Abstract
Campylobacter jejuni and Campylobacter coli are the most common cause of bacterial gastroenteritis worldwide. Additionally, C. jejuni is the most common bacterial etiological agent in the autoimmune Guillain-Barré syndrome (GBS). Ganglioside mimicry by C. jejuni lipooligosaccharide (LOS) is the triggering factor of the disease. LOS-associated genes involved in the synthesis and transfer of sialic acid (glycosyltranferases belonging to family GT-42) are essential in C. jejuni to synthesize ganglioside-like LOS. Despite being isolated from GBS patients, scarce genetic evidence supports C. coli role in the disease. In this study, through data mining and bioinformatics analysis, C. coli is shown to possess a larger GT-42 glycosyltransferase repertoire than C. jejuni. Although GT-42 glycosyltransferases are widely distributed in C. coli population, only a fraction of C. coli strains (1%) are very likely able to express ganglioside mimics. Even though the activity of C. coli specific GT-42 enzymes and their role in shaping the bacterial population are yet to be explored, evidence presented herein suggest that loss of function of some LOS-associated genes occurred during agriculture niche adaptation.
Introduction
Glycan mimicry is a strategy utilized by pathogens to evade detection by the host innate immune system1,2. Campylobacter jejuni, the most commonly reported cause of gastroenteritis in the world, boasts a large repertoire of human glycans3. Molecular mimicry between sialylated C. jejuni lipooligosaccharides (LOS) and gangliosides may result in the onset of Guillain-Barré syndrome (GBS)4,5; an autoimmune acute progressive polyradiculoneuropathy with approximately 5% mortality rate6. To express ganglioside-like LOS7–9, C. jejuni synthesizes cytidine-5′-monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) from uridine-5′-diphosphate-N-acetylglucosamine (UDP-GlcNAc) by the consecutive actions of an N-acetylglucosamine-6-phosphate 2-epimerase (NeuC), a sialic acid synthase (NeuB), and a CMP-Neu5Ac synthase (NeuA)10. Then, CMP-Neu5Ac is transferred by either of the LOS associated sialyltransferases; CstII (α2,3/8-sialyltransferase) or CstIII (α2,3-sialyltransferase)11. Both sialyltransferases belong to the, so far, monospecific CAZy (Carbohydrate-active enzymes database)12–14 glycosyltransferase (GT) family 4215–17. Although the presence of GT-42 and N-acetylneuraminate biosynthesis genes (neuABC) is insufficient for expressing molecular mimics, all C. jejuni strains containing this set of genes8,18–20 (LOS locus classes A, B, C, M, and R) have been shown to synthesize ganglioside-like structures3,7–9,21. Therefore, the presence of GT-42 and neuABC genes has been used as proxy for identifying C. jejuni strains capable of producing human glycan mimics9,22.
Campylobacter coli is the second most common cause of campylobacteriosis contributing, depending on the geographical region, to as many as 25% of all the infections23. Although C. coli has also been isolated from GBS patients24,25, its role in promoting this autoimmune disease remains controversial26. Additionally, despite the pervasive introgression with C. jejuni27, C. coli containing C. jejuni-like LOS classes linked to ganglioside mimicry have not been reported so far. Based on genomic data analysed hitherto, C. coli LOS locus appears to be marginally affected by horizontal gene transfer (HGT) or homologous recombination28.
Discovery of alternative orthologues of GT-42 encoding genes and associated LOS locus classes has been hindered by the very limited availability of genomic data. Consequently, it was only recently that C. coli LOS locus classes containing putative sialyltransferases, distantly related to those found in C. jejuni, were described28,29. The C. coli LOS locus class IX contains a GT-42 (cstV) and neuABC genes, LOS class II harbours an orphan GT-42 (cstIV), and LOS class III has a pseudogenized orphan GT-4220,28,29.
At present, the decreasing costs of next generation sequencing has driven a mass production of genomic sequences of several bacterial pathogens including Campylobacter spp. At the time of writing, the approximately 12,000 C. jejuni and 3,000 C. coli genome sequences found in public repositories offer unforeseeable opportunities. Thus, we took advantage of the large number of sequenced Campylobacter spp. strains to comprehensively investigate presence, frequency, and distribution of the molecular machinery for the biosynthesis of sialylated LOS structures in C. coli population.
Results
C. coli GT42 genes
Of the 45 C. coli GT42 protein sequences retrieved from NCBI nr database, six were partial sequences (i.e. incomplete coding sequences). Thus, they were excluded from further analysis (Supplementary Table S1). Based on BlastP Score Ratio (BSR), the remaining 39 sequences clustered into 7 different groups (Supplementary Tables S2 and S3), with average BSR values ranging from 0.80 to 0.98 (Table 1). Group 1, 2, and 4 contain proteins showing the highest similarity to CstI, CstIII, and CstII, respectively, while the other groups show limited homology to C. jejuni GT-42 enzymes (Table 1). Group 5 comprises orthologues to the previously described CstV in LOS class IX of C. coli 7633929, while Group 7 includes CstIV, the GT-42 within C. coli LOS locus class II20,28. Group 6 contains a novel group of orthologous proteins (named herein CstVI) showing high similarity to the pseudogenizised GT-42 described as part of LOS locus class III20,28. Similarly, Group 3 includes a single novel protein sequence named herein CstVII.
Table 1.
Average Blastp Score Ratio (BSR) of the C. coli GT-42 homologs.
| Groups | Generef | BSR | CstI (Q9RGF1) | CstII (Q9F0M9) | CstIII (Q7BP25) | Id (%) | Cov (%) | Id (%) |
|---|---|---|---|---|---|---|---|---|
| Cov (%) | Id (%) | Cov (%) | ||||||
| 1 | cstI 21 | 0.87 | 93 | 70 | 89 | 51 | 83 | 50 |
| 2 | cstIII 21 | 0.80 | 56 | 54 | 98 | 51 | 100 | 88 |
| 3a | cstVII c | 1 | 61 | 51 | 90 | 52 | 86 | 53 |
| 4 | cstII 21 | 0.87 | 60 | 52 | 99 | 89 | 95 | 52 |
| 5 | cstV 29 | 0.98 | −b | — | 98 | 48 | 94 | 43 |
| 6 | cstVI c | 0.95 | — | — | 97 | 37 | 96 | 35 |
| 7 | cstIV 29 | 0.86 | — | — | 97 | 40 | 94 | 37 |
aSingleton.
bNo significant hits.
cGene name proposed in this study.
Furthermore, evolutionary analysis revealed that the 7 BSR groups form monophyletic clades and are divided into two clusters (Fig. 1). Cluster A is comprised of cstI, cstII, cstIII, and cstVII, while cluster B includes cstIV, cstV and cstVI.
Figure 1.
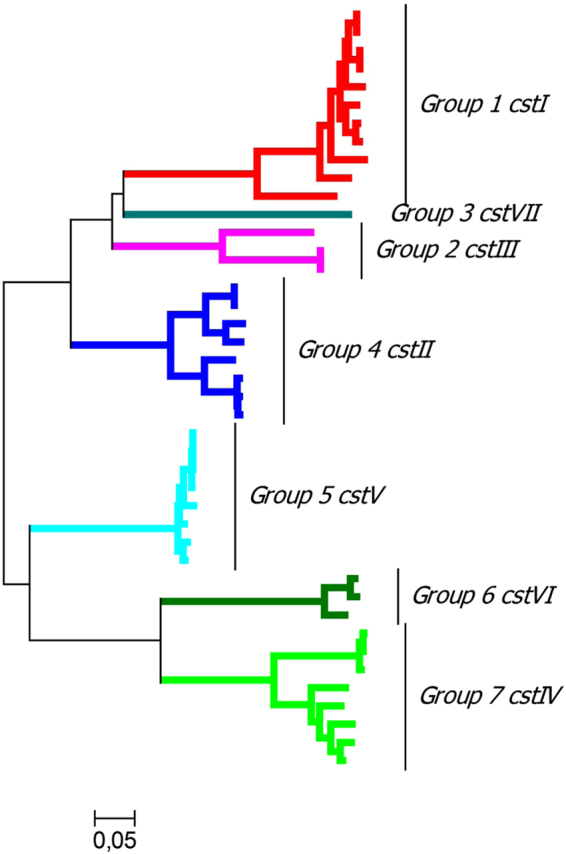
Evolutionary analyses of C. coli GT-42. Evolutionary analysis of 45 C. coli GT-42 sequences and three C. jejuni sequences (cstI, cstII and cstIII) was conducted in MEGA7 and the evolutionary history was inferred using the Minimum Evolution method calculating the distance using Maximum Composite Likelihood. The tree is drawn to scale, with branch lengths in the same units (number of base substitutions per site) as those of the evolutionary distances used to infer the phylogenetic tree.
Prevalence of GT-42 encoding genes in C. coli population
Raw reads from 2,582 genomes submitted as C. coli were retrieved from European Nucleotide Archive (ENA) and classified into one of the three major C. coli phylogenetic clades based on atpA phylogeny and hierBAPS clustering (Supplementary Fig. S1). A total of 29 genomes were excluded from further analyses, as atpA phylogenetic analysis confirmed them to be C. jejuni. Altogether, 2,432 (95%) genomes belonging to Clade 1, 40 (1.6%) to Clade 2 and 81 (3.2%) to Clade 3 were mapped against all the sequences classified into the 7 C. coli GT-42 groups. A total of 818 (32%) C. coli genomes were positive for at least one GT-42 encoding gene (Table 2; Supplementary Table S4). GT-42 genes were found in approximately one third of C. coli Clade 1 (774/2,432; 31.8%). Furthermore, GT-42 genes were underrepresented in C. coli Clade 2 (2/40; 5%; P < 0.0001), while overrepresented in Clade 3 (42/81; 52%; P < 0.001). Overall, cluster B GT-42 genes (cstIV, cstV and cstVI) were the most abundant GT-42 detected in the C. coli population, accounting for 84.2% of the alleles. Conversely, cluster A GT-42 genes (cstI, cstII, cstIII and cstVII) only represented 15.8% of the alleles (Table 2). The most abundant C. coli GT-42 was cstVI, whereas cstIII was the rarest. C. coli Clade 1 strains were overrepresented in cstVII and cstVI, and underrepresented in cstV (P < 0.01). Conversely, Clade 3 strains were underrepresented in cstVII and cstVI, and overrepresented in cstV (P < 0.01).
Table 2.
Distribution of GT-42 genes among C. coli clades.
| C. coli | GT-42 Cluster A | GT-42 Cluster B | Total | |||||
|---|---|---|---|---|---|---|---|---|
| cstI (G1) | cstII (G4) | cstIII (G2) | cstVII (G3) | cstIV (G7) | cstV (G5) | cstVI (G6) | ||
| Clade 1 | 2 | 22 | 2 | 91 | 267 | 0 | 414 | 798 (94%) |
| Clade 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 (0.2%) |
| Clade 3 | 10 | 5 | 0 | 0 | 14 | 15 | 5 | 49 (5.8%) |
| Total | 12 (1.41%) | 27 (3.19%) | 4 (0.47%) | 91 (10.73%) | 281 (33.10%) | 15 (1.77%) | 419 (49.35%) | 849 (100%) |
C. coli LOS classes contain GT-42 encoding genes
To predict the LOS locus composition of GT-42 positive strains, genomes were mapped against all genes from all known LOS locus classes28. Results are available in Supplementary Table S4. The presence of GT-42 gene alleles from cluster B was strongly concordant with predicted LOS locus classes. For cstVI positive strains, 99% were predicted to have a LOS locus class III-like. Similarly, 93% of cstV positive C. coli possessed a LOS locus class IX-like, and 68% of cstIV positive strains harboured a LOS locus class II-like. Contrastingly, genomes exclusively positive for cluster A GT-42 genes had no significant match to any of the previously defined LOS locus classes (Supplementary Table S4).
To determine the exact genetic composition and synteny of the LOS loci, 261 GT-42 positive genomes were assembled and manually inspected. The data set included all Clade 2 and 3 strains and a selection of Clade 1 strains comprising all cstI, cstII, cstIII, and cstVII positive strains, and a subset of randomly selected cstIV and cstVI positive strains (Supplementary Table S4). Annotation of the identified LOS locus classes is available in Supplementary Table S5. Apart from cstI and cstVII, all GT-42 genes were found within the LOS locus. Among assembled genomes, 61.3% (160) were found to contain a LOS-associated GT-42 gene. Besides the three previously described LOS locus classes containing GT-42 genes (i.e. classes II, III, and IX), 23 novel classes were identified (Fig. 2). LOS class III was the most abundant accounting for 72 isolates, followed by II (39), XXIII (9), XXV (8), XXVI (5), XXX (4), XVI (2), XXVIII (2), and XXXIII (2). The rest of the classes (17) were represented by a single strain. A strong association between LOS locus composition, C. coli Clade, and GT-42 gene alleles, was observed. In general, C. coli Clade 1 exhibited lower LOS locus diversity compared to the other clades. In Clade 1, genomes positive for cstIV and cstVI (88.9% of the total) possess LOS locus classes II and III, respectivelly, with 99% nt sequence identity. In all cases cstVI was present as a pseudogene. Contrastingly, Clade 3 C. coli evince a larger genetic variability in LOS locus classes containing cstIV (8 classes), cstV (3), or cstVI (2). Interestingly, no pseudogenes were found.
Figure 2.
C. coli LOS classes containing GT-42 genes. Arrows represent open reading frames. White arrows: genes putatively unrelated to biosynthesis and transfer of Neu5Ac. Grey arrows: conserved genes. Purple arrows: sialic acid biosynthesis genes always present neuB, neuC, and neuA order. Pink arrows: GT-42 orthologues from group 2. Dark blue arrows: GT-42 orthologues from group 4. Light blue arrows: GT-42 orthologues from group 5. Dark green arrows: GT-42 orthologues from group 6. Light green arrows: GT-42 orthologues from group 7. Striped genes are fragmented. Representation of LOS class II was adapted to reflect origin from LOS class XXXIV. Gene size is not drawn to scale.
Albeit the rarity of LOS associated cluster A GT-42 genes (cstII and cstIII) in C. coli population (1.2%), several distinct LOS locus classes were identified (Fig. 2a). Out of the ten LOS classes containing cstII (Fig. 2a), only XXIII was detected in multiple Clade 1 (7) and Clade 3 (3) strains. Meanwhile, cstIII was located in two different LOS locus classes in C. coli Clade 2 strains.
All LOS locus classes containing cstII, cstIII, or cstV were positive for neuABC genes. Contrastingly, only 6.44% and 4.51% of cstIV and cstVI positive strains, respectively, contained neuABC genes which were invariably located outside the LOS locus and frequently in association with cstI or cstVII.
Gene flow and evolution of GT-42 containing LOS locus classes in C. coli
Based on orthologue group delineation by Roary (>95% amino acid identity), strains belonging to different C. coli clades were shown to share LOS-associated orthologues (Fig. 3). Hence, suggesting gene flow of LOS genes across C. coli clades. Interestingly, most of the share orthologues between clades encode proteins putatively involved in sugar biosynthesis or sugar modification (Table 3).
Figure 3.
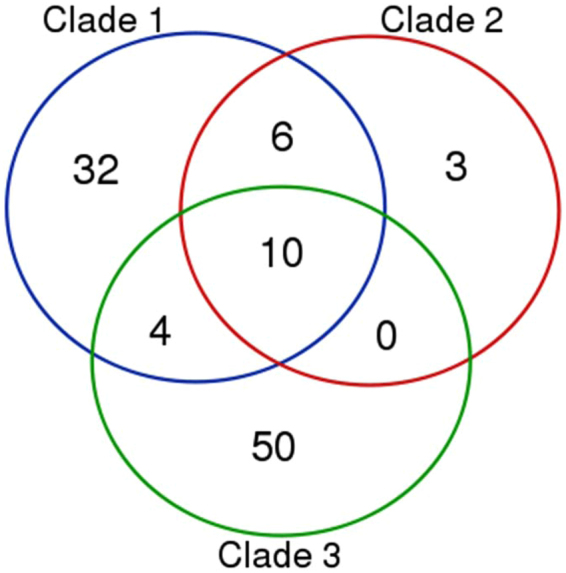
Venn diagram illustrating the number of orthologues shared between C. coli major clades. 10 orthologue were found in all three C. coli clades.
Table 3.
Group of orthologues shared among C. coli clades.
| Roary Orthologue | Prokka annotation | 1 | 2 | 3 |
|---|---|---|---|---|
| 4026 | dTDP-glucose 4,6-dehydratase | + | + | + |
| 4027 | Glucose-1-phosphate thymidylyltransferase | + | + | + |
| 4078 | TDP-4-oxo-6-deoxy-alpha-D-glucose-3,4-oxoisomerase | + | + | + |
| 4014 | UDP-N-acetylglucosamine 2-epimerase | + | + | + |
| 4155 | N,N′-diacetyllegionaminic acid synthase | + | + | + |
| 4077 | Polysialic acid O-acetyltransferase | + | + | + |
| 4042 | UDP-glucose 6-dehydrogenase | + | + | + |
| 4041 | UDP-glucose 4-epimerase | + | + | + |
| 4076 | UDP-glucose 4-epimerase | + | + | + |
| 4075 | UDP-galactopyranose mutase | + | + | + |
| 4003 | General stress protein A | + | + | − |
| 4002 | GalNAc-alpha-(1->4)-GalNAc-alpha-(1->3)-diNAcBac-PP-undecaprenol alpha-1,4-N-acetyl-D-galactosaminyltransferase | + | + | − |
| 4004 | putative glycosyltransferase EpsJ | + | + | − |
| 4051 | putative glycosyltransferase EpsJ | + | + | − |
| 4059 | N-acylneuraminate cytidylyltransferase | + | + | − |
| 4052 | N-acylneuraminate cytidylyltransferase | + | + | − |
| 4127 | hypothetical protein | + | − | + |
| 4126 | hypothetical protein | + | − | + |
| 4132 | hypothetical protein | + | − | + |
| 4157 | hypothetical protein | + | − | + |
Insights into the evolution of C. coli GT-42 containing LOS classes were gain by comparing Clade 1 with Clade 3 LOS classes. Reciprocal blastn analysis between LOS locus classes II (Clade 1) and class XXXIV (Clade 3) showed ~88% nucleotide identity over ~99% of length. Likewise, the terminal part of LOS class III showed high similarity (>90% nucleotide identity) to LOS classes XXVII and XXVIII. Notably, in both Clade 1 LOS locus classes gene pseudogenization was observed: the phosphoethanolamine transferase genes (eptC) in class II, and cstVI in class III. Thus, LOS locus classes II and III plausibly originated from Clade 3 LOS classes and underwent a diversification process (including pseudogenization) and clonally expanded as a consequence of adaptation to the agricultural niche.
Prevalence of GT-42 genes in C. jejuni
Prevalence of GT-42 homologues in C. jejuni was investigated by mapping 12,391 genome sequences deposited as C. jejuni against the 7 C. coli GT-42 groups. A total of 61.15% of the putative C. jejuni genomes were positive for at least one gene. Unsurprisingly, cstII and cstIII were the most abundant representing 95.75% of the GT-42 sequences detected. The remaining gene groups were either present in a minority of the tested genomes (cstIV, 101; cstVI 211; cstVII, 2) or non-detected (cstV). Genomes positive to GT-42 sequences other than cstII and cstIII were assembled for species verification and to manually inspect the LOS locus gene composition. Only 52 (16.6%) genomes were confirmed as C. jejuni by INNUca (Supplementary Table S6), 40 of which (77%) were positive for cstIV, 10 to cstVI (19.2%), and 2 to cstVII (3.8%). Similarly to C. coli, C. jejuni cstVII was located outside the LOS locus and downstream from neuABC genes.
Introgression between C. jejuni and C. coli affect GT-42 containing LOS classes
High similarity between C. coli and C. jejuni cstII-associated LOS locus was observed (i.e. >80% gene lengths and >95% of nucleotide identity), implying recent gene flow between the two species. In fact, C. coli LOS classes XVI, XVII, XVIII, XIX, XX, XXI, and XXII are a mosaic of C. jejuni LOS classes A, B, S/F, and I/D. C. coli LOS class XXIV is further evidence of admixture between the two species, as it includes the C. coli specific cstV and neuB orthologues, as well as the C. jejuni cstII and neuB copies (Supplementary Table S4, Fig. 2). Finally, cross-species mobilization of an entire LOS locus classes was also encountered. C. coli strain SRR5152313 carries C. jejuni 11168 LOS class C, and 35 out of 40 cstIV positive C. jejuni strains, 37.1% of which from MLST sequence type 459, harbour a C. coli LOS locus class II.
Discussion
The small number of GBS associated C. coli isolates and the supposedly absence of molecular machinery for ganglioside mimicry are the main reasons, so far, supporting the idea of no link between C. coli and GBS. In 1994 von Wulffen and colleagues reported the first C. coli isolated from a GBS patient in a comparative seroreactivity study30. The C. coli strain in question exhibited a Lior type 11 phenotype, which had also been found in GBS-associated C. jejuni strains. Thus, in the following years C. coli was considered as a plausible GBS causing organism30. However, after recognition of C. jejuni expressing ganglioside-like LOS as the infectious agent triggering GBS, testing for cross-reactivity with anti-ganglioside autoantibodies became critical in understanding GBS aetiology. So far, insufficient evidence supporting a causal relationship between C. coli and GBS has been found since studied GBS-associated C. coli strains have been unreactive to monoclonal anti-ganglioside antibodies24,25. Furthermore, although the GBS-associated C. coli strain 664H2004 has been shown to carry a cstII orthologue and di-sialylated LOS, no further evidence suggesting expression of ganglioside mimics was attained, as authors failed to genetically and structurally characterize C. coli 664H2004 LOS26.
In the present study, 16 C. coli LOS locus classes (Fig. 2) were shown to contain the essential molecular machinery to potentially express sialylated LOS (i.e. a cst homologue and neuABC). While genotype is generally insufficient to predict LOS structure3,8,19, considerable evidence supporting the expression of ganglioside-like LOS in C. coli was found. In contrast to previous reports27,28, C. coli LOS locus may be substantially affected by introgression with C. jejuni. Herein, 10 C. coli LOS locus classes containing a cstII were demonstrated to be mosaics of C. jejuni LOS classes. C. jejuni strains carrying cstII containing LOS classes have hitherto rarely being found to express non-ganglioside sialylated LOS3,8,9,18,21. Furthermore, extreme introgression resulted in acquisition of the entire C. jejuni LOS class C in C. coli SRR5152313 (100% homology). Consequently, this strain, isolated from turkey in US in 2016, could potentially trigger GBS as most likely expresses a GM1a- or GM2-like LOS9.
However, it is to be noted that strains carrying C. jejuni-like LOS locus are a minority in the C. coli population (approximately 1% of sequenced strains). Most of the C. coli possessing GT-42 genes (i.e. cstIV and cstVI) carry LOS classes lacking neuABC genes (approximately 27% of the sequenced strains). Furthermore, genome-wise analysis failed to identify genes potentially linked to the synthesis of CstIV and CstVI sugar donors. Even though functional studies are needed to clarify the activity of CstIV and CstVI, it seems plausible to believe that these elements are not involved in LOS ganglioside mimicry based on the results presented here and the absence of Neu5Ac in the LOS of cstIV positive strains28. Thus, the infrequency of the genetic structures related to ganglioside mimicry in the population might be the reason behind C. coli little contribution to GBS incidence26.
Beside ganglioside mimicry and the pathogenesis of GBS, expression of sialylated structures has a strong impact on host-bacteria interaction10. In our broad-gauge screening, we have shown that a considerable proportion of C. coli strains carry GT-42 genes within the LOS locus (29% of C. coli deposited in ENA at the time of writing). Overall, 23 new GT-42 associated LOS classes were described, 15 of which were present exclusively in the non-agriculture C. coli belonging to Clade 3. Thus, underrepresentation of non-agricultural C. coli strains27 in studies characterizing the LOS loci of extensive strain collections20,28,29 probably hampered earlier identification of a wider diversity of LOS classes with GT-42 genes.
We also discovered that LOS locus classes II and III28, the most predominant among agriculture-adapted Clade 1 C. coli, most likely originated from non-agriculture Clade 3 LOS classes. Moreover, few genes in both classes, including the GT-42 cstVI, lost their function in Clade 1. Cell surface structural changes as result of natural selection is a dominant phenomenon in microbial evolution. In pneumococcus, for example, natural selection as a consequence of vaccination programs targeting polysaccharide structures has resulted in shifts in the population of nonvaccine-type strains31. Outer membrane or wall-associated structures in bacteria (i.e. oligo and polysaccharides and proteins) play also a fundamental role in host interaction. Thus, they are subjected to diversifying selective pressure to conform to distinct receptors in different host species32. Moreover, reductive evolution leading to functional loss of several genes through e.g. pseudogenization is a common feature of bacterial undergoing niche adaptation32. For example, a single naturally occurring nucleotide mutation responsible for the inactivation of a gene essential for D-alanylation of teichoic acids, has been shown to be sufficient to convert a human-specific Staphylococcus aureus strain into one that could infect rabbits33. Introduction of the agricultural niche was key in the evolution of C. coli clades34: clade 1 expanded within this niche and underwent an extensive genome introgression with C. jejuni27.
Therefore, it is tempting to speculate that gene loss within imported LOS classes II and III, may have played a significant role in the expansion of C. coli in the agricultural niche by shaping the outer membrane composition. This hypothesis is supported by two pieces of evidence: (i) the predominance of LOS classes II and III in C. coli Clade 1 generalist (i.e. multihost) strains28 and (ii) the strong purifying selection resulting in limited nucleotide variability in these LOS locus classes (>99% identity. The importance of LOS locus classes II and III in adaptation to the agricultural niche is further evidenced by the flow of these genetic elements between C. coli and agricultural C. jejuni. Although introgression between C. jejuni and C. coli has been considered to be unilateral until now27, we identified several C. jejuni strains carrying LOS classes typically detected in C. coli Clade 1. Most of the C. jejuni strains carried a LOS class II with 99% identity, while some other presented a mosaic of Clade 3 LOS classes containing cstIV. As described previously22, a strong association between MLST type and LOS class was observed, being the bovine associated ST-45935 the most prevalent among the C. jejuni carrying C. coli LOS class II.
Conclusion
Although at extremely low frequencies, bacterial factors implicated in GBS aetiology can cross clade and species barriers. Furthermore, spreading of these factors in the population could potentially result in C. coli playing a more prominent role in GBS. C. coli also presents a larger GT-42 enzyme repertoire than C. jejuni. Nevertheless, the activity of these enzymes and their role shaping C. coli population is yet to be explored. Overall, C. coli glycobiology is largely unknown in spite of being a major foodborne pathogen.
Methods
Genome sequences mining, genes detection and allele calling
All whole genome raw sequence reads of entries deposited in the ENA as either Campylobacter coli or Campylobacter jejuni at the time of analysis (August 2017) were mapped against a set of reference genes (see below) for performing variant calling and inferring presence or absence using the ReMatCh framework v3.2 (https://github.com/B-UMMI/ReMatCh)36. Briefly, ReMatCh interacts with ENA for extracting and downloading all publicly available raw IlluminaTM reads in fastq format for a given taxon. Then, it maps the reads onto the desired target loci using Bowtie237, and performs variant calling with Samtools/Bcftools38 and ReMatCh Single Nucleotide Polymorphism call criteria. The minimum coverage depth to consider a position to be present in the alignment was fixed at 5 reads, and to perform allele calling the threshold was 10 reads. A locus was considered to be present if 1) at least 70% of the target reference gene sequence was successfully mapped and 2) if the consensus sequence was ≥80% identity at nucleotide level. When needed, the consensus sequence alignment was extracted using the script combine_alignment_consensus.py available in ReMatCh utilities.
Identification and frequency of C. coli GT-42 homologues
To collect a set of C. coli reference genes homologous to C. jejuni GT-42 encoding genes, amino acid sequences of CstI (Uniprot Q9RGF1), CstII (Uniprot Q9F0M9) and CstIII (Uniprot Q7BP25) were used to search non-redundant (nr) NCBI protein sequences collection using blast + V 2.7.139 for best C. coli blastp hits (>30% of amino acid identity; >50% query coverage). Partial sequences were discarded and the remaining ones were used for an all-versus-all blastp analysis. Sequences were then categorized in separate groups having >0.7 of BSR40. A Minimum Evolution phylogenetic tree based on the back-translated nucleotide sequence alignments (built with MUSCLE41 with default parameters) of all detected C. coli GT-42 proteins and C. jejuni cstI, cstII, and cstIII was inferred using MEGA742. Finally, the detected C. coli GT-42 nucleotide sequences were used as reference for calling orthologues in all C. coli and C. jejuni strains using ReMatCh as described above.
Identification of Campylobacter coli clades
To assign C. coli samples to one of the three previously described major phylogenetic clades27,29, population structure analysis and inferred phylogenetic relationships based on atpA gene43 were performed. The atpA sequence of C. coli strain RM2228 (KF855277) was used for allele calling in all C. coli strains using ReMatCh as described above. Based on the ReMatCh atpA consensus sequence alignment, samples were clustered using hierBAPS44 at first level and a Neighbor joining phylogenetic tree was inferred using MEGA742. Representative strains from each C. coli clade27,29 were used as reference for classifying the clusters, and a set of C. jejuni strains were used as outgroup. The generated tree was visualized in iTOL45.
Classification into LOS classes
To assign samples to one of the previously described C. coli LOS locus classes, nucleotide sequences of loci located between the “conserved putative two-domain glycosyltransferase” (orthologue 16 as described previously20,28) and the “LOS biosynthesis glycosyltransferase waaV” (orthologue 10 described previously20,28) from C. coli LOS locus classes I to XII28 were used for calling orthologues in all GT-42 positive C. coli using ReMatCh, as described above. Results were reported as percentage of genes present for a given LOS locus class.
Pangenome analysis and gene flow investigation of LOS loci
For a set of C. coli strains of interest, raw sequencing data were retrieved from ENA with getSeqENA (https://github.com/B-UMMI/getSeqENA). Then, the paired-end raw reads were assembled using the INNUca pipeline (https://github.com/INNUENDOCON/INNUca), which consists of several modules and QA/QC steps. In brief, INNUca starts by calculating if the sample raw data fulfill the expected coverage (min 15×). After subjecting reads to quality analysis using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and cleaning with Trimmomatic46, INNUca proceeds to de novo draft genome assembly with SPAdes 3.1147 and checking assembly depth of coverage (min 30×). Finally, Pilon48 improves the draft genome by correcting bases, fixing misassembles, and filling gaps, prior species confirmation and MLST prediction with mlst software (https://github.com/tseemann/mlst).
Draft genomes passing INNUca QA/QC were annotated with Prokka49, and pangenome analysis was executed using Roary50 (default parameters). To annotate novel LOS locus classes, assemblies were manually inspected with Artemis51.
Horizontal Gene Transfer (HGT) among C. coli clades was inferred by mapping presence/absence of LOS associated group of orthologues into the atpA tree (see above). To infer possible gene transfer between C. coli and C. jejuni, representative sequences of LOS associated group of orthologues were blastn against nt NCBI database and HGT was detected if the best blast hit for C. jejuni was >90% nucleotide identity over >70% of the C. coli query length.
Statistical analysis
Fisher’s exact test was used to assess clade and GT-42 associations. P values of ≤0.05 were considered significant.
Data availability
Data are available in Supplementary Information.
Electronic supplementary material
Acknowledgements
This study was funded by the following grants; University of Helsinki three years research grant 313/51/2013, ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI - “Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020” and by national funds from FCT - “Fundação para a Ciência e a Tecnologia” and BacGenTrack (TUBITAK/0004/2014) [FCT/ Scientific and Technological Research Council of Turkey (Türkiye Bilimsel ve Teknolojik Araşrrma Kurumu, TÜBITAK)]”. A. C was supported by the Microbiology and Biotechnology graduate program from the University of Helsinki. The authors wish to thank CSC- Tieteen tietotekniikan keskus Oy for providing access to cloud computing resources.
Author Contributions
A.C. designed and coordinated the study. A.C. and M.R. performed data analysis, prepared figures, and wrote the manuscript. J.A.C. and M.P.M. design and developed INNUca and ReMatCh. All authors have contributed to data interpretation, have critically reviewed the manuscript, and approved the final version as submitted.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21438-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cress BF, et al. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol. Rev. 2014;38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlin AF, et al. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houliston RS, et al. Lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J. Biol. Chem. 2011;286:12361–12370. doi: 10.1074/jbc.M110.181750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tam CC, et al. Incidence of Guillain-Barré syndrome among patients with Campylobacter infection: a general practice research database study. J. Infect. Dis. 2006;194:95–97. doi: 10.1086/504294. [DOI] [PubMed] [Google Scholar]
- 5.Yuki N. Carbohydrate mimicry: a new paradigm of autoimmune diseases. Curr. Opin. Immunol. 2005;17:577–582. doi: 10.1016/j.coi.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Goodfellow JA, Willison HJ. Guillain-Barre syndrome: a century of progress. Nat. Rev. Neurol. 2016;12:723–731. doi: 10.1038/nrneurol.2016.172. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert M, et al. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384 identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-MHz 1H AND 13C NMR analysis. J. Biol. Chem. 2000;275:3896–3906. doi: 10.1074/jbc.275.6.3896. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert M, et al. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 2002;277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]
- 9.Godschalk PCR, et al. The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré syndrome. J. Clin. Invest. 2004;114:1659–1665. doi: 10.1172/JCI200415707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis AL, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. 2009;106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, et al. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell J, Davies G, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1998;329:719. doi: 10.1042/bj3290719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 15.Blixt O, et al. Chemoenzymatic synthesis of 2-azidoethyl-ganglio-oligosaccharides GD3, GT3, GM2, GD2, GT2, GM1, and GD1a. Carbohydr. Res. 2005;340:1963–1972. doi: 10.1016/j.carres.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Zuccolo AJ, Li W, Zheng RB, Ling C-C. Probing a sialyltransferase’s recognition domain to prepare α(2,8)-linked oligosialosides and analogs. Chem. Commun. 2009;0:4233–4235. doi: 10.1039/b908933k. [DOI] [PubMed] [Google Scholar]
- 17.Yu C-C, et al. A plate-based high-throughput activity assay for polysialyltransferase from Neisseria meningitidis. Anal. Biochem. 2014;444:67–74. doi: 10.1016/j.ab.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Parker CT, et al. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 2005;43:2771–2781. doi: 10.1128/JCM.43.6.2771-2781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J. Bacteriol. 2008;190:5681–5689. doi: 10.1128/JB.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards VP, Lefébure T, Pavinski Bitar PD. & Stanhope, M. J. Comparative characterization of the virulence gene clusters (lipooligosacharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013;14:200–213. doi: 10.1016/j.meegid.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert, M., Parker, C. T. & Moran, A. P. Campylobacter jejuni lipooligosaccharides: structures and biosynthesis. 483–504, 10.1128/9781555815554.ch27 (2008).
- 22.Revez J, Hänninen M-L. Lipooligosaccharide locus classes are associated with certain Campylobacter jejuni multilocus sequence types. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:2203–2209. doi: 10.1007/s10096-012-1556-3. [DOI] [PubMed] [Google Scholar]
- 23.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funakoshi K, Koga M, Takahashi M, Hirata K, Yuki N. Campylobacter coli enteritis and Guillain-Barré syndrome: no evidence of molecular mimicry and serological relationship. J. Neurol. Sci. 2006;246:163–168. doi: 10.1016/j.jns.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Bersudsky M, Rosenberg P, Rudensky B, Wirguin I. Lipopolysaccharides of a Campylobacter coli isolate from a patient with Guillain–Barré syndrome display ganglioside mimicry. Neuromuscul. Disord. 2000;10:182–186. doi: 10.1016/S0960-8966(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 26.van Belkum A, et al. Can Campylobacter coli induce Guillain-Barré syndrome? Eur. J. Clin. Microbiol. Infect. Dis. 2009;28:557–560. doi: 10.1007/s10096-008-0661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheppard SK, et al. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol. Ecol. 2013;22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culebro A, et al. Large sequence diversity within the biosynthesis locus and common biochemical features of Campylobacter coli lipooligosaccharides. J. Bacteriol. 2016;198:2829–2840. doi: 10.1128/JB.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skarp-de Haan CP, et al. Comparative genomics of unintrogressed Campylobacter coli clades 2 and 3. BMC Genomics. 2014;15:129. doi: 10.1186/1471-2164-15-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wulffen H, Hartard C, Scharein E. Seroreactivity to Campylobacter jejuni and gangliosides in patients with Guillain-Barré syndrome. J. Infect. Dis. 1994;170:828–833. doi: 10.1093/infdis/170.4.828. [DOI] [PubMed] [Google Scholar]
- 31.Cohen T, Colijn C, Murray M. Modeling the effects of strain diversity and mechanisms of strain competition on the potential performance of new tuberculosis vaccines. Proc. Natl. Acad. Sci. USA. 2008;105:16302–16307. doi: 10.1073/pnas.0808746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guinane CM, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2010;2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viana D, et al. A single natural nucleotide mutation alters bacterial pathogen host-tropism. Nat. Genet. 2015;47:361–366. doi: 10.1038/ng.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard SK, et al. Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from national surveillance data in Scotland. PLOS ONE. 2010;5:e15708. doi: 10.1371/journal.pone.0015708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha W, et al. Comparing the genetic diversity and antimicrobial resistance profiles of Campylobacter jejuni recovered from cattle and humans. Front. Microbiol. 2017;8:818. doi: 10.3389/fmicb.2017.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machado MP, Ribeiro-Gonçalves B, Silva M, Ramirez M, Carriço JA. Epidemiological surveillance and typing methods to track antibiotic resistant strains using high throughput sequencing. Methods Mol. Biol. Clifton NJ. 2017;1520:331–355. doi: 10.1007/978-1-4939-6634-9_20. [DOI] [PubMed] [Google Scholar]
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller WG, Yee E, Jolley KA, Chapman MH. Use of an improved atpA amplification and sequencing method to identify members of the Campylobacteraceae and Helicobacteraceae. Lett. Appl. Microbiol. 2014;58:582–590. doi: 10.1111/lam.12228. [DOI] [PubMed] [Google Scholar]
- 44.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker BJ, et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 50.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinforma. Oxf. Engl. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in Supplementary Information.



