Significance
We report in this article that c-Myc-mediated repression of lncRNA IDH1-AS1 sustains activation of the Warburg effect by HIF1α under normoxic conditions. IDH1-AS1 would otherwise enhance IDH1 enzymatic activity through promoting its homodimerization, leading to increased production of α-KG, which, along with decreases in ROS levels similarly resulting from increased IDH1 activity, causes down-regulation of HIF1a and a reduction in glycolysis. Collectively, our results have identified a signaling axis c-Myc-(IDH1-AS1)-IDH1-αKG/ROS-HIF1α that is important for activation of the Warburg effect under normoxia. Moreover, the results reveal IDH1 as a member of c-Myc-responsive metabolic enzymes and demonstrate that c-Myc plays an important part in balancing mitochondrial respiration and glycolysis to ensure glycolysis be executed efficiently in cancer cells under normoxia.
Keywords: IDH1-AS1, c-Myc, HIF1α, IDH1, Warburg effect
Abstract
The oncoprotein c-Myc plays an important role in regulating glycolysis under normoxia; yet, in cancer cells, HIF1α, which is essential for driving glycolysis under hypoxia, is often up-regulated even in the presence of oxygen. The relationship between these two major regulators of the Warburg effect remains to be fully defined. Here we demonstrate that regulation of a long noncoding RNA (lncRNA), named IDH1-AS1, enables c-Myc to collaborate with HIF1α in activating the Warburg effect under normoxia. c-Myc transcriptionally repressed IDH1-AS1, which, upon expression, promoted homodimerization of IDH1 and thus enhanced its enzymatic activity. This resulted in increased α-KG and decreased ROS production and subsequent HIF1α down-regulation, leading to attenuation of glycolysis. Hence, c-Myc repression of IDH1-AS1 promotes activation of the Warburg effect by HIF1α. As such, IDH1-AS1 overexpression inhibited cell proliferation, whereas silencing of IDH1-AS1 promoted cell proliferation and cancer xenograft growth. Restoring IDH1-AS1 expression may therefore represent a potential metabolic approach for cancer treatment.
A hallmark of cancer is metabolic reprogramming to generate energy through increased glycolysis even in the presence of oxygen, known as the Warburg effect (1, 2), which is essential for providing energy and biosynthesis building blocks to sustain high proliferation rates of cancer cells. Under normoxic conditions, the Warburg effect is commonly driven by c-Myc, the protein product of the oncogene MYC, which is frequently deregulated in cancer cells (3–5). c-Myc promotes glycolysis through effects on glycolytic enzymes (6)—for example, it up-regulates glucose transporters (GLUTs), lactate dehydrogenase A (LDHA), and some key switches of glucose metabolism, such as hexokinase 2 (HK2) and pyruvate dehydrogenase kinase 1 (PDK1) (7–9).
Another key regulator of glycolysis is the transcription factor hypoxia-inducible factor 1α (HIF1α), which is rapidly up-regulated under hypoxic conditions through increased transcription and protein stabilization (10, 11). HIF1α is critical for hypoxic survival through transcriptional activation of its target genes to promote glycolysis (12, 13). Under normoxic conditions, the oxygen sensors prolyl hydroxylases (PHDs) hydroxylate HIF1α, leading to its degradation through the von Hippel-Lindau (VHL)-mediated ubiquitin-proteasome pathway (14). HIF1α expression is often up-regulated in cancer cells (15). Although this is closely associated with the hypoxic microenvironment of solid tumors (16), hypoxia-independent up-regulation of HIF1α has also been described (17, 18). Of interest, while c-Myc and HIF1α regulate glycolysis independently (19), they can also cooperatively function to activate glycolytic genes when c-Myc is overexpressed (20). However, the mechanisms involved remain elusive, although overexpression of c-Myc has been reported to stabilize HIF1α under normoxic and hypoxic conditions (21).
Isocitrate dehydrogenase (IDH) 1 and 2, which act in the cytoplasm and mitochondrion, respectively, are NADPH-dependent enzymes that metabolize isocitrate to α-ketoglutarate (α-KG), an intermediary product in the TCA cycle that is involved in various metabolic pathways (22–25). In particular, α-KG functions as an electron donor to PHDs for prolyl hydroxylation, which is important for hydroxylation and degradation of HIF1α (26). Indeed, whereas increased α-KG alone is sufficient to suppress hypoxia-induced HIF1α, leading to inhibition of glycolysis, specific mutations of IDH1 (IDH1R132H) and IDH2 (IDH2R172H) cause a shift in substrate specificity where IDH catabolizes α-KG into 2-hydroxyglutarate (2-HG) (27–30).
An increasing number of noncoding RNAs have been found to be involved in regulation of metabolism (31). For example, the long noncoding RNA (lncRNA) ceruloplasmin (NRCP) alters glycolysis through up-regulating glucose-6-phosphate isomerase (32), whereas lincRNA-21 promotes stabilization of HIF1α and thus enhances glycolysis under hypoxic conditions (33). Moreover, the lncRNA urothelial cancer-associated 1 (UCA1) up-regulates hexokinase 2 and promotes glycolysis (34). In this report, we demonstrate that c-Myc-mediated repression of the lncRNA IDH1 antisense RNA1 (IDH1-AS1; HUGO Gene Nomenclature Committee: 40292) sustains activation of the Warburg effect by HIF1α under normoxic conditions. IDH1-AS1 is shown to enhance IDH1 enzymatic activity through facilitating its homodimerization, leading to increased production of α-KG, which, along with reduced ROS production (35), causes down-regulation of HIF1α and a reduction in glycolysis. Moreover, we show that IDH1-AS1 inhibits cell proliferation and retards cancer xenograft growth, with implications of its restoration in the treatment of cancer.
Results
LncRNA IDH1-AS1 Enhances IDH1 Enzymatic Activity.
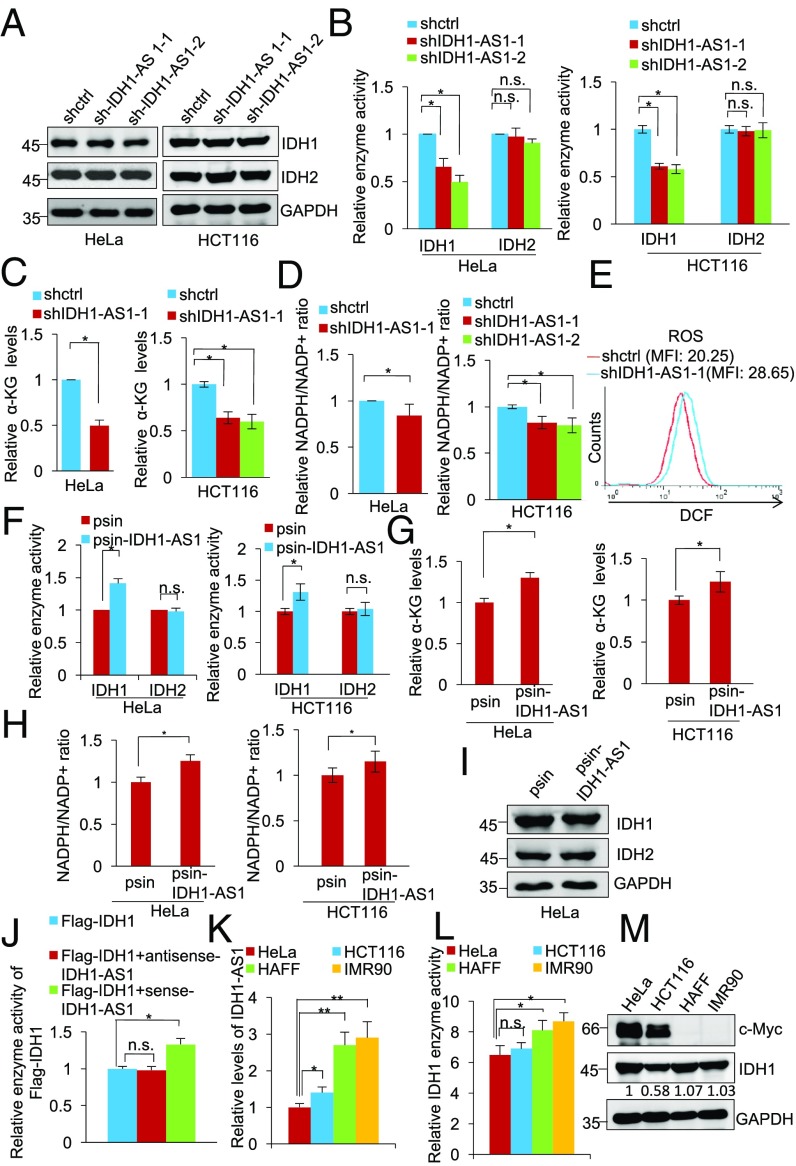
The gene encoding the lncRNA IDH1-AS1 is located head to head with the IDH1 gene on chromosome 2 separated by a short distance of 90 bp (Fig. S1A). This suggests that IDH1-AS1 may have a role in regulating the expression of IDH1 (36). However, shRNA silencing of IDH1-AS1 did not impinge on IDH1 expression (Fig. 1A and Fig. S1B) but rather reduced its enzymatic activity in HeLa and HCT116 cells (Fig. 1B and Fig. S1B). In accordance, silencing of IDH1-AS1 decreased the levels of α-KG, the product converted from isocitrate by IDH1, and NADPH, the specific by-product of isocitrate conversion in HeLa and HCT116 cells (Fig. 1 C and D and Fig. S1B). Moreover, silencing of IDH1-AS1, similar to deficiency in IDH1 (37), resulted in an increase in cellular ROS levels (Fig. 1E). These results suggest that IDH1-AS1 promotes IDH1 activity without affecting its expression levels. In support of this, introduction of an IDH1-AS1-expressing construct (psin-IDH1-AS1) caused an increase in IDH1 activity and up-regulation of α-KG and NADPH but did not alter the expression levels of IDH1 (Fig. 1 F–I and Fig. S1C). Furthermore, incubation of purified Flag-IDH1 with in vitro-transcribed sense but not antisense IDH1-AS1 enhanced IDH1 activity (Fig. 1J). Of note, neither knockdown nor overexpression of IDH1-AS1 caused significant changes in IDH2 activity (Fig. 1 B and F and Fig. S1 B and C), indicating that the effect of IDH1-AS1 was selective for IDH1.
Fig. 1.
See also Fig. S1. IDH1-AS1 enhances IDH1 enzymatic activity. (A) Whole cell lysates from HeLa and HCT116 cells transduced with shctrl, shIDH1-AS1-1, or shIDH1-AS1-2 were subjected to Western blot analysis. n = 3. (B–D) Whole cell lysates from HeLa and HCT116 cells transduced with shctrl, shIDH1-AS1-1, or shIDH1-AS1-2 were analyzed for IDH1 and IDH2 activity (B), α-KG levels (C), and NADPH/NADP+ ratios (D). IDH1 and IDH2 activity, α-KG levels, or NADPH/NADP+ ratios in cells transduced with shctrl were arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (E) Representative flow cytometry histograms of analysis of ROS levels in HeLa cells transduced with shctrl or shIDH1-AS1-1. MFI, mean fluorescence intensity. n = 3. (F) Whole cell lysates from HeLa and HCT116 cells transduced with the psin vertor or psin-IDH1-AS1 were analyzed for IDH1 and IDH2 enzyme activity. The activity of IDH1 and IDH2 in cells transduced with the vector alone was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (G and H) Whole cell lysates from HeLa and HCT116 cells transduced with the psin vertor or psin-IDH1-AS1 were analyzed for α-KG levels (G) and NADPH/NADP+ ratios (H). The α-KG level (G) and NADPH/NADP+ ratios (H) in cells transduced with the vector alone were arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (I) Whole cell lysates from HeLa cells transduced with the psin vector alone or psin-IDH1-AS1 were subjected to Western blot analysis. n = 3. (J) In vitro-transcribed sense or antisense IDH1-AS1 was incubated with purified Flag-IDH1 for 1 h before analysis of IDH1 enzyme activity. The activity of IDH1 in samples without incubation with in vitro-transcribed sense or antisense IDH1-AS1 was arbitrarily designated as 1. Values are mean ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (K) Total RNA from HeLa, HCT116, HAFF, and IMR90 cells was analyzed using qPCR for IDH1-AS1 expression. The relative abundance of IDH1 mRNA in HeLa cells was arbitrarily designated as 1. Values are means ± SEMs; n = 3. (*P < 0.05, **P < 0.01, two-tailed paired Student’s t test). (L) Whole cell lysates from HeLa, HCT116, HAFF, and IMR90 cells were analyzed for IDH1 activity. The activity of IDH1 in HeLa cells was arbitrarily designated as 1. Values are means ± SEMs; n = 3. (*P < 0.05, two-tailed paired Student’s t test). (M) c-Myc and IDH1 expression in HeLa, HCT116, HAFF, and IMR90 cells detected using Western blot analysis of whole cell lysates. n = 3. n.s., not significant; shc-Myc, c-Myc shRNA; shctrl, control shRNA.
Cursory screening of human cell lines for the expression of IDH1-AS1 showed that normal human HAFF and IMR90 cells expressed relatively high levels of IDH1-AS1, whereas the cancer cell lines HeLa and HCT116 displayed significantly lower levels (Fig. 1K). Consistent with the finding that IDH1-AS1 promotes IDH1 activity, there was a trend correlating higher IDH1-AS1 expression with increased IDH1 activity, although no association was observed between IDH1 and IDH1-AS1 expression levels (Fig. 1 K and M). Intriguingly, the expression levels of c-Myc, which is encoded by the MYC gene that is amplified in both HeLa and HCT116 cells (38), were negatively associated with IDH1-AS1 expression levels and IDH1 activity (Fig. 1 K–M). However, there was no association between c-Myc and IDH1 expression (Fig. 1M). Collectively, these results point to a potential role of c-Myc in regulating IDH1 activity through IDH1-AS1. In support of this, analysis of publicly available gene expression datasets revealed that IDH1-AS1 expression in colon and lung cancer tissues was negatively correlated with the expression of the MYC gene (Expression Project for Oncology, https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) (Fig. S1 D and E).
c-Myc Inhibits IDH1 Activity Through Repression of IDH1-AS1.
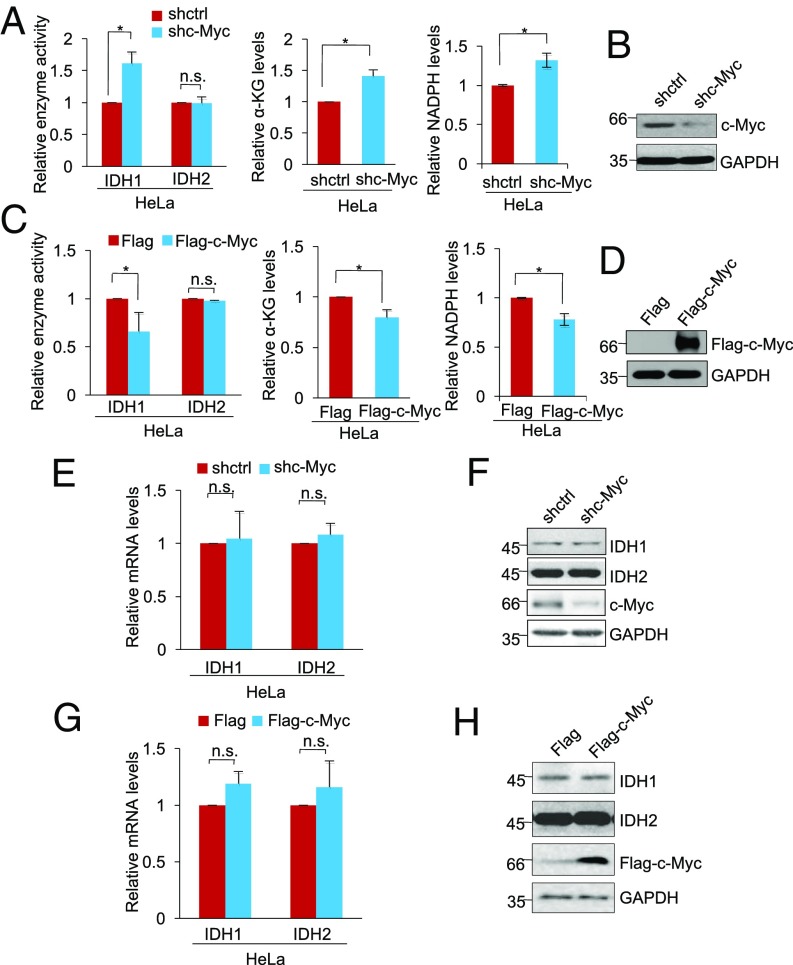
We sought to substantiate the potential role of c-Myc in regulating IDH1 activity. ShRNA silencing of c-Myc increased the activity of IDH1 but not IDH2 and caused increases in α-KG and NADPH in HeLa, HCT116, and H1299 cells (Fig. 2 A and B and Fig. S2 A–D). Conversely, introduction of Flag-tagged c-Myc reduced IDH1 but not IDH2 activity, concomitant with decreased cellular levels of α-KG and NADPH (Fig. 2 C and D and Fig. S2 E–H). Neither knockdown nor overexpression of c-Myc caused significant changes in the expression of IDH1 (Fig. 2 E–H), supporting the notion that c-Myc regulates IDH1 activity without affecting its expression.
Fig. 2.
See also Fig. S2. c-Myc inhibits IDH1 enzyme activity. (A) Whole cell lysates from HeLa cells transduced with shctrl or shc-Myc were analyzed for IDH1 and IDH2 activity (Left), α-KG levels (Middle), and NADPH levels (Right). The relative IDH1 and IDH2 activity and α-KG abundance and NADPH/NADP+ ratio in cells transduced with shctrl were arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (B) ShRNA knockdown efficiency of c-Myc in HeLa cells as shown in Western blot analysis of whole cell lysates. n = 3. (C) Whole cell lysates from HeLa cells transduced with Flag-vector or Flag-tagged c-Myc were analyzed for IDH1 and IDH2 activity (Left), α-KG levels (Middle), and NADPH levels (Right). The relative IDH1 and IDH2 activity and α-KG abundance and NADPH/NADP+ ratio in cells transduced with Flag-vector were arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (D) Overexpression of c-Myc in HeLa cells transduced Flag-tagged c-Myc as measured by Western blot analysis of whole cell lysates. n = 3. (E) Total RNA from HeLa cells transduced with shctrl or shc-Myc was subjected to qPCR analysis of IDH1 and IDH1 mRNA expression. The relative abundance of IDH1 and IDH2 mRNA in cells transduced with shctrl was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (two-tailed paired Student’s t test). (F) Whole cell lysates from HeLa cells transduced with shctrl or shc-Myc were subjected to Western blot analysis. n = 3. (G) Total RNA from HeLa cells transduced with Flag-vector or Flag-tagged c-Myc was subjected to qPCR analysis of IDH1 and IDH1 mRNA expression. The relative abundance of IDH1 and IDH2 mRNA in cells transduced with shctrl was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (two-tailed paired Student’s t test). (H) Whole cell lysates from HeLa cells transduced with Flag-vector or Flag-tagged c-Myc were subjected to Western blot analysis. n = 3. n.s., not significant; shctrl, control shRNA.
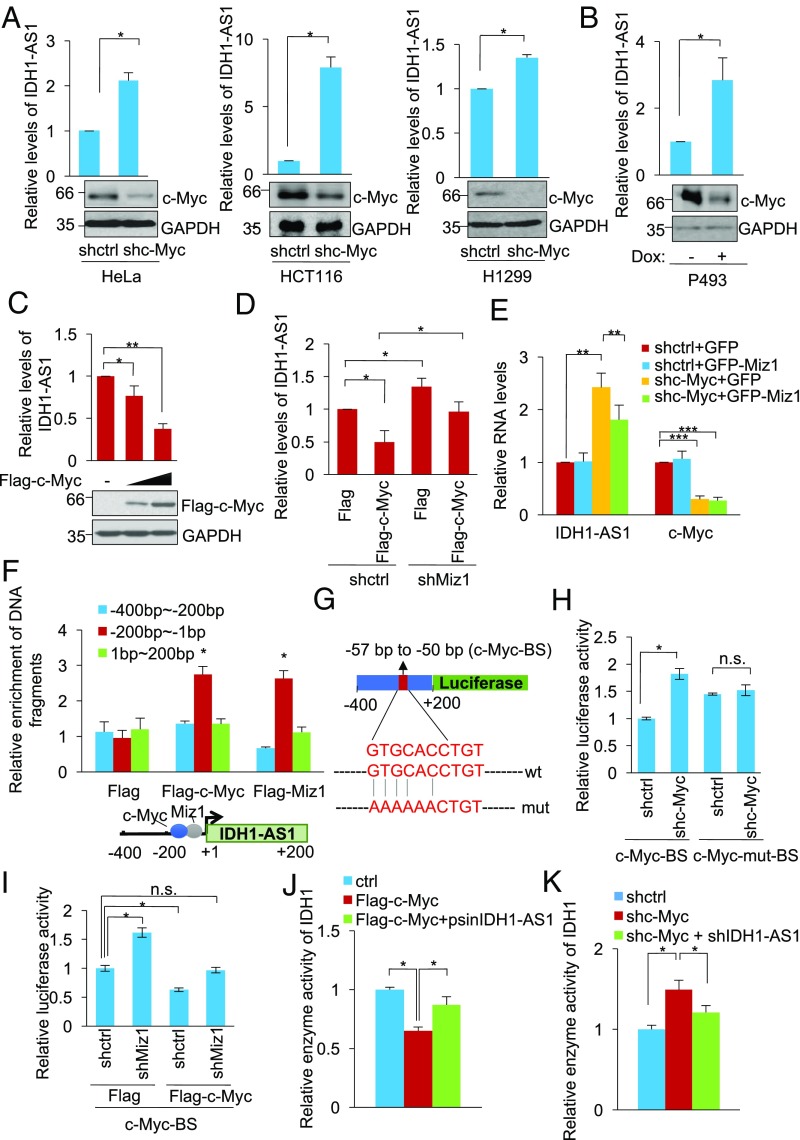
Remarkably, c-Myc silencing up-regulated IDH1-AS1 in HeLa, HCT116, and H1299 cells (Fig. 3A), and similarly, tetracycline-induced inhibition of c-Myc expression in P493 cells carrying a Tet-Off c-Myc expression system resulted in an increase in IDH1-AS1 expression (Fig. 3B). In contrast, IDH1-AS1 expression levels were reduced in proportion to the levels of exogenous c-Myc expressed in HeLa cells (Fig. 3C). Moreover, shRNA silencing of Miz1, which is required for c-Myc-mediated transcriptional repression of gene expression (39), was able to reverse the down-regulation of IDH1-AS1 caused by c-Myc overexpression (Fig. 3D). Consistently, overexpression of Miz1 reversed, at least partially, the up-regulation of IDH1-AS1 resulting from silencing of c-Myc (Fig. 3E). Therefore, c-Myc transcriptionally represses the expression of IDH1-AS1. On the other hand, IDH1-AS1 did not appear to have a role in regulating c-Myc expression (Fig. S3).
Fig. 3.
See also Fig. S3. c-Myc transcriptionally represses IDH1-AS1. (A) Total RNA from HeLa (Left), HCT116 (Middle), and H1299 (Right) cells transduced with shctrl or shc-Myc was analyzed using qPCR for the expression of IDH1-AS1, whereas whole cell lysates were subjected to Western blot analysis. The relative abundance of IDH1-AS1 in cells transduced with shctrl was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (B) Total RNA and whole cell lysates from P493 cell lysates from P493 cells carrying a Tet-Off c-Myc expression system treated with doxycycline (1 μM) were respectively subjected to qPCR Western blot analysis. The relative abundance of IDH1-AS1 in cells without doxycycline treatment was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). Dox, doxycycline. (C) Total RNA and whole cell lysates from HeLa cells transfected with Flag-vector or Flag-tagged c-Myc at increasing concentrations were subjected to qPCR and Western blot analysis, respectively. The relative abundance of IDH1-AS1 in cells transduced with Flag-vector was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, **P < 0.01; two-tailed paired Student’s t test). (D) Total RNA from HeLa cells cotransduced with shctrl or shMiz1 and Flag-vector or Flag-tagged c-Myc was subjected to qPCR analysis. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (E) Total RNA from HeLa cells cotransduced with GFP-vector or GFP-tagged Miz1 and shctrl or shc-Myc was subjected to qPCR analysis. Values are means ± SEMs; n = 3 (**P < 0.01, ***P < 0.001; two-tailed paired Student’s t test). (F) Whole cell lysates from HeLa cells transduced with Flag-vector, Flag-tagged c-Myc, or Flag-tagged Miz1 were subjected to ChIP assays. ChIP products were analyzed by qPCR using primers directed to the indicated fragments of the IDH1-AS1 promoter. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (G) Schematic illustration of luciferase reporter constructs containing the WT or mutant c-Myc binding site in the promoter of IDH1-AS1. BS, binding site; mut, mutant. (H) HeLa cells transduced with shctrl or shc-Myc were cotransfected with reporter constructs containing the WT (c-Myc-BS) or mutant (c-Myc-mut-BS) c-Myc binding site as illustrated in G and Renilla luciferase plasmids. Transcriptional activity was determined by luciferase assays. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (I) HeLa cells transduced with shctrl or shMiz1 were cotransfected with Flag-vector or Flag-tagged c-Myc and Renilla luciferase plasmids. Transcriptional activity was determined by luciferase assays. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (J) Whole cell lysates from HeLa cells transduced with Flag-vector, Flag-tagged c-Myc, or Flag-tagged c-Myc plus psin-IDH1-AS1 were analyzed for IDH1 activity. The activity of IDH1 in cells transduced with Flag-vector was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (K) Whole cell lysates from HeLa cells transduced with shctrl, shc-Myc, or shc-Myc plus shIDH1-AS1 were analyzed for IDH1 activity. The activity of IDH1 in cells transduced with shctrl was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). ctrl, control; n.s., not significant; shctrl, control shRNA.
To determine the region of the IDH1-AS1 promoter subject to repression by c-Myc, we carried out ChIP assays using an anti-Flag antibody in HeLa cells introduced with Flag-tagged c-Myc or Miz1. Both Flag-c-Myc and Flag-Miz1 bound to the −200/+1 (numbers relative to the transcriptional start site) fragment of the IDH1-AS1 promoter but not to the −400/−200 or +1/+200 fragment of the gene (Fig. 3F). Using bioinformatic analyses, we identified a c-Myc response element (c-Myc-RE) (CACGTG)-like sequence (GTGCACCTGT) at the −200/+1 fragment (−57/−50) of the IDH1-AS1 promoter (Fig. 3G). Mutation of the c-Myc-RE-like sequence in a luciferase construct containing −400 and +200 (pGL3-IDH1-AS1 −400/+200) abolished the increased reporter activity caused by c-Myc knockdown (Fig. 3H), suggesting that binding of c-Myc to the GTGCACCTGT motif is essential for transcriptional repression of IDH1-AS1. Consistent with the requirement of Miz1, the luciferase activity driven by the IDH1-AS1 promoter also increased after Miz1 shRNA and this increase was blunted when c-Myc was overexpressed (Fig. 3I).
To confirm the role of IDH1-AS1 in c-Myc-mediated regulation of IDH1 activity, we manipulated the levels of both IDH1-AS1 and c-Myc in HeLa cells. c-Myc overexpression decreased IDH1 activity, while co-overexpression of IDH1-AS1 reversed the inhibition of IDH1 activity resulting from overexpression of c-Myc (Fig. 3J). Conversely, silencing of c-Myc increased IDH1 activity, but this increase could be prevented through cosilencing of IDH1-AS1 (Fig. 3K). Together, these findings indicate that c-Myc inhibits IDH1 activity through negative regulation of IDH1-AS1.
IDH1-AS1 Interacts with WT IDH1 and Promotes Its Homodimerization.
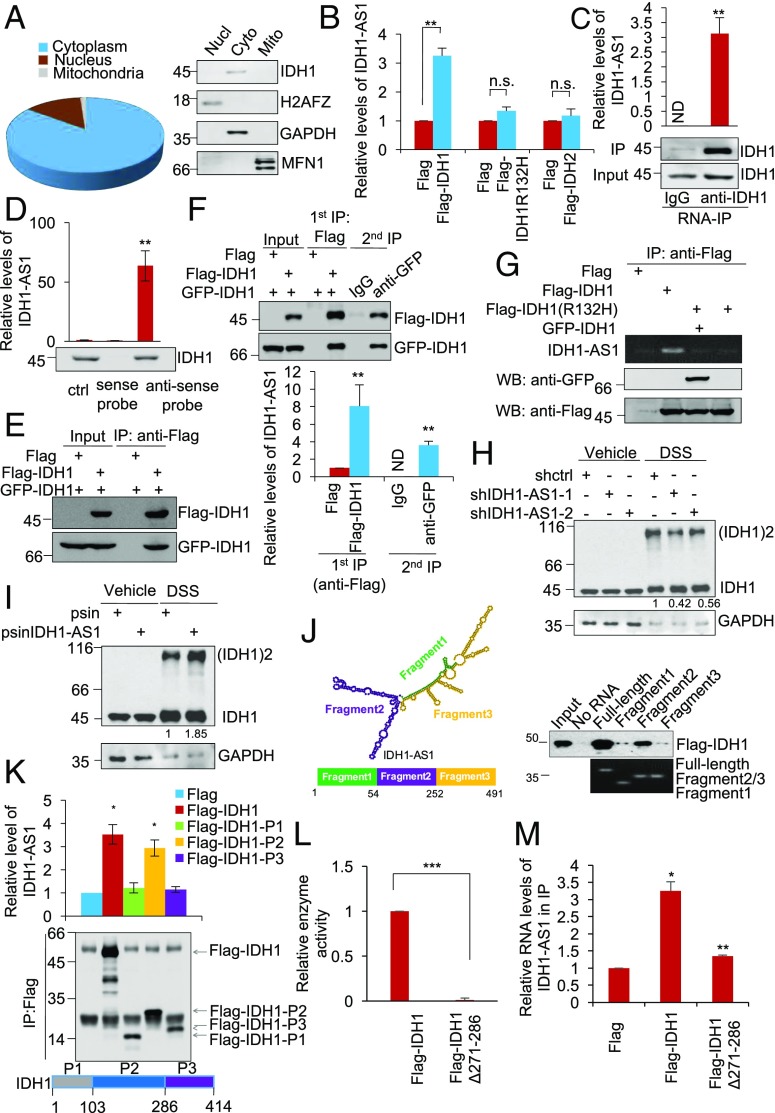
To understand the mechanism by which IDH1-AS1 promotes the enzymatic activity of IDH1, we first examined its subcellular localization. Similar to IDH1, IDH1-AS1 was predominantly located to the cytoplasm (Fig. 4A). RNA immunoprecipitation (RIP) assays demonstrated that Flag-tagged WT IDH1, but not the IDH1R132H mutant or the WT IDH2, was associated with IDH1-AS1 (Fig. 4B). Similarly, endogenous IDH1-AS1 was coprecipitated with IDH1 (Fig. 4C). Moreover, IDH1 was co-pulled down with IDH1-AS1 by using antisense but not sense probes directed against IDH1-AS1 (Fig. 4D). Thus, IDH1-AS1 physically associates with WT IDH1 within the cytoplasm but does not bind to the mutant form of IDH1.
Fig. 4.
See also Fig. S4. IDH1-AS1 interacts with IDH1 and promotes its dimerization. (A) Total RNA from the cytoplasmic, nuclear, or mitochondrial fraction of HeLa cells was subjected to qPCR analysis of IDH1-AS1 expression (Left), whereas total protein from each fraction was subjected to Western blot analysis. The overall relative abundance of IDH1-AS1 cytoplasmic, nuclear and mitochondrial fractions was designated as 100%. Values are means ± SEMs; n = 3. Cyto, cytoplasmic; Mito, mitochondrial; Nucl, nuclear. (B) Whole cell lysates from HeLa cells transduced with Flag-vector, Flag-tagged IDH1, Flag-tagged IDH1R132H, or Flag-tagged IDH2 were subjected to RNA-IP with an anti-Flag antibody. The resulting precipitates were then subjected to qPCR analysis. The relative abundance of IDH1-AS1 in cells transduced with Flag-vector was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (**P < 0.01, two-tailed paired Student’s t test). (C) Whole cell lysates from HeLa cells were subjected to RNA-IP using an anti-IDH1 antibody. The resulting precipitates were then subjected to qPCR analysis. Values are mean ± SEMs; n = 3 (**P < 0.01, two-tailed paired Student’s t test). ND, not detectable. (D) Whole cell lysates from HeLa cells were incubated with in vitro-synthesized, biotin-labeled sense or antisense IDH1-AS1 probes followed by incubation with streptavidin-conjugated beads. The eluted fractions were then subjected to Western blot analysis. Values are means ± SEMs; n = 3 (**P < 0.01, two-tailed paired Student’s t test). ctrl, control. (E) Whole cell lysates from HeLa cells ectopically expressing GFP-tagged IDH1 and Flag-tagged IDH1 were subjected to IP using an anti-Flag antibody. The resulting precipitates were then subjected to Western blot analysis using an anti-Flag or anti-GFP antibody. n = 3. (F) Whole cell lysates from HeLa cells transduced with Flag-Vector and GFP-IDH1, Flag-IDH1 plus GFP-IDH1, and Flag-Vector plus GFP-IDH1 were subjected to IP using an anti-Flag antibody. The resulting precipitates were then eluted using Flag peptides. Ten percent of the eluents were subjected to Western blot analysis using an anti-Flag or anti-GFP antibody. Ten percent of the eluents were subjected to qPCR analysis of IDH1-AS1 expression. Eighty percent of the eluents were subjected to a secondary phase of IP using an anti-GFP antibody. Ten percent of the resulting precipitates were subjected to Western blot analysis using an anti-Flag or anti-GFP antibody. The rest of the eluents were subject to qPCR analysis of IDH1-AS1 expression. Values are means ± SEMs; n = 3 (**P < 0.01, two-tailed paired Student’s t test). (G) Whole cell lysates from HeLa cells ectopically expressing Flag tagged-IDH1 or Flag-tagged IDH1R132H mutant were incubated with in vitro-transcribed IDH1-AS1. The resulting precipitates were subjected to either RT-PCR or Western blot analysis. n = 3. (H) Whole cell lysates from HeLa cells transduced with shctrl, shIDH1-AS1-1, or shIDH1-AS1-2 with or without treatment with DSS were subjected to Western blot analysis. n = 3. (I) Whole cell lysates from HeLa cells overexpressing IDH1-AS1 with or without treatment with DSS were subjected to Western blot analysis. n = 3. (J) Whole cell lysates from HeLa cells ectopically expressing Flag-tagged IDH1 were incubated with in vitro-transcribed full-length IDH1-AS1, IDH1-AS1 Fragment 1, IDH1-AS1 Fragment 2, or IDH1-AS1 Fragment 3 as schematically illustrated. The resulting precipitates were subjected to either RT-PCR or Western blot analysis. n = 3. (K) Whole cell lysates from HeLa cells transfected with Flag-tagged IDH1, Flag-tagged IDH1-P1, Flag-tagged IDH1-P2, or Flag-tagged IDH1-P3 as schematically illustrated were subjected to RNA-IP using an anti-Flag antibody, whereas total RNA was subjected to qPCR analysis. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (L) Whole cell lysates from HeLa cells transduced with Flag-tagged IDH1 or Flag-tagged IDH1 with deletion of residue 271–286 (Flag-IDH1Δ271–286) were analyzed for IDH1 activity. The activity of IDH1 in cells transduced with Flag-tagged IDH1 was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (***P < 0.001, two-tailed paired Student’s t test). (M) Whole cell lysates from HeLa cells transfected with Flag-tagged IDH1 or Flag-tagged IDH1 with deletion of residue 271–286 (Flag-IDH1Δ271–286) were subjected to RNA-IP using an anti-Flag antibody, whereas total RNA was subjected to qPCR analysis. Values are means ± SEMs; n = 3 (*P < 0.05, **P < 0.01; two-tailed paired Student’s t test). DSS, disuccinimidyl suberate; IP, immunoprecipitation; shctrl, control shRNA; WB, Western blot.
The enzymatically active conformation of IDH1 is a homodimer (40). Indeed, ectopically expressed GFP-IDH1 was coprecipitated with ectopically expressed Flag-tagged IDH1 in HeLa cells (Fig. 4E). To characterize the relationship between IDH1-AS1 and IDH1 dimerization, we employed a two-step immunoprecipitation (IP) assay in HeLa cells expressing GFP- and Flag-IDH1. The first-phase IP using anti-Flag antibody captured high levels of GFP-IDH1 and IDH1-AS1 in addition to Flag-IDH1 (Fig. 4F), whereas the second IP using anti-GFP IgG coprecipitated Flag-IDH1 and IDH1-AS1 (Fig. 4F). These results suggest that IDH1-AS1 and dimerized IDH1 form a ternary structure. Indeed, in vitro-transcribed IDH1-AS1 could bind to purified Flag-IDH1 in a cell-free system (Fig. 4G). However, it did not bind to Flag-IDH1R132H or to Flag-IDH1R132H/GFP-IDH1 under the same conditions (Fig. 4G). Furthermore, shRNA silencing of IDH1-AS1 decreased, whereas IDH1-AS1 overexpression increased, the amount of IDH1 dimers detected in HeLa cell total protein extracts captured through chemical cross-linking with DSS (Fig. 4 H and I). Taken together, these data indicate that IDH1-AS1 promotes homodimerization of WT IDH1. Intriguingly, IDH1-AS1 consists of two similarly sized exons with pairwise BLAST analysis indicating ∼40% sequence similarity (Fig. S4), suggesting that it has the potential to form a symmetrical molecule. Consistent with this notion, plotting its secondary structure based on minimum free energy also predicted a symmetrical structure, with each exon contributing one pole of the molecule (Fig. 4J) (41). Deletion of the fragment that formed the predicted symmetric structure diminished binding of IDH1-AS1 with IDH1 (Fig. 4J), implicating that this symmetry may be important for IDH1-AS1 to promote dimerization of IDH1.
To better understand which domain of WT IDH1 physically interacts with IDH1-AS, we used a domain mapping strategy. Either Flag-tagged IDH1 or one of three truncated IDH1 constructs, namely IDH1-P1 (aa 1–103), IDH1-P2 (aa 103–286), or IDH1-P3 (aa 286–414), was separately introduced into IDH1 KO HeLa cells (Fig. 4K) (42). RIP assays demonstrated that IDH1-P2 but not IDH1-P1 or IDH1-P3 was able to bind IDH1-AS1 to the same extent as full-length Flag-IDH1 (Fig. 4K), suggesting that the intermediate part of IDH1 (IDH1-P2) is responsible for its interaction with IDH1-AS1.
To clarify the mechanism by which IDH1-AS1 is disabled in binding to the IDH1R132H mutant, we interrogated the crystal structures of WT IDH1 and IDH1R132H (42). Strikingly, a helix structure formed by aa 271–286 was located within IDH1-P2 in WT IDH1 but not IDH1R132H (42), suggesting that the inability of IDH1-AS1 to bind IDH1R132H is associated with this alteration in protein structure. Indeed, deletion of residues 271–286 abolished binding of WT IDH1 with IDH1-AS1 and inhibited its enzymatic activity (Fig. 4 L and M), consolidating the importance of this segment for the physical association between IDH1 and IDH1-AS1.
IDH1-AS1 Down-Regulation Promotes c-Myc-Mediated Glycolysis.
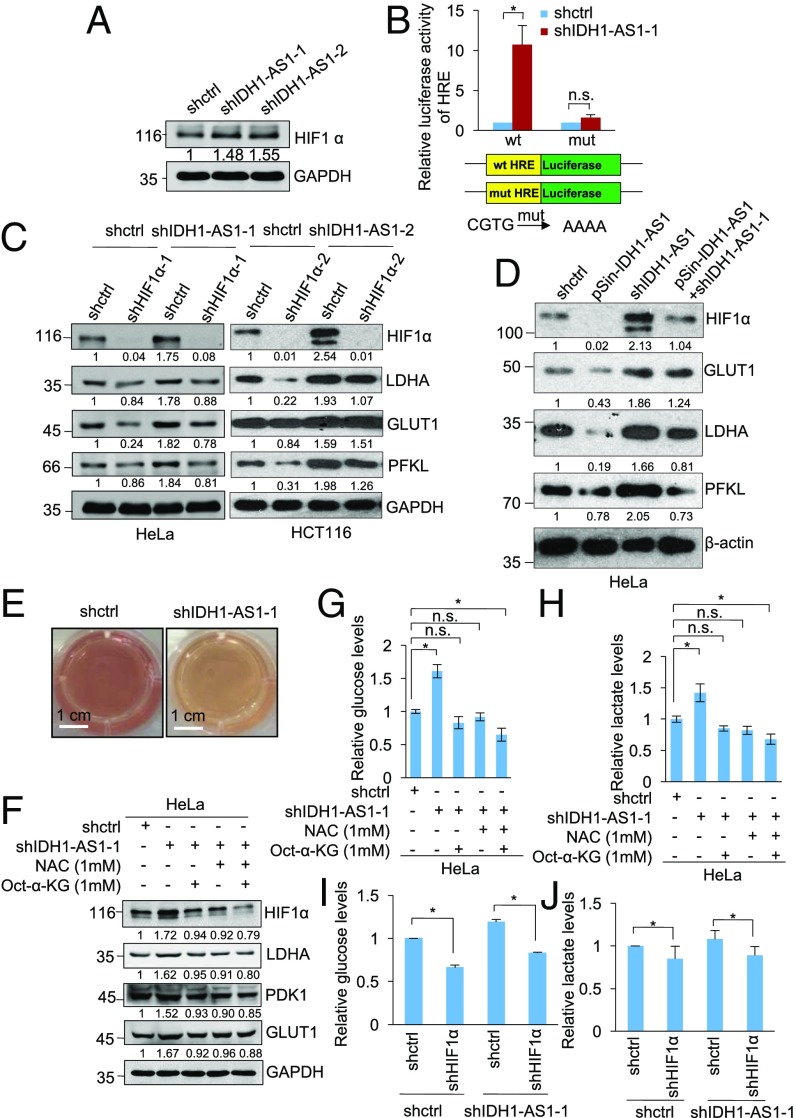
Since IDH1-AS1 promotes the enzymatic activity of IDH1 leading to increased production of α-KG, which is important for hydroxylation and degradation of HIF1α (Fig. 1 C and D) (27), we examined the effect of IDH1-AS1 on HIF1α expression and function. While silencing of IDH1-AS1 (i) up-regulated HIF1α, (ii) enhanced the reporter activity of luciferase constructs containing the WT HIF1α RE (HRE) (pGL3-HRE), and (iii) increased the expression of a number of transcriptional targets of HIF1α (including LDHA, GLUT1, PFKL, and PDK1) in cells under normoxia (Fig. 5 A–C), the introduction of a construct expressing shRNA-resistant IDH1-AS1 transcript reversed up-regulation of HIF-1α and its downstream transcriptional targets caused by IDH1-AS1 knockdown in HeLa cells (Fig. 5D), demonstrating the specificity of the IDH1-AS1 shRNAs and substantiating the role of IDH1-AS1 in the regulation of HIF1α expression. In accordance with the role of IDH1-AS1 in the regulation of HIF1α under normoxic conditions, unbiased RNA-seq analysis of the global transcriptome revealed that IDH1-AS1 silencing caused up-regulation of dozens of known transcriptional targets of HIF1α, inclusive of a large proportion of glycolytic genes (Fig. S5A). This impact on the global HIF1α gene expression signature suggested that the regulatory effects of IDH1-AS1 on glycolysis are mediated through HIF1α. Indeed, silencing of HIF1α nullified changes in its target genes observed when IDH1-AS1 levels were inhibited (Fig. 5C). Moreover, IDH1-AS1 silencing led to the acceleration of acidification of the culture medium (Fig. 5E). Consistently, overexpression of IDH1-AS1 reduced HIF1α expression, inhibited HRE-driven reporter activity, decreased the expression of the HIF1α downstream targets, and inhibited glucose uptake and production of lactate (Fig. S5 B–G).
Fig. 5.
See also Fig. S5. IDH1-AS1 negatively regulates glycolysis through suppressing HIF1α. (A) Whole cell lysates from HeLa cells transduced with shctrl, shIDH1-AS1-1, or shIDH1-AS1-2 were subjected to Western blot analysis. n = 3. (B) HeLa cells transduced with shctrl or shIDH1-AS1-1 were cotransfected with the indicated reporter constructs and Renilla luciferase plasmids. Transcriptional activity was determined by luciferase assays. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). mut, mutant. (C) Whole cell lysates from HeLa and HCT116 cells cointroduced with shctrl or shIDH1-AS1 and shHIF-1α were subjected to Western blot analysis. n = 3. (D) Whole cell lysates from HeLa cells cointroduced with shctrl or shIDH1-AS1-1 and psin-IDH1-AS1 were subjected to Western blot analysis. n = 3. (E) IDH1-AS1 silencing accelerated acidification of the culture medium of HeLa cells. n = 3. (F) Whole cell lysates from HeLa cells transduced with shctrl or shIDH1-AS1-1 with or without treatment with Octyl-α-KG (1 mM) and/or NAC (1 mM) were subjected to Western blot analysis. n = 3. (G and H) Whole cell lysates from HeLa cells transduced with shctrl or shIDH1-AS1-1 with or without treatment with Octyl-α-KG and/or NAC were analyzed for glucose (G) and lactate (H) levels. The relative abundance of glucose or lactate in cells transduced with shctrl without treatment was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (I and J) Whole cell lysates from HeLa cells cointroduced with shctrl or shIDH1-AS1-1 and shHIF-1α were analyzed for glucose (I) and lactate (J) levels. The relative abundance of glucose or lactate in cells transduced with shctrl only was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). n.s., not significant; shctrl, control shRNA. (Scale bars, 1 cm.)
Treatment with the cell-permeable α-KG analog Octyl-α-KG, similar to treatment with the ROS scavenger N-acetyl-l-cysteine (NAC) (43), reversed the increase in LDHA, GLUT1, and PDK1 resulting from IDH1-AS1 silencing (Fig. 5F). When cells were cotreated with Octyl-α-KG and NAC, the expression of HIF1α and its transcriptional targets was further reduced (Fig. 5F), suggesting that down-regulation of α-KG together with the increase in ROS are cooperatively responsible for IDH1-AS1 knockdown-caused up-regulation of HIF1α (27, 44). In support of this, silencing of IDH1-AS1 promoted glucose uptake and lactate production and these changes could be reversed by treatment with Octyl-α-KG or NAC, and were further inhibited by cotreatment with Octyl-α-KG and NAC or HIF1α silencing (Fig. 5 G–J).
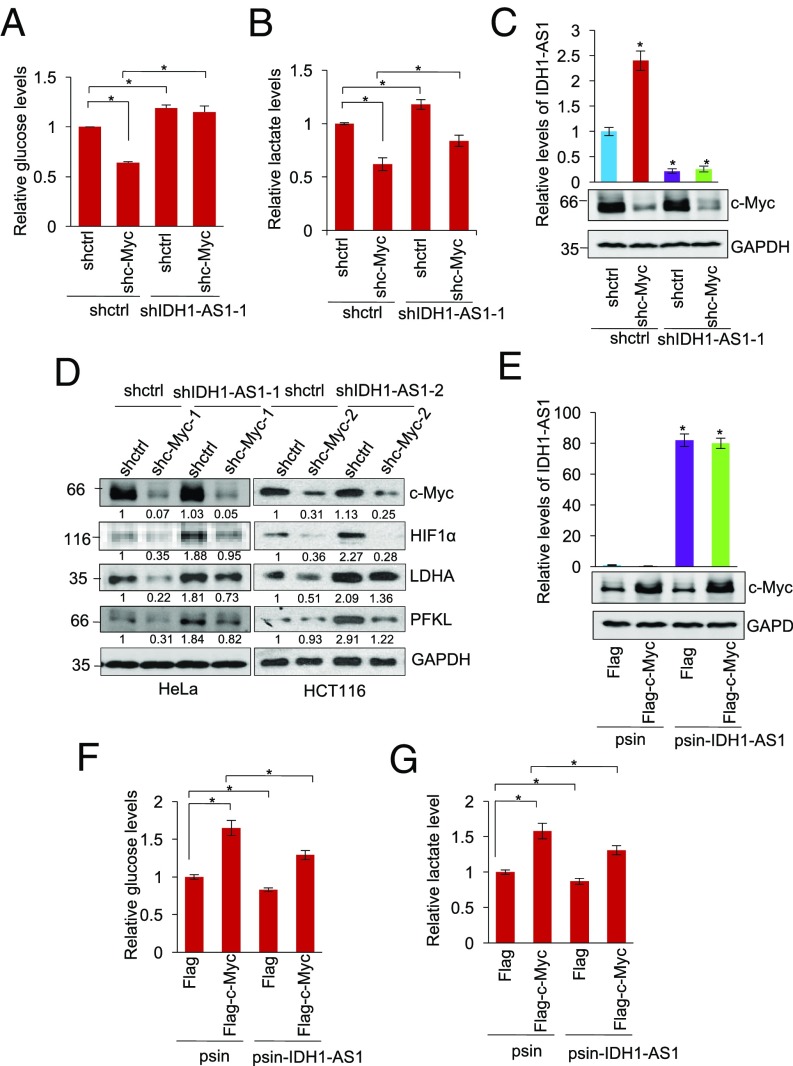
We next investigated the role of IDH1-AS1 down-regulation in c-Myc-mediated glycolysis (45). Cosilencing of IDH1-AS1 reversed, albeit partially, the reduction in glucose uptake and lactate production caused by c-Myc silencing (Fig. 6 A–C). In addition, silencing of IDH1-AS1 also diminished the down-regulation of HIF1α observed in response to c-Myc knockdown along with changes to the HIF1α transcriptional targets LDHA and PFKL (Fig. 6D). Conversely, co-overexpression of IDH1-AS1 inhibited, albeit partially, the increase in glucose uptake and lactate production caused by c-Myc overexpression (Fig. 6 E–G). Therefore, suppression of IDH1-AS1 plays an important role in c-Myc-mediated glycolysis through the IDH1-α-KG-HIF1α axis under normoxic conditions.
Fig. 6.
IDH1-AS1 inhibition contributes to c-Myc-mediated glycolysis. (A and B) Whole cell lysates from HeLa cells cointroduced with shctrl or shc-Myc and shIDH1-AS1-1 were analyzed for glucose (A) and lactate (B) levels. The relative abundance of glucose or lactate in cells transduced with shctrl only was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (C) shRNA knockdown efficiency of IDH1-AS1 and c-Myc in HeLa cells as tested using qPCR and Western blot analysis, respectively. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (D) Whole cell lysates from HeLa and HCT116 cells cointroduced with shctrl or shHIF-1α and shIDH1-AS1 were subjected to Western blot analysis of the expression. n = 3. (E) Successful overexpression of IDH1-AS1 and c-Myc in HeLa cells as tested using qPCR and Western blot analysis, respectively. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (F and G) Whole cell lysates from HeLa cells cointroduced with the psin vector alone or psin-IDH1-AS1 and Flag-vector or Flag-tagged c-Myc were analyzed for glucose (F) and lactate (G) levels. The relative abundance of glucose or lactate in cells transduced with the psin vector plus Flag-vector was arbitrarily designated as 1. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). shctrl, control shRNA.
IDH1-AS1 Inhibits Cell Proliferation and Retards Cancer Xenograft Growth.
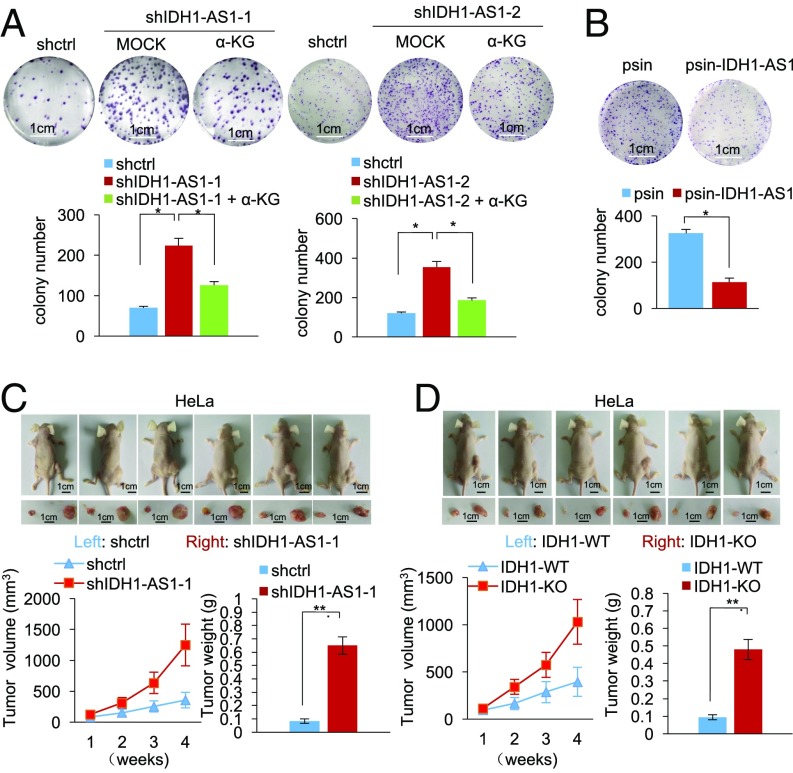
Finally, we examined the functional significance of IDH1-AS1 dysregulation in cancer cells under normoxia. Silencing of IDH1-AS1 enhanced, whereas its overexpression inhibited, HeLa cell proliferation as shown in carboxyfluorescein succinimidyl ester (CFSE) proliferation and clonogenic assays (Fig. 7 A and B and Fig. S6A). Moreover, IDH1-AS1 silencing or IDH1 deficiency promoted the growth of HeLa cell xenografts in nu/nu mice (Fig. 7 C and D). Strikingly, silencing of IDH1-AS1 did not significantly alter proliferation of HeLa cells under hypoxic conditions (Fig. S6B). Taken together, these results suggest that the IDH1-AS1-IDH1 axis may have a tumor-suppressive role under normoxic conditions through the inhibition of cell proliferation.
Fig. 7.
See also Fig. S6. IDH1-AS1 inhibits cell proliferation and shows a tumor-suppressive effect. (A) IDH1-AS1 silencing promoted clonogenicity, which was partially reversed by treatment with α-KG, in HeLa cells. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (B) IDH1-AS1 overexpression inhibited clonogenicity in HeLa cells. Values are means ± SEMs; n = 3 (*P < 0.05, two-tailed paired Student’s t test). (C) IDH1-AS1 silencing promoted HeLa cell xenograft growth in nu/nu mice. HeLa cells (2 × 106) transduced with shctrl or shIDH1-AS1-1 were xenografted s.c. into flanks of nu/nu mice. Four weeks after injection, mice were killed and photographed and tumors were measured and weighed. Values are means ± SEMs; n = 6 (**P < 0.01, two-tailed paired Student’s t test). (D) IDH1 deficiency promoted HeLa cell xenograft growth in nu/nu mice. HeLa cells (2 × 106) carrying WT IDH1 (WT) or those with IDH1 knocked out (IDH1-KO) were xenografted s.c. into flanks of nu/nu mice. Four weeks after injection, mice were killed and photographed and tumors were measured and weighed. Values are means ± SEMs; n = 6 (**P < 0.01, two-tailed paired Student’s t test). shctrl, control shRNA. (Scale bars, 1 cm.)
Discussion
As two major regulators of glycolysis, c-Myc and HIF1α have long been known to crosstalk, either directly or indirectly, and in a cooperative or antagonistic manner (9, 46–49). On one hand, c-Myc promotes mitochondrial respiration, whereas HIF1α actively inhibits this process (50–52). On the other, both c-Myc and HIF1α activate virtually all glycolytic genes (53). Mechanistically, HIF1α inhibits c-Myc functions through various mechanisms, such as direct binding to Max blocking c-Myc/Max interaction or induction of MXI1, which competes with c-Myc for binding MAX that is required for c-Myc-mediated transcriptional activation (52, 54). However, in cancer cells in which c-Myc is commonly deregulated, c-Myc and HIF1α often cooperate to promote glycolysis through up-regulating common downstream targets such as PDK1 and HK2 (9). Moreover, while overexpression c-Myc improves the stability of HIF1α under normoxic and hypoxic conditions (21), knockdown of IDH1 has been reported to result in stabilization of HIF1α in HeLa and several other cell types under normoxia (27). Nevertheless, the mechanisms responsible for these remain to be fully defined.
Herein we demonstrate that c-Myc stabilizes HIF1α through transcriptional repression of the lncRNA IDH1-AS1, a mechanism that otherwise promotes IDH1 activity and thus increases α-KG production and decreases ROS levels leading to degradation of HIF1α (27, 44). Notably, this mechanism appeared to be only functional in cells under normoxia, as IDH1-AS1 did not have a role in regulating HIF1α expression and its function in cells under hypoxic conditions. It is not clear why this difference occurs but one possibility is that, under hypoxia, the inhibitory effect afforded by reduced prolyl hydroxylation signals on VHL-mediated ubiquitin-proteasome degradation of HIF1α is overwhelming (14), which would override the IDH1-AS1-mediated mechanism for HIF1α degradation. Regardless, our results reveal a signaling axis c-Myc-(IDH1-AS1)-IDH1-α-KG/ROS-HIF1α, which plays a role in activation of the Warburg effect under normoxia (Fig. S7). This bears practical implications, in that cancer cells in solid tumors commonly encounter complex microenvironments with hypoxic and normoxic conditions depending on the vasculature status (55, 56). While hypoxia-stimulated HIF1α is essential for metabolism of cancer cells in a hypoxic microenvironment (57), it is conceivable that decreased α-KG production and increased levels of ROS ensure the Warburg effect to provide energy and biosynthesis building blocks required by fast proliferation of cancer cells under normoxic conditions (1).
Noticeably, it has been previously reported that knockdown of IDH1 does not affect the NADPH/NADP+ ratio in 293T cells (58). Indeed, we found that manipulation of c-Myc or IDH1-AS1 similarly did not induce HIF1α expression (Fig. S2 I and J). Nevertheless, knockdown of IDH1-AS1 and overexpression of c-Myc resulted in a reduction in IDH1 activity, whereas knockdown of c-Myc increased the activity of IDH1 (Figs. 1B and 2 A and C). These results suggest that the c-Myc-(IDH1-AS1)-IDH1- HIF1α pathway is not functional in 293T cells, perhaps obstructed by dysregulated IDH1 function, and are informative that further cell-type differences are likely to be found.
Although c-Myc regulates the vast majority of enzymes involved in the TCA cycle (45), its relation to the activity of IDH enzymes is largely uncharacterized. Therefore, the finding that c-Myc attenuates the enzymatic activity of IDH1 highlights IDH1 as a member of c-Myc-responsive metabolic enzymes. However, the negative regulatory effect of c-Myc on IDH1 activity seems paradoxical, as this role of c-Myc diverged from its conventional function as an activator of the TCA cycle (50). Nevertheless, attenuation of IDH1 activity contributes to c-Myc-mediated glycolysis. Thus, these results identify a role of c-Myc in balancing mitochondrial respiration and glycolysis, which may be important for ensuring glycolysis to be executed efficiently in fast-proliferating cancer cells under normoxia (59).
c-Myc attenuated IDH1 activity through repression of lncRNA IDH1-AS1, whose gene is located head to head at close proximity with the IDH1 gene. It is therefore not surprising that IDH1-AS1 plays a role in regulating IDH1; yet, the manner of this regulation appears to be unusual, since IDH1-AS1 did not affect the expression of IDH1 as did some other endogenous antisense lncRNAs that regulated the expression of their neighboring genes, such as BACE1-AS and APOA1-AS (60, 61). Rather, IDH1-AS1 was involved in the facultative activation of IDH1 where it acted to facilitate IDH1 homodimerization (23, 40). Supporting this conclusion, we found that IDH1-AS1 formed a ternary structure with the IDH1 enzyme and that manipulating the levels of IDH1-AS1 affected the quanta of IDH1 dimers. Therefore, IDH1-AS1 is a de novo RNA component of the IDH1 dimer critical for its enzymatic activity. Although our results proposed a likely structural symmetry of IDH1-AS1 based on minimum free energy, further clarification of how binding of IDH1-AS1 to IDH1 enhances its dimerization is awaiting the resolution of the crystal structure of the complex. Regardless, our report indeed showed that homodimerization of a protein is enhanced by an lncRNA.
An important yet intriguing finding of this study was that IDH1-AS1 did not bind to the IDH1R132H mutant. Although there has been no demonstration that the R132H mutation has a significant impact on the structure of the mutant IDH1 protein, we identified that the segment of aa 271–286 that formed a helix structure in WT IDH1 but not IDH1R132H was important for the binding of IDH1-AS1 to WT IDH1. These results indicate that the interaction between IDH1-AS1 and IDH1 does not involve the R132 residue that is frequently mutated in selected types of cancers, including glioma and acute myeloid leukemia (62, 63). Interestingly, the aa 271–286 region of IDH1 has been highlighted in early work as a regulatory segment of the enzyme (40). Resolving the crystal structure of the IDH1-AS1 and WT IDH1 complex is needed to fully understand the basis responsible for the inability of IDH1-AS1 to bind to the IDH1R132H mutant.
As an isocitrate dehydrogenase, IDH1 catalyzes isocitrate to produce α-KG (23). Indeed, α-KG levels increased when IDH1 activity was enhanced by overexpression of IDH1-AS1. Among the diverse functions of α-KG is its role as an electron donor to PHDs, which enables hydroxylation of HIF1α at its oxygen-dependent degradation domain that is required for VHL-mediated HIF1α polyubiquitination and subsequent proteasomal degradation (14). Interestingly, PHD also appears to be the effector responsible for down-regulation of HIF1α caused by the inhibition of ROS (64). In line with this, treatment with exogenous α-KG or a ROS scavenger reversed the increased expression of HIF1α and enhanced glycolysis caused by IDH1-AS1 knockdown. Nevertheless, cotreatment with exogenous α-KG and the ROS scavenger caused only a moderate further inhibitory effect on the expression of HIF1α and glycolysis activity. This was conceivably related to the saturation of PHD activity, which would limit its response to additional stimulation. Therefore, the increase in α-KG production and decrease in ROS levels mediated by the IDH1-AS1-IDH1 axis provide two cooperative yet likely redundant mechanisms that regulate HIF1α expression and glycolysis activity.
The glycolytic effects of IDH1-AS1 were tightly controlled by c-Myc, in that c-Myc silencing and overexpression recapitulated, respectively, the effect of IDH1-AS1 overexpression and silencing. Moreover, silencing of IDH1-AS1 reversed, in part, the impact of c-Myc depletion on glycolysis, whereas overexpression of IDH1-AS1 also partially reversed the promotion of glycolysis observed when c-Myc was overexpressed. Hence, our results indicate collaborative interplay between c-Myc with HIF1α in the regulation of glycolysis, with effects mediated through IDH1-AS1.
The functional significance of the IDH1-AS1-IDH1 axis was demonstrated by the findings that IDH1-AS1 or IDH1 silencing promoted cancer xenograft growth. Thus, consistent with the finding that c-Myc represses the expression of IDH1-AS1, our data suggest that the IDH1-AS1-IDH1 axis may have a tumor-suppressive role. Intriguingly, IDH1 has been reported to promote anchorage-independent growth through mitigating ROS production and to enhance cell survival under conditions of systemic inflammation similarly through inhibiting the production of ROS (35). Nevertheless, these studies were carried out under conditions where additional ROS production was induced. The difference between these studies and our findings may be related to the levels of ROS production involved and the ensuing cellular responses. Cancer cells are known to have elevated ROS levels resulting from oncogenic stimulation (65). On one hand, ROS promote cancer cell survival and proliferation (66). On the other, high levels of ROS suppress tumor growth through the inhibition of proliferation and induction of cell death (67). It is conceivable that when cellular ROS levels are above a threshold, IDH1-mediated down-regulation of ROS protects cells from detrimental effects of excessive oxidative stress (37). However, when cellular ROS levels are below the threshold, down-regulation of ROS by IDH1-AS1-IDH1 does not exert a protective role. Instead, the inhibition of glycolysis by the axis triggers the inhibition of cell proliferation as shown by our results.
Experimental Procedures
Reagents and Antibodies.
The following antibodies used in this study were purchased from the indicated sources: anti-HIF-1α and anti-GAPDH (Santa Cruz); rabbit IgG, anti-GFP, and anti-Flag (Sigma); anti-β-actin, anti-GLUT1, anti-c-Myc, anti-IDH2, anti-H2AFZ, anti-MFN1, and anti-LDHA (Cell Signaling Technology); and anti-IDH1, anti-PFKL, and anti-PDK1 (proteintech). Octyl-α-KG was obtained from Cayman Chemical. DSS was obtained from Sigma. The primers and oligo DNA used in this study are listed in Table S1.
Cell Culture and Transfection.
The cell lines HeLa, HCT116, H1299, and 293T were purchased from ATCC. The cell line P493 was provided by Dr. Ping Gao (University of Science and Technology of China). The IDH1-KO HeLa cells generated with CRISPR/Cas9 system were provided by Prof. Zhiyin Song (Wuhan University, Wuhan, China). Cells were cultured in DMEM supplemented with 10% FBS, 1 mM sodium pyruvate, and 100 mg/mL streptomycin (Invitrogen) (68). Transfection was performed as previously described (68).
ChIP Assay.
The ChIP assay was performed as previously described (69). Briefly, HeLa cells individually expressing Flag, Flag-c-Myc, or Flag-Miz1 were first cross-linked with 1% formaldehyde for 10 min at room temperature. Glycine (0.125 M) was added to stop the reaction. Treated cells were suspended in lysis buffer [50 mM Tris⋅HCl (pH 8.1), 1% SDS, 10 mM EDTA, and protease inhibitors]. Protein A/G-Sepharose beads conjugated with either anti-GFP antibody or control IgG were used for incubation with cell lysates. After elution, the resulting reactants were amplified by PCR.
Glucose Uptake Assay.
HeLa cells expressing control shRNA or indicated shRNAs were cultured in glucose-free DMEM for 16 h, and then incubated with high-glucose DMEM for an additional 24 h. After removal of the culture medium, the levels of intracellular glucose were assessed using a fluorescence-based glucose assay kit (BioVision) according to manufacturer’s instructions.
Lactate Production Assay.
HeLa cells expressing control shRNA or indicated shRNAs were cultured. The levels of lactate in the medium were quantitated with a lactate assay kit that is based on fluorescence activity (BioVision) according to manufacturer’s instructions.
NADPH and α-KG Assay.
HeLa cells expressing control shRNA or indicated shRNAs, psin, or psin-IDH1-AS1 were cultured. NADPH or α-KG levels in the cell lysates were determined using a fluorescence-based lactate assay kit (AAT Bioques for NADPH, BioVision for α-KG) according to the manufacturers’ instructions.
RNA-IP.
RNA-IP was performed as described previously (70). Briefly, 1 ∼ 3 × 107 cells were lysed in hypotonic buffer supplemented with RNase A inhibitor and DNase I before centrifugation. Cell lysates were precleared with protein A/G beads (Pierce) before they were incubated with protein A/G beads coated with the indicated antibodies at 4 °C for 3 h. After extensive washing, the bead-bound immunocomplexes were eluted using elution buffer [50 mM Tris (pH 8.0), 1% SDS, and 10 mM EDTA] at 65 °C for 10 min. To isolate protein-associated RNAs from the eluted immunocomplexes, samples were treated with proteinase K, and RNAs were extracted by phenol/chloroform. Purified RNAs were then subjected to RT-PCR analysis.
Biotin Pull-Down Assay.
All processes were performed in the RNase-free conditions. For the antisense oligomer affinity pull-down assay, sense or antisense biotin-labeled DNA oligomers corresponding to human IDH1-AS1 (3 mg) were incubated with lysates from 2 ∼ 3 × 107 HeLa cells. One hour after incubation, streptavidin-coupled Dynabeads (Invitrogen) were added to isolate the RNA-protein complex. For the in vitro RNA pull-down assay, 3 μg in vitro-synthesized biotin-labeled IDH1-AS1 was incubated with purified Flag or Flag-IDH1 protein for 3 h. Streptavidin-coupled Dynabeads (Invitrogen) were then added to the reaction mix to isolate the RNA-protein complex.
IDH Enzymatic Activity Assays.
IDH enzyme assays were carried out with 100 μM NADP+ and 2 mM MnCl2 in 30 mM Tris⋅HCl buffer (pH 7.5), as described previously (27). Protein extracts from the cytoplasmic fraction were used for analysis of IDH1, whereas protein extracts from the mitochondrial fraction were used for analysis of IDH2 activity.
Xenograft Mouse Model.
HeLa cells (2 × 106) expressing control shRNA or IDH1-AS1 shRNA were s.c. injected into the dorsal flanks of 4-week-old female athymic nude mice (Shanghai SLAC Laboratory Animal Co. Ltd.; n = 6 mice per group). After 4 weeks, mice were killed, and tumors were excised and weighed. The tumors were homogenized, and RNAs were extracted for quantitative real-time RT-PCR analysis. Studies on animals were conducted with approval from the Animal Research Ethics Committee of the University of Science and Technology of China.
Reproducibility.
All of the data analyses were repeated at least three times. The Western blot and ChIP analyses were representative of three independent experiments.
Statistical Analysis.
Analysis was performed using Microsoft Excel and GraphPad Prism to assess differences between varying groups. Statistical significance was defined by Student’s t test and expressed as a P value. P values <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Prof. Zhiyin Song (Wuhan University, Wuhan, China) and Prof. Ping Gao (University of Science and Technology of China, Hefei, China) for providing us with IDH1-KO HeLa and P493 cells, respectively. This work was supported by grants from the National Key R&D Program of China (2016YFC1302302) and the National Natural Science Foundation of China (81430065 and 31371388).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711257115/-/DCSupplemental.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shou Y, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim H, et al. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osthus RC, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 9.Kim JW, Gao P, Liu Y-C, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semenza GL, Rue EA, Iyer NV, Pang MG, Kearns WG. Assignment of the hypoxia-inducible factor 1α gene to a region of conserved synteny on mouse chromosome 12 and human chromosome 14q. Genomics. 1996;34:437–439. doi: 10.1006/geno.1996.0311. [DOI] [PubMed] [Google Scholar]
- 11.Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenza GL. HIF-1: Upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaakkola P, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 15.Masoud GN, Li W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 17.Lee BL, et al. A hypoxia-independent up-regulation of hypoxia-inducible factor-1 by AKT contributes to angiogenesis in human gastric cancer. Carcinogenesis. 2008;29:44–51. doi: 10.1093/carcin/bgm232. [DOI] [PubMed] [Google Scholar]
- 18.Gottschald OR, et al. TIAR and TIA-1 mRNA-binding proteins co-aggregate under conditions of rapid oxygen decline and extreme hypoxia and suppress the HIF-1α pathway. J Mol Cell Biol. 2010;2:345–356. doi: 10.1093/jmcb/mjq032. [DOI] [PubMed] [Google Scholar]
- 19.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 20.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 21.Doe MR, Ascano JM, Kaur M, Cole MD. Myc posttranscriptionally induces HIF1 protein and target gene expression in normal and cancer cells. Cancer Res. 2012;72:949–957. doi: 10.1158/0008-5472.CAN-11-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corpas FJ, et al. Peroxisomal NADP-dependent isocitrate dehydrogenase. Characterization and activity regulation during natural senescence. Plant Physiol. 1999;121:921–928. doi: 10.1104/pp.121.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu N, Giles RE, Kucherlapati RS, Shimizu Y, Ruddle FH. Somatic cell genetic assignment of the human gene for mitochondrial NADP-linked isocitrate dehydrogenase to the long arm of chromosome 15. Somatic Cell Genet. 1977;3:47–60. doi: 10.1007/BF01550986. [DOI] [PubMed] [Google Scholar]
- 25.Bruns GA, Eisenman RE, Gerald PS. Human mitochondrial NADP-dependent isocitrate dehydrogenase in man-mouse somatic cell hybrids. Cytogenet Cell Genet. 1976;17:200–211. doi: 10.1159/000130713. [DOI] [PubMed] [Google Scholar]
- 26.MacKenzie ED, et al. Cell-permeating α-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27:3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paschka P, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 30.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornfeld J-W, Brüning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupaimoole R, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–2402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–955. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 37.Itsumi M, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ. 2015;22:1837–1845. doi: 10.1038/cdd.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peter S, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staller P, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 41.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Suppl 2):W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010;20:1188–1200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 43.Kerksick C, Willoughby D. The antioxidant role of glutathione and N-acetyl-cysteine supplements and exercise-induced oxidative stress. J Int Soc Sports Nutr. 2005;2:38–44. doi: 10.1186/1550-2783-2-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Podar K, Anderson KC. A therapeutic role for targeting c-Myc/Hif-1-dependent signaling pathways. Cell Cycle. 2010;9:1722–1728. doi: 10.4161/cc.9.9.11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mongiardi MP, et al. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget. 2016;7:33257–33271. doi: 10.18632/oncotarget.8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Liu C, Cao S, Xu Y. Elucidation of drivers of high-level production of lactates throughout a cancer development. J Mol Cell Biol. 2015;7:267–279. doi: 10.1093/jmcb/mjv031. [DOI] [PubMed] [Google Scholar]
- 50.Morrish F, Hockenbery D. MYC and mitochondrial biogenesis. Cold Spring Harb Perspect Med. 2014;4:a014225. doi: 10.1101/cshperspect.a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang HJ, et al. Hypoxia inducible factors modulate mitochondrial oxygen consumption and transcriptional regulation of nuclear-encoded electron transport chain genes. Biochemistry. 2015;54:3739–3748. doi: 10.1021/bi5012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis: The seventh hallmark of cancer. Cell Mol Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corn PG, et al. Mix1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- 55.Brown JM, Giaccia AJ. The unique physiology of solid tumors: Opportunities (and problems) for cancer therapy. Cancer Res. 1998;58:1408–1416. [PubMed] [Google Scholar]
- 56.Less JR, Skalak TC, Sevick EM, Jain RK. Microvascular architecture in a mammary carcinoma: Branching patterns and vessel dimensions. Cancer Res. 1991;51:265–273. [PubMed] [Google Scholar]
- 57.Harris AL. Hypoxia: A key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 58.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation. Oncol Lett. 2012;4:1151–1157 (review). doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halley P, et al. Epigenetic regulation of the apolipoprotein gene cluster by a long non-protein-coding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Mutation analysis of isocitrate dehydrogenase in acute lymphoblastic leukemia. Genet Test Mol Biomarkers. 2012;16:991–995. doi: 10.1089/gtmb.2011.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balss J, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 64.Pan Y, et al. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol. 2007;27:912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liou G-Y, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 67.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 68.Huang X, Wu Z, Mei Y, Wu M. XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 2013;32:2204–2216. doi: 10.1038/emboj.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin L, et al. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci USA. 2011;108:15840–15845. doi: 10.1073/pnas.1019312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.