Summary
Functional genomics has transformed from futuristic concept to well‐established scientific discipline during the last decade. Cotton functional genomics promise to enhance the understanding of fundamental plant biology to systematically exploit genetic resources for the improvement of cotton fibre quality and yield, as well as utilization of genetic information for germplasm improvement. However, determining the cotton gene functions is a much more challenging task, which has not progressed at a rapid pace. This article presents a comprehensive overview of the recent tools and resources available with the major advances in cotton functional genomics to develop elite cotton genotypes. This effort ultimately helps to filter a subset of genes that can be used to assemble a final list of candidate genes that could be employed in future novel cotton breeding programme. We argue that next stage of cotton functional genomics requires the draft genomes refinement, re‐sequencing broad diversity panels with the development of high‐throughput functional genomics tools and integrating multidisciplinary approaches in upcoming cotton improvement programmes.
Keywords: genome sequencing, cotton databases, gene discovery tools, CRISPR/Cas9, cotton improvement, biotic and abiotic stresses
Introduction
Cotton (Gossypium hirsutum) is a foundation of the global economy, prized for its important renewable fibre resource. It serves as an ideal plant for different biological studies such as genome evolution, polyploidization and single‐celled biological processes (Qin and Zhu, 2011; Shan et al., 2014). Decoding cotton's genome provides useful understanding about the agronomic and functional importance of polyploidy and genome size variations within the genus Gossypium (Chen et al., 2007). However, evolution and function of allopolyploid cotton genome is complicated by the presence of two subgenomes (A T and D T) in its nucleus. About 5–10 million years ago (MYA), the African‐derived ‘A’ diploid genome diverged from the eudicot progenitor simultaneously with the diploid genome ‘D’ which was native to Mexican (Wendel, 1989; Wendel and Albert, 1992). Then around 1–2 MYA, these two species were reunited together by the transoceanic dispersal of an A‐genome ancestor (Gossypium arboreum) to the New World and hybridized with a D‐genome ancestor (Gossypium raimondii) followed by chromosome doubling, which produced the allotetraploid cotton (Wendel, 1989). These well‐established relationships between the allotetraploid and diploid cotton genomes help us to explore the evolution of gene expression, because most of the gene functions are highly conserved between wild as well as diploid and tetraploid cotton species.
Whole‐genome sequencing is a fundamental component for comprehensive molecular analysis, and for several thousands of plant species, genome sequencing projects are now complete or underway. Compared with model plants, that is Arabidopsis, rice and maize, the whole‐genome sequencing of cotton was lacking behind. During the last decade, sequenced genomes of tetraploid cotton (Li et al., 2015; Liu et al., 2015; Yuan et al., 2015; Zhang et al., 2015b) and their diploid progenitors (Li et al., 2014b; Paterson et al., 2012; Wang et al., 2012) have been released that provide critical understanding of the evolution and differentiation of genome structures. Though, knowledge of the precise sequences and position of all the genes of an organism is an initial step to explore how biological systems work together. Previously, various studies have performed to compare the structural variations in genomes, which showed the differences in the expression pattern rather than in the absence and presence of genes (Gingle et al., 2006). In this respect, functional genomics is the main approach which is generally referred as ‘development and application of global experimental approaches to evaluate gene functions by using the information and reagents obtained from structural genomics’. It helps us to understand the basic plant biology and exploit the genomic information for cotton improvement, which is a vital step for manipulating cotton genes in agriculture. However, in cotton functional genomics, a persistent challenge is the absence of genetic and molecular tools partly due to large genome size, low transformation efficiency and long growth cycle. In this article, we provide an over‐review of the currently available tools and resources for cotton functional genomics with its recent advances for different important traits. Ultimately, this overview helps in assembling a final list of candidate genes that might be employed in future novel cotton breeding programme.
Tools and resources for cotton functional genomics
Cotton has become a system of choice for functional genomics studies. Here, we overview the available resources and tools for functional genomics studies in cotton and also discuss the ways (Figure 1) in which existing resources or tools can be used to further support large‐scale functional studies in cotton.
Figure 1.
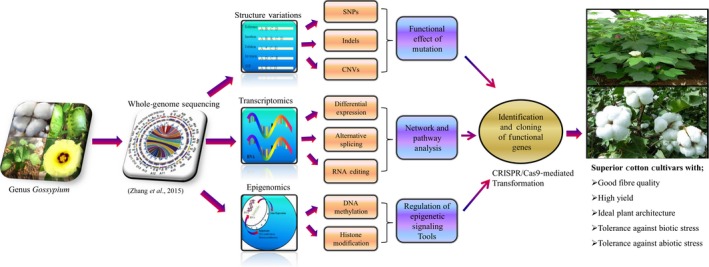
A scheme of the sequential research processes from whole‐genome sequencing to practical functional genomics in cotton. This figure shows the integrative approach of comprehensive information flows from the whole‐genome sequencing to practical functional genomics in cotton. It systematically represents the typical data evaluation path with bioinformatics tools in genomics, transcriptomics and epigenomics technologies to analyse the genomic mutations, differential gene expression and regulation of epigenetic signalling tools. It also incorporates protein expression data into appropriate genes and functional networks which ultimately facilitate the identification and cloning of functional genes. SNPs (single nucleotide polymorphisms), Indels (insertion/deletion) and CNVs (copy number variations)
Cotton genome sequencing: progress and implications
Efforts towards increased efficiency of functional genomics are demonstrated by the advancements that initiated from genome sequencing. In the last 20 years, rapid and impressive progress has been made in developing genetically modified cotton cultivars against resistance to insects and herbicides (Guo et al., 2015b; Yu et al., 2016). Conversely, slow advances have been made in genetic improvements of cotton for plant architecture, flowering, fibre quality, yield and resistance against biotic and abiotic stresses. The successful implementation and accessibility of well‐established whole‐genome sequences of Arabidopsis and rice has facilitated the consortium‐based cotton genome research. In 2007, the Cotton Genome Consortium (Chen et al., 2007) set a strategic plan to sequence cotton genomes that they primarily target less‐complicated diploid genomes that can be directly applied to the tetraploid cotton. For the persistent objective of cotton genome sequencing, the D‐genome species G. raimondii was prioritized for complete sequencing. A major initial source of cotton genome sequencing was released in 2012 by two independent studies that released the draft genome sequence of G. raimondii (Paterson et al., 2012; Wang et al., 2012), which was a rational step to characterize the larger ‘A’ diploid and ‘AD’ tetraploid cotton genomes.
Two years later, the same research group sequenced the 1694‐Mb genome of G. arboreum, which is a supposed donor species for the A chromosome group (Li et al., 2014b) in tetraploid cotton. However, the genomes of two existing progenitors (G. raimondii and G. arboreum) have been sequenced; the precise species that directs the development of the tetraploid cotton species about 1–2 MYA currently not exists (Wendel, 1989). Further, G. hirsutum revealed important variations in plant morphology and economic characteristics as compared to diploid cotton species, showing that precise natural and artificial selection has happened during evolution. Therefore, it was essential to sequence the allotetraploid species of cotton to understand the evolutionary history and gain insights into fibre biology. Using the genome sequences of A and D progenitor species, Li et al. (2015) and Zhang et al. (2015b) simultaneously but independently sequenced the genome of allotetraploid G. hirsutum. Besides G. hirsutum, Sea Island cotton (G. barbadense) is prized due to its superior quality and extra‐long fibre for the fabrication of high‐quality textiles. Considering its importance, genome of the G. barbadense was sequenced (Liu et al., 2015; Yuan et al., 2015), which covered 2470 and 2570 Mb of the genome, respectively.
At present, reference genome sequences for diploid and tetraploid cotton species are released by different groups, but the researchers assumed that some of these sequenced genomes contain assembly errors. For example, differences have been observed in the sequenced and assembled draft genomes of the G. raimondii (Paterson et al., 2012; Wang et al., 2012) and G. hirsutum (Li et al., 2015; Zhang et al., 2015b) by two independent groups in terms of chromosome length and their annotated genes (Figure 2a–c). These differences might be due to errors in their assemblies (Zhang et al., 2015b), at least at large scale, which in turn also affects the tremendous amount of genome analysis among different cotton species. Currently, we need to devote more effort in capturing these genome assemblies with a more sceptical eye for careful comparison, evaluation and fixing their misassemblies by developing quality control standards. In addition, re‐sequencing the genome for which there is a reference genome available permits exploration of the association between sequence variations. Recent comprehensive genome assessment by genome‐wide re‐sequencing of 34 (Page et al., 2016), 318 (Fang et al., 2017b), 147 (Fang et al., 2017a) and 352 (Wang et al., 2017b) cotton accessions represented extensive collections in order to identify genome regions that are signature of selection. These studies provide new genomic resources that significantly advance molecular breeding in cotton. Particularly, under the guidance of sequence information, the favourable genes that are linked with high yield, wide adaptation and fibre quality can be introgressed between different gene pools to further improve cotton production.
Figure 2.
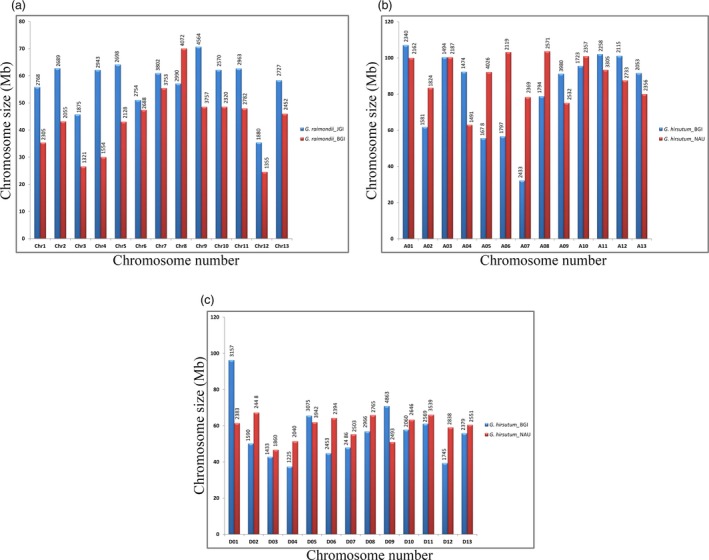
Chromosome size distribution (y‐axis) and number of annotated genes (above each bar) of different Gossypium species. Differences in chromosome size and number of annotated genes (above each bar) by two independent studies between the sequenced genomes of (a) G. raimondii (Paterson et al., 2012 (blue); Wang et al., 2012 (red)); (b) At‐subgenome of G. hirsutum (Li et al., 2015 (blue); Zhang et al., 2015b (red)), and (c) Dt‐subgenome of G. hirsutum (Li et al., 2015 (blue); Zhang et al., 2015b (red)). These differences might be due to errors in their assemblies, which in turn also affects the various genome analyses among different cotton species. Currently, we need to devote more efforts in capturing, evaluating and fixing their misassemblies by developing quality control standards.
Gene discovery tools recently available
Sequenced and re‐sequenced cotton genomes are simply the foundation; the main challenge is to discover the features of the genome that explain the biology. The next stage of cotton genomics will entirely expose these biologically active states of DNA, as has been made for other model crop plants where high‐density genetic and fine maps, SNP array platforms, epigenetic modifications and transcript abundance are studied across multiple species and tissues. Previous to the release of the draft genome sequences of four Gossypium species, limited ultra‐precision genetic maps were the main obstruction that prevented the intense genetic research and breeding of improved cotton cultivars. Currently, few comparatively dense linkage maps of cotton are available (Guo et al., 2008; John et al., 2012; Li et al., 2016; Wang et al., 2015), which provide a platform for high‐throughput marker development, gene mapping, gene isolation and cloning. Moreover, during the last decade, at least 1075 QTLs from 58 studies of intraspecific G. hirsutum and 1059 QTLs from interspecific populations of G. hirsutum × G. barbadense have been published for fibre quality, yield, seed quality and resistance against biotic and abiotic stresses (Said et al., 2015). However, these identified QTLs reside at large genomic regions that may contain several genes, provide only coarse resolution for marker‐assisted selection. Therefore, it is also crucial to fine map the genomic regions with large number of markers that will enhance the efficiency of selection, which ultimately helps to clone the genes present at the target loci. In cotton, fine mapping of few important genes and QTLs has been reported, that is fine mapping of glandless gene (Cheng et al., 2016b), leaf shape (Andres et al., 2014) and fibre quality‐related QTLs (Fang et al., 2017c; Liu et al., 2016a; Xu et al., 2017).
The rising efficacy of NGS technique and advanced in silico methods has permitted the development of single nucleotide polymorphisms (SNPs) at the whole‐genome level, even for the 2.5‐Gb genome of allotetraploid cotton. In cotton, SNP63K has been developed that contains assays for 45 104 and 17 954 putative intraspecific and interspecific SNP markers (Ashrafi et al., 2015). This initial effort for developing SNP63K array of cotton provides a standard high‐throughput genotyping tool and a base for the genetic analysis of economically and agronomically important traits. As a large proportion of the genome was affected by copy number variations (CNVs) rather than SNPs, they may help to explore several phenotypic variations that are not captured by SNPs. Many evidences support that CNVs are prevalent in plant genomes that can change gene structure, dosage and gene regulation (Mills et al., 2011), and mainly CNV‐affected genes are related to important traits. In cotton, 989 CNV‐affected genes have been identified (Fang et al., 2017b), which are related to cell wall organization, plant type and translational regulation.
Recently, transcriptome profiling has evolved into the most important tool demonstrating how information obtained from sequence data can be transformed into an extensive knowledge of gene function. In this regard, RNA‐Seq has revealed strong potential for whole‐genome transcriptome profiling as it allows the direct sequencing of transcripts by high‐throughput sequencing technologies. The recent transcriptome assembly of the G. hirsutum inbred line TM‐1 together with an assembly of all publicly available ESTs (expressed sequence tags) (Ashrafi et al., 2015) served as a reference for RNA‐Seq‐based SNP identification of cotton. Also, the application of the diploid and tetraploid genome sequence and NGS technology to practise RNA‐Seq analysis of large‐scale gene expression in cotton has been published by many reports. For instance, transcriptome analysis for leaf senescence (Lin et al., 2015), fibre development (Islam et al., 2016a; Naoumkina et al., 2015; Yoo and Wendel, 2014), biotic stress (Artico et al., 2014; Xu et al., 2011) and abiotic stress (Bowman et al., 2013; Zhang et al., 2016a) has been reported. However, RNA‐Seq technique faces some challenges such as library construction, development of efficient methods to store and process large amounts of data (Wang et al., 2009). It is anticipated that once these obstacles to the extensive use of RNA‐Seq are overcome, this technique will become the major tool for transcriptome analysis (Zhao et al., 2014).
Besides genetic factors, many traits in living things are controlled by other processes known as epigenetic modifications that resolve whether, when and how many genes are expressed. There are many epigenetic signalling tools that controlled gene expression, but most common is the DNA methylation (Phillips, 2008), which has appeared to play an important role in evolution and morphological diversity in crop plants (Cubas et al., 1999; Suzuki and Bird, 2008). In cotton, DNA methylation changes are related to seasonal variation in the development of fibres (Jin et al., 2013) and different tissues (Osabe et al., 2014). For example, dynamic role of methylation in ovule and fibre development showed that RdDM (RNA‐directed DNA methylation)‐dependent CHH methylation is related to gene activation in ovules, while CMT2 (chromomethylase2)‐dependent methylation guides to the silencing of genes in fibres (Song et al., 2015). Subsequently, 519 cotton genes are epigenetically modified between wild and domesticated cotton cultivars, some of which are related to agronomic and domesticated traits (Song et al., 2017). This finding gives insights into epigenetic regulation for the development of different traits and polyploid evolution of cotton. So, knowing how the methylome changed during evolution and domestication helps to bring this technology one step closer to reality.
Functional genomics databases for cotton
A comprehensive study of any genome depends on the availability of information regarding genome sequence, map position, mRNA and protein expression, metabolism and allelic variation. Hence, with the development of enormous omics data sets, it is important to have functional genomics database that permits users to easily obtain and visualize genomic information. Currently, many functional genomics databases are available for the cotton research community: the CottonGen (https://www.cottongen.org), Cotton Functional Genomic Database (CottonFGD; https://cottonfgd.org), Cotton Genome Resource Database (CGRD; http://cgrd.hzau.edu.cn/index.php), Cotton Genome Database (CottonDB; http://www.cottondb.org), Cotton Genome Project (CGP; http://cgp.genomics.org.cn/page/species/index.jsp), Platform of Functional Genomics Analysis in Gossypium raimondii (GraP; http://structuralbiology.cau.edu.cn/GraP/about.html), Comparative Evolutionary Genomics of Cotton (http://cottonevolution.info/), Join Genome Institute (JGI; http://jgi.doe.gov) and Database for Co‐expression Networks with Function Modules (ccNET; http://structuralbiology.cau.edu.cn/gossypium/).
CottonGen is the most important curated web‐based intellectual database, offering easy access to available genomic and genetic data of cotton. It contains annotated whole‐genome sequences of different cotton species, unigenes from ESTs, genetic maps, markers trait loci, genes and germplasm resources. Similarly, CottonFGD also provides a quick and easy access to genome sequences, functional annotations, transcriptome and genome re‐sequencing data for all of the sequenced Gossypium genomes. However, ccNET displays co‐expression networks and identified functional modules from diploid and polyploid cotton species including 1155 and 1884 modules in G. arboreum and G. hirsutum, respectively.
Potential of CRISPR/Cas9 in cotton gene editing
Clustered regularly interspaced short palindromic repeat (CRISPR)‐associated protein 9 (Cas9) from Streptococcus pyogenes (Sapranauskas et al., 2011) is a fast developing genome editing technology that has been effectively employed in many model plants (Belhaj et al., 2015). A distinctive feature of CRISPR/Cas9 is that DNA cleavage sites are recognized through Watson–Crick base pairing (Lowder et al., 2015) by three components: Cas9 protein, CRISPR‐RNA (crRNA) and trans‐activating crRNA (trancrRNA) (Karvelis et al., 2013; Mei et al., 2016). The utilization of the CRISPR/Cas9 system as a genome engineering tool came out when it was revealed that the target DNA sequence could be simply re‐programmed by altering 20 nucleotides in the CRISPR‐RNA (Jinek et al., 2012). Further, multiple gRNAs with diverse sequences could also be used to get multiplex genome engineering at various loci at the same time. This milestone study established that the CRISPR/Cas9 is a simple, efficient, economical and multipurpose tool for gene mutation, gene expression repression or activation and genome editing.
In plant biology, the first application of CRISPR/Cas9‐based genome editing (Li et al., 2013c; Shan et al., 2013) demonstrated its vast adaptability in the model species Arabidopsis as well as in crop plant rice. Subsequently, it has been applied in other crop plants, that is maize (Liang et al., 2014) and wheat (Wang et al., 2014b). In cotton, the application of CRISPR/Cas9 is still at its infancy. Most recently, multiple sites genome editing through CRISPR/Cas9 system in allotetraploid cotton by targeting arginase (GhARG), discosoma red fluorescent protein2 (DsRed2) and chloroplast development (GhCLA1) genes proves that it is highly reliable and effective for cotton genome editing (Wang et al., 2017c,2017d). It is expected that the potential and applications of CRISPR/Cas9 in cotton genome editing are certain to be further developed over time. In future, improvements will continue to increase its use from mutant generation to accurate gene regulation at noncoding enhancer regions in cotton.
Functional genomics for different traits
With the success of whole‐genome sequencing of cotton, its annotated genes were assigned some degree of functions by comparing them with the sequences of genes with known function and RNA‐Seq analysis. For example, the functional allocation of G. hirsutum genes (Zhang et al., 2015b) was shown by a Venn diagram. The RNA‐Seq data in fragments per kilobase of exon per million fragments mapped (FPKM) of each G. hirsutum gene were downloaded from CottonFGD. The venn diagram was constructed by online analysis tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The distributions of these genes (Figure 3) highlight that 52 854 differentially expressed genes were commonly identified during fibre, organ and ovule development and resistance against abiotic stress. Interestingly, there were several more differentially expressed genes during the stress resistance (1115) than during fibre and organ development, implying that the stress resistance is more complicated transcript regulation. However, knowing gene functions by comparative analysis and RNA‐Seq mainly does not give an insight into their specific role. In this regard, large‐scale functional genomics mainly helps in which all of the genes will be assigned functions on the basis of experimental verification.
Figure 3.
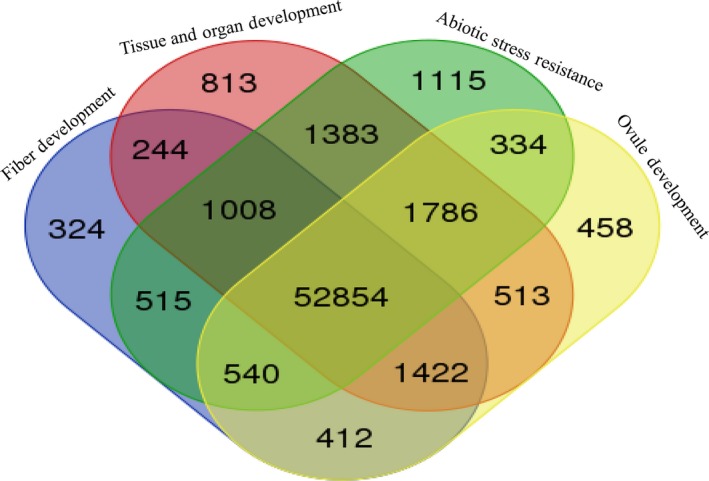
A venn diagram of the differentially expressed genes during fibre development, tissue and organ development, abiotic stress resistance and ovule development. The functional distribution of annotated genes from G. hirsutum (Zhang et al., 2015b) highlights that 52 854 differentially expressed genes were commonly identified during fibre, organ, tissue and ovule development and resistance against abiotic stress. However, more genes were differentially expressed during stress resistance than during fibre and organ development.
Fibre quality
Cotton fibres are single‐celled trichomes that originate from outer integument cells of the ovular surface. The cotton fibre undergoes a complex developmental programme that can be divided into four overlapping stages: fibre cell initiation, elongation, secondary cell wall biosynthesis and maturation (Manik and Ravikesavan, 2009; Wilkins and Jernstedt, 1999). Fibre initiation occurs around the time of anthesis during which about 30% of fibre primordia differentiate into mature fibres (Tiwari and Wilkins, 1995), while cell exhibits highly emphasized polarized expansion during fibre elongation (0–25 day postanthesis) (John and Keller, 1996). The secondary cell wall is primarily comprised of cellulose, which generally occurs between 24 and 27 day postanthesis (Wilkins and Jernstedt, 1999). The last stage of fibre development, maturation is related to the accumulation of minerals and a concurrent decrease in water potential (John and Keller, 1996). Although the above‐mentioned developmental stages are coinciding, each stage has its own specific features of physiological and cellular states. The genetic complexity of the cotton fibre transcriptome lies in the involvement of ~18 000 and 36 000 genes in diploid and allotetraploid cotton genomes (Arpat et al., 2004).
Currently, cotton fibre has become a trait of primary interest and several efforts have been made (Table 1) to identify fibre‐related genes and their functions to improve fibre quality such as E6 (John and Crow, 1992), GhExp1 (Harmer et al., 2002), GhSusA1 (Jiang et al., 2012), PIP2s (Li et al., 2013b) and GA20ox (Bai et al., 2014). Additionally, actin cytoskeleton (Li et al., 2005), polysaccharide biosynthesis, signal transduction and protein translocation (Sun et al., 2017)‐related genes are also preferentially expressed in various fibre developmental pathways. Among these cotton fibre genes, some are predominantly expressed during fibre initiation (Deng et al., 2012; Hu et al., 2016), some are highly expressed during secondary cell wall biosynthesis (Brill et al., 2011; Harmer et al., 2002), and some show high expression during fibre elongation (Shi et al., 2006; Yang et al., 2014). For instance, a cotton protodermal factor1 gene (GbPDF1) is preferentially expressed during fibre initiation by the HDZIP2ATATHB2 core cis element (Deng et al., 2012). However, alpha‐expansins (GhExp1) gene highly expressed in developing fibres encodes a cell wall protein and regulates cell wall loosening (Harmer et al., 2002). During fibre elongation, many genes related to osmosis regulation are highly expressed. Previously, Ruan et al. (2003) reported that antisense suppression of a sucrose synthase (SuSy) gene disturbed the fibre elongation, signifying the contribution of SuSy in osmosis regulation. In contrast, proline‐rich protein‐coding gene (GhPRP5) worked as a negative regulator during cotton fibre development (Xu et al., 2013). Cellulose synthesis is a principal event in fibre cells during the secondary cell wall biosynthesis. Previously, many efforts have been made to explore that how the cotton fibre regulates and supports the strong irreversible carbon sink characterized by secondary wall cellulose synthesis (Brill et al., 2011; Haigler et al., 2007). It has been shown that suppression of Sus gene expression affects the cellulose deposition (Ruan, 2007), emphasizing the importance of this enzyme in cellulose synthesis. Subsequently, Brill et al. (2011) identified and characterized a novel Sus isoform (SusC), which was up‐regulated during secondary wall cellulose synthesis in cotton fibre. Besides secondary wall cellulose synthesis, maturation stage of fibre development begins. During fibre maturation, the majority of the expressed genes belong to cellular respiration (Kim et al., 2013).
Table 1.
Functional genomics for fibre traits
| Functional study aspect | Specific fibre trait | Cotton species (cultivar) | References |
|---|---|---|---|
| Jasmonate ZIM‐domain protein‐encoding (GhJAZ2) gene | Fibre initiation | G. hirsutum (TM‐1, YZ1 & Xu142) | Hu et al. (2016) |
| Bulb biogenesis 1 (GhRBB1_A07) gene | Fibre quality | G. hirsutum | Islam et al. (2016b) |
| Receptor‐like kinase (RLK) gene | Fibre strength | G. hirsutum (MD52ne & MD90ne) | Islam et al. (2016c) |
| Phytohormone‐related (PHYA1) gene | Fibre length | G. hirsutum (Coker 312) | Abdurakhmonov et al. (2014) |
| Homeodomain‐leucine zipper (GhHOX3) gene | Fibre elongation | G. hirsutum (R15), G. arboreum (Qinyangxiaozi), G. herbaceum & G. raimondii | Shan et al. (2014) |
| Calcium sensor (GhCaM7) gene | Fibre elongation | G. hirsutum | Tang et al. (2014) |
| Brassinosteroid catabolism (PAG1) gene | Fibre elongation | G. hirsutum (CCRI24) | Yang et al. (2014) |
| LIM domain‐encoding (WLIM1a) gene | Fibre elongation and secondary wall synthesis | G. hirsutum (R15) | Han et al. (2013) |
| Annexins (AnnGh3) gene | Fibre initiation and elongation | G. hirsutum (Xuzhou 142, Emian 9,10 & Coker 312) | Li et al. (2013a) |
| Plasma membrane intrinsic protein 2s encoding (PIP2s) gene | Fibre elongation | G. hirsutum (Xuzhou 142, Emian 9 & Coker 312) | Li et al. (2013b) |
| Proline‐rich proteins (PRP5) gene | Fibre length | G. hirsutum (Coker 312) | Xu et al. (2013) |
| Protodermal factor1 (GbPDF1) gene | Fibre initiation | G. barbadense (3‐79) & G. hirsutum (Xu142, Xu142 fl & YZ1) | Deng et al. (2012) |
| TCP transcription factor (GbTCP) gene | Fibre elongation | G. barbadense (3‐79) & G. hirsutum (YZ1) | Hao et al. (2012) |
| Sucrose synthase (GhSusA1) gene | Fibre length and strength | G. hirsutum (TM‐1 & 7235) | Jiang et al. (2012) |
| Homeodomain‐leucine zipper (GhHD‐1) gene | Fibre initiation | G. hirsutum (Acala Maxxa) | Walford et al. (2012) |
| MADS box protein‐coding (GhMADS11) gene | Fibre elongation | G. hirsutum (Coker312 & Xuzhou 142) | Li et al. (2011) |
| RAD‐like (GbRL1) gene | Fibre initiation | G. barbadense (Pima‐90) | Zhang et al. (2011a) |
| Auxin biosynthesis (iaaM) gene | Fibre initiation | G. hirsutum (Jimian 14) | Zhang et al. (2011c) |
| Xyloglucan endotransglycosylase/hydrolase (GhXTH) gene | Fibre elongation | G. hirsutum (Coker 312) | Lee et al. (2010) |
| Gibberellin 20‐oxidase (GhGa20ox1‐3) gene | Fibre initiation and elongation | G. hirsutum (Jimian 14) | Xiao et al. (2010) |
| Peroxidase‐encoding (GhPOX1) gene | Fibre elongation | G. hirsutum (Xuzhou 142) | Mei et al. (2009) |
| Calcium‐dependent protein kinase (GhCPK1) gene | Fibre elongation | G. hirsutum (TM‐1) | Huang et al. (2008b) |
| Steroid 5a‐reductase (GhDET2) gene | Fibre initiation and elongation | G. hirsutum (Jimian 14) | Luo et al. (2007) |
| Ethylene biosynthesis (ACO) genes | Fibre elongation | G. hirsutum (Xuzhou 142) | Shi et al. (2006) |
| GhMyb25 and the homeodomain genes | Fibre initiation | G. hirsutum (Xu 142 or XZ 142) & six lintless lines | Wu et al. (2006) |
| WDT‐repeat (GhTTG1‐GhTTG4) genes | Fibre initiation | Different cotton species | Humphries et al. (2005) |
| Actin cytoskeleton (ACTIN) genes | Fibre elongation | G. hirsutum (Coker 312) | Li et al. (2005) |
Many genes encoding transcription factors, that are MYB, C2H2, bHLH, WRKY and HD‐ZIP families, were also expressed during cotton fibre development. Previously, various studies indicated that MYB‐related genes have high expression during fibre development in G. hirsutum (Machado et al., 2009; Pu et al., 2008). For example, expression studies of six MYB‐related genes in G. hirsutum indicated that GhMYB6 has high expression in cotton fibre (Loguercio et al., 1999), while R2R3 MYB‐like transcription factor‐encoding gene ‘GhMYB109’ is expressed particularly in fibre initiation and elongation (Suo et al., 2003). The RAD‐like GbRL1 was also highly expressed in cotton ovules during fibre initiation (Zhang et al., 2011a). TCP transcription factor has played a significant role in fibre and root hair development by controlling the jasmonic acid biosynthesis, ethylene signalling, calcium channel and reactive oxygen species (Hao et al., 2012). Though, GhHOX3 from class IV homeodomain‐leucine zipper (HD‐ZIP) family showed strong expression during early fibre elongation (Shan et al., 2014). Besides transcription factors, phytohormones such as ethylene, auxins and brassinosteroids (BR) also play a critical role during fibre development. Ethylene plays a vital function in fibre elongation by stimulating the pectin biosynthesis network (Qin and Zhu, 2011), while gibberellins (GA) and indole‐3‐acetic acid (IAA) are required for fibre initiation and elongation in cotton (Xiao et al., 2010; Zhang et al., 2011c). In contrast, the persistent high concentration of jasmonic acid (JA) inhibits fibre elongation (Tan et al., 2012).
Although several gene expression studies have been reported on cotton fibre development, some issues are illustrated here. First, most of the differentially expressed genes identified by the comparative analysis are associated with variations between species rather than related to fibre traits. Second, in some cases, the use of the protein‐coding gene sequences from G. raimondii and G. arboreum may not be accurate enough for gene annotation in tetraploid cotton. Third, it is unknown whether any of the expressed genes recognized from earlier reports had sequence variations between a cotton fibre mutant and its wild type, because only the differentially expressed genes having sequence differences and colocalization with target fibre traits are possible candidates for advanced cotton studies.
Plant architecture and flowering
The productivity of the cotton plant is mainly affected by the plant architectural traits such as the shape, position of branches and distribution of reproductive structures (Wang et al., 2006; Ye and Zhu, 2001). Flowering and terminal loci such as single flower truss (SFT) and self‐pruning (SP) genes regulate the balance between monopodial and sympodial growth habits in woody perennial plants (Lifschitz et al., 2006; McGarry et al., 2016; Shalit et al., 2009). In cotton, GhSP gene is required to maintain both monopodial and sympodial branches and is also vital to ascertain cambial activity (McGarry et al., 2016). However, GhSFT stimulates the quick onset of sympodial branching and flowering inside the shoots of day neutral and wild photoperiodic accessions (McGarry et al., 2016). The floricaula/leafy homologs of cotton also play an important role in the flower initiation, LFY (GhLFY) gene from G. hirsutum was expressed in the shoot apex (Li et al., 2013d) with extensive up‐regulation at the third stage of true leaf expansion, and it might function downstream of MADS box GhSOC1 gene.
The time of floral initiation is one of the most important factors related to early maturation of cotton. Many genes have been differentially expressed during floral initiation (Table 2), including those encoding the B3, MADS and MYB domain transcription factors (Wu et al., 2015). MADS box genes are an important class of transcription factors in plants, involved in various cellular processes particularly in floral developmental processes, that is GhMADS3 (Guo et al., 2007b) and GhMADS9 (Shao et al., 2010). Despite these efforts, little is known about the mechanism underlying plant architecture and floral development in cotton. Nevertheless, it is expected that recent advances in cotton genome sequencing and transformation techniques will increase applications of various molecular biology approaches in cotton, which may help to explore the role of different genes during plant architecture and floral development.
Table 2.
Functional genomics for plant architecture and flowering
| Functional study aspect | Specific trait | Cotton species (cultivar) | References |
|---|---|---|---|
| Late meristem identity1‐D1b (GhLMI1‐D1b) gene | Leaf shape | G. hirsutum | Andres et al. (2017) |
| Single flower truss (GhSFT) and self‐pruning (GhSP) genes | Monopodial and sympodial branches | G. hirsutum (TX701 & DP61) | McGarry et al. (2016) |
| MADs box (GhSOC1 and GhMADS42) genes | Flowering | G. hirsutum (CCRI36) | Zhang et al. (2016b) |
| Flowering‐promoting factor 1 (GhFPF1) gene | Flowering time control and shade avoidance | G. hirsutum (TM1 & CCRI36) | Wang et al. (2014a) |
| Leafy (GhLFY) gene | Shoot apex | G. hirsutum (CCRI36) | Li et al. (2013d) |
| Florigen‐encoded flowering locus T (FT) gene | Determinate growth | G. hirsutum (TX701 & DP61) | McGarry et al. (2013) |
| Sepallata (GhSEP) gene | Squares or flowers | G. hirsutum | Lai et al. (2011) |
| Mitogen‐activated protein kinase (GhMAPK7) gene | Plant growth and development | G. hirsutum (Lumian 22) | Shi et al. (2010) |
| MADS box (GhMADS9) gene | Anther/pollen development | G. hirsutum (Coker312) | Shao et al. (2010) |
| MADS box (GhMADS3) gene | Stamens and carpels | G. hirsutum (Xuzhou 142 & Chuanmian 239) | Guo et al. (2007b) |
Abiotic stresses
Cotton's production is limited by various abiotic stresses, which cause about 73% yield loss worldwide (Saranga et al., 2009). Among different abiotic stresses, drought and salinity are the two main factors that affect the cotton production and it has become a challenging task to improve tolerance in cotton against these stresses. Previously, few stress‐related genes such as GhCIPK6 (He et al., 2013), GbRLK (Zhao et al., 2013), GhMKK1 (Lu et al., 2013) and GhSnRK2 (Bello et al., 2014) have been reported in cotton (Table 3). A cotton Raf‐like MAP3K (GhMAP3K40) gene positively regulates defence response but mediates reduced tolerance to biotic and abiotic stresses in transgenic Nicotiana benthamiana (Chen et al., 2015). However, the overexpression of annexin‐encoding (GhAnn1) gene showed higher chlorophyll content, increased peroxidase activities and lower lipid peroxidation levels, which ultimately increase the salt and drought stress tolerance in transgenic cotton (Zhang et al., 2015a). Previous studies have also reported that CBL‐interacting protein kinase (GhCIPK6) and sucrose nonfermenting 1‐related protein kinase 2 (SnRK2) genes are also involved in abiotic stress tolerance in cotton (Bello et al., 2014; He et al., 2013). Genes related to ethylene, abscisic acid and jasmonic acid signalling pathways have also played a significant role in drought tolerance (Chen et al., 2013). Further, 1528 and 1128 leaf‐ and root‐related genes with 28 biological pathways have been identified in response to water‐deficient conditions (Ranjan and Sawant, 2015), which signifies that leaves are distinct from roots for molecular mechanisms of drought tolerance in cotton.
Table 3.
Functional genomics for abiotic stress
| Functional study aspect | Abiotic stress | Cotton species (cultivar) | References |
|---|---|---|---|
| ERF‐encoding (GhERF38) gene | Salinity, drought and abscisic acid | G. hirsutum (Coker 312) | Ma et al. (2017) |
| bZIP‐encoding (GhABF2) gene | Drought and salinity | G. hirsutum (Simian 3) | Liang et al. (2016) |
| WRKY transcription factor‐encoding (GhWRKY25) gene | Drought and salinity | G. hirsutum (Lumian 22) | Liu et al. (2016b) |
| Trehalose‐6‐phosphate synthase (GhTPS11) gene | Heat, drought, salinity, gibberellin and abscisic acid | G. hirsutum (ZM19) | Wang et al. (2016a) |
| NAC domain‐encoding (GbNAC1) gene | Abscisic acid, mannitol and NaCl | G. barbadense (Xinhai 15 & Xinhai 16) | Wang et al. (2016b) |
| Mitogen‐activated protein kinase (GhMAP3K40) gene | Drought and salinity | G. hirsutum (Lumian 22) | Chen et al. (2015) |
| WRKY transcription factor‐encoding (GhWRKY41) gene | Drought and salinity | G. hirsutum (Lumian 22) | Chu et al. (2015) |
| Annexin gene (GhAnn1) | Salinity | G. hirsutum (7235) | Zhang et al. (2015a) |
| Sucrose nonfermenting 1‐related protein kinase 2 (GhSnRK2) gene | Drought, cold, abscisic acid and salinity | G. hirsutum (CCRI24) | Bello et al. (2014) |
| Mitogen‐activated protein kinase (GbMPK3) gene | Drought | G. barbadense (7124) | Long et al. (2014) |
| WRKY transcription factor (GhWRKY39‐1) gene | Salinity | G. hirsutum (Lumian 22) | Shi et al. (2014a) |
| WRKY transcription factor‐encoding (GhWRKY39) gene | Salinity | G. hirsutum (Lumian 22) | Shi et al. (2014b) |
| CBL‐interacting protein kinase (GhCIPK6) gene | Salinity, drought and abscisic acid | G. hirsutum (YZ‐1) | He et al. (2013) |
| NAC domain protein (GhNAC7‐GhNAC13) genes | Cold, abscisic acid, drought and salinity | G. hirsutum (Coker 312) | Huang et al. (2013) |
| Mitogen‐activated protein kinase (GhMPK6a) gene | Salinity and drought | G. hirsutum (Lumian 22) | Li et al. (2013e) |
| Mitogen‐activated protein kinase kinases (GhMKK1) gene | Salinity and drought | G. hirsutum (Lumian 22) | Lu et al. (2013) |
| Receptor‐like kinase (GbRLK) gene | Salinity and drought | G. barbadense (Hai 7124) | Zhao et al. (2013) |
| Mitogen‐activated protein kinase (GhMKK5) gene | Salinity and drought | G. hirsutum (Lumian 22) | Zhang et al. (2012) |
| Mitogen‐activated protein kinase (GhMPK16) gene | Drought | G. hirsutum (Lumian 22) | Shi et al. (2011) |
| Mitogen‐activated protein kinase (GhMPK2) gene | Salinity and drought | G. hirsutum | Zhang et al. (2011b) |
| Ethylene responsive (GhERF2, GhERF3, GhERF6) genes | Ethylene, abscisic acid, salt, cold and drought | G. hirsutum (Zhongmian 12) | Jin et al. (2010) |
| DRE‐binding transcription factor (GhDREB) gene | Drought, salinity and cold | Cotton (Simian 3) | Gao et al. (2009) |
| CCCH‐type zinc finger protein‐encoding (GhZFP1) gene | Salinity | G. hirsutum (ZMS19) | Guo et al. (2009) |
| CBF/DREB1‐encoding (GhDREB1) gene | Freezing, salinity and osmotic | G. hirsutum | Huang et al. (2009) |
| NAC transcription factor (GhNAC1‐GhNAC6) genes | Drought, salinity, cold and abscisic acid | G. hirsutum (Jinmian 19) | Meng et al. (2009) |
| DRE‐binding protein‐encoding (GhDBP2) gene | Drought, low temperature and abscisic acid | G. hirsutum (Zhongmian 12) | Huang et al. (2008a) |
| Ethylene response factors (GhERF1) gene | Ethylene, abscisic acid, salinity, cold and drought | G. hirsutum (Zhongmian 12) | Qiao et al. (2008) |
| DREB1/CBF‐like (GhDREB1L) gene | Low temperature, drought and salinity | G. hirsutum (Zhongmian 35) | Huang et al. (2007) |
Comparative analysis of genome‐wide expression profile reveals that different genes, transcription factors and physiological processes work together to induce stress tolerance (Ranjan et al., 2012). Transcription factors could be used as candidate genes to increase stress tolerance in cotton as they act in response to stress signals by regulating the expression of various downstream genes involved in response to high salt, drought and cold stresses (Guo et al., 2015a). The WRKY is one of the largest families of transcription factors in plants that bind to particular DNA sequences to repress or activate the transcription of various genes (Dou et al., 2014). To date, various WRKY‐based studies have been conducted in cotton against abiotic stresses (Shi et al., 2014b; Zhou et al., 2014). Additionally, NAC is also an important class of the transcription factors and its proteins are distinguished by a highly conserved N‐terminal (DNA‐binding) and highly divergent C‐terminal regions (Puranik et al., 2011), which is valuable for the diversity in the transcriptional activities. In cotton, putative NAC genes ‘GhNAC1–GhNAC6’ (Meng et al., 2009) and ‘GhNAC7–GhNAC13’ (Huang et al., 2013) have been highly expressed in leaves and roots and distinctively regulate under high salt, drought, cold and ABA conditions. The basic region leucine zipper (bZIP) and ethylene response factors (ERF) are among the largest and most diverse transcription factor families involved in stress tolerance in many plant species. However, in cotton, few members of these families, that are GhABF2 (Liang et al., 2016), GhERF2, GhERF3, GhERF6 (Jin et al., 2010) and GhERF38 (Ma et al., 2017), have been characterized for stress tolerance (Abid et al., 2017). Evidence from transgenic plants has demonstrated that C‐repeat/dehydration‐responsive element binding factor (GhDREB1) gene could function as positive regulators to enhance abiotic stress tolerance in cotton (Huang et al., 2009).
The development of stress tolerance cotton cultivars has become more feasible in recent years; though, it is still a difficult task that needs extensive interdisciplinary research efforts. Wild Gossypium species offer genetic diversity related to stress tolerance (Saranga et al., 2009), which can be employed in future cotton improvement programmes. More recently, different genomic tools have also become available to identify underlying genes and pathways during stress tolerance and to transfer them into different cotton cultivars.
Biotic stresses
Globally, biotic stresses such as insects, weeds and diseases occur with different levels of intensity, which may not be relevant in a particular year but they generally reduce plant yield in most years (Fritsche‐Neto and Borem, 2012). Among the different biotic factors, cotton breeding against disease resistance remains the primary objective. The shortage of resistant cotton germplasms makes Verticillium wilt the most serious disease to influence cotton production (Chang et al., 2008). The molecular mechanisms of resistance to V. dahliae reported that cotton phenylpropanoid pathway (Xu et al., 2011), terpenoid pathway (Luo et al., 2001), salicylic acid, reactive oxygen species and jasmonic acid signalling pathways (Xu et al., 2014) are important contributors to the pathogen response (Table 4). Plant mitogen‐activated protein kinase (MAPK) cascades have also been shown to regulate a number of stress responses. In cotton, GhMPK16 from D‐MAPK group has been characterized, which is involved in disease resistance (Shi et al., 2011). Additionally, transgenic cotton plants expressing the synthetic antimicrobial peptide (D4E1) gene showed the significant resistance to disease and mycotoxin causing fungal pathogens (Rajasekaran et al., 2005). Many WRKY proteins have also played a regulatory function in response to different pathogen infections either by regulating itself or due to their proximity to well‐characterized genes that play a central role in cotton defence (Zhou et al., 2014). Cotton leaf curl virus (CLCuV) is also one of the important rising threats to cotton production in different countries. It has been reported that resistance against CLCuV is conferred by two dominant and one suppressor gene (Rahman et al., 2005). In other studies, antisense coat protein gene (AV1) and truncated AC1 gene were targeted for restricting viral replication and movement in transgenic cotton (Amudha et al., 2013; Hashmi et al., 2011).
Table 4.
Functional genomics for biotic stress
| Functional study aspect | Biotic stress | Cotton species (cultivar) | References |
|---|---|---|---|
| Jasmonate ZIM‐domain (GhJAZ2) gene | Verticillium dahliae | G. hirsutum | He et al. (2017) |
| GR79‐EPSPS and N‐acetyltransferase (GAT) genes | Resistant to glyphosate | G. hirsutum (R18) | Liang et al. (2017) |
| Jasmonate ZIM‐domain interactor (NINJA) gene | Verticillium dahliae | G. hirsutum (BD18) | Wang et al. (2017a) |
| Ve homologous (Gbvdr3) gene | Verticillium dahliae | G. barbadense (7124) | Chen et al. (2016) |
| MYB transcription factor (GhMYB108) gene | Verticillium dahliae | G. hirsutum (BD18) | Cheng et al. (2016a) |
| Tectaria macrodonta (Tma12) gene | Cotton leaf curl virus and whitefly | G. hirsutum (Coker 312) | Shukla et al. (2016) |
| NAC transcription factor (GbNAC1) gene | Verticillium dahliae | G. barbadense (Xinhai 15 & Xinhai 16) | Wang et al. (2016c) |
| WRKY transcription factor (GbWRKY1) gene | Botrytis cinerea and V. dahliae | G. barbadense (7124) & G. hirsutum (YZ1) | Li et al. (2014a) |
| WRKY transcription factor (GhWRKY39‐1) gene | R. solanacearum and R. solani | G. hirsutum (Lumian 22) | Shi et al. (2014a) |
| Mitogen‐activated protein kinase (GhMPK6a) gene | Ralstonia solanacearum | G. hirsutum (Lumian 22) | Li et al. (2013e) |
| WRKY transcription factor (GhWRKY15) gene | Viral and fungal pathogens | G. hirsutum (Lumian 22) | Yu et al. (2012) |
| Mitogen‐activated protein kinase (GhMKK5) gene | Ralstonia solanacearum | G. hirsutum (Lumian 22) | Zhang et al. (2012) |
| Disease resistance (GhNDR1) and MAP kinase kinase 2 (GhMKK2) genes | Verticillium dahliae | G. hirsutum (Deltapine 90, R135, Phytogen 480WR, Phytogen 425RF, FM 832, PSC 355 & FM 9160B2F) | Gao et al. (2011) |
| WRKY transcription factor (GhWRKY3) gene | R. solani, Colletotrichum gossypii and F. oxysporeum | G. hirsutum | Guo et al. (2011) |
| Mitogen‐activated protein kinase (GhMPK16) gene | X. campestris pv. malvacearum, R. solani and C. gossypii | G. hirsutum (Lumian 22) | Shi et al. (2011) |
| Lignin‐related genes | Verticillium dahliae | G. barbadense (7124) & G. hirsutum (YZ‐1) | Xu et al. (2011) |
| Mitogen‐activated protein kinase (GhMPK7) gene | R. solani, C. gossypii and F. oxysporum f. sp. vasinfectum | G. hirsutum (Lumian 22) | Shi et al. (2010) |
| CCCH‐type zinc finger protein (GhZFP1) gene | R. solani | G. hirsutum (ZMS19) | Guo et al. (2009) |
| CP4 5‐enolpyruvylshikimate‐3‐phosphate synthase (CP4 EPSPS) gene | Resistant to glyphosate | G. hirsutum (Coker 312 & Coker130) | Chen et al. (2006) |
| Nonsymbiotic haemoglobin (GhHb1) gene | Verticillium dahliae | G. hirsutum (BD18) | Qu et al. (2005) |
| Synthetic antimicrobial peptide (D4E1) gene | F. verticillioides, V. dahlia, A. flavus and T. basicola | G. hirsutum (Coker 312) | Rajasekaran et al. (2005) |
| (+)‐δ‐Cadinene synthase (cdn1‐C4) gene | Bacterial blight | G. hirsutum | Townsend et al. (2005) |
Insect herbivores and cotton plant have waged war from millions of years. In cotton plant, transgenic technology has been mainly used to induce the resistance against insect herbivores. Among the different transgenic approaches against insects, Cry gene encoding Bacillus thuringiensis toxin has gained a fabulous success against bollworms (Guo et al., 2007a; Rashid et al., 2008). Recently, tma‐12 gene encoding insecticidal protein has been identified that gives substantial results against whitefly and cotton leaf curl viral disease (Shukla et al., 2016). Many secondary metabolites in cotton such as gossypol and related sesquiterpene aldehydes form phytoalexin chemicals that facilitate it to escape from herbivores. For example, expression of a P450 monooxygenase gene (CYP6AE14) is correlated with larval growth and its expression was induced by gossypol (Mao et al., 2007, 2011, 2013). Besides insect, cotton yield has also been largely affected by weeds throughout the growing season, which is generally managed by the application of several classes of herbicides. One such herbicide is the glyphosate which has become the most valuable herbicide due to its low cost and broad‐spectrum weed control. Initially, few genetically modified herbicide‐tolerant cotton lines have been developed by transferring gene encoding the 5‐enolpyruvylshikimate‐3‐phosphate synthase isolated from Agrobacterium sp. CP4 (CP4 EPSPS) (Nida et al., 1996). Lately, it was reported that CP4 EPSPS gene has played an important role in vegetative tolerance to glyphosate; however, its expression is critical for the development of male reproductive organ in response to high glyphosate application during late developmental stage (Chen et al., 2006). Additionally, highly glyphosate‐resistant cotton plants have also been developed by pyramiding the glyphosate resistance and detoxification genes (Liang et al., 2017), which presents attractive promise for developing highly herbicide‐resistant cotton cultivars.
Future perspectives
For determining the entire set of genes with their functions, genome sequencing of an organism is an important prerequisite resource. At present, the sequenced and re‐sequenced genomes of diploid and allotetraploid (Fang et al., 2017a,2017b; Li et al., 2014b, 2015; Liu et al., 2015; Paterson et al., 2012; Wang et al., 2012; Yuan et al., 2015; Zhang et al., 2015b) cotton have been available, which presents valuable information for cotton genomes. However, large knowledge gaps still persist as compared to Arabidopsis and rice, concerning with the molecular regulation of the fundamental biological processes. Due to which, characterization and cloning of more essential genes controlling complex traits is a major challenge for current and future cotton functional genomics studies. Currently, there is a dire need to further analyse multiple cotton cultivars which will improve the depth and pave a better way that will lead to more optimized marker applications and automated genotyping platforms for CNV determination (Rasheed et al., 2017).
Additionally, the development of a well‐organized system for molecular breeding by various functional components is necessary. Previously, few efforts have been made to develop a multiscale crop system for high‐throughput association studies of composite traits, that is the ePlant model (Zhu et al., 2013). Moreover, a revolution is underway in cotton functional genomics which is spearheaded by the CRISPR/Cas9 system due to its several valuable features. Also, there is a need to understand the composite connections among genes related to different cotton traits under control as well as diverse environmental conditions which ultimately boost our capability to adapt cotton plants appropriate for improvement in various traits.
Harnessing the full potential of functional genomics requires a multidisciplinary approach and integrated knowledge of the molecular and other biological processes underlying different traits because gene functions cannot be inferred by only one approach. The addition of information obtained from genomics, transcriptomics, proteomics and epigenomics studies of cotton will help us to critically explore and investigate the different regulatory pathways underlying different traits. Also, new user‐friendly bioinformatics tools and software with better resolving power and technological improvements need to be developed to increase the potential offered by functional genomics. The resulting huge amount of data from different high‐throughput techniques should, in turn, be further organized, stored and interconnected into fundamental timely updated databases in order to let easy extraction and comparison that will increase the understanding and opportunities for future functional genomics advancements in cotton.
Conclusion
The strong background of cotton genetics and the great efforts of the cotton genome consortium led to the start of cotton genome sequencing in 2007. With the wealth of cotton genome sequence information, cotton genomics research has entered the phase of fast functional characterization of all genes. However, despite great efforts in whole‐genome sequencing and re‐sequencing of cotton, large knowledge gaps still persist as compared to model plants Arabidopsis and rice. Therefore, next stage of cotton genomics requires draft genome refinement, re‐sequencing broad diversity panels and diverse wild relatives to better understand its genome. However, for taking the full benefits of the available genomic information on cotton genes, only the multidisciplinary integrated approach allows their functional characterization. So, advances in functional genomics of cotton will depend on developing high‐throughput technologies and integrating multidisciplinary approaches including genomics, transcriptomics, proteomics, epigenomics and bioinformatics in upcoming cotton improvement programmes.
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this manuscript.
Author contributions
JA and GS conceived and designed the experiments; all authors performed data analysis and interpretation; JA, WM, MAA and GS wrote the manuscript.
References
- Abdurakhmonov, I.Y. , Buriev, Z.T. , Saha, S. , Jenkins, J.N. , Abdukarimov, A. and Pepper, A.E. (2014) Phytochrome RNAi enhances major fibre quality and agronomic traits of the cotton Gossypium hirsutum L. Nat. Commun. 5, 3062. [DOI] [PubMed] [Google Scholar]
- Abid, M.A. , Liang, C. , Malik, W. , Meng, Z. , Tao, Z. , Ashraf, J. , Guo, S. et al (2017) Cascades of ionic and molecular networks involved in expression of genes underpin salinity tolerance in cotton. J. Plant Growth Regul. https://doi.org/org/10.1007/s00344-017-9744-0. [Google Scholar]
- Amudha, J. , Balasubramani, G. , Malathi, V. , Monga, D. and Kranthi, K. (2013) Cotton leaf curl virus resistance transgenics with antisense coat protein gene (AV1). Curr. Sci. 104, 1542. [Google Scholar]
- Andres, R.J. , Bowman, D.T. , Kaur, B. and Kuraparthy, V. (2014) Mapping and genomic targeting of the major leaf shape gene (L) in Upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 127, 167–177. [DOI] [PubMed] [Google Scholar]
- Andres, R.J. , Coneva, V. , Frank, M.H. , Tuttle, J.R. , Samayoa, L.F. , Han, S.W. , Kaur, B. et al (2017) Modifications to a late meristem identity1 gene are responsible for the major leaf shapes of upland cotton (Gossypium hirsutum L.). Proc. Natl Acad. Sci. 114, E57–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat, A. , Waugh, M. , Sullivan, J.P. , Gonzales, M. , Frisch, D. , Main, D. , Wood, T. et al (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol. Biol. 54, 911–929. [DOI] [PubMed] [Google Scholar]
- Artico, S. , Ribeiro‐Alves, M. , Oliveira‐Neto, O.B. , de Macedo, L.L.P. , Silveira, S. , Grossi‐de‐Sa, M.F. , Martinelli, A.P. et al (2014) Transcriptome analysis of Gossypium hirsutum flower buds infested by cotton boll weevil (Anthonomus grandis) larvae. BMC Genom. 15, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi, H. , Hulse‐Kemp, A.M. , Wang, F. , Yang, S.S. , Guan, X. , Jones, D.C. , Matvienko, M. et al (2015) A long‐read transcriptome assembly of cotton (L.) and intraspecific single nucleotide polymorphism discovery. Plant Genome, 8, 1–14. [DOI] [PubMed] [Google Scholar]
- Bai, W.Q. , Xiao, Y.H. , Zhao, J. , Song, S.Q. , Hu, L. , Zeng, J.Y. , Li, X.B. et al (2014) Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS ONE, 9, e96537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj, K. , Chaparro‐Garcia, A. , Kamoun, S. , Patron, N.J. and Nekrasov, V. (2015) Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. [DOI] [PubMed] [Google Scholar]
- Bello, B. , Zhang, X. , Liu, C. , Yang, Z. , Yang, Z. , Wang, Q. , Zhao, G. et al (2014) Cloning of Gossypium hirsutum sucrose non‐fermenting 1‐related protein kinase 2 gene (GhSnRK2) and its overexpression in transgenic Arabidopsis escalates drought and low temperature tolerance. PLoS ONE, 9, e112269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, M.J. , Park, W. , Bauer, P.J. , Udall, J.A. , Page, J.T. , Raney, J. , Scheffler, B.E. et al (2013) RNA‐Seq transcriptome profiling of upland cotton (Gossypium hirsutum L.) root tissue under water‐deficit stress. PLoS ONE, 8, e82634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill, E. , van Thournout, M. , White, R.G. , Llewellyn, D. , Campbell, P.M. , Engelen, S. , Ruan, Y.L. et al (2011) A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol. 157, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. , Guo, W.Z. , Li, G.Y. , Gao, F. , Lin, S.S. and Zhang, T.Z. (2008) QTLs mapping for verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci. 174, 290–298. [Google Scholar]
- Chen, Y.C.S. , Hubmeier, C. , Tran, M. , Martens, A. , Cerny, R.E. , Sammons, R.D. and CaJacob, C. (2006) Expression of CP4 EPSPS in microspores and tapetum cells of cotton (Gossypium hirsutum) is critical for male reproductive development in response to late‐stage glyphosate applications. Plant Biotechnol. J. 4, 477–487. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J. , Scheffler, B.E. , Dennis, E. , Triplett, B.A. , Zhang, T. , Guo, W. , Chen, X. et al (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Liu, Z.H. , Feng, L. , Zheng, Y. , Li, D.D. and Li, X.B. (2013) Genome‐wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS ONE, 8, e80879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Wang, J. , Zhu, M. , Jia, H. , Liu, D. , Hao, L. and Guo, X. (2015) A cotton Raf‐like MAP3K gene, GhMAP3K40, mediates reduced tolerance to biotic and abiotic stress in Nicotiana benthamiana by negatively regulating growth and development. Plant Sci. 240, 10–24. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Kan, J. , Yang, Y. , Ling, X. , Chang, Y. and Zhang, B. (2016) A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae . Plant Physiol. Biochem. 98, 101–111. [DOI] [PubMed] [Google Scholar]
- Cheng, H.Q. , Han, L.B. , Yang, C.L. , Wu, X.M. , Zhong, N.Q. , Wu, J.H. , Wang, F.X. et al (2016a) The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67, 1935–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. , Lu, C. , Yu, J.Z. , Zou, C. , Zhang, Y. , Wang, Q. , Huang, J. et al (2016b) Fine mapping and candidate gene analysis of the dominant glandless gene Gl2 e in cotton (Gossypium spp.). Theor. Appl. Genet. 129, 1347–1355. [DOI] [PubMed] [Google Scholar]
- Chu, X. , Wang, C. , Chen, X. , Lu, W. , Li, H. , Wang, X. , Hao, L. et al (2015) The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana . PLoS ONE, 10, e0143022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P. , Vincent, C. and Coen, E. (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature, 401, 157–161. [DOI] [PubMed] [Google Scholar]
- Deng, F. , Tu, L. , Tan, J. , Li, Y. , Nie, Y. and Zhang, X. (2012) GbPDF1 is involved in cotton fiber initiation via the core cis‐element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, L. , Zhang, X. , Pang, C. , Song, M. , Wei, H. , Fan, S. and Yu, S. (2014) Genome‐wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics, 289, 1103–1121. [DOI] [PubMed] [Google Scholar]
- Fang, L. , Gong, H. , Hu, Y. , Liu, C. , Zhou, B. , Huang, T. , Wang, Y. et al (2017a) Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 18, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L. , Wang, Q. , Hu, Y. , Jia, Y. , Chen, J. , Liu, B. , Zhang, Z. et al (2017b) Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 49, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Fang, X. , Liu, X. , Wang, X. , Wang, W. , Liu, D. , Zhang, J. , Liu, D. et al (2017c) Fine‐mapping qFS07. 1 controlling fiber strength in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 130, 795–806. [DOI] [PubMed] [Google Scholar]
- Fritsche‐Neto, R. and Borem, A. (2012) Plant Breeding for Biotic Stress Resistance. New York: Springer. [Google Scholar]
- Gao, S.Q. , Chen, M. , Xia, L.Q. , Xiu, H.J. , Xu, Z.S. , Li, L.C. , Zhao, C.P. et al (2009) A cotton (Gossypium hirsutum) DRE‐binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Wheeler, T. , Li, Z. , Kenerley, C.M. , He, P. and Shan, L. (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingle, A.R. , Yang, H. , Chee, P.W. , May, O.L. , Rong, J. , Bowman, D.T. , Lubbers, E.L. et al (2006) An integrated web resource for cotton. Crop Sci. 46, 1998–2007. [Google Scholar]
- Guo, X. , Huang, C. , Jin, S. , Liang, S. , Nie, Y. and Zhang, X. (2007a) Agrobacterium‐mediated transformation of Cry1C, Cry2A and Cry9C genes into Gossypium hirsutum and plant regeneration. Biol. Plant. 51, 242–248. [Google Scholar]
- Guo, Y. , Zhu, Q. , Zheng, S. and Li, M. (2007b) Cloning of a MADS box gene (GhMADS3) from cotton and analysis of its homeotic role in transgenic tobacco. J. Genet. Genomics, 34, 527–535. [DOI] [PubMed] [Google Scholar]
- Guo, W. , Cai, C. , Wang, C. , Zhao, L. , Wang, L. and Zhang, T. (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum . BMC Genom. 9, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y.H. , Yu, Y.P. , Wang, D. , Wu, C.A. , Yang, G.D. , Huang, J.G. and Zheng, C.C. (2009) GhZFP1, a novel CCCH‐type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. [DOI] [PubMed] [Google Scholar]
- Guo, R. , Yu, F. , Gao, Z. , An, H. , Cao, X. and Guo, X. (2011) GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol. Biol. Rep. 38, 49–58. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Shi, G. , Guo, X. , Zhang, L. , Xu, W. , Wang, Y. , Su, Z. et al (2015a) Transcriptome analysis reveals that distinct metabolic pathways operate in salt‐tolerant and salt‐sensitive upland cotton varieties subjected to salinity stress. Plant Sci. 238, 33–45. [DOI] [PubMed] [Google Scholar]
- Guo, S. , Wang, Y. , Sun, G. , Jin, S. , Zhou, T. , Meng, Z. and Zhang, R. (2015b) Twenty years of research and application of transgenic cotton in China. Sci. Agi. Sin. 48, 3372–3387. [Google Scholar]
- Haigler, C.H. , Singh, B. , Zhang, D. , Hwang, S. , Wu, C. , Cai, W.X. , Hozain, M. et al (2007) Transgenic cotton over‐producing spinach sucrose phosphate synthase showed enhanced leaf sucrose synthesis and improved fiber quality under controlled environmental conditions. Plant Mol. Biol. 63, 815–832. [DOI] [PubMed] [Google Scholar]
- Han, L.B. , Li, Y.B. , Wang, H.Y. , Wu, X.M. , Li, C.L. , Luo, M. , Wu, S.J. et al (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell, 25, 4421–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J. , Tu, L. , Hu, H. , Tan, J. , Deng, F. , Tang, W. , Nie, Y. et al (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S. , Orford, S. and Timmis, J. (2002) Characterisation of six a‐expansin genes in Gossypium hirsutum (upland cotton). Mol. Genet. Genomics, 268, 1–9. [DOI] [PubMed] [Google Scholar]
- Hashmi, J.A. , Zafar, Y. , Arshad, M. , Mansoor, S. and Asad, S. (2011) Engineering cotton (Gossypium hirsutum L.) for resistance to cotton leaf curl disease using viral truncated AC1 DNA sequences. Virus Genes, 42, 286–296. [DOI] [PubMed] [Google Scholar]
- He, L. , Yang, X. , Wang, L. , Zhu, L. , Zhou, T. , Deng, J. and Zhang, X. (2013) Molecular cloning and functional characterization of a novel cotton CBL‐interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 435, 209–215. [DOI] [PubMed] [Google Scholar]
- He, X. , Zhu, L. , Wassan, G.M. , Wang, Y. , Miao, Y. , Shaban, M. , Hu, H. et al (2017) GhJAZ2 attenuates cotton resistance to biotic stresses via inhibiting the transcriptional activity of GhbHLH171. Mol. Plant Pathol. https://doi.org/10.1111/mpp.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , He, X. , Tu, L. , Zhu, L. , Zhu, S. , Ge, Z. and Zhang, X. (2016) GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3‐MYB transcription factor GhMYB25‐like. Plant J. 88, 921–935. [DOI] [PubMed] [Google Scholar]
- Huang, B. , Jin, L. and Liu, J. (2007) Molecular cloning and functional characterization of a DREB1/CBF‐like gene (GhDREB1L) from cotton. Sci. China, Ser. C Life Sci. 50, 7–14. [DOI] [PubMed] [Google Scholar]
- Huang, B. , Jin, L. and Liu, J.Y. (2008a) Identification and characterization of the novel gene GhDBP2 encoding a DRE‐binding protein from cotton (Gossypium hirsutum). J. Plant Physiol. 165, 214–223. [DOI] [PubMed] [Google Scholar]
- Huang, Q.S. , Wang, H.Y. , Gao, P. , Wang, G.Y. and Xia, G.X. (2008b) Cloning and characterization of a calcium dependent protein kinase gene associated with cotton fiber development. Plant Cell Rep. 27, 1869. [DOI] [PubMed] [Google Scholar]
- Huang, J.G. , Yang, M. , Liu, P. , Yang, G.D. , Wu, C.A. and Zheng, C.C. (2009) GhDREB1 enhances abiotic stress tolerance, delays GA‐mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant Cell Environ. 32, 1132–1145. [DOI] [PubMed] [Google Scholar]
- Huang, G.Q. , Li, W. , Zhou, W. , Zhang, J.M. , Li, D.D. , Gong, S.Y. and Li, X.B. (2013) Seven cotton genes encoding putative NAC domain proteins are preferentially expressed in roots and in responses to abiotic stress during root development. Plant Growth Regul. 71, 101–112. [Google Scholar]
- Humphries, J.A. , Walker, A.R. , Timmis, J.N. and Orford, S.J. (2005) Two WD‐repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57, 67–81. [DOI] [PubMed] [Google Scholar]
- Islam, M.S. , Fang, D.D. , Thyssen, G.N. , Delhom, C.D. , Liu, Y. and Kim, H.J. (2016a) Comparative fiber property and transcriptome analyses reveal key genes potentially related to high fiber strength in cotton (Gossypium hirsutum L.) line MD52ne. BMC Plant Biol. 16, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.S. , Thyssen, G.N. , Jenkins, J.N. , Zeng, L. , Delhom, C.D. , McCarty, J.C. , Deng, D.D. et al (2016b) A MAGIC population‐based genome‐wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genom. 17, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.S. , Zeng, L. , Thyssen, G.N. , Delhom, C.D. , Kim, H.J. , Li, P. and Fang, D.D. (2016c) Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes. Theor. Appl. Genet. 129, 1071–1086. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Guo, W. , Zhu, H. , Ruan, Y.L. and Zhang, T. (2012) Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 10, 301–312. [DOI] [PubMed] [Google Scholar]
- Jin, L.G. , Li, H. and Liu, J.Y. (2010) Molecular characterization of three ethylene responsive element binding factor genes from cotton. J. Integr. Plant Biol. 52, 485–495. [DOI] [PubMed] [Google Scholar]
- Jin, X. , Pang, Y. , Jia, F. , Xiao, G. , Li, Q. and Zhu, Y. (2013) A potential role for CHH DNA methylation in cotton fiber growth patterns. PLoS ONE, 8, e60547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, M.E. and Crow, L.J. (1992) Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc. Natl Acad. Sci. USA, 89, 5769–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, M.E. and Keller, G. (1996) Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc. Natl Acad. Sci. USA, 93, 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, Z.Y. , Kohel, R.J. , Fang, D.D. , Cho, J. , Van Deynze, A. , Ulloa, M. , Hoffman, S.M. et al (2012) A high‐density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. G3 Genes Genomes Genet. 2, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis, T. , Gasiunas, G. , Miksys, A. , Barrangou, R. , Horvath, P. and Siksnys, V. (2013) crRNA and tracrRNA guide Cas9‐mediated DNA interference in Streptococcus thermophilus . RNA Biol. 10, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. , Tang, Y. , Moon, H.S. , Delhom, C.D. and Fang, D.D. (2013) Functional analyses of cotton (Gossypium hirsutum L.) immature fiber (im) mutant infer that fiber cell wall development is associated with stress responses. BMC Genom. 14, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, D. , Li, H. , Fan, S. , Song, M. , Pang, C. , Wei, H. , Liu, J. et al (2011) Generation of ESTs for flowering gene discovery and SSR marker development in upland cotton. PLoS ONE, 6, e28676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Burns, T.H. , Light, G. , Sun, Y. , Fokar, M. , Kasukabe, Y. , Fujisawa, K. et al (2010) Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta, 232, 1191–1205. [DOI] [PubMed] [Google Scholar]
- Li, X.B. , Fan, X.P. , Wang, X.L. , Cai, L. and Yang, W.C. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell, 17, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Ning, H. , Zhang, Z. , Wu, Y. , Jiang, J. , Su, S. , Tian, F. et al (2011) A cotton gene encoding novel MADS‐box protein is preferentially expressed in fibers and functions in cell elongation. Acta Biochim. Biophys. Sin. 43, 607. [DOI] [PubMed] [Google Scholar]
- Li, B. , Li, D.D. , Zhang, J. , Xia, H. , Wang, X.L. , Li, Y. and Li, X.B. (2013a) Cotton AnnGh3 encoding an annexin protein is preferentially expressed in fibers and promotes initiation and elongation of leaf trichomes in transgenic Arabidopsis. J. Integr. Plant Biol. 55, 902–916. [DOI] [PubMed] [Google Scholar]
- Li, D.D. , Ruan, X.M. , Zhang, J. , Wu, Y.J. , Wang, X.L. and Li, X.B. (2013b) Cotton plasma membrane intrinsic protein 2s (PIP2s) selectively interact to regulate their water channel activities and are required for fibre development. New Phytol. 199, 695–707. [DOI] [PubMed] [Google Scholar]
- Li, J.F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al (2013c) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Fan, S.L. , Song, M.Z. , Pang, C.Y. , Wei, H.L. , Li, W. , Ma, J.H. et al (2013d) Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Rep. 32, 1675–1686. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, L. , Wang, X. , Zhang, W. , Hao, L. , Chu, X. and Guo, X. (2013e) Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J. 280, 5128–5144. [DOI] [PubMed] [Google Scholar]
- Li, C. , He, X. , Luo, X. , Xu, L. , Liu, L. , Min, L. , Jin, L. et al (2014a) Cotton WRKY1 mediates the plant defense‐to‐development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM‐DOMAIN1 expression. Plant Physiol. 166, 2179–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Fan, G. , Wang, K. , Sun, F. , Yuan, Y. , Song, G. , Li, Q. et al (2014b) Genome sequence of the cultivated cotton Gossypium arboreum . Nat. Genet. 46, 567–572. [DOI] [PubMed] [Google Scholar]
- Li, F. , Fan, G. , Lu, C. , Xiao, G. , Zou, C. , Kohel, R.J. , Ma, Z. et al (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM‐1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. [DOI] [PubMed] [Google Scholar]
- Li, X. , Jin, X. , Wang, H. , Zhang, X. and Lin, Z. (2016) Structure, evolution, and comparative genomics of tetraploid cotton based on a high‐density genetic linkage map. DNA Res. 23, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Zhang, K. , Chen, K. and Gao, C. (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics, 41, 63–68. [DOI] [PubMed] [Google Scholar]
- Liang, C. , Meng, Z. , Meng, Z. , Malik, W. , Yan, R. , Lwin, K.M. , Lin, F. et al (2016) GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C. , Sun, B. , Meng, Z. , Meng, Z. , Wang, Y. , Sun, G. , Zhu, T. et al (2017) Co‐expression of GR79 EPSPS and GAT yields herbicide‐resistant cotton with low glyphosate residues. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz, E. , Eviatar, T. , Rozman, A. , Shalit, A. , Goldshmidt, A. , Amsellem, Z. , Alvarez, J.P. et al (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl Acad. Sci. USA, 103, 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M. , Pang, C. , Fan, S. , Song, M. , Wei, H. and Yu, S. (2015) Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA‐Seq. BMC Plant Biol. 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhao, B. , Zheng, H.J. , Hu, Y. , Lu, G. , Yang, C.Q. , Chen, J.D. et al (2015) Gossypium barbadense genome sequence provides insight into the evolution of extra‐long staple fiber and specialized metabolites. Sci. Rep. 5, 14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Zhang, J. , Liu, X. , Wang, W. , Liu, D. , Teng, Z. , Fang, X. et al (2016a) Fine mapping and RNA‐Seq unravels candidate genes for a major QTL controlling multiple fiber quality traits at the T 1 region in upland cotton. BMC Genom., 17, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Song, Y. , Xing, F. , Wang, N. , Wen, F. and Zhu, C. (2016b) GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana . Protoplasma, 253, 1265–1281. [DOI] [PubMed] [Google Scholar]
- Loguercio, L.L. , Zhang, J.Q. and Wilkins, T.A. (1999) Differential regulation of six novel MYB‐domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol. Gen. Genet. 261, 660–671. [DOI] [PubMed] [Google Scholar]
- Long, L. , Gao, W. , Xu, L. , Liu, M. , Luo, X. , He, X. , Yang, X. et al (2014) GbMPK3, a mitogen‐activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco. Plant Cell, Tissue Organ Cult. 116, 153–162. [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. , Tang, X. , Zheng, X. , Voytas, D.F. et al (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Chu, X. , Li, Y. , Wang, C. and Guo, X. (2013) Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana . PLoS ONE, 8, e68503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, P. , Wang, Y.H. , Wang, G.D. , Essenberg, M. and Chen, X.Y. (2001) Molecular cloning and functional identification of (+)‐δ‐cadinene‐8‐hydroxylase, a cytochrome P450 mono‐oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28, 95–104. [DOI] [PubMed] [Google Scholar]
- Luo, M. , Xiao, Y. , Li, X. , Lu, X. , Deng, W. , Li, D. , Hou, L. et al (2007) GhDET2, a steroid 5α‐reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 51, 419–430. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Longxing, H. , Jibiao, F. , Erick, A. , Khaldun, A.B.M. , Yong, Z. and Liang, C. (2017) Cotton GhERF38 gene is involved in plant response to salt/drought and ABA. Ecotoxicology, 26, 841–854. [DOI] [PubMed] [Google Scholar]
- Machado, A. , Wu, Y. , Yang, Y. , Llewellyn, D.J. and Dennis, E.S. (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. [DOI] [PubMed] [Google Scholar]
- Manik, N. and Ravikesavan, R. (2009) Emerging trends in enhancement of cotton fiber productivity and quality using functional genomics tools. Biotechnol. Mol. Biol. Rev. 4, 11–28. [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. et al (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Mao, Y.B. , Tao, X.Y. , Xue, X.Y. , Wang, L.J. and Chen, X.Y. (2011) Cotton plants expressing CYP6AE14 double‐stranded RNA show enhanced resistance to bollworms. Transgenic Res. 20, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y.B. , Xue, X.Y. , Tao, X.Y. , Yang, C.Q. , Wang, L.J. and Chen, X.Y. (2013) Cysteine protease enhances plant‐mediated bollworm RNA interference. Plant Mol. Biol. 83, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, R.C. , Prewitt, S. and Ayre, B.G. (2013) Overexpression of FT in cotton affects architecture but not floral organogenesis. Plant Signal. Behav. 8, e23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry, R.C. , Prewitt, S.F. , Culpepper, S. , Eshed, Y. , Lifschitz, E. and Ayre, B.G. (2016) Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF‐PRUNING orthologs. New Phytol. 212, 244–258. [DOI] [PubMed] [Google Scholar]
- Mei, W. , Qin, Y. , Song, W. , Li, J. and Zhu, Y. (2009) Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics, 36, 141–150. [DOI] [PubMed] [Google Scholar]
- Mei, Y. , Wang, Y. , Chen, H. , Sun, Z.S. and Ju, X.D. (2016) Recent progress in CRISPR/Cas9 technology. J. Genet. Genomics, 43, 63–75. [DOI] [PubMed] [Google Scholar]
- Meng, C. , Cai, C. , Zhang, T. and Guo, W. (2009) Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 176, 352–359. [Google Scholar]
- Mills, R.E. , Walter, K. , Stewart, C. , Handsaker, R.E. , Chen, K. , Alkan, C. , Abyzov, A. et al (2011) Mapping copy number variation by population scale genome sequencing. Nature, 470, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina, M. , Thyssen, G.N. and Fang, D.D. (2015) RNA‐seq analysis of short fiber mutants Ligon‐lintless‐1 (Li 1) and–2 (Li 2) revealed important role of aquaporins in cotton (Gossypium hirsutum L.) fiber elongation. BMC Plant Biol. 15, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nida, D.L. , Kolacz, K.H. , Buehler, R.E. , Deaton, W.R. , Schuler, W.R. , Armstrong, T.A. , Taylor, M.L. et al (1996) Glyphosate‐tolerant cotton: genetic characterization and protein expression. J. Agric. Food Chem. 44, 1960–1966. [Google Scholar]
- Osabe, K. , Clement, J.D. , Bedon, F. , Pettolino, F.A. , Ziolkowski, L. , Llewellyn, D.J. , Finnegan, E.J. et al (2014) Genetic and DNA methylation changes in cotton (Gossypium) genotypes and tissues. PLoS ONE, 9, e86049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, J.T. , Liechty, Z.S. , Alexander, R.H. , Clemons, K. , Hulse‐Kemp, A.M. , Ashrafi, H. , Van Deynze, A. et al (2016) DNA sequence evolution and rare homoeologous conversion in tetraploid cotton. PLoS Genet. 12, e1006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H. , Wendel, J.F. , Gundlach, H. , Guo, H. , Jenkins, J. , Jin, D. , Llewellyn, D. et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature, 492, 423–427. [DOI] [PubMed] [Google Scholar]
- Phillips, T. (2008) The role of methylation in gene expression. Nat. Educ. 1, 116. [Google Scholar]
- Pu, L. , Li, Q. , Fan, X. , Yang, W. and Xue, Y. (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics, 180, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik, S. , Bahadur, R.P. , Srivastava, P.S. and Prasad, M. (2011) Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Mol. Biotechnol. 49, 138–150. [DOI] [PubMed] [Google Scholar]
- Qiao, Z.X. , Huang, B. and Liu, J.Y. (2008) Molecular cloning and functional analysis of an ERF gene from cotton (Gossypium hirsutum). Biochim. Biophys. Acta Gene Regul. Mech. 1779, 122–127. [DOI] [PubMed] [Google Scholar]
- Qin, Y.M. and Zhu, Y.X. (2011) How cotton fibers elongate: a tale of linear cell‐growth mode. Curr. Opin. Plant Biol. 14, 106–111. [DOI] [PubMed] [Google Scholar]
- Qu, Z.L. , Wang, H.Y. and Xia, G.X. (2005) GhHb1: a nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae . Biochim. Biophys. Acta Gene Struct. Expr. 1730, 103–113. [DOI] [PubMed] [Google Scholar]
- Rahman, M. , Hussain, D. , Malik, T. and Zafar, Y. (2005) Genetics of resistance to cotton leaf curl disease in Gossypium hirsutum . Plant. Pathol. 54, 764–772. [Google Scholar]
- Rajasekaran, K. , Cary, J.W. , Jaynes, J.M. and Cleveland, T.E. (2005) Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol. J. 3, 545–554. [DOI] [PubMed] [Google Scholar]
- Ranjan, A. and Sawant, S. (2015) Genome‐wide transcriptomic comparison of cotton (Gossypium herbaceum) leaf and root under drought stress. 3 Biotech, 5, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, A. , Nigam, D. , Asif, M.H. , Singh, R. , Ranjan, S. , Mantri, S. , Pandey, N. et al (2012) Genome wide expression profiling of two accession of G. herbaceum L. in response to drought. BMC Genom. 13, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed, A. , Hao, Y. , Xia, X. , Khan, A. , Xu, Y. , Varshney, R.K. and He, Z. (2017) Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant. 10, 1047–1064. [DOI] [PubMed] [Google Scholar]
- Rashid, B. , Saleem, Z. , Husnain, T. and Riazuddin, S. (2008) Transformation and inheritance of Bt genes in Gossypium hirsutum . J. Plant Biol. 51, 248–254. [Google Scholar]
- Ruan, Y. (2007) Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single‐celled cotton fibre. Funct. Plant Biol. 34, 1–10. [DOI] [PubMed] [Google Scholar]
- Ruan, Y.L. , Llewellyn, D.J. and Furbank, R.T. (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell, 15, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said, J.I. , Song, M. , Wang, H. , Lin, Z. , Zhang, X. , Fang, D.D. and Zhang, J. (2015) A comparative meta‐analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol. Genet. Genomics, 290, 1003–1025. [DOI] [PubMed] [Google Scholar]
- Sapranauskas, R. , Gasiunas, G. , Fremaux, C. , Barrangou, R. , Horvath, P. and Siksnys, V. (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in E. coli . Nucleic Acids Res. 39, 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saranga, Y. , Paterson, A.H. and Levi, A. (2009) Bridging classical and molecular genetics of abiotic stress resistance in cotton In Genetics and Genomics of Cotton (Paterson A.H., ed), pp. 337–352. New York: Springer. [Google Scholar]
- Shalit, A. , Rozman, A. , Goldshmidt, A. , Alvarez, J.P. , Bowman, J.L. , Eshed, Y. and Lifschitz, E. (2009) The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl Acad. Sci. USA, 106, 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. , Zhang, Y. , Chen, K. , Liang, Z. , Zhang, K. et al (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Shan, C.M. , Shangguan, X.X. , Zhao, B. , Zhang, X.F. , Chao, L.M. , Yang, C.Q. , Wang, L.J. et al (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5, 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, S.Q. , Li, B.Y. , Zhang, Z.T. , Zhou, Y. , Jiang, J. and Li, X.B. (2010) Expression of a cotton MADS‐box gene is regulated in anther development and in response to phytohormone signaling. J. Genet. Genomics, 37, 805–816. [DOI] [PubMed] [Google Scholar]
- Shi, Y.H. , Zhu, S.W. , Mao, X.Z. , Feng, J.X. , Qin, Y.M. , Zhang, L. , Cheng, J. et al (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell, 18, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , An, H.‐L. , Zhang, L. , Gao, Z. and Guo, X.‐Q. (2010) GhMPK7, a novel multiple stress‐responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Zhang, L. , An, H. , Wu, C. and Guo, X. (2011) GhMPK16, a novel stress‐responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W. , Hao, L. , Li, J. , Liu, D. , Guo, X. and Li, H. (2014a) The Gossypium hirsutum WRKY gene GhWRKY39‐1 promotes pathogen infection defense responses and mediates salt stress tolerance in transgenic Nicotiana benthamiana . Plant Cell Rep. 33, 483–498. [DOI] [PubMed] [Google Scholar]
- Shi, W. , Liu, D. , Hao, L. , Wu, C.‐A. , Guo, X. and Li, H. (2014b) GhWRKY39, a member of the WRKY transcription factor family in cotton, has a positive role in disease resistance and salt stress tolerance. Plant Cell, Tissue Organ Cult. 118, 17–32. [Google Scholar]
- Shukla, A.K. , Upadhyay, S.K. , Mishra, M. , Saurabh, S. , Singh, R. , Singh, H. , Thakur, N. et al (2016) Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 34, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Song, Q. , Guan, X. and Chen, Z.J. (2015) Dynamic roles for small RNAs and DNA methylation during ovule and fiber development in allotetraploid cotton. PLoS Genet. 11, e1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q. , Zhang, T. , Stelly, D.M. and Chen, Z.J. (2017) Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 18, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z. , Wang, X. , Liu, Z. , Gu, Q. , Zhang, Y. , Li, Z. , Ke, H. et al (2017) Genome‐wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol. J. 15, 982–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo, J. , Liang, X. , Pu, L. , Zhang, Y. and Xue, Y. (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim. Biophys. Acta 1630, 25–34. [DOI] [PubMed] [Google Scholar]
- Suzuki, M. and Bird, A. (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. [DOI] [PubMed] [Google Scholar]
- Tan, J. , Tu, L. , Deng, F. , Wu, R. and Zhang, X. (2012) Exogenous jasmonic acid inhibits cotton fiber elongation. J. Plant Growth Regul. 31, 599–605. [Google Scholar]
- Tang, W. , Tu, L. , Yang, X. , Tan, J. , Deng, F. , Hao, J. , Guo, K. et al (2014) The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 202, 509–520. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.C. and Wilkins, T.A. (1995) Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can. J. Bot. 73, 746–757. [Google Scholar]
- Townsend, B.J. , Poole, A. , Blake, C.J. and Llewellyn, D.J. (2005) Antisense suppression of a (+)‐δ‐cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiol. 138, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford, S.A. , Wu, Y. , Llewellyn, D.J. and Dennis, E.S. (2012) Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD‐1. Plant J. 71, 464–478. [DOI] [PubMed] [Google Scholar]
- Wang, B.H. , Wu, Y.T. , Huang, N.T. , Zhu, X.F. , Guo, W.Z. and Zhang, T.Z. (2006) QTL mapping for plant architecture traits in upland cotton using RILs and SSR markers. Acta Genet. Sin. 33, 161–170. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Gerstein, M. and Snyder, M. (2009) RNA‐Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Wang, Z. , Li, F. , Ye, W. , Wang, J. , Song, G. , Yue, Z. et al (2012) The draft genome of a diploid cotton Gossypium raimondii . Nat. Genet. 44, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Fan, S. , Song, M. , Pang, C. , Wei, H. , Yu, J. , Ma, Q. et al (2014a) Upland cotton gene GhFPF1 confers promotion of flowering time and shade‐avoidance responses in Arabidopsis thaliana . PLoS ONE, 9, e91869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014b) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Chen, J. , Zhang, W. , Hu, Y. , Chang, L. , Fang, L. , Wang, Q. et al (2015) Sequence‐based ultra‐dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 16, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.L. , Zhang, S.C. , Qi, S.D. , Zheng, C.C. and Wu, C.A. (2016a) Delayed germination of Arabidopsis seeds under chilling stress by overexpressing an abiotic stress inducible GhTPS11. Gene, 575, 206–212. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Yuan, Y. , Yang, C. , Geng, S. , Sun, Q. , Long, L. , Cai, C. et al (2016b) Characterization, expression, and functional analysis of a novel NAC gene associated with resistance to verticillium wilt and abiotic stress in cotton. G3 Genes Genomes Genet. 6, 3951–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Yuan, Y. , Yang, C. , Geng, S. , Sun, Q. , Long, L. , Cai, C. et al (2016c) Characterization, expression, and functional analysis of a novel NAC gene associated with resistance to verticillium wilt and abiotic stress in cotton. Genes Genomes Genet. 6, 3951–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wu, S.M. , Zhu, Y. , Fan, Q. , Zhang, Z.N. , Hu, G. , Peng, Q.Z. et al (2017a) Functional characterization of a novel jasmonate ZIM‐domain interactor (NINJA) from upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 112, 152–160. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Tu, L. , Lin, M. , Lin, Z. , Wang, P. , Yang, Q. , Ye, Z. et al (2017b) Asymmetric subgenome selection and cis‐regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Zhang, J. , Sun, L. , Ma, Y. , Xu, J. , Liang, S. , Deng, J. et al (2017c) High efficient multi‐sites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. https://doi.org/10.1111/pbi.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Meng, Z. , Liang, C. , Meng, Z. , Wang, Y. , Sun, G. , Zhu, T. et al (2017d) Increased lateral root formation by CRISPR/Cas9‐mediated editing of arginase genes in cotton. Sci. China Life Sci. 60, 524–527. [DOI] [PubMed] [Google Scholar]
- Wendel, J.F. (1989) New World tetraploid cottons contain Old World cytoplasm. Proc. Natl Acad. Sci. 86, 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F. and Albert, V.A. (1992) Phylogenetics of the cotton genus (Gossypium L.): character‐state weighted parsimony analysis of chloroplast DNA restriction site data and its systematic and biogeographic implications. Syst. Bot. 17, 115–143. [Google Scholar]
- Wilkins, T.A. and Jernstedt, J.A. (1999) Molecular genetics of developing cotton fibers, pp. 231–267. New York: Cotton Fibers. Haworth Press. [Google Scholar]
- Wu, Y. , Machado, A.C. , White, R.G. , Llewellyn, D.J. and Dennis, E.S. (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol. 47, 107–127. [DOI] [PubMed] [Google Scholar]
- Wu, M. , Li, J. , Fan, S. , Song, M. , Pang, C. , Wei, J. , Yu, J. et al (2015) Gene expression profiling in shoot apical meristem of Gossypium hirsutum . Russ. J. Plant Physiol. 62, 684–694. [Google Scholar]
- Xiao, Y.H. , Li, D.M. , Yin, M.H. , Li, X.B. , Zhang, M. , Wang, Y.J. , Dong, J. et al (2010) Gibberellin 20‐oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 167, 829–837. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Zhu, L. , Tu, L. , Liu, L. , Yuan, D. , Jin, L. , Long, L. et al (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA‐Seq‐dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W.L. , Zhang, D.J. , Wu, Y.F. , Qin, L.X. , Huang, G.Q. , Li, J. , Li, L. et al (2013) Cotton PRP5 gene encoding a proline‐rich protein is involved in fiber development. Plant Mol. Biol. 82, 353–365. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Zhang, W. , He, X. , Liu, M. , Zhang, K. , Shaban, M. , Sun, L. et al (2014) Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 65, 6679–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Gao, J. , Cao, Z. , Chee, P.W. , Guo, Q. , Xu, Z. , Paterson, A.H. et al (2017) Fine mapping and candidate gene analysis of qFL‐chr1, a fiber length QTL in cotton. Theor. Appl. Genet. 130, 1309–1319. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Zhang, C. , Yang, X. , Liu, K. , Wu, Z. , Zhang, X. , Zheng, W. et al (2014) PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 203, 437–448. [DOI] [PubMed] [Google Scholar]
- Ye, Z.H. and Zhu, J. (2001) Genetic analysis on flowering and boll setting in upland cotton (Gossypium hirsutum L.) 11. The genetic behavior of different fruiting sites. Actu Agron. Sin. 27, 243–252. [PubMed] [Google Scholar]
- Yoo, M.J. and Wendel, J.F. (2014) Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome. PLoS Genet. 10, e1004073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Huaxia, Y. , Lu, W. , Wu, C. , Cao, X. and Guo, X. (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.H. , Wu, S.J. , Peng, Y.S. , Liu, R.N. , Chen, X. , Zhao, P. , Xu, P. et al (2016) Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, D. , Tang, Z. , Wang, M. , Gao, W. , Tu, L. , Jin, X. , Chen, L. et al (2015) The genome sequence of Sea‐Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5, 17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Liu, X. , Zuo, K. , Zhang, J. , Sun, X. and Tang, K. (2011a) Molecular cloning and characterization of a novel Gossypium barbadense L. RAD‐like gene. Plant Mol. Biol. Rep. 29, 324–333. [Google Scholar]
- Zhang, L. , Xi, D. , Li, S. , Gao, Z. , Zhao, S. , Shi, J. , Wu, C. et al (2011b) A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 77, 17–31. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Zheng, X. , Song, S. , Zeng, Q. , Hou, L. , Li, D. , Zhao, J. et al (2011c) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 29, 453–458. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, Y. , Lu, W. , Meng, F. , Wu, C.A. and Guo, X. (2012) Cotton GhMKK5 affects disease resistance, induces HR‐like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana . J. Exp. Bot. 63, 3935–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Li, S. , Yang, S. , Wang, L. and Guo, W. (2015a) Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol. Biol. 87, 47–67. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Hu, Y. , Jiang, W. , Fang, L. , Guan, X. , Chen, J. , Zhang, J. et al (2015b) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM‐1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang, F. , Zhu, G. , Du, L. , Shang, X. , Cheng, C. , Yang, B. , Hu, Y. et al (2016a) Genetic regulation of salt stress tolerance revealed by RNA‐Seq in cotton diploid wild species, Gossypium davidsonii . Sci. Rep. 6, 20582. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang, X. , Fan, S. , Song, M. , Pang, C. , Wei, H. , Wang, C. and Yu, S. (2016b) Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.). Plant Sci. 242, 178–186. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Gao, Y. , Zhang, Z. , Chen, T. , Guo, W. and Zhang, T. (2013) A receptor‐like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought‐stress tolerance in Arabidopsis. BMC Plant Biol. 13, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. , Fung‐Leung, W.P. , Bittner, A. , Ngo, K. and Liu, X. (2014) Comparison of RNA‐Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE, 9, e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Wang, N.N. , Kong, L. , Gong, S.Y. , Li, Y. and Li, X.B. (2014) Molecular characterization of 26 cotton WRKY genes that are expressed differentially in tissues and are induced in seedlings under high salinity and osmotic stress. Plant Cell, Tissue Organ Cult. 119, 141–156. [Google Scholar]
- Zhu, X.G. , Wang, Y. , Ort, D.R. and Long, S.P. (2013) e‐photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant Cell Environ. 36, 1711–1727. [DOI] [PubMed] [Google Scholar]


