EG et al. High-throughput gene expression analysis at the level of single proteins using a microfluidic turbidostat and automated cell tracking. Philos Trans R Soc Lond B Biol Sci 2013;368:20120025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara S, Wakamoto Y, Inoue I et al. On-chip single-cell microcultivation assay for monitoring environmental effects on isolated cells. Biochem Biophys Res Commun 2003;305:534–40. [DOI] [PubMed] [Google Scholar]
- Unthan S, Gruenberger A, Van Ooyen J et al. Beyond growth rate 0.6: What drives Corynebacterium glutamicum to higher growth rates in defined medium. Biotechnol Bioeng 2014;111:359–71. [DOI] [PubMed] [Google Scholar]
- Uphoff S, Kapanidis AN. Studying the organization of DNA repair by single-cell and single-molecule imaging. DNA Repair 2014;20:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban PL, Jefimovs K, Amantonico A et al. High-density micro-arrays for mass spectrometry. Lab Chip 2010;10:3206–9. [DOI] [PubMed] [Google Scholar]
- Valkonen M, Mojzita D, Penttila M et al. Noninvasive high-throughput single-cell analysis of the intracellular pH of Saccharomyces cerevisiae by ratiometric flow cytometry. Appl Environ Microbiol 2013;79:7179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkonen M, Penttila M, Bencina M. Intracellular pH responses in the industrially important fungus Trichoderma reesei. Fungal Genet Biol 2014;70:86–93. [DOI] [PubMed] [Google Scholar]
- Valli M, Sauer M, Branduardi P et al. Intracellular pH distribution in Saccharomyces cerevisiae cell populations, analyzed by flow cytometry. Appl Environ Microbiol 2005;71:1515–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli M, Sauer M, Branduardi P et al. Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl Environ Microbiol 2006;72:5492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE. Fluorescent biosensors of protein function. Curr Opin Chem Biol 2008;12:60–5. [DOI] [PubMed] [Google Scholar]
- Vasdekis AE, Stephanopoulos G. Review of methods to probe single cell metabolism and bioenergetics. Metab Eng 2015;27:115–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeant MA, Ford RM, Wagner M et al. Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl Environ Microbiol 2002;68:2794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram A, Jesudhasan PR, Jayaprakasha GK et al. Citrus limonoids interfere with Vibrio harveyi cell-cell signalling and biofilm formation by modulating the response regulator LuxO. Microbiology 2011;157:99–110. [DOI] [PubMed] [Google Scholar]
- Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng 2006;8:425–54. [DOI] [PubMed] [Google Scholar]
- Von Stockar U, Briou B. The heat generated by yeast cultures with a mixed metabolism in the transition between respiration and fermentation. Biotechnol Bioeng 1989;34:86–101. [DOI] [PubMed] [Google Scholar]
- Von Stockar U, Gustafsson L, Larrson C et al. Thermodynamic considerations in constructing energy balances for cellular growth. Biochim Biophys Acta 1993;1183:221–40. [Google Scholar]
- Von Stockar U, Vojinovic V, Maskow T et al. Can microbial growth yield be estimated using simple thermodynamic analogies to technical processes? Chem Eng Process 2008;47:980–90. [Google Scholar]
- Wachtler V, Rajagopalan S, Balasubramanian MK. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci 2003;116:867–74. [DOI] [PubMed] [Google Scholar]
- Wagner M, Nielsen PH, Loy A et al. Linking microbial community structure with function: Fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr Opin Biotechnol 2006;17:83–91. [DOI] [PubMed] [Google Scholar]
- Wallden M, Fange D, Lundius EG. et al, The synchronization of replication and division cycles in individual E. coli cells. Cell 2016;166:729–39. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen L, Chen Z et al. RNA-seq based transcriptomic analysis of single bacterial cells. Integr Biol 2015;7:1466–76. [DOI] [PubMed] [Google Scholar]
- Wang P, Robert L, Pelletier J et al. Robust growth of Escherichia coli. Curr Biol 2010a;20:1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Arellano-Santoyo H, Combs PA et al. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci U S A 2010b;107:9182–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Moffitt JR, Dempsey GT et al. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci U S A 2014;111:8452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Li GW, Chen CY et al. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 2011a;333:1445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen S, Kong M et al. Enhanced cell sorting and manipulation with combined optical tweezer and microfluidic chip technologies. Lab Chip 2011b;11:3656–62. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 2005;21:319–46. [DOI] [PubMed] [Google Scholar]
- Wegner AS, Alexeeva S, Odijk T et al. Characterization of Escherichia coli nucleoids released by osmotic shock. J Struct Biol 2012;178:260–9. [DOI] [PubMed] [Google Scholar]
- Weibel DB, Di Luzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol 2007;5:209–18. [DOI] [PubMed] [Google Scholar]
- Weigert C, Steffler F, Kurz T et al. Application of a short intracellular pH method to flow cytometry for determining Saccharomyces cerevisiae vitality. Appl Environ Microbiol 2009;75:5615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland M, Bumann D. Optimization of GFP levels for analyzing Salmonella gene expression during an infection. FEBS Lett 2002;521:105–8. [DOI] [PubMed] [Google Scholar]
- Weng Y, Delgado FF, Son S et al. Mass sensors with mechanical traps for weighing single cells in different fluids. Lab Chip 2011;11:4174–80. [DOI] [PubMed] [Google Scholar]
- Westerwalbesloh C, Gruenberger A, Stute B et al. Modeling and CFD simulation of nutrient distribution in picoliter bioreactors for bacterial growth studies on single-cell level. Lab Chip 2015;15:4177–86. [DOI] [PubMed] [Google Scholar]
- Westerwalbesloh C, Grunberger A, Wiechert W et al. Coarse-graining bacteria colonies for modelling critical solute distributions in picolitre bioreactors for bacterial studies on single-cell level. Microb Biotechnol 2017;10:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 1999;4:683–94. [DOI] [PubMed] [Google Scholar]
- Winderickx J, De Winde JH, Crauwels M. et al, Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol Gen Genet 1996;252:470–82. [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Ostuni E, Takayama S et al. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng 2001;3:335–73. [DOI] [PubMed] [Google Scholar]
- Wimpenny J, Manz W, Szewzyk U. Heterogeneity in biofilms. FEMS Microbiol Rev 2000;24:661–71. [DOI] [PubMed] [Google Scholar]
- Woodward JR, Cirillo VP, Edmunds LN Jr. Light effects in yeast: Inhibition by visible light of growth and transport in Saccharomyces cerevisiae grown at low temperatures. J Bacteriol 1978;133:692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FB, Van Schie BGC, Keymer JE et al. Symmetry and scale orient Min protein patterns in shaped bacterial sculptures. Nat Nanotechnol 2015a;10:719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Moser C, Wang HZ et al. Strategies for combating bacterial biofilm infections. Int J Oral Sci 2015b;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma H, Kawai S, Tabata KV et al. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci Rep 2014;4:6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JW, Locke JCW, Altinok A et al. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat Protoc 2012;7:80–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JW, Locke JC, Elowitz MB. Rate of environmental change determines stress response specificity. Proc Natl Acad Sci U S A 2013;110:4140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S et al. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev 2014;28:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 1999;32:315–26. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang T, Zhou Q et al. Development of a facile droplet-based single-cell isolation platform for cultivation and genomic analysis in microorganisms. Sci Rep 2017;7:41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangle TA, Teitell MA. Live-cell mass profiling: An emerging approach in quantitative biophysics. Nat Methods 2014;11:1221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zare RN, Kim S. Microfluidic platforms for single-cell analysis. Annu Rev Biomed Eng 2010;12:187–201. [DOI] [PubMed] [Google Scholar]
- Zeng G, Mueller T, Meyer RL. Single-cell force spectroscopy of bacteria enabled by naturally derived proteins. Langmuir 2014;30:4019–25. [DOI] [PubMed] [Google Scholar]
- Zenobi R. Single-cell metabolomics: Analytical and biological perspectives. Science 2013;342:6163. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu KK. Optical tweezers for single cells. J R Soc Interface 2008;5:671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PR, Ren LH, Zhang X et al. Raman-activated cell sorting based on dielectrophoretic single-cell trap and release. Anal Chem 2015;87:2282–9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Khodakov DA, Ellis AV et al. Surface modification for PDMS-based microfluidic devices. Electrophoresis 2012;33:89–104. [DOI] [PubMed] [Google Scholar]
- Zipfel WR, Williams RM, Webb WW. Nonlinear magic: Multiphoton microscopy in the biosciences. Nat Biotechnol 2003;21:1369–77. [DOI] [PubMed] [Google Scholar]