ABSTRACT
The earthworm gut is an anoxic, saccharide-rich microzone in aerated soils. The apparent degradation of diverse saccharides in the alimentary canal of the model earthworm Lumbricus terrestris is concomitant with the production of diverse organic acids, indicating that fermentation is an ongoing process in the earthworm gut. However, little is known about how different gut-associated saccharides are fermented. The hypothesis of this investigation was that different gut-associated saccharides differentially stimulate fermentative microorganisms in gut contents of L. terrestris. This hypothesis was addressed by (i) assessing the fermentation profiles of anoxic gut content microcosms that were supplemented with gut-associated saccharides and (ii) the concomitant phylogenic analysis of 16S rRNA sequences. Galactose, glucose, maltose, mannose, arabinose, fucose, rhamnose, and xylose stimulated the production of fermentation products, including H2, CO2, acetate, lactate, propionate, formate, succinate, and ethanol. Fermentation profiles were dependent on the supplemental saccharide (e.g., glucose yielded large amounts of H2 and ethanol, whereas fucose did not, and maltose yielded large amounts of lactate, whereas mannose did not). Approximately 1,750,000 16S rRNA sequences were affiliated with 37 families, and phylogenic analyses indicated that a respective saccharide stimulated a subset of the diverse phylotypes. An Aeromonas-related phylotype displayed a high relative abundance in all treatments, whereas key Enterobacteriaceae-affiliated phylotypes were stimulated by some but not all saccharides. Collectively, these results reinforce the likelihood that (i) different saccharides stimulate different fermentations in gut contents of the earthworm and (ii) facultative aerobes related to Aeromonadaceae and Enterobacteriaceae can be important drivers of these fermentations.
IMPORTANCE The feeding habits of earthworms influence the turnover of elements in the terrestrial biosphere. The alimentary tract of the earthworm constitutes an anoxic saccharide-rich microzone in aerated soils that offers ingested microbes a unique opportunity for anaerobic growth. The fermentative activity of microbes in the alimentary tract are responsible for the in situ production of (i) organic compounds that can be assimilated by the earthworm and (ii) H2 that is subject to in vivo emission by the earthworm and can be trophically linked to secondary microbial events in soils. To gain insight on how fermentative members of the gut microbiome might respond to the saccharide-rich alimentary canal, this study examines the impact of diverse gut-associated saccharides on the differential activation of fermentative microbes in gut contents of the model earthworm L. terrestris.
KEYWORDS: anaerobes, fermentation, earthworms, invertebrate microbiology
INTRODUCTION
Earthworms are subsurface dwellers that go largely unseen but nonetheless have profound impact on the terrestrial biosphere (1–5). For example, their feeding and burrowing habits affect soil porosity, gas exchange between soil and air, nutrient availability, plant growth, and the annual turnover of litter fall (2–4, 6–14). As a major soil macrofauna, earthworm populations can reach extraordinary numbers, and up to 2,000 individuals per square meter have been documented (4, 13, 14), a number that could theoretically yield approximately 500 ml earthworm gut content per square meter of soil (15). The alimentary canal of the earthworm is anoxic and rich in organic compounds (2, 14, 16, 17). These properties (i) are in marked contrast to the aerated soil that is ingested by the earthworm and (ii) augment anaerobiosis in the alimentary canal (14, 16, 18–20). Gut-associated anaerobic microbial processes can lead to the in vivo emission of diverse gases from the earthworm, including molecular hydrogen (H2), nitrous oxide (N2O), molecular nitrogen (N2), and methane (CH4) (17, 20–25). Thus, the alimentary canal of the earthworm constitutes an important anoxic microzone in soils that are inhabited by earthworms.
Dissimilatory microbial activities in the earthworm gut are dependent in part upon organic-carbon-derived reductant, and potential sources of organic carbon include ingested plant material and ingested microbial cells that become ruptured in the crop/gizzard (9, 14, 16, 17, 26, 27). In addition to these ingested sources of organic carbon, earthworm-produced mucus in the alimentary canal is conceived to be an important source of saccharides, and mucus-derived saccharides have been proposed to augment fermentation and denitrification in the alimentary canal of the model earthworm Lumbricus terrestris (14, 16, 17, 28–30) and to also facilitate methanogenic food webs in the gut of the CH4-emitting earthworm Eudrilus eugeniae (15, 24).
Independent of the origin of organic carbon in the alimentary canal, diverse saccharides occur in gut contents of L. terrestris, and hydrolysis is conceived to increase the availability of these saccharides (17). Indeed, over 100 mM polymeric saccharide equivalents can occur in the aqueous phase of the alimentary canal (14, 16, 17), and the in situ hydrolysis of this localized enrichment of high-quality organic carbon offers an “anoxic sugar buffet” for nutritionally starved ingested microbes capable of anaerobiosis (16, 17). The apparent in situ degradation of these saccharides is concomitant with the occurrence of diverse fermentation products, including organic acids and H2 (17), suggesting that saccharides in the gut might drive diverse fermentations along the alimentary canal. Indeed, glucose, as a model gut-associated saccharide that can exceed 10 mM in the aqueous phase of the alimentary canal (16, 17), stimulates fermentation in gut contents (15, 29). However, the large number of different saccharides in the alimentary canal suggests that competition might lead to the differential engagement of fermentative taxa in response to each specific saccharide. Thus, the hypothesis of this investigation was that different gut-associated saccharides differentially stimulate fermentative microorganisms in the earthworm gut. The main objective, therefore, was to determine which members of the microbial community in gut contents respond to a specific gut-associated saccharide.
RESULTS
Saccharide-dependent stimulation of fermentation.
Arabinose, fucose, galactose, glucose, maltose, mannose, and rhamnose are found in the alimentary canal of L. terrestris (17). Plant-derived hemicellulose is a source of xylose (31, 32), and earthworms such as L. terrestris that ingest plant material could theoretically have this saccharide in the alimentary canal. These saccharides were subjected to consumption in anoxic gut content microcosms (Table 1). Certain saccharides stimulated the production of carbon dioxide (CO2) and H2 without appreciable delay (Fig. 1), indicating that fermentative microbes in gut content were poised to respond rapidly to nutrient availability under anoxic conditions. Neither N2O nor CH4 was detected, and the pH did not vary much in the different treatments with the exception of the maltose treatment, in which the pH decreased a bit more than in the other treatments (see Table S2 in the supplemental material).
TABLE 1.
Fermentation profiles of different saccharide treatmentsa
| Treatment | Time (h) | Amt of fermentation product (μmol/g gut contentFW) |
||||||
|---|---|---|---|---|---|---|---|---|
| Saccharide | Succinate | Formate | Propionate | Ethanol | Lactate | Acetate | ||
| None (control)b | 0 | 0 | 0 | 0 | 0 | 0 | 3.9 ± 0.1 | 0.2 ± 0.3 |
| 30 | 0 | 0 | 1.5 ± 0.4 | 7.0 ± 0.5 | 2.4 ± 3.1 | 0.8 ± 0.1 | 13.5 ± 0.8 | |
| Arabinose | 0 | 59.4 ± 1.8 | 0 | 0 | 0 | 0 | 3.4 ± 0.3 | 0.3 ± 0.4 |
| 30 | 0 | 8.9 ± 0.7 | 3.1 ± 0.2 | 9.5 ± 1.7 | 41.5 ± 0.3 | 0 | 44.6 ± 2.0 | |
| Fucose | 0 | 56.0 ± 3.2 | 0 | 0 | 0 | 0 | 2.3 ± 0.5 | 0.7 ± 0.3 |
| 30 | 35.9 ± 8.1 | 1.6 ± 1.4 | 3.3 ± 0.3 | 16.2 ± 1.8 | 0 | 0 | 22.6 ± 2.4 | |
| Galactose | 0 | 48.3 ± 10.0 | 0 | 0 | 0 | 0 | 2.2 ± 0.0 | 0.8 ± 0.2 |
| 30 | 0 | 9.7 ± 0.8 | 2.3 ± 0.5 | 9.7 ± 0.5 | 33.5 ± 4.9 | 0 | 38.1 ± 2.1 | |
| Glucose | 0 | 54.3 ± 3.4 | 0 | 0 | 0 | 0 | 2.4 ± 0.2 | 3.1 ± 2.0 |
| 30 | 0 | 0 | 0 | 10.6 ± 1.5 | 36.4 ± 4.2 | 12.5 ± 2.1 | 43.3 ± 5.4 | |
| Maltose | 0 | 51.8 ± 4.3 | 0 | 0 | 0 | 0 | 2.7 ± 0.1 | 4.4 ± 0.3 |
| 30 | 0 | 0 | 0 | 15.5 ± 3.1 | 71.2 ± 21.7 | 38.7 ± 4.7 | 60.5 ± 11.0 | |
| Mannose | 0 | 52.4 ± 24.5 | 0 | 0 | 0 | 0 | 2.9 ± 0.3 | 1.3 ± 0.1 |
| 30 | 0 | 12.4 ± 0.7 | 0.3 ± 0.5 | 13.7 ± 0.8 | 42.5 ± 10.4 | 0 | 41.6 ± 1.9 | |
| Rhamnose | 0 | 47.6 ± 1.6 | 0 | 0 | 0 | 0 | 2.7 ± 0.1 | 3.3 ± 1.6 |
| 30 | 0 | 3.6 ± 0.1 | 0.3 ± 0.5 | 28.3 ± 2.7 | 0 | 5.0 ± 0.7 | 31.2 ± 5.9 | |
| Xylose | 0 | 56.8 ± 6.2 | 0 | 0 | 0 | 0 | 2.9 ± 0.2 | 2.6 ± 2.2 |
| 30 | 11.0 ± 4.3 | 2.8 ± 2.2 | 14.6 ± 2.4 | 0 | 27.0 ± 5.6 | 0.4 ± 0.7 | 33.8 ± 4.4 | |
Values were calculated from three replicate analyses; values and standard deviations are rounded to the nearest first decimal place. The gaseous product profile of each treatment is shown in Fig. 1. Statistical P values are shown in Table S1. FW, fresh weight.
Control lacked supplemental saccharides.
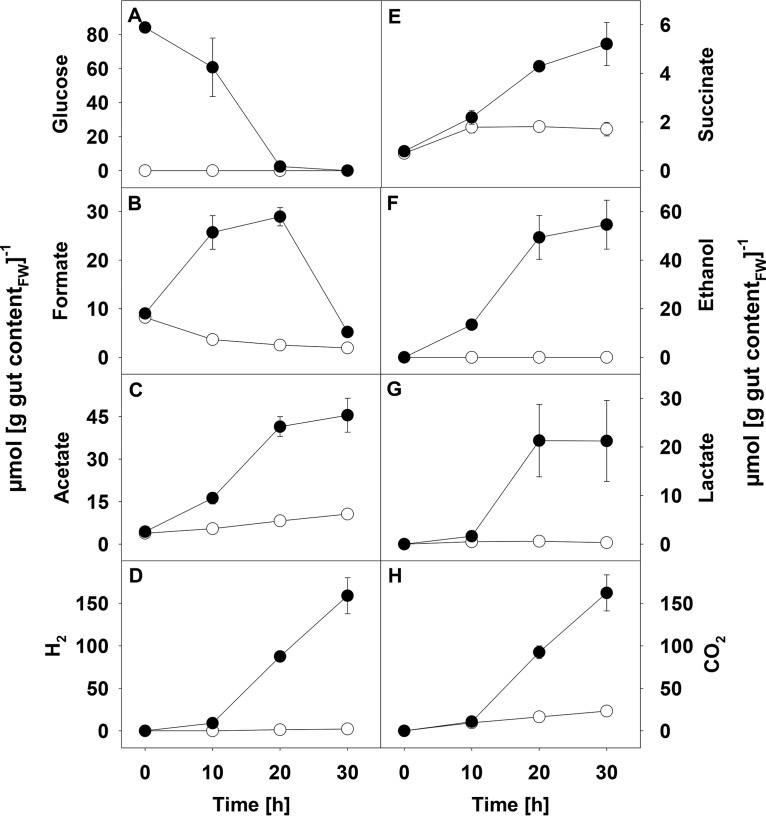
FIG 1.
Production of H2 and CO2 in different saccharide treatments. Triplicate anoxic microcosms with gut content (A to D) or soil (E to H) were supplemented with the following saccharides: arabinose (●), fucose (⬥), galactose (◇), glucose (○), maltose (▽), mannose (△), rhamnose (▲), and xylose (▼). The gray squares represent the unsupplemented controls. Incubation was at 15°C in the dark. Additional information on the substrate/product profiles of the treatments is shown in Table 1. Error bars indicate standard deviations. Statistical P values are shown in Table S1 in the supplemental material.
In most cases, supplemental saccharides stimulated the production of ethanol and numerous organic acids, particularly acetate (Table 1). However, fucose was poorly utilized and did not yield appreciable amounts of H2 (Table 1; Fig. 1). Rhamnose and fucose did not yield ethanol, and high concentrations of lactate occurred only in glucose- and maltose-supplemented microcosms. Formate was more abundant in xylose treatments than in other treatments. Thus, the apparent production of a particular fermentation product was saccharide dependent. The accumulation of fatty acids in the microcosms indicated that syntrophic fermentation was minimal or absent.
The recovery of carbon and reductant for most saccharide-supplemented incubations ranged between 73 and 87% (see Table S3 in the supplemental material), suggesting that most fermentation products were detected. However, recoveries for xylose-, rhamnose-, and fucose-supplemented microcosms were lower, indicating that additional undetected products (e.g., 2,3-butanediol) may have been produced from these saccharides (Table S3). Nitrate concentrations in microcosms with gut contents were below the detection limit of 0.08 mg nitrate liter−1; traces of nitrate were detected in the anoxic soil microcosms at the beginning of the incubations (data not shown). As illustrated by the very limited effect of supplemental saccharides on the production of H2 and CO2 by preingested soil (Fig. 1), the consumption of saccharides and the production of fatty acids and alcohols in soil microcosms were very marginal compared to that of gut content microcosms (data not shown).
Saccharide-dependent stimulation of microbes in gut content microcosms.
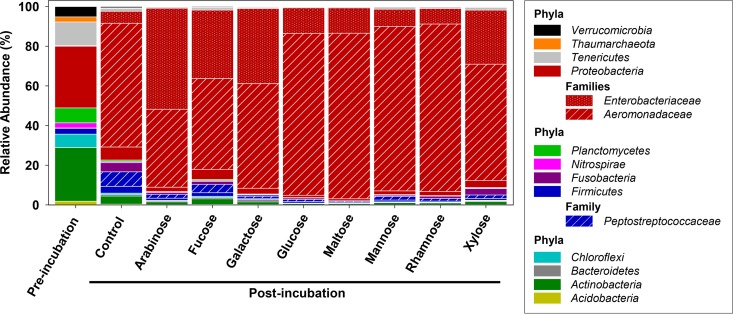
A total of 890,072 16S rRNA sequences were obtained from the aforementioned experiment, and rarefaction analyses indicated that the most abundant taxa were effectively targeted (see Fig. S1 in the supplemental material). The high phylum and family level diversities detected in gut content prior to incubation decreased at the end of the incubation period (Fig. 2), whereas the number of phylotypes increased from 117 to 134 to 136 at the end of the incubation period relative to those detected prior to incubation (Fig. S1). Thus, anoxic conditions and supplemental saccharides stimulated certain but not all gut content microbes.
FIG 2.
Relative abundances of phylum and family level 16S rRNA sequences obtained from different gut content saccharide treatments. Number of sequences for each treatment: preincubation, 86,762; control postincubation, 110,496; arabinose, 89,649; fucose, 73,148; galactose, 85,325; glucose, 93,192; maltose, 98,364; mannose, 88,619; rhamnose, 96,928; and xylose, 67,589.
The phyla Actinobacteria, Chloroflexi, Planctomycetes, Proteobacteria, Tenericutes, and Verrucomicrobia displayed the highest relative initial abundances (Fig. 2). The relative abundance of the Actinobacteria became very low postincubation, whereas the Proteobacteria displayed the highest relative abundances in both unsupplemented and saccharide-supplemented gut content microcosms postincubation (Fig. 2). Aeromonadaceae and Enterobacteriaceae constituted the most-represented Proteobacteria-affiliated families postincubation (Fig. 2). However, these two families were differentially distributed in the saccharide treatments. For example, the arabinose-supplemented treatment displayed a high relative abundance of Enterobacteriaceae, whereas this family was of minor importance in rhamnose treatments, in which Aeromonadaceae became the dominant family. Sequences belonging to the Aeromonas-affiliated phylotype were 100% similar to 16S rRNA sequences of Aeromonas lacus, Aeromonas aquatica, and Aeromonas hydrophila (Aeromonadaceae) (Table 2). This phylotype displayed high relative abundances in all saccharide treatments and in the unsupplemented control at the end of the incubation period. In addition to the Aeromonas-affiliated phylotype, (i) the Enterobacteriaceae-affiliated phylotype E (100% similarity to Klebsiella pneumoniae, Enterobacter aerogenes, and Enterobacter xiangfangensis) displayed a high relative abundance in arabinose-supplemented microcosms, (ii) the Enterobacteriaceae-affiliated phylotype C (100% similarity to Citrobacter freundii, Citrobacter braakii, and Raoultella terrigena) displayed a high relative abundance in fucose- and galactose-supplemented microcosms, and (iii) the Enterobacteriaceae-affiliated phylotype R (100% similarity to Rosenbergiella australoborealis, Rosenbergiella epipactidis, and Raoultella planticola) was an abundant phylotype in all supplemented microcosms (Table 2).
TABLE 2.
Relative abundance of the most abundant phylotypes of different saccharide treatmentsa
| Taxonomy of phylotype | Closest cultured microorganism | S (%)b | Relative abundance (%)c |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre | C | A | F | Ga | Gl | Mt | Mn | R | X | |||
| Proteobacteria | ||||||||||||
| Enterobacteriaceae | ||||||||||||
| Enterobacteriaceae-affiliated phylotype E [LT838793] | Klebsiella quasipneumoniae [NR_134062] | 100 | <1 | 2 | 33 | 7 | 5 | 3 | 2 | 3 | 3 | 4 |
| Enterobacter aerogenes [NR_102493] | 100 | |||||||||||
| Enterobacter xiangfangensis [NR_126208] | 100 | |||||||||||
| Enterobacteriaceae-affiliated phylotype C [LT838794] | Citrobacter freundii [NR_117752] | 100 | <1 | 1 | 13 | 16 | 28 | <1 | <1 | <1 | 1 | 1 |
| Citrobacter braakii [NR_117750] | 100 | |||||||||||
| Raoultella terrigena [NR_113703] | 100 | |||||||||||
| Enterobacteriaceae-affiliated phylotype R [LT838795] | Rosenbergiella australoborealis [NR_126305] | 100 | <1 | 3 | 4 | 10 | 4 | 9 | 10 | 5 | 4 | 21 |
| Rosenbergiella epipactidis [NR_126303] | 100 | |||||||||||
| Raoultella planticola [NR_113701] | 100 | |||||||||||
| Aeromonadaceae | ||||||||||||
| Aeromonas-affiliated phylotype A [LT838791] | Aeromonas lacus [NR_136831] | 100 | <1 | 58 | 37 | 43 | 50 | 77 | 78 | 77 | 79 | 55 |
| Aeromonas aquatica [NR_136829] | 100 | |||||||||||
| Aeromonas hydrophila [NR_074841] | 100 | |||||||||||
| Bradyrhizobiaceae | ||||||||||||
| Bradyrhizobium-affiliated phylotype [LT838788] | Bradyrhizobium japonicum [NR_119191] | 100 | 5 | 1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Bradyrhizobium neotropicale [NR_133987] | 100 | |||||||||||
| Bradyrhizobium ingae [NR_133985] | 100 | |||||||||||
| Hyphomicrobiaceae | ||||||||||||
| Rhodoplanes-affiliated phylotype [LT838789] | Rhodoplanes oryzae [NR_134156] | 97 | 5 | 1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Rhodoplanes piscinae [NR_115060] | 97 | |||||||||||
| Rhodoplanes elegans [NR_029125] | 97 | |||||||||||
| Fusobacteria | ||||||||||||
| Fusobacteriaceae | ||||||||||||
| Fusobacteriaceae-affiliated phylotype [LT838792] | Fusobacterium mortiferum [NR_117734] | 96 | <1 | 5 | 1 | 1 | 1 | 1 | <1 | 1 | 1 | 3 |
| Cetobacterium somerae [NR_025533] | 96 | |||||||||||
| Fusobacterium varium [NR_113384] | 95 | |||||||||||
| Verrucomicrobia | ||||||||||||
| Unclassified family | ||||||||||||
| Unclassified phylotype [LT838790] | Terrimicrobium sacchariphilum [NR_133878] | 92 | 5 | 1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Chthoniobacter flavus [NR_115225] | 90 | |||||||||||
| Verrucomicrobium spinosum [NR_026266] | 84 | |||||||||||
| Tenericutes | ||||||||||||
| Unclassified family | ||||||||||||
| Unclassified phylotype [LT838787] | “Candidatus Lumbricincola sp.” [FM165581] | 100 | 11 | 1 | <1 | 1 | 1 | <1 | <1 | 1 | 1 | 1 |
| “Candidatus Bacilloplasma” mollicute [DQ485975] | 91 | |||||||||||
| Staphylococcus massiliensis [KF600533] | 85 | |||||||||||
Accession numbers are in brackets. The table contains all phylotypes that had a relative abundance of at least 4% in at least one treatment. The fermentation profiles of the treatments are displayed in Fig. 1 and Table 1. All phylotypes are shown in Table S4.
S, similarity of 16S rRNA sequence.
Abbreviations: Pre, preincubation; C, unsupplemented control; A, arabinose supplemented; F, fucose supplemented; Ga, galactose supplemented; Gl, glucose supplemented; Mt, maltose supplemented; Mn, mannose supplemented; R, rhamnose supplemented; X, xylose supplemented.
In general, Proteobacteria increased in relative abundance by the end of the incubation. However, certain phylotypes of this phylum (i.e., Bradyrhizobium- and Rhodoplanes-affiliated phylotypes) decreased in relative abundance (Table 2). Other taxa displayed high relative abundances preincubation (e.g., an unclassified phylotype belonging to the Tenericutes) or in the unsupplemented control postincubation (i.e., a Fusobacteriaceae-affiliated phylotype) (Table 2), illustrating that the different incubations did not yield a uniform stimulation of the microbial taxa, but rather fostered a selective stimulation of specific taxa.
The apparent saccharide-dependent stimulation of these contrasting phylotypes suggests that they had the capacity to be differentially engaged in the fermentation of the diverse saccharides found in the earthworm gut. Archaea-affiliated phylotypes were not abundant in either the saccharide treatments or the unsupplemented control (Fig. 2).
Time-dependent changes of the bacterial community.
The experiment described above provided an overview of the saccharide-dependent responses of fermentative taxa in gut contents. To more closely examine temporal shifts of the microbial community, time-resolved fermentative processes and associated taxa were evaluated with glucose as a model gut-associated saccharide.
Glucose was degraded within the first 20 h, and a variety of organic acids and ethanol were concomitantly produced without apparent delay (Fig. 3), reaffirming the ability of gut content microbes to respond rapidly to nutrient input. Formate was initially produced but subsequently consumed, suggesting that certain fermentation products were subject to secondary dissimilation. In this regard, H2 and CO2 were produced in nearly equal amounts during the consumption of formate, suggesting that formate might have been dissimilated by formate hydrogenlyase-containing taxa. However, H2 and CO2 production exceeded the amount that could have been derived from the detected formate and thus may have also been produced from other processes. The recoveries of glucose-derived carbon and reductant were 79% and 76%, respectively (values were similar to that given in Table S3 for the glucose treatment [data not shown]), suggesting that most fermentation products were detected.
FIG 3.
Time-dependent product profiles of glucose treatment. Triplicate anoxic gut content microcosms contained (●) or did not contain (○) supplemental glucose. Incubation was at 15°C in the dark. The glucose concentration at the beginning of incubation represents the value of only one replicate due to the occurrence of unknown interfering peaks in the HPLC profile. Error bars indicate standard deviations. Statistical P values are shown in Table S1.
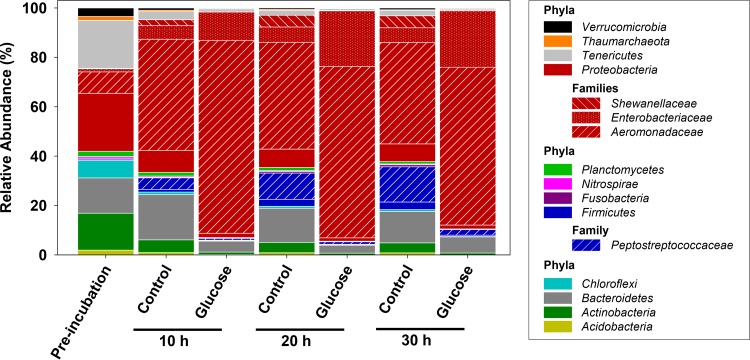
A total of 863,108 16S rRNA sequences were obtained, and rarefaction analyses indicated that the most abundant taxa were effectively targeted (see Fig. S2 in the supplemental material). A high diversity of phyla, including Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Nitrospirae, Planctomycetes, Proteobacteria, Tenericutes, Thaumarchaeota, and Verrucomicrobia, were detected at the beginning of the incubation (Fig. 4). The relative abundances of the different RNA-based phylotypes in both the unsupplemented control and glucose-supplemented treatments were greatly altered in the first 10 h of incubation, demonstrating that anoxia, as well as nutrient input, had pronounced effects on the relative abundance of 16S rRNA of gut content microbes.
FIG 4.
Effect of time on the relative abundances of phylum and family level 16S rRNA sequences obtained from control and glucose treatments. Number of sequences for each treatment: preincubation, 78,847; 10-h control, 107,853; 10-h glucose, 144,612; 20-h control, 98,655; 20-h glucose, 155,353; 30-h control, 118,099; 30-h glucose, 159,689.
Proteobacteria and Firmicutes increased in relative abundance within the first 10 h in the unsupplemented control (Fig. 4). At the end of the incubation period, the relative abundances of Bacteroidetes and Firmicutes were lower in the glucose treatments than in the unsupplemented controls (Fig. 4). A Flavobacterium-affiliated phylotype (Bacteroidetes) displayed the second-highest relative abundance in the unsupplemented control, and its relative abundance remained fairly stable during the anoxic incubation (Table 3). Bacteroidetes-related phylotypes displayed lower relative abundances in the first glucose experiment (Fig. 2), a difference likely due to differences in the soil used to feed worms and also differences in worm batches. The relative abundance of Firmicutes increased with time in the unsupplemented control, and this phylum was dominated by the family Peptostreptococcaceae. Independent of the differences observed with Firmicutes-affiliated phylotypes, Proteobacteria-affiliated phylotypes displayed the highest relative abundance in all treatments and at all sampling intervals, with Aeromonadaceae- and to a lesser extent Enterobacteriaceae-affiliated family level phylotypes displaying the highest relative abundances in this phylum (Fig. 4). Similar observations were made in the first experiment (Table 2, glucose treatment and control; see Materials and Methods), underlining the reproducibility of these findings. The apparent dominance of the Aeromonas-related phylotype in both the unsupplemented control and glucose treatment indicated that microbes associated with this phylotype were participants in the fermentations observed in these treatments. Sequences affiliated to the Enterobacteriaceae-related phylotype R increased in apparent response to glucose (Table 3). The modest time-dependent increase in the relative abundance of Shewanellaceae-affiliated phylotypes occurred only in the unsupplemented control.
TABLE 3.
Shifts in the relative abundance of the most abundant phylotypes during the fermentation of glucosea
| Taxonomy of phylotype | Closest cultured microorganism | S (%)b | Relative abundance (%)c |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre | 10 h |
20 h |
30 h |
||||||
| C | G | C | G | C | G | ||||
| Proteobacteria | |||||||||
| Enterobacteriaceae | |||||||||
| Enterobacteriaceae-affiliated phylotype R [LT838795] | For details, see Table 2 | 1 | 4 | 10 | 3 | 21 | 4 | 21 | |
| Aeromonadaceae | |||||||||
| Aeromonas-affiliated phylotype A [LT838791] | For details, see Table 2 | 8 | 42 | 74 | 40 | 66 | 39 | 60 | |
| Shewanellaceae | |||||||||
| Shewanella-affiliated phylotype [LT838797] | Shewanella putrefaciens [NR_113582] | 100 | <1 | 2 | <1 | 5 | <1 | 5 | <1 |
| Shewanella profunda [NR_104770] | 100 | ||||||||
| Shewanella xiamenensis [NR_116732] | 100 | ||||||||
| Bacteroidetes | |||||||||
| Flavobacteriaceae | |||||||||
| Flavobacterium-affiliated phylotype [LT838796] | Flavobacterium succinicans [NR_118478] | 100 | 11 | 14 | 4 | 11 | 3 | 10 | 5 |
| Flavobacterium spartansii [NR_133748] | 99 | ||||||||
| Flavobacterium granuli [NR_114020] | 99 | ||||||||
| Firmicutes | |||||||||
| Peptostreptococcaceae | |||||||||
| Peptostreptococcaceae-affiliated phylotype [LT838798] | Romboutsia sedimentorum [NR_134800] | 100 | <1 | 2 | <1 | 4 | <1 | 5 | 1 |
| Clostridium bifermentans [NR_113323] | 98 | ||||||||
| Clostridium dakarense [NR_133028] | 97 | ||||||||
| Firmicutes | |||||||||
| Peptostreptococcaceae | |||||||||
| Peptostreptococcaceae-affiliated phylotype [LT838799] | Romboutsia sedimentorum [NR_134800] | 99 | <1 | 2 | <1 | 4 | <1 | 5 | 1 |
| Clostridium bifermentans [NR_113323] | 99 | ||||||||
| Romboutsia lituseburensis [NR_118728] | 98 | ||||||||
| Tenericutes | |||||||||
| Unclassified family | |||||||||
| Unclassified phylotype [LT838787] | For details, see Table 2 | 18 | 3 | 1 | 2 | 1 | 2 | <1 | |
Accession numbers are in brackets. The table contains all phylotypes that had a relative abundance of at least 4% in at least one treatment at a given time interval. The glucose-dependent fermentation profiles are displayed in Fig. 3. All phylotypes are shown in Table S5.
S, similarity of 16S rRNA sequence.
Abbreviations: Pre, preincubation; C, unsupplemented control; G, glucose treatment.
DISCUSSION
The microorganisms found in the alimentary canal of the earthworm are conceived to be derived primarily from ingested material (16, 19, 33, 34). The anoxic organic-carbon-rich environment of the earthworm gut augments the activities of ingested microbes that are capable of anaerobiosis, and this activation leads to an apparent increase in both the detectability and activities of certain ingested microbes (e.g., denitrifiers) (16–18, 20, 33, 34). As such, the anaerobic microbial activity of preingested soil is marginal compared to that of ingested soil, i.e., gut contents (16–18, 20, 33). Dietary intake of litter and soil by earthworms introduces diverse sources of organic carbon into the alimentary canal. However, saccharide-rich worm-derived mucus can constitute up to 80% of gut contents on a dry weight basis (27, 28, 30, 35) and is considered to be an important source of saccharides during gut passage of ingested microbes (14, 16, 17). Given the potential importance of fermentation to the metabolic activities in the alimentary canal, the present investigation assessed how microbes in gut contents might respond to different gut-associated saccharides.
Aeromonadaceae and Enterobacteriaceae are important fermentative families.
Aerated soils contain highly complex microbial communities and include Aeromonadaceae- and Enterobacteriaceae-affiliated bacteria (36–39). Ingestion of soil introduces this complex microbiome into the alimentary canal of the earthworm, and phylotypes affiliated with Aeromonadaceae and Enterobacteriaceae were detected in gut contents and, based on the relative abundance of detected 16S rRNA sequences, became highly active under anoxic conditions with supplemental saccharides (Fig. 2). Likewise, fermentation profiles were indicative of mixed-acid fermentations catalyzed by these facultatively aerobic taxa (Fig. 1; Table 1).
A time-dependent shift in the 16S rRNA profile of the gut microbiome occurred in the absence of supplemental saccharides (Fig. 4; Table 3), indicating that naturally occurring substrates in gut contents and anoxia fostered an alteration of the microbial community. The high diversity of 16S rRNA-based phylotypes based on the phylum and family levels initially detected decreased during the anoxic incubation. Although secondary in relative abundance to Aeromonadaceae, 16S rRNA profiles indicated that Peptostreptococcaceae, Enterobacteriaceae, and Fusobacteriaceae became highly active families postincubation in unsupplemented controls. The occurrence of those families in gut contents of different earthworm species has been previously reported (15, 29, 40, 41).
The highest relative abundance of the Fusobacteriaceae-affiliated phylotype was detected in the control lacking saccharides and xylose-supplemented treatment postincubation (Table 2; Fig. 2). Genera of Fusobacteriaceae are capable of fermenting carbohydrates as well as amino acids and peptides (42). Although this family is well known for its fermentative metabolism and was active in the unsupplemented control, it did not dominate in saccharide-supplemented treatments (Fig. 2; Table 2), a finding also observed with gut contents of the methane-emitting earthworm Eudrilus eugeniae (15).
Peptostreptococcaceae-affiliated 16S rRNA sequences increased in relative abundance during the incubation period in the presence and absence of supplemental saccharides (Fig. 2 and 4). The family Peptostreptococcaceae occurs in various habitats, including soil, and contains diverse fermentative obligate anaerobes or facultative aerobes (43, 44). Based on the relative abundances of 16S rRNA sequences detected in the different treatments, phylotypes affiliated with this family were less competitive than Aeromonadaceae- and Enterobacteriaceae-affiliated phylotypes relative to their comparative responses to supplemental saccharides.
Dominant phylotypes.
A phylotype related to “Candidatus Lumbricincola sp.” (Table 2) that was abundant preincubation has been previously detected in earthworm casts, tissue, and gut contents and may have a symbiotic lifestyle in earthworms (45).
Anoxic incubation of gut contents with or without supplemental saccharides yielded large alterations of the relative abundances of specific 16S rRNA sequences, with sequences of an Aeromonas-related phylotype displaying a high relative abundance in all treatments (Tables 2 and 3). This genus was previously detected in earthworm gut contents and is a member of the family Aeromonadaceae (15, 29, 33, 40, 41). Aeromonas is a metabolically robust genus, and affiliated species utilize formate hydrogenlyase in various fermentations and are capable of anaerobic respiration and dissimilatory metal reduction (46–48), properties that enhance the competitiveness of this genus under anoxic conditions. Aeromonas-affiliated species ferment saccharides to ethanol and organic acids (including succinate, acetate, and formate), but to our knowledge, the production of propionate has not been reported for this taxon (47–50). Based on its relative abundance of 16S rRNA, this phylotype responded very quickly (i.e., in the first 10 h of incubation) to anoxia, and glucose augmented this response (Table 3), illustrating the marked anaerobic ability of this phylotype.
Although a single Aeromonadaceae-affiliated phylotype appeared to be dominant in all treatments, three Enterobacteriaceae-related phylotypes, E, C, and R, displayed strong responses to certain saccharides (Table 2). The Enterobacteriaceae use a fermentative metabolism for the dissimilation of saccharides, yielding a variety of products, including organic acids and H2 (37, 51–54). Enterobacteriaceae-related species are widely distributed in various environments, including soils (36, 37, 51). Some related genera, including Enterobacter, have been investigated for their potential to produce H2 (53–55), suggesting that they might be associated with the microbial production of H2 in the earthworm gut that is associated with novel hydrogenases (17, 41).
Diverse genera belonging to the family Enterobacteriaceae ferment substrates by either mixed-acid or butanediol fermentations (52, 56). The Enterobacteriaceae-affiliated phylotype E was associated mainly with Enterobacter- and Klebsiella-related species and displayed a high relative abundance in the arabinose-supplemented treatment (Table 2). Certain species of these genera utilize butanediol fermentation and produce large amounts of H2 (52, 56, 57). Some Enterobacter-related species are notable for their capacity to ferment diverse saccharides, yielding large amounts of H2, ethanol, 2,3-butanediol, and lactate (53, 57, 58).
The Enterobacteriaceae-affiliated phylotype C responded strongly to the hexose galactose (Table 2; Fig. 2) and is related to Citrobacter. Citrobacter-related species are fermentative facultative aerobes that can produce a variety of fermentation products, including H2, acetate, formate, ethanol, and lactate (56, 59, 60), products observed in the product profiles of this study (Tables 1 and 2). Citrobacter-related species are noted for their formate hydrogenlyase activity (61–63), an activity that might have contributed to the formation of CO2 and H2 in treatments in which Citrobacter-affiliated phylotype C increased in relative abundance and formate was detected (Fig. 1; Table 2).
The Enterobacteriaceae-related phylotype R is related to the genus Rosenbergiella (Table 2). Rosenbergiella-related species are facultative aerobes and capable of fermenting hexoses and pentoses (64). Rosenbergiella australoborealis and Rosenbergiella epipactidis are closely related to phylotype R, which displayed high relative abundances in various hexose (glucose and maltose) and pentose (fucose and xylose) treatments (Tables 2 and 3). Strains of the genus Rosenbergiella display a broad temperature range of 4 to 37°C (64, 65), making the genus well suited for soils inhabited by earthworms. To our knowledge, propionate production has not been reported for Rosenbergiella-related species, suggesting that the Enterobacteriaceae-affiliated phylotype R was not the source of propionate in certain fermentations (e.g., fucose, rhamnose).
A Flavobacterium-affiliated phylotype that was abundant preincubation and also in the unsupplemented control postincubation became less abundant in the glucose treatment (Table 3). In addition, the relative abundance of a Shewanella-affiliated phylotype increased in the unsupplemented control but did not increase in the glucose treatment (Table 3). Most Flavobacterium-related species are obligate aerobes (66), and most Shewanella-affiliated species are respiratory and not capable of fermentation (67, 68). The lack of a strong response of the Shewanella- and Flavobacterium-affiliated phylotypes in saccharide-supplemented treatments indicated that these phylotypes were less competitive than the aforementioned fermentative phylotypes under these conditions.
Conclusions and outlook.
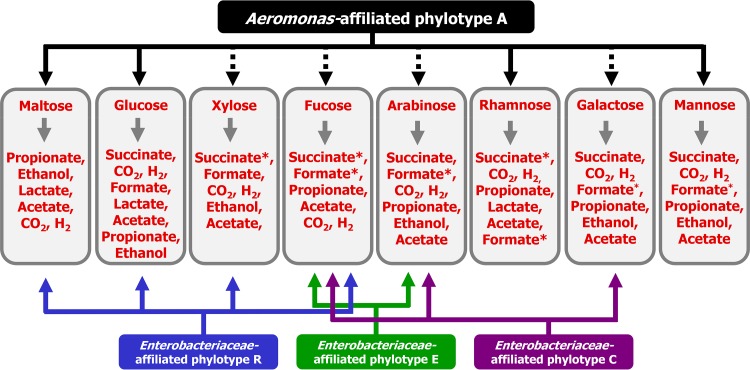
The findings of this study indicate that microbes in the earthworm gut have the capacity to respond rapidly to diverse gut-associated saccharides under anoxic conditions and that the response to a specific saccharide is not uniform across all microbial taxa in gut content (Fig. 5). Aeromonas-affiliated phylotypes can occur in gut content of L. terrestris and E. eugeniae (15, 29, 40), and it is noteworthy that an Aeromonas-affiliated phylotype was stimulated by several saccharides and also responded strongly to anoxia in the absence of supplemental saccharides. Most fucose-derived electrons were recovered in propionate, and the Aeromonas-affiliated phylotype as well as the Enterobacter-affiliated phylotypes C and R displayed high relative abundances in this treatment. Likewise, the Aeromonas-affiliated phylotype displayed a high relative abundance in the rhamnose treatment, in which most electrons were also recovered in propionate. Such taxa might therefore be important in the production of propionate in the alimentary canal. In this regard, strains related to the Aeromonas-affiliated phylotype and Enterobacteriaceae-affiliated phylotype C produce propionate (60, 69). Although the quantitative differences observed for the contrasting phylotypes cannot be directly extended to in situ conditions, the findings qualitatively illustrate the potential competitiveness of subsets of the fermentative taxa that could respond to saccharide availability and thus contribute to in situ gut-associated fermentations.
FIG 5.
Hypothetical model illustrating the main phylotypes that responded to saccharide consumption in anoxic treatments of gut contents of L. terrestris. A broken line indicates that the response was marginal, and an asterisk (*) indicates that only traces were detected.
The dominate saccharide-stimulated phylotypes are related to facultative aerobes that facilitate H2-forming mixed-acid fermentations under anoxic conditions, a finding that reinforces earlier work indicating that the anaerobic activity of facultative aerobes in the gut might be linked to the in vivo emission of H2 by the earthworm (17), an activity potentially linked to secondary H2-driven processes in soil (70, 71). Although the current study did not resolve the composition of the microbial community in different regions of the alimentary canal, the in situ concentrations of fermentation products along the alimentary canal are highly variable and peak in the midgut (e.g., over 30 mM fatty acids can occur in the aqueous phase of the midgut of L. terrestris [17]). Likewise, saccharide availability decreases sharply along the alimentary canal, with the largest amounts occurring in the crop/gizzard (17). Thus, it seems likely that the saccharide-dependent stimulation of fermentative taxa occurs both temporally and spatially during gut passage. In this regard, dietary uptake and thus earthworm feeding guilds likely affect saccharide availability in the gut.
Ingested material is presumed to be the primary source of microbes in the earthworm alimentary canal (14, 16, 17, 26, 27), and the comparative inactiveness of soil under anoxic conditions (Fig. 1) highlights the unique potential of the earthworm gut to augment anaerobic processes in aerated soils, a microbe-invertebrate interaction that contributes to the overall turnover dynamics of organic carbon in the terrestrial biosphere. Future studies focused on specific saccharide-derived fermentation profiles and the associated microbiota along the alimentary canal of different earthworm feeding guilds would provide further insight on microbe-mediated transformation of organic carbon in the earthworm gut, activities that are likely interfaced to the utilization of gut-derived nutrients by the earthworm.
MATERIALS AND METHODS
Experimental setup and earthworms.
Two experiments were conducted. The first experiment analyzed the stimulation of gut content bacteria by diverse saccharides (the earthworms for this experiment were obtained in November 2014). The second experiment analyzed the time-dependent changes of the gut content bacterial community during the fermentation of glucose (the earthworms for this experiment were obtained in March 2015). Adult specimens of L. terrestris were obtained from distributor ANZO in Bayreuth (Germany). Earthworms were incubated in soil for at least 7 days in the dark and at 15°C and provided with composted leaves for dietary needs. Soil was collected from the meadow Trafo-Wiese near Bayreuth (for details, see reference 16). Earthworms were numbed with carbon dioxide and washed with filtered water to remove soil particles, and gut content was obtained under anoxic conditions in an O2-free chamber (Mercaplex, Grenchen, Switzerland) (29).
Anoxic microcosms.
Soil was derived from the soil in which earthworms were incubated. Gut content or soil was diluted 1:10 (wt/vol) with sodium phosphate buffer (pH 7 [29]) and placed into anoxic sterile 125 ml-serum bottles under anoxic conditions in an O2-free chamber. Gas-tight serum bottles were sealed with sterile butyl rubber stoppers and flushed with sterile 100% N2 (29). Anoxic microcosms and anoxic solutions were prepared by a modified Hungate technique (72, 73). Saccharides were supplemented to a final concentration of 5 mM (50 μmol g−1 fresh weight). Unsupplemented controls and saccharide-supplemented treatments were set up in triplicates. Anoxic microcosms were incubated in the dark at 15°C, and sterile and gas-flushed syringes were used to sample microcosms (29). Liquid samples for chemical analysis were stored at −20°C. Liquid samples for molecular analysis were stored at −80°C.
Chemical analyses.
CO2, N2O, CH4, and H2 were measured by gas chromatography (using SRI 8610C from SRI Instruments, Torrance, CA, and Hewlett Packard 5890 Series II from Hewlett Packard, Palo Alto, CA) as previously described, with the following modifications for quantification of N2O with the SRI 8610C instrument (74, 75): the oven temperature was 80°C, the flow rate was 25 ml carrier gas min−1, the injector temperature was 60°C, the temperature of the column oven was 80°C, and the temperature of the electron capture detector was 300°C. Gas concentrations were calculated from the ideal gas law as previously described (76, 77).
Sugars, ethanol, and organic acids were quantified by high-performance liquid chromatography (HPLC; HP 1090 Series II; Hewlett Packard, Palo Alto, CA, USA) with a refractive index detector and a wavelength detector (Agilent Series 1200; Agilent Technologies, Santa Clara, CA) (17). The column used was Rezex ROA-Organic Acid H+ (300 by 7.8 mm; Phenomenex, Torrance, CA, USA). The mobile phase was 4 M H3PO4 (17) and had a flow rate of 0.4 ml min−1.
The pH was measured with a pH electrode (InlabSemi-Micro pH; Mettler Toledo, Gießen, Germany) and a digital pH meter (WTW pH 323; Xylem Analytics Germany Sales GmbH and Co. KG, Weilheim, Germany). Nitrate was analyzed by exclusion chromatography (Central Analytic, University of Bayreuth, Bayreuth, Germany).
Extraction of nucleic acids.
The extraction of nucleic acids was by published protocol (78) and included (i) mechanical lysis of cells with glass beads, (ii) extraction using organic solvents, and (iii) precipitation of nucleic acids with polyethylene glycol 6000.
Preparation and quantification of RNA.
DNA was removed from the nucleic acid extract with the DNase I, RNase-free kit (Thermo Fisher Scientific, Waltham, MA, USA). The DNase I, RNase-free kit was utilized according to the manufacturer's protocol with the following modifications: 26 μl of the nucleic acid extract was mixed with 3 μl of reaction buffer (100 mM Tris-HCl [pH 7.5], 25 mM MgCl2, 1 mM CaCl2) and 1 μl of RNase-free DNase (1 U μl−1). The DNA-free RNA extract yielded no PCR signal with primers 907RM (5′-CCGTCAATTCMTTTGAGTTT-3′) and 27f (5′-AGAGTTTGATCMTGGCTC-3′) and a Taq DNA polymerase (79). The amplification of the bacterial 16S rRNA gene was prepared as follows: (i) initial denaturation at 95°C for 5 min, (ii) 5 precycles of denaturation at 95°C for 60 s, annealing at 40°C for 60 s, and elongation 72°C for 70 s, (iii) 30 subsequent cycles of denaturation at 95°C for 30 s, annealing at 43°C for 30 s, and elongation at 72°C for 70 s, and (iv) terminal elongation at 72°C for 5 min (modified from reference 75). PCR was facilitated with 5 Prime Mastermix (5 Prime, Hamburg, Germany); the 25-μl PCR mixture contained 0.6 U Taq DNA polymerase, 0.2 mM each deoxynucleoside triphosphate (dNTP), 4 mM MgCl2, and 1.2 μM each primer (modified from reference 73). The Quant-iT RiboGreen RNA assay kit (Molecular Probes; Thermo Fisher Scientific, Waltham, MA, USA) was used for quantification of single-stranded RNA.
Reverse transcription.
As noted above, each treatment was replicated three times, equal amounts of RNA from each replicate were pooled after digestion, and the pooled sample was used for further analysis. The reverse transcription of pooled RNA to cDNA was performed with SuperScript Reverse Transcriptase III (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's protocol with the following modifications: 5 μl pooled RNA was used for reverse transcription, 1 μl of RNase-free water was used instead of 1 μl of RNaseOUT, and reverse transcription ran for 120 min. cDNA was precipitated for approximately 12 h with 100% isopropyl alcohol and 5 mM RNase-free NaCl at −20°C. Pellets were washed with 70% DNase-free ethanol and stored at −80°C until further analysis.
Illumina sequencing.
Sequencing was performed by Microsynth (Balgach, Switzerland). The primer pair 515f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806r (5′-GGACTACHVGGGTWTCTAAT-3′) was used to target archaeal and bacterial 16S rRNA at the V4 region and yielded a fragment of approximately 250 bp (80). The QIIME software package was used (81). Sequences were demultiplexed, stitched, and quality filtered. Chimeric sequences (23,390 sequences) were identified with ChimeraSlayer software (82) and excluded from further analysis. Nonchimeric sequences (2,172,982 sequences) were clustered in phylotypes with a 99% similarity threshold without singletons (2,114,028 sequences) (83, 84). For further analysis, only phylotypes that contained 1,000 sequences or more were evaluated, resulting in a data set that contained 1,753,180 sequences. This analysis yielded 136 phylotypes (see Tables S4 and S5 in the supplemental material). Phylotypes were classified with SILVA (85) and aligned by SINA (86). Phylotypes were assigned to genera or families and not to single species due to the short Illumina-based sequence reads (87–89). The classification of the most abundant taxa was done according to LPSN (List of prokaryotic names with standing in nomenclature [www.bacterio.net]). The diversity of the microbial community was determined by rarefaction analysis (90, 91). The protocol outlined above with pooled replicates of gut content microcosms has been shown in additional studies to be highly reproducible, yielding highly similar taxa in the analysis of nonpooled replicates.
Accession number(s).
Sequencing data can be found at European Nucleotide Archive (ENA) under study number ERP022175. 16S rRNA sequences representative of phylotypes are available at ENA under accession numbers LT838787 to LT838799.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter S. Depkat-Jakob, Oliver Schmidt, and Ralf Mertel for technical assistance.
Support for this study was provided by the Deutsche Forschungsgemeinschaft (DR310/4-2) and the University of Bayreuth.
Funding Statement
The University of Bayreuth also provided resources.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01851-17.
REFERENCES
- 1.Darwin CR. 1881. The formation of vegetable mould through the action of worms, with observations on their habits. Murray, London, United Kingdom. [Google Scholar]
- 2.Lee KE. 1985. Earthworms: their ecology and relationships with soils and land use. Academic Press, Sydney, Australia. [Google Scholar]
- 3.Lavelle P. 1988. Earthworm activities and the soil system. Biol Fertil Soils 6:237–251. doi: 10.1007/BF00260820. [DOI] [Google Scholar]
- 4.Edwards CA, Bohlen PJ. 1996. Biology and ecology of earthworms, 3rd ed Chapman & Hall, London, United Kingdom. [Google Scholar]
- 5.Makeschin F. 1997. Earthworms (Lumbricidae: Oligochaeta): important promoters of soil development and soil fertility, p 173–223. In Benckiser G. (ed), Fauna in soil ecosystems. Marcel Dekker, Inc., New York, NY. [Google Scholar]
- 6.Lee KE. 1992. Some trends and opportunities in earthworm research or: Darwin's children—the future of our discipline. Soil Biol Biochem 24:1765–1771. doi: 10.1016/0038-0717(92)90185-Z. [DOI] [Google Scholar]
- 7.Scheu S. 2003. Effects of earthworms on plant growth: patterns and perspectives. Pedobiologia 47:846–856. [Google Scholar]
- 8.Dávalos A, Nuzzo V, Stark J, Blossey B. 2013. Unexpected earthworm effects on forest understory plants. BMC Ecol 13:48. doi: 10.1186/1472-6785-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curry JP, Schmidt O. 2007. The feeding ecology of earthworms—a review. Pedobiologia 50:463–477. doi: 10.1016/j.pedobi.2006.09.001. [DOI] [Google Scholar]
- 10.Zaller JG, Arnone JA. 1999. Earthworm responses to plant species' loss and elevated CO2 in calcareous grassland. Plant Soil 208:1–8. doi: 10.1023/A:1004424720523. [DOI] [Google Scholar]
- 11.Zaller JG, Arnone JA. 1999. Earthworm and soil moisture effects on the productivity and structure of grassland communities. Soil Biol Biochem 31:517–523. doi: 10.1016/S0038-0717(98)00126-6. [DOI] [Google Scholar]
- 12.Zaller JG, Heigl F, Grabmaier A, Lichtenegger C, Piller K, Allabashi R, Frank T, Drapela T. 2011. Earthworm-mycorrhiza interactions can affect the diversity, structure and functioning of establishing model grassland communities. PLoS One 6:e29293. doi: 10.1371/journal.pone.0029293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards CA. 2004. The importance of earthworms as key representatives of the soil fauna, p 3–11. In Edwards CA. (ed), Earthworm ecology, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 14.Drake HL, Horn MA. 2007. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu Rev Microbiol 61:169–189. doi: 10.1146/annurev.micro.61.080706.093139. [DOI] [PubMed] [Google Scholar]
- 15.Schulz K, Hunger S, Brown GG, Tsai SM, Cerri CC, Conrad R, Drake HL. 2015. Methanogenic food web in the gut contents of methane-emitting earthworm Eudrilus eugeniae from Brazil. ISME J 9:1778–1792. doi: 10.1038/ismej.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn MA, Schramm A, Drake HL. 2003. The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl Environ Microbiol 69:1662–1669. doi: 10.1128/AEM.69.3.1662-1669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wüst PK, Horn MA, Drake HL. 2009. In situ hydrogen and nitrous oxide as indicators of concomitant fermentation and denitrification in the alimentary canal of the earthworm Lumbricus terrestris. Appl Environ Microbiol 75:1852–1859. doi: 10.1128/AEM.02745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karsten GR, Drake HL. 1995. Comparative assessment of the aerobic and anaerobic microfloras of earthworm guts and forest soils. Appl Environ Microbiol 61:1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drake HL, Horn MA. 2006. Earthworms as a transient heaven for terrestrial denitrifying microbes: a review. Eng Life Sci 6:261–265. doi: 10.1002/elsc.200620126. [DOI] [Google Scholar]
- 20.Karsten GR, Drake HL. 1997. Denitrifying bacteria in the earthworm gastrointestinal tract and in vivo emission of nitrous oxide (N2O) by earthworms. Appl Environ Microbiol 63:1878–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthies C, Grießhammer A, Schmittroth M, Drake HL. 1999. Evidence for involvement of gut-associated denitrifying bacteria in emission of nitrous oxide (N2O) by earthworms obtained from garden and forest soils. Appl Environ Microbiol 65:3599–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn MA, Mertel R, Gehre M, Kastner M, Drake HL. 2006. In vivo emission of dinitrogen by earthworms via denitrifying bacteria in the gut. Appl Environ Microbiol 72:1013–1018. doi: 10.1128/AEM.72.2.1013-1018.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wüst PK, Horn MA, Henderson G, Janssen PH, Rehm BHA, Drake HL. 2009. Gut-associated denitrification and in vivo emission of nitrous oxide by the earthworm families Megascolecidae and Lumbricidae in New Zealand. Appl Environ Microbiol 75:3430–3436. doi: 10.1128/AEM.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depkat-Jakob PS, Hunger S, Schulz K, Brown GG, Tsai SM, Drake HL. 2012. Emission of methane by Eudrilus eugeniae and other earthworms from Brazil. Appl Environ Microbiol 78:3014–3019. doi: 10.1128/AEM.07949-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depkat-Jakob PS, Brown GG, Tsai SM, Horn MA, Drake HL. 2013. Emission of nitrous oxide and dinitrogen by diverse earthworm families from Brazil and resolution of associated denitrifying and nitrate-dissimilating taxa. FEMS Microbiol Ecol 83:375–391. doi: 10.1111/j.1574-6941.2012.01476.x. [DOI] [PubMed] [Google Scholar]
- 26.Brown GG, Doube BM. 2004. Functional interactions between earthworms, microorganisms, organic matter, and plants, p 213–239. In Edwards CA. (ed), Earthworm ecology, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 27.Brown GG, Barois I, Lavelle P. 2000. Regulation of soil organic matter dynamics and microbial activity in the drilosphere and the role of interactions with other edaphic functional domains. Eur J Soil Biol 36:177–198. doi: 10.1016/S1164-5563(00)01062-1. [DOI] [Google Scholar]
- 28.Martin A, Cortez J, Barois I, Lavelle P. 1987. Les mucus intestinaux de Ver de Terre, moteur de leurs interactions avec la microflore. Rev Ecol Biol Sol 24:549–558. [Google Scholar]
- 29.Wüst PK, Horn MA, Drake HL. 2011. Clostridiaceae and Enterobacteriaceae as active fermenters in earthworm gut content. ISME J 5:92–106. doi: 10.1038/ismej.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trigo D, Barois I, Garvín MH, Huerta E, Irisson S, Lavelle P. 1999. Mutualism between earthworms and soil microflora. Pedobiologia 43:866–873. [Google Scholar]
- 31.Saddler JN, Yu EKC, Mes-Hartree M, Levitin N, Brownell HH. 1983. Utilization of enzymatically hydrolyzed wood hemicelluloses by microorganisms for production of liquid fuels. Appl Environ Microbiol 45:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha BC. 2003. Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 33.Ihssen J, Horn MA, Matthies C, Gößner A, Schramm A, Drake HL. 2003. N2O-producing microorganisms in the gut of the earthworm Aporrectodea caliginosa are indicative of ingested soil bacteria. Appl Environ Microbiol 69:1655–1661. doi: 10.1128/AEM.69.3.1655-1661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Depkat-Jakob PS, Hilgarth M, Horn MA, Drake HL. 2010. Effect of earthworm feeding guilds on ingested dissimilatory nitrate reducers and denitrifiers in the alimentary canal of the earthworm. Appl Environ Microbiol 76:6205–6214. doi: 10.1128/AEM.01373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahemtulla F, Løvtrup S. 1975. The comparative biochemistry of invertebrate mucopolysaccharides—III. Oligochaeta and Hirudinea. Comp Biochem Physiol Part B 50:627–629. doi: 10.1016/0305-0491(75)90101-7. [DOI] [PubMed] [Google Scholar]
- 36.Degelmann DM, Kolb S, Dumont M, Murrell JC, Drake HL. 2009. Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol Ecol 68:312–319. doi: 10.1111/j.1574-6941.2009.00681.x. [DOI] [PubMed] [Google Scholar]
- 37.Octavia S, Lan R. 2014. The family Enterobacteriaceae, p 225–286. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed, vol 9 Springer, Berlin, Germany. [Google Scholar]
- 38.Hayase N, Kouno K, Ushio K. 2000. Isolation and characterization of Aeromonas sp. B-5 capable of decolorizing various dyes. J Biosci Bioeng 90:570–573. doi: 10.1016/S1389-1723(01)80044-X. [DOI] [PubMed] [Google Scholar]
- 39.Bashir AH. 2012. Diversity and morphology of bacterial community characterized in topsoil samples from the Gaza Strip, Palestine. Res J Microbiol 7:309–318. doi: 10.3923/jm.2012.309.318. [DOI] [Google Scholar]
- 40.Byzov BA, Nechitaylo TY, Bumazhkin BK, Kurakov AV, Golyshin PN, Zvyagintsev DG. 2009. Culturable microorganisms from the earthworm digestive tract. Microbiology 78:360–368. doi: 10.1134/S0026261709030151. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt O, Wüst PK, Hellmuth S, Borst K, Horn MA, Drake HL. 2011. Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl Environ Microbiol 77:5842–5850. doi: 10.1128/AEM.05432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen I. 2014. The family Fusobacteriaceae, p 109–132. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed, vol 9 Springer, Berlin, Germany. [Google Scholar]
- 43.Ezaki T. 2009. Family VII. Peptostreptococcaceae, p 1008–1008. In Whitman WB. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 3 Springer, New York, NY. [Google Scholar]
- 44.Slobodkin A. 2014. The family Peptostreptococcaceae, p 291–302. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed, vol 9 Springer, Berlin, Germany. [Google Scholar]
- 45.Nechitaylo TY, Timmis KN, Golyshin PN. 2009. “Candidatus Lumbricincola”, a novel lineage of uncultured Mollicutes from earthworms of family Lumbricidae. Environ Microbiol 11:1016–1026. doi: 10.1111/j.1462-2920.2008.01837.x. [DOI] [PubMed] [Google Scholar]
- 46.Knight V, Blakemore R. 1998. Reduction of diverse electron acceptors by Aeromonas hydrophila. Arch Microbiol 169:239–248. doi: 10.1007/s002030050567. [DOI] [PubMed] [Google Scholar]
- 47.Huys G. 2014. The family Aeromonadaceae, p 27–57. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed, vol 9 Springer, Berlin, Germany. [Google Scholar]
- 48.Martin-Carnahan A, Joseph SW. 2005. Genus I. Aeromonas, p 557–578. In Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 49.Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Kim J, Shin SG, Hwang S. 2008. Biokinetic parameters and behavior of Aeromonas hydrophila during anaerobic growth. Biotechnol Lett 30:1011–1016. doi: 10.1007/s10529-008-9660-2. [DOI] [PubMed] [Google Scholar]
- 51.Brenner DJ, Farmer JJ III. 2005. Family I. Enterobacteriaceae, p 587–607. In Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 52.Ciani M, Comitini F, Mannazzu I. 2008. Fermentation, p 1548–1557. In Jorgensen SE, Fath B (ed), Encyclopedia of ecology. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 53.Rachman MA, Furutani Y, Nakashimada Y, Kakizono T, Nishio N. 1997. Enhanced hydrogen production in altered mixed acid fermentation of glucose by Enterobacter aerogenes. J Ferment Bioeng 83:358–363. doi: 10.1016/S0922-338X(97)80142-0. [DOI] [Google Scholar]
- 54.Jayasinghearachchi HS, Sarma PM, Singh S, Aginihotri A, Mandal AK, Lal B. 2009. Fermentative hydrogen production by two novel strains of Enterobacter aerogenes HGN-2 and HT 34 isolated from sea buried crude oil pipelines. Int J Hydrog Energy 34:7197–7207. doi: 10.1016/j.ijhydene.2009.06.079. [DOI] [Google Scholar]
- 55.Long C, Cui J, Liu Z, Liu Y, Long M, Hu Z. 2010. Statistical optimization of fermentative hydrogen production from xylose by newly isolated Enterobacter sp. CN1. Int J Hydrog Energy 35:6657–6664. doi: 10.1016/j.ijhydene.2010.04.094. [DOI] [Google Scholar]
- 56.Müller V. 2008. Bacterial fermentation. In Encyclopedia of life sciences. John Wiley & Sons, Ltd, Chichester, United Kingdom. doi: 10.1002/9780470015902.a0001415.pub2. [DOI] [Google Scholar]
- 57.Grimont PAD, Grimont F. 2005. Genus XII. Enterobacter, p 661–669. In Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 58.Tajima Y, Kaida K, Hayakawa A, Fukui K, Nishio Y, Hashiguchi K, Fudou R, Matsui K, Usuda Y, Sode K. 2014. Study of the role of anaerobic metabolism in succinate production by Enterobacter aerogenes. Appl Microbiol Biotechnol 98:7803–7813. doi: 10.1007/s00253-014-5884-3. [DOI] [PubMed] [Google Scholar]
- 59.Frederiksen W. 2005. Genus X. Citrobacter, p 651–656. In Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 60.Mangayil R, Santala V, Karp M. 2011. Fermentative hydrogen production from different sugars by Citrobacter sp. CMC-1 in batch culture. Int J Hydrog Energy 36:15187–15194. doi: 10.1016/j.ijhydene.2011.08.076. [DOI] [Google Scholar]
- 61.Sawers G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek 66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 62.Kim S, Seol E, Mohan Raj S, Park S, Oh Y, Ryu D. 2008. Various hydrogenases and formate-dependent hydrogen production in Citrobacter amalonaticus Y19. Int J Hydrog Energy 33:1509–1515. doi: 10.1016/j.ijhydene.2007.09.029. [DOI] [Google Scholar]
- 63.McDowall JS, Murphy BJ, Haumann M, Palmer T, Armstrong FA, Sargent F. 2014. Bacterial formate hydrogenlyase complex. Proc Natl Acad Sci U S A 111:E3948–E3956. doi: 10.1073/pnas.1407927111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Halpern M, Fridman S, Atamna-Ismaeel N, Izhaki I. 2013. Rosenbergiella nectarea gen. nov., sp. nov., in the family Enterobacteriaceae, isolated from floral nectar. Int J Syst Evol Microbiol 63:4259–4265. doi: 10.1099/ijs.0.052217-0. [DOI] [PubMed] [Google Scholar]
- 65.Lenaerts M, Álvarez-Pérez S, de Vega C, Van Assche A, Johnson SD, Willems KA, Herrera CM, Jacquemyn H, Lievens B. 2014. Rosenbergiella australoborealis sp. nov., Rosenbergiella collisarenosi sp. nov. and Rosenbergiella epipactidis sp. nov., three novel bacterial species isolated from floral nectar. Syst Appl Microbiol 37:402–411. doi: 10.1016/j.syapm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol 61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 67.Satomi M. 2014. The family Shewanellaceae, p 597–625. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes, 4th ed, vol 9 Springer, Berlin, Germany. [Google Scholar]
- 68.Bernardet J-F, Bowman JP. 2011. Genus I. Flavobacterium, p 112–154. In Whitman WB. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 4 Springer, New York, NY. [Google Scholar]
- 69.Namdari H, Cabelli VJ. 1989. The suicide phenomenon in motile aeromonads. Appl Environ Microbiol 55:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Osborne CA, Peoples MB, Janssen PH. 2010. Detection of a reproducible, single-member shift in soil bacterial communities exposed to low levels of hydrogen. Appl Environ Microbiol 76:1471–1479. doi: 10.1128/AEM.02072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khdhiri M, Piché-Choquette S, Tremblay J, Tringe SG, Constant P. 2017. The tale of a neglected energy source: elevated hydrogen exposure affects both microbial diversity and function in soil. Appl Environ Microbiol 83:e00275-17. doi: 10.1128/AEM.00275-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hungate RE. 1969. A roll tube method for cultivation of strict anaerobes, p 117–132. In Norris JR, Ribbons DW (ed), Methods in microbiology. Academic Press, New York, NY. [Google Scholar]
- 73.Daniel SL, Drake HL. 1993. Oxalate- and glyoxylate-dependent growth and acetogenesis by Clostridium thermoaceticum. Appl Environ Microbiol 59:3062–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Küsel K, Drake HL. 1995. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl Environ Microbiol 61:3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hunger S, Schmidt O, Hilgarth M, Horn MA, Kolb S, Conrad R, Drake HL. 2011. Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl Environ Microbiol 77:3773–3785. doi: 10.1128/AEM.00282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blachnik R. 1998. Elemente, anorganische Verbindungen und Materialien, Minerale, 4th ed Springer, Berlin, Germany. [Google Scholar]
- 77.Krichevsky IR, Kasarnosky JS. 1935. Thermodynamical calculations of solubilities of nitrogen and hydrogen in water at high pressures. J Am Chem Soc 57:2168–2171. doi: 10.1021/ja01314a036. [DOI] [Google Scholar]
- 78.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lane DJ. 1996. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 80.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, The Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 84.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155. [Google Scholar]
- 85.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caro-Quintero A, Ochman H. 2015. Assessing the unseen bacterial diversity in microbial communities. Genome Biol Evol 7:3416–3425. doi: 10.1093/gbe/evv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hurlbert SH. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 91.Heck KL, van Belle G, Simberloff D. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459–1461. doi: 10.2307/1934716. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.