Far-red light alleviates cold-induced photoinhibition and enhances photoprotection in shade leaves via activation of phyA-dependent HY5-ABI5-RBOH1 signaling pathways.
Abstract
Photoreceptor-mediated light signaling plays a critical role in plant growth, development, and stress responses but its contribution to the spatial regulation of photoinhibition and photoprotection within the canopy remains unclear. Here, we show that low-red/far-red (L-R/FR) ratio light conditions significantly alleviate PSII and PSI photoinhibition in the shade leaves of tomato (Solanum lycopersicum) plants. This protection is accompanied by a phytochrome A-dependent induction of LONG HYPOCOTYL5 (HY5). HY5 binds to the promoter of ABA INSENSITIVE5 (ABI5), triggering RESPIRATORY BURST OXIDASE HOMOLOG1 (RBOH1)-dependent H2O2 production in the apoplast. Decreased levels of HY5, ABI5, and RBOH1 transcripts increased cold-induced photoinhibition and abolished L-R/FR-induced alleviation of photoinhibition. L-R/FR illumination induced nonphotochemical quenching (NPQ) of chlorophyll a fluorescence and increased the activities of Foyer-Halliwell-Asada cycle enzymes and cyclic electron flux (CEF) around PSI. In contrast, decreased HY5, ABI5, and RBOH1 transcript levels abolished the positive effect of L-R/FR on photoprotection. Loss of PROTON GRADIENT REGULATION5-dependent CEF led to increased photoinhibition and attenuated L-R/FR-dependent NPQ. These data demonstrate that HY5 is an important hub in the cross talk between light and cold response pathways, integrating ABA and reactive oxygen species signaling, leading to the attenuation of photoinhibition by enhanced induction of photoprotection in shade leaves.
Low temperatures are a major factor limiting the productivity and geographical distribution of plant species. Tropical and subtropical plants are generally sensitive to chilling because of lack of the capacity for cold acclimation (Zhu et al., 2007). Many economically important species such as maize (Zea mays), rice (Oryza sativa), and tomato (Solanum lycopersicum) are unable to survive long-term exposures of temperatures below 12°C. In addition to interspecific differences in chilling sensitivity, the tolerance of a given species to low growth temperatures varies between organs and according to developmental stage. Within the canopy, the upper “sun” leaves often exhibit a higher sensitivity to chilling than the shade leaves. However, little is known about the mechanisms that contribute to the spatial differences in chilling tolerance.
In many situations, leaves absorb more light than can be effectively utilized in photosynthesis, especially when plants are exposed to stress. The excess light energy has to be dissipated because overexcitation has the potential to damage the photosynthetic machinery, particularly PSII, in the process called “photoinhibition” (Kingston-Smith et al., 1997; Kingston-Smith and Foyer, 2000; Foyer et al., 2017). This process is characterized by the decreases in the maximal photochemical efficiency of PSII (Fv/Fm) and in maximal P700 oxidation (ΔP700max) in PSI. Meanwhile, plants have evolved a range of photoprotective mechanisms to decrease the probability of damage to the PSII and PSI reaction centers. Photoprotection involves diverse processes such as chloroplast avoidance movement, dissipation of absorbed light energy as thermal energy (NPQ), pseudocyclic electron flow coupled to reactive oxygen species (ROS) scavenging systems (Foyer-Halliwell-Asada cycle), cyclic electron flow (CEF) around PSI, and the photorespiratory pathway (Takahashi and Badger, 2011). The dominant component of NPQ is the energy-dependent nonphotochemical quenching (qE), which is induced by an increase in the proton gradient across the thylakoid membrane (ΔpH) under excess light conditions (Munekage et al., 2004). PSII subunit S (PsbS) protein acts as a sensor of lumen pH and may activate qE through conformational changes of LHCII (Li et al., 2002; Ahn et al., 2008). The mechanisms that contribute to NPQ are not completely understood, but it is widely accepted that two distinct xanthophyll-dependent quenching mechanisms involving xanthophyll cycle pigments and lutein1, respectively, participate in the ΔpH-triggered, PsbS-mediated conformational changes of LHCII (Ruban et al., 2007; Ahn et al., 2008). In the xanthophyll cycle, violaxanthin (V) is converted into zeaxanthin (Z) under high light, via the intermediate antheraxanthin (A), a reaction that is catalyzed by the enzyme violaxanthin de-epoxidase (VDE). The presence of Z activates thermal dissipation of the excess energy (Niyogi et al., 1997, 1998). The de-epoxidation state of the xanthophyll cycle pigments is thought to regulate qE-dependent NPQ (Kromdijk et al., 2016). Alterations in VDE activity influence the extent of PSII photoinhibition (Niyogi et al., 1998; Han et al., 2010).
CEF around PSI is considered to involve NAD(P)H dehydrogenase (NDH) complex-dependent and PROTON GRADIENT REGULATION5 (PGR5)/PGRL1 complex-dependent pathways (Shikanai, 2007), the latter being responsible for most of the required additional ΔpH generation across the thylakoid membrane (Munekage et al., 2004). The generation of an increased trans-thylakoid ΔpH gradient by CEF is important for the activation of qE (Munekage et al., 2004). PGR5-PGRL1-dependent CEF pathway is regulated by the chloroplastic redox state and is activated under stress conditions (Okegawa et al., 2008; Strand et al., 2015). Decreases in both CEF and qE resulted in an inhibition of the synthesis of the D1 protein (Takahashi et al., 2009). Moreover, loss of function of proteins involved in CEF around PSI increased the sensitivity of plants to photoinhibition of PSII and also PSI (Munekage et al., 2002). Furthermore, suppression of Foyer-Halliwell-Asada cycle enzymes increased photoinhibition, whereas an overexpression of the genes encoding these enzymes tended to decrease photoinhibition (Foyer et al., 1995; Maruta et al., 2010).
The effects of high light intensities or fluctuations in light intensity on the extent of photoinhibition of PSII and PSI have been intensively studied over the past 40 years (Kim and Tokura, 1995). In contrast, relatively little is known about the effects of light quality on photoinhibition or photoprotection. Plants have developed a set of sophisticated photoreceptors, including phytochromes, cryptochromes, phototropins (PHOTs), and UV-B light photoreceptors (e.g. UVR8) to perceive changes in light quality (Möglich et al., 2010). Of these, blue-light photoreceptors (e.g. PHOT) have been reported to activate chloroplast avoidance movements in sessile plants under excess light conditions (Kasahara et al., 2002). Energy dissipation in green algae is also controlled by the PHOT and UVR8 photoreceptors, which are activated by blue and UV light (Petroutsos et al., 2016; Allorent et al., 2016; Allorent and Petroutsos, 2017). PhyA and phyB, which are the photoreceptors for far-red light (FR) and red light (R), respectively, play a central role in regulating the expression of a large number of light-responsive genes that are involved in regulation of a wide range of processes from photomorphogenesis to stress responses (Quail, 2002a, 2002b; Franklin and Quail, 2010; Wang et al., 2016), but the role of phytochromes in the regulation of photoinhibition has not been well characterized. LONG HYPOCOTYL5 (HY5), a basic Leu zipper (bZIP) transcription factor, acts downstream of multiple photoreceptors, in the signal transduction pathway that links various signaling pathways including light and phytohormone signaling (Cluis et al., 2004; Jiao et al., 2007; Lau and Deng, 2010). HY5 is also important in the regulation of cold acclimation responses, promoting the expression of a large number of cold-inducible genes (Catalá et al., 2011). Interestingly, low temperatures lead to the stabilization of HY5 through exclusion of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) from the nucleus. COP1 is an E3 ubiquitin ligase targeting HY5 for proteasome-mediated degradation in response to light (Catalá et al., 2011). Whether HY5 is involved in the regulation of photoprotection and photoinhibition is as yet unknown. However, HY5 is known to regulate the expression of several ROS and anthocyanin-related genes (Catalá et al., 2011).
Spatial variations in the chilling sensitivity of leaves at different positions in a canopy have been reported under field conditions, the upper sun-exposed leaves being more easily injured by a cold episode than the shade leaves. Shading not only decreases the light intensity arriving the leaf surface, but also reduces the R/FR of the light available to the shade leaves (Sasidharan et al., 2009). However, the role of changes in R/FR on cold tolerance within a canopy remains largely unexplored, particularly with regard to effects on photoinhibition and photoprotection. Here we show that spatial differences in cold tolerance and in photoinhibition are linked to light-quality-regulated photoprotection. Data are presented showing that low R/FR ratios induce an accumulation of HY5 transcripts in a phyA-dependent manner. The increased expression of HY5 leads to improved cold tolerance by enhancing ABA signaling through direct binding of HY5 to the ABI5 promoter and induction of RBOH1-dependent apoplastic H2O2 generation. ABI5 participates in the regulation of photoprotection by inducing a strong antioxidant response, as well as enhancing PGR5-dependent NPQ. Taken together, these data show that phytochrome-mediated HY5-ABA-ROS signaling plays a key role in avoiding cold-induced photoinhibition by inducing photoprotection within the canopy in response to variations in light quality.
RESULTS
Spatial Variation in Photoinhibition Is Partially Attributable to the Changes in Light Quality Conditions
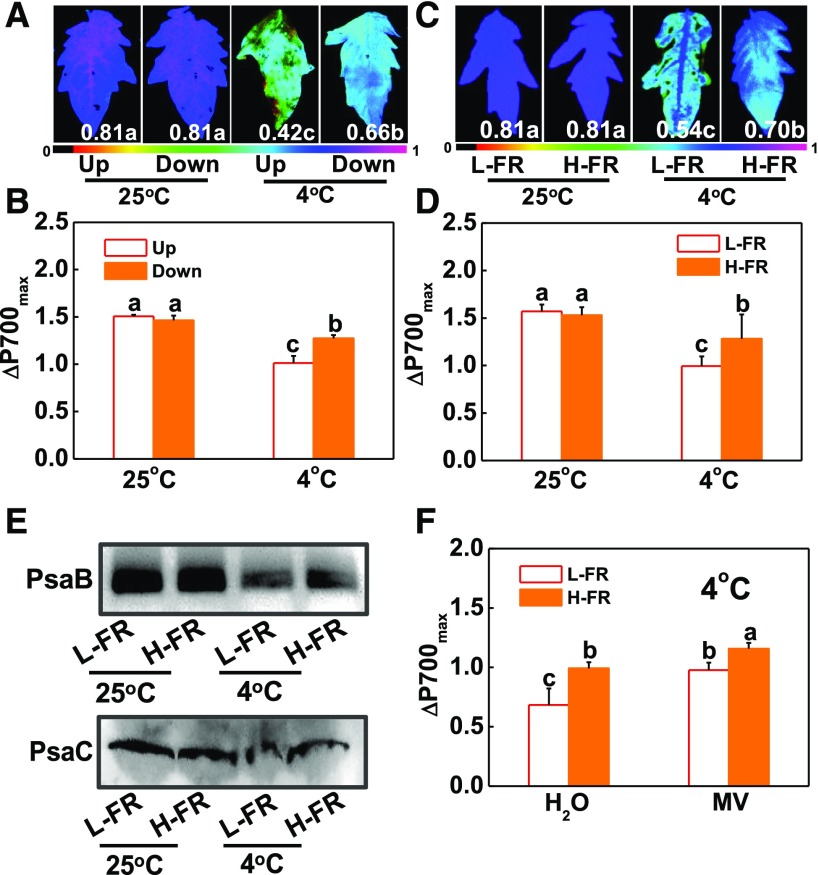
Tomato plants were grown to the 11th leaf stage and then exposed to a cold treatment at 4°C under white light for 7 d. The degree of photoinhibition of PSII and PSI was then compared in leaves at the 9th and 5th ranks from the base. Leaves at the 9th rank had lower Fv/Fm and lower maximum P700 photooxidation level (ΔP700max), together with higher levels of relative electrolyte leakage (REL) than the 5th leaves (Fig. 1, A and B; Supplemental Fig. S1). Light quality analysis revealed that the R/FR was decreased from 1.3 at the 9th leaf rank to 0.5 at 5th leaf rank.
Figure 1.
Spatial variation in photoinhibition is partially attributable to the changes in light quality conditions. A and B, Maximum photochemical efficiency of PSII (Fv/Fm, A), maximum P700 photooxidation level (ΔP700max, B) in leaves at the 9th (Up) and 5th (Down) ranks from the base in plants at 11-leaf stage under white light conditions after exposure to 4°C for 7 d. C and D, Fv/Fm (C) and ΔP700max (D) at 4th leaves of the tomato plants at 6-leaf stage grown in temperature-controlled chambers at 25°C or 4°C under l-FR or H-FR light conditions for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple) represents the level of damage in leaves. E, Immunoblot detection of thylakoid proteins (PsaB and PsaC) separated by SDS-PAGE. Detached leaves were exposed to 25°C or 4°C for 3 d under l-FR or H-FR. F, Effect of MV on the ΔP700max under cold stress in different light quality. After treated with 25 μM MV for 3 h in darkness at 25°C, leaves were transferred to 4°C for 6 h under different light quality conditions. For the l-FR and H-FR, R/FR values at 1.5 and 0.5, respectively, plants were kept at R conditions (200 μmol m−2 s−1) supplemented with different intensities of FR (133 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates except for Fv/Fm, which was the mean for 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test.
We then examined the effects of light quality on cold-induced photoinhibition by exposing tomato plants at the 6-leaf stage to a cold treatment at 4°C under high R/FR, i.e. low FR intensity (L-FR; 133 μmol m−2 s−1) or low R/FR, i.e. high FR intensity (H-FR; 400 μmol m−2 s−1) light conditions, respectively, using monochromatic LEDs. In these experiments, the R light intensity was maintained at 200 μmol m−2 s−1 under both light quality treatment regimes. Chilling-induced decreases in Fv/Fm and in ΔP700max were lower under H-FR than the values determined in plants exposed to the L-FR conditions (Fig. 1, C and D; Supplemental Fig. S2, A and B). NPQ values, PsbS protein accumulation, and de-epoxidation state of the xanthophyll cycle, i.e. (A + Z)/(V + A + Z) ratio, were increased after cold stress, especially under H-FR light conditions (Supplemental Fig. S2, C to E). Western-blot analysis revealed that chilling stress induced a decrease in the accumulation of the PsaB and PsaC proteins, especially under L-FR conditions (Fig. 1E). We also tested the acceptor-side limitation in chilled leaves by application of 25 μm methyl viologen (MV). Results showed that chilling-induced decrease of ΔP700max was mostly relieved in the MV-treated leaves, especially under H-FR conditions (Fig. 1F). These results suggest that the main cause of the chilling-induced decrease in ΔP700max is the degradation of PSI submits resulting in an acceptor side limitation in PSI.
RNA-seq analysis was performed on the 4th leaves after exposure of plants to a cold treatment at 4°C under either l- or H-FR light conditions. This generated a total of 103,463,126 reads, which were aligned to the S. lycopersicum reference genome (https://solgenomics.net/). Compared with the plants grown under L-FR conditions, a total of 6312 transcripts (3607 increased in abundance and 2705 decreased in abundance) were differentially changed under the H-FR (Supplemental Table S1). An examination of the levels of transcripts encoding photosynthetic proteins and proteins involved in light and ABA signaling revealed a subset of mRNAs that showed differential changes in response to the light quality, having higher levels in leaves exposed to H-FR light compared to l-FR. These transcripts encoded proteins are that involved in PSII (PHOTOSYSTEM II LIGHT HARVESTING COMPLEX GENE2.1 and PHOTOSYSTEM II REACTION CENTER PROTEIN A), PSI (PHOTOSYSTEMI SUBUNITI and PHOTOSYSTEMI, PsaA/PsaB PROTEIN), cyclic electron flux (PGR5 and PGR5-LIKE A), plastoquinone cytochrome b6f complex (CYTOCHROME b561/FERRIC REDUCTASE TRANSMEMBRANE PROTEIN FAMILY and CYTOCHROME B5 ISOFORM B), FAD/NAD(P)-binding oxidoreductase family protein [NAD(P)-BINDING ROSSMANN-FOLD SUPERFAMILY PROTEIN and FAD/NAD(P)-BINDDING OXIDOREDUCTASE FAMILY PROTEIN], oxidoreductase proteins (PEROXIDASE2 and ASCORBATE PEROXIDASE3), and thermal energy dissipation (CAROTENOID CLEAVAGE DIOXYGENASE1 and ZEAXANTHIN EPOXIDASE; Supplemental Fig. S2F; Supplemental Table S2).
PhyA Acts as a Positive Regulator for Light Quality-Dependent Regulation of Photoinhibition
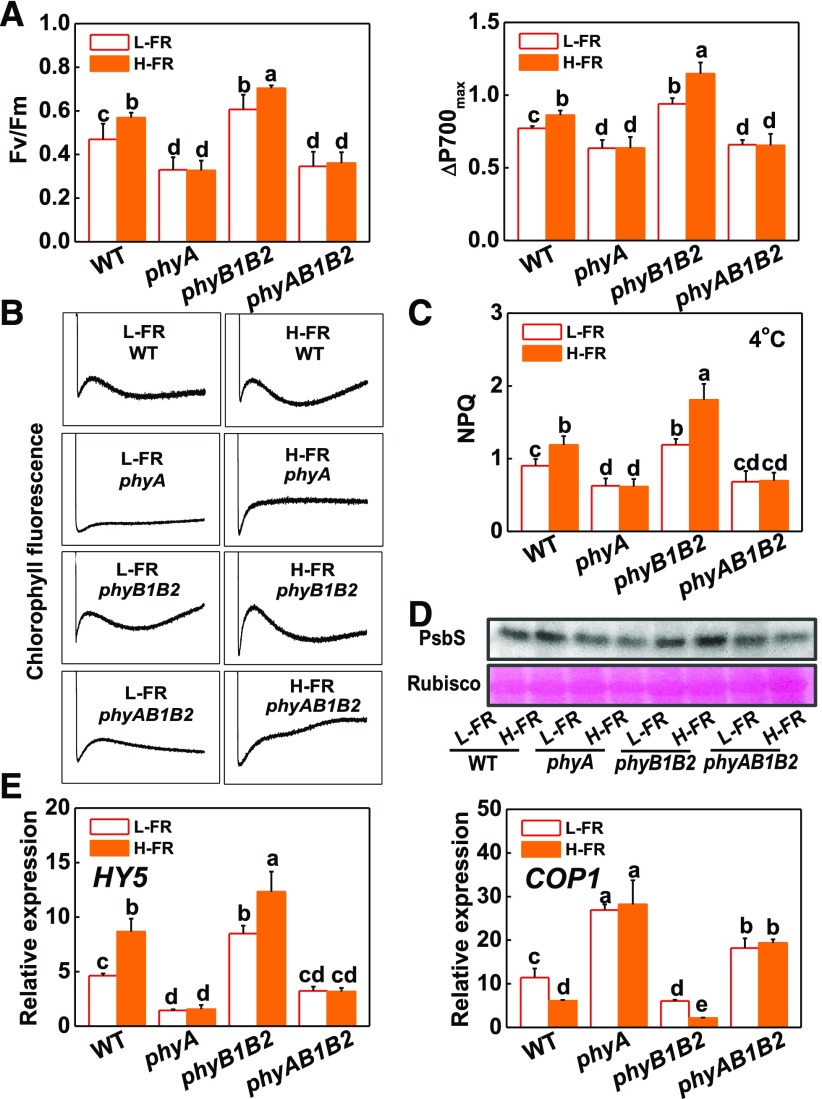
Cold-induced photoinhibition was compared in wild-type tomato leaves and in mutants deficient in phytochrome A (phyA and phyAB1B2) or phytochrome B (phyB1B2) grown under either L-FR or H-FR conditions. The phyA mutants had lower Fv/Fm and ΔP700max levels than the wild type after exposure to a cold treatment at 4°C for 7 d (Fig. 2A). In contrast, the phyB1B2 plants showed higher Fv/Fm and ΔP700max values than the wild type under these conditions. Moreover, the phyA and phyAB1B2 mutants had lower NPQ values, with less PsbS protein accumulation and lower CEF rates compared to wild-type plants (Fig. 2, B to D). In contrast, the phyB1B2 plants had higher NPQ values, PsbS protein accumulation, and CEF rates than the wild type. In addition, H-FR significantly induced increases in Fv/Fm, ΔP700max values, NPQ values, PsbS protein accumulation, and CEF rates in wild type and phyB1B2 mutant, but not in phyA and phyAB1B2 mutants after the cold treatment (Fig. 2, A to D).
Figure 2.
Roles for tomato phytochromes in the light quality regulation of photoinhibition and the expression of light signaling genes (HY5 and COP1). A, Fv/Fm and ΔP700max of the tomato phytochrome mutant plants after exposure to cold at 4°C under l-FR or H-FR light conditions for 7 d. B, Postillumination chlorophyll fluorescence (CEF around PS I) in tomato plants after exposure to cold at 4°C for 3 d under l-FR and H-FR conditions. C and D, Changes of NPQ (C) and PsbS protein (D) in wild-type and phytochrome mutant plants exposed for either 3 d or 1 d to cold stress (at 4°C) under l-FR and H-FR light conditions. E, Levels of HY5 and COP1 transcripts at 6 h after tomato phytochrome mutants were exposed to 4°C under l-FR or H-FR light conditions. For the l-FR and H-FR, R/FR treatments of 1.5 or 0.5, respectively, plants were kept under R (200 μmol m−2 s−1) light conditions, supplemented with different intensities of FR (133 μmol m−2 s−1 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates except for Fv/Fm ratios, which are the means of 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test. WT, wild type.
The levels of HY5 transcripts were increased in response to a cold treatment in wild-type tomato leaves grown under H-FR compared to L-FR conditions (Fig. 2E). The H-FR growth regime also resulted in significant chilling-induced increases in the levels of HY5 transcripts in the phyB1B2 mutant leaves but not in those of the phyA or phyAB1B2 mutants. However, an inverse pattern of response to change in FR intensity was observed for COP1 transcripts. Growth under the H-FR light regime decreased the levels of COP1 mRNAs in the wild type and in the phyB1B2 mutants under cold stress. In contrast, differences in the FR intensity had little effect on the levels of COP1 transcripts in phyA and phyAB1B2 mutants after the cold treatment.
FR-Induced HY5 Alleviated Photoinhibition by Induction of Photoprotection
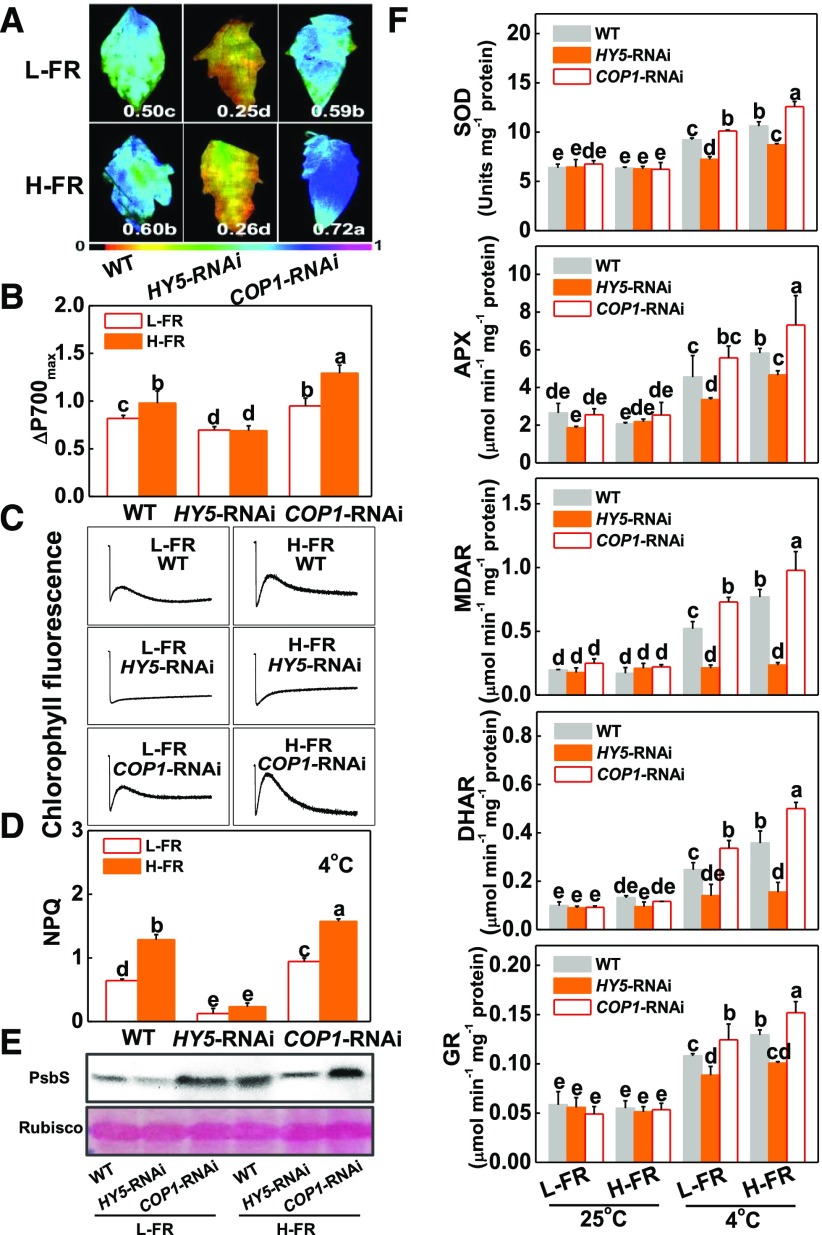
The levels of HY5 and COP1 transcripts were decreased by 80% and 70%, respectively, in HY5-RNAi and COP1-RNAi plants used for the study (Supplemental Fig. S3A). Cold and FR intensity-induced changes in Fv/Fm, ΔP700max, survival rates, REL, and the levels of oxidized protein, as determined by the presence of protein carbonyl groups, were measured in the HY5-RNAi and COP1-RNAi plants (Fig. 3, A and B; Supplemental Fig. S3, B and C). Cold-induced increases in REL and in the levels of oxidized proteins were higher in the HY5-RNAi plants compared to the wild-type and COP1-RNAi plants regardless of FR intensity (Supplemental Fig. S3C). In contrast, the Fv/Fm and the ΔP700max values were much lower in the leaves of the HY5-RNAi plants compared to the wild-type and COP1-RNAi plants under both FR light conditions (Fig. 3, A and B). The chilling-induced decreases in the Fv/Fm and the ΔP700max values were significantly less in the COP1-RNAi plants than the wild type (Fig. 3, A and B). Significantly, H-FR treatment induced increases in Fv/Fm, ΔP700max values, survival rates, and decreases in REL or the levels of oxidized proteins in the wild-type and the COP1-RNAi plants, but had little effects on these parameters in the HY5-RNAi plants (Fig. 3, A and B; Supplemental Fig. S3, B and C). Moreover, the HY5-overexpressing plants showed an increased tolerance to the cold treatment compared to the wild type (Supplemental Fig. S4). Taken together, these results indicate that HY5 is required for the light quality-mediated regulation of chilling tolerance in tomato and that COP1 negatively regulates this process.
Figure 3.
HY5 alleviated photoinhibition by induction of photoprotection. A and B, Fv/Fm (A) and ΔP700max (B) of the wild-type, HY5-RNAi, and COP1-RNAi tomato plants after exposure to a cold stress at 4°C under l-FR or H-FR light conditions for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple) represents the level of damage in leaves. C and D, Postillumination chlorophyll fluorescence (CEF around PSI, C) and NPQ (D) in wild-type, HY5-RNAi, and COP1-RNAi tomato plants after exposure to 4°C for 3 d under l-FR and H-FR conditions. E, Immunoblot analysis of PsbS in wild-type, HY5-RNAi, and COP1-RNAi tomato plants after exposure to 4°C for 1 d under l-FR and H-FR conditions. Samples were loaded at equal total proteins amounts based on Coomassie Blue staining. F, Activity of antioxidant enzymes (SOD, APX, MDAR, DHAR, and GR) involved in Foyer-Halliwell-Asada cycle after the wild-type, HY5-RNAi, and COP1-RNAi tomato plants exposure to 25°C or 4°C under l-FR or H-FR light conditions for 3 d. For the l-FR and H-FR, R/FR at 1.5 and 0.5, respectively, plants were kept at R conditions (200 μmol m−2 s−1) supplemented with different intensities of FR (133 μmol m−2 s−1 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates except for Fv/Fm, which are the means of 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test. WT, wild type.
NPQ, cyclic electron flux (CEF), and Foyer-Halliwell-Asada cycle all play important roles in preventing the photosystems from photodamage or photoinhibition (Foyer et al., 1995; Takahashi et al., 2009; Chen and Gallie, 2012). In comparison to the wild-type plants, the HY5-RNAi plants showed decreased levels of NPQ, PsbS protein accumulation, antioxidant enzyme activities, and CEF rates (Fig. 3, C to F; Supplemental Fig. S5). These parameters were increased in the COP1-RNAi plants relative to the wild type. An increase in the FR intensity significantly increased the level of NPQ and the accumulation of PsbS protein, and the abundance of transcripts encoding Foyer-Halliwell-Asada cycle enzymes (Cu/Zn-SUPEROXIDE DISMUTASE, Cu/Zn-SOD; ASCORBATE PEROXIDASE, APX; MONODEHYDROASCORBATE REDUCTASE, MDAR; DEHYDROASCORBATE REDUCTASE, DHAR; and GLUTATHIONE REDUCTASE1, GR1), as well as the activities of these enzymes, and the rate of CEF in wild-type and COP1-RNAi plants (Fig. 3, C to F; Supplemental Fig. S5). The effects of the FR intensity were more pronounced in the COP1-RNAi plants. However, the H-FR treatment had little effect on the level of NPQ, the activities of antioxidant enzymes, or the rates of CEF in the HY5-RNAi plants, suggesting that HY5 is essential for the H-FR regulation of photoprotection.
HY5 Is a Transcriptional Activator of ABI5
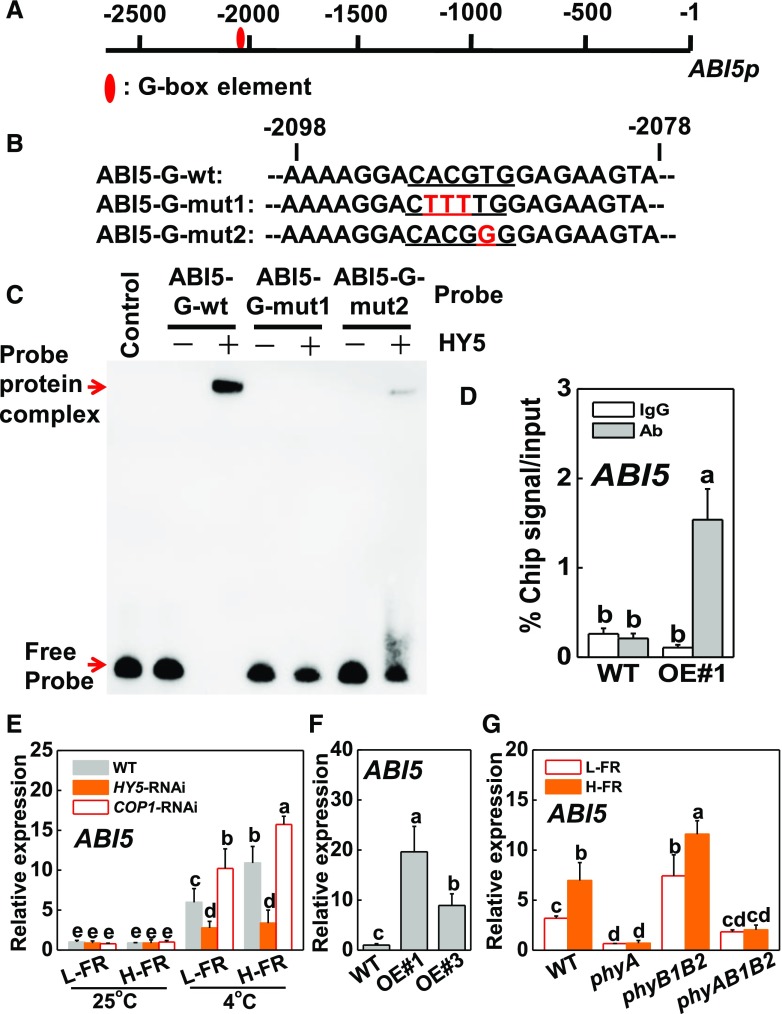
Although exposure to cold stress had no effect on stomatal movements in HY5-RNAi plants, this treatment caused a decrease in stomatal aperture in COP1-RNAi leaves, especially under H-FR light conditions (Supplemental Fig. S6, A and B). Given that ABA signaling positively regulates stomatal movement, we examined whether HY5 could bind to the promoters of any of the ABA signaling genes. For this analysis, we inspected 2.5-kb sequences upstream of the transcriptional start sites of a set of tomato ABA INSENSITIVE (ABI) genes. Of these, the promoters of three ABA signaling genes (ABI3-1, ABI3-2, and ABI5) contain the G-box sequences: CACGTG (Fig. 4A; Supplemental Fig. S6C). Electrophoretic mobility shift assays (EMSAs) were used to analyze whether HY5 binds directly to these promoters in vitro. The probe-protein complex was not detected using ABI3-1 and ABI3-2 probes. However, HY5 directly bound to the promoter probe of ABI5 (Fig. 4C; Supplemental Fig. S6D). When the core sequence of G-box element motif in ABI5 probe was mutated in a single base (ABI5-G-mut2) or multiple bases (ABI5-G-mut1; Fig. 4B), the binding to the complexes was decreased, or even totally lost (Fig. 4C). Based on these observations, we conclude that HY5 protein binds specifically to the G-box element sequences of the synthesized probes for the ABI5 promoters in vitro.
Figure 4.
HY5-induced transcript level of ABI5 by binding to promoter of ABI5. A and B, G-box elements in the promoter of tomato ABI5 gene (A) and oligonucleotide used in the EMSAs (B). Numbering is from predicted transcriptional start sites. The ABI5 probe contains one G-box (ABI5-G-wt), whereas in the ABI5-G-mut1 and ABI5-G-mut2 probes, the G-box core sequence was mutated. The wild-type and mutated G-box sequences are underlined. The mutated bases were indicated in red. C, HY5 directly binds to the G-box of ABI5 promoter in vitro. Recombinant HY5 was purified from E. coli cells and used for DNA binding assays with probes of ABI5-G-wt, ABI5-G-mut1, and ABI5-G-mut2. The protein purified from empty vector was used as the negative control. D, Direct binding of HY5 to the ABI5 promoter was analyzed using ChIP-qPCR in 35S-HY5-3HA-overexpressing (HY5-OE#1) tomato plants. HY5-OE#1 plants at 6-leaf stage were exposed to 4°C under H-FR condition and input chromatin was isolated from leaf samples at 6 h. The epitope-tagged HY5-chromatin complex was immunoprecipitated with an anti-HA antibody. A control reaction was processed side-by-side using mouse IgG. Input- and ChIP-DNA samples were quantified by qRT-PCR using primers specific for the promoter of the ABI5 gene. The ChIP results are presented as a percentage of the input DNA. #1, line of HY5-OE plants. E and F, Transcript level of ABI5 gene at 6 h after HY5-RNAi and COP1-RNAi tomato plants exposed to 25°C or 4°C under different R/FR light regimes (E), and two independent HY5 overexpressing transgenic tomato lines (HY5-OE#1, OE#3) exposed to 4°C under H-FR conditions (F). G, Transcript level of ABI5 gene at 6 h after wild-type and phytochrome mutants of tomato exposed to 4°C under different R/FR light regimes. For the l-FR and H-FR, R/FR values at 1.5 and 0.5, respectively, plants were kept at R conditions (200 μmol m−2 s−1) supplemented with different intensities of FR (133 and 400 μmol m−2 s−1). Four independent experiments were performed yielding similar results. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test. OE, overexpressing; WT, wt, wild type.
To further determine whether the tomato HY5 protein binds directly to the promoter of ABI5 in vivo under cold stress, we performed ChIP-qPCR assays. As shown in Figure 4D, the ABI5 promoter sequence was substantially enriched in fractions using the anti-HA antibody that immunoprecipitates the 3HA-tagged HY5 transgene product in the HY5 overexpressing (OE) lines but not the wild type after 6 h of cold stress under H-FR. However, the IgG control antibody failed to pull down the ABI5 gene promoter DNA segment (Fig. 4D). We then assessed the levels of ABI5 transcripts in wild type, HY5-RNAi, and COP1-RNAi plants exposed to H- and l-FR conditions under cold stress conditions (Fig. 4E). No changes in ABI5 transcript levels were detected in the HY5-RNAi plants in relation to the FR intensity. In contrast, an increase in the FR gradually induced increase the abundance of ABI5 transcripts in wild type and COP1-RNAi plants, the induction being more significant in the COP1-RNAi plants than the wild type. The induction of ABI5 expression was greater in the HY5 overexpressing lines (OE#1 and OE#3, expressing high HY5 protein levels; Supplemental Fig. S4A) than the wild type after 6 h of cold stress under the H-FR irradiance regime (Fig. 4F). These results indicate that HY5 binds directly the promoter of ABI5 and activates its expression, subsequently regulating cold tolerance of tomato in response to light quality.
When wild-type and phytochrome mutant plants were exposed to l-FR and H-FR light conditions at 4°C, higher levels of ABI5 transcripts were maintained in phyB1B2 mutants compared to the wild-type, phyA, or phyAB1B2 plants under both light quality conditions (Fig. 4G). Moreover, the higher FR intensity increased the levels of ABI5 transcripts in wild-type and phyB1B2 plants, but not in phyA or phyAB1B2 mutants.
Role of ABI5 in Light Quality-Regulated Photoinhibition and Photoprotection
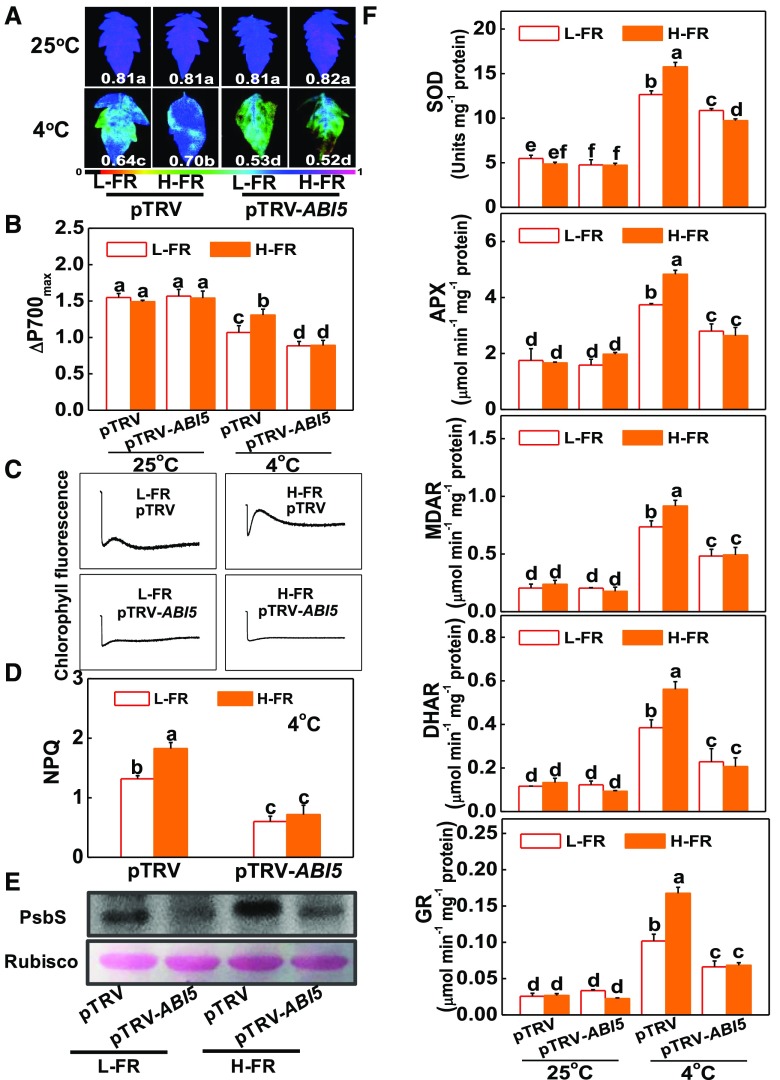
Lines of ABI5-silenced tobacco rattle virus (TRV; pTRV-ABI5) plants were generated, using a virus-induced gene silencing (VIGS). These lines showed a reduction in ABI5 transcript levels of 75% (Supplemental Fig. S7, A and B). Tomato pTRV-ABI5 plants showed an increased sensitivity to cold-induced photoinhibition compared to the pTRV plants, as measured by a decrease in the Fv/Fm ratio and in ΔP700max, as well as an increase in REL (Fig. 5, A and B; Supplemental Fig. S7, C and D). Interestingly, the H-FR-induced cold tolerance and alleviation of photoinhibition observed in the pTRV plants was completely lost in the ABI5-silenced plants, which showed no significant differences in Fv/Fm and ΔP700max under cold stress at both light quality regimes. These observations clearly indicate that loss of ABI5 function compromised the H-FR-induced alleviation of chilling-dependent photoinhibition in tomato. In support of this hypothesis, we observed that the H-FR-induced changes in NPQ, PsbS protein accumulation, and the activities of antioxidant enzymes, as well as the rate of CEF, were abolished or attenuated in the pTRV-ABI5 plants (Fig. 5, C to F). These results show that ABI5 functions as a down-stream of HY5 in light-regulated photoprotection.
Figure 5.
Role of ABI5 in light quality-regulated photoinhibition and photoprotection. A and B, Fv/Fm (A) and ΔP700max (B) of the nonsilenced (pTRV) and silenced (pTRV-ABI5) tomato plants grown in temperature-controlled chambers at 25°C or 4°C under l-FR or H-FR light conditions for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple), representing the level of damage in leaves. C and D, Postillumination chlorophyll fluorescence (CEF around PSI, C) and NPQ (D) in the pTRV and pTRV-ABI5 tomato plants after exposure to 4°C for 3 d under l-FR and H-FR conditions. E, Immunoblot analysis of PsbS in pTRV and pTRV-ABI5 tomato plants after exposure to 4°C for 1 d under l-FR and H-FR conditions. Samples were loaded at equal total proteins amounts based on Coomassie Blue staining. F, Activity of antioxidant enzymes (SOD, APX, MDAR, DHAR, and GR) involved in Foyer-Halliwell-Asada cycle after the pTRV and pTRV-ABI5 tomato plants’ exposure to 25°C or 4°C under l-FR or H-FR light conditions for 3 d. For the l-FR and H-FR, R/FR values at 1.5 and 0.5, respectively, plants were kept at R conditions (200 µmol m−2 s−1) supplemented with different intensities of FR (133 μmol m−2 s−1 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates except for Fv/Fm values, which are the means of 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test.
RBOH1-Dependent ROS Production Prevents Photoinhibition by Activation of Photoprotection
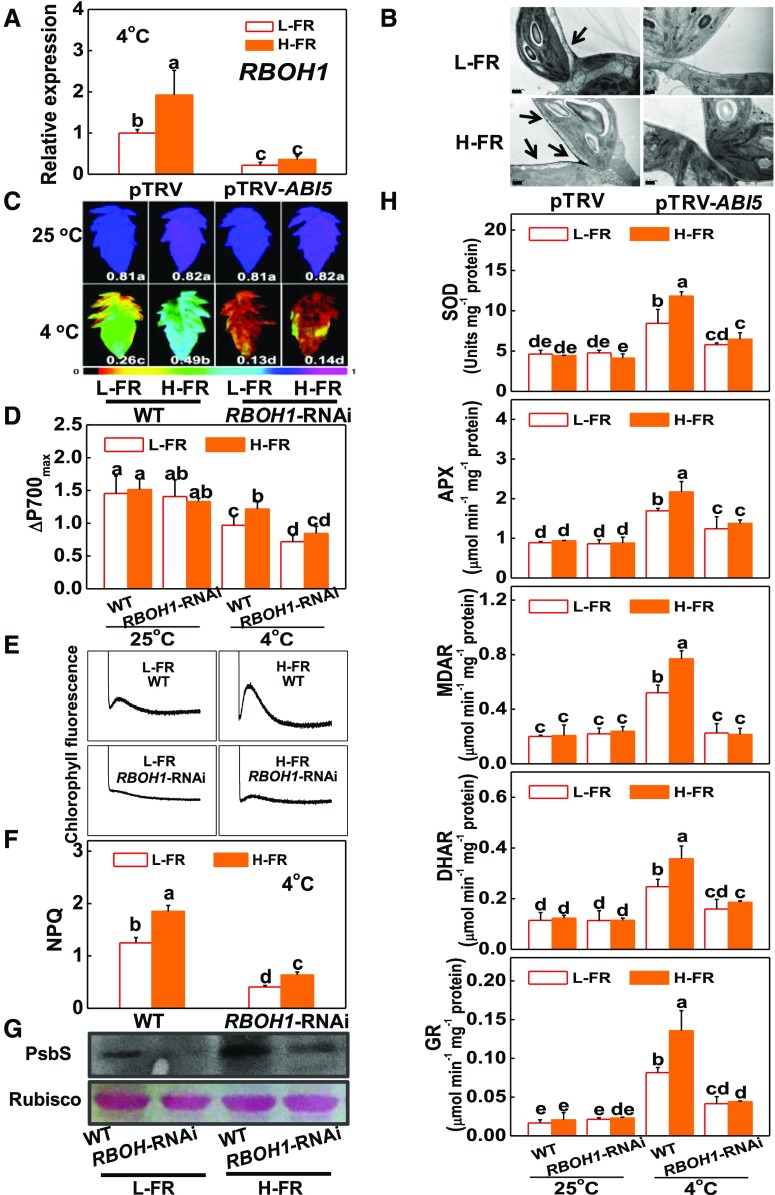
ABA signaling is linked to the up-regulation of RBOH-dependent ROS production in response to stress (Murata et al., 2001; Xing et al., 2008; Zhou et al., 2014). The cold treatment used in this study increased the levels of RBOH1 transcripts and apoplastic H2O2 accumulation (Fig. 6, A and B). Moreover, silencing ABI5 (pTRV-ABI5) abolished the H-FR-dependent induction of RBOH1 expression and apoplastic H2O2 accumulation.
Figure 6.
RBOH1-dependent ROS production prevents plants from photoinhibition by activating photoprotection. A and B, Transcript level of RBOH1 gene at 6 h (A) and cytochemical localization of H2O2 accumulation in leaf mesophyll cells at 1 d as visualized by CeCl3 staining and transmission electron microscopy (B) after pTRV and pTRV-ABI5 tomato plants exposed to 4°C under different R/FR light regimes. The arrows indicate CeCl3 precipitates. Scale bars = 0.5 μm. C and D, Fv/Fm (C) and ΔP700max (D) of the wild-type and RBOH1-RNAi tomato plants were exposed to 25°C or 4°C under l-FR or H-FR light conditions for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple) represents the level of damage in leaves. E and F, Postillumination chlorophyll fluorescence (CEF around PSI, E) and NPQ (F) in the wild-type and RBOH1-RNAi tomato plants after exposure to 4°C for 3 d under l-FR and H-FR conditions. G, Immunoblot analysis of PsbS in WT and RBOH1-RNAi tomato plants after exposure to 4°C for 1 d under l-FR and H-FR conditions. Samples were loaded at equal total proteins amounts based on Coomassie Blue staining. H, Activity of antioxidant enzymes (SOD, APX, MDAR, DHAR, and GR) involved in Foyer-Halliwell-Asada cycle after the wild-type and RBOH1-RNAi tomato plants’ exposure to 25°C or 4°C under l-FR or H-FR light conditions for 3 d. For the l-FR and H-FR, R/FR values at 1.5 and 0.5, respectively, plants were kept at R conditions (200 µmol m−2 s−1) supplemented with different intensities of FR (133 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates except for Fv/Fm values, which are the means of 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test. WT, wild type.
RBOH1-RNAi plants were used to examine whether RBOH1 plays a role in the regulation of cold-induced photoinhibition and photoprotection under L- and H-FR conditions. The RBOH1-RNAi plants showed lower apoplastic H2O2 accumulation and an increased sensitivity to photoinhibition compared to the wild type (Fig. 6, C and D; Supplemental Fig. S8D). The response to changes in FR intensities was also compromised in terms of survival rates, wilting symptoms and changes in the Fv/Fm, ΔP700max, and REL (Fig. 6, C and D; Supplemental Fig. S8, A to C). In addition, ABA-induced alleviation of photoinhibition was compromised by treatment with dimethylthiourea (a ROS scavenger). This effect was also not observed in the RBOH1-RNAi plants (data not shown). Moreover, H-FR-induced changes in NPQ, PsbS protein accumulation, and the activities of antioxidant enzymes, as well as the rate of CEF, were abolished in the RBOH1-RNAi plants (Fig. 6, E to H). These results suggest that the ABI5-dependent production of H2O2 plays a pivotal role in HY5-regulated photoprotection by functioning as a critical downstream component in light signaling.
Light-Activated CEF Plays Dual Roles in Preventing Plants from Photoinhibition
The roles of NPQ and antioxidants in photoprotection are well established (Foyer et al., 1995; Niyogi et al., 1997, 1998; Chen and Gallie, 2012). However, relatively little is known about the role of PGR5-PGRL1-dependent and NDH-dependent CEF in photoprotection (Shikanai, 2007). Here, we show that chilling stress increased the accumulation of PGR5 transcripts by more than 5-fold but had less effect on the levels of PGRL1A and ORANGE RIPENING (ORR) transcripts (Supplemental Fig. S9). ORR encodes an NDH-M subunit in the tomato NDH complex (Nashilevitz et al., 2010). Chilling-induced increases in PGR5, PGRL1A, and ORR transcripts were greater after exposure of plants to H-FR light conditions. In comparison, PGRL1B transcripts were decreased by the chilling treatment and they were not affected by FR levels. Moreover, PHYA deficiency or silencing of HY5, ABI5, and RBOH1 abolished the H-FR-dependent induction of PGR5, PGRL1A, and ORR transcripts. These results suggested the potential involvement of the PGR5-PGRL1-dependent and NDH-dependent CEF in the photoprotection in response to the cold stress.
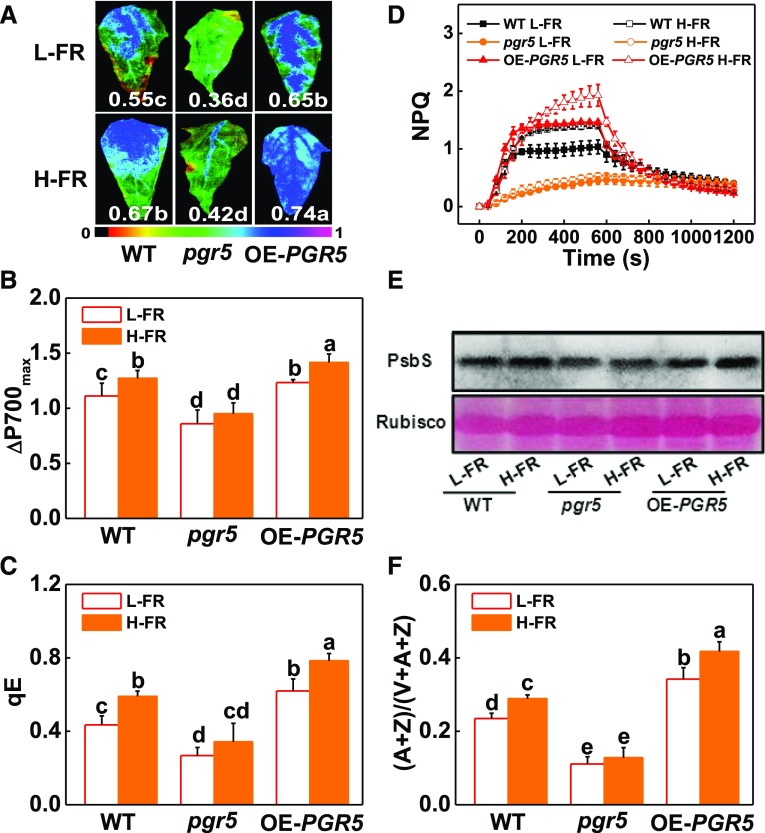
We then generated pgr5 mutants by using a Crisp/cas9 technique and also PGR5-overexpressing (PGR5-OE) tomato plants (Supplemental Fig. S10). The pgr5 plants showed decreased CEF rates whereas the PGR5-OE plants had increased CEF rates under cold stress (Supplemental Fig. S11C). Moreover, the cold-mediated induction of CEF under H-FR light conditions was lower in the pgr5 plants than the wild type. In contrast, this parameter was higher in the PGR5-OE plants exposed to cold stress under H-FR light conditions. Significantly, the cold treatment led to greater decreases in the Fv/Fm and in ΔP700max, an increase in REL, and more severe necrosis and wilting symptoms in the pgr5 plants and the wild type (Fig. 7, A and B; Supplemental Fig. S11, A and B), whereas PGR5-overexpressing significantly increased Fv/Fm ratios and ΔP700max values after a cold stress. H-FR induced increases in qE, NPQ, PsbS protein acclimation, and (A + Z)/(V + A + Z) ratio in the wild-type and PGR5-OE plants but not in the pgr5 plants (Fig. 7, C to F). These results suggest that PGR5-dependent CEF is essential for light-regulated photoprotection. To provide further evidence for the roles of HY5, ABI5, and RBOH1 in PGR5-dependent photoprotection, we silenced HY5, ABI5, and RBOH1 in PGR5-OE plants (Supplemental Fig. S12). As observed in wild-type plants, silencing of these genes in PGR5-OE plants significantly decreased ΔP700max and compromised H-FR-induced increase in ΔP700max. These findings show that HY5-ABI5-RBOH1 cascades play a critical role in FR-induced and PGR5-dependent photoprotection in the plants.
Figure 7.
PGR5-dependent CEF plays dual roles in preventing plants from photoinhibition. A and B, Fv/Fm (A) and ΔP700max (B) of the wild type, pgr5 mutant (pgr5#5), and PGR5-overexpressing (OE-PGR5#3) transgenic plants grown at 4°C under L-FR or H-FR light conditions for 7 d. The false color code depicted at the bottom of the image ranges from 0 (black) to 1.0 (purple) represents the level of damage in leaves. C and D, qE (C) and NPQ (D) in the wild-type, pgr5#5 mutant, and OE-PGR5#3 tomato plants after exposure to 4°C for 3 d under l-FR and H-FR conditions. E and F, PsbS protein (E) and de-epoxidation state of the xanthophyll cycle (F) in the wild-type, pgr5#5 mutant, and OE-PGR5#3 tomato plants after exposure to 4°C for 1 d and 3 d, respectively, under L-FR and H-FR conditions. For the L-FR and H-FR, R/FR values at 1.5 and 0.5, respectively, plants were kept at R conditions (200 μmol m−2 s−1) supplemented with different intensities of FR (133 μmol m−2 s−1 and 400 μmol m−2 s−1). Data are the means (±sd) of four biological replicates, except for Fv/Fm, which are the means for 15 leaves from independent plants. Different letters indicate significant differences (P < 0.05) according to the Tukey’s test. WT, wild type.
DISCUSSION
The management of light energy usage in photosynthesis is a key concept of photosynthetic regulation (Foyer et al., 2017). A wide range of mechanisms have evolved to protect the photosystems from the potentially damaging effects of the high irradiances that occur in the natural environments. Although photoinhibition may not be such a common phenomenon in nature as was once thought, because recovery without damage is facilitated by the protective component of NPQ (Foyer et al., 2017), understanding the regulation of NPQ at different leaf ranks within the plant canopy is crucial to plant productivity. The recent demonstration that acceleration of the NPQ relaxation can lead to significant increases in crop yield (Kromdijk et al., 2016) highlights the importance of understanding how photosynthetic efficiency is regulated. The data presented here provides new information concerning the regulation of leaves to shading. We show that spatial variations in susceptibility to cold-induced photoinhibition are attributable to differences in the R/FR experienced by the leaves. In particular, phyA-mediated induction of HY5 under different R/FR regimes plays a critical role in photoprotection. Through binding to the promoter of ABI5, HY5 triggers enhanced photoprotection through induction of an apoplastic H2O2 burst that influences antioxidant status, CEF, and NPQ. This enhanced photoprotection allows shade leaves to avoid photoinhibition.
Chlorophyll-containing cells absorb blue light and R light, whereas FR photons are either transmitted or reflected. This leads to a decrease in the R/FR experienced deep leaves within the vegetative canopy compared to leaves exposed to full sunlight (Sasidharan et al., 2009). The data presented here show that the upper leaves experiencing a high R/FR (i.e. low FR intensity, L-FR) growth environment have a higher degree of photoinhibition compared to shade leaves that experience a low R/FR (i.e. high FR intensity, H-FR) growth environment (Fig. 1, A and B; Supplemental Fig. S1). To exclude the potential effects of other parameters such as leaf developmental stage and light intensity on photoinhibition and cold tolerance, plants were exposed to R at the same light intensity and only FR intensities were changed. Photoinhibition and electrolyte leakage were significantly decreased, whereas NPQ values, PsbS protein accumulation, and (A + Z)/(V + A + Z) ratio were increased under the low-R/FR value light growth regime (Fig. 1, C to F; Supplemental Fig. S2, A to E). Therefore, spatial differences in sensitivity to photoinhibition and in cold tolerance are largely attributable to differences in R/FR within the growth environment. Moreover, the low R/FR experience by the leaves plays a positive role in tolerance to excess light such that shade leaves are less sensitive to photoinhibition. By using western blotting against PSI subunits PsaB and PsaC, and suppling of MV (an artificial electron acceptor from PSI), we found FR plays a critical role in photoprotection by alleviating the degradation of PSI submits and the release of acceptor side limitation of PSI (Fig. 1, E and F).
It is widely accepted that the primary function of phytochromes is to detect environmental fluctuations in the relative proportions of R and FR radiation (Chen et al., 2004). Data presented here show that the FR receptor phyA and R receptor phyB are, respectively, positive and negative regulators of photoinhibition (Fig. 2, A to D). These findings are in agreement with earlier studies demonstrating that phyA- and phyB1B2-deficient tomato plants had increased and decreased sensitivities to chilling (Wang et al., 2016). It is of interest to note that the higher tolerance to cold observed in shade leaves contrasts markedly with the reported decreased resistance to herbivory and pathogens. Numerous studies have reported that the low R/FR experienced by shade leaves increase plant population densities, and increase herbivory and disease (Xie et al., 2011; Ballaré, 2014). In contrast to the increases in cold tolerance observed here in phyB tomato plants, similar mutants in Arabidopsis were reported to be more sensitive to Pseudomonas syringae pv. tomato DC3000 (de Wit et al., 2013). Recent studies have demonstrated that high R/FR values have important effects on plant defenses through effects on JA signaling and other defense pathways (Cerrudo et al., 2012; Nagata et al., 2015). Therefore, plants appear to have evolved different mechanisms for coping with biotic stress and abiotic stresses through the integration of light signaling pathways with those involving the perception of other environmental stimuli.
Multiple photoreceptors promote the accumulation of HY5 in response to changing light conditions. As a member of the bZIP transcription factor family, HY5 plays a critical role in different plant processes such as hormone-, nutrient-, abiotic stress-, and redox-signaling pathways (Gangappa and Botto, 2016). This places HY5 at the center of the transcriptional network hub that regulates plant responses to environmental change. One mechanism by which this is achieved is through regulation of the nuclear abundance of COP1, an E3 ubiquitin ligase that targets HY5 for proteasome-mediated degradation in darkness (Osterlund et al., 2000; Yi and Deng, 2005). Exposure to cold stress induced HY5 expression in wild type and phyB1B2. However, mutation in phyA abolished cold-induced transcript of HY5 under both L-FR and H-FR light conditions (Fig. 2E). In addition, changes in the COP1 transcript levels were in contrast with those in HY5 transcript levels in wild-type leaves and in phytochrome-deficient mutants. These results suggest that the induction of HY5 and COP1 in response to the cold stress is phyA-and phyB-dependent, respectively. To date, our knowledge of the role of HY5 in plant cold responses was limited to the regulation of anthocyanin accumulation in Arabidopsis (Catalá et al., 2011). The data presented here show that HY5 and COP1 are positive and negative regulators for the plant cold response, leading to the regulation of photoinhibition (Fig. 3; Supplemental Figs. S3, S4, and S5).
ABA signaling is known to play an important role in responses to various environmental stresses (Zhao et al., 2013). Analysis using EMSAs and ChIP-qPCR assays revealed that HY5 binds to the G-box element of the ABI5 promoter in vitro and in vivo with a high specificity (Fig. 4, A to D; Supplemental Fig. S6, C and D). In the absence of HY5, the ability of H-FR-induced signals to increase ABI5 transcript levels was impaired (Fig. 4E). The induction of ABI5 expression was also found to be phyA-dependent and significantly increased in HY5-overexpressing plants compared to wild-type plants after exposure to cold stress (Fig. 4, F and G). ABI5 had been shown to be involved in the regulation of seed germination and responses to drought (Chen et al., 2008). ABI5 involvement in drought stress responses has been assessed through adjustments in ROS scavenging and in osmotic potential in cotton (Mittal et al., 2014). ABA signaling, like brassinosteroid signaling, is known to have a role in the induction of apoplastic H2O2 accumulation in plants (Zhou et al., 2014). The data presented here show that silencing ABI5 compromised the H-FR-induced alleviation of photoinhibition (Fig. 5), as well as the induction of RBOH1 expression and H2O2 accumulation in the leaf apoplast (Fig. 6, A and B). Taken together, these findings strongly suggest that ABI5 not only functions as a downstream component of the light-regulated cold tolerance pathway in a HY5-dependent manner, but that it is also linked to ROS signaling. Furthermore, RBOH1-RNAi plants failed to respond to changes in FR intensities in terms of effects on Fv/Fm , ΔP700max, survival rates and REL (Fig. 6, C to H; Supplemental Fig. S8, A and C). These findings suggest that RBOH1-dependent H2O2 production plays an essential role in the adjustment of the photosynthetic processes to changes in light quality. Taken together, our results demonstrate that light quality signaling participates in the regulation of the responses of photosynthesis to chilling by regulation of HY5-ABI5-RBOH1 signaling pathways.
Plants absorb sunlight to power the photochemical reactions of photosynthesis with the generation of ROS, a process that is increased under stress (Foyer et al., 2012). Whereas ROS are highly reactive and have been proposed to accelerate photoinhibition through direct oxidative damage to PSII/PSI (Nishiyama et al., 2006), they are also vital signals relaying information concerning changes in the redox status of the chloroplast to the nucleus stress (Foyer et al., 2012). Plants have developed diverse photoprotection mechanisms to limit light-induced damage to the photosynthetic apparatus (Takahashi and Badger, 2011). Thermal energy dissipation, cyclic electron flow, and the direct transfer of energy and electrons to oxygen in pseudocyclic electron flow fulfill crucial roles in photosynthetic regulation and photoprotection (Foyer et al., 2012). The data presented here show that the increased sensitivity to cold-induced photoinhibition observed in the HY5-RNAi, pTRV-ABI5, and RBOH1-RNAi plants was linked to decreased capacity of photoprotection (Figs. 3, 5, and 6; Supplemental Figs. S3, S4, S5, S7, and 8). These findings suggest that the HY5-ABI5-RBOH1 signaling pathway plays a critical role in the induction of the photoprotection mechanisms that serve to avoid cold-induced photoinhibition.
The data presented here show that exposure to H-FR intensities induce NPQ, PsbS protein accumulation, and CEF, as well as increasing the activities of five enzymes involved in antioxidant reactions in plants experiencing cold stress. Moreover, loss of HY5, ABI5, or RBOH1 functions compromised the H-FR-induced NPQ, CEF, and the increases in antioxidant enzyme activities at low temperatures (Figs. 3, C to E; 5, C to F; and 6, D to H). We conclude that the HY5-ABI5-RBOH1 pathway is required for the FR induction of photoprotection in response to cold stress. It is worth noting that phyA, HY5-RNAi, pTRV-ABI5, and RBOH1-RNAi plants all showed reduced accumulation of the NPQ effector protein PsbS relative to wild type and they showed little response to increases in FR light intensities (Figs. 2D, 3E, 5E, and 6G). Reduced accumulation of PsbS and the insufficient trans-thylakoid ΔpH, caused by severely damaged CEF, may contribute to the impaired NPQ in these plants (Figs. 2, B to D; 3, C to E; 5, C to E; and 6, E to G). In addition, an FR-induced increase in (A + Z)/(V + A + Z) ratios was not observed in these plants (data not shown). It is plausible that the FR-activated and phyA-mediated HY5-ABI5-RBOH1-dependent signaling pathway is linked to a NPQ-specific effect on photoprotection. Whereas transcript of PGR5 was under the regulation by HY5, ABI5, and RBOH1 in response to the change in FR intensity, silencing of HY5, ABI5, and RBOH1 in PGR5-OE plants also compromised H-FR-induced increase in ΔP700max. (Supplemental Fig. S12). In this case, ΔP700max was influenced, respectively, by both the inherent PGR5 (which could be modified by light conditions) and 35S promoter-driven PGR5 (which is insensitive to the changes in light conditions). This is why H-FR altered the ΔP700max in wild-type and PGR5-OE plants to a similar degree. However, we could not exclude the possibility for the involvement of other regulatory mechanisms. HY5 is also required for the suppression of excessive ROS accumulation during acclimation to low temperatures (Catalá et al., 2011). Similarly, ABA signaling also plays a role in the expression and/or activities of antioxidant enzymes, a role that is dependent to a large extent on the induction of apoplastic H2O2 production (Zhang et al., 2007). We have previously reported that apoplastic H2O2 production plays a critical role in cold acclimation by protection of PSII (Zhou et al., 2012). Therefore, FR-induces photoprotection and suppresses excessive ROS accumulation in a HY5-, ABI5-, and RBOH1-dependent manner. The role of SOD, APX, MDAR, DHAR, and GR as well as NPQ in photoprotection has been well established in plants including tomato (Foyer et al., 1995; Chen and Gallie, 2012; Duan et al., 2012). The results presented here show that PGR5-dependent CEF is important in photoprotection in tomato leaves experiencing cold stress (Fig. 7; Supplemental Figs. S9 and S10). Similar to the apoplastic H2O2-dependent induction of the antioxidant response, the induction of CEF was also shown to be dependent on apoplastic H2O2 production (Fig. 6E). These observations are in agreement with earlier findings showing that H2O2 participates in the induction of CEF (Strand et al., 2015; Guo et al., 2016). In agreement with the role of CEF in the activation of ATP production and qE (Munekage et al., 2004; Guo et al., 2016; Yamori et al., 2016), we show that loss of PGR5 functions in the pgr5 mutant impaired H-FR-induced qE during NPQ and PsbS protein accumulation and increases in (A + Z)/(V + A + Z) ratios (Fig. 7, C to F). These results not only demonstrate the involvement of apoplastic H2O2 in the induction of ROS scavenging, CEF, and NPQ, but also emphasize the roles of CEF in photoprotection.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type tomato (Solanum lycopersicum) cv “Ailsa Craig” and cv “Moneymaker”, and the phyA, phyB1B2, and phyAB1B2 mutants in the cv Moneymaker background were obtained from the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu). HY5-RNAi, COP1-RNAi, and RBOH1-RNAi plants were generated as described previously (Liu et al., 2004; Guo et al., 2016). These transgenic plants were identified by resistance to Basta and then by quantitative real-time (qRT)-PCR analysis for the transgene. For the generation of HY5 overexpressing transgenic plants, a 474-bp full-length HY5 cDNA fragment was obtained by RT-PCR using the primer pair HY5-OE-F with an AscI site and HY5-OE-R with a SalI site (Supplemental Table S3). The PCR product was cloned into pFGC1008-HA vector behind the CaMV 35S promoter to generate the HY5-OE-HA clone. The tobacco rattle virus (TRV)-based vectors (pTRV1/2) were used for the virus-induced gene silencing (VIGS) of tomato HY5, ABI5, and RBOH1 genes with the specific PCR-amplified primers listed in Supplemental Table S3 (Liu et al., 2002). VIGS was performed as described previously (Xia et al., 2014).
PGR5 CRISPR/Cas9 vector was constructed as described by Pan et al. (2016). The target sequence (TTGGAAAGGCAGTGAGATCA) was designed using a web tool from CRISPR-P (Lei et al., 2014). The synthesized sequences were annealed and inserted into the BbsI site of AtU6-sgRNA-AtUBQ-Cas9 vector, and the AtU6-sgRNA-AtUBQ-Cas9 cassette was inserted into the HindIII and KpnI sites of the pCAMBIA1301 binary vector. To obtain the tomato PGR5 overexpressing construct, the 357-bp full-length coding DNA sequence was amplified with the primers PGR5-OE-F and PGR5-OE-R (Supplemental Table S3) using tomato cDNA as the template. The PCR product was digested with AscI and KpnI and inserted behind the CaMV 35S promoter in the plant transformation vector pFGC1008-HA. The resulting plasmids (HY5-OE-HA, PGR5 CRISPR/Cas9 vector, and PGR5-OE-HA) were transformed into Agrobacterium tumefaciens strain EHA105, and then introduced into tomato seeds of Ailsa Craig via a method as previously described (Fillatti et al., 1987). Two independent homozygous lines of the F2 generation were used for the study. Two independent pgr5 lines, pgr5#4 and pgr5#5, mutated at the first base of the protospacer adjacent motif (PAM) and stopped translation immediately (Supplemental Fig. S10, A to C).
Seedlings were grown in pots with a mixture of three parts peat to one part vermiculite, receiving Hoagland nutrient solution. The growth conditions were as follows: 12-h photoperiod, temperature of 25/20°C (day/night), and photosynthetic photo flux density (PPFD) of 600 μmol m−2 s−1.
Cold, Light, and Chemical Treatments
Plants at the 11-leaf stage were used for the determination of spatial variation in photoinhibition. Experiments were carried out in growth rooms with a 12-h photoperiod, and a PPFD of 200 μmol m−2 s−1 by providing white light from directly above the plants. Light quality analysis revealed that R/FR decreased from 1.3 at the 9th leaf rank to 0.5 at the 5th leaf rank. Growth room temperatures were controlled at either 25°C (optimal growth temperatures) or 4°C (cold stress). Other light quality treatments were carried out in controlled environment growth chambers (model no. E15; Conviron) on plants at the 6-leaf stage. Plants were grown under a 12-h/12-h light/dark cycle, with 85% humidity. For these light quality treatments, plants were exposed to cold stress at 4°C under either high R/FR (1.5), i.e. low FR intensity (L-FR, 133 μmol m−2 s−1) or low R/FR (0.5), i.e. high FR intensity (H-FR, 400 μmol m−2 s−1) light conditions. R light, supplied by LED (λmax = 660 nm; Philips), was maintained at 200 μmol m−2 s−1. FR was supplied by a FR LED (λmax = 735 nm; Philips). R/FR values were calculated via the quantum flux densities measured between 655 nm and 665 nm divided by the quantum flux densities measured between 730 nm and 740 nm.
To determine the cause of light-induced changes in photooxidizable P700 in plants exposed to low growth temperatures, fully expanded leaves were excised from the plants at 6-leaf stage and put onto petri dishes containing either water or 25 μM MV. Leaves were allowed to float on either 100 mL of 25 µM MV or water for 3 h in darkness at 25°C. The petri dishes were then transferred to the 4°C chambers and exposed to different light quality (L-FR or H-FR) conditions (R/FR, 1.5 or 0.5) for 6 h. The maximum level of P700 photooxidation (ΔP700max) was then determined in the MV-treated leaves and water-treated controls using the Dual-PAM-100 system (Heinz Walz).
Cold Tolerance Assays
Cellular membrane permeability, measured as REL, was determined after 7-d exposure to the cold stress, as described previously (Cao et al., 2007). Levels of oxidized leaf proteins were assayed by immunoblot detection as described previously (Wang et al., 2016). Plant death was recorded after 6-d recovery from the cold treatment, i.e. after return to optimal temperatures (25°C) with a 12-h/12-h light/dark cycle (PPFD of 600 μmol m−2 s−1) and 85% humidity.
Chlorophyll Fluorescence Measurements
Plants were dark-adapted for 30 min to measurement. The maximum quantum yield of PSII (Fv/Fm) and NPQ were determined with the Imaging-PAM (IMAG-MAXI; Heinz Walz) as previously described (Jin et al., 2014). qE was simultaneously measured with the Dual-PAM-100 system (Heinz Walz). Fluorescence quenching was induced by 10 min of actinic illumination with white light. The maximal fluorescence in the dark-adapted state (Fm) and in the light-adapted state (Fm′) and after 10 min of dark relaxation after actinic illumination (Fm′′) were determined using a saturating pulse of light applied at 2 min intervals. Energy-dependent quenching (qE) was calculated according to the equations qE = Fm/Fm′ – Fm/Fm′′ (Liu and Last, 2015).
P700 values were measured simultaneously with the Dual-PAM-100 system (Heinz Walz) after leaves had dark-adapted for 30 min (to obtain open reaction centers). The maximum P700 photooxidation level (ΔP700max) was determined using a saturation pulse (100 ms; 10,000 μmol m−2 s−1) under an FR background (720 nm; approximately 0.3 μmol m−2 s−1) according to the method of Klughammer and Schreiber (2008). The decrease in ΔP700max is an indicator of PSI photoinhibition.
Postillumination chlorophyll fluorescence (CEF around PSI) was monitored by the transient increase of dark-level chlorophyll fluorescence after actinic light illumination (250 μmol m−2 s−1 for 3 min) had been turned off by using a Dual-PAM-100 instrument (Heinz Walz; Nashilevitz et al., 2010).
Activity of Antioxidant Enzymes and Pigment Analysis
Frozen leaf segments (0.3 g) were ground with 2 mL ice-cold buffer containing 50 mm PBS (pH 7.8), 0.2 mm EDTA, 2 mm AsA, and 2% (w/v) polyvinylpolypyrrolidone. The homogenates were centrifuged at 4°C for 20 min at 12,000 g, and the resulting supernatants were used for the determination of enzymatic activity. The protein concentration was determined with bovine serum albumin as standard (Bradford, 1976). The activity of SOD, APX, MDAR, DHAR, and GR was measured following the protocol used as previously described (Xia et al., 2009).
Total pigments were extracted as previously described (Xu et al., 2006). Xanthophyll cycle pigments (V, violaxanthin; A, antheraxanthin; Z, zeaxanthin) were analyzed using a C30 column (YMC) equipped for HPLC (Waters) as described previously (Xu et al., 2006), with the following modification to the elution program. Mobile phases A (90% methanol), and B (tert-butyl methyl ether) were applied as follows: 92% A, 8% B, a linear gradient to 75% A and 25% B by 30 min, and gradient changed to 30% A, 70% B by 35 min; then held until 50 min, changed to 92% A and 8% B by 50.01 min, and held to the end of analysis (60 min). The de-epoxidation state of the xanthophyll cycle pigments is defined as the (A + Z)/(V + A + Z) ratio, where A, Z, and V are the concentrations of antheraxanthin, zeaxanthin, and violaxanthin, respectively.
Determination of Stomatal Aperture and Visualization of Cellular H2O2 Accumulation
Tomato stomatal apertures were measured as described previously (Xia et al., 2014) by peeling off the abaxial epidermises with forceps and floating it on a buffer containing 30 mm KCl, 10 mm 2-(N-morpholino)-ethanesulfonic acid (pH 6.15). All images were captured using a light microscope equipped with a digital camera (Leica Microsystems).
The localization of H2O2 accumulation in leaves was visualized at the subcellular level using cytochemical CeCl3 staining and transmission electron microscopy (H7650; Hitachi) as described previously (Xia et al., 2009).
Thylakoid Isolation and Immunoblot Analysis
Total protein was extracted from tomato leaves after exposure to a cold stress at 4°C under either H-FR or l-FR light conditions for 1 d as described by Wang et al. (2016). After quantification of total protein concentrations, samples of 50 µg protein were separated by SDS-PAGE electrophoresis, and immuno-labeled with primary antibodies raised against PsbS (AS09533; Agrisera). After incubation with secondary anti-rabbit antibodies (Invitrogen, Sweden), enhanced chemical luminescence (ECL) was performed to detect labeled proteins.
Fractions of intact chloroplasts were prepared from (10 g) leaves harvested from tomato plants that had been grown at either 25°C or 4°C for 3 d under either H-FR or L-FR conditions as described by Hertle et al. (2013). Thylakoid fractions were prepared from isolated chloroplasts by osmotic rupture. After centrifugation (4°C, 14,000 g, 3 min), the pellet containing the thylakoid membranes was resuspended in a buffer containing 10 mm Tris/HCl (pH 6.8), 10 mm MgCl2, and 20 mm KCl. The chlorophyll (Chl) concentration of the membranes was quantified spectrophotometrically as described by Porra et al. (1989). The thylakoid membranes (15 μg Chl at 1 mg Chl/mL) were solubilized using 2% (w/t) n-dodecyl-β-d-maltoside (Anatrace), as described by Kromdijk et al. (2016). After incubation at 30 min at 4°C with gentle agitation, insoluble fractions were removed by centrifugation (15,000 g) for 10 min at 4°C. The solubilized membrane proteins were subjected to SDS-PAGE (15% polyacrylamide) electrophoresis. Proteins were then transferred onto nitrocellulose membranes (Bio-Rad), which were then incubated with antibodies against PsaB (AS10695; Agrisera) or PsaC (AS10939; Agrisera). Secondary antibodies used in these studies were anti-rabbit (Invitrogen). Signal detection was by ECL.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from tomato leaves using an RNAprep Pure Plant Kit (Tiangen Biotech) according to the manufacturer’s instruction. Residual DNA was removed with RNase Mini Kit (Qiagen). The extracted RNA was reverse transcribed using a ReverTra Ace qPCR RT Kit (Toyobo), following the manufacturer’s recommendations. qRT-PCR experiments were performed using a Power SYBR Green PCR Master Mix kit (Takara). qRT-PCR was performed with 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, and 1 min at 72°C. The tomato ACTIN2 gene was used as an internal control. Primers sequence can be found in Supplemental Table S4. The relative gene expression was calculated following previously described formulae (Livak and Schmittgen, 2001).
RNA-seq Analysis
For tomato RNA-seq analysis, leaf tissues from 6-leaf stage tomato seedlings were collected from l-FR and H-FR treatments after 6 h under 4°C to conduct the RNA-seq analysis. Total RNA was isolated using TRIzol reagent (Biotopped) and RNA integrity was evaluated using a Bioanalyzer 2100 (Agilent). The RNA samples were then subjected to RNA sequencing by LC Sciences. Genes with P value < 0.05 and fold change ≥ 2 were regarded as differentially expressed genes.
Recombinant Protein and EMSA
The full-length coding region of HY5 was first PCR-amplified using the primers in Supplemental Table S3, then, the product was digested with BamHI and SacI and ligated into the same sites of pET-32a vector. The recombinant vector was transformed into Escherichia coli strain BL21 (DE3). The recombinant His-tagged HIS-HY5 proteins were induced by isopropyl β-d-1-thiogalactopyranoside and purified following the instructions of the Novagen pET purification system.
For binding assay, probes were biotin end-labeled following the instructions of the Biotin 3′ End DNA Labeling Kit (Cat. no. 89818; Pierce) and annealed to double-stranded probe DNA by incubating sequentially at 95°C for 5 min, then the temperature decreased from 95°C to 55°C by 40 cycles (−1°C/cycle, 1 cycle/min), 55°C for 30 min, from 55°C to 25°C by 30 cycles (−1°C/cycle, 1 cycle/min), finally, 4°C for 5 min. EMSAs of the HY5-DNA complexes were performed using biotin-labeled probes according to the instructions of the Light Shift Chemiluminescent EMSA Kit (Cat. no. 20148; Thermo Fisher Scientific). Briefly, 0.5 μg of HY5 fusion proteins were incubated together with biotin-labeled probes in 20 μL reaction mixtures containing 10 mm Tris-HCl, 1 mm DTT, 150 mm KCl, 100 mm ZnCl2, 50 ng μL−1 poly (dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, and 0.5 μg mL−1 BSA for 20 min at room temperature and separated on 6% native polyacrylamide gels in Tris-Gly buffer at 100 V. After electrophoresis, the gel was dried and autoradiographed as described previously (Xu et al., 2014).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed following the instructions of the EpiQuik Plant ChIP Kit (Cat. no. P-2014; Epigentek) as described previously (Li et al., 2011). Approximately 1 g of leaf tissue was harvested from 35S-HY5-HA and wild-type plants after cold stress under H-FR conditions. Chromatin was immunoprecipitated with an anti-HA antibody (Cat. no. 26183; Pierce), and the goat anti-mouse IgG (Cat. no. AP124P; Millipore) was used as the negative control. Both immunoprecipitated DNA and input DNA were analyzed by qRT-PCR (Light Cycler; Roche). Primers for ChIP-qPCR of the ABI5 promoters were listed in Supplemental Table S5. Each ChIP value was normalized to its respective input DNA value. All ChIP-qPCR experiments were independently performed in triplicate.
Statistical Analysis
The experimental design was a completely randomized block design with four replicates. Each replicate contained 6 to 12 plants. ANOVA was used to test for significance. When interaction terms were significant (P < 0.05), differences between means were analyzed using Turkey comparisons. Significant differences between treatment means are indicated by different letters.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S4.
Accession Numbers
Sequence data from this article can be found in Supplementary Tables. S1, S2, S4, and S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of spatial variation on the cold tolerance of tomato.
Supplemental Figure S2. Effect of FR intensity on the cold tolerance and photoprotective response of tomato.
Supplemental Figure S3. Silencing efficiency and cold tolerance of HY5-RNAi and COP1-RNAi tomato plants.
Supplemental Figure S4. Cold tolerance of HY5-overexpressing transgenic tomato lines.
Supplemental Figure S5. Transcripts of genes involved in Foyer-Halliwell-Asada cycle in wild-type, HY5-RNAi, and COP1-RNAi tomato plants.
Supplemental Figure S6. HY5 regulated ABA-mediated stomatal movement and directly binds to the G-boxes of the ABI promoters in vitro.
Supplemental Figure S7. Silencing efficiency and cold tolerance of ABI5-silenced plants.
Supplemental Figure S8. Cold tolerance of wild-type and RBOH1-RNAi transgenic tomato plants.
Supplemental Figure S9. Relative expression of CEF-related genes in response to cold stress and far red light.
Supplemental Figure S10. Transgenic tomato of pgr5 mutant and PGR5-overexpressing plants.
Supplemental Figure S11. Cold tolerance and CEF around PSI in pgr5 mutant and OE-PGR5 plants.
Supplemental Figure S12. Changes of ΔP700max in wild-type and OE-PGR5 tomato plants as altered by the silencing of HY5, ABI5, or RBOH1.
Supplemental Table S1. Differentially expressed genes of tomato plants after exposure to a cold at 4°C under H-FR and l-FR light conditions.
Supplemental Table S2. Differentially expressed genes in the photosystems and photoprotection of tomato plants after exposure to cold at 4°C under H-FR and l-FR light conditions.
Supplemental Table S3. PCR primer sequences used for vector construction.
Supplemental Table S4. List of primer sequences used for qRT-PCR analysis.
Supplemental Table S5. Primers used for ChIP-qPCR assays.
Acknowledgments
We are grateful to Prof. Jim Giovannoni of Cornell University and the Tomato Genetics Resource Center at the California University for tomato seeds. We thank Prof. Gang Lu (Zhejiang University) for denoting the CRISP/Cas9 vectors.
References
- Ahn TK, Avenson TJ, Ballottari M, Cheng Y-C, Niyogi KK, Bassi R, Fleming GR (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320: 794–797 [DOI] [PubMed] [Google Scholar]
- Allorent G, Lefebvre-Legendre L, Chappuis R, Kuntz M, Truong TB, Niyogi KK, Ulm R, Goldschmidt-Clermont M (2016) UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 113: 14864–14869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allorent G, Petroutsos D (2017) Photoreceptor-dependent regulation of photoprotection. Curr Opin Plant Biol 37: 102–108 [DOI] [PubMed] [Google Scholar]
- Ballaré CL. (2014) Light regulation of plant defense. Annu Rev Plant Biol 65: 335–363 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen Z, Gallie DR (2012) Violaxanthin de-epoxidase is rate-limiting for non-photochemical quenching under subsaturating light or during chilling in Arabidopsis. Plant Physiol Biochem 58: 66–82 [DOI] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38: 332–347 [DOI] [PubMed] [Google Scholar]
- de Wit M, Spoel SH, Sanchez-Perez GF, Gommers CMM, Pieterse CMJ, Voesenek LACJ, Pierik R (2013) Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J 75: 90–103 [DOI] [PubMed] [Google Scholar]
- Duan M, Ma NN, Li D, Deng YS, Kong FY, Lv W, Meng QW (2012) Antisense-mediated suppression of tomato thylakoidal ascorbate peroxidase influences anti-oxidant network during chilling stress. Plant Physiol Biochem 58: 37–45 [DOI] [PubMed] [Google Scholar]
- Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology (N Y) 5: 726–730 [Google Scholar]
- Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J (2012) Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 63: 1637–1661 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Ruban AV, Noctor G (2017) Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem J 474: 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Souriau N, Perret S, Lelandais M, Kunert KJ, Pruvost C, Jouanin L (1995) Overexpression of glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance to photoinhibition in poplar trees. Plant Physiol 109: 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang F, Xiang X, Ahammed GJ, Wang M, Onac E, Zhou J, Xia X, Shi K, Yin X, et al. (2016) Systemic induction of photosynthesis via illumination of the shoot apex is mediated sequentially by phytochrome B, auxin and hydrogen peroxide in tomato. Plant Physiol 172: 1259–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Gao S, Li B, Dong XC, Feng HL, Meng QW (2010) Overexpression of violaxanthin de-epoxidase gene alleviates photoinhibition of PSII and PSI in tomato during high light and chilling stress. J Plant Physiol 167: 176–183 [DOI] [PubMed] [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jin H, Liu B, Luo L, Feng D, Wang P, Liu J, Da Q, He Y, Qi K, Wang J, Wang HB (2014) HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26: 1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002) Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Kim HE, Tokura H (1995) Influence of different light intensities during the daytime on evening dressing behavior in the cold. Physiol Behav 58: 779–783 [DOI] [PubMed] [Google Scholar]
- Kingston-Smith AH, Foyer CH (2000) Bundle sheath proteins are more sensitive to oxidative damage than those of the mesophyll in maize leaves exposed to paraquat or low temperatures. J Exp Bot 51: 123–130 [PubMed] [Google Scholar]
- Kingston-Smith AH, Harbinson J, Williams J, Foyer CH (1997) Effect of chilling on carbon assimilation, enzyme activation, and photosynthetic electron transport in the absence of photoinhibition in maize leaves. Plant Physiol 114: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U (2008) Saturation pulse method for assessment of energy conversion in PSI. PAM Appl Notes 1: 11–14 [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354: 857–861 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li XP, Muller-Moule P, Gilmore AM, Niyogi KK (2002) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA 99: 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Liu J, Last RL (2015) A land plant-specific thylakoid membrane protein contributes to photosystem II maintenance in Arabidopsis thaliana. Plant J 82: 731–743 [DOI] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔΔCTmethod. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maruta T, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S (2010) Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol 51: 190–200 [DOI] [PubMed] [Google Scholar]
- Mittal A, Gampala SSL, Ritchie GL, Payton P, Burke JJ, Rock CD (2014) Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J 12: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Yang X, Ayers RA, Moffat K (2010) Structure and function of plant photoreceptors. Annu Rev Plant Biol 61: 21–47 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Murata Y, Pei ZM, Mori IC, Schroeder J (2001) Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata M, Yamamoto N, Shigeyama T, Terasawa Y, Anai T, Sakai T, Inada S, Arima S, Hashiguchi M, Akashi R, Nakayama H, Ueno D, et al. (2015) Red/far red light controls arbuscular mycorrhizal colonization via jasmonic acid and strigolactone signaling. Plant Cell Physiol 56: 2100–2109 [DOI] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Izkovich Y, Rogachev I, Osorio S, Itkin M, Adato A, Pankratov I, Hirschberg J, Fernie AR, Wolf S, Usadel B, et al. (2010) An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 22: 1977–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim Biophys Acta 1757: 742–749 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR (1997) The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA 94: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y, Kagawa Y, Kobayashi Y, Shikanai T (2008) Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol 49: 825–834 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6: 24765.10.1038/srep24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Tokutsu R, Maruyama S, Flori S, Greiner A, Magneschi L, Cusant L, Kottke T, Mittag M, Hegemann P, Finazzi G, Minagawa J (2016) A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls A and B extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Quail PH. (2002a) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180–188 [DOI] [PubMed] [Google Scholar]
- Quail PH. (2002b) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Berera R, Ilioaia C, van Stokkum IHM, Kennis JTM, Pascal AA, van Amerongen H, Robert B, Horton P, van Grondelle R (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Voesenek LACJ, Pierik R (2009) A molecular basis for the physiological variation in shade avoidance responses: a tale of two ecotypes. Plant Signal Behav 4: 528–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Strand DD, Livingston AK, Satoh-Cruz M, Froehlich JE, Maurino VG, Kramer DM (2015) Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc Natl Acad Sci USA 112: 5539–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Badger MR (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Milward SE, Fan DY, Chow WS, Badger MR (2009) How does cyclic electron flow alleviate photoinhibition in Arabidopsis? Plant Physiol 149: 1560–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Guo Z, Li H, Wang M, Onac E, Zhou J, Xia X, Shi K, Yu J, Zhou Y (2016) Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol 170: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Gao CJ, Song LX, Zhou YH, Shi K, Yu JQ (2014) Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ 37: 2036–2050 [DOI] [PubMed] [Google Scholar]
- Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ (2009) Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol 150: 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XZ, Xue YJ, Zhou JJ, Zhang B, Chang H, Takano M (2011) Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol Plant 4: 688–696 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J 54: 440–451 [DOI] [PubMed] [Google Scholar]
- Xu CJ, Fraser PD, Wang WJ, Bramley PM (2006) Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J Agric Food Chem 54: 5474–5481 [DOI] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M (2014) Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Makino A, Shikanai T (2016) A physiological role of cyclic electron transport around photosystem I in sustaining photosynthesis under fluctuating light in rice. Sci Rep 6: 20147 10.1038/srep20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1—from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Zhang A, Jiang M, Zhang J, Ding H, Xu S, Hu X, Tan M (2007) Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol 175: 36–50 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Xing L, Liu X, Hou YJ, Chinnusamy V, Wang P, Duan C, Zhu JK (2013) The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res 23: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Li X, Xia XJ, Zhou YH, Shi K, Chen Z, Yu JQ (2014) H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J Exp Bot 65: 4371–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Shi K, Xia XJ, Zhou YH, Yu JQ (2012) Hydrogen peroxide is involved in the cold acclimation-induced chilling tolerance of tomato plants. Plant Physiol Biochem 60: 141–149 [DOI] [PubMed] [Google Scholar]
- Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10: 290–295 [DOI] [PubMed] [Google Scholar]