Abstract
Centromeric identity and chromosome segregation are determined by the precise centromeric targeting of CENP-A, the centromere-specific histone H3 variant. The significance of the amino-terminal domain (NTD) of CENP-A in this process remains unclear. Here, we assessed the functional significance of each residue within the NTD of CENP-A from Schizosaccharomyces pombe (SpCENP-A) and identified a proline-rich ‘GRANT’ (Genomic stability-Regulating site within CENP-A N-Terminus) motif that is important for CENP-A function. Through sequential mutagenesis, we show that GRANT proline residues are essential for coordinating SpCENP-A centromeric targeting. GRANT proline-15 (P15), in particular, undergoes cis–trans isomerization to regulate chromosome segregation fidelity, which appears to be carried out by two FK506-binding protein (FKBP) family prolyl cis–trans isomerases. Using proteomics analysis, we further identified the SpCENP-A-localizing chaperone Sim3 as a SpCENP-A NTD interacting protein that is dependent on GRANT proline residues. Ectopic expression of sim3+ complemented the chromosome segregation defect arising from the loss of these proline residues. Overall, cis–trans proline isomerization is a post-translational modification of the SpCENP-A NTD that confers precise propagation of centromeric integrity in fission yeast, presumably via targeting SpCENP-A to the centromere.
INTRODUCTION
The centromere is a critical locus linking the chromosome with the spindle microtubule to control precise chromosome segregation (1,2). CENP-A is a centromere-specific, histone H3 variant that is essential for organizing centromeric chromatin and for epigenetically defining centromeric identity (1,3,4). CENP-A bears significant homology to other histone H3 variants; albeit, the αN helix in CENP-A is shorter than that of the canonical histone H3 structure, which may affect the flexibility of the N-terminal domain and the various DNA conformations that occur at the entrance and exit of the CENP-A nucleosome (4).
Consistent with its epigenetic role, in vitro cell-free reconstitution experiments have shown that CENP-A incorporation is sufficient for centromere and kinetochore assembly (5,6). The presence of CENP-A determines the functionality of neocentromeres formed on extra-centromeric loci (7). Consistently, disruptions in CENP-A chromatin architecture cause the loss of centromeric integrity and the formation of a chromosomal missegregation phenotype (2,8). Furthermore, recent studies have implicated CENP-A as a prognostic marker for many cancers, with reduced CENP-A function noted as an inhibitor of neoplastic proliferation (9–11).
In Drosophila, the CENP-A assembly factor CAL1 is sufficient to mediate CENP-A deposition (12,13), whereas, in humans, amphibians, fungi, and budding yeast, the deposition of new centromeric nucleosomes requires the Holliday junction recognition protein (HJURP)/Scm3 chromatin assembly factors (14–16). HJURP/Scm3 is a CENP-A-specific chaperone that mediates CENP-A deposition at centromeres (17). Histone chaperones are involved in the stepwise formation of nucleosomes and affect chromatin assembly and disassembly by depositing, replacing, or removing histones (18). In Schizosaccharomyces pombe, Scm3 binds CENP-ACnp1 to maintain chromatin integrity. However, the association between Scm3 and chromatin is uncoupled from its links with CENP-ACnp1 and it is thus unlikely to be a vital component of the CENP-ACnp1 nucleosome (19). Another histone chaperone, Sim3, bears significant homology to the histone binding proteins NASP (human), N1/N2 (Xenopus) and Hif1 (Saccharomyces cerevisiae) (20). Sim3 is necessary for the proper deposition of CENP-A at fission yeast chromosomes (20) and plays a role in the maintenance of chromatin structure (21).
Early events in centromere establishment differ from those involved in maintaining centromeric identity, with centromere establishment requiring both the N-terminal tail and the CENP-A targeting domain (CATD) of CENP-A (22). The cis-acting CATD, which is nested within the histone fold of CENP-A, offers unique conformational rigidity to CENP-A in its heterotetrameric conformation as well as to the centromere (23–25). However, recent studies have shown that the CATD is not sufficient for centromeric localization in Drosophila, and thus the role of the CATD may not be universal across species (26). In fission yeast, various other factors are reported to act in trans to orchestrate centromeric positioning of HJURP/Scm3 and of CENP-A itself, including the Mis16-Mis18 complex (27,28), Mis6 (29), the Ams2 GATA factor (30,31), and Sim3 (20,21).
The CENP-A amino (N)-terminal domain (NTD), which controls microtubule–kinetochore attachment (32,33), also epigenetically regulates centromeric identity (24,34). CENP-A in human and fission yeast also regulates the localization of CENP-B, another centromere-localizing protein that controls de novo centromeric chromatin establishment of human artificial chromosomes (24,35), as well as Cnp20/CENP-T, an inner kinetochore protein of the Constitutive Centromere-Associated Network (CCAN) family, which interacts with the outer kinetochore (34). Interestingly, the NTD is involved in a proteolysis-linked mechanism that results in the degradation of mis-targeted CENP-A in budding and fission yeast (36–38); however, the exact role of the NTD in this epigenetic control of CENP-A remains elusive (34).
Here, we sought to assess the functional significance of the NTD of S. pombe CENP-A (SpCENP-A). We identified a proline-rich ‘GRANT’ (Genomic stability-Regulating site within CENP-A N-Terminus) motif within the NTD that is essential for ensuring precise chromosome segregation. Using site-directed mutagenesis of the proline residues within this domain (GRANT-prolines), we confirmed that the proline residues indeed coordinate a CATD-independent regulation of SpCENP-A centromeric targeting. The GRANT motif coordinates the localization of SpCENP-A via its interaction with Sim3 to ensure faithful propagation of centromeric identity in a novel mode of epigenetic regulation. Further, we show that cis–trans isomerization of an essential proline residue within this GRANT domain is carried out by two FKBP-like peptidyl prolyl isomerases.
MATERIALS AND METHODS
Fission yeast manipulation and microscopy (observation of progenies)
YEA, EMM2 and SPA growth media were used (39). C-terminal gene tagging was performed as previously described (40) using strains and primers listed in Supplementary Tables S1 and S2, respectively. The correct integration of the tag sequence was ensured by PCR, and the expression of the epitope-fused gene was confirmed by immunoblotting. Individual yeast cells were separated with a dissection microscope MSM400 (Singer Instruments, Somerset, UK).
Peptide pull-down assay and co-immunoprecipitation
The peptide pull-down and co-immunoprecipitation (co-IP) assays were performed in HB buffer (22) prior to immunoblotting or mass spectrometry. Log-phase yeast cell extracts were prepared in HB buffer (25 mM Tris–HCl pH 7.5, 60 mM β-glycerol-2-phosphate, 15 mM disodium-ρ-nitrophenylphosphate hexahydrate, 0.1 mM sodium fluoride, 0.5 mM sodium orthovanadate, 5% Triton X-100, 15 mM EDTA, 15 mM MgCl2, 1 mM PMSF, 1 mM DTT and complete protease inhibitor [EDTA-free; Roche, Basel, Switzerland]). Cells were disrupted by homogenization with glass beads (Fastprep, MP Biomedicals, Santa Ana, CA, USA) in five rounds of beating for 40 s interspersed with 5 min of rest on ice. Whole cell lysates were subsequently clarified via centrifugation at 21 000 × g for 5 min at 4°C, before pre-clearing with streptavidin beads (Life Technologies, Carlsbad, CA, USA) or protein A or G sepharose beads (Life Technologies) on a rotating nutator for 1 h at 4°C. For co-IP, the antibody (mouse anti-GFP or -FLAG; Roche, Basel, Switzerland) was crosslinked in bis(sulfosuccinimidyl)suberate (Sigma-Aldrich, St Louis, MO) for 30 min, quenched with 50 mM Tris–HCl, pH 7.5 for 15 min, and then washed twice in HB buffer. The pre-cleared cell extracts were then incubated with crosslinked beads for 3 h at 4°C. For peptide IP, the streptavidin beads were removed and the whole cell extracts (WCEs) subsequently incubated with biotin-linked peptides (Mimotopes, Victoria, Australia; see Supplementary Table S3) on the nutator for 2 h at 4°C. Extracts were incubated for an additional 1 h with streptavidin beads and rotated at 4°C.
For both assays, protein-bound beads were recovered by centrifugation at 400 × g for 2 min, washed four times in 1 × HB buffer, resuspended in 50 μl 2 × Laemmli buffer containing 1/20 (v/v) β-mercaptoethanol, and denatured at 96°C for 5 min. For silver staining, one-third of the samples were resolved on 4–12% Bis–Tris NUPAGE Novex polyacrylamide gels in MES-SDS (NuPAGE) buffer (Life Technologies). The whole gel was subjected to silver staining, according to standard protocols. Protein bands of interest were excised and digested for mass spectrometry. The peptide sequences are listed in Supplementary Table S3.
Cnp1 amino acid substitution, 6 × His-tagged protein constructs, and single-copy gene replacement
Single amino acid substitution and 6 × His-tagged constructs were amplified from wild-type (WT) genomic DNA with appropriate primers and high-fidelity pfu polymerase (Bioatlas, Tartu, Estonia) as per the manufacturer's instructions. PCR products were then precipitated, gel-purified, digested with appropriate restriction enzymes (New England Biolabs, Ipswich, MA, USA), ligated into pREP41 plasmid or pET-32a plasmid (for 6 × His tag), and amplified in competent TOP10 bacterial cells (Life Technologies) using selection on LB media containing ampicillin (Sigma-Aldrich). Plasmids were recovered with a QIAprep Miniprep Plasmid extraction kit (Qiagen, Limburg, Netherlands), verified by DNA sequencing (Axil Scientific, Singapore), and then transformed into selected yeast strains. 6 × His-tagged constructs were transformed and selected using BL21 competent cells (Sigma-Aldrich) under ampicillin selection. Constructs for gene replacement in the construction of the endogenous cnp1 point mutants (cnp1-E9A to Y22A) and the cnp1–4PA strain were amplified as mentioned above except when using a set of overlapping primers harboring the mutation. The resultant construct was transformed into a yeast strain bearing an endogenous copy of the cnp1+ gene replaced with a ura4+ gene at the original locus and cnp1+HA or cnp1–1 at the extragenic lys locus. Replacement of the ura4+ gene was selected using EMM media containing 5-fluoroorotic acid (5-FOA) (MP Biomedicals). The extragenic copy of cnp1+HA or cnp1–1 was removed by backcrossing with the wild-type strain. Sequences of all strains and primers used are listed in the Supplementary Tables S1 and S2, respectively.
Mass spectrometry analysis, and protein abundance with emPAI scores
Mass spectrometry (MS) analysis was performed by the Protein and Proteomics Center (National University of Singapore). The relative abundance of an identified protein between different samples was calculated with respective emPAI scores provided by the MASCOT search results. First, the %emPAI scores of each individual protein identified was calculated by dividing its emPAI score with the sum of the emPAI scores of all the identified proteins within a sample. Then, the %emPAI scores of the same protein across different samples were directly compared to derive the relative abundance.
Expression and purification of 6 × His-tagged native proteins
BL21 cells harboring 6 × His-tagged constructs were induced with 1 mM IPTG for 4.5 h for protein expression, and then lysed as per the manufacturer's instructions using the Ni-NTA Fast-Start kit (Qiagen). Lysates were then incubated with wild-type S. pombe WCE and Ni-NTA beads and then eluted. Eluted proteins were concentrated and buffer-exchanged with 20 mM NaH2PO4, pH 6.5, using a PES concentrator 30K MWCO (Pierce, Thermo Scientific, Rockford, IL, USA).
Microscopy
For nuclear phenotype observation, log-phase cells were fixed with 1/10 (v/v) glutaraldehyde on ice for 15 min, and then rinsed thrice with cold 1 × PBS before mounting onto glass slides with an equal volume of 100 μg/ml DAPI stain (Sigma-Aldrich). For direct observation of GFP signals, log-phase cells were concentrated by filtration through GF/C glass microfiber filters (GE Healthcare, Little Chalfont, UK) and then fixed for 2 h in 100% methanol at −80°C. Cells were progressively rehydrated before mounting with 1 μg/ml DAPI for microscopy with a Nikon Eclipse Ti-E inverted microscope using Plan-Apo 100 × objective lens with oil immersion (Nikon, Tokyo, Japan). Image intensities were measured using the Nikon NIS-Elements AR software and ImageJ.
Chromatin immunoprecipitation (ChIP)
Log-phase yeast cultures were fixed in 3% paraformaldehyde (Sigma-Aldrich) for 30 min on ice. The reaction was quenched with the addition of 0.0625 (v/v) of 2.5 M glycine (Thermo Fisher Scientific, Wilmington, DE, USA) and the samples washed twice in PBS, sonicated, and lysed. The samples were then immunoprecipitated with specific antibodies, which were later bound to protein A agarose (Life Technologies, Carlsbad, CA) and protein G agarose (Roche Diagnostic, Indianapolis, IN) (1:1). The agarose was washed and reverse crosslinked by incubating for >8 h at 65°C. The immunoprecipitants were then digested with 20 mg/ml Proteinase K (Roche Diagnostics, Basel, Switzerland) for 2 h at 37°C and extracted with 1 vol phenol:chloroform:isoamyl alcohol (25:24:1) (Nacalai Tesque, Kyoto, Japan). The aqueous layer was then ethanol precipitated, dried, and analyzed with PCR using centromere-specific primers (see Supplementary Table S2). The previously published sonication method and buffer composition were closely followed (30,41).
Cell growth measurement and serial dilution assay
For single-gene copy strains, the log-phase yeast culture was diluted to an OD600 of 0.1 and incubated at 30°C with shaking. Subsequent measurements at OD600 were monitored hourly. In monitoring cell division, single yeast cells were separated via micro-manipulation with the dissection microscope MSM400 (Singer Instruments, Somerset, UK). The number of cells and divisions (per single cell) were then monitored at regular time intervals. For monitoring of cell growth, 0.2 ml of log-phase yeast culture at OD600 of 0.5 was serial diluted in multiples of 5 in the respective growth medium, followed by spotting 2.1 μl of each serial dilution onto the agar plates. Growth of cells on the plates was tracked at 30 or 33°C, over 2–7 days.
Protein turnover rate
pRep41 plasmids bearing GFP-tagged WT or GRANT-prolines substitution mutant copies of SpCENP-A were transformed into wild-type HF123 cells and selected on EMM-leu plates with thiamine repression. Gene expression was induced with the removal of the thiamine repressor for 18 h at 30°C followed by 3 h treatment with 100 μg/ml cycloheximide (Sigma-Aldrich). Cell pellet collections were performed every 30 min from the point of treatment and, thereafter, proteins were extracted and subjected to western blot analysis.
Whole cell protein extract preparation
Whole protein extracts from yeast cultures were recovered using trichloroacetic acid (TCA; Sigma-Aldrich). Log-phase yeast cultures were harvested by centrifugation, reconstituted in 400 μl of 20% TCA, and then homogenized with glass beads at 4°C for 2 min. WCEs were collected and precipitated on ice for 10 min, then subsequently pelleted at 14 000 rpm for 5 min at 4°C. Protein pellets were then washed once with 100% ethanol and resuspended in 150 μl of 1 M Tris, pH 8.8 (Sigma-Aldrich). An equal volume of 2 × Laemmli buffer and 1/20 (v/v) of β-mercaptoethanol were added to the extract, which was then heated at 96°C for 5 min before western blotting. All primary and secondary antibodies were diluted at 1:500 and 1:4000, respectively.
Nuclear magnetic resonance (NMR) spectroscopy
All NMR experiments were performed on a Bruker 800 Hz NMR spectrometer (Bruker Biospin GmbH, Ettlingen, Germany) at 25°C with 2 mM peptide (Mimotopes, Victoria, Australia) dissolved in 20 mM phosphate buffer (90% H2O + 10% D2O, pH 6.5) in the presence or absence of 0.03 mM bacterially produced Ani1 and Ani2 proteins. 256 × 512 complex points were acquired with spectral widths of 7200 × 9600 Hz in t1 × t2 dimensions and a relaxation delay of 1 s. Rotating frame nuclear Overhauser effect spectroscopy (ROESY) spectra (42) were acquired in series over a mixing time of 39, 45, 60, 75, 90 and 110 ms, with a spin-lock field strength of 4 kHz and 16 scans.
Isothermal titration calorimetry (ITC)
ITC experiments were performed on a Microcal ITC200 (Northampton, MA). Proteins and peptides were prepared in the same buffer of 20 mM Tris (pH 8.0) and 200 mM NaCl. All samples were centrifuged and degassed at room temperature before the measurement. The sample cell was filled with 100 μM Sim3 protein and the syringe was filled with 1 mM WT or 4PA peptides. Each ITC titration run was performed to saturation with 19 injections of 2 μl of peptide into the sample cell (0.206 ml) containing the Sim3 at a stirring rate of 1000 rpm at 25°C. Intervals were at 150 s, with an initial delay of 60 s. Reference power was 5. All data was analyzed by MicroCal Origin version 7.0 program.
RESULTS
A critical region within the SpCENP-A NTD ensures SpCENP-A localization for precise chromosome segregation
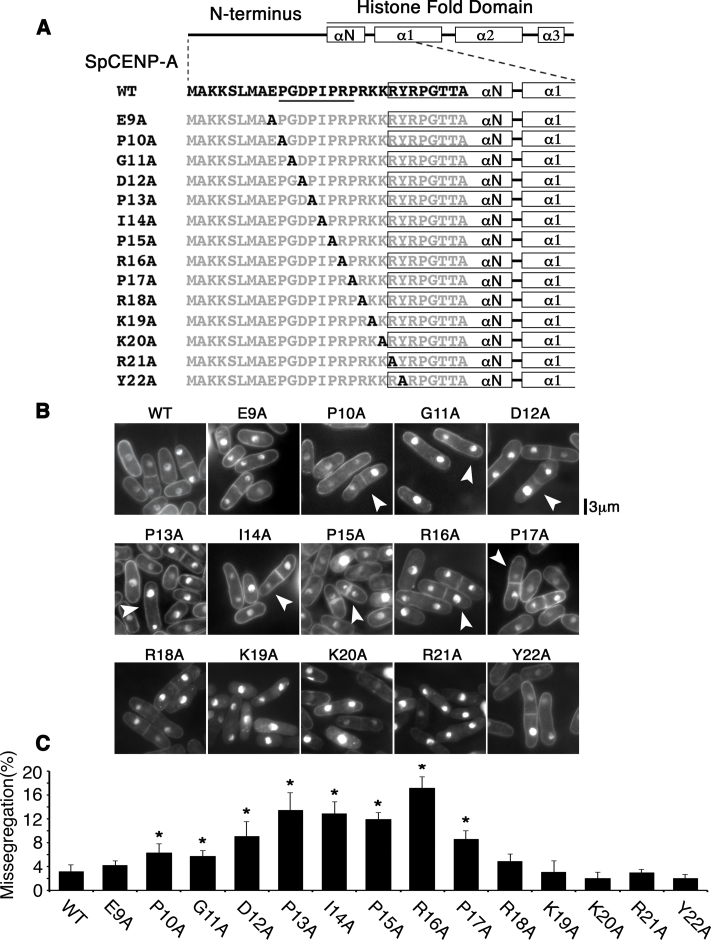
We adopted an unbiased approach by interrogating the functional significance of all amino acids (a.a.) in the SpCENP-A NTD. First, we generated 24 mutant cnp1 genes, replacing each a.a. from position 3 to 27 with alanine, and ectopically expressed the genes in wild-type (WT) cells to assess for a dominant-negative effect. We found that the ectopic expression of P10A to R16A mutant cnp1 induced chromosome missegregation and some growth retardation (Supplementary Figure S1A and B). No observable growth defects were seen for mutant genes outside this region (Supplementary Figure S1A–C).
To further ascertain the physiological implication of the mutagenesis results—especially pertaining to the residues that showed a functional link to chromosome segregation fidelity—we integrated these mutations into the endogenous copy of cnp1+ gene. We constructed 14 alanine-replacement mutants spanning residues 9 to 22 of the SpCENP-A NTD in the endogenous copy of the cnp1+ gene, so that cells only expressed the mutant proteins (Figure 1A). Because the N-terminus of the SpCENP-A gene stretches from residues 1 to 20, the 21st and 22nd residues were thus situated at the boundary between the N-terminus and the αN-helix (Figure 1A) (34). WT cells were employed as a control (Figure 1A; ‘WT’). Similar to the ectopic expression results (Supplementary Figure S1), point mutations affecting residues 10 through 17 led to significant chromosomal missegregation defects, whereas strains with mutations outside this region (cnp1-E9A, cnp1-R18A to cnp1-Y22A) showed WT-like growth and proper chromosome segregation (Figure 1B and C). From these results, we surmised that the stretch from a.a. 10 to 17 is an important regulatory region for safeguarding precise chromosome segregation. This region is hereafter referred to as the ‘Genomic stability-Regulating site within CENP-A N-Terminus’ or ‘GRANT’ motif.
Figure 1.
Proline-rich region within the SpCENP-A NTD is essential for ensuring the fidelity of chromosome segregation. (A) Schematic of the 14 site-directed single alanine substitution mutations incorporated into the endogenous copy of the cnp1+ gene. ‘A’ in black indicates the position of the alanine mutation, whereas the non-mutated residues are in gray. The GRANT residues (P10 to P17) are underlined. (B) Phenotype of mutant cells shown in (A). Numerous cells show unequal chromosome segregation. Arrowhead: Unequal chromosome segregation. (C) Quantification of phenotype in (B). Two-tailed t-test, *P < 0.05, N > 220. Bar: S.D. Bar plot indicates the mean of three experimental replicates.
Proline residues within the GRANT motif are functionally critical for the maintenance of chromosome segregation fidelity
The GRANT motif comprises four proline residues (PGDPIPRP) at positions P10, P13, P15 and P17. We performed a sequence similarity search and found that GRANT is highly conserved in CENP-A proteins, especially within the fungal kingdom, with a consensus motif of PGxPyPz(P), where x denotes D or E; y denotes I, V or L; and z denotes any amino acid. P17 and P10 appeared to be least conserved, whereas P13 and P15 appeared to be present in most of the CENP-A proteins (Supplementary Figure S2).
To understand the physiological significance of these proline residues, we constructed a cnp1 gene with quadruple P-to-A mutations (cnp1–4PA-GFP). We noticed that the introduction of this mutation affected neither the overall protein level nor the stability of the protein with respect to that of WT SpCENP-A upon ectopic expression in WT cells (Supplementary Figure S3A and B), suggesting that the GRANT-prolines do not coordinate proteolysis of the SpCENP-A protein as do proline residues in the budding yeast Cse4 NTD (36–38). Furthermore, we found that ectopic expression of cnp1–4PA-GFP could retard cell growth and result in mitotic chromosome missegregation, similar to those changes observed for SpCENP-A with a disrupted GRANT region (Supplementary Figure S3C and D).
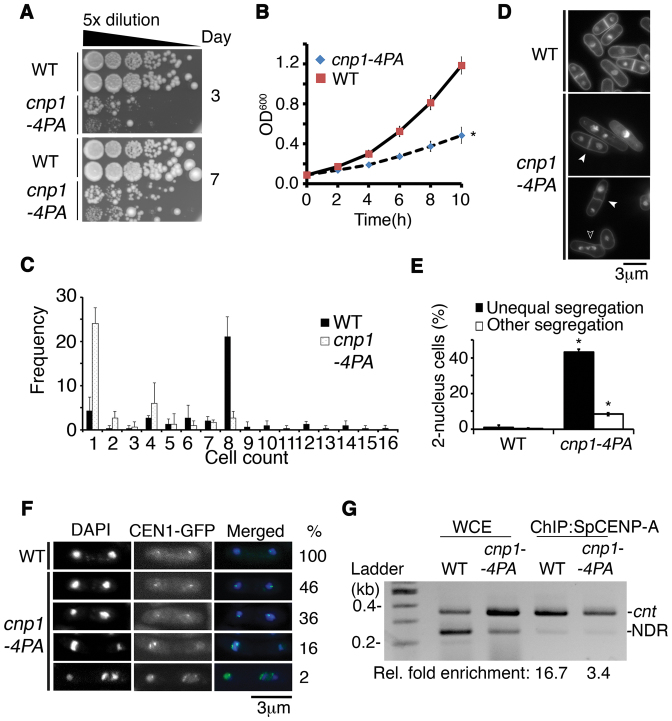
To further investigate the role of GRANT-prolines in genomic stability, we integrated a cnp1–4PA mutation into the genomic copy of cnp1+ gene at the original chromosomal locus; this mutant replaced the original gene and became the only cnp1 copy in the mutant cell (Figure 2). This cnp1–4PA mutation caused a slow-growth phenotype, with numerous cells failing to engage in cell division (Figure 2A–C). Furthermore, 43.2% of cnp1–4PA cells displayed unequal chromosome segregation, of which 16% of 2N cells harbored a missegregated chromosome 1 and exhibited two CEN1-GFP spots in a single segregated nucleus (8) (Figure 2D–F).
Figure 2.
Proline residues in endogenous SpCENP-A are essential for safeguarding precise chromosome segregation. (A) Growth retardation in the cnp1–4PA mutant at 30°C on agar plates as compared with wild type (WT). (B) Growth retardation in the cnp1–4PA mutant at 30°C in liquid media as compared with WT. Two tailed t-test, *P < 0.05. Error: S.D. Plot indicates the mean of three experimental replicates. (C) Quantification of the number of progeny cells derived from the mitotic divisions of individual cnp1–4PA cells as compared with WT cells over 10 h at 30°C. Mode of 1 and 8 were obtained for cnp1–4PA (black) and WT (dotted), respectively. N = 40. Error: S.D. Bar plot indicates the mean of three experimental replicates. (D) Chromosomal missegregation (white arrowhead) and multi-nucleated cells (open arrowhead) in cnp1–4PA as compared with WT cells. (E) Quantification of unequal nuclei segregation and other types of missegregation in cnp1–4PA as compared with WT cells. Two tailed t-test, *P < 0.05, N = 200. Error: S.D. Bar plot indicates the mean of three experimental replicates. (F) Missegregation of chromosome 1 indicated from segregation assays of GFP-labeled centromere 1 (CEN1-GFP) in cnp1–4PA strains. Percentages to the right list the frequencies of the types of missegregated CEN1-GFP spots in bi/multi-nucleated cells as a percentage of the total number (100%) of missegregated cells. N = 110. (G) ChIP assay for WT and 4PA-mutated (cnp1–4PA) cnt: SpCENP-A at the central core region of centromere 1. WCE: Whole cell extract. NDR: nucleosome-depleted region as normalization control. Representative image and average enrichment of two independent experimental replicates.
We performed chromatin immunoprecipitation (ChIP) against endogenous SpCENP-A using an antibody that recognizes SpCENP-A protein (43). Compared with WT SpCENP-A protein, we observed a 79.6% reduction in the localization of SpCENP-A-4PA at the central centromeric core region (Figure 2G). We next constructed strains hosting a GFP-fused WT cnp1+ (WT) or mutant cnp1–4PA gene at the original locus on the genome and incorporated an additional copy of HA-tagged WT cnp1+ gene at the extragenic lys1+ locus (Supplemental Figure S4A) to examine centromeric incorporation of the SpCENP-A protein. As expected, we observed a reduction in chromatin incorporation of the mutant Cnp1–4PA-GFP at the inner centromere relative to that of WT cells (Supplemental Figure S4B). In contrast, WT SpCENP-A-HA in both strains exhibited comparable binding to the inner centromeric sequence. Consistent with the reduction in ChIP signal at the centromere, we found reduced localization of the mutant Cnp1–4PA protein (30.8% reduction in mean GFP intensity; Supplementary Figure S5A–C). Taken together, these localization studies demonstrated that the disruption of the GRANT motif resulted in the partial mislocalization of SpCENP-A from centromeric chromatin, despite an intact CATD domain.
Cis–trans isomerization of the SpCENP-A NTD
To further understand the regulation of the SpCENP-A NTD, we used affinity purification and mass spectrometry to identify potential NTD-binding proteins that would bind to a biotinylated peptide of the NTD (a.a. 1 to 20 of the SpCENP-A NTD). We co-purified a protein annotated as a FK506-binding protein 39 kDa (FKBP39) peptidyl prolyl isomerase (Supplementary Figure S6A). Since proline residues can undergo cis–trans isomerization, we assessed the functional implication of this isomerase. A sequence alignment revealed high similarity between this putative FKBP39 (SPBC1347.02) protein and another fission yeast coding sequence of unknown function (SPAC27F1.06c), and both showed close sequence homology to the budding yeast proteins, which were recently linked to cis–trans prolyl isomerization in budding yeast (38,44–46) (Supplementary Figure S6B).
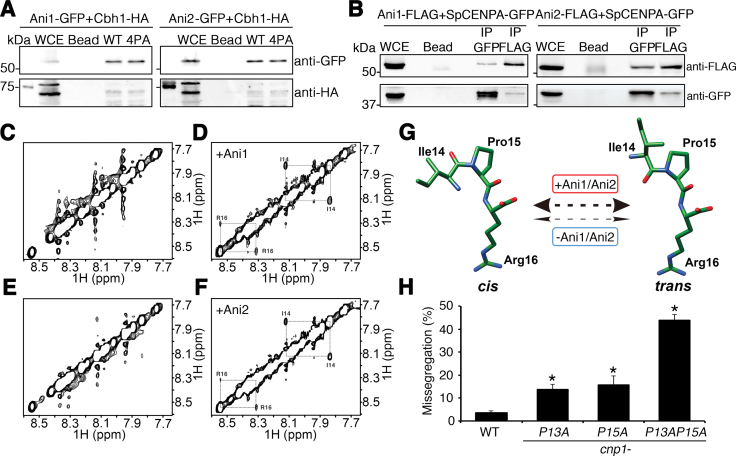
Using a peptide pulldown experiment with GFP-tagged SPBC1347.02 and SPAC27F1.06c, we confirmed their physical associations with the SpCENP-A NTD (Figure 3A), and this occurred in a GRANT-proline–independent manner. Previous studies have shown that CENP-B interacts with human CENP-A NTD (24), and this interaction was used as a positive control for the assay (Figure 3A, Supplemental Figure S7). These isomerases were hereafter referred to as SpCENP-A NTD isomerases 1 and 2 (Ani1 and Ani2), respectively. Reciprocal in vivo co-immunoprecipitation (co-IP) experiments with endogenous full-length SpCENP-A-GFP (expressed from the authentic chromosomal locus) and the FLAG-tagged isomerases confirmed the binding (Figure 3B; left Ani1, right Ani2).
Figure 3.
Cis–trans conformational change of SpCENP-A NTD by Ani1 and Ani2 peptidyl prolyl isomerases. (A) Binding of Ani1-GFP (left) and Ani2-GFP (right) to wild type (WT) and 4PA SpCENP-A amino-terminal domain (NTD) peptides. Ani1 and Ani2 were detected by immunoblotting with anti-GFP. WCE, whole cell extract; Bead, beads only control; Cbh1-HA, positive IP and loading control. (B) Co-immunoprecipitation of Ani1-FLAG (left) or Ani2-FLAG (right) to wild type SpCENP-A-GFP in vivo. IP GFP, immunoprecipitation performed with anti-GFP; IP FLAG, immunoprecipitation performed with anti-FLAG. Representative blot of three experimental replicates. (C–F) Two-dimensional 1H rotating frame Overhauser effect spectroscopy (ROESY) nuclear magnetic resonance (NMR) profiles of SpCENP-A NTD peptides in the absence (C, E) and presence (D, F) of recombinant Ani1 (C, D) and Ani2 (E, F). Diagonal cis–trans cross-peaks at Isoleucine (I)-14 and Arginine (R)-16 were obtained for both Ani1 and Ani2. (G) Structural model depicting isomerization catalyzed by Ani1 and Ani2 (thick double-headed arrow) centering on Proline (Pro)15 in the SpCENP-A NTD. The rate of isomerization is negligible in the absence of the enzyme (thin double-headed arrows). (H) Chromosome missegregation frequency of WT and cnp1-P13A, cnp1-P15A and cnp1-P13AP15A mutants. Two tailed t-test, *P < 0.05, N = 300. Error: S.D. Bar plot indicates the mean of three biological replicates.
We used NMR spectroscopy to assess whether Ani1 and Ani2 indeed possessed peptidyl prolyl cis–trans isomerase activity. 1H signals in the ROESY spectra were obtained for a 14-a.a peptide (SLMAEPGDPIPRPR) that spanned the four GRANT-proline residues. In the absence of Ani1 or Ani2, no cross-peaks were observed, indicating that the speed of the cis- and trans-conformational switch was too slow to be detected under the NMR time scale (<0.1 s−1; Figure 3C and E). In contrast, we detected two prominent and reproducible exchange cross-peaks corresponding to Ile14 and Arg16 in the presence of Ani1 or Ani2, confirming that both isomerases could regulate the conformation of the SpCENP-A NTD (Figure 3D and F). No cross peaks were found corresponding to P10 or P13. Preserving the aromatic and basic nature of the original a.a., we further showed that a peptide bearing P17FR18K mutations—but not P15FR16K mutations—produced cross-peaks (Supplementary Figure S8A–D). These observations indicate that P15 as a key residue on which the isomerases induce a conformational change (Figure 3G). These NMR analyses showed that P15 is the major isomerized proline within the GRANT motif, confirming the data observed within the detection limit of the one-dimensional ROESY analysis.
GRANT-proline residues synergistically safeguard precise mitotic chromosome segregation
Next, we sought to investigate the in vivo cooperation among the GRANT-prolines. Since P15 and P13 were among the most conserved residues, we examined the chromosomal segregation defects associated with single-point mutations in these proline resides (cnp1-P15A and cnp1-P13A) relative to double-point mutations (cnp1-P13AP15A). These mutations were incorporated into the endogenous cnp1+ gene at the authentic chromosomal locus (Figure 3H). We observed a cumulative increase in the proportion of chromosomal segregation defects in the double mutant (43.8%) over the single mutants (cnp1-P13A, 13.6%; cnp1-P15A, 15.7%), indicating that these two prolines indeed have synergistic action. Interestingly, the severity of the chromosome missegregation phenotype observed for the double mutant approximated that of cnp1–4PA, suggesting that the function of the GRANT motif is indeed mediated by synergism between the isomerized P15 and another GRANT proline(s) (Figure 3H).
Ani1 and Ani2 safeguard chromosome segregation through isomerase activity in vivo
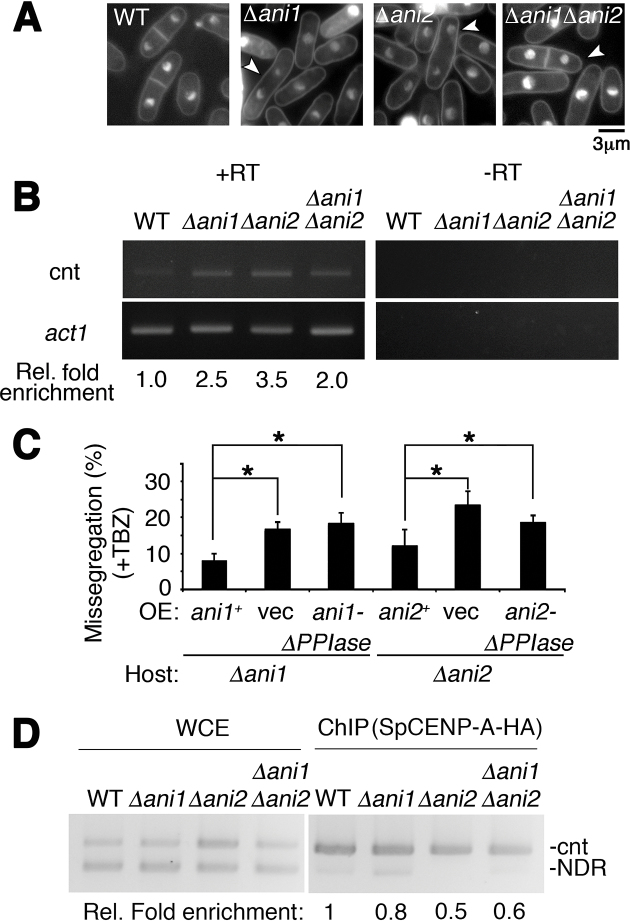
To determine whether Ani1 and Ani2 are required for chromosome segregation, we disrupted the genes encoding these two prolyl isomerases. Null mutants of ani1 (Δani1) and ani2 (Δani2) resulted in a similar chromosome missegregation phenotype as cnp1–4PA (Figure 4A); the mutants also caused a de-repression of the inner centromeric DNA sequence that is associated with a disruption to centromeric chromatin architecture (47) (Figure 4B).
Figure 4.
Catalytic function of Ani1 and Ani2 is required for regulation of precise chromosome segregation. (A) Nuclear phenotypes of wild type (WT), Δani1, Δani2 and Δani1Δani2. Cells harboring Δani1, Δani2 and Δani1Δani2 all showed unequal chromosome segregation. White arrowhead indicates missegregated bi-nucleated cells. Bar: 3 μm. (B) Transcription de-repression of centromeric DNA following the concurrent disruption of ani1 and ani2. WT, wild type; Ani, SpCENP-A NTD-isomerase. +RT, –RT: with and without reverse transcription, respectively; cnt, central core region of centromere 1; Act1, actin normalizing control. Representative gel image of two biological replicates. Rel. fold enrichment: fold enrichment relative to WT. (C) Mutant ani1 and ani2 genes containing mutation in catalytic domains cannot suppress chromosome segregation defects of Δani1 and Δani2. Frequency of the chromosome missegregation phenotype of strains expressing vector only (vec), full-length ani1+ and ani2+, and isomerase domain null (-ΔPPIase) proteins in ani1 or ani2 deletion strains. Assay was performed 18 h post-induction in EMM-leucine after thiamine wash-out at 26°C followed with 3 h treatment in thiabendazole (TBZ). Missegregation phenotypes were suppressed in Full-length but not in the isomerase domain null mutant. Vec, empty vector control. N = 150. Error: S.D. Mean of three biological replicates.Two tailed t-test. *P < 0.05. (D) ChIP assay for SpCENP-A-HA in WT, Δani1, Δani2 and Δani1Δani2 cells. cnt: SpCENP-A at the central core region of centromere 1. WCE, Whole cell extract; NDR, nucleosome-depleted region as normalization control. Representative image and average enrichment of two independent experimental replicates.
The budding yeast Fpr3 has been shown to suppress a mutant of aurora B kinase in an isomerase domain-independent manner (48). Hence, we next investigated the functional dependency of the isomerization event to mitotic chromosome segregation in vivo. We transformed Δani1 and Δani2 with WT ani1+ and ani2+ genes (under the control of the nmt41 promoter), or genes lacking the C-terminal isomerase domain (ani1ΔPPIase, ani2ΔPPIase) in the presence of thiabendazole (TBZ) (Figure 4C) (38,44). Δani1 with the empty vector showed 16.9% chromosome missegregation, which could be partially recovered (to 9.1%) by WT ani1+ overexpression. Interestingly, ani1ΔPPIase could not suppress the effects of Δani1, indicating that the isomerase activity is functionally important for safeguarding precise chromosome segregation. Similar results were also observed for Δani2 (Figure 4C). Consistent with this view, in vivo localization of SpCENP-A-HA at the inner centromere was greatly reduced in the absence of both ani1 and ani2 (Figure 4D).
Taken together, these results suggest that a defect in the isomerization of P15 by Ani1/FKBP39 or Ani2 leads to a disruption in the centromeric locus and a chromosome missegregation phenotype.
GRANT-prolines promote binding of the Sim3 SpCENP-A-loading chaperone onto SpCENP-A NTD
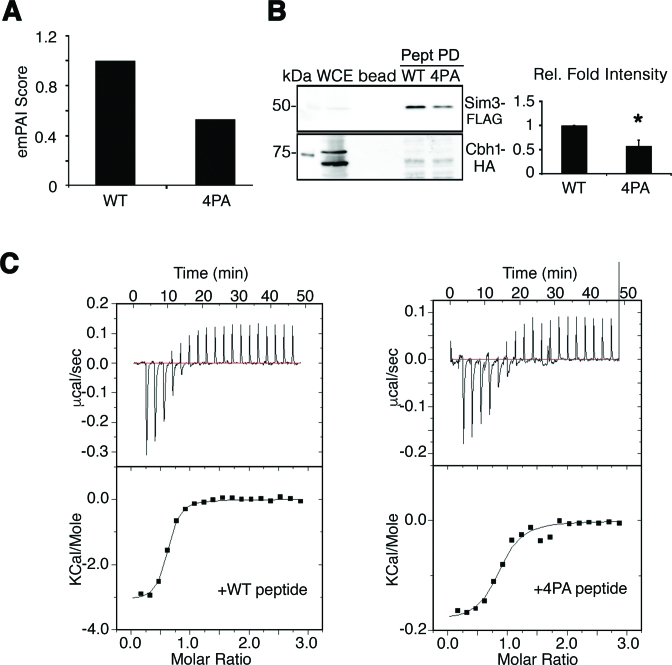
To identify other proteins that interact with GRANT-prolines in the SpCENP-A NTD, we further interrogated the affinity purification results to find a preferential association with the WT peptide and not the 4PA peptide. We identified Sim3, a human NASP-like histone chaperone (49) (Figure 5A) previously reported to incorporate SpCENP-A into centromeric chromatin (20). Further tests using exponentially modified protein abundance index (emPAI) analyses (50) confirmed that Sim3 preferentially associates with the WT over mutant SpCENP-A (normalized emPAI of 4PA = 0.54; Figure 5A) and this physical interaction was further confirmed with a biotinylated peptide pulldown assay. Cells expressing C-terminal FLAG-tagged Sim3 (Sim3-FLAG) showed preferential binding to the WT SpCENP-A NTD over that of the 4PA mutant (Figure 5B). In contrast, the peptides bearing the sequence of the histone H3 NTD failed to associate (Figure 5B, Supplementary Figure S9A); this contrasts with the findings for Ani1 and Ani2 (Supplementary Figure S9B). Finally, to ascertain a direct physical interaction between Sim3 and SpCENP-A NTD, we performed isothermal titration calorimetry (ITC) on bacterially purified recombinant Sim3 protein with SpCENP-A NTD peptides. Consistent with the co-IP results, we observed a direct binding with WT SpCENP-A NTD and Sim3 protein, with a 3-fold lower dissociation constant (Kd= 1.5μM) as compared with the 4PA mutant peptide (Kd= 4.5 μM) (Figure 5C). It is worth noting that, because these peptide pull-down assays were performed using the soluble part of the cell lysate, it is possible that the observed interactions occurred in the non-chromatin fraction.
Figure 5.
SpCENP-A loading chaperone Sim3 requires GRANT prolines for binding. (A) Normalized emPAI scores of co-precipitated Sim3 by the Cnp1-4PA (4PA) peptide relative to wild type (WT) identified through mass spectrometric quantification. Numbers are the mean of two experiments (Normalized emPAI of 4PA = 0.54). (B) Sim3-GFP physically interacts with the SpCENP-A NTD peptide but the interaction is reduced for Cnp1–4PA. Cbh1-HA is used as a positive control and loading control. Representative blot of three experimental replicates. Graph depicts quantitative band intensities. Relative fold intensities are obtained via normalization with WT. Two tailed t-test, Error: S.D., *P < 0.05. Pept PD, peptide pull down (C) Isothermal titration calorimetry (ITC) profiles between Sim3-His and WT (left) or Cnp1-4PA peptides (right).
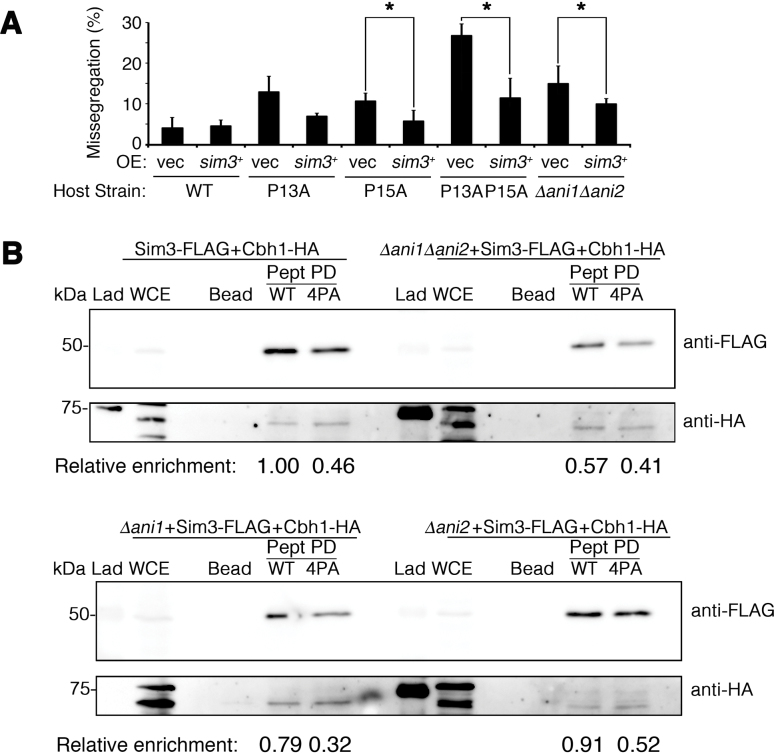
To determine what effect Sim3 binding has on the chromosome missegregation phenotype in the SpCENP-A GRANT-proline mutants, we transformed WT, cnp1-P13A (P13A), cnp1-P15A (P15A) and cnp1-P13A,P15A (P13A P15A) strains with empty vector (vec) or sim3+ under the control of the nmt41 promoter and scored the chromosome missegregation frequency. Interestingly, sim3+ overexpression could restore chromosome segregation fidelity in the cnp1 mutant strains lacking the critical GRANT-prolines. Similarly, overexpression of sim3+ was able to suppress the chromosome missegregation defects that arose from the loss of the ani1 and ani2 isomerases (Figure 6A).
Figure 6.
Sim3 is the effector for the GRANT motif under the regulation of cis–trans isomerases in safeguarding chromosome segregation. (A) Suppression of chromosome missegregation defects in mutants of GRANT motif and isomerases—P13A, P15A, P13AP15A and Δani1Δani2 by ectopic expression of sim3+His using pRep41. N > 150, two tailed t-test, *P < 0.05. Bar plot indicates the mean of three biological replicates. Vec, empty vector. WT, wild type (B) Physical interaction between Sim3 and wild type (WT) or Cnp1-4PA (4PA) NTDs in the presence or absence of Ani1 and/or Ani2 isomerases (Δani1, Δani2, Δani1Δani2). Extracts were prepared from WT and single and double mutants of Δani1 and Δani2 cells expressing Sim3-FLAG; and Cbh1-HA, Bead, beads only control; Pept PD, peptide pull down; Lad, molecular weight ladder. Cbh1-HA, positive and loading control. Representative image of experimental duplicates. Relative enrichment intensity measured against WT peptide pull down in WT cells, adjusted for loading with Cbh1-HA.
Finally, we investigated the composite effect of the isomerases and the GRANT-prolines on the interaction of Sim3 with the SpCENP-A NTD. We performed peptide pull-down assays using WT and 4PA peptides with Sim3-FLAG in WT, single and double mutants of Δani1 and Δani2. Consistent with previous observations, the interaction of Sim3-FLAG with the SpCENP-A NTD was reduced 54% (relative fold binding, 1–0.46) by the 4PA mutations. A similar reduction of ∼43% (1–0.57) was detected for Sim3-FLAG with WT SpCENP-A NTD in the absence of both ani1 and ani2 (Figure 6B, top gel and section on Δani1Δani2, WT). The 4PA peptide showed a similar level of Sim3-FLAG binding to the SpCENP-A NTD relative to the WT in the double isomerase deletion background (0.57–0.41) (Figure 6B, top gel and section on Δani1Δani2, 4PA). Single null mutation of the isomerases did not grossly affect the binding of Sim3-FLAG to the WT peptide, supporting the view that these isomerases show redundancy in conferring this binding (Figure 6B, bottom gel). Taken together, these results strongly suggest that the Ani1 and Ani2 isomerases synergistically cooperate to isomerize SpCENP-A NTD, thereby regulating Sim3 binding to the SpCENP-A NTD.
DISCUSSION
Proline-rich regions are commonly found in proteins of both prokaryotes and eukaryotes. These regions are variable in length, often repeated, and essential for driving specific signaling interactions. Indeed, one of the classical proline-rich stretches in cellular proteins is the PxxP motif, which binds to SH3 domains in proteins, such as tyrosine kinases, which are important in various cell functions (51). Proline residues are unusual amino acids because of their unique structure, and they tend to act as a helix breaker, causing a few degrees of rotation (52). Peptide bonds to proline residues can adopt both cis and trans configurations, and proline cis–trans isomerization plays a pivotal role in protein folding.
Here, we characterized the functional implications of a proline-rich stretch within the NTD of SpCENP-A (GRANT) that preserves the fidelity of chromosome segregation. We found that P15 within this domain undergoes cis–trans isomerization by two previously uncharacterized FKBP-type isomerases—denoted as Ani1 and Ani2—and show that P15 acts synergistically with the nearby P13 to ensure precise chromosome segregation, probably by regulating its interaction with the SpCENP-A loading factor, Sim3. Thus, the NTD acts as a module to localize SpCENP-A to the centromere, and this occurs alongside the canonical control of SpCENP-A localization by the CATD within the histone fold domain. Overall, our results describe a new and complementary mechanism for ensuring centromeric identity.
The NTD of CENP-A has been the focus of much recent investigation. Simulation experiments were recently used to interrogate and compare DNA flexibility between CENP-A and the canonical histone H3 variant (3,4). They confirmed previous suppositions that the ends of the CENP-A nucleosome show greater flexibility than those of the H3 nucleosome (3,4). The CENP-A NTD has also been implicated in the epigenetic regulation of centromeric identity (24,34) and in the regulation of CENP-B localization (24,35). Exactly how the NTD contributes to the epigenetic role of CENP-A, nevertheless, remained elusive (34). Although we show that GRANT prolines in the SpCENP-A NTD are essential for chromosome segregation, our site-directed mutagenesis studies show that non-proline residues within the GRANT motif may also have functional implications. For example, I14 and R16 may safeguard the structural integrity of the isomerized NTD. Alternatively, these residues may act as additional sites of regulation; although, this is not necessarily mutually exclusive. Indeed, the Arg nested in the XP(R/K) motif is methylated by the N-terminal methyltransferase NRMT1 in human and fly CENP-A (53). Whether fission yeast R16 may be similarly regulated needs to be explored in future studies.
We noted a synergistic interaction among the four proline residues of the GRANT motif, with disruptions to multiple residues resulting in more severe chromosomal missegregation phenotypes. It is possible that proline isomerization cooperatively coordinates the interactions with centromeric factors, with P15 constituting the major isomerized residue to modulate conformation of the SpCENP-A NTD. Also worth noting is that P10 and P13 comprise a PxxP motif, which, as mentioned above, is a well-characterized, interacting interface or docking site for SH3 domain-containing proteins (51). It is thus tempting to postulate that these two prolines may also act as a platform to dynamically regulate the interactions of other factors in SpCENP-A localization, of which Sim3 may be a possibility.
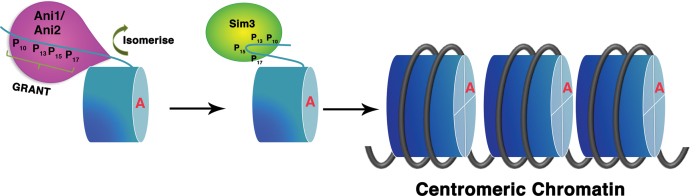
The physical interaction between SpCENP-A and its own loading factor Sim3 points to a self-propagating positive feedback mechanism: the NTD mediates the recruitment of Sim3, which, in turn, acts to incorporate SpCENP-A to safeguard the epigenetic inheritance of the centromeric chromatin. Sim3 facilitates the accumulation of newly synthesized SpCENP-A at the centromere both in the S- and G2-phases of the cell cycle (20). Since no insoluble chromatin fraction was included in the peptide pull-down experiments, the interaction of SpCENP-A and Sim3 is likely to occur preferentially in the soluble, non-chromatin-bound fraction. It is possible that the NTD may facilitate the co-accumulation of SpCENP-A and its chaperone to a ‘peri-centromeric’ space surrounding the centromeric chromatin. We speculate that, in doing so, the GRANT motif-dependent process may precede and facilitate the subsequent rapid CATD-dependent deposition of SpCENP-A by chaperones such as Sim3, Scm3 and Mis16–18 (20,27,28). Such biphasic regulation, differentially controlled by two discrete regions within SpCENP-A (GRANT and CATD) can efficiently and accurately incorporate CENP-A into the centromere within a brief window in the cell cycle of fission yeast (41,54) (Figure 7). In this scenario, the conformational change in the NTD of SpCENP-A acts as a temporal switch to promote precise inheritance of the centromeric epigenetic state. These suppositions need to be verified in future tests.
Figure 7.
Model showing isomerization-dependent targeting of SpCENP-A and Sim3 to the centromeric chromatin to facilitate the efficient deposition of SpCENP-A. Deposition of SpCENP-A into the centromeric chromatin is facilitated by the ‘GRANT’ (Genomic stability-Regulating site within CENP-A N-Terminus) motif within its amino-terminal domain (NTD) via the interaction with the SpCENP-A chaperone Sim3.
Overall, our findings point to a novel epigenetic regulation of centromeric identity. Proline-rich stretches within the GRANT motif of the N-terminus synergize to ensure precise chromosome segregation in a CATD-independent manner. This is made possible by the cis–trans isomerization of the SpCENP-A NTD to control the physical association of the SpCENP-A-loading chaperone, Sim3, which, in turn, regulates SpCENP-A centromeric localization to ensure faithful epigenetic propagation of centromeric identity.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rebecca Jackson for editing the manuscript; Robin Allshire for α-SpCENP-A antibody, Mitsuhiro Yanagida, Yasushi Hiraoka and National BioResource Project for yeast strains; Keith Gull for anti-tubulin antibody; Ron Ng and Nikon Imaging Center (Biopolis, Singapore) for help with deconvolution microscopy, and Jocelyn Shumei Wu for advice and help on protein work. Mass spectrometry was performed at the Protein & Proteomics Centre, Department of Biological Sciences, NUS, Singapore; and NMR was performed at the NMR Laboratory of Faculty of Science, NUS, Singapore.
Author Contributions: H.L.T. made all of the cnp1 constructs. H.L.T., L.S.L. and B.R. constructed the strains and performed the microscopy. H.L.T., K.K.L., B.R. and E.S.C. performed the peptide pull-down and the ChIP. H.L.T. performed the Co-IP. T.K.L and Q.L. performed proteomics. Q.L. performed the emPAI analysis. Y-C.L., Q.Y., H.L.T. and J.F. performed the NMR experiments. A.M.M.S., Y-K. M. and H.L.T. performed the ITC. E.S.C. and H.L.T. wrote the manuscript. E.S.C. conceived the study and coordinated the work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Singapore Ministry of Education Academic Research Fund (AcRF) [MOE2010-T2-1-111 to E.S.C]. Funding for open access charge: Singapore Ministry of Education.
Conflict of interest statement. None declared.
REFERENCES
- 1. Allshire R.C., Karpen G.H.. Epigenetic regulation of centromeric chromatin: old dogs, new tricks. Nat. Rev. Genet. 2008; 9:923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sekulic N., Black B.E.. Molecular underpinnings of centromere identity and maintenance. Trends Biochem. Sci. 2012; 37:220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kurumizaka H., Horikoshi N., Tachiwana H., Kagawa W.. Current progress on structural studies of nucleosomes containing histone H3 variants. Curr. Opin. Struct. Biol. 2013; 23:109–115. [DOI] [PubMed] [Google Scholar]
- 4. Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka T., Oda T., Sato M., Park S.Y. et al. . Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011; 476:232–235. [DOI] [PubMed] [Google Scholar]
- 5. Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F.. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011; 477:354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mendiburo M.J., Padeken J., Fulop S., Schepers A., Heun P.. Drosophila CENH3 is sufficient for centromere formation. Science. 2011; 334:686–690. [DOI] [PubMed] [Google Scholar]
- 7. Burrack L.S., Berman J.. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012; 20:607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takahashi K., Chen E.S., Yanagida M.. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000; 288:2215–2219. [DOI] [PubMed] [Google Scholar]
- 9. Li Y., Zhu Z., Zhang S., Yu D., Yu H., Liu L., Cao X., Wang L., Gao H., Zhu M.. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011; 6:e17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu J.J., Guo J.J., Lv T.J., Jin H.Y., Ding J.X., Feng W.W., Zhang Y., Hua K.Q.. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumour Biol. 2013; 34:2971–2975. [DOI] [PubMed] [Google Scholar]
- 11. Wu Q., Qian Y.M., Zhao X.L., Wang S.M., Feng X.J., Chen X.F., Zhang S.H.. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012; 77:407–414. [DOI] [PubMed] [Google Scholar]
- 12. Chen C.C., Dechassa M.L., Bettini E., Ledoux M.B., Belisario C., Heun P., Mellone B.G.. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 2014; 204:313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F.. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008; 183:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R.. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011; 194:229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G.. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009; 137:485–497. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Z., Feng H., Zhou B.R., Chirlando R., Hu K., Zwolak A., Miller-Jenkins L.M., Xiao H., Tiandra N., Wu C. et al. . Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011; 472:234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perpelescu M., Hori T., Toyoda A., Misu S., Monma N., Ikeo K., Obuse C., Fujiyama A., Fukagawa T.. HJURP is involved in the expansion of centromeric chromatin. Mol. Biol. Cell. 2015; 26:2742–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Das C., Tyler J.K., Churchill M.E.. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 2010; 35:476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kaganski A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X. et al. . Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009; 33:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dunleavy E.M., Pidoux A.L., Monet M., Bonilla C., Richardson W., Hamilton G.L., Ekwall K., McLaughlin P.J., Allshire R.C.. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell. 2007; 28:1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanae K., Horiuchi T., Yamakawa T., Matsuo Y., Kawamukai M.. Sim3 shares some common roles with the histone chaperone Asf1 in fission yeast. FEBS Lett. 2012; 586:4190–4196. [DOI] [PubMed] [Google Scholar]
- 22. Logsdon G.A., Barrey E.J., Bassett E.A., DeNizio J.E., Guo L.Y., Panchenko T., Dawicki-McKenna J.M., Heun P., Black B.E.. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 2015; 208:521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Black B.E., Jansen L.E., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W.. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007; 25:309–322. [DOI] [PubMed] [Google Scholar]
- 24. Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E. et al. . A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013; 15:1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. 3rd, Bassett E.A., Wood S., Black B.E., Cleveland D.W.. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009; 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moreno-Moreno O., Medina-Giro S., Torras-Llort M., Azorin F.. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr. Biol. 2011; 21:1488–1493. [DOI] [PubMed] [Google Scholar]
- 27. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M.. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004; 118:715–729. [DOI] [PubMed] [Google Scholar]
- 28. Williams J.S., Hayashi T., Yanagida M., Russell P.. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 2009; 33:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saitoh S., Takahashi K., Yanagida M.. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997; 90:131–143. [DOI] [PubMed] [Google Scholar]
- 30. Chen E.S., Saitoh S., Yanagida M., Takahashi K.. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003; 11:175–187. [DOI] [PubMed] [Google Scholar]
- 31. Takayama Y., Sato H., Saitoh S., Ogiyama Y., Masuda F., Takahashi K.. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell. 2008; 19:682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunitoku N., Sasayama T., Marumoto T., Zhang D., Honda S., Kobayashi O., Hatakeyama K., Ushio Y., Saya H., Hirota T.. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell. 2003; 5:853–864. [DOI] [PubMed] [Google Scholar]
- 33. Samel A., Cuomo A., Bonaldi T., Ehrenhofer-Murray A.E.. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9029–9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Folco H.D., Campbell C.S., May K.M., Espinoza C.A., Oegema K., Hardwick K.G., Grewal S.I., Desai A.. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 2015; 25:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W.R., Larionov V., Masumoto H.. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007; 131:1287–1300. [DOI] [PubMed] [Google Scholar]
- 36. Au W.C., Dawson A.R., Rawson D.W., Taylor S.B., Baker R.E., Basrai M.A.. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013; 194:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gonzalez M., He H., Dong Q., Sun S., Li F.. Ectopic centromere nucleation by CENP–a in fission yeast. Genetics. 2014; 198:1433–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohkuni K., Abdulle R., Kitagawa K.. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl Cis-Trans isomerase. Genetics. 2014; 196:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cam H.P., Noma K., Ebina H., Levin H.L., Grewal S.I.. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008; 451:431–436. [DOI] [PubMed] [Google Scholar]
- 40. Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. 3rd, Steever A.B., Wach A., Philippsen P., Pringle J.R.. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998; 14:943–951. [DOI] [PubMed] [Google Scholar]
- 41. Chen E.S., Zhang K., Nicolas E., Cam H.P., Zofall M., Grewal S.I.. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008; 451:734–737. [DOI] [PubMed] [Google Scholar]
- 42. Bax A., Davis D.G.. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 1985; 63:207–213. [Google Scholar]
- 43. Castillo A.G., Mellone B.G., Partridge J.F., Richardson W., Hamilton G.L., Allshire R.C., Pidoux A.L.. Plasticity of fission yeast CENP-A chromoatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007; 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hochwagen A., Tham W.H., Brar G.A., Amon A.. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005; 122:861–873. [DOI] [PubMed] [Google Scholar]
- 45. Monneau Y.R., Soufari H., Nelson C.J., Mackereth C.D.. Structure and activity of the peptidyl-prolyl isomerase domain from the histone chaperone Fpr4 toward histone H3 proline isomerization. J. Biol. Chem. 2013; 288:25826–25837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nelson C.J., Santos-Rosa H., Kouzarides T.. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006; 126:905–916. [DOI] [PubMed] [Google Scholar]
- 47. Lim K.K., Ong T.Y.R., Tan Y.R., Yang E.G., Ren B., Seah K.S., Yang Z., Tan T.S., Dymock B.W., Chen E.S.. Mutation of histone H3 serine 86 disrupts GATA factor Ams2 expression and precise chromosome segregation in fission yeast. Sci. Rep. 2015; 5:14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghosh A., Cannon J.F.. Analysis of protein phosphatase-1 and aurora protein kinase suppressors reveals new aspects of regulatory protein function in Saccharomyces cerevisiae. PLoS One. 2013; 8:e69133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finn R.M., Ellard K., Eirin-Lopez J.M., Ausio J.. Vertebrate nucleoplasmin and NASP: egg histone storage proteins with multiple chaperone activities. FASEB J. 2012; 26:4788–4804. [DOI] [PubMed] [Google Scholar]
- 50. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M.. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics. 2005; 4:1265–1272. [DOI] [PubMed] [Google Scholar]
- 51. Kay B.K., Williamson M.P., Sudol M.. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000; 14:231–241. [PubMed] [Google Scholar]
- 52. Lu K.P., Finn G., Lee T.H., Nicholson L.K.. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 2007; 3:619–629. [DOI] [PubMed] [Google Scholar]
- 53. Wu R., Yue Y., Zheng X., Li H.. Molecular basis for histone N-terminal methylation by NRMT1. Genes Dev. 2015; 29:2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mellone B.G., Zhang W., Karpen G.H.. Frodos found: Behold the CENP-a “Ring” bearers. Cell. 2009; 137:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.