Abstract
Xenotransplantation is a promising strategy to alleviate the shortage of organs for human transplantation. In addition to the concern on pig-to-human immunological compatibility, the risk of cross-species transmission of porcine endogenous retroviruses (PERVs) has impeded the clinical application of this approach. Earlier, we demonstrated the feasibility of inactivating PERV activity in an immortalized pig cell line. Here, we confirmed that PERVs infect human cells, and observed the horizontal transfer of PERVs among human cells. Using CRISPR-Cas9, we inactivated all the PERVs in a porcine primary cell line and generated PERV-inactivated pigs via somatic cell nuclear transfer. Our study highlighted the value of PERV inactivation to prevent cross-species viral transmission and demonstrated the successful production of PERV-inactivated animals to address the safety concern in clinical xenotransplantation.
The shortage of human organs and tissues for transplantation represents one of the most significant unmet medical needs (1). Xenotransplantation holds great promise. Porcine organs are considered favorable resources for xenotransplantation since they are similar to human organs in size and function, and can be bred in large numbers (2).
However, the clinical use of porcine organs has been hindered by immunological incompatibilities (2) and by the potential risk of PERV transmission (3). PERVs are gamma retroviruses found in the genome of all pig strains and can be vertically transferred through inheritance (4). Although to date, no study has shown PERV transmission to humans in clinical settings, it has been demonstrated that PERVs can infect human cells (3,5) and integrate into human genome in cell culture (6). PERV integration could potentially lead to immunodeficiency and tumorigenesis, as reported with other retroviruses (7, 8).
We recently demonstrated a method to inactivate all 62 copies of PERVs in an immortalized porcine cell line (PK15) and eliminated PERV transmission to human cells (5). In the present study, we adopted a strategy to conduct multiplexed genome engineering to inactivate PERV activity in a primary porcine fibroblast cell line; then we used the modified fibroblasts to produce embryos through somatic cell nuclear transfer (SCNT) and transferred the SCNT embryos into surrogate sows. Using such an approach, we successfully generated PERV-inactivated pigs.
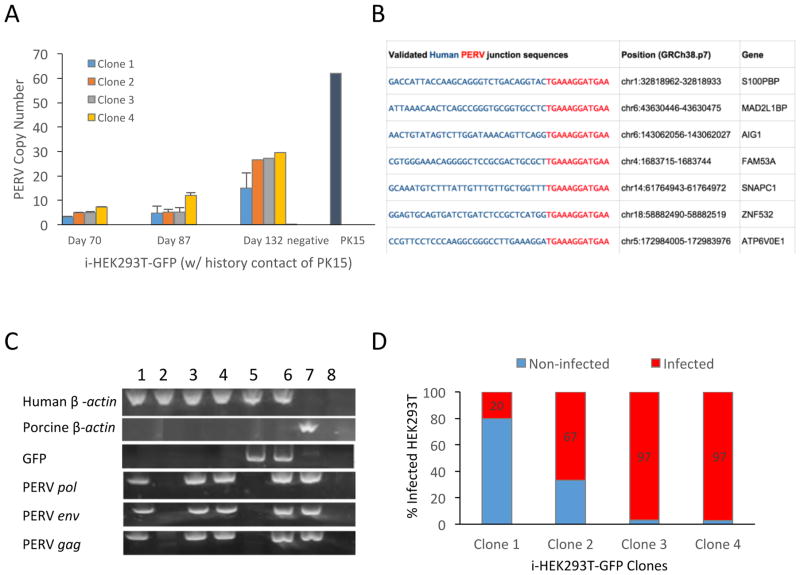
We previously demonstrated the transmission of PERVs from an immortalized pig epithelial cell line, PK15, to GFP-labeled human embryonic kidney cells (HEK293T-GFP), after co-culturing them for one week (5). We wondered whether PERVs remain active and propagate in human cells. To detect this, we monitored PERV copy number both in a population and in clones of PERV-infected HEK293T-GFP cells (i-HEK293T-GFP) for more than 4 months and observed that PERV copy number increased over time (Fig. 1A, S1B), as determined by droplet digital PCR (ddPCR). Consistent with previous reports (3, 9), we detected that both subtypes PERV-A and PERV–B were present in the infected human cells (Fig. S1B), confirming that they are human-tropic. We did not detect PERV-C in either PK15 or i-HEK293T-GFP cells (Fig. S1A, S1B). To determine whether the PERVs integrate into the human genome or stay episomal in the infected human cells, we performed junction capture sequencing of the infected clonal i-HEK293T-GFP cells. We detected novel PERV junctions in the human genome and observed that they are overrepresented in intragenic regions and in active chromatin areas (Fig. 1B, S2A, S2B).
Figure 1. Pig-to-human and human-to-human PERVs transmission.
A) PERVs copy number in infected human cells increases over time when co-cultured with PK15 cells. Human HEK293T-GFP cells were co-cultured with equivalent numbers of pig PK15 cells for one week. HEK293T-GFP without any contact of PK15 cells were used as negative control (negative). B) Detection of PERVs insertion sites in human genome. Among the 22 PERV insertion sites detected by inverse PCR, 15 were mapped to the intragenic region. We tested a portion of the intragenic hits and validated 7 out of 12 by junction PCR (shown here). The 30bp human genomic sequences are shown in blue, whereas the PERV LTRs are shown in red. C) Detection of human-to-human PERVs transmission. Individual clones of HEK293T were grown from the single cells isolated from the co-culture of i-HEK293T-GFP with HEK293T through flow cytometry. The PCR gel image showed that 3 out of 4 randomly tested HEK293T clones were infected, which contained PERVs sequences (PERV pol, env, and gag), but no sequence of GFP or pig genomic DNA (tested by pig specific GGTA). Sample orders are: 1) HEK293T clone 1; 2) HEK293T clone 2; 3) HEK293T clone 3; 4) HEK293T clone 4; 5) HEK293T-GFP control; 6) i-HEK293T-GFP; 7) PK15 WT; and 8) negative. D) Four different i-HEK293T-GFP clones have different infectious potential. Four infected parental i-HEK293T-GFP clones are co-cultured with WT HEK293T. The PERV copy number of the 4 parental i-HEK293T-GFP clones are 15, 28, 27 and 28, respectively. The percentages of the infected WT HEK293T clones from the co-culture of i-HEK293T-GFP and WT HEK293T varied from 20% to 97%. Primers used are listed in the Methods Table 1.
The increased copy number of PERVs in i-HEK293T-GFP clones can be caused by intracellular transposition or by intercellular PERV transmission among human cells. To clarify this, we then examined whether infected human cells could transmit PERVs to wild-type (WT) human cells. We co-cultured clonal i-HEK293T-GFP cells with WT HEK293T cells for two weeks, and subsequently checked PERV elements in the co-cultured WT clones via PCR. We detected the robust presence of PERV elements in WT HEK293T cells with no history of contact with porcine cells (Fig. 1C, S2C). The percentage of infected WT HEK293T cells varies from 20% to 97% (Fig. 1D) depending on different parental i-HEK293T-GFP clones in the co-culture. We concluded from our observations that infected human cells can transmit PERVs to previously unexposed human cells.
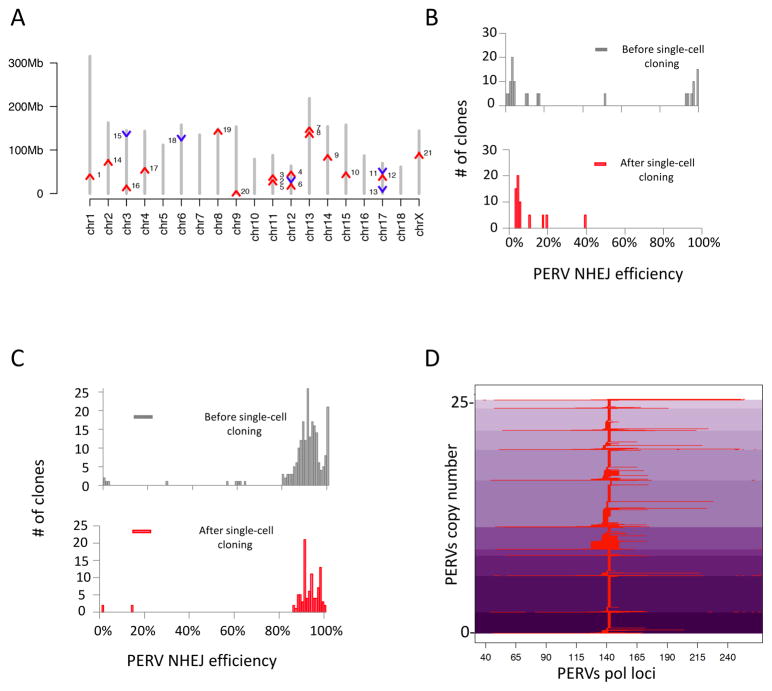
Generating PERV-inactivated pigs involves construction of primary porcine cells devoid of PERV activity, which can be cloned via SCNT to produce porcine embryos. We tested whether we could use the same strategy we used previously in PK15 (5) to inactivate the PERV activities in a primary porcine fetal fibroblast cell line (denoted as FFF3 hereafter). We first aimed to map and characterize the PERVs present in the FFF3 genome. We detected 25 copies of functional PERVs, as determined by ddPCR on the reverse transcriptase (pol) gene (Fig. S3). The detected PERV copy number was close to the sum of 10 copies of the PERV-A env gene, 14 copies of the PERV-B env gene, and 0 copies of the PERV-C env gene that we identified in the genome (Fig. S3) by ddPCR. Using whole genome sequencing, we further detected one additional copy of truncated PERV-B not detectable by ddPCR. We used hybridization capture followed by sequencing to map PERVs copies into the genome (Fig. 2A, Fig. S4). To target these PERVs for inactivation, we designed two CRISPR guide RNAs (gRNAs) specific to the catalytic core of the PERV pol gene (Fig. S5). After treating a population of FFF3 cells with CRISPR-Cas9 and the two gRNAs for 12 days, we observed 37% PERV pol inactivation. Notably, we observed a bimodal distribution of targeting efficiency among single FFF3 cells after treatment, resembling our previous results obtained with PK15 cells (5). About 35% single cells had high editing efficiency (>90û) and 61% single cells had low editing efficiency (<20%) (Fig. 2B). Unfortunately, despite the presence of highly modified cells in the population, we could not grow up the single-cell clones with >90% PERV editing efficiency.
Figure 2. PERVs insertion sites mapping, and genome-wide inactivation.
A) Chromosome mapping of PERVs locations in FFF3. Chromosomal scaffolds are in grey. Red arrows represent PERVs in the forward or positive chain of chromosome. Blue arrows represent PERVs in the reverse or negative chain. Y-axis represents chromosomal coordinates. Two additional copies were mapped to repetitive regions, and two could not be mapped to the current pig genome assembly and are not shown (11% gaps, Sus scrofa build 10.2) (10). B) Failure to obtain 100% PERV-inactivated FFF3 clones using CRISPR-Cas9. After targeting the PERVs in FFF3, single cells were sorted and immediately genotyped. We observed a bimodal distribution of PERV targeting frequencies among single cells (upper panel), similar to that seen in the PK15 clones (5). 100% PERV-inactivated FFF3 cells were present among the single cells directly genotyped. However, this pattern changed after expansion of the single cells (bottom panel). Among the single cell clones, we only obtained the ones with lower efficiency (≤39%, the average targeting efficiency in the population was 37%), but not the ones with 100% PERV inactivation (lower panel). C) Treatment with PFTα and bFGF sustained the growth of highly modified FFF3 clones. The combined use of a p53 inhibitor, PFTα, and a growth factor, bFGF, rescued the highly modified cells. A population of FFF3 was treated with PFTα and bFGF during the gene editing experiment (Methods); then, single cells were sorted for direct genotyping and for colony growth followed by genotyping. Both the single cells and expanded clones showed similar distribution in PERV targeting efficiency, and highly modified clones survived under this condition. D) Genotype of 100% PERV inactivated clones. Several 100% PERV-inactivated clones were achieved from the PFTα and bFGF treated FFF3 population. The figure shows haplotypes of one of the 100% PERV-inactivated clones at PERV pol loci, after CRISPR-Cas9 treatment. The y-axis indicates the edited PERVs loci. The x-axis indicates the relative locations of the indels within the PERV loci. Aligned indel events in the PERV pol sequence are represented in red. Shades of purple indicate different haplotypes of PERVs.
We hypothesized that simultaneous DNA cleavages by Cas9 at multiple PERV sites in the FFF3 genome trigger DNA damage-induced senescence or apoptosis; hence, we could not obtain the highly modified FFF3 clones. Through screening of different anti-apoptotic strategies (Fig. S6, S7), we observed that the application of a cocktail containing p53 inhibitor, pifithrin alpha (PFTα), and basic fibroblast growth factor, bFGF, during genetic modification, significantly increased the average targeting efficiency of the resulting FFF3 populations (Fig. S6A (ANOVA, p=0.00002), Fig. S6B). Using this optimized cocktail, we were able to grow up 100% PERV-inactivated FFF3 cells (PERV-inactivated FFF3) from the population treated with CRISPR-Cas9 (Fig. 2C, 2D).
Having confirmed that we genetically mutated PERV pol in the genome, we performed RNA-seq (Fig. S8) on PERV-inactivated FFF3 clones and confirmed that all pol transcripts had been mutated. Furthermore, we examined whether the genome-wide disruption of PERV pol would eliminate in vitro production of PERVs from FFF3. We could not detect reverse transcriptase (RT) activity of PERVs in the cell culture supernatant of the PERV-inactivated FFF3 (Fig. S9), suggesting that there is no viral particle secreted by the modified cells.
We next sought to examine off-target effects of CRISPR-Cas9 in the PERV-inactivated FFF3. We carried out karyotyping of 8 PERV-inactivated FFF3 and observed that 5 carried chromosomal abnormalities. Of note, the translocation sites in the genome tend to correlate with presence of PERV cutting sites, suggesting that CRISPR-Cas9 on-target toxicity may contribute to the translocations observed (Fig. S10A). The remaining 3 PERV-inactivated FFF3 clones carry normal chromosomal structures (Fig. S10B). To examine chromosomal integrity with higher resolution, we performed PERV genomic junctions sequencing on the 3 normal karyotype clones to examine potential deletions between Cas9-induced double-strand breaks (DSBs). All 21 tested junctions remained intact (Fig. S11), which indicates no Cas9-induced macrodeletions in these regions. Therefore, we concluded that we could obtain PERV-inactivated FFF3 without detectable structural variations.
Having obtained PERV-inactivated FFF3 cells, we attempted to produce PERV-inactivated embryos via SCNT. For every round of embryogenesis, ~20–40% of the constructed PERV-inactivated embryos reached blastocyst stage after being cultured for 6 days (Methods), which is within the normal range of porcine SCNT efficiency. We observed normal 64-cell stage blastocyst structure and validated the pluripotency of inner cell mass (detected by SOX2 antibody) on day 6 (Fig. S12A). We performed genomic deep sequencing to check the PERV pol genotypes in embryos, and confirmed 100% PERV-inactivation efficiency (Fig. S12B).
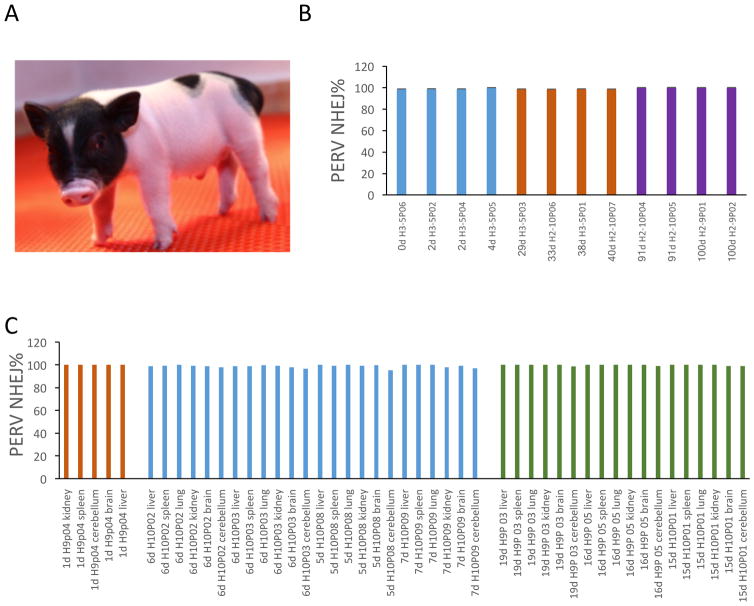
Next, we transferred the PERV-inactivated embryos into surrogate sows which are PERV-C free and present a total PERV copy number of 12–30 (Fig. S13A). We detected pregnancy rates of 75% (33/44), 63% (28/44), and 52% (23/44) for PERV-inactivated FFF3 cells at day 23, 51 and 70, respectively, and 100% (5/5) for WT cells at all the three time points. Fetuses at pregnancy date 50 were analyzed, all PERV-inactivated fetuses showed 100% PERV inactivation (Fig. S12C, S12D) and similar PERV copy number as WT FFF3 (Fig. S13B). Despite the lower pregnancy efficiency, which is commonly observed in transgenic pig production (11), we successfully produced putative PERV- inactivated pigs (Fig. 3A, Fig. S14). We did not observe difference between PERV-inactivated and WT cells on the SCNT efficiency. The ratios of piglets born/number of embryos transferred are similar for PERV-inactivated cells (0.9%) and WT cells (0.8%). To test PERV inactivation in these pigs, we isolated genomic DNA from both the bulk cells and single cell clones derived from these pigs and observed that all the pigs exhibited ~100% PERV inactivation at the genomic DNA level (Fig. 3B, S15A, S15B). In addition, we observed that the copy number of PERVs in the generated pigs stays close to 25, reconfirming that there is no reinfection (Fig. S15C). We further isolated total RNAs from a variety of tissues of the pigs and confirmed ~100% PERV inactivation at the mRNA level (Fig. 3C). We performed karyotyping and did not detect abnormal structural changes in the PERV-inactivated pigs (Fig. S16). The PERV-inactivated pig production is robust that we have so far produced 37 PERV-inactivated piglets from 17 sows (200–300 embryo transferred/sow), 15 piglets remain alive, and the oldest healthy animals are 4-month old. We are conducting long term studies to monitor the impact of PERV-inactivation and gene editing on big animals.
Figure 3. PERV-inactivated pigs.
A) Image of the first born PERV-inactivated pig. This picture showed the first born pig (Laika) at day 2 after birth. B) PERV inactivation at genomic DNA level. We genotyped PERV-inactivated pigs at different ages (up to 100 days) by deep sequencing of the PERV pol loci. All examined pigs showed ~100% PERV inactivation efficiency, which demonstrates that there is no detectable PERV reinfection from surrogate sows to cloned pigs. C) PERV inactivation at mRNA level. Total mRNA generated cDNA was used to detect the PERV inactivation efficiency of the PERV-ko pig transcripts. All pigs exhibited ~100% PERV eradication efficiency at mRNA level.
In summary, we observed in our studies that PERVs can be transmitted from pig to human cells and transmitted among human cells in vitro. These results substantiate the risk of cross-species viral transmission in the context of xenotransplantation. To work towards eliminating this risk, we generated PERV-inactivated primary porcine cell lines using a combination of CRISPR-Cas9, apoptosis inhibitor and growth factor, and with these cell lines, we produced PERV-inactivated porcine embryos, fetuses, and live pigs.
In this study, we discovered that treatment with p53 inhibitor can mitigate the stress from multiplex DNA damage during multiplexible genome engineering and support clonal expansion of 100% PERV-inactivated cells. Although we have focused in this paper on the applications to xenotransplantation, we envision, more generally, that the synergistic combination of CRISPR-Cas technology with anti-apoptosis treatment may also be used to enable large-scale genome engineering in primary cells for a broad range of applications, including pathway engineering and modifications of other genetic repetitive elements of biological interest.
Although it is still unclear whether PERVs infect humans in vivo, our study shows that PERV-infected human cells pass the PERVs robustly to fresh human cells that have no prior-exposure to pig cells. Therefore, our data substantiates the value of PERV-inactivation for safe xenotransplantation practice. The physiological functions of endogenous retrovirus, which exists in all mammalian species, remain largely unknown. Further studies on our PERV-inactivated pigs will shed light on the endogenous retrovirus functionalities in related to the hosts. Most importantly, the PERV-inactivated pig can serve as a foundation pig strain, which can be further engineered to provide safe and effective organ and tissue resources for xenotransplantation.
Supplementary Material
Acknowledgments
We thank J. Markmann from Massachusetts General Hospital and P. O’connell from Sydney University for providing advices on xenotransplantation; J. Hu from the Alt lab at Boston Children’s’ Hospital for assistance with junction capture sequencing; Y. Takeuchi from University College London for his insightful comments on virus interference; T. Ferrante from WYSS Institute, Harvard University for his assistance with embryo imaging and members of the Seidman lab at Harvard Medical School for advice regarding ddPCR. This study is mainly funded by eGenesis Inc. and was funded by NIH grant P50 HG005550. Y.L. was funded by Danish Research Council for Independent Research (DFF-1337-00128) and Sapere Aude Young Research Talent Prize (DFF-1335-00763). M.G. was funded by a Human Frontiers Science Program Long Term fellowship. Some of the pig production was funded by Major Program on Basic Research Projects of Yunnan Province, China (Grant No. 2014FC006). PERV elements genotyping illumina miseq data have been uploaded to the European Nucleotide Archive (ENA) hosted by the European Bioinformatics Institute (EBI) with the submission reference PRJEB11222.
References
- 1.Shafran D, Kodish E, Tzakis A. Organ shortage: the greatest challenge facing transplant medicine. World J Surg. 2014;38:1650–7. doi: 10.1007/s00268-014-2639-3. [DOI] [PubMed] [Google Scholar]
- 2.Deschamps JY, Roux FA, Saï P, Gouin E. History of xenotransplantation. Xenotransplantation. 2005;12:91–109. doi: 10.1111/j.1399-3089.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 3.Patience C, Takeuchi Y, Weiss RA. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–6. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 4.Denner J. How active are porcine endogenous retroviruses (PERVs)? Viruses. 2016;8:216. doi: 10.3390/v8080215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–4. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 6.Moalic Y, et al. Porcine endogenous retrovirus integration sites in the human genome: features in common with those murine leukemia virus. J Virol. 2006;80:10980–8. doi: 10.1128/JVI.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendinelli M, Matteucci D, Friedman H. Retrovirus-induced acquired immunodeficiencies. Adv Cancer Res. 1985;45:125–81. doi: 10.1016/s0065-230x(08)60268-7. [DOI] [PubMed] [Google Scholar]
- 8.Wegman-Points LJ, et al. Retroviral-infection increases tumorigenic potential of MDA-MB-231 breast carcinoma cells by expanding an aldehyde dehydrogenase (ALDH1) positive stem-cell like population. Redox Biol. 2:847–54. doi: 10.1016/j.redox.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patience C, et al. Multiple groups of novel retroviral genomes in pigs and related species. J Virol. 2001;75:2771–5. doi: 10.1128/JVI.75.6.2771-2775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenen MaM, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–8. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, et al. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 2010;341(1):13–21. doi: 10.1007/s00441-010-1000-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.