Arabidopsis VPS38 is required for the intracellular localization of PI3P, an important lipid regulator of endosomal and vacuolar trafficking.
Abstract
Phosphatidylinositol 3-P (PI3P) is a signaling molecule that controls a variety of processes in endosomal, autophagic, and vacuolar/lysosomal trafficking in yeasts and mammals. Vacuolar protein sorting 34 (Vps34) is a conserved PI3K present in multiple complexes with specific functions and regulation. In yeast, the PI3K complex II consists of Vps34p, Vps15p, Vps30p/Atg6p, and Vps38p, and is essential for vacuolar protein sorting. Here, we describe the Arabidopsis (Arabidopsis thaliana) homolog of yeast Vps38p and human UV radiation resistance-associated gene protein. Arabidopsis VPS38 interacts with VPS30/ATG6 both in yeast and in planta. Although the level of PI3P in Arabidopsis vps38 mutants is similar to that in wild type, vps38 cells contain enlarged multivesicular endosomes and fewer organelles enriched in PI3P than the wild type. The vps38 mutants are defective in the trafficking of vacuolar cargo and its receptor VACUOLAR SORTING RECEPTOR2;1. The mutants also exhibit abnormal cytoplasmic distributions of endocytic cargo, such as auxin efflux carriers PINFORMED1 (PIN1) and PIN2. Constitutive autophagy is normal in the mutants but starvation-induced autophagy was slightly inhibited. We conclude that Arabidopsis VPS38 is dispensable for autophagy but essential for efficient targeting of biosynthetic and endocytic cargo to the vacuole.
Vacuoles are the most conspicuous compartments in mature plant cells (Marty, 1999). Their biogenesis and maintenance are not well understood, partly because cargo destined for the vacuole is heterogeneous in function and origin. Soluble or membrane cargo can be biosynthetic (e.g. newly synthesized structural proteins, transporters, and resident vacuolar hydrolases) or degradative (i.e. macromolecules that will be degraded in the vacuole). Most of the biosynthetic cargo is synthesized at the endoplasmic reticulum (ER), reaches the trans-Golgi network (TGN) via the Golgi apparatus, and is delivered into the multivesicular endosomes (MVEs), which partially derives from the TGN (Scheuring et al., 2011). Various soluble vacuolar proteins, such as seed storage proteins (SSPs), bind VACUOLAR SORTING RECEPTORs (VSRs; De Marcos Lousa et al., 2012) to be properly sorted into the vacuole. The recycling of these receptors for a new round of sorting is mediated by the retromer complex and is essential for continuous delivery of soluble cargo to the vacuole (Shimada et al., 2006; Oliviusson et al., 2006).
The TGN also functions as early endosome (EE) in plant cells, receiving plasma membrane (PM) proteins internalized by endocytosis and acting as a merging point for the biosynthetic and endosomal trafficking routes (Viotti et al., 2010). At the TGN/EE, a fraction of the endocytosed PM cargo is recycled back to the cell surface, whereas cargo proteins targeted for degradation are recognized by the Endosomal Sorting Complex Required for Transport (ESCRT) machinery and internalized into intraluminal vesicles (ILVs) in MVEs. The limiting membrane of the MVE fuses with the tonoplast, the vacuolar membrane, releasing its contents into the vacuolar lumen where ILVs are degraded. Thus, MVEs act as late endosomes (LEs) in plant cells.
Another trafficking route that delivers cargo to the vacuole is autophagy. Here, cytoplasmic constituents are sequestered into organelles called autophagosomes and directly delivered to the vacuole (Kim et al., 2012). When the outer membrane of the autophagosome fuses with the tonoplast, a vesicle containing the autophagic cargo is released as an autophagic body into the vacuolar lumen, where it is rapidly degraded by vacuolar hydrolases.
Polyphosphoinositides (PPIs) play critical roles in intracellular signaling pathways and membrane trafficking (Munnik and Nielsen, 2011; Gerth et al., 2017). In particular, phosphatidylinositol 3-P (PI3P) is important for endosomal, vacuolar/lysosomal, and autophagic trafficking through recruitment of effector proteins (Schink et al., 2013). PI3P effectors typically bind to PI3P through their Fab1, YOTB/ZK632.12, Vac1, and EEA1 (FYVE) or Phox homology domains, such as those found in some subunits of the retromer, ESCRT, and autophagy machinery. In yeast and plants, PI3P is produced through phosphorylation of phosphatidylinositol (PI) by PI 3-kinase activity of VPS34. In yeast, two types of Vps34-containing complexes are found, sharing three common proteins (Vps34p, Vps15p, and Vps30p/Atg6p) but differing in the fourth. Vps34 complex I contains Atg14p as additional factor, which participates in the formation of autophagosomes, whereas complex II contains Vps38p, which is required for vacuolar protein sorting (Kihara et al., 2001). Both types of VPS34 complexes have also been found in mammals and are presumed to exhibit similar functions (Itakura et al., 2008), except that ATG14L and the UV radiation resistance-associated gene protein (UVRAG) appear to be the mammalian counterparts of Atg14p and Vps38p, respectively. Whether UVRAG homologs participate in autophagy remains controversial (Levine et al., 2015), and there is evidence that UVRAG and ATG14L can promote membrane fusion, independently of their roles in the PI 3-kinase complex (Liang et al., 2008; Diao et al., 2015).
PI3P is a very minor membrane lipid. Typically, PPIs represent less than a percent of all phospholipids, and PI3P only accounts for approximately 2.5% to 5% of all PPIs (Munnik et al., 1994a, 1994b; Vermeer et al., 2006). Its turnover, however, is relatively rapid compared to structural phospholipids as judged by 32Pi-labeling experiments in vivo (Munnik et al., 1994a, 1994b; Vermeer et al., 2006; Leprince et al., 2015). Important topological information has come from studies using the expression of a genetically encoded-PI3P biosensor, which is a tandem FYVE domain fused to a fluorescent protein (FP) of any color (Vermeer et al., 2006; Simon et al., 2014). Coexpression with FP-tagged membrane markers revealed that PI3P is highly enriched at the LE membrane and the tonoplast (Vermeer et al., 2006; Simon et al., 2014).
Biosynthesis of PI3P appears to be essential for normal plant development, as gene silencing dramatically retards plant growth and development (Welters et al., 1994). Moreover, no viable homozygous null mutants have been found for vps34, vps15, or atg6 (Fujiki et al., 2007; Lee et al., 2008). Limited information is available from genetic studies on PI3P effectors in Arabidopsis (Arabidopsis thaliana; van Leeuwen et al., 2004), including the ESCRT component FREE1 (Gao et al., 2014) and the retromer subunits SNX1 (Pourcher et al., 2010) and SNX2b (Phan et al., 2008). Pharmacological approaches using PI 3-kinase inhibitors like Wortmannin (Wm) have been shown to affect vacuolar sorting (Matsuoka et al., 1995) and MVE morphology (Haas et al., 2007), but because Wm may also affect downstream PPI pools and inhibit PI 4-kinases and other kinases, putative functions of PI3P must be confirmed by genetic approaches (Nováková et al., 2014).
Here we describe a plant homolog of the yeast Vps38p and mammalian UVRAG. We show that Arabidopsis VPS38 interacts with ATG6 in yeast and in planta. Arabidopsis vps38 mutants show an altered distribution of PI3P, are defective in the vacuolar trafficking of biosynthetic and endocytic cargo, and exhibit enlarged MVEs. In contrast, no drastic effects on autophagy are found. We propose that VPS38 in plants is involved in efficient vacuolar and endosomal sorting.
RESULTS
Identification of Plant Homologs of ATG14L/Atg14p and UVRAG/Vps38p
The Atg14 domain (pfam10186) is found in both ATG14L and UVRAG, the subunits of mammalian Vps34 complexes I and II, respectively. To identify plant homologs, we searched the GenBank RefSeq database, and found three Arabidopsis genes containing Atg14-domain-like sequences: At1g77890, At4g08540, and At2g32760. For the phylogenetic analysis of the resultant Arabidopsis proteins, we searched RefSeq protein databases using At2g32760 protein as a query in DELTA-BLAST. Amino acid sequence alignment of representative DELTA-BLAST hits (Supplemental Fig. S1) revealed that these sequences could be classified into two groups, based on conserved Cys repeats at their N terminus (Matsunaga et al., 2010). The Cys repeats are present in yeast Atg14p, human ATG14L, and their putative plant homologs At1g77890 and At4g08540. In contrast, yeast Vps38p, metazoan UVRAG homologs, and At2g32760 protein lack these Cys repeats (Fig. 1A).
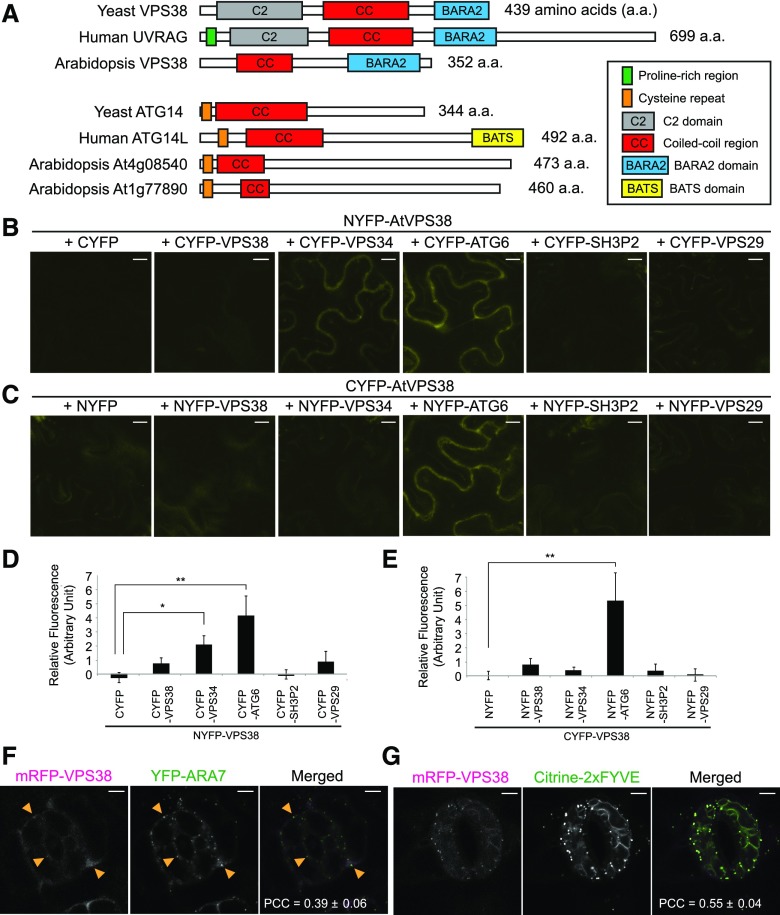
Figure 1.
Conserved domains, protein interactions, and subcellular localization of Arabidopsis VPS38. A, Diagram showing conserved domains/motifs in VPS38/UVRAG and ATG14/ATG14L homologs of yeast, human, and Arabidopsis. B to E, In planta interaction assay using BiFC in tobacco leaf epidermis. BiFC combinations of NYFP-VPS38 and various CYFP fusions (B) and of various NYFP fusions and CYFP-VPS38 (C) were tested, and fluorescence quantified from the combinations is shown in (D) and (E), respectively (mean ± se; n = 6 leaf areas of inoculation or more). Background fluorescence from a control pair of NYFP and CYFP was set to zero. Columns marked with asterisks represent significantly different means (*, 0.01 < P < 0.05; **, P < 0.01; t test). F and G, Colocalization of mRFP-VPS38 with YFP-ARA7 (F) and citrine-2xFYVE (G) in guard cells of Arabidopsis cotyledon epidermis. Arrowheads indicate small puncta of mRFP-VPS38 overlapping with larger YFP-ARA7 puncta. Pearson correlation coefficients were calculated from 4 (F) and 8 (G) pairs of guard cells and presented as mean ± se. Bars = 5 μm. PCCs, Pearson correlation coefficients.
Using sequence alignment of the putative Atg14 and Vps38p/UVRAG proteins to construct a phylogenetic tree, we found that At2g32760 clustered together with the Vps38/UVRAG proteins (Supplemental Fig. S2). Based on this sequence homology and the molecular genetic evidence presented below, we referred to At2g32760 as Arabidopsis VPS38.
VPS38 Interacts with Components of the PI 3-Kinase Complex
A crystal structure of the yeast Vps34 complex II (Rostislavleva et al., 2015) indicated that Vps38p interacts with Atg6p/Vps30p via their coiled-coil regions, whereas only the N-terminal region of Vps38p is adjacent to Vps34p. Based on this structural information, we tested the interactions between Arabidopsis VPS38 protein and putative components of the PI 3-kinase complex. Our yeast two-hybrid (Y2H) assay revealed interaction of VPS38 with ATG6 but not with VPS34 (Supplemental Fig. S3A). VPS38-ATG6 interaction in planta was verified by pairwise bimolecular fluorescence complementation (BiFC; Fig. 1, B to E). We detected positive BiFC signals in the pair of NYFP-VPS38 and CYFP-VPS34 (Fig. 1, B and D) but not in the reciprocal pair of NYFP-VPS34 and CYFP-VPS38 (Fig. 1, C and E). This could be due to the fact that the orientation of the two interacting partners, VPS34 and VPS38, only favors reconstitution of YFP in one of the two combinations or that in the pair NYFP-VPS34 and CYFP-VPS38, one or both halves of YFP block interaction between VPS34 and VPS38.
To investigate the subcellular localization of VPS38, we generated Arabidopsis plants expressing either VPS38-GFP or mRFP-VPS38 fusion proteins under the Cauliflower Mosaic Virus 35S (CaMV35S) promoter. In both cases, mostly diffuse fluorescence signal was observed in the cytoplasm of guard cells (Supplemental Fig. S3, B and C), but very weak or no fluorescence was detected in other plant parts, including root tips (Supplemental Fig. S3D). The levels of VPS38-GFP (n = 19 T1 plants) and mRFP-VPS38 (n = 20 T1 plants) in all transgenic lines were much lower than we expected based on the use of a strong promoter such as CaMV35S. Transgenic seedlings treated with the proteasome inhibitor MG132 contained a higher level of mRFP-VPS38 (Supplemental Fig. S3E), suggesting that overexpressed mRFP-VPS38 is unstable in Arabidopsis.
The subcellular localization of mRFP-VPS38 was compared with LE markers YFP-ARA7/RabF2b and with citrine-2xFYVE (Fig. 1, F and G). The FYVE domain, which binds specifically to PI3P in membranes, has been used as a biosensor for PI3P in Arabidopsis with preferential localization to LEs but also to tonoplast in specific cells (Simon et al., 2014; Vermeer et al., 2006). Although punctate signals of mRFP-VPS38 were weak in transgenic plants, they overlapped with a small fraction of endosomal puncta (Fig. 1F, arrowheads). Interestingly, mRFP-VPS38 puncta were easily observed when citrine-2xFYVE was coexpressed, and mRFP-VPS38 was colocalized with citrine-2xFYVE signal (Fig. 1G). The increased abundance of mRFP-VPS38 puncta in the Citrine-2xFYVE background likely resulted from elevated levels of mRFP-VPS38 (Supplemental Fig. S3E). These data indicate that mRFP-VPS38 is associated with organelles enriched in PI3P.
VPS38 Is Required for Seedling Development and Plant Growth
To analyze the function of VPS38 in Arabidopsis, three T-DNA mutant alleles designated as vps38-1, vps38-2, and vps38-3 (Fig. 2A) were investigated. The location of the insertion within the VPS38 gene was confirmed by sequencing, and viable homozygous individuals from each allele were isolated. We found that vps38-1 and vps38-2 did not accumulate full-length VPS38 transcripts (Fig. 2B). Instead, vps38-1 produced two VPS38 transcripts (asterisks in Fig. 2B), representing aberrant splice variants encoding exon-skipped and truncated VPS38 polypeptides. The vps38-3 plants accumulated full-length wild-type VPS38 transcripts but only at 1% of the wild-type level (Supplemental Table S1; Fig. 2B).
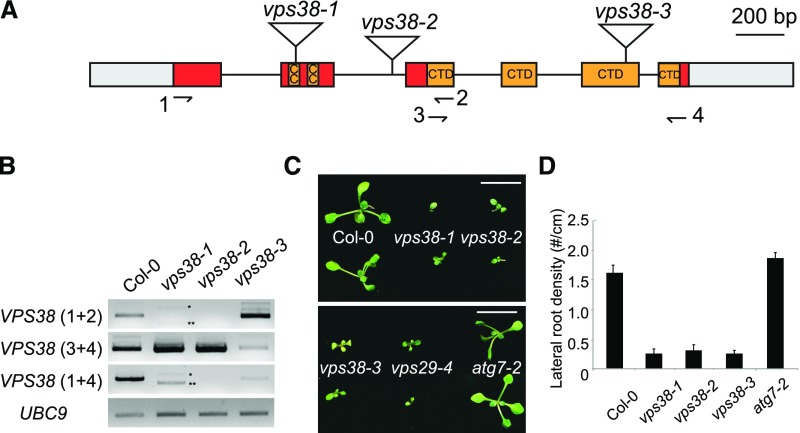
Figure 2.
Arabidopsis vps38 mutants show phenotypic similarity to retromer mutants but not to autophagy mutants. A, Diagram of the VPS38 gene. Exons (boxes) and introns (connecting lines) are shown with T-DNA insertion sites (triangles). Gray boxes illustrate untranslated regions, whereas colored boxes represent coding regions. Coding sequences for coiled coil regions and a conserved C-terminal BARA2 domain (see Supplemental Fig. S1) are indicated. B, Transcript analysis of vps38 mutants. RT-PCR was performed to determine VPS38 transcript levels using three pairs of primers (see arrows in the gene diagram for primer positions) and UBC9 transcript level (internal control). Asterisks indicate vps38-1-specific RT-PCR products representing two aberrant splice variants that either skip exon 2 (**) or contain additional nucleotides derived from T-DNA vector sequence (*). C, Picture of 10-d-old seedlings with indicated genotypes. Bars = 10 mm. D, Lateral root density of seedlings with indicated genotypes. Seedlings were vertically grown for 9 d. Each bar represents the mean value ± se from more than 16 seedlings. CC, coiled coil; CTD, conserved C-terminal domain.
The vps38 homozygous seedlings showed reduced growth compared to wild type (Fig. 2C). Primary root growth and lateral root development were also inhibited (Fig. 2D; Supplemental Fig. S4A). Interestingly, growth on Suc-lacking medium was severely retarded (Supplemental Fig. S4B), which is characteristic of mutants defective in LE functions too (Silady et al., 2008; Kleine-Vehn et al., 2008). Although vps38 adult plants exhibited stunted growth (Supplemental Fig. S4C), they produced viable homozygous seeds. A leaf-curling phenotype was frequently observed in vps38 mutants (Supplemental Fig. S4D). Expression of VPS38-GFP or mRFP-VPS38 transgenes rescued developmental phenotypes of vps38-1 (Supplemental Fig. S4E), indicating that the fusion proteins were functional (Supplemental Fig. S3, B to D).
PI3P Is Aberrantly Distributed in vps38 Mutant Cells
To examine the role of VPS38 in PI3P signaling, the subcellular localization of PI3P was investigated in VPS38/+ and vps38 root cells using the PI3P biosensor, citrine-2xFYVE. This biosensor predominantly localizes to the MVE/LE and, to a much lesser extent, with the TGN/EE (Simon et al., 2014; Vermeer et al., 2006). Wm, a PI 3-kinase inhibitor, is known to induce the formation of ring-shaped YFP-2xFYVE signals (Vermeer et al., 2006). As expected, citrine-2xFYVE was found to decorate small and abundant organelles in control, young root cells (Fig. 3A). In vps38 cells, however, citrine-2xFYVE was detected on enlarged organelles, sometimes in a ringlike pattern, mimicking the organelles found in Wm-treated control seedlings (Fig. 3, A and B). Moreover, the number of citrine-2xFYVE puncta was significantly decreased in vps38 seedlings as was found after Wm treatment (Fig. 3C). The number of the citrine-2xFYVE puncta in vps38 was further reduced by Wm, suggesting either residual PI3K activity in vps38 or a PI3K-independent action of Wm on PI3P-enriched organelles.
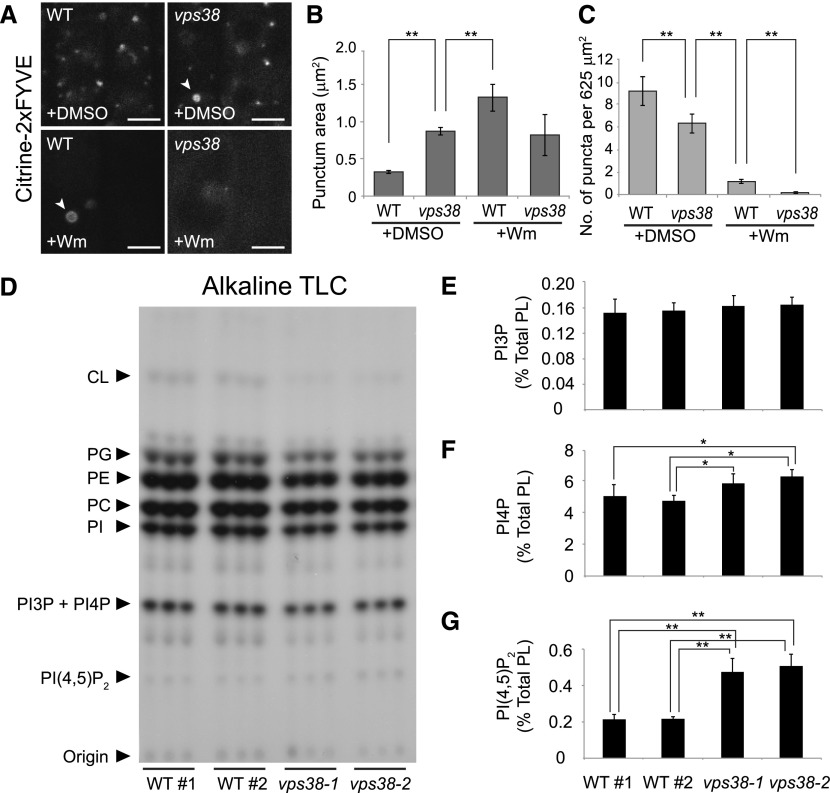
Figure 3.
vps38 affects the intracellular distribution of PI3P but not its total levels. A, Root tip images of wild-type and vps38 expressing the PI3P biosensor, citrine-2xFYVE. Wild-type and vps38 seedlings expressing the sensor were incubated with either 30 μm Wm or DMSO for 1 h before confocal imaging. Arrowheads indicate ring-shaped compartments. Bars = 5 μm. B and C, Graphs illustrate area (B) and density (C) of the citrine-2xFYVE puncta in DMSO- or Wm-treated wild type and vps38-1 (mean ± se; n > 46 regions of interest; *, 0.01 < P < 0.05; **, P < 0.01; two-way ANOVA followed by Tukey’s test). D, Autoradiograph of TLC of 32P-labeled lipid extracts, which were prepared from three biological replicates, each consisting of three seedlings selected from four seed populations of wild type and two vps38 alleles that were 32Pi-labeled in vivo. E to G, Levels of PI3P (E), PI4P (F), and PI(4,5)P2 (G) were quantified by phosphoimaging the phosphoinositide spots in (D) and determining the ratios of PI3P to PI4P by HPLC analysis (see “Materials and Methods”). Columns (mean ± sd) marked with asterisks represent significantly different means (*, 0.01 < P < 0.05; **, P < 0.01; one-way ANOVA followed by Tukey’s test). CL, cardiolipin; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; WT, wild type; WT#1 and WT#2, wild-type seed populations.
To investigate how the vps38 mutation affects PI3P levels, wild-type and vps38 seedlings were 32Pi-labeled in vivo and their phosphoinositide levels determined through TLC and HPLC analysis (Munnik, 2013; Munnik and Zarza, 2013). We did not detect statistically significant changes in the amount of 32P-labeled PI3P (Fig. 3, D and E), but we did find significant increases in the levels of PI(4,5)P2 and PI4P in vps38 seedlings (Fig. 3, F and G). Together, these results indicate that VPS38 plays a role in the subcellular distribution of PI3P-enriched organelles, although it does not seem to affect the total PI3P content.
VPS38 Is Dispensable for Autophagy
Wm has inhibitory effects on vacuolar transport and autophagy (Shin et al., 2014; Zhuang et al., 2013; Matsuoka et al., 1995). To test whether VPS38 is required for autophagy, we employed an autophagy marker, GFP-ATG8a (Kim et al., 2013). Upon activation of autophagy, cytosolic GFP-ATG8a becomes lipidated and associated to autophagic membranes (Chung et al., 2010). As a negative control, we used atg7 mutants that cannot lipidate ATG8 (Chung et al., 2010) and therefore, only accumulate soluble GFP-ATG8a in the cytosol.
To induce autophagy, we subjected the GFP-ATG8a-expressing seedlings to nitrogen starvation and treated them with or without concanamycin A (ConA), an inhibitor of H+-ATPase at the TGN and the vacuole (Matsuoka et al., 1997; Dröse et al., 1993). ConA impairs vacuolar hydrolytic activity and thereby stabilizes autophagic bodies decorated with GFP-ATG8a. In vps38 cells, the distribution of the GFP-ATG8a signal was similar to wild-type cells, and GFP-ATG8a fluorescence was weaker than in atg7 root cells (Fig. 4A). Autophagic bodies, detected by ConA treatment, were observed in both wild-type and vps38 vacuoles but not in atg7 vacuoles (Fig. 4A). Similarly, the autophagic delivery of GFP-ATG8a to the vacuole was normal in vps38 seedlings that were grown with sufficient nitrogen (Supplemental Fig. S5A).
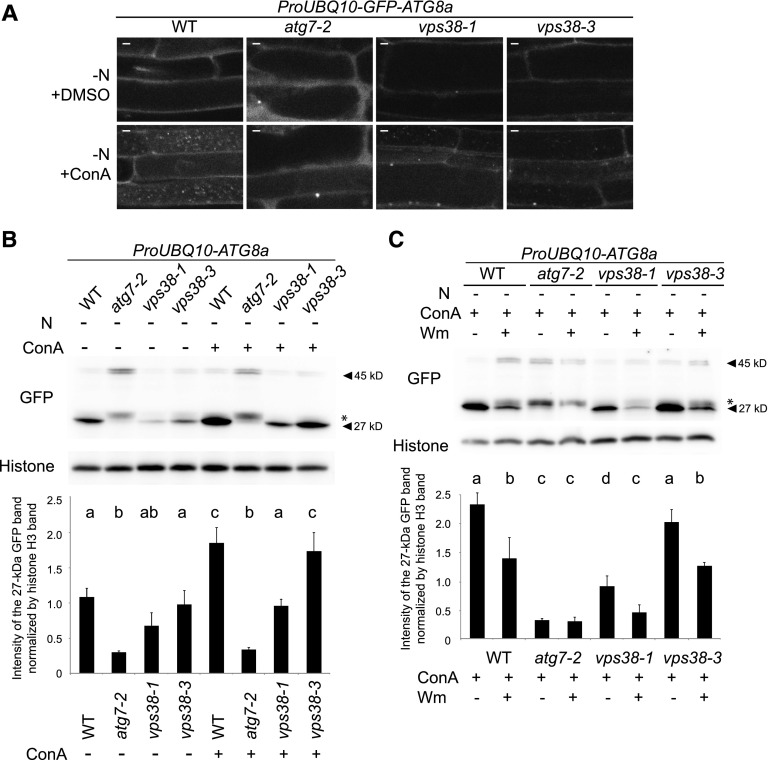
Figure 4.
Autophagy flux under N starvation is reduced but not fully inhibited in vps38-1. A, Images of the autophagy marker line GFP-ATG8a in the genetic backgrounds of wild type, atg7-2, vps38-1, and vps38-3. Seven-d-old seedlings were starved for nitrogen for 32 h, and subsequently treated with DMSO or 1 μm ConA for 16 h. Bar = 5 μm. B and C, GFP-ATG8a processing assay of wild type, atg7, vps38-1, and vps38-3. Where applicable, seedlings were treated with N starvation for 2 d, 30 μm Wm for 16 h, or 1 μm ConA for 16 h before protein extraction. A protein band indicated by an asterisk is an autophagy-independent cleavage product of GFP-ATG8a (Shin et al., 2014). Graphs below illustrate the quantification of 27-kD GFP band intensities normalized by those of histone H3. Mean ± se. n = 6 (B) and 5 (C). Columns marked with different letters represent significantly different means according to two-way ANOVA followed by Tukey’s test (P < 0.01). WT, wild type. −N, nitrogen starvation.
Autophagic flux was quantified by immunoblot analysis of GFP-ATG8a and its cleaved GFP fragment, which is generated by the degradation of GFP-ATG8a upon autophagic delivery to the vacuole (Shin et al., 2014). Autophagic flux was slightly reduced in vps38-1 under nitrogen-deficient conditions (Fig. 4B). In contrast, autophagic flux in vps38-3 was indistinguishable from that in wild type (Fig. 4B; Supplemental Fig. S5B). Importantly, in both wild type and vps38, Wm treatment inhibited the accumulation of the 27-kD GFP fragment (Fig. 4C; Supplemental Fig. S5C; Shin et al., 2014). These results suggest that in plants, VPS38 only plays a minor, VPS34-independent role in autophagy.
Consistent with the dispensability of VPS38 in autophagy, vps38 mutants did not show the accelerated leaf senescence typically observed in atg mutants (Supplemental Fig. S5D). Similarly, unlike atg7, vps38 mutants did not exhibit pexophagy-related phenotypes, like overaccumulation of catalase (Supplemental Fig. S5E; Shibata et al., 2013). Finally, no punctate signal of mRFP-VPS38 was detected at GFP-ATG8a puncta (Supplemental Fig. S3F).
VPS38 Is Required for Proper Vacuolar Trafficking of SSPs
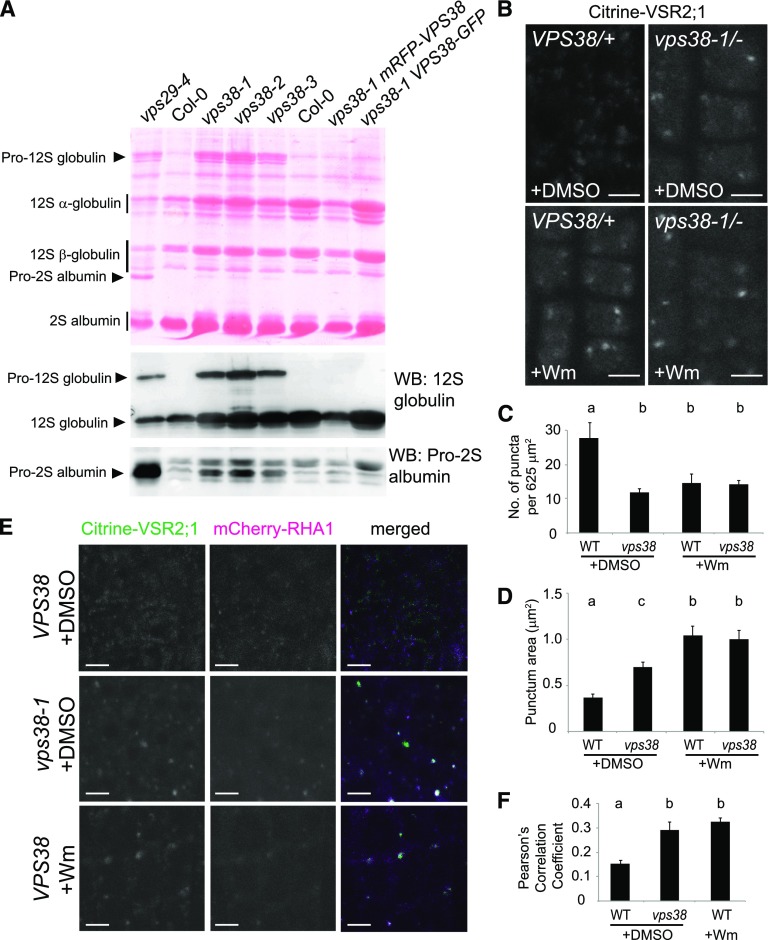
Yeast VPS38 was originally identified from a genetic screen for mutations affecting vacuolar protein sorting (Rothman et al., 1989). To determine whether vacuolar sorting is compromised in Arabidopsis vps38 mutants, we monitored the proteolytic processing of the SSP precursor. In Arabidopsis seeds, two abundant classes of SSPs are the 12S globulins and 2S albumins. Their precursor forms, pro-12S globulins and pro-2S albumins, are synthesized at the ER, move through the Golgi apparatus, and are processed to mature forms at the LEs and the vacuole (Otegui et al., 2006). We found that vps38 mutant seeds accumulated pro-12S globulin, similar to many Arabidopsis mutants with defective vacuolar trafficking including the retromer mutant vps29 (Fig. 5A; Shimada et al., 2003, 2006). However, accumulation of pro-2S albumin in vps38 was not as pronounced as in vps29 (Fig. 5A). Moreover, the overaccumulation of SSP precursors in vps38-1 was completely suppressed by the expression of mRFP-VPS38 or VPS38-GFP (Fig. 5A).
Figure 5.
Vacuolar protein sorting is compromised in vps38 mutants. A, Seed protein profile of the indicated mutants and transgenic plants. A protein blot stained with Ponceau S is shown in the upper panel with seed storage protein species indicated by arrowheads (precursors) and vertical bars (mature forms). The two immunoblots below were generated with antibodies against 12S globulin (middle panel) and pro-2S albumin (lower panel). Each lane contains the protein extract of five dry seeds. B, Confocal microscopy images of Citrine-VSR2;1 transgenic plants with indicated genotypes. Root tips of seedlings treated for 1 h with either DMSO or 30 μm Wm were observed. C and D, Graphs illustrate the quantification of citrine-VSR2;1 puncta in wild-type and vps38-1 mutant cells (mean ± se; n > 11 regions of interest). E, Confocal images of wild-type and vps38-1 root cells expressing both citrine-VSR2;1 and mCherry-RHA1. Roots were treated with DMSO or Wm as in (B). F, Quantification of colocalization using images similar to one shown in (E) (mean ± se; n > 23 regions of interest). Columns marked with different letters represent significantly different means according to two-way ANOVA followed by Tukey’s test (C and D) or t test (F) (P < 0.01). Bars = 5 μm. WB, western blot; WT, wild type.
Trafficking of SSPs into the vacuole is mediated by VSRs. We examined the effect of vps38 on the subcellular distribution of a fluorescently tagged VSR isoform, VSR2;1::citrine-VSR2;1, which restored an early flowering phenotype of vsr2;1 (Supplemental Fig. S4F; Avila et al., 2008). In wild-type Arabidopsis, VSR isoforms have been detected at various endomembrane compartments but predominantly localized at the LEs (De Marcos Lousa et al., 2012). In control (VPS38/+) root cells, citrine-VSR2;1 exhibited a punctate pattern in the cytoplasm, whereas in vps38 root tip cells, citrine-VSR2;1 associated with fewer and larger compartments (Fig. 5, B to D). Wm is known to induce the enlargement of LEs (Jaillais et al., 2006; Haas et al., 2007). The citrine-VSR2;1-decorated compartments in vps38 cells were bigger than those in wild-type cells and somewhat smaller than the enlarged endosomes resulting from Wm treatment (Fig. 5D). The strong citrine-VSR2;1 signals on enlarged compartments of vps38 cells are consistent with the hypothesis that the recycling of citrine-VSR2;1 is compromised in the mutants. Notably, Wm did not cause a further reduction in the number of citrine-VSR2;1 puncta in vps38 mutant (Fig. 5C). Taken together, our results indicate that VPS38 plays a critical role for vacuolar trafficking.
To further investigate the effect of VPS38 on the localization of citrine-VSR2;1, we crossed a vps38-1 mutant expressing citrine-VSR2;1 with the LE marker line mCherry-RHA1, and observed F2 seedling roots. In wild-type control, citrine-VSR2;1 signals largely separated from the endosome marker, but either Wm treatment or vps38 mutation significantly increased the tendency of colocalization (Fig. 5, E and F). This result is consistent with our hypothesis that VPS38 is essential for recycling of the vacuolar sorting receptors from endosomes.
vps38 Cells Contain Enlarged Late Endosomes and Abnormal Vacuoles
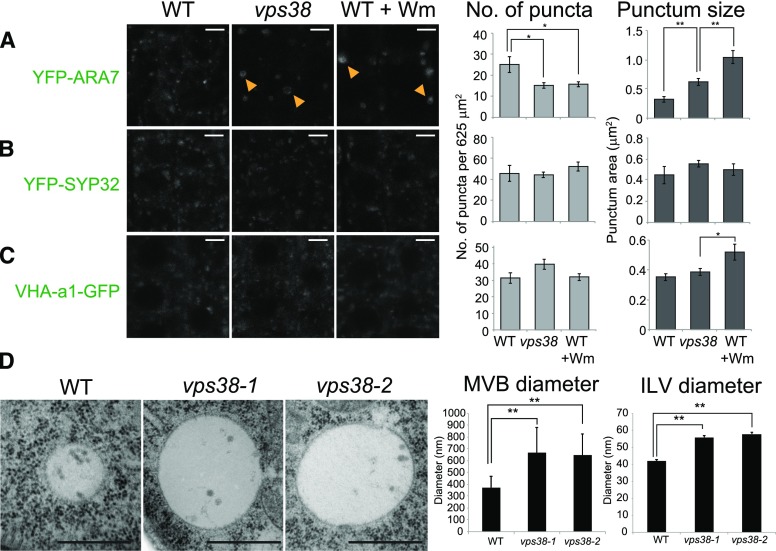
We investigated the subcellular distribution of various organelle markers in vps38 mutants. Transgenic plants carrying fluorescent markers were crossed to the vps38 mutants. Using confocal imaging, we found that, in vps38 cells, the LE marker YFP-ARA7 decorated enlarged organelles when compared to wild-type root cells (Fig. 6A). YFP-ARA7 puncta were even larger and often ring-shaped, like in Wm-treated wild-type controls, consistent with the pattern observed for the PI3P biosensor (Fig. 3A). The number of YFP-ARA7 puncta was reduced in both vps38 and Wm-treated wild-type cells (Fig. 6A). In contrast, the distribution of the Golgi (Fig. 6B) and TGN marker (Fig. 6C; Dettmer et al., 2006) in vps38 was similar to wild-type cells.
Figure 6.
MVE morphology is altered in vps38 mutants. A to C, Root tip images of wild-type and vps38-expressing MVE marker, YFP-ARA7 (A); Golgi marker, YFP-SYP32 (B); and TGN marker, VHA-a1-GFP (C). Wild-type seedlings expressing each marker were incubated with either DMSO or 30 μm Wm for 1 h before observation under confocal microscope. Arrowheads indicate ring-shaped compartments. Graphs on the right illustrate the quantities of each marker in DMSO-treated wild type and vps38, and Wm-treated wild type (mean ± se; n > 11 regions of interest; *, 0.01 < P < 0.05; **, P < 0.01 by t test). D, Transmission electron micrograph showing MVEs and their ILVs of wild type, vps38-1, and vps38-2. Between 34 and 46 MVEs and between 36 and 47 ILVs from two independent roots were analyzed. Bars = 5 μm (A to C) or 500 nm (D). WT, wild type.
Upon Wm treatment, MVEs become enlarged and produce ILVs (Wang et al., 2009; Haas et al., 2007), suggesting a role for PI3P in the formation of ILVs during the sorting of endocytosed proteins for vacuolar degradation. To assess this possibility, we performed transmission electron microscopy of vps38 roots (Fig. 6D; Supplemental Fig. S6, A to C). MVEs in vps38 root cells were easily discernible by the presence of ILVs, and they were significantly larger than wild-type MVEs. The ILVs in both vps38-1 and vps38-2 were also significantly larger than wild-type endosomes, further suggesting that Wm treatment mimics disruption of VPS38 function. In addition, we often found in differentiating vps38 root cells abnormal vacuoles with highly convoluted tonoplasts (Supplemental Fig. S6, D to F), further supporting a role of VPS38 in endosomal and vacuolar function.
VPS38 Is Crucial for Efficient Vacuolar Degradation of Endocytosed Cargo
Because endocytosis and endosomal trafficking play critical roles in plant development by controlling the homeostasis of various PM proteins (Reyes et al., 2011), we investigated whether the abnormal growth and development of vps38 mutants (Fig. 2) could be explained by defects in the endocytic pathway.
First, we assessed endocytic uptake and endosomal recycling by analyzing the dynamics of the endocytic tracer FM4-64 and the plasma membrane-localized PINFORMED2 (PIN2) protein fused to GFP. We incubated seedlings in FM4-64 (Supplemental Fig. S7A), but found no differences in the timing it took the tracer to reach the vacuolar membrane in wild-type and vps38 root cells (Supplemental Fig. S7, B and C). Brefeldin A (BFA) causes the formation of the BFA compartment, an aggregate of the Golgi and TGN membranes that blocks the recycling of endocytosed proteins back to the PM (Geldner et al., 2003; Naramoto et al., 2014). In both wild-type and vps38 cells, FM4-64 accumulated in BFA bodies, which dissipated again upon washing out the BFA (Supplemental Fig. S7D). Thus, VPS38 does not globally affect the uptake and dynamics of endocytic tracer FM4-64.
To test whether vps38 mutation affects recycling of the endocytosed proteins, we carried out BFA washout experiments for PIN2-GFP (Supplemental Fig. S7E). De novo synthesis of PIN2-GFP was blocked by cycloheximide treatment, BFA was added to block recycling of PIN2-GFP, and BFA was washed out in the presence of cycloheximide to reinitiate the recycling to the PM. The PIN2-GFP signal at the PM of both wild-type and vps38 root cells was increased 2 h after BFA washout (Supplemental Fig. S7, F and G), indicating that VPS38 is not required for the endosomal recycling of PIN2-GFP. Notably, vps38 seemed to show enhanced recycling of PIN2-GFP at both 2 h and 4 h after washout. This could relate to the higher intensity of intracellular PIN2-GFP signal at 0 h in vps38 than in wild type (Supplemental Fig. S7, E and G; see below).
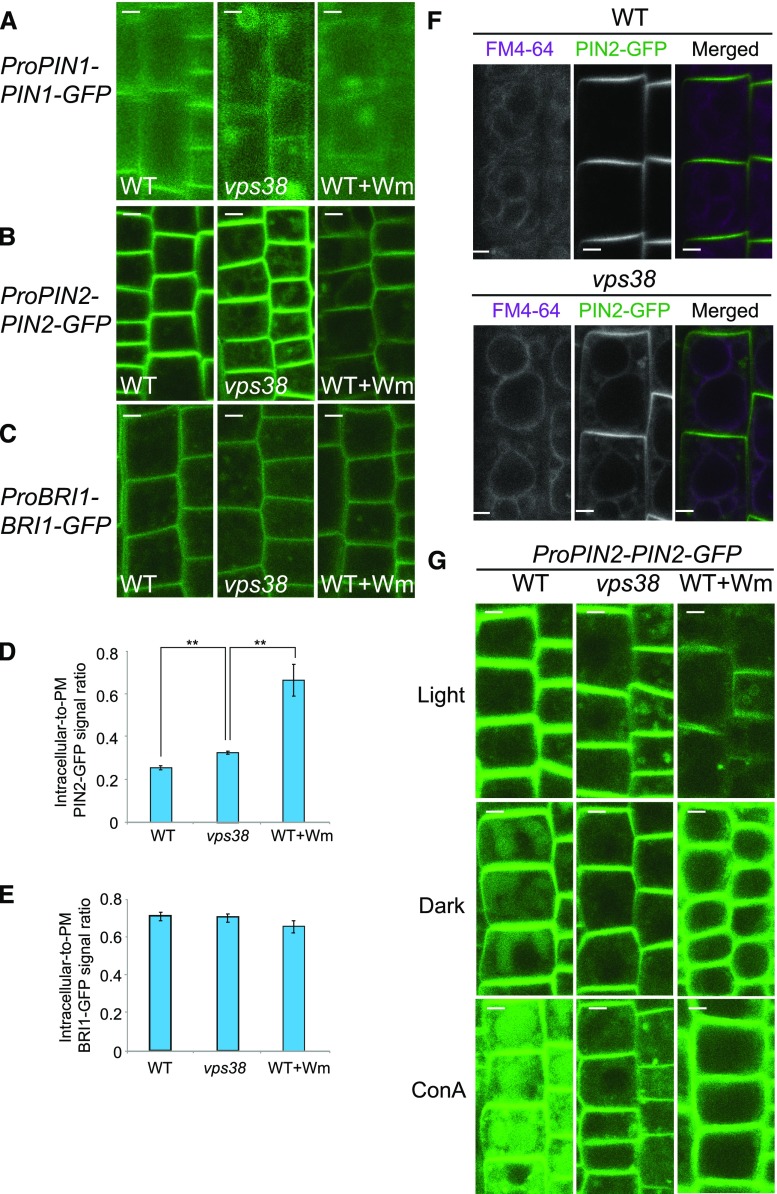
Next, we assessed the function of VPS38 in endosomal degradative trafficking of the plasma membrane proteins PIN1, PIN2, and BRASSINOSTEROID INSENSITIVE1 (BRI1) fused to GFP. In vps38, PIN1-GFP accumulated in large (up to 5 μm in diameter) cytoplasmic compartments, likely enlarged endosomes or small vacuoles, similar to those observed in control root cells treated with Wm (Fig. 7A; Supplemental Fig. S7H; Kleine-Vehn et al., 2008; Jaillais et al., 2007). vps38 root cells also had more intracellular puncta of PIN2-GFP (Fig. 7, B and D) than control, but ratios of intracellular-to-PM signals of BRI1-GFP did not differ between control and vps38 (Fig. 7, C and E).
Figure 7.
Trafficking of endocytic cargo to the vacuole is hindered in vps38 mutants. A to C, Confocal microscopic images of wild-type and vps38 roots expressing PIN1-GFP (A), PIN2-GFP (B), or BRI1-GFP (C). Seedlings expressing each marker were incubated with either DMSO or 30 μm Wm for 1 h before observation under confocal microscope. D and E, Graphs illustrating the quantification of PIN2-GFP (D) and BRI1-GFP (E) signal intensities in the PM and intracellular region (mean ± se; n > 11; *, 0.01 < P < 0.05; **, P < 0.01 by t test). F, Images of wild-type and vps38 roots expressing PIN2-GFP in the presence of the endocytic tracer FM4-64. Seven-d-old seedlings were pulse-labeled with 5 μΜ FM4-64 for 2 h, washed 3 times, and incubated for 4 h before observation. G, Images of dark- or ConA-treated wild type or vps38 seedlings expressing PIN2-GFP. Seedlings were incubated with either DMSO, 1 μm ConA, or 30 μm Wm for 12 h before observation. Dark treatment was performed by incubating seedlings for 12 h in the dark. Bars = 5 μm. WT, wild type.
We observed that some of the intracellular PIN2-GFP signal in vps38 was associated with the tonoplast (Fig. 7F). Accumulation of PM proteins in the tonoplast usually results from a failure to sort proteins targeted for vacuolar degradation into ILVs, as in the case of ESCRT mutants (Spitzer et al., 2009; Buono et al., 2016). Therefore, to investigate the potential role of VPS38 in the degradation of endocytosed membrane proteins, we incubated PIN2-GFP- and BRI1-GFP-expressing seedlings in the dark and in the presence of ConA, both of which were used to monitor GFP fluorescence in the vacuolar lumen (Kleine-Vehn et al., 2008). As expected, either dark or ConA treatment led to the accumulation of diffuse GFP signal in the vacuolar lumen of cells expressing PIN2-GFP (Fig. 7G) and BRI1-GFP (Supplemental Fig. S7I). Both Wm treatment and the vps38 mutation suppressed the accumulation of diffuse PIN2-GFP (Fig. 7G) and BRI1-GFP (Supplemental Fig. S7I) signal in the vacuolar lumen. These results indicate that VPS38 plays a role in the targeting of endocytosed cargo to the vacuole.
DISCUSSION
In this study, we identified VPS38 as the Arabidopsis homolog of yeast Vps38 and mammalian UVRAG. This argument is supported by similarities in their amino acid sequence (Supplemental Fig. S1), conserved protein interactions (Fig. 1; Supplemental Fig. S3), and vps38 phenotypes relating to vacuolar and endosomal trafficking (Figs. 5 and 7). Abnormal subcellular structures observed in vps38 cells include enlarged MVEs (Fig. 6). Transport of biosynthetic cargo (e.g. SSPs) to the vacuole is less efficient in vps38, likely because its receptor VSR cannot be fully retrieved from the endosomes (Fig. 5). The targeting of endocytosed membrane cargo to the vacuole is also inhibited (Fig. 7; Supplemental Fig. S7). The combination of these defective processes in vacuolar sorting may result in irregular plant development and poor responses to nutrient limitation (Fig. 2; Supplemental Fig. S4).
The biochemical functions of VPS38 are not known. Yeast vps38 mutants showed only a slight decrease in PI 3-kinase activity (Kihara et al., 2001). We found that PI3P levels in vps38 were comparable to that of wild type (Fig. 3), suggesting that PI3P levels are robustly regulated in plants (Vermeer et al., 2006). Several mechanisms for maintaining a PI3P level in vps38 are possible. For example, ATG14 homologs and/or unidentified PI 3-kinases may act redundantly in PI3P production, compensating for the lack of VPS38. Alternatively, the activity of PI3P 3-phosphatases or 5-kinases may be reduced in vps38. Interestingly, D4-phosphorylated phosphoinositides PI(4,5)P2 and PI4P accumulated in vps38 (Fig. 3), implying a shift in phosphoinositide metabolism toward the D4-phosphorylation pathway (Munnik and Nielsen, 2011; Nováková et al., 2014). Besides a possible redirection effect on phosphoinositide metabolism, vps38 mutations cause defective endosomal and vacuolar trafficking, which may in turn lead to a compensatory remodeling of secretory trafficking, endocytosis, and/or metabolism of PI4P and PI(4,5)P2. Conversely, it is also possible that the change in PI(4,5)P2 and PI4P affects the dynamics of some PM proteins and is partially responsible for some of the abnormal vps38 phenotypes. These possibilities remain to be tested in the future.
Molecular interactions and subcellular localization of VPS38 fusion proteins (Fig. 1) led us to speculate that VPS38 recruits a putative plant PI 3-kinase complex II to endosomal compartments via direct interaction with ATG6. It remains to be seen whether the complex is poorly targeted to vps38 endosomes, which may explain fewer citrine-2xFYVE puncta (Fig. 3) and enlarged MVEs (Fig. 6) and other vps38 phenotypes that are mimicked by Wm treatment (Figs. 5, 6, and 7).
It is well established that PI3P is crucial for membrane dynamics and sorting events on endosomes through the actions of PI3P effectors (Schink et al., 2013). Based on the known PI3P effectors in mammalian and yeast cells and the effects of Wm on the plant vacuolar trafficking (Tse et al., 2004; daSilva et al., 2005), we proposed at least three classes of potential PI3P effectors in the plant vacuolar trafficking: (1) retromer-dependent retrograde transport of vacuolar sorting receptors; (2) ESCRT-dependent ILV formation; and (3) MVE maturation, which is mediated by Rab5-to-Rab7 conversion (Ebine et al., 2014; Singh et al., 2014). Our genetic analysis indicated that vps38 is most similar to mutants that are defective in retromer and ESCRT. Like vps38, both retromer-defective vps29 and ESCRT-related free1 mutants showed accumulation of PM proteins in endosomal compartments (Fig. 7, A and B; Spitzer et al., 2009; Gao et al., 2014; Jaillais et al., 2007). In contrast, abnormal targeting of endocytic cargo to the vacuole was seen in vps38 and ESCRT-related mutants (Fig. 7, F and G; Spitzer et al., 2009; Gao et al., 2014), but not in retromer mutants (Jaillais et al., 2007). The difference suggests that VPS38 is required for two separated PI3P-dependent processes, i.e. vacuolar sorting of biosynthetic and endocytic cargo.
Another potential PI3P effector is ATG18a, which is involved in autophagy. However, our data indicated that vps38 mutation did not block autophagy (Fig. 4; Supplemental Fig. S5). It is possible that two Atg14-like proteins, At1g77890 and At4g08540 (Fig. 1A), replace VPS38, as in the autophagy-specific PI 3-kinase complex I in yeast and mammals (Kihara et al., 2001; Itakura et al., 2008). These proteins may also be responsible for the sensitivity of vps38 mutants to Wm in both our autophagic flux (Fig. 4C; Supplemental Fig. S5C) and PI3P biosensor assays (Fig. 3C).
Our data are consistent with recent findings that challenge direct involvement of UVRAG/VPS38 in autophagy (Takáts et al., 2014; Farré et al., 2010; Jiang et al., 2014). However, it is still possible that a role of VPS38 in autophagy is masked by its greater impact on vacuolar protein sorting. For example, mutations in VPS38 may block autophagic flux, but this effect would be canceled out if the mutant vacuole had weaker proteolytic activity due to inefficient vacuolar sorting.
Clearly, the use of vps38 mutants provides, to our knowledge, a new tool to elucidate the in vivo roles of PI3P in plant vacuolar/endosomal sorting pathways, complementing the pharmacological approach of PI 3-kinase inhibitors, which are less specific. For example, Wm has been reported to deplete TGN marker proteins and induce heterotypic fusion of the TGN vesicles with the MVE (Takáč et al., 2012). Treatment with another pan-PI3K inhibitor LY294002 also results in a slight increase in the size of the TGN (Fujimoto et al., 2015). Our data that Wm, but not vps38 mutation, was associated with an increase in the punctum size of a TGN marker (Fig. 6C) indicate that inhibition of a non-VPS34 target by Wm and LY294002 is responsible for the apparent enlargement of the TGN. Future investigation on vps38 mutants using pan-PI3K inhibitors and more specific VPS34 inhibitors (Ronan et al., 2014; Dowdle et al., 2014; Bago et al., 2014) may reveal additional major players in PI3P metabolism in plants.
MATERIALS AND METHODS
DNA Constructs and Transgenic Plants
The Arabidopsis Biological Resource Center (ABRC; Columbia, OH) provided cDNA entry clones for Arabidopsis (Arabidopsis thaliana VPS29; ABRC stock no. G20137), VPS38 (G82505), VPS34 (GC105027), ATG6 (G12843), and SH3P2 (G11736). We amplified cDNA for VPS38(NS) by PCR using primers listed in Supplemental Table S2 and cloned them into pENTR/D-TOPO (Invitrogen).
For yeast two-hybrid (Y2H) constructs, the entry clones were reacted with the destination vectors pDEST22 and pDEST32 using LR Clonase (Invitrogen). For BiFC, entry clones were recombined with pSITE-nEYFP-C1 or with pSITE-cEYFP-C1 (Martin et al., 2009) via the LR Clonase reaction.
For Pro-35S::VPS38-GFP and Pro-35S::mRFP-VPS38, entry clones for VPS38(NS) and VPS38 were reacted with the destination vectors pMDC83 (Curtis and Grossniklaus, 2003) and pSITE-4CA (Chakrabarty et al., 2007), respectively.
To generate stable transformants of Arabidopsis, expression clones were introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium transformants were used to infect Arabidopsis by the floral dip method (Clough and Bent, 1998). Multiple T1 events were analyzed for initial analysis, and homozygous T3 populations from a representative line were used for quantitative analyses.
We used the fusion protein citrine-VSR2;1 as previously described in Saint-Jean et al. (2010) except that the 35S promoter was replaced by the promoter of AtVSR2;1 (VSR4). The fluorescent tag was put in frame between the signal peptide of VSR2;1 and the sequence of mature VSR2;1. The promoter region starting from −1397 to −3 bp before the start codon was provided by Jan Zouhar (Zouhar et al., 2010). The citrine-VSR2;1 coding region was amplified using primers VSR2;1_F and VSR2;1_R and inserted between XhoI and AscI of pENTR/D-TOPO behind the promoter. The final construct was transferred by recombination into pGWB1. The citrine-VSR2;1 construct was introduced into vsr2;1 mutant plants (SALK_094467) by the floral dip method. Selection was made using hygromycin plus kanamycin and homozygous T2 plants were selected on the basis of fluorescent progeny.
Plant Materials and Growth
Mutant seeds of vps38-1 (SAIL_552_F02), vps38-2 (SAIL_164_D09), vps38-3 (SALK_094540C), vps29-4 (Jaillais et al., 2007), atg7-2 (Chung et al., 2010), and transgenic seeds of ProUBQ10::YFP-ARA7, ProUBQ10::YFP-SYP32, ProUBQ10::mCherry-RHA1 (Geldner et al., 2009), ProUBQ10::Citrine-2xFYVE (Simon et al., 2014), and ProPIN1-PIN1-GFP (Benková et al., 2003) were obtained from the ABRC. ProVHA-a1::VHA-a1-GFP (Dettmer et al., 2006), ProPIN2::PIN2-GFP (Xu and Scheres, 2005), ProBRI1::BRI1-GFP (Nam and Li, 2002), and ProUBQ10::GFP-ATG8a (Kim et al., 2013) were previously described. Seeds were germinated on solid Murashige & Skoog (MS) medium with 1% (w/v) Suc and 0.25% (w/v) to 0.6% Phytagel (Sigma-Aldrich). For liquid culture, seeds were germinated in 1 mL of liquid MS medium with 1% Suc. For nitrogen starvation, 7-d-old seedlings were washed and supplemented with liquid MS-N with 1% Suc (Chung et al., 2009).
Sequence Analysis
For phylogenetic analyses, putative VPS38 and ATG14-like protein sequences were identified by the software DELTA-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROGRAM=blastp&BLAST_PROGRAMS=deltaBlast) using Arabidopsis VPS38 amino acid sequence as a query. Sequences of BLAST hits were aligned using the software MUSCLE (Edgar, 2004), and maximum-likelihood trees were generated by the software MEGA 6 (Tamura et al., 2013) using the Jones/Taylor/Thornton matrix-based model, partial deletion of gaps, uniform rates among sites, and 500 bootstrap replicates. Coiled coil regions were predicted by COILS (https://embnet.vital-it.ch/software/COILS_form.html) with more than 30% probability and 28 amino acid window width.
RNA Analysis
Seeds were germinated on the solid medium and total RNA was extracted from 10-d-old seedlings using Trizol (Invitrogen). Total RNA was treated with DNase I (New England BioLabs) and used for cDNA synthesis by RevertAid H Minus reverse transcriptase (Thermo Fisher Scientific) with oligo(dT) primers. cDNA was amplified for 35 (for VPS38) or 26 cycles (for UBC9 as an internal control). For real-time quantitative PCR, real-time PCR mix (SolGent) including Evagreen was used. Amounts of transcripts were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) normalizing against UBC9 expression according to the Rotor Gene Q system (Qiagen). The primers used for Reverse transcriptase PCR (RT-PCR) are presented in Supplemental Table S2.
Protein Interaction
An activation domain fusion construct and a DNA binding domain construct were transformed into yeast strain MaV203 using the Yeastmarker Yeast Transformation System 2 (Clontech). Y2H was performed with ProQuest (Invitrogen), using the manufacturer’s instructions. Expression of both constructs was tested on medium lacking Leu and Trp and protein interaction was identified on medium lacking Leu, Trp, and His and containing 25 mm 3-amino-1,2,4-triazole.
For BiFC, transgenes were introduced into the Agrobacterium strain GV3101 and then used to infiltrate 6-week-old Nicotiana benthamiana leaves as described in Martin et al. (2009). p19 genes were coinfiltrated to repress gene silencing. Leaf epidermal cells were analyzed by confocal microscopy 36 h to 48 h after infiltration.
Immunoblot Analysis
For immunoblotting, samples were homogenized in Laemmli buffer and centrifuged at 16,000g for 10 min. The supernatants were analyzed by SDS-PAGE after boiling. Western blotting was performed as described in Kim et al. (2013). Antibodies used were as follows: anti-GFP (Roche), anti-mCherry (Clontech), and anti-Catalase (Agrisera). The antibodies against Arabidopsis 12S (Shimada et al., 2003), and 2S propeptides (Otegui et al., 2006) were previously described. Band intensity was determined using the software ImageJ (National Institutes of Health).
Transmission Electron Microscopy
Root tips from 5-d-old wild-type Col-0, vps38-1, and vps38-2 seedlings were high-pressure-frozen in a model no. HP-010 (Bal-Tec), substituted overnight at −80°C in acetone containing 2% (w/v) osmium tetroxide and embedded in Epon resin (Electron Microscopy Sciences). Samples were sectioned, and stained with 2% uranyl acetate in 70% methanol and lead citrate (2.6% lead nitrate and 3.5% sodium citrate, pH 12). For the MVE morphometric analysis, two independent roots for each genotype were analyzed using the software FIJI (https://fiji.sc/). In total, we analyzed 46 (Col-0), 34 (vps38-1), and 38 (vps38-2) MVEs and 36 (Col-0), 42 (vps38-1), and 47 (vps38-2) ILVs.
Confocal Microscopy and Image Analysis
In most cases, images were acquired using a model no. 510 laser scanning confocal microscope (Carl Zeiss). For GFP, YFP, and citrine, a 488-nm laser and BP500-530IR emission filter were used. For mRFP, mCherry, and FM4-64, a 543-nm laser and BP565-615IR emission filter were used. For colocalization of mRFP-VPS38 with YFP-ARA7 or with citrine-2xFYVE, a model no. 710 AxioObserver (Carl Zeiss) was used. mRFP-VPS38 was excited by a 594-nm laser, and 599-nm to 638-nm fluorescence was collected. YFP-ARA7 and citrine-2xFYVE were excited by a 488-nm laser and 493-563-nm fluorescence was collected. For each quantitative analysis, more than 12 regions of interest were analyzed by the software ImageJ. The number and the size of puncta were measured by the Analyze Particles function. For the quantification of PM and intracellular (IC) signals of PIN2-GFP and BRI1-GFP, PM and IC regions per one cell were manually selected using the Area Selection Tools, and the mean brightness value was measured. To calculate the Pearson correlation coefficient (r), the Coloc 2 Plug-in for ImageJ (https://imagej.net/Coloc_2) was used.
Inhibitors and Endocytic Tracer Analysis
Inhibitors were added to liquid medium in which seedlings were hydroponically grown for 7 d. Wm (Sigma-Aldrich), ConA (Santa Cruz Biotechnology), BFA (Molecular Probes), MG132 (Sigma-Aldrich), and cycloheximide (Sigma-Aldrich) were dissolved in DMSO. For pulse-labeling with endocytic tracer FM4-64 (Molecular Probes), 7-d-old seedlings grown on the MS liquid medium were incubated with cold 5 μm FM4-64 for 5 min. Roots were then washed three times with the liquid MS medium at room temperature, and observed at various times. To detect vacuolar membrane, roots were treated with 5 μm FM4-64 for 2 h, then washed and incubated in the liquid MS medium at room temperature for 4 h. For the BFA washout experiments (Martins et al., 2015), roots expressing PIN2-GFP were preincubated in liquid medium containing 50 μm cycloheximide for 30 min, and subsequently incubated in medium with 50 μm BFA and 50 μm cycloheximide for 2 h, before being washed out with medium containing 50 μm cycloheximide.
Phosphoinositide Analysis
Five-d-old wild-type and vps38 seedlings were labeled overnight using [32P]PO43− as detailed in Munnik and Zarza (2013). Next day, fresh label (5 to 10 μCi/sample) was added for another 60 min, before lipid extraction. Extracts were separated by alkaline TLC and the radioactivity visualized and quantified by PImaging (Munnik and Zarza, 2013). The experiment was performed in triplicate and three seedlings were used per sample. To distinguish PI3P from PI4P, PIP spots were scraped from the TLC, deacylated and desalted, and the resulting GPIPs separated by HPLC using a Parisil SAX column (Sigma-Aldrich) and a gradient of NaH2PO4 pH 3.7 (Munnik, 2013; Munnik et al., 1994a, 1994b). The final results for the different PIP isomers were obtained by quantifying the PIP spots from the TLC and normalizing the results with respect to the ratios of PI3P to PI4P obtained from the HPLC analysis.
Accession Numbers
Sequence data from this article can be found in the TAIR (http://www.arabidopsis.org/) database: ATG6 (At3g61710), ATG7 (At5g45900), ATG8a (At4g21980), BRI1 (At4g39400), PIN1 (At1g73590), PIN2 (At5g57090), RABF2b/ARA7 (At4g19640), RHA1 (At5g45130), SH3P2 (At4g34660), SYP32 (At3g24350), VHA-a1 (At2g28520), VPS29 (At3g47810), VPS34 (At1g60490), VPS38 (At2g32760), VSR2;1 (At2g14720), and UBC9 (At4g27960).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of amino acid sequences predicted from 33 proteins containing the ATG14 domain.
Supplemental Figure S2. A phylogenetic analysis of ATG14 and VPS38/UVRAG homologs.
Supplemental Figure S3. VPS38 interactions and subcellular localization.
Supplemental Figure S4. Phenotypes of Arabidopsis vps38 mutants and complementation tests in this study.
Supplemental Figure S5. vps38 mutants show reduced autophagic flux under nitrogen-sufficient (+N) conditions but retain sensitivity to Wm.
Supplemental Figure S6. vps38 cells have enlarged MVE and abnormal vacuoles.
Supplemental Figure S7. vps38 mutants show normal endocytic uptake but defective trafficking of PM proteins into the vacuole.
Supplemental Figure S8. Immunoblots used for the quantification in Figure 4B.
Supplemental Figure S9. Immunoblots used for the quantification in Figure 4C.
Supplemental Table S1. Quantitative RT-PCR to estimate VPS38 transcript level in vps38-3.
Supplemental Table S2. Primers used for plasmid construction or RT-PCR.
Acknowledgments
We thank Karin Schumacher for providing VHA-a1-GFP; Ben Scheres for PIN2-GFP; Kyoung Hee Nam for BRI1-GFP; and Ikuko Hara-Nishimura for anti-12S globulin antiserum.
Footnotes
This work was supported by grants no. NRF-2014R1A1A1A05003740 and “Cooperative Research Program for Agriculture Science & Technology Development (PJ01110801), RDA, Korea” to T.C., the Netherlands Organization for Scientific Research (NWO 867.15.020 and 711.017.005) to T.M., and National Science Foundation (NSF) MCB1614965 to M.S.O.
Articles can be viewed without a subscription.
References
- Avila EL, Brown M, Pan S, Desikan R, Neill SJ, Girke T, Surpin M, Raikhel NV (2008) Expression analysis of Arabidopsis vacuolar sorting receptor 3 reveals a putative function in guard cells. J Exp Bot 59: 1149–1161 [DOI] [PubMed] [Google Scholar]
- Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR (2014) Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 463: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Buono RA, Paez-Valencia J, Miller ND, Goodman K, Spitzer C, Spalding EP, Otegui MS (2016) Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol 171: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M (2007) PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana-virus interactions. Mol Plant Microbe Interact 20: 740–750 [DOI] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T, Suttangkakul A, Vierstra RD (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva LL, Taylor JP, Hadlington JL, Hanton SL, Snowden CJ, Fox SJ, Foresti O, Brandizzi F, Denecke J (2005) Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17: 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcos Lousa C, Gershlick DC, Denecke J (2012) Mechanisms and concepts paving the way towards a complete transport cycle of plant vacuolar sorting receptors. Plant Cell 24: 1714–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, et al. (2015) ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520: 563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, et al. (2014) Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 16: 1069–1079 [DOI] [PubMed] [Google Scholar]
- Dröse S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K (1993) Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32: 3902–3906 [DOI] [PubMed] [Google Scholar]
- Ebine K, Inoue T, Ito J, Ito E, Uemura T, Goh T, Abe H, Sato K, Nakano A, Ueda T (2014) Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr Biol 24: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Mathewson RD, Manjithaya R, Subramani S (2010) Roles of Pichia pastoris UVRAG in vacuolar protein sorting and the phosphatidylinositol 3-kinase complex in phagophore elongation in autophagy pathways. Autophagy 6: 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143: 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Suda Y, Vernhettes S, Nakano A, Ueda T (2015) Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol 56: 287–298 [DOI] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L (2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24: 2556–2563 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth K, Lin F, Menzel W, Krishnamoorthy P, Stenzel I, Heilmann M, Heilmann I (2017) Guilt by association: a phenotype-based view of the plant phosphoinositide network. Annu Rev Plant Biol 68: 349–374 [DOI] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martínez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS (2007) The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Santambrogio M, Rozier F, Fobis-Loisy I, Miège C, Gaude T (2007) The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N (2014) The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 25: 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kwon C, Lee JH, Chung T (2012) Genes for plant autophagy: functions and interactions. Mol Cells 34: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J (2008) Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, Lee Y (2008) The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol 147: 1886–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince AS, Magalhaes N, De Vos D, Bordenave M, Crilat E, Clément G, Meyer C, Munnik T, Savouré A (2015) Involvement of Phosphatidylinositol 3-kinase in the regulation of proline catabolism in Arabidopsis thaliana. Front Plant Sci 5: 772 10.3389/fpls.2014.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Liu R, Dong X, Zhong Q (2015) Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol 25: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N, Vert G (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151 10.1038/ncomms7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F. (1999) Plant vacuoles. Plant Cell 11: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T (2010) Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 190: 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K (1995) Different sensitivity to Wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol 130: 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Higuchi T, Maeshima M, Nakamura K (1997) A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T. (2013) Analysis of D3-,4-,5-phosphorylated phosphoinositides using HPLC. Methods Mol Biol 1009: 17–24 [DOI] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A (1994a) Rapid turnover of phosphatidylinositol 3-phosphate in the green alga Chlamydomonas eugametos: signs of a phosphatidylinositide 3-kinase signalling pathway in lower plants? Biochem J 298: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Musgrave A, de Vrije T (1994b) Rapid turnover of polyphosphoinositides in carnation flower petals. Planta 193: 89–98 [Google Scholar]
- Munnik T, Nielsen E (2011) Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol 14: 489–497 [DOI] [PubMed] [Google Scholar]
- Munnik T, Zarza X (2013) Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol Biol 1009: 3–15 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Otegui MS, Kutsuna N, de Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H, Nakano A, Friml J (2014) Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková P, Hirsch S, Feraru E, Tejos R, van Wijk R, Viaene T, Heilmann M, Lerche J, De Rycke R, Feraru MI, Grones P, van Montagu M, et al. (2014) SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci USA 111: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviusson P, Heinzerling O, Hillmer S, Hinz G, Tse YC, Jiang L, Robinson DG (2006) Plant retromer, localized to the prevacuolar compartment and microvesicles in Arabidopsis, may interact with vacuolar sorting receptors. Plant Cell 18: 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan NQ, Kim SJ, Bassham DC (2008) Overexpression of Arabidopsis sorting nexin AtSNX2b inhibits endocytic trafficking to the vacuole. Mol Plant 1: 961–976 [DOI] [PubMed] [Google Scholar]
- Pourcher M, Santambrogio M, Thazar N, Thierry AM, Fobis-Loisy I, Miège C, Jaillais Y, Gaude T (2010) Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Buono R, Otegui MS (2011) Plant endosomal trafficking pathways. Curr Opin Plant Biol 14: 666–673 [DOI] [PubMed] [Google Scholar]
- Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, Marquette JP, El-Ahmad Y, et al. (2014) A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 10: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350: aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Howald I, Stevens TH (1989) Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J 8: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jean B, Seveno-Carpentier E, Alcon C, Neuhaus JM, Paris N (2010) The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 22: 2825–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, Schumacher K (2011) Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink KO, Raiborg C, Stenmark H (2013) Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays 35: 900–912 [DOI] [PubMed] [Google Scholar]
- Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M (2013) Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25: 4967–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Koumoto Y, Li L, Yamazaki M, Kondo M, Nishimura M, Hara-Nishimura I (2006) AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, Kobayashi M, Kondo M, et al. (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J Biol Chem 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Shin KD, Lee HN, Chung T (2014) A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells 37: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silady RA, Ehrhardt DW, Jackson K, Faulkner C, Oparka K, Somerville CR (2008) The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J 53: 29–41 [DOI] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y (2014) A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 77: 322–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JEM, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, Jürgens G (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS (2009) The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáč T, Pechan T, Samajová O, Ovečka M, Richter H, Eck C, Niehaus K, Samaj J (2012) Wortmannin treatment induces changes in Arabidopsis root proteome and post-Golgi compartments. J Proteome Res 11: 3127–3142 [DOI] [PubMed] [Google Scholar]
- Takáts S, Pircs K, Nagy P, Varga Á, Kárpáti M, Hegedűs K, Kramer H, Kovács AL, Sass M, Juhász G (2014) Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell 25: 1338–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L (2004) Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Okrész L, Bögre L, Munnik T (2004) Learning the lipid language of plant signalling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr., Munnik T (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, Jürgens G, de Vries SC, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Miao Y, Lam SK, Jiang L (2009) Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters P, Takegawa K, Emr SD, Chrispeels MJ (1994) AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci USA 91: 11398–11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Muñoz A, Rojo E (2010) Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J 64: 577–588 [DOI] [PubMed] [Google Scholar]