Abstract
Metals are essential for life, and they play a central role in the struggle between infecting microbes and their hosts. In fact, an important aspect of microbial pathogenesis is the ‘nutritional immunity’, in which metals are actively restricted (or, in an extended definition of the term, locally enriched) by the host to hinder microbial growth and virulence. Consequently, fungi have evolved often complex regulatory networks, uptake and detoxification systems for essential metals such as iron, zinc, copper, nickel and manganese. These systems often differ fundamentally from their bacterial counterparts, but even within the fungal pathogens we can find common and unique solutions to maintain metal homeostasis. Thus, we here compare the common and species-specific mechanisms used for different metals among different fungal species—focusing on important human pathogens such as Candida albicans, Aspergillus fumigatus or Cryptococcus neoformans, but also looking at model fungi such as Saccharomyces cerevisiae or A. nidulans as well-studied examples for the underlying principles. These direct comparisons of our current knowledge reveal that we have a good understanding how model fungal pathogens take up iron or zinc, but that much is still to learn about other metals and specific adaptations of individual species—not the least to exploit this knowledge for new antifungal strategies.
Keywords: transition metals, pathogenic fungi, nutritional immunity, metal homeostasis, host–pathogen interactions, regulatory networks
Pathogenic fungi require metals to survive and cause disease in the host. Their complex regulatory, uptake and detoxification systems are often uniquely adapted to conditions in vivo. This review compares and contrasts metal homeostasis mechanisms of human fungal pathogens.
INTRODUCTION
Fungi are frequently underestimated as causes of disease and death worldwide—by the public, by health practitioners, and even by national and global health organizations (Brown et al. 2012). Because of their often high mortality rates, infections with invasive fungi from genera as diverse as Candida, Aspergillus, Cryptococcus, Histoplasma, Paracoccidioides or Blastomyces are responsible for about one and a half million deaths per year (Brown et al. 2012), and non-fatal infections will affect most people at least once in their lifetime, with correspondingly high costs for healthcare systems worldwide. The search for fungal virulence factors and thus potential new drug targets in these eukaryotic pathogens is therefore all the more important.
Metals play a surprisingly central role in infection processes, as they serve as cofactors in a multitude of enzymes—including many with direct and indirect roles in virulence, such as metal-dependent superoxide dismutases (SODs), metalloproteases or melanin-producing laccases. Especially the first-row transition metals—manganese (Mn), iron (Fe), cobalt (Co), nickel (Ni) and copper (Cu)—provide the necessary redox and catalytic activity for many important biological processes. Their ionization energies increase slowly both over the row and for subsequent ionization events in the same metal. In the case of first-row transition metals, this is due to the shielding effect of their 3d-electrons on the 4s-electrons, and these are first lost during ionization. In fact, all these transition metals thus have a stable +2 oxidation state (lacking the 4s-electrons) and generally many additional stable states (up to seven in the case of Mn), which allows them to readily change their oxidation states in biological reactions. Zinc (Zn), with its single oxidation state (+2) and its filled d-orbital, is a notable exception, but nonetheless plays important roles especially in eukaryotic gene regulation.
The host is similarly dependent on metals, and should theoretically present a near optimal, metal-rich environment for infecting microbes. However, this is counterintuitively not the case, a fact that helps our intact immune system to fend off pathogenic fungi and bacteria. This is due to a process aptly named ‘nutritional immunity’, where the host actively sabotages and counteracts metal uptake by microorganisms (Weinberg 1975) and to make matters worse—as seen from the pathogen's side—can also fight invaders by deploying toxic levels of certain metals (Hood and Skaar 2012). Iron, copper and manganese, for example, are intrinsically toxic via Fenton chemistry (Fenton 1894), the metal-catalyzed generation of oxygen radical species from hydrogen peroxide, which at high metal concentrations results in oxidative damage to the microbes (Higson, Kohen and Chevion 1988; Touati 2000). Furthermore, many of the common biological metals have similar divalent cation properties in binding ligands, but strikingly different catalytic functions. Mismetallation, i.e. the replacement of an enzyme's metal cofactor by a different metal by host-induced metal excess and oxidative stress (reviewed in Imlay 2014), could thus inhibit the function of microbial enzymes that require defined metals as cofactors (Macomber and Imlay 2009; McDevitt et al. 2011; Veyrier et al. 2011). Consequently, the pathogens must keep these essential metals within strict homeostatic boundaries even when moving through rapidly changing metal microenvironments within the host. Finally, in biologically relevant pH ranges, these metals are frequently more soluble under acidic conditions, which results in often pH-dependent systems of metal homeostasis, many of which are described below.
Many of the metal conditions in microbial organisms still reflect the environment that we envision to have existed during the emergence of life. Then, iron was mainly present in its ferrous form (Fe2+)—due to the anoxic environment, which also led to copper and other soft metals to be trapped away in sulfide minerals. Especially eukaryotes, like fungi, later learned to include zinc and, to a certain extent, copper into the spectrum of biologically useful metals. Still, the profound differences between the evolutionary inherited patterns of metal use and the modern lower availability of iron (mostly ferric (Fe3+) rather than ferrous, due to the newly oxic conditions), and the relative abundance of soft metals, like copper, presents a continuing challenge to microbes, which nonetheless may have ‘trained’ the microorganisms to better deal with the metal-based nutritional immunity of mammals.
In fact, pathogenic fungi have developed often complex and advanced detection and signaling networks to upregulate the import of specific metals in times of need. Frequently, biological processes that rely on these metals are downregulated by dedicated regulators, reducing the consumption and liberating the bound metal. Under metal excess, often (but not always) a different regulator stops the expression of importers and initiates the sequestration of surplus metal to special proteins like metallothioneins (MTs) or to the vacuole, which serves as an overflow basin and emergency reservoir for many different metals. Many transporters have evolved that allow the transport of the charged metal ions over the plasma or vacuolar membranes, but unspecific transport of several metals by the same transporter is not uncommon—bringing with it the danger of the loss of full control over the metals that enter the cell and possibly leaving the microbe vulnerable to metal toxicity (Liu et al. 1997; Li and Kaplan 1998; Viau et al. 2012; Caetano et al. 2015).
Excellent recent reviews exist on many aspects of bacterial metal use, and among those we highly recommend (Palmer and Skaar 2016) for readers interested in non-fungal systems. On the topic of nutritional immunity, we recommend (Hood and Skaar 2012) for an outstanding overview of metal-related bacteria–host interactions, and (Crawford and Wilson 2015) for a view on common fungal pathogens. For an in-depth view on individual metals and their role in microbial pathogenesis, we refer the reader to Garcia-Santamarina and Thiele (2015) for copper, and for iron to Ganz and Nemeth (2015) and Soares and Weiss (2015) for a host view and Bairwa, Hee Jung and Kronstad (2017) for the fungal side.
In this review, we compile and compare strategies that fungi employ to obtain metals during pathogenesis, and we provide examples for different homeostatic mechanisms, and how they connect to fungal virulence. To this end, we summarize here the basic principles of homeostatic regulation in pathogenic fungi for iron, zinc, copper and manganese—metals for which a sufficiently large body of literature exists. The direct comparisons of known mechanisms among fungi will, we hope, allow the reader to discover common principles and identify open questions in order to complete our picture of the role of metals in fungal infections.
IRON
Most texts on microbial metal homeostasis start with a focus on iron. This is for good reason, as iron is the most abundant of the trace metals in organisms and arguably the one with the most diverse roles in cellular processes. These include central metabolic pathways such as oxygen transport, the tricarboxylic acid (TCA) cycle or electron transport chains, mostly via incorporation of iron or the iron-containing prosthetic group heme into the active centers of key enzymes. For these reasons, iron is an essential metal in nearly all organisms (Borrelia burgdorferi, the causative agent of Lyme disease, is one of the rare and notable exceptions; Posey and Gherardini 2000). While the ubiquity of iron is related to its chemical redox properties, namely the capacity to readily switch between the ferric and the ferrous form, this same quality is also at the root of the problems that can be caused by iron in many biological systems. For instance Fe3+, the prevalent form under aerobic conditions, is essentially insoluble in water and hence inaccessible to most microbes. Fe2+ in contrast is much more soluble, but at the same time more prone to elicit iron-induced toxicity mediated by the formation of radicals via the Fenton reaction. Additionally, iron, similar to copper, has a high affinity to replace other metals in enzymatic reactive centers, a mismetallation that usually results in a disruption of the enzymatic function (Vance and Miller 1998; Martin and Imlay 2011).
Accordingly, vertebrates and microorganisms alike have developed sophisticated strategies to ensure solubility, distribution and steady supply of iron while keeping its homeostatic levels sufficiently low to prevent toxicity. In vertebrates, this includes the almost complete binding of iron via a plethora of transport and storage proteins, such as hemoglobin, transferrin, lactoferrin and ferritin (reviewed in Wang and Pantopoulos 2011). During infection, microbial access to iron (and other metals) is actively restricted even further by nutritional immunity mechanisms (Weinberg 1975). This occurs at the systemic level by hepcidin-induced reduction of circulating iron (Nemeth et al. 2004) and at the tissue level by the active redistribution of iron away from sites of infection (Potrykus et al. 2013). In these processes, iron is shuttled to intracellular stores to keep it out of reach of invading pathogens—predominantly in macrophages, which also act as natural heme recycling sites via phagocytosis of senescent erythrocytes (reviewed in Wang and Pantopoulos 2011).
However, a range of microbial pathogens have adopted an intracellular lifestyle and use macrophages as hiding places from the immune system, or even as a source of nutrients and metals for their own growth. This includes many pathogenic fungi such as the dimorphic ascomycete Histoplasma capsulatum (Newman et al. 1994; Hwang et al. 2008), the basidiomycete Cryptococcus neoformans (Levitz et al. 1997), the yeast-like ascomycete Candida glabrata (Nevitt and Thiele 2011; Seider et al. 2014) and other dimorphic ascomycetes e.g. Paracoccidioides brasiliensis (Cano et al. 1994) or Blastomyces dermatitidis (Sterkel et al. 2015). All these species are able to survive phagocytosis and replicate inside macrophages, and they use diverse strategies in order to exploit the intracellular iron stores of macrophages, not all of which have yet been elucidated (Hilty, Smulian and Newman 2008, 2011; Nevitt and Thiele 2011; Hu et al. 2015).
Iron homeostasis and uptake
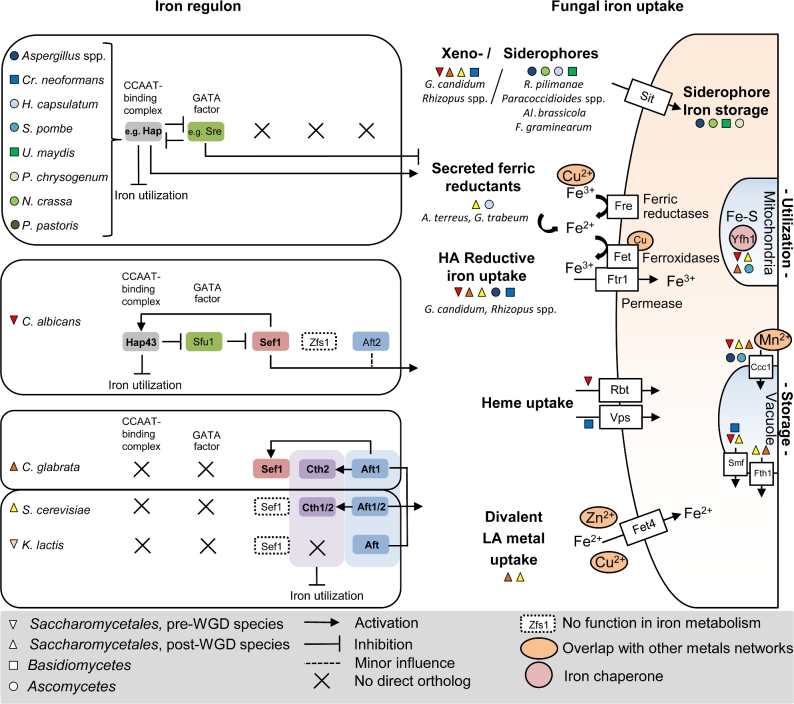
Pathogens have evolved elaborate systems to acquire iron from their environment (Fig. 1). A common theme in iron uptake is the utilization of siderophores, a heterogeneous class of small molecules, which are secreted by bacteria and fungi to bind extracellular ferric iron with extremely high affinity. This is achieved by coordinating Fe3+ by normally six oxygen ligands per molecule in an octahedral geometry, although siderophores with less donor atoms per molecule can bind in stochiometries different from 1:1 or use water as an additional oxygen donor. Siderophore–iron complexes are then either taken up directly or they deliver their precious load to receptors of the microbe's surface for uptake via specific transporters (reviewed for fungi in Haas, Eisendle and Turgeon 2008). Like in bacteria, many different classes of fungal siderophores are known, such as the most commonly produced hydroxamates [triacetylfusarinine C (Charlang et al. 1981; Oide et al. 2006; Schrettl et al. 2007), coprogens (Matzanke et al. 1987), ferrichromes (Neilands 1952), rhodotorulic acid (Muller, Barclay and Raymond 1985)], polycarboxylates produced by zygomycetes (Thieken and Winkelmann 1992) and phenolates-catecholates, which are present in wood-rotting fungi (Fekete, Chandhoke and Jellison 1989). Some fungal siderophores have highly specialized roles: Aspergillus fumigatus and A. nidulans ferricrocins, for example, are found inside the fungus rather than being secreted, and are involved in intracellular iron homeostasis and storage (Eisendle et al. 2006; Schrettl et al. 2007; Gsaller et al. 2012). Similarly, ferrichromes of the plant-pathogenic fungi Ustilago sphaerogena and U. maydis can be secreted or store iron intracellularly (Ecker, Lancaster and Emery 1982; Budde and Leong 1989). Importantly, Fe3+ bound to siderophores, due to their strongly negative redox potential, is not readily reduced to Fe2+ and hence will not generate hydroxyl radicals (Cornish and Page 1998). By this mechanism, intracellular siderophores can help to protect microbes from the toxic effects of iron (Eisendle et al. 2006).
Figure 1.
Fungal iron homeostasis. Regulation of iron homeostasis (left panel side) is shown for different fungal species (species is color coded, shape defines phylogenetic ancestry according to Gabaldon et al. 2013). Major transcription factors upregulated during iron starvation to initiate fungal iron uptake (right panel side) are written in bold. Functional orthologs are color shaded and aligned vertically, X indicates lack of ortholog and a white box with dashed borders indicates that an ortholog is present but not involved in iron homeostasis. HA, high affinity; LA, low affinity.
Overall, siderophore producers are widespread in the fungal kingdom and include animal and human pathogens such as Aspergillus spp. (Zähner et al. 1963; Nilius and Farmer 1990; Gressler et al. 2015), H. capsulatum (Howard et al. 2000), Rhodotorula pilimanae (Carrano and Raymond 1978), Neurospora crassa (Horowitz et al. 1976), Paracoccidioides spp. (Silva-Bailao et al. 2014) and the plant pathogens U. maydis (Budde and Leong 1989) and Alternaria brassicicola (Oide et al. 2006), among many others. In fact, siderophores are essential for the virulence of most fungal pathogens producing them. Deletion mutants lacking siderophore synthesis genes show severe virulence defects in A. fumigatus (Schrettl et al. 2004; Hissen et al. 2005), and also in H. capsulatum (Hwang et al. 2008). Consequently, the host has been shown to sequester fungal (and bacterial) siderophores via siderocalins, special siderophore-binding lipocalins (Goetz et al. 2002; Leal et al. 2013). Notably, the cellular energy cost to sustain siderophore synthesis is rather high for the microbe. Hence, biosynthesis is generally tightly controlled and activated solely upon significant iron shortage (Mei, Budde and Leong 1993; Oberegger et al. 2001). In addition, many fungal species, including C. albicans, C. glabrata or Saccharomyces cerevisiae, as well as Cr. neoformans, Geotrichum candidum and Rhizopus spp., lack the key enzyme L-ornithine N5-oxygenase (Sid1/SidA), which is needed for the initiation of hydroxamate siderophore biosynthesis, and they thus do not produce their own siderophores (reviewed in Haas, Eisendle and Turgeon 2008). Controversially, siderophore production was reported for C. albicans (Ismail, Bedell and Lupan 1985), but no putative biosynthesis genes were subsequently found in the genome.
Lacking their own biosynthetic machinery, these species often rely on xenosiderophores, i.e. siderophores produced by other fungi or bacteria. Dedicated xenosiderophore transporters with different substrate specificities have evolved, e.g. Sit1 homologs for hydroxamate-type fungal siderophores in C. glabrata (Nevitt and Thiele 2011), C. albicans (Heymann et al. 2002; Lesuisse et al. 2002), Cr. neoformans (Tangen et al. 2007) and S. cerevisiae [Arn1–4, with Arn3 and Arn4 specific for bacterial ferroxamines and Enterobactin B, respectively (Heymann, Ernst and Winkelmann 2000a,b; Yun et al. 2000)] and in many other fungi. Candida glabrata Sit1 enhances fungal survival in macrophages (Nevitt and Thiele 2011), and C. albicans Sit1 is required for invasion of human epithelial cells in vitro (Heymann et al. 2002); in the absence of xenosiderophores, these observations seem puzzling, and although mammals were recently found to produce siderophores (Devireddy et al. 2010), these are similar to enterobactin und thus unlikely to be taken up via Sit1. Accordingly, SIT1 deletion causes no attenuation in virulence of C. albicans in a systemic mouse model of infection (Hu et al. 2002). Similarly, Cr. neoformans Sit1 deletion mutants showed changes in melanin and capsule formation and in cell wall density, but were not reduced in virulence (Tangen et al. 2007)—however, there are six more potential siderophore transporters encoded in the Cr. neoformans genome (Jung and Kronstad 2008).
Overall, the ability to use a broad spectrum of xenosiderophores likely reflects microbial competition for iron. This would make such a strategy advantageous when close interspecies contacts are frequent, such as in biofilms in the oral cavity, gut or vagina, as well as generally in co-infections. However, in the absence of any evident producer, the role of xenosiderophore binding during dissemination in blood or host tissue remains unclear at best. In these environments, it seems more important that many fungi have developed multiple mechanisms to directly exploit iron-binding molecules of the host. Candida albicans shows an impressive versatility in using host sources and can directly or indirectly obtain iron from hemoglobin (Moors et al. 1992), hemin (Santos et al. 2003), ferritin (Almeida et al. 2008) and transferrin (Knight et al. 2005). Similarly, Cr. neoformans can use transferrin (Jung et al. 2008), heme and hemin (Jung et al. 2008; Cadieux et al. 2013; Hu et al. 2015), and H. capsulatum is known to obtain iron from transferrin and hemin (Timmerman and Woods 1999; Foster 2002), but Aspergillus spp. appear to be unable to acquire iron from heme (Vaknin et al. 2014).
In hemoglobin, iron is incorporated in heme in its ferrous form and can be acquired by C. albicans and Cr. neoformans with specific heme uptake mechanisms. The former relies on a family of heme receptors [Rbt51 (Moors et al. 1992; Weissman and Kornitzer 2004)] and hemophores [Rbt5, Pga7, Csa2 (Weissman and Kornitzer 2004; Weissman et al. 2008; Kuznets et al. 2014; Nasser et al. 2016)] for initial uptake followed by ESCRT complex-mediated internalization into the vacuole via the endocytic pathway (Weissman et al. 2008). In Cr. neoformans, the ESCRT complex similarly has a pronounced role in heme utilization [Vps23, Vps22, Snf7 (Hu et al. 2013, 2015)] along with the putative hemophore Cig1 (Cadieux et al. 2013). The internalized heme-bound iron is then released by a heme oxygenase, which has been described in many Candida species and in S. cerevisiae to recycle self-generated heme (Santos et al. 2003; Kim et al. 2006). Other host iron sources containing Fe3+ can also be taken up directly, or, more commonly, the bound Fe3+ is first extracted from host molecules (or siderophores) on the cell surface via ferric reductases. Fe2+ is then oxidized again by permease-coupled multicopper ferroxidases followed by trans-membrane transport of Fe3+ via high-affinity permeases to complete the uptake process. This system is especially important for virulence in non-siderophore producing fungi such as Cr. neoformans (Jung et al. 2009; Han, Do and Jung 2012), C. albicans (Ramanan and Wang 2000; Fang and Wang 2002; Knight et al. 2005; Cheng et al. 2013) and C. glabrata (Srivastava, Suneetha and Kaur 2014), which heavily rely on the reductive pathway for iron uptake to facilitate growth and virulence (Srivastava, Suneetha and Kaur 2014; Gerwien et al. 2016, 2017). In contrast, while A. fumigatus siderophore synthesis mutants were dramatically attenuated in virulence (Hissen et al. 2005), defects in reductive iron assimilation had no significant effect (Schrettl et al. 2004). Similarly, other siderophore producers, such as Fusarium graminearum (Greenshields et al. 2007) or Al. brassicicola (Oide et al. 2006), cannot fully compensate the loss of siderophore-mediated iron uptake by the reductive uptake system alone.
As described above, the reductive uptake system comprises reductase and linked permease/ferroxidase functions. Pathogenic fungi commonly have large families of cell-surface NAD(P)H-dependent ferric reductases at their disposal, such as Cr. neoformans (eight known reductases) (Saikia et al. 2014), C. albicans (18 putative) (Jeeves et al. 2011; Xu et al. 2014b) or A. fumigatus (15 putative) (Blatzer, Binder and Haas 2011)—with no number currently available for H. capsulatum. Candida albicans Fre2, Fre5/Frp1 and Fre9 (Bensen et al. 2004; Baek, Li and Davis 2008) are expressed under alkaline conditions, and there are indications that Fre2 might be secreted or shedded under azole treatment (Sorgo et al. 2011). In Cr. neoformans, transcription levels of Fre3 seem to be associated with virulence: RNAi suppression of FRE3 decreased survival in macrophages, while artificial upregulation led to increased virulence in mice (Hu et al. 2014).
Ferric reductases are best characterized in S. cerevisiae, where, despite obvious redundancy, the nine known members each play specific roles in siderophore-Fe reduction (Fre1, Fre2, Fre3, Fre4) (Martins et al. 1998; Yun et al. 2001), copper reduction (Fre1, Fre2, Fre7) (Martins et al. 1998) and presumably in intracellular transmembrane shuttling at the vacuole (Fre6) (Huh et al. 2003). In C. albicans, similar specific functions have been attributed to Fre7 and Fre10 as cupric reductases (Jeeves et al. 2011). Candida glabrata (Srivastava, Suneetha and Kaur 2014) and the fission yeast Schizosaccharomyces pombe (Roman et al. 1993) are notable exceptions, since they each possess only two ferric reductase genes. In C. glabrata, the lack of FRE6 has been associated with attenuated virulence in a Drosophila model (Brunke et al. 2015) and slightly decreased kidney fungal burdens in mice (Srivastava, Suneetha and Kaur 2014). However, our own work has shown that both Fre6 and Fre8 might have roles other than ferric or cupric reduction in C. glabrata, since this fungus does not exhibit evident surface ferric reductase activity (Gerwien et al. 2017). Finally, low-affinity broad-spectrum metal transporters for iron, copper and zinc have been identified in S. cerevisiae (Fet4) (Dix et al. 1994) and in C. glabrata (Fet4) (Srivastava, Suneetha and Kaur 2014; Gerwien et al. 2016) with possible orthologs in Cr. neoformans (Jacobson, Goodner and Nyhus 1998; Jung et al. 2008) and in Sc. pombe (Dainty et al. 2008).
Non-siderophore secreted molecules with the capacity to bind and reduce iron are also known in fungi. For instance, H. capsulatum uses the glutathione-dependent γ-glutamyltransferase Ggt1 to extracellularly reduce ferric iron from siderophores, transferrin and hemin (Timmerman and Woods 1999; Timmerman and Woods 2001; Zarnowski et al. 2008). Non-enzymatic ferric reductants are also excreted by this fungus (Timmerman and Woods 1999), although their exact nature is still unknown. In Cr. neoformans, 3-hydroxyanthranilate has been identified as an extracellular ferric reductant, but additional active compounds seem to exist (Nyhus, Wilborn and Jacobson 1997; Jacobson, Goodner and Nyhus 1998; Jung et al. 2008). As melanized Cr. neoformans cells reduce iron at a much higher rate than non-melanized cells, ferric reduction activity may be associated with this polymer (Nyhus, Wilborn and Jacobson 1997). In S. cerevisiae, excretion of anthranilate and 3-hydroxyanthranilate correlates with ferric reduction capacity in the extracellular medium, although, counterintuitively, cells grown in iron-rich medium show a higher secretion than those in iron-poor medium (Lesuisse et al. 1992). Likewise, culture supernatants of C. albicans, C. glabrata and S. cerevisiae show ferric reduction activity, which depends on a so far unknown low-molecular-weight compound (Gerwien et al. 2017), and A. terreus has recently been shown to secrete terrein under iron starvation, which acts as a ferric reductant and can partially rescue strains defective in siderophore biosynthesis (Gressler et al. 2015).
In a similar fashion, the active lowering of the environmental pH can increase iron bioavailability, either by increasing the overall solubility or via pH-dependent release of iron from host molecules such as transferrin (Lestas 1976). Histoplasma capsulatum is known to exploit this strategy inside macrophages, keeping the intraphagosomal pH at 6.5 (Eissenberg, Goldman and Schlesinger 1993). This is alkaline enough to inhibit phagolysosome function, but acidic enough to keep iron accessible and possibly even release it from host transferrin (Newman et al. 1994; Hilty, Smulian and Newman 2008). In fact, this strategy was found to be essential for intracellular growth and virulence of H. capsulatum (Hilty, Smulian and Newman 2008). Similar mechanisms are probably also used by other fungi with the ability to manipulate phagosomal pH, like, for example, C. glabrata (Kasper et al. 2014).
Excess iron is stored both as a stockpile for times of need and to avoid its toxicity at high concentrations. Storage is mediated either by vacuolar polyphosphates or by intracellular siderophores (see above); with the exception of zygomycetes, ferritin-like molecules with this purpose are so far unknown in fungi (Carrano, Bohnke and Matzanke 1996). In S. cerevisiae, the transporter Ccc1 mediates vacuolar iron (and manganese) import (Lapinskas, Lin and Culotta 1996; Li et al. 2001), while export is controlled by Smf3 (Portnoy, Liu and Culotta 2000) or a complex consisting of Fth1/Fet5 coupled to a ferric reductase, resembling the reductive uptake system of the plasma membrane (Urbanowski and Piper 1999). Ccc1 orthologs with similar roles in iron storage exist in C. glabrata (Gerwien et al. 2016), C. albicans (Xu et al. 2014a), A. fumigatus (Gsaller et al. 2012), A. nidulans (Eisendle et al. 2006) and Sc. pombe (Mercier, Pelletier and Labbe 2006), indicating that vacuolar iron storage is important in both siderophore producers and non-producers. Similarly, Smf3 has been associated with intracellular iron homeostasis in S. cerevisiae (Portnoy, Jensen and Culotta 2002) and in C. albicans (Xu et al. 2014a), and an ortholog is present in C. glabrata. Deletion of C. glabrata Fth1 or Fet5 does not cause sensitivity to the iron chelator bathophenanthroline disulfonate (Srivastava, Suneetha and Kaur 2014), although FTH1 was found to be iron regulated (Gerwien et al. 2016). Aspergillus nidulans and Sc. pombe finally lack orthologs for both genes—however, in the latter, Abc3 has been suggested to have a similar role in vacuolar iron mobilization (Pouliot et al. 2010).
The organelles with the highest need for iron are mitochondria. Here iron-sulfur (Fe-S) clusters are synthesized as prosthetic group for respiratory chain complexes, the TCA cycle and various other metabolic processes. Consequently, a highly conserved short-term storage molecule has evolved in fungi and mammals: the mitochondrial matrix iron chaperone Yfh1 (Huynen et al. 2001), which has been found in S. cerevisiae (Babcock et al. 1997; Wilson and Roof 1997), Sc. pombe (Fxn1) (Wang et al. 2014), C. albicans (Santos et al. 2004) and C. glabrata (Srivastava, Suneetha and Kaur 2014).
Iron-sensing and transcriptional regulation
Regulation of fungal iron homeostasis has mostly been studied in the model yeast S. cerevisiae. However, baker's yeast is barely representative of other fungi, since it employs a rather unusual regulation system, which among the pathogenic fungi has so far only been found in the closely related C. glabrata (Gerwien et al. 2016). In both species, an Aft transcription activator (Aft1 and Aft2 in S. cerevisiae) upregulates genes involved in Fe uptake under iron limitation (Yamaguchi-Iwai, Dancis and Klausner 1995; Ueta et al. 2012). Mechanistically, this is mediated by Fe-S clusters produced in the mitochondria, which—when present—bind the glutaredoxins Grx3 and Grx4 and enable them to interact with Aft1 to remove it from its promoter targets (Rutherford et al. 2005; Ueta et al. 2012). Such Fe-S clusters also play a role in adaptation to high iron, as they can activate the high iron-responsive regulator Yap5 (Li et al. 2012). Its limited range of target genes includes CCC1 (Li et al. 2008), coding for the vacuolar iron importer (Li et al. 2001), and CUP1 (Pimentel et al. 2012), encoding a copper-binding MT.
During Fe starvation, iron-requiring processes are post-transcriptionally further downregulated via degradation of mRNAs that carry the target sequence 5΄-(U)UAUUUAU(U)-3΄ in their 3΄UTR region. This process is mediated by the combined action of the RNA-binding proteins Cth1 and Cth2 in S. cerevisiae (Shakoury-Elizeh et al. 2004; Puig, Askeland and Thiele 2005; Puig, Vergara and Thiele 2008; Martinez-Pastor et al. 2013) and by a single Cth2 ortholog in C. glabrata (Gerwien et al. 2016). Thus, C. glabrata and S. cerevisiae (and likely their closest relatives) uniquely share the Aft/Cth iron regulatory system, although their opportunistic pathogenic and environmental lifestyles would at first glance suggest the need for vastly different iron homeostasis mechanisms. Interestingly, further Aft orthologs with roles in iron homeostasis have been identified in Kluyveromyces lactis (Conde e Silva et al. 2009), also a part of the Saccharomycetaceae clade (Gabaldon et al. 2013), and surprisingly in the evolutionary more distant yeast C. albicans (Liang et al. 2010; Xu et al. 2013). However, K. lactis lacks any Cth2 ortholog, whereas the one present in C. albicans (Zfs1) has no function in iron homeostasis, but influences biofilm formation (Wells et al. 2015). Notably also, C. albicans Aft2 has only a very minor function in iron homeostasis regulation (Liang et al. 2010; Xu et al. 2013), since, like most other fungi, C. albicans relies on a different iron regulation strategy.
This other system has so far been found (often with slight variations) in C. albicans, Cr. neoformans, both A. fumigatus and A. nidulans, and Sc. pombe. It usually comprises two repressors: a GATA transcription factor for the downregulation of iron acquisition (called Sfu1, Cir1, SreA, or Fep1 in these fungi) and a CCAAT-binding complex to downregulate iron consumption pathways (Hap43, HapX, HapX, or Php4) (Haas et al. 1999; Oberegger et al. 2001; Tuncher et al. 2005; Mercier, Pelletier and Labbe 2006; Hortschansky et al. 2007; Schrettl et al. 2008; Jung et al. 2010; Schrettl et al. 2010; Chen et al. 2011; Hsu, Yang and Lan 2011; Kronstad, Hu and Jung 2013). In H. capsulatum (Hwang et al. 2012), N. crassa (Zhou, Haas and Marzluf 1998), Penicillium chrysogenum (Haas, Angermayr and Stoffler 1997) and U. maydis (Voisard et al. 1993), a GATA factor (Sre1, Sre, SreP, Urbs1) with an iron-regulatory function has been characterized, but in these fungi, a complete iron-related CCAAT-binding complex has not been described yet. It is likely to be present, though, as both components play complementary roles for the efficient adaption to varying iron levels: under iron depletion, which is frequently encountered during active infections, the CCAAT-binding complex represses the iron-consuming cellular processes. At the same time, it indirectly induces iron acquisition by repressing the GATA transcription factor to alleviate its repressive effect on iron uptake. The latter function of the GATA transcription factor is in turn important under iron-replete conditions likely encountered by C. albicans cells commensally growing in the mammalian gut (Chen et al. 2011). In these environments, it also downregulates the CCAAT-binding complex, increasing the iron-consuming cellular processes. In A. fumigatus, HapX was recently shown to be important under both iron starvation and excess. Through different domains, this factor can either repress consumption or activate vacuolar sequestration of iron, depending on the current concentration of the metal (Gsaller et al. 2014). With these central roles, it is not surprising that a deletion of the CCAAT-binding complex results in attenuated virulence in Cr. neoformans (Jung et al. 2010), C. albicans (Hsu, Yang and Lan 2011; Singh et al. 2011) and A. fumigatus (Schrettl et al. 2010). Deletion of the GATA transcription factor leads to more varied outcomes, from complete avirulence in Cr. neoformans (Jung et al. 2006) to unchanged, wild-type level virulence in mouse infections for A. fumigatus ΔsreA (Schrettl et al. 2008) and C. albicans sfu1Δ/Δ (Chen et al. 2011). Notably, however, the C. albicans sfu1Δ/Δ mutant is severely defective in GI tract colonization, where iron is abundant (Chen et al. 2011).
Candida albicans adds a twist to this established system, as this fungus has incorporated a third regulator into the GATA/CCAAT partnership. Sef1 is an activator of Hap43 expression (Chen et al. 2011) and is required for full virulence (Chen and Noble 2012). Possibly, the two lifestyles of C. albicans—both as a pathogen and as a commensal in the gut where iron levels can change rapidly through food intake and microbial competition—require an additional stabilizing element in iron homeostasis regulation (Chen et al. 2011). Interestingly, a Sef1 ortholog is also present in C. glabrata, like C. albicans a commensal of mucosal surfaces, but with a vastly different regulatory network, and this has been shown to play an (albeit less pronounced) role in iron homeostasis (Gerwien et al. 2016).
Finally, with the close connection between pH and metal solubility, some fungi, such as C. albicans and Cr. neoformans, use the pH-responsive factor Rim101 to detect alkaline pH as a marker for iron starvation and signal to upregulate the iron acquisition systems (Bensen et al. 2004). Consequently, a C. albicans rim101Δ/Δ mutant is attenuated in virulence (Davis et al. 2000). Similarly, a Cr. neoformans rim101Δ/Δ mutant is unable to utilize heme (Cadieux et al. 2013), but was found to be hypervirulent (O’Meara et al. 2010) likely because of an (unrelated) defective shedding of capsule polysaccharides, which results in a hyperactivation of the host immune response (O’Meara et al. 2010, 2013).
The evolution of these various and partly redundant systems for iron homeostasis throughout fungal pathogens reflects the importance of this particular micronutrient. Adaptations occurred in response to host-induced scarcity, to conditions of varying pH, and to the changing availability of host iron sources. Our current knowledge on these adaptations is already being used to develop new therapeutic approaches, for example, by supporting the host in its iron restriction during fungal infections (reviewed in Bruhn and Spellberg 2015; Lamb 2015) or by using fungal iron acquisition systems as targets for potential vaccines—as has been done with C. albicans Als3, although its involvement in iron uptake was not known at that time (Spellberg et al. 2006, 2008). It is therefore noteworthy that, beyond well-researched examples such as C. albicans or A. fumigatus, many fungal iron acquisition strategies are likely still unknown to us.
ZINC
Zinc is a structural and catalytic co-factor for many proteins, including the ubiquitous zinc finger DNA-binding proteins. Recently, zinc was also shown to be an intracellular second messenger in various transduction signaling pathways (Yamasaki et al. 2007). In fact, zinc is the second most abundant trace metal in the human body: there are more than 300 zinc-dependent enzymes, and ≈10% of human genes code for zinc-binding proteins (Andreini et al. 2006a). The importance of zinc is sadly evident in the two billion people who suffer from zinc deficiency, especially in developing countries: A lack of zinc leads to thymic atrophy and lymphopenia, and weakens both the innate and adaptive immune responses: phagocytosis, cytokine production by macrophages, host defense by neutrophils and natural killer cells, and antibody secretion of both T and B cells are all impaired under zinc deficiency (reviewed in Prasad 2012).
Like for humans, zinc is of high importance for microorganisms. Within the fungi, zinc homeostasis has been investigated mainly in S. cerevisiae: Following the pattern of a high proportion of zinc-binding proteins in eukaryotes, about 8% of the yeast proteome is thought to bind zinc (Andreini et al. 2006b) and more than 400 yeast genes are involved in growth under zinc limitation (North et al. 2012). These include genes essential for zinc homeostasis, but also endoplasmic reticulum (ER) function, oxidative stress resistance, protein folding, vesicular trafficking and chromatin modification. Moreover, SODs, which are essential for the detoxification of reactive oxygen species (ROS) generated by host cells, are copper-, manganese- and zinc-dependent enzymes (Huang et al. 2009).
Consequently, zinc is vital for growth and metabolism in both the host and pathogens. Thus, like for iron, there is a constant competition for zinc during infections, and zinc sequestration is another aspect of the vertebrates' nutritional immunity (Corbin et al. 2008). The frequently near-neutral pH in the host lowers the solubility of zinc and therefore restricts its accessibility for microorganisms. In the oral cavity, antimicrobial peptides within saliva, the histatins, are able to bind zinc and copper, which adds to their inhibitory effect on the growth of C. albicans (Gusman et al. 2001). Intracellularly, stimulated T cells, macrophages and dendritic cells decrease their lysosomal zinc content via the expression of the zinc transporter ZIP8, inducing zinc limitation for pathogens in the phagolysosome (Begum et al. 2002; Aydemir et al. 2009). Similarly, stimulated dendritic cells reduce their cytoplasmic zinc concentration by upregulating zinc exporters and downregulating zinc importers (Kitamura et al. 2006). Cytokine-activated macrophages restrict the intracellular growth of H. capsulatum by diminishing intracellular zinc availability (Winters et al. 2010) via binding to MTs and by sequestering labile zinc into the Golgi apparatus (Vignesh et al. 2013).
The host protein calprotectin inhibits bacterial and fungal growth by chelating transition metals, including zinc (Lulloff, Hahn and Sohnle 2004; Corbin et al. 2008). In fact, calprotectin is the most abundant cytosolic protein of neutrophils and is released mainly during the formation of neutrophil extracellular traps (NETs) as their key antifungal effector (Urban et al. 2009; Bianchi et al. 2011). In vitro the stimulation of neutrophils with phorbol myristate acetate triggers NET formation, which leads to the reduction of the supernatant zinc content, while no changes were detected for Fe, Cu and Mn concentrations (Niemiec et al. 2015). NET-dependent inhibition of fungal growth is consequently reversible in vitro by zinc supplementation (Urban et al. 2009; McCormick et al. 2010; Bianchi et al. 2011).
Fungi have developed sophisticated countermeasures to this host-imposed zinc limitation, including the expression of high-affinity membrane zinc importers and specialized secreted zinc uptake proteins, known as zincophores, in order to obtain zinc from the host environment (Citiulo et al. 2012). However, excessive zinc levels can also be toxic for cells—mainly due to competition with other metals for metal-binding sites in enzymes (McDevitt et al. 2011; Gu and Imlay 2013), as zinc does not participate in Fenton chemistry. Vertebrates use this to their advantage and are able to accumulate zinc to toxic levels in certain niches. As an example from bacteria, a drastic increase of the intraphagosomal zinc level leads to an impaired growth of Mycobacterium tuberculosis, although the bacterium can partially cope with this metal excess by the expression of metal efflux ATPases (Botella et al. 2011).
Zinc homeostasis and uptake
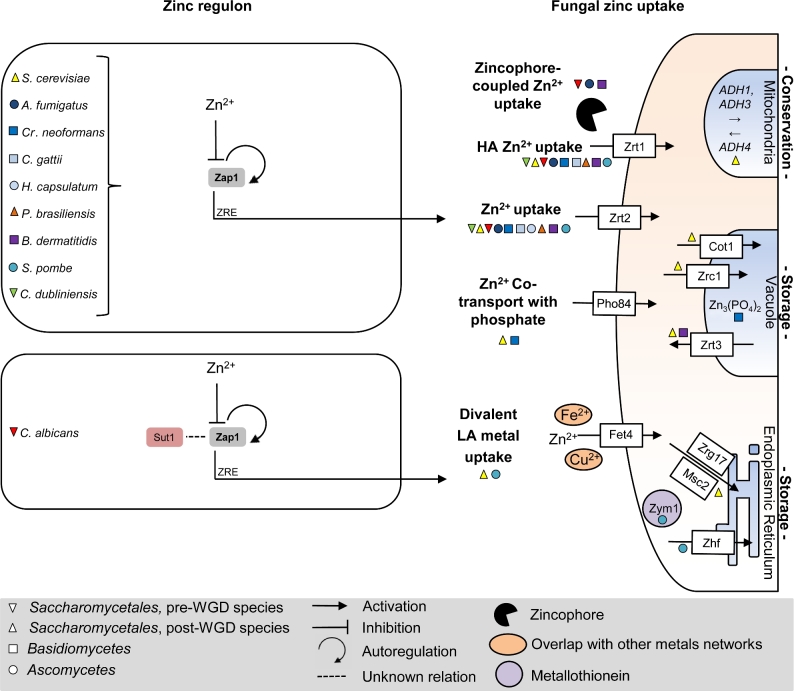
Our knowledge of zinc transporters, their transcriptional regulation and zinc trafficking mechanisms within the cell (Fig. 2) is (again) based, for a good part, on studies in S. cerevisiae—all these were first described in baker's yeast. There are two known classes of eukaryotic zinc transporters: ZRT-IRT-like proteins (ZIP) (Grotz et al. 1998), which include S. cerevisiae Zrt1, Zrt2 and Zrt3 (MacDiarmid, Gaither and Eide 2000), and the cation diffusion facilitators (Paulsen and Saier 1997), represented by Zrc1, Cot1, Msc2 (Li and Kaplan 2001) and Zrg17 (Ellis, Macdiarmid and Eide 2005).
Figure 2.
Fungal zinc homeostasis. Regulation of zinc homeostasis (left panel side) is shown for different fungal species (species is color coded, shape defines phylogenetic ancestry according to Gabaldon et al. 2013). Major transcription factors upregulated during zinc starvation to initiate fungal zinc uptake (right panel side) are written in bold. Orthologs are color shaded and aligned vertically. ZRE, recognition of target genes via zinc responsive elements. HA, high affinity; LA, low affinity.
The uptake of zinc from the extracellular milieu takes place mainly via two ZIP transporters in S. cerevisiae, the high-affinity Zrt1 (Zhao and Eide 1996a) and the low-affinity Zrt2 membrane transporters (Zhao and Eide 1996b). Under severe zinc limitation, ZRT1 expression increases 30-fold (Zhao and Eide 1996a) compared to optimal zinc conditions, while ZRT2 is usually expressed only under mild zinc limitation. In addition, under conditions of low zinc, S. cerevisiae also expresses the low-affinity metal transporter Fet4 that imports zinc, iron and copper into the cell (Li and Kaplan 1998). An additional system that exists is the phosphate/H+ symporter family member Pho84, a known phosphate transporter, which is also able to import zinc complexed with phosphate (Jensen, Ajua-Alemanji and Culotta 2003).
Aspergillus fumigatus is able to robustly grow under a broader range of pH values than S. cerevisiae, especially in alkaline environments (Wheeler, Hurdman and Pitt 1991; Amich et al. 2010) where metal solubility is low (Martinez and Motto 2000). Of its eight putative ZIP transporters, ZrfA and ZrfB have functions in zinc uptake that resemble S. cerevisiae Zrt1, although ZrfB appears to be the main transporter (Vicentefranqueira et al. 2005). Interestingly, and in contrast to baker's yeast, this system is active only under acidic pH (Vicentefranqueira et al. 2005). In neutral to alkaline low zinc environments, resembling host tissue, A. fumigatus instead employs the ZrfC zinc transporter, which does not have a S. cerevisiae ortholog (Amich et al. 2010). Its ability to acquire zinc in alkaline environments seems to depend on its long N-terminus (not present in ZrfA and ZrfB), which contains four putative zinc-binding motifs (Amich et al. 2010). Consequently, this N-terminal sequence was found to be important for zinc uptake during lung infections, and it enables growth even in the presence of zinc-binding calprotectin (Amich et al. 2014).
The Cr. neoformans and Cr. gattii zinc uptake systems comprise the ZIP transporters Zip1 and Zip2, orthologs of S. cerevisiae Zrt1 and Zrt2, respectively (Do et al. 2016). In both fungi, the high-affinity membrane transporter Zip1 is the main (pH-independent) zinc importer, while Zip2 seems to contribute little, if anything, to zinc uptake in vitro (de Oliveira Schneider et al. 2015; Do et al. 2016). In Cr. gattii, both transporters must be deleted for a visible effect on virulence (de Oliveira Schneider et al. 2015), while in Cr. neoformans, deletion of Zip1 already results in attenuation in a mouse model of cryptococcosis (Do et al. 2016). However, residual virulence even in a Cr. neoformans zip1Δzip2Δ double deletion mutant hints towards additional, still undetected zinc uptake mechanisms in this fungus and possibly, Cr. gattii. Interestingly, a connection between phosphate uptake and zinc homeostasis was shown for Cr. neoformans (Kretschmer et al. 2014), which could imply a role of its Pho84 homologs in zinc uptake similar to S. cerevisiae. Further zinc-regulated homologs of Zrt1 and/or Zrt2 have been described in H. capsulatum (Dade et al. 2016), P. brasiliensis (Parente et al. 2013) and B. dermatitidis (Muñoz et al. 2015), generally in connection to virulence—indicating the central role of zinc and this conserved acquisition system in fungal diseases.
Not surprisingly, C. albicans follows the same pattern of transport via Zrt1 and Zrt2 ZIPs (Kim et al. 2008), and again zinc uptake was found to be upregulated in the early stages of C. albicans infection in mice (Xu et al. 2015). However, the C. albicans zinc uptake system was shown to additionally include a ‘zincophore’ (Citiulo et al. 2012). In response to alkaline pH and to zinc limitation, C. albicans releases the metalloprotease-like Pra1 into the medium, where it is able to bind zinc ions with high affinity. Zinc-loaded Pra1 can then bind back to Zrt1, in a manner reminiscent of siderophores used by other fungi for iron (Citiulo et al. 2012). Interestingly, PRA1 and ZRT1 are co-expressed (Ihmels et al. 2005), as they share the same upstream intergenic region, and both were found to be upregulated on epithelial cells and in a liver infection model (Thewes et al. 2007; Zakikhany et al. 2007). So far, the C. albicans Pra1-Zrt1 pairing is the only proven zincophore system in fungi, but a similar locus structure is conserved in A. fumigatus: ASPF2-ZRFC is orthologous to PRA1-ZRT1 (Amich et al. 2010), and like Pra1, AspF2 is secreted in high amounts during infections (Segurado et al. 1999). Not surprisingly, a possible zincophore function has recently been suggested (Amich et al. 2014). In B. dermatitidis mice infections, BDFG_05357 is one of the most highly expressed genes. Like Pra1, it encodes an HRXXH domain-containing secreted protein, and has also been predicted to function as a zincophore (Muñoz et al. 2015). It seems that research into zincophores and their role in fungal pathogenesis is still gathering momentum.
High zinc levels can pose the opposite problem, and surplus zinc must be dealt with swiftly by the microorganism. In fungi, the vacuole serves as an organelle for zinc sequestration, storage and detoxification. Vacuolar zinc homeostasis has been investigated in some detail in S. cerevisiae, where it depends—among others—on the Zrc1 and Cot1 zinc importers of the vacuolar membrane (MacDiarmid, Gaither and Eide 2000). Surprisingly, ZRC1 transcription is also induced under low zinc concentration, likely in anticipation of a possible sudden zinc excess: as all zinc importers are fully active, they will immediately relay any environmental increase in zinc abundance (MacDiarmid, Milanick and Eide 2003). Inside the vacuole, zinc is likely bound to polyphosphates, as shown for Cr. neoformans (Kretschmer et al. 2014). In contrast, Sc. pombe does not rely on the vacuole as a zinc sink; instead, the zinc homeostasis factor, Zhf, transports excess zinc into the ER (Borrelly et al. 2002; Clemens et al. 2002)—a function derived maybe from its S. cerevisiae counterpart, Msc2, which in a heterodimer with Zrg17 imports zinc into the ER for proper protein processing (Li and Kaplan 2001; Ellis et al. 2004). Schizosaccharomyces pombe strikingly also uses the metallothionein Zym1 to sequester zinc, similar to higher eukaryotes, but in contrast to other fungi, where MTs mainly sequester copper (Borrelly et al. 2002).
The vacuole not only serves as an emergency disposal site, but can also replenish cellular zinc in times of need. Zinc mobilization under starvation occurs via the Zrt3 vacuole zinc exporter in S. cerevisiae (MacDiarmid, Gaither and Eide 2000). Its orthologs have been found upregulated during co-incubation of B. dermatitidis with macrophages (Muñoz et al. 2015) and during zinc starvation in C. dubliniensis (Böttcher et al. 2015). Another approach to deal with low zinc is to conserve the metal by decreasing its use. S. cerevisiae reduces the expression of major zinc-dependent enzymes and induces expression of alternative proteins of identical function, which either require less zinc or different metals. For example, the alcohol dehydrogenases Adh1 and Adh3 (which bind two zinc ions each) are replaced under zinc limitation by Adh4, which only requires one zinc ion, allowing cells to continue fermentation even under zinc deficiency (Bird et al. 2006). Important infection-associated extracellular SODs of C. albicans (Sod4–6) and H. capsulatum (Sod3) uniquely use a single copper instead of the otherwise nearly universal Cu and Zn cofactors of SODs, likely reflecting the copper-rich, zinc-poor host environment (Gleason et al. 2014a)—a factor we will come back to in the section on copper.
Zinc sensing and transcriptional regulation
In contrast to iron and copper, zinc is a redox-inactive metal and does not damage cells via ROS. However, it avidly binds to many metallation sites of proteins, replacing the native metal and interfering with their function. Hence, like for the other metals, zinc homeostasis must be precisely regulated. In yeast, the zinc responsive activator protein 1 (Zap1) is the major transcription factor regulating zinc homeostasis genes (Zhao and Eide 1997). It binds to conserved zinc responsive elements in the promoters of more than 80 genes, including ZRT1, ZRT2, ZRT3, FET4 and ZRC1 (Wu et al. 2008). Moreover, Zap1 positively autoregulates its own expression to ensure a robust response to zinc limitation (Zhao and Eide 1997; Wu et al. 2008). The structure of Zap1 was analyzed in detail in S. cerevisiae: it contains two activation domains, AD1 and AD2, which are evolutionary conserved within the fungal species (Frey and Eide 2011); AD1 is responsible for the induction of most Zap1 target genes, while AD2 regulates genes when zinc deficiency appears in concert with other stresses (Frey and Eide 2011). The intracellular zinc level is sensed via direct interaction of metal and protein: under a sufficient cytosolic zinc concentration, zinc ions directly bind AD1 and AD2 to inhibit the expression of Zap1 targets (Frey and Eide 2011). Overall, this system is highly conserved within fungi and can be found with few variations throughout the non-pathogenic and pathogenic species, including Cr. gattii (Zap1, de Oliveira Schneider et al. 2012) and A. fumigatus (ZafA, Moreno et al. 2007), and in both it was found important for full virulence.
For a fast downregulation of the importers during zinc repletion, post-translational effects come into play. Zrt1 is a stable membrane protein under low environmental zinc levels; however, the presence of zinc leads to its rapid ubiquitination and internalization for vacuolar degradation (Gitan et al. 1998). Moreover, under low zinc, Zap1 activates the expression of PIS1, encoding a phosphatidylinositol synthase, and DTT1, encoding a diacylglycerol pyrophosphate phosphatase, which results in increased levels of phosphatidylinositol and decreased levels of phosphatidylethanolamine in the membrane (Carman and Han 2007). This change in the membrane phospholipid composition is thought to influence the function and the localization of membrane zinc transporters.
The C. albicans Zap1 ortholog, also called Csr1, controls zinc homeostasis including Pra1 expression (Nobile et al. 2009), but is, of note, also involved in filamentation and biofilm matrix elaboration (Kim et al. 2008; Nobile et al. 2009)—two important contributors to C. albicans virulence. However, the virulence defect of a csr1Δ mutant likely depends not only on these morphological effects, but also directly on defective zinc homeostasis in the host. In support of this, a csr1Δ mutant of the closely related species C. dubliniensis shows no such filamentation defects, but still exhibits reduced virulence (Böttcher et al. 2015). Interestingly, in C. albicans, an additional transcription factor, Sut1, was recently implicated in controlling Csr1 expression in vivo, but surprisingly not in vitro (Xu et al. 2015). No functional relationship between the two S. cerevisiae counterparts is known (Xu et al. 2015), which suggests that this seemingly host-specific interaction is an adaptation to the pathogenic lifestyle of C. albicans. It will be interesting to see whether any other pathogen exhibits a similar departure from S. cerevisiae’s zinc regulation template.
A final twist is the pH-dependency of zinc uptake. As mentioned before, A. fumigatus switches from zinc uptake via ZrfA and ZrfB to ZrfC (and possibly AspF2) depending on the environment's alkalinity. While the Zap1 ortholog ZafA activates all transporters under zinc limitation independent of pH, the pH-dependent transcription factor PacC represses ZrfA and ZrfB under alkaline pH (Amich, Leal and Calera 2010) and ZrfC/AspF2 under acidic conditions (Amich et al. 2010). In C. albicans, expression of the Zrt1/Pra1 zincophore is similarly alkaline specific via the PacC ortholog Rim101 (Bensen et al. 2004; Citiulo et al. 2012; Xu et al. 2015), mirroring the Rim101-dependent expression of iron uptake-related genes. It seems likely that these expression patterns evolved as highly effective systems to deal with the low solubility of metals under alkaline conditions.
COPPER
Copper is in many ways a different beast than iron or zinc (Fig. 3). Like those metals, it is required as an essential trace element in many biochemical reactions, but it rapidly becomes highly toxic at increased levels (reviewed in Festa and Thiele 2011). Copper started to be bioavailable at a large scale only after the great oxidation event ≈2.4 billion years ago, when earth's atmosphere became oxidizing. Eukaryotes, which evolved after these events, consequently harbor many more Cu-containing proteins than the more ancient bacteria (Dupont et al. 2010). For the same reason, many Cu-containing enzymes have oxygen-related functions. For instance, the mitochondrial cytochrome c oxidase requires Cu for its function in the respiratory electron transport chain. Cytoplasmic or cell-wall associated Cu/Zn-SODs (like their mostly mitochondrial manganese-dependent counterparts) can protect fungal cells from externally and internally generated oxidative stress. Again, the C. albicans SODs are unusual: C. albicans is the only known organism to contain both Cu/Zn- and Mn-SOD enzymes in the cytosol (Lamarre et al. 2001)—in addition to the Cu-only variety of extracellular SODs mentioned above. The Mn-dependent Sod3 is expressed to replace the Cu-dependent counterparts under copper starvation, for example during infections of the murine kidney (Li et al. 2015). This flexibility probably tells as much about the necessity of SODs for pathogens as about the diverse metal environments C. albicans is facing during infections. In addition, copper has an important helper role as a cofactor in multicopper ferroxidases to allow the uptake of iron via the reductive pathway (see above). Finally, it also has an important function as a cofactor of laccases and tyrosinases (Shaw and Kapica 1972; Williamson 1994), which are required for the biosynthesis of melanin—an important virulence factor of pigmented fungi.
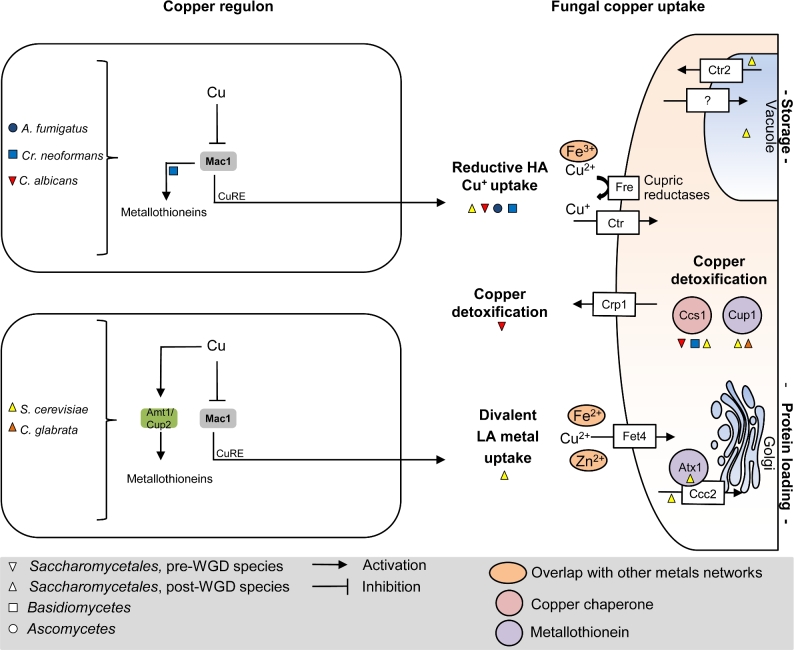
Figure 3.
Fungal copper homeostasis. Regulation of copper homeostasis (left panel side) is shown for different fungal species (species is color coded, shape defines phylogenetic ancestry according to Gabaldon et al. 2013). Major transcription factors upregulated during copper starvation to initiate fungal copper uptake (right panel side) are written in bold. Orthologs are color shaded and aligned vertically. CuRE, recognition of target genes via copper responsive elements. HA, high affinity; LA, low affinity.
However, due to its toxicity, copper has also been used as an antimicrobial agent for much of human civilization. As a fungicide against plant pathogens, it is part of the Bordeaux mixture used in vineyards, and copper surfaces show promise as a weapon against pathogens in hospitals (Casey et al. 2010). Part of its toxic effects derives from the ability of Cu+ (under anaerobic, reducing conditions) to disrupt Fe-S clusters (Macomber and Imlay 2009) and from its high capacity to displace other metals from their coordination sites, as, according to the Irving-Williams series, Cu2+ forms the most stable complexes of the divalent transition metals (Irving and Williams 1948). Furthermore, like iron, it can also readily form ROS by the Fenton reaction by Cu+/Cu2+ redox cycling under aerobic conditions, although the precise role of this for microbes is somewhat disputed (Macomber, Rensing and Imlay 2007), and in fact copper seems even more toxic under anaerobic than under aerobic conditions both for bacteria (Evans et al. 1986) and fungi like S. cerevisiae and C. albicans (Strain and Culotta 1996; Weissman, Shemer and Kornitzer 2000).
Given this comparatively high toxicity, the host and fungal strategies during infections differ significantly from the Fe-based nutritional immunity: instead of limiting access, the host seems to actively pump copper into microbe-containing phagosomes via the P-type ATPase ATP7A (Wagner et al. 2005; White et al. 2009). In fact, Cr. neoformans copper detoxification is activated during murine pulmonary infections, and the relevant MTs are required for virulence in this model (Ding et al. 2013). According to some reports, copper limitation may also play a role as an immune defense mechanism. A Cr. neoformans copper transporter was seen to be upregulated after phagocytosis by macrophage-like cells and during human cryptococcosis (Waterman et al. 2007, 2012), and the C. albicans copper transporter similarly shows upregulation upon phagocytosis (Lorenz, Bender and Fink 2004). Whether these observations represent a bona fide copper limitation or a loss of bioavailability due to the oxidative phagosomal environment (Waterman et al. 2007) remains to be seen. However, overlapping regulation of Cu uptake and resistance pathways (Ding et al. 2011), as well as possible confounding effects of the deletion and detection systems, seem to call for further investigation into the matter (Ding et al. 2013). Thus, the jury is still out whether both Cu ion overload and withholding are complementary strategies employed by the host, possibly depending on the microenvironment the fungus is facing.
Copper homeostasis and uptake
Similar to iron, copper is usually reduced from Cu2+ to Cu+ (in part by the same cell-surface metalloreductases as for Fe) for efficient uptake and then imported via high-affinity Cu+ importers—Ctr1 in C. albicans (Marvin, Williams and Cashmore 2003), the functionally redundant Ctr1 and Ctr4 in Cr. neoformans (Ding et al. 2011), and at least two importers (CtrA2 and CtrC) in A. fumigatus (Park et al. 2014). In contrast to iron, no oxidase is involved in this process. In S. cerevisiae at least, the iron transporter Fet4 also imports copper with low affinity (Hassett et al. 2000). Another source of copper in addition to the surrounding medium is the vacuolar storage. In S. cerevisiae, the transmembrane copper transporter Ctr2, a homolog of Ctr1, allows copper mobilization from this organelle (Rees, Lee and Thiele 2004) with the help of a metalloreductase in the vacuolar membrane (Rees and Thiele 2007), mimicking the cytoplasmic membrane setup. Pathogenic fungi like C. albicans possess orthologs of these proteins, but their role in virulence has not been investigated so far.
Once intracellular, the potentially toxic Cu+ is immediately bound by different specific chaperones, which allow its quick and targeted transport to Cu-requiring enzymes. For example, Ccs1 proteins deliver copper to the Cu/Zn-SODs of C. albicans (Gleason et al. 2014b), Cr. neoformans, S. cerevisiae (Liu et al. 1997) and in fact nearly all eukaryotes (Leitch et al. 2009). Similarly, Atx1 homologs escort copper to Ccc2 Cu-transporting ATPases of the Golgi membrane (Lin et al. 1997; Huffman and O’Halloran 2000). These then pump the metal into late secretory vesicles to serve as a cofactor, for example, in the aforementioned Fe multicopper oxidases or laccases. This also intimately links copper to iron homeostasis, as multicopper oxidases are required for efficient iron uptake in fungi like yeast or C. albicans (Askwith et al. 1994; Eck et al. 1999; Cheng et al. 2013).
A similar binding mechanism prevents toxicity under high copper conditions. MTs, small proteins rich in cysteine residues, can sequester Cu (and, especially in non-fungal organisms, other metals) to render it biologically inactive. They are also present in plants and animals, but in very few bacteria—one example being specifically the pathogenic mycobacteria (Gold et al. 2008). Characteristically, the genes coding for MTs vary strongly in numbers between species: in pathogenic fungi, some C. glabrata strains harbor more than 30 copies of the MT-IIa gene, in addition to one copy each of MT-IIb and MT-I (Mehra, Garey and Winge 1990; Mehra et al. 1992). Similarly, S. cerevisiae can increase its copy number of the CUP1 metallothionein gene and thereby obtain higher Cu resistance (Fogel and Welch 1982). No such mechanism has been described for C. albicans with its three known MTs or C. neoformans with its two (Ding et al. 2011) so far. Similarly, it seems that in S. cerevisiae copper is also detoxified, like other metals, via the vacuolar storage (Szczypka et al. 1997; Jo et al. 2008), but little is known about this process in other fungi.
In S. cerevisiae (and likely other fungi), high intracellular Cu levels furthermore rapidly block the Ctr1 Cu importer by direct binding and subsequent conformational changes to restrict copper influx (Wu et al. 2009). However, C. albicans achieves its high intrinsic Cu resistance (when compared to S. cerevisiae) also by active outward transport over the plasma membrane by Crp1, a P-type ATPase (Riggle and Kumamoto 2000; Weissman et al. 2000), in a process functionally resembling the copper transport by Ccc2 ATPase into the Golgi (Weissman, Shemer and Kornitzer 2002)—or even into the phagosome by the host's ATP7A, in an interesting example of a molecular-level arms race using the same mechanism on both sides. This export mechanism, although common in bacteria (reviewed in Samanovic et al. 2012) and present in other eukaryotes, has so far been found only in C. albicans and—very recently—in A. nidulans (Antsotegi-Uskola, Markina-Inarrairaegui and Ugalde 2017).
Copper sensing and transcriptional regulation
Low copper levels lead to an activation of the transcription factor Mac1 in S. cerevisiae (Jungmann et al. 1993), and the same is true for its orthologs in C. albicans (Mac1; Marvin, Mason and Cashmore 2004), A. fumigatus (Afmac1; Kusuya et al. 2017) and most likely also C. glabrata. The Mac1 activator comprises a copper fist DNA-binding domain to recognize copper response elements, and a Cu-binding domain to gauge the intracellular copper concentration and inhibit DNA binding under copper replete conditions (Graden and Winge 1997). Under copper starvation, Mac1 binding leads to the expression of the dedicated copper transporter and metalloreductase genes via their upstream regulatory elements (Yamaguchi-Iwai et al. 1997). Under copper excess, Mac1 is quickly degraded to avoid copper toxicity (Zhu et al. 1998), and in contrast to copper-depleted conditions, MAC1 mRNA exists in a readily degradable isoform when copper is present (Peccarelli et al. 2016). This Cu-dependent regulation directly influences virulence: deletion of the Mac1 ortholog Cuf1 reduces dissemination of C. neoformans to the mouse brain, and abolishes transcription of the copper-dependent laccase (Jiang et al. 2009). In C. albicans, Mac1 is—among other functions—responsible for shifting from the Cu-dependent Sod1 to the Cu-independent Sod3, by repressing the former and activating the latter (Li et al. 2015).
The Cr. neoformans Cuf1 (Lin et al. 2006; Waterman et al. 2007) is not only responsible for upregulation of copper uptake under starvation, but also positively regulates MTs under Cu excess (Ding et al. 2011). In fact, Cuf1 seems to be a hybrid factor, as in C. glabrata and S. cerevisiae these roles are separated, and in C. glabrata another transcription factor, called Amt1 [homologous to Cup2 or Ace1 in S. cerevisiae (Buchman et al. 1989; Szczypka and Thiele 1989)], is activated under high copper levels by the binding of four Cu+ ions to its N-terminal domain (Thorvaldsen et al. 1994). Active Amt1 then induces the transcription of all three MT genes and itself, leading to a positive autoregulatory loop and thus a robust copper resistance response (Zhou et al. 1992; Zhou and Thiele 1993; Koch et al. 2001). The role of its homolog in C. albicans is not well investigated so far (although it likely has similar functions), but the cAMP pathway has been implicated in copper resistance in this fungus. A deletion of C. albicans GPA2 (encoding the G-protein α subunit upstream of protein kinase A) decreases Cu uptake, increases MT expression and hence renders the fungus more resistant to copper (Schwartz et al. 2013). Overall, the typical fungal response to high copper thus seems to be determined by a fast inactivation and degradation of the Mac1 activator homologs, and copper sequestration via upregulation of MTs by different mechanisms. However, our knowledge of these regulatory systems still lacks behind what we have learned about zinc and especially iron homeostasis in fungal pathogenesis.
NICKEL
Nickel is a comparatively rare metal, but an efficient fungicide that seems to exert its effects mainly by interfering with the carbohydrate metabolism and DNA repair, by production of ROS (albeit less than copper or iron), and by membrane damage (reviewed in Macomber and Hausinger 2011). Many of these effects are exerted by nickel replacing the original metal in metalloenzymes—and as nickel is rather stable in the Ni2+ state, this replacement abolishes the redox function of the metal cofactor (Macomber and Hausinger 2011). At high external concentrations, nickel can non-specifically enter the microbial cell via the magnesium transport system. Still, dedicated uptake systems for this mostly toxic transition metal also exist, especially in bacteria (Zhang et al. 2009), and a functional Ni permease with high similarity to its bacterial co-family members has, for example, been found in Sc. pombe (Eitinger et al. 2000). So why would microbes, and especially fungi, actively import nickel? In Sc. pombe, this seems to be related to its urease activity (Eitinger et al. 2000), which requires Ni to allow the use of urea as a nitrogen source and the concomitant alkalization of the environment. For pathogens, ureases (and with them, most likely dedicated nickel permeases) often play important roles as virulence factors, for example, in Coccidioides immitis and in Cr. neoformans (Singh et al. 2013). With no known Ni metalloenzymes in vertebrates, nickel homeostasis has thus been suggested as a promising avenue for fighting infections (Morrow and Fraser 2013). However, the Saccharomycetes—like S. cerevisiae, C. albicans and C. glabrata—do not employ a Ni-requiring urease (Navarathna et al. 2010), and consequently seem to lack Ni permeases—instead, these fungi use non-nickel, biotin-requiring urea amidolyases to metabolize urea (Navarathna et al. 2010). In A. fumigatus, a nickel permease homolog can be found in the genome, but little is known so far about its potential role in virulence.
Excess nickel, as is so often the case with toxic metals, is sequestered into the vacuole by S. cerevisiae (Nishimura, Igarashi and Kakinuma 1998)—in this case with the help of the avid nickel binder, histidine (Pearce and Sherman 1999). It seems likely that pathogenic fungi have similar mechanisms at their disposal, paralleling the existence of nickel resistance mechanisms in many bacteria. Overall, however, little is currently known about the role of nickel in fungal pathogenesis, and we may yet be surprised by unexpected findings in the future.
MANGANESE
Manganese is required in the function of polymerases, sugar transferases of the Golgi and of course for the Mn-SODs especially of the mitochondria (reviewed for baker's yeast in Reddi, Jensen and Culotta 2009). Its intracellular concentration has been shown to vary significantly, over nearly two orders of magnitude (Reddi, Jensen and Culotta 2009). One reason may be that—in contrast to most of the other metals described here—manganese acts as an anti-oxidant at high concentrations, rather than a ROS producer. In fact, at high intracellular concentrations Mn-containing complexes can take the role of SODs in certain bacteria and in yeast SOD deletion mutants (Reddi et al. 2009). Excessive levels are nonetheless toxic to yeasts leading to the induction of apoptosis (Liang and Zhou 2007).
External manganese is taken up in baker's yeast via the Nramp transporters, Smf1 and Smf2 (Supek et al. 1996; Cohen, Nelson and Nelson 2000; Portnoy, Liu and Culotta 2000), and a possible ortholog in C. neoformans has been described to transport Mn and other metals (Agranoff et al. 2005). It has been suggested that Smf1 is responsible for keeping up the intracellular Mn levels required for its anti-oxidant action, while Smf2 imports manganese for the Mn-requiring enzymes (Luk and Culotta 2001; Reddi et al. 2009). These transporters are continuously expressed and regulated mainly post-translationally, and at sufficiently high (physiological) Mn levels they are continually targeted for vacuolar degradation (Reddi et al. 2009). Furthermore and in a manner similar to zinc, high extracellular manganese can be imported by yeast in complex with phosphate via the Pho84 transmembrane transporter (Jensen, Ajua-Alemanji and Culotta 2003). Once inside the cell, it can then be transported by the Golgi P-type Ca2+/Mn2+ ATPase, Pmr1, to serve as a cofactor in the secretory pathway (Dürr et al. 1998). In fact, a Pmr1 homolog is required for full C. albicans virulence due to this cofactor role in glycosylation (Bates et al. 2005). Finally, in S. cerevisiae at least, excess manganese is excreted via the secretory pathway, but also sequestered to the vacuole (like iron via Ccc1; Li et al. 2001), and in C. albicans its complexation with polyphosphate has been shown (Ikeh et al. 2016). If and how manganese can leave the vacuole again is still an open question, as no dedicated exporter has been described so far.
Due to these biological functions, the host employs Mn starvation to fight bacteria and possibly fungi. Macrophage phagosomes are severely limited for manganese (Jabado et al. 2000), and the host-defense protein, calprotectin, chelates manganese in addition to zinc (Corbin et al. 2008) and—as shown recently—iron (Nakashige et al. 2015). In vitro at least, Mn chelation by calprotectin reduces growth of A. fumigatus (Amich et al. 2014; Clark et al. 2016), and Mn withdrawal may thus play a role in fungal infections—although in contrast to bacteria, the effects of manganese limitation on fungal virulence are probably eclipsed by the removal of zinc and iron. As with nickel, research into the role of manganese in fungi may yet reveal some unexpected connections to pathogenesis, as our knowledge so far is comparatively incomplete.
CONCLUSIONS
Metals clearly play a central role during fungal pathogenesis. This is shown by the sheer number and diversification of the regulatory, uptake and detoxification systems in fungal pathogens, and of course by the host's many efforts to efficiently withhold metals. We seem to have a good concept of how iron and—with a few gaps—zinc are acquired by fungi during infections, but for many of the metals that are experimentally more difficult to address, our knowledge is still quite limited. The protection mechanisms against many metals with toxic effects are not well established, nor are the uptake systems for those which are required only in minute amounts—from cobalt to silver or cadmium. It seems likely that fungal research can learn a lot from the bacterial field, as even though the molecular mechanisms may differ, the basic problems the microbes are facing are essentially the same, and analogous solutions may have been found by both groups of pathogens.
Metal homeostasis also presents a largely untapped resource for potential treatment options. The natural response of the host already indicates the effectiveness of targeting the microbial requirement for metals. Strategies that may be worthwhile to follow in the future include a knowledge-guided combination of deprivation and excess: withholding one metal to induce a partially unspecific uptake response, which is exploited to introduce toxic levels of another. It seems that the immune system may already follow this strategy inside the phagosome, as described above e.g. for copper. Metal-based drugs were found highly effective against parasites like Leishmania spp. or Plasmodium spp. (reviewed in Navarro et al. 2010), and it seems at least possible that a similar approach may prove useful for fungi as well. We hope that with this review, we have enabled the reader to see the connections and similarities between metals and among fungi, maybe forming the kernel of a new hypothesis. The potential and the need for many more findings still exist in this growing field.
Acknowledgements
We thank Selene Mogavero for the critical reading of this manuscript and Maria Joanna Niemiec for her valuable input on NETs.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft (Collaborative Research Centre/Transregio FungiNet project C1) [grant number TR/CRC 124]; the Jena School for Microbial Communication [grant number GSC 214]; the Bundesministerium für Bildung und Forschung (Infect-ERA FunComPath & Center for Sepsis Control and Care) [Grant Numbers 031L0001A, 01E01002]; the European Union (Marie Sklodowska-Curie grant agreement OPATHY [From Omics to Patient: Improving Diagnostics of Pathogenic Yeasts]) [Grant number 642095]; and the Leibniz Association (International Leibniz Research School, ILRS).
Conflict of interest. None declared.
REFERENCES
- Agranoff D, Collins L, Kehres D et al. . The Nramp orthologue of Cryptococcus neoformans is a pH-dependent transporter of manganese, iron, cobalt and nickel. Biochem J 2005;385:225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RS, Brunke S, Albrecht A et al. . The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 2008;4:e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amich J, Leal F, Calera JA. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int Microbiol 2010;12:39–47. [PubMed] [Google Scholar]
- Amich J, Vicentefranqueira R, Leal F et al. . Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot Cell 2010;9:424–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amich J, Vicentefranqueira R, Mellado E et al. . The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn‐chelating protein calprotectin. Cell Microbiol 2014;16:548–64. [DOI] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I et al. . Counting the zinc-proteins encoded in the human genome. J Proteome Res 2006a;5:196–201. [DOI] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I et al. . Zinc through the three domains of life. J Proteome Res 2006b;5:3173–8. [DOI] [PubMed] [Google Scholar]
- Antsotegi-Uskola M, Markina-Inarrairaegui A, Ugalde U. Copper resistance in Aspergillus nidulans Relies on the PI-Type ATPase CrpA, Regulated by the Transcription Factor AceA. Front Microbiol 2017;8:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C, Eide D, Van Ho A et al. . The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 1994;76:403–10. [DOI] [PubMed] [Google Scholar]
- Aydemir TB, Liuzzi JP, McClellan S et al. . Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-γ expression in activated human T cells. J Leukocyte Biol 2009;86:337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M, de Silva D, Oaks R et al. . Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 1997;276:1709–12. [DOI] [PubMed] [Google Scholar]
- Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell 2008;7:1168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Hee Jung W, Kronstad JW. Iron acquisition in fungal pathogens of humans. Metallomics 2017;9:215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, MacCallum DM, Bertram G et al. . Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem 2005;280:23408–15. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kobayashi M, Moriwaki Y et al. . Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics 2002;80:630–45. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Martin SJ, Li M et al. . Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol 2004;54:1335–51. [DOI] [PubMed] [Google Scholar]
- Bianchi M, Niemiec MJ, Siler U et al. . Restoration of anti-Aspergillus defense by neutrophil extracellular traps in human chronic granulomatous disease after gene therapy is calprotectin-dependent. J Allergy Clin Immunol 2011;127:1243–52. e1247. [DOI] [PubMed] [Google Scholar]
- Bird AJ, Gordon M, Eide DJ et al. . Repression of ADH1 and ADH3 during zinc deficiency by Zap1‐induced intergenic RNA transcripts. EMBO J 2006;25:5726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M, Binder U, Haas H. The metalloreductase FreB is involved in adaptation of Aspergillus fumigatus to iron starvation. Fungal Genet Biol 2011;48:1027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelly GP, Harrison MD, Robinson AK et al. . Surplus zinc is handled by Zym1 metallothionein and Zhf endoplasmic reticulum transporter in Schizosaccharomyces pombe. J Biol Chem 2002;277:30394–400. [DOI] [PubMed] [Google Scholar]
- Botella H, Peyron P, Levillain F et al. . Mycobacterial P 1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 2011;10:248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher B, Palige K, Jacobsen ID et al. . Csr1/Zap1 maintains Zinc homeostasis and influences virulence in Candida dubliniensis but is not coupled to morphogenesis. Eukaryot Cell 2015;14:661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA et al. . Hidden killers: human fungal infections. Sci Transl Med 2012;4:165rv113. [DOI] [PubMed] [Google Scholar]
- Bruhn KW, Spellberg B. Transferrin-mediated iron sequestration as a novel therapy for bacterial and fungal infections. Curr Opin Microbiol 2015;27:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunke S, Quintin J, Kasper L et al. . Of mice, flies–and men? Comparing fungal infection models for large-scale screening efforts. Dis Model Mech 2015;8:473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman C, Skroch P, Welch J et al. . The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol Cell Biol 1989;9:4091–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde AD, Leong SA. Characterization of siderophores from Ustilago maydis. Mycopathologia 1989;108:125–33. [DOI] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G et al. . The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J Infect Dis 2013;207:1339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano SM, Menezes R, Amaral C et al. . Repression of the low affinity iron transporter gene FET4: A novel mechanism against cadmium toxicity orchestrated by YAP1 via ROX1. J Biol Chem 2015;290:18584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano LE, Gomez B, Brummer E et al. . Inhibitory effect of deferoxamine or macrophage activation on transformation of Paracoccidioides brasiliensis conidia ingested by macrophages: reversal by holotransferrin. Infect Immun 1994;62:1494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han G-S. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. BBA-Mol Cell Biol L 2007;1771:322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano CJ, Bohnke R, Matzanke BF. Fungal ferritins: the ferritin from mycelia of Absidia spinosa is a bacterioferritin. FEBS Lett 1996;390:261–4. [DOI] [PubMed] [Google Scholar]
- Carrano CJ, Raymond KN. Coordination chemistry of microbial iron transport compounds: rhodotorulic acid and iron uptake in Rhodotorula pilimanae. J Bacteriol 1978;136:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AL, Adams D, Karpanen TJ et al. . Role of copper in reducing hospital environment contamination. J Hosp Infect 2010;74:72–7. [DOI] [PubMed] [Google Scholar]
- Charlang G, Ng B, Horowitz NH et al. . Cellular and extracellular siderophores of Aspergillus nidulans and Penicillium chrysogenum. Mol Cell Biol 1981;1:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Noble SM. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 2012;8:e1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Pande K, French SD et al. . An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 2011;10:118–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Xu N, Yu Q et al. . Novel insight into the expression and function of the multicopper oxidases in Candida albicans. Microbiology 2013;159:1044–55. [DOI] [PubMed] [Google Scholar]
- Citiulo F, Jacobsen ID, Miramón P et al. . Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 2012;8:e1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HL, Jhingran A, Sun Y et al. . Zinc and manganese chelation by neutrophil S100A8/A9 (Calprotectin) limits extracellular Aspergillus fumigatus hyphal growth and corneal infection. J Immunol 2016;196:336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Bloss T, Vess C et al. . A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and differentially affects transition metal tolerance. J Biol Chem 2002;277:18215–21. [DOI] [PubMed] [Google Scholar]
- Cohen A, Nelson H, Nelson N. The family of SMF metal ion transporters in yeast cells. J Biol Chem 2000;275:33388–94. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N, Goncalves IR, Lemaire M et al. . KlAft, the Kluyveromyces lactis ortholog of Aft1 and Aft2, mediates activation of iron-responsive transcription through the PuCACCC Aft-type sequence. Genetics 2009;183:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A et al. . Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 2008;319:962–5. [DOI] [PubMed] [Google Scholar]
- Cornish AS, Page WJ. The catecholate siderophores of Azotobacter vinelandii: their affinity for iron and role in oxygen stress management. Microbiology 1998;144:1747–54. [DOI] [PubMed] [Google Scholar]
- Crawford A, Wilson D. Essential metals at the host–pathogen interface: nutritional immunity and micronutrient assimilation by human fungal pathogens. FEMS Yeast Res 2015;15:fov071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dade J, DuBois JC, Pasula R et al. . HcZrt2, a zinc responsive gene, is indispensable for the survival of Histoplasma capsulatum in vivo. Med Mycol 2016;54:865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dainty SJ, Kennedy CA, Watt S et al. . Response of Schizosaccharomyces pombe to zinc deficiency. Eukaryot Cell 2008;7:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Edwards JE Jr, Mitchell AP et al. . Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun 2000;68:5953–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Schneider R, Diehl C, Dos Santos FM et al. . Effects of zinc transporters on Cryptococcus gattii virulence. Sci Rep 2015;5:10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Schneider R, de Souza Süffert Fogaça N, Kmetzsch L et al. . Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii. PLoS One 2012;7:e43773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy LR, Hart DO, Goetz DH et al. . A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell 2010;141:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Festa RA, Chen YL et al. . Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 2013;13:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Yin J, Tovar EM et al. . The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol Microbiol 2011;81:1560–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DR, Bridgham JT, Broderius MA et al. . The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem 1994;269:26092–9. [PubMed] [Google Scholar]
- Do E, Hu G, Caza M et al. . The ZIP family zinc transporters support the virulence of Cryptococcus neoformans. Med Mycol 2016;54:605–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Butcher A, Valas RE et al. . History of biological metal utilization inferred through phylogenomic analysis of protein structures. P Natl Acad Sci USA 2010;107:10567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr G, Strayle J, Plemper R et al. . The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol Biol Cell 1998;9:1149–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck R, Hundt S, Hartl A et al. . A multicopper oxidase gene from Candida albicans: cloning, characterization and disruption. Microbiology 1999;145(pt 9):2415–22. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Lancaster JR Jr, Emery T. Siderophore iron transport followed by electron paramagnetic resonance spectroscopy. J Biol Chem 1982;257:8623–6. [PubMed] [Google Scholar]
- Eisendle M, Schrettl M, Kragl C et al. . The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans. Eukaryot Cell 2006;5:1596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg LG, Goldman WE, Schlesinger PH. Histoplasma capsulatum modulates the acidification of phagolysosomes. J Exp Med 1993;177:1605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitinger T, Degen O, Bohnke U et al. . Nic1p, a relative of bacterial transition metal permeases in Schizosaccharomyces pombe, provides nickel ion for urease biosynthesis. J Biol Chem 2000;275:18029–33. [DOI] [PubMed] [Google Scholar]
- Ellis CD, Macdiarmid CW, Eide DJ. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J Biol Chem 2005;280:28811–8. [DOI] [PubMed] [Google Scholar]
- Ellis CD, Wang F, MacDiarmid CW et al. . Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J Cell Biol 2004;166:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SL, Tolbert C, Arceneaux JE et al. . Enhanced toxicity of copper for Streptococcus mutans under anaerobic conditions. Antimicrob Agents Ch 1986;29:342–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang HM, Wang Y. Characterization of iron-binding motifs in Candida albicans high-affinity iron permease CaFtr1p by site-directed mutagenesis. Biochem J 2002;368:641–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete FA, Chandhoke V, Jellison J. Iron-binding compounds produced by wood-decaying basidiomycetes. Appl Environ Microb 1989;55:2720–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton HJH. Oxidation of tartaric acid in presence of iron. J Chem Soc Trans 1894;65:899–910. [Google Scholar]
- Festa RA, Thiele DJ. Copper: an essential metal in biology. Curr Biol 2011;21:R877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Welch JW. Tandem gene amplification mediates copper resistance in yeast. P Natl Acad Sci USA 1982;79:5342–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster LA. Utilization and cell-surface binding of hemin by Histoplasma capsulatum. Can J Microbiol 2002;48:437–42. [DOI] [PubMed] [Google Scholar]
- Frey AG, Eide DJ. Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem 2011;286:6844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Martin T, Marcet-Houben M et al. . Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics 2013;14:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol 2015;15:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Santamarina S, Thiele DJ. Copper at the fungal pathogen-host axis. J Biol Chem 2015;290:18945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwien F, Safyan A, Wisgott S et al. . A novel hybrid iron regulation network combines features from pathogenic and nonpathogenic yeasts. MBio 2016;7:e01782–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwien F, Safyan A, Wisgott S et al. . An unusual ferric reductase-independent iron acquisition system in a fungal pathogen. Front Microbiol 2017;8:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitan RS, Luo H, Rodgers J et al. . Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J Biol Chem 1998;273:28617–24. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Galaleldeen A, Peterson RL et al. . Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. P Natl Acad Sci USA 2014a;111:5866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JE, Li CX, Odeh HM et al. . Species-specific activation of Cu/Zn SOD by its CCS copper chaperone in the pathogenic yeast Candida albicans. J Biol Inorg Chem 2014b;19:595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N et al. . The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 2002;10:1033–43. [DOI] [PubMed] [Google Scholar]
- Gold B, Deng H, Bryk R et al. . Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol 2008;4:609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graden JA, Winge DR. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. P Natl Acad Sci USA 1997;94:5550–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenshields DL, Liu G, Feng J et al. . The siderophore biosynthetic gene SID1, but not the ferroxidase gene FET3, is required for full Fusarium graminearum virulence. Mol Plant Pathol 2007;8:411–21. [DOI] [PubMed] [Google Scholar]
- Gressler M, Meyer F, Heine D et al. . Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals. Elife 2015;4:e07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotz N, Fox T, Connolly E et al. . Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. P Natl Acad Sci USA 1998;95:7220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsaller F, Eisendle M, Lechner BE et al. . The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus. Metallomics 2012;4:1262–70. [DOI] [PubMed] [Google Scholar]
- Gsaller F, Hortschansky P, Beattie SR et al. . The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J 2014;33:2261–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Imlay JA. Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol 2013;89:123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusman H, Lendenmann U, Grogan J et al. . Is salivary histatin 5 a metallopeptide? BBA-Protein Struct Mol Enzymol 2001;1545:86–95. [DOI] [PubMed] [Google Scholar]
- Haas H, Angermayr K, Stoffler G. Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 1997;184:33–7. [DOI] [PubMed] [Google Scholar]
- Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol 2008;46:149–87. [DOI] [PubMed] [Google Scholar]
- Haas H, Zadra I, Stoffler G et al. . The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem 1999;274:4613–9. [DOI] [PubMed] [Google Scholar]
- Han K, Do E, Jung WH. A human fungal pathogen Cryptococcus neoformans expresses three distinct iron permease homologs. J Microbiol Biotechnol 2012;22:1644–52. [DOI] [PubMed] [Google Scholar]
- Hassett R, Dix DR, Eide DJ et al. . The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem J 2000;2(351 Pt):477–84. [PMC free article] [PubMed] [Google Scholar]
- Heymann P, Ernst JF, Winkelmann G. A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1p) in Saccharomyces cerevisiae. Biometals 2000a;13:65–72. [DOI] [PubMed] [Google Scholar]
- Heymann P, Ernst JF, Winkelmann G. Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol Lett 2000b;186:221–7. [DOI] [PubMed] [Google Scholar]
- Heymann P, Gerads M, Schaller M et al. . The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun 2002;70:5246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higson FK, Kohen R, Chevion M. Iron enhancement of ascorbate toxicity. Free Radic Res Commun 1988;5:107–15. [DOI] [PubMed] [Google Scholar]
- Hilty J, Smulian AG, Newman SL. The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages and virulence in a murine model of histoplasmosis. Mol Microbiol 2008;70:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty J, Smulian AG, Newman SL. Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Med Mycol 2011;49:633–42. [DOI] [PubMed] [Google Scholar]
- Hissen AH, Wan AN, Warwas ML et al. . The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence. Infect Immun 2005;73:5493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 2012;10:525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz NH, Charlang G, Horn G et al. . Isolation and identification of the conidial germination factor of Neurospora crassa. J Bacteriol 1976;127:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortschansky P, Eisendle M, Al-Abdallah Q et al. . Interaction of HapX with the CCAAT-binding complex–a novel mechanism of gene regulation by iron. EMBO J 2007;26:3157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DH, Rafie R, Tiwari A et al. . Hydroxamate siderophores of Histoplasma capsulatum. Infect Immun 2000;68:2338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot Cell 2011;10:207–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Bai C, Zheng XD et al. . Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem 2002;277:30598–605. [DOI] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B et al. . Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun 2013;81:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B et al. . The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol Microbiol 2015;96:973–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chen SH, Qiu J et al. . Microevolution during serial mouse passage demonstrates FRE3 as a virulence adaptation gene in Cryptococcus neoformans. MBio 2014;5:e00941–00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Canadien V, Lam GY et al. . Activation of antibacterial autophagy by NADPH oxidases. P Natl Acad Sci USA 2009;106:6226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman DL, O’Halloran TV. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J Biol Chem 2000;275:18611–4. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC et al. . Global analysis of protein localization in budding yeast. Nature 2003;425:686–91. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Snel B, Bork P et al. . The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum Mol Genet 2001;10:2463–8. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Mayfield JA, Rine J et al. . Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog 2008;4:e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Seth E, Gilmore SA et al. . SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell 2012;11:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Berman J et al. . Comparative gene expression analysis by a differential clustering approach: application to the Candida albicans transcription program. PLoS Genet 2005;1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeh MA, Kastora SL, Day AM et al. . Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol Biol Cell 2016;27:2784–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. The mismetallation of enzymes during oxidative stress. J Biol Chem 2014;289:28121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving H, Williams RJP. Order of stability of metal complexes. Nature 1948;162:746–7. [Google Scholar]
- Ismail A, Bedell GW, Lupan DM. Siderophore production by the pathogenic yeast, Candida albicans. Biochem Bioph Res Co 1985;130:885–91. [DOI] [PubMed] [Google Scholar]
- Jabado N, Jankowski A, Dougaparsad S et al. . Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med 2000;192:1237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson ES, Goodner AP, Nyhus KJ. Ferrous iron uptake in Cryptococcus neoformans. Infect Immun 1998;66:4169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeves RE, Mason RP, Woodacre A et al. . Ferric reductase genes involved in high-affinity iron uptake are differentially regulated in yeast and hyphae of Candida albicans. Yeast 2011;28:629–44. [DOI] [PubMed] [Google Scholar]
- Jensen LT, Ajua-Alemanji M, Culotta VC. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J Biol Chem 2003;278:42036–40. [DOI] [PubMed] [Google Scholar]
- Jiang N, Sun N, Xiao D et al. . A copper-responsive factor gene CUF1 is required for copper induction of laccase in Cryptococcus neoformans. FEMS Microbiol Lett 2009;296:84–90. [DOI] [PubMed] [Google Scholar]
- Jo WJ, Loguinov A, Chang M et al. . Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants. Toxicol Sci 2008;101:140–51. [DOI] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W et al. . Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot Cell 2009;8:1511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Kronstad JW. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol 2008;10:277–84. [DOI] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G et al. . HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog 2010;6:e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T et al. . Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog 2008;4:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R et al. . Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 2006;4:e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Reins HA, Lee J et al. . MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J 1993;12:5051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper L, Seider K, Gerwien F et al. . Identification of Candida glabrata genes involved in pH modulation and modification of the phagosomal environment in macrophages. PLoS One 2014;9:e96015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Yukl ET, Moenne-Loccoz P et al. . Fungal heme oxygenases: Functional expression and characterization of Hmx1 from Saccharomyces cerevisiae and CaHmx1 from Candida albicans. Biochemistry 2006;45:14772–80. [DOI] [PubMed] [Google Scholar]
- Kim M-J, Kil M, Jung J-H et al. . Roles of Zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic Yeast Candida albicans. J Microbiol Biot 2008;18:242–7. [PubMed] [Google Scholar]
- Kitamura H, Morikawa H, Kamon H et al. . Toll-like receptor–mediated regulation of zinc homeostasis influences dendritic cell function. Nat Immunol 2006;7:971–7. [DOI] [PubMed] [Google Scholar]
- Knight SA, Vilaire G, Lesuisse E et al. . Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect Immun 2005;73:5482–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KA, Allard S, Santoro N et al. . The Candida glabrata Amt1 copper-sensing transcription factor requires Swi/Snf and Gcn5 at a critical step in copper detoxification. Mol Microbiol 2001;40:1165–74. [DOI] [PubMed] [Google Scholar]
- Kretschmer M, Reiner E, Hu G et al. . Defects in phosphate acquisition and storage influence virulence of Cryptococcus neoformans. Infect Immun 2014;82:2697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, Jung WH. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol 2013;21:457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuya Y, Hagiwara D, Sakai K et al. . Transcription factor Afmac1 controls copper import machinery in Aspergillus fumigatus. Curr Genet 2017;63:777–89. [DOI] [PubMed] [Google Scholar]
- Kuznets G, Vigonsky E, Weissman Z et al. . A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 2014;10:e1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre C, LeMay JD, Deslauriers N et al. . Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J Biol Chem 2001;276:43784–91. [DOI] [PubMed] [Google Scholar]
- Lamb AL. Breaking a pathogen's iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim Biophys Acta 2015;1854:1054–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas PJ, Lin SJ, Culotta VC. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol 1996;21:519–28. [DOI] [PubMed] [Google Scholar]
- Leal SM Jr, Roy S, Vareechon C et al. . Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog 2013;9:e1003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch JM, Jensen LT, Bouldin SD et al. . Activation of Cu,Zn-superoxide dismutase in the absence of oxygen and the copper chaperone CCS. J Biol Chem 2009;284:21863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestas AN. The effect of pH upon human transferrin: selective labelling of the two iron-binding sites. Brit J Haematol 1976;32:341–50. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Knight SA, Camadro JM et al. . Siderophore uptake by Candida albicans: effect of serum treatment and comparison with Saccharomyces cerevisiae. Yeast 2002;19:329–40. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Simon M, Klein R et al. . Excretion of anthranilate and 3-hydroxyanthranilate by Saccharomyces cerevisiae: relationship to iron metabolism. J Gen Microbiol 1992;138:85–9. [DOI] [PubMed] [Google Scholar]
- Levitz SM, Harrison TS, Tabuni A et al. . Chloroquine induces human mononuclear phagocytes to inhibit and kill Cryptococcus neoformans by a mechanism independent of iron deprivation. J Clin Invest 1997;100:1640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Gleason JE, Zhang SX et al. . Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. P Natl Acad Sci USA 2015;112:E5336–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bagley D, Ward DM et al. . Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 2008;28:1326–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen OS, McVey Ward D et al. . CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 2001;276:29515–9. [DOI] [PubMed] [Google Scholar]
- Li L, Kaplan J. Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 1998;273:22181–7. [DOI] [PubMed] [Google Scholar]
- Li L, Kaplan J. The yeast gene MSC2, a member of the cation diffusion facilitator family, affects the cellular distribution of zinc. J Biol Chem 2001;276:5036–43. [DOI] [PubMed] [Google Scholar]
- Li L, Miao R, Bertram S et al. . A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J Biol Chem 2012;287:35709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Zhou B. Copper and manganese induce yeast apoptosis via different pathways. Mol Biol Cell 2007;18:4741–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Wei D, Wang H et al. . Role of Candida albicans Aft2p transcription factor in ferric reductase activity, morphogenesis and virulence. Microbiology 2010;156:2912–9. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Pufahl RA, Dancis A et al. . A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 1997;272:9215–20. [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG et al. . Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα; allele enhances filamentation. PLoS Genet 2006;2:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Supek F, Nelson N et al. . Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J Biol Chem 1997;272:11763–9. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 2004;3:1076–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk EE, Culotta VC. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J Biol Chem 2001;276:47556–62. [DOI] [PubMed] [Google Scholar]
- Lulloff SJ, Hahn BL, Sohnle PG. Fungal susceptibility to zinc deprivation. J Lab Clin Med 2004;144:208–14. [DOI] [PubMed] [Google Scholar]
- McCormick A, Heesemann L, Wagener J et al. . NETs formed by human neutrophils inhibit growth of the pathogenic mold Aspergillus fumigatus. Microbes Infect 2010;12:928–36. [DOI] [PubMed] [Google Scholar]
- McDevitt CA, Ogunniyi AD, Valkov E et al. . A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog 2011;7:e1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J 2000;19:2845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid CW, Milanick MA, Eide DJ. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J Biol Chem 2003;278:15065–72. [DOI] [PubMed] [Google Scholar]
- Macomber L, Hausinger RP. Mechanisms of nickel toxicity in microorganisms. Metallomics 2011;3:1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. P Natl Acad Sci USA 2009;106:8344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 2007;189:1616–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 2011;80:319–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez CE, Motto HL. Solubility of lead, zinc and copper added to mineral soils. Environ Pollut 2000;107:153–8. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, de Llanos R, Romero AM et al. . Post-transcriptional regulation of iron homeostasis in Saccharomyces cerevisiae. Int J Mol Sci 2013;14:15785–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LJ, Jensen LT, Simon JR et al. . Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J Biol Chem 1998;273:23716–21. [DOI] [PubMed] [Google Scholar]
- Marvin ME, Mason RP, Cashmore AM. The CaCTR1 gene is required for high-affinity iron uptake and is transcriptionally controlled by a copper-sensing transactivator encoded by CaMAC1. Microbiology 2004;150:2197–208. [DOI] [PubMed] [Google Scholar]
- Marvin ME, Williams PH, Cashmore AM. The Candida albicans CTR1 gene encodes a functional copper transporter. Microbiology 2003;149:1461–74. [DOI] [PubMed] [Google Scholar]
- Matzanke BF, Bill E, Trautwein AX et al. . Role of siderophores in iron storage in spores of Neurospora crassa and Aspergillus ochraceus. J Bacteriol 1987;169:5873–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra RK, Garey JR, Winge DR. Selective and tandem amplification of a member of the metallothionein gene family in Candida glabrata. J Biol Chem 1990;265:6369–75. [PubMed] [Google Scholar]
- Mehra RK, Thorvaldsen JL, Macreadie IG et al. . Disruption analysis of metallothionein-encoding genes in Candida glabrata. Gene 1992;114:75–80. [DOI] [PubMed] [Google Scholar]
- Mei B, Budde AD, Leong SA. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. P Natl Acad Sci USA 1993;90:903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A, Pelletier B, Labbe S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell 2006;5:1866–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moors MA, Stull TL, Blank KJ et al. . A role for complement receptor-like molecules in iron acquisition by Candida albicans. J Exp Med 1992;175:1643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno MÁ, Ibrahim‐Granet O, Vicentefranqueira R et al. . The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol Microbiol 2007;64:1182–97. [DOI] [PubMed] [Google Scholar]
- Morrow CA, Fraser JA. Is the nickel-dependent urease complex of Cryptococcus the pathogen's Achilles' heel? MBio 2013;4:e00408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller G, Barclay SJ, Raymond KN. The mechanism and specificity of iron transport in Rhodotorula pilimanae probed by synthetic analogs of rhodotorulic acid. J Biol Chem 1985;260:13916–20. [PubMed] [Google Scholar]
- Muñoz JF, Gauthier GM, Desjardins CA et al. . The dynamic genome and transcriptome of the human fungal pathogen Blastomyces and close relative Emmonsia. PLoS Genet 2015;11:e1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashige TG, Zhang B, Krebs C et al. . Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol 2015;11:765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser L, Weissman Z, Pinsky M et al. . Structural basis of haem-iron acquisition by fungal pathogens. Nat Microbiol 2016;1:16156. [DOI] [PubMed] [Google Scholar]
- Navarathna DH, Harris SD, Roberts DD et al. . Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res 2010;10:209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Gabbiani C, Messori L et al. . Metal-based drugs for malaria, trypanosomiasis and leishmaniasis: recent achievements and perspectives. Drug Discov Today 2010;15:1070–8. [DOI] [PubMed] [Google Scholar]
- Neilands JB. A crystalline organo-iron pigment from a rust fungus (Ustilago sphaerogena). J Am Chem Soc 1952;74:4846–7. [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J et al. . Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004;306:2090–3. [DOI] [PubMed] [Google Scholar]
- Nevitt T, Thiele DJ. Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog 2011;7:e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SL, Gootee L, Brunner G et al. . Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J Clin Invest 1994;93:1422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiec MJ, De Samber B, Garrevoet J et al. . Trace element landscape of resting and activated human neutrophils on the sub-micrometer level. Metallomics 2015;7:996–1010. [DOI] [PubMed] [Google Scholar]
- Nilius AM, Farmer SG. Identification of extracellular siderophores of pathogenic strains of Aspergillus fumigatus. J Med Vet Mycol 1990;28:395–403. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Igarashi K, Kakinuma Y. Proton gradient-driven nickel uptake by vacuolar membrane vesicles of Saccharomyces cerevisiae. J Bacteriol 1998;180:1962–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD et al. . Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 2009;7:e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North M, Steffen J, Loguinov AV et al. . Genome-wide functional profiling identifies genes and processes important for zinc-limited growth of Saccharomyces cerevisiae. PLoS Genet 2012;8:e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhus KJ, Wilborn AT, Jacobson ES. Ferric iron reduction by Cryptococcus neoformans. Infect Immun 1997;65:434–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Holmer SM, Selvig K et al. . Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio 2013;4:e00522–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Norton D, Price MS et al. . Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 2010;6:e1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberegger H, Schoeser M, Zadra I et al. . SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol Microbiol 2001;41:1077–89. [DOI] [PubMed] [Google Scholar]
- Oide S, Moeder W, Krasnoff S et al. . NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 2006;18:2836–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LD, Skaar EP. Transition metals and virulence in bacteria. Annu Rev Genet 2016;50:67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente AFA, de Rezende TCV, De Castro KP et al. . A proteomic view of the response of Paracoccidioides yeast cells to zinc deprivation. Fungal Biol 2013;117:399–410. [DOI] [PubMed] [Google Scholar]
- Park YS, Lian H, Chang M et al. . Identification of high-affinity copper transporters in Aspergillus fumigatus. Fungal Genet Biol 2014;73:29–38. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Saier MH Jr.. A novel family of ubiquitous heavy metal ion transport proteins. J Membr Biol 1997;156:99–103. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Sherman F. Toxicity of copper, cobalt, and nickel salts is dependent on histidine metabolism in the yeast Saccharomyces cerevisiae. J Bacteriol 1999;181:4774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccarelli M, Scott TD, Steele M et al. . mRNAs involved in copper homeostasis are regulated by the nonsense-mediated mRNA decay pathway depending on environmental conditions. Fungal Genet Biol 2016;86:81–90. [DOI] [PubMed] [Google Scholar]
- Pimentel C, Vicente C, Menezes RA et al. . The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One 2012;7:e37434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy ME, Jensen LT, Culotta VC. The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem J 2002;362:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy ME, Liu XF, Culotta VC. Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Mol Cell Biol 2000;20:7893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science 2000;288:1651–3. [DOI] [PubMed] [Google Scholar]
- Potrykus J, Stead D, Maccallum DM et al. . Fungal iron availability during deep seated candidiasis is defined by a complex interplay involving systemic and local events. PLoS Pathog 2013;9:e1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot B, Jbel M, Mercier A et al. . abc3+ encodes an iron-regulated vacuolar ABC-type transporter in Schizosaccharomyces pombe. Eukaryot Cell 2010;9:59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AS. Discovery of human zinc deficiency: 50 years later. J Trace Elem Med Bio 2012;26:66–9. [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 2005;120:99–110. [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab 2008;7:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science 2000;288:1062–4. [DOI] [PubMed] [Google Scholar]
- Reddi AR, Jensen LT, Culotta VC. Manganese homeostasis in Saccharomyces cerevisiae. Chem Rev 2009;109:4722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi AR, Jensen LT, Naranuntarat A et al. . The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic Biol Med 2009;46:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees EM, Lee J, Thiele DJ. Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J Biol Chem 2004;279:54221–9. [DOI] [PubMed] [Google Scholar]
- Rees EM, Thiele DJ. Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J Biol Chem 2007;282:21629–38. [DOI] [PubMed] [Google Scholar]
- Riggle PJ, Kumamoto CA. Role of a Candida albicans P1-type ATPase in resistance to copper and silver ion toxicity. J Bacteriol 2000;182:4899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DG, Dancis A, Anderson GJ et al. . The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol 1993;13:4342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Ojeda L, Balk J et al. . Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem 2005;280:10135–40. [DOI] [PubMed] [Google Scholar]
- Saikia S, Oliveira D, Hu G et al. . Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect Immun 2014;82:839–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic MI, Ding C, Thiele DJ et al. . Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 2012;11:106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Buisson N, Knight S et al. . Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology 2003;149:579–88. [DOI] [PubMed] [Google Scholar]
- Santos R, Buisson N, Knight SA et al. . Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol Microbiol 2004;54:507–19. [DOI] [PubMed] [Google Scholar]
- Schrettl M, Beckmann N, Varga J et al. . HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog 2010;6:e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Bignell E, Kragl C et al. . Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 2004;200:1213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Bignell E, Kragl C et al. . Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 2007;3:1195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M, Kim HS, Eisendle M et al. . SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol 2008;70:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JA, Olarte KT, Michalek JL et al. . Regulation of copper toxicity by Candida albicans GPA2. Eukaryot Cell 2013;12:954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Lopez-Aragon R, Calera JA et al. . Zinc-regulated biosynthesis of immunodominant antigens from Aspergillus spp. Infect Immun 1999;67:2377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seider K, Gerwien F, Kasper L et al. . Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot Cell 2014;13:170–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, Tiedeman J, Rashford J et al. . Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell 2004;15:1233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CE, Kapica L. Production of diagnostic pigment by phenoloxidase activity of Cryptococcus neoformans. Appl Microbiol 1972;24:824–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Bailao MG, Bailao EF, Lechner BE et al. . Hydroxamate production as a high affinity iron acquisition mechanism in Paracoccidioides spp. PLoS One 2014;9:e105805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Panting RJ, Varma A et al. . Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans. MBio 2013;4:e00220–00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Prasad HK, Sinha I et al. . Cap2-HAP complex is a critical transcriptional regulator that has dual but contrasting roles in regulation of iron homeostasis in Candida albicans. J Biol Chem 2011;286:25154–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MP, Weiss G. The Iron age of host-microbe interactions. EMBO Rep 2015;16:1482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgo AG, Heilmann CJ, Dekker HL et al. . Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot Cell 2011;10:1071–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Ibrahim AS, Lin L et al. . Antibody titer threshold predicts anti-candidal vaccine efficacy even though the mechanism of protection is induction of cell-mediated immunity. J Infect Dis 2008;197:967–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg BJ, Ibrahim AS, Avanesian V et al. . Efficacy of the anti-Candida rAls3p-N or rAls1p-N vaccines against disseminated and mucosal candidiasis. J Infect Dis 2006;194:256–60. [DOI] [PubMed] [Google Scholar]
- Srivastava VK, Suneetha KJ, Kaur R. A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem J 2014;463:103–14. [DOI] [PubMed] [Google Scholar]
- Sterkel AK, Mettelman R, Wuthrich M et al. . The unappreciated intracellular lifestyle of Blastomyces dermatitidis. J Immunol 2015;194:1796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain J, Culotta VC. Copper ions and the regulation of Saccharomyces cerevisiae metallothionein genes under aerobic and anaerobic conditions. Mol Gen Genet 1996;251:139–45. [DOI] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H et al. . A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. P Natl Acad Sci USA 1996;93:5105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Thiele DJ. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol Cell Biol 1989;9:421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Zhu Z, Silar P et al. . Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 1997;13:1423–35. [DOI] [PubMed] [Google Scholar]
- Tangen KL, Jung WH, Sham AP et al. . The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 2007;153:29–41. [DOI] [PubMed] [Google Scholar]
- Thewes S, Kretschmar M, Park H et al. . In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol 2007;63:1606–28. [DOI] [PubMed] [Google Scholar]
- Thieken A, Winkelmann G. Rhizoferrin: a complexone type siderophore of the Mucorales and entomophthorales (Zygomycetes). FEMS Microbiol Lett 1992;73:37–41. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Sewell AK, Tanner AM et al. . Mixed Cu+ and Zn2+ coordination in the DNA-binding domain of the AMT1 transcription factor from Candida glabrata. Biochemistry 1994;33:9566–77. [DOI] [PubMed] [Google Scholar]
- Timmerman MM, Woods JP. Ferric reduction is a potential iron acquisition mechanism for Histoplasma capsulatum. Infect Immun 1999;67:6403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman MM, Woods JP. Potential role for extracellular glutathione-dependent ferric reductase in utilization of environmental and host ferric compounds by Histoplasma capsulatum. Infect Immun 2001;69:7671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys 2000;373:1–6. [DOI] [PubMed] [Google Scholar]
- Tuncher A, Sprote P, Gehrke A et al. . The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A.nidulans and human. J Mol Biol 2005;352:517–33. [DOI] [PubMed] [Google Scholar]
- Ueta R, Fujiwara N, Iwai K et al. . Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol 2012;32:4998–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban CF, Ermert D, Schmid M et al. . Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 2009;5:e1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem 1999;274:38061–70. [DOI] [PubMed] [Google Scholar]
- Vaknin Y, Shadkchan Y, Levdansky E et al. . The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fungal Genet Biol 2014;63:55–64. [DOI] [PubMed] [Google Scholar]
- Vance CK, Miller AF. A simple proposal that can explain the inactivity of metal-substituted superoxide dismutases. J Am Chem Soc 1998;120:461–7. [Google Scholar]
- Veyrier FJ, Boneca IG, Cellier MF et al. . A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog 2011;7:e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau CM, Cardone JM, Guecheva TN et al. . Enhanced resistance of yeast mutants deficient in low-affinity iron and zinc transporters to stannous-induced toxicity. Chemosphere 2012;86:477–84. [DOI] [PubMed] [Google Scholar]
- Vicentefranqueira R, Moreno MA, Leal F et al. . The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot Cell 2005;4:837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignesh KS, Figueroa JAL, Porollo A et al. . Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity 2013;39:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisard C, Wang J, McEvoy JL et al. . urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol Cell Biol 1993;13:7091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B et al. . Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol 2005;174:1491–500. [DOI] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J 2011;434:365–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Marcus S et al. . The role of frataxin in fission yeast iron metabolism: implications for Friedreich's ataxia. Biochim Biophys Acta 2014;1840:3022–33. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Hacham M, Hu G et al. . Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest 2007;117:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Park YD, Raja M et al. . Role of CTR4 in the virulence of Cryptococcus neoformans. MBio 2012;3:e00285–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 1975;231:39–41. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Berdicevsky I, Cavari BZ et al. . The high copper tolerance of Candida albicans is mediated by a P-type ATPase. P Natl Acad Sci USA 2000;97:3520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol 2004;53:1209–20. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Conibear E et al. . An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol 2008;69:201–17. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Kornitzer D. Deletion of the copper transporter CaCCC2 reveals two distinct pathways for iron acquisition in Candida albicans. Mol Microbiol 2002;44:1551–60. [DOI] [PubMed] [Google Scholar]
- Wells ML, Washington OL, Hicks SN et al. . Post-transcriptional regulation of transcript abundance by a conserved member of the tristetraprolin family in Candida albicans. Mol Microbiol 2015;95:1036–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler KA, Hurdman BF, Pitt JI. Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium. Int J Food Microbiol 1991;12:141–9. [DOI] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T et al. . A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 2009;284:33949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol 1994;176:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat Genet 1997;16:352–7. [DOI] [PubMed] [Google Scholar]
- Winters MS, Chan Q, Caruso JA et al. . Metallomic analysis of macrophages infected with Histoplasma capsulatum reveals a fundamental role for zinc in host defenses. J Infect Dis 2010;202:1136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-Y, Bird AJ, Chung LM et al. . Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics 2008;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sinani D, Kim H et al. . Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J Biol Chem 2009;284:4112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Cheng X, Yu Q et al. . Aft2, a novel transcription regulator, is required for iron metabolism, oxidative stress, surface adhesion and hyphal development in Candida albicans. PLoS One 2013;8:e62367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Dong Y, Cheng X et al. . Cellular iron homeostasis mediated by the Mrs4-Ccc1-Smf3 pathway is essential for mitochondrial function, morphogenesis and virulence in Candida albicans. Biochim Biophys Acta 2014a;1843:629–39. [DOI] [PubMed] [Google Scholar]
- Xu N, Qian K, Dong Y et al. . Novel role of the Candida albicans ferric reductase gene CFL1 in iron acquisition, oxidative stress tolerance, morphogenesis and virulence. Res Microbiol 2014b;165:252–61. [DOI] [PubMed] [Google Scholar]
- Xu W, Solis NV, Ehrlich RL et al. . Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol 2015;13:e1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Dancis A, Klausner RD. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 1995;14:1231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Serpe M, Haile D et al. . Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J Biol Chem 1997;272:17711–8. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Sakata-Sogawa K, Hasegawa A et al. . Zinc is a novel intracellular second messenger. J Cell Biol 2007;177:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CW, Bauler M, Moore RE et al. . The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J Biol Chem 2001;276:10218–23. [DOI] [PubMed] [Google Scholar]
- Yun CW, Tiedeman JS, Moore RE et al. . Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J Biol Chem 2000;275:16354–9. [DOI] [PubMed] [Google Scholar]
- Zähner H, Keller-Schierlein W, Hütter R et al. . Stoffwechselprodukte von Mikroorganismen: Sideramine aus Aspergillaceen. Arch Mikrobiol 1963;45:119–35. [Google Scholar]
- Zakikhany K, Naglik JR, Schmidt‐Westhausen A et al. . In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 2007;9:2938–54. [DOI] [PubMed] [Google Scholar]
- Zarnowski R, Cooper KG, Brunold LS et al. . Histoplasma capsulatum secreted gamma-glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol Microbiol 2008;70:352–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodionov DA, Gelfand MS et al. . Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 2009;10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. P Natl Acad Sci USA 1996a;93:2454–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem 1996b;271:23203–10. [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol 1997;17:5044–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LW, Haas H, Marzluf GA. Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol Gen Genet 1998;259:532–40. [DOI] [PubMed] [Google Scholar]
- Zhou P, Thiele DJ. Rapid transcriptional autoregulation of a yeast metalloregulatory transcription factor is essential for high-level copper detoxification. Gene Dev 1993;7:1824–35. [DOI] [PubMed] [Google Scholar]
- Zhou P, Szczypka MS, Sosinowski T et al. . Expression of a yeast metallothionein gene family is activated by a single metalloregulatory transcription factor. Mol Cell Biol 1992;12:3766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Labbe S, Pena MM et al. . Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J Biol Chem 1998;273:1277–80. [DOI] [PubMed] [Google Scholar]