Significance
Gene expression is governed by the opposing forces of transcription and decay. How RNA decay modulates the transcriptome is largely unknown, including whether the three major cytoplasmic degradative pathways (mRNA decapping, the RNA exosome, and SOV/DIS3L2) act on specific sets of RNAs. We used maximum likelihood mathematical modeling and RNAseq to analyze mRNA decay in wild-type and Arabidopsis RNA decay mutants. We demonstrate that mRNA decapping contributes broadly to mRNA decay (68% of RNAs), and is especially important for short-lived RNAs. SOV deficiency caused slower decay of few transcripts, but also elicited broad feedback: Many RNAs decayed faster, and required mRNA decapping for this faster rate. Despite altered turnover, these RNAs were maintained at near-wild-type abundance.
Keywords: RNA degradation, posttranscriptional regulation, SOV/DIS3L2, RNA turnover, mathematical modeling
Abstract
The decay of mRNA plays a vital role in modulating mRNA abundance, which, in turn, influences cellular and organismal processes. In plants and metazoans, three distinct pathways carry out the decay of most cytoplasmic mRNAs: The mRNA decapping complex, which requires the scaffold protein VARICOSE (VCS), removes a protective 5′ cap, allowing for 5′ to 3′ decay via EXORIBONUCLEASE4 (XRN4, XRN1 in metazoans and yeast), and both the exosome and SUPPRESSOR OF VCS (SOV)/DIS3L2 degrade RNAs in the 3′ to 5′ direction. However, the unique biological contributions of these three pathways, and whether they degrade specialized sets of transcripts, are unknown. In Arabidopsis, the participation of SOV in RNA homeostasis is also unclear, because Arabidopsis sov mutants have a normal phenotype. We carried out mRNA decay analyses in wild-type, sov, vcs, and vcs sov seedlings, and used a mathematical modeling approach to determine decay rates and quantify gene-specific contributions of VCS and SOV to decay. This analysis revealed that VCS (decapping) contributes to decay of 68% of the transcriptome, and, while it initiates degradation of mRNAs with a wide range of decay rates, it especially contributes to decay of short-lived RNAs. Only a few RNAs were clear SOV substrates in that they decayed more slowly in sov mutants. However, 4,506 RNAs showed VCS-dependent feedback in sov that modulated decay rates, and, by inference, transcription, to maintain RNA abundances, suggesting that these RNAs might also be SOV substrates. This feedback was shown to be independent of siRNA activity.
Gene expression is a highly regulated process with major research focus on transcriptional control, and its resulting impact on RNA abundance. However, RNA steady-state levels arise through balanced activity of transcription and decay, and, despite strong advances in understanding regulatory roles of transcription factors (1, 2), we have only a rudimentary understanding of how cytoplasmic RNA decay pathways shape the transcriptome. Specialized decay pathways, e.g., miRNA-specified cleavage, target specific mRNA substrates (3), but most mRNAs do not decay by specialized pathways, and instead undergo deadenylation followed by either 3′-to-5′ decay [via the RNA exosome or the exoribonuclease SUPPRESSOR OF VCS (SOV)/DIS3L2], or by 5′-to-3′ decay, which removes the 7-methyl guanosine cap (decapping) followed by exoribonucleolytic decay by EXORIBONUCLEASE (XRN1 or XRN4) (4). The extent to which these three cytoplasmic pathways contribute to overall decay, whether they have specialized functions, and whether transcripts show decay pathway specificity are not known.
Decapping of mRNA is executed by a DCP2−DCP1 heterodimer (5, 6). In metazoans and plants, decapping activity also requires a scaffold protein, known as VARICOSE (VCS) in plants and HEDLS/Ge-1/EDC4 in metazoans (7–10). Decapping efficiency is also influenced by accessory factors, including Lsm proteins and DExD/H-box RNA helicase Dhh1 (11–16). Analyses of mRNA decapping and 5′-to-3′ decay substrates carried out in yeast suggested that most of the transcriptome decays via the 5′-to-3′ pathway; however, contributions from other decay pathways were not assessed (17). Potential roles for mRNA decapping in shaping the transcriptome have been suggested in studies of osmotic stress responses (18, 19), yet we have little understanding of this pathway’s decay substrates.
RNA decay in the 3′-to-5′ direction is carried out by the RNA exosome, a large multisubunit complex with both nuclear and cytoplasmic functions, and also by SOV/DIS3L2 (20, 21). RNA substrates of the exosome were analyzed in Arabidopsis using knockdowns of exosome subunits and microarrays, which revealed miRNA intermediates and noncoding RNAs, yet few mRNAs (22). Nevertheless, the Arabidopsis RNA exosome is essential in Arabidopsis, as loss-of-function mutations in the catalytic subunit (RRP44) fail to produce female gametophytes (23).
We previously showed that SOV is a conserved 3′-to-5′ exoribonuclease that suppresses the phenotype of vcs mutants by contributing to decay of decapping substrates (23). In metazoans, the SOV ortholog, DIS3L2, has emerged as an important ribonuclease, as mutants show an overgrowth condition called Perlman syndrome in humans, and a similar overgrowth condition in Drosophila (24–26). Perlman arises because DIS3L2 is required to degrade the uridylated precursor to the let-7 miRNA (25–28). DIS3L2 has also been shown to degrade mRNAs (e.g., during apoptosis and Drosophila development) and noncoding RNAs (24, 29, 30). Normal functions for this enzyme in Arabidopsis, however, are not obvious, as the sov mutation was discovered in the widely used wild-type Arabidopsis strain, Col-0 (23). One goal for this study was to investigate the impact of SOV on the Arabidopsis transcriptome.
Here we describe a genome-wide analysis of cytoplasmic mRNA decay using single and double mutants with defects in decapping and SOV. Maximum likelihood modeling allowed us to determine the decay rates of 17,293 transcripts, and to quantify the contributions of VCS and SOV to these rates. Simple patterns of additive genotypic effects were found, but also more complicated patterns suggesting activation of feedback pathways in decay mutants. An analysis of the identified RNA substrates indicated some specializations of the three decay pathways, and an especially important role for mRNA decapping in decay of short-lived RNAs.
Results
Strategy and Modeling.
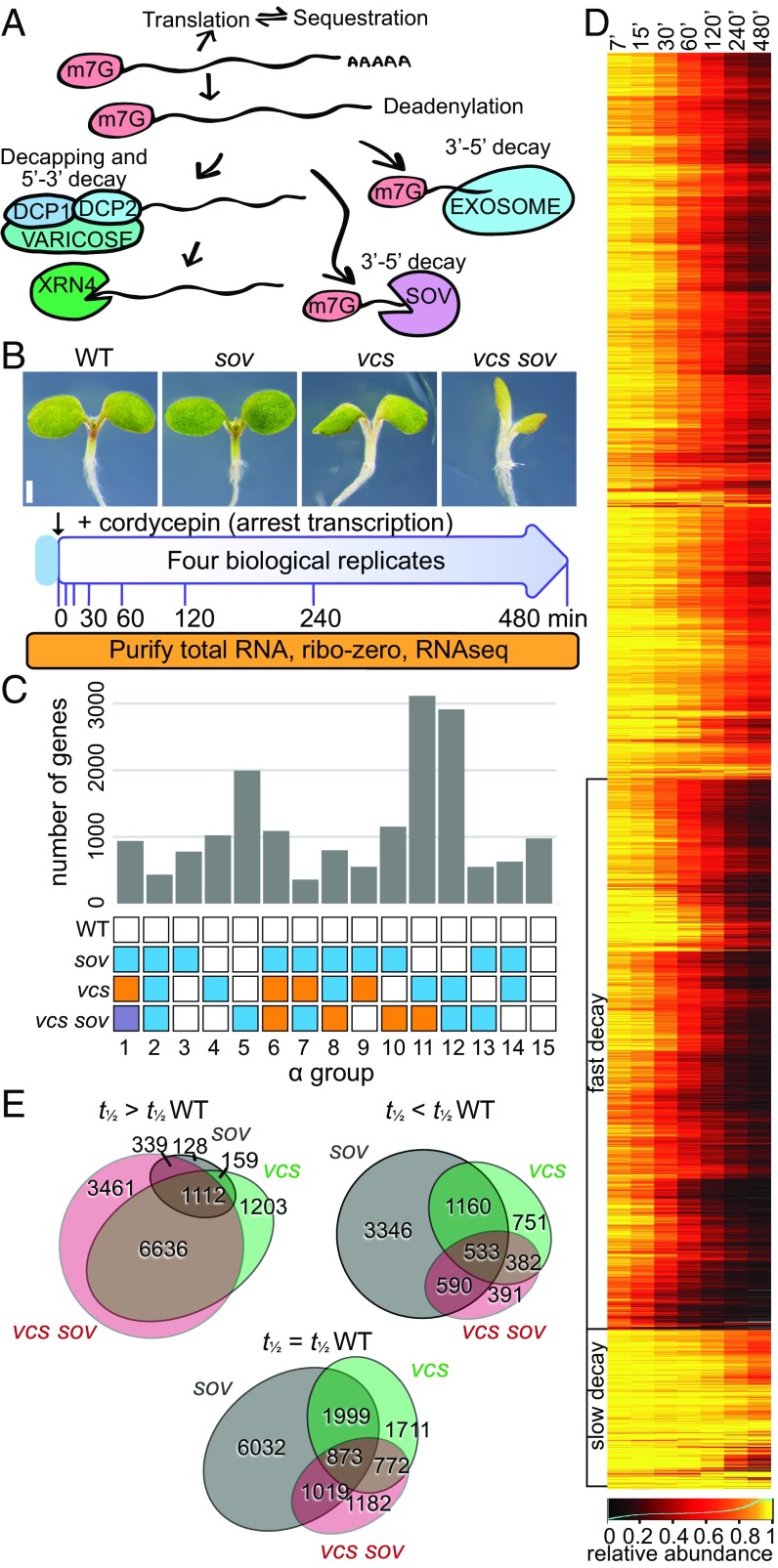
To identify the mRNA substrates of SOV and mRNA decapping (Fig. 1A), we carried out genome-wide mRNA decay analyses using four Arabidopsis genotypes (Fig. 1B and Table 1), expecting RNA substrates to decay more slowly in RNA decay mutants. We analyzed mRNA decapping using vcs-7, a null allele lacking the scaffold for the mRNA decapping complex (7–9). Because vcs-7 was isolated in the Col-0 accession, and Col-0 is a sov mutant (23), we refer to this double mutant as vcs sov. The vcs single mutant was analyzed using the previously described transgenic line carrying the functional SOV allele from Landsberg erecta expressed from its endogenous promoter (8, 23). We also used a synthetic wild type (WT), the Col-0 accession carrying this same transgene, and the sov single mutant (the Col-0 accession). To measure RNA decay, we treated 5-d-old seedlings with a transcription inhibitor (cordycepin), and collected samples across an 8-h time course (higher temporal resolution at earlier time points; Fig. 1B and Fig. S1A). To include mRNA decay intermediates in our analysis, including deadenylated mRNAs and decay intermediates, we isolated total RNA, subjected it to ribosomal RNA depletion, and then carried out RNAseq.
Fig. 1.
RNA decay experimental strategy and outcomes. (A) Schematic of cytoplasmic RNA decay pathways in Arabidopsis. (B) Experimental design. Five-day-old Arabidopsis seedlings were treated as presented. (C) Distribution of RNAs in RNA decay α groups indicating genotypic influence as a histogram. Colored boxes within each group represent identically modeled decay rates. (D) Depiction of RNA decay. WT transcript abundance changes following inhibition of transcription were hierarchically clustered and are presented as a heatmap. Clusters of transcripts with fast and slow decay rates are indicated. (E) Proportional Venn diagrams show overlap in transcripts that were stabilized (t1/2 > t1/2 WT), unaffected (t1/2 = t1/2 WT), or destabilized (t1/2 < t1/2 WT) in mutants. Gray circles, sov; green circles, vcs; pink circles, vcs sov.
Table 1.
Genotypes used in this study
Multiplexed sequence libraries averaged 28 million reads, and 98 to 99% mapped to the Arabidopsis genome. Each set of four bioreplicates showed high reproducibility (mean Pearson correlation = 0.993 between biological replicates) (Fig. S1B). Read counts for each gene were normalized to library size, and libraries were scaled to 1 × 106. Because stable RNAs become an increasingly large proportion of library-normalized read counts as RNA decay proceeds, we scaled read count values using a decay factor for each library based on levels of 30 abundant and stable RNAs (Dataset S1). To further facilitate decay rate comparisons, data were normalized to the mean expression of the respective initial (T0) sample. After removing low abundance transcripts [0 reads in the T0 of any genotype, and those with a sum of the mean T0 reads per million (RPM) from all genotypes ≤4], we initiated analysis of mRNA decay data for 18,674 genes.
To identify the decay pathways used by each mRNA, we used likelihood functions to estimate mRNA decay rates in each genotype, and to assign each gene a statistically supported genotypic effect. Modeling considered all 15 combinations of genotypic influences: An mRNA might show a different decay rate in each genotype, a few genotypes might share decay rates, or all genotypes could share the same decay rate (Fig. 1C). Typically, RNA decay studies assume a single exponential decay curve (31–33); however, the decay rate of some transcripts appeared to slow over the time course. To accurately quantify these more complicated decay kinetics, we compared exponential decay and decaying decay models, and estimated parameters for the initial primary decay rate (α) and a secondary decay of decay rate (β) for each transcript. As with the α groups, we reasoned that the decay-of-decay rate might have the same 15 potential impacts of genotype on the slowing of decay rate, or decay rate might remain steady throughout the time course (β = 0) (Fig. S2A). We assessed data quality using the estimated parameter for variance (σ2). This revealed a high-quality data set of 17,293 genes, which was used for all subsequent analyses, and a lower-quality set (1,381 genes, σ2 ≥ 0.0625) (Fig. S2 B–D). All data, and their quality scores, are available in Dataset S1.
The mRNA Decay Landscape in Arabidopsis.
An analysis of gene-specific RNA abundance over the time course showed a diverse range of decay rates, including groups with fast and slow decay (Fig. 1D). Decay rates ranged from inestimably slow to very fast, with the most rapidly degraded mRNA (DREB1B, At4g25490) having a decay rate of 0.185 (t1/2 3.7 min). These data compared well to a previous genome-wide analysis of RNA decay in Arabidopsis (32) (Fig. S2E), which found a similar range of decay rates, although their shortest half-life was 12 min, and median mRNA half-life also differed for the two studies (107 min for WT in our study, vs. 228 min). These differences are likely due to sampling strategies, methodologies, and tissues analyzed.
The identified contributions of mRNA decapping and SOV to decay are depicted in Figs. 1E and 2A. Gene-specific requirements for these pathways were evident because RNAs of some genes decayed at the same rate in WT and mutants, and others showed longer half-lives in the mutants, suggesting they were VCS and SOV substrates. In vcs, the median t1/2 was 30 min slower (107 min to 137 min), and the 168-min median half-life in vcs sov (31 min longer than vcs) indicated that SOV contributed to decay of many of these mRNAs in vcs mutants (Fig. S2F). However, a striking and unexpected finding was that some RNAs decayed faster in mutants. Surprisingly, a large majority of the decay rates altered in sov were faster, with a median half-life of 97 min, 10 min shorter than the WT.
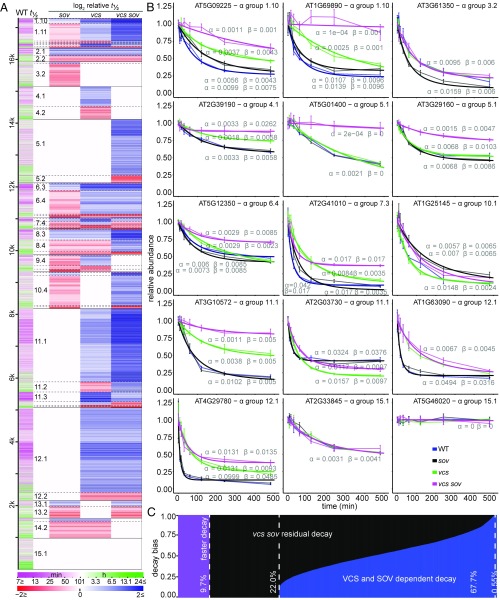
Fig. 2.
Substrates for mRNA Decapping and SOV. (A) Genotypic effects on decay rate. Predominant patterns were revealed by genotype half-life comparisons presented as a heatmap. Transcript half-life in WT is represented in the first column. Relative stabilization or destabilization is presented in the remaining four columns as log2 half-life relative to WT (WT, sov, vcs, vcs sov). Transcripts (rows) are ordered by α groups, which are divided by solid black lines. Subgroups are defined by dashed lines, and large subgroups are labeled in the second column. (B) Representative transcript decay profiles: relative RNA abundance following inhibition of transcription (thin lines, mean ± SE, n = 4; thick line, modeled values). The α and β parameters are listed near each model line (blue, WT; black, sov; green, vcs; pink, vcs sov). (C) Genome-wide contributions of VCS and SOV to mRNA stability based on the relative decay rate decrease (blue) and residual decay rate (black) in vcs sov. Transcripts with faster decay in vcs sov are represented in purple.
More detail about the patterns of VCS and SOV contributions to RNA decay came from analysis of the α groups (Fig. 1C), which, because of the presence of faster decay rates, were divided into subgroups to reflect whether RNAs showed slower or faster decay in the mutants; these are labeled numerically with the α group number following a decimal as in Fig. 2A; e.g., subgroup 12.1 is the largest subgroup (2,657 mRNAs). The three most populated α groups, 5, 11, and 12, which together represented 46.4% of analyzed RNAs, demonstrated three important patterns of cytoplasmic mRNA decay. The largest (18%) was α group 11; these genes required VCS for normal decay of their RNA, and, because they decayed more slowly in vcs sov than vcs, they were also SOV substrates. The second-most populated was α group 12 (16.8%); RNAs from these genes required VCS, but not SOV, for normal decay rates. Third-most populated was α group 5 (11.5%); this group’s RNAs were only affected in vcs sov seedlings, indicating functional redundancy between VCS and SOV, as either pathway was sufficient to sustain normal decay rates. These findings indicated that SOV has the potential to make broad contributions to the transcriptome, but that, in 5-d Arabidopsis seedlings, mRNA decapping was typically sufficient to sustain most normal decay rates. Representative examples of these and other decay patterns are also depicted in graphs of actual and modeled data (Fig. 2B).
Contributions of SOV to decay were much more complex. When examined in the vcs mutant background, of the 9,112 genes whose RNA required VCS for decay, 3,374 showed even slower decay in vcs sov, and, genome-wide, 77.7% of seedling-expressed genes required VCS and/or SOV for normal decay (Fig. 2C). However, SOV substrates that followed the predicted slower decay in sov mutants were rare. RNAs that were substrates for SOV, but not decapping, were identified in α group 13 (Fig. 2A). The slower RNA decay rates for sov in subgroup 13.1 support a role for SOV in cytoplasmic mRNA decay, albeit RNAs of only 197 different genes were in this group. RNAs with slower decay rates in sov and that were also substrates for mRNA decapping were found in α subgroups 1.10 (145 genes), 6.3 (212 genes), 8.3 (335 genes), and 10.3 (103 genes), which, together with 13.1, comprised 5.7% of the analyzed genes. However, each of these α groups contained much larger subgroups with RNAs that showed faster decay in sov (subgroups 1.11, 6.4, 8.4, 10.4, and 13.2); these 2,953 genes (17% of modeled RNAs) decayed ∼43% faster in sov than in WT. These faster RNA decay rates demonstrate a complicated influence of SOV on mRNA decay; siRNAs and feedback as possible explanations are discussed in Elevated 21- to 22-nt siRNAs in RNA Decay Mutants Do Not Generally Associate with Faster Decay Rates.
RNA decay that was independent of VCS or SOV, which we call residual decay, arose from the combined activity of other RNA decay pathways, and most activity is likely from the exosome. RNAs decaying exclusively by residual pathways were found in α group 15 (975 RNAs; 5.6%); because this group contained some very stable RNAs, it is possible that modest SOV or VCS contributions to normal decay were not resolved. Residual decay was apparent in most RNA substrates of VCS and SOV, as only 0.55% of RNAs decayed exclusively by VCS and SOV (i.e., no degradation in vcs sov was detected) (Fig. 2C).
Elevated 21- to 22-nt siRNAs in RNA Decay Mutants Do Not Generally Associate with Faster Decay Rates.
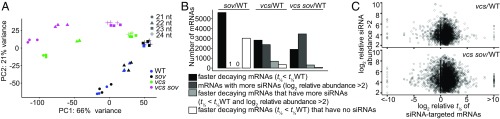
The common faster RNA decay rates in sov, and other less common patterns of faster decay, including in vcs and vcs sov, were surprising, and led us to test whether these arose from siRNAs, which have previously been reported as elevated in mRNA decay mutants (21, 34). We sequenced small RNAs (<200 nt) from all four genotypes, using seedlings age-matched to those of the decay analysis. Multiplexed sequencing yielded an average of 10.5 million genome-aligned 21- to 24-nt reads. A principal component analysis revealed clustering of read lengths for each genotype, indicating high reproducibility of bioreplicates (Fig. 3A). The 21- to 22-nt reads, which have ascribed roles in posttranscriptional gene silencing (3, 35), were widely separated for vcs and vcs sov, and clustered for sov and WT.
Fig. 3.
Widespread increases in siRNA do not drive faster decay. (A) Principal component analysis of DESeq2 normalized read counts of 21- to 24-nt small RNAs. PC1 and PC2 are principal components 1 and 2. (B) Genotype comparisons of shorter half-life mRNAs (t1/2 < t1/2 WT), mRNAs with elevated relative siRNA abundance (>2), shorter half-life mRNAs (t1/2 < t1/2 WT) with elevated siRNAs, and mRNAs with no siRNAs. (C) Relative siRNA abundance compared with relative t1/2 of corresponding mRNAs in vcs and vcs sov compared with WT.
We identified siRNAs as 21- to 22-nt reads aligning antisense to exons, indicating that they likely arose through the activity of an RNA-dependent RNA polymerase, and quantified differential expression using DESeq2 (Padj < 0.05, |log2 rel. abundance| > 2) (Fig. 3B) (36). As described previously, siRNA levels were higher in vcs mutants than the WT (Fig. S3A) (21, 34). In sov, where faster decay was most common, only a single gene had elevated siRNAs compared with WT (At2g01008), and this gene’s mRNA decayed at the same rate in WT and sov. The lack of elevated siRNAs in sov suggested that either the extensive faster mRNA decay rates in sov arose from a mechanism independent of siRNAs or that sov mutants might have heightened siRNA sensitivity. To assess this latter possibility, we identified transacting siRNAs (tasiRNAs) (37) and tested for heightened sensitivity by comparing decay rates of their mRNA targets (Fig. S3 B and C). However, most tasiRNA targets showed identical decay rates; only three showed faster decay, a ratio similar to the genome-wide distribution of faster decay rates.
We also assessed siRNA levels and activity in vcs and vcs sov, where we found genes with elevated 21- to 22-nt siRNAs (|log2 FC| > 2, Padj < 0.05) (2,235 and 3,244, respectively). Although vcs and vcs sov had 2,826 and 1,896 genes that encoded faster-decaying RNAs, faster decay rates did not appear to be explained by siRNA-induced cleavage and decay. Most genes encoding faster-decaying RNAs did not have elevated siRNAs (75.5% and 83.8%, in vcs and vcs sov, respectively) (Fig. 3B). Indeed, a comparison of relative decay rates with relative siRNA levels showed no significant relationship (Fig. 3C). These data indicate that, in our system, faster decay rates in mutant backgrounds, including those in sov, largely arose independently of siRNAs.
Feedback on mRNA Decay Defects.
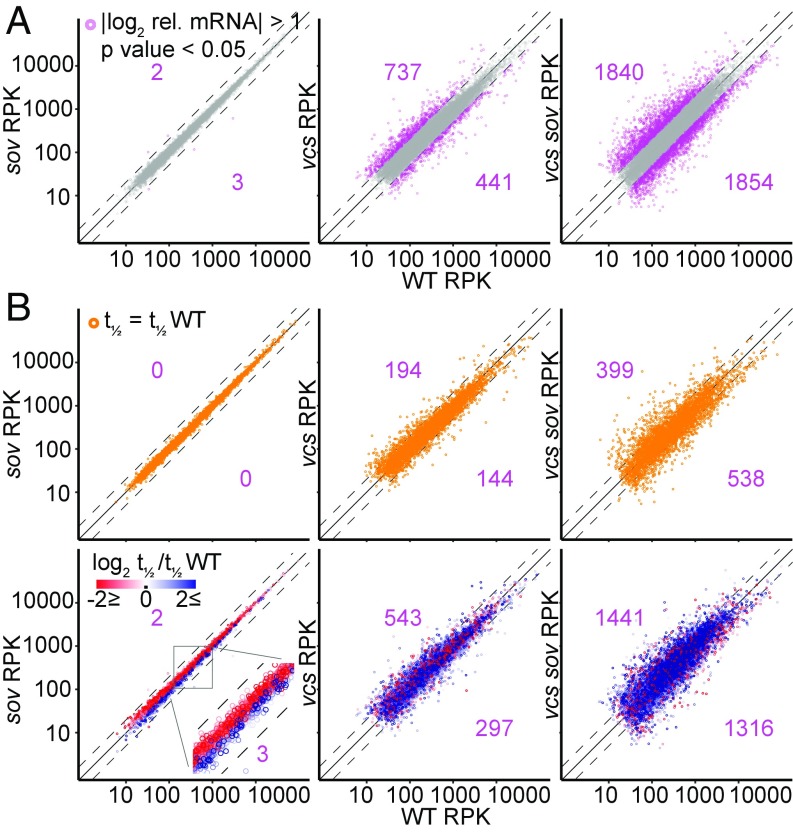
To assess whether the observed faster decay rates might be feedback responses, we investigated the relationship between altered decay rates and mRNA abundance. A comparison of T0 RNA abundances in sov, vcs, and vcs sov, to those in the WT revealed tight correlations for sov (only five mRNAs with significantly altered steady-state levels; Fig. 4A). By contrast, vcs and vcs sov mutants had many RNAs with significantly altered steady-state levels, including ones elevated or depleted relative to the WT.
Fig. 4.
RNA decay feedback in sov maintains WT-like RNA levels. (A) Comparison of DESeq normalized transcript abundance scaled to 1-kb mRNA length (RPK) from T0 samples of sov, vcs, and vcs sov to WT. Differentially expressed genes (|log2 relative abundance| > 1, Padj < 0.05) are pink. Numbers indicate counts of higher (upper left) and lower (lower right) expressed genes compared with WT. (B) Comparisons of transcript abundances for the subset of RNAs which decay at the same rate as in WT (Upper, orange), and for the subset of RNAs which decay faster (red) or slower (blue) than in WT (Lower). Numbers indicate counts of higher (upper left) and lower (lower right) expressed genes compared with WT. Dashed lines, |log2 rel. abundance| >1; solid line, equal expression.
To distinguish between RNA abundance differences that arose as a direct result of altered mRNA decay and those that resulted from indirect responses, we analyzed abundances of RNAs that decayed at the same rate as WT separately from those that had differing decay rates (Fig. 4B). In vcs and vcs sov, many RNAs that decayed with the same kinetics as WT showed altered abundances, which suggested either altered transcription rates or changes in populations of some cell types. The comparisons of vcs and vcs sov RNAs with faster or slower decay rates also showed many RNA abundance differences. We anticipated that slower-decaying RNAs would be more abundant, and faster-decaying RNAs would be less abundant; however, we saw no such pattern. To determine whether the abundance differences reflected altered morphology in vcs and vcs sov, we tested the differentially expressed genes for enrichment of genes previously identified as showing cell type specificity (38, 39). Modest enrichment of some cell types, e.g., xylem-related gene sets, in the differentially expressed RNAs was found (Dataset S1), suggesting an influence on some of the altered transcript abundances in vcs and vcs sov.
By contrast, despite RNA abundances in sov resembling those of WT, decay rates differed widely. Moreover, RNAs with modestly higher abundance showed faster decay rates, and RNAs with modestly lower abundance showed slower decay rates (Fig. 4B). This pattern extended to RNAs with small decay rate differences (Fig. S4), and is the opposite of the expectation for a simple decay rate/abundance relationship. RNAs that are both more abundant and faster decaying are likely to have elevated transcription rates. Similarly, RNAs that are both less abundant and slower decaying are likely to have slower transcription rates. These findings suggest that SOV loss leads to feedback that maintains RNAs at WT levels by adjusting both decay and transcription rates, reminiscent of the feedback system between transcription and RNA degradation that has been identified in yeast, called RNA buffering (17, 40).
A possible role for mRNA decapping in sov’s faster decay rates was suggested by the heat map (Fig. 2A), as faster decay was generally eliminated in vcs sov. Faster-decaying RNAs were concentrated in subgroups 1.11, 3.2, 6.4, 8.4, and 10.4, and the faster decay rates of all 3,320 of these RNAs were eliminated in vcs sov. The decay profile of subgroup 1.11 is particularly interesting; VCS was required to maintain normal decay rates of these 551 RNAs, and, because their decay rates were even slower in vcs sov, they are also SOV substrates. A similar case can be made for subgroup 10.4, where 939 RNAs showed faster decay in sov mutants, but, in vcs sov, they degraded more slowly than WT. These groups are similar because the faster decay in sov was negated when decapping was also lost, but they differ in that VCS is necessary to sustain normal decay rates for subgroup 1.11, but not 10.4. Inspection of the heat map suggests similar explanations for other cohorts of fast-decaying RNAs in sov mutants.
We also observed RNAs with faster decay rates in vcs that suggested the existence of SOV-mediated feedback. For example, subgroup 11.2’s 315 RNAs decayed at the same rate in WT and sov, faster in vcs, but then slower in vcs sov, suggesting that SOV was required for the faster decay in vcs (Fig. 2A). A general moderating influence of SOV on mRNA levels was also revealed in comparisons of T0 RNA abundances in vcs (relative to WT) and vcs sov (Fig. 4B and Fig. S4B). Participation of both SOV and VCS in feedback decay might also explain subgroup 8.4, whose 361 RNAs have identical faster decay rates in sov and vcs, but slower decay in vcs sov. Finally, not all of the faster decay rates observed can be explained by SOV- and/or VCS-mediated feedback. For example, RNAs in subgroup 2.2 (194) decay faster in sov, vcs, and vcs sov, and RNAs in subgroup 12.2 (255) decay faster in both vcs and vcs sov. This suggests additional decay pathways might also participate in feedback.
Specialized Functions of mRNA Decay Pathways.
To determine whether the different cytoplasmic RNA decay machinery degrades functionally distinct mRNAs, we searched the α-subgroups (Fig. 2A) for overrepresented gene ontologies (GOs) (41). The strongest enrichment was chloroplast part in subgroup 15.1 (VCS- and SOV-independent decay) (Padj 5E-31), and this subgroup also showed strong enrichment for mitochondrial membrane (Padj 2E-12). Structural constituent of ribosome was strongly overrepresented in subgroup 12.1 (VCS-dependent decay), with Padj 2E-24, and this same subgroup showed overrepresentation of RNA methylation (Padj 2E-19). Subgroups containing RNAs that use both VCS and SOV also had overrepresented GOs, e.g., subgroup 5.2 included plastid stroma (Padj 1E-14) and glyceraldehyde-3-phosphate metabolism (Padj 1E-14), and subgroup 11.3 included transcription factor activity (Padj 1E-08) and response to chitin (Padj 3E-11). The distinct GO enrichments suggest these mRNA decay pathways play distinct roles in shaping the transcriptome. The complete set of GO enrichments for the subgroups is provided in Dataset S1.
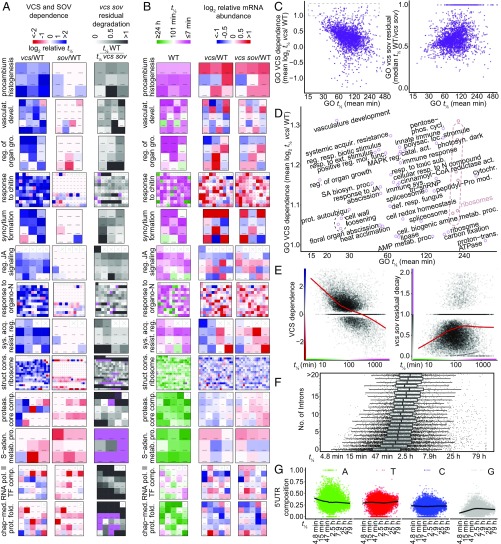
As a complementary approach, we calculated RNA decay pathway dependence, which was the extent to which decapping, SOV, or residual decay pathways accounted for each gene’s RNA decay rate. For example, a gene was classified as highly VCS-dependent if its RNA half-life was much longer in vcs mutants [log2 (t1/2 vcs/t1/2 WT)] or with high usage of residual decay pathways if its RNA half-life in vcs sov was close to that of the WT (t1/2 WT/t1/2 vcs sov). We then calculated the mean pathway dependence (VCS, SOV, and residual) for each GO, and identified GOs whose RNAs were highly VCS-dependent or had high residual decay pathway usage, but saw no strong SOV dependence (Fig. 5A). For example, most genes in procambium histogenesis, vasculature development, regulation of organ growth, and response to chitin GOs were strongly VCS-dependent, and showed little dependence on SOV or residual decay in vcs sov, whereas the RNA polII TF complex and chaperone-mediated protein folding GOs showed high residual decay pathway usage and little SOV or VCS dependence. These functionally related mRNAs with similar decay pathway preferences are candidates for posttranscriptional coregulation.
Fig. 5.
RNA decay pathway preferences and decay rates partition among functional groups. (A) Analysis of VCS and SOV dependence (log2 relative t1/2), and levels of residual decay in vcs sov mutants (t1/2 WT/t1/2 vcs sov) by GOs and displayed as heatmaps. All genes in each GO are depicted; empty squares contain crosses (x). (B) RNA half-life (WT) and log2 relative RNA abundance in vcs and vcs sov compared with WT are displayed as heatmaps for the same GOs in A. (C) Scatter plots comparing mean half-life (WT) with overall GO VCS dependence (Left) and residual decay (Right) for all GOs. (D) Enlargement of box in C with GO labels for GOs with high VCS dependence. (E) Scatterplots comparing VCS dependence (Left) and vcs sov residual decay (Right) to half-life for all RNAs with decay rate estimates. Red line indicates a Loess smoothing function. (F) Short mRNA half-life is limited by larger intron numbers. Boxplot of mRNA half-life (WT) for subsets of RNAs with different numbers of introns. (G) Short mRNA half-life is correlated with increased A and decreased G content in the 5′ UTR. Black line indicates a Loess smoothing function. vasculat., vasculature; devel., development; reg., regulation; gro., growth; JA, jasmonic acid; sys., systemic; acq. or acquir., acquired; resist., resistance; struct., structural; cons., constituent; proteas., proteasome; comp., complex; S-aden., S-adenosyl methionine; metab., metabolic; proc., process; pol., polymerase; TF, transcription factor; chap-med., chaperone-mediated; prot., protein; fold., folding; resp., response; ext., external; mol., molecular; func., function; MAPK, mitogen associated protein kinase; SA, salicylic acid; biosyn., biosynthetic; autoubiqu., autoubiquitination; phos., phosphate; cycl., cycle; polysac., polysacharide; catal., catalytic; act., activity; sub., substance; def., defense; cell., cellular; photosyn., photosynthesis; loc., location; cytochr., cytochrome; mod., modification; trans., transport; AMP, adenosine monophosphate.
To find other shared characteristics of VCS-dependent RNAs or RNAs that decay by residual pathways, we expanded the GO analysis to include half-lives and relative T0 abundances in vcs and vcs sov (Fig. 5B). Most VCS-dependent GOs tended to also have short-lived RNAs (Fig. 5 A and B). However, an exception was the structural constituent of ribosome GO, whose RNA decay showed generally strong VCS dependence, and the RNAs were also typically long-lived. Nevertheless, highly VCS-dependent GOs had a wide distribution of decay rates (Fig. 5 C and D). Most GOs with high residual decay, by contrast, had generally long-lived RNAs. For example, RNAs in the S-adenosyl methionine metabolic process GO were strongly degraded by residual decay pathways, and were generally long-lived. To see if these selected patterns represented a genome-wide trend, we compared VCS dependence and residual decay to WT decay rates (Fig. 5E), and found VCS dependence to be strongly correlated with short half-life (r = −0.237), whereas high residual decay was correlated with long half-life (r = 0.079). A similar analysis examining mean GO dependence showed a similar trend (Fig. 5C).
We also noted an increased relative abundance for many RNAs in the GOs with fast decay and high VCS dependence (Fig. 5B). This was unexpected, given the lack of a genome-wide correlation between decay rate and abundance (Fig. 4B). Closer inspection demonstrated that very rapidly decaying (t1/2 < 15 min) VCS-dependent RNAs were more abundant, but that this trend diminished quickly as decay rates slowed (Fig. S5).
To identify physical attributes that correlated with short RNA half-lives, we examined intron number, which has been noted in other studies, and intron presence interpreted as stabilizing the resulting mRNA (32, 42, 43). We also found intron number to correlate with half-life (Fig. 5F). Intron number was inversely correlated with decay rate (r = −0.22), and the shortest half-life RNAs were transcribed from intronless genes (39 of the 50 shortest t1/2, including the five shortest half-lives: AT4G25490, AT2G21210, AT4G38840, AT2G17070, and AT2G17080). However, RNAs from intronless genes also had the widest half-life distribution. Moreover, RNAs with very long half-lives were represented in all intron number classes, but there was a marked loss of short half-life RNAs as intron number increased. Thus, introns appear to limit an RNA’s capacity to show rapid decay rates, rather than low intron number being a determinant of rapid decay. How the presence of introns slows the half-life for the resulting mRNAs is unknown.
We also examined 5′ and 3′ UTRs for features that correlated with decay rates. Although neither the 5′ nor 3′ UTR lengths, nor coding region lengths, appeared to broadly influence decay rates, we identified a strong 5′ UTR nucleotide composition bias (Fig. 5G). Increased A content and decreased G content were strongly correlated with fast decay rates. This contrasted with nt content of the coding sequence (CDS) and 3′ UTRs, where no correlations were found (Fig. S5).
RNA Half-Life and Functions of Encoded Proteins.
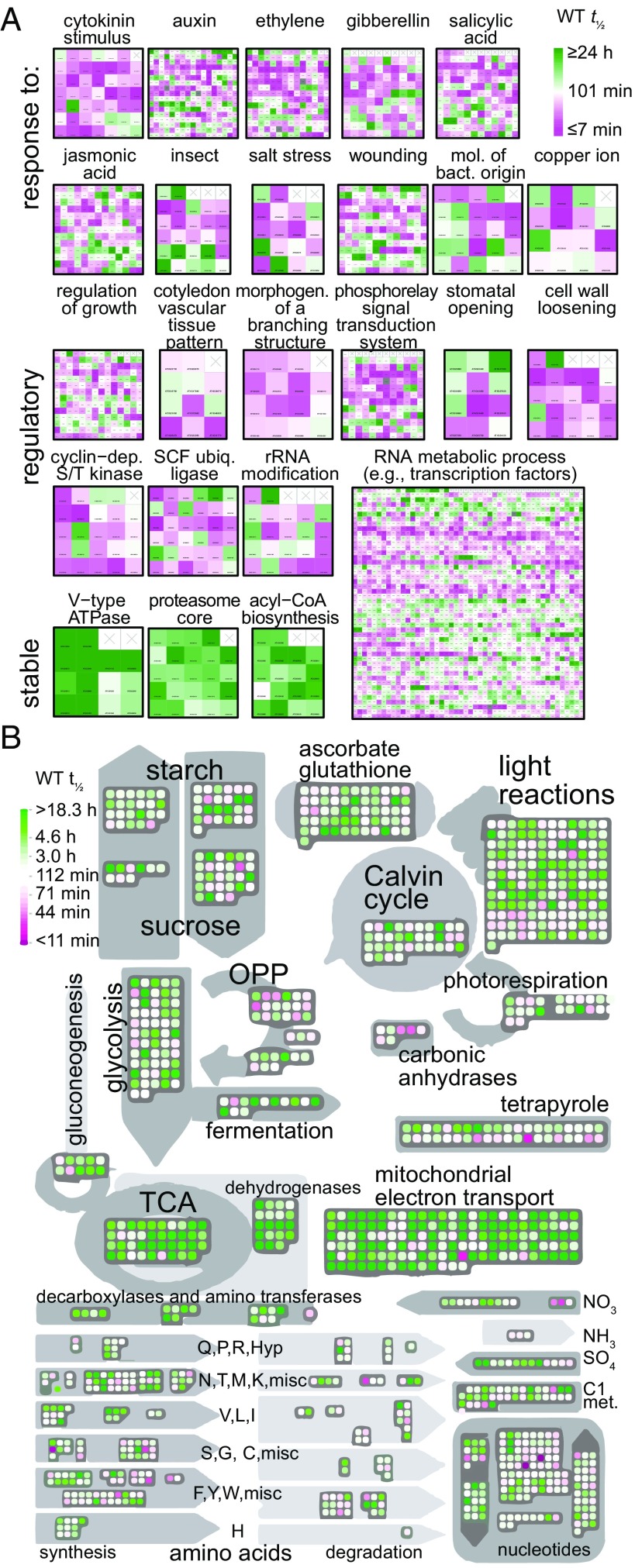
The analysis of GOs containing RNAs with high VCS dependence or residual decay and comparison with decay rates (Fig. 5) suggested that short half-life RNAs encoded proteins that respond rapidly to signaling or that encoded signal transduction components. To extend this analysis, we examined the 150 GOs with shortest mean WT half-life (range 17.8 min to 43.1 min); these included seven of the nine “response to” hormone GOs, and the other two (responses to ABA and to brassinosteroid) were still within the fastest quartile (mean half-lives of 56 min and 58 min, respectively) (Fig. 6A). Similarly, many biotic and abiotic stress-related GOs were among those with short half-lives. By contrast, the 150 GOs with longest median half-life revealed many encoding enzymes with metabolic and housekeeping functions (Fig. 6A). To see if this was a general trend, we used MapMan to display half-life for RNAs encoding enzymes in basic metabolic pathways; while not universal, this analysis revealed a strong tendency for very long half-lives (Fig. 6B). Chloroplast RNAs identified in this analysis might have decayed slowly because cordycepin, our transcription inhibitor, is also likely to disrupt polyadenylation, which, in plastids, activates RNA decay (44).
Fig. 6.
High-flux RNAs control many responsive and regulatory processes. (A) RNA half-lives of each gene in a GO are displayed as a heatmap. Selected GOs illustrate that RNAs responsive to hormone and environmental stimuli, and encoding regulatory or signaling function, are short-lived (WT). Highly stable GOs are shown for comparison. (B) MapMan (57) display of half-life (WT) of RNAs coding for proteins involved in central metabolic pathways. mol., molecules; bact., bacterial; dep., dependent; ubiq., ubiquitin.
β Analysis Suggests Diverse Constraints on Decay Rates.
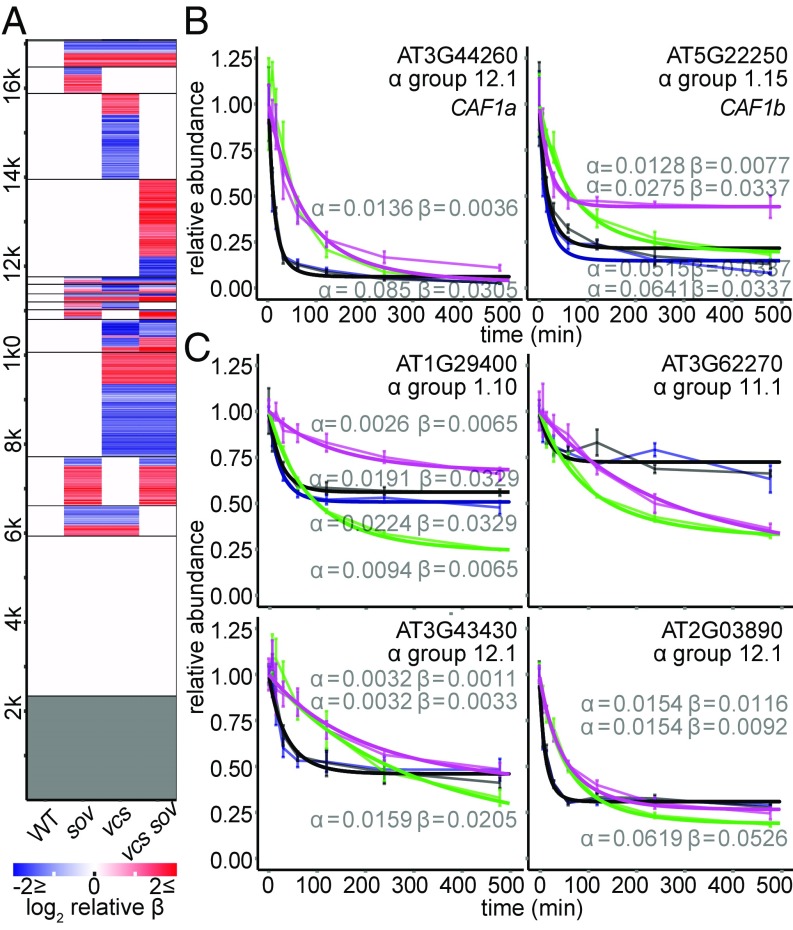
Our decay modeling included a nuisance factor, β, to account for the slowing of RNA decay rates. This was common in our dataset: Only about 14% of transcripts had no slowing of decay rates, decay rates of many transcripts slowed uniformly in all genotypes (21%), and 65% showed genotype-specific β (Fig. 7A). Slowing of decay rates could arise for a variety of reasons. One possibility is that transcription was not uniformly inhibited, e.g., if cordycepin failed to penetrate the interior-most cell types. We evaluated this possibility by examining β-values for sets of cell type-specific marker genes (38, 39), and found them to be distributed similarly for different marker gene sets in WT (Fig. S7), suggesting that β is unlikely to arise from a cell type-specific phenomenon. Alternatively, RNA decay rates could slow if the RNA decay machinery itself was not stable through the time course. We found that AtCAF1a and AtCAF1b, which encode important components of the CCR4−CAF1 deadenylation complex (45–47), had short RNA half-lives (t1/2 of 8.2 min and 9.3 min, respectively) (Fig. 7B and Dataset S1), but, in vcs mutants, these RNAs were more stable. The β-analysis also identified RNAs whose decay slowed substantially, commensurate with the loss of AtCAF1a and AtCAF1b RNAs in the WT, but only in genotypes where decapping was intact (Fig. 7C). Together, these observations suggest that these RNAs might require CAF1a and/or CAF1b deadenylation to initiate decay. Thus, a detailed analysis of β-patterns might allow discovery of additional RNA decay pathway substrates, and reveals the added value that can arise from using mathematical approaches.
Fig. 7.
Decay of decay. (A) Modeling of decay included β, which accounted for the slowing of decay rates across the time course. Heatmap depicting the effects of genotype on β. RNAs sharing the same β in all genotypes are white, and those with no β (single exponential models) are gray. (B) RNAs encoding CAF1a and CAF1b have fast VCS-dependent decay. (C) Representative decay plots showing strong VCS-dependent decay of decay.
Discussion
In this study, we applied mathematical modeling to genome-wide mRNA decay analyses that used Arabidopsis mRNA decay mutants, which revealed feedback pathways, and identified specialized roles for the mRNA decapping complex. An underlying assumption of our study was that, in mutants with decay defects, mRNAs would either decay at the same rate as the WT, indicating that the affected enzyme was not necessary, or they would decay more slowly, indicating they were normally substrates of the affected enzyme. While we did observe both of these outcomes in all three of our mutant genotypes, we also observed some RNAs that decayed faster in the mutants, suggesting a strong overcompensating feedback.
Feedback on RNA decay defects has been investigated in yeast, where mutants with mRNA decay rate defects led to reduced transcription rates, and RNA polymerase II defects resulted in reduced rates of mRNA decay (48). This feedback pathway has been called RNA buffering, because it maintains RNAs at near-WT levels, and it appears to work through the XRN1 enzyme, which is a cytoplasmic 5′-to-3′ exoribonuclease that also functions as a general transcription repressor (17, 40, 49). In our system, RNA levels in sov mutants appeared to also be buffered; despite approximately one-third of the transcriptome showing significantly faster decay rates, all but five RNAs were maintained at near-WT levels, with adjustments of both RNA decay and, presumably, transcription rates that maintained RNAs at a near-WT steady state. However, whether these two systems are mechanistically similar is less clear. The yeast XRN1 and plant XRN4 are not orthologs (50), and there is no evidence of nuclear roles for XRN4, and yet it is tantalizing that both feedback systems require components of the 5′-to-3′ decay pathway.
Why the loss of SOV evoked these changes is less clear. We favor the idea that loss of the SOV decay pathway led to increased levels of substrate mRNAs, causing recruitment of decapping for feedback. Alternatively, because the sov mutant is the Col-0 accession, it is also possible that this strain has accumulated compensatory mutations that were unlinked to SOV. We consider this unlikely, as the faster decay rates were not found in the same Col-0 accession that contained the active SOV transgene. Further support for SOV being an important RNA decay pathway in Arabidopsis is its widespread conservation among the sequenced accessions; only 14 accessions match the nonfunctional Col-0 sequence at SOV’s amino acid 705 (Arg), whereas 1,033 accessions match the active L.er sequence (Pro) (51). Other faster decay rates, e.g., observed in vcs, or in all three mutants, suggest the possibility of more-widespread feedback. In our system, we found no evidence that faster mRNA decay rates arose from elevated siRNAs. Whether this is general, or specific for genotypes with defects in VCS, and thus indicating a requirement for VCS for siRNA-induced RNA cleavage and decay, is unknown.
RNAs with short half-lives, or high flux, have strong regulatory potential, as decreases in transcription or decay rates can result in rapid changes in RNA abundance. Our study allowed us to identify high-flux RNAs; these largely decayed via the mRNA decapping pathway, while long-lived RNAs tended to degrade by pathways that did not include decapping or SOV. VCS-dependent short-lived RNAs included many genes known to have rapid RNA-level responses to environmental or developmental signals. This is consistent with short-lived RNAs functioning in circadian control (52), and with the recent findings of enhanced flux and XRN4-mediated decay playing roles in Arabidopsis gene expression responses to heat and cold (53–55). Studies demonstrating phosphorylation of mRNA decay components in response to osmotic stress, and subsequent changes in mRNA half-life, suggests that regulation could arise from direct modulation of mRNA decapping machinery (18, 19), or through upstream pathways, such as the LSM proteins (12). An understanding of how stress perception leads to selective changes in mRNA decay rates is an important area of future research.
The GOs with highest VCS dependence and shortest mean half-life include procambium histogenesis, vasculature development, and regulation of organ growth. These GOs are consistent with the vcs phenotype, which we described as a mutant with cotyledon and leaf vascular development defects. The eponymous varicose veins contain ectopic tracheary elements (7), indicating compromised spatial regulation of vascular development-associated genes, and a role for VCS-mediated decay in establishing tight boundaries during development.
Materials and Methods
Plant Growth.
The Arabidopsis thaliana genotypes used were Col-0, Col-0 [SOVL.er], vcs-7, and vcs-7 [SOVL.er] (7, 8, 23). Seedlings were grown vertically on growth media [0.216% (wt/vol) Murashige and Skoog salts, 4% (wt/vol) sucrose, 23 mM Mes, pH 5.8, 0.8% agar] at 22 °C in a Conviron growth chamber under ∼75 µEinstein⋅m−2⋅s−1 constant illumination. Homozygous mutants were sorted from segregating siblings and placed on fresh growth media before transcriptional arrest treatment. WT seedlings were similarly handled to standardize genotype treatment.
Transcriptional Arrest Time Course.
Seedlings were preincubated in 3 mL of incubation buffer (15 mM sucrose, 1 mM Pipes pH 6.25, 1 mM KCl, 1 mM sodium citrate) with aeration (swirling 100 rpm) in 35 mm × 10 mm Petri dishes for 15 min (23, 56). Initial (T0) samples were frozen immediately, while the rest were subjected to transcriptional inhibition by addition of 3 mL of fresh buffer containing 1 mM cordycepin. Vacuum infiltration was performed three times. Tissue was collected 7.5, 15, 30, 60, 120, 240, and 480 min following the first vacuum release, frozen in liquid nitrogen, and stored at −80 °C until RNA extraction.
Modeling Decay of Decay.
For each genotype, RNA abundance [c(t)] as a function of time after inhibition of transcription (t) was modeled as , where the parameters α and β are the initial decay rate and the decay of decay rate, respectively. Using an optimization algorithm described in Supporting Information, we modeled genotype effects on decay kinetics of RNAs from each gene by combinatorially constraining equivalence of α and β parameters across genotypes. These constraints yielded 240 different models with α and β estimates for each genotype for all genes (Fig. S2A). We present RNA half-life throughout as a more interpretable proxy for α, as it is inversely proportional to decay rate, calculated as t1/2 = ln(2)/α. For details of the modeling implementation and model selection, see SI Materials and Methods.
RNA extraction, library preparation, and sequencing methods are described in SI Materials and Methods. For details of RNA sequencing data treatment, small RNA analysis, and GO analysis, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Richard Clark and Robert Greenhalgh for bioinformatics advice and manuscript comments. We thank Thuy Le for help learning C++ and Jason Griffiths for helpful suggestions. This work was supported by National Science Foundation (NSF) Grants NSF 2010 1022435 and MCB-1616779. M.J.D. was supported by National Institutes of Health (NIH) Genetics Training Grant T32 GM07464, R.S.S. was supported by NIH Developmental Biology Training Grant 5T32 HD07491, K.J. was supported by NSF-funded Research Training Group in Mathematical and Computational Biology (NSF-DMS 1148230) and NIH-funded Interdisciplinary Training Grant T32 Program in Computational Approaches to Diabetes and Metabolism Research (1T32DK11096601), and The University of Utah core facility (NCI P30CA042014).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE86361).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712312115/-/DCSupplemental.
References
- 1.Taylor-Teeples M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–575. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz-Gutiérrez E, et al. A dynamic gene regulatory network model that recovers the cyclic behavior of Arabidopsis thaliana cell cycle. PLOS Comput Biol. 2015;11:e1004486. doi: 10.1371/journal.pcbi.1004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 4.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 6.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deyholos MK, et al. VARICOSE, a WD-domain protein, is required for leaf blade development. Development. 2003;130:6577–6588. doi: 10.1242/dev.00909. [DOI] [PubMed] [Google Scholar]
- 8.Goeres DC, et al. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Yang J-Y, Niu Q-W, Chua N-H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Golisz A, Sikorski PJ, Kruszka K, Kufel J. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013;41:6232–6249. doi: 10.1093/nar/gkt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perea-Resa C, et al. The LSM1-7 complex differentially regulates Arabidopsis tolerance to abiotic stress conditions by promoting selective mRNA decapping. Plant Cell. 2016;28:505–520. doi: 10.1105/tpc.15.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radhakrishnan A, et al. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016;167:122–132.e9. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tharun S, et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 15.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J. LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell. 2012;24:4930–4947. doi: 10.1105/tpc.112.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haimovich G, et al. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Chua N-H. Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 2012;31:1975–1984. doi: 10.1038/emboj.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soma F, et al. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat Plants. 2017;3:16204. doi: 10.1038/nplants.2016.204. [DOI] [PubMed] [Google Scholar]
- 20.Januszyk K, Lima CD. The eukaryotic RNA exosome. Curr Opin Struct Biol. 2014;24:132–140. doi: 10.1016/j.sbi.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015;348:120–123. doi: 10.1126/science.aaa2618. [DOI] [PubMed] [Google Scholar]
- 22.Chekanova JA, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci USA. 2010;107:15981–15985. doi: 10.1073/pnas.1007060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Towler BP, Jones CI, Harper KL, Waldron JA, Newbury SF. A novel role for the 3′-5′ exoribonuclease Dis3L2 in controlling cell proliferation and tissue growth. RNA Biol. 2016;13:1286–1299. doi: 10.1080/15476286.2016.1232238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Astuti D, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44:277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 26.Morris MR, Astuti D, Maher ER. Perlman syndrome: Overgrowth, Wilms tumor predisposition and DIS3L2. Am J Med Genet C Semin Med Genet. 2013;163C:106–113. doi: 10.1002/ajmg.c.31358. [DOI] [PubMed] [Google Scholar]
- 27.Ustianenko D, et al. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang H-M, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirouz M, Du P, Munafò M, Gregory RI. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 2016;16:1861–1873. doi: 10.1016/j.celrep.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas MP, et al. Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Reports. 2015;11:1079–1089. doi: 10.1016/j.celrep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedel CC, Dölken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narsai R, et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pai AA, et al. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 2012;8:e1003000. doi: 10.1371/journal.pgen.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez de Alba AE, et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015;43:2902–2913. doi: 10.1093/nar/gkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Chen X. RNA quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol Plant. 2016;9:826–836. doi: 10.1016/j.molp.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Li G, Zhu S, Zhang S, Fang J. tasiRNAdb: A database of ta-siRNA regulatory pathways. Bioinformatics. 2014;30:1045–1046. doi: 10.1093/bioinformatics/btt746. [DOI] [PubMed] [Google Scholar]
- 38.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 39.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun M, et al. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao C, Hamilton T. Introns regulate the rate of unstable mRNA decay. J Biol Chem. 2007;282:20230–20237. doi: 10.1074/jbc.M700180200. [DOI] [PubMed] [Google Scholar]
- 43.Wang H-F, Feng L, Niu D-K. Relationship between mRNA stability and intron presence. Biochem Biophys Res Commun. 2007;354:203–208. doi: 10.1016/j.bbrc.2006.12.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kudla J, Hayes R, Gruissem W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 45.Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010;152:866–875. doi: 10.1104/pp.109.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang W, et al. The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 2009;19:307–316. doi: 10.1038/cr.2008.317. [DOI] [PubMed] [Google Scholar]
- 47.Sarowar S, et al. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J. 2007;51:792–802. doi: 10.1111/j.1365-313X.2007.03174.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun M, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medina DA, et al. Cytoplasmic 5′-3′ exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front Genet. 2014;5:1–26. doi: 10.3389/fgene.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arae T, et al. Co-ordinated regulations of mRNA synthesis and decay during cold acclimation in Arabidopsis cells. Plant Cell Physiol. 2017;58:1090–1102. doi: 10.1093/pcp/pcx059. [DOI] [PubMed] [Google Scholar]
- 54.Merret R, et al. XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 2013;5:1279–1293. doi: 10.1016/j.celrep.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 55.Sidaway-Lee K, Costa MJ, Rand DA, Finkenstadt B, Penfield S. Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response. Genome Biol. 2014;15:R45. doi: 10.1186/gb-2014-15-3-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usadel B, et al. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.