Abstract
Endosymbiosis is a widespread phenomenon and hosts of bacterial endosymbionts can be found all-over the eukaryotic tree of life. Likely, this evolutionary success is connected to the altered phenotype arising from a symbiotic association. The potential variety of symbiont’s contributions to new characteristics or abilities of host organisms are largely unstudied. Addressing this aspect, we focused on an obligate bacterial endosymbiont that confers an intraspecific killer phenotype to its host. The symbiosis between Paramecium tetraurelia and Caedibacter taeniospiralis, living in the host’s cytoplasm, enables the infected paramecia to release Caedibacter symbionts, which can simultaneously produce a peculiar protein structure and a toxin. The ingestion of bacteria that harbor both components leads to the death of symbiont-free congeners. Thus, the symbiosis provides Caedibacter-infected cells a competitive advantage, the “killer trait.” We characterized the adaptive gene expression patterns in symbiont-harboring Paramecium as a second symbiosis-derived aspect next to the killer phenotype. Comparative transcriptomics of infected P. tetraurelia and genetically identical symbiont-free cells confirmed altered gene expression in the symbiont-bearing line. Our results show up-regulation of specific metabolic and heat shock genes whereas down-regulated genes were involved in signaling pathways and cell cycle regulation. Functional analyses to validate the transcriptomics results demonstrated that the symbiont increases host density hence providing a fitness advantage. Comparative transcriptomics shows gene expression modulation of a ciliate caused by its bacterial endosymbiont thus revealing new adaptive advantages of the symbiosis. Caedibacter taeniospiralis apparently increases its host fitness via manipulation of metabolic pathways and cell cycle control.
Keywords: ciliate, fitness advantage, mutualist, parasite, R-body, RNA-Seq
Introduction
New phenotypes and abilities arise from symbioses. Some of those have been studied at the functional and transcriptome level. Examples comprise nutritional symbioses where the symbionts substitute the diet of their host by provision of nutrients or by cooperation in the digestion of complex compounds, for example, in cellulose-digesting termites (Tartar et al. 2009; He et al. 2013), phloem-feeding insects (Sloan et al. 2014), and mice (Sonnenburg et al. 2006). Defensive symbioses provide another kind of altered phenotype, for example, immune system activation by murine and human microbiomes (Anahtar et al. 2015; Zelante et al. 2013) or resistance to chytridiomycosis in amphibians (Rebollar et al. 2016). More “exotic” cases report developmental and behavioral modifications (Stilling et al. 2015; Heijtz et al. 2011; Shin et al. 2011).
The association between the eukaryote Paramecium tetraurelia (Ciliophora, Alveolata) and the cytoplasmic endosymbiont Caedibacter taeniospiralis (Gammaproteobacteria) might be considered a less complex symbiosis as it includes only two participants. However, complexity is not necessarily restricted to the number and variety of involved partners. The Paramecium-Caedibacter symbiosis exhibits a rather peculiar phenotypic change: Paramecia carrying these obligate intracellular bacteria are termed killer paramecia because they become toxic for uninfected Paramecium cells, a phenomenon called killer trait (Schrallhammer and Schweikert 2009; Pond et al. 1989; Preer et al. 1974). The underlying mechanism is not yet completely understood. Caedibacter taeniospiralis residing in the cytoplasm of P. tetraurelia occur in two morphologically different forms: Normal bacterial rods (“nonbright”) and slightly bigger “bright” cells. The first multiply by standard cell division whereas the latter are incapable of division and contain a microscopically visible (up to 20 µm in length, 0.4 µm in width), proteinaceous structure termed Refractile body (R-body) due to its bright appearance in phase contrast microscopy. The R-body is build-up from a large, tightly coiled protein ribbon. As a prerequisite for the killer trait, bright bacteria need to be released and ingested by Caedibacter-free and thus sensitive paramecia (Preer et al. 1974; Schrallhammer et al. 2012). Phagocytosis represents the normal feeding behavior of bacterivorous paramecia. Incorporation in a phagosome followed by acidification triggers the R-body to unroll and thereby to destroy bacterial and vacuolar membranes. This action is probably connected with the release of an unidentified toxin, which ultimately leads to the death of the Caedibacter-free Paramecium cell. Infected paramecia are protected from the killer trait by an unknown mechanism provided by the symbiotic bacteria. Thus, they gain a selective advantage compared with symbiont-free competitors (Schrallhammer and Schweikert 2009).
While environmental paramecia can be found either free of symbiotic bacteria or infected by one or more bacterial endosymbionts from diverse phylogenetic origins (Görtz and Fokin 2009), C. taeniospiralis reproduces exclusively in the cytoplasm of P. tetraurelia and is vertically transmitted. Hence, its survival, reproduction, and evolution are coupled to its host. At the first glance, all benefits of expressing the killer trait are gained by Paramecium while Caedibacter pays the costs. It devotes a part of its population to R-body production, an evolutionary dead-end because those bacteria lose the ability to divide. Finally, those cells die when the R-body unrolls, albeit once inside a phagosome Caedibacter would be digested in any case. However, by sacrificing a proportion of individual members, the host and therewith the rest of the symbiont population can thrive.
Clearly, the killer trait and the corresponding resistance are beneficial phenotype modifications provided by Caedibacter (Görtz et al. 2009). Interestingly, the alphaproteobacterial symbiont Caedimonas varicaedens (basonym Caedibacter varicaedens, Schrallhammer et al. 2018) also confers its Paramecium host the killer trait. The question regarding the nature of additional effects of these symbionts is controversial. Preer et al. (1974) state that under laboratory conditions Caedibacter generally do not harm their hosts. Experimental studies performing growth assays with genetically identical lines either infected by Caedibacter or symbiont-free highlight the complexity of this question (Kusch et al. 2002; Dusi et al. 2014). While both studies arrive at the conclusion that C. taeniospiralis is a parasite, they present contradicting results: One observes a significant difference in population growth rates (Kusch et al. 2002), the other exclusively during at carrying capacity but not during exponential growth (Dusi et al. 2014). Thus, cultivation conditions (e.g., food abundance, food quality, medium composition, temperature) could have a strong influence on the outcome of the interaction between Caedibacter and Paramecium.
Among ciliate-bacteria symbioses, the Paramecium-Caedibacter system is exceptional because it is relatively well-studied (reviewed by Schrallhammer and Schweikert 2009; Preer et al. 1974; Pond et al. 1989; Schweikert et al. 2013). We introduce a new perspective on this interaction by applying mRNA-Seq to reveal differential gene expression in Caedibacter-carrying paramecia compared with symbiont-free Paramecium cells. Fitness assays are used as a methodological independent confirmation of transcriptome results. This approach aims to elucidate so far hidden aspects of host’s phenotype modification caused by its bacterial symbionts.
Materials and Methods
Organisms and Cultivation
Paramecium tetraurelia 51K infected by the bacterial symbiont C. taeniospiralis (Preer et al. 1974) was obtained from the Culture Collection of Algae and Protozoa (CCAP Scotland, UK; CCAP 1660/3F). “K” stands for “Killer” indicating that this strain exhibits the killer trait and thus harbors the causative symbiont. The line P. tetraurelia 51 (genetically identical P. tetraurelia background as 51K) was generated from 51K cells by curing them from C. taeniospiralis infection via antibiotic treatment. From now on, the two lines are referred to as 51K and 51.
Paramecium cells were maintained at 24 °C in Wheat Grass Powder (WGP, Pines International Co., Lawrence, KS, USA) infusion medium supplemented with 0.8 µg ml−1 stigmasterol (Sigma–Aldrich, Seelze, Germany). As food source Klebsiella pneumoniae (Cheaib et al. 2015) inoculated in WGP was provided twice a week.
For the elimination of the bacterial symbionts, 51K cells were incubated two times with 100 µg ml−1 streptomycin (Carl Roth GmbH, Karlsruhe, Germany). Briefly, 30 cells of 51K were placed in antibiotic solution (100 µg ml−1 streptomycin in sterile Volvic) at 24 °C for 24 h. Thereafter, cells were washed individually four times and individual cells were incubated in antibiotic solution as described. Then the cells were washed as before and transferred to 0.5 ml of bacterized CM diluted in a ratio 1:2 with sterile Volvic. Cultivation was continued as described above until >100 cells per batch were available to control for the successful elimination of C. taeniospiralis by Fluorescence In Situ Hybridization (FISH, see below). The line 51 was established circa seven months prior to the preparation for RNA extraction and fitness experiments by pooling two batches of Caedibacter-free paramecia.
For analysis of the potential killer effect of 51K and 51 an additional line with reported susceptibility to this phenomenon (P. tetraurelia 51S, “S” stands for “Sensitive”) was used (Schrallhammer et al. 2012). Furthermore, to exclude potential undesired side effects of streptomycin on paramecia, 51S was also subjected to the above-described antibiotic treatment as control.
FISH
Presence or absence of endosymbionts was monitored regularly (before and after antibiotic treatment and fitness assays, before killer tests, cell age synchronization, and harvesting for RNA extraction). Therefore, bacteria were visualized and identified inside the Paramecium cells by FISH applying the universal probe EUB338 (Amann 1990) and the species-specific probe Ctaenio-998 (Beier et al. 2002). For each FISH experiment, at least 20 Paramecium cells were collected and fixed in 2% paraformaldehyde (w/v) on an adhesion slide (Superfrost Ultra Plus, Thermo Scientific, Braunschweig, Germany). Hybridizations were carried out in hybridization buffer (900 mM NaCl, 20 mM Tris–HCl, 0.01% SDS, 35% formamide) at 46 °C for 18 h, followed by washing (70 mM NaCl, 20 mM Tris–HCl, 0.01% SDS, 5 mM EDTA) at 48 °C for 20 min. The slides were examined with an epifluorescence microscope (Axio Imager M2, Carl Zeiss Microscopy GmbH, Jena, Germany). Pictures were taken (AxioCam MRM, Zeiss Microscopy GmbH, Jena, Germany) and processed using the implemented ZEN software package (blue edition).
Killer Tests
Caedibacter-bearing paramecia confer the killer trait to their host and hence kill sensitive cells while infected cells are resistant. Without the symbionts, the Paramecium cells become susceptible. The occurrence of this characteristic behavior was assessed for 51K and 51 using the symbiont-free and reportedly sensitive line P. tetraurelia 51S (Schrallhammer et al. 2012) as susceptibility control. Ten cells of 51S were exposed to a lysate corresponding to ∼25 cells of 51K or 51, respectively. The lysate was prepared by vortexing the Paramecium suspension with glass beads until no intact paramecia were observed. Thus, potentially present R-body containing Caedibacter, the lethal agents of the killer trait, were released at least in case of 51K. The number of living 51S was determined at several time points. Six replicates were included for each experiment. A one-way ANOVA with repeated measures followed by a multiple comparison test was performed using the standard settings of PRISM 6 (version 6, GraphPad Software, Inc., La Jolla, USA).
RNA Extraction, cDNA Library Generation, and Illumina Sequencing
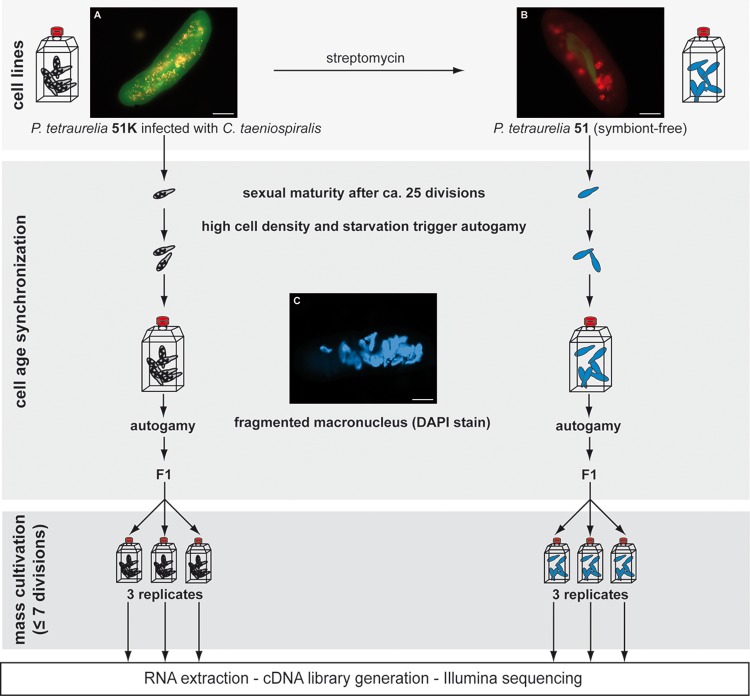
For the identification of Differentially Expressed Genes (DEGs) in P. tetraurelia as response to C. taeniospiralis, the transcriptomes of the genetically identical lines 51K (fig. 1A) and 51 (fig. 1B) were analyzed. The symbiont-free 51 was used as reference.
Fig. 1.
—Experimental set-up. Fluorescence In Situ hybridizations of used cell lines Paramecium tetraurelia 51K infected by Caedibacter taeniospiralis (A) and symbiont-free P. tetraurelia 51 (B) generated by treatment with streptomycin. Applied probes are the universal EUB338 (labeled with Cy3) and the symbiont-specific Ctaenio-998 (fluorescein). Merged images show the presence of numerous cytoplasmic bacteria (A) respectively bacteria in digestive vacuoles (B). For visualization of the macronuclear fragmentation during autogamy, DAPI staining was performed (C). Bars represent 20 µm. To obtain cell age synchronized cells, paramecia were cultivated for circa 25 divisions and then triggered to undergo autogamy, which was confirmed by the observation of fragmented macronuclei after DAPI staining (C). F1 of lines with > 95% autogamous cells were split into three replicates, cultivated for max. seven divisions and harvested for subsequent steps.
In order to avoid differences in gene expression resulting from sexual recombination (autogamy), 51K and 51 were subjected to cell age synchronization (fig. 1). Therefore, the paramecia were triggered to undergo autogamy, a self-fertilization process that results in homozygote cells which remain immature (unable to undergo a new round of autogamy) for about seven cell divisions (Ishikawa et al. 1998). The F1 paramecia were then cultivated to obtain the desired cell number before reaching maturity again.
Starting from single cells, both 51K and 51 were grown in bacterized 0.2× WGP until the cells completed ∼25 divisions and thus should be fully sexually mature. The cells were harvested by centrifugation and resuspended in exhausted 0.2× WGP to obtain a final cell density of 2,000 cells ml−1. Exhausted medium is the cell-free supernatant of bacterized 0.2× WGP obtained after centrifugation (6,000 × g for 20 min). High cell density and starvation triggered the cells to undergo autogamy as microscopically confirmed by visualization of macronuclear fragmentation after DAPI staining (fig. 1C). Mass cultivation of F1 paramecia were performed with Paramecium lines that contained >95% autogamous cells. The respective cells were transferred to 1× WGP and incubated for three days (approximately max. seven divisions) till the cell number exceeded 200,000. After harvesting, the collected paramecia were resuspended in sterile Volvic water and incubated for 20 min at 24 °C to reduce bacteria in Paramecium food vacuoles. Total RNA of ∼200,000–400,000 cells per line was isolated (modified from Chomczynski and Sacchi 1987) using TriReagent (Sigma–Aldrich, Seelze, Germany). After DNAse I treatment and subsequent purification by acid phenol, DNA-free total RNA integrity was checked by denaturing gel electrophoresis. For the generation of cDNA libraries, poly-A RNA was enriched using the NEBNext Poly(A) mRNA Magnetic Isolation Module followed by library preparation with NEBNext Ultra Directional RNA Library Prep Kit for Illumina. Both steps were carried out according to manufacturer’s (NEB, Frankfurt a. M., Germany) recommendations using 11 PCR cycles of library enrichment. DNA quantity of the prepared cDNA libraries was measured using the Qubit 1.0 Fluorometer (Life Technologies, United States). Illumina sequencing was carried out on a HiSeq2500 platform generating 2 × 100 nt paired end reads. Read numbers were: 51-1, 115.094.792; 51-2, 19.897.636; 51-3, 24.146.344; 51K-1, 16.888.465; 51K-2, 15.131.427; 51K-3, 13.962.278 (study accession PRJEB21163). Reads were demultiplexed with bcl2fastq (v1.8.4) and trimmed for adaptor contamination and low quality bases with the cutadapt (v1.4.1) wrapper trim_galore (v0.3.3).
Bioinformatics
Gene expression levels for each individual library were quantified using Sailfish (version 0.9.2), an alignment-free quantification algorithm (Patro et al. 2014). The expression values were normalized using Transcripts Per Million (TPM) values reported by Sailfish. However, differential expression of genes was determined by DESeq2 (version 3.3; Love et al. 2014) using read counts per gene not TPM. DESeq2 uses negative binomial linear models for dispersion estimates. After performing multiple testing correction, genes with a False Discovery Rate (FDR) of ≤0.01 were considered as DEGs.
Gene Ontology (GO) enrichment was performed via Ontologizer (Bauer et al. 2008), using the Java webstart application (version 2.1). Thereby, the DEGs were categorized into different GO terms. Those are concepts to describe gene molecular functions or biological processes. The GO annotation file was obtained from the Gene Ontology Consortium webpage (http://geneontology.org, accessed December 15, 2017). The GO term association file has been created according to Cheaib et al. (2015) using their gene association file as template. Gene enrichment was determined for up- and down-regulated genes using the Parent–Child-Union method with Benjamini–Hochberg multiple testing correction (Bauer et al. 2008). Multidimensional scaling scatter plots of GO terms exhibiting a P-value of ≤0.001 were generated using the REViGO tool (Supek et al. 2011). This visualization summarizes biological processes according to their similarity and GO enrichment P-values.
Fitness Assays
The impact of Caedibacter on P. tetraurelia was functionally analyzed. Thus, the exponential growth rate r (division rate during the exponential growth phase) and the carrying capacity k (maximal cell density in the stationary phase) were used as indicative fitness parameters for 51K and symbiont-free 51 (modified from Bella et al. 2016). In preparation for the fitness assay, cells were washed and adjusted to exponential growth. Therefore, ten single cells per line were individually washed three times and cultivated until a density of ∼50 cells ml−1 was reached. In order to adapt the cells for the assays, nine cultures (for 51K as well as 51) were pooled and each day provided with ¼ volume of bacterized WGP to ensure exponential growth for three days. For the actual assay, five replicates per line were included. Each replicate started with identical volumes of bacterized medium and ∼80 Paramecium cells ml−1. The total volume of each replicate was set to 30 ml. In order to obtain growth curves, samples for density counts (Castelli et al. 2015) were collected at regular time points and fixed with 2.9% Bouin’s solution (Sigma–Aldrich, Seelze, Germany). The exponential growth rate r and carrying capacity k were calculated by fitting a nonlinear parametric regression based on a logistic growth model as described elsewhere (Bella et al. 2016). Differences of those two fitness parameters between symbiont-bearing 51K and symbiont-free 51 were determined by unpaired t-test with Welch’s correction (PRISM 6, version 6, GraphPad Software, Inc., La Jolla, USA).
Results
Phenotypes of Infected and Symbiont-free P. tetraurelia
Caedibacter taeniospiralis was removed from infected cells via antibiotic treatment enabling subsequent comparisons between killer and symbiont-free paramecia. Ideally, cured cells should be re-infected and used in parallel for transcriptome or fitness analyses. However, C. taeniospiralis is strictly vertically transmitted and experimental infections are not feasible. By streptomycin treatment of C. taeniospiralis-bearing 51K, the genetically identical endosymbiont-free line P. tetraurelia 51 was successfully generated (fig. 1B). In analogously treated 51S cells no physiological effects (abnormal cell morphology or division rate, increased mortality) caused by the antibiotics were observed.
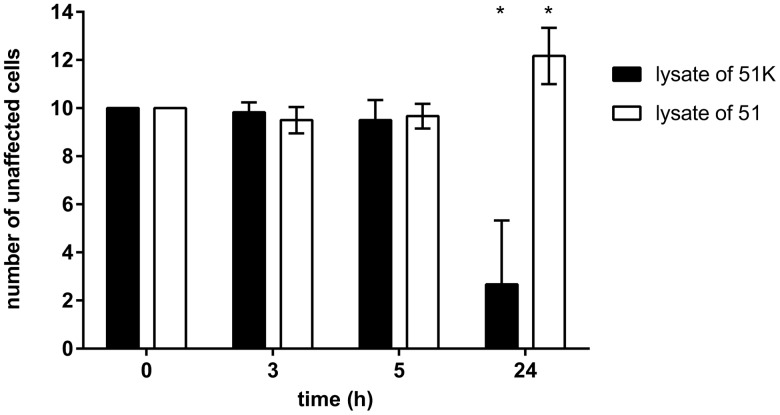
The prominent feature of Caedibacter symbionts is the alteration of their host’s phenotype into so-called killer paramecia. We performed killer tests to verify the presence of the symbiont and hence if 51K can cause the death of sensitive 51S. Vice versa, in case of 51 we tested for the loss of this ability together with the elimination of C. taeniospiralis. Therefore, 51K and 51 were mechanically lysed to maximize the number of potentially released bacteria. Exposing 51S cells to these lysates confirmed toxic effects of 51K (fig. 2): After 24 h, the cell number of 51S mixed with this lysate had decreased significantly (P-value < 0.001). Those exposed to 51 lysate did not show reduced cell numbers or any prelethal symptoms. On the contrary, 51S multiplied in these replicates (fig. 2).
Fig. 2.
—Survival rates of Paramecium tetraurelia 51S in killer tests. Ten cells of 51S were exposed to lysate corresponding to ∼25 paramecia up to 24 h. Lysates were prepared from P. tetraurelia 51K infected with Caedibacter taeniospiralis (black) and Caedibacter-free 51 (white). After exposure, the number of unaffected 51S cells was reported at different time points. Statistically significant differences to initial cell number were observed (* P-value ≤0.001; repeated measurements one-way ANOVA followed by a multiple comparison test). Bars represent the mean of six replicates ± SD.
Analysis of Differential Gene Expression in Infected and Symbiont-Free Cells
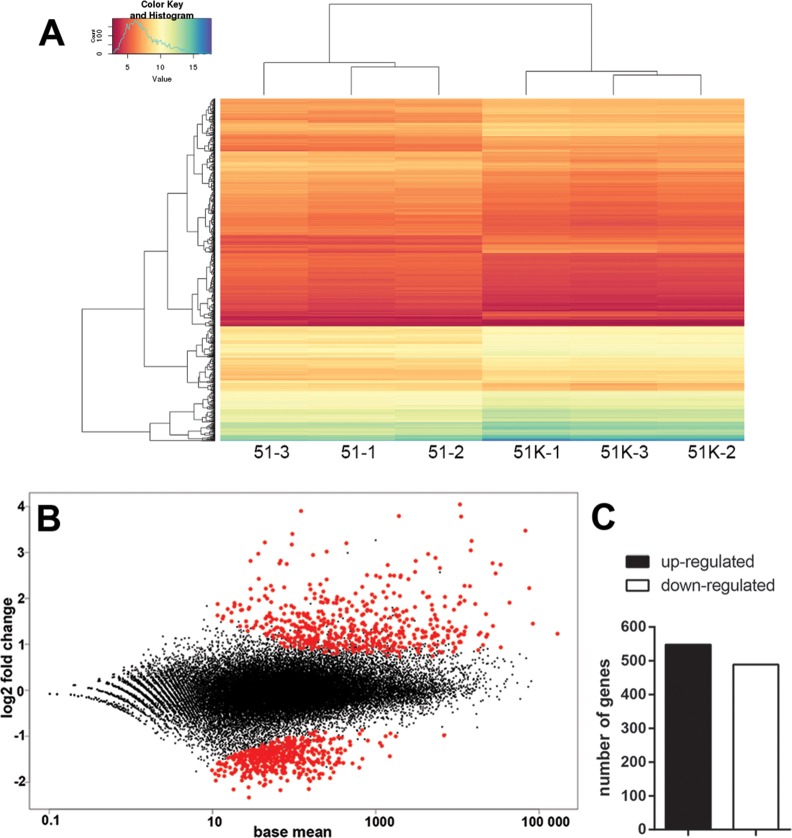
RNA sequencing with three biological replicates was carried out to determine the genome-wide gene expression patterns for C. taeniospiralis-infected 51K and symbiont-free 51. A hierarchical clustering analysis of these transcriptomes revealed major reproducible differences between both lines as the replicates of infected and symbiont-free cells cluster clearly together (fig. 3A). Using the DESeq2 approach (Love et al. 2014) with an adjusted P-value of ≤0.01 as cut-off, we identified 1,037 genes to be differentially expressed between C. taeniospiralis-carrying and symbiont-free paramecia (fig. 3B). Considering symbiont-free 51 as reference, out of 39,495 annotated genes, slightly more were significantly up- (548) than down-regulated (489) in Caedibacter-infected paramecia (fig. 3C). Up-regulated genes show more variation than down-regulated genes concerning both their mean expression values and absolute fold changes (fig. 3B).
Fig. 3.
—Gene expression in Paramecium tetraurelia 51K infected with Caedibacter taeniospiralis compared with symbiont-free line 51. (A) Hierarchical clustering of gene expression values (logTPM) in rows and columns for all six samples. (B) MA plot of log2 fold change of gene expression level (y-axis) against mean gene expression level (x-axis). Each point represents a gene; those with significant differential expression (FDR ≤0.01) are indicated in red. (C) Bar plot of DEGs in 51K with the number of up- and down-regulated genes, in black and white respectively.
GO Term Enrichment in Infected P. tetraurelia
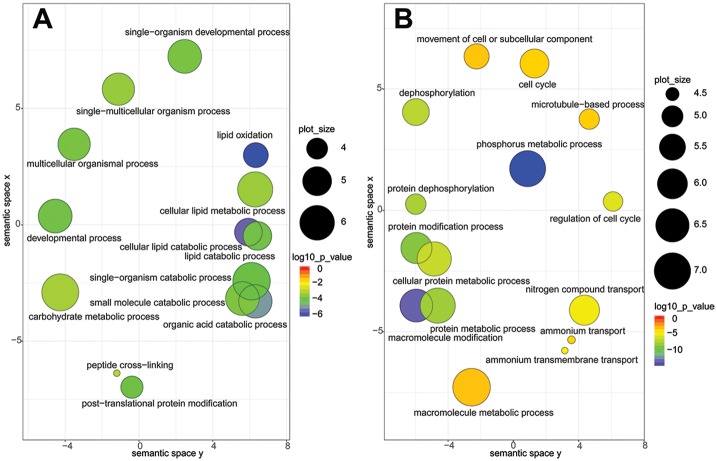
DEGs with a P-value of ≤0.001 were subjected to GO enrichment analysis with the Ontologizer (Bauer et al. 2008) using the Biological Process subontology. Enriched GO terms are specified (supplementary tables S1 and S2, Supplementary Material online) and depicted by REViGO (fig. 4; Supek et al. 2011). The most significantly up-regulated GO term in infected paramecia (fig. 4A) is lipid oxidation (including cellular lipid metabolism, cellular lipid catabolism, and lipid catabolism). It comprises six DEGs (table 1) with sequence homologies to peroxisomal acyl-CoA oxidases, important enzymes for kinetics and substrate specificity in the β-oxidation (Poirier et al. 2006). Further up-regulated processes (fig. 4A) are organic acid catabolism (included DEGs almost identical with those in lipid oxidation, table 1) and carbohydrate metabolism. The latter comprises mostly transcripts involved in citrate cycle, glycolysis and glycogenesis and therefore indicates an increased energy metabolism (table 1).
Fig. 4.
—Visualization of Gene Ontology (GO) terms representing biological processes. Up- (A) and down-regulated (B) GO terms (FDR ≤0.01) are depicted as circles; the distance between those indicates the relationship between terms: closer distance means higher similarity. Color indicates significance of differential expression of an individual GO term (red low and blue high); size (in log10 P-value) indicates the percentage of genes annotated with a term in the reference database (UniProt) and thus indicates more general terms (large) and more specific ones (small).
Table 1.
Metabolic Pathways and Cellular Functions of DEGs Included in Most Significantly Enriched GO Terms
| GO Term | Transcript Stable ID | Protein | Involved In |
|---|---|---|---|
| Up-regulated DEGs | |||
| Lipid oxidation | CAK94090, CAK59990, CAK90106, CAK92996, CAK77450, CAK79917 | Acyl-coenzyme A oxidase | Peroxisomal β-oxidation |
| Organic acid catabolic process | CAK94090, CAK59990, CAK90106, CAK92996, CAK77450, CAK79917 | Acyl-coenzyme A oxidase | Peroxisomal β-oxidation |
| CAK76270 | Homogentisate 1, 2-dioxygenase | Catabolization of aromatic amion acids | |
| Carbohydrate metabolic process | CAK89181, CAK72962, CAK86438, CAK94271 | Fructose-1, 6-bisphosphatase | Glycolysis and glycogenesis |
| CAK61489, CAK90992 | Citrate synthase | Citrate cycle | |
| CAK60798 | Glycosyl hydrolase family 31 protein | Hydrolysis of complex sugars | |
| CAK89297 | Malate dehydrogenase | Citrate cycle | |
| CAK93624 | Amylo-alpha-1, 6-glucosidase | Glycogen degradation | |
| CAK74015 | Poly [ADP-ribose] polymerase | DNA repair and programmed cell death | |
| CAK69096 | Lipid A-disaccharide synthase | Lipid A biosynthesis | |
| CAK88889 | Proteobacterial glycerol kinase-like protein | Triglyceride and glycerophospholipid synthesis or degradation | |
| CAK94394 | Hypothetical protein | Unknown | |
| Down-regulated DEGs | |||
| Phosphorus metabolic process 1 | CAK74095, CAK82985, CAK63555, CAK77180, CAK77847, CAK89645, CAK93119, CAK59145, CAK65257, CAK69124 | Dual specificity phosphatase domain protein | Protein phosphorylation |
| CAK92571, CAK72540, CAK70776, CAK75609, CAK77917 | Protein kinase domain protein | Protein phosphorylation | |
| CAK81054, CAK71834, CAK84440 | Hypothetical protein | Unknown | |
| CAK71484 | Serine/threonine kinase domain protein | Protein phosphorylation | |
| CAK92511 | Cyclin-dependent kinase-like serine/threonine kinase family protein | Cell cycle regulation | |
Only the 20 DEGs with the lowest P-values comprised by this GO term are listed (in total: 82).
The most significantly down-regulated process (fig. 4B) is phosphorus metabolism (82 DEGs), directly followed by macromolecule modification (79 DEGs, all also included in phosphorus metabolism). These DEGs show homologies either to phosphatases or protein kinases (supplementary table S2, Supplementary Material online). The four transcripts comprised by the GO term nitrogen compound belong to ammonium transporters (supplementary table S2, Supplementary Material online), which are generally involved in nitrogen uptake for nutritional purposes and ammonia excretion for waste removal (Kirsten et al. 2008). Additionally, the GO term cell cycle contains four genes annotated as cyclin-dependent kinases (CDKs).
Caedibacter taeniospiralis Infection Affects Host Fitness
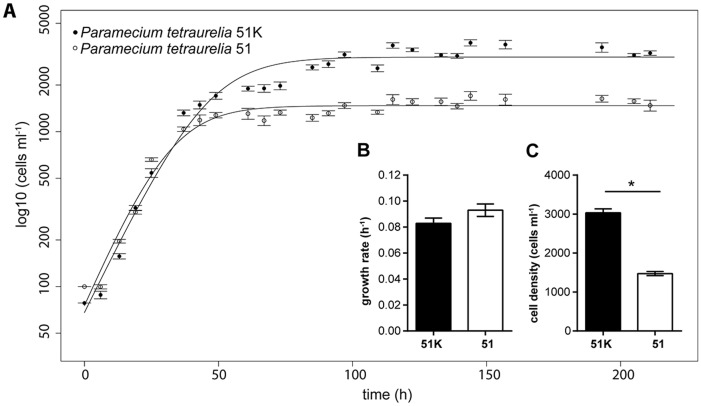
Comparative transcriptomics of infected and symbiont-free P. tetraurelia indicated that 51K carrying C. taeniospiralis has an increased energy metabolism. In parallel growth assays with both lines the possible consequences of Caedibacter infection on Paramecium fitness were tested. FISH was applied at the beginning and end of the fitness assay to determine the infection level of C. taeniospiralis in 51K and confirmed a constant infection rate (100% infection prevalence). After application of a logistic growth model to fit a nonlinear regression (fig. 5A), exponential growth rate and carrying capacity (cell density that is reached and remains constant during stationary phase) were used for comparisons. Symbiont-bearing 51K reached a significantly higher carrying capacity (P-value < 0.001) than genetically identical symbiont-free 51, whereas no relevant difference was observed during exponential growth (fig. 5B). Thus, in the stationary phase infected cells had a fitness advantage (fig. 5C).
Fig. 5.
—Fitness parameters of Paramecium tetraurelia infected with Caedibacter and symbiont-free cells. (A) Data points represent the mean cell density of five replicates ± SD at different time points (in hours). A nonlinear parametric regression model fits the regressions (lines). Filled circles indicate P. tetraurelia infected by Caedibacter taeniospiralis 51K, empty circles indicate symbiont-free 51 cells. Bar plots depict exponential growth rate (B) and carrying capacity (C) of infected 51K (black) and symbiont-free 51 (white). Statistically significant differences were only observed at carrying capacity (* P-value ≤0.001; unpaired t-test with Welch’s correction). Bars represent the mean of five replicates ± SD.
Infection Causes Permanent Activation of Individual HSP70 Isoforms
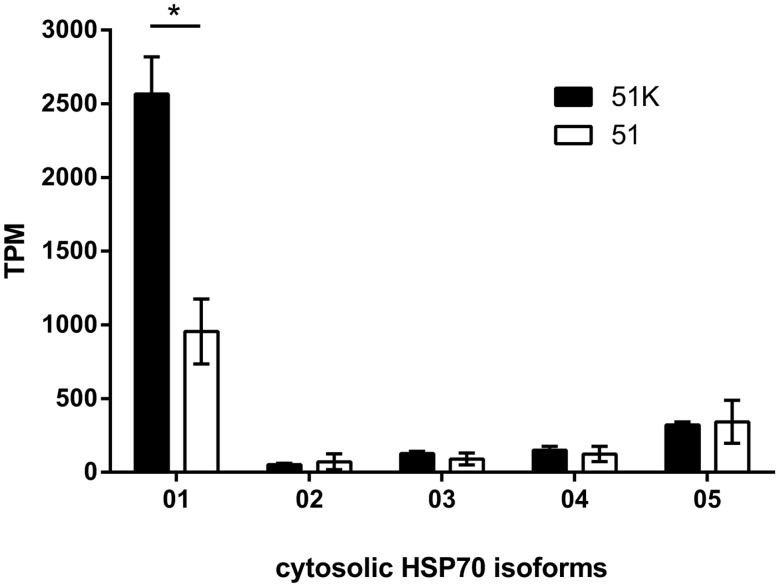
Expression of Heat Shock Protein (HSP70) genes is not only related to classical heat shock situations, it furthermore involves constitutively expressed isoforms and additionally those up-regulated in response to different stressors. In P. tetraurelia, five cytosolic HSP70 isoforms have been identified (HSP70Pt01-05; Cheaib et al. 2015). HSP70Pt01 is the major constitutively expressed in P. tetraurelia and shows the highest total abundance of transcripts in response to heat shock. This gene is significantly up-regulated in Caedibacter-harboring 51K (fig. 6).
Fig. 6.
—Cytosolic HSP70 expression of Paramecium tetraurelia. Transcripts Per Million (TPM) normalized expression values for the five cytosolic heat shock (HSP70) genes in Caedibacter-infected P. tetraurelia 51K (black) and symbiont-free 51 (white). Statistically significant differences were observed (* P-value ≤0.001; unpaired t-test with Welch’s correction).
Discussion
Comparative Transcriptomics Reveal Modulation of the Host Transcriptome Caused by Caedibacter
Differential gene expression analysis comparing the transcriptome of Caedibacter-harboring to symbiont-free cells as reference reveals that the presence of the bacterial endosymbiont causes a rather discrete modification of the host transcriptome with 2.6% of the sequenced genes classified as DEGs. In comparison, a study analyzing P. tetraurelia gene expression alterations in response to environmental changes revealed a much higher number of DEGs (starvation: ca. 11.4%, cold adaptation: ca. 8.2%) using the same significance criteria (Cheaib et al. 2015). Apparently, a discrete number of genes are involved in the host’s response to the presence of C. taeniospiralis rather than a massive modulation of the transcriptome taking place.
Up- and down-regulated DEGs were analyzed by GO enrichment analysis, revealing that up-regulated GO terms relate to several metabolic and catabolic pathways (supplementary table S1, Supplementary Material online). These observations suggest that Caedibacter-bearing 51K probably possess an increased fatty acid catabolism by peroxisomal β-oxidation as well as increased glycolysis and citrate cycle activity. Enzymes involved in each of these pathways were detected in up-regulated processes by GO enrichment analysis (table 1). Those pathways are connected by substrates and cofactors and are used, also via the electron transport chain, to generate energy. Thus, our findings suggest that Caedibacter-harboring paramecia have an increased energy metabolism by catabolizing the available resources faster and/or activating storage compounds such as fatty acids. This additional (or faster) available energy could be used by infected paramecia to sustain their cytoplasmic symbionts and their higher cell density compared with symbiont-free cells (fig. 5).
Prominent among the down-regulated DEGs (supplementary table S2, Supplementary Material online) are kinases and phosphatases comprised in the significantly enriched GO terms phosphorus metabolism (table 1) and macromolecule modification. Both enzyme types are expected to play major roles in various signaling pathways (Patterson et al. 2009). The observed down-regulation of kinases and phosphatases in infected cells compared with symbiont-free ones amounts to ∼1/6 of all DEGs with reduced expression levels. Down-regulation of kinases and phosphatases can result in a modified activity in phosphorylation-dependent signaling cascades. This suggests that symbiont-bearing 51K have altered signaling and regulatory functions compared with symbiont-free 51.
A specific group of kinases, CDKs, are comprised in the GO term cell cycle which is also enriched in down-regulated DEGs. Like in other eukaryotes, P. tetraurelia CDKs are major regulators of the cell cycle (Zhang et al. 1999; Zhang and Berger 1999; Berger 2001). Thus, down-regulated cell cycle signaling pathways could result in altered density-dependent regulation of the cell cycle via CDKs and cause modified cell division inhibition, which might partially explain the higher cell number of Caedibacter-bearing 51K (compared with symbiont-free 51) at carrying capacity as observed in the fitness assays.
Caedibacter Induces Multiple Modifications of Its Host Phenotype
The killer tests demonstrate that the analyzed lines represent killer paramecia (51K) and cells that have lost this ability (51) together with the bacterial symbionts. Paramecium may serve as a model for comparative studies addressing the function of various symbionts, as it can harbor a large variety of bacteria (Görtz and Fokin 2009) or algae (Kodama et al. 2014). Symbiont-free genetically identical lines are easily established as well as – in case of infective symbionts – infected Paramecium lines (Bella et al. 2016). Additional to the killer trait, fitness assays reveal the significant impact of Caedibacter on the cell density of its host. The obtained results differ from those of previous studies and hence stress the influence of biotic and abiotic factors such as cultivation medium and food organisms on the outcome of the Paramecium-Caedibacter symbiosis. Presumably, these factors not only alter the magnitude of the fitness impact but also shift a mutualistic interaction (fig. 5) into a parasitic one (Kusch et al. 2002; Dusi et al. 2014). Accordingly, C. taeniospiralis can cause a broad spectrum of effects ranging from harmful to beneficial.
The obligate cytoplasmic symbiont C. taeniospiralis induces a permanent up-regulation of the HSP70Pt01 isoform in its host. Similarly, HSP70Pt01 was found to be permanently up-regulated in P. tetraurelia expressing a specific surface antigen (51D, Cheaib et al. 2015). Both cases do not represent classic short-term stress situations. Thus, elevated HSP70Pt01 expression might be indicative for adaptations of the P. tetraurelia proteome in situations such as specific serotypes or infections considering the chaperone nature of HSPs. Interestingly, a similar up-regulation of HSP70 genes was observed in Paramecium bursaria and Paramecium caudatum carrying endosymbionts: The transcriptome alteration analysis of P. bursaria carrying the microalgae Chlorella sp. compared with symbiont-free P. bursaria revealed an up-regulation of individual HSP70 genes in the symbiosis (Kodama et al. 2014). Hori and Fujishima (2003) observed by RT-PCR that P. caudatum infected with the alphaproteobacterial symbiont Holospora obtusa express higher levels of HSP70 mRNA already at physiological cultivation temperature (25 °C) which was further enhanced when transferred to 35 °C. This may indicate that HSP70Pt01 is involved in the host–symbiont interaction in P. tetraurelia or it could represent a general adaptation of the host to exogenous intracellular particles.
Symbiont-Induced Modulation of Host Energy Metabolism and Evolutionary Implications
The combination of comparative transcriptomics and comparative functional analyses provides new insights into the interactions of bacteria with their unicellular host. Killer paramecia represent an evolutionary outstanding symbiosis as the symbiont provides its host with a phenotype to kill other paramecia. Although the host’s killer phenotype is well-studied in terms of the bacterial protein structure involved in intraspecific competition, the complexity of the host’s adaptation processes to infection remains hardly understood.
Here, comparative transcriptomics reveals discrete but significant gene expression differences in C. taeniospiralis-harboring 51K cells relative to symbiont-free 51. Most strikingly, enzymes of several connected metabolic pathways (β-oxidation, glycolysis, gluconeogenesis, and citrate cycle) are up-regulated in infected paramecia. The energy gained from 51K up-regulated metabolism is apparently used to increase the host cell number at carrying capacity. Caedibacter should profit from an increased amount of infected paramecia as it is vertically transmitted. Thus, the increased host cell number implies an increase also in Caedibacter population size. Modified expression of kinases and cell cycle regulatory genes could assist in increasing the host population density. As the genome of C. taeniospiralis (Zaburannyi et al. 2018) encodes various transporters, the cytoplasmic symbiont might profit even more directly from the up-regulated metabolism of its host by importing additionally available metabolites. Accordingly, 51K maintains C. taeniospiralis in exchange for the killer advantage and the symbionts maintain their intracellular niche.
Possibly, these obligate endosymbionts have evolved an indirect response to react to environmental changes by manipulating their host’s metabolism. In our fitness assays they act as mutualists and cause an increased host cell density. Further investigations will show which factors can trigger a switch from mutualistic to parasitic behavior and how those are sensed by the symbiont. Currently the true nature of this symbiosis in the environment is still undisclosed.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Authors’ Contributions
M.Sch. and M.S. designed the study. K.G. conducted the experiments and prepared the cDNA libraries. Bioinformatics and statistical analyses were performed by K.G., P.R., A.D.A., and M.H.S.
K.G., P.R., M.H.S., and M.Sch. prepared graphs and figures. G.G. performed clustering, sequencing, and data processing. M.H.S., M.S., and M.Sch. contributed to the interpretation and discussion of results. M.Sch. wrote the manuscript with contributions of K.G. and M.S. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Miriam Cheaib for kind assistance in the lab and Dr. Simon Neumann for his technical assistance in photographic artwork. This work was supported by the Research Innovation Fund of the Albert-Ludwigs University of Freiburg. The article processing charge was funded by the German Research Foundation (DFG) and the University of Freiburg in the funding programme Open Access Publishing.
Literature Cited
- Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 56(6):1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anahtar MN, et al. 2015. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42(5):965–976.http://dx.doi.org/10.1016/j.immuni.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Grossmann S, Vingron M, Robinson PN.. 2008. Ontologizer 2.0 – a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24(14):1650–1651.http://dx.doi.org/10.1093/bioinformatics/btn250 [DOI] [PubMed] [Google Scholar]
- Beier CL, et al. 2002. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl Environ Microbiol. 68(12):6043–6050.http://dx.doi.org/10.1128/AEM.68.12.6043-6050.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella C, et al. 2016. Fitness impact of obligate intranuclear bacterial symbionts depends on host growth phase. Front Microbiol. 7:2084.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JD. 2001. Riding the ciliate cell cycle – a thirty-five-year prospective. J Eukaryot Microbiol. 48(5):505–518.http://dx.doi.org/10.1111/j.1550-7408.2001.tb00186.x [DOI] [PubMed] [Google Scholar]
- Castelli M, Lanzoni O, Fokin SI, Schrallhammer M, Petroni G.. 2015. Response of the bacterial symbiont Holospora caryophila to different growth conditions of its host. Eur J Protistol. 51(1):98–108. [DOI] [PubMed] [Google Scholar]
- Cheaib M, Dehghani Amirabad A, Nordström KJV, Schulz MH, Simon M.. 2015. Epigenetic regulation of serotype expression antagonizes transcriptome dynamics in Paramecium tetraurelia. DNA Res. 22(4):293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N.. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 162(1):156–159. [DOI] [PubMed] [Google Scholar]
- Dusi E, et al. 2014. Vertically transmitted symbiont reduces host fitness along temperature gradient. J Evol Biol. 27(4):796–800.http://dx.doi.org/10.1111/jeb.12336 [DOI] [PubMed] [Google Scholar]
- Görtz HD, et al. 2009. Microbial symbionts for defense and competition among ciliate hosts In: White J, Torres M, editors. Defensive mutualism in microbial symbiosis. Boca Raton (FL: ): CRC Press; pp. 45–65. [Google Scholar]
- Görtz HD, Fokin SI.. 2009. Diversity of endosymbiotic bacteria in Paramecium In: Fujishima M, editor. Endosymbionts in Paramecium. Microbiology Monographs. Berlin, Heidelberg: Springer; pp. 131–160. [Google Scholar]
- He S, et al. 2013. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS ONE. 8(4):e61126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtz RD, et al. 2011. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 108(7):3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori M, Fujishima M.. 2003. The endosymbiotic bacterium Holospora obtusa enhances heat-shock gene expression of the host Paramecium caudatum. J Eukaryot Microbiol. 50(4):293–298.http://dx.doi.org/10.1111/j.1550-7408.2003.tb00137.x [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Suzuki A, Takagi Y.. 1998. Factors controlling the length of autogamy-immaturity in Paramecium tetraurelia. Zool Sci. 15(5):707–712.http://dx.doi.org/10.2108/zsj.15.707 [Google Scholar]
- Kirsten JH, Xiong Y, Davis CT, Singleton CK.. 2008. Subcellular localization of ammonium transporters in Dictyostelium discoideum. BMC Cell Biol. 9:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, et al. 2014. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics. 15:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J, et al. 2002. Competitive advantages of Caedibacter-infected paramecia. Protist 153(1):47–58.http://dx.doi.org/10.1078/1434-4610-00082 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550..http://dx.doi.org/10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C.. 2014. Sailfish enables alignment-free isoform quantification from RNA- seq reads using lightweight algorithms. Nat Biotechnol. 32(5):462–464.http://dx.doi.org/10.1038/nbt.2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KI, Brummer T, O’Brien PM, Daly RJ.. 2009. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 418(3):475–489. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK.. 2006. Peroxisomal β-oxidation—a metabolic pathway with multiple functions. Mol Cell Res. 1763(12):1413–1426. [DOI] [PubMed] [Google Scholar]
- Pond FR, Gibson I, Lalucat J, Quackenbush RL.. 1989. R-body-producing bacteria. Microbiol Rev. 53(1):25–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preer JR, Preer LB, Jurand A.. 1974. Kappa and other endosymbionts in Paramecium aurelia. Bacteriol Rev. 38(2):113–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollar EA, et al. 2016. Using ‘omics’ and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front Microbiol. 7:68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrallhammer M, et al. 2012. Tracing the role of R-bodies in the killer trait: absence of toxicity of R-body producing recombinant E. coli on paramecia. Eur J Protistol. 48(4):290–296.http://dx.doi.org/10.1016/j.ejop.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Schrallhammer M, Castelli M, Petroni G.. Forthcoming 2018. Phylogenetic relationships among endosymbiotic R-body producer: bacteria providing their host the killer trait. Syst Appl Microbiol. [DOI] [PubMed] [Google Scholar]
- Schrallhammer M, Schweikert M.. 2009. The killer effect of Paramecium and its causative agents. In: Fujishima M, editor. Endosymbionts in Paramecium, Microbiology Monographs. Berlin, Heidelberg: Springer; pp. 227–249. [Google Scholar]
- Schweikert M, Fujishima M, Görtz HD.. 2013. Symbiotic associations between ciliates and prokaryotes In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: prokaryotic biology and symbiotic associations. Berlin, Heidelberg: Springer: pp. 427–463. [Google Scholar]
- Shin SC, et al. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334(6056):670–674.http://dx.doi.org/10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31(4):857–871.http://dx.doi.org/10.1093/molbev/msu004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI.. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4(12):e413.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, et al. 2015. Microbes & neurodevelopment – absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 50:209–220.http://dx.doi.org/10.1016/j.bbi.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 6(7):e21800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartar A, et al. 2009. Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol Biofuels. 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaburannyi N, et al. 2018. Draft genome sequence and annotation of the obligate bacterial endosymbiont Caedibacter taeniospiralis, causative agent of the killer phenotype in Paramecium tetraurelia. Genome Announc. 6:e01418–e01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, et al. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39(2):372–385.http://dx.doi.org/10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zhang H, Adl SM, Berger JD.. 1999. Two distinct classes of mitotic cyclin homologues, Cyc1 and Cyc2, are involved in cell cycle regulation in the ciliate Paramecium tetraurelia. J Eukaryot Microbiol. 46(6):585–596.http://dx.doi.org/10.1111/j.1550-7408.1999.tb05134.x [DOI] [PubMed] [Google Scholar]
- Zhang H, Berger JD.. 1999. A novel member of the cyclin-dependent kinase family in Paramecium tetraurelia. J Eukaryot Microbiol. 46(5):482–491.http://dx.doi.org/10.1111/j.1550-7408.1999.tb06065.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.