ABSTRACT
Integrative conjugative elements (ICEs) are chromosomal elements that are widely distributed in bacterial genomes, hence contributing to genome plasticity, adaptation, and evolution of bacteria. Conjugation requires a contact between both the donor and the recipient cells and thus likely depends on the composition of the cell surface envelope. In this work, we investigated the impact of different cell surface molecules, including cell surface proteins, wall teichoic acids, lipoteichoic acids, and exopolysaccharides, on the transfer and acquisition of ICESt3 from Streptococcus thermophilus. The transfer of ICESt3 from wild-type (WT) donor cells to mutated recipient cells increased 5- to 400-fold when recipient cells were affected in lipoproteins, teichoic acids, or exopolysaccharides compared to when the recipient cells were WT. These mutants displayed an increased biofilm-forming ability compared to the WT, suggesting better cell interactions that could contribute to the increase of ICESt3 acquisition. Microscopic observations of S. thermophilus cell surface mutants showed different phenotypes (aggregation in particular) that can also have an impact on conjugation. In contrast, the same mutations did not have the same impact when the donor cells, instead of recipient cells, were mutated. In that case, the transfer frequency of ICESt3 decreased compared to that with the WT. The same observation was made when both donor and recipient cells were mutated. The dominant effect of mutations in donor cells suggests that modifications of the cell envelope could impair the establishment or activity of the conjugation machinery required for DNA transport.
IMPORTANCE ICEs contribute to horizontal gene transfer of adaptive traits (for example, virulence, antibiotic resistance, or biofilm formation) and play a considerable role in bacterial genome evolution, thus underlining the need of a better understanding of their conjugative mechanism of transfer. While most studies focus on the different functions encoded by ICEs, little is known about the effect of host factors on their conjugative transfer. Using ICESt3 of S. thermophilus as a model, we demonstrated the impact of lipoproteins, teichoic acids, and exopolysaccharides on ICE transfer and acquisition. This opens up new avenues to control gene transfer mediated by ICEs.
KEYWORDS: integrative conjugative element, Streptococcus thermophilus, biofilm, cell surface molecules, conjugation, exopolysaccharides, gene transfer, lipoteichoic acids, mutant, wall teichoic acids
INTRODUCTION
Horizontal gene transfer (HGT) significantly impacts bacterial evolution by driving genome plasticity. Comparison of the increasingly available prokaryotic genome sequences enables estimation of the intraspecies diversity and reveals a high dynamic of gene exchanges (1). Conjugation is a mechanism through which the DNA is transferred following the establishment of a cell contact between a donor and a recipient bacterium (2, 3). This mechanism has been found to be the primary contributor to HGT by allowing an effective DNA transfer to a large spectrum of hosts. Mobile genetic elements (MGEs) are essential actors in HGT. Among them, integrative conjugative elements (ICEs), whose sizes range from 18 to 600 kb, are widely distributed in bacterial genomes regardless of the species or any other classification (4–6). ICEs are chromosomal mobile elements that are able to excise from the donor chromosome, transfer by conjugation, and integrate into the recipient chromosome. Like many other MGEs, ICEs owe their evolutionary success in part to the adaptive genes they carry, which can significantly contribute to the competitiveness of their hosts. Hence, ICEs are notably contributing to the spread of antimicrobial resistance (AMR) and the emergence of multidrug-resistant strains, which constitute a serious threat to global public health, as restated recently by the World Health Organization (4, 7). This highlights the necessity of drawing a more comprehensive picture of the conjugative mechanism employed by ICEs. Since conjugation needs a physical contact between the donor and recipient cells, it likely depends on the cell surface composition and on the donor-recipient interactions. Previous studies on ICEs have focused mainly on the mechanisms of transfer and regulation encoded by these elements, whereas the impact of host factors on ICE transfer, including donor and recipient cell surface components, are still poorly described. This has been studied for ICEBs1, an ICE from the Gram-positive bacterium Bacillus subtilis, for which the authors reported an impact of the phospholipid biosynthesis pathway on ICEBs1 transfer (8, 9). The impact of this pathway has also been tested for Tn916 by the same group in order to extrapolate the ICEBs1 results, demonstrating that some impacts are specific to ICEBs1, whereas others could be generalized to other ICEs (8, 9). A recent study has also showed a role of common polysaccharide antigen, a homopolymer of d-rhamnose attached on lipopolysaccharide, in the initiation of PAPI-1 ICE transfer in Pseudomonas aeruginosa (10). In order to provide more information about this topic, we investigated the impact of various cell surface molecules, including surface-exposed proteins, wall teichoic acids (WTA), lipoteichoic acids (LTA), and exopolysaccharides (EPS), on the conjugative transfer of ICESt3 of Streptococcus thermophilus.
ICESt3 of S. thermophilus is a 28-kb element inserted in the 3′ end of the fda gene, encoding a 1.6-disphosphate aldolase. It transfers at a frequency of 10−6 transconjugants per donor to other S. thermophilus strains but also to Streptococcus pyogenes and Enterococcus faecalis (11). The frequency of transfer of ICESt3 can be increased 25-fold after exposure of donor cells to mitomycin C treatment (11). ICESt3 does not encode any known aggregation factor or cell surface-exposed molecule; thus, its transfer depends on successful donor-recipient contacts and likely relies on host factors, as already suggested by a previous study (12).
The Gram-positive bacterium S. thermophilus is a clonal species that has recently emerged from a commensal ancestor of the Streptococcus salivarius group, which also includes the closely related species Streptococcus vestibularis and Streptococcus salivarius (13). S. thermophilus has evolved mainly by loss of gene functions unnecessary for its adaptation to a narrow and well-defined ecological niche, milk (14–16). This notably includes loss of functions linked with the cell surface composition, making S. thermophilus a simple model of cell surface envelope suitable for the purpose of this study.
LPXTG-containing proteins are cell surface proteins covalently linked to the peptidoglycan through the action of sortase enzymes. These proteins are known to fulfill functions mainly linked to the interactions of pathogenic strains with their host (17). Thus, it is not surprising that only rare S. thermophilus strains harbor LPXTG proteins at their surface (14).
Lipoproteins (Lpp) are surface proteins covalently linked to the plasma membrane through the sequential action of several enzymes, including the lipoprotein signal peptidase II (LspA) (18).
The bacterial cytoplasmic membrane is a bilayer composed of complex lipids which vary not only in the length and modifications of their acylated fatty acids but also in the composition of their head groups (19). Some of them are positively charged (e.g., lysylphosphatidylglycerol, synthesized by the MprF protein) (19).
Wall teichoic acids (WTA) are major components of the Gram-positive bacterial envelope (20). Their exposure at the cell surface depends on the action of TagO or its homolog TarO, described in B. subtilis and Staphylococcus aureus as essential for the initiation of WTA biosynthesis (21, 22). LTA are covalently attached to the plasma membrane through their bond to a glycolipid inserted in the membrane (22). In S. aureus, LtaS polymerizes the carbon backbone of LTA, whereas three homologs of LtaS, LtaSBS, YfnI, and YqgS, are involved in the biosynthesis of LTA in B. subtilis (21, 23, 24). WTA and LTA backbones are maintained with phosphodiester bounds that confer a negative global charge to the whole polymers. For both components, a d-alanylation driven by DltABCD can neutralize the negative charges of the WTA and LTA polymers (21, 25, 26). A dlt cluster has been described in S. thermophilus LMG 18311 (16).
Exopolysaccharides (EPS) are long chains of polysaccharides that are branched with repeated units of sugar (e.g., glucose, galactose, rhamnose, and derivatives) and are transiently linked to the plasma membrane before their secretion in the neighborhood environment (27, 28). EPS of S. thermophilus are well-studied components because of the texture they form, which is useful for the dairy industry. In S. thermophilus LMG 18311, a large cluster of genes encodes all the proteins involved in the formation of the repeated sugar units and the export and polymerization of the EPS chain (29–31). The EpsE phosphogalactosyltransferase is essential for EPS production in S. thermophilus, since strains lacking this enzyme do not show a detectable amount of EPS (32).
In this study, cell surface mutants of S. thermophilus strain LMG 18311 were constructed, characterized (by growth, microscopic observations, and biofilm formation), and used to test their ability to transfer ICESt3 compared to that of the wild-type (WT) strain. Mating experiments were carried out with either mutated recipients or mutated donors and with both mutated donors and recipients. Whereas some mutations led to an increase, up to 400-fold, of the ICESt3 transfer frequency, others led to a decrease of the transfer frequency compared to that for the WT. However, none of the tested molecules appeared to be essential for ICESt3 transfer.
RESULTS
Genes involved in cell surface composition in S. thermophilus LMG 18311 and homologs in other bacterial genomes.
Eight S. thermophilus LMG 18311 mutants were constructed to target cell surface proteins (ΔlspA mutant), teichoic acids (ΔtagO-like, ΔyfnI-like, and ΔdltA mutants), lysyl-phosphatidylglycerol biosynthesis (ΔmprF-like mutant), and polysaccharide production, including exopolysaccharides (ΔepsE and Δeps9 Δeps10 Δeps11 mutants) and rhamnose-glucose polysaccharides (Δstu1482 ΔrgpX2 mutant). Genes encoding some of the targeted functions have already been described (16) or clearly annotated, such as the EPS and dlt operons and the lspA gene. Identification of genes linked to LTA and WTA functions in S. thermophilus required a preliminary in silico analysis to detect possible homologs of known coding genes from other bacterial species. Analysis of the available LMG 18311 genome (14) revealed homologs of B. subtilis proteins involved in LTA, WTA, and lysyl-phosphatidylglycerol biosynthesis: Stu0163, Stu0636, and Stu1256 were found to be homologous to TagO (33) (41% identity with 86% sequence coverage, E = 2e−69), YfnI (24) (42% identity with 96% sequence coverage, E = 7e−158), and MprF (19) (31% identity with 96% sequence coverage, E = 2e−121), respectively. We searched the LMG 18311 proteome for homologs of the two other proteins, LtaSBS and YqgS, involved in LTA synthesis in B. subtilis (24). Stu0636 was the sole protein detected, suggesting that as observed in S. aureus (23) and Listeria monocytogenes (34), only one LTA synthase is involved in LTA biosynthesis in S. thermophilus LMG 18311. Only one LTA synthase was also found in other S. thermophilus strains as revealed by in silico analyses (e.g., in the TH1435, CNRZ1066, TH1477, and JIM8232 genomes).
Homologs of the 8 genes or clusters of genes selected for mutant construction in S. thermophilus LMG18311 were searched in the other available genomes of S. thermophilus and the closely related species S. salivarius, S. pyogenes, and E. faecalis. These latter species were chosen since we previously demonstrated transfer of ICESt3 to a strain of S. pyogenes (ATCC 12202) and a strain of E. faecalis (JH2-2) (11). Four proteins (Stu0163/TagO, Stu0521/LspA, Stu0636/YfnI, and Stu0761/DltA) appeared to be highly conserved in the 4 species (encoded by 94 to 100% of the strains, with amino acid sequence identities higher than 99%, 91%, 59%, and 45% for the other S. thermophilus genomes and the S. salivarius, S. pyogenes, and E. faecalis genomes, respectively). Stu1256/MprF was found in all the genomes of S. thermophilus (99 to 100% identity) and S. salivarius (88 to 92% identity) and in 98% of the strains of E. faecalis (40 to 42% identity). Stu1108/EpsE (with at least 50% of query cover and more than 40% identity with LMG 18311) was found in the majority of the genomes of S. thermophilus (>78% with >94% identity) and in all S. salivarius genomes (with 76 to 97% identity). In contrast, the rhamnose synthesis cluster was found in only one-third of the S. thermophilus and S. salivarius genomes, and sequence conservation was lower (from 62% to 98% amino acid identity). The stu1097 stu1098 stu1099 Eps synthesis cluster was found to be specific to LMG 18311, since no homolog (with >40% amino acid identity) was found in the other genomes examined.
Characterization of cell surface mutants. (i) Growth properties of mutants.
None of the cell surface mutants displayed drastic growth defects compared to the WT during growth in M17 broth complemented with 0.5% lactose. We noted, however, that the ΔyfnI-like and Δstu1482 ΔrgpX2 mutants showed a higher generation time (46 ± 1 min and 62 ± 1 min, respectively) than the WT (30 ± 1 min), while the ΔlspA, ΔtagO-like, ΔepsE, and Δeps9 Δeps10 Δeps11 mutants displayed generation times (28 ± 1 min, 32 ± 1 min, 32 ± 1 min, and 32 ± 1 min, respectively) close to those of the WT.
(ii) Morphological characteristics of mutants.
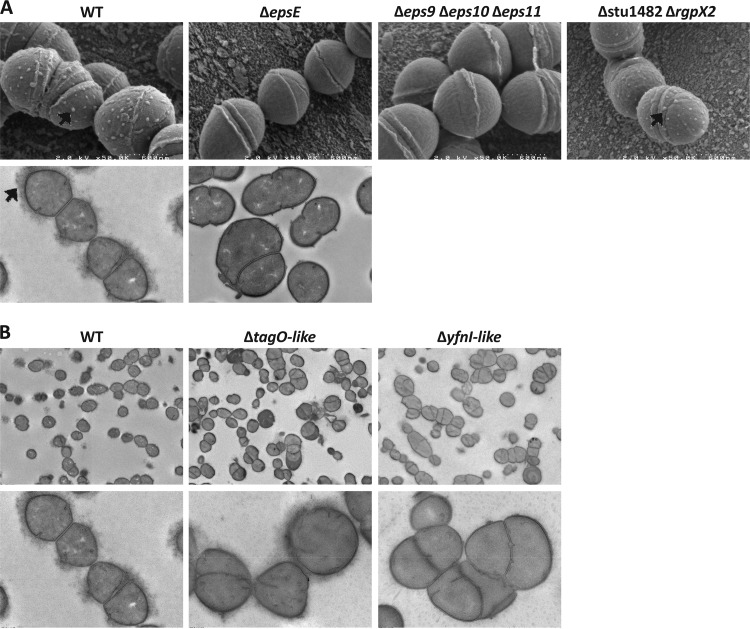
The LMG 18311 WT strain and cell surface mutants were observed with scanning electron microscopy (SEM) and transmission electron microscopy (TEM) in order to characterize the cell, surface, and chain morphology. LMG 18311 WT cells displayed a homogenous size and were assembled in typical ovococci chains, with septa formed in successive parallel planes perpendicular to the chain axis (Fig. 1). In WT cells, exopolysaccharides appeared as disseminated white spots at the cell surface when using SEM (Fig. 1A, upper panels, black arrows) and as amorphous structures surrounding cells when using TEM (Fig. 1A, lower panels, black arrows). All the mutants except the ΔepsE and Δeps9 Δeps10 Δeps11 mutants shared these characteristics (Fig. 1A), thus confirming that the latter mutants lack exopolysaccharides. It also appeared that wall teichoic and lipoteichoic acid mutants displayed an aberrant pattern of septation and cell separation during division (Fig. 1B). Furthermore, two mutants (the ΔtagO-like and ΔdltA mutants) formed aggregates compared to the WT (see Fig. S3 in the supplemental material for the ΔtagO-like mutant).
FIG 1.
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) observations of WT LMG 18311 and mutants. (A) SEM observations (original magnification, ×50,000) (upper panels) and TEM observations (original magnification, ×10,000) (lower panels) for the indicated strains. Black arrows indicate EPS. (B) TEM observations (original magnification, ×2,500 [upper panels] and ×10,000 [lower panels]) for the epsE mutant compared to the WT.
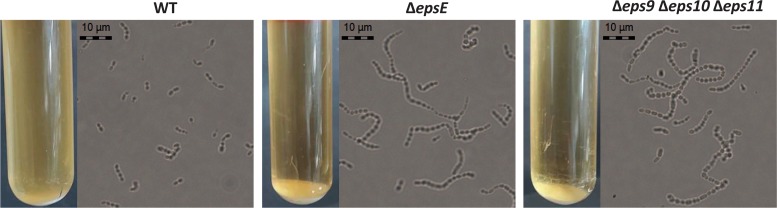
Finally, the two mutants linked to EPS biosynthesis (ΔepsE and Δeps9 Δeps10 Δeps11 mutants) exhibited a distinguishable phenotype of sedimentation in liquid culture compared to the WT (Fig. 2). This phenotype was likely linked to the number of cells per chain of these mutants, which was greater than in the WT, as observed by phase-contrast microscopy (Fig. 2).
FIG 2.
Sedimentation and chain length of WT LMG 18311 and EPS mutants. The picture shows standing LM17 cultures and phase-contrast microscopy of the LMG 18311 WT, ΔepsE, and Δeps9 Δeps10 Δeps11 strains. Photographs were taken after 8 h of growth at 42°C in LM17 medium, corresponding to the conditions where sedimentation is visible. Original magnification, ×400.
(iii) Cell surface charge of mutants.
In order to characterize the impact of mutations on the global charge of the cell surface envelope, the zeta potential of mutants was compared to that of the WT at various pHs. At very acidic pH (around pH 2), the zeta potentials of the ΔepsE and Δeps9 Δeps10 Δeps11 mutants differed from those of the WT and other mutants with a more positively charged zeta potential (close to 10 mV compared to less than 5 mV) (see Fig. S1 in the supplemental material). The zeta potentials of the WT and mutants were negative at the three tested pHs, pH 5, 7, and 9. At the two latter pHs, the ΔepsE and Δeps9 Δeps10 Δeps11 mutants displayed a more negative zeta potential than the WT and other mutants (Fig. S1).
(iv) Biofilm-forming abilities of mutants.
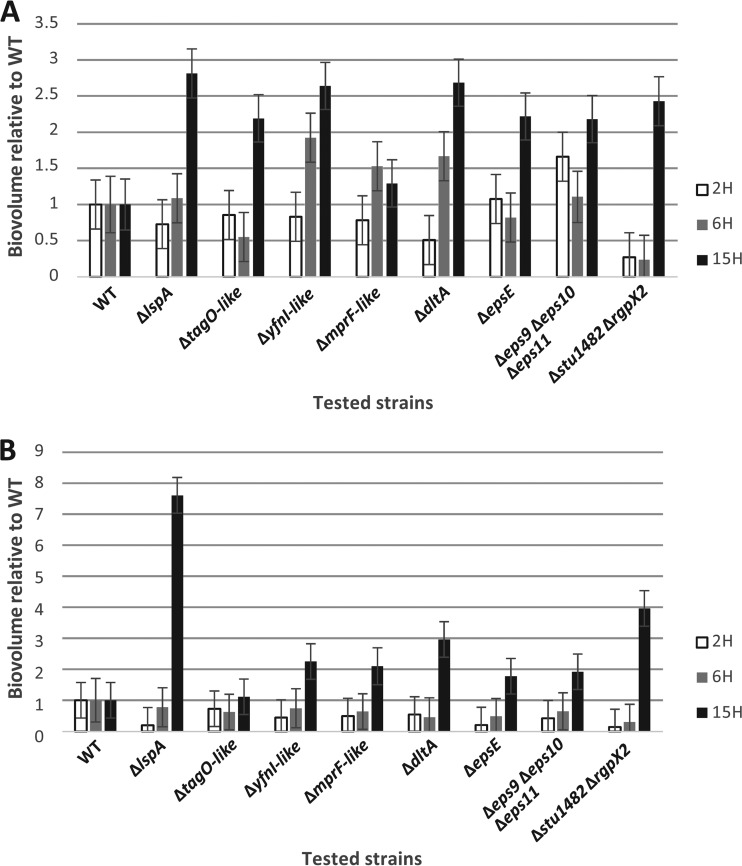
Biofilm formation was evaluated after different time lapses (2 h, 6 h, and 15 h) and measured two times: before and after rinsing the 96-well polystyrene plates to test the robustness of the formed biofilms. The quantitative comparison of biofilm biovolumes showed diversity in biofilm formation compared to that of the WT. Before rinsing, the ΔyfnI-like mutant showed a significant higher biofilm biovolume than the WT after 6 h of growth (Fig. 3A). After 15 h of growth, all mutants except the ΔmprF-like mutant showed at least a 2-fold higher biofilm biovolume than the WT (Fig. 3A). After rinsing of the well plates, the ΔlspA mutant stood out from the WT and other mutants as shown by the robustness of its biofilm structure, with a 7-fold higher biofilm biovolume after 15 h of growth than the WT (Fig. 3B). Two other mutants (the ΔdltA, and Δstu1482 ΔrgpX2 mutants) also displayed a robust biofilm structure compared to that of the WT after 15 h of growth (Fig. 3B).
FIG 3.
Biofilm biovolumes of LMG 18311 mutants relative to the WT after 2, 6, and 15 h of growth, before (A) and after (B) rinsing of the polystyrene microplates. Error bars show standard errors of the means.
Conjugative transfer of ICESt3 using mutated recipients.
To test whether the cell surface properties impact the transfer of ICESt3, mating experiments were first carried out using mutated recipient cells as detailed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| LMG 18311 | Wild-type strain | |
| LMG 18311(ICESt3) | LMG 18311 carrying ICESt3 tagged with a chloramphenicol resistance cassette; Cmr | 11 |
| LMG 18311(ICESt3 GFP) | LMG 18311 carrying ICESt3 tagged with a chloramphenicol resistance cassette and with a GFP gene under the control of the pLDH promoter of S. thermophilus LMG 18311; Cmr | This work |
| LMG 18311 Eryr | LMG 18311 carrying an erythromycin resistance cassette in its chromosome between stu0627 and stu0629 genes; Eryr | This work |
| LMG 18311 Spcr | LMG 18311 carrying a spectinomycin resistance cassette in its chromosome between stu0627 and stu0629 genes; Spcr | This work |
| LMD-9(pMG36e) | LMD-9 carrying the pMG36e plasmid conferring erythromycin resistance; Eryr | This work |
| Mutated recipient cells for mating with WT donor cells | ||
| LMG 18311 ΔlspA | LMG 18311 with lspA (stu0521) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 ΔtagO-like | LMG 18311 with tagO-like (stu0163) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 ΔdltA-like | LMG 18311 with dltA (stu0761) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 ΔyfnI-like | LMG 18311 with yfnI-like (stu0636) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 ΔmprF-like | LMG 18311 with mprF-like (stu1256) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 ΔepsE | LMG 18311 with epsE (stu1108) gene deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 Δeps9 Δeps10 Δeps11 | LMG 18311 with eps9 to eps11 (stu1097 to stu1099) genes deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMG 18311 Δstu1482 ΔrgpX2 | LMG 18311 with stu1482 and rgpX2 (stu1482 and stu1473) genes deleted by insertion of an erythromycin cassette; Eryr | This work |
| LMD-9 ΔsrtA | LMD-9 with srtA gene deleted by insertion of an erythromycin cassette; Eryr | 43 |
| Mutated donor cells for mating with WT recipient cells | ||
| LMG 18311(ICESt3) ΔlspA | LMG 18311 ΔlspA carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) ΔtagO-like | LMG 18311 ΔtagO-like carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) ΔdltA-like | LMG 18311 ΔdltA-like carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) ΔyfnI-like | LMG 18311 ΔyfnI-like carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) ΔmprF-like | LMG 18311 ΔmprF-like carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) ΔepsE | LMG 18311 ΔepsE carrying ICESt3; Eryr Cmr | This work |
| LMG 18311(ICESt3) Δeps9 Δeps10 Δeps11 | LMG 18311 Δeps9 Δeps10 Δeps11 carrying ICESt3; Eryr Cmr | This work |
| Mutated recipient cells used in mutant/mutant experiments | ||
| LMG 18311 ΔlspA | LMG 18311 ΔlspA with spectinomycin resistance cassette; Eryr Spcr | This work |
| LMG 18311 ΔtagO-like | LMG 18311 ΔtagO-like with spectinomycin resistance cassette; Eryr Spcr | This work |
| LMG 18311 ΔyfnI-like | LMG 18311 ΔyfnI-like with spectinomycin resistance cassette; Eryr Spcr | This work |
| LMG 18311 ΔepsE | LMG 18311 ΔepsE with spectinomycin resistance cassette; Eryr Spcr | This work |
| Plasmids | ||
| pMG36e | 3.4 kb, replication origin from pWV01; Eryr | 44 |
| pG+host9 | 4.6 kb, thermosensitive derivative of pWV01; Eryr | 45 |
| pSL1180 spec lox | 4.5 kb, derivative of pBR322; Spcr | This work |
| pSET4s | 4.5 kb, thermosensitive derivative of pWV01; Spcr | 46 |
| pOri23 | 5.8 kb, ColE1 replication origin; Eryr | 47 |
| pOri23-pLDH | pOri23 with the promoter of the l-lactate dehydrogenase gene (stu1280) of S. thermophilus LMG18311 instead of the P23 promoter | This work |
| pSW4-GFPopt | 7.3 kb, Gram-positive/Gram-negative shuttle vector with GFPopt gene; Kmr Apr | 42 |
| pOri23-pLDH-gfp | pOri23 vector carrying the GFP gene of pSW4-GFPopt under the control of the pLDH promoter of S. thermophilus LMG18311 | This work |
Abbreviations: Cmr, chloramphenicol resistance; Eryr, erythromycin resistance; Spcr, spectinomycin resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance.
(i) Mating experiments using recipient cells affected in protein exposure (ΔsrtA and ΔlspA mutants).
Strain LMG 18311 harbors a truncated srtA gene and three pseudogenes of LPXTG proteins. An efficient transfer was observed using this strain as either the donor or recipient in conjugation experiments. This indicates that LPXTG proteins are not essential for transfer and acquisition of ICESt3. Experiments were also carried out using the LMD-9 strain of S. thermophilus, which harbors a functional sortase A protein and has three LPXTG proteins exposed at its surface. The LMD-9 ΔsrtA mutant is a strain with a sortase A gene that was interrupted to inhibit the synthesis of SrtA (32). No significant difference in ICESt3 acquisition was observed when using WT LMG 18311(ICESt3) Cmr as a donor and LMD-9 ΔsrtA Eryr or LMD-9(pMG36e) Eryr as a recipient (see Fig. S2 in the supplemental material).
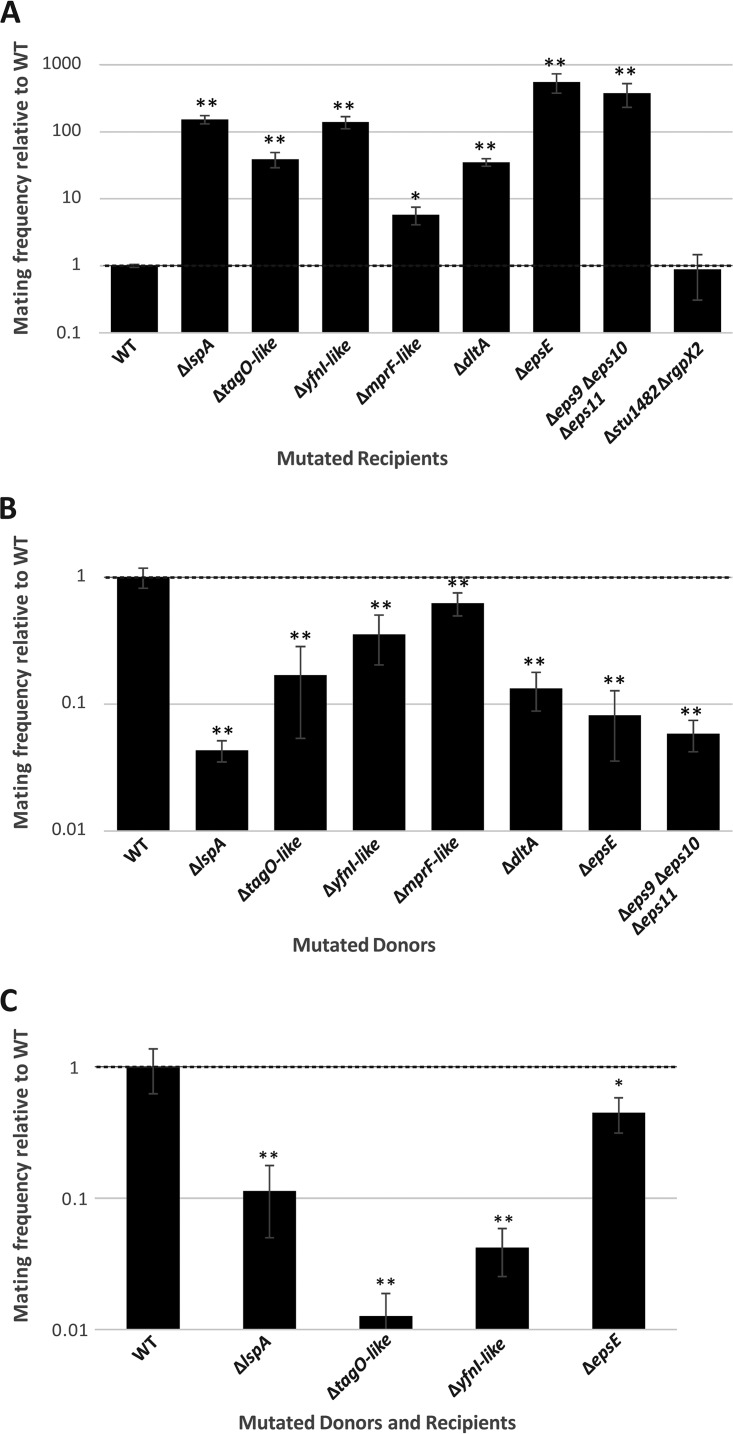
In contrast, an lspA mutation in LMG 18311 led to a 100-fold increase of ICESt3 acquisition compared to that for a WT recipient strain (i.e., LMG 18311 Eryr) (P < 0.01, t test) (Fig. 4A).
FIG 4.
Conjugative transfer of ICESt3 from and/or to mutants. Conjugation frequencies of LMG 18311 mutants relative to the WT are shown. (A) Conjugative transfer of ICESt3 using a WT donor and a mutated recipient; (B) conjugative transfer of ICESt3 using a mutated donor and a WT recipient; (C) conjugative transfer of ICESt3 using a mutated donor and a mutated recipient. For each experiment, means and standard deviations from at least 3 biological repetitions on 2 independent clones of mutants are shown. * and **, the conjugation frequency is statistically different from that of the WT (P < 0.05 and P < 0.01, respectively [t test]).
(ii) Mating experiments using recipient cells affected in WTA (ΔtagO-like mutant) and LTA (ΔyfnI-like mutant) exposure.
The ΔtagO-like and ΔyfnI-like mutants showed 40- and 100-fold increases of ICESt3 acquisition (P < 0.01, t test), respectively, compared to that of a WT recipient strain (Fig. 4A).
(iii) Mating experiments using recipient cells that could be affected in cell surface charge (ΔmprF-like and ΔdltA mutants).
An increase of ICESt3 acquisition, albeit lower than those observed for the other mutants (approximately 5-fold [P < 0.05, t test]), was observed using the ΔmprF-like mutant compared to the WT (Fig. 4A). For the ΔdltA mutant, an increase similar to the one observed for the WTA mutant (approximately 40-fold [P < 0.01, t test]) was observed (Fig. 4A).
(iv) Mating experiments using recipient cells affected in cell surface polysaccharide production.
A 400-fold increase of ICESt3 acquisition was obtained for mutants lacking genes or a group of genes belonging to the same cluster and involved in glycosyltransferase activity (epsE, eps11, and eps10), or pyruvyltranferase activity (eps9) (P < 0.01, t test) (Fig. 4A). Under these conditions, the frequency of ICESt3 transfer was close to 10−1 transconjugants per donor. In contrast, a mutant affected in rhamnose-glucose polysaccharide production (Δstu1482 ΔrgpX2), showed no significant difference in ICESt3 acquisition compared to WT recipient cells (Fig. 4A).
Conjugative transfer of ICESt3 using mutated donor cells.
In order to investigate whether the phenotypes observed for the mutants are specific to the recipient cells and/or to ICESt3 activity, mating experiments were also carried out using mutated donor cells. Mating experiments were carried out using selected LMG 18311 mutants (ΔlspA, ΔtagO-like, ΔyfnI-like, ΔmprF-like, ΔdltA, ΔepsE, and Δeps9 Δeps10 Δeps11) carrying ICESt3 as donor cells (obtained in the experiments mentioned in the preceding paragraph) and the WT LMG 18311 Spcr strain as recipient cells (see Table 1). While the mutation of these cell surface molecules in the recipient led to a 5- to 400-fold increase of ICESt3 acquisition compared to that for the WT, the same mutations in the donor cells led to a significant decrease of ICESt3 transfer (ranging from less than 10- to 25-fold) compared to that for the WT (P < 0.01, t test) (Fig. 4B).
Conjugative transfer of ICESt3 using a mutant/mutant mating pair.
To test whether the observed effects of mutations in the donor or recipient cells were additive, mating experiments using mutated donor and recipient cells were also carried out for the ΔlspA, ΔtagO-like, ΔyfnI-like, and ΔepsE mutants. The results showed a decrease of ICESt3 transfer when lipoproteins, teichoic acids, lipoteichoic acids, or exopolysaccharides were affected in both the donor and recipient cells. These results were close to the ones obtained when only the donor strain was mutated. Hence, no additive effect between the donor mutation and the recipient mutation was observed (Fig. 4C).
DISCUSSION
Our results highlight a significant impact of cell surface composition on the conjugation of ICESt3 of S. thermophilus. Indeed, deletion of the lspA, tagO-like, yfnI-like, dltA, epsE, and eps9 to -11 genes in recipient S. thermophilus LMG 18311 cells led to an increase of ICESt3 acquisition. Except for the eps9 to -11 cluster, which was found to be specific to S. thermophilus LMG 18311, these genes appeared to be highly conserved in S. thermophilus and the closely related species S. salivarius but also in the more distantly related species S. pyogenes and E. faecalis (except for epsE). It is thus possible that the observed impact on conjugation of these surface molecules could apply to other bacterial species. This indicates that the presence of mature lipoproteins, teichoic acids, and exopolysaccharides at the cell surface can be a hindrance to ICESt3 acquisition by recipient cells. Taken as a whole, this suggests that the fewer surface molecules, the better conjugation efficiency is, at least when considering the recipient cell. Bearing in mind the important proportion and networks of teichoic acids and exopolysaccharides in the Gram-positive cell surface envelope (20), the lack or decrease of these components could significantly reduce the steric hindrance in the cell envelope. This less congested surface may be suitable for the peptidoglycan hydrolysis activity of the ICESt3 hydrolase OrfA or for the establishment or activity of the conjugation machinery required for DNA transport in the recipient cell. Furthermore, we cannot exclude that the observed effect of deletions can be linked to the modification of physical-chemical properties of the cell envelope impacting cell-cell interactions and/or DNA transport across the membranes.
Characterization of the mutants revealed some phenotypes that could contribute to the observed impact of mutations on ICESt3 conjugation. The ΔlspA mutant, but also the ΔtagO-like, ΔyfnI-like, ΔdltA, ΔepsE, and Δeps9 Δeps10 Δeps11 mutants, showed in particular higher biovolumes and ability of interactions in biofilm assays than the WT. Furthermore, the LMG 18311 ΔlspA mutant stood out from the WT and derivatives by the robustness of its biofilm-forming ability, thus suggesting better cell-to-cell interactions for this mutant.
Lpp.
LMG 18311 ΔlspA is expected to be affected in its lipoprotein (Lpp) content, as observed in a Streptococcus sanguinis ΔlspA mutant, whose Lpp show partial activity (35). In Streptococcus uberis, the type I signal peptidase Eep seems to replace the cleavage activity of LspA (36), suggesting that an alternative pathway takes place in a ΔlspA mutant. Genome analysis of LMG 18311 predicts the presence of two type I signal peptidases (16), but it is not known whether these proteins could be involved in Lpp maturation in a ΔlspA mutant context. LMG 18311 ΔlspA showed a large increase (up to 100-fold) of ICESt3 acquisition. LMG 18311 harbors 24 predicted lipoproteins whose presence could interfere with the conjugation machinery assembly. Among these 24 predicted lipoproteins, 15 are involved in the binding of substrates of ABC transporters, whereas others are involved in unknown functions (16). We cannot exclude that a molecular interaction between one or several of these lipoproteins and components of the conjugation machinery could impair ICESt3 transfer.
WTA.
TagO and TarO homologs are described as essential for wall teichoic acid (WTA) biosynthesis in B. subtilis (33) and S. aureus (37). The LMG 18311 ΔtagO-like mutant is thus expected to lack WTA. The ΔtagO-like mutant showed an approximately 40-fold increase of ICESt3 acquisition by recipient cells. TEM analysis revealed that the tagO mutant is affected in its septation (this is also true for the yfnI mutant). However, it is unclear whether and how this phenotype could contribute to better ICESt3 acquisition. Contrast and confocal microscopies also indicated that it forms aggregates, as described in other species such as S. aureus (37). This phenotype can contribute to better cell-cell interactions.
LTA.
The yfnI-like gene product is the only lipoteichoic acid (LTA) synthase identified in our in silico analysis. Therefore, the disruption of this gene is expected to abrogate, or at least to reduce, LTA presence in the LMG 18311 cell envelope. As in B. subtilis for ICEBs1 acquisition (9), the ΔyfnI-like mutant of S. thermophilus LMG 18311 showed an increased propensity for ICESt3 acquisition. The much higher impact of the yfnI-like mutation on ICESt3 transfer (approximately 100-fold) than on ICEBs1 acquisition (less than 10-fold) could be explained by the fact that two additional enzymes, LtaSBS and YqgS, are involved with YfnI in LTA biosynthesis in B. subtilis (24). Thus, yfnI deletion in these species is not expected to trigger the same phenotypes. This can also be linked to the difference of transfer frequency between ICEBs1 and ICESt3, thus making ICEBs1 more prone to saturation.
We also investigated the impact of an mprF mutation on ICESt3 acquisition by the recipient cells. Unlike the case for ICEBs1 and Tn916 (8), the ΔmprF-like mutant showed an increase of ICESt3 acquisition, although to a lesser extent than for the other mutants. As suggested by Johnson and Grossman, MprF, which impacts the level of lysyl-phosphatidylglycerol, a positively charged molecule, could be involved in cell buffering under the various environmental conditions encountered by the bacteria (9). Specific environmental growth conditions and cell surface charge of S. thermophilus could explain these observed differences, suggesting a species-dependent impact of lysyl-phosphatidylglycerol on conjugation.
d-Alanylation of teichoic acid content.
WTA and LTA are known to be negatively charged polymers; however, little is known about the cell envelope composition of S. thermophilus regarding its teichoic acid content. One hypothesis could be that the observed impacts on ICESt3 transfer and acquisition are partly linked to the negative charges carried by the WTA and LTA polymers. The absence of these molecules would make the cell envelope less negatively charged, hence contributing to better interactions with the donor that is negatively charged. To test whether WTA and LTA impacts are linked to cell surface charge, ICESt3 transfers were assessed using a ΔdltA mutant as the recipient. The dltA gene belongs to the dlt operon, which is responsible for the d-alanylation of both WTA and LTA, thus neutralizing their negative charge and making the whole polymers zwitterionic, or at least reducing them. It has been described that a dltA mutation is sufficient to impair d-alanylation of WTA and LTA in B. subtilis (26). Thus, LMG 18311 ΔdltA is expected to have fully negatively charged WTA and LTA polymers exposed at its surface. However, the ΔdltA mutant showed an increase of ICESt3 acquisition similar to that for a ΔtagO-like mutant (approximately a 40-fold increase for both compared to the WT) but lower than that for the ΔyfnI-like mutant (approximately a 100-fold increase compared to the WT), which is not consistent with this hypothesis. However, we cannot exclude that the ΔdltA effect could be due to the lack of d-alanylation of another cell envelope component. Furthermore, no measurable difference of zeta potential was detected between the WT, ΔdltA, ΔtagO-like, and ΔyfnI-like strains under the tested conditions, suggesting that the change of surface charge is minor in these mutants, which can be explained by the following hypotheses: (i) LTA and WTA in S. thermophilus LMG 18311 could harbor a carbon backbone poorly loaded in phosphoglycerol repeat units, and (ii) LMG 18311 could display small amounts of LTA and WTA at its surface, thus explaining the absence of a significant change in the cell surface net charge. However, this could also indicate that not all teichoic acids were removed by these mutations. These results also highlight that the increase of ICESt3 acquisition observed when recipient is affected in WTA or LTA biosynthesis is not linked to the negative charges that both these polymers could confer to the bacteria. Moreover, the same impact on ICESt3 acquisition observed for both the ΔdltA and ΔtagO-like mutants could be linked to the formation of aggregates that could improve cell-to-cell interactions of both mutants. Indeed, these mutants form mixed aggregates with donor cells carrying a green fluorescent protein (GFP) gene-labeled ICE when examined by confocal microscopy (see Fig. S3 in the supplemental material for the ΔtagO-like mutant).
EPS.
Several genes belonging to a large gene cluster involved in exopolysaccharide (EPS) biosynthesis were mutated in LMG 18311. These mutants showed the greatest increase of ICESt3 acquisition, with a transfer frequency reaching 10−1 transconjugants per donor. Unlike S. thermophilus teichoic acids, EPS are well documented, and it has been described that the lack of the EpsE glycosyltransferase leads to a lack of EPS at the S. thermophilus cell surface (32). We confirmed this observation by SEM and TEM, since EPS is absent from the cell surfaces of ΔepsE and Δeps9 Δeps10 Δeps11 mutants. Liquid culture of these mutants showed sedimentation following growth, which could be a consequence of the cell chain length increase of these mutants compared to the WT. This increase in chain length likely facilitates ICE retransfer within the same chain of recipient cells, as described for B. subtilis (38), and could thus artificially increase the frequency of ICESt3 transfer. However, we found, by counting CFU before and after vortexing, that the vortexing step used in the conjugation experiments is not sufficient to disrupt cell chains of S. thermophilus LMG 18311. This indicates that, in our experiments, retransfer events are not counted in the conjugation frequency and do not explain the observed increase of ICESt3 transfer. EPS mutants show a significant change in cell surface charge compared to the WT and the other mutants. This phenotype could originate from the presence of phosphate and acetate components that may be attached to the EPS backbone. It could be hypothesized that these changes in electrostatic cell interactions favor ICESt3 acquisition.
LPXTG proteins.
the mutation of srtA in LMD-9 did not have any significant impact regarding ICESt3 acquisition compared to that of the WT, thus suggesting that the covalent linkage of LPXTG proteins to the cell membrane does not interfere with DNA transport inside the recipient cell.
Mutations affecting cell surface composition were also tested in a donor context (except for the Δstu1482 ΔrgpX2 mutant) instead of a recipient one. ΔlspA, ΔtagO-like, ΔyfnI-like, ΔepsE, and Δeps9 Δeps10 Δeps11 donor mutants were not able to efficiently transfer ICESt3 toward WT recipient cells. Our results indicate that the impact of mutations is different when the donor cell is targeted. Since the same strain was used, this difference in behavior is likely linked to the donor or recipient “status” in link with ICESt3 activity. The transfer frequencies of ICESt3 using ΔlspA, ΔtagO-like, ΔyfnI-like, and ΔepsE mutant donors also decreased when using recipient cells carrying the same mutation (mutant/mutant pairs).
No obvious additive effect between donor and recipient mutations was observed, thus suggesting that (i) the effect of a donor mutation is dominant and (ii) the adverse effect occurs prior to ICESt3 entry in the recipient cell. These results suggest that lipoproteins, wall teichoic acids, lipoteichoic acids, and exopolysaccharides are important for the proper positioning/assembly or activity of the conjugation machinery in the donor cell.
This study contributes to a better understanding of the impact of host factors on conjugation, but further studies are needed to decipher precisely how these factors interfere with the transfer of conjugative elements.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. S. thermophilus LMG 18311, LMD-9, and their derivatives were grown in M17 broth supplemented with 0.5% lactose (LM17) at 42°C without shaking. When required, cultures were supplemented with antibiotics at the following concentrations: chloramphenicol, 6 μg ml−1; erythromycin, 5 μg ml−1; and spectinomycin, 500 μg ml−1.
Genome analysis.
A search for homologs of B. subtilis proteins involved in WTA (33), LTA (24), and lysyl-phosphatidylglycerol (19) biosynthesis was carried out on the S. thermophilus LMG 18311 genome using the blastp program (with default parameters and low-complexity filter disabled). This genome was also screened for the presence of annotated clusters encoding polysaccharides and exopolysaccharides. Eight genes or clusters of genes were selected for mutant construction: stu0163 (homolog of tagO of B. subtilis), stu0521 (lspA gene), stu0636 (homolog of yfnI of B. subtilis), stu0761 (dltA gene), stu1256 (homolog of mprF of B. subtilis), stu1108 (encoding EpsE, the major glycosyltransferase involved in exopolysaccharide synthesis in S. thermophilus), 1 cluster encoding exopolysaccharides (stu1097, stu1097, and stu1099), and a cluster encoding rhamnose (stu1473, stu1480, stu1481, and stu1482).
All the genomes available in GenBank (last interrogation, 3 November 2017) for S. thermophilus (19 complete and 18 draft genomes), the closely related S. salivarius species (9 complete and 33 draft genomes), and the two species that successfully acquired ICESt3, S. pyogenes (60 complete and 297 draft genomes) and E. faecalis (15 complete and 533 draft genomes), were searched for homologs of these genes or clusters of genes with the tBLASTn program (with default parameters and low-complexity filter disabled). Hits were retained when they showed more than 50% of query cover and more than 40% of amino acid identity with the query proteins of S. thermophilus LMG 18311.
Construction of mutants affected in cell surface composition.
S. thermophilus LMG 18311 (WT) was used to construct cell surface mutants. For each targeted molecule, the gene sequence was deleted by insertion of an erythromycin resistance cassette by overlap PCR as described previously (39). PCRs were performed with 50 ng of genomic DNA, 200 μM each deoxynucleoside triphosphate (dNTP), 0.5 μM each primer (primer sequences are given in Table 2), and 0.02 U μl−1 of Phusion high-fidelity DNA polymerase (Thermo Scientific) in appropriate buffer per 50-μl reaction volume. Cycling conditions for the overlap PCR were 3 min at 98°C, 30 s at the annealing temperature (with a 1°C increment at each cycle), and 30 s/kb at 72°C, followed by 30 additional cycles with an annealing temperature of 55°C and a final extension of 10 min at 72°C. PCR products were then used for natural transformation of LMG 18311. The same steps were followed for chromosomal tagging of strain LMG 18311 with two different resistance genes (erythromycin or spectinomycin resistance gene) (Table 1). These resistance cassettes were inserted in an intergenic region between two convergent open reading frames (ORFs) in the S. thermophilus genome (14), stu0627 and stu0629. LMD-9(pMG36e) was obtained by natural transformation of LMD-9 with purified extract of pMG36e using a previously described protocol (39).
TABLE 2.
Primers used in this work
| Primer use | Primer name | Sequence (5′→3′) |
|---|---|---|
| LMG 18311 Eryr construction | MchromLMG I_Fwd | GAGGAACTCGGATTGGTAG |
| MchromLMG I_rev_ery | AGCCATCCGGAAGATCTGGGACCATTTCGTTTGCAC | |
| MchromLMG II_fwd_ery | AACACGAACCGTCTTATCTCCCAACTTAATTGAAGGCCAT | |
| MchromLMG II_Rev | CAGGGCTAGCTATTGTTTC | |
| Prom-Ery-For | GGAGATAAGACGGTTCGTGTT | |
| pSL1180 ery rev | CAGATCTTCCGGATGGCT | |
| LMG 18311 Spcr construction | MchromLMG I_Fwd | GAGGAACTCGGATTGGTAG |
| MchromLMG I_rev_spc | GGGAAATATTCATTCTAATTGGGGACCATTTCGTTTGCAC | |
| MchromLMG II_fwd_spc | ATTTATAGATTTCATTGGCTTCCAACTTAATTGAAGGCCAT | |
| MchromLMG II_Rev | CAGGGCTAGCTATTGTTTC | |
| SpecFwd | TAGAAGCCAATGAAATCTAT | |
| Speclox66 Rev | CCAATTAGAATGAATATTTCCC | |
| LMG 18311 ΔlspA construction | EG1885 | ACGAGTACTTCTTGACAGACAAATCAGA |
| EG1886 | TCATGTAATCACTCCTTCTTAATTACACATAAGTCCTCCTATGGTTTATAAGTATCA | |
| EG1887 | TATTTAACGGGAGGAAATAATTCTACATTAAAGAGGCGGGAAACCGTCTGGACAAGT | |
| EG1888 | AGAACATCCGTTGGATGACTATTAAGCT | |
| LMG 18311 ΔtagO-like construction | EG1873 | ATAGGCAGTCATGGTTGTTACCTCC |
| EG1874 | TCATGTAATCACTCCTTCTTAATTACACATGTCCTAGCTCCATTTCGTTGCTTGT | |
| EG1875 | TATTTAACGGGAGGAAATAATTCTAAAATAAACATTTGAAAAGCCAAGCAATGGCT | |
| EG1876 | AGATCTTGCAACCAGAGTGGCTCTGCT | |
| LMG 18311 ΔyfnI-like construction | EG1897 | TCCATACTAAAGCCATTAGCTTCAAA |
| EG1898 | TCATGTAATCACTCCTTCTTAATTACACAAAATAATACTTCCTTTGATTTCATATTA | |
| EG1899 | TATTTAACGGGAGGAAATAATTCTAGAATAATCCTAAAAAGACTGTTCTAAT | |
| EG1900 | ACTTGACTGTGCATCATCTGAATTCTA | |
| LMG 18311 ΔmprF-like construction | EG1951 | TCATTTATGGTATCTAAGCTTGTCCGT |
| EG1902 | TCATGTAATCACTCCTTCTTAATTACACATGCCACCACCTCTTTTTGACTAATTCTA | |
| EG1903 | TATTTAACGGGAGGAAATAATTCTAAAGTAAATACGACAAAAAAAGTGACCCTCCAGGGTT | |
| EG1904 | AGCATTCTCGATATGGATATTCCTGA | |
| LMG 18311 ΔdltA construction | EG1947 | AGTGCTTTAGCCTGTGCTGATCGTCTA |
| EG1914 | TCATGTAATCACTCCTTCTTAATTACACATTATTCTTCCTAAAATTCGTTATAGATA | |
| EG1915 | TATTTAACGGGAGGAAATAATTCTACGATGATAGACTTCTTGAAACAGCTTCCCC | |
| EG1916 | TCGCATGAGTACTATGACTAAGCGCATA | |
| LMG 18311 ΔepsE construction | EG1945 | ACTAAGGTTGATAAGAACAATATCGAGA |
| EG1946 | TCATGTAATCACTCCTTCTTAATTACACACTTATTTTTCCTCCATCAGATTTTTGAT | |
| EG1923 | TATTTAACGGGAGGAAATAATTCTAAAATGATAACTTCAAAGATGATTAGATGAG | |
| EG1924 | AGACCTGTAATTCCTGGCTTGAAGCT | |
| LMG 18311 Δeps9 Δeps10 Δeps11 construction | EG1941 | AGGAATGTCAAGATTTAGGAATTACA |
| EG1942 | TCATGTAATCACTCCTTCTTAATTACACATCTTCTCATCACCTAAATATTGATTTTT | |
| EG1943 | TATTTAACGGGAGGAAATAATTCTACAATAAATTCAATGATAATATAAGAGTTGC | |
| EG1944 | TTGCTAAATGCTGAGTAAATCCATTCCA | |
| LMG 18311 Δstu1482 ΔrgpX2 construction | EG1937 | TGGTATTGATAGTATCGAAAGTAGAGA |
| EG1938 | TCATGTAATCACTCCTTCTTAATTACACATTTTTATACGTAGTTTCTCCTGAAAACT | |
| EG1939 | TATTTAACGGGAGGAAATAATTCTAAAATAATATTTTATTAATAGCAGTCCCCTG | |
| EG1940 | ATCAGTTTGTGCCATAGCCTCCAGTA | |
| Ery resistance cassette used for mutant construction | EG940 | TGTAATTAAGAAGGAGTGA |
| EG941 | TAGAATTATTTCCTCCCGT | |
| pSL1180 spec lox vector construction | Spec-lox71-SpeI F | TTTTTACTAGTTCGTACCGTTCGTATAGCATACATTATACGAAGTTATCGTAACGTGACTGGCAAGA |
| Spec-lox66-SpeI R | TTTTTACTAGTCGTACCGTTCGTATAATGTATGCTATACGAAGTTATCCAATTAGAATGAATATTTCCC | |
| pOri23-pLDH-gfp vector construction | PLDHthermo-fwd-EcoRI | TTTTTGAATTCTTTCAATCAAATTATTCC |
| PLDHthermo-rev-BamHI-SacI | TTTTTGGATCCGTTGCAGTCATGAGCTCAACATCTC | |
| gfp-fwd-SacI | TTTTTGAGCTCATGTCATGTCAAAAGAATTA | |
| gfp-rev-PstI | GATAAGCTTGGCTGCAGGT | |
| pldh_oe_fwd | GGAGATTGAGCATACCTAGGGGAATTCTTTCAATCAAATTATTCC | |
| gfp-oe-rev | CGGTGACTAGTTATCTACACGGATAAGCTTGGCTGCAGG |
The pSL1180 spec vector was obtained by cloning an SpeI-SpeI spectinomycin resistance cassette at the AvrII site of pSL1180. The SpeI-SpeI spectinomycin resistance cassette was amplified from the pSET4S plasmid using the Spec-lox71-SpeI F and Spec-lox66-SpeI R primers (Table 2), which introduce SpeI sites upstream and downstream the resistance cassette.
Mating experiments.
S. thermophilus LMG 18311 was chosen for mating experiments. This strain was previously successfully used as a recipient for ICESt3 using the original donor strain CNRZ385 (at a frequency of 10−6 transconjugants per donor), but it can also act as a donor transferring ICESt3 at the same frequency (11). In order to avoid interference with host factors, LMG 18311 was used as both donor and recipient in mating experiments. Depending on the mating pair used, derivatives of strain LMG 18311, tagged with either an erythromycin or a spectinomycin resistance gene, were used as recipients (Table 1). The mating pair using the LMG 18311(ICESt3) donor strain with these recipient strains was considered a WT mating pair (with an ICESt3 transfer frequency of 10−4 transconjugants per donor), and the term mutant was used for the cells affected in their cell surface composition.
Donor and recipient strains were grown overnight with an appropriate antibiotic. Fifteen milliliters of broth medium was inoculated with 150 μl of donor or recipient stationary-phase cultures. Cultures were grown until mid-exponential phase (optical density at 600 nm [OD600] of 0.4) and then were mixed and centrifuged for 15 min in a prewarmed centrifuge at 4,200 × g to pellet cells. The pellet was resuspended in 1 ml of LM17 broth, and 150 μl was spread on 0.45-μm-pore-size cellulose nitrate filters (Millipore) deposited on LM17 soft agar (0.8%) plates. The plates were then incubated at 42°C. After an overnight incubation, the filters were removed from the agar plates and placed into 10 ml of LM17 liquid medium. Bacteria were recovered by vortexing for 30 s. By counting CFU before and after vortexing, we showed that such a short vortexing step is not sufficient to disrupt cell chains of S. thermophilus LMG 18311 (data not shown). The suspension was then directly spread on agar plates supplemented with the appropriate antibiotic to enable counting the CFU of the donor, recipient, and transconjugant cells after a 24-h incubation.
Mating frequencies were calculated by dividing the number of transconjugants by the number of donor CFU. At least three independent biological repetitions were done on two independent transformants. Statistical analysis was carried out by using Student's t test.
Sedimentation tests.
Sedimentation of the LMG 18311 ΔepsE and Δeps9 Δeps10 Δeps11 mutants was visualized after 8 h of growth in LM17 liquid culture and was compared to that of WT LMG 18311. The cultures were observed by phase-contrast microscopy with an original magnification of ×400.
Determination of bacterial cell wall zeta potential.
Bacterial cells from overnight cultures were harvested by centrifugation (5 min at 7,000 × g), washed twice with demineralized water, and suspended in demineralized water. To break bacterial chains, cells were vortexed for 3 min. Electrophoretic mobility measurements were done by suspending the bacterial strains in 3 ml of 10 mM potassium phosphate buffer with the pH ranging from 2 to 9 to obtain an OD600 of 0.07. The electrophoretic mobility at 150 V of the suspended bacteria was then measured using the ZetaSizer Nano ZS apparatus (Malvern Instruments Ltd., Malvern, UK). Electrophoretic mobilities were converted to the zeta potentials using the Helmholtz-Smoluchowski equation. At least three independent biological replicates were done.
TEM.
For each strain, 50 ml of planktonic bacteria grown until the end of exponential growth was pelleted at 2,000 rpm for 10 min at 4°C. Samples were then fixed for 1 h at room temperature in a 0.1 M cacodylate buffer containing 2% (vol/vol) glutaraldehyde (pH 7.2). Samples were kept overnight at 4°C in a 0.1 M cacodylate and 0.2 M sucrose buffer. Bacteria were then washed one time during 5 min with 0.1 M cacodylate buffer, contrasted during 1 h with 0.5% Oolong tea extract (OTE) in 0.1 M cacodylate buffer, and washed 2 times during 5 min with 0.1 M cacodylate buffer. Samples were postfixed for 1 h at room temperature in 0.1 M cacodylate buffer containing 1% (vol/vol) osmium tetroxide with 1.5% potassium cyanoferrate and then washed twice for 5 min with distilled water. Thereafter, cells were dehydrated in a gradual ethanol series (30%, 50%, 70%, and 90% [vol/vol] with distilled water and 3 times with 100% ethanol, 10 min for each step, except overnight for 70% ethanol). A 10-min intermediate bath in propylene oxide was performed. The bacteria were then impregnated at room temperature in successive mixes of propylene oxide and Epon (2:1; 1:1, and 1:2; each step for 2 h), in pure Epon overnight, and under vacuum conditions. A final inclusion bath with pure Epon and dimethylaminoethanol (DMAE) (an accelerator) was performed, and polymerization was allowed by incubating for 48 h at 60°C. Ultrathin sections of 70 nm were cut with an ultramicrotome (UC6; Leica, Germany) and deposited on 200-mesh copper-platinum grids. Sections were stained for 2 min in Reynold's lead citrate and rinsed in distilled water. Observations were performed using an HT7700 transmission electron microscope (Hitachi, Japan) equipped with an 8-million-pixel format charge-coupled device (CCD) camera driven by the image capture engine software AMT, version 6.02, at the INRA MIMA2 microscopy platform (Jouy-en-Josas, France). Images were made at 80 kV in high-contrast mode with an objective aperture adjusted for each sample and magnification.
SEM.
Bacterial suspensions, collected at the end of exponential growth, were immersed in a fixative solution (2.5% glutaraldehyde in 0.2 M sodium cacodylate buffer, pH 7.4), deposited on sterile cover glass discs (Marienfeld, VWR, France), and stored for 1 h at room temperature and overnight at 4°C. The fixative was removed, and samples were rinsed three times for 10 min each in the sodium cacodylate solution (pH 7.4). The samples underwent progressive dehydration by soaking in a gradual ethanol series (50 to 100%) before critical-point drying under CO2. Samples were mounted on aluminum stubs (10-mm diameter) with carbon adhesive discs (Agar Scientific; Oxford Instruments SAS, Gometz-La-Ville, France) and sputter coated with platinum (Polaron SC7640; Elexience, Verrières-le-Buisson, France) for 200 s at 10 mA. Samples were visualized by field emission gun scanning electron microscopy (SEM). They were viewed as secondary electron images (2 kV) with a Hitachi S4500 instrument (Elexience, Verrières-le-Buisson, France). Scanning electron microscopy analyses were performed at the Microscopy and Imaging Platform MIMA2 (INRA, Jouy-en-Josas, France).
Biofilms assays by laser scanning confocal microscopy.
Biofilms were measured in polystyrene 96-well microtiter plates with a μclear base (Greiner Bio-One, France), enabling high-resolution fluorescence imaging, as previously described (40). A volume of 200 μl of an overnight culture in LM17 (adjusted to an optical density at 600 nm of 0.01) was added to the wells of a microtiter plate. The microtiter plate was then kept at 42°C for 60 min to allow the bacteria to adhere to the bottom of the wells. After this adhesion step, the wells were rinsed with the growth medium to eliminate any nonadherent bacteria and then refilled with 200 μl of LM17. The microtiter plate was then incubated at 42°C for 2, 6, or 15 h and rinsed or not with a microplate autowasher (Thermo Fisher Wellwash) before microscopic evaluation. Bacterial cells were fluorescently stained in green with the nucleic acid marker SYTO9 (1:500 dilution in LM17 from a SYTO9 stock solution at 5 mM in dimethyl sulfoxide [DMSO]; Invitrogen, France). After 20 min of incubation in the dark to enable fluorescent labeling of the bacteria, the plate was mounted on the motorized stage of the confocal microscope (Leica SP8 AOBS inverter confocal laser scanning microscope at the MIMA2 platform, http://www6.jouy.inra.fr/mima2_eng/). The microtiter plates were scanned using a 63×/1.2-numerical-aperture (NA) water immersion objective lens and scanned at excitation wavelengths of 488 nm (argon laser; 3% intensity), with emission wavelengths collected from 493 to 550 nm using hybrid detectors (HyD Leica Microsystems, Germany). Three-dimensional (3D) projections of the biofilm structures before and after the washing step were reconstructed using the Easy 3D function of the IMARIS software (Bitplane, Switzerland). The biofilm biovolume is defined as the number of biomass pixels in all images of a stack multiplied by the voxel size and divided by the substratum area of the image stack (41). The biofilm biovolume (in μm3) was automatically extracted from image series using the dedicated ImageJ COMSTAT2 plugin (www.comstat.dk) (41).
Analysis of aggregates by laser scanning confocal microscopy.
Cells of S. thermophilus LMG 18311 carrying ICESt3 were discriminated by labeling ICESt3 with a GFP gene. A pOri23-pLDH vector was first constructed by cloning the promoter of the l-lactate dehydrogenase gene from S. thermophilus LMG 18311 (stu1280) in the pOri23 plasmid using the PLDHthermo-fwd-EcoRI and PLDHthermo-rev-BamHI-SacI primers (Table 2). The GFP gene, encoding a green fluorescent protein that was codon optimized for low-GC Gram-positive bacteria, was amplified by PCR with the gfp-fwd-SacI and gfp-rev-PstI primers using the pSW4-GFPopt plasmid constructed by Sastalla et al. (42) and then cloned downstream of the pLDH promoter in the pOri23-pLDH vector after digestion by the SacI and PstI restriction enzymes to give pOri23-pLDH-gfp (Table 1). The pLDH-gfp fragment was then amplified by PCR using primers that introduce extensions matching sequences of ICESt3 (pldh_oe_fwd and gfp-oe-rev). This enables synthesis of an overlap PCR fragment carrying the pLDH-gfp cassette flanked by sequences of ICESt3. After induction of natural competence of S. thermophilus cells, the overlap PCR product was then added for transformation. The crossover events, upstream and downstream from the tagged region, were positively selected by the newly acquired fluorescence of the transformed clones.
Cells carrying ICESt3-gfp (LMG 18311 ICESt3 pLDH-gfp) were mixed with LMG 18311 WT or mutants (ΔtagO-like or ΔdltA) and incubated for 4 to 6 h at 42°C before observation. Cells were counterlabeled using the syto61 red fluorescent nucleic acid stain before observation by confocal microscopy using a Leica SP8 AOBS inverter confocal laser scanning microscope at the MIMA2 platform (http://www6.jouy.inra.fr/mima2_eng/).
Supplementary Material
ACKNOWLEDGMENTS
We thank Sophie Bobet and Johan Staub for the construction of mutant strains, Alexis Canette for TEM observations, Steve Leppla of the Laboratory of Bacterial Diseases of the National Institute of Allergy and Infectious Diseases (Bethesda, MD, USA) for providing the pSW4-GFPopt vector, and UR AFPA laboratory for providing the srtA mutant.
N.D. is the recipient of a scholarship funded by INRA and Région Grand Est. This work received financial support from the Région Lorraine and Université de Lorraine (2011 to 2013) and from ANR (MATICE project, ANR-15-CE21-0007).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02109-17.
REFERENCES
- 1.Koonin EV. 2016. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res 5:F1000 Faculty Rev-1805. doi: 10.12688/f1000research.8737.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Aljaro C, Balleste E, Muniesa M. 2017. Beyond the canonical strategies of horizontal gene transfer in prokaryotes. Curr Opin Microbiol 38:95–105. doi: 10.1016/j.mib.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 4.Bellanger X, Payot S, Leblond-Bourget N, Guedon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 5.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraro N, Burrus V. 2014. Biology of three ICE families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol Spectr 2:1–20. doi: 10.1128/microbiolspec.MDNA3-0008-2014. [DOI] [PubMed] [Google Scholar]
- 8.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. doi: 10.1111/mmi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CM, Grossman AD. 2016. The composition of the cell envelope affects conjugation in Bacillus subtilis. J Bacteriol 198:1241–1249. doi: 10.1128/JB.01044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong TP, Carter MQ, Struffi P, Casonato S, Hao Y, Lam JS, Lory S, Jousson O. 2017. Conjugative type IVb pilus recognizes lipopolysaccharide of recipient cells to initiate PAPI-1 pathogenicity island transfer in Pseudomonas aeruginosa. BMC Microbiol 17:31. doi: 10.1186/s12866-017-0943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol 191:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraro N, Libante V, Morel C, Decaris B, Charron-Bourgoin F, Leblond P, Guedon G. 2011. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol 11:238. doi: 10.1186/1471-2180-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delorme C, Abraham AL, Renault P, Guedon E. 2015. Genomics of Streptococcus salivarius, a major human commensal. Infect Genet Evol 33:381–392. doi: 10.1016/j.meegid.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol 22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couvigny B, Therial C, Gautier C, Renault P, Briandet R, Guedon E. 2015. Streptococcus thermophilus biofilm formation: a remnant trait of ancestral commensal life? PLoS One 10:e0128099. doi: 10.1371/journal.pone.0128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guedon E, Monnet V, Renault P, Kleerebezem M. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev 29:435–463. [DOI] [PubMed] [Google Scholar]
- 17.Siegel SD, Reardon ME, Ton-That H. 2017. Anchoring of LPXTG-like proteins to the Gram-positive cell wall envelope. Curr Top Microbiol Immunol 404:159–175. doi: 10.1007/82_2016_8. [DOI] [PubMed] [Google Scholar]
- 18.Buddelmeijer N. 2015. The molecular mechanism of bacterial lipoprotein modification—how, when and why? FEMS Microbiol Rev 39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 19.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgain J, Scher J, Francius G, Borges F, Corgneau M, Revol-Junelles AM, Cailliez-Grimal C, Gaiani C. 2014. Lactic acid bacteria in dairy food: surface characterization and interactions with food matrix components. Adv Colloid Interface Sci 213:21–35. doi: 10.1016/j.cis.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Chapot-Chartier MP, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schade J, Weidenmaier C. 2016. Cell wall glycopolymers of Firmicutes and their role as nonprotein adhesins. FEBS Lett 590:3758–3771. doi: 10.1002/1873-3468.12288. [DOI] [PubMed] [Google Scholar]
- 23.Grundling A, Schneewind O. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wormann ME, Corrigan RM, Simpson PJ, Matthews SJ, Grundling A. 2011. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol Microbiol 79:566–583. doi: 10.1111/j.1365-2958.2010.07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyauchi E, Morita M, Rossi M, Morita H, Suzuki T, Tanabe S. 2012. Effect of d-alanine in teichoic acid from the Streptococcus thermophilus cell wall on the barrier-protection of intestinal epithelial cells. Biosci Biotechnol Biochem 76:283–288. doi: 10.1271/bbb.110646. [DOI] [PubMed] [Google Scholar]
- 26.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270:15598–15606. [DOI] [PubMed] [Google Scholar]
- 27.Rehm BH. 2010. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 28.Schmid J, Sieber V, Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol 6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Vuyst L, Weckx S, Ravyts F, Herman L, Leroy F. 2011. New insights into the exopolysaccharide production of Streptococcus thermophilus. Int Dairy J 21:586–591. doi: 10.1016/j.idairyj.2011.03.016. [DOI] [Google Scholar]
- 30.Pachekrepapol U, Lucey JA, Gong Y, Naran R, Azadi P. 2017. Characterization of the chemical structures and physical properties of exopolysaccharides produced by various Streptococcus thermophilus strains. J Dairy Sci 100:3424–3435. doi: 10.3168/jds.2016-12125. [DOI] [PubMed] [Google Scholar]
- 31.Ren W, Xia Y, Wang G, Zhang H, Zhu S, Ai L. 2016. Bioactive exopolysaccharides from a S. thermophilus strain: screening, purification and characterization. Int J Biol Macromol 86:402–407. doi: 10.1016/j.ijbiomac.2016.01.085. [DOI] [PubMed] [Google Scholar]
- 32.Minic Z, Marie C, Delorme C, Faurie JM, Mercier G, Ehrlich D, Renault P. 2007. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J Bacteriol 189:1351–1357. doi: 10.1128/JB.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb AJ, Karatsa-Dodgson M, Grundling A. 2009. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol Microbiol 74:299–314. doi: 10.1111/j.1365-2958.2009.06829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, Kanamoto T, Ge X, Xu P, Unoki T, Munro CL, Kitten T. 2009. Contribution of lipoproteins and lipoprotein processing to endocarditis virulence in Streptococcus sanguinis. J Bacteriol 191:4166–4179. doi: 10.1128/JB.01739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denham EL, Ward PN, Leigh JA. 2008. Lipoprotein signal peptides are processed by Lsp and Eep of Streptococcus uberis. J Bacteriol 190:4641–4647. doi: 10.1128/JB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, Lasa I. 2008. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154:865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auchtung JM, Aleksanyan N, Bulku A, Berkmen MB. 2016. Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 86:14–25. doi: 10.1016/j.plasmid.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Dahmane N, Libante V, Charron-Bourgoin F, Guedon E, Guedon G, Leblond-Bourget N, Payot S. 2017. Diversity of integrative and conjugative elements of Streptococcus salivarius and their intra- and interspecies transfer. Appl Environ Microbiol 83:e00337-17. doi: 10.1128/AEM.00337-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridier A, Dubois-Brissonnet F, Boubetra A, Thomas V, Briandet R. 2010. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J Microbiol Methods 82:64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 42.Sastalla I, Chim K, Cheung GY, Pomerantsev AP, Leppla SH. 2009. Codon-optimized fluorescent proteins designed for expression in low-GC gram-positive bacteria. Appl Environ Microbiol 75:2099–2110. doi: 10.1128/AEM.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kebouchi M, Galia W, Genay M, Soligot C, Lecomte X, Awussi AA, Perrin C, Roux E, Dary-Mourot A, Le Roux Y. 2016. Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Appl Microbiol Biotechnol 100:3667–3679. doi: 10.1007/s00253-016-7322-1. [DOI] [PubMed] [Google Scholar]
- 44.van de Guchte M, van der Vossen JM, Kok J, Venema G. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 55:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takamatsu D, Osaki M, Sekizaki T. 2001. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 46:140–148. doi: 10.1006/plas.2001.1532. [DOI] [PubMed] [Google Scholar]
- 47.Que YA, Haefliger JA, Francioli P, Moreillon P. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect Immun 68:3516–3522. doi: 10.1128/IAI.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.