Abstract
Background:
Leptomonas pyrrhocoris is a parasite of the firebug Pyrrhocoris apterus. This flagellate has been recently proposed as a model species for studying different aspects of the biology of monoxenous trypanosomatids, including host – parasite interactions. During its life cycle L. pyrrhocoris never tightly attaches to the epithelium of the insect gut. In contrast, its dixenous relatives (Leishmania spp.) establish a stable infection via attachment to the intestinal walls of their insect hosts.
Material and Methods:
This process is mediated by chemical modifications of the cell surface lipophosphoglycans. In our study we tested whether the inability of L. pyrrhocoris to attach to the firebug’s midgut is associated with the absence of these glycoconjugates. We also analyzed evolution of the proteins involved in proper lipophosphoglycan assembly, cell attachment and establishment of a stable infection in L. pyrrhocoris, L. seymouri, and Leishmania spp. Our comparative analysis demonstrated differences in SCG/L/R repertoire between the two parasite subgenera, Leishmania and Viannia, which may be related to distinct life strategies in various Leishmania spp. The genome of L. pyrrhocoris encodes 6 SCG genes, all of which are quite divergent from their orthologs in the genus Leishmania. Using direct probing with an antibody recognizing the β-Gal side chains of lipophosphoglycans, we confirmed that these structures are not synthesized in L. pyrrhocoris.
Conclusion:
We conclude that either the SCG enzymes are not active in this species (similarly to SCG5/7 in L. major), or they possess a different biochemical activity.
Keywords: LPG, Insect gut's attachment, Host-parasite interaction, Monoxenous trypanosomatids, L. pyrrhocoris, SCG enzymes
1. Introduction
Trypanosomatids are obligatory parasitic protists of the class Kinetoplastea [1]. Some of them (dixenous = having two hosts in their life cycle: Trypanosoma, Leishmania, and Phytomonas spp.) were intensely studied because they cause diseases in humans, domestic animals, and cultured plants. The vast majority of trypanosomatids is represented by monoxenous (with one host, insect, in their life cycle) species [2, 3]. For decades, these parasites were neglected and only sporadically used in physiological, biochemical, or molecular studies [4-6]. The situation has dramatically improved in recent years with realization that the evolutionary history of dixenous pathogens can be traced back to their monoxenous relatives [7], and with appreciation of the roles these parasites can play in insect communities [8, 9]. Trypanosomatids exhibit numerous unusual molecular and biochemical traits, such as RNA editing, trans-splicing, compartmentalized glycolysis, to name the most prominent ones [10-12].
The life cycles of dixenous Trypanosoma and Leishmania spp. are characterized in fine detail [13-15], while much less is known about the life cycle and molecular features of Phytomonas spp. [16, 17]. In contrast, the life cycles of monoxenous Trypanosomatidae remain virtually unknown. So far, they have been studied only in a handful of species (Leptomonas pyrrhocoris, Crithidia brevicula, C. fasciculata, Herpetomonas nabiculae, Blastocrithidia triatomae, B. papi, and few others, mainly using light microscopy techniques [18-22]. One outstanding question unifying these studies was to understand how trypanosomatids establish a stable infection, which in absolute majority of cases, seems to be achieved via the attachment of flagellates to the intestinal walls of their insect hosts. Several mechanisms were proposed which included simple physical anchoring of the parasite's flagellum, or specific interactions of modified flagella with microvilli or microvilli-free epithelial cuticle [23].
Leptomonas pyrrhocoris is a parasite of the cosmopolitan firebug, Pyrrhocoris apterus. The life cycle of this trypanosomatid is well understood and consists of developmental stages in the insect's midgut, followed by the occasional invasion of the host's hemolymph and salivary glands [24]. It is transmitted by coprophagy [18] and, supposedly, by necrophagy [25]. Importantly, at any stage of its life cycle, L. pyrrhocoris is never tightly attached to the cells, neither in the firebug’s gut nor in hemocoel [23]. The genome of this flagellate has been recently sequenced and assembled almost to chromosomal level [26]. Therefore, L. pyrrhocoris has been proposed as a model species for monoxenous trypanosomatids. Taken together, knowledge of the life cycle and the available genomic information provide us with a unique opportunity to analyze physiology of this organism and to compare it with its trypanosomatid relatives. The best candidates for this would be Leishmania spp. and Leptomonas seymouri [27]. The latter is an emerging opportunistic pathogen with sequenced genome [28-30] although the details of its life cycle and the specific host remain unknown. Such a comparison between these representatives of two distinct life styles is well justified because: i) the dixenous life cycle of Leishmania apparently evolved from a Leptomonas-like ancestor [7, 31, 32]; ii) mechanisms governing the attachment of Leishmania to the insect's gut are known in considerable detail [33-35]; iii) genomes of numerous Leishmania spp. were sequenced [27, 36] and a comprehensive comparative analysis is thus feasible.
The development of Leishmania spp. within the alimentary tract of the sand fly's host is mediated by chemical modifications of the lipophosphoglycan (LPG) on the cell surface [37-40]. This molecular complex consists of an 1-O-alkyl-2-lyso-phosphatidylinositol anchor, a conserved Gal(α1,6)Gal(α1,3)Galf(β1,3)[Glc(α1)-PO4]Man(α1,3)Man (α1,4)-GlcN(α1) glycan core region, a conserved repetitive element Gal(β1,4)Man(α1)-PO4, and a terminal cap [41]. However, the repetitive units and caps may incorporate additional stage- and species-specific modifications. For example, in L. major and L. turanica, the repetitive units are branched-off with β-1,3-galactosyl side chains (scβ1,3-Gal) [37, 42]. This modification mediates attachment of the procyclic promastigotes to the midgut of the vector Phlebotomus papatasi [34]. Upon differentiation into infectious metacyclic promastigotes, scGal is further capped with arabinose, enabling parasite's release and subsequent migration to the foregut [43]. The scβ1,3-Gal is added by members of the scGal transferase family (SCG/L/R, 14 enzymes in L. major), while the α1,2-Ara attachment to the scβGal is facilitated by two scAra transferases, SCA1/2 [44, 45].
In this work, we tested whether the inability of L. pyrrhocoris to attach to the firebug’s midgut is associated with the absence of surface lipophosphoglycans. In addition, we analyzed evolution of the SCG/L/R and SCA-encoding genes in L. pyrrhocoris, L. seymouri, and Leishmania spp.
2. Materials and Methods
2.1. Cultivation of Trypanosomatids and Experimental Infection of Pyrrhocoris apterus with Leptomonas pyrrhocoris
Leptomonas pyrrhocoris isolates PP1 [46] and ATCC 30974 were cultivated in Brain Heart Infusion (BHI) medium (Sigma-Aldrich, St. Louis, USA) supplemented with 10 µg/ml hemin, 100 µg/ml ampicillin, 100 µg/ml chloramphenicol and 50 µg/ml tetracycline at 25 °C as described previously [47]. Due to the low number of passages of the primary culture preserved in the collection, the isolate PP1 was chosen for experimental infection. Non-infected firebugs Pyrrhocoris apterus were starved for 4 days and then provided with a medium containing cultivated trypanosomatids [22]. The smears prepared from experimentally infected insects were prepared on day 10 post-infection.
For extraction and purification of GPI-anchored glycoconjugates, starter cultures represented by promastigotes of Leishmania major strain MHOM/IL/80/Friedlin and Leptomonas pyrrhocoris ATCC 30974 were grown in M199 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 50 mg/ml streptomycin, 12.5 mM glutamine, 0.1 M adenine, 5 µg/ml hemin, and 40 mM HEPES, pH 7.4 at 25 °C. Two liters of BHI medium supplemented with 27 µg/ml adenosine and 5 µg/ml hemin were seeded with 200 ml of starter cultures. Cells were grown for two days at 25 °C with agitation to a density of 5 - 7 x 107 cells/ml, as described previously [39].
2.2. Light and Transmission Electron Microscopy
Smears on slides containing life stages of either species were air-dried, fixed with ethanol and stained with Giemsa. Cells were examined using Leica DM 2500 light microscope (Leica Microsystems, Wetzlar, Germany) equipped with UCMOS14000KPA 14-Mpx camera. For transmission electron microscopy (TEM), the insect gut sections were fixed with 2.5% glutaraldehyde in 0.1 М cacodylate buffer (pH 7.4) for 1 h on ice and processed as described elsewhere [16].
2.3. Genomic Analysis and Phylogenetic Inferences
For the identification of the SCG/L/R and SCA gene family members, a BLASTP search was performed using these proteins of Leishmania major Friedlin as a query [44, 48] and annotated proteins of L. infantum JPCM5 (MCAN/ES/98/LLM-877), L. (Viannia) braziliensis M2904 (MHOM/BR/75M2904) [49], Leptomonas pyrrhocoris H10 [26] and L. seymouri ATCC 30220 [28] as a database with E-value cut-off of 10-20 and the minimal coverage of 60%. Amino acid sequences of all identified proteins were aligned using Muscle v. 3.8.31 with default parameters [50]. Poorly aligned regions of the alignment were removed using Gblocks [51] with the following parameters: -b3 = 10; -b4 = 2; -b5 = h. The SCA and SCA-like proteins were analyzed separately from the SCG/L/R family members due to the notable divergence of their sequences. The average identities within the SCA/SCA-like and SCG/L/R groups were 37.53% and 27.39%, respectively, whereas the same value between them was only 5.09%. Maximum likelihood phylogenetic trees were inferred using the IQTREE v.1.5.3 web-server with JTTDCMut + G4 model for SCG/L/R and LG + F + G4 model for SCA/SCA-like proteins, as selected by the built-in module [52]. Branch supports were assessed by bootstrapping with 100 standard replicates. Bayesian inference of phylogeny was accomplished in MrBayes 3.2.6 with analysis run for 1 million generations, sampling every 100th of them and other parameters of MCMC left as default [53]. The model of site heterogeneity was set based on IQTREE analyses, while the models of amino acid substitutions were assessed by the MrBayes (mixed amino acid model prior). The resulting models were WAG/Jones + F + G4 for SCG/L/R and Jones + F + G4 for SCA/SCA-like.
2.4. Extraction and Purification of Glycoconjugates
Glycoconjugates (LPG-like structures) from parasites were subjected to organic extraction in solvent E (H2O/ethanol/diethyl ether/pyridine/NH4OH, 15:15:5:1: 0.017). The extract was dried by N2 evaporation, dissolved in 0.1 M acetic acid/0.1 M NaCl, and applied to a column of phenyl-Sepharose (2 ml), equilibrated in the same buffer. LPG was eluted using solvent E as described elsewhere [54].
2.5. Stains-all and Immunoblotting
In order to confirm purification, purified GPI-anchored glycoconjugates (~20 µg per lane) eluted from the Phenyl-Sepharose column were resolved by SDS-PAGE and subjected to stains-all technique as described previously [55]. Samples were also transferred to nitrocellulose membrane and subjected to immunoblotting with the WIC 79.3 monoclonal antibody (1:1,000), which recognizes terminal β1,3Gal sequences that branches off the Galβ1,4Manα1-PO4 repeat units in the L. major LPG [56].
3. Results
3.1. Leptomonas pyrrhocoris Does not Attach to the Insect's Gut
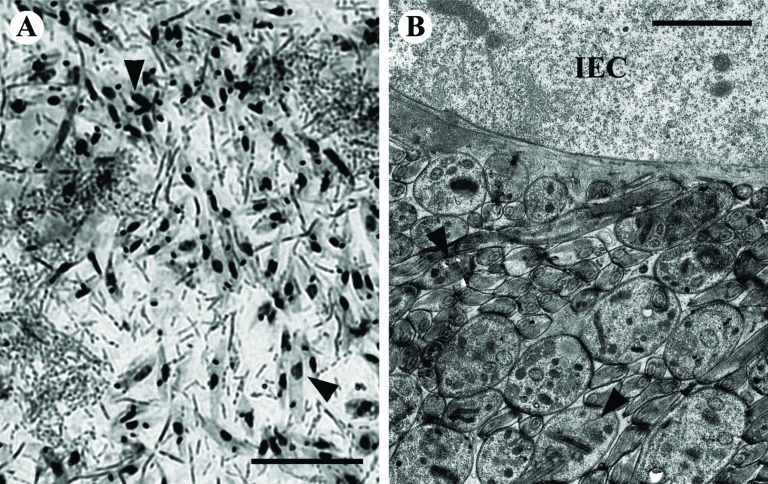
It was observed that during its life cycle L. pyrrhocoris never tightly attaches to the epithelium of the insect gut [18, 23]. To test this, we prepared smears and ultrathin sections of the P. apterus gut of specimens experimentally infected with L. pyrrhocoris, and examined them by light and electron microscopy (Fig. 1). Consistent with previous observations, we have never detected L. pyrrhocoris cells attached to the host midgut epithelium.
Fig. (1).
Leptomonas pyrrhocoris does not attach to the cells of insect's gut. Light- (A, Giemsa-stained) and transmission electron microscopy (B) of the insect's gut. Selected trypanosomatid cells are marked by arrowheads. The intestinal epithelium cell is labeled IEC. Scale bars are 20 µm (A) and 2 µm (B).
3.2. Leptomonas pyrrhocoris Encodes Almost Complete Set of Divergent SCG/L/R and SCA Enzymes
In order to analyze genes potentially involved in the biosynthesis of LPG in L. pyrrhocoris, we first identified a set of orthologs to the well-characterized SCA and SCG/L/R families of L. major Friedlin [44].
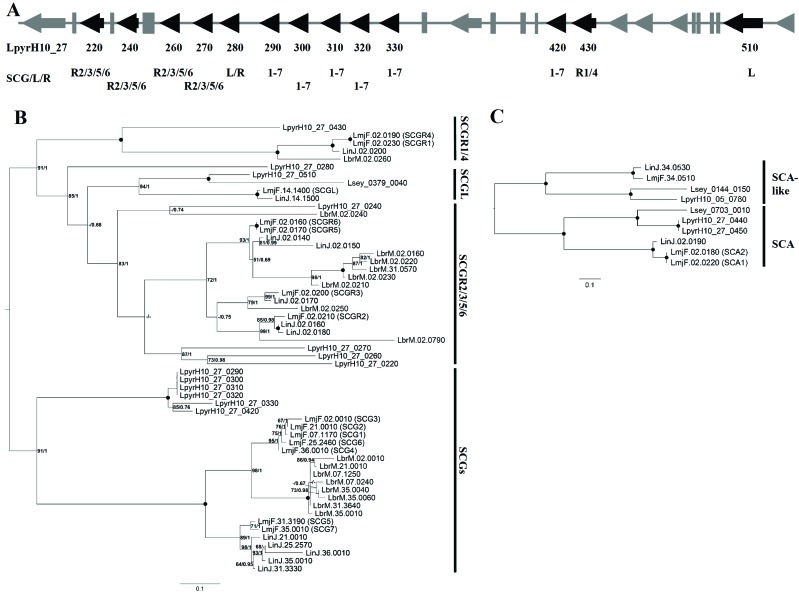
Thirteen of the SCG/L/R family genes found in L. pyrrhocoris (compared to 14 in L. major Friedlin, 12 in L. infantum JPCM5, and 17 in L. braziliensis M2904) cluster together in the same assembled contig (LpyrH10_27) and span about 90 kb (Fig. 2A). This contrasts with the situation in all Leishmania spp. analyzed so far, in which these genes are located on different chromosomes. Of note, only a single gene of this family was identified in our analysis of L. seymouri genome.
Fig. (2).
SCG/L/R orthologs in Leptomonas pyrrhocoris. (A) Schematic organization of the genomic locus containing identified SCG/L/R genes (black arrows). Grey arrows and rectangles are genes not involved in LPG biosynthesis. Numbering within LpyrH10_27 contig and predicted SCG/L/R affiliation are shown below the scheme. (B and C) Midpoint-rooted phylogenetic trees of Leishmania major SCG/L/R (B) and SCA (C) proteins along with their orthologs from Leptomonas pyrrhocoris, Leptomonas seymouri, Leishmania braziliensis, and Leishmania infantum. Numbers at nodes indicate bootstrap percentage/posterior probability. Dots mark branches with maximal statistical support. Dashes (-) indicate bootstrap support below 50%, posterior probability below 0.5, or different topology. The scale bar denotes the number of substitutions per site.
To assess relationships within this group of proteins, we built a phylogenetic tree with all sequences of the SCG/L/R family members of L. major, L. infantum, L. braziliensis, L. pyrrhocoris, and L. seymouri (Fig. 2B). There are 6 orthologs of SCG proteins in L. pyrrhocoris (LpyrH10 _27_290 – LpyrH10_27_330, and LpyrH10_27_420) compared to 7 (SCG1 – 7) in L. major. These protein sequences form a divergent monophyletic group sister to all Leishmania SCG sequences. Confirming a previous observation [44], SCG5/7 differ from other SCG proteins. Interestingly, all 8 SCG orthologs from L. braziliensis cluster together with SCG1-4/6, while 4 SCG orthologs in L. infantum belong to the SCG5/7 group (Fig. 2B). It appears that separation of SCG5/7 from the rest of the SCG proteins occurred before radiation of Leishmania spp. The species-specific clusters on the tree suggest that each species has experienced an independent expansion of these genes.
The SCG5/7 enzymes are inactive in L. major [45], implying that L. infantum uses mechanisms of attachment to the insects' gut that differ from those of L. major and L. braziliensis. Indeed, a structural analysis of the LPG molecules from L. infantum and L. braziliensis demonstrated that they are different from their L. major counterparts [39, 40, 57]. However, whether divergent L. pyrrhocoris orthologs play the same functional role in the biosynthesis of LPG (addition of the scβ1,3-Gal) as they do in L. major cannot be assessed from the genomic analysis.
The situation with other members of the SCG/L/R family is more complicated. LpyrH10_27_280 is sister to the SCGL+ SCGR2/3/5/6 cluster and therefore it cannot be assigned to any of the known groups of these proteins. Meanwhile, LpyrH10_27_220, LpyrH10_27_240, LpyrH10_ 27_260, and LpyrH10_27_270 are phylogenetically related to SCGR2/3/5/6, while LpyrH10_27_430 and LpyrH10_ 27_510 cluster together with SGR1/4 and SCGL, respectively (Fig. 2B). Again, we observed evidence of independent multiplication of these genes, similar to the case of SCGs. A single gene of the whole SCG/L/R family present in L. seymouri (Lsey_0379_0040) belongs to the SCGL group along with LpyrH10_27_510 of L. pyrrhocoris, LinJ.14.1500 of L. infantum, and LmjF.14.1400 of L. major (Fig. 2B).
Three orthologs of the SCA proteins identified in L. pyrrhocoris are phylogenetically related to the SCA1/2 (LpyrH10_27_440 and LpyrH10_27_450) and SCA-like protein (LpyrH10_05_780) of L. major (Fig. 2C). While orthologs of SCA or SCA-like proteins are present in L. seymouri (Lsey_0703_010 and Lsey_0144_150, respectively) and L. infantum (LinJ.02.190 and LinJ.34.530, respectively), they are absent in Leishmania braziliensis.
3.3. Leptomonas pyrrhocoris Does not Express LPG with β-Gal Side Chains
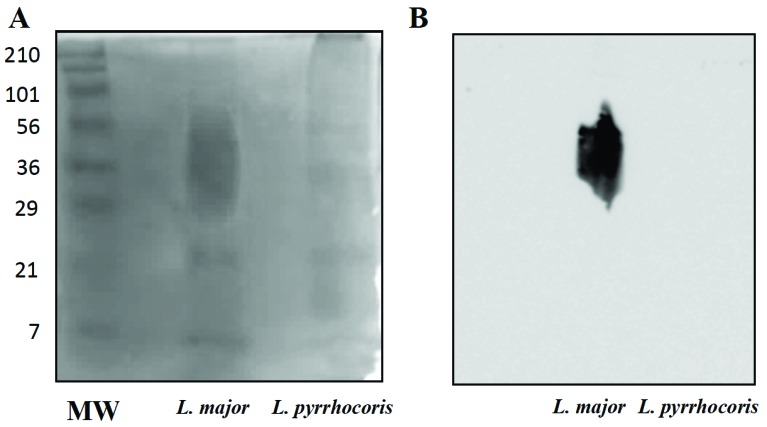
GPI-anchored LPG-like structures were purified from L. major and L. pyrrhocoris as confirmed by stains-all technique (Fig. 3A). After probing the membrane with the WIC 79.3 antibody only the LPG of L. major (positive control) was recognized (Fig. 3B). These data suggest that LPG-like structures from L. pyrrhocoris are devoid of the β-Gal side chains.
Fig. (3).
Stains-all (A) and Western blotting (B) analysis of purified GPI-anchored glycoconjugates from Leishmania major (positive control) and Leptomonas pyrrhocoris parasites. Glycoconjugates (20 µg per lane) from late-log stage cultures were probed with the WIC 79.3 monoclonal antibody (1:1000), specific for β1,3Gal residues of LPG. Protein ladder sizes on the left are in kDa.
Discussion and conclusion
The attachment of Leishmania spp. to the insects' gut is governed by lipophosphoglycans on their cell surface [41, 58]. Thus, enzymes involved in biosynthesis of LPG are important for understanding the physiology and behavior of the parasitic protists in their hosts. It is a family of side chain galactosyltransferases (SCGs) that in Leishmania determines proper LPG assembly, cell attachment and establishment of a stable infection [45]. These side chains are further modified with the arabinose moiety ensuring detachment and subsequent transmission of the Leishmania species to the vertebrate host [34, 37, 43]. The enzymes involved in this process are side chain arabinosyltransferases (SCAs) [59]. The extended SCG/L/R family in L. major Friedlin contains 14 members [44]. Our comparative analysis demonstrated that there are 12 and 17 SCG/L/R enzymes in L. infantum JPCM5 and L. braziliensis M2904, respectively. The expansion of this family in L. braziliensis (8 SCG orthologs compared to 6 in L. major and 7 in L. infantum; 5 SCGR5/6 orthologs compared to 2 in L. major and L. infantum), combined with the absence of SCAs or SCA-like proteins in this species, may be related to the differences in various Leishmania spp. life strategies. The New World leishmanias of the subgenus Viannia, such as L. braziliensis are peripylarian parasites that parasitize in the hindgut before migrating forward into the midgut. In contrast, most Leishmania species (subgenus Leishmania) are suprapylarian parasites and they develop in the midgut of their insect host [60]. Of note, enzymes of this family can be detected only within Leishmaniinae and not in other trypanosomatids with sequenced and annotated genomes. This can be explained either by high divergence of these proteins or by their absence in other species.
Another interesting aspect of our comparative analysis is clustering of SCG orthologs of L. infantum. They appear different from the related enzymes in L. major or L. braziliensis and group together with presumably inactive SCG5/7 sequences. The molecular mechanisms of LPG synthesis and modification in L. infantum remain to be investigated further. In most cases, they are governed by β-glucosyl transferases [39, 61, 62]. The tree topology demonstrating the independent expansion of SCGs implies that these proteins may be responsible for species-specific functions.
L. pyrrhocoris encodes 6 SCG enzymes (Fig. 2B), all of which are only distantly related to their SCGs orthologs in the genus Leishmania. Direct probing with an antibody recognizing the β-Gal side chains of LPG confirmed that these structures are not synthesized (Fig. 3B). There are two alternative explanations for these observations. Either the SCG enzymes are not active in L. pyrrhocoris (similarly to SCG5-7 in L. major), or they possess a different biochemical activity. We favor the second scenario because these genes in L. pyrrhocoris are quite divergent from those of Leishmania and in many cases expanded independently.
In conclusion, trypanosomatids exhibit a wide range of adaptations to the different hosts and environments that they circulate in. The survival of these parasites depends on glycoconjugates enabling them to circumvent adverse conditions they face in their hosts. Genomic and functional studies of the enzymes responsible for the assembly of those molecules may illuminate the mechanisms used by trypanosomatids for interactions with their hosts.
Ethics Approval and Consent to Participate
Not applicable.
Human and Animal Rights
No Animals/Humans were used for studies that are base of this research.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
We thank the members of our laboratories for stimulating discussions and Dr. d'Avila-Levy (Fundação Oswaldo Cruz, Rio de Janeiro, Brazil) for providing the Leptomonas pyrrhocoris ATCC 30974 strain (COLPROT-026). Support from the Czech Grant Agency awards 17-10656S to V.Y., 16-18699S to J.L. and V.Y., and 15-16406S to A.B. and Moravskoslezský kraj research initiative 01211/2016/RRC to V.Y. and A.Y.K., and the EU COST Action CM1307 to F.R.O. and J.L. is kindly acknowledged. The contribution of A.O.F. (experimental infection of insects, light and electron microscopical studies) was supported by the Russian Foundation for Basic Research grant 15-29-02734. R.P.S. and T.S.V. were supported by CNPq (305065/2016-5) and FAPEMIG (PPM-00102-16).This work was also financially supported by the Ministry of Education, Youth and Sports of the Czech Republic in the “National Feasibility Program I”, project LO1208 “TEWEP” and the EU structural funding Operational Programme “Research and Development for Innovation”, project CZ.1.05/2.1.00/19.0388.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Lumsden W.H., Evans D.A. Biology of Kinetoplastida. Vol. 1 London: Academic Press; 1976. [Google Scholar]
- 2.Podlipaev S.A. The more insect trypanosomatids under study - the more diverse Trypanosomatidae appears. Int. J. Parasitol. 2001;31(5-6):648–652. doi: 10.1016/s0020-7519(01)00139-4. [DOI] [PubMed] [Google Scholar]
- 3.Maslov D.A., Votýpka J., Yurchenko V., Lukeš J. Diversity and phylogeny of insect trypanosomatids: All that is hidden shall be revealed. Trends Parasitol. 2013;29(1):43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Bacchi J., Lambros C., Goldberg B., Hutner S.H., de Carvalho G.D. Susceptibility of an insect Leptomonas and Crithidia fasciculata to several established antitrypanosomatid agents. Antimicrob. Agents Chemother. 1974;6(6):785–790. doi: 10.1128/aac.6.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan H.F., Coombs G.H. A comparative study of the purine- and pyrimidine-metabolising enzymes of a range of trypanosomatids. Comp. Biochem. Physiol. B. 1986;84(2):219–223. [PubMed] [Google Scholar]
- 6.Schaub G.A., Jensen C. Developmental time and mortality of the reduviid bug Triatoma infestans with differential exposure to coprophagic infections with Blastocrithidia triatomae (Trypanosomatidae). J. Invertebr. Pathol. 1990;55(1):17–27. doi: 10.1016/0022-2011(90)90027-4. [DOI] [PubMed] [Google Scholar]
- 7.Lukeš J., Skalický T., Týč J., Votýpka J., Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 2014;195(2):115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Kozminsky E., Kraeva N., Ishemgulova A., Dobáková E., Lukeš J., Kment P., Yurchenko V., Votýpka J., Maslov D.A. Host-specificity of monoxenous trypanosomatids: Statistical analysis of the distribution and transmission patterns of the parasites from Neotropical Heteroptera. Protist. 2015;166(5):551–568. doi: 10.1016/j.protis.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton P.T., Votýpka J., Dostalova A., Yurchenko V., Bird N.H., Lukeš J., Lemaitre B., Perlman S.J. Infection dynamics and immune response in a newly described Drosophila-trypanosomatid association. MBio. 2015;6(5):e01356–e01315. doi: 10.1128/mBio.01356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart K., Panigrahi A.K. RNA editing: Complexity and complications. Mol. Microbiol. 2002;45(3):591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- 11.Opperdoes F.R., Butenko A., Flegontov P., Yurchenko V., Lukeš J. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J. Eukaryot. Microbiol. 2016;63(5):657–678. doi: 10.1111/jeu.12315. [DOI] [PubMed] [Google Scholar]
- 12.Záhonová K., Kostygov A., Ševčíková T., Yurchenko V., Eliáš M. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr. Biol. 2016;26(17):2364–2369. doi: 10.1016/j.cub.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 13.Fenn K., Matthews K.R. The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 2007;10(6):539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton J. Chagas disease 101. Nature. 2010;465(7301):S4–S5. doi: 10.1038/nature09220. https://www.nature.com/articles/nature09220 [DOI] [PubMed] [Google Scholar]
- 15.Bates P.A., Rogers M.E. New insights into the developmental biology and transmission mechanisms of Leishmania. Curr. Mol. Med. 2004;4(6):601–609. doi: 10.2174/1566524043360285. [DOI] [PubMed] [Google Scholar]
- 16.Frolov A.O., Malysheva M.N., Yurchenko V., Kostygov A.Y. 2016 http://www.sciencedirect.com/science/article/pii/S09324739
- 17.Jankevicius J.V., Jankevicius S.I., Campaner M., Conchon I., Maeda L.A., Teixeira M.M., Freymuller E., Camargo E.P. Life cycle and culturing of Phytomonas serpens (Gibbs), a trypanosomatid parasite of tomatoes. J. Protozool. 1989;36(3):265–271. [Google Scholar]
- 18.Frolov A.O. The life cycle of Leptomonas pyrrhocoris (Kinetoplastida, Trypanosomatidae). Zool. zhurnal. 1987;66(1):5–11. [Google Scholar]
- 19.Frolov A.O. Life cycle of Blastocrithidia miridarum (Kinetoplastida, Trypanosomatidae). Zool. zhurnal. 1987;66(5):655–661. [Google Scholar]
- 20.Mehlhorn H., Schaub G.A., Peters W., Haberkorn A. 1971 (Trypanosomatidae). Tropenmed. Parasitol. 1979;30(3):289–300. [PubMed] [Google Scholar]
- 21.Alcolea P.J., Alonso A., Garcia-Tabares F., Torano A., Larraga V. An insight into the proteome of Crithidia fasciculata choanomastigotes as a comparative approach to axenic growth, peanut lectin agglutination and differentiation of Leishmania spp. promastigotes. PLoS One. 2014;9(12):e113837. doi: 10.1371/journal.pone.0113837. journals.plos.org/plosone/article?id=10.1371/journal.pone.0113837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frolov A.O., Malysheva M.N., Ganyukova A.I., Yurchenko V., Kostygov A.Y. Life cycle of Blastocrithidia papi sp. n. (Kinetoplastea, Trypanosomatidae) in Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Eur. J. Protistol. 2017;57:85–98. doi: 10.1016/j.ejop.2016.10.007. http://www.sciencedirect.com/science/article/pii/S0932473916301249 [DOI] [PubMed] [Google Scholar]
- 23.Frolov A.O., Skarlato S.O. Fine structure and mechanisms of adaptation of lower trypanosomatids in Hemiptera. Tsitologiia. 1995;37(7):539–560. [Google Scholar]
- 24.Frolov A.O., Malysheva M.N., Kostygov A.Y. Homoxenous trypanosomatids from true bugs Pyrrhocoris apterus (L.) in the North of the Pskov region. Parazitologiia. 2014;48(6):461–471. [PubMed] [Google Scholar]
- 25.Teodoro G. Osservazioni sul Pyrrhocoris apterus L., con particolare riguardo alla flagellosi. Redia (Firenze) 1927;16:45–50. [Google Scholar]
- 26.Flegontov P., Butenko A., Firsov S., Kraeva N., Eliáš M., Field M.C., Filatov D., Flegontova O., Gerasimov E.S., Hlaváčová J., Ishemgulova A., Jackson A.P., Kelly S., Kostygov A., Logacheva M.D., Maslov D.A., Opperdoes F.R., O’Reilly A., Sádlová J., Ševčíková T., Venkatesh D., Vlček Č., Volf P., Votýpka J., Záhonová K., Yurchenko V., Lukeš J. Genome of Leptomonas pyrrhocoris: A high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci. Rep. 2016;6:23704. doi: 10.1038/srep23704. https://www.nature.com/ articles/srep23704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantacessi C., Dantas-Torres F., Nolan M.J., Otranto D. The past, present, and future of Leishmania genomics and transcriptomics. Trends Parasitol. 2015;31(3):100–108. doi: 10.1016/j.pt.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraeva N., Butenko A., Hlaváčová J., Kostygov A., Myškova J., Grybchuk D., Leštinová T., Votýpka J., Volf P., Opperdoes F., Flegontov P., Lukeš J., Yurchenko V. Leptomonas seymouri. 2015 doi: 10.1371/journal.ppat.1005127. journals.plos.org/plospathogens/article?id=10.1371/journal.ppat [DOI] [PMC free article] [PubMed]
- 29.Singh N., Chikara S., Sundar S. SOLiD sequencing of genomes of clinical isolates of Leishmania donovani from India confirm Leptomonas co-infection and raise some key questions. PLoS One. 2013;8(2):e55738. doi: 10.1371/journal.pone.0055738. journals.plos.org/plosone /article?id=10.1371/journal.pone.0055738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh S., Banerjee P., Sarkar A., Datta S., Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J. Clin. Microbiol. 2012;50(8):2774–2778. doi: 10.1128/JCM.00966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandes A.P., Nelson K., Beverley S.M. Evolution of nuclear ribosomal RNAs in kinetoplastid protozoa: Perspectives on the age and origins of parasitism. Proc. Natl. Acad. Sci. USA. 1993;90(24):11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jirků M., Yurchenko V.Y., Lukeš J., Maslov D.A. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J. Eukaryot. Microbiol. 2012;59(6):537–547. doi: 10.1111/j.1550-7408.2012.00636.x. [DOI] [PubMed] [Google Scholar]
- 33.Spath G.F., Epstein L., Leader B., Singer S.M., Avila H.A., Turco S.J., Beverley S.M. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc. Natl. Acad. Sci. USA. 2000;97(16):9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamhawi S., Ramalho-Ortigao M., Pham V.M., Kumar S., Lawyer P.G., Turco S.J., Barillas-Mury C., Sacks D.L., Valenzuela J.G. A role for insect galectins in parasite survival. Cell. 2004;119(3):329–341. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Dostálová A., Volf P. Leishmania development in sand flies: Parasite-vector interactions overview. Parasit. Vectors. 2012;5:276. doi: 10.1186/1756-3305-5-276. http://www.academia.edu/28877718/ [Leishmania_development _in_sand_flies_parasite-vector_interactions_overview]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Sayed N.M., Myler P.J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E.A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D.C., Haas B.J., Tran A.N., Wortman J.R., Alsmark U.C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J.M., Cerqueira G.C., Creasy T., Delcher A.L., Djikeng A., Embley T.M., Hauser C., Ivens A.C., Kummerfeld S.K., Pereira-Leal J.B., Nilsson D., Peterson J., Salzberg S.L., Shallom J., Silva J.C., Sundaram J., Westenberger S., White O., Melville S.E., Donelson J.E., Andersson B., Stuart K.D., Hall N. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. doi: 10.1126/science.1112181. science.sciencemag.org /content/sci/309/5733/404.full.pdf [DOI] [PubMed] [Google Scholar]
- 37.McConville M.J., Turco S.J., Ferguson M.A., Sacks D.L. Developmental modification of lipophosphoglycan during the differentiation of Leishmania major promastigotes to an infectious stage. EMBO J. 1992;11(10):3593–3600. doi: 10.1002/j.1460-2075.1992.tb05443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacks D.L., Pimenta P.F., McConville M.J., Schneider P., Turco S.J. Stage-specific binding of Leishmania donovani to the sand fly vector midgut is regulated by conformational changes in the abundant surface lipophosphoglycan. J. Exp. Med. 1995;181(2):685–697. doi: 10.1084/jem.181.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soares R.P., Macedo M.E., Ropert C., Gontijo N.F., Almeida I.C., Gazzinelli R.T., Pimenta P.F., Turco S.J. Leishmania chagasi: lipophosphoglycan characterization and binding to the midgut of the sand fly vector Lutzomyia longipalpis. Mol. Biochem. Parasitol. 2002;121(2):213–224. doi: 10.1016/s0166-6851(02)00033-6. [DOI] [PubMed] [Google Scholar]
- 40.Soares R.P., Cardoso T.L., Barron T., Araujo M.S., Pimenta P.F., Turco S.J. Leishmania braziliensis: A novel mechanism in the lipophosphoglycan regulation during metacyclogenesis. Int. J. Parasitol. 2005;35(3):245–253. doi: 10.1016/j.ijpara.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 41.de Assis R.R., Ibraim I.C., Nogueira P.M., Soares R.P., Turco S.J. Glycoconjugates in New World species of Leishmania: Polymorphisms in lipophosphoglycan and glycoinositolphospholipids and interaction with hosts. Biochim. Biophys. Acta. 2012;1820(9):1354–1365. doi: 10.1016/j.bbagen.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Volf P., Nogueira P.M., Myšková J., Turco S.J., Soares R.P. Structural comparison of lipophosphoglycan from Leishmania turanica and L. major, two species transmitted by Phlebotomus papatasi. Parasitol. Int. 2014;63(5):683–686. doi: 10.1016/j.parint.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Sacks D.L., Saraiva E.M., Rowton E., Turco S.J., Pimenta P.F. The role of the lipophosphoglycan of Leishmania in vector competence. Parasitology. 1994;108(Suppl.):S55–S62. doi: 10.1017/s0031182000075727. journals.cambridge.org/article_S0031182000075727 [DOI] [PubMed] [Google Scholar]
- 44.Dobson D.E., Scholtes L.D., Myler P.J., Turco S.J., Beverley S.M. Genomic organization and expression of the expanded SCG/L/R gene family of Leishmania major: Internal clusters and telomeric localization of SCGs mediating species-specific LPG modifications. Mol. Biochem. Parasitol. 2006;146(2):231–241. doi: 10.1016/j.molbiopara.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Dobson D.E., Scholtes L.D., Valdez K.E., Sullivan D.R., Mengeling B.J., Cilmi S., Turco S.J., Beverley S.M. Functional identification of galactosyltransferases (SCGs) required for species-specific modifications of the lipophosphoglycan adhesin controlling Leishmania major-sand fly interactions. J. Biol. Chem. 2003;278(18):15523–15531. doi: 10.1074/jbc.M301568200. [DOI] [PubMed] [Google Scholar]
- 46.Malysheva M.N., Mamkaeva M.A., Kostygov A.Y., Frolov A.O., Karpov S.A. Culture collection of parasitic protists at the Zoological Institute RAS (CCPP ZIN RAS). Protistology. 2016;10(1):26–42. [Google Scholar]
- 47.Kostygov A.Y., Grybchuk-Ieremenko A., Malysheva M.N., Frolov A.O., Yurchenko V. Molecular revision of the genus Wallaceina. Protist. 2014;165(5):594–604. doi: 10.1016/j.protis.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Dobson D.E., Kamhawi S., Lawyer P., Turco S.J., Beverley S.M., Sacks D.L. Leishmania major survival in selective Phlebotomus papatasi sand fly vector requires a specific SCG-encoded lipophosphoglycan galactosylation pattern. PLoS Pathog. 2010;6(11):e1001185. doi: 10.1371/journal.ppat.1001185. journals.plos.org/plospathogens/ article?id=10.1371/journal.ppat.1001185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M.A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S.L., Faulconbridge A., Jeffares D., Depledge D.P., Oyola S.O., Hilley J.D., Brito L.O., Tosi L.R., Barrell B., Cruz A.K., Mottram J.C., Smith D.F., Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 52.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232-235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu A.C., Freitas M.A., Silva Sde O., Nogueira P.M., Soares R.P., Pesquero J.B., Gomes M.A., Pesquero J.L., Melo M.N. Genetic differences between two Leishmania major-like strains revealed by suppression subtractive hybridization. Mol. Biochem. Parasitol. 2015;203(1-2):34–38. doi: 10.1016/j.molbiopara.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Mookherjee N., Pearson T.W. Trypanosoma simiae and Trypanosoma congolense: Surface glycoconjugates of procyclic forms-the same coats on different hangers? Exp. Parasitol. 2002;100(4):257–268. doi: 10.1016/s0014-4894(02)00023-1. [DOI] [PubMed] [Google Scholar]
- 56.Kelleher M., Bacic A., Handman E. Identification of a macrophage-binding determinant on lipophosphoglycan from Leishmania major promastigotes. Proc. Natl. Acad. Sci. USA. 1992;89(1):6–10. doi: 10.1073/pnas.89.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soares R.P., Margonari C., Secundino N.C., Macedo M.E., da Costa S.M., Rangel E.F., Pimenta P.F., Turco S.J. 2010 doi: 10.1155/2010/439174. https://www.hindawi.com/journals/bmri/2010/439174/abs/ [DOI] [PMC free article] [PubMed]
- 58.Sacks D.L., Modi G., Rowton E., Spath G., Epstein L., Turco S.J., Beverley S.M. The role of phosphoglycans in Leishmania-sand fly interactions. Proc. Natl. Acad. Sci. USA. 2000;97(1):406–411. doi: 10.1073/pnas.97.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobson D.E., Mengeling B.J., Cilmi S., Hickerson S., Turco S.J., Beverley S.M. Identification of genes encoding arabinosyltransferases (SCA) mediating developmental modifications of lipophosphoglycan required for sand fly transmission of Leishmania major. J. Biol. Chem. 2003;278(31):28840–28848. doi: 10.1074/jbc.M302728200. [DOI] [PubMed] [Google Scholar]
- 60.Lainson R., Ward R.D., Shaw J.J. Leishmania in phlebotomid sandflies: VI. Importance of hindgut development in distinguishing between parasites of the Leishmania mexicana and L. braziliensis complexes. Proc. R. Soc. Lond. B Biol. Sci. 1977;199(1135):309–320. doi: 10.1098/rspb.1977.0141. rspb.royalsocietypublishing.org/content/199/1135/309 [DOI] [PubMed] [Google Scholar]
- 61.Mahoney A.B., Sacks D.L., Saraiva E., Modi G., Turco S.J. Intra-species and stage-specific polymorphisms in lipophosphoglycan structure control Leishmania donovani-sand fly interactions. Biochemistry. 1999;38(31):9813–9823. doi: 10.1021/bi990741g. [DOI] [PubMed] [Google Scholar]
- 62.Coelho-Finamore J.M., Freitas V.C., Assis R.R., Melo M.N., Novozhilova N., Secundino N.F., Pimenta P.F., Turco S.J., Soares R.P. Leishmania infantum: Lipophosphoglycan intraspecific variation and interaction with vertebrate and invertebrate hosts. Int. J. Parasitol. 2011;41(3-4):333–342. doi: 10.1016/j.ijpara.2010.10.004. [DOI] [PubMed] [Google Scholar]