Significance
Intracellular pathogens hijack many host processes, including the vesicle trafficking pathway, to help their survival and replication. Legionella pneumophila is the causative agent of Legionnaires’ disease, which utilizes numerous effector proteins, including RidL during infection. RidL suppresses retromer transport activity, but the exact mechanism for this has remained unclear. Using a combination of structural, biochemical, and cellular approaches, we show how retromer specifically targets to retromer subunit VPS29. The RidL–VPS29 interaction is necessary for the proper localization and function of RidL. Mechanistically, RidL inhibits retromer activity by direct competition with TBC1d5, an essential retromer regulator. Our study highlights the beauty of studying host–pathogen interaction in the discovery of novel aspects of host cell biology.
Keywords: vesicular trafficking, host–pathogen interaction, endosomal sorting, retromer
Abstract
Retrograde vesicle trafficking pathways are responsible for returning membrane-associated components from endosomes to the Golgi apparatus and the endoplasmic reticulum (ER), and they are critical for maintaining organelle identity, lipid homeostasis, and many other cellular functions. The retrograde transport pathway has emerged as an important target for intravacuolar bacterial pathogens. The opportunistic pathogen Legionella pneumophila exploits both the secretory and recycling branches of the vesicle transport pathway for intracellular bacterial proliferation. Its Dot/Icm effector RidL inhibits the activity of the retromer by directly engaging retromer components. However, the mechanism underlying such inhibition remains unknown. Here we present the crystal structure of RidL in complex with VPS29, a subunit of the retromer. Our results demonstrate that RidL binds to a highly conserved hydrophobic pocket of VPS29. This interaction is critical for endosomal recruitment of RidL and for its inhibitory effects. RidL inhibits retromer activity by direct competition, in which it occupies the VPS29-binding site of the essential retromer regulator TBC1d5. The mechanism of retromer inhibition by RidL reveals a hotspot on VPS29 critical for recognition by its regulators that is also exploited by pathogens, and provides a structural basis for the development of small molecule inhibitors against the retromer.
Targeted protein transport is critical for almost every aspect of cell functions (1). Endosomes are major sorting compartments in cells and lie in the center of a variety of trafficking pathways (1). Molecules or ligands taken up during endocytosis can be sent to lysosomes for degradation via endosomes, or they can be transported from the endosome to the trans-Golgi network (TGN) or the plasma membrane. Thence, endosomes sorting is essential for many important physiological processes, including maintaining metabolite homeostasis, establishing hormone-mediated signal transduction, and preserving immune surveillance.
One of the key protein machineries controlling endosomal sorting is the evolutionarily conserved retromer complex (1, 2). Retromer mediates the trafficking from endosomes to the TGN or the plasma membrane. Established retromer cargoes include sorting receptors, α5β1 integrin, glucose and metal transporters, amyloid precursor protein receptor, and the list is still growing (1). Retromer has been shown to be critical for many physiological and developmental processes (1, 3, 4). The core of the retromer complex is the trimeric assembly consisting of VPS35, VPS26, and VPS29 (5). Key regulators of retromer include various sorting nexin (SNX) proteins (6), TBC1d5 (7, 8), VARP (9, 10), and the WASH actin regulatory complex (11–13), which impose different regulatory effects on retromer-mediated transport. Importantly, dysregulation of retromer activity has been linked with multiple neurological disorders, in particular Alzheimer’s disease and Parkinson’s disease (3, 14).
Because of its essential role in vesicle trafficking, retromer is coopted by intracellular pathogens to promote survival and replication (15). Several toxins, such as bacterial Shiga toxins and the ricin toxin of plant origin, hijack the retromer pathway for transport within the cell (16, 17). Hence, small-molecule inhibitors that selectively block endosomal trafficking have been used to protect against toxin exposure; however, development of such molecules is relatively limited, in part due to the complexity of retromer transport (18). More recently, the Chlamydial effector protein IncE has been shown to recognize a highly conserved region on SNX5/SNX6, resulting in the inhibition of retromer activity (19–23). Legionella pneumophila, the causative agent of Legionnaires’ disease, exploits host functions, particularly vesicle transport, by numerous effectors delivered into infected cells by its Dot/Icm type IV secretion system (24). Among these, the effector RidL appears to block retrograde transport, which has been shown to restrict intracellular growth of L. pneumophila (25). RidL binds the retromer as well as phosphoinositol 3-phosphate [PtdIns(3)P], a signaling lipid associated with the endosome (25). However, the molecular mechanism by which RidL blocks retromer transport remains elusive.
To gain mechanistic insights into the regulation of retromer by RidL, we performed a combination of biochemical, structural, and cellular studies. We show that RidL, through its N-terminal region, interacts with both the VPS29 subunit of the retromer complex and PtdIns(3)P. The crystal structure of a VPS29–RidL complex reveals that RidL contacts a conserved hydrophobic pocket of VPS29, which is also the binding site of TBC1d5 and VARP, two key regulators of retromer activity. In addition, overexpression of RidL displaces TBC1d5 from endosomes and thus inhibits retromer- and TBC1d5-dependent cargo transport. In combination, our study suggests that RidL inhibits retromer transport through directly competing with TBC1d5 and VARP, illustrating how host and pathogen contend in controlling cellular trafficking and providing insights into the regulation of endosomal sorting.
Results
Interaction Between RidL and Retromer or PtdIns(3)P.
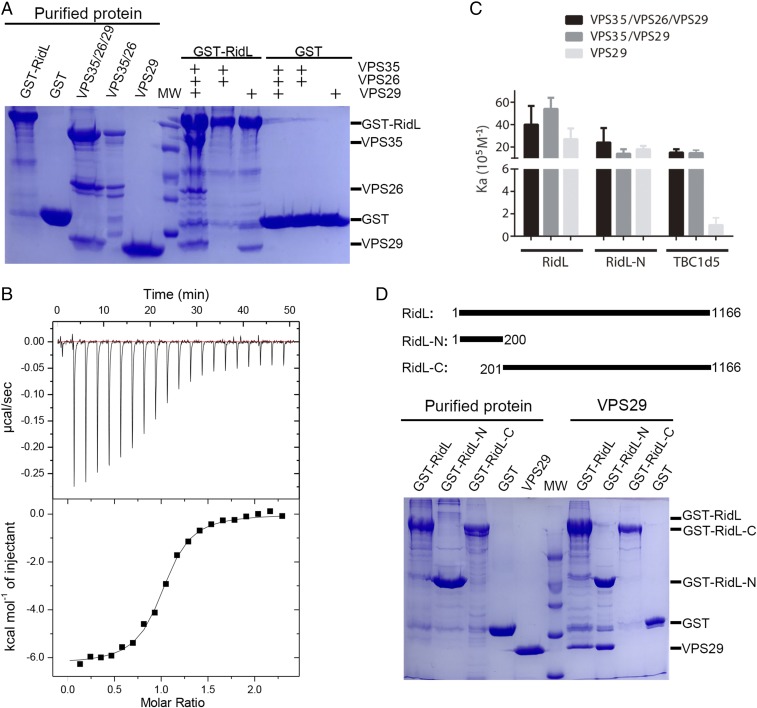
A previous study suggests that RidL interacts with the retromer subunit VPS29 (25). To test whether this is the only retromer component engaged by RidL, we used GST pull-down and isothermal titration calorimetry (ITC) assays to examine the binding of RidL to other retromer subunits. GST–RidL specifically retained VPS35/VPS26/VPS29 or VPS29, but not the VPS35/VPS26 subcomplex (Fig. 1A). RidL bound to VPS35/VPS26/VPS29, VPS35/VPS29, or VPS29 with similar affinities, with a dissociation constant (Kd) value of 200∼500 nM (Fig. 1 B and C). These results suggest that RidL solely contacts VPS29, in contrast to TBC1d5, which requires both VPS35 and VPS29 to achieve maximal binding affinity (Fig. 1C) (8).
Fig. 1.
The RidL–retromer interaction is mediated by the N terminus of RidL and VPS29 subunit of retromer. (A) GST–RidL or GST pull-down of purified retromer VPS35/VPS26/VPS29, VPS35/VPS26, and VPS29. Shown are Coomassie blue stained SDS/PAGE gels of purified proteins used (Left) and bound samples (Right). (B) Isothermal titration calorimetry (ITC) of RidL titrated into VPS35/VPS26/VPS29 in a buffer containing 20 mM Tris⋅HCl, pH 8.0, 200 mM NaCl, 5 mM βME at 20 °C. Top and Bottom show raw and integrated heat from injections, respectively. The black curve at Bottom represents a fit of the integrated data to a single-site binding model. (C) Affinity between retromer proteins and full-length RidL, RidL-N, or TBC1d5, determined by ITC. Association constant (Ka) are shown together with errors from data fitting. (D) GST–RidL, RidL-N, RidL-C, or GST pull-down of purified VPS29. Shown are Coomassie blue stained SDS/PAGE gels of purified proteins used (Left) and bound samples (Right).
To determine the region of RidL essential for the binding, we next truncated RidL into various lengths. The N-terminal portion of RidL (RidL-N) harboring the first 200 residues retained the ability to bind VPS29, whereas the carboxyl portion (RidL-C, 201-C terminus) did not (Fig. 1D). Furthermore, full-length RidL and RidL-N bound to VPS29 or VPS29-containing complex with similar affinity (Fig. 1C).
When measured for the ability to bind PtdIns(3)P by the liposome flotation assay, we found that full-length RidL and RidL-N, but not RidL-C, are specifically associated with this lipid (SI Appendix, Fig. S1) (26, 27). Consistent with the fact that RidL-N makes contact with both retromer and PtdIns(3)P, RidL-N displayed a punctate cellular localization similar to that of the early endosomal marker EEA1; RidL-C, on the other hand, appeared cytosolic (SI Appendix, Fig. S2). The majority of full-length RidL distributed diffusely in cells, similar to that of RidL-C; however, a small portion of proteins displayed a punctate localization (SI Appendix, Fig. S2).
Complex Structure Between VPS29 and RidL.
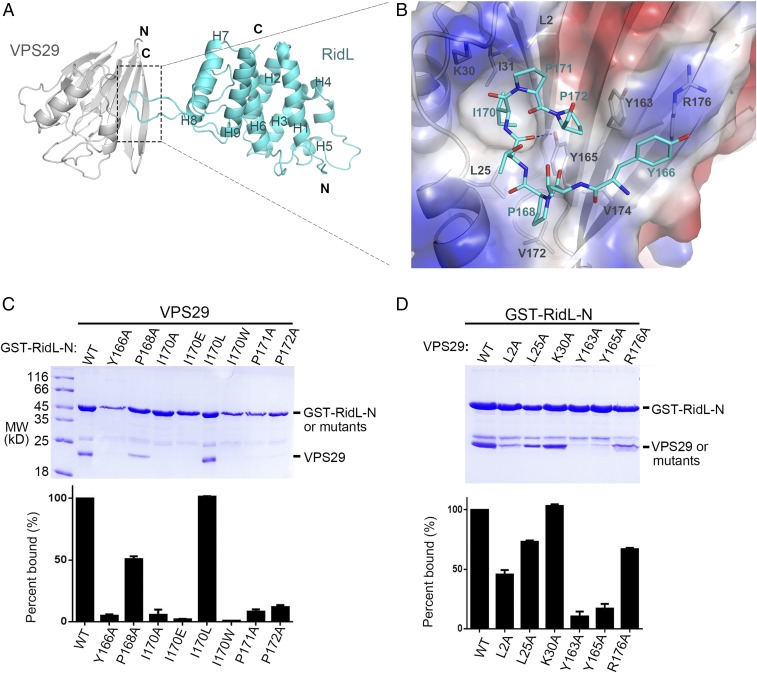
To understand the binding mechanism between RidL and VPS29, we determined the crystal structure of VPS29:RidL-N complex at 2.5 Å (Fig. 2; see SI Appendix, Fig. S3 and Table S1; PDB ID code 5WYH). VPS29 is highly similar to previous published VPS29 structures (0.50 Å rmsd for all atoms aligned in PDB 1W24) (28, 29). DALI search identified no structures with considerable similarity to that of RidL, indicating that RidL-N possesses a novel fold (30). The protein contains nine helices that formed two intimately packed helix bundles (H1–6 and H6–9), with two loops inserted between H4 and H5 as well as H8 and H9, respectively (SI Appendix, Fig. S4).
Fig. 2.
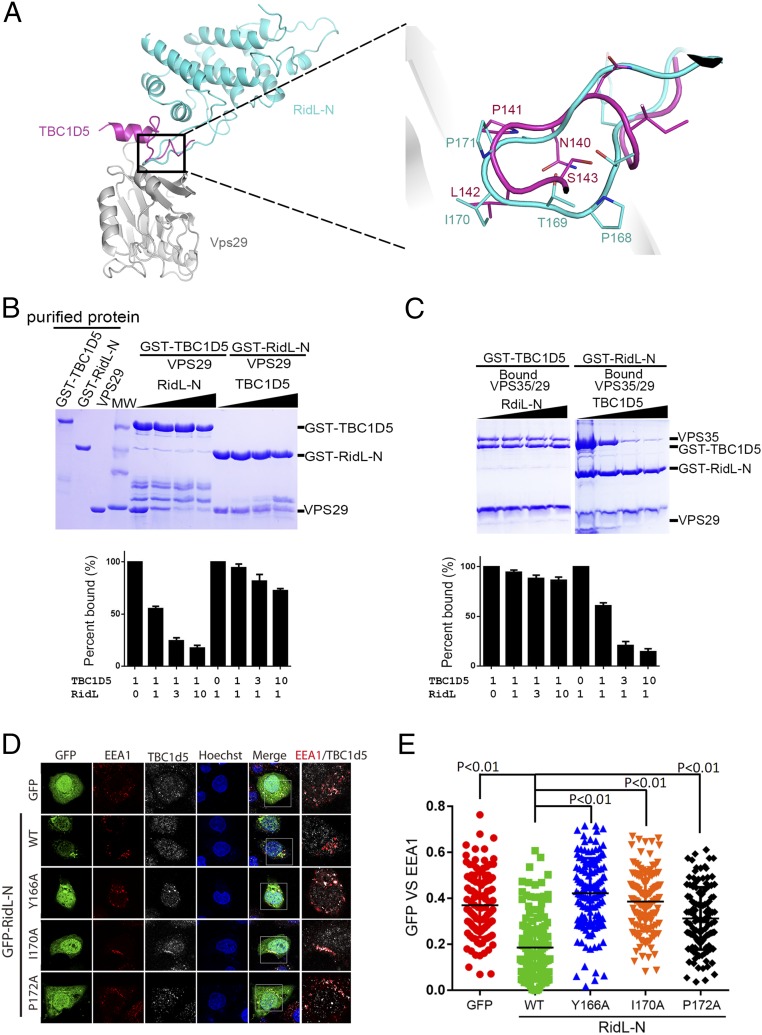
Structural mechanism of VPS29 recognition by RidL-N. (A) Ribbon diagram of the VPS29/RidL complex (VPS29, gray; RidL, cyan). N and C terminus of proteins are labeled. The secondary structure elements of RidL are labeled. (B) VPS29–RidL interactions in detail. Critical VPS29 and RidL residues are shown in stick modes. A partially transparent electrostatic surface potential map is presented for VPS29. Blue dash represents hydrogen bond between RidL and VPS29. (C and D) Coomassie blue stained SDS/PAGE gels of bound proteins are shown. Results are representative of three independent experiments. Amount of VPS29 retained was expressed relative to the amount of GST–RidL in the bound sample and then normalized to the amount of wild-type protein. All values are presented as mean ± SD, derived from three independent experiments. In C, immobilized GST–RidL-N or its mutants (Y166A, P168A, I170A, I170E, I70L, I170W, P171A, and P172A) was used to pull down VPS29. In D, immobilized GST–RidL-N was used to pull down VPS29 WT or mutants (L2A, L25A, K30A, Y163A, Y165A, and R176A).
The extended loop between H8 and H9 in RidL-N forms a hairpin structure, which binds to a conserved hydrophobic patch on VPS29 (Fig. 2 A and B). Although the buried surface area is only 427 Å2, its interacting site is rich in components capable of hydrophobic interactions (Fig. 2B, dash lines). The hydrophobic interactions are formed by side chain from Y166, P168, I170, P171, and P172 from RidL and L2, L25, K30, I31, Y163, Y165, V172, V174, and R176 from VPS29. There is only one intramolecular hydrogen bond observed, which is formed by the phenol oxygen of Y165VPS29 and the amide oxygen of T169RidL.
In the center of the VPS29–RidL contact surface, the side chain of I170RidL is deeply inserted into a hydrophobic pocket formed by L2, L25, I31, and the hydrophobic portion of K30 side chain from VPS29. The observation that conversion of I170 to a small (I170A) or large (I170W) hydrophobic residue, or a charged residue (I170E), significantly disrupted the interaction with VPS29, further indicating the importance of I170RidL (Fig. 2C). In contrast, replacement of I170RidL with an isometric amino acid (I170L) did not affect the binding. In addition to I170RidL, several surrounding hydrophobic residues are critical for binding as well. Y166ARidL, I170ARidL, P171ARidL, and P172ARidL mutations completely abolished the binding between RidL and VPS29. P168ARidL also reduced the interaction, but to a lesser extent. In comparison, mutation on VPS29 residues generally had lower impact on the binding (Fig. 2D). Whereas Y163AVPS29 and Y165AVPS29 almost completely abolished the interactions, several other mutations (L2AVPS29, L25AVPS29, and R176AVPS29) only partially reduced the binding affinity.
The Conformation of the RidL Loop Is Essential for Binding.
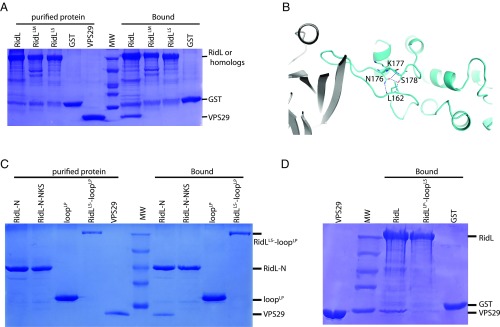
Sequence alignment of RidL from closely related Legionella species revealed that residues in the interacting hairpin loop are highly variable, including the key residue I170RidL (SI Appendix, Fig. S4). Consistent with these variations, RidL from L. pneumophila (RidL), but not related Legionella moravica (RidLLM) or Legionella shakspearei (RidLLS) bound to VPS29 (Fig. 3A).
Fig. 3.
The sequence and conformation of the RidL loop are essential for the binding to VPS29. (A) GST–RidL from related Legionella species pull-down of purified VPS29. Shown are Coomassie blue stained SDS/PAGE gels of purified proteins used (Left) and bound samples (Right). (B) Detailed VPS29–RidL interactions highlight RidL residues that are distant from the binding sites of VPS29, but critical for the conformation of the binding loop. Residues L162, N176, K177, and S178 are shown in stick representation. Hydrogen bonds are denoted with blue dash lines. (C) GST–RidL-N, RidL-N-NKS, loopLP, RidLLS-loopLP pull-down of purified VPS29. Shown are Coomassie blue stained SDS/PAGE gels of purified proteins used (Left) and bound samples (Right). (D) GST–RidL, RidLLP-loopLS, and GST pull-down of purified VPS29. Shown are Coomassie blue stained SDS/PAGE gels of purified VPS29 protein (Left) and bound samples (Right).
TBC1d5 contacts VPS29 through a short motif that is unique to TBC1d5 and its orthologs among the TBC family (8). Since the interaction between VPS29 and RidL also involves a short motif from RidL, we were interested in determining whether this motif/loop is sufficient for the interaction. Side chains of N176RidL and S178RidL form an extensive hydrogen bond network together with the main chain of L162RidL, which seems to stabilize the conformation of the VPS29-binding loop (Fig. 3B). Whereas RidL-N retained VPS29 in a pull-down assay, the RidL hairpin from L. pneumophila (loopLP) on its own or a chimera protein, RidLLS-loopLP (the hairpin loop of L. shakspearei RidL is replaced by an equivalent sequence from L. pneumophila) did not bind to VPS29 (Fig. 3C). Furthermore, although none of three residues (N176/K177/S178) directly contact VPS29, a mutant (N176L/K177D/S178A, RidL-N-NKS) failed to retain VPS29 as wild-type (WT) protein (Fig. 3C). Finally, we substituted the RidL loop with the corresponding loop from RidLLS and found that the chimera protein, RidLLP-loopLS, failed to retain VPS29 (Fig. 3D). Hence, both the key residues from the hairpin loop and its correct conformation are required for the binding to VPS29.
RidL Localizes to Endosomes and Inhibits Shiga Toxin Transport Through Its Interaction with Retromer.
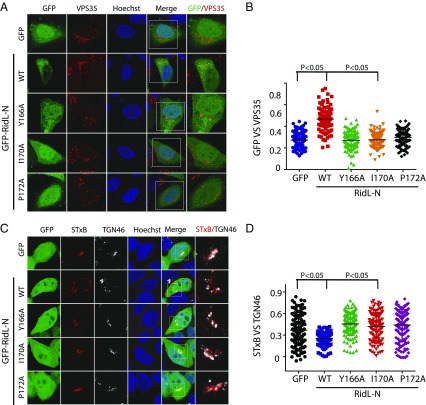
Having demonstrated that RidL directly contacts VPS29 and PtdIns(3)P, we next asked whether these interactions are important for the endosomal localization and inhibitory function of RidL. RidL-N WT extensively colocalized with VPS35 or EEA1 (Fig. 4A and SI Appendix, Fig. S2). The VPS29-interacting deficient mutants (Y166A, I170A, and P172A) became predominantly cytosolic, suggesting that interactions with retromer are essential for the membrane association of RidL (Fig. 4 A and B). The structure of the RidL–VPS29 complex reveals two positive patches on RidL that are distal from the VPS29-binding site (SI Appendix, Fig. S5A). To test whether these regions are involved in PtdIns(3)P binding, we mutated several positively charged residues in the two regions and examined the PtdIns(3)P-binding activity of these mutants in a liposome flotation assay (SI Appendix, Fig. S5B). Whereas RidL-N K56D interacted with PtdIns(3)P similar to wild type protein, mutations in the second patch (K27A/R33A/K122A, KRR) abolished the lipid-binding activity. In contrast, the mutants deficient in interaction with VPS29 (Y166A, I170A, and P172A) bound to the PtdIns(3)P-containing liposome similar to RidL WT (SI Appendix, Fig. S5C). To examine the potential role of lipid binding in cellular localization of RidL, we constructed relevant mutants and determined their cellular localization (SI Appendix, Fig. S5 D and E). The RidL-N-KRR mutant showed lower levels of colocalization with EEA1 than the wild type protein, but its rates of colocalization were higher than that of RidL-N-Y166A, the mutant deficent for VPS29 binding. Therefore, interactions with retromer and lipid binding contribute to the endosomal localization of RidL, with the retromer interaction playing a more significant role.
Fig. 4.
The RidL localizes to endosomes and inhibits Shiga toxin transport through its interaction with retromer. (A) HeLa cells were transfected with GFP, or GFP–RidL-N WT, Y166A, I170A, P172A (green), and then fixed and labeled with anti-VPS35 (red) antibody. (B) Quantitation of GFP colocalization with VPS35 in cells in A. Each dot represents Pearson’s correlation coefficients from one cell. P values shown are the result of one-way ANOVA, post hoc Tukey’s test. (C) RidL inhibits retrograde transport of STxB through its interaction with the retromer. Cells were first transformed for 24 h with GFP, GFP–RidL-N WT, or mutants (green), and then fed with purified STxB protein. The trafficking of STxB was analyzed by staining STxB with antibody (red) and determining the colocalization with the trans-Golgi marker, TGN46 (white). (D) Quantitation of STxB colocalization with TGN46 in cells in C. Each dot represents Pearson’s correlation coefficients from one cell. P values shown are the result of one-way ANOVA, post hoc Tukey’s test.
To examine the impact of RidL on retromer-mediated trafficking, we chose to study the trafficking of subunit B of Shiga toxin (STxB), which enters the cells and then transports from endosomes to the Golgi network in a retromer-dependent manner (16, 17). Cells were first transfected with GFP, GFP–RidL-N WT, or mutants, and then treated with purified STxB protein. In control cells expressing GFP, STxB was efficiently transported to the Golgi apparatus as indicated by its extensive colocalization with TGN46, a trans-Golgi marker. In contrast, in cells producing GFP–RidL-N WT, the transportation was delayed and colocalization with TGN46 was significantly lower than that in control cells expressing GFP (Fig. 4 C and D). GFP–RidL-N mutants appeared unable to interfere with the transport of STxB, since STxB colocalized with TGN46 in cells expressing GFP–RidL-N mutants to a similar extent as in control cells (Fig. 4 C and D). Thus, RidL inhibits STxB transport through its interaction with the retromer.
VARP, TBC1d5, and RidL Recognize VPS29 Through Similar Modes.
Our study herein and previous studies by us and by others indicate that RidL, TBC1d5, and VARP recognize the same surface on VPS29, opposite to the binding surface of VPS35 (8, 9, 31). Structural comparison between VPS29–RidL and VPS29–TBC1d5–Ins1 revealed the sites for binding RidL and TBC1d5 are almost identical (Fig. 5A) (8). RidL and TBC1d5 insert an isoleucine or leucine residue in the hyhrophobic pocket of VPS29. Remarkably, these two proteins display little similarity except for the leucine/isoleucine residue essential for binding. In fact, the main chain direction of RidL is opposite to that of TBC1d5. Despite these differences, in both proteins the leucine/isoleucine site is surrounded with proline or small amino acids, which may allow the proteins to make sharp turns and to properly position leucine/isoleucine.
Fig. 5.
RidL and TBC1d5 compete with each other for binding to retromer. (A) Structural comparison of VPS29/RidL and VPS29/TBC1d5-Ins1 overlaid by VPS29 (Left). (VPS29, gray; RidL, cyan; and TBC1d5, magenta). (Right) Zoomed-in view of the VPS29-interacting regions of TBC1d5 and RidL. Key interacting residues from TBC1d5 and RidL are shown in stick representation. (B and C) Coomassie blue stained SDS/PAGE gels of bound proteins are shown. Results are representative of three independent experiments. Amount of VPS29 (B) or VPS35 (C) retained was expressed relative to the amount of immobilized GST-fusion proteins in the bound sample and then normalized to control sample (no competing protein). All values are presented as mean ± SD, derived from three independent experiments. The molar ratio of immobilized protein and competing protein is indicated at the Bottom of the table. (B) GST–RidL or TBC1d5 pull-down of purified VPS29 in the absence or presence of competing TBC1d5 or RidL. (C) GST–RidL or TBC1d5 pull-down of purified VPS35/VPS29 in the absence or presence of competing TBC1d5 or RidL. (D and E) RidL inhibits endosomal localization of TBC1d5 through its interaction with the retromer. Cells were first transformed with GFP, GFP–RidL-N WT, or mutants (green), then fixed and stained with anti-TBC1d5 (white) and EEA1 (red) antibodies. Each dot represents Pearson’s correlation coefficients from one cell.
VARP possesses two cysteine-rich motifs that can bind to VPS29 independently (9, 10). Sequence alignment between cysteine-rich motifs of VARP and Ins1 of TBC1d5 suggests that the two proteins share a conserved leucine residue (L434 and L714 in VARP, and L142 in TBC1d5) and a preceding proline residue (SI Appendix, Fig. S6A). RidL is not included in the sequence alignment as its main chain direction is different from that of TBC1d5 and possibly VARP. In addition, the cysteine-rich motif is also known to feature variable loops within a short amino acid sequence (32). To examine the importance of the corresponding leucine of VARP in contacting VPS29, we generated a VARP fragment (amino acids 1–450) harboring the first cysteine-rich motif and examined its interaction with VPS29 in a GST pull-down assay. Consistent with published studies, this fragment associated with VPS29 (9, 10). Replacement of L434VARP to alanine (L434A), but not isoleucine (L434I), greatly weakened the interaction between VARP and VPS29 (SI Appendix, Fig. S6B). Thus, with little similarity in the primary sequence or tertiary structure, two host proteins and one pathogen protein interact with VPS29 through similar modes: all three proteins insert a leucine/isoleucine into the hydrophobic pocket of VPS29 that matches the size of leucine/isoleucine, and the VPS29-contacting motifs are primarily sharp turns.
RidL and TBC1d5 Compete with Each Other in Vitro and in Vivo.
Our structural analysis indicates that RidL and TBC1d5 bind to the same site of VPS29, suggesting that the two may compete with each other (8). Indeed, RidL-N inhibited the binding of GST–TBC1d5 to VPS29 in a dose-dependent manner; in contrast, TBC1d5 only slightly outcompeted RidL-N for VPS29 binding (Fig. 5B). This is consistent with our measurement that the binding affinity between VPS29 and RidL is at least one order of magnitude higher than that of VPS29 and TBC1d5 (Fig. 1B). Intriguingly, in similar experiments with VPS35/VPS29, TBC1d5 pronouncedly inhibited the binding of GST–RidL to VPS35/VPS29, but not vice versa (Fig. 5C). Since RidL and TBC1d5 bound to VPS35/VPS29 with a similar affinity, the difference in pull-down experiments was likely due to the different binding mechanisms. TBC1d5 contacts both VPS35 and VPS29, whereas RidL only interacts with VPS29, resulting in distinct association/dissociation behaviors. Furthermore, the binding between TBC1d5 or RidL with several VPS29 mutants (L2A, Y163A, or Y165A) is greatly reduced or abolished when comparing with VPS29 WT (Fig. 2D and SI Appendix, Fig. S7) (8).
Since TBC1d5 is necessary for the transport of retromer cargoes such as CI-MPR and Integrin a5b1, we asked whether RidL inhibits retromer transport through competing with TBC1d5 in cells (8). Transfection of GFP–RidL–WT efficiently displaced TBC1d5 from endosomes, leading to a less TBC1d5:EEA1 colocalization (Fig. 5 D and E). In contrast, the RidL mutants (Y166A, I170A, and P172A) defective in binding VPS29 failed to dislodge TBC1d5. Since VARP binds to retromer with an affinity lower than that of TBC1d5, it is most likely that RidL could outcompete with VARP in cells as well (9).
Discussion
In summary, our biochemical, structural, and cellular studies reveal that RidL binds to a conserved surface on VPS29 that is also targeted by its endogenous regulators such as TBC1d5 and VARP, and demonstrate that competing with these regulators is the primary mechanism underlying RidL-mediated retromer inhibition (Fig. 6). It remains to be determined how many other proteins, either endogenous or from pathogens, bind to the same surface of VPS29. The lack of sequence similarity among these known VPS29-binding proteins makes it difficult to predict additional partners by bioinformatics tools. The RidL–VPS29 complex structure also suggests small molecules mimicking the interaction could function as inhibitors for the retromer transport. These inhibitors will be important probes to investigate cellular trafficking and could potentially be used to protect against ricin exposure or Shigella infection. In this regard, lysine-mimic small molecules have been shown to occupy a small hydrophobic pocket on the surface of Cdc20 and to block Cdc20-dependent function (33). Finally, although our study herein and previous studies have begun to reveal the distinct strategies utilized by pathogenic bacteria to control retrograde transport, it remains to be determined how retrograde trafficking restricts or promotes bacterial pathogens. Since both the retromer and TBC1d5 regulate autophagy, further studies will be necessary to address whether RidL plays a role in the formation of autophagosome, a well-known mechanism to curtail intracellular bacterial pathogens (34–36). While our manuscript was under final stage of review, Bärlocher et al. (37) reported the crystal structure of the N terminus of RidL. Our complex structure between RidL and VPS29, and detailed biochemical and cellular studies extended their findings, and more importantly, provided a molecular basis for the displacement of TBC1d5 by RidL.
Fig. 6.
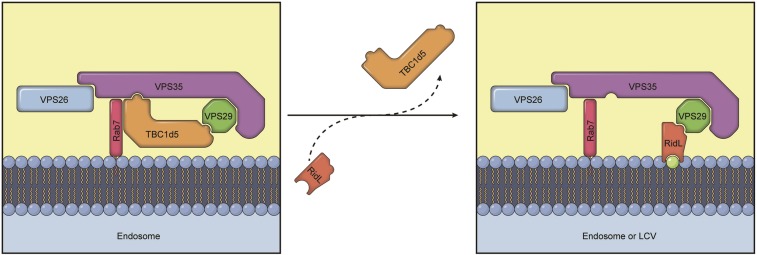
Model showing how RidL inhibits retromer-dependent transport. (Left) Retromer requires TBC1d5 for endosomal transport. TBC1d5 forms a tight complex with retromer through interacting both VPS35 and VPS29 and may function to regulate retromer assembly and turnover on endosomal membranes. Retromer cargoes, Sortin Nexins, VARP, and other known regulators are omitted for simplicity. (Right) During L. pneumophila infection or ectopic expression of RidL, RidL replaces TBC1d5. Upon ectopic expression of RidL, RidL is recruited to endosomes through interaction with VPS29 and endosomal lipid PtdIns(3)P (green dot). RidL replaces TBC1d5, and likely VARP, to block retromer-mediated trafficking. During L. pneumophila infection, retromer subunits are recruited to Legionella-containing vacuoles (LCVs) through their interaction with RidL and potentially other bacterial effector proteins.
Methods
Antibodies and Plasmids.
DNA constructs and antibodies used in this paper are listed in SI Appendix, Tables S2 and S3, respectively.
Cloning, Expression, and Purification.
The RidL cDNAs from L. pneumophila, L. moravica, or L. shakespearei were cloned into a pGEX-4T1–based expression vector incorporating a TEV-cleavable N-terminal GST-tag fusion. Expression of GST–RidL was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the culture was grown overnight at 20 °C in LB broth (Miller). Cells were harvested and sonicated in lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl and 1 mM PMSF). Proteins were purified on a GST column and eluted after TEV cleavage in a buffer containing: 50 mM Tris pH 8.0, 200 mM NaCl, followed by a Superdex 200 increase gel filtration column on the Äkta Pure (GE Healthcare) using the gel filtration buffer (20 mM Tris pH 8.0, 200 mM NaCl). Purification of retromer cargo-selective complex and subcomplexes was described previously (38).
Crystallization and Data Collection.
RidL-N and VPS29 proteins were mixed at 1:1.5 molar ratio and purified by Superdex 200 increase gel filtration column on the Äkta Pure (GE Healthcare). Fractions containing 1:1 complex were concentrated to 3.7∼5 mg/mL. Diffraction-quality crystals were obtained using hanging-drop vapor-diffusion methods at 18 °C, by mixing equal amounts of protein solution and reservoir solution containing 0.1 M Tris PH 8.5, 20∼25% wt/vol polyethylene glycol 3350. The 20% (vol/vol) glycerol was supplemented with crystallization condition as the cryoprotectant. X-ray diffraction data were collected at Shanghai Synchrotron Radiation facility beamline BL17U1 (39). The data collection statistics are given in SI Appendix, Table S1.
Structure Solution and Refinement.
Coordinates of VPS29 (PDB code 1W24) were used as the search model, and the program MolRep found two copies of VPS29 per asymmetric unit. The resulting phase was accurate enough to allow modeling of two RidL molecules through the program Autobuild (40) and manual building using the program COOT (41). Refinement was performed using the program Refmac5 (42). Translation/libration/screw (TLS) refinement (43) and noncrystallographic symmetry (NCS) restraints were used in the refinement process. Data in the interval 50.00- to 2.5-Å resolution were used and at the end of the refinement, the R value was 0.188 (Rfree = 0.224) for all reflections (SI Appendix, Table S1).
ITC.
ITC experiments were conducted at 20 °C using ITC200 (Microcal) in gel filtration buffer (20 mM Tris pH 8.0, 200 mM NaCl). RidL, RidL-N, or TBC protein (100∼200 μM) were titrated into the sample cell containing retromer subunit or complex (8–16 μM). Data were analyzed with the Origin 7.0 software package (OriginLab) by fitting the “one set of sites” model.
Pull-Down Experiments.
GST pull-down experiments were performed as previous studies (38). The mixture contained 20 μg of GST or GST-tagged protein, and 200 μg of bait proteins. The proteins were mixed with glutathione Sepharose 4B resin in 1 mL of pull-down buffer (PB: 20 mM Tris, pH 8.0, 200 mM NaCl, 0.005% Triton X-100). After extensive washing with PB buffer, bound proteins were separated by SDS/PAGE and visualized by Coomassie staining. For the competition experiment, 400 pmol of GST-tagged protein was mixed with 800 pmol of VPS35/VPS29 or 2,500 pmol of VPS29, and indicated amount of competitor protein. Each experiment was repeated at least once and checked for consistency. Band intensity quantification was performed using the program ImageJ (44).
Cell Culture, Immunofluorescent Staining, and Confocal Microscopy.
HeLa cells were maintained and analyzed as previously described (8, 19). Cells were maintained in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% (vol/vol) FBS (Sangon Biotech), and transfected with TurboFect transfection reagent (Thermo Fisher Scientific). Confocal images were acquired by Zeiss LSM 780 and Olympus FV-1000 confocal microscopes and analyzed using NIH ImageJ software. All cellular experiments were duplicated at least once.
Retrograde Trafficking Assay of STxB.
Purification of subunit B of Shiga toxin (STxB) and retrograde trafficking assay of STxB were performed according to established protocol (45, 46). Briefly, the plasmid encoding recombinant STxB was transformed into Escherichia coli DH5α cells. The bacteria were grown overnight at 30 °C, and then shifted to 42 °C for 4 h. Recombinant STxB was expressed in the periplasm, first isolated by osmosis shock, and further purified by Q-Sepharose Fast Flow and Mono Q chromatography.
To assay the intracellular trafficking of STxB, HeLa cells were transiently transformed for 24 h with plasmid encoding GFP, GFP–RidL-N WT, or mutants. Transformed cells were incubated with purified STxB on 16.5 °C for 1 h. After washing three times with cold PBS, the cells were shifted for 20 min to 37 °C in serum-free DMEM. After fixation, the cells were staining with STxB and TGN46 antibodies. STxB trafficking was determined by calculating Pearson’s correlation coefficient for colocalization between STxB and TGN46.
Liposome Flotation Assay.
Liposome flotation assays were performed as previously described (26, 27). Briefly, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and phosphatidylinositol-3-phosphate [PtdIns(3)P] (all from Avanti Polar Lipids) were mixed with a ratio of 78:20:2. Lipid mixtures in glass tubes were then dried under nitrogen flow in a 42 °C waterbath and brought to complete dryness in a vacuum desiccator for at least 3 h. Dried lipid films were resuspended by a detergent-free buffer containing 25 mM Hepes-K+ pH 7.60, 150 mM KCl 10% glycerol (vol/vol) and 0.2 mM TCEP (buffer H) and vortexed for 5 min. The mixtures were then frozen and thawed five times and extruded through a 200-nm polycarbonate filter with a miniextruder (Avanti Polar Lipids) 17 times. Qualities of the prepared liposomes were checked using dynamic light scattering. Solution containing 2 mM liposomes (total lipids) was mixed with 5 µM proteins. After incubation at room temperature for 1 h, the samples were loaded into a Histodenz density gradient (40%, 30%, and 0%) and centrifuged at 240,000 × g, 20 °C for 1 h in a SW55Ti rotor (Beckman Coulter). Samples from the top of the gradient (30 µL) were taken and analyzed by SDS/PAGE followed by immunoblotting using rabbit anti-GST polyclonal antibody (Proteintech).
Supplementary Material
Acknowledgments
We thank Drs. Dan Billadeau and Guihua Tai for reagents, Drs. Bo Sun and Huan Zhou (Shanghai Synchrotron Radiation Facility) for help with diffraction data collection, and members of our laboratories for helpful discussions. This research is supported by Natural Science Foundation of China Grants 80502629 (to Q.S.) and 31671477 (to D.J.) and by NIH National Institute of Allergy and Infectious Diseases Grant 1R01AI127465 (to Z.-Q.L.). D.J. is a “One Thousand Talents” program scholar, supported by the Chinese Central Government and Sichuan Province.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.R.R. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5WYH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717383115/-/DCSupplemental.
References
- 1.Burd C, Cullen PJ. Retromer: A master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMillan KJ, Korswagen HC, Cullen PJ. The emerging role of retromer in neuroprotection. Curr Opin Cell Biol. 2017;47:72–82. doi: 10.1016/j.ceb.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas M, Hierro A. Retromer. Curr Biol. 2017;27:R687–R689. doi: 10.1016/j.cub.2017.05.072. [DOI] [PubMed] [Google Scholar]
- 5.Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas M, et al. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–1635.e14. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia D, et al. Structural and mechanistic insights into regulation of the retromer coat by TBC1d5. Nat Commun. 2016;7:13305. doi: 10.1038/ncomms13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hesketh GG, et al. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev Cell. 2014;29:591–606. doi: 10.1016/j.devcel.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGough IJ, et al. Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma-membrane recycling. J Cell Sci. 2014;127:4940–4953. doi: 10.1242/jcs.156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia D, et al. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Small SA, Petsko GA. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat Rev Neurosci. 2015;16:126–132. doi: 10.1038/nrn3896. [DOI] [PubMed] [Google Scholar]
- 15.Personnic N, Bärlocher K, Finsel I, Hilbi H. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol. 2016;24:450–462. doi: 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Bujny MV, Popoff V, Johannes L, Cullen PJ. The retromer component sorting nexin-1 is required for efficient retrograde transport of Shiga toxin from early endosome to the trans Golgi network. J Cell Sci. 2007;120:2010–2021. doi: 10.1242/jcs.003111. [DOI] [PubMed] [Google Scholar]
- 17.Popoff V, et al. The retromer complex and clathrin define an early endosomal retrograde exit site. J Cell Sci. 2007;120:2022–2031. doi: 10.1242/jcs.003020. [DOI] [PubMed] [Google Scholar]
- 18.Stechmann B, et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell. 2010;141:231–242. doi: 10.1016/j.cell.2010.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, et al. Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE. Signal Transduct Target Ther. 2017;2:17030. doi: 10.1038/sigtrans.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Z-Q. Catch and arrest: Exploiting the retromer by a Chlamydial effector. Signal Transduct Target Ther. 2017;2:17039. doi: 10.1038/sigtrans.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul B, et al. Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis. eLife. 2017;6:e22311. doi: 10.7554/eLife.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu Y, et al. Ablation of SNX6 leads to defects in synaptic function of CA1 pyramidal neurons and spatial memory. eLife. 2017;6:e20991. doi: 10.7554/eLife.20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirrashidi KM, et al. Global mapping of the Inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu J, Luo ZQ. Legionella and Coxiella effectors: Strength in diversity and activity. Nat Rev Microbiol. 2017;15:591–605. doi: 10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- 25.Finsel I, et al. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe. 2013;14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Li Y, Ma C. Synaptotagmin-1 C2B domain interacts simultaneously with SNAREs and membranes to promote membrane fusion. eLife. 2016;5:e14211. doi: 10.7554/eLife.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins BM, Skinner CF, Watson PJ, Seaman MN, Owen DJ. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat Struct Mol Biol. 2005;12:594–602. doi: 10.1038/nsmb954. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, et al. Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. J Biol Chem. 2005;280:22962–22967. doi: 10.1074/jbc.M500464200. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hierro A, et al. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 33.Sackton KL, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–649. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy S, Leidal AM, Ye J, Ronen SM, Debnath J. Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol Cell. 2017;67:84–95.e5. doi: 10.1016/j.molcel.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popovic D, et al. Rab GTPase-activating proteins in autophagy: Regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruzs T, et al. Retromer ensures the degradation of autophagic cargo by maintaining lysosome function in Drosophila. Traffic. 2015;16:1088–1107. doi: 10.1111/tra.12309. [DOI] [PubMed] [Google Scholar]
- 37.Bärlocher K, et al. Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5. Nat Commun. 2017;8:1543. doi: 10.1038/s41467-017-01512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, et al. Automatic crystal centring procedure at the SSRF macromolecular crystallography beamline. J Synchrotron Radiat. 2016;23:1323–1332. doi: 10.1107/S160057751601451X. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Painter J, Merritt EA. TLSMD web server fpr the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallard F, et al. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J Cell Biol. 1998;143:973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D, et al. The effects of the carboxyl-terminus amino acids of the Shiga toxin B-subunit on retrograde transport. Mol Med Rep. 2012;6:220–226. doi: 10.3892/mmr.2012.885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.