Significance
Ancient DNA metabarcoding of coprolites (ancient dung) has greatly improved our ability to investigate the behavior and ecology of extinct species. We use coprolites from extinct New Zealand birds to show how this approach can reveal aspects of dietary behavior, such as the consumption of ectomycorrhizal fungi, and both the distribution and coextinction of parasites. We show how this approach can identify lost ecological interactions, which have key implications for understanding, conserving, and restoring currently threatened ecosystems.
Keywords: ancient DNA, metabarcoding, coprolites, moa, New Zealand
Abstract
Over the past 50,000 y, biotic extinctions and declines have left a legacy of vacant niches and broken ecological interactions across global terrestrial ecosystems. Reconstructing the natural, unmodified ecosystems that preceded these events relies on high-resolution analyses of paleoecological deposits. Coprolites are a source of uniquely detailed information about trophic interactions and the behaviors, gut parasite communities, and microbiotas of prehistoric animal species. Such insights are critical for understanding the legacy effects of extinctions on ecosystems, and can help guide contemporary conservation and ecosystem restoration efforts. Here we use high-throughput sequencing (HTS) of ancient eukaryotic DNA from coprolites to reconstruct aspects of the biology and ecology of four species of extinct moa and the critically endangered kakapo parrot from New Zealand (NZ). Importantly, we provide evidence that moa and prehistoric kakapo consumed ectomycorrhizal fungi, suggesting these birds played a role in dispersing fungi that are key to NZ’s natural forest ecosystems. We also provide the first DNA-based evidence that moa frequently supplemented their broad diets with ferns and mosses. Finally, we also find parasite taxa that provide insight into moa behavior, and present data supporting the hypothesis of coextinction between moa and several parasite species. Our study demonstrates that HTS sequencing of coprolites provides a powerful tool for resolving key aspects of ancient ecosystems and may rapidly provide information not obtainable by conventional paleoecological techniques, such as fossil analyses.
The late Quaternary (last 50,000 y) has seen global declines and extinctions of terrestrial biota on an unprecedented scale, characterized by the widespread extinction of charismatic megafauna, such as the woolly mammoth (Mammuthus primigenius) (1). Advances in ecological network theory have highlighted the extent to which the effects of extinction can reverberate throughout an ecosystem due to loss of interactions (e.g., propagule dispersal, pollination, parasitism) and processes (e.g., nutrient cycling) (2–5). Thus, assessing the real-world effects of late Quaternary extinctions and declines on terrestrial ecosystems requires detailed information about the ecological interactions and ecosystem processes that were provided by the lost species. While such details are seldom preserved in the fossil record, coprolites (ancient dung) provide a key resource for obtaining these data (6–16).
Most of the work performed on late Quaternary coprolites to date has focused on microscopic analyses and morphological identification of visible remains. While such analyses are valuable, they lack the ability to confirm the identity of the depositing species or to resolve well-digested remains. Ancient DNA (aDNA) techniques offer a tool for addressing these issues. While several studies have sequenced aDNA from coprolites, the majority have used cloning and Sanger sequencing methods to assess the diversity of dietary or parasitic taxa (6–14).
High-throughput sequencing (HTS) offers clear advantages over these methods. By indexing and pooling amplicons of key barcoding loci, such as the 16S (bacterial) or 18S (eukaryote) ribosomal RNA (rRNA) genes, multiple taxonomic groups can be identified at depths of tens of thousands of sequences from many individual samples simultaneously (17). Despite these clear advantages, however, thus far aDNA metabarcoding of coprolites has been used only to characterize ancient human gut bacteria (18, 19), dietary archaeological data from Polynesian domestic dogs (20), and seed plants consumed by Pleistocene Arctic megafauna (21). Metabarcoding has been underutilized on coprolites to resolve the paleoecology of extinct species.
Here we used HTS metabarcoding approaches on coprolites to explore lost species interactions in the large (268,021 km2) South Pacific archipelago of New Zealand (NZ). NZ is an excellent locality for aDNA studies, as it was the last large landmass to be colonized by humans (during the 13th century A.D.) (22), and the ecological record of its unique, prehuman ecosystems are exceptional (23). Due to NZ’s large size and long isolation (ca. 55 Ma) from the supercontinent Gondwana (24), NZ’s indigenous biota has older lineages, and greater phylogenetic diversity and ecological complexity, than most oceanic islands (25). NZ’s recent indigenous fauna lacks terrestrial mammals (except bats) and was dominated by birds - the largest of which were nine species of extinct herbivorous moa (Dinornithiformes) (up to 250 kg in body mass and 3.6 m tall) (23, 26). NZ’s terrestrial ecosystems experienced dramatic and rapid changes following human settlement, including the loss of 41% of indigenous bird species (including all moa species within 200 y of human arrival) (23, 26–28); however, NZ’s temperate climate, widespread cave systems, and recent extinction ages have resulted in exceptional preservation of prehuman faunal remains. Included in these are thousands of avian coprolites, up to >7,000 y old (14), of which nearly 100 have been assigned (using aDNA) to five moa species (7–11, 13) and the critically endangered kakapo parrot (Strigops habroptilus) (12). The plant and parasite contents of these coprolites have been identified using microfossil and Sanger sequencing methods (7–13, 15, 16), but these approaches are labor-intensive and have not addressed the full diversity of potential lost ecological interactions. For example, fossil analyses are unable to elucidate the full diversity of gastrointestinal parasites or microbes, or foods in the diet that are unlikely to be preserved, such as nonfibrous plant tissues or fungi. Thus, there is little information on foods that were regularly exploited by moa, other than flowering or coniferous plants. It has also been hypothesized that the many NZ fungi with bright-colored fruiting bodies are adapted for dispersal by native ground-dwelling birds (29, 30), but this hypothesis has not been tested, as most of the relevant birds are now extinct.
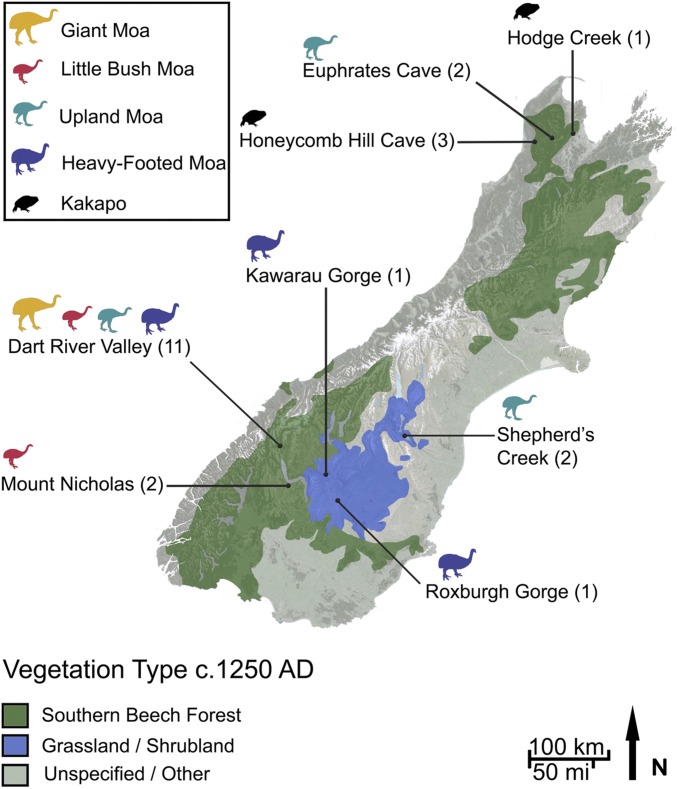
In this study, we examined whether an HTS survey of NZ’s remarkable avian coprolite assemblages can provide insight into NZ’s prehuman ecosystems. Using a fragment of the 18S rRNA gene, which is amplifiable across a wide variety of eukaryotic taxa (20, 31), we assessed 23 coprolites, dated to between 124 and 1,557 y in age, deposited by kakapo, little bush moa (Anomalopteryx didiformis), South Island giant moa (Dinornis robustus), upland moa (Megalapteryx didiformis), and heavy-footed moa (Pachyornis elephantopus) (7, 10, 12, 13). For a modern comparison, fecal metabarcoding data from two captive, extant ratite species (North Island brown kiwi Apteryx mantelli and ostrich Struthio camelus), and a range of captive and wild mammals from Parfrey et al. (32), were included in our analysis. The coprolites originate from eight localities that were prehistorically vegetated by southern beech (Nothofagaceae) forest or semiarid shrubland/grassland (Fig. 1 and SI Appendix, Tables S1 and S2). Ecologically, kakapo were found throughout NZ’s forested habitats, and their broad herbivorous diet is known to include fruit, leaves, and fungi (15, 33). Conversely, subfossil distributions, cranial/bill morphology, and coprolite/gizzard contents of different moa taxa clearly demonstrate interspecific niche partitioning, allowing several large herbivores to exploit a broad range of habitats and food sources (7, 9–11, 13, 23, 34–36). Based on this evidence, the current research supports the idea that little bush moa browsed nearly exclusively on fibrous, forest vegetation, whereas heavy-footed moa browsed and grazed on vegetation characteristic of open habitats. Upland moa and giant moa appear to have been widespread dietary generalists, with upland moa populating higher altitudes. Due to the differing ecological and geographic ranges of different moa species, most coprolite localities contain coprolites of single moa species, but one site near a forest margin (Dart River Valley) contains coprolites from all four moa species, allowing for examination of niche partitioning (Fig. 1 and SI Appendix, Tables S1 and S2).
Fig. 1.
Map of New Zealand’s South Island with localities of the coprolites used in this study. Relevant paleoenvironments are shown.
Results
Sequence Diversity.
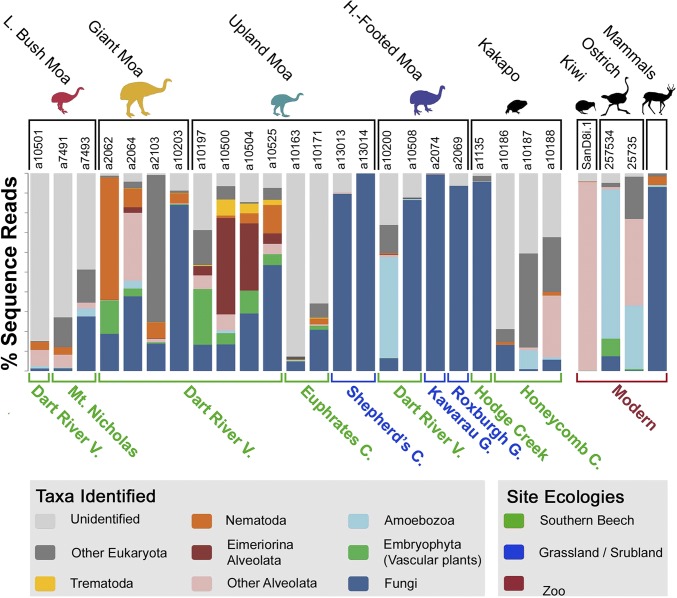
Between 26,102 and 257,186 sequence reads (after quality filtering) were obtained from each sample (SI Appendix, Table S2), with read depth greatest in upland and giant moa from southern beech sites. Taxonomic identities assigned to operational taxonomic units (OTUs) included a diversity of fungi, land plants, parasites, and environmentally common microbial eukaryotes (Fig. 2 and SI Appendix, Figs. S1–S4 and Dataset S1). Alpha diversity analyses (SI Appendix, Figs. S5 and S6 and Dataset S2) identified that southern beech site coprolites had significantly greater diversity than semiarid site coprolites, possibly reflecting poorer DNA preservation in the latter environment (as supported by very low read counts of expected taxa, such as plants or parasites from semiarid coprolites). Beta diversity analyses clustered bird samples distinct from mammal samples and bird samples clustered into groups representative of modern birds, southern beech site coprolites, and semiarid site coprolites (SI Appendix, Figs. S7–S12). Although PERMANOVA analyses identified some beta-diversity signal related to vegetation type or depositor species, collection locality was identified as the major determinant of coprolite diversity (SI Appendix, Table S3). Kruskal–Wallis and G-test statistics, as well as principal coordinates analyses, suggested that fungi (mostly common microfungi) were responsible for the majority of geographically driven beta diversity, although green plants and several microbial taxa also contributed unique signals (Fig. 2 and SI Appendix, Figs. S1 and S9–S12). These analyses indicated that taphonomy may be more important than depositing species or local ecology in determining coprolite DNA diversity, highlighting the need for a conservative approach when examining coprolite sequence taxa.
Fig. 2.
Proportions of reads per sample to taxonomic and ecological groups as determined using QIIME. All OTUs present in extraction blanks are filtered. Note the unequal total read counts among samples.
Fungal aDNA.
Fungi (predominantly Ascomycota and Basidiomycota) dominated the coprolite sequences (Figs. 2 and 3 and SI Appendix, Figs. S1 and S2). Ascomycota and yeast-forming taxa (Saccharomycetes, Microbotryomycetes, and Tremellomycetes) were common in most samples, although none could be confidently supported as endogenous or dietary. For example, all yeast-forming OTUs (including species of Cryptococcus) could not be differentiated from environmentally common taxa, and the only definite macrofungal (and potentially edible) Ascomycota taxa (Lasiobolus, Ascobolus) are coprophilous (37) (Fig. 3). Therefore, Ascomycota and yeast fungi were not analyzed further.
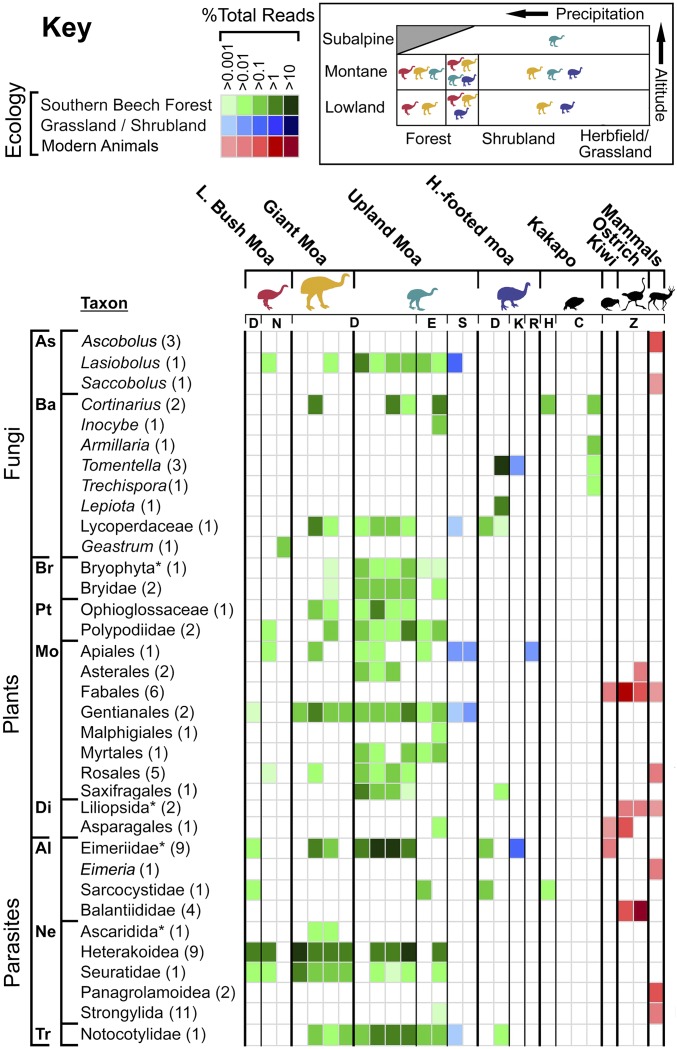
Fig. 3.
Distribution of select taxa among samples, identified to the lowest possible taxonomic rank. *Designates a taxon inclusive of the taxon immediately following. The number of OTUs is in brackets. (Top Right) An approximation and overlap of the different moa species distributions relative to precipitation/aridity and altitude. Taxon groups: Al, Alveolata; As, Ascomycota; Ba, Basidiomycota; Br, Bryophyta; Di, dicots; Mo, monocots; Ne, Nematoda; Pt, Pteridiophyta (ferns); Tr, Trematoda. Sites: C, Honeycomb Cave; D, Dart River; E, Euphrates Cave; H, Hodge Creek; K, Kawarau Gorge; N, Mt. Nicholas; R, Roxburgh Gorge; S, Shepherd’s Creek; Z, zoo.
Nonetheless, likely dietary macrofungi were detected among Basidiomycota reads, which were dominated by Agaricomycetes (which include “true” mushrooms, bracket fungi, and puffballs). Agaricomycetes were abundant only in coprolites from southern beech forest sites, which might be expected as a rich diversity of mycorrhizal mushroom species occur in this forest type (38) (SI Appendix, Fig. S2). Notable mushroom-forming genera identified exclusively in coprolites from the southern beech forest sites include the ectomycorrhizal Cortinarius (two OTUs) and Inocybe (one OTU), and the plant parasite Armillaria (one OTU). Importantly, these fungi are not saprobic (decomposers) and are dependent on a symbiosis with plant roots. Therefore, these plant-symbiotic fungi are very unlikely to have colonized the coprolites postdeposition in the dry sediment where they were found (especially for cave samples) (39). Moreover, their DNA presence is unlikely to be explained by settlement of ambient spores on consumed foods or on the coprolites postdeposition, as these taxa were not widespread among coprolites within collection sites, they had high-read counts when present, and coprolite surfaces were removed before laboratory analysis. Finally, these plant-symbiotic fungi were only found in coprolites of kakapo (a bird known to consume fungi) (33) and upland and giant moa (generalist moa species that frequently fed on the forest floor) (7, 9, 10). Along with plant-symbiotic macrofungi, saprobic Agaricomycete macrofungi with the potential to be dietary were identified as well (Lepiota, Geastrum, Lycoperdaceae sp.). Overall, we infer that this observation provides direct evidence for mycophagy in the extinct moa.
Plant aDNA.
Plants were detected in all samples except the kakapo coprolites (Figs. 2 and 3 and SI Appendix, Figs. S1 and S3), which were a notable exception. The absence of plant DNA in kakapo may be due to a preservation bias, possibly related to the intensive food-processing characteristic of this species, with food plants thoroughly masticated and fibrous material rarely swallowed (33). Such factors could potentially result in poorly preserved plant materials and bias enzymatic amplification to better-preserved fungi DNA (which may be bound in spores) that dominated the kakapo coprolite sequences (Fig. 2 and SI Appendix, Figs. S1, S2, and S4). Plant reads were most diverse and abundant in moa coprolites from southern beech forest sites and closely reflected local vegetation communities. Although taxon resolution was generally limited to the ordinal level due to the low evolutionary rate of 18S rRNA in plants (20, 40), we identified the first DNA sequences of nonangiosperm plants from moa coprolites of both ferns (Ophioglossaceae and Polypodiidae) and mosses (Bryidae and an unassigned Bryophyta) (Fig. 3). Mosses were found only in upland moa samples from southern beech forest sites (except for extremely low numbers of reads from a single giant moa), while ferns were found in coprolites of all moa species except heavy-footed moa (Fig. 3 and SI Appendix, Fig. S3). This nonrandom distribution of fern and moss DNA is inconsistent with background environmental contamination and is also consistent with fossil evidence (Dataset S4). The remaining majority of identified plant taxa were angiosperms and included native shrubs, trees, and herbs, many representing plant groups detected in previous studies of moa coprolites (Dataset S4) (7, 9–11, 13). Specific identified taxa included Saxifragales (aquatic Myriophyllum and the shrub Gonocarpus in NZ) from upland moa and heavy-footed moa and Myrtales (comprising the tree/climber genus Metrosideros and the shrub Neomyrtus) from upland moa only (9, 10, 16).
Parasite aDNA.
Obligate parasitic taxa were detected in most coprolites (Figs. 2 and 3 and SI Appendix, Figs. S2 and S16–S20) and included all taxa found from previously found samples via cloning Sanger sequencing evidence (Dataset S4) (8). The most abundant moa parasites were those previously detected, including apicomplexans (Eimeriidae), nematodes (Ascaridida: Heterakoidea), and trematode (Notocotylidae) sequences. However, Eimeriidae and Heterakoidea were found to be unexpectedly diverse, containing nine OTUs each (SI Appendix, Fig. S20). This diversity of OTUs cannot be explained by sequencing error or aDNA damage, as genetic diversity between these OTUs was high (up to an 8% pairwise difference between Heterakoidea OTUs; SI Appendix, Figs. S16 and S18), and their distribution between coprolites was complex (e.g., upland moa and giant moa had high counts of Heterakoidea OTUs not found in other moas; SI Appendix, Fig. S20). Therefore, these Eimeriidae and Heterakoidea OTUs likely represent several different species. In addition, we identified parasites not detected previously, including an additional apicomplexan (Sarcocystis) and two Ascaridida nematode OTUs (Seuratidae and an unassigned Ascaridida).
Parasite distributions rarely supported a specific relationship to a single moa species. Although upland moa had a Strongylida nematode OTU not found in other moa species, this OTU also has been found in some ruminant mammals, suggesting this nematode occurs across a wide variety of vertebrates and is less likely to be specific to upland moa or moa in general. Several Eimeriidae OTUs were also distributed between several moa species and the extant kiwi (SI Appendix, Figs. S16 and S20). Where different moa species co-occurred (i.e., Dart River Valley), little bush, giant, and upland moa had similar parasite communities, suggesting regular parasite transmission across different moa taxa. Finally, the single trematode OTU detected was abundant and widespread among upland moa coprolites, but was present in very low counts in some heavy-footed and giant moa coprolites, possibly reflecting differences in host behavior instead of host-specificity to upland moa (Fig. 3).
A few parasite OTUs were found only in moa as a group or in single moa species (i.e., an unidentified Ascaridida and one heterakoid to giant moa and two heterakoids and several Eimeriidae to upland moa). Furthermore, the high interspecific diversity and phylogenetic uniqueness of some moa parasites suggest a long history of coevolution between these parasites and their hosts; for example, heterakoid nematodes were found only in moa, were very genetically diverse, and had no close relatives. Therefore, several of these parasites were likely moa-specific and have experienced coextinction.
Discussion
Advantages of HTS Metabarcoding for Coprolite Studies.
This report presents the largest DNA-based dataset yet obtained from the coprolites of NZ’s ancient avifauna and reveals that metabarcoding can obtain a diversity of paleoecological information from coprolites. Although moa coprolites have been thoroughly studied using microfossil analysis, DNA cloning, and Sanger sequencing, our HTS analysis of moa coprolites has provided several novel results (Dataset S4). Our approach has (i) characterized a wide taxonomic diversity (i.e., fungi, plants, and parasites) in a single analysis, allowing for assessment of several previously understudied groups; (ii) identified rare taxa, including previously unrecognized dietary components and parasite lineages; (iii) provided finer taxonomic resolution within certain parasitic taxa (e.g., Eimeriidae, Heterakoidea); and (iv) identified the degree to which parasites are shared between host species. Taken together, these results have several important implications for NZ paleoecology and provide insights into niche partitioning, ecological roles, behavior, and parasitic communities of the extinct moa.
New Dietary Taxa, Niche Partitioning, and Ecological Roles.
We have found direct evidence from moa and kakapo coprolites of the consumption of fungi, and report DNA-based evidence to support the consumption of mosses and ferns by several moa species. We also observed considerable differences in plant taxa between the coprolites of each moa taxon, especially where they occurred in sympatry (i.e., Dart River Valley). Moreover, only two moa species—upland and South Island giant moa—showed evidence of eating fungi and moss which, based on other evidence, are considered dietary generalists (9). Our data indicate that the upland moa had the broadest diet, consuming relatively higher amounts and a greater diversity of mosses, ferns, and fungi. Unique foraging behavior in upland moa is also supported by the near-universal presence of trematode (Notocotylidae) DNA in their coprolites. Notocotylidae are usually transmitted via consumption of aquatic vegetation or pond snails (41), suggesting that upland moa regularly consumed aquatic plants (8–10, 16). Taken together, these data indicate that the relatively small (14–63 kg) upland moa often foraged on small and relatively inaccessible foods, suggesting how the species may have coexisted with the similarly generalist, but much larger (maximum 74 kg m/250 kg f) South Island giant moa (23, 26).
Mosses were widespread in upland moa coprolites and nearly absent in other species, which may contradict some fossil studies. Congruent with our data, moss fragments were found in nearly one-half (17 of 35) of upland moa coprolites from Euphrates Cave examined in a previous study (10). However, another study of 51 little bush, giant, upland, and heavy-footed moa coprolites from the Dart River Valley identified moss spores or fragments in just four coprolites, although these represented all four species found at this site (9). Moss structures also have been identified in preserved gizzard contents from heavy-footed and giant moa, as well as the eastern moa Emeus crassus (not represented in our study), although these may have represented a taphonomic artifact (e.g., anomalous behavior of trapped moa, postmortem introduction of microfossils) (34). However, compared with our data, these observations of moss remains in moa coprolites are rare, suggesting that moss structures are often not physically preserved and require aDNA for their detection. Thus, our metabarcoding data provide evidence that while several moa species occasionally consumed mosses, only upland moa fed on them regularly and in quantity.
Today, many herbivorous birds frequently consume fungi, including kakapo (15, 33) and extant ratites (42, 43). Evidence that moa (and historical kakapo) consumed fungi is a key finding, as southern beech trees, which compose more than one-half of NZ’s forest canopies, are codependent on an ectomycorrhizal network (38). The close association between southern beech trees and mycorrhizal fungi may underpin broad-scale biogeographic patterns in NZ, such as several enigmatic gaps in the distribution of beech trees across the country (38, 44). A fungi genus (Cortinarius) identified in moa coprolites is also one of several NZ mycorrhizal lineages to have independently evolved “sequestrate” mushrooms (i.e., enclosed caps preventing spore release), which typically rely on consumption by animals for dispersal and germination (generally mammals) (29, 30, 43, 45). NZ’s sequestrate mushrooms tend to be uniquely colorful, a classic adaptation for bird dispersal, even though the majority of NZ’s ground-foraging avifauna are extinct or endangered (23, 26). DNA data, spore identification, and video evidence confirm that introduced mammals in NZ, including red deer (Cervus elaphus) and brushtail possum (Trichosurus vulpecula), consume and disperse both native and introduced mycorrhizal fungi today (46); however, only introduced fungi in NZ, many of which are symbiotic with invasive tree species, have been confirmed to survive digestion by mammals and germinate. Our findings suggest that moa and other native birds may have provided a potentially irreplaceable dispersal role for fungi essential for a dominant forest type in NZ. Furthermore, as the extinct moa and the nearly extinct kakapo consumed both seeds and fungi (7, 10, 16, 33, 34), they may have provided a unique mechanism for codispersal of native trees and mycorrhizal fungi that is now lost. This is potentially alarming in light of current species reductions and local extinctions, and further illustrates that NZ’s ancient bird lineages have no ecological analogs among introduced species.
Evidence for Moa Interactions and Migratory Behavior.
Gaining insight into the behavior (e.g., migration movements) or social interactions of extinct animals tends to be extremely difficult (47). In our study, some details of moa behavior may be inferred from patterns of parasite prevalence among moa species. In the Dart River Valley, coprolites of all moa species except heavy-footed moa shared similar communities of Ascaridida nematodes and Eimeriidae (Fig. 3 and SI Appendix, Figs. S12 and S20). These parasites are usually transmitted via food that has been contaminated by infected fecal material (48); thus, these three moa species were likely coexisting in the same habitat (which may have been the forest, explaining why the heavy-footed moa, which inhabited nonforest habitats, did not follow this pattern). Despite sharing several parasite taxa with other moa species and kiwi, upland moa had the most unique parasite communities, including unusually high numbers of reads from a trematode (Notocotylidae) across all localities. Notocotylidae typically infest semiaquatic animals, such as waterfowl, and generally require ingestion of larvae (cercariae) encysted on water plants or snails found in stillwater ponds or lakes (41). As noted, this finding indicates that upland moa consumed more aquatic plants compared with other moa (16). It is also possible that the more alpine-dwelling upland moa were uniquely inoculated by interactions with tarns (i.e., enclosed alpine ponds or small lakes) (8, 16), which in NZ provide one of the few reliable sources of fresh water in the alpine zone, contain abundant aquatic plants, and are frequented by large numbers of wetland birds (49). Given the finding of trematodes in non-alpine upland moa coprolites, an alpine tarn hypothesis is compatible with the theory that upland moa exhibited seasonal altitudinal migration (10, 16).
Coextinction of Parasites.
The coextinction of hosts and parasites is now recognized as perhaps one of the most important mechanisms of biodiversity loss (50, 51). This is particularly important as parasites are being increasingly recognized as important components of healthy ecosystems and can reveal a great deal about host ecology and evolutionary history (51–53). Nonetheless, little empirical evidence remains on the extent and nature of host-parasite coextinction. Although we have confirmed that several Eimeriidae species infested moa, as previously suggested by past evidence (8), we identified that some of these species may still occur in the extant kiwi, which was previously confirmed to host an unidentified Eimeria (54). This result perhaps is unsurprising, as some Eimeriidae species can infect a broad range of host taxa (55). In contrast, Heterakoidea nematodes identified in this study were restricted to moa. The moa heterakoids were also genetically distinct from the cosmopolitan genus Ascaridia, which in NZ infect parakeets and migratory cuckoos (54). Outside Ascaridia, the only known native NZ heterakoids are two rare species of the relictual family Kiwinematidae host-specific to either the kiwi or the tuatara (Sphenodon punctatus, the sole surviving rhynchocephalian reptile) (8). Like moa, these hosts are phylogenetically distinct and endemic to NZ. Although DNA sequences of extant Kiwinematidae species are unavailable for comparison, putative heterakoid eggs from moa coprolites also share morphological similarities with described Kiwinematidae eggs (8). We demonstrate that the moa heterakoids were common in their hosts and comprised several taxa, patterns not shared with extant Kiwinematidae (SI Appendix, Fig. S18). Furthermore, the divergent physiology and ecology of kiwi, tuatara, and moa makes it extremely unlikely that nematode species could be shared among them without being widespread in other hosts as well. Instead, this host distribution, as well as the phylogenetic distinctiveness of moa heterakoids, is consistent with the hypothesis that these parasites have had long evolutionary histories in NZ. As a result, the heterakoid species that we have identified were likely unique to moa and may demonstrate that two phylogenetically deep clades of taxa became coextinct.
Conclusions
Our HTS metabarcoding analysis of coprolites has provided insight into the diet, behavior, and parasites of extinct and endangered birds and provides a unique window into the presettlement ecosystems of NZ. Our study has identified several potential cascade effects from the loss of native bird species impacting ecosystem processes, including the dispersal of ecologically important fungi. We also provide evidence supporting widespread parasite-host coextinction, as well as niche partitioning among the extinct moa. Although we show that 18S rRNA sequences may provide a useful “snapshot” of coprolite content, additional information may be obtained by using deeper sequencing, longer read lengths (depending on the severity of ancient DNA fragmentation), or barcoding loci with improved resolution and reference databases for specific taxa (e.g., ITS for fungi, rbcL or trnL for plants). HTS metabarcoding of animal coprolites from other late Quaternary ecosystems will provide valuable insight into the wider ecological consequences of extinctions. As extinction and biodiversity change continues, the need for relevant and detailed paleoecological information is increasingly urgent. High-resolution data, such as that provided by HTS metabarcoding of coprolites, is vital to better understanding the past and hence identifying the risks to ecosystems posed by future extinctions.
Methods
Sampling, DNA Sequencing, and Dataset Compilation.
A total of 23 coprolites from eight sites (Fig. 1 and SI Appendix, Table S1) were subsampled following published procedures (56) in a sterile subsampling room at Landcare Research (Lincoln, New Zealand), and had DNA extracted following the procedure described by Wood et al. (7) in a fully-integrated aDNA laboratory at the Australian Centre for Ancient DNA (ACAD), Adelaide. One kiwi and two ostrich fecal samples were obtained from the San Diego and Houston Zoos, respectively, and extracted using standard Earth Microbiome Project protocols (press.igsb.anl.gov/earthmicrobiome/protocols-and-standards/dna-extraction-protocol/). For each set of DNA extractions performed, an “extraction blank” control was produced by omitting sample material from one reaction. The identity of the depositing species was confirmed by sequencing of moa or kakapo DNA from each coprolite, as described previously (7, 9–12). AMS radiocarbon dates were obtained for nine coprolites at the Waikato Radiocarbon Dating Laboratory, Waikato University, Hamilton, New Zealand (SI Appendix, Table S2). Extracts were amplified by PCR in triplicate by the 18S rRNA eukaryote primers Euk1391f and EukBr (31). Pooled, quantified, and purified PCR sets were sequenced on a single lane of the Illumina HiSeq platform at the BioFrontiers Institute, University of Colorado, Boulder. Raw data and supporting metadata from this study are available on both Qiita, https://qiita.ucsd.edu (study ID 11507) and EBI, https://www.ebi.ac.uk (accession no. ERP106282). Published metabarcoding 18S rRNA data from feces of captive zoo animals (32) were also included in our analyses.
QIIME Analyses.
Combined reads were analyzed using the QIIME software package (57), with barcoded sequences demultiplexed and quality filtered using default parameters. Sequences were open-reference clustered to 97% into OTUs using an RDP classifier toward the PR2 database (58) modified to be QIIME-compatible. OTUs that clustered with the reference database inherited the reference taxonomy. Nonassigned hits were clustered de novo and were blasted to the PR2 database using an e-value cutoff of 1e−25. For OTUs that fell below the threshold, the RDP classifier was retrained on PR2 using >50% confidence. In all instances, the most abundant sequence for each clustered OTU was selected as the representative sequence.
The dataset was subjected to additional filtering, which included removal of all OTUs present in extraction blanks to control for in vitro contaminants, and removal of reads with fewer than five reads per OTU per sample (maximum 0.015% of total reads, considered a sequencing error risk). Finally, all OTUs that could not be identified to any eukaryotic kingdom were removed from downstream analyses. Rarefactions of this dataset were used to calculate Shannon’s diversity and t test statistics. Jackknifed beta-diversity statistics were calculated (on the entire dataset and several subsets), using an unweighted pair group method with arithmetic mean (UPGMA) tree of representative OTU sequences from each dataset to calculate UniFrac distance matrices. These matrices were used in the calculation of PcoA plots, UPGMA distance-matrix plots, and beta-diversity statistics (e.g., Adonis test, G-test, Kruskal–Wallis test).
BLASTn and Phylogenetic Identifications.
High-resolution IDs for each OTU sequence (Dataset S3) were improved using BLASTn searches (maximum target sequences, 25; minimum percent identity match, 80%) against a custom, curated database of sequences obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The output file was opened in MEGAN5 (ab.inf.uni-tuebingen.de/software/megan5/), and was collapsed into reliable taxonomic identification using custom parameters (minimum score, 150; top 5%; minimum percent support, 0.1%; minimum support, 5%). In addition, coprolite OTUs identified as plant-symbiotic fungi or parasites had their identity confirmed by phylogenetic analyses. This was achieved by building seven independent gene alignments representing three fungal and four parasite taxa, composing all OTU representative sequences for each group (SI Appendix, Table S4). All sequences of the 18S rRNA barcoding region of each taxon available in GenBank and a known outgroup taxon were also included in these alignments (SI Appendix). Appropriate substitution models were selected using jModelTest2 (59), and phylogenetic trees were estimated using MrBayes 3.2 (60), with the first 25% of trees used as a burn-in to estimate posterior probabilities. Model selection and generations used are summarized in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank L. Wegener Parfrey for assisting with sequencing and bioinformatics analyses of this dataset, and members of the ACAD research group for advising on the analyses, writing, and presentation of this research project, particularly K. Mitchell and J. Breen. We also thank the Houston and San Diego Zoos for providing fecal samples. Funding for this study was provided by the Royal Society of New Zealand Marsden Fund, the Australian Research Council, and the Templeton Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data and supporting metadata from this study are available on both Qiita, https://qiita.ucsd.edu (study ID 11507) and EBI, https://www.ebi.ac.uk (accession no. ERP106282).
See Commentary on page 1411.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712337115/-/DCSupplemental.
References
- 1.Burney DA, Flannery TF. Fifty millennia of catastrophic extinctions after human contact. Trends Ecol Evol. 2005;20:395–401. doi: 10.1016/j.tree.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Hansen DM, Galetti M. Ecology: The forgotten megafauna. Science. 2009;324:42–43. doi: 10.1126/science.1172393. [DOI] [PubMed] [Google Scholar]
- 3.Gill JL, Williams JW, Jackson ST, Lininger KB, Robinson GS. Pleistocene megafaunal collapse, novel plant communities, and enhanced fire regimes in North America. Science. 2009;326:1100–1103. doi: 10.1126/science.1179504. [DOI] [PubMed] [Google Scholar]
- 4.Johnson CN. Ecological consequences of Late Quaternary extinctions of megafauna. Proc Biol Sci. 2009;276:2509–2519. doi: 10.1098/rspb.2008.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petchey OL, Eklöf A, Borrvall C, Ebenman B. Trophically unique species are vulnerable to cascading extinction. Am Nat. 2008;171:568–579. doi: 10.1086/587068. [DOI] [PubMed] [Google Scholar]
- 6.Poinar HN, et al. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science. 1998;281:402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 7.Wood JR, et al. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes) Quat Sci Rev. 2008;27:2593–2602. [Google Scholar]
- 8.Wood JR, et al. A megafauna’s microfauna: Gastrointestinal parasites of New Zealand’s extinct moa (Aves: Dinornithiformes) PLoS One. 2013;8:e57315. doi: 10.1371/journal.pone.0057315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood JR, et al. Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes) Proc Natl Acad Sci USA. 2013;110:16910–16915. doi: 10.1073/pnas.1307700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood JR, et al. High-resolution coproecology: Using coprolites to reconstruct the habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus) PLoS One. 2012;7:e40025. doi: 10.1371/journal.pone.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood JR, Wilmshurst JM, Worthy TH, Cooper A. First coprolite evidence for the diet of Anomalopteryx didiformis, an extinct forest ratite from New Zealand. N Z J Ecol. 2012;36:164–170. [Google Scholar]
- 12.Wood JR, Wilmshurst JM, Worthy TH, Holzapfel AS, Cooper A. A lost link between a flightless parrot and a parasitic plant and the potential role of coprolites in conservation paleobiology. Conserv Biol. 2012;26:1091–1099. doi: 10.1111/j.1523-1739.2012.01931.x. [DOI] [PubMed] [Google Scholar]
- 13.Wood JR, Wilmshurst JM. Pollen analysis of coprolites reveals dietary details of heavy-footed moa (Pachyornis elephantopus) and coastal moa (Euryapteryx curtus) from Central Otago. N Z J Ecol. 2013;37:151–155. [Google Scholar]
- 14.Wood JR, Wilmshurst JM. Late Quaternary terrestrial vertebrate coprolites from New Zealand. Quat Sci Rev. 2014;98:33–44. [Google Scholar]
- 15.Horrocks M, et al. Plant microfossil analysis of coprolites of the critically endangered kakapo (Strigops habroptilus) parrot from New Zealand. Rev Palaeobot Palynol. 2008;149:229–245. [Google Scholar]
- 16.Horrocks M, D’Costa D, Wallace R, Gardner R, Kondo R. Plant remains in coprolites: Diet of a subalpine moa (Dinornithiformes) from southern New Zealand. Emu. 2004;104:149–156. [Google Scholar]
- 17.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol. 2012;21:2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- 18.Cano RJ, et al. Paleomicrobiology: Revealing fecal microbiomes of ancient indigenous cultures. PLoS One. 2014;9:e106833. doi: 10.1371/journal.pone.0106833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tito RY, et al. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood JR, Crown A, Cole TL, Wilmshurst JM. Microscopic and ancient DNA profiling of Polynesian dog (kurī) coprolites from northern New Zealand. J Archaeol Sci Rep. 2016;6:496–505. [Google Scholar]
- 21.Willerslev E, et al. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature. 2014;506:47–51. doi: 10.1038/nature12921. [DOI] [PubMed] [Google Scholar]
- 22.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc Natl Acad Sci USA. 2008;105:7676–7680. doi: 10.1073/pnas.0801507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worthy TH, Holdaway RN. The Lost World of the Moa: Prehistoric Life of New Zealand. Indiana Univ Press; Bloomington, IN: 2002. [Google Scholar]
- 24.Schellart WP, Lister GS, Toy VG. A Late Cretaceous and Cenozoic reconstruction of the Southwest Pacific region: Tectonics controlled by subduction and slab rollback processes. Earth Sci Rev. 2006;76:191–233. [Google Scholar]
- 25.Wallis GP, Trewick SA. New Zealand phylogeography: Evolution on a small continent. Mol Ecol. 2009;18:3548–3580. doi: 10.1111/j.1365-294X.2009.04294.x. [DOI] [PubMed] [Google Scholar]
- 26.Tennyson AJD, Martinson P. Extinct Birds of New Zealand. Te Papa Press; Wellington, New Zealand: 2006. [Google Scholar]
- 27.Perry GLW, Wheeler AB, Wood JR, Wilmshurst JM. A high-precision chronology for the rapid extinction of New Zealand moa (Aves, Dinornithiformes) Quat Sci Rev. 2014;105:126–135. [Google Scholar]
- 28.Holdaway RN, Worthy TH, Tennyson AJ. A working list of breeding bird species of the New Zealand region at first human contact. N Z J Zool. 2001;28:119–187. [Google Scholar]
- 29.Johnston PR. Causes and consequences of changes to New Zealand’s fungal biota. N Z J Ecol. 2010;34:175–184. [Google Scholar]
- 30.Bougher NL, Lebel T. Sequestrate (truffle-like) fungi of Australia and New Zealand. Aust Syst Bot. 2001;14:439–484. [Google Scholar]
- 31.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parfrey LW, et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front Microbiol. 2014;5:298. doi: 10.3389/fmicb.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powlesland R, Merton DV, Cockrem JF. A parrot apart: The natural history of the kakapo (Strigops habroptilus), and the context of its conservation management. Notornis. 2006;53:3–26. [Google Scholar]
- 34.Wood JR. Moa gizzard content analyses: Further information on the diets of Dinornis robustus and Emeus crassus, and the first evidence for the diet of Pachyornis elephantopus (Aves: Dinornithiformes) Rec Canterb Mus. 2007;21:27–39. [Google Scholar]
- 35.Attard MR, et al. Moa diet fits the bill: Virtual reconstruction incorporating mummified remains and prediction of biomechanical performance in avian giants. Proc Biol Sci. 2016;283:20152043. doi: 10.1098/rspb.2015.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worthy TH, Scofield RP. Twenty-first century advances in knowledge of the biology of moa (Aves: Dinornithiformes): A new morphological analysis and moa diagnoses revised. N Z J Zool. 2012;39:87–153. [Google Scholar]
- 37.Bezerra JL, Kimbrough J. The genus Lasiobolus (Pezizales, Ascomycetes) Can J Bot. 1975;53:1206–1229. [Google Scholar]
- 38.McKenzie EHC, Buchanan PK, Johnston PR. Checklist of fungi on Nothofagus species in New Zealand. N Z J Bot. 2000;38:635–720. [Google Scholar]
- 39.Bastian F, Alabouvette C, Saiz-Jimenez C. The impact of arthropods on fungal community structure in Lascaux Cave. J Appl Microbiol. 2009;106:1456–1462. doi: 10.1111/j.1365-2672.2008.04121.x. [DOI] [PubMed] [Google Scholar]
- 40.Soltis P, Doyle JJ. Molecular Systematics of Plants II: DNA Sequencing. Kluwer Academic; Dordrecht, The Netherlands: 2012. [Google Scholar]
- 41.Flores V, Brugni N. Catatropis hatcheri n. sp. (Digenea: Notocotylidae) from Heleobia hatcheri (Prosobranchia: Hydrobiidae) and notes on its life-cycle in Patagonia, Argentina. Syst Parasitol. 2006;63:111–118. doi: 10.1007/s11230-005-9004-8. [DOI] [PubMed] [Google Scholar]
- 42.Wright DD. Diet, keystone resources and altitudinal movement of dwarf cassowaries in relation to fruiting phenology in a Papua New Guinean rainforest. In: Dew JL, Boubli JP, editors. Tropical Fruits and Frugivores. Springer; Dordrecht, The Netherlands: 2005. pp. 205–236. [Google Scholar]
- 43.Reddell P, Spain AV, Hopkins M. Dispersal of spores of mycorrhizal fungi in scats of native mammals in tropical forests of northeastern Australia. Biotropica. 1997;29:184–192. [Google Scholar]
- 44.Orlovich DA, Cairney JG. Ectomycorrhizal fungi in New Zealand: Current perspectives and future directions. N Z J Bot. 2004;42:721–738. [Google Scholar]
- 45.Peintner U, et al. Multiple origins of sequestrate fungi related to Cortinarius (Cortinariaceae) Am J Bot. 2001;88:2168–2179. [PubMed] [Google Scholar]
- 46.Wood JR, et al. Novel interactions between non-native mammals and fungi facilitate establishment of invasive pines. J Ecol. 2015;103:121–129. [Google Scholar]
- 47.Hone D, Faulkes C. A proposed framework for establishing and evaluating hypotheses about the behaviour of extinct organisms. J Zool. 2014;292:260–267. [Google Scholar]
- 48.Anderson RC. Nematode Parasites of Vertebrates: Their Development and Transmission. Centre for Agriculture and Bioscience International; Wallingford, United Kingdom: 2000. [Google Scholar]
- 49.Walls G. 1999 Visitor impacts on freshwater avifauna in New Zealand. Available at: doc.org.nz/documents/science-and-technical/casn240.pdf. Accessed November 16, 2016.
- 50.Dunn RR, Harris NC, Colwell RK, Koh LP, Sodhi NS. The sixth mass coextinction: Are most endangered species parasites and mutualists? Proc Biol Sci. 2009;276:3037–3045. doi: 10.1098/rspb.2009.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. Colloquium paper. Homage to Linnaeus: How many parasites? How many hosts? Proc Natl Acad Sci USA. 2008;105:11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hudson PJ, Dobson AP, Lafferty KD. Is a healthy ecosystem one that is rich in parasites? Trends Ecol Evol. 2006;21:381–385. doi: 10.1016/j.tree.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Whiteman NK, Kimball RT, Parker PG. Co-phylogeography and comparative population genetics of the threatened Galápagos hawk and three ectoparasite species: Ecology shapes population histories within parasite communities. Mol Ecol. 2007;16:4759–4773. doi: 10.1111/j.1365-294X.2007.03512.x. [DOI] [PubMed] [Google Scholar]
- 54.McKenna P. An updated checklist of helminth and protozoan parasites of birds in New Zealand. WebmedCentral Parasit. 2010;1:WMC00705. [Google Scholar]
- 55.Vrba V, Pakandl M. Host specificity of Turkey and chicken Eimeria: Controlled cross-transmission studies and a phylogenetic view. Vet Parasitol. 2015;208:118–124. doi: 10.1016/j.vetpar.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 56.Wood JR, Wilmshurst JM. A protocol for subsampling Late Quaternary coprolites for multi-proxy analysis. Quat Sci Rev. 2016;138:1–5. [Google Scholar]
- 57.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guillou L, et al. The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012;41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.