Abstract
Melanoma treatment with the BRAF V600E inhibitor vemurafenib (VMF) provides therapeutic benefits but the common emergence of drug resistance remains a challenge. We generated A375 melanoma cells resistant to VMF with the goal of investigating changes in miRNA expression patterns that might contribute to resistance. Increased expression of miR-204-5p and miR-211-5p occurring in VMF-resistant cells was determined to impact VMF response. Their expression was rapidly affected by VMF treatment through RNA stabilization. Similar effects were elicited by MEK and ERK inhibitors but not AKT or Rac inhibitors. Ectopic expression of both miRNA in drug-naive human melanoma cells was sufficient to confer VMF resistance and more robust tumor growth in vivo. Conversely, silencing their expression in resistant cells inhibited cell growth. Joint overexpression of miR-204-5p and miR-211-5p durably stimulated Ras and MAPK upregulation after VMF exposure. Overall, our findings show how upregulation of miR-204-5p and miR-211-5p following VMF treatment enables the emergence of resistance, with potential implications for mechanism-based strategies to improve VMF responses.
Keywords: Melanoma, miRNA, BRAF inhibitor, resistance, MAP kinases
Introduction
The BRAF V600E mutation is the most prevalent genetic alteration in malignant melanoma, and the focus of recently-developed BRAF inhibitors (BRAFi), such as vemurafenib (VMF) and dabrafenib (1-3). Both agents have provided substantial benefits for melanoma patients, but a major challenge in melanoma treatment with mitogen-activated protein kinase (MAPK)-targeted therapy is an almost universal emergence of resistance that leads to patient relapse. The most frequent mechanisms involved in BRAFi resistance of melanoma cells converge in the reactivation of the MAPK pathway usually following NRAS mutations (4), alterations in BRAF splicing (5) as well as BRAF amplification (6,7). Another signaling route mediating melanoma resistance to BRAFi is the PI3K-Akt pathway, which becomes hyperactivated in some patients(8). Yet, a significant portion (40%) of tumors displays unknown resistance mechanisms (9) that cannot be accounted for genetic alterations (10).
The class of small non-coding RNAs called microRNAs (miRNAs) has emerged as key post-transcriptional regulators in tumor progression. Mature miRNAs are 20-30 nucleotide-long RNAs that by targeting mRNA transcripts keep the transcriptome under tight control. miRNAs base-pair to partially complementary motifs in target mRNAs, usually in the 3′ UTR, leading to translational repression or exonucleolytic mRNA decay (11). The first indication that miRNAs play important roles in cancer came from an early study showing that the miR-15/16 cluster is frequently deleted in chronic lymphocytic leukemia, therefore implicating miRNAs as tumor suppressors (12). Moreover, transgenic expression of miR-21 initiates lymphomagenesis in mice (13). Despite a more frequent pattern of reduction in the levels of miRNAs in cancer, several miRNAs are upregulated and play oncogenic roles, which have led to call them oncomiRs, such as the miR-17/92 cluster, which is upregulated in several cancer cell types (14).
Large-scale expression profiling and deep-sequencing approaches have revealed that miRNAs play pivotal roles in melanoma progression. Some of these miRNAs have tumor suppressor roles, such as let-7b and miR-137 (15,16), whereas other act as oncomiRs, including miR155, miR-30b/30d and miR-182 (17-19). Importantly, miR-137 expression correlates with melanoma's patient clinical outcome, with lower miR-137 levels associated to shorter survival of Stage IV patients (20). Various miRNAs control melanoma cell invasion and metastasis, including the miR-211 (21).
Several miRNAs have been linked to resistance responses in different cancers (22), but only few recent studies have so far addressed the possible involvement of miRNAs in BRAFi resistance of melanoma. Thus, miR-200c and miR-7 have been shown to be reduced in BRAFi-resistant cells (23,24). In the present study we performed RNA-seq analyses comparing miRNA expression in parental and VMF-resistant melanoma cells, and identified and characterized selected miRNAs which contribute to BRAFi resistance.
Materials and Methods
Cells and antibodies
The human melanoma cell line A375 was latest authenticated in August 2017 at Secugen (Madrid, Spain) by short tandem repeat analysis. The melanoma cell lines SK-Mel-103, SK-Mel-28 and SK-Mel-147 were gifts from Dr. Marisol Soengas (Centro Nacional de Investigaciones Oncológicas, Madrid; April 2014), and were not authenticated in our laboratory. All cell lines were used within 5-50 passages of thawing the original stocks, were tested every 3 months for mycoplasma contamination, and cultured in DMEM medium supplemented with 10% fetal bovine serum (Gibco, Paisley, UK) (complete medium). Vemurafenib-resistant A375 cells (A375-VR) were derived from parental A375 cells by treatment with sequential increases of vemurafenib (Selleckchem, Houston, TX) concentrations, from 10 nM to 1.3 μM, and were finally maintained as an uncloned resistant cell population in complete medium with 1.3 μM of VMF. We also obtained A375 cells growing with the MEK inhibitor trametinib (Selleckchem) (40 nM; A375-TR).
Vectors and lentiviral-mediated gene transfer
Lentiviral vectors carrying miRNA precursor transcripts (H-miR-204-5p or H-miR-211-5p) (System Biosciences, Palo Alto, CA), or antisense miRNA sequences (Zip-mIR-140-3p; System Biosciences) were used to stably overexpress mature microRNAs or inhibit the endogenous microRNAs, respectively. Pre-miR and anti-miR-scramble sequences (H-scr and Zip-scr) were used as negative controls (System Biosciences). For virus production, HEK-293FT cells were transfected with H-miR or Zip-miR vectors, pPAX2 and pMD2G using lipofectamine 2000 (Invitrogen, Carlsbad, CA). Viruses were collected and filtered 48h after transfection, and infection of A375 or SK-Mel-28 cells was performed for 72h using polybrene (Millipore). Cells were subsequently assessed by real-time quantitative PCR (qPCR). Triple-miRNA transductants were derived from H-miR-204 cells that were co-infected with H-miR-211 and Zip-miR-140 viruses using polybrene. SK-Mel-28 double-miRNA transductants were derived from SK-Mel-28 H-miR-211 cells that were infected with H-miR-204 viruses.
Oligonucleotide and siRNA transient transfection
miRIDIAN microRNA Hairpin Inhibitors (Dharmacon, Lafayette, CO) for miRNA-204-5p and miRNA-211-5p (40 nM) were transfected using lipofectamine 2000 according to manufacturer's protocol. miRIDIAN microRNA Hairpin Inhibitor Negative Control #1 (Dharmacon) was used as negative control. siRNAs were transfected using Interferin (Polyplus Transfection, Illkirch, France), following manufacter's instructions. PAX6 and MITF SMARTpool siRNAs were purchased from Dharmacon. Sequences for control and STAT3 siRNAs (Sigma-Aldrich) are provided in Supplementary Table S1A. Transfection efficiency was monitored by qPCR at 48 h.
Cell proliferation
Cells were seeded in complete medium into 96- or 6-well plates, treated for different times with VMF, and analyzed by staining with crystal violet and measurement of absorbance at 590 nm after being dissolved with 15% acetic acid. For MTT assays, 8000 or 5000 cells were seeded with or without serum in triplicates the day before treatment, and after 24h or 48h, respectively, 10 μl of MTT reagent (Sigma-Aldrich; 5 mg/ml) were added for 1.5 h. MTT was solubilized using DMSO and plates were read at 540 nm.
Western blotting and GTPase assays
Following cell lysis (25), proteins were resolved by SDS/PAGE, transferred to PVDF membranes and detected by chemiluminescence. Densitometry of the resulting bands was performed using ImageJ software. GTPase assays to detect the active forms of Ras were performed as described (25), using GST-RAF-RBD and immunoblotting with anti-Ras antibodies.
Real-time quantitative PCR
Total RNA was extracted using miRNeasy Mini Kit (Qiagen, Valencia, CA). To analyze miRNA expression, we used the miRNA-specific Taqman MicroRNA Assay Kits (Applied Biossystems, Foster City, CA). miRNA-enriched RNA was first reverse-transcribed with the Taqman microRNA Reverse Transcription Kit (Applied Biossystems), and quantitave PCR was performed in triplicate with corresponding Taqman PCR primers (Applied Biosystems), and Taqman Universal Master Mix, according to manufacturer's instructions. The RNU44 small RNA was used for normalization. For mRNA expression analyses, reverse transcription was performed using M-MLV-RT (Promega, Madison, WI), and quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). Assays were performed in triplicate and results normalized for the expression levels of TBP (TATA-binding protein). Quantitative PCR was analyzed using the LightCycler 480 (Roche). Oligonucleotide sequences are provided in Supplementary Table S1B.
Small RNA-Seq and data analysis
RNA was extracted using the mirVana miRNA Isolation kit (Thermo Fisher Scientific, Waltham, MA). Small RNA sequencing was performed in duplicate at the Genomics Unit of the Centro Nacional de Investigaciones Cardiovasculares (Madrid), using Next Generation Sequencing (NGS, Illumina HiSeq 2500, Illumina, San Diego, CA). For NGS data analysis, 3′ adapters were trimmed from sequencing reads using CutAdapt 1.7.1.Software. The remaining reads were mapped to known human mature miRNAs database (miRBase21) using Bowtie version 1.0.0 (global alignment), where 100% identity between reads and known miRNAs sequences was required. The total read count for each sample was scaled relative to the library size. Differential expression analysis between samples was performed using DESeq2 R package (26) or BIOSAGE library (27). Differentially-expressed miRNAs between samples were considered using absolute Log2FC (fold change) criteria of 0.9. p-values for multiple tests were calculated using Benjamini-Hochberg correction and only those miRNAs with p<0.05 values were selected for further analyses. GEO (Gene Expression Omnibus) accession number for the RNA sequencing: GSE107576.
Animal studies
For xenografting studies we followed the described method (28). The Consejo Superior de Investigaciones Científicas Ethics Committee approved the protocols used for experiments with mice. Briefly, NOD/SCID/IL2gR-/- (NSG) mice were subcutaneously or intravenously inoculated with 1×106 parental or A375-VR cells in 0.2 ml PBS. Subcutaneous tumor growth was inspected on a daily basis and tumor volumes measured until day 36, when all mice were sacrificed. Intravenously-injected mice were sacrificed when signs of respiratory stress were noted and/or when weight sharply decreased. Lung and liver metastases were excised, minced and filtered through 40 μm filters (BD Biosciences). Isolated melanoma cells were cultured for 10-15 days before testing them in MTT assays. For experiments involving VMF administration, 1.5 × 106 H-Scr or triple-miR A375 cells were subcutaneously inoculated, and after 20 days (tumor volume approximately 100 mm3), NSG mice were randomized in four groups and daily treated intra-peritoneally with vehicle (5% DMSO+10% 2-hydroxypropyl-β-cyclodextrin) or VMF (25 mg/kg). Tumor volume was measured three times per week.
Statistical analyses
Unless otherwise indicated, mean values and standard deviations are representative of one of three independent experiments. Data were analyzed by Student's T test or one-way ANOVA, followed by Tukey–Kramer multiple comparison test. In all analyses the minimum acceptable level of significance was p<0.05.
Results
Characterization of vemurafenib-resistant melanoma cells
As reported (1), A375 melanoma cells carrying the BRAF V600E mutation were sensitive to vemurafenib as well as to the MEK inhibitor trametinib (TMT) (Supplementary Fig. S1A). Unlike A375, SK-Mel-103 cells expressing wild type (wt) BRAF and Q61R N-Ras were insensitive to VMF concentrations up to 1 μM, while they were sensitive to TMT. Accordingly, Erk1/2 activation in A375 cells was gradually lost at increasing concentrations of VMF or TMT, enhanced in SK-Mel-103 cells exposed to VMF, and blocked when these cells were incubated with TMT (Supplementary Fig. S1B).
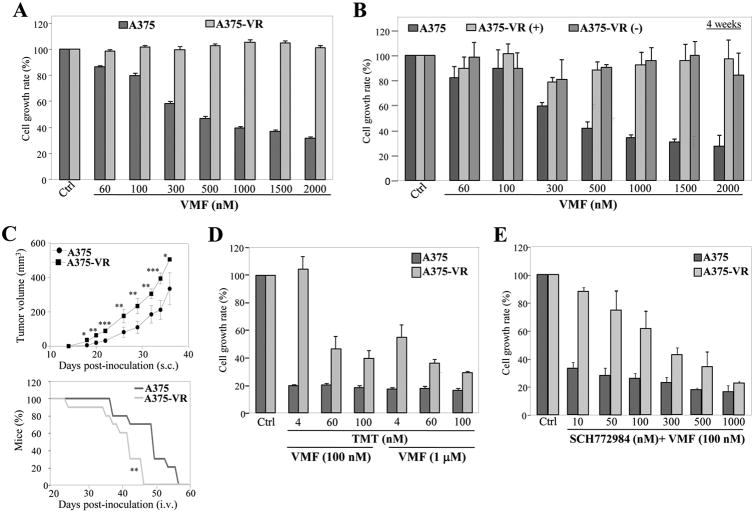
A375 cells were exposed to sequential VMF increases to a final concentration of 1.3 μM, rendering a cell population resistant to up to 2 μM VMF (Fig. 1A). When VMF-resistant cells (A375-VR) were cultured without VMF for up to 4 weeks and then re-plated in the presence of VMF, they remained fully resistant (Fig. 1B; Supplementary Fig. S1C). Determination of in vivo tumor cell growth using subcutaneously-inoculated NSG mice revealed that A375-VR tumors grew faster than parental cell tumors (Fig. 1C, top). Furthermore, based on signs of respiratory stress and/or sharply decrease of weight, mice intravenously injected with A375-VR cells were sacrificed significantly sooner than those inoculated with parental cells (Fig. 1C, bottom). A375 and VMF-resistant cells displayed similar metastasis degrees to both lung and livers (66+/-11 and 16+/-5 for A375, and 57+/-16 and 18+/-6 for A375-VR, respectively). Cell proliferation assays showed that cells extracted from lung and liver metastases from mice injected with A375-VR cells retained resistance to VMF relative to cells from metastases of mice inoculated with parental cells (Supplementary Fig. S1D).
Figure 1. Characterization of vemurafenib-resistant melanoma cells.
(A) Cells were tested in MTT assays (48 h) in the absence (Ctrl) or presence of the indicated concentrations of vemurafenib (VMF) (n=4). (B) A375-VR cells were incubated for 4 weeks without (-) or with (+) VMF (1.3 μM), and subsequently subjected for 48 h to MTT assays as in (A). Parental A375 are shown as control. (C) Cells were subcutaneously (top) or intravenously (bottom) inoculated into NSG mice in the absence of drug treatment, and tumor growth and percentage of alive mice, respectively, were assessed. (n=9-10 mice/condition; ***p<0.001, **p<0.01, *p<0.05). (D, E) Cells were incubated for 48 h without (Ctrl) or with the indicated concentrations of VMF, TMT or SCH772984 (n=3).
Cells resistant to VMF also displayed partial resistance to TMT and to the Erk1/2 inhibitor SCH772984, whereas they were sensitive to the Akt inhibitor triciribine (Supplementary Fig. S2A-C). Importantly, A375-VR cells displayed resistance to VMF combined either with TMT or SCH772984 (Fig. 1D and E).
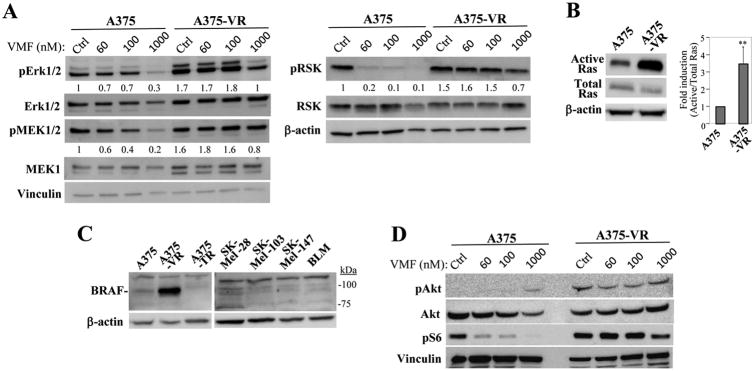
Although basal Erk1/2 and MEK levels are slightly increased in A375-VR cells relative to parental ones, nevertheless VMF-resistant cells showed higher levels of activation of MEK, Erk1/2 and its downstream effector RSK than parental counterparts, which was linked to stimulation of the GTPase Ras (Fig. 2A, B). Furthermore, we detected overexpression of 90 kD BRAF in VMF-resistant cells which was not observed in A375, A375-TR, the BRAF wt SK-Mel-103 and SK-Mel-147, and in BRAF wt/N-Ras wt BLM melanoma cells (Fig. 2C). Sequence analyses confirmed that resistant cells remained mutant V600E for BRAF and wt for NRAS (not shown). We did not detect diminished levels in A375-VR cells of SPRED, an inhibitor of MAPK activation (29), or increased expression of COT, whose overexpression is associated to resistance to BRAFi (7) (Supplementary Fig. S2D). The partial resistance of A375-VR cells to TMT and to SCH772984 was linked with sustained Erk1/2 and MEK activation (Supplemental Figs. S2E and S2F). In addition to enhanced MAPK activation, the PI3-K pathway was also activated in A375-VR cells, as revealed by increased Akt and S6 phosphorylation (Fig. 2D).
Figure 2. Analysis of Ras-MAPK and PI3-K/Akt activation in vemurafenib-resistant melanoma cells.
(A) Cells were incubated for 48 h in medium (Ctrl) or VMF, as indicated, and subsequently tested by immunoblotting using antibodies to the shown proteins. Numbers below gels indicate values from densitometric analyses (n=3). (B, left) Cells were subjected to Ras GTPase assays. (Right) Densitometric analyses of protein gel bands show the mean ±SD of five independent experiments (**p<0.01). (C) Cells were tested by western blotting using anti-BRAF antibodies. (D) Cells were subjected to immunoblotting to test for Akt and S6 activation.
The A375-VR cells displayed a blockade of invasion across Matrigel relative to parental counterparts (Supplemental Fig. S3A), which was linked to increased Rac and RhoA activation in A375-VR cells (Supplementary Fig. S3B). These results raise the possibility that excessive Rho GTPase activation could impair resistant cell migration and invasion. As increased Rac activation involving the RAC P29S mutation has been associated to resistance to BRAFi (30), we tested whether A375-VR cell resistance could be affected by the Rac inhibitor NSC23766. Both parental and A375-VR cells showed similar sensitivity to increasing NSC23766 concentrations (Supplementary Fig. S3C), and combination of the Rac inhibitor together with increasing VMF concentrations did not cause significant reduction in A375-VR cell proliferation relative to that intrinsically caused by NSC23766 (Supplementary Fig. S3D). We detected a tendency, albeit not statistically significant of a small increase in A375-VR cell viability in the presence of 25 μM of the Rac inhibitor starting at 500 nM of VMF. The molecular mechanistics of the potential counteraction between NSC23766 and VMF are not known. In addition, NSC23766 did not affect the increased Erk1/2 phosphorylation shown by the resistant cells (Supplemental Fig. S3E), suggesting that resistance to VMF was independent of the increased Rac activation exhibited by A375-VR cells.
Identification of miRNAs differentially expressed in A375-VR cells
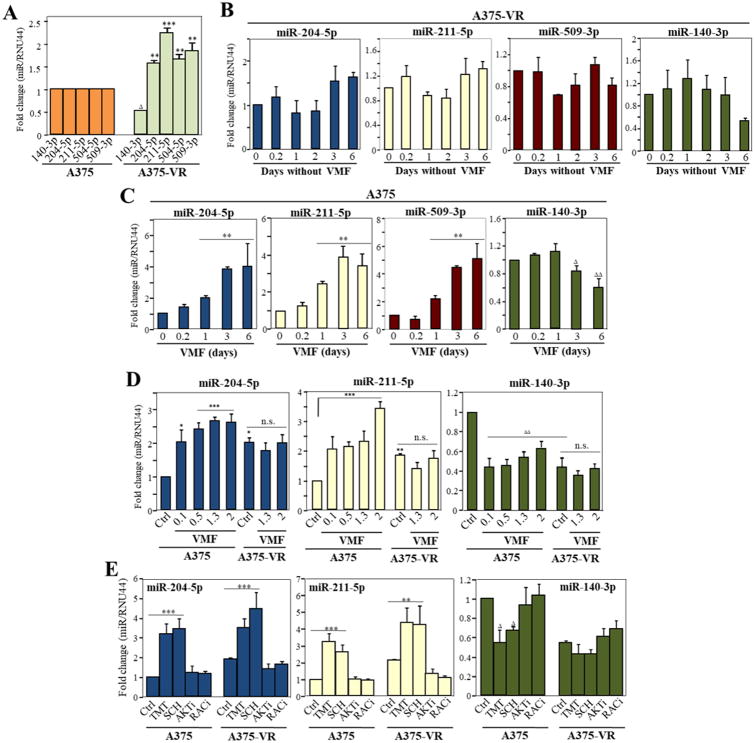
We performed small RNA-seq from parental and A375-VR cells with the aim of identifying miRNAs whose altered expression could potentially contribute to melanoma resistance to VMF. Sequences were aligned against the miRBase21 database, and following DESeq2 and BioSAGE analyses, the expression of four miRNAs, miR-204-5p, miR-211-5p, miR-504-5p and miR-509-3p was found to be significantly upregulated in A375-VR cells compared to A375, whereas miR-4454, miR-140-3p and miR-210-3p were reduced (Supplementary Table S2). Expression of miR-204-5p, miR-211-5p, miR-509-3p, miR-140-3p and miR-504-5p was detected in samples from two different cohorts of melanoma patients (not shown), whereas no expression of miR-4454 and miR-210-3p was found, which led us to discontinue their study.
Subsequent qPCR experiments validated the changes for miR-204-5p, miR-211-5p, miR-504-5p, miR-509-3p and miR-140-3p expression in A375-VR cells (Fig. 3A). Since miR-504-5p expression was very low in A375 and A375-VR cells, we decided to also cease its study. To analyze that the observed miRNA alterations in A375-VR cells are independent of VMF presence, we assessed their miRNA levels by incubation in culture medium without VMF for up to 6 days. The altered expression levels of miR-204-5p, miR-211-5p, miR-509-3p and miR-140-3p in A375-VR cells were preserved in the absence of VMF (Fig. 3B), supporting the stability of these miRNA alterations. We next investigated the dynamics of VMF-dependent miRNA changes by incubating A375 cells with VMF. Notably, significant increases in miR-204-5p, miR-211-5p and miR-509-3p were already detected after 24 h of VMF exposure, reaching a plateau between 3-6 days (Fig. 3C). In addition, cell incubation with VMF led to decreased miR-140-3p expression, although with slower kinetics. These results indicate that alterations in the expression of these miRNAs occur early after melanoma cell treatment with VMF. We decided then to focus our studies on miR-204-5p, miR-211-5p and miR-140-3p, leaving miR-509-3p for future analyses. Dose-response experiments with A375 cells revealed substantial changes in miR-204-5p, miR-211-5p and miR-140-3p expression already at VMF concentrations of 100 nM (Fig. 3D), whereas only minor but non-significant alterations in miRNA expression in A375-VR cells were observed upon increasing VMF amounts. To study if changes in miRNA expression can be detected in an additional VMF-resistant BRAF V600E melanoma cell line, we used several resistant clones of SK-Mel 239 cells (31). Augmented levels of miR-204-5p and miR-211-5p were detected in 5 out of 6 clones, whereas reduced expression of miR-140-3p was observed in 2 out of the 6 clones (Supplementary Fig. S4A).
Figure 3. Identification of miRNAs differentially expressed in VMF-resistant A375 melanoma cells.
(A) qPCR validation of miRNA expression changes between parental and VMF-resistant cells (n=5). (B, C) A375-VR or parental A375 cells were incubated for the indicated times in the absence (B) or presence of VMF (1.3 μM) (C), and subsequently subjected to qPCR analyses for miRNA expression (n=3). (D) Dose-response analyses of miRNA expression in A375 and A375-VR cells cultured for 48 h in the absence (Ctrl) or presence of the indicated concentrations (0.1 to 1.3 μM) of vemurafenib. miRNA expression was significantly upregulated, ***p<0.001, **p<0.01, *p<0.05, or significantly reduced, ΔΔΔp<0.001, ΔΔp<0.01, Δp<0.05. (E) Cells were incubated for 48 h in medium (Ctrl) or in the presence of TMT (4 nM), SCH772984 (50 nM), triciribine (5 μM), NSC23766 (10 μM), and subsequently analyzed by qPCR for expression of the shown miRNAs (n=3-4).
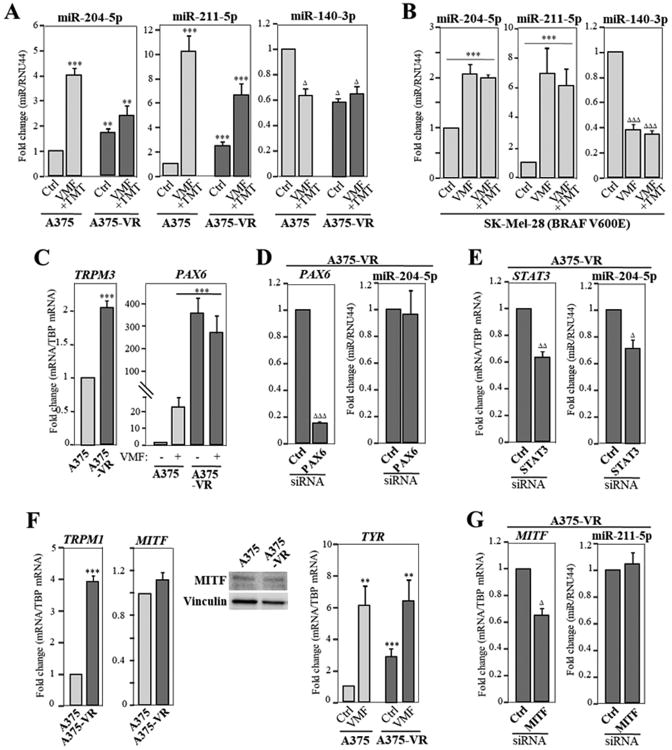
Interestingly, upregulation of miR-204-5p and miR-211-5p was also detected in parental and A375-VR cells exposed to TMT, to SCH772984, or to the combination of VMF and TMT, whereas reduced levels of miR-140-3p were observed in A375 cells incubated with these inhibitors, but not in resistant cells (Figs. 3E; 4A). Meanwhile, the same miRNAs were unaltered in response to AKT or Rac inhibitors (Fig. 3E). Moreover, increase in miR-204-5p and miR-211-5p, and reduction in miR-140-3p expression were also observed in VMF/TMT-double resistant (VR/TR) A375 cells (Supplementary Fig. S4B, C). Of note, BRAF V600E mutant SK-Mel-28 melanoma cells also showed enhanced miR-204-5p and miR-211-5p expression and decreased levels of miR-140-3p in response to incubation with VMF or with combined VMF and TMT (Fig. 4B), suggesting that these miRNA changes might represent a common event in melanoma cells displaying this mutation. Together, these data point out to a strong correlation between changes in miR-204-5p, miR-211-5p and miR-140-3p expression in melanoma and BRAF/MEK/Erk1/2 inhibition, but not PI3-K/Akt or Rac blockade. Normal foreskin melanocytes were also tested for potential alterations in miRNA expression after exposing them to VMF and TMT. Whereas miR-204-5p expression was upregulated by VMF, no significant changes in miR-211-5p and miR-140-3p expression were detected (Supplemental Fig. S4D). The molecular basis for VMF-triggered increase in miR-204-5p levels in normal melanocytes and its functional consequences in melanocyte biology have not been addressed in the present study.
Figure 4. Characterization of miRNAs differentially expressed in VMF-resistant melanoma cells.
(A, B) Cells were incubated for 48 h in medium (Ctrl), or in the presence of VMF alone (1.3 μM) or combined VMF (100 nM) and TMT (4 nM), and subsequently analyzed by qPCR for expression of the shown miRNAs (n=3-4). Expression was significantly upregulated, ***p<0.001, **p<0.01, or significantly reduced, ΔΔΔp<0.001, ΔΔp<0.01, Δp<0.05. (C) Cells were analyzed by qPCR for TRPM3 and PAX6 mRNA expression. (D, E) Cells were transfected with control, PAX6 or STAT3 siRNA, and transfectants tested by qPCR for PAX6, STAT3 and miR-204-5p expression (n=3). (F) Cells were analyzed by qPCR for TRPM1, MITF or TYR mRNA expression, or by immunoblotting for MITF protein levels (n=2-4). (G) Cells were transfected with control or MITF siRNA, and transfectants tested by qPCR for MITF and miR-211-5p expression (n=3).
mir-204 is located in intron 6 of the TRPM3 (Transient Receptor Potential Melastatin 3) gene, and it has been proposed that expression of miR-204 and TRPM3 has common regulatory mechanisms (32,33). Correlating with increased miR-204-5p expression in A375-VR cells, we found upregulated TRPM3 mRNA in the resistant cells compared to parental ones (Fig. 4C, left). Furthermore, the PAX6 transcription factor promotes expression of miR-204-5p (34), and we observed a strong PAX6 induction in A375-VR cells, as well as in A375 cells treated with VMF (Fig. 4C, right). In spite of the strong PAX6 upregulation in A375-VR cells, its silencing did not alter miR-204-5p expression (Fig. 4D). Recent data revealed a role for STAT3 in regulation of miR-204-5p expression in melanoma (35). We then silenced STAT3 in A375-VR cells (30-35% depletion) and found a significant decrease in the expression of miR-204-5p (Fig. 4E), suggesting that STAT3 is involved in the increased miR-204-5p expression in A375-VR cells.
miR-211 is expressed from the intron 6 of the melanoma marker TRPM1 (Transient receptor potential cation channel subfamily M member 1; melastatin) (36,37), and we found that the increased miR-211-5p expression in A375-VR cells directly correlated with enhanced expression TRPM1 relative to parental cells (Fig. 4F, left). As MITF (microphthalmia-associated transcription factor) regulates the transcription of TRPM1, we analyzed MITF expression and found no significant differences between A375 and A375-VR cells (Fig. 4F, middle panels). However, expression of tyrosinase (TYR), a marker of MITF activation (38), was enhanced in resistant cells both with and without VMF, as well as in parental cells exposed to the inhibitor (Fig. 4F, right), together opening the possibility that increased miR-211-5p expression in A375-VR cells could be dependent of MITF activity. However, MITF silencing in A375-VR cells did not cause detectable alterations in miR-211-5p expression (Fig. 4G), suggesting that upregulated miR-211-5p expression in A375-VR cells is independent of MITF activation.
Role of miR-204-5p, miR-211-5p and miR-140-3p in melanoma resistance to vemurafenib
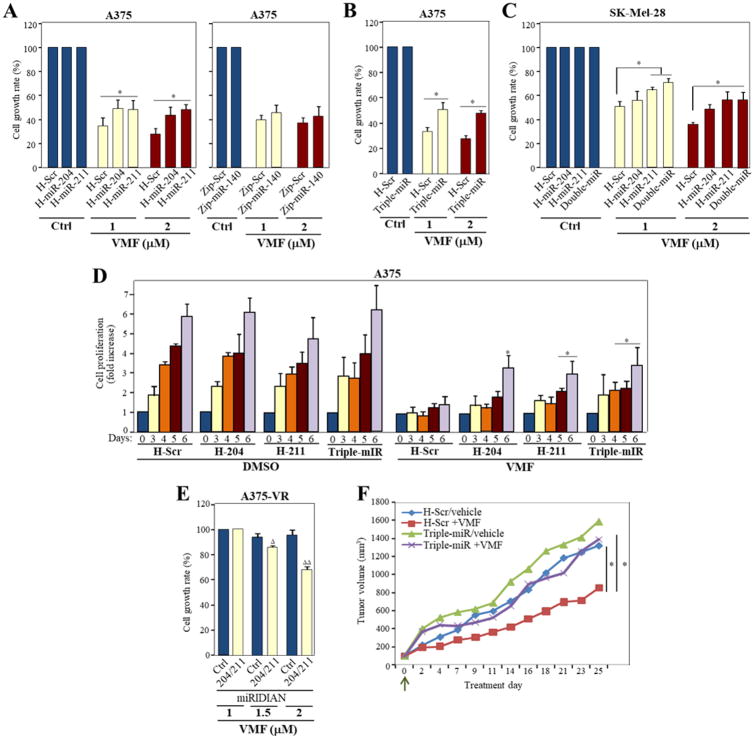
To test the potential involvement of the miRNAs in resistance to VMF, A375 cells were infected with lentiviral vectors to either overexpress miR-204-5p or miR-211-5p, or to silence miR-140-3p (Supplementary Fig. S5A). MTT assays revealed a moderate but consistent increase in resistance to VMF in cells overexpressing miR-211-5p or miR-204-5p relative to control transfectants (from 28-34% cell viability in H-Scr control cells to 45-49% in H-miR-211 or -miR-204) (Fig. 5A, left). Instead, no significant variations in proliferation were observed in cells silenced for miR-140-3p (Fig. 5A, right).
Figure 5. Role of miR-204-5p, miR-211-5p and miR-140-3p in melanoma resistance to vemurafenib.
(A-C) The indicated control or miRNA A375 and SK-Mel 28 transductants were tested in MTT assays in medium without serum, in the absence (Ctrl) or presence of the indicated concentrations of VMF. Proliferation was significantly augmented, *p<0.05 (n=3-4). (D) Control (H-Scr cells), transductants over-expressing miR-204-5p or miR-211-5p, or triple-miR transductants were cultured in complete medium for the indicated times in the absence (DMSO samples) or presence of VMF (100 nM), and cell proliferation determined. Data is shown as fold increase in cell proliferation from three independent experiments. *Proliferation was significantly augmented (p<0.05), relative to the corresponding time points obtained with H-Scr control cells exposed to VMF. (E) A375-VR cells were transiently transfected with a combination of miRNA-204-5p and miRNA-211-5p miRIDIAN microRNA hairpin inhibitors or with a negative control, and transfectants tested by quadriplicate in cell proliferation assays in the absence of serum (n=2; ΔΔp<0.01, Δp<0.05). (F) Cells were subcutaneously inoculated into NSG mice and after 20 days, mice were randomized in four groups and treated with vehicle or VMF, as stated in the Methods. Shown are tumor volume determinations. Arrow denotes starting of treatments. (n=5 mice/condition, *p<0.05).
We then generated A375 triple transductants that overexpress miR-204-5p and miR-211-5p, and that are knocked-down for miR-140-3p (triple-miR; Supplementary Fig. S5B). These triple-miR transductants also displayed enhanced resistance to VMF relative to control cells (Fig. 5B; Supplementary Fig. S5C), overall indicating that miR-204-5p and miR-211-5p overexpression, but not depletion of miR-140-3p in A375 cells provides a survival advantage against vemurafenib. Supporting a role for miR-204-5p and miR-211-5p in providing resistance to VMF, SK-Mel-28 melanoma cells overexpressing either one or both miRNAs (double-miR) (Supplemental Fig. S5D, E) exhibited a moderate and consistent increase in growth rate in the presence of the inhibitor compared to control counterparts (Fig. 5C).
H-miR-204, H-miR-211 and triple-miR, but not Zip-miR-140 A375 cells, also showed increased resistance to TMT and to combined VMF and TMT treatments, as compared to control cells (Supplementary Fig. S6A). Control H-Scr SK-Mel-28 transductants displayed close to two-fold higher intrinsic resistance to TMT than H-Scr A375 counterparts, and H-miR-211 and double-miR, but not H-miR-204 SK-Mel-28 transductants exhibited higher resistance to TMT and to combined VMF and TMT treatments than control cells (Supplementary Fig. S6B). Contrary to VMF or TMT, neither miR-204-5p nor miR-211-5p overexpression conferred resistance to SCH772984 (not shown), and only triple-miR cells displayed a moderate increase in growth rate with VMF and SCH772984 (Supplementary Fig. S6C), but not with triciribine (Supplementary Fig. S6D).
Further confirmation of miR-204-5p and miR-211-5p contribution to resistance to VMF came from cell proliferation assays for 6 days in the presence of VMF, which showed that H-miR-204, H-miR-211 and triple-miR stable cells have significantly higher proliferation than control H-Scr counterparts (Fig. 5D). Instead, Zip-miR-140-3p transductants lacked proliferation advantage when exposed to the inhibitor (not shown). To add further significance to these data, we silenced both miR-204-5p and miR-211-5p in A375-VR cells (Supplementary Fig. S6E), and tested cell viability in the presence of vemurafenib. Transfectants displayed decreased growth at 1.5-2 μM of VMF (Fig. 5E), highlighting the involvement of these miRNAs in resistance to VMF. Moreover, NSG mice subcutaneously inoculated with triple-miR cells displayed significantly larger tumor volumes following VMF administration than mice inoculated with H-Scr control cells (Fig. 5F). No significant differences in tumor growth were observed in mice injected with triple-miR or H-Scr cells upon vehicle administration, or in mice inoculated with triple-miR cells and exposed to vehicle or VMF.
We next used SK-Mel-103 cells to test the effect of VMF in an inherently VMF-resistant melanoma cell line. miR-204-5p expression was augmented by VMF, but reduced when cells were incubated with both VMF and TMT (Supplementary Fig. S7A). No miR-211-5p expression was detected in these cells (not shown), and expression of miR-140-3p was unaltered in the presence of VMF, and increased with combined VMF and TMT. We then silenced miR-204-5p in SK-Mel-103 cells and tested cell resistance after exposing them to increasing VMF concentrations. The results revealed no significant differences in growth between miR-204-5p-knockdown cells and control counterparts (Supplementary Fig. S7B), overall indicating that miR-204-5p does not influence proliferation in inherently VMF-resistant SK-Mel-103 cells.
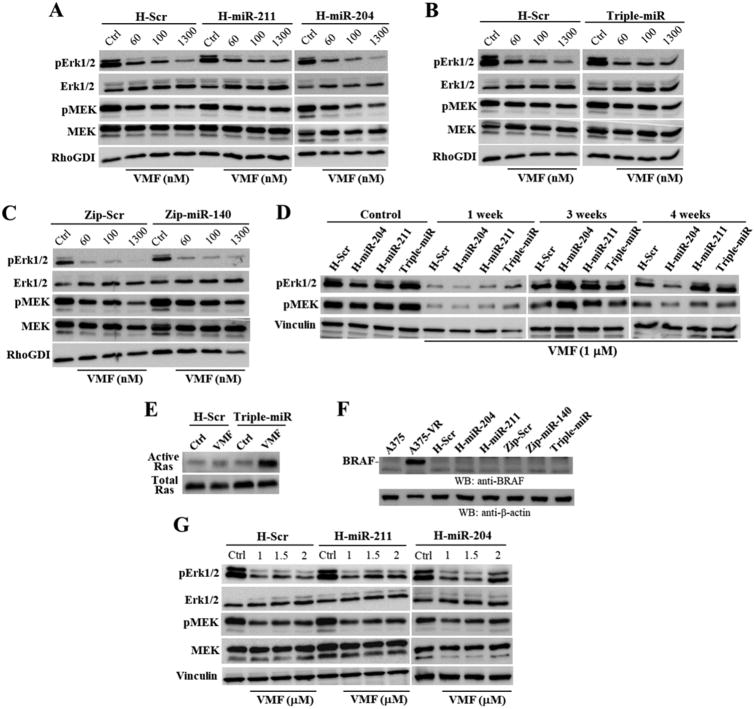
Analyses of Erk1/2 activation revealed that VMF-induced decrease in pErk1/2 was more attenuated in H-miR-211 and in triple-miR A375 cells than in control cells (Fig. 6A-C). Furthermore, reduced pMEK due to VMF was more mitigated in H-miR-211 cells. As expected, when we extended cell exposure to VMF to 1 week, pErk1/2 and pMEK levels sharply diminished, but higher phosphorylation of Erk1/2 and MEK in triple-miR cells than in control cells was observed (Fig. 6D). By week 3 of treatment with VMF, pErk1/2 and pMEK levels were recovering in H-Scr cells, but triple-miR, as well as cells overexpressing miR-204-5p or miR-211-5p still showed remarkably higher pErk1/2 and pMEK. After 4 weeks with VMF, H-miR-211 and triple-miR, but not H-miR-204 cells still exhibited higher pErk1/2 and pMEK1/2 than control cells (Fig. 6D). Furthermore, VMF-exposed triple-miR transductants displayed substantially higher Ras activation levels than control cells (Fig. 6E). None of the different miRNA transductants exhibited BRAF overexpression shown by A375-VR cells (Fig. 6F). The attenuated decrease in Erk1/2 activation upon VMF treatment was not only observed in A375 cells overexpressing miR-211-5p, but H-miR-211 as well as H-miR-204 SK-Mel 28 transductants also displayed mitigated reduction in pErk1/2 relative to control cells (Fig. 6G).
Figure 6. Analysis of Erk1/2 and MEK activation in miRNA transductants.
(A-C) A375 transductants were incubated for 24 h in the absence (Ctrl) or presence of the indicated concentrations of VMF, and subsequently subjected to immunoblotting. (D) Cells were left untreated (Control) or incubated for the times shown with VMF, and analyzed by western blotting. (E) Cells were incubated for 24 h without (Ctrl) or with VMF (1.3 μM) and subsequently subjected to Ras GTPase assays. (F) Parental and A375-VR cells, or the indicated transductants were analyzed by immunoblotting using anti-BRAF antibodies. (G) SK-Mel-28 transductants were incubated for 24 h with or without VMF, and subsequently subjected to immunoblotting. Loading controls were assessed with antibodies to RhoGDI, vinculin or β-actin.
Examination of miR-204-5p and miR-211-5p target expression
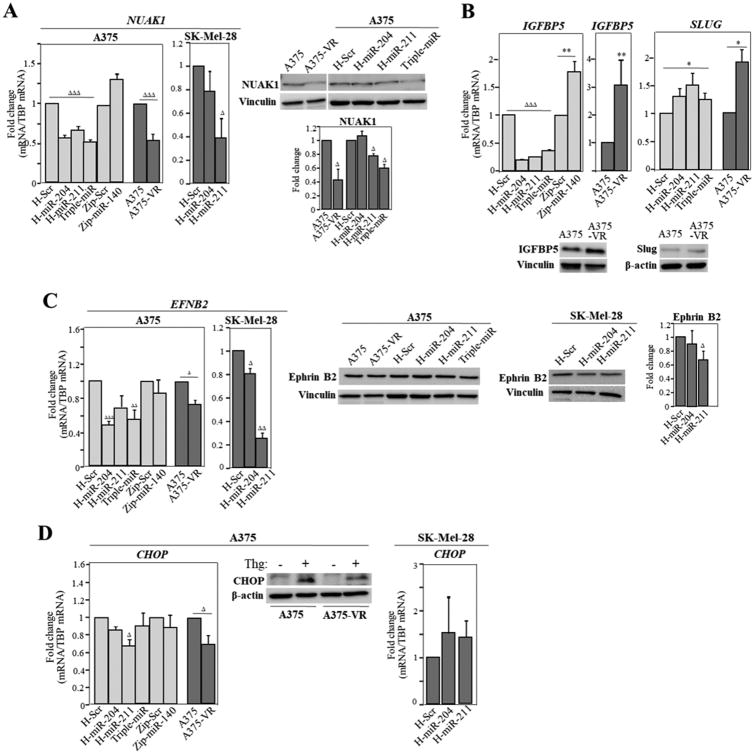
Data from previous published work and from available miRNA databases indicate that miR-204-5p and miR-211-5p share some common targets, including NUAK1/ARK5, IGFBP5, TGF-βRII, Slug and CHD5 (21,37,39,40). qPCR analyses showed that NUAK1 mRNA was downregulated in A375-VR cells and in H-miR-204, H-miR-211 and triple-miR cells, but not in Zip-miR-140 counterparts (Fig. 7A, left). Moreover, NUAK1 expression was also reduced in SK-Mel-28 cells overexpressing miR-211-5p. Decreased NUAK1 protein expression was confirmed in immunoblot analyses in A375-VR, H-miR-211 and triple-miR A375 cells, but not in H-miR-204 counterparts (Fig. 7A, right). Although IGFBP5 and TGF-βRII mRNAs were reduced in cells overexpressing miR-204-5p or miR-211-5p, confirming that they represent targets for these miRNAs, their levels were increased in A375-VR cells (Fig. 7B, right; Supplementary Fig. S8A, left), indicating that other mechanisms overall upregulated their expression in the resistant cells. Interestingly, IGFBP5 mRNA was enhanced in Zip-miR-140 transductants, suggesting that it might represent a miRNA target. Contrary to what it was expected, SLUG expression was increased in H-miR-204, H-miR-211 and in triple-miR transductants, as well as in A375-VR cells (Fig. 7B, right). No significant alterations in CHD5 expression was detected in the miRNA transductants or in A375-VR cells (Supplementary Fig. S8A, right).
Figure 7. Analyses of miRNA target expression in cells overexpressing miR-204-5p and miR-211-5p and in A375-VR cells.
mRNA and protein levels for NUAK1 (A), IGFBP5 and Slug (B), Ephrin B2 (C) and CHOP were determined by qPCR or immunoblotting, respectively. Relative fold change values from the blots are also displayed. ΔΔΔExpression was significantly reduced, p<0.001, ΔΔp<0.01, Δp<0.05, or **stimulated, p<0.01, *p<0.05 (n=3-5).
The miR-204-5p target ephrin B2 (EFNB2) (41) was reduced in H-miR-204, triple-miR, and to a lower extent in A375-VR cells relative to their controls (Fig. 7C, left). However, we were unable to detect significant alterations in ephrin B2 protein expression in these cells (Fig. 7C, middle). EFNB2 mRNA expression was also decreased in SK-Mel-28 cells overexpressing miR-204-5p or miR-211-5p, and furthermore, we could detect reduced ephrin B2 expression in H-mir-211 SK-Mel-28 cells (Fig. 7C). Although the expression of another putative miR-204-5p target, SNAIL, was moderately diminished in resistant cells, we did not observe variations in its levels in the miRNA transductants (Supplementary Fig. S8B).
The pro-apoptotic transcription factor CHOP/GADD153 and IGF-2R are well-known miR-211-5p targets (42,43). We found that CHOP mRNA expression was reduced both in A375-VR and H-miR-211cells, but not in SK-Mel-28 cells overexpressing miR-211-5p (Fig. 7D, left and right). To efficiently visualize CHOP protein expression in A375 cells, we incubated them with thapsigargin, an agent which elicits endoplasmic reticulum (ER) stress. We found decreased CHOP expression (25-30%) in thapsigargin-exposed A375-VR cells compared to parental counterparts (Fig. 7D, middle). IGF-2R levels were not significantly altered in A375-VR cells relative to parental ones, or in the different miRNA transductants (Supplementary Fig. S8C). Finally, the expression of additional miR-204-5p and miR-211-5p targets, such as RAB22A, CREB5, SOX4, BCL2, NFAT5 and POU3F2 was unaltered in VMF-resistant cells (not shown).
Whereas NUAK1, EFNB2 and CHOP expression is reduced in A375-VR relative to A375 cells (Fig. 7A, C, D), NUAK1 and EFNB2 were found to be increased in SK-Mel-103 compared to A375 cells, and no significant differences between A375 and SK-Mel-103 cells were detected for CHOP expression (Supplementary Fig. S8D).
Discussion
Resistance to BRAFi in melanoma involves genetic alterations that lead to reactivation of the MAPK pathway or activation of PI3-K/AKT signalling (4-7,44,45). In addition, there can be other non-genetic alterations conferring melanoma resistance. Here we have generated A375 melanoma cells displaying resistance to vemurafenib with the goal of identifying changes in miRNA expression that could provide survival advantage against this inhibitor. Resistant A375-VR cells displayed Ras-MAPK pathway reactivation, as well as activation of PI3-K/AKT, consistent with previous data (1). Activation of these pathways likely contributed to increased tumor growth in the A375-VR xenograft models. VMF-resistant cells also showed partial resistance to trametinib, as well as to combined VMF and TMT. We have not addressed whether additional mechanisms independently of Ras-MAPK re-activation, including Notch1 pathway activation (46) or increased expression of PDGFβ or EGF receptors (4,47) contribute to VMF resistance in A375-VR cells. Stability of the resistance was demonstrated both in in vitro and in vivo assays, such that lung and liver A375-VR metastases retained full resistance to VMF after culturing tumor cells with the inhibitor.
Analyses of small RNAseq data and subsequent qPCR studies revealed increased miR-204-5p and miR-211-5p levels in A375-VR cells relative to parental counterparts, whereas miR-140-3p expression was reduced. Upregulation of miR-204-5p and miR-211-5p was already evident after 24 h of cell exposure to VMF, and was detected at low concentrations of the inhibitor. miRNA alterations persisted for at least 6 days in the absence of VMF, overall suggesting that miRNAs are rapidly increased by the inhibitor, but do not require its continuous presence. Of note, enhanced miR-204-5p and miR-211-5p expression was not exclusive of A375-VR cells, as it was also detected in BRAF V600E-mutant SK-Mel-239-VR melanoma cells. Together, these results strongly suggest that increased miR-204-5p and miR-211-5p expression could represent a common event during development of melanoma resistance to vemurafenib. Our results are in line with previous data showing that VMF induces the expression of these miRNAs in A375 cells (35).
The rapid elevation of miR-204-5p and miR-211-5p levels was not only detected in A375 cells exposed to VMF, but also to TMT, to an ERK inhibitor and to combined VMF and TMT treatment. Moreover, BRAF V600E-mutant SK-Mel-28 cells also showed miR-204-5p and miR-211-5p upregulation after incubation with VMF or with combined VMF and TMT. The fact that the increased levels of these miRNAs were not observed in cells incubated with AKT or Rac inhibitors suggests that it might represent a specific response to MAPKi.
We studied the impact of miRNA alterations on melanoma resistance to VMF, by ectopic expression of miR-204-5p or miR-211-5p, or by silencing miR-140-3p in parental A375 cells. The data revealed that miR-204-5p and miR-211-5p, but not miR-140-3p contribute to cell resistance to VMF. Moreover, overexpression of miR-204-5p and miR-211-5p in another BRAF V600E melanoma cell line, SK-Mel-28, also conferred VMF resistance. Further supporting the involvement of these miRNAs in VMF resistance, their depletion in A375-VR cells led to decreased cell growth in the presence of VMF. It is noteworthy that as miR-204-5p silencing in inherently VMF-resistant SK-Mel-103 cells does not affect their proliferation in the presence of VMF, the results suggest that inherent resistant cells and BRAF V600E cells that have developed VMF resistance behave differently in the context of miR-204-5p involvement. Underlining the miR-204-5p and miR-211-5p contribution to VMF resistance, we show that triple-miR cells exhibit higher in vivo resistance to VMF than control cells, resulting in larger tumors in NSG mice.
Notably, overexpression of miR-204-5p and miR-211-5p provided MAPK reactivation during the first 4 weeks of cell exposure to VMF, earlier and on top of that observed in control cells. One of the possible mechanisms behind the increased MAPK reactivation detected in triple-miR cells could be their remarkable enhancement of Ras activation when exposed to VMF. Therefore, based on the fast kinetics of miR-204-5p and miR-211-5p upregulation, the data overall suggest that these miRNAs might provide initial survival advantage supported by their early contribution to maintain sufficient Ras/MEK/ERK activation levels before further activation caused by genetic or non-genetic alterations of components of this pathway lead to resistance to VMF. Furthermore, the reduced viability of A375-VR cells depleted for miR-204-5p and miR-211-5p indicates that these miRNAs facilitate resistance to VMF not only at initial but also at more advanced stages of treatment.
mir-204 is located in the TRPM3 gene, and we show here that, similarly to miR-204-5p, TRPM3 mRNA expression is also enhanced in A375-VR cells, highlighting early proposal that miR-204 and TRPM3 have common regulatory mechanisms (32,33). PAX6 promotes expression of this miRNA (34). Although we found a strong induction of PAX6 expression in A375-VR cells, the increased miR-204-5p expression was independent of PAX6, as PAX6 silencing did not alter the miRNA levels. On the contrary, knocking-down the expression of STAT3, another transcription factor regulating miR-204-5p expression in melanoma (35), led to a significant reduction in the levels of this miRNA in A375-VR cells, suggesting the involvement of STAT3 in the upregulation of miR-204-5p in the VMF-resistant cells.
mir-211 is found in an intron of TRPM1, a target gene of MITF, and therefore MITF could indirectly drive mir-211 expression by transcriptionally enhancing TRPM1 levels (37). A375-VR cells display higher expression of both miR-211-5p and TRPM1 than parental cells, as well as enhanced MITF activity based on increased TYR expression, together raising the possibility that MITF could contribute to miR-211 upregulation in A375-VR cells. Nevertheless, MITF silencing in resistant cells did not significantly alter the expression of this miRNA, suggesting that MITF plays minor or no roles in the regulation of miR-211-5p expression in A375-VR cells, and that other mechanisms stimulate the expression of this miRNA in these cells.
miR-204 and miR-211 have very similar nucleotide sequences with only two different nucleotides in the whole sequence and the same seed region, a likely explanation for sharing some common targets. Examination of target expression in A375-VR cells and in cells overexpressing miR-204-5p or miR-211-5p revealed that NUAK1/ARK5, EFNB2 and CHOP mRNA levels were significantly reduced. However, only NUAK1/ARK5 protein expression was consistently diminished in the VMF-resistant cells and in H-miR-211 and triple-miR transductants. NUAK1 is a kinase related to AMP-activated protein kinase (AMPK) that can be phosphorylated and activated by the tumor suppressor LKB1 (48). Interestingly, LKB1 can be phosphorylated by both ERK and p90RSK in melanoma cell lines harboring the BRAF V600E mutation, leading to inhibition of LKB1 binding to and activation of AMPK, and ultimately causing increased cell proliferation (49). It is currently unknown whether the BRAF V600E mutation could also lead to LKB1-NUAK1 uncoupling and upregulation of cell growth. The present data opens up the possibility that NUAK1 could be involved in VMF resistance in melanoma cells, an hypothesis that will be the focus of future investigations.
Although ephrin B2 is a target for miR-204-5p, its expression did not significantly decrease in A375-VR or in H-miR-204 cells, suggesting no apparent roles for ephrin B2 in resistance to VMF in A375-VR cells. miR-211-5p elicits pro-survival cell responses by downregulating its target gene CHOP (42), a pro-apoptotic transcription factor involved in the unfolded protein response. CHOP mRNA and protein expression was reduced in A375-VR cells, which raised the possibility that it could contribute to early viability against VMF. However, this hypothesis becomes weakened by the reported increase in CHOP expression in melanoma cells exposed to BRAFi (50). It is worth mentioning that NUAK1/ARK5 and EFNB2 expression was increased in SK-Me-103 relative to A375 cells, which might suggest that inherent resistant cells or cells that developed resistance to VMF behave differently in the context of miRNA involvement.
Both oncogenic and tumor suppressor roles have been linked to miR-204-5p and miR-211-5p. Thus, their ectopic expression leads to colorectal and breast cancer cell proliferation (40,51). In melanoma, these miRNAs act as tumor suppressors and also inhibit cell invasion (21,37,52,53). This is consistent with our observation of A375-VR in vitro cell invasion blockade, which could potentially translate into reduced metastatic capacity. However, parental and A375-VR cells display similar metastatic capacities, and thus our data would not support that the decreased A375-VR cell invasion would result in lower metastasis, at least in later steps of this process. Yet, we do not know whether initial invasion and dissemination events of parental cells might be more efficient than those of VMF-resistant cells. Anyhow, as A375-VR tumors grew faster than parental cell tumors, this suggests that a possible initial lower invasion efficiency of A375-VR cells might be later counteracted by their higher proliferation rate in vivo. The involvement of miR-204-5p and miR-211-5p in melanoma resistance to VMF reported here represents a novel function for these miRNAs, and functional identification of their targets will shed mechanistic view of their implication in melanoma resistance. The upregulation of miR-204-5p and miR-211-5p expression could provide early viability in the presence of VMF that might allow subsequent resistance. Our study together with recent work addressing the role of miRNAs in BRAFi resistance of melanoma (23,24,54,55) highlights the contribution of non-genetic alterations in melanoma resistance to BRAFi.
Supplementary Material
Statement of significance.
Identification of miRNAs that enable resistance to BRAF inhibitors in melanoma suggests a mechanism-based strategy to limit resistance and improve clinical outcomes.
Acknowledgments
We thank Nohemi Arellano-Sánchez for expert technical support. Drs. Angeles García-Pardo, Marisol Soengas, Ignacio Casal, Emily Bernstein, Poulikos Poulikakos, Erica Riveiro-Falkenbach, José Luis Rodríguez-Peralto and Agustin Zapata, are acknowledged for providing useful reagents, and Dr. Iván Marquez and Imanol Arozarena for helpful discussions. We also thank Olivia Barcón-Minchán for assistance in the Animal Facility.
Financial support: This work was supported by grants SAF2014-53059-R and RD12/0036/0061 to J. Teixidó, and NIH/NCI grants R01CA155234-03 and R01CA163891-03 to E. Hernando.
Footnotes
Conflict of interest disclosure statement: The authors declare no potential conflicts of interest
References
- 1.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19:1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 9.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–77. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 10.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 14.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–8. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 17.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–73. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 19.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–18. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–75. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 21.Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol. 2014;134:441–51. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 22.Migliore C, Giordano S. Resistance to targeted therapies: a role for microRNAs? Trends Mol Med. 2013;19:633–42. doi: 10.1016/j.molmed.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Tetzlaff MT, Wang T, Yang R, Xie L, Zhang G, et al. miR-200c/Bmi1 axis and epithelial-mesenchymal transition contribute to acquired resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res. 2015;28:431–41. doi: 10.1111/pcmr.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Li J, Sun Y, Zhang Y, Dong L, Shen C, et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7:53558–70. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Varas P, Colo GP, Bartolome RA, Paterson A, Medrano-Fernandez I, Arellano-Sanchez N, et al. Rap1-GTP-interacting adaptor molecule (RIAM) protein controls invasion and growth of melanoma cells. J Biol Chem. 2011;286:18492–504. doi: 10.1074/jbc.M110.189811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 28.Bartolome RA, Wright N, Molina-Ortiz I, Sanchez-Luque FJ, Teixido J. Activated G(alpha)13 impairs cell invasiveness through p190RhoGAP-mediated inhibition of RhoA activity. Cancer Res. 2008;68:8221–30. doi: 10.1158/0008-5472.CAN-08-0561. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5:441–50. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 30.Watson IR, Li L, Cabeceiras PK, Mahdavi M, Gutschner T, Genovese G, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–52. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koetz-Ploch L, Hanniford D, Dolgalev I, Sokolova E, Zhong J, Diaz-Martinez M, et al. MicroRNA-125a promotes resistance to BRAF inhibitors through suppression of the intrinsic apoptotic pathway. Pigment Cell Melanoma Res. 2017 doi: 10.1111/pcmr.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall DP, Cost NG, Hegde S, Kellner E, Mikhaylova O, Stratton Y, et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26:738–53. doi: 10.1016/j.ccell.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, et al. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitiello M, Tuccoli A, D'Aurizio R, Sarti S, Giannecchini L, Lubrano S, et al. Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget. 2017;8:25395–417. doi: 10.18632/oncotarget.15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004;64:509–16. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- 37.Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010;40:841–9. doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–70. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PLoS One. 2013;8:e73473. doi: 10.1371/journal.pone.0073473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, Liu Y, et al. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS One. 2012;7:e29750. doi: 10.1371/journal.pone.0029750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–9. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48:353–64. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol. 2016;18:1006–17. doi: 10.1038/ncb3399. [DOI] [PubMed] [Google Scholar]
- 44.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–81. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal. 2014;7:ra121. doi: 10.1126/scisignal.aaa1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 48.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. Embo J. 2004;23:833–43. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33:237–47. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–17. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee H, Lee S, Bae H, Kang HS, Kim SJ. Genome-wide identification of target genes for miR-204 and miR-211 identifies their proliferation stimulatory role in breast cancer cells. Sci Rep. 2016;6:25287. doi: 10.1038/srep25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazar J, Qi F, Lee B, Marchica J, Govindarajan S, Shelley J, et al. MicroRNA 211 Functions as a Metabolic Switch in Human Melanoma Cells. Mol Cell Biol. 2016;36:1090–108. doi: 10.1128/MCB.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J, et al. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. Onco Targets Ther. 2017;10:1237–46. doi: 10.2147/OTT.S128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget. 2015;6:17753–63. doi: 10.18632/oncotarget.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vergani E, Di Guardo L, Dugo M, Rigoletto S, Tragni G, Ruggeri R, et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. 2016;7:4428–41. doi: 10.18632/oncotarget.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.