Abstract
DNA methylation patterns are disrupted in various malignancies, suggesting a role in the development of cancer, but genetic aberrations directly linking the DNA methylation machinery to malignancies were rarely observed, so this association remained largely correlative. Recently, however, mutations in the gene encoding DNA methyltransferase 3A (DNMT3A) were reported in patients with acute myeloid leukaemia (AML), and subsequently in patients with various other haematological malignancies, pointing to DNMT3A as a critically important new tumour suppressor. Here, we review the clinical findings related to DNMT3A, tie these data to insights from basic science studies conducted over the past 20 years and present a roadmap for future research that should advance the agenda for new therapeutic strategies.
Previous interest in DNA methylation in cancer focused on the role of DNA methyltransferase 1 (DNMT1) because mutations in this protein have been described in colorectal, prostate and haematological malignancies, albeit rarely1, and reduced DNMT1 activity has been shown to promote cancer in mouse models2,3. Recent cancer genome sequencing efforts exposed DNMT3A as one of the most frequently mutated genes across a range of haematological malignancies, raising questions concerning how these lesions promote malignancies. Basic research has examined the role of DNMT3A in gene repression, but the insights gained from these fundamental studies have not been placed in context with the recent findings suggesting that DNMT3A mutations play a prominent part in clonal and malignant haematopoiesis. The intention of this Review is to analyse the clinical findings related to DNMT3A and survey its role in normal stem cell biology, highlighting the large conceptual gaps that remain to be filled to understand and target DNMT3A mutation-associated malignancies. Although DNMT3A has been implicated in several types of cancer4–6, we focus primarily on the role of DNMT3A in haematological malignancies.
DNA methylation
DNA methylation is an epigenetic modification that is important in development, imprinting, stem cell regulation, X-chromosome inactivation and several diseases7. Methylation of DNA refers to the addition of a methyl (CH3) group to the C5 position of the pyrimidine ring of cytosines to form 5-methylcytosine (5mC)8, usually in the context of a CpG dinucleotide pair. Although 60–80% of individual CpGs are methylated, clusters called CpG islands in gene regulatory regions tend to be unmethylated. In general, high DNA methylation is associated with the silencing of gene expression9. DNA methylation is particularly concentrated on repetitive elements, and it may limit their genomic activity.
In cancer, aberrant DNA methylation has been observed for more than two decades, with interest initially focused on promoter hypermethylation and the consequent silencing of tumour suppressor genes10,11. Global hypomethylation was also observed and proposed to be associated with genomic instability12. Regions of lower-density methylation near CpG islands, known as ‘shores’, exhibit great variation in methylation, including hypomethylation and hypermethylation, across different types of cancers13. Importantly, the mechanisms that drive aberrant methylation and its pathological importance are mostly unknown.
DNA methylation is mediated by a family of DNA methyltransferase enzymes, including DNMT1, DNMT3A and DNMT3B14. The related member DNMT3-like (DNMT3L) lacks a catalytic domain and functions as an accessory protein to DNMT3A during embryonic development and genomic imprinting15,16. DNMT1 primarily maintains pre-existing DNA methylation patterns17, whereas DNMT3A and DNMT3B carry out de novo DNA methylation14. DNMT2 is considered an RNA methyltransferase18. The methylcytosine dioxygenase proteins (TET1, TET2 and TET3), convert 5mC to 5-hydroxymethylcytosine (5hmC)19. 5hmC is not maintained by DNMT1 but the mark leads to passive demethylation during cell division20,21 and thus provides a mechanism for DNA demethylation.
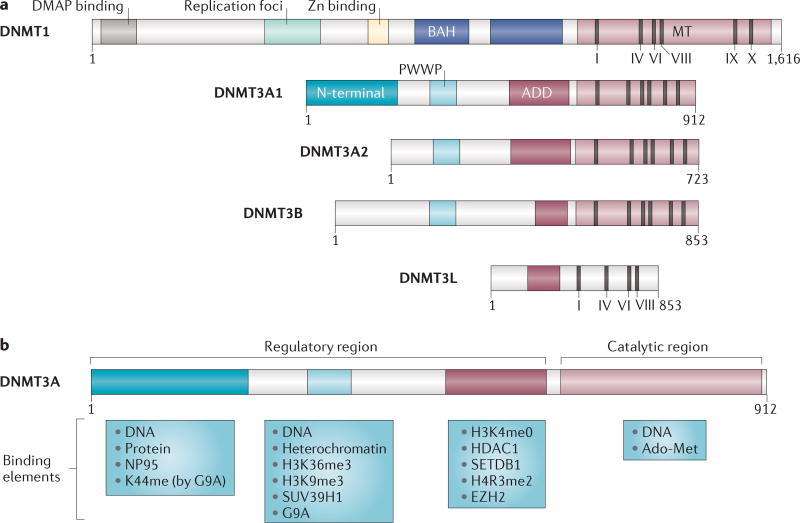
Structure of DNMT3A
DNMT3A is a 130 kDa protein encoded by 23 exons on human chromosome 2p23 (REF. 22). The protein is highly conserved across mammals, with 98% homology between human and murine homologues23. There are two major splice isoforms of the murine RNA: Dnmt3a1 (long) and Dnmt3a2 (short). Dnmt3a2 lacks the first six exons of the amino-terminal domain24, and its expression is restricted to embryonic stem cells (ESCs), testes, ovaries, spleen and thymus24,25. Dnmt3a1 is widely expressed and transcribed from two different promoter regions11,26. The basis for the cell type specificity and significance of the isoforms is not known.
The major protein domains are the Pro-Trp-Trp-Pro (PWWP) domain, the ATRX-DNMT3-DNMT3L (ADD) domain (also known as the plant homeodomain (PHD)) and the catalytic methyltransferase domain (FIG. 1). Although they have not been exhaustively studied, the ADD and PWWP domains are known to interact with proteins that are involved in transcriptional repression, and the N terminus is thought to be involved in DNA binding27–31. The ADD domain also inhibits the DNMT3A catalytic domain by forming an auto-inhibitory loop that is released by interaction with the unmodified lysine 4 of histone H3 (H3K4me0), linking DNMT3A and H3 chromatin marks32.
Figure 1. The structure of DNA methyltransferase proteins and their binding partners.
a | Domain architecture of DNA methyltransferase 1 (DNMT1), DNMT3A1, DNMT3A2, DNMT3B and DNMT3-like (DNMT3L) and major DNMT3A splice isoforms. Note that DNMT3B has a number of additional isoforms that are not depicted here36,37. Protein length is indicated (length given as number of amino acids). Domain abbreviations: ADD, ATRX-DNMT3-DNMT3L (related to the plant homology (PHD)-like domain of regulator ATRX); BAH, Bromo adjacent homology domain; DMAP, DNMT1-associated protein; PWWP, Pro-Trp-Trp-Pro. MT is the catalytic methyltransferase domain, and I, IV, VI, IX and X are motifs in the catalytic domain: motif I allows the binding of the methyl group donor AdoMet (S-adenosyl methionine). Motifs I and X are for cofactor binding, and motifs VIII and IX are for DNA binding. The catalysis of DNA methylation occurs at the IV, VI and VIII motifs155. b | DNMT3A contains an amino-terminal domain that is unique to the long isoform and exhibits DNA-binding capability27,156. It may also interact with transcription factors such as OCT3 (also known as OCT4 and POU5F1) in embryonic stem cells157. Lysine 44 (K44) of the N-terminal domain is dimethylated by G9A and is important for interactions with G9A and/or EHMT1 (also know as GLP)50,51; this interaction is necessary for the DNA methylation of some loci, such as the OCT3 promoter, which is involved in embryonic stem cell pluripotency. PWWP has diverse capabilities, including DNA156 and heterochromatin binding40,158, as well as interactions with the histone H3 lysine 36 trimethylation (H3K36me3) and H3K9me3 marks28. The ADD domain has strong affinity for histone deacetylase 1 (HDAC1)29 and for unmodified H3K4 (REFS 30–32), and is thought to positively affect DNA methyltransferase activity159.This domain binds to the methyltransferase domain to act as an auto-inhibition loop in the presence of unmodified histone H3 binding32; it also interacts with histone modifiers involved in gene repression, such as the histone-lysine N-methyltransferase SUV39H1 (REF. 53). Furthermore, the ADD domain binds to the histone-lysine N-methyltransferase enhancer of zeste homologue 2 (EZH2); EZH2 is part of Polycomb repressive complex 2 (PRC), which is responsible for H3K27 methylation52,53.
The catalytic domain of DNMT3A is highly conserved, even within prokaryotic DNA methyltransferases33, implying that the specificity of DNMT3A originates in its regulatory domains. X-ray crystallography indicates that the murine protein acts as a heterotetramer, with the catalytic core consisting of a mixed β-sheet of seven strands, among which the methyl donor and DNA binding functions are distinguished34. Interaction with DNMT3L substantially increases enzyme processivity15,35. DNMT3B, which is usually co-expressed with DNMT3A, is expressed in multiple isoforms with different activities36,37, some of which may also influence DNMT3A activity, particularly in cells that lack DNMT3L38.
Methylation activities of DNMT3A
DNMT3A is reported to methylate DNA at unique sites as well as at repetitive elements. In mouse ESCs, DNMT3A localizes to discrete nuclear foci and, together with DNMT3B, to pericentromeric heterochromatin39, suggesting that the DNMT3A–DNMT3B dimer methylates major satellite repeats at these sites40. Furthermore, in Dnmt3a and Dnmt3b double-knockout (Dnmt3a−/−Dnmt3b−/−) ESCs, transfection of Dnmt3a restores methylation at specific repetitive element families36. In genomic studies, DNMT3A methylates specific regions of the genome overlapping with those methylated by DNMT3B36,41,42, suggesting some degree of functional redundancy. DNMT3A also methylates free or linker DNA with higher efficiency than nucleosome-bound DNA43,44.
Some of the gene-silencing activities of DNMT3A are intimately tied to chromatin modifications through interactions with specific proteins (FIG. 1). As described above, DNMT3A interacts with H3K4me0, a mark of inactive gene transcription30,32. DNMT3A also interacts with histone modifiers involved in gene repression, such as the histonelysine N-methyltransferases SUV39H1, SETDB1 and G9A (also known as EHMT2)45–51, which are linked to H3K9 methylation. Similarly, the histonelysine N-methyltransferase enhancer of zeste homologue 2 (EZH2), which is the catalytic component of Polycomb repressive complex 2 (PRC2), is required for DNA methylation of EZH2 target promoters and interacts with DNMT3A, DNMT3B and DNMT1. This interaction, however, is insufficient for de novo DNA methylation, suggesting that other factors are needed to initiate methyltransferase activity52,53. Overall, most studies have been limited to specific cells and loci, and we still have a poor understanding of how DNMT3A activity is regulated more broadly. How is DNMT3A recruited to chromatin? How does it target particular sites? And how is the protein regulated through interactions with other protein partners? These are crucial questions that need to be answered if we are to understand cancer development and to develop targeted therapies.
Unique methylation patterns set by DNMT3A have also been investigated. In haematopoietic stem cells (HSCs), DNMT3A probably regulates methylation at the edges of large regions of hypomethylation called DNA methylation canyons54 (or valleys55). DNMT3A is also involved in gene body DNA methylation56.
Although there is no known consensus sequence that DNMT3A methylates57, biochemical studies show that DNMT3A prefers to methylate sequences rich in T, C and A around CpG sites, methylating one strand at a time15,58–60. Methylation at CpGs is the primary target, but DNMT3A is also active in other contexts, such as CpA61–63 (also termed CpH methylation, where H can be A, C or T). This non-CpG methylation occurs at extremely low levels in most tissues, except in ESCs and the postnatal brain64–66, and its potential function is a matter of controversy67. The relative importance of different DNA methylation sites, and how DNMT3A activity is directed to different types of loci, is unclear. These multiple targets of DNMT3A underscore the complexity of its functions and the major gaps in our understanding of its regulation.
Role of DNMT3A in development
Mouse models have offered important insights into the function of the DNMT family. Dnmt3a−/− mice survive for approximately 1 month after birth but die with an uncharacterized failure to thrive, whereas Dnmt3b−/− mice die mid-gestation. Dnmt3a−/−Dnmt3b−/− embryos show limited embryonic development68. Both Dnmt3a and Dnmt3b are highly expressed in mouse ESCs and are downregulated during differentiation14. Although deleting either gene alone has minimal impact, eliminating both together in ESCs resulted in persistent self-renewal and inefficient differentiation, as measured by teratoma formation assay after serial passage36. This phenotype was associated with DNA hypomethylation36 and incomplete repression of pluripotency genes after differentiation69.
Recently, Dnmt3a was also found to have a role in somatic stem cell differentiation. When Dnmt3a was conditionally deleted in HSCs, self-renewal was markedly favoured over differentiation. This increased self-renewal caused Dnmt3a-null HSCs dramatically to outcompete their wild-type counterparts and accumulate in the bone marrow70. The Dnmt3a-null HSCs exhibited a surface phenotype that was indistinguishable from that of their wild-type counterparts, and they differentiated into all types of downstream progeny, such as myeloid cells, B cells and T cells; however, the output of differentiated cells per HSC was reduced relative to that of wild-type cells70. Similar to wild-type HSCs, they were mostly quiescent, suggesting that their competitive advantage lay in the proportion of cell divisions that generated stem cells versus differentiated cells. When Dnmt3b was ablated, HSCs showed only minor differences from wild-type cells, perhaps because the predominant splice isoform generates a catalytically inactive protein. However, its ablation along with Dnmt3a (Dnmt3a−/−Dnmt3b−/− HSCs) greatly exacerbated the stem cell expansion and led to almost completely blocked differentiation71.
To date, the mechanisms accounting for increased self-renewal in the absence of DNMT3A have been studied by analysing changes in gene expression and DNA methylation in mutant cells. Dnmt3a-null HSCs exhibit a net loss of DNA methylation, particularly at the edges of large hypomethylated canyon regions54, which are enriched for genes associated with stem cell self-renewal and cancer, such as homeobox A9 (Hoxa9), Meis homeobox 1 (Meis1), and MDS1 and EVI1 complex locus (Mecom; which encodes the transcriptional regulator ecotropic virus integration site 1 protein homologue (EVI1)). In Dnmt3a−/− mice, many genes associated with HSC self-renewal increase in expression, and some fail to be appropriately repressed during differentiatiation70. These data suggest that the absence of DNMT3A abrogates the ability to switch from a self-renewal to a differentiation programme, ultimately resulting in more self-renewal cell divisions.
Analysis of the mechanisms leading to the differentiation block of Dnmt3a−/−Dnmt3b−/− HSCs suggested a contribution of β-catenin to this phenotype. The promoter region of Ctnnb1 (which encodes β-catenin) became hypomethylated, and both gene and target gene expression increased. Knockdown of Ctnnb1 partially rescued the differentiation block, consistent with a role for β-catenin in HSC self-renewal71, similar to its role in ESC self-renewal72. However, other unknown factors are also likely to contribute.
Together, the sustained expression of self-renewal-associated genes is consistent with the expansion of Dnmt3a−/− and Dnmt3a−/−Dnmt3b−/− HSCs, but many questions remain to be answered. For example, the mechanisms that dictate the activity of DNMT3A at specific genomic loci are obscure. Similarly, although the levels of DNA methylation change at canyons and other regions, how these changes contribute to the phenotype is unclear. More broadly, whether de novo DNA methylation can initiate or merely enforce differentiation decisions is unknown, as is a possible role for DNMT3A in maintaining ‘stemness’ beyond differentiation.
The role of de novo DNA methylation in the function of ESCs and HSCs raises the possibility that DNMT3A and/or DNMT3B may be involved in the differentiation of other somatic stem cell types. Indeed, DNMT3A has been implicated in neural stem cell differentiation56,73. In addition, germline mutations in DNMT3A cause a human overgrowth syndrome resulting in extreme height74, perhaps through the modulation of tissue-resident stem cell activity.
DNMT3A mutations in blood malignancies
Despite the long-recognized aberrant DNA methylation in cancer, the first mutations in DNMT3A associated with malignancy were only identified in 2010, with three groups reporting mutations in acute myeloid leukaemia (AML) with frequencies of up to 22%75–77. A mutational hotspot at arginine 882 (R882) was highlighted, but other mutations throughout the gene were also seen. Mutations in DNMT3A have now been found in most types of haematological malignancy with varying frequency (TABLE 1; Supplementary information S1 (table)). Although a few mutations in DNMT1 and DNMT3B have also been reported78,79, the overwhelming prevalence of DNMT3A mutations across a range of diseases demonstrates the special role of this gene in preventing malignancy.
Table 1.
Frequency of DNMT3A mutations in haematological disorders
| Disease | Patient population | Frequency (%) | Refs |
|---|---|---|---|
| AML | De novo AML | 62/281 (22.1) | 76 |
| CN AML | 44/120 (36.7) | ||
| De novo AML | 70/500 (14) | 147 | |
| CN AML | 51/223 (22.9) | ||
| <60 years CN AML | 64/181 (35.5) | 123 | |
| ≥60 years CN AML | 77/234 (33.3) | ||
| sAML | 13/37 (35) (10/27 (37) from MDS; 3/10 (33) from MPN) | 126 | |
| tAML | 10/59 (17) | ||
| Paediatric AML | 3/140 (2.1) | 115 | |
| MDS | Adult de novo MDS | 127/944 (13.5) | 121 |
| Paediatric MDS, tMDS | 0/44* | 117 | |
| MPN | MPN (entire cohort) | 10/155 (9) | 98 |
| PMF | 1/16 (6) | ||
| PV | 2/30 (7) | ||
| ET | 0/30 (0) | ||
| Blast phase MPN | 5/35 (14) | ||
| Systemic mastocytosis | 3/26 (12) | 162 | |
| CML | 0/79 | 163 | |
| CMML and JMML | CMML | 5/227 (2)* | 164 |
| CMML-1 | 1/48 (2) | 100,134 | |
| CMML-2 | 1/16 (6.3) | ||
| sAML from CMML | 6/23 (26) | ||
| JMML | 1/113 (0.8) | 165 | |
| T cell leukaemia and lymphoma | T cell lymphoma | 11/96 (11) | 147 |
| PTCL | 21/79 (26.6) | 103 | |
| PTCL-NOS | 9/33 (27.3) | ||
| AITL | 12/46 (26.1) | ||
| Adult T-ALL (entire cohort) | 16/83 (18) | 104 | |
| Early (pro- and pre-leukaemic) T cell leukaemia | 10/38 (26.3) | ||
| Cortical T-ALL | 5/39 (12.8) | ||
| Adult ETP ALL | 11/68 (16) | 137 | |
| Paediatric T-ALL | 0/91 | 79 | |
| Paediatric ETP ALL | 0/12 | 119 | |
| MPAL | Adult T cell and myeloid MPAL | 10/18 (55.6) | 166 |
| Paediatric mixed lineage leukaemia | 0/20 | 79 |
The range of frequencies reported within a given disease type depends on the subset of patients examined as well as whether the entire DNMT3A gene or just the hotspots were examined. Also see the comprehensive online Supplementary information S1 (table). AITL, angioimmunoblastic T cell lymphoma; ALL, acute lymphoblastic leukaemia; AML, acute myeloid leukaemia; CML, chronic myeloid leukaemia; CMML, chronic myelomonocytic leukaemia; CN AML, cytogenetically normal AML; ET, essential thrombocytopaenia; ETP, early T cell precursor; JMML, juvenile myelomonocytic leukaemia; MDS, myelodysplastic syndrome; MPAL, mixed phenotype acute leukaemia; MPN, myeloproliferative neoplasm; PMF, primary myelofibrosis; PTCL, peripheral T cell lymphoma; PTCL-NOS, PTCL-not otherwise specified; PV, polycythaemia vera; sAML, secondary AML; T-ALL, T cell ALL; tAML, therapy-related AML (AML that develops after exposure to cytotoxic chemotherapy and/or radiation therapy administered for a prior neoplastic or non-neoplastic disorder); tMDS, therapy-related MDS.
Only sequenced exon 23.
Across haematological malignancies, DNMT3A mutations are distributed throughout all of the functional domains (FIG. 2a), some of which have been shown by biochemical studies to govern specific functions and interactions (TABLE 2; Supplementary information S2 (table)). For example, mutation of R878 in the catalytic domain of the murine protein (equivalent to R882 of the human protein; discussed below) has been shown to abrogate catalytic activity and to result in reduced DNA binding60. However, most of the specific mutations found in cancer have not been functionally characterized. Although the impact of some mutations may be predicted by the domain interactions, their specific relevance for the cancer phenotype has not been explored.
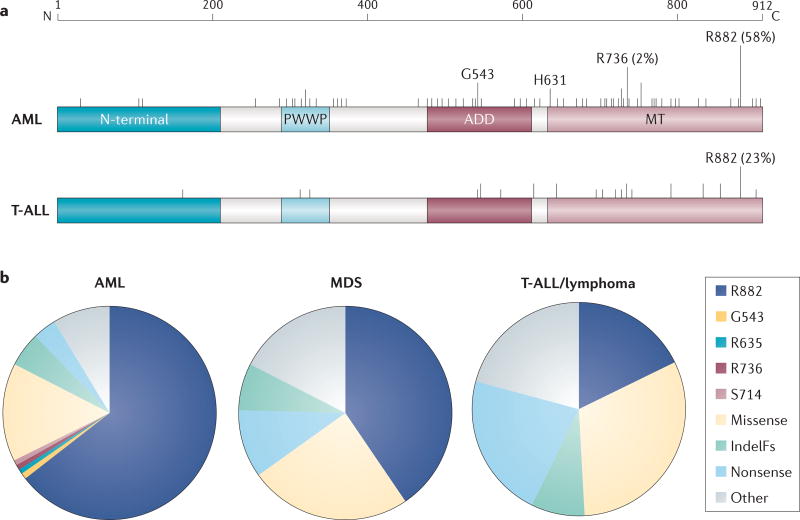
Figure 2. Distribution of DNMT3A mutation frequency in myeloid and lymphoid leukaemia.
a | Structural diagram of the location of mutations in DNA methyltransferase 3A (DNMT3A). The top band indicates amino acid position and scale. The vertical lines show the mutations mapped to DNMT3A in acute myeloid leukaemia (AML) and T cell acute lymphoblastic leukaemia (T-ALL) in the subset of patients in which the entire gene has been sequenced, as curated from the published literature. Long lines represent >0.03% frequency of mutation. Also shown is the distinct frequency of mutation at arginine 882 (R882) in AML versus T-ALL, and the minor hotspot in AML G543 (1.4%) and R736 (2.1%) residues. b | Frequency of mutations in AML, myelodysplastic syndrome (MDS) and T-ALL/lymphoma, as curated from the Catalogue of Somatic Mutations in Cancer (COSMIC) database (see Further information). ADD, ATRX-DNMT3-DNMT3L; IndelFs, insertion–deletion frameshift mutations; MT, methyltransferase; PWWP, Pro-Trp-Trp-Pro.
Table 2.
Biochemical impact of mutations in DNMT3A found in cancer patients
| Residue (murine) | AA | Disease | Domain | Consequence | Refs |
|---|---|---|---|---|---|
| 308 (304) | Q | AML | PWWP | H3 | 159 |
| 664 (660) | E | AML | Motif II | Cat | 60 |
| 710 (706) | C | sAML from MDS | Motif IV | Cat, DNA, AdoMet (−) | 60 |
| 714 (710) | C | AML | Following motif IV | AdoMet | 60 |
| 720 (716) | N | T-ALL | Following motif IV | Cat, DNA | 60 |
| 729 (725) | R | AML | Catalytic | DNA | 106 |
| 733 (729) | E | AML, CMML | Catalytic | DNA | 106 |
| 771 (767) | R | AML, MDS, SM | Catalytic | DNA | 106 |
| 792 (788) | R | T-ALL | Motif VIII | Cat, DNA (+) | 60 |
| 826 (822) | K | MDS | Catalytic | DNAMulti | 167 |
| 841 (837) | K | AML | Catalytic | DNAMulti | 167 |
| 856 (852) | E | T-ALL | Catalytic | DNA (+) | 167 |
| 860 (856) | W | AML | Catalytic | Cat | 105 |
| 879 (875) | N | AML | Catalytic | Cat | 105 |
| 882 (878) | R | AML | In front of motif X | Cat, DNA | 60 |
AA, amino acid; AdoMet, S-adenosyl methionine reduced AdoMet binding; AdoMet (−), no AdoMet binding; AML, acute myeloid leukaemia; Cat, reduced catalytic activity; CMML, chronic myelomonocytic leukaemia; DNA, reduced DNA binding; DNA (+), enhanced DNA binding; DNAMulti, loss of ability to multimerize on DNA; DNMT3A, DNA methyltransferase 3A; H3, insensitive to H3 peptide; MDS, myelodysplastic syndrome; PWWP, Pro-Trp-Trp-Pro; sAML, secondary AML; SM, systemic mastocytosis; T-ALL, T cell acute lymphoblastic leukaemia.
DNMT3A mutations as a pre-leukaemic lesion
Deep sequencing of haematological malignancies showed that DNMT3A mutations were typically found at higher variant-allele frequencies (VAFs) than other accompanying mutations, suggesting that they were among the first to arise80,81. If, as in mice, DNMT3A-mutant (DNMT3Amut) human HSCs enjoy a self-renewal advantage, this could lead to their expansion over time, potentially serving as a pre-leukaemic lesion (FIG. 3). Indeed, this concept has been substantiated. Two independent studies found that human HSCs purified from patients with AML could harbour DNMT3A mutations in the absence of other common leukaemia-associated mutations82,83. The DNMT3A mutations also appeared in ostensibly normal lymphoid progeny from the same patients. Furthermore, as in mice, human DNMT3Amut HSCs seemed to have a marked advantage relative to wild-type HSCs, at least in xenograft models. Together, these studies indicate that human HSCs can harbour DNMT3A mutations and still contribute to multiple blood lineages, existing in a pre-leukaemic state prior to the acquisition of additional mutations that lead to leukaemia.
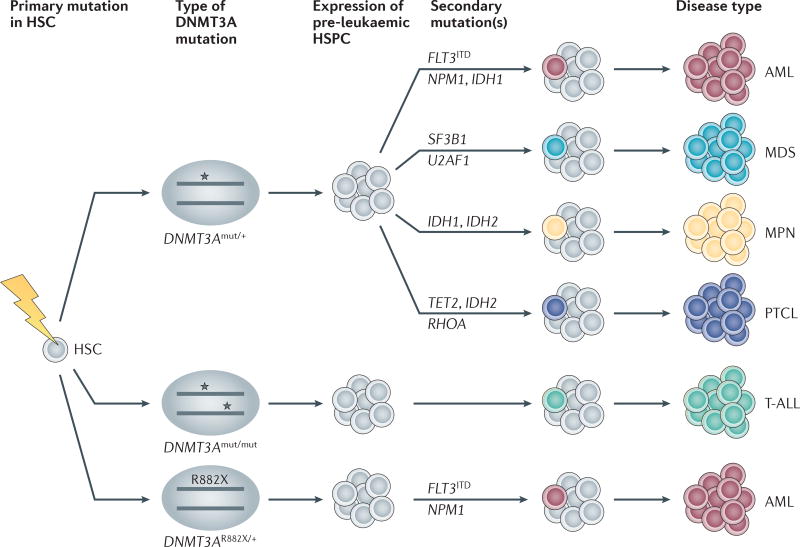
Figure 3. DNMT3A mutation allele and gene dosage, combined with secondary mutations, are likely to dictate the type of haematological disease.
DNA methyltransferase 3A (DNMT3A) mutations (indicated by stars) are likely to arise in the pre-leukaemic haematopoietic stem cell (HSC) compartment, in which heterozygous mutations predispose the occurrence of myeloid disease and peripheral T cell lymphoma (PTCL), whereas homozygous mutations are likely to occur in T cell disease. Certain mutations in R882X, where X is an amino acid other than R, lead to the acquisition of co-mutations; that is, internal tandem duplication in the gene encoding the receptor tyrosine kinase FLT3 (FLT3ITD) and mutations in the gene encoding nucleophosmin (NPM1). Acquisition of a secondary mutation in myeloid disease is associated with distinct myeloid neoplasms, including acute myeloid leukaemia (AML), myelodysplastic syndrome (MDS) and myeloproliferative neoplasms (MPNs). HSPC, haematopoietic stem and progenitor cell; IDH, isocitrate dehydrogenase; mut, mutant; RHOA, RAS homologue family member A; SF3B1, splicing factor 3b, subunit 1; T-ALL, T cell acute lymphoblastic leukaemia; U2AF1, U2 small nuclear RNA auxiliary factor 1.
Because HSCs are maintained in vivo for decades, these findings raised the possibility that DNMT3Amut HSCs might arise months, or even years, before the development of disease. Now, extensive analysis of genome sequencing data from more than 42,000 individuals without haematological malignancies across three studies has incontrovertibly established this concept84–86. In multiple cohorts, haematopoiesis was shown to frequently become clonally derived with age: 5–10% of 70-year-olds derived almost all of their peripheral blood cells from a single HSC. Genome variants associated with this remarkable state were enriched for a number of cancer-associated mutations, with mutations in DNMT3A overwhelmingly the most common. Although most individuals harbouring such mutations did not develop haematological malignancies in the time frame examined, the somatic mutations (collectively) were associated with an increased risk of leukaemia as well as all-cause mortality86. This striking clonal haematopoiesis demonstrates the Darwinian nature of the bone marrow, in which a small advantage of variant stem cells can lead to their dominance over time, and highlights the particular fitness benefit conferred on stem cells by even partial loss of DNMT3A function.
These studies established the existence of preleukaemic stem cells, in which DNMT3A mutations are common. These mutations can predispose to, but are insufficient for, the development of leukaemia. Accordingly, these studies underscore the importance of understanding the mechanisms that endow mutant HSCs with their competitive advantage, the features that may promote or inhibit the edge that mutant HSCs have over their wild-type counterparts and the factors that influence the lag time for disease development. The observation that loss of DNMT3A in murine HSCs also confers a selective advantage, allows HSCs to persist for months in the absence of transformation70, and predisposes to multiple types of haematological disease87,88 indicates that murine models may be valuable for investigating these crucial questions.
Distribution of DNMT3A mutations
Interestingly, the domains of DNMT3A that are enriched for mutations and the frequency of heterozygous versus homozygous (or compound heterozygous) mutations varies among different malignancies (FIG. 2). In the myeloid lineage, DNMT3A mutations are most prevalent in adults with AML, with most studies reporting a mutation frequency of 20–25% in DNMT3A in de novo disease76,78,89–94. Many studies examining all or most of the coding region in AML reported that around 60% of DNMT3A mutations are found at the residue R882 in the methyltransferase domain76,92,93. Across other myeloid malignancies, including myelodysplastic syndrome (MDS), myeloproliferative neoplasms (MPNs) and chronic myelomonocytic leukaemia (CMML), the R882 position is the most frequently mutated, although less frequently than in AML95–100. In all of these diseases, DNMT3A mutations are typically heterozygous, with biallelic involvement essentially confined to non-R882 mutants.
T lymphoid lineage malignancies also frequently harbour DNMT3A mutations, although the domain distribution of mutations is more diverse than that in myeloid lineages (FIG. 2). In peripheral T cell lymphoma (PTCL), DNMT3A mutations are clustered in the methyltransferase domain, but less than 20% affect the R882 position101–103. A similar proportion of mutations in T cell acute lymphoblastic leukaemia (T-ALL) affect the R882 position96,104, and the frequency of biallelic involvement is very high, occurring in up to 62% of patients104. Biallelic mutation suggests a more complete loss of function, which is consistent with classic tumour suppressor activity.
The prevalence of the R882 variant has made it of special interest. Recent data indicate that R882 mutation results in a hypomorphic protein60,105,106 that acts in a dominant-negative manner, inhibiting the methyltransferase activity of the remaining wild-type DNMT3A (DNMT3AWT)107,108. In the heterozygous state, some DNMT3AWT function remains (estimated at 20%), either through homodimeric interactions of remaining DNMT3AWT or through other protein interactions that have not yet been determined105. By contrast, heterozygous mutation of DNMT3A at other sites may lower activity of the wild-type protein to only 50% and thus be insufficient to drive malignancy. Therefore, selection for a second mutation (or loss of heterozygosity) is more common in the context of non-R882 mutations. The level of DNMT3A activity in these cases remains to be examined.
The prevalence of R882 mutations in myeloid malignancies, and the predominance of biallelic mutations in lymphoid malignancies, is intriguing and not understood. We conjecture that the varying levels of residual DNMT3A activity that are present in different scenarios have distinct effects. Possibly, some DNMT3A activity is needed for myeloid lineage choice and cancer development by action at specific gene targets, whereas the development of lymphoid malignancies may tolerate (or require) more complete functional DNMT3A loss. Alternatively, interaction with accessory proteins, including DNMT3B, may vary among the lineages, leading to different outcomes. A better understanding of the functional impact of other mutant alleles would provide valuable insight into how these malignancies develop.
The frequency of the R882 mutation may also have therapeutic implications. Patients heterozygous for the R882 mutation have some DNMT3AWT, the function of which may be improved if the R882 form was selectively inhibited, whereas patients with null-like biallelic mutations may need alternative strategies to regain normal DNA methylation.
DNMT3A mutations and aberrant DNA methylation
The mechanisms through which mutations in DNMT3A drive leukaemia are unclear. The R882 mutation in patients with AML correlates with global hypomethylation, especially at CpG islands, shores and promoters108,109, although some promoter hypermethylation is also reported77,109. Although the hypomethylation of genes previously implicated in AML, such as homeobox-containing transcription factors, can be observed77,109, the role of these events in disease development is unknown. More large-scale and unbiased methylation studies are needed to fully assess the impact of R882 and non-R882 mutations on DNA methylation.
In Dnmt3a−/− HSCs, hypomethylation was predominantly found when whole-genome DNA methylation analysis was carried out54. In murine leukaemias generated from Dnmt3a−/− HSCs, loss of methylation at intergenic regions was associated with AML, whereas hypermethylation, often at CpG islands, was observed in T-ALL87. The mechanism that leads to an increase in methylation when a DNA methyltransferase is disrupted in mice (or mutated in humans) is enigmatic; it might originate partly from residual DNMT3B, although the predominant DNMT3B transcript present in samples from patients with AML108, as well as murine HSCs71, is thought to encode a catalytically inactive protein. Moreover, the relative contribution of hypermethylation versus hypomethylation to the pathological state and leukaemia development is not known.
Further impeding a mechanistic understanding is the relative lack of correlation between changes in DNA methylation and gene expression56,70,110. Emerging data from other studies suggest that changes in enhancer DNA methylation, in addition to changes in promoters, may be important111,112. Further studies of aberrant DNA methylation should be linked with topological studies of the genome, including the three-dimensional structure that links enhancers and gene expression by DNA looping113.
The lack of understanding of how DNMT3A mutations affect DNA methylation and the poor correlation of genetic changes with gene expression have led to speculation that mutations in DNMT3A may be disrupting functions that are distinct from DNA methylation. Although no activities of DNMT3A that are unrelated to DNA methylation have been reported, this possibility certainly merits further study.
DNMT3A-related disease features
In some studies, the prevalence of DNMT3A mutations is associated with age, ethnicity and gender. Additionally, a number of clinical studies have reported possible associations between DNMT3A mutational status and clinical features such as blood cell counts and the percentage of blasts in the blood and bone marrow at the time of cancer diagnosis (see Supplementary information S1 (table)). In both myeloid and lymphoid malignancies, an increased incidence of DNMT3A mutations with advanced age has been reported (see Supplementary information S1 (table)). This relationship with age is consistent with the recent findings of DNMT3A mutations linked to clonally derived haematopoiesis that is progressively more common with age84–86. In a subset of individuals with such clonal haematopoiesis, malignant transformation may occur after a subsequent deleterious lesion is acquired following a lag time that may be in the order of years. In accordance with this observation, DNMT3A mutations are exceedingly rare in paediatric haematological malignancies114–119 (TABLE 1; see Supplementary information S1 (table)). Alternatively, fundamental differences in the pathogenic process may account for the dearth of DNMT3A mutations in paediatric compared to adult haematological malignancies.
Specific co-mutation patterns of DNMT3A
DNTM3A mutations occur non-randomly in association with other genetic abnormalities, including cytogenetic aberrations (FIG. 4). For example, in AML, DNMT3A mutations are essentially never present in patients with the chromosomal translocations t(15;17), inv(16) and t(8;21)76,78,89,91,92,94, despite their association with specific DNA methylation patterns78,120. Similarly, DNMT3A mutations are almost never found concurrently with rearrangements involving the histone-lysine N-methyltransferase KMT2A (also known as MLL) in acute leukaemias, and are negatively correlated with mutations in the transcriptional regulator additional sex combs-like transcriptional regulator 1 (ASXL1), an enzyme that is important in H3K27 methylation in MDS95,121,122. Thus, the mutual exclusion of DNMT3A mutations and these abnormalities suggests possible convergence on similar epigenetic perturbations. Of the positive correlations, there are some striking patterns of co-mutation that seem to dictate disease outcome (FIG. 4) and that may have implications for how DNMT3A functions.
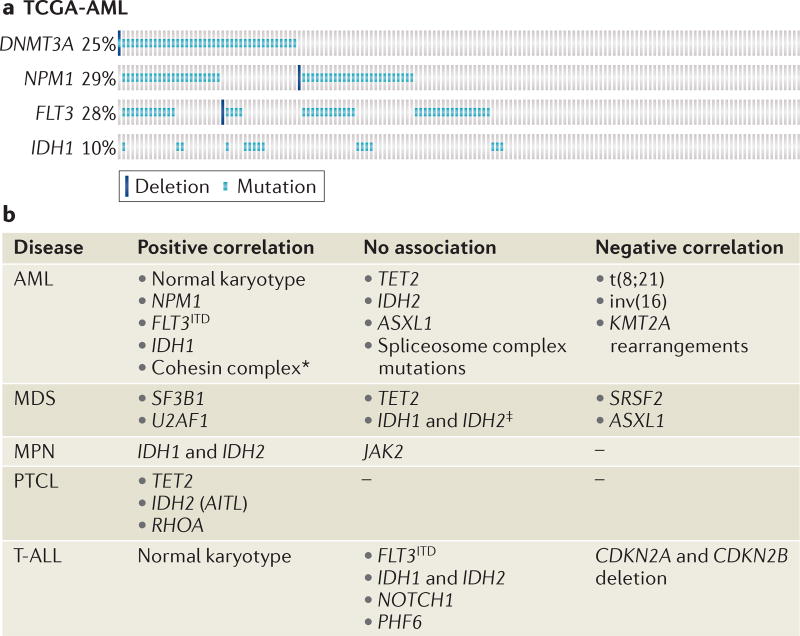
Figure 4. Co-mutations with DNMT3A in acute myeloid leukaemia and other diseases.
a | Mutations in nucleophosmin (NPM1) and internal tandem duplication in the gene encoding the receptor tyrosine kinase FLT3 (FLT3ITD) occur more frequently in DNA methyltransferase 3A (DNMT3A)-mutant acute myeloid leukaemia (AML) than in non-DNMT3A-mutant AML. Each column represents a patient from The Cancer Genome Atlas (TCGA) database. Each coloured mark represents a mutation (light blue) or deletion (dark blue). The frequency of patients with each mutation is indicated on the left-hand side. The figure was made using cBioPortal160,161. b | Frequent co-mutations with DNMT3A. AITL, angioimmunoblastic T cell lymphoma; ASXL1, additional sex combs-like transcriptional regulator 1; CDKN, cyclin-dependent kinase inhibitor; IDH, isocitratedehydrogenase; JAK2, Janus kinase 2; KMT2A, histone-lysine N-methyltransferase KMT2A (also known as MLL); MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; PHF6, PHD finger protein 6; PTCL, peripheral T cell lymphoma; RHOA, RAS homologue family member A; SF3B1, splicing factor 3b, subunit 1; SRSF2, serine/arginine-rich splicing factor 2; T-ALL, T cell acute lymphoblastic leukaemia; U2AF1, U2 small nuclear RNA auxiliary factor 1.*One large study showed significant association, but another study found no association. ‡One study found a significant association with IDH2 mutations that was not confirmed in two additional large studies.
Two of the most striking examples of co-occurrence are in AML: that is, the association between DNMT3A mutations and mutations in the gene encoding nucleophosmin (NPM1), and the co-occurrence of DNMT3A mutations with internal tandem duplication in the gene encoding the receptor tyrosine kinase FLT3 (FLT3ITD) (FIG. 4). A remarkable 60% of patients with DNMT3A mutations also carry an NPM1 mutation, whereas only 13% of patients with DNMT3AWT harbour an NPM1 mutation76,78,89,91,92,96,123,124. Similarly, FLT3ITD mutations are specifically enriched in patients with DNMT3A mutations76,78,89,91,94,96,123,125 (FIG. 4). The known association between NPM1 and FLT3ITD mutations raises the possibility that the high frequency of co-occurring FLT3ITD and DNMT3A mutations is merely a reflection of the high frequency of concomitant FLT3ITD and NPM1 mutations. However, extensive genomic and epigenomic analysis performed by The Cancer Genome Atlas Research Network found that samples with all three mutations formed distinct mRNA, microRNA and DNA methylation clusters, suggesting that the occurrence of all three mutations is non-random, and that NPM1mutFLT3ITDDNMT3Amut AML is a distinct entity78. How these mutations interact to cause leukaemia is unclear and warrants further investigation.
In haematological malignancies, an interesting pattern exists of co-mutations of DNMT3A with genes encoding enzymes that are important in 5-hydroxymethylation, including isocitrate dehydrogenase 1 (IDH1), IDH2 and TET2. Mutations in IDH1 and IDH2 are thought to contribute to leukaemogenesis by the accumulation of 2-hydroxyglutarate, which inhibits TET2, the enzyme responsible for DNA hydroxymethylation. IDH1 mutations are enriched in patients with AML who harbour DNMT3A mutations76,78,91,124 (FIG. 4). In MDS, the correlation between mutations in IDH1 and IDH2 and mutations in DNMT3A is still uncertain; however, three of ten patients with DNMT3Amut secondary acute myeloid leukaemia (sAML) derived from MDS also had mutations in IDH1 or IDH2, whereas none of the patients with DNMT3AWT sAML derived from MDS exhibited IDH1 or IDH2 mutations126,127. This suggests that the combination of DNMT3A mutation with mutations in IDH1 and IDH2 may contribute to progression from MDS to AML. The evidence suggests that an important interaction exists between mutations in DNMT3A and mutations in IDH1 and IDH2 in myeloid malignancies, implying that aberration of DNA methylation and DNA hydroxymethylation contribute to leukaemogenesis. Curiously, no specific association between DNMT3A and TET2 mutations exists, perhaps suggesting an impact of IDH mutations on histone, rather than DNA, demethylation128. Nevertheless, in T cell lymphoma a strong and interesting association between mutations in DNMT3A, TET2 and IDH exists that may have important mechanistic implications (BOX 1).
Box 1. DNMT3A, TET2 and IDH mutations in lymphoma.
In peripheral T cell lymphomas (PTCLs), combined aberration of DNA methylation and hydroxymethylation is likely to contribute to lymphomagenesis. However, unlike in myeloid disease, mutations in DNA methyltransferase 3A (DNMT3A) are highly correlated with concomitant mutations in the gene encoding a methylcytosine dioxygenase (TET2), with 70–100% of DNMT3A-mutant patients also harbouring mutations in TET2 (REFS 101–103,147). In addition, in angioimmunoblastic T cell lymphoma (AITL; a PTCL subset), nearly two-thirds of patients with TET2 mutations have two or three different TET2 mutations, whereas such mutations are predominantly heterozygous in myeloid disease. Further, whereas in myeloid disease mutations of TET2 and isocitrate dehydrogenase 1 (IDH1) and/or IDH2 are mutually exclusive, they can co-occur in AITL, mostly in patients with only one TET2 mutation101. These data strongly suggest that a substantial impairment of DNA hydroxymethylation — either from the loss of both normal copies of TET2 or through a combination of TET2 and IDH2 mutation — is required for this disease to develop. Although the interaction between DNMT3A mutation and mutations in TET2 and IDH1 and/or IDH2 in PTCL is distinct from that in myeloid malignancies, importantly PTCL has a significantly different pattern of mutations when compared with T cell acute lymphoblastic leukaemia (T-ALL). In both T-ALL and PTCL, non-R882 DNMT3A mutations predominate; however, biallelic DNMT3A mutations are rarely encountered in PTCL, whereas most patients with T-ALL have them. There are at least two possible explanations for these interesting findings: first, with such profound loss of DNA hydroxymethylation in PTCL, less impact on DNA methylation is necessary for disease development; or second, retention of at least partial DNMT3A function is necessary for these diseases to develop.
In MDS there is a strong association between DNMT3A mutations and mutations of the spliceosome factor SF3B1 (splicing factor 3b, subunit 1), with 50–56% of patients with MDS and DNMT3A mutations also harbouring mutations of SF3B1, compared with 12–17% of patients with DNMT3AWT (REFS 95,121,122) (FIG. 4). Co-mutation with the spliceosome factor U2AF1 (U2 small nuclear RNA auxiliary factor 1) has also been reported129. By contrast, a negative correlation between mutations of serine/arginine-rich splicing factor 2 (SRSF2) and DNMT3A is seen121, indicating differential genetic interactions between mutant DNMT3A and the spliceosomal machinery, and a potential mechanistic convergence (BOX 2).
Box 2. DNMT3A and alternative splicing.
Epigenetic regulation through histone modifications has been linked to alternative splicing148,149, perhaps also implicating DNA methylation. Gene body DNA methylation is associated with transcriptionally active genes150, the exons of which are preferentially marked by histone H3 lysine 36 trimethylation (H3K36me3)151. DNA methyltransferase 3A (DNMT3A) physically interacts with H3K36me3 (REF. 28), linking DNMT3A to exon methylation. Interestingly, a spike of DNA methylation is observed at the 5′ splice site or exon–intron junction, followed by a sharp plummet at the 3′ splice site152. Moreover, in maize, high levels of DNA methylation at the 5′ spice site correlate with higher rates of alternative splicing153, linking DNA methylation to alternative splicing. With these observations, we can speculate that DNMT3A might have a role at the 5′ splice site. Intriguingly, a number of spliceosome factors that are co-mutated with DNMT3A in myelodysplastic syndrome are assigned specifically to the 3′ splice junction154. The implications of DNA methylation for splicing, and its role in diseases in which mutations in spliceosome factors are found, warrant further exploration.
Prognostic impact of DNMT3A mutations
Most AML studies have found no difference in the rate of complete remission between patients with and without DNMT3A mutations; however, analysis of the impact of DNMT3A mutations on survival has generated differing results. Ley et al. reported that DNMT3A mutations had a highly significant negative impact on survival76, a finding supported by a number of large studies90,91,93,123,124,130 but not uniformly corroborated89,92,96. Comparing these reports is challenging because patient populations, treatment regimens and outcome measures vary widely from study to study. The prognostic impact of R882 versus non-R882 mutations is also inconclusive92,94,123,124.
As in AML, reports vary regarding the impact of DNMT3A mutations on outcome in MDS, with some studies finding a significantly worse overall and event-free survival and a higher rate of progression to AML97,131, but others finding no significant association between outcome and mutation of DNMT3A95,96,121,122,132. Interestingly, patients with SF3B1WT and DNMT3Amut had an inferior overall survival and higher risk of AML transformation compared with other related mutation combinations (SF3B1mutDNMT3Amut, SF3B1WTDNMT3AWT and SF3B1mutDNMT3AWT)121. Because over 50% of patients with MDS who have DNMT3A mutations have a concomitant SF3B1 mutation — and these mutations are associated with superior outcomes — perhaps the prognostic significance of DNMT3A mutation is diluted when only looking at these mutations as a whole group. Future studies could evaluate the prognostic impact of the SF3B1–DNMT3A ‘risk group’ to better stratify patients.
Despite the lack of clarity regarding the impact of DNMT3A mutation on outcome, evidence in MDS, MPN and CMML suggests that the presence of a DNMT3A mutation may facilitate the transition from myeloproliferation and/or myelodysplasia to frank myeloid leukaemia. In all of these neoplasms, there is a striking enrichment for DNMT3A mutations in sAML compared with the frequency in de novo acute myeloid leukaemia (de novo AML)127,133,134. In addition, the order of acquiring a mutation in DNMT3A relative to other genes may affect disease progression. In MDS and myelofibrosis, the DNMT3A mutations found in sAML can be traced back to the original MDS clone, which is consistent with the concept that DNMT3A mutations are likely to instigate a pre-leukaemic state. However, in the MPNs polycythaemia vera and essential thrombocythaemia, patients harbouring mutations in Janus kinase 2 (JAK2) may acquire DNMT3A mutations later135, indicating that DNMT3A mutations can be acquired through the evolution of a JAK2mut clone and that this may contribute to progression in these specific diseases.
As in myeloid diseases, a number of studies have reported significantly worse overall survival for patients with T-ALL who have DNMT3A mutations96,104,136. It is not clear whether this is cause or correlation because DNMT3A mutations are enriched in the more primitive and/or immature T-ALL subtypes, which tend to have a worse prognosis than mature T-ALL104,136,137. However, the sum of available data suggests that the presence of a DNMT3A mutation is a negative prognostic marker independent of disease phenotype. Thus, it is reasonable to consider screening patients with T-ALL for mutations of DNMT3A as a way to refine risk stratification.
Therapeutic implications
DNA-damaging anthracyclines are a key component of most AML treatment regimens. Some studies suggest that better outcomes for patients with AML who harbour DNMT3A mutations have correlated with intensified treatment with DNA-damaging anthracycline therapy89,138. If confirmed, this may indicate that AML cells with DNMT3A mutations are relatively resistant to this class of agents, perhaps due to DNMT3A-linked resistance to DNA damage-induced cell death or to other unknown mechanisms. Therefore, patients with DNMT3A mutations may require higher anthracycline doses compared with patients who have DNMT3AWT. Intensification through the administration of higher daunorubicin doses or the use of a more potent anthracycline, idarubicin, may overcome the negative prognostic impact of DNMT3A mutations, but this strategy would come at the cost of increased toxicity; therefore, more targeted approaches are desirable.
The use of the DNA methyltransferase inhibitor 5-azacytidine and its deoxy derivative, decitabine, has become increasingly common for the treatment of MDS and is being explored for the treatment of AML. These agents are thought to work by inhibiting DNMT1, leading to the demethylation of aberrantly hypermethylated genes, such as cyclin-dependent kinase inhibitor 2B (CDKN2B)139,140, MDR1 (also known as ABCB1) and syndecan 4 (SDC4)141, which in turn results in the expression of these genes being reinstated.
Importantly, use of these agents was adopted before the incidence of DNMT3A mutations was appreciated. Given the variable response to these agents, even among patients with similar disease phenotypes, a number of clinical studies were set up to examine the relationship between hypomethylating agent response and the mutational status of DNMT3A and other epigenetic modifiers. One study of 92 patients with MDS, MDS/MPN and sAML showed that patients with a DNMT3A mutation, a TET2 mutation or both were more likely to have a favourable response to hypomethylating therapy142. Additionally, one small AML study found that decitabine treatment resulted in a higher complete remission rate and a trend towards increased overall survival in patients with DNMT3A mutations123. However, in vitro treatment with decitabine of primary samples from patients with AML did not lead to different responses in samples with DNMT3A mutations compared to those without143. Because DNMT3A mutations are likely to cause a reduction in DNA methyltransferase activity, the mechanism of therapeutic benefit provided by a drug that inhibits DNA methylation is puzzling. Studies including larger cohorts of patients are necessary to determine whether a true correlation exists between DNMT3A status and response to hypomethylating agents. Furthermore, there is poor correlation between changes in DNA methylation and gene expression after treatment with hypomethylating therapy, indicating that the mechanism of action of these agents is likely to be complex143. More work to delineate the mechanism of action of these drugs may improve our ability to predict which patients are most likely to benefit from treatment.
Given that DNMT3A mutations seem to have no clear impact on ability to achieve complete remission, the poor survival rate associated with such mutations is probably attributable to a high relapse rate91,93,125,144. Because DNMT3A mutations show remarkable stability during disease evolution, it is likely that the high relapse rate is due to the presence of DNMT3A mutations in ostensibly normal-appearing patient HSCs that persist even after chemotherapy and during relapse82,83. These mutant HSCs would be available to reinitiate disease after new oncogenic hits. With DNMT3Amut-associated disease, long-term disease surveillance should be based on molecular, rather than histological, criteria.
The concept that DNMT3A-mutant progenitors are difficult to eradicate points to an urgent need to identify new approaches to specifically target these HSCs. The challenge will be the similarity of the mutant HSCs to their wild-type counterparts. In mice, Dnmt3a−/− HSCs retain most characteristics of normal stem cells, including relative quiescence70,71. One strategy may be to identify mechanisms to enforce mutant clone silence. For example, some patients with CML who are taken off tyrosine kinase inhibitor therapy after long treatment periods remain in remission despite the continued presence of breakpoint cluster region (BCR)–ABL1-positive cells, indicating that complete eradication of the leukaemic clone is not always necessary145. Alternatively, improved mechanistic understanding may lead to new approaches, such as the upregulation of catalytically active DNMT3B splice isoforms. Considering the large number of patients with DNMT3A mutations across many haematological diseases, novel therapeutic approaches are greatly needed.
Conclusions
DNMT3A has recently emerged as one of the most important tumour suppressors in haematological malignancies. Its exceptional role is rooted in its crucial function in stem cells, in which it enables the first steps of haematopoietic differentiation. The observation that Dnmt3a−/− HSCs have a marked selective advantage over their normal counterparts in bone marrow underscores the Darwinian nature of competing HSCs in the haematopoietic system and suggests that certain mutations can have large effects on the stem cell pools over time. Indeed, the increasing prevalence of clonal haematopoiesis with age146, which is highly associated with DNMT3A mutations84–86, strongly suggests that stem cells that have acquired a DNMT3A mutation early in life may slowly accumulate, finally appearing in large numbers many years later.
In addition to age, we expect that other factors — including haematological stress, such as that caused by infection — could positively or negatively affect the proportion of normal versus mutant HSCs. Thus, the way that mutations in DNMT3A affect normal haematopoiesis, as well as the propensity of mutant clones to contribute to disease, will be of considerable future interest. These questions also have a crucial bearing on risk stratification and the choice of therapeutic modalities for patients.
On a mechanistic level, despite nearly two decades of basic research, we understand only poorly how DNMT3A carries out its duties with regard to DNA methylation and gene expression, and we have no inkling of any DNA methylation-independent functions.
In summary, DNMT3A has a crucial biological role in self-renewing cells, enabling their differentiation. When lost or reduced in activity, the balance is shifted, resulting in a predisposition to cancer and other pathological consequences. Further study of the different facets of this molecule, including basic research and more clinical data, should generate new insights, leading to new therapeutic opportunities.
Supplementary Material
Acknowledgments
The authors thank C. Gillespie for critical comments on the manuscript. L.Y. is an M.D./Ph.D. McNair Scholar. This work was supported by US National Institutes of Health grants CA183252 and DK092883, the Sam Waxman Cancer Research Foundation and the Edward P. Evans Foundation.
Glossary
- Imprinting
A genetic phenomenon in which the expression status of each allele of a gene is dictated by the parent from which the allele was inherited. DNA methylation plays a part in the crucial parent-specific regulation of gene expression.
- CpG
A dinucleotide pair on the same DNA strand consisting of cytosine (C) and guanine (G) nucleotides joined by a phosphodiester bond (p); CpGs are the predominant target of DNA methylation.
- Repetitive elements
Stretches of DNA that are found in multiple copies (often many thousands) throughout the genome. Most represent types of transposable elements, the activity of which is partly repressed by DNA methylation.
- Pericentromeric heterochromatin
Regions of compact genomic DNA and chromatin that are located near centromeres. Pericentromeric heterochromatin is associated with repressive chromatin marks and inactive gene transcription.
- Teratoma formation assay
A stringent test of pluripotency in which embryonic (or other) stem cells are transplanted into a mouse and examined for their ability to differentiate into all three germ layers.
- Variant-allele frequencies (VAFs)
The relative proportions of sequencing reads from variant alleles. Variants found at a VAF of 50% usually represent heterozygous mutations that are present in all cells within the sample (the founding clone). Lower VAFs suggest that the variant occurs in only a fraction of the cells (possible subclone).
- Compound heterozygous
The presence of two different mutant alleles at a particular gene locus, on each chromosome of a pair.
- Myelodysplastic syndrome (MDS)
A group of myeloid disorders characterized by clonal and ineffective haematopoiesis, dysplasia in one or more of the myeloid cell lines, cytopaenia(s) and increased risk of the development of acute myeloid leukaemia.
- Myeloproliferative neoplasms (MPNs)
A heterogeneous group of clonal haematopoietic stem cell disorders characterized by the abnormal proliferation of one or more of the myeloid lineages.
- Chronic myelomonocytic leukaemia (CMML)
A clonal haematological malignancy with features of both a myeloproliferative neoplasm and a myelodysplastic syndrome. CMML is characterized by persistent monocytosis, dysplasia of one or more myeloid lineages, and <20% blasts in the blood and bone marrow. By definition, CMML lacks the breakpoint cluster region (BCR)–ABL1 fusion gene and platelet-derived growth factor receptor-α (PDGFRA) or PDGFRB rearrangements.
- Enhancers
Distal regulatory regions of the genome up to 1 megabase upstream of transcription start sites defined by chromatin marks such as monomethylation of histone H3 lysine 4 (H3K4me1) and acetylation of H3K27 (H3K27ac). Enhancers are often bound by transcription factors.
- Secondary acute myeloid leukaemia (sAML)
A documented myelodysplastic syndrome or myeloproliferative neoplasm that transforms into AML. This subset of AML is now included in the category ‘AML with myelodysplasia-related changes’.
- Spliceosome factor
A member of the large and complex molecular machinery known as the spliceosome, which functions to remove introns from a transcribed precursor mRNA.
- De novo acute myeloid leukaemia (De novo AML)
Initial diagnosis of AML, not preceded by myelodysplastic syndrome or myeloproliferative neoplasm, and not associated with prior chemotherapy or radiation therapy.
- Polycythaemia vera
A chronic myeloproliferative neoplasm characterized by aberrantly increased red blood cell production independent of the mechanisms that normally regulate erythropoiesis. Polycythaemia vera is molecularly characterized by activating mutations of the tyrosine kinase Janus kinase 2 (JAK2), which are present in nearly all patients with polycythaemia vera.
- Essential thrombocythaemia
A chronic myeloproliferative neoplasm characterized by increased platelet count in the peripheral blood and megakaryocyte proliferation with large and mature morphology in the bone marrow. Essential thrombocytosis is characterized molecularly by activating mutations of the tyrosine kinase Janus kinase 2 (JAK2), which are present in 40–50% of patients with essential thrombocytosis.
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
COSMIC: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
References
- 1.Peters SL, et al. Essential role for Dnmt1 in the prevention and maintenance of MYC-induced T-cell lymphomas. Mol. Cell. Biol. 2013;33:4321–4333. doi: 10.1128/MCB.00776-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laird PW, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 3.Broske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nature Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. Apmis. 2013;121:85–94. doi: 10.1111/j.1600-0463.2012.02940.x. [DOI] [PubMed] [Google Scholar]
- 5.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 8.Holliday R, Grigg GW. DNA methylation and mutation. Mutat. Res. 1993;285:61–67. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- 9.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Weisenberger DJ, et al. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene. 2002;298:91–99. doi: 10.1016/s0378-1119(02)00976-9. [DOI] [PubMed] [Google Scholar]
- 12.Schoofs T, Müller-Tidow C. DNA methylation as a pathogenic event and as a therapeutic target in AML. Cancer Treat. Rev. 2011;37(Suppl. 1):1–6. doi: 10.1016/j.ctrv.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 15.Wienholz BL, et al. DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet. 2010;6:e1001106. doi: 10.1371/journal.pgen.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 17.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nature Rev Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 18.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front. Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson KD, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie S, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 24.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 25.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagisawa Y, Ito E, Yuasa Y, Maruyama K. The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents. Biochim. Biophys. Acta. 2002;1577:457–465. doi: 10.1016/s0167-4781(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 27.Suetake I, et al. Characterization of DNA-binding activity in the N-terminal domain of the DNA methyltransferase Dnmt3a. Biochem. J. 2011;437:141–148. doi: 10.1042/BJ20110241. [DOI] [PubMed] [Google Scholar]
- 28.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2014 doi: 10.1038/nature13899. http://dx.doi.org/10.1038/nature13899 References 30 and 32 demonstrate that the auto-inhibitory function of the ADD domain of DNMT3A provides the link between H3K4 methylation and DNMT3A. [DOI] [PubMed]
- 33.Xu F, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr. Med. Chem. 2010;17:4052–4071. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holz-Schietinger C, Reich NO. The inherent processivity of the human de novo methyltransferase 3A (DNMT3A) is enhanced by DNMT3L. J. Biol. Chem. 2010;285:29091–29100. doi: 10.1074/jbc.M110.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. This article deeply explores the phenotype in ESCs of ablation of Dnmt3a and Dnmt3b, establishing the paradigm of extended self-renewal and DNA methylation loss, foreshadowing the somatic stem cell phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostler KR, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon CA, Hartono SR, Chedin F. Inactive DNMT3B splice variants modulate de novo DNA methylation. PLoS ONE. 2013;8:e69486. doi: 10.1371/journal.pone.0069486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 40.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hattori N, et al. Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells. Genome Res. 2004;14:1733–1740. doi: 10.1101/gr.2431504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi SH, et al. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Res. 2011;39:104–118. doi: 10.1093/nar/gkq774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson AK, Geiman TM, Sankpal UT, Hager GL, Robertson KD. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun. 2004;322:110–118. doi: 10.1016/j.bbrc.2004.07.083. [DOI] [PubMed] [Google Scholar]
- 44.Takeshima H, et al. Distinct DNA methylation activity of Dnmt3a and Dnmt3b towards naked and nucleosomal DNA. J. Biochem. 2006;139:503–515. doi: 10.1093/jb/mvj044. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene. 2013;34:104–118. doi: 10.1038/onc.2013.522. [DOI] [PubMed] [Google Scholar]
- 46.Weissmann F, et al. DNA hypermethylation in Drosophila melanogaster causes irregular chromosome condensation and dysregulation of epigenetic histone modifications. Mol. Cell. Biol. 2003;23:2577–2586. doi: 10.1128/MCB.23.7.2577-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muramatsu D, Singh PB, Kimura H, Tachibana M, Shinkai Y. Pericentric heterochromatin generated by HP1 protein interaction-defective histone methyltransferase Suv39h1. J. Biol. Chem. 2013;288:25285–25296. doi: 10.1074/jbc.M113.470724. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Karimi MM, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nature Struct. Mol. Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang Y, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nature Commun. 2011;2:533. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rush M, et al. Targeting of EZH2 to a defined genomic site is sufficient for recruitment of Dnmt3a but not de novo DNA methylation. Epigenetics. 2009;4:404–414. doi: 10.4161/epi.4.6.9392. [DOI] [PubMed] [Google Scholar]
- 53.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 54.Jeong M, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nature Genet. 2014;46:17–23. doi: 10.1038/ng.2836. This paper defines ‘DNA methylation canyons’, which are large, under-methylated regions of the genome that are particularly affected by the loss of DNMT3A. It is also suggested that TET proteins and histone marks interplay at canyons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin IG, Han L, Taghva A, O’Brien LE, Hsieh CL. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 2002;22:704–723. doi: 10.1128/MCB.22.3.704-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 60.Gowher H, et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 2006;357:928–941. doi: 10.1016/j.jmb.2006.01.035. This paper describes a mutational analysis that reveals the key role of specific residues in the catalytic domain of DNMT3A. [DOI] [PubMed] [Google Scholar]
- 61.Suetake I, Miyazaki J, Murakami C, Takeshima H, Tajima S. Distinct enzymatic properties of recombinant mouse DNA methyltransferases Dnmt3a and Dnmt3b. J. Biochem. 2003;133:737–744. doi: 10.1093/jb/mvg095. [DOI] [PubMed] [Google Scholar]
- 62.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 63.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo JU, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nature Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziller MJ, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt CS, et al. Global DNA hypomethylation prevents consolidation of differentiation programs and allows reversion to the embryonic stem cell state. PLoS ONE. 2012;7:e52629. doi: 10.1371/journal.pone.0052629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genet. 2012;44:23–31. doi: 10.1038/ng.1009. This article shows that ablation of Dnmt3a in HSCs leads to stem cell expansion and inhibited differentiation, providing the conceptual framework for understanding the role of DNMT3A mutations in human haematological malignancies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Challen GA, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. This article shows the roles of DNMT3B and DNMT3A in HSC differentiation, implicating the β-catenin pathway, upregulation of self-renewal genes and attendant epigenetic changes in the molecular and cellular phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 73.Wu Z, et al. Dnmt3a regulates both proliferation and differentiation of mouse neural stem cells. J. Neurosci. Res. 2012;90:1883–1891. doi: 10.1002/jnr.23077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tatton-Brown K, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nature Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamashita Y, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 76.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. This seminal paper reports the frequency and clinical significance of DNMT3A mutations in a large cohort of patients with acute myeloid leukaemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 78.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. This work is a comprehensive genetic and epigenetic analysis of patients with AML that provides a valuable database of patients with DNMT3A and other commonly occurring mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huether R, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nature Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. This study identifies DNMT3Amut HSCs as the pre-leukaemic founding clones in AML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. This study demonstrates the existence of pre-leukaemic HSCs in AML and at remission, and the predominance of mutations in epigenetic modifiers, including DNMT3A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. References 84, 85 and 86 demonstrate the striking age-related increase of clonal haematopoiesis and highlight DNMT3A mutations as being highly correlated with this state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2014;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Celik H, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2014;125:619–628. doi: 10.1182/blood-2014-08-594564. References 87 and 88 show that ablation (knockout) of Dnmt3a in mouse stem cells can recapitulate the range of haematological malignancies seen in patients, despite the distinct genetic lesions observed in such malignancies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. This study reports the frequency and prognostic relevance of DNMT3A mutations and other recurrent mutations in a large, uniformly treated cohort of adults with AML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen Y, et al. Gene mutation patterns and their prognostic impact in a cohort of 1,185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 91.Ribeiro AFT, et al. Mutant DNMT3A: a new marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- 92.Gaidzik VI, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–4777. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- 93.Hou HA, et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28:50–58. doi: 10.1038/leu.2013.236. [DOI] [PubMed] [Google Scholar]
- 94.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 95.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roller A, et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia. 2013;27:1573–1578. doi: 10.1038/leu.2013.65. [DOI] [PubMed] [Google Scholar]
- 97.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stegelmann F, et al. DNMT3A mutations in myeloproliferative neoplasms. Leukemia. 2011;25:1217–1219. doi: 10.1038/leu.2011.77. [DOI] [PubMed] [Google Scholar]
- 99.Abdel-Wahab O, et al. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia. 2011;25:1219–1220. doi: 10.1038/leu.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature Genet. 2014;46:171–175. doi: 10.1038/ng.2872. References 101–103 demonstrate the high frequency of DNMT3A mutations and their striking association with IDH and TET2 mutations in peripheral T cell lymphomas. [DOI] [PubMed] [Google Scholar]
- 104.Grossmann V, et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer. 2013;52:410–422. doi: 10.1002/gcc.22039. [DOI] [PubMed] [Google Scholar]
- 105.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J. Biol. Chem. 2012;287:30941–30951. doi: 10.1074/jbc.M112.366625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J. Biol. Chem. 2011;286:41479–41488. doi: 10.1074/jbc.M111.284687. References 105 and 106 show the crucial role of DNMT3A oligomerization in maintaining high DNA methylation activity in addition to the mechanistic effect of specific AML-associated mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim SJ, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. References 107 and 108 provide insights into the common DNMT3A R882 mutation, showing that it can act as a dominant-negative mutation in mouse ESCs and human AML cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qu Y, et al. Differential methylation in CN-AML preferentially targets non-CGI regions and is dictated by DNMT3A mutational status and associated with predominant hypomethylation of HOX genes. Epigenetics. 2014;9:1108–1119. doi: 10.4161/epi.29315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raddatz G, Gao Q, Bender S, Jaenisch R, Lyko F. Dnmt3a protects active chromosome domains against cancer-associated hypomethylation. PLoS Genet. 2012;8:e1003146. doi: 10.1371/journal.pgen.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 113.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ho PA, et al. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children’s Oncology Group. Pediatr. Blood Cancer. 2011;57:204–209. doi: 10.1002/pbc.23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hollink IHIM, et al. Low frequency of DNMT3A mutations in pediatric AML, and the identification of the OCI-AML3 cell line as an in vitro model. 2011;26:371–373. doi: 10.1038/leu.2011.210. [DOI] [PubMed] [Google Scholar]
- 116.Liang DC, et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood. 2013;121:2988–2995. doi: 10.1182/blood-2012-06-436782. [DOI] [PubMed] [Google Scholar]
- 117.Shiba N, et al. DNMT3A mutations are rare in childhood acute myeloid leukaemia, myelodysplastic syndromes and juvenile myelomonocytic leukaemia. Br. J. Haematol. 2012;156:413–414. doi: 10.1111/j.1365-2141.2011.08879.x. [DOI] [PubMed] [Google Scholar]
- 118.Thol F, et al. DNMT3A mutations are rare in childhood acute myeloid leukemia. Haematologica. 2011;96:1238–1240. doi: 10.3324/haematol.2011.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Damm F, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- 123.Marcucci G, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Renneville A, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26:1247–1254. doi: 10.1038/leu.2011.382. [DOI] [PubMed] [Google Scholar]
- 125.Hajkova H, et al. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk. Res. 2012;36:1128–1133. doi: 10.1016/j.leukres.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 126.Fried I, et al. Frequency, onset and clinical impact of somatic DNMT3A mutations in therapy-related and secondary acute myeloid leukemia. Haematologica. 2012;97:246–250. doi: 10.3324/haematol.2011.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zebisch A, Hoefler G, Quehenberger F, Wolfler A, Sill H. Mutant DNMT3A in acute myeloid leukemia: guilty of inducing genetic instability? Leukemia. 2013;27:1777–1778. doi: 10.1038/leu.2013.48. [DOI] [PubMed] [Google Scholar]
- 128.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thol F, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 130.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J. Clin. Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 131.Thol F, et al. Rare occurence of DNMT3A mutations in myelodysplastic syndromes. Haematologica. 2011;96:1870–1873. doi: 10.3324/haematol.2011.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lin CC, et al. IDH mutations are closely associated with mutations of DNMT3A, ASXL1 and SRSF2 in patients with myelodysplastic syndromes and are stable during disease evolution. Am. J. Hematol. 2014;89:137–144. doi: 10.1002/ajh.23596. [DOI] [PubMed] [Google Scholar]
- 133.Fried I, et al. Mutations in DNMT3A and loss of RKIP are independent events in acute monocytic leukemia. Haematologica. 2012;97:1936–1937. doi: 10.3324/haematol.2012.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kar SA, et al. Spliceosomal gene mutations are frequent events in the diverse mutational spectrum of chronic myelomonocytic leukemia but largely absent in juvenile myelomonocytic leukemia. Haematologica. 2012;98:107–113. doi: 10.3324/haematol.2012.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Quintas-Cardama A, et al. Molecular analysis of patients with polycythemia vera or essential thrombocythemia receiving pegylated interferon α-2a. Blood. 2013;122:893–901. doi: 10.1182/blood-2012-07-442012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Vlierberghe P, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neumann M, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 138.LaRochelle O, et al. Do AML patients with DNMT3A exon 23 mutations benefit from idarubicin as compared to daunorubicin? A single center experience. Oncotarget. 2011;2:850–861. doi: 10.18632/oncotarget.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Herman JG, et al. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–841. [PubMed] [Google Scholar]
- 140.Paul TA, Bies J, Small D, Wolff L. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood. 2010;115:3098–3108. doi: 10.1182/blood-2009-07-233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Toyota M, et al. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–2829. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 142.Traina F, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28:78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 143.Klco JM, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2013;121:1633–1643. doi: 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ostronoff F, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: a SWOG report. Leukemia. 2013;27:238–241. doi: 10.1038/leu.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ross DM, et al. Patients with chronic myeloid leukemia who maintain a complete molecular response after stopping imatinib treatment have evidence of persistent leukemia by DNA PCR. Leukemia. 2010;24:1719–1724. doi: 10.1038/leu.2010.185. [DOI] [PubMed] [Google Scholar]
- 146.Busque L, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl. J. Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 148.Brown SJ, Stoilov P, Xing Y. Chromatin and epigenetic regulation of pre-mRNA processing. Hum. Mol. Genet. 2012;21:R90–R96. doi: 10.1093/hmg/dds353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 151.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Regulski M, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 155.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 156.Purdy MM, Holz-Schietinger C, Reich NO. Identification of a second DNA binding site in human DNA methyltransferase 3A by substrate inhibition and domain deletion. Arch. Biochem. Biophys. 2010;498:13–22. doi: 10.1016/j.abb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 157.Kotini AG, Mpakali A, Agalioti T. Dnmt3a1 upregulates transcription of distinct genes and targets chromosomal gene clusters for epigenetic silencing in mouse embryonic stem cells. Mol. Cell. Biol. 2011;31:1577–1592. doi: 10.1128/MCB.01093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ge YZ, et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 159.Li BZ, et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Traina F, et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS ONE. 2012;7:e43090. doi: 10.1371/journal.pone.0043090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lin J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE. 2011;6:e26906. doi: 10.1371/journal.pone.0026906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Itzykson R, Kosmider O, Fenaux P. Somatic mutations and epigenetic abnormalities in myelodysplastic syndromes. Best Pract. Res. Clin. Haematol. 2013;26:355–364. doi: 10.1016/j.beha.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 165.Wlodarski MW, et al. Abnormal promoter DNA methylation in juvenile myelomonocytic leukemia is not caused by mutation in DNMT3A. Blood. 2011;118:4490–4491. doi: 10.1182/blood-2011-07-370684. [DOI] [PubMed] [Google Scholar]
- 166.Kern W, et al. Mixed phenotype acute leukemia, T/myeloid, NOS (MPAL-TM) has a high DNMT3A mutation frequency and carries further genetic features of both AML and T-ALL: results of a comprehensive next-generation sequencing study analyzing 32 genes. Blood. 2012;120:403. [Google Scholar]
- 167.Rajavelu A, Jurkowska RZ, Fritz J, Jeltsch A. Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation. Nucleic Acids Res. 2012;40:569–580. doi: 10.1093/nar/gkr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.