Abstract
Comparative genomics analysis of conserved gene cassettes demonstrated resemblance between a recently described cassette of genes involved in sulphoquinovose degradation in Escherichia coli K-12 MG1655 and a Bacilli cassette linked with lactose degradation. Six genes from both cassettes had similar functions related to carbohydrate metabolism, namely, hydrolase, aldolase, kinase, isomerase, transporter, and transcription factor. The Escherichia coli sulphoglycolysis cassette was thus predicted to be associated with lactose degradation. This prediction was confirmed experimentally: expression of genes coding for aldolase (yihT), isomerase (yihS), and kinase (yihV) was dramatically increased during growth on lactose. These genes were previously shown to be activated during growth on sulphoquinovose, so our observation may indicate multi-functional capabilities of the respective proteins. Transcription starts for yihT, yihV and yihW were mapped in silico, in vitro and in vivo. Out of three promoters for yihT, one was active only during growth on lactose. We further showed that switches in yihT transcription are controlled by YihW, a DeoR-family transcription factor in the Escherichia coli cassette. YihW acted as a carbon source-dependent dual regulator involved in sustaining the baseline growth in the absence of lac-operon, with function either complementary, or opposite to a global regulator of carbohydrate metabolism, cAMP-CRP.
Introduction
Catabolism and synthesis of carbohydrate molecules are important segments of bacterial metabolism. Many bacterial species can assimilate a wide range of carbohydrate substrates, adjusting to environmental changes, and are able to quickly switch between pathways. With multiple energy sources in the environment, a bacterial cell goes through metabolic assessment and, in most cases, selects a specific preferred energy source, e.g. glucose1,2.
Genes encoding proteins responsible for carbohydrate transport, degradation, and synthesis are very diverse. According to the IMG database3, over a hundred thousand genes related to carbohydrate metabolism have been described for over six hundred genomes of various bacterial species. Moreover, some of these genes are known to encode proteins that have multiple functions and can be involved in different metabolic pathways. For example, thermophilic glycoside hydrolase CoGH1A from Caldicellulosiruptor owensensis has a high capability of saccharification, being able to hydrolyze various substrates4, polysaccharide lyase Smlt1473 from Stenotrophomonas maltophilia is able to catalyze chemical bond cleavage in a variety of carbohydrate molecules5, and hexokinase Glk/TM1469 from Thermotoga maritima switches between substrates under different temperature conditions6.
Carbohydrate metabolism in most bacteria is controlled by the carbon catabolite repression (CCR) mechanism7,8. CCR allows the cell to select a preferred source over a non-preferred one, maintaining a proper energy balance9. In a glucose-free environment or under stress conditions bacteria switch to the next best carbon source available10,11. In Escherichia coli and related species, this switch is often controlled by cAMP-CRP as a global regulator, and a local regulator encoded by a gene usually located either in the same operon with genes encoding enzymes and/or transporters necessary for growth on alternative carbohydrates, or in a divergent orientation12–14.
cAMP-CRP is one of the most important transcription factors that regulates transcription initiation for more than a hundred genes of carbohydrate catabolism15,16. It functions as a dimer in the form of the CRP-cAMP complex and may activate or repress transcription. In most cases, CRP downregulates transcription when its operator region is situated upstream of the transcription start site17,18. CRP-mediated activation occurs when cAMP-CRP binds upstream of the target promoter and directly contacts RNA polymerase19. It can also bind DNA in a cAMP-independent manner, resulting in a weak target repression17. An additional, substrate-specific layer of regulation is provided by the binding sites of local transcription factors from six major families (LacI, ROK, DeoR, AraC, GntR, and TetR) that are located close to the CRP binding sites, or even overlap them. In most cases, these factors play a role opposite to CRP and act as dimers. Dimerisation caused by the ligand binding leads to a metabolic switch20.
Here, we identify and explore the second lactose catabolism system in E. coli, supplementing the lac operon described in the pioneer work of François Jacob and Jacques Monod21. The lac operon consists of three genes, encoding β-galactosidase, β-galactoside transacetylase, and β-galactoside permease. Until now, no other pathways for lactose utilisation in E. coli have been known.
In a recent biochemical study, Denger et al.22 described the E. coli gene cassette consisting of 10 yih genes and, according to that study, completing the biogeochemical cycle of sulphur. Four reactions of sulphoquinovose (SQ) catabolism were demonstrated using purified, heterologously expressed enzymes; other genes from this cassette were predicted to be involved in regulation, transport, hydrolysis, and other processes of sulphoglycolysis. The yih genes have not been linked to the lactose catabolism, and their transcription profile has not been characterised.
Unknown functions of a gene can be predicted using comparative genomics analysis, based on gene proximity in the chromosome and functional similarity of their protein products23–29. We compared functional composition of the yih cassette with other carbohydrate metabolism gene cassettes and found a match with a cassette involved in the lactose catabolism of Bacilli. Six genes from these two cassettes encoded proteins with similar functions, namely, hydrolase, aldolase, kinase, isomerase, transporter, and transcription factor, allowing us to suggest involvement of the E. coli yih-cassette in the lactose catabolism. We then confirmed it experimentally using expression analysis. We also mapped promoters for the yih genes, and studied how the switch to lactose utilization is regulated, thus describing a possible additional pathway for lactose utilization in E. coli and suggesting multi-functional features for the respective proteins.
Results
The functions of genes in the E. coli cassette encoding enzymes of sulfoquinovose degradation are similar to those in the lactose cassette in Bacilli
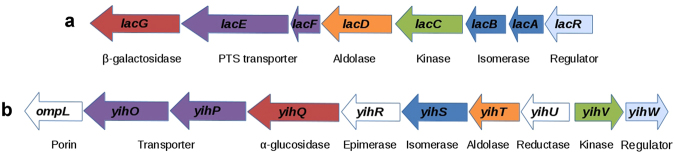
Comparison of the combinations of gene functions in cassettes related to bacterial carbohydrate metabolism revealed a six-gene match between a cassette linked with SQ degradation in E. coli and present in some other Enterobacteriaceae, and a lactose catabolism cassette in several Bacilli species (Fig. 1). Namely, the functional content of the yih cassette in a number of strains of E. coli, Enterobacter cloacae, Salmonella enterica, Cronobacter turicensis, and Pantoea anantis, all belonging to Enterobacteriaceae, matched the content in the lactose degradation cassette of the Streptococcus spp. (S. gallolyticus, S. suis, S. pyogenes, S. agalactiae, S. uberis, S. equi, S. mutans, and S. sanguinis) and Staphylococcus spp. (S. aureus, S. epidermidis, S. haemolyticus, and S. lugdunensis).
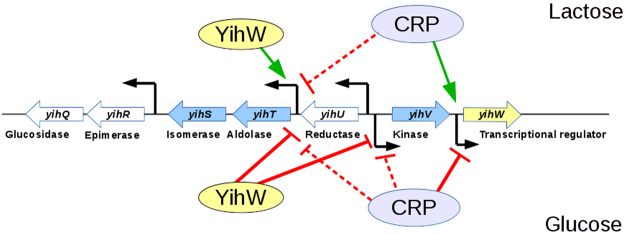
Figure 1.
The lactose catabolism cassette of Bacilli (a) and the yih cassette of Enterobacteriaceae (b). Genes encoding proteins with similar functions are marked with the same colour. Genes marked white have no matches between the cassettes.
These matching genes encoded enzymes converting sugar substrates which shared at least three first numbers of the Enzyme Commission nomenclature, thus having similar chemical reaction types catalysed, namely hydrolases (3.2.1), aldolases (EC 4.1.2), kinases (EC 2.7.1), and isomerases (2.3.1), and also transporters and transcription factors (Fig. 2)3. The genes coding for aldolases also belonged to one and the same cluster of orthologous genes (COG3684). Such co-localization of genes linked with functions similar to those required for the Bacilli lactose pathway (Fig. 2) in the Enterobacteriaceae cassette allowed us to predict lactose degrading capabilities of the latter.
Figure 2.
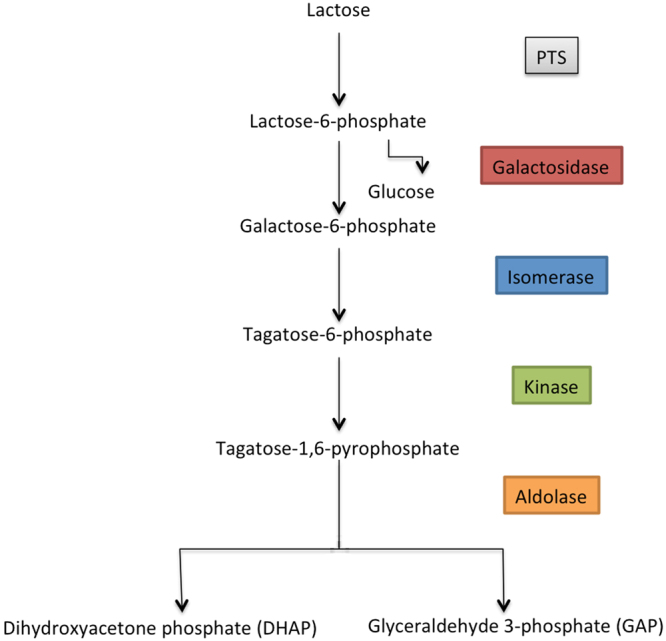
The lactose catabolism pathway encoded by the Bacilli cassette.
Our next goal was to test whether this E. coli cassette is actually involved in the lactose metabolism. Since no experimentally confirmed data on positioning of transcription start points for the genes of the yih cassette were available, the first step was to map promoters for the main genes encoding the key enzymes of the suggested catabolic pathway.
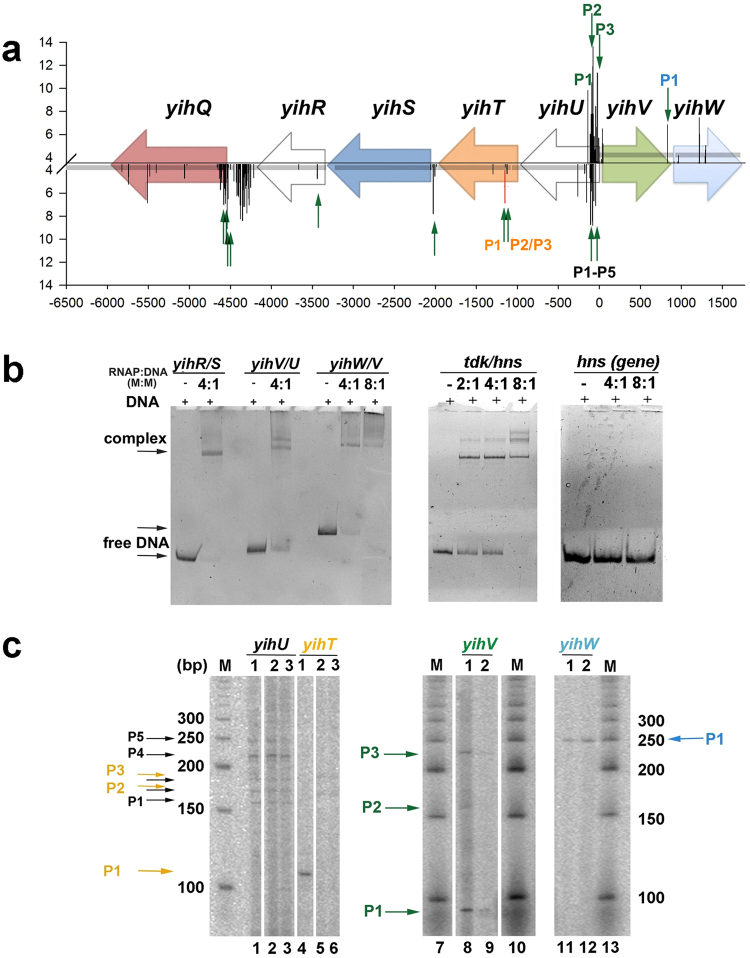
Promoter mapping
Computational analysis of the intergenic regions by phylogenetic footprinting and promoter search algorithm PlatProm30 allowed us to predict candidate promoters for the genes encoding aldolase (yihT), kinase (yihV), isomerase (yihS), alpha-glucosidase (yihQ), and a local transcription factor (yihW). To obtain a comprehensive view on the transcription pattern in this locus, the yihU and yihQ genes coding for aldolase and aldose epimerase that are not associated with the suggested pathway, were also incorporated in the analyses (Fig. 3a). Computational predictions were then compared with the starts detected in 5′-end specific RNA-seq31 (Fig. 3a), and transcription starts for the mRNAs of the key enzymes were further confirmed experimentally using band-shift assays (Fig. 3b) and primer extension (Fig. 3c). Band-shift assays with σ70-RNA polymerase (RNAP) revealed the presence of promoters capable of efficient binding with RNAP in the intergenic regions yihU/V, yihV/W, and yihS/R (Fig. 3b). Formation of two complexes with almost identical efficiency suggested the presence of at least two equally strong promoters between yihU and yihV. Specificity of the RNAP binding was confirmed by its strong interaction with known σ70-promoter of the hns gene and the absence of complex formation with the intergenic region of hns where no promoters had been mapped (Fig. 3b). To map the exact transcription start points and to test whether these promoters are lactose-sensitive, primer extension was done with RNA isolated from cells growing for 6 hours on M9 + 10% LB supplemented with 0.2% lactose, glucose, or glycerol. Reverse transcription with primer yihU_RT revealed several products (black arrows in Fig. 3c) corresponding to the transcription starts at positions −15 (PlatProm score 6.83), −25 (5.04), −35 (3.52), −62/−63 (8.75), −82/−92 (a large cluster with scores 8.75–13.61, yihU promoters yihUP1-yihUP5) relative to the annotated ATG codon. RNA synthesis from yihUP1, yihUP2, yihUP4, and yihUP5 was also confirmed with primer yihU/yihV_F (data not shown, Supplementary Table 1) and 5′-end specific RNA-seq31 (green arrows in Fig. 3a). Note, however, that none of these promoters was dependent on a carbon source suggesting that the protein product of yihU (putative reductase) is not involved in the lactose metabolism. For yihV (putative kinase), several transcription starts were also mapped, with the major one located at –25/–27 relative to the starting ATG (a large cluster of predicted promoters with the highest score of 11.35 at position 4071737 in U00096.2). This promoter was activated during growth on lactose (Fig. 3c). For yihT, three possible starts were detected (orange arrows in Fig. 3c). Two of them, corresponding to yihTP2 and P3, located +35/+45 relative to the putative ATG codon, were predicted with a relatively low score of 3.67–4.33 and, in line with this, had low transcriptional activity. Nonetheless, these products were not detected during cell growth on lactose (lane 1 in Fig. 3c), while another promoter (yihTP1) was switched on, with transcription start located at +93/+94 relative to ATG (lane 4 in Fig. 3c).
Figure 3.
Promoter mapping within the yih cassette. (a) Schematic representation of the yih genes and candidate promoters predicted in silico. Horizontal arrows show gene positioning. Bars represent the transcription start points predicted by PlatProm on both DNA strands, their scores are indicated on the Y axis. Score assigned to the yihTP1 promoter by the unified algorithm, PlatPromU, is marked with red. X axis represents positioning related to the yihV ATG start codon. Green vertical arrows point out the 5′-end of RNA detected by the 5′-end-specific RNA-Seq31. (b) Band-shift assays with σ70-RNA polymerase suggests the presence of single promoters in the intergenic regions yihV/W and yihS/R, and two promoters in yihU/V. Molar ratios are indicated above the lanes. All samples were run on one gel. Known σ70-promoter of the hns gene was used as a positive control; intergenic region within hns where no promoters were predicted was used as a negative control (c) Primer extension analysis revealed transcription start point for yihW and several starts for the yihU, yihT, and yihV genes, some of which were activated during growth on lactose. (1) Growth on lactose, (2) growth on glucose, (3) growth on glycerol. The yihU and yihT samples, as well as the DNA ladder on the left, were run on one gel, while yihV and yihW were run on another gel, together with the DNA ladders displayed. Lanes that were taken from different parts of the gels are divided by spacing.
This promoter was predicted only with the universal algorithm, PlatPromU, suggesting its recognition by an alternative sigma factor. The existence of transcriptional switch is also supported by the stringent control discriminator GCGC motif present between the transcription start and the −10 element of yihTP1. All mapped promoters for yihT are located within the proposed ORF for this gene suggesting either incorrect identification of the translational start (at least four additional candidate start-codons were detected, 51, 78, 84 and 87 nt downstream), or their regulatory role in the yihT transcription32. For yihW, only one promoter was mapped, with transcription start located at −27/−28 relative to the ATG codon (Fig. 3a–c).
Expression of the yih genes during growth on various carbon sources
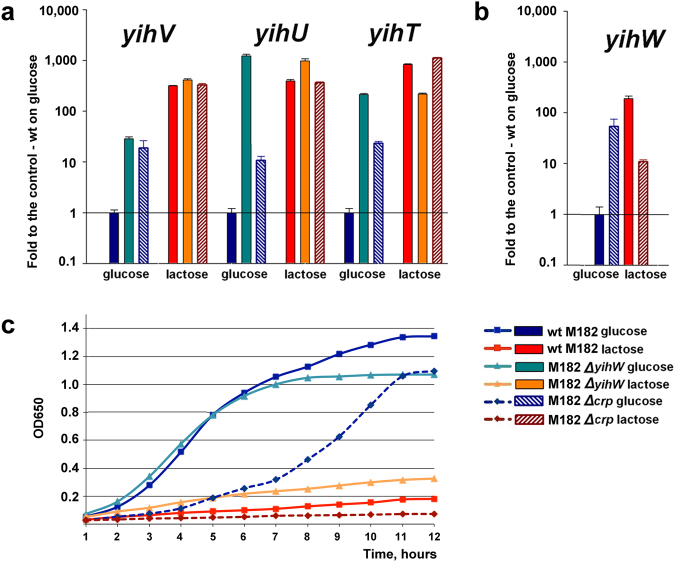
To further check whether transcription of the yih genes is lactose-dependent, we tested their mRNA levels in cultures growing in the same conditions as used for primer extension, and also during growth on galactose. Glucose was taken as a standard rich carbon source, glycerol as a standard poor carbon source, and galactose as an intermediate of the possible pathway shown in Fig. 2. Results of qRT-PCR tests for cultures grown on various carbon sources for 6 hours are shown in Fig. 4. Genes encoding kinase (yihV), aldolase (yihT), and isomerase (yihS) were activated during growth on galactose and lactose, and this activation was more pronounced during growth on lactose. yihW expression, conversely, was approximately 2-fold repressed during growth with lactose suggesting either its function as a repressor for the yih genes at this time point, or a self-repressor function. The yihU gene, encoding a putative reductase, did not respond to differences in carbon sources (Fig. 4).
Figure 4.
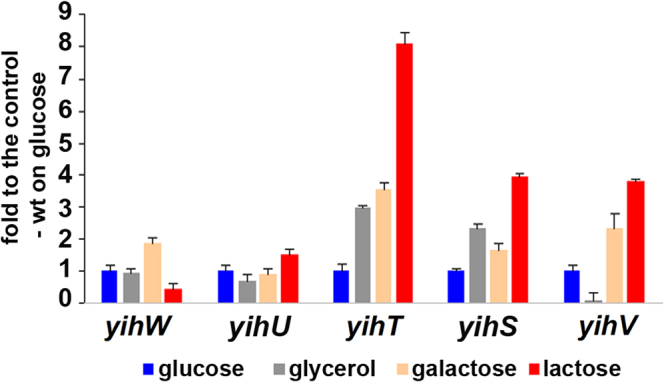
mRNA levels of the yih genes measured by qRT-PCR. Growth conditions are indicated at the bottom. No changes were detected in the hns-mRNA and ysaA-aRNA levels that were used as controls. mRNA levels are expressed relative to their levels during growth on glucose. Standard deviations were calculated on the basis of three biological and three technical replicates.
Expression of yihR and yihQ (data not shown), encoding epimerase and glucosidase, respectively, was also independent (yihR) or barely dependent (yihQ) on the carbohydrates used; this result supports our previous assumption about their independent transcription (Fig. 3) and suggests that these genes are not related to the lactose metabolism. Comparative analysis showed that a number of Enterobacteriaceae bacteria, including Escherichia albertii and Citrobacter koseri strains, have orthologs only for the yihSTUVW genes, while the rest of this cassette, including yihR and yihQ, is missing, which also indicates that different parts of the 10-gene cassette may function separately.
cAMP-CRP and YihW are involved in the regulation of the yih genes
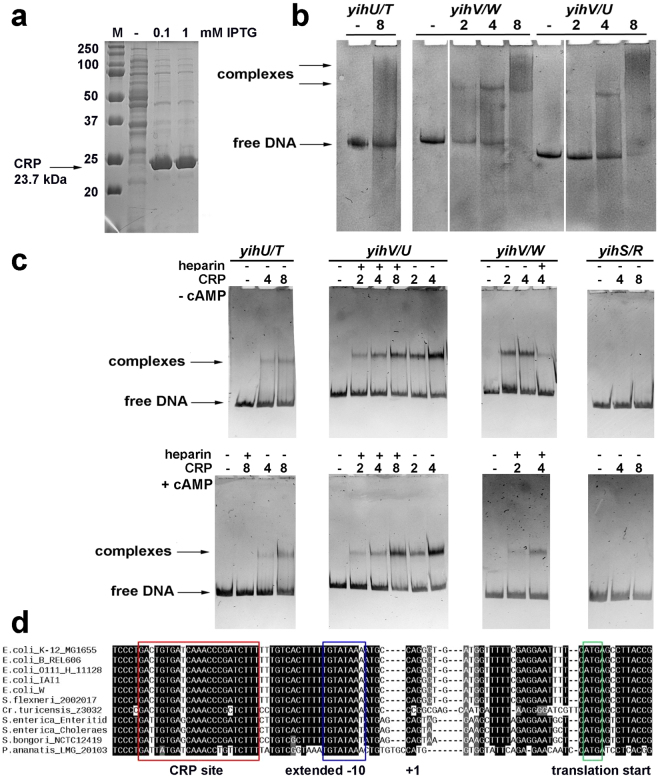
A moderate decrease of the yihW-mRNA level in the presence of lactose suggested that the YihW transcription factor may be involved in down-regulation of the yih genes as a local regulator. If so, it may be complemented by cAMP-CRP as a global regulator. Thus, we searched for potential CRP binding sites near the mapped promoters and tested whether these promoters are CRP-dependent.
The sites were found in the yihT/U, yihU/V, and yihV/W intergenic regions and were further confirmed by phylogenetic footprinting and band-shifts assays with native CRP from the cell lysate (Fig. 5b; CRP expression level is shown in Fig. 5a). The presence of CRP in the formed complexes was confirmed by Western blot analysis. Strong specific binding of CRP to the yihV/yihU and yihW promoters was also detected with the purified protein, and interaction with both promoter regions was ~30% enhanced in the presence of cAMP (Fig. 5c). In line with this, a highly conserved site was found in the yihV/W intergenic region at the position −41.5 relative to the start site of the yihW promoter, that is typical for the Class II CRP-dependent promoters (Fig. 5d).
Figure 5.
(a) The level of cAMP-CRP synthesized in the cell from pET_CRP after IPTG induction. (b) cAMP-CRP interacted with the yihU/yihT, yihV/yihW, and yihV/yihU intergenic regions, with most efficient binding being registered for yihV/W. The protein:DNA ratio is indicated above the lanes. Control lane for the yihV/yihW intergenic region was run on a separate gel, as well as the sample with 1:1 protein:DNA ratio (this ratio gave no relevant binding and the corresponding lanes were deleted from the final figure). (c) Binding of purified CRP to the studied regions with/without cAMP. The protein:DNA ratio and the presence of heparin in the sample is indicated above the lanes. (d) Multiple alignment of the yihV/W intergenic region in several bacteria containing orthologs for these genes revealed high conservation of the mapped yihW promoter and the CRP binding motifs.
Assuming that CRP acts as a global regulator for all yih genes, and YihW is their local regulator, we investigated the growth rates of K-12 MG1655 cells with deleted yihW and crp33 on glucose and lactose.
The wild type and the yihW deletion strains grew equally well in the presence of glucose, with slightly retarded growth of Δcrp reflecting the key role of CRP in the regulation of glucose metabolism. The parent strain grew more slowly on lactose than on glucose as the carbon source, but deletion of yihW resulted in more rapid growth on lactose (Fig. 6). In contrast, growth of the crp mutant on lactose almost stopped as soon as the LB had been consumed. These results reveal that YihW is likely involved in regulation of the lactose metabolism and may play the role opposite to CRP. The next aim, therefore, was to uncover the roles of these proteins in the transcription of the yih genes.
Figure 6.
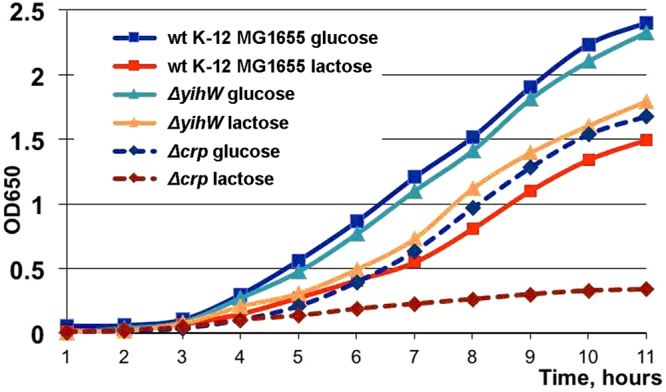
Effects of yihW and crp deletions on growth in the presence of 0.2% glucose or lactose. Squares: the parent strain; triangles: the yihW mutant. Dashed lines correspond to the cultures with deleted crp. Each growth curve is an average based on three independent measurements.
Effects of cAMP-CRP and YihW on transcription of the yih genes
Transcription of the yihT, yihS, and yihV genes was activated after 6 hours of growth on lactose (Fig. 4), while yihW was slightly repressed. Together with affected growth (Fig. 6), this suggested involvement of YihW in the control of the yih genes that was confirmed with qRT-PCR (Fig. 7). At that, RNA was isolated from the lac-deficient E. coli M182 cells34,35 exponentially growing in the minimal medium containing twice less LB (5%) to get more pronounced effects of the carbon source. The experiment showed that yihT expression on both glucose and lactose is totally controlled by YihW that functions as a sugar-dependent dual regulator (Fig. 7a), and repressed by CRP during growth on glucose. Expression of yihW itself is up-regulated by CRP in the presence of lactose, and down-regulated on glucose (Fig. 7b). Finally, both YihW and CRP function as repressors for yihV (Fig. 7a).
Figure 7.
Effects of yihW and crp deletion on mRNA levels the yih genes (a,b), and culture growth on glucose or lactose (c). Growth conditions are indicated at the bottom right. The mRNA levels are expressed relative to their levels in the parent strain during growth on glucose. No changes were detected in the hns-mRNA and ysaA-aRNA levels that were used as controls. Standard deviations were calculated on the basis of three biological and three technical replicates. Growth curve in (c) is an average based on three independent measurements. Squares: the parent strain; triangles: the yihW mutant. Dashed lines correspond to cultures with deleted crp.
Discussion
Since Jacob and Monod described the lac operon in late 1950s21, no other pathway for lactose utilisation has been observed in Escherichia coli. Here, we demonstrate that the yih cassette of E. coli, previously linked with SQ degradation22, may be also involved in lactose catabolism, thus suggesting the existence of an alternative pathway of further lactose degradation following initial hydrolysis. This hypothesis was initially stated based on the comparative genomics analysis, and then confirmed in a number of experiments.
The lactose catabolism pathway of some Bacilli species, such as Streptococcus, Staphylococcus, and Lactococcus spp., differs from the common lactose degrading pathway, which involves initial hydrolytic cleavage of lactose to glucose and galactose by β-galactosidase. The Bacilli species do not hydrolyse lactose directly; they transport it into the cell by a phosphoenolpyruvate-dependent phosphotransferase system that phosphorylates lactose to lactose-6-phosphate36. Then, phospho-β-galactosidase hydrolytically cleaves the phosphorylated derivative of lactose into glucose and galactose-6-phosphate37,38. Finally, D-galactose-6-phosphate is degraded via the D-tagatose-6-phosphate pathway comprised of D-galactose-6-phosphate isomerase38, D-tagatose-6-phosphate kinase39, and D-tagatose-1,6-diphosphate aldolase40 (Fig. 1).
Based on the concordance between functions encoded in the yih cassette and the Bacilli cassette, which is linked with the lactose catabolism, we suggested that the proteins encoded by the yih cassette could be involved in a similar lactose degradation pathway. To validate this prediction, we performed a detailed analysis of gene expression in different growth conditions. As one of quantitative characteristics indicating involvement of a particular enzyme in conversion of a particular substrate is changing expression of its gene in response to this substrate, we first compared expression of genes encoding the key enzymes of the predicted pathway during growth on glucose, galactose, and lactose. Following initial lactose hydrolysis, it should further be degraded by kinase (YihV), aldolase (YihT), and isomerase (YihS) (Figs 1 and 2). Initial hydrolysis is possibly performed by β-galactosidase from the lac-operon; however, even in the absence of lac-operon, all strains, except ∆crp, were capable of slow growth with lactose and galactose, thus suggesting possible existence of an alternative way of lactose cleavage (Fig. 7, Supplementary Figs S2 and S3).
Indeed, qRT-PCR demonstrated that all three respective genes were substantially activated on both lactose and galactose, the next intermediate of the pathway. As shown in Figs 4 and 7, these genes were activated after either four or six hours of growth. Note, however, that the yihW gene, encoding a predicted DeoR-family transcriptional regulator, was also lactose-activated during exponential growth but repressed upon transition to starvation where the substrates start depleting. This observation led us to an assumption that YihW may act as a dual regulator for the yih genes, activating them in the presence of extra lactose and repressing when lactose is lacking. Stimulated growth of lac-minus cultures with lactose in the absence of YihW, that is even more pronounced on galactose (Fig. 7 and Supplementary Fig. S3), is of particular interest. This observation might support involvement of YihW and the whole yih cassette in sustaining the baseline E. coli growth with lactose in the absence of lac-operon.
Effects of local regulators like YihW are usually opposed or supported, depending on growth conditions, by the action of global regulators; in the case of sugar metabolism of E. coli, that is cAMP-CRP15,19. Figures 6 and 7 clearly show that YihW may play a role opposite to that of CRP.
To further check this assumption and to reconstruct regulatory events that take place in the yih locus upon switching the carbon source from glucose to lactose and vice versa, we examined how expression of the key genes would change in the absence of CRP and YihW. The results are shown in Fig. 7a,b and summarized in Fig. 8. Activation of yihW itself during exponential growth with lactose was CRP-dependent, due to direct binding of CRP at −41.5 relative to the yihW promoter mapped here (Figs 3c and 5b). When lactose was absent, CRP repressed the yihW expression.
Figure 8.
Regulation of the studied yih genes during growth on lactose (top) and glucose (bottom) by the CRP and YihW transcription factors. Green arrows indicate activation of transcription, red lines indicate repression. Dashed lines indicate cases where inhibition is moderate and may be due to indirect effects.
YihW, in turn, is critical for the balanced expression of the aldolase gene yihT; it is responsible for the lactose-driven activation and for tight repression in the absence of lactose. Most probably, the same is also true for the isomerase gene yihS, because its transcription profile is almost identical to that of yihT (Fig. 4). When yihW was deleted, the yihT expression became totally independent on the carbon source (Fig. 7). CRP here plays a role of sugar-independent repressor due to its weak binding in a cAMP-independent manner16. This was consistent with positioning of its binding sites found by phylogenetic footprinting17 and was further confirmed by band-shift assays for the yihT/U intergenic region (Fig. 5b,c).
Interestingly, YihT predicted to be 6-deoxy-6-sulphofructose-1-phosphate aldolase is homologous to tagatose 1,6-diphosphate aldolase LacD present both in various Salmonella and Shigella species, and in several E. coli strains such as E. coli APEC O1 and E. coli O157:H7. The nucleotide sequence identity is 80% or even higher, with most substitutions occurring in the third codon positions, yielding 96% conservation of the respective amino acid sequences. Organization of the genomic neighborhood of these genes in Enterobacteriaceae strains differs, but in most cases yihT and lacD are accompanied by divergently transcribed genes homologous to yihW.
The yihV gene expression is also activated during growth on lactose and repressed on glucose, controlled by both YihW and CRP (Fig. 8).
This case is an example of a successful prediction of gene functions based on their co-localization patterns. The revealed complexity of the yih locus in E. coli seems to represent a case where, depending on environmental conditions, bacteria use the same set of multifunctional enzymes in different pathways, exploiting a finely-tuned regulatory mechanism.
Demonstrating possible involvement of the yih genes, previously known for only a very narrow specialization in SQ degradation, in lactose metabolism, our study raises a number of further questions concerning biochemical specialization of the proteins encoded by the yih genes, particularly, their specificity and affinity to lactose and SQ, and the regulation of their expression during growth on SQ.
Materials and Methods
Cassette database and comparison
We previously compiled a database41 comprising over a hundred and forty-five thousand genes linked to carbohydrate metabolism according to the IMG database3 from over six hundred bacterial species. Gene functions were identified based on the Enzyme Nomenclature numbers provided in the IMG annotation. Cassettes were assembled based on co-localization of these genes in bacterial chromosomes. Genes were assigned to the same cassette if a distance between adjacent genes was less than 300 bp; the order of genes and strands were not taken into consideration. The database contains over twenty-six thousand cassettes. Tools developed ad hoc were used to search the database for conserved function combinations within cassettes belonging to different species.
Strains, plasmids and growth conditions
All strains and plasmids used in this study are listed in Supplementary Table S1 online. The yihW and crp genes were disrupted in E. coli BW25113 using recombineering42. The gene::kan mutations were then transferred by P1-transduction into E. coli K-12 MG165543 or M182, which is a lac-minus derivative of K-1234. Cells were grown in the minimal salts (MS) medium supplemented with 5% or 10% (v/v) LB, 0.2% (w/v) of the appropriate carbon source - D-glucose, D-galactose, lactose or glycerol. Cultures were grown aerobically at 37 °C under constant shaking. Cells were harvested after 4.5 hours of growth (mid-log phase with OD~0.2–0.4, depending on the strain). To express the cAMP-CRP protein, E. coli BL21* (DE3) cells44 were chemically transformed with the pET_CRP plasmid constructed based on pET28b (Invitrogen), with the crp gene inserted between the NdeI and Bpu1102 sites. Purified transformants were grown on LB or Terrific broth (TB) at 37 °C till OD650 = 0.3, and the protein synthesis was induced by addition of a very low IPTG concentration (20 μM) to avoid the effects of potential protein toxicity. The induction rate of CRP after 3 hours of IPTG induction was ~70% of the total protein (See Supplementary Fig. S1).
CRP purification
To obtain pure CRP, BL21*(DE3) cells with overproduced CRP (16 hours of induction with 20 μM IPTG in 200 ml TB, Amp100) were harvested by centrifugation at +4 °C, washed with 1xPBS and lysed with 2 ml BugBuster protein extraction reagent (Novagen) for 20 minutes at +4 °C. Due to the very high induction level, all protein was in insoluble fraction even when very low IPTG concentration and low temperatures were used. To extract CRP from inclusion bodies, two-step lysis was performed: after initial lysis with BugBuster, 6 ml of ice-cold Lysis buffer15,19 (50 mM KPO4 buffer pH 7.5, 2 mM EDTA, 0.2 M NaCl, 5% w/v glycerol, 2 mM DTT, 50 μg/ml PMSF) was added, and cells were further sonicated 3 × 30 seconds on ice. Then, the lysate was cleared by 20 min centrifugation at 15,000 rpm, +4 °C, and loaded onto cAMP-agarose (Sigma). Then, the protocol described in15,19 was applied. Final yield of purified native CRP was ~20 mg of protein for 200 ml culture. All steps of purification are illustrated in Supplementary Fig. S1.
Promoter mapping
Four intergenic regions with lengths allowing for promoter or transcription factor site mapping, e.g., exceeding 40–50 nucleotides, were selected for genes of the yih cassettes from the Escherichia coli, Enterobacter cloacae, Salmonella enterica, Cronobacter turicensis, and Pantoea anantis (namely, the yihR/S, yihT/U, yihU/V, and yihV/W regions). Regions were then extended in both directions by 100 nucleotides allowing for possible errors in the original open reading frame assignments. The resulting nucleotide sequences were obtained from GenBank45 and aligned using the T-Coffee multiple alignment tool46. Promoter-like motifs in the E. coli K-12 MG1655 genome (U00096.2) were found by the software PlatProm30 and its unified version PlatPromU47 where the impact of a sigma factor in promoter recognition by RNAP was depleted. Transcription starts were localized by band-shift assays with σ70-RNA polymerase, single-round transcription in vitro as previously described48 and by measuring the lengths of primer extension products in vivo. For primer extension, cells were grown in MS in the presence of 10% LB and 0.2% glucose, lactose, or glycerol, and harvested at OD650 = 0.4. RNA was extracted from 10 ml of cell culture with TRIzol reagent (Invitrogen, USA) and treated with DNAse I (New England Biolabs, USA) following the manufacturers’ instructions. 10 µg of total RNA was incubated with 4 pmol of the [γ-32P]ATP-labeled appropriate primer (Supplementary Table S2) and SuperScript II reverse transcriptase (Invitrogen, USA) according to the manufacturer’s protocol. The resulting samples were preheated and loaded onto a 6% denaturing polyacrylamide gel calibrated with [γ-32P]ATP-labeled 50 bp DNA ladder (New England Biolabs, USA). Gels were scanned using PMI Molecular Imager (Bio-Rad, USA) and autoradiographed.
Search for transcription factor binding sites
Candidate binding sites for transcription factors were identified in the intergenic regions using phylogenetic footprinting and the Virtual Footprint database49, with respect to the mapped transcription start sites, within the region −250/+50. The resulting sites were ranked according to their scores and conservation among the Enterobacteriaceae species. Highly conserved sites with best scores were retained for further analysis.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSA, band-shift assays) were used to test the efficiency of the RNA polymerase binding to the regulatory regions of the genes belonging to the yih cassette. DNA fragments containing potential promoter regions of separate genes were PCR amplified (primers marked as R and F for each gene, Supplementary Table S2), extracted from the gel and used for subsequent experiments. One pmol of DNA was incubated at 37 °C in 1× Transcription buffer50, for 10 min, then 1 to 4 pmol of purified σ70-RNA polymerase (obtained as described in ref.50) was added. After further 30 min at 37 °C, complexes were loaded onto a 5% polyacrylamide gel that had been pre-warmed to 37 °C. Protein complexes were separated from free DNA by electrophoresis in 1xTBE buffer. The DNA fragment containing known σ70-promoter of the hns gene amplified with primers hns_Bgl_263 and hns_Xba (Table S2) was used as a positive control; intergenic region within hns (primers RT-PCR) where no promoters were predicted was used as a negative control.
The same approach, but with cell lysate with the overproduced protein, was used to test the ability of cAMP-CRP to bind the regulatory regions of the yih genes. This approach allowed us to skip addition of extracellular cAMP in the sample in vitro. CRP was produced in BL21*(DE3) cells, then cells (10 ml) were harvested, washed and resuspended in 3 ml of 0.5× Transcription buffer with 10 mM PMSF. Cell lysate was prepared using standard 3 × 30 seconds rounds of sonication, and a total protein concentration was determined using the Bradford reagent (Sigma, USA). Approximate molar concentration of CRP in the lysate was calculated, and 1–8 molar excess of the protein was used in band-shifts.
To estimate the efficiency of binding, free DNA fragments and proteins were loaded on separate lanes. Gels were stained by ethidium bromide to reveal bands containing DNA. Western blotting was used for specific detection of CRP in the complexes. In brief, band-shifts were performed as above in duplicate, one of which was stained with ethidium bromide and the other one was transferred onto a PVDF membrane (Immobilon, Sigma-Aldrich, USA) using the Mini trans-blot protocol (Bio-rad, USA). The membrane was then blocked with 0.5% skimmed milk (Oxoid, UK) and incubated with anti-CRP (T14) antibodies (Cell Signalling, USA) in Tris-buffered saline (TBS) supplemented with 0.1% Tween-20 and 0.5% skimmed milk for 2 hours at 37 °C. After 3 × 10 min washes with TBS-T, secondary antibodies (A3687, anti-rabbit IgG, Sigma, USA) were added and incubation was allowed for 1 hour at room temperature. Membranes were stained with Western-blue stabilized substrate for alkaline phosphatase (Promega, USA) and scanned. To prove specific binding of CRP, band-shifts were also made with the purified protein with or without addition of cAMP. Band-shifts were made exactly as above, but 200 μM cAMP was added to the samples, to the gel and the running 1xTBE buffer15, and 20 μg/ml heparin was added to some samples to prove or disprove binding specificity.
Quantitative PCR
DT-322 thermocycler (DNA-Technology, Russia) and SYBR Green I as a fluorescent dye (Invitrogen, USA) were used for quantitative PCR (qRT-PCR). Primers used for reverse transcription (−RT) and amplification (−PCR) are listed in Supplementary Table S2. No PCR products were detected in negative controls in the absence of reverse transcriptase. To avoid influence of the changed growth, both hns and ysaA-aRNA51 were used as controls. Data obtained from at least three biological samples and analyzed in three statistical replicates were calculated by the ΔΔCt method. The error bars indicate the standard deviations of the respective mean values.
Availability of data and material
Data generated or analyzed during this study are included in the present manuscript, additional datasets analyzed during the current study are available from the corresponding author by request. All strains and plasmids are also available by request.
Software used for figure preparation
Schemes in Figs 1, 2 and 8 were prepared with Libre Office Draw and Multiple Align Show (http://www.bioinformatics.org/sms/multi_align.html). Graph in Fig. 3a was first built in SigmaPlot 8 and finalized in Adobe Photoshop CS5. Gels shown in Figs 3b, 5a–c were photographed, converted to a greyscale, and the text was added in Adobe Photoshop CS5. Images shown in Fig. 3c were taken from the PMI Molecular Imager, then resolution was increased and the text was added in Adobe Photoshop CS5. Graphs for Figs 4, 6 and 7c were prepared in Microsoft Excel 2003, for Fig. 7a,b – in SigmaPlot 8. Headings, legends and axes legends were then unified using Adobe Photoshop CS5.
Electronic supplementary material
Acknowledgements
The project was partially performed at the Summer Schools of Molecular and Theoretical Biology for high school students, supported by the Dynasty Foundation and the Zimin Foundation. This study was supported by the Russian Science Foundation under grant 14-50-00150. CRP purification was made as the part of the Russian Foundation for Basic Research grant 15-04-08716.
Author Contributions
A.Kaz. and M.S.G. conceived the project; A.Kaz. performed bioinformatic analysis with A.E., E.B. and P.S.; M.T. and U.S. conceived the experiments; U.S., E.B., D.B., V.E. and A.Kor. performed the experiments. A.Kaz. and M.T. wrote the manuscript. A.Kaz., M.T. and M.S.G. reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21534-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stülke J, Hillen W. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften. 1998;85:583–592. doi: 10.1007/s001140050555. [DOI] [PubMed] [Google Scholar]
- 2.Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. doi: 10.1023/A:1020628909429. [DOI] [PubMed] [Google Scholar]
- 3.Chen I-MA, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng X, Su H, Mi S, Han Y. A multifunctional thermophilic glycoside hydrolase from Caldicellulosiruptor owensensis with potential applications in production of biofuels and biochemicals. Biotechnol. Biofuels. 2016;9:98. doi: 10.1186/s13068-016-0509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald LC, Berger BW. Insight into the role of substrate-binding residues in conferring substrate specificity for the multifunctional polysaccharide lyase Smlt1473. J. Biol. Chem. 2014;289:18022–18032. doi: 10.1074/jbc.M114.571299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodionova IA, et al. Diversity and versatility of the Thermotoga maritima sugar kinome. J. Bacteriol. 2012;194:5552–5563. doi: 10.1128/JB.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PloS One. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lulko AT, Buist G, Kok J, Kuipers OP. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 2007;12:82–95. doi: 10.1159/000096463. [DOI] [PubMed] [Google Scholar]
- 9.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 10.Bernhardt J, Weibezahn J, Scharf C, Hecker M. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 2003;13:224–237. doi: 10.1101/gr.905003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang D-E, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mironov AA, Koonin EV, Roytberg MA, Gelfand MS. Computer analysis of transcription regulatory patterns in completely sequenced bacterial genomes. Nucleic Acids Res. 1999;27:2981–2989. doi: 10.1093/nar/27.14.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aidelberg, G. et al. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst. Biol. 8 (2014). [DOI] [PMC free article] [PubMed]
- 14.Bren A, et al. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 2016;6:24834. doi: 10.1038/srep24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 16.Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee DJ, Busby SJW. Repression by cyclic AMP receptor protein at a distance. mBio. 2012;3:e00289–212. doi: 10.1128/mBio.00289-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano M, Ogasawara H, Shimada T, Yamamoto K, Ishihama A. Involvement of cAMP-CRP in transcription activation and repression of the pck gene encoding PEP carboxykinase, the key enzyme of gluconeogenesis. FEMS Microbiol. Lett. 2014;355:93–99. doi: 10.1111/1574-6968.12466. [DOI] [PubMed] [Google Scholar]
- 19.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 20.Khoroshkin MS, Leyn SA, Van Sinderen D, Rodionov DA. Transcriptional regulation of carbohydrate utilization pathways in the Bifidobacterium genus. Front. Microbiol. 2016;7:120. doi: 10.3389/fmicb.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob F, Perrin D, Sanchez C, Monod J. Operon: a group of genes with the expression coordinated by an operator. Comptes Rendus Hebd. Seances Acad. Sci. 1960;250:1727–1729. [PubMed] [Google Scholar]
- 22.Denger K, et al. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature. 2014;507:114–117. doi: 10.1038/nature12947. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, et al. Genome evolution reveals biochemical networks and functional modules. Proc. Natl. Acad. Sci. USA. 2003;100:15428–15433. doi: 10.1073/pnas.2136809100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity determining residues. Genome Biol. 2002;3:PREPRINT0002. doi: 10.1186/gb-2002-3-3-preprint0002. [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Hagelsieb G. The power of operon rearrangements for predicting functional associations. Comput. Struct. Biotechnol. J. 2015;13:402–406. doi: 10.1016/j.csbj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Pellegrini M, Eisenberg D. Detection of parallel functional modules by comparative analysis of genome sequences. Nat. Biotechnol. 2005;23:253–260. doi: 10.1038/nbt1065. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Vitkup D. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome Biol. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shavkunov KS, Masulis IS, Tutukina MN, Deev AA, Ozoline ON. Gains and unexpected lessons from genome-scale promoter mapping. Nucleic Acids Res. 2009;37:4919–4931. doi: 10.1093/nar/gkp490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dornenburg, J. E., Devita, A. M., Palumbo, M. J. & Wade, J. T. Widespread antisense transcription in Escherichia coli. mBio1 (2010). [DOI] [PMC free article] [PubMed]
- 32.Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014;12:647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- 33.Tutukina MN, Potapova AV, Cole JA, Ozoline ON. Control of hexuronate metabolism in Escherichia coli by the two interdependent regulators, ExuR and UxuR: derepression by heterodimer formation. Microbiology. 2016;162:1220–1231. doi: 10.1099/mic.0.000297. [DOI] [PubMed] [Google Scholar]
- 34.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 35.Busby SJW, Kotlarz D, Buc H. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J. Mol. Biol. 1983;167:259–274. doi: 10.1016/S0022-2836(83)80335-0. [DOI] [PubMed] [Google Scholar]
- 36.Hengstenberg W, Penberthy WK, Morse ML. Purification of the staphylococcal 6-phospho-beta-D-galactosidase. Eur. J. Biochem. 1970;14:27–32. doi: 10.1111/j.1432-1033.1970.tb00256.x. [DOI] [PubMed] [Google Scholar]
- 37.Hengstenberg W, Egan JB, Morse ML. Carbohydrate transport in Staphylococcus aureus. V. The accumulation of phosphorylated carbohydrate derivatives, and evidence for a new enzyme-splitting lactose phosphate. Proc. Natl. Acad. Sci. USA. 1967;58:274–279. doi: 10.1073/pnas.58.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bissett DL, Wenger WC, Anderson RL. Lactose and D-galactose metabolism in Staphylococcus aureus. II. Isomerization of D-galactose 6-phosphate to D-tagatose 6-phosphate by a specific D-galactose-6-phosphate isomerase. J. Biol. Chem. 1980;255:8740–8744. [PubMed] [Google Scholar]
- 39.Bissett DL, Anderson RL. Lactose and D-galactose metabolism in Staphylococcus aureus. III. Purification and properties of D-tagatose-6-phosphate kinase. J. Biol. Chem. 1980;255:8745–8749. [PubMed] [Google Scholar]
- 40.Bissett DL, Anderson RL. Lactose and D-galactose metabolism in Staphylococcus aureus. IV. Isolation and properties of a class I D-ketohexose-1,6-diphosphate aldolase that catalyzes the cleavage of D-tagatose 1,6-diphosphate. J. Biol. Chem. 1980;255:8750–8755. [PubMed] [Google Scholar]
- 41.Kaznadzey A, Shelyakin P, Gelfand MS. Sugar Lego: gene composition of bacterial carbohydrate metabolism genomic loci. Biol. Direct. 2017;12:28. doi: 10.1186/s13062-017-0200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blattner FR, et al. & other authors. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 44.Studier FW. Constructing T7 based protein expression system with reduced T7 RNA polymerase thus allow toxic proteins to be expressed in bacteria. The system is included in current version of BL21 strains from Invitrogen. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-Z. [DOI] [PubMed] [Google Scholar]
- 45.Benson DA, et al. GenBank. Nucleic Acids Res. 2014;42:D32–37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 47.Panyukov VV, Shavkunov KS, Masulis IS, Kiselev SS, Ozoline ON. Mixed promoter islands as genomic regions with specific structural and functional properties. Mat. Biolog. Bioinform. 2013;8:432–448. doi: 10.17537/2013.8.432. [DOI] [Google Scholar]
- 48.Ozoline ON, Fujita N, Ishihama A. Mode of DNA-protein interaction between the C-terminal domain of Escherichia coli RNA polymerase alpha subunit and T7D promoter UP element. Nucleic Acids Res. 2001;29:4909–4919. doi: 10.1093/nar/29.24.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Münch R, et al. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinforma. Oxf. Engl. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 50.Ozoline ON, Fujita N, Murakami K, Ishihama A. Monitoring of RNA polymerase-DNA UP element interaction by a fluorescent probe conjugated to alpha subunit. Eur. J. Biochem. 1998;253:371–381. doi: 10.1046/j.1432-1327.1998.2530371.x. [DOI] [PubMed] [Google Scholar]
- 51.Purtov YA, et al. Promoter islands as a platform for interaction with nucleoid proteins and transcription factors. J Bioinform Comput Biol. 2014;12:1441006. doi: 10.1142/S0219720014410066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.