Stage- and cell type-specific gene expression profiling reveals dynamics and characteristics of the mitosis to meiosis transition in male germinal cells of maize.
Abstract
In flowering plants, germ lines are induced from somatic meristems within reproductive organs. Within anthers, germinal cell initials first undergo several rounds of mitotic proliferation before synchronously entering meiosis. Our understanding of the progression and the molecular basis of this mitosis to meiosis transition is still limited. Taking advantage of the correlation between anther length and premeiotic germinal cell development in maize (Zea mays), we studied the transcriptome dynamics of germinal cells at three sequential stages, mitotic archesporial cells, enlarging pollen mother cells at the premeiosis interphase, and pollen mother cells at the early prophase of meiosis, using laser microdissection-based expression profiling. Our analysis showed that cells undergoing the mitosis-meiosis switch exhibit robust transcriptional changes. The three stages are distinguished by the expression of genes encoding transcription factor subsets, meiotic chromosome recombination proteins, and distinct E3 ubiquitin ligases, respectively. The transcription level of genes encoding protein turnover machinery was significantly higher in these three stages of germinal cells than in mature pollen, parenchyma cells, or seedlings. Our experimental results further indicate that many meiotic genes are not only transcribed, but also translated prior to meiosis. We suggest that the enlarging pollen mother cells stage represents a crucial turning point from mitosis to meiosis for developing germinal cells.
The transition from the mitotic cycle to the meiosis cycle in germ cells is a key step in sexual reproduction. Understanding how germ cells initiate and execute this program holds great potential for both medical and agricultural applications (Kimble, 2011). Unlike animals, plants do not have primordial germ cells that are set aside early in embryonic development. Rather, they are induced from somatic cells during the development of reproductive organs (Ma and Sundaresan, 2010). Compared to conserved meiosis-specific events (Marston and Amon, 2004), the mitosis to meiosis transition is less understood.
Anthers of maize (Zea mays) are an excellent model system for studying microsporogenesis and cell differentiation (Ma, 2005; Hamant et al., 2006; Kelliher and Walbot, 2011). One advantage is that the developmental events of microsporogenesis in maize correlate closely with anther length (Kelliher and Walbot, 2011). In the maize inbred lines W23 and B73, for example, the specification of the male germinal cell, archesporial (AR) cell, starts in anthers that are shorter than 0.2 mm (Ma et al., 2008; Kelliher and Walbot, 2014). Later, in anthers up to 0.8 to 1 mm, AR cells undergo two phases of rapid mitotic proliferation, while the surrounding somatic cells gradually form four layers by mitotic divisions and differentiation (epidermis, endothecium, middle layer, and tapetum, from outer to inner). In 1- to 2-mm anthers, AR cells stop mitotic divisions. They enlarge considerably and mature into cells competent for meiosis, named pollen mother cells (PMCs). Meiosis then begins at the 2-mm stage and ends by the 3-mm stage. In addition, maize anther development spans nearly 1 month (Ma et al., 2008). Therefore, it is possible to further resolve the stage of the mitosis to meiosis transition within the range of 0.3- to 2-mm anthers.
Transcriptome analysis has been extensively used to help address fundamental questions in maize anther development. In addition to analyzing whole anthers at various developmental stages (Ma et al., 2007, 2008; Skibbe et al., 2009; Wang et al., 2010; Nan et al., 2011; Sekhon et al., 2011; Zhang and Yang, 2014; Zhang et al., 2014), cell type-specific transcriptomes have also been reported for AR cells undergoing the first phase of mitotic proliferation in 0.30- to 0.35-mm and 0.7-mm anthers (Kelliher and Walbot, 2014; Zhang et al., 2014) and prophase I meiocytes in anthers about 2 mm long (Dukowic-Schulze et al., 2014a). Rapid progress has been made in understanding the mechanisms of AR cell fate specification and meiotic progression. However, stage- and cell type-specific gene expression profiling of developing germinal cells after the first phase of mitotic proliferation and before meiosis has not been reported. Consequently, there is a gap in our understanding of the transition from mitosis to meiosis in premeiotic germinal cells. Notably, it has been established that germinal cells express transcripts for meiotic proteins long before they actually start meiosis (Tang et al., 2010; Kelliher and Walbot, 2014), but it is unclear whether these early-expressed meiotic gene transcripts are translated into proteins before meiosis.
Here, we studied the correlation between anther length and premeiotic germinal cell development in the maize inbred B73 then, using laser microdissection, conducted transcriptome analysis on mitotic ARs in 0.7- to 0.9-mm anthers, enlarging pollen mother cells (ePMC) in 1.2- to 1.5-mm anthers, and late pollen mother cells (PMC) just starting meiosis in 1.8- to 2.0-mm anthers. These captured samples are male germinal cells at three stages: before, undergoing, and after the mitosis to meiosis transition. The expression features of transcription factor subsets and E3 ubiquitin ligases in these three stages of germinal cells indicate that the ePMC stage is a critical turning point for the mitosis-to-meiosis transition.
RESULTS AND DISCUSSION
Identification of Germinal Cell Stages in Developing Anthers of Maize Inbred B73
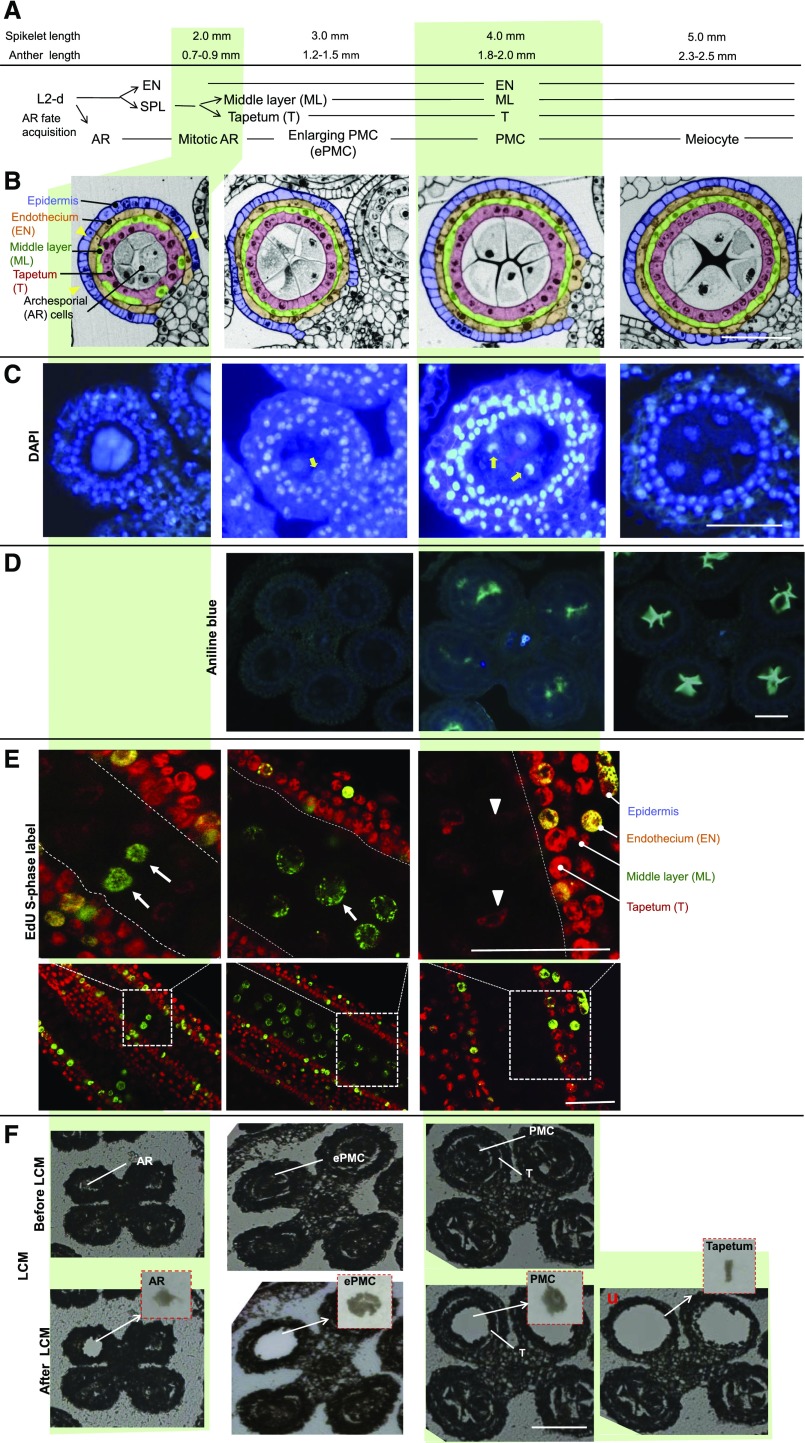
To understand the development of germinal cells during the mitosis-meiosis transition, anther lobe morphology and organization were scrutinized in maize B73 and W23 anthers of different lengths. Gene expression profiling of AR cells at the first and second phases of mitotic proliferation in 0.30- to 0.35-mm anthers and 0.7-mm anthers, respectively, have been reported for maize inbred W23 (Kelliher and Walbot, 2014; Zhang et al., 2014). We began with 0.7- to 0.9-mm anthers of inbred B73 (Fig. 1, left). Cross section of anthers at this stage showed that, in each anther lobe, three or four concentric cell layers surrounding the central germinal cells could be observed (Fig. 1B, left), indicating the differentiation of tapetum and the middle layer from the secondary parietal cells had not been completed. Longitudinal sections of anthers stained with 5-ethynyl-2'-deoxyuridine (EdU), a thymidine analog that can be incorporated into DNA during S phase in these cells, showed that many cells, including germinal cells, were proliferating at this stage (Fig. 1E, left). Therefore, we judged the anthers at this stage contained AR cells that were undergoing the second phase of mitotic proliferation before meiosis.
Figure 1.
Morphological and histochemical analysis of maize B73 anthers in 2- to 5-mm spikelets and the isolation of male germinal cells using laser microdissection. A, Developmental progression of L2-d cells in maize anthers in spikelets of given lengths. B, Cross sections of anther lobes at the stages analyzed. Yellow arrowheads point to the positions where only three concentric cell layers can be observed. AR, Archesporial cells; EN, endothecium (yellow); ePMC, enlarging pollen mother cells; ML, middle layer (green); PMC, pollen mother cells; SPL, secondary parietal layer; T, tapetum (pink). C, Cross sections of anthers stained with DAPI. Yellow arrows point to nucleolus. D, Cross sections of anthers stained with aniline blue. E, Longitudinal sections of anthers stained with EdU (green signal). Arrows indicate EdU-labeled S-phase cells. Arrowheads point to cells without EdU signal. F, Isolation of male germinal cells and tapetal cells by the Zeiss PALM microdissection system. Representative pictures of anther sections before and after laser capture were shown. Insets: Cells captured. Bars = 50 μm.
B73 AR cells accomplish mitotic proliferation by the 1-mm anther stage (Kelliher and Walbot, 2011). We next analyzed 1.2- to 1.5-mm anthers. Cross-sections showed that at this stage, the differentiation of the tapetum and middle layer had been completed (Fig. 1B). No callose deposition was observed in cell walls of germinal cells at this stage, indicated by aniline blue staining (Fig. 1D). The size of germinal cells had also increased. Noticeably, with DAPI staining (Fig. 1C), centrally positioned nucleoli (i.e. the weak staining region in DAPI stained nuclei) could readily be seen in germinal cells, suggesting that the cells were at interphase (Luck and Jordan, 1977; Yang et al., 2006). Moreover, EdU was incorporated into some germinal cells at this stage (Fig. 1E), indicating that most of the cells were at the S phase when the synchronization of meiosis had not yet been achieved (Heslop-Harrison, 1966; Boavida et al., 2005). We therefore defined germinal cells at this stage as ePMC.
In 1.8- to 2.0-mm anthers, four complete layers of somatic cells surrounding the central germinal cells were observed in the anther lobe (Fig. 1B). The central germinal cells showed aniline blue staining (Fig. 1D), indicating the start of callose deposition preceding the synchronous entry into meiosis (Abramova et al., 2003). The lack of EdU staining at this stage indicated no DNA replication. Also, DAPI staining showed that the nucleolus had moved to the side of nucleus of the germinal cell (Fig. 1C). Therefore, we considered that the germinal cells in 1.8- to 2-mm anthers were PMCs starting meiosis. This is consistent with a previous report that meiosis begins in 2-mm anthers (Kelliher and Walbot, 2011).
In 2.3- to 2.5-mm anthers, condensed chromatin was observed in germinal cells stained with DAPI, callose deposition was abundant in germinal cells stained with aniline blue, and germinal cells had started to separate from each other (Fig. 1 right). These germinal cells were in the first prophase of meiosis.
Altogether, in 0.7- to 2.0-mm maize anthers, germinal cells progressed from mitosis to meiosis, and the three stages along this transition were mitotic AR, ePMC, and PMC.
Gene Profiling of Germinal Cells in the Progress of Mitosis-to-Meiosis Transition with a Fine Spatiotemporal Resolution
Based on the correlation between anther length and germinal cell developmental stages, three homogenous germinal cell populations that cover the mitosis to meiosis transition were obtained by laser capture microdissection from maize anthers of three lengths: 0.7 to 0.9 mm, 1.2 to 1.5 mm, and 1.8 to 2.0 mm (Fig. 1F). These three cell populations were named mitotic AR cells, ePMC, and PMC, respectively.
The tapetum cells surrounding the PMCs in 1.8- to 2.0-mm anthers were also laser captured (Fig. 1F, right). These neighboring somatic cells serve as a good control for spatial resolution. Four other samples (Supplemental Fig. S1) were used for comparison: mature pollen; the final product of microsporogenesis, which is haploid and distinct from other cell types examined; 2-week-old seedlings, which contain diverse vegetative cell types and in which many of the cells are dividing or expanding; and parenchyma cells, which were isolated from the stem pith tissue of maize that had just entered the reproductive growth phase (2 months old). Parenchyma cells in stem tissue that were nondividing were also obtained by laser microdissection (Supplemental Fig. S1). In total, seven samples, including six types of cells and the seedling, were used for maize microarray hybridization.
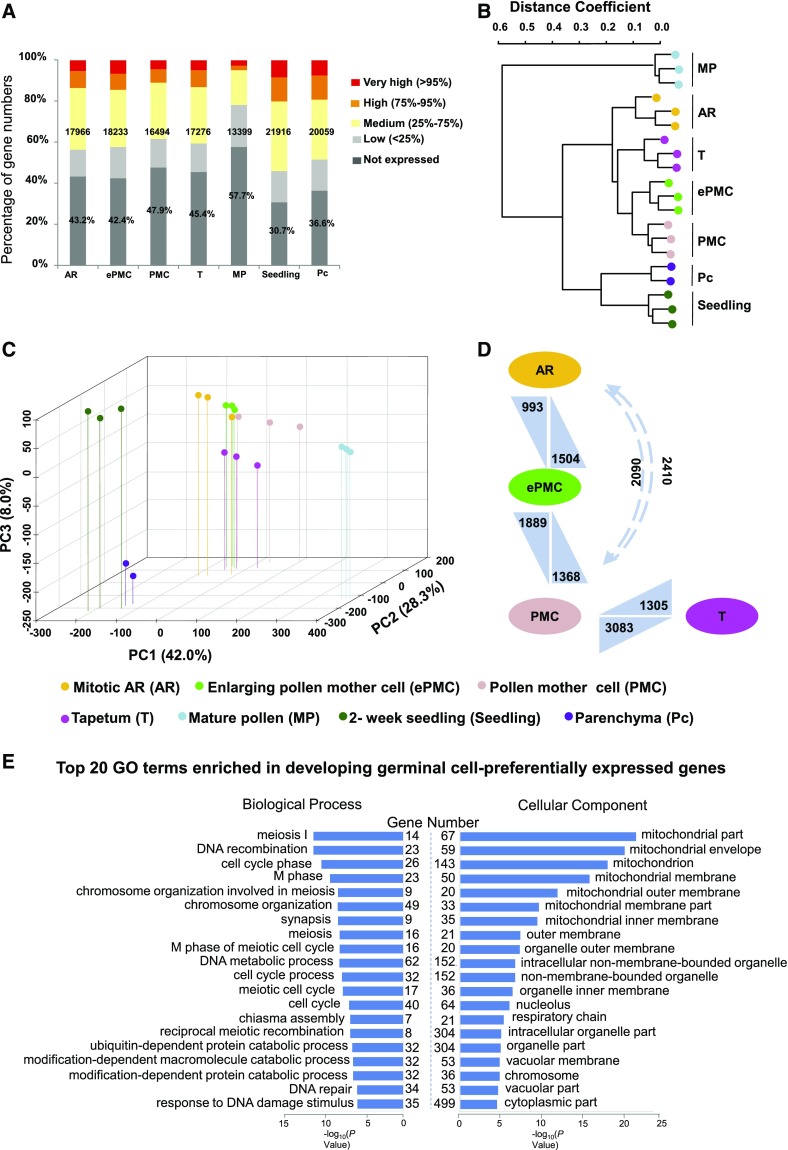
Microarray results showed that the number of expressed genes in AR, ePMC, PMC, and tapetum were similar, ranging from 16,494 to 18,233 (52.1% to 57.6% of the total 31,642 unigenes on the microarray). 21,916 (69.3%) and 20,059 (63.4%) genes were expressed in seedling or parenchyma cells, respectively. In contrast, only 42.3% of the genes (13,399) were expressed in mature pollen (Fig. 2A). In two reports on maize pollen transcriptome, one estimated that there were 10,500 pollen-expressed genes based on microarray data (Ma et al., 2008), while the other detected a total of 13,418 genes expressed in maize pollen using RNA sequencing (Davidson et al., 2011). Our data are in line with these two studies and support the notion that mature pollen maintains a transcriptome with reduced complexity (Honys and Twell, 2004).
Figure 2.
Global expression levels, principal component analysis, and hierarchical clustering for gene expression data of seven samples. A, Percentages of genes in different categories according to their expression levels. Numbers indicate expressed genes in each sample. Percentages refer to genes that are not expressed. B, Hierarchical clustering dendrogram using Pearson correlation coefficients. C, Principle component analysis for gene expression data of the seven samples. Spatial distribution of the first three principle components is shown. D, Numbers of differentially expression genes. Arrows point to the sample with up-regulated genes. E, GO enrichment analysis. The x axis indicates the significance level (scored as –log(P value)) from Fisher’s exact test and y axis represents Numbers of genes preferentially expressed in developing germinal cells fall in the indicated GO terms are listed.
We further grouped expressed genes into different categories according to their expression levels (Fig. 2A; Supplemental Dataset 1): the bottom 25th percentile was considered low-expressed, the 25th to 75th percentile medium-expressed, the 75th to 95th percentile highly expressed, and above the 95th percentile very highly expressed. In particular, genes that had signal intensities >50,000 were among the top-expressed genes.
Unsupervised hierarchical clustering of transcriptomes showed that all the biological replicates grouped together with high correlations (Fig. 2B), indicating reproducibility of our experiments. Hierarchical clustering and principal component analysis both showed that the three developing germinal cell samples and the tapetum samples were more similar to each other than to mature pollen, stem parenchyma cells, or seedling (Fig. 2, B and C), consistent with our current understanding that developing germinal cells and tapetum cells are all derived from L2 layer cells of anther primordia (Kelliher and Walbot, 2011).
Quantitative PCR and in Situ Hybridization
As a part of an effort to assess the fidelity of the microarray results, we also performed quantitative PCR after reverse transcription analysis of 20 randomly selected genes and 4 meiotic genes in the 7 samples (Supplemental Fig. S2). Expression patterns of 21 of these genes were highly consistent with microarray data (r > 0.8), and the other three were moderately consistent with microarray data (0.6 < r < 0.8).
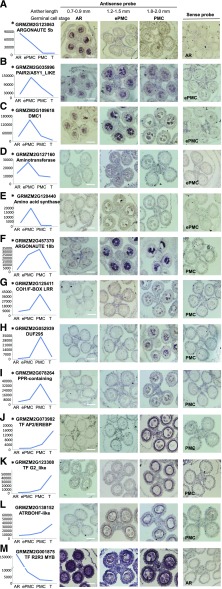
We also selected 13 genes of interest and performed in situ hybridization on developing anthers. These were genes encoding two Argonaute proteins (Fig. 3, A and F), two meiotic proteins (Fig. 3, B and C), two enzymes involved in amino acid metabolism (Fig. 3, D and E), two F-box proteins (Fig. 3, G and H), three transcription factors (Fig. 3, J, K, and M), a PPR-domain containing protein (Fig. 3I), and a NADPH oxidase (Fig. 3L). All 13 antisense probes yielded positive signal, while no hybridization signal was detected with control sense probes (Fig. 3 right). Twelve of the 13 genes showed time-course expression patterns well matched with the microarray data. Among these 12 transcripts, 8 were detected specifically in developing germinal cells and 3 exclusively in the tapetum. One probe, corresponding to a R2R3-type MYB transcription factor (GRMZM2G001875), showed a strong in situ signal in AR and lower signals in ePMC and PMC, consistent with microarray hybridization results on developing germinal cells. The only inconsistency is that in situ hybridization showed strong signal in tapetum, but the microarray showed very low expression in tapetum (Fig. 3M). This might be caused by cross-hybridization to other related MYB transcripts, as subsets of MYB transcription factors are highly expressed in premeiotic anthers and some of them share high sequence identity with the probe (Supplemental Fig. S3). Overall, our gene profiling using laser microdissection has high spatiotemporal resolution.
Figure 3.
In situ hybridizations of candidate genes in developing maize anthers at different developmental stages. Left: expression levels of candidate genes on microarray. *Developing germinal cell-preferential genes. Representative images of sense RNA probe hybridization were shown in the right-most panels, with germinal cell stage indicated. Bars = 50 μm.
Differential Expression Analysis Suggest Protein Turnover Pathway Genes Are Highly Expressed in ePMC
We then applied differential expression analysis using the Significance Analysis of Microarrays (SAM) method (Tusher et al., 2001); a 3-fold change with a false discovery rate of 0.05 was used as cutoff to select for differentially expressed genes (DEGs; Supplemental Dataset 2 A). Although the three developing germinal cells were isolated within a comparatively short time window (3 d), we identified 1,504 and 993 genes expressed significantly higher and lower, respectively, in ePMC than in AR (Fig. 2D), and 1,368 and 1,889 genes expressed significantly higher and lower respectively in PMC than in ePMC (Fig. 2D). The numbers of DEGs between two adjacent stages (AR to ePMC, and ePMC to PMC) varied from 2,497 to 3,257 (8 to 10% of total unigenes in the microarray), and a total of 4,500 DEGs (14%) were identified between AR and PMC, indicating robust transcriptional changes in developing germinal cells at these stages.
The overall correlation between PMC and tapetum cell transcriptomes was low (r = 0.44). A total of 4,388 DEGs were identified between PMC and tapetum based on microarray data (Fig. 2D). In situ hybridization verified four DEGs whose expression in PMC was significantly higher than in the tapetum (Fig. 3, F–I) and two DEGs whose expression in tapetum was significantly higher than in PMC (Fig. 3, J and K). Furthermore, there were 407 genes that were not detected in the tapetum but were expressed at a medium level or higher in PMC, and 106 genes that were not expressed in PMC but were expressed above the medium level in the tapetum (Supplemental Dataset 2B). These differences indicate effective separation of the two neighboring cell types in our sampling.
Forty-six genes showed significantly higher expression in AR than in any one of the other six samples we examined by microarray, termed AR-preferential genes (Supplemental Dataset 3). Fifty-three genes showed significantly higher expression in ePMC than in the other six samples, termed ePMC-preferential genes. A total 300 genes showed significantly higher expression in PMC than in the other six samples, termed PMC-preferential genes.
There are 2,452 genes whose expression in at least one examined germinal cell sample (AR, ePMC, or PMC) was significantly higher than their expression in mature pollen, stem parenchyma cells, and seedling (Supplemental Dataset 3). These were termed developing germinal cell-preferentially expressed genes. Gene ontology (GO) enrichment analysis showed that genes in biological processes such as meiosis I, DNA recombination, and cell cycle phase were significantly enriched in these 2,452 developing germinal cell-preferentially expressed genes (Fig. 2E). These meiosis-related processes enrichment are consistent with the expected major functions of these germinal cells. GO enrichment analysis also showed that various mitochondrial parts are the most enriched cell component categories in these 2,452 genes (Fig. 2E).
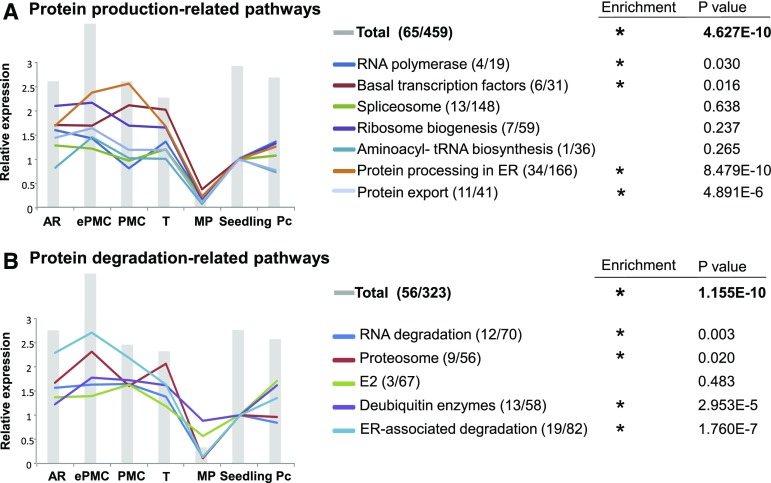
Based on annotations in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa and Goto, 2000), 459 (i.e. 1.45%) of 31,642 maize genes on the microarray are grouped into genetic information processing pathways toward protein production (including RNA polymerases, basal transcription factors, spliceosome, ribosome biogenesis, aminoacyl-tRNA biosynthesis, ER protein processing, etc). Significantly, 65 (2.65%) of those 2,452 developing germinal cell-preferentially expressed genes are grouped into genetic information processing pathways toward protein production (Supplemental Dataset 4). Genes in genetic information processing pathways toward protein product accumulation, particularly those categories of RNA polymerases, basal transcription factors, ER protein processing, and protein export, were significantly enriched in these premeiotic germinal cells-preferentially expressed genes (Fig. 4A). Interestingly, the genetic information processing pathways toward protein degradation (including RNA degradation, proteasome, ER-associated degradation, etc.) were also enriched in these germinal cell-preferentially expressed genes (Fig. 4B).
Figure 4.
Protein turnover (both production and degradation) are maintained at high levels in premeiotic germinal cells. Relative total intensities of genes in each KEGG pathway leading to protein production (A) or degradation (B). Gray bars represent total intensities of all the genes included in protein production (A) or protein degradation (B). The relative total expression of genes in each subcategory in different sample compared to that of seedling are shown in lines. The first and second numbers after each pathway indicated the number of genes present on developing germinal cell-preferential and genes present on microarray, respectively. For details, see Supplemental Dataset 4.
Furthermore, the aggregate (i.e. total combined) expression of 459 genes in pathways toward protein production or of 323 genes in pathways toward protein degradation was highest in ePMC among the seven samples (Fig. 4), and the aggregate expression for individual subcategories in these pathways also showed a similar trend: expression in ePMC was 80% to 90% higher compared to those in seedling or in stem parenchyma cells. This suggests that ePMCs may increase protein turnover (both synthesis and degradation) to exit mitotic proliferation and switch to meiosis.
We also observed extremely low expression activities in mature pollen. Mature pollen is in metabolically less active state. This is consistent with reports showing that dividing or growing cells maintain higher rates of protein turnover than cells in periods of steady state (Doherty and Whitfield, 2011; Hinkson and Elias, 2011). The highest expression level of protein turnover-related genes suggests that ePMCs are in a highly active state. The continual protein synthesis and degradation may underlie the stage transition in cells.
E3 Ubiquitin-Ligase Genes Are Enriched in ePMC- and PMC-Preferential Genes
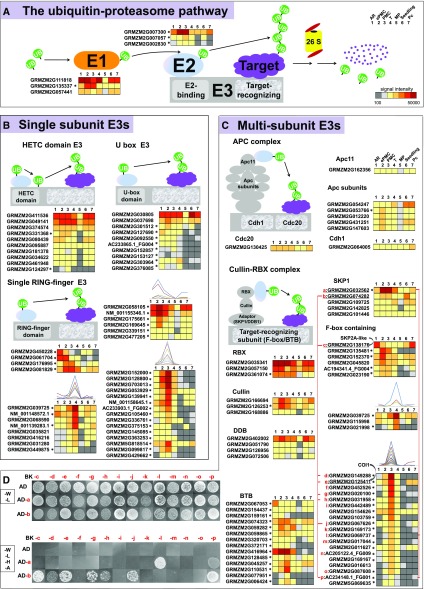
The highly activated protein turnover machinery dictates the importance of progressive protein degradation in germinal cell development. The regulation of protein degradation, which affects nearly every aspect of cell function and development, is carried out predominantly by the ubiquitin-proteasome system (Song et al., 2011). Ubiquitination of target proteins depends on the combination and sequential action of three classes of enzymes: ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2), and ubiquitin protein ligases (E3). Using the annotation of Database of Plants Ubiquitin Proteasome System (plantsUPS), 1 E1, 3 E2s, and 120 E3s are developing germinal cell-preferential genes (Fig. 5, indicated by * after the gene name).
Figure 5.
Expression of genes in ubiquitin-proteasome systems, and interactions between SKP1 and F-box proteins coexpressed in germinal cells. A, Diagram of plant ubiquitin-proteasome pathway. B and C, Expression patterns of identified single subunit E3s (B) or multisubunit E3s (C). *Developing germinal cell-preferential genes. For details, see Supplemental Table S5. D, Yeast two-hybrid assay. The coding sequences of two highly expressed SKP1-like proteins were cloned into the pray expression vector (pGADT7, AD) to test their interactions with various F-box proteins used as baits (in pGBKT7, BK). Transformants were spotted on SD/-Leu-Trp (-W –L) or SD/-Leu-Trp-His-Ade (-W –L –H -A) and grew for 1 week before imaging. Corresponding gene IDs for a top in red are provided in C.
A total of 1,156 E3 encoding genes are present on the maize microarray, that is 3.65% of total unigenes on the microarray. Furthermore, 5 of the 53 ePMC-preferential genes and 39 of the 300 PMC-preferential genes encode E3 ubiquitin ligases (Supplemental Datasets 3 and 5). Genes encoding the E3 ubiquitin ligases were significantly enriched in ePMC-preferential genes and PMC-preferential genes.
We further checked expression of three major classes of E3 ligases in plants: HECT-domain proteins and monomeric RING/U-box proteins function as single subunit E3s, and cullin-RING ligases as multisubunit E3s. Two of the 11 HETC E3s present on the microarray and eight of the U-box E3s were preferentially expressed in germinal cells (as marked with * in Figure 5B; see also Supplemental Dataset 5). Among monomeric RING type E3s, 15 members were preferentially expressed in PMCs (Fig. 5B).
Multisubunit E3s can be further divided into different subgroups: the APC complex (for Anaphase Promoting Complex), the SCF complex (for Skp-Cullin1-F-box), and the CUL3-BTB complex (for Cullin3 and Broad-complex, Tramtrack, Bric-a-Brac). Only one subunit of the APC complex, GRMZM2G053766, was developing germinal cell-preferential expressed (Fig. 5C). The SCF complex and the CUL3-BTB complex both consist of a RING-finger-containing subunit (RBX) that binds E2s, a scaffold-like cullin, a adaptor proteins (SKP1) and a target recognizing subunit (F-box protein or BTB protein, respectively). One RBX isoform, one cullin, and one SKP1 (GRMZM2G032562) were preferentially expressed in premeiotic germinal cells (Fig. 5C). GRMZM2G032562 and GRMZM2G074282 are homologs of Arabidopsis AtSKP1 (ASK1), which is essential for male gametophyte meiosis (Yang et al., 1999). Furthermore, 14 BTB proteins and 32 F-box proteins were identified as developing germinal cell-preferential expressed. Among which, 18F-box proteins were preferentially expressed in PMCs (Fig. 5C). This is consistent with our previous finding in rice (Oryza sativa) that F-box proteins were enriched in genes preferentially expressed in PMCs (Tang et al., 2010). Therefore, the appearance of numerous E3s seems to be a feature for germinal cells at the PMC stage.
Two LRR domain-containing F-box proteins are of special interest given their expression features and knowledge from homologous genes. One is GRMZM2G138176, the homolog of Arabidopsis SKP2A. SKP2A forms a SCF complex in vivo and positively regulates cell division by degrading the cell division E2FC-DPB transcription factor in Arabidopsis (Jurado et al., 2008). GRMZM2G138176 is highly expressed in AR (Fig. 5C, labeled as “c”). The other LRR-type F-box is GRMZM2G125411, the closest homolog of Arabidopsis CORONATINE INSENSITIVE (COI)-1, the receptor for jasmonates. COI1 is required for jasmonate-regulated defense and fertility (Xie et al., 1998). It is well established that as the receptor for jasmonates, the activated SCF COI1 degrades JA-ZIM (JAZ) transcriptional regulators, thus releasing JAZ-binding MYC or MYB transcription factors to direct JA-dependent transcription (Chini et al., 2007; Thines et al., 2007; Song et al., 2011). The maize homolog of COI1, GRMZM2G125411, was preferentially expressed in PMC (Fig. 5C, labeled “e”).
Yeast Two-Hybrid to Test Interactions between SKP1 and F-Box Proteins
SKP1 adaptor proteins pair with a target recognizing subunit (F-box protein) in the SCF complex to function. Our expression data highlight probable roles of the highly expressed SKP1 proteins (GRMZM2G032562 and GRMZM2G074282, labeled “a” and “b,” respectively) in the mitosis to meiosis transition. We conducted yeast two-hybrid assays to further narrow down functional SKP1-F-box protein pairs (Fig. 5D). Among the 14 F-box proteins tested (labeled c to p), homologs of SKP2A and COI1 (i.e. GRMZM2G138176 and GRMZM2G125411), along with 5 other F-box proteins may form SCF complex with one of the SKP1 proteins, GRMZM2G074282. The SKP1 protein GRMZM2G032562 interacted with two F-box proteins preferentially expressed in PMC GRMZM2G069737 and AC234148.1_FG001. AC234148.1_FG001 can interact with both SKP1 proteins. Our expression profiling and yeast two-hybrid analysis thus provided probable SKP1-F-box combinations that may function in ePMC and PMC.
Transcription Factors Are Enriched in AR-Preferential Genes
The Plant Transcription Factor Database (Pérez-Rodriguez et al., 2010) and the Maize Genetics and Genomics Database (Schaeffer et al., 2011) list approximately 3600 transcription factors (TFs) in the maize genome. However, our knowledge of which transcription factors might regulate the germinal cell mitosis to meiosis transition is still limited. Unique oligonucleotide probes corresponding to 2,000 of these TFs are present on the microarray (Supplemental Dataset 6). Among 2,452 developing germinal cell-preferential genes, there are 126 encodes putative TFs. Among these 126 TFs, only 6 have been previously identified in anthers at premeiotic stages, including one, GRMZM2G104078, that was previously identified in 0.3-mm anther when AR fate specification events are completed (Kelliher and Walbot, 2014), and 5 TFs were identified in 0.7-mm anther (Zhang et al., 2012; Supplemental Dataset 6).
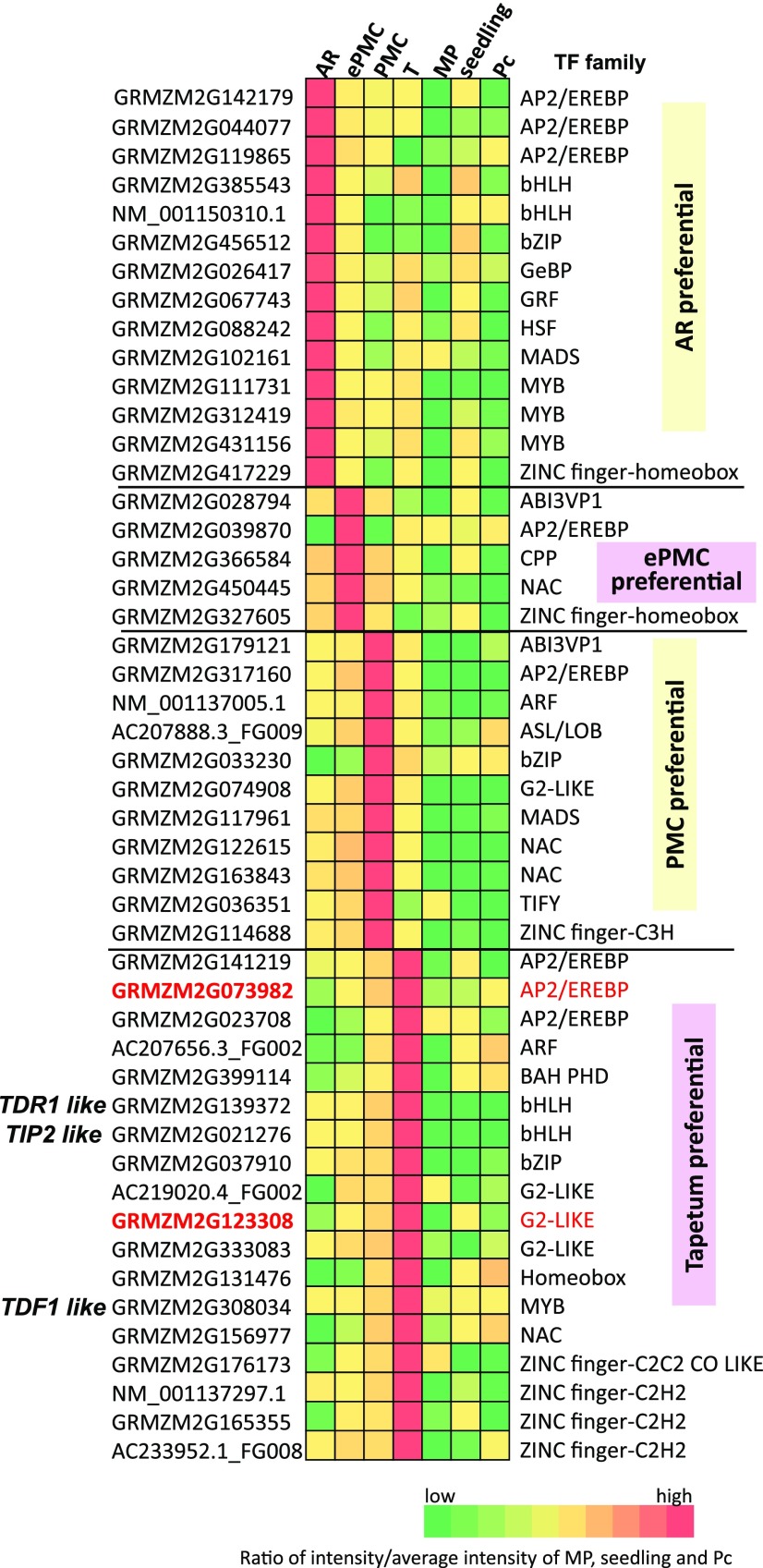
Fourteen of the 46 AR-preferential expressed genes encode putative TFs, including 3 MYBs, 3 APETALA 2/Ethylene response element binding proteins (AP2/EREBP), and 2 helix-loop-helix (bHLH) proteins. The transcription factor family is significantly enriched in AR-preferential genes (Fig. 6; Supplemental Dataset 3).
Figure 6.
A heatmap presentation of relative expression levels of transcription factors from various families that are preferentially expressed in developing germinal or tapetal cells. See also Supplemental Table S6 for detailed information. The two genes analyzed in in situ hybridization are in red.
The MYB TF family is known to play important roles in anther development. Four MYB TFs were identified from our DEGs, and three of them were specifically highly expressed in AR while the other was preferentially expressed in the tapetum (Fig. 6; Supplemental Dataset 6). Four Arabidopsis MYB-TFs, including Tapetal Development and Function 1 (TDF1; Zhu et al., 2008), AtMYB33, AtMYB65 (Millar and Gubler, 2005), and AtMYB80 (Phan et al., 2011), are essential for proper tapetal development and microspore maturation. In maize, the closest homolog for TDF1 (GRMZM2G308034) represents one of the top-expressed genes in tapetum and GRMZM2G139688, homologous to both AtMYB33 and AtMYB65, was also highly expressed in tapetum (Fig. 6). Another well-known MYB protein, DUO1 POLLEN1, regulates generative cell cycle progression, a postmeiosis event that is essential for sperm cell differentiation (Durbarry et al., 2005; Borg et al., 2011). It is not surprising that expression of its maize homolog (GRMZM2G105137) was not detected in premeiotic germinal cells (Supplemental Dataset 6).
Four rice bHLH proteins, including UNDEVELOPED TAPETUM1 (a homolog of Arabidopsis DYT1; Jung et al., 2005; Zhang et al., 2006), TAPETUM DEGENERATION RETARDATION (TDR) (Li et al., 2006), ETERNAL TAPETUM1 (EAT1; Niu et al., 2013), and TDR INTERACTING PROTEIN2 (TIP2/ bHLH142; Fu et al., 2014; Ko et al., 2014), are known to regulate tapetal cell differentiation and tapetal programmed cell death. A model suggested that TIP2 acts upstream of TDR and EAT1 and they form a regulatory cascade to control programmed cell death in tapetum. Consistent with a role for their maize homologs in tapetum development, ZmTDR1 (GRMZM2G139372), ZmEAT1 (AC233960.1_FG005), and ZmTIP2 (GRMZM2G021276) identified from our enrichment analysis, were highly expressed in the tapetum. Intriguingly, these three genes were also highly expressed in AR or PMCs. The expression of ZmEAT1, for example, reached its highest level at ePMC, was relatively low level at PMC, but maintained a high level at tapetal cells of the same stage.
Several members of plant-specific TF families have been identified as tapetum-preferential expressed, including three AP2/EREBP, three G2-like proteins, and one NAC proteins. With in situ hybridization we verified expression of an AP2/EREBP GRMZM2G073982 and a G2-like GRMZM2G123308 (Fig. 3, J and K).
In total, 14 genes encoding TF were preferentially expressed in AR, 5 TF genes were preferentially expressed in ePMC, 11 TF genes were preferentially expressed in PMC, and18 TF genes were tapetum preferentially expressed (Fig. 6). Among these TFs, only 7 are common with the 69 TFs reported as up-regulated in maize meiocytes (Dukowic-Schulzeet al., 2014b). The identification of 48 TFs preferentially expressed in AR, ePMC, PMC, or tapetum should help unravel the complex transcriptional network during early anther development.
Expression Patterns of Meiotic Genes
Transcriptome profiling of rice PMC showed that many meiotic genes were preferentially expressed in premeiotic PMCs, suggesting that at the mRNA level, the molecular machinery for meiosis was installed in advance before the cells actually carried out meiosis (Tang et al., 2010). Later, Kelliher and Walbot (2014) clearly demonstrated that in maize, meiotic factors were actively transcribed early in AR (0.3–0.35 mm), an even earlier stage prior to PMC. However, it is still not clear how the expression of these meiotic genes changes during AR specification and PMC maturation and when their mRNAs are translated. To address these questions, we looked at the expression patterns of meiosis-related genes in proliferating AR, enlarging PMC and PMC.
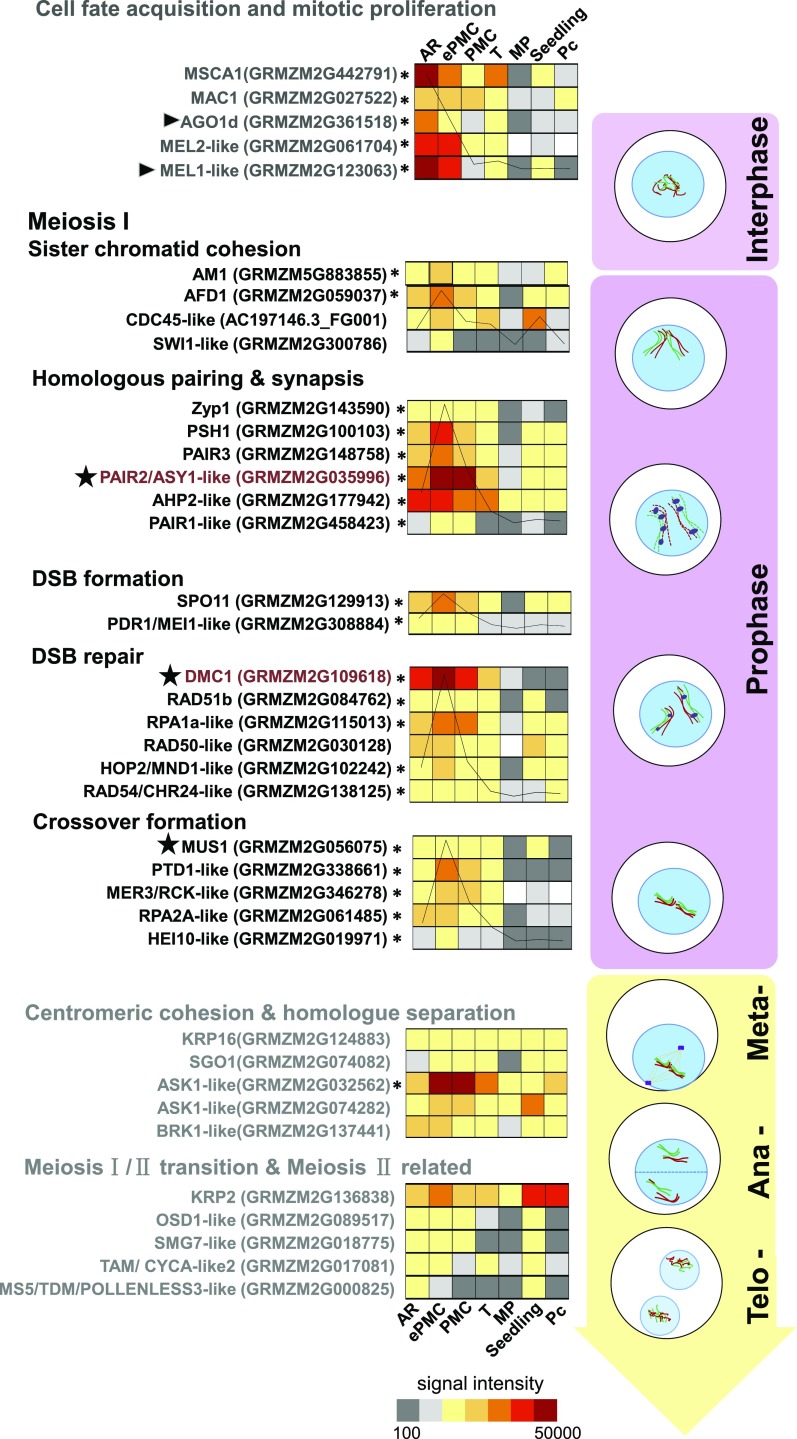
The male sterile converted anther 1 (MSCA1) glutaredoxin functions to trigger germinal cell (i.e. AR cell) initiation in maize anther primordia by hypoxia (Chaubal et al., 2003; Kelliher and Walbot, 2012). MSCA1 was highly expressed in AR, then down-regulated in ePMC and PMC (Fig. 7), consistent with its role in early steps of germinal fate determination. Rice MEL1, an ARGONAUTE (AGO) family protein, also regulates the progression of premeiotic mitosis and meiosis (Nonomura et al., 2007; Zhai et al., 2014). Two of its maize homologs (AGO1d GRMZM2G361518 and GRMZM2G123063) showed an expression pattern similar to that of MSCA1. In situ hybridization (Fig. 3A) verified that GRMZM2G123063 was preferentially expressed in AR cells. MEL2, a RNA-recognition-motif protein in rice, is required for premeiotic G1/S-phase transition (Nonomura et al., 2011), and its homolog in maize (GRMZM2G061704) was highly expressed in AR and ePMC but then markedly down-regulated in PMC (45-fold). The expression patterns of the above-mentioned genes are in agreement with their roles in early meiosis initiation.
Figure 7.
The heatmap presentation of meiotic gene expression positioned in their function events in meiosis. Signal intensity was shown for each gene. Aggregate intensities of genes within the group are charted and mounted on the heatmap. Arrowheads indicate proteins identified from premeiotic anthers at AR and ePMC stages. Stars indicate proteins identified from ePMC anthers. *Developing germinal cell-preferential genes. Genes analyzed in in situ hybridization are in red.
We also found that GRMZM2G300786 is expressed higher in AR than in later stages in germinal cells. It is a homolog of the Arabidopsis protein SWITCH1, which functions in meiotic chromosome remodeling (Mercier et al., 2003). The maize multiple archesporial cells 1 (MAC1) gene controls the proliferation of AR cells (Wang et al., 2012a). The expression of MAC1 was sustained from AR to PMC.
Ameiotic 1 (am1, GRMZM5G883855), a plant-specific gene, affects the switch from mitotic to meiotic cell cycle (Pawlowski et al., 2009) and showed higher expression in ePMC than in AR or in PMC. Genes encoding the synaptonemal complex protein Zyp1 (GRMZM2G143590) and absence of first division 1 (AFD1; GRMZM2G059037) both showed ePMC preferential expression. Furthermore, the meiosis-specific DMC1 promotes strand exchange that is required for the recombination repair of double strand breaks (Li et al., 1997). OsDMC1 (Tang et al., 2010) and ZmDMC1 (Kelliher and Walbot, 2014) were preferentially expressed in rice PMC and maize AR (0.3–0.35 mm), respectively. In three replicates, the average signal intensity (SI) for ZmDMC1 (GRMZM2G109618) in AR was 35,184, while in ePMC, it rose to 168,001 and then went down to 38,195 in PMC. For another example, GRMZM2G035996, the maize homolog of PAIR2/ASY1, which functions in meiotic chromosome synapsis (Armstrong et al., 2002; Nonomura et al., 2006), also showed highest expression in ePMC (Fig. 7). In conclusion, we found that the majority of meiotic process genes showed a similar expression pattern: they were expressed in AR and their expression levels peaked at ePMC then went down at PMC (Fig. 7, detailed description of the meiotic genes is in Supplemental Dataset 7).
We found that several genes functioning in later stages of meiosis did not show highest expression in ePMC. Arabidopsis ASK1, as an essential component of the SCF E3 complex, is thought to participate in cohesion distribution during late prophaseI (Zhao et al., 2006) and control homolog separation at anaphaseI (Yang et al., 1999). The expression of maize ASK1 homologs (GRMZM2G032562 and GRMZM2G074282) increased significantly in ePMC and PMC. STUD/TETRASPORE/AtNACK2, which encodes a kinesin, is required for male meiotic cytokinesis in meiosisII (Spielman et al., 1997; Yang et al., 2003; Tanaka et al., 2004). The expression of the gene encoding the putative kinesin-related protein2 (KRP2, GRMZM2G136838) was low in AR, and increased in ePMC then remained at a comparable level in PMC. Other genes, including a kinesin KRP16 (ATK1 homolog, Chen et al., 2002), a spindle checkpoint kinase (BRK1 homolog; Wang et al., 2012b), a centromeric cohesion protein (ZmSGO1), an A-type cyclin (TAM/CYCA1;2 homolog), a component of nonsense-mediated RNA decay machinery (SMG7 homolog; Riehs et al., 2008), and OSD1, which is essential for the meiosisI/ meiosisII transition in Arabidopsis (d’Erfurth et al., 2009), did not show specific up-regulation at the ePMC stage.
Zhang et al. (2014) annotated 222 maize genes as putative meiotic genes based on mis-regulated genes in am1 mutants and most of them were constitutively expressed in anthers <1 mm. There are 179 annotated genes included in our microarray but not included in the above list of known or putative meiotic genes. Supplemental Dataset 7D provides detailed expression data of these 179 genes in our samples; 61% of these genes were expressed higher in premeiotic germinal cells than in seedling, parenchyma cells, or tapetum cells. These two gene sets for putative meiotic genes are likely complementary and both should be useful for identifying genes directly involved in meiosis.
Mass Spectrometry Detected Protein Products of Several Meiotic Genes in AR and/or ePMC
The cell-type and -stage specific transcriptomic results provided strong evidence that many meiotic genes are transcribed before meiosis, but it remained a question whether these meiosis-associated transcripts were also translated before meiosis. Zhang et al. (2014) tried to investigate whether high-abundance transcripts would be translated at the same stage by comparing microarray data with proteomics data of maize anthers. However, they did not find a good correlation between transcript abundance and protein abundance at a given developmental stage. We argue that a correlation might not be readily observable for transcriptome data and proteomic data from mixed tissues, given that the sensitivity of protein detection is much more limited than transcript detection. To address this question with increased sensitivity, proteins from premeiotic anthers at the AR stage or the ePMC stage were isolated and then size-fractionated by SDS-PAGE. Proteins within three gel sections, 30 to 50 kD, 60 to 80 kD, and 100 to 120 kD, were subjected to trypsin digestion and sequenced by electrospray ionization-mass spectrometry. From three biological replicates, a total of 2,027 and 2,818 proteins were reproducibly detected in maize anthers at AR and ePMC stages, respectively (Supplemental Dataset 8). Among these, 56 known or putative meiotic proteins were identified in per-meiotic stage anthers (Supplemental Dataset 8). For example, the MEL1-like protein (GRMZM2G123063) was detected in AR anthers and in ePMC anthers, consistent with their expression patterns and putative functions in promoting premeiotic mitosis. Other known or putative meiotic proteins detected in ePMC anthers include DMC1, MUS1, PAIR2/ASY2-like, RAD51a2, consistent with the idea that these meiotic proteins are translated in ePMC. In line with this idea, the meiotic protein ASY1 in Arabidopsis was detected in pollen mother cells during meiotic interphase (Armstrong et al., 2002). Protein products of 56 genes possibly functioning in meiosis were identified in premeiotic anthers (Supplemental Dataset 8), indicating that these meiotic genes were translated before meiosis.
Potential Role for Phenylpropanoid Metabolism in Germinal Cell Development
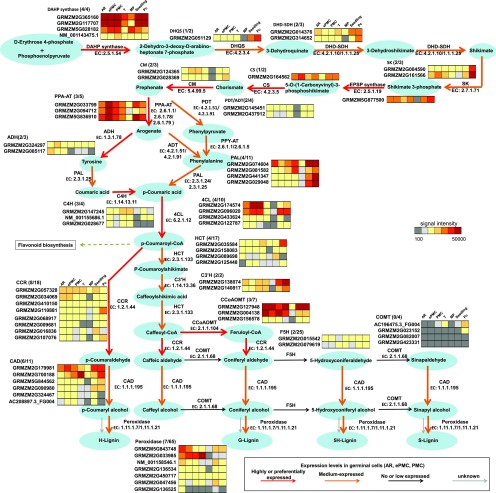
Regulation at the mRNA and protein levels ultimately changes cell metabolic homeostasis. The regulation of specific metabolic pathways is important to cell behaviors. For example, in AR initials, the germinal cells acclimate to hypoxia by using alternative metabolic pathways to ensure ATP and reduce power generation without respiration (Kelliher and Walbot, 2014). We mapped our gene expression data to metabolic pathways. It is interesting that 35 genes annotated to function in the shikimate or phenylpropanoid pathways were highly or very highly expressed in examined germinal cells, and 14 of these pathway genes were developing germinal cell-preferentially expressed (Supplemental Dataset 9). The genes highly or preferentially expressed in developing germinal cells include 3-deoxy-7-phosphoheptulonate synthase and chorismate synthase, the first and the last enzyme in the common shikimate pathway that leads to the biosynthesis of Phe, Tyr, and Trp. This set also includes l-Phe ammonia-lyase, 4-Coumarate: Coenzyme A ligase and caffeoyl-CoA O-methyltransferase, three core enzymes in the phenylpropanoid pathway, which generates the secondary metabolites lignin and flavonoids (Fig. 8, red arrows).
Figure 8.
Phenylpropanoid pathway gene expression in developing germinal cells. Arrow colors indicate the aggregate expression of genes encoding enzymes responsible for the reaction. The first number in the brackets indicates number of genes expressed in developing germinal cells. The second number indicates number of genes on our microarray. ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; CAD, cinnamyl-alcohol dehydrogenase; CCR, cinnamoyl-CoA reductase; C3′H, 5-O-(4-coumaroyl)-d-quinate 3′-monooxygenase; C4H, transcinnamate 4-monooxygenase; 4CL, 4-coumarate CoA ligase; CM, Chorismate mutase; CS, chorismate synthase; DAHP synthase, 3-deoxy-7-phosphoheptulonate synthase; DHD-SDH, shikimate dehydrogenase; DHQS, 3-dehydroquinate synthase; EPSP synthase, enolpyruvylshikimate phosphate synthase; F5H, ferulate-5- hydroxylase; HCT, shikimate O-hydroxycinnamoyltransferase; HPP-AT, histidinol phosphate aminotransferase; PAL, plasmalogen synthase; PDH, prephenate dehydrogenase; PDT, prephenate dehydratase; PPA-AT, Asp-prephenate aminotransferase; PPY-AT, Asp transaminase; SK, shikimate kinase.
In contrast, the transcripts of three core enzymes responsible for flavonoid biosynthesis, namely chalcone synthase, chalcone isomerase, and chalcone reductase, were either absent or maintained at low levels (Supplemental Dataset 9), suggesting that active Phe metabolism in germinal cells does not result in flavonoid production.
We further found that genes responsible for the final steps of monolignol synthesis are very low or not expressed (Fig. 8, black arrows). For example, Ferulate-5- hydroxylase (F5H) and caffeate O-methyltransferase (COMT) catalyze hydroxylation or further O-methylation of coniferyl aldehyde or coniferyl alcohol, which produces of 5H-Lignin or S-Lignin. Thus, our expression data suggest that the final product, lignin, may not be highly synthesized in developing germinal cells. This is consistent with the presumption that developing germinal cells are not producing secondary cell walls.
Based on these expression data, we suggest that certain intermediates of phenylpropanoid metabolism (such as ferulic acid and caffeic acid) may accumulate in developing germinal cells. This may help in understanding why genetic engineering in the phenylpropanoid pathway aiming to reduce total lignin content in plants for biofuel production has been hampered by defects in pollen development. For example, over-expression of F5H in COMT-deficient Arabidopsis causes ectopic enrichment of 5H-substituted phenolic precursors and leads to male sterility (Weng et al., 2010). Mutation in the Cinnamate 4-hydroxylase gene (C4H) also results in male sterility (Schilmiller et al., 2009). Interestingly, in reduced epidermal fluorescence 3 (ref3) mutants, which harbor mis-sense mutations in C4H, pollen was not present in open flowers or in anthers at earlier developmental stages. It was not known whether male germinal cells in these mutants carry out meiosis normally. Our results suggest that meiosis abnormality to be a possible reason for causing ref3 male sterility. The significance of phenylpropanoid metabolism in germinal cell development thus deserves further investigation.
The intermediates of phenylpropanoid metabolism (such as ferulic acid and caffeic acid) are known to have antioxidant activity (Kanski et al., 2002; Xue et al., 2015). Redox status is a determinant of maize germ cell fate, as hypoxia in anthers triggers germinal cell fate acquisition (Kelliher and Walbot, 2012). It might be worthwhile to explore whether increasing partial phenylpropanoid metabolism to produce antioxidant intermediates is related to maintaining cell fate in maize germinal cells at the mitosis to meiosis transition.
MATERIALS AND METHODS
Plant Material
Maize (Zea mays) inbred line B73 plants were grown in the greenhouse under 14 h light and 10 h dark per day in a growth chamber at 22°C to 26°C with 65% relative humidity. For spikelet collection, only spikelets around the central region of a tassel stalk were collected to ensure a good correlation between the spikelet length and anther length. To define the exact sampling region, several spikelets were first dissected to make sure the anthers within would fall in certain length ranges. Based on our observation, 2-mm spikelets (anther length 0.7–0.9 mm), 3-mm spikelets (anther length 1.2–1.5 mm), 4-mm spikelets (anther length 1.8–2.0 mm), and 5-mm spikelets (anther length 2.3–2.5 mm) were collected for following analysis. To discriminate anther developmental stages in maize B73, 2-mm spikelet (anther length 0.7–0.9 mm), 3-mm spikelet (anther length 1.2–1.5 mm), 4-mm spikelet (anther length 1.8–2.0 mm), and 5-mm spikelet (anther length 2.3–2.5 mm) were made into semithin sections, stained with 1% toluidine blue (0.1%) or 4’,6-diamino-2-phenylindole solution (1 μg/mL, with 0.2% [v/v] Tween 20) and examined.
Aniline BLUE and EdU staining
For callose staining, transverse anther sections were stained with 0.1% (w/v) aniline blue in 0.1 m K2HPO4 (pH 10) for 5 min and visualized under UV illumination using a fluorescence microscope (Olympus BX51).
EdU and PI (propidium iodide) staining were performed according to Kotogány et al., (2010). Anthers of each stage were incubated in N6 culture medium with 20 μg/ml EdU (5-ethynyl-2'-deoxyuridine, catalog no. A10044, Invitrogen) at room temperature for 6 h to allow EdU incorporation. Then anthers were fixed in phosphate buffered saline (PBS) with 4% (w/v) formaldehyde and 0.1% Triton X-100 for 15 min, washed twice by PBS with 2% BSA. The samples were further permeabilized in 10% KOH at 85°C for 2 min and washed twice again by PBS with 2% BSA. The detection of the incorporated EdU using click-it color reaction cocktail was performed according to manufacturer’s instruction (catalog no. C10350, Life Technologies). The whole anthers were then incubated in PI (20 μg/mL, catalog no. 40711ES10, Yeasen) staining solution for 30 min, rinsed twice in PBS, and kept at 4°C in the dark for up to 2 weeks until used for observation. Slides were observed using a Leica TSC SP8 with excitation/emission spectra of 559/570-620 nm for PI and 495/519 nm for EdU. Seedlings used in this study were collected 14 d after planting in soil; parenchyma cells were obtained from 2-month-old B73 plants while mature pollen grains were collected from 75-d-old maize tassels.
Laser Microdissection and Microarray Hybridization
For laser microdissection, germinal cells or tapetal cells we isolated from 2-mm, 3-mm, or 4-mm spikelets. Spikelets or stem sections were fixated by microwave-accelerated acetone method and embedded by paraffin as described by Tang et al. (2006). Then 0.10-μm-thick cross-sections were cut using a paraffin-tape transfer system (Leica) on a rotary microtome (Leica RM2235_CN, Leica Biosystems Nussloch). The target cells were isolated and captured using ZEISS PALM Microbeam laser microdissection system. For each cell type, around 800 cells were captured per biological replicate. Total RNA from Laser microdissection samples were extracted with the PicoPure RNA isolation kit (catalog no. 12204, Applied Biosystems). An average of 8 ng of total RNA can be obtained per sample from 800 cells. The concentration and integrity of RNA samples were measured by Agilent Bioanalyzer 2100 (Agilent Technologies) as previously described (Tang et al., 2006; 2010). Amino-ally1 aRNA was recovered through 2-round amplification using a Target Amp 2-Round Aminoallyl-aRNA Amplification Kit (catalog no. TAA2R4924, Epicentre Biotechnologies) and a Qiagen RNeasy MinElute Cleanup Kit (catalog no. 74204, Qiagen), starting from approximately 1 ng of total RNA. Total RNA of seedling and mature pollen were extracted using EasyPure Plant RNA Kit (catalog no. DP432, Tiangen Biotech) and amplified as above. Three biological replicates were obtained for each sample except for the parenchyma cells, which had two biological replicates. cRNA was hybridized in a 4 x 44K format two-color microarray based on ZmB73 4a.53-filtered cDNA data (http://ftp.maizesequence.org/release-4a.53/filtered-set/) and NCBI B73 cDNA data (design ID: 026609; Agilent Technologies) at ShanghaiBiotechnology Corporation. Probes for 31,642 unigenes were designed by Agilent eArray (https://earray.chem.agilent.com/earray/).
Microarray Data Processing and Analysis
Microarray data were processed as described (Tang et al., 2010). Based on the 719 negative probes on our microarray chip, a signal intensity (SI) of 101 that represented the 99th percentile of their intensities was defined as the threshold for significant gene expression (Jiao et al., 2009). The genes are considered as expressed in a sample if SI of at least two replicates and the average SI are all greater than the threshold. In the case of parenchyma cells where only two replicates exist, only the average SI is considered. To identify differentially expressed genes, we used the web-enabled and cross-platform Significance Analysis of Microarrays (SAM 5.0) software package via Shiny (http://statweb.stanford.edu/∼tibs/SAM/). Differentially expressed genes were extracted using a 3-fold and a false discovery rate of 0.05 cutoff for significance.
GO enrichment analysis was performed with the AgriGO portal (http://bioinfo.cau.edu.cn/agriGO/analysis.php) (Du et al., 2010). The input sample list was 2,452 developing germinal cell-preferentially expressed genes and the background was the total 31,642 unigenes on our microarray data. The enrichment criteria include FDR < 0.05.
Real-Time PCR
For quantitative PCR, approximately 300 ng aRNA was used to synthesize cDNA with EasyScript Fisrt-Strand cDNA Synthesis SuperMix Kit (catalog no. AT301, Transgen Biotech) using oligo dT. Quantitative PCR was performed with about 10 ng cDNA using the Transcript Green One-Step qRT-PCR SuperMix Kit (catalog no. AQ211, Transgen Biotech). Primers used were listed in Supplemental Dataset 10. A Bio-Rad IQ double-color real time PCR detection system was used in conjunction with the Bio-Rad CFX Manage Software (Bio-Rad). The program used was as follows: an initial denaturation step at 95°C for 30s followed by 40 amplification cycles at 95°C for 5 s and 55°C for 30 s. All above steps were performed according to the manufacturer’s instructions.
In Situ Hybridization
RNA in situ hybridizations were performed on paraffin-embedded cross-sections (10 μm) of 2-mm, 3-mm, and 4-mm spikelets. Targeting cDNAs were subcloned into pBluescript SK+ vector. Thereby, sense and antisense probes were synthesized by in vivo transcription using T7 and T3 RNA polymerase, respectively, using DIG RNA labeling kit (catalog no. 11175025910, Roche). The details of primer sequences can be found in Supplemental Dataset 10. Hybridization procedure closely followed the protocol previously described (Tang et al., 2010).
Yeast Two-Hybrid Assays
Coding sequences of SKP1-like genes or F-box-like genes were cloned and subcloned to pGADT7 or pGBKT7 (MATCHMAKER GAL4 Two-Hybrid System, catalog no. PT3247, Clontech), respectively, using CloneExpressII One Step Cloning Kit (catalog no. C112, Vazyme). Yeast strain AH109 harboring appropriate plasmids were spotted on SD medium lacking Trp/Leu (-W, -l) or Trp/Leu/His/adenine (-W, -l, -H, -A) and imaged 5 d after growth.
Protein Extraction and Peptide Analysis
Three biological replicate samples of 0.7- to 0.9-mm anthers (around 200 anthers per sample), 1.2- to 1.5-mm anthers (around 200 anthers per sample), mature pollen (200 mg per sample), and 2-week-old seedlings (3 seedlings per sample) of maize B73 were collected and homogenized in liquid nitrogen with extraction buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 10% [v/v] glycerol, 0.1% Nonidet P-40, 1 mm DTT, and 1XProtease Inhibitor Cocktail Complete [Boehringer Mannheim]). The protein samples were immediately mixed with equal volume of 2x SDS-PAGE sample loading buffer (Mybiosource, MBS355551), boiled for 5 min, and loaded on 10% SDS-PAGE gels. Proteins were separated by SDS-PAGE. Gel sections containing protein bands ranging between 30 and 50 kD, 60 and 80 kD, and 100 and 120 kD were excised and digested in-gel with trypsin. Peptide analysis was carried out as described (Huang et al., 2014). Tandem mass spectrometry spectra were searched against an in-house database constructed with the FASTA protein sequences (proteome MAIZE GDB.REVERSED.fasta) downloaded from http://www.plantgdb.org/download/download.php?dir=/PublicPlantSeq/Dump/Z/Zea_mays/FASTA. Proteome of mature pollen was also analyzed according to the above procedure, except that the whole gel lane rather than gel sections were analyzed.
Accession Numbers
The Agilent microarray data from this study have been deposited at GEO and are available under the accession number GSE70215.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative images of three samples used for comparison.
Supplemental Figure S2. Quantitative RT-PCR analysis of 20 randomly selected genes and 4 meiotic genes.
Supplemental Figure S3. Nucleotide sequence of GRMZM2G001875 and alignment details within the probe region.
Supplemental Dataset 1. Detailed expression data of 31,642 unigenes in seven samples.
Supplemental Dataset 2. Lists of differentially expressed genes.
Supplemental Dataset 3. Detailed expression data of various preferentially expressed genes.
Supplemental Dataset 4. Microarray data for genes involved in protein turnover.
Supplemental Dataset 5. Detailed expression data of genes in the ubiquitin-proteasome pathway.
Supplemental Dataset 6. Detailed expression data of known or putative transcription factor genes.
Supplemental Dataset 7. Detailed expression data of known and putative meiosis genes.
Supplemental Dataset 8. Proteins identified from anthers at AR or ePMC stage by electrospray ionization-mass spectrometry.
Supplemental Dataset 9. Expression data of phenylpropanoid pathway genes.
Supplemental Dataset 10. Primers used in this study.
Acknowledgments
We thank Drs. Hong Ma, Yufeng Wang, and Xuan Li for discussion and Sheila McCormick for editing the manuscript.
Footnotes
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences “Molecular Mechanism of Plant Growth and Development,” the National Key Research and Development Program of China (2016YFD0100600), the National GMO project (2016ZX08009-003), and the Natural Science Foundation of China (Grant 31570318).
Articles can be viewed without a subscription.
References
- Abramova LI, Avalkina NA, Golubeva EA, Pyzhenkova ZS, Golubovskaya IN (2003) Synthesis and deposition of callose in anthers and ovules of meiotic mutants of maize (Zea mays). Russ J Plant Physiol 50: 324–329 [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Boavida LC, Becker JD, Feijó JA (2005) The making of gametes in higher plants. Int J Dev Biol 49: 595–614 [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D (2011) The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23: 534–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubal R, Anderson JR, Trimnell MR, Fox TW, Albertsen MC, Bedinger P (2003) The transformation of anthers in the msca1 mutant of maize. Planta 216: 778–788 [DOI] [PubMed] [Google Scholar]
- Chen C, Marcus A, Li W, Hu Y, Calzada JPV, Grossniklaus U, Cyr RJ, Ma H (2002) The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin HN, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. (2011) Utility of RNA Sequencing for Analysis of Maize Reproductive Transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R (2009) Turning meiosis into mitosis. PLoS Biol 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty MK, Whitfield PD (2011) Proteomics moves from expression to turnover: update and future perspective. Expert Rev Proteomics 8: 325–334 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze S, Sundararajan A, Mudge J, Ramaraj T, Farmer AD, Wang M, Sun Q, Pillardy J, Kianian S, Retzel EF, et al. (2014a) The transcriptome landscape of early maize meiosis. BMC Plant Biol 14: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze S, Harris A, Li J, Sundararajan A, Mudge J, Retzel EF, Pawlowski WP, Chen C (2014b) Comparative transcriptomics of early meiosis in Arabidopsis and maize. J Genet Genomics 41: 139–152 [DOI] [PubMed] [Google Scholar]
- Durbarry A, Vizir I, Twell D (2005) Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol 137: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The Rice Basic Helix-Loop-Helix Transcription Factor TDR INTERACTING PROTEIN2 Is a Central Switch in Early Anther Development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ (2006) Genetics of meiotic prophase I in plants. Annu Rev Plant Biol 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1966) Cytoplasmic connections between angiosperm meiocytes. Oxford JournalsScience & Mathematics Annals of Botany 30: 221–222 [Google Scholar]
- Hinkson IV, Elias JE (2011) The dynamic state of protein turnover: it’s about time. Trends Cell Biol 21: 293–303 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WJ, Liu HK, McCormick S, Tang WH (2014) Tomato pistil factor STIG1 promotes in vivo pollen tube growth by binding to phosphatidylinositol 3-phosphate and the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 26: 2505–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Díaz-Triviño S, Abraham Z, Manzano C, Gutierrez C, del Pozo C (2008) SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J 53: 828–841 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanski J, Aksenova M, Stoyanova A, Butterfield DA (2002) Ferulic acid antioxidant protection against hydroxyl and peroxyl radical oxidation in synaptosomal and neuronal cell culture systems in vitro: structure-activity studies. J Nutr Biochem 13: 273–281 [DOI] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2011) Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol 350: 32–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2014) Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J 77: 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. (2011) Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol 3: a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Sun-Ben Ku M, Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC, et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotogány E, Dudits D, Horváth GV, Ayaydin F (2010) A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Golub EI, Gupta R, Radding CM (1997) Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci USA 94: 11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck BT, Jordan EG (1977) The nucleolus and meiosis during microsporogenesis in Endymion non-scriptus (L.). J Cell Sci 25: 111–123 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Ma H, Sundaresan V (2010) Development of flowering plant gametophytes. Curr Top Dev Biol 91: 379–412 [DOI] [PubMed] [Google Scholar]
- Ma J, Duncan D, Morrow DJ, Fernandes J, Walbot V (2007) Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell types. Plant J 50: 637–648 [DOI] [PubMed] [Google Scholar]
- Ma J, Skibbe DS, Fernandes J, Walbot V (2008) Male reproductive development: gene expression profiling of maize anther and pollen ontogeny. Genome Biol 9: R181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A (2004) Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FCH (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130: 3309–3318 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan GL, Ronceret A, Wang RC, Fernandes JF, Cande WZ, Walbot V (2011) Global transcriptome analysis of two ameiotic1 alleles in maize anthers: defining steps in meiotic entry and progression through prophase I. BMC Plant Biol 11: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Nonomura K, Eiguchi M, Nakano M, Takashima K, Komeda N, Fukuchi S, Miyazaki S, Miyao A, Hirochika H, Kurata N (2011) A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N (2007) A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Wang CJ, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ (2009) Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc Natl Acad Sci USA 106: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K (2008) Arabidopsis SMG7 protein is required for exit from meiosis. J Cell Sci 121: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Schaeffer ML, Harper LC, Gardiner JM, Andorf CM, Campbell DA, Cannon EK, Sen TZ, Lawrence CJ (2011) MaizeGDB: curation and outreach go hand-in-hand. Database (Oxford) 2011: bar022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Skibbe DS, Fernandes JF, Medzihradszky KF, Burlingame AL, Walbot V (2009) Mutator transposon activity reprograms the transcriptomes and proteomes of developing maize anthers. Plant J 59: 622–633 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D (2011) The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman M, Preuss D, Li FL, Browne WE, Scott RJ, Dickinson HG (1997) TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124: 2645–2657 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishikawa M, Kitamura S, Takahashi Y, Soyano T, Machida C, Machida Y (2004) The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9: 1199–1211 [DOI] [PubMed] [Google Scholar]
- Tang W, Coughlan S, Crane E, Beatty M, Duvick J (2006) The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola. Mol Plant Microbe Interact 19: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Tang X, Zhang ZY, Zhang WJ, Zhao XM, Li X, Zhang D, Liu QQ, Tang WH (2010) Global gene profiling of laser-captured pollen mother cells indicates molecular pathways and gene subfamilies involved in rice meiosis. Plant Physiol 154: 1855–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ (2012a) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Oses-Prieto JA, Li KH, Fernandes JF, Burlingame AL, Walbot V (2010) The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis-regulation of metabolic functions. Plant J 63: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tang D, Luo Q, Jin Y, Shen Y, Wang K, Cheng Z (2012b) BRK1, a Bub1-related kinase, is essential for generating proper tension between homologous kinetochores at metaphase I of rice meiosis. Plant Cell 24: 4961–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Mo H, Chapple C (2010) Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64: 898–911 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xue J, Luo D, Xu D, Zeng M, Cui X, Li L, Huang H (2015) CCR1, an enzyme required for lignin biosynthesis in Arabidopsis, mediates cell proliferation exit for leaf development. Plant J 83: 375–387 [DOI] [PubMed] [Google Scholar]
- Yang CY, Spielman M, Coles JP, Li Y, Ghelani S, Bourdon V, Brown RC, Lemmon BE, Scott RJ, Dickinson HG (2003) TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J 34: 229–240 [DOI] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie WR, Ma H (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci USA 96: 11416–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Timofejeva L, Ma H, Makaroff CA (2006) The Arabidopsis SKP1 homolog ASK1 controls meiotic chromosome remodeling and release of chromatin from the nuclear membrane and nucleolus. J Cell Sci 119: 3754–3763 [DOI] [PubMed] [Google Scholar]
- Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z (2014) Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J Integr Plant Biol 56: 1042–1052 [DOI] [PubMed] [Google Scholar]
- Zhang D, Yang L (2014) Specification of tapetum and microsporocyte cells within the anther. Curr Opin Plant Biol 17: 49–55 [DOI] [PubMed] [Google Scholar]
- Zhang H, Egger RL, Kelliher T, Morrow D, Fernandes J, Nan GL, Walbot V (2014) Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda) 4: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang XW, Jia LJ, Zhang Y, Jiang G, Li X, Zhang D, Tang WH (2012) In planta stage-specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell 24: 5159–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Yang X, Quan L, Timofejeva L, Rigel NW, Ma H, Makaroff CA (2006) ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol Biol 62: 99–110 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]