Significance
Exogenous metabolites have been documented to potentiate antibiotics to kill multidrug-resistant pathogens, but the mechanisms are largely unknown. The work presented here shows that intermetabolites from the TCA cycle and the phosphoenolpyruvate (PEP)-pyruvate-AcCoA pathway have the potential to improve targeting of these resistant microorganisms. The enzymes that connect the PEP-pyruvate-AcCoA pathway to the TCA cycle are essential for this potentiation, indicating both pathway and cycle can be merged to be considered a pyruvate cycle (P cycle). The P cycle operates routinely as a general mechanism for energy production and regulating the TCA cycle in Escherichia coli and Edwardsiella tarda. The results reported here provide insights into metabolite-facilitated targeting by antibiotics as well as bacterial energy metabolism and homeostasis.
Keywords: P cycle, TCA cycle, energy metabolism, metabolites, antibiotic resistance
Abstract
The emergence and ongoing spread of multidrug-resistant bacteria puts humans and other species at risk for potentially lethal infections. Thus, novel antibiotics or alternative approaches are needed to target drug-resistant bacteria, and metabolic modulation has been documented to improve antibiotic efficacy, but the relevant metabolic mechanisms require more studies. Here, we show that glutamate potentiates aminoglycoside antibiotics, resulting in improved elimination of antibiotic-resistant pathogens. When exploring the metabolic flux of glutamate, it was found that the enzymes that link the phosphoenolpyruvate (PEP)-pyruvate-AcCoA pathway to the TCA cycle were key players in this increased efficacy. Together, the PEP-pyruvate-AcCoA pathway and TCA cycle can be considered the pyruvate cycle (P cycle). Our results show that inhibition or gene depletion of the enzymes in the P cycle shut down the TCA cycle even in the presence of excess carbon sources, and that the P cycle operates routinely as a general mechanism for energy production and regulation in Escherichia coli and Edwardsiella tarda. These findings address metabolic mechanisms of metabolite-induced potentiation and fundamental questions about bacterial biochemistry and energy metabolism.
The emergence and spread of antibiotic-resistant bacteria is a major threat to human health and could also have significant impacts on the health and survival of other species (1–3). Conventional approaches to antibacterial development are based on the inhibition of essential processes of antibiotic-resistant mechanisms, which seem to have reached the point of diminishing returns (4). The discovery that bacterial environments and pathways confound antibiotic efficacy represents a fundamental shift in our understanding of bactericidal antibiotic modes of action (4, 5). A bacterium acquires antibiotic resistance as a response to the antibiotic environment, including the horizontal acquisition of new genes and spontaneous mutations within chromosomally located genes and the exhibition of intrinsic resistance. Conversely, the reversibility of the resistance could be a result of environmental interventions such as metabolic modulation, which leads to elevated sensitivity to antibiotics (4). Therefore, environmental interventions can lead to the development of new antimicrobial therapies.
Recent reports indicate that a high abundance of endogenous metabolites, such as indole, nitric oxide, hydrogen sulfide, and gaseous ammonia, correlate with higher prevalence of bacterial resistance to antibiotics (5–9), whereas increased abundance of an exogenous carbon source enhances inactivation by aminoglycoside antibiotics (10–12), which suggests that the microbial metabolic environment modulates antibiotic uptake and/or efficacy (8, 13). Exogenous carbon-mediated inactivation by aminoglycoside antibiotics is attributed to elevated NADH and proton motive force (PMF) generation and increased antibiotic uptake (10–13), which is supported by metabolic modulation, including promotion of the tricarboxylic acid (TCA) cycle (11, 12). Thus, better understanding of the metabolic flux could lead to effective therapeutic or preventive interventions for elimination of antibiotic-resistant bacteria (14–16).
The TCA cycle (also known as the Krebs cycle or citric acid cycle) was identified 80 y ago and is now understood to be the final pathway in aerobic organisms for oxidation of carbohydrates, fatty acids, and amino acids (17). The evolutionary origin of the TCA cycle is an important question in biochemical research (18, 19). As a new metabolic pathway is established, it could opportunistically exploit enzymes from existing pathways in the cell (9). With this strategy in mind, several alternatives of the TCA cycle have been proposed and theoretically tested (18–20), including the phosphoenolpyruvate (PEP)-glyoxylate cycle in Escherichia coli (21, 22). However, such speculation on the origin of a metabolic pathway has limitations to understanding its metabolic reaction network (23). Conversely, both stable and radioactive isotopes represent an objective approach to tracing these molecules as they pass through the metabolic reaction network in a relatively objective manner, including techniques such as isotope-ratio mass spectrometry (IRMS) and nontargeted isotope fate detection (NTFD) (23, 24). Using IRMS, the PEP-glyoxylate cycle has been demonstrated as an alternative to the TCA cycle in E. coli (25, 26). However, IRMS neither provides quantitative information about mass isotopomer distributions (MIDs) nor quantifies the particular ion fragments present in mass spectra. Comparatively, NTFD allows for the tracing of labeled atoms present as an externally supplied compound. Thus, metabolic pathways coupled to the applied tracer can be elucidated without a priori knowledge of the pathway and downstream identification of enzymes (23).
In a previous study, we characterized the metabolomes of kanamycin-susceptible Edwardsiella tarda, LTB4-S, and isogenic kanamycin-resistant E. tarda LTB4-R, and identified depressed alanine and glucose as two crucial metabolic biomarkers that are related to antibiotic resistance. We demonstrated that exogenous alanine and glucose increase flux through the TCA cycle and promote antibiotic uptake, thereby eliminating antibiotic-resistant bacteria (11). In the current study, we further showed that glutamate, another depressed biomarker, also promotes the inactivation of drug-resistant bacteria by kanamycin. The detailed study of the underlying mechanism revealed a previously unknown prevalent pathway, which we call the pyruvate cycle (P cycle). We found that the P cycle is a common pathway of respiration and energy production in E. tarda and E. coli grown under various conditions. The P cycle also prioritizes use of oxaloacetate (OAA), which regulates the TCA cycle, and together with the TCA cycle provides respiratory energy in E. tarda and E. coli.
Results
Exogenous Glutamate Fluxes to the TCA Cycle Through the PEP-Pyruvate-AcCoA Pathway.
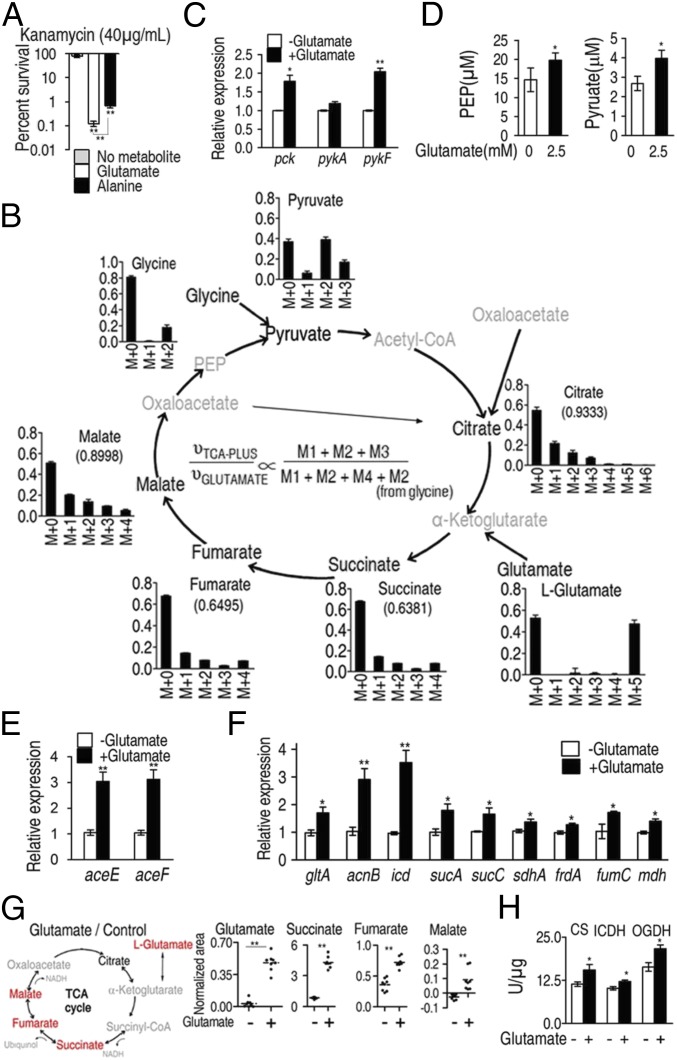
We previously showed that for the E. tarda LTB4-R metabolome, alanine, aspartate, and glutamate metabolism is the most important metabolic pathway and depressed alanine, glutamate, and aspartate are crucial metabolites. Exogenous alanine or/and glucose restored susceptibility to antibiotics in antibiotic-resistant bacteria by increasing TCA flux, NADH production, and PMF generation to enhance kanamycin uptake and thereby efficiently kill the multidrug-resistant pathogens (11). Alanine and glutamate are reversibly converted to pyruvate and α-ketoglutarate, respectively, by glutamic-pyruvic transaminase (GPT), which then feed the TCA cycle (SI Appendix, Fig. S1B). Thus, the present study further explored whether glutamate has the same potential as alanine. Results showed that glutamate also elevates NADH and PMF to promote antibiotic uptake and thereby restores susceptibility of drug-resistant E. tarda to kanamycin (SI Appendix, Text S1, and Figs. S1 and S2). Furthermore, 2.5 mM exogenous glutamate facilitates cell death of the multidrug-resistant bacteria by kanamycin with higher efficiency than that of 40 mM alanine (Fig. 1A), suggesting differential modes of action between these two metabolites. Additionally, variable uptake efficiencies of the two metabolites by the bacteria could be another possibility.
Fig. 1.
The P cycle in E. tarda. (A) Percent survival of E. tarda EIB202. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM) or alanine (40 mM) plus kanamycin (40 μg/mL). (B) Mass isotopomer distributions in EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with 13C-labeled glutamate (1.25 mM) and unlabeled glutamate (1.25 mM). Please see SI Appendix, Text S2 for detailed interpretation of this result. (C) Expression of pck, pykA, and pykF of EIB202, which was determined by qRT-PCR. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM). (D) PEP and pyruvate abundance of EIB202, which was determined by UPLC/MS. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM). (E and F) Expression of aceE and aceF (E) and genes in the TCA cycle (F) of EIB202, which was determined by qRT-PCR. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM). (G) Scatterplot of abundance of the indicated metabolites, which were analyzed by GC-MS. Red, increased metabolites; black, unchanged metabolite; gray, undetected metabolites. Each dot represents a biological or technical replicate. (H) Activity of citrate synthase (CS), isocitrate dehydrogenase (ICDH), and α-oxoglutarate dehydrogenase (OGDH). Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM). Information on these genes and their encoding enzymes is described as follows: aceE and aceF, pyruvate dehydrogenase complex E1 and E2, respectively; acnB, aconitase; frdA, fumarate reductase subunit; fumC, fumarase; mdh, malate dehydrogenase; gltA, citrate synthase; icd, isocitrate dehydrogenase; pck, phosphoenolpyruvate carboxykinase; pykA and pykF, pyruvate kinases 2 and 1, respectively; sdhA, succinate dehydrogenase subunit; sucA, α-oxoglutarate dehydrogenase subunit; and sucC, succinyl-CoA synthetase. Results (A and C–H) are displayed as mean ± SEM; at least three biological repeats were carried out. Statistically significant values are indicated with asterisk (*P < 0.05, **P < 0.01) and determined by Student’s t test.
To explore the metabolic mechanisms performed by exogenous glutamate, we used NTFD to analyze metabolite fate (23, 24) in EIB202, a multidrug-resistant E. tarda, cultured in the presence of 1.25 mM 13C-labeled glutamate and 1.25 mM unlabeled glutamate. This analysis identified 21 13C-labeled metabolites, including succinate, fumarate, malate, and citrate (TCA cycle intermetabolites), glycine, threonine, alanine, and some precursors of biosynthesis of fatty acids (SI Appendix, Tables S1 and S2). 13C-labeled pyruvate was also detected by ultra performance liquid chromatography-mass spectrometry (UPLC-MS/MS). These results showed that the labeled alanine, glycine, and threonine were from 13C-OAA through 13C-PEP rather than 13C-pyruvate, since 13C-pyruvate is not irreversibly converted to PEP (www.genome.jp/kegg), but 13C-alanine, -glycine, and -threonine could be reversibly transferred to pyruvate through PEP. Another interesting finding is the low abundance of M4-labeled citrate, indicating the transformation of M4-labeled OAA with unlabeled AcCoA to citrate was limited by the TCA cycle (Fig. 1B). (Please see SI Appendix, Text S2 for detailed interpretation of this result). In summary, the tracer experiments suggest that exogenous glutamate is converted to OAA and that OAA preferentially enters the PEP-pyruvate-AcCoA pathway, and then enters the TCA cycle, which we refer to as the P cycle (Fig. 2A).
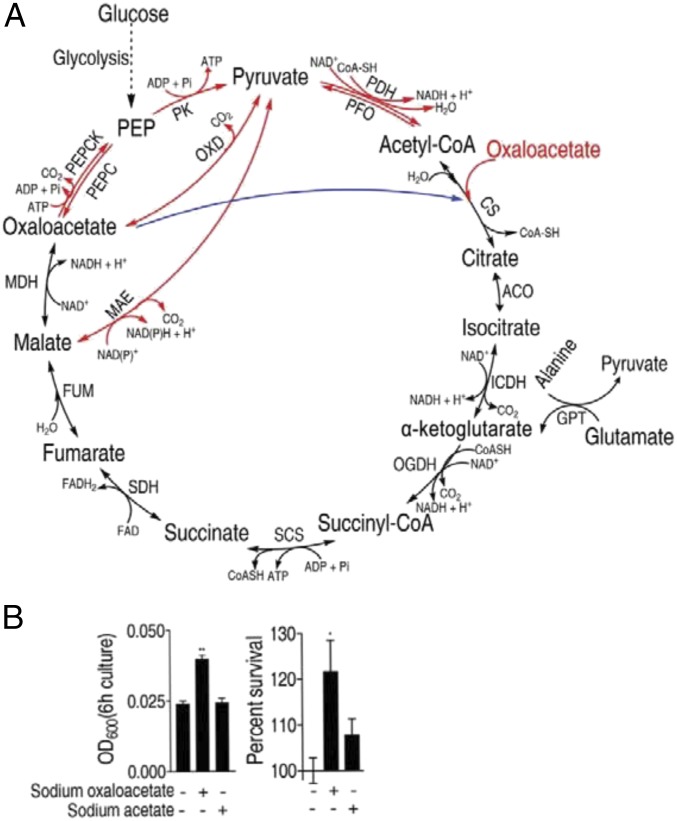
Fig. 2.
The P cycle and the TCA cycle in E. tarda. (A) Overview of the P cycle. Black line, overlapped pathway between the P cycle and the TCA; blue, a step of the TCA cycle, which does not exist in the P cycle; and red, pathways of the P cycle, which do not exist in the TCA cycle. ACO, aconitase; CS, citrate synthase; FUM, fumarase; GPT, glutamic-pyruvic transaminase; ICDH, isocitric dehydrogenase; MAE, malic enzyme (NAD-dependent malic enzyme and NADP-dependent malic enzyme); MDH, malate dehydrogenase; OGDH, α-oxoglutarate dehydrogenase; OXD, oxaloacetate decarboxylase; PDH, pyruvate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase; PEPCK, phosphoenolpyruvate carboxykinase; PFO, pyruvate-flavodoxin oxidoreductase; PK, pyruvate kinase; SCS, succinyl-CoA synthetase; and SDH, succinic dehydrogenase. (B) Optical density (OD) (Left) and percent survival (Right) of EIB202. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) or oxaloacetate (10 mM). Result (B) is displayed as mean ± SEM; at least three biological repeats were carried out. Statistically significant values are indicated with asterisk (*P < 0.05, **P < 0.01) and determined by Student’s t test.
To examine flux through the P cycle more carefully, the expression of pck and pykA, pykF, which encode for PEP carboxykinase (PEPCK) and pyruvate kinase (PK), respectively, were analyzed by qRT-PCR. The results show that glutamate stimulates expression of both pck and pykF, (Fig. 1C), which suggests increased flux through the P cycle, leading to transformation of OAA to PEP and pyruvate (Fig. 1D). However, exogenous glutamate does not increase production of lactate (SI Appendix, Fig. S3), which is another possible product from this pathway. Glutamate also stimulated expression of aceE and aceF, encoding two components of the pyruvate dehydrogenase complex (PDH) that converts pyruvate to AcCoA (Fig. 1E) as well as nine other genes in the TCA cycle (Fig. 1F). Exogenous glutamate increased the abundance of succinate, fumarate, and malate (Fig. 1G) and the activity of citrate synthase (CS), isocitrate dehydrogenase (ICDH), and α-oxoglutarate dehydrogenase (OGDH) (Fig. 1H). Therefore, these results indicate that glutamate increases flux through the TCA cycle preferentially through the OAA-PEP-pyruvate-AcCoA-CIT(citrate) pathway (i.e., the P cycle).
An Energy-Generating Cycle: The P Cycle.
The difference between the P cycle and the TCA cycle is that the former consumes OAA, while the latter consumes AcCoA; both cycles reduce NAD+ to NADH, produce carbon dioxide, and fuel ATP production via oxidative phosphorylation. The equation of the P cycle is: OAA + 4 NAD+ + FAD + ADP + Pi+ H2O → 3 CO2 + 4 NADH + 4 H+ + FADH2 + ATP. The equation of the TCA cycle is: AcCoA + 3 NAD+ + FAD + ADP + Pi + 2 H2O → CoA-SH + 3 NADH + 3 H+ + FADH2 + ATP + 2 CO2 (Fig. 2A). Note that the P cycle generates one more NADH than the TCA cycle, which could explain why bacteria grow faster in M9 mineral medium (M9 medium) with OAA than in M9 medium with acetate (Fig. 2B).
The P Cycle Is a Prevalent Cycle.
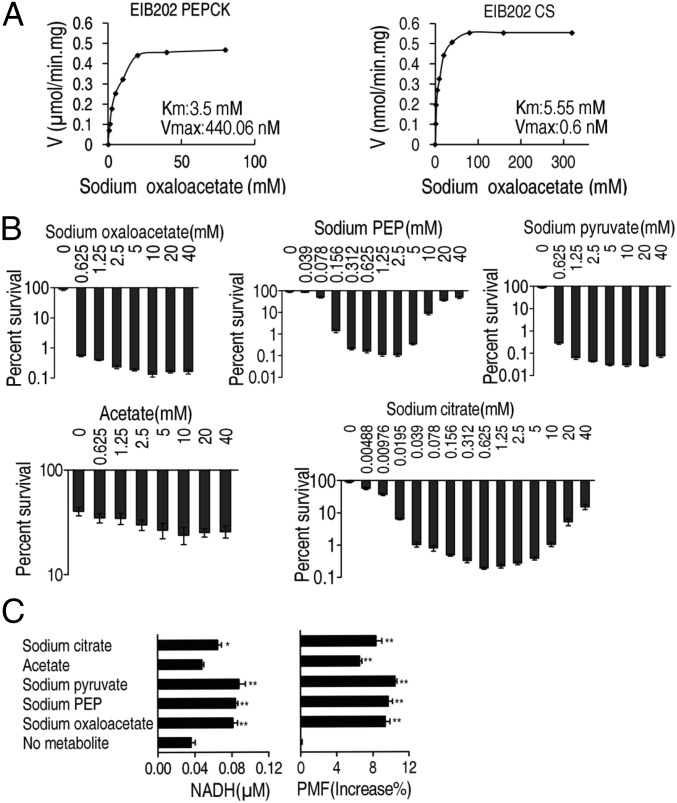
Our results raised the question of whether the P cycle is glutamate dependent. We speculated that the P cycle was a primary cycle rather than a secondary pathway initiated by glutamate due to the reduced transformation of OAA and AcCoA into citrate by the TCA cycle. To demonstrate this idea, Michaelis–Menten kinetics were used to determine the rate of PEPCK and citrate synthase (CS) reactions to OAA concentrations in a glutamate-independent manner. PEPCK was shown to have a lower Km value than citrate synthase, indicating that PEPCK was more competitive than citrate synthase for OAA utilization (Fig. 3A), which suggests OAA prefers the P cycle to the TCA cycle.
Fig. 3.
Oxaloacetate favors PEPCK to citrate synthesis, and the P cycle generates more NADH and PMF than the TCA cycle in E. tarda. (A) Michaelis–Menten kinetics of phosphoenolpyruvate carboxykinase (PEPCK) and citrate synthase (CS) in oxaloacetate (0.625, 1.25, 2.5, 5, 10, 20, 40, 80 mM and 1.25, 2.5, 5, 10, 20, 40, 80, 160, 320 mM, respectively). Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM). (B) Percent survival of EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of the indicated concentrations of metabolites plus kanamycin (30 μg/mL). (C) NADH and PMF of EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of the indicated metabolites sodium citrate (0.625 mM), sodium pyruvate (10 mM), sodium PEP (1.25 mM), or sodium oxaloacetate (10 mM). Result (B and C) is displayed as mean ± SEM, and three biological repeats were carried out. Significant differences are identified (*P < 0.05, **P < 0.01) as determined by Student’s t test. P < 0.01 in B except for acetate.
Kanamycin-mediated bacterial cell death is highly dependent on PMF, which is associated with the buildup of protons (10). Compared with the TCA cycle, the P cycle generated one more NADH through PDH. Therefore, the P cycle can generate more PMF than the TCA cycle and, therefore, would be more efficient in kanamycin-mediated bacterial inactivation. To validate our hypothesis, the following experiments were performed. The additions of OAA, PEP, pyruvate, acetate (it is converted to AcCoA in vivo), and citrate were assessed for bactericidal effects for E. tarda exposed to 30 μg/mL kanamycin. OAA, PEP, and pyruvate showed stronger inactivation (670- to 3,200-fold) than that of citrate (480-fold) and acetate (1.6-fold) in comparison with a control not given additional substrate, which may be due to the additional molecule of NADH generated by PDH. Both PEP and citrate increased the inactivation efficiency in a dose-dependent manner when at low concentration (<2.5 mM of PEP and <0.625 mM of citrate), but the efficiency decreased gradually at high concentration (Fig. 3B), implying that high concentration of PEP impedes the transformation of OAA to PEP, and high concentration of citrate decreases the condensation of OAA and AcCoA to citrate in unidentified manners. Meanwhile, the increased intracellular concentrations of NADH and PMF were consistent with the bactericidal effects imposed by OAA, PEP, and pyruvate and to a lesser extent citrate, due to more proton generation (Fig. 3C). These findings provide evidence for the P cycle. Note that acetate also increased PMF, although somewhat lower than that induced by the other intermetabolites, but failed to potentiate the bactericidal effect, suggesting the threshold is required for effective results.
The TCA Cycle Is a Part of the P Cycle.
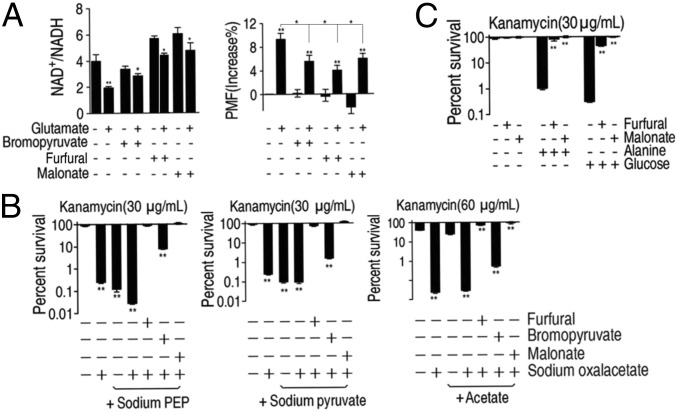
The above results motivated us to explore the relationship between the TCA cycle and the P cycle. One difference between the proposed P cycle and the TCA cycle was that the P cycle had three alternative metabolic steps before following the TCA cycle. To validate the three steps that represent essential components of the P cycle, and understand the effect of these steps on the TCA cycle, we inactivated enzymes with inhibitors to see whether they would abolish the glutamate-triggered effects. Bromopyruvate and furfural, active site-directed and noncompetitive inhibitor for pyruvate dehydrogenase El, respectively, which converts pyruvate to AcCoA, impaired the ability of glutamate to stimulate the production of NADH, and to increase PMF (Fig. 4A). Similar results were obtained with malonate, a competitive inhibitor of the enzyme succinate dehydrogenase (SDH) in the TCA cycle, as a control (Fig. 4A). These data indicate that inhibition of the P cycle or the TCA cycle leads to the similar increase of NAD+/NADH and decrease of PMF. Seemingly, the TCA cycle is suppressed when PDH is inhibited. Thus, inhibition of either PDH (not in the TCA cycle) or SDH (in the TCA cycle) abolished the potency of glutamate in bacterial killing, suggesting that the P cycle rather than the TCA cycle led the functional link. Meanwhile, two other inhibitors, namely Na2-ATP (inhibitor for PEPCK, CS, ICDH, and OGDH) and furfural (noncompetitive inhibitor for PDH) were used to further define the role of the three steps (SI Appendix, Text S3, and Fig. S4A). All of these inhibitors were shown to reduce glutamate-triggered effects regardless of the cycles these enzymes belong to, while they did not affect the growth of EIB202. Furfural inhibited glutamate-mediated bactericidal effects most efficiently while the other three reduced inactivation promoted by glutamate in a dose-dependent manner (SI Appendix, Fig. S4B). The inhibition of the conversion from pyruvate to AcCoA completely abolished glutamate-triggered effects, implying a critical role for the related alternative three steps and subsequent effects on the TCA cycle. Thus, the above data strongly implied the TCA cycle is a part of the P cycle in E. tarda energy generation.
Fig. 4.
Inhibition of the P cycle abrogates metabolite-mediated killing. (A) NAD+/NADH and PMF of EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without glutamate (2.5 mM) in the presence or absence of bromopyruvate (2.5 mM), furfural (10 mM), or malonate (20 mM), as indicated. (B) Percent survival of EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of oxaloacetate (10 mM) and PEP (1.25 mM), pyruvate (10 mM), or acetate (10 mM) with or without furfural (10 mM), bromopyruvate (2.5 mM), or malonate (20 mM). (C) Percent survival of EIB202. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of alanine (40 mM) or glucose (10 mM) and effect of inhibitors furfural (10 mM) or malonate (20 mM). Results are displayed as mean ± SEM, and three biological repeats were carried out. Statistically significant differences are indicated by asterisk (*P < 0.05, **P < 0.01) and were determined by Student’s t test.
Further tests demonstrated that inhibition of the OAA-PEP-pyruvate-AcCoA-CIT pathway could attenuate or block the TCA cycle, while confirming that inhibition of PDH does not down-regulate the TCA cycle indirectly by decreasing the abundance of AcCoA. To test this idea, cells were grown in the presence of OAA plus PEP, pyruvate, or acetate. These metabolites restored susceptibility to antibiotics, but this effect was blocked by furfural, bromopyruvate, or malonate agents, that inhibited the P cycle or TCA cycle, respectively (Fig. 4B), and similar effects were observed with glucose and alanine instead of glutamate (Fig. 4C). Therefore, we suggest that the P cycle, rather than TCA cycle, is the fundamental energy-generating cycle in E. tarda cells.
Demonstration of the P Cycle in E. coli.
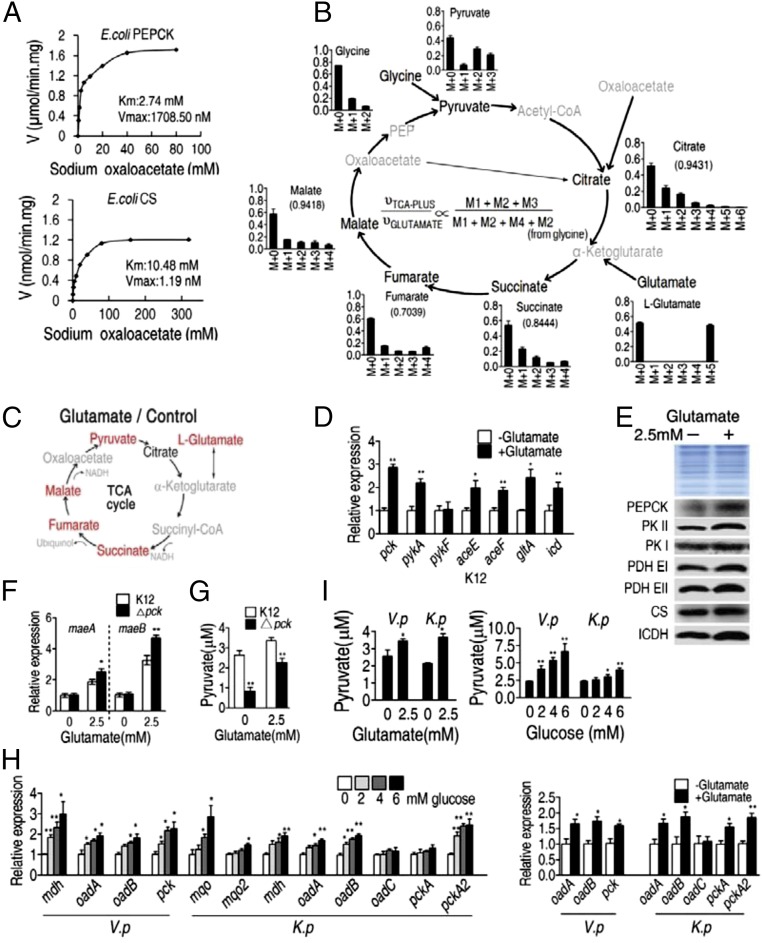
To determine whether the P cycle is unique to E. tarda, additional experiments were performed in E. coli K12 BW25113. Michaelis–Menten kinetic analysis showed that, the Km of PEPCK for OAA was 2.74 mM with a Vmax of 1708.5 nM⋅min−1⋅mg−1. The Km of citrate synthase for OAA was 10.48 mM with a Vmax of 1.19 nM⋅min−1⋅mg−1 (Fig. 5A), suggesting higher affinity of OAA for PEPCK. Nontargeted 13C5-glutamate isotope tracer analysis was carried out when E. coli K12 BW25113 cells were cultured in Luria-Bertani (LB) medium and then M9 medium with acetate plus the labeled metabolite. The obtained results were similar in results described above in E. tarda EIB202. Specifically, 13C labeled alanine, glycine, and threonine were detected and the abundance of M4-labeled citrate was low, which supported the P cycle and low flux through the TCA cycle, respectively (Fig. 5B and SI Appendix, Table S3), and exogenous glutamate elevated the relative abundance of malate, pyruvate, succinate, and fumarate (Fig. 5C). Similar results were obtained in M9 medium cultures with 0.3% glucose, as well as super optimal broth with catabolite repression (SOC medium) with subsequent transfer to M9 medium with acetate plus the labeled metabolite, and shorter incubation periods did not significantly change the results (SI Appendix, Fig. S5, and Tables S4–S7). qRT-PCR and Western blot data, similar to that presented for E. tarda, also confirmed the presence of an active P cycle in E. coli K12 BW25113 (i.e., elevated mRNA and protein expression of pck, pykA, aceE aceF, gltA, and icd in the presence of exogenous glutamate) (Fig. 5 D and E).
Fig. 5.
The P cycle in E. coli. (A) Kinetic analysis of E. coli PEPCK and E. coli citrate synthase (CS) with oxaloacetate (0.625, 1.25, 2.5, 5, 10, 20, 40, 80 mM and 1.25, 2.5, 5, 10, 20, 40, 80, 160, 320 mM, respectively) as substrate at the indicated concentration. Cells were grown in LB medium and then incubated in M9 plus acetate (10 mM). (B) Mass isotopomer distributions in E. coli. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with 13C-labeled glutamate (1.25 mM) and unlabeled glutamate (1.25 mM). Black, 13C-labeled metabolites; gray, undetected metabolites. (C) Metabolites with increased abundance (in red) in the TCA cycle, which were detected by GC-MS. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without glutamate (2.5 mM). Gray, undetected metabolites. Each dot represents a biological or technical replicate. (D and E) Expression of the indicated genes (D) and proteins (E) determined by qRT-PCR and Western blot, respectively. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without glutamate (2.5 mM). pck, pykA, pykF, aceE aceF, gltA, icd encode PEPCK, PKII, PKI, PDH E1, PDH EII, CS, ICDH, respectively. (F and G) Expression of malic enzyme (F) and intracellular pyruvate (G) in ∆pck and E. coli K12. Genes maeA and maeB encode NAD-dependent malic enzyme and NADP-dependent malic enzyme, respectively. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without glutamate (2.5 mM). (H) Expression of malic enzyme, oxaloaceate decarboxylase and PEPCK genes in V. parahaemolyticus or K. pneumonia. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without the indicated concentrations of glucose. (I) Intracellular pyruvate in V. parahaemolyticus (V. p) or K. pneumonia (K. p). Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) with and without glutamate (2.5 mM) or the indicated glucose concentrations. Results (D and F–I) are displayed as mean ± SEM, and three biological repeats were carried out. Statistically significant values are indicated by asterisk (*P < 0.05, **P < 0.01) and were determined by Student’s t test.
Two alternative metabolic pathways for malate/OAA to pyruvate have been reported previously in some bacterial species (27, 28), namely, OAA decarboxylase conversion of OAA to pyruvate (OAA-pyruvate), as well as enzymatic conversion of malate to pyruvate (MAL-pyruvate) (Fig. 2A). To test whether these pathways respond to exogenous glutamate, expression of maeA and maeB, which encode NAD-dependent malic enzyme and NADP-dependent malic enzyme, respectively, were examined in E. coli K12 BW25113 and Δpck (pck encodes PEPCK). It was found that maeA and maeB expression increased in the presence of glutamate, and that the loss of pck led to higher expression of maeA and maeB (Fig. 5F). Furthermore, exogenous glutamate was observed to lead to higher pyruvate, while loss of pck led to lower pyruvate (Fig. 5G). OAA decarboxylase is absent in E. coli, but present in Vibrio parahaemolyticus (oadA and oadB) and Klebsiella pneumonia (oadA, oadB, and oadC). Results in these bacteria showed increased expression of oadA, oadB, pck, mdh, mqo, and mqo2 (Fig. 5H) and increased pyruvate in the presence of glutamate or glucose (Fig. 5I). These results suggest that MAL-pyruvate or/and OAA-pyruvate alternative pathways function in E. coli, V. parahaemolyticus, and K. pneumonia, which supports evidence for the P cycle.
Loss of the OAA-PEP-Pyruvate-AcCoA-CIT Pathway Loses the TCA Cycle in the Presence or Absence of Glutamate.
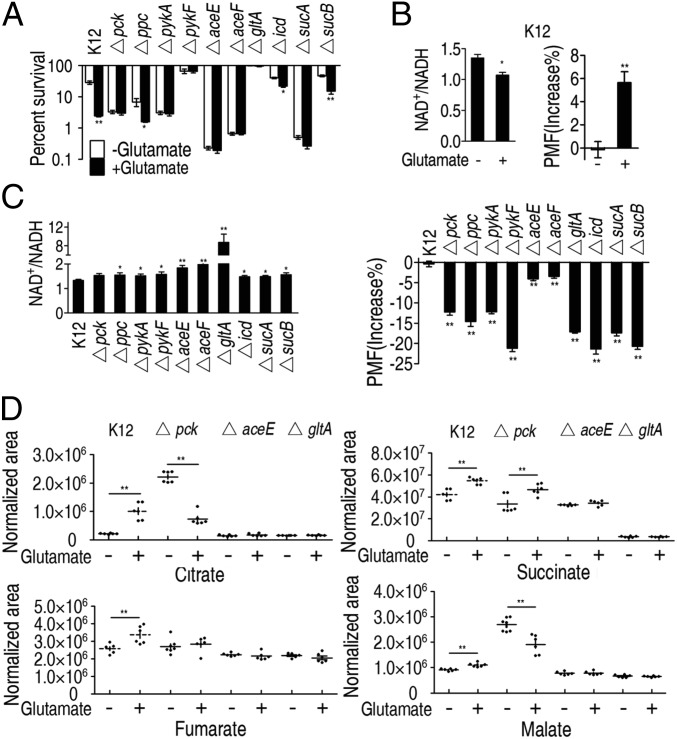
To understand the effect of OAA-PEP-pyruvate-AcCoA-CIT on the TCA cycle, we also analyzed the effect of glutamate on gentamicin susceptibility of 19 strains of E. coli K12, each carrying a deletion mutation in one enzyme of the P cycle with kanamycin resistance. E. coli K12 was susceptible to gentamicin in the presence of glutamate, showing decreased survival and intracellular NAD+/NADH and increased PMF (Fig. 6 A and B). In contrast, the 19 mutant strains with increased NAD+/NADH and decreased PMF (Fig. 6C) were not susceptible or were weakly susceptible to killing by gentamicin in the presence of glutamate (Fig. 6A and SI Appendix, Fig. S6A). When the mutation in each strain was corrected by genetic complementation, susceptibility to antibiotics in the presence of glutamate was restored (SI Appendix, Fig. S6B).
Fig. 6.
Effect of exogenous glutamate on the P cycle and the TCA cycle in E. coli. (A) Percent survival of E. coli K12 and selected P cycle mutants. Cells were grown in LB medium and then incubated in M9 medium with acetate (10 mM) in the presence or absence of glutamate (2.5 mM) plus gentamicin (1.2 μg/mL). (B and C) NAD+/NADH (Left) and PMF (Right) of E. coli K 12 (B) and selected P cycle mutants (C). Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of glutamate (2.5 mM). (D) Effect of pck, aceE, and gltA deficiency on abundance of the indicated metabolites. Cells grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of glutamate (2.5 mM). Results (A–D) are mean ± SEM of three biological repeats. Statistically significant values are indicated with an asterisk (*P < 0.05, **P < 0.01) and were determined by Student’s t test.
Interestingly, among the 19 mutants, Δpck, Δppc, ΔpykA, ΔaceE, and ΔaceF play no role in the TCA cycle. The loss of these genes and sucA in the TCA cycle led to decreased PMF but increased killing, indicating gentamicin-mediated killing and PMF level are not tightly correlated. Thus, the mechanism by which the PMF-dependent increased susceptibility to gentamicin is unclear (10–12), where the deleted genes may have additional effects. Intriguingly, ΔaceF cells had high intracellular oleate and intracellular gentamicin, and exogenous oleate enhanced both uptake and bactericidal effects of gentamicin (SI Appendix, Fig. S6 C–F), indicating that oleate promotes gentamicin uptake in the lower PMF, but the significance of this observation is not yet understood. It was also shown that exogenous glutamate promoted gentamicin to kill E. coli K12 but not ΔaceE and ΔaceF strains in mouse urinary tracts (SI Appendix, Fig. S6G).
Metabolite profiling was conducted in Δpck, ΔaceE (within the OAA-PEP-pyruvate-AcCoA-CIT pathway), ΔgltA (within TCA cycle), and E. coli K12 grown without glutamate. Lower citrate, fumarate, succinate, and malate were detected in ΔaceE and ΔgltA, and lower succinate was detected in Δpck. However, exogenous glutamate led to higher levels of citrate, fumarate, succinate, and malate in E. coli K12 but not in Δpck, ΔaceE, or ΔgltA cells, except for succinate in Δpck (Fig. 6D). These results suggest that the OAA-PEP-pyruvate-AcCoA-CIT pathway affects the TCA cycle in the presence or absence of glutamate.
Loss of the OAA-PEP-Pyruvate-AcCoA-CIT Pathway Interrupts the TCA Cycle in the Presence or Absence of Glucose or Fructose.
When antibiotic-resistant E. coli and E. tarda were grown in the presence of glutamate, glucose, or fructose, these metabolites had similar effects, stimulating uptake and bactericidal effects of aminoglycoside antibiotics (10–12). Our previous study emphasized that the TCA cycle was the critical pathway for glucose- and fructose-enabled bacterial inactivation (11, 12). However, given the current results, we arrived at expanded conclusions, as described below.
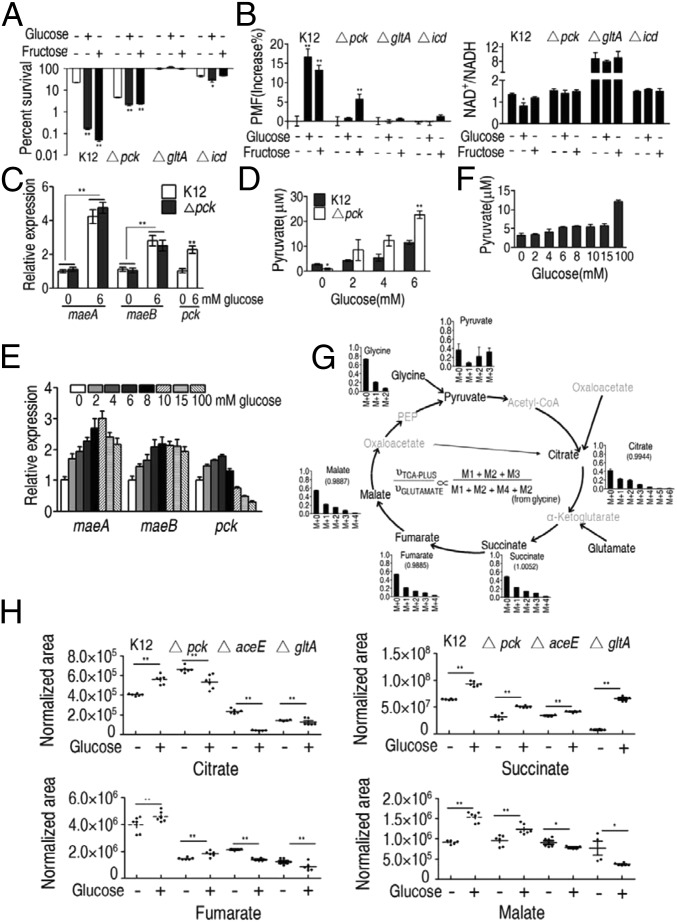
Because PEPCK is involved in the P cycle but not the TCA cycle, experiments were performed in ∆pck cells. Results were compared with those in ∆gltA or ∆icd cells (within the TCA cycle). These mutations abrogated glucose- and fructose-enabled inactivation in the presence of gentamicin (Fig. 7A), which was attributed to decreased PMF and increased NAD+/NADH (Fig. 7B). Thus, the P cycle is an important part of glucose- and fructose-enabled bactericidal activity. One noticeable finding is that similarly increased PMF was detected in E. tarda cultured with exogenous acetate and in E. coli ∆pck cultured with exogenous glucose, but low killing and strong killing were found in E. tarda and in E. coli ∆pck, respectively. This discrepancy suggests that PMF-dependent killing thresholds could be different for different bacteria, and the deleted gene possibly plays an unidentified role in increasing PMF-dependent susceptibility to gentamicin. Comparatively, stronger action was detected in ΔgltA and Δicd than Δpck, which should be attributed to the MAL-pyruvate pathway. To demonstrate this, qRT-PCR was used to examine the expression of maeA and maeB in E. coli K12 and Δpck cells grown in M9 medium with or without glucose. Exogenous glucose promoted maeA and maeB expression (Fig. 7C) and increased pyruvate in a dose-dependent manner in Δpck and E. coli K12 (Fig. 7D). Interestingly, higher pyruvate was detected in Δpck than the parental strain (Fig. 7D), suggesting that the TCA cycle is obstructed when the P cycle is inactive. Studies have indicated that high doses of glucose depress pck expression (29), which is consistent with results reported here for cells grown in LB with variable glucose. As shown in earlier studies (29), pck was negatively regulated by glucose, in the 10- to 100-mM range (Fig. 7E). However, maeA and maeB expression increased in high glucose (Fig. 7E), which was consistent with the elevated pyruvate (Fig. 7F).
Fig. 7.
Effect of exogenous glucose on the P cycle and the TCA cycle in E. coli. (A) Percent survival of the indicated strains. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of glucose (10 mM) or fructose (2.5 mM) plus gentamicin (1.2 μg/mL). (B) As in A showing intracellular PMF (Left) and NAD+/NADH (Right). (C) Relative expression of pck and maeA and maeB determined by qRT-PCR in Δpck or control cells. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of glucose (6 mM). (D) Intracellular pyruvate determined by UPLC-MS in Δpck or control cells. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence of 0, 2, 4, or 6 mM glucose, as indicated. (E and F) Relative expression of maeA, maeB, and pck determined by qRT-PCR (E) and intracellular pyruvate determined by UPLC-MS (F) in E. coli K12. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence of the indicated concentrations of glucose. (G) Mass isotopomer distributions in E. coli. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of 13C-labeled glucose (5 mM) and unlabeled glucose (5 mM). Black, 13C-labeled metabolites; gray, undetected metabolites. (H) Scatterplot of the abundance of the indicated metabolites in Δpck, ∆aceE, and ∆gltA E. coli. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of glucose (10 mM). Results (A–E and H) are mean ± SEM of three biological repeats. Statistically significant values are indicated with an asterisk (*P < 0.05, **P < 0.01) and were determined by Student’s t test. P < 0.01 in gradient of D–F.
When 13C5-glutamate was replaced with 13C6-glucose in the nontargeted isotope tracer analysis, a similar pattern of labeled metabolites existed for both, with M4-labeled metabolites and M6-labeled citrate detected in low levels (Fig. 7G and SI Appendix, Table S8). These results support evidence that the P cycle is a prevalent metabolic pathway regardless of nutrient sources.
GC-MS analysis was used to quantify metabolites of the P cycle in Δpck, ΔaceE, and ΔgltA in the presence or absence of glucose. Exogenous glucose increased citrate, fumarate, succinate, and malate in wild-type variants, while not affecting citrate in Δpck cells, or citrate, fumarate, or malate in ΔaceE and ΔgltA cells (Fig. 7H).
Loss of the OAA-PEP-Pyruvate-AcCoA-CIT Pathway Interrupts the TCA Cycle in the Presence of Ample Fuels for the TCA Cycle.
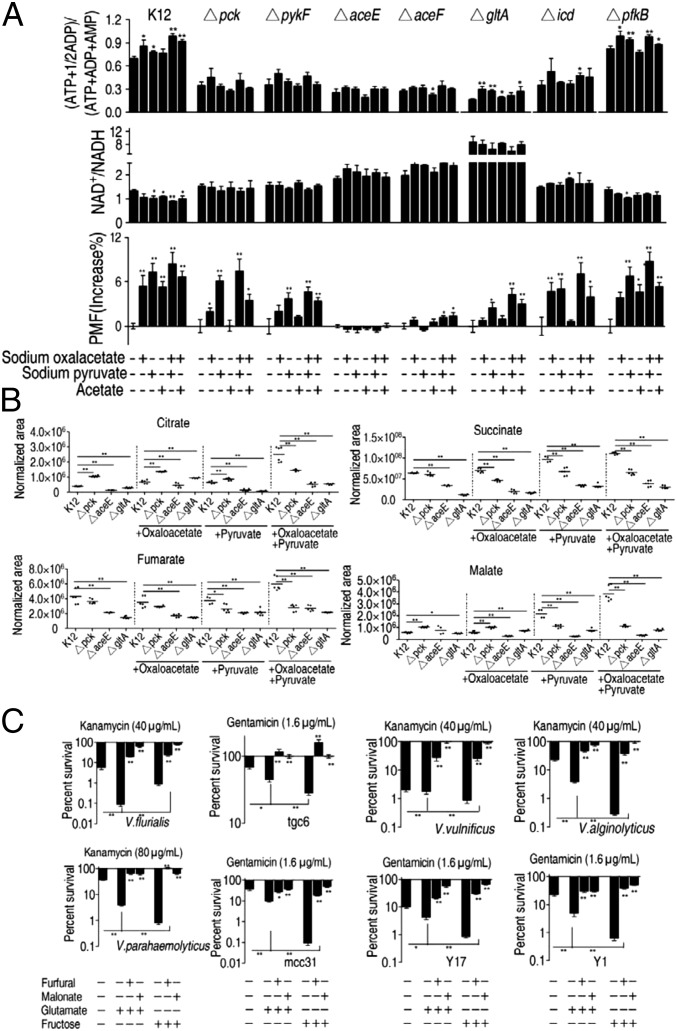
We showed that exogenous OAA, pyruvate, and acetate, which fuel the TCA cycle, elevated the adenylate energy charge and PMF, and decreased the NAD+/NADH ratio in E. coli K12, but not in ΔaceE or ΔaceF (Fig. 8A), two genes that transform pyruvate into AcCoA. Similar results were detected in Δpck, ΔpykE, ΔgltA, and Δicd in some mutants unless complemented with exogenous pyruvate. Generally, high NAD+/NADH and low adenylate energy charge and PMF levels were found in ΔaceE, ΔaceF, and ΔgltA, which may have resulted from the additional NADH generated while transforming pyruvate into AcCoA in ΔaceE and ΔaceF (Fig. 8A). It is also possible that the gltA deletion affected the transformation, due to accumulation of PDH production (AcCoA) that was transformed by citrate synthase.
Fig. 8.
The P cycle and the TCA cycle in media containing excess oxaloacetate, pyruvate, or acetate in E. coli. (A) Adenylate energy charge, NAD+/NADH, and PMF of E. coli K12 and the indicated mutants. Cells were grown in LB medium and then incubated in M9 medium with and without acetate (10 mM) in the presence or absence of the indicated oxaloacetate (10 mM) and pyruvate (10 mM), as indicated. (B) Scatterplot of the abundance of the indicated metabolites in ∆pck, ∆aceE, and ∆gltA or control cells. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of oxaloacetate (10 mM), pyruvate (10 mM), or both. (C) Percent survival of the indicated bacterial species/strains. Cells were grown in LB medium and then incubated in M9 medium plus acetate (10 mM) in the presence or absence of the indicated compounds: glutamate (2.5 mM) or fructose (2.5 mM), furfural (10 mM) or malonate (20 mM), and kanamycin (40 or 80 μg/mL) or gentamicin (1.6 μg/mL). Strains tgc6, mcc31, and Y1, Y17 are clinically isolated E. coli from infected chicken and humans, respectively. Results (A–C) are mean ± SEM of three biological repeats. Statistically significant values are indicated with an asterisk (*P < 0.05, **P < 0.01) and were determined by Student’s t test.
In addition, NAD+/NADH levels were higher, and the adenylate energy charge and PMF levels were lower in these mutants compared with control cultures of E. coli K12 cultured in medium without exogenous metabolites, indicating the importance of the whole P cycle (Fig. 8A). GC-MS analysis of metabolite abundance in Δpck, ΔaceE, and ΔgltA cells showed lower or similar amounts of metabolites (but not citrate and malate in Δpck). In the presence of OAA or/and pyruvate, succinate, fumarate, and malate were substantially lower or similar in all of the mutants but not citrate and malate in Δpck with OAA and citrate in Δpck with pyruvate (Fig. 8B), indicating that depletion of these genes shuts down the TCA cycle even in the presence of excess carbon sources.
Finally, experiments were performed in ΔpfkB cells to test the effect of a mutation that plays no role in the P cycle or TCA cycle on cell growth and energy production in the presence of exogenous metabolites. ΔpfkB encodes ATP-dependent 6-phosphofructokinase isozyme 2, which catalyzes the phosphorylation of d-fructose 6-phosphate to fructose 1,6-bisphosphate by ATP. Loss of this gene had little or no effect on the intracellular adenylate energy charge, NAD+/NADH ratio, and PMF levels in the presence of exogenous metabolites (Fig. 8A).
The P Cycle Is Present in Other Bacterial Pathogens.
Similar experiments were performed in E. coli strains from humans and chicken, Vibrio anguillarum, Vibrio alginolyticus, V. parahaemolyticus, Photobacterium damsel, Vibrio vulnificus, and Vibrio fluvialis. Glutamate and fructose increased susceptibility to kanamycin in bacteria tested here, but this was inhibited by furfural or malonate (Fig. 8C).
Discussion
During investigation of metabolite suppression in kanamycin-resistant bacteria, it was demonstrated that exogenous glutamate conferred distinct efficacy to the biocidal effects of kanamycin. We further found that OAA, PEP, pyruvate, and citrate also showed potentiation; however, higher induced efficacy was detected in glutamate, OAA, PEP, and pyruvate compared with citrate. To understand the metabolic flux, we performed nontargeted tracer fate detection to determine the fate of glutamate carbon atoms after uptake. The results show that glutamate enters the TCA cycle through the OAA-PEP-pyruvate-AcCoA-CIT pathway, which we call here the P cycle.
This raises questions regarding whether OAA-PEP-pyruvate-AcCoA-CIT was a causal pathway initiated by glutamate or a prevalent metabolic pathway in energy generation, and the details of the relationship between this pathway and the TCA cycle. In the current study, we performed a series of knockout and inhibition experiments and found that the bioconversion OAA-PEP-pyruvate-AcCoA-CIT was an essential pathway for glutamate-mediated bacterial inactivation by aminoglycoside antibiotics, and it also represents an integrated, glutamate-independent, energy metabolism pathway. Our results showed that loss of the pathway interrupts action of the TCA cycle, indicating that the P cycle controls the TCA cycle. We also characterized two alternative opinions of OAA-pyruvate-AcCoA and MAL-pyruvate-AcCoA in the P cycle. The P cycle is similar to pyruvate recycling common in mammalian cells based on homologous cycle steps (30). However, the pyruvate recycling is distinctly different from the P cycle, in that pyruvate recycling is a causal pathway resulting from excess metabolites (30), whereas our experiments with or without excess metabolites identified the P cycle as a prevalent cycle. Thus, while pyruvate recycling is a bypass of the TCA cycle, OAA favored PEPCK over citrate synthase, indicating that the TCA cycle is a bypass of the P cycle. Additionally, the effect of pyruvate recycling on the TCA cycle is not known, but the P cycle appears to regulate the TCA cycle. Furthermore, pyruvate recycling occurs in specific cells under specific conditions (30–33), whereas the P cycle can occur in different culture conditions including LB, M9, with acetate plus glucose or SOC medium, over different incubation times. Furthermore, the P cycle is different from the PEP-glyoxylate cycle in E. coli, which integrates OAA-PEP-pyruvate-AcCoA-CIT with the glyoxylate cycle rather than the TCA cycle, in which two PEP molecules are oxidized sequentially from AcCoA, citrate, glyoxylate, and OAA to CO2, and one PEP is regenerated (25). The present study found that the integration of OAA-PEP-pyruvate-AcCoA-CIT, OAA-pyruvate-AcCoA, and/or MAL-pyruvate-AcCoA pathways with the TCA cycle operates routinely as a general mechanism for energy production.
The elucidation of the P cycle leads to an understanding of the TCA cycle and the integrated basic energy mechanism. Our demonstration that the TCA cycle does not work when a step in the P cycle is perturbed, such as loss of aceE and inhibition by furfural, indicates that the TCA cycle is only a component of the P cycle, regardless of whether the origin is glycolysis (such as PEP) or alanine, aspartate, and glutamate metabolism (such as α-ketoglutaric acid). Our finding that the synergistic action of citrate synthase substrates, such as OAA, AcCoA, or its upstream metabolites, did not drive the TCA cycle when PDH activity was inhibited or a key gene of the enzyme complex (aceE) or pck was deleted, suggests that the P cycle, rather than the TCA cycle, generates respiratory energy in E. tarda and E. coli.
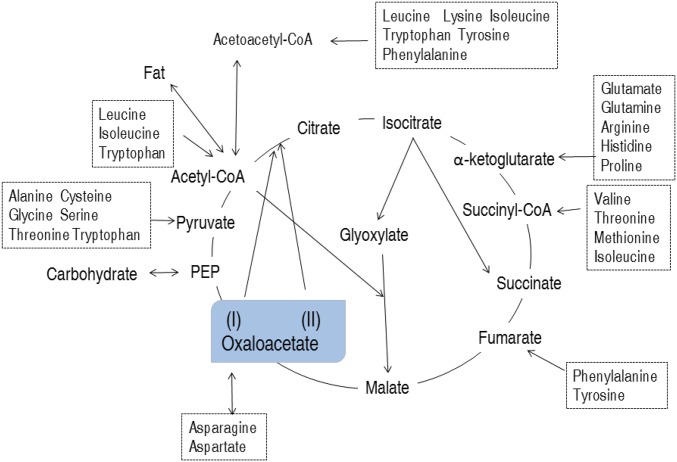
Michaelis–Menten kinetics demonstrated that PEPCK has higher affinity than citrate synthase for OAA, which supports the conclusion that PEPCK prioritizes metabolism of OAA through the P cycle. It is noteworthy that nontargeted tracer fate analysis showed that OAA plays many roles in and beyond the TCA cycle. The OAA function beyond the TCA cycle could represent a main source of the P cycle due to higher detection of M1 and M2 labels and lower detection of M4 labels, which is metabolized in the P cycle. OAA is a metabolic intermediate involved in processes such as gluconeogenesis, the glyoxylate cycle, amino acid synthesis, and fatty acid synthesis. Therefore, OAA arises from rich energy sources, such as carbohydrates through glycolysis, AcCoA through the glyoxylate cycle, and amino acids through protein metabolism (Fig. 9) (28, 34–36). Comparatively, carbohydrates have similar metabolic pathways to form OAA or AcCoA (28), while amino acids and fatty acids may be transformed and degraded to OAA and AcCoA, respectively (37, 38). AcCoA is then synthesized to OAA in the glyoxylate cycle (39). Clearly, the P cycle permits carbohydrates, amino acids, and fatty acids to directly enter energy metabolism pathways, whereas the TCA cycle requires indirect entrance for carbohydrates and fatty acids by AcCoA (Fig. 9). Thus, the P cycle is more suitable than the TCA cycle for the utilization of all three potential energy inputs. These data indicate the importance of the P cycle in regulating energy metabolism, and further investigations are required to understand metabolic regulation among the carbohydrates, amino acids, and fatty acids in context of the P cycle.
Fig. 9.
Model showing proposed metabolic pathways for oxaloacetate. See text for Discussion. Oxaloacetate (I) and (II) represent sources within and beyond the TCA cycle, respectively.
Reports have indicated that cells have a transport system with high affinity for acetate and with lower affinity for pyruvate, and both pyruvate and acetate are not limited in the TCA cycle and the PEP-pyruvate-AcCoA pathway (27). Among the five metabolites, OAA, pyruvate, and acetate showed that the highest inactivation efficiency reached a plateau at 10 mM. When the concentration was elevated by 400%, inactivation was unaffected, and similar results were obtained for PEP and citrate. Thus, the differences in inactivation efficacy among the five metabolites were not significantly related to intermembrane or off-target effects. In summary, these results indicate that the P cycle is a prevalent cycle that acts in a glutamate-independent manner in E. tarda and E. coli.
The present study indicates that exogenous glutamate stimulates metabolic flux through the P cycle. The idea that carbon metabolism in the TCA cycle is central to antibiotic resistance is not new (10–12, 40–48), but the reported impact that exogenous nitrogen sources, such as glutamate, as well as OAA, PEP, and pyruvate can exert on energy homeostasis, PMF, and drug uptake is, as far as we know, unprecedented, and indicates the role that the P cycle plays in this process. These findings also support previous work (11), confirming that these pathways can be exploited to combat antibiotic resistance in naturally occurring and laboratory-derived bacterial pathogens. Our results demonstrate that P cycle gene knockouts display decreased resistance to aminoglycosides, regardless of whether these genes are components of the TCA cycle. This is not consistent with the finding that aminoglycoside uptake is related to PMF, which is attributed to increased NADH generated by promotion of the TCA cycle (10, 11). A possible mechanism could be that the cell reallocates metabolites during glycolysis and/or pyruvate metabolism due to abrogation of the P cycle or the TCA cycle to replenish the stores of other amino acids and/or intermediates. This was demonstrated by the finding that loss of aceF leads to elevated oleate concentrations, which promotes gentamicin uptake in a PMF-independent manner. These results support the conclusion that the microbial metabolic environment has a strong influence on antibiotic efficacy (5). As quickly as new antibiotics are being discovered (49, 50), bacterial strains are emerging with acquired drug resistance to the new reagents. Our strategy provides an alternative approach by restoring sensitivity to previously effective antibiotics through metabolic modulation. In addition, both acetate and glucose were used together in the glucose-enabled bacterial inactivation. Recently, acetate has been shown to block the flow of glucose carbons into AcCoA and the TCA cycle (51), suggesting that further optimization of reaction conditions may promote the potential of metabolites such as glucose.
In summary, the most critical results of this study are: (i) the P cycle plays a critical role in energy homeostasis in E. coli and E. tarda; (ii) the TCA cycle can be a bypass pathway that provides OAA for the P cycle; (iii) the P cycle regulates the TCA cycle; and (iv) exogenous glutamate, OAA, PEP, and pyruvate revert the phenotype of antibiotic resistance and modulate flux through the P cycle. The results provide insights into metabolite-enabled cell death via administration of antibiotics as well as bacterial energy metabolism and homeostasis.
Materials and Methods
In all experiments, bacterial cells were cultured in 50 mL LB broth for 24 h at 30 °C (E. tarda) and 16 h (E. coli) at 37 °C, 200 rpm and 80% humidity in 250-mL flasks. E. coli K12 BW25113 and its knockout strains from the Keio collection, and other clinical isolated strains were obtained from the collection of our laboratory. Unless otherwise noted, overnight bacterial cultures were harvested by centrifugation and washed for GC-MS and bacterial survival experiments in M9 medium plus acetate or other metabolites indicated. GC-MS analysis was carried out with a variation on the two-stage technique and the resulting data were processed using pattern recognition methods as described previously (52, 53). For bacterial survival detection, bacterial cells were resuspended in M9 medium, adjusted OD600 to 0.6, and supplied glutamate and/or antibiotic as desired. Measurement of percentage survival was performed by serially diluting and spot plating onto LB agar plates to determine colony-forming units per milliliter and survival as described previously with a few modifications (10, 11). NADH, PMF, ATP, enzyme activity and antibiotic concentration were measured with commercially available assay kits according to the manufacturer’s manual. The isotope tracer experiment was carried out as previously described (23, 54). qPCR, SDS/PAGE, Western blot, and gene complementation assays were carried out according to routine procedures (53–55). For full details of all these processes, please see SI Appendix.
Supplementary Material
Acknowledgments
This work was sponsored by National Natural Science Foundation of China Grants 31572654, U1701235, 31672656, and 31700119.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714645115/-/DCSupplemental.
References
- 1.Band VI, et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol. 2016;1:16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakradhar S. Reservoirs of resistance: To understand why antibiotics fail, geneticists chase the ‘resistome’. Nat Med. 2016;22:1069–1071. doi: 10.1038/nm1016-1069. [DOI] [PubMed] [Google Scholar]
- 3.King AM, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2011;8:6–9. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen NR, et al. A role for the bacterial GATC methylome in antibiotic stress survival. Nat Genet. 2016;48:581–586. doi: 10.1038/ng.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitko NP, Spahich NA, Richardson AR. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. MBio. 2015;6:e00045-e15. doi: 10.1128/mBio.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: A universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 10.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng B, et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015;21:249–261. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Su YB, Peng B, Han Y, Li H, Peng XX. Fructose restores susceptibility of multidrug-resistant Edwardsiella tarda to kanamycin. J Proteome Res. 2015;14:1612–1620. doi: 10.1021/pr501285f. [DOI] [PubMed] [Google Scholar]
- 13.Vilchèze C, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114:4495–4500. doi: 10.1073/pnas.1704376114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhargava P, Collins JJ. Boosting bacterial metabolism to combat antibiotic resistance. Cell Metab. 2015;21:154–155. doi: 10.1016/j.cmet.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Peng B, Li H, Peng XX. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell. 2015;6:628–637. doi: 10.1007/s13238-015-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen XH, et al. Exogenous L-valine promotes phagocytosis to kill multidrug-resistant bacterial pathogens. Front Immunol. 2017;8:207. doi: 10.3389/fimmu.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchanan BB, Arnon DI. A reverse KREBS cycle in photosynthesis: Consensus at last. Photosynth Res. 1990;24:47–53. [PubMed] [Google Scholar]
- 18.Korla K, Mitra CK. Modelling the Krebs cycle and oxidative phosphorylation. J Biomol Struct Dyn. 2014;32:242–256. doi: 10.1080/07391102.2012.762723. [DOI] [PubMed] [Google Scholar]
- 19.Meléndez-Hevia E, Waddell TG, Cascante M. The puzzle of the Krebs citric acid cycle: Assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. J Mol Evol. 1996;43:293–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]
- 20.Egri-Nagy A, Nehaniv CL, Rhodes JL, Schilstra MJ. Automatic analysis of computation in biochemical reactions. Biosystems. 2008;94:126–134. doi: 10.1016/j.biosystems.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Kaleta C, de Figueiredo LF, Schuster S. Can the whole be less than the sum of its parts? Pathway analysis in genome-scale metabolic networks using elementary flux patterns. Genome Res. 2009;19:1872–1883. doi: 10.1101/gr.090639.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster S, de Figueiredo LF, Kaleta C. Predicting novel pathways in genome-scale metabolic networks. Biochem Soc Trans. 2010;38:1202–1205. doi: 10.1042/BST0381202. [DOI] [PubMed] [Google Scholar]
- 23.Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G. Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem. 2010;82:6621–6628. doi: 10.1021/ac1011574. [DOI] [PubMed] [Google Scholar]
- 24.Hiller K, Metallo C, Stephanopoulos G. Elucidation of cellular metabolism via metabolomics and stable-isotope assisted metabolomics. Curr Pharm Biotechnol. 2011;12:1075–1086. doi: 10.2174/138920111795909096. [DOI] [PubMed] [Google Scholar]
- 25.Fischer E, Sauer U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J Biol Chem. 2003;278:46446–46451. doi: 10.1074/jbc.M307968200. [DOI] [PubMed] [Google Scholar]
- 26.Haverkorn van Rijsewijk BR, Nanchen A, Nallet S, Kleijn RJ, Sauer U. Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol Syst Biol. 2011;7:477. doi: 10.1038/msb.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolkver E, et al. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J Bacteriol. 2009;191:940–948. doi: 10.1128/JB.01155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou SY, Chao YP, Liao JC. A mutant phosphoenolpyruvate carboxykinase in Escherichia coli conferring oxaloacetate decarboxylase activity. J Bacteriol. 1995;177:1620–1623. doi: 10.1128/jb.177.6.1620-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Goldie H. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: Studies with pck-lacZ operon fusions. J Bacteriol. 1984;159:832–836. doi: 10.1128/jb.159.3.832-836.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JG, et al. Measurement of gluconeogenesis and pyruvate recycling in the rat liver: A simple analysis of glucose and glutamate isotopomers during metabolism of [1,2,3-(13)C3]propionate. FEBS Lett. 1997;412:131–137. doi: 10.1016/s0014-5793(97)00764-3. [DOI] [PubMed] [Google Scholar]
- 32.McKenna MC. Glutamate pays its own way in astrocytes. Front Endocrinol (Lausanne) 2013;4:191. doi: 10.3389/fendo.2013.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen GM, Sonnewald U. Glutamate: Where does it come from and where does it go? Neurochem Int. 2015;88:47–52. doi: 10.1016/j.neuint.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Waagepetersen HS, Qu H, Hertz L, Sonnewald U, Schousboe A. Demonstration of pyruvate recycling in primary cultures of neocortical astrocytes but not in neurons. Neurochem Res. 2002;27:1431–1437. doi: 10.1023/a:1021636102735. [DOI] [PubMed] [Google Scholar]
- 35.Bauzá A, Quiñonero D, Deyà PM, Frontera A. Long-range effects in anion-π interactions: Their crucial role in the inhibition mechanism of Mycobacterium tuberculosis malate synthase. Chemistry. 2014;20:6985–6990. doi: 10.1002/chem.201304995. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Bommareddy RR, Frank D, Rappert S, Zeng AP. Deregulation of feedback inhibition of phosphoenolpyruvate carboxylase for improved lysine production in Corynebacterium glutamicum. Appl Environ Microbiol. 2014;80:1388–1393. doi: 10.1128/AEM.03535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 38.Sawers G. The anaerobic degradation of L-serine and L-threonine in enterobacteria: Networks of pathways and regulatory signals. Arch Microbiol. 1998;171:1–5. doi: 10.1007/s002030050670. [DOI] [PubMed] [Google Scholar]
- 39.Riedel SL, Lu J, Stahl U, Brigham CJ. Lipid and fatty acid metabolism in Ralstonia eutropha: Relevance for the biotechnological production of value-added products. Appl Microbiol Biotechnol. 2014;98:1469–1483. doi: 10.1007/s00253-013-5430-8. [DOI] [PubMed] [Google Scholar]
- 40.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 41.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: From targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin X, Kang L, Li H, Peng X. Fluctuation of multiple metabolic pathways is required for Escherichia coli in response to chlortetracycline stress. Mol Biosyst. 2014;10:901–908. doi: 10.1039/c3mb70522f. [DOI] [PubMed] [Google Scholar]
- 44.Purohit HJ, Cheema S, Lal S, Raut CP, Kalia VC. In search of drug targets for Mycobacterium tuberculosis. Infect Disord Drug Targets. 2007;7:245–250. doi: 10.2174/187152607782110068. [DOI] [PubMed] [Google Scholar]
- 45.Belenky P, et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobritz MA, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meylan S, et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasr T, Bondock S, Eid S. Design, synthesis, antimicrobial evaluation and molecular docking studies of some new thiophene, pyrazole and pyridone derivatives bearing sulfisoxazole moiety. Eur J Med Chem. 2014;84:491–504. doi: 10.1016/j.ejmech.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 50.Spellberg B. The future of antibiotics. Crit Care. 2014;18:228. doi: 10.1186/cc13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enjalbert B, Millard P, Dinclaux M, Portais JC, Létisse F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci Rep. 2017;7:42135. doi: 10.1038/srep42135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du CC, et al. Metabolic mechanism for L-Leucine-induced metabolome to eliminate Streptococcus iniae. J Proteome Res. 2017;16:1880–1889. doi: 10.1021/acs.jproteome.6b00944. [DOI] [PubMed] [Google Scholar]
- 53.Zeng ZH, et al. Glucose enhances tilapia against Edwardsiella tarda infection through metabolome reprogramming. Fish Shellfish Immunol. 2017;61:34–43. doi: 10.1016/j.fsi.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 54.Walther JL, Metallo CM, Zhang J, Stephanopoulos G. Optimization of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Metab Eng. 2012;14:162–171. doi: 10.1016/j.ymben.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YL, et al. C-terminal domain of hemocyanin, a major antimicrobial protein from litopenaeus vannamei: Structural homology with immunoglobulins and molecular diversity. Front Immunol. 2017;8:611. doi: 10.3389/fimmu.2017.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.