Abstract
The nuclear receptor family of transcription factor proteins mediate endocrine function and play critical roles in development, physiology and pharmacology. Malfunctioning nuclear receptors are associated with several disease states. The functional activity of nuclear receptors is regulated by small molecular hormonal and synthetic molecules. Multiple sources of evidence have identified and distinguished between the different allosteric pathways initiated by ligands, DNA and cofactors such as co-activators and co-repressors. Also, these biophysical studies are attempting to determine how these pathways that regulate co-activator and DNA recognition can control gene transcription. Thus, there is a growing interest in determining the genome-scale impact of allostery in nuclear receptors. Today, it is accepted that a detailed understanding of the allosteric regulatory pathways within the nuclear receptor molecular complex will enable the development of efficient drug therapies in the long term.
Keywords: allostery, nuclear receptor, transcription, drug design, endocrine, genome-scale
1. Introduction
Nuclear receptors (also nuclear hormone receptors and abbreviated as NR) are a family of transcription factors whose transcriptional activity is controlled by lipophilic hormone molecules and the ensuing recruitment of coactivator molecules (Evans, 1988; Evans & Mangelsdorf, 2014). There is a specific classification of nuclear receptors proposed by the International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification (NC-IUPHAR) (Alexander, et al., 2015; Auwerx, et al., 1999). It is based on a phylogenetic tree that connects all known nuclear receptor sequences. This nomenclature also accounts for the evolution of the two well-conserved domains of nuclear receptors (described below).
2. Nuclear receptors, human disease and pharmacology
Nuclear receptors are widely expressed in all human tissue (Bookout, et al., 2006; Kumar, et al., 2013; McKenna & O'Malley,). For instance, the peroxisome proliferator-activated receptor (PPAR) and liver X receptor (LXR) isoforms are expressed in tissues as diverse as the epidermis (Schmuth, Jiang, Dubrac, Elias, & Feingold, 2008) and in adipose tissue (Michalik, et al., 2006). The estrogen receptor (ER) and PPAR are also expressed in neural tissue (Couse, Lindzey, Grandien, Gustafsson, & Korach, 1997; Cullingford, et al., 1998) and ER is also expressed within the reproductive tract, hypothalamus and the lungs (Couse, et al., 1997). The constitutive androstane receptor (CAR) whose activity is linked to xeno and endobiotic metabolism (Huang, et al., 2003; Sonoda, Rosenfeld, Xu, Evans, & Xie, 2003; Sueyoshi & Negishi, 2001; Wei, Zhang, Egan-Hafley, Liang, & Moore, 2000; Zhang, Huang, Chua, Wei, & Moore, 2002; Zhang, Huang, Qatanani, Evans, & Moore, 2004) is most abundantly expressed in the liver and intestine (Bertilsson, et al., 1998; Lamba, et al., 2004) and is also expressed in the testis, adrenal tissue and the brain (Lamba, et al., 2004). Multiple reviews on the many disease states associated with nuclear receptor malfunction already exist (McKenna & O'Malley). For instance, the PPARs are associated with diseases as diverse as diabetes (Cipolletta, et al., 2012) and Alzheimer’s (Moutinho & Landreth, 2017; Prakash & Kumar, 2014). Signaling through the glucocorticoid receptor (GR) has been linked with cardiovascular disease, psychiatric disorders and hyperglycemia, among many other ailments (Kadmiel & Cidlowski, 2013). Also, nuclear receptors have been linked to the progression of multiple cancers (McKenna & O'Malley; Tang, et al., 2011), by the fatty liver disease and liver tumors by the farnesoid X (FXR) (Neuschwander-Tetri, et al., 2015) and the constitutive androstane receptors (CAR) (Yamamoto, Moore, Goldsworthy, Negishi, & Maronpot, 2004).
Consequently, nuclear receptors are vital targets of therapeutic drugs (Alexander, et al., 2015; Burris, et al., 2013; Evans & Mangelsdorf, 2014; Moore, Collins, & Pearce, 2006; Safe, Jin, Hedrick, Reeder, & Lee, 2014). Multiple small-molecule scaffolds have been designed as pharmaceutical nuclear receptor ligands that function as agonists or antagonists. These include therapeutic drugs such as bicalutamide that bind to the androgen receptor (AR) and target prostate cancer (Blackledge, 1996), tamoxifen for ERs (that target breast cancer) (Ward, 1973), thiazolidinediones against PPARγ that target type II diabetes (Lehmann, et al., 1995) and corticosteroids such as dexamethasone which targets the GR when treating ailments associated with inflammation (Madretsma, Dijk, Tak, Wilson, & Zijlstra, 1996).
3. Nuclear receptor structural topology, assembly and signaling
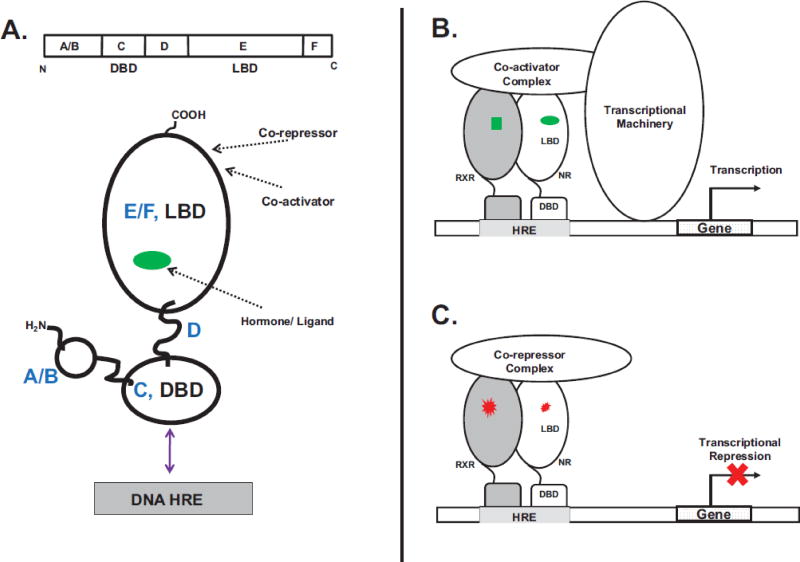
Nuclear receptors have common modular structural features that include an N-terminal domain (A/B domain, Figure 1A). This A/B domain is of variable length and amino acid sequence and is critical for regulating transactivation (Dieken & Miesfeld, 1992; Kato, et al., 1995; O'Malley, et al., 1995; Tora, et al., 1989; Werman, et al., 1997). With a few exceptions, the A/B domain encompasses a ligand-independent transactivation function (AF1) domain (Tora, et al., 1989; Tsai & O'Malley, 1994). Following the A/B domain is a highly conserved DNA-binding domain (DBD) (C domain, Figure 1B) that binds palindromic or direct repeat DNA sequences (six nucleotide segments of varied arrangements), or response elements (RE). A short ‘hinge’ sequence (D domain) connects the DBD to a C-terminal ligand-binding domain (LBD) (E & F domain, Figure 1A). Upon binding agonist-ligands the LBD undergoes conformational changes and recruits coactivator molecules to the ligand-dependent transactivation function (AF2) domain within the LBD (Suino, et al., 2004; Wright, et al., 2011; Wright, Vincent, & Fernandez, 2007; Xu, et al., 2004). Inverse agonists disrupt the ‘active’ AF2 conformation and the resulting LBD conformation functions as a docking site for co-repressors (Dussault, et al., 2002; Shan, et al., 2004).
Figure 1. Nuclear Receptor (NR) Mode of Action and Molecular Topology.
A. The nuclear receptor topology and functional organization consists of distinct N-terminal A/B, a DNA-binding (C, DBD), linker D and C-terminal ligand-binding (EF, LBD) domains. Arrows show locations of the binding sites for ligand, co-activators/co-repressors and the DNA HRE. B. Ligand agonists (green) interact with the receptor (heterodimer of nuclear receptor (NR):retinoid X receptor, (RXR)). Ligand binding is accompanied by the recruitment of co-activators and the basal transcriptional machinery. C. In the absence of agonists or when bound to antagonists (red) the nuclear receptor is maintained in an inactive transcriptional state by co-repressor molecules.
These receptors function as monomers, homodimers and as heterodimers, most commonly in a bimolecular complex with the nuclear receptor, the retinoid X receptor (RXR) (Auwerx, et al., 1999; Evans & Mangelsdorf, 2014; Kliewer, Umesono, Noonan, Heyman, & Evans, 1992; D. D. Moore, et al., 2006). When activated, nuclear receptors bind specific DNA sequences called hormone response elements (HRE) which are usually labeled to signify the activity-initiating hormone. For instance, the estrogen response elements (ERE) are HREs that bind the estrogen hormone receptor (ER) (Klock, Strahle, & Schutz, 1987), the androgen response elements (ARE) bind the androgen hormone receptor (AR) (Cato, Henderson, & Ponta, 1987), glucocorticoid response elements (GRE) bind the glucocorticoid receptor (Klock, et al., 1987), and the thyroid hormone response elements (TRE) bind the thyroid hormone receptor (Figure 1B) (Umesono, Giguere, Glass, Rosenfeld, & Evans, 1988), among others (Evans, 1988; Olefsky, 2001). This DNA/nuclear receptor complex recruits and binds to transcriptional coactivator proteins such as the steroid receptor coactivators (SRC) (A. B. Johnson & O'Malley, 2012), TIF-2/GRIP-1/NcoA-2 (transcriptional intermediary factor 2/glucocorticoid receptor interacting protein 1/nuclear receptor coactivator 2) (Min, Kemper, & Kemper, 2002), peroxisome proliferator-activated receptor ɣ coactivator 1 α (PGC-1α) (Ding, Lichti, Kim, Gonzalez, & Staudinger, 2006; Shiraki, Sakai, Kanaya, & Jingami, 2003), Activating signal cointegrator-2 (ASC-2) (Choi, et al., 2005) and others (Arnold, Eichelbaum, & Burk, 2004). Coactivator recruitment can be accompanied by histone acetylation, recruitment of the RNA polymerase II complex and gene expression (Dasgupta, Lonard, & O'Malley, 2014; Glass & Rosenfeld, 2000; Kamei, et al., 1996; Rastinejad, Huang, Chandra, & Khorasanizadeh, 2013; Yao, Ku, Zhou, Scully, & Livingston, 1996). Within the nucleus, the transcriptional activity of nuclear receptors can be maintained in a repressed state by antagonists and inverse agonists (Weatherman, Fletterick, & Scanlan, 1999) which promote the recruitment of transcriptional co-repressors (Lonard & O'Malley, 2012) such as the silencing mediator of retinoid and thyroid-hormone receptors (SMRT) (J. D. Chen & Evans, 1995; Sande & Privalsky, 1996) and the nuclear receptor co-repressor (NCoR) (Figure 1C) (Horlein, et al., 1995; Seol, Mahon, Lee, & Moore, 1996).
4. Noncanonical nuclear receptor signaling
Several additional mechanisms can also control the action of nuclear receptors and alter target gene expression. For instance, RNA-seq studies on CAR in hepatocyte-like (HepaRG) cell lines have shown distinct ligand (CITCO)-dependent and ligand-independent (phenobarbital, PB, activated) gene expression profiles (Li, et al., 2015; Mutoh, et al., 2009). Likewise, diverse mechanisms can control the DNA-binding-site specificity of nuclear receptors. Genome-scale studies with the estradiol (E2)-activated ER using ChIP-seq data in breast cancer tissue cell lines have been reported to bind multiple, non-overlapping ER-binding DNA sites (Welboren, Sweep, Span, & Stunnenberg, 2009; W. J. Welboren, et al., 2009). Also, different thiazolidinedione (TZDs)-agonists, the type 2 diabetes directed medications, that activate the PPARγ elicit overlapping but distinct in vivo gene expression profiles (Camp, et al., 2000; Lehmann, et al., 1995; Sears, et al., 2007). ChIP-seq studies using a border pattern-based motif recognition approach in multiple prostate cancer cell lines show that agonist dihydrotestosterone (DHT)-liganded human androgen receptor (AR) and antagonist bicalutamide and enzalutamide-liganded AR bind to distinctly different DNA ARE motifs (Z. Chen, et al., 2014). Furthermore, these motifs can be linked to distinct prostate cancer-relevant transcriptional outcomes.
Conversely, different DNA HRE sequences can also alternately activate or repress nuclear receptor transactivation. ‘Negative’ GREs are GR HREs that effectively repress transcription of agonist-liganded glucocorticoid (GR) (Surjit, et al., 2011). Indeed, these negative GREs promote the recruitment of transcriptional repression-associated SMRT and NCoR proteins to the negative GRE-GR(+agonist) molecular complex. Such negative HRE have also been reported to repress the thyroid receptors (TR) (Sharma, Thakran, Deng, Elam, & Park, 2013). By binding to TREs upstream, the agonist-ligand TRβ actively represses transcription of the secretory phospholipase A2 group IIa (PLA2g2a) gene. As with GR (Surjit, et al., 2011), the TRE-TR(+agonist) molecular complex also recruits co-repressor molecules SMRT and NCoR (Sharma, et al., 2013). Thus, noncanonical mechanisms can direct DNA-recognition for target gene selection by liganded nuclear receptors and a comprehensive analysis of these mechanisms will be essential to explain the overall in vivo significance of nuclear receptor ligands.
5. Allostery in biology
Allosteric coupling of distinct sites on proteins and DNA is fundamental to many biological processes (Monod, Changeux, & Jacob, 1963). Within nuclear receptors, allostery is increasingly recognized as a common regulatory process (Forman, Umesono, Chen, & Evans, 1995; Hilser & Thompson, 2011; Q. R. Johnson, Lindsay, Nellas, Fernandez, & Shen, 2015; Kojetin, et al., 2015; Mangelsdorf & Evans, 1995; Pavlin, Brunzelle, & Fernandez, 2014; Putcha, Wright, Brunzelle, & Fernandez, 2012; Shulman, Larson, Mangelsdorf, & Ranganathan, 2004; Wright, et al., 2011; Wright, et al., 2007). Structural and biophysical tools have shown that ligand binding and even minor perturbations (such as non-binding-site mutations) can be detected at distal regions of nuclear receptors. There are significant structural changes associated with allostery which are observed with crystallography (Osz, et al., 2012; Putcha, et al., 2012), hydrogen-deuterium exchange mass spectrometry (HDX MS) (Wright, et al., 2011) and NMR spectroscopy (Kojetin, et al., 2015).
6. Linking ligand and ligand through allostery
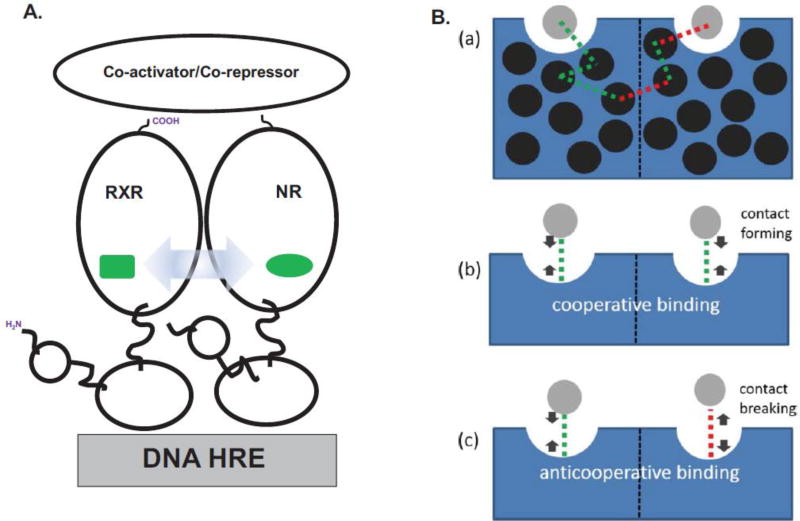
Since both receptors within nuclear receptor heterodimers can bind small-molecule agonist ligands in the simplest model for transactivation, agonist binding to either receptor can generate comparable transcriptional levels of downstream genes (Figure 1B) (Evans, 1988; Forman & Samuels, 1990). Such model systems are exemplified by the PPAR:RXR; CAR:RXR and LXR:RXR heterodimers (Clark, et al., 2016; Shulman, et al., 2004; Wright, et al., 2011). Yet, there are other nuclear receptor heterodimers that exhibit transcriptional responses that are distinct from this model (Forman, et al., 1995; Shulman, et al., 2004). For instance, transactivation by RAR:RXR, VDR:RXR and TR:RXR only occurs in the presence of the RAR, VDR and TR agonists, respectively. However, when these agonists are applied exogenously in combination with the agonist for the heterodimeric partner RXR (9-cis retinoic acid, 9c), transactivation levels are either enhanced, unaffected or are repressed, respectively (Forman, et al., 1995; Kurokawa, et al., 1994; Putcha, et al., 2012; Shulman, et al., 2004; Yao, et al., 1993). Such ligand-ligand allostery has been observed to link ligand bound to one nuclear receptor subunit to ligand and co-activator binding to the associated dimeric partner as observed in the RAR:RXR (Schulman, Li, Schwabe, & Evans, 1997) and TR:RXR (Putcha, et al., 2012) heterodimeric complexes (Figure 2A). These allostery-initiated conformational changes are significantly large and have easily identifiable local conformational pathways that can be characterized through multiply-liganded crystal structure analyses of the RAR:RXR and VDR:RXR heterodimers (Rochel, et al., 2011). With the TR:RXR heterodimer complex, the corresponding conformational changes are less obvious (Putcha, et al., 2012). The ‘frustrated fit’ mechanism is a recent approach to understanding how subtle conformational changes associated with ligand-ligand allostery are propagated throughout the molecular complex and across the heterodimer interface (Clark, et al., 2016; Q. R. Johnson, et al., 2015). The propagation of these allosteric signals can also be ligand-specific as observed in the murine CAR:RXR heterodimer with the agonists tcpobop and meclizine (Huang, Zhang, Wei, Schrader, & Moore, 2004; Wright, et al., 2011). Although these ligands elicit comparable transcriptional activity, studies with fluorescence spectroscopy and HDX MS show that they induce discrete conformational changes across the CAR:RXR dimer interface (Wright, et al., 2011).
Figure 2. Ligand-to-Ligand Allostery.
A. In nuclear receptor heterodimers, the conformational changes (blue arrow) induced within one receptor subunit upon binding its cognate ligand (green) are transmitted to the ligand-binding pocket of the second receptor (green). B. (a) The allosteric pathway from one ligand-binding pocket to the second includes the formation of both new interactions and the breaking of old interactions, as determined by the program CAMERRA (Q. R. Johnson, et al., 2015). (b) The result of these allosteric conformational changes are cooperative implying that the binding of ligand to one ligand-binding pocket can facilitate the binding of ligand to the second binding pocket in the nuclear receptor heterodimer. (c) Conversely, anticooperative binding occurs when the binding of ligand to one binding pocket diminishes the binding-affinity of the second binding pocket for ligand. Adapted with permission from (Clark, et al., 2016). Copyright (2016) American Chemical Society.
The immediate molecular consequence of this ligand-ligand allostery results in unique nuclear receptor heterodimer:co-activator molecular stoichiometries: 1:2 nuclear receptor↔co-activator stoichiometries for CAR:RXR (Pavlin, et al., 2014) and RARβ (Osz, et al., 2012) and ER homodimers (Yi, et al., 2015), reflecting the ligand response of these receptor systems, the 1:1 nuclear receptor↔co-activator stoichiometries for RAR:RXR and VDR:RXR (Rochel, et al., 2011) and the ‘phantom ligand effect’ where binding of ligand to RXR within the RAR:RXR heterodimer results in a linked conformational change within RAR (Schulman, et al., 1997).
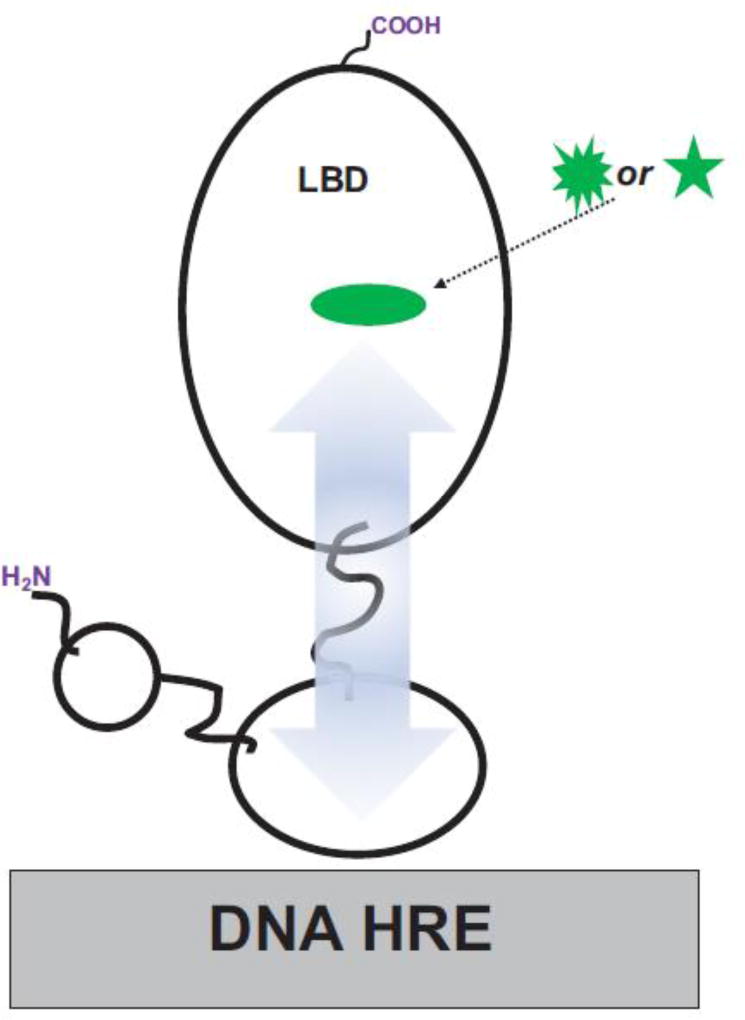
7. Linking ligand and DNA through allostery
Ligand-DNA allostery is a potent mechanism for ligand-dependent gene expression (Meijsing, Elbi, Luecke, Hager, & Yamamoto, 2007; Meijsing, et al., 2009). It has been shown that ligand binding can also affect the DNA-binding-site specificity of the nuclear receptor (ligand-DNA allostery) (Forman, et al., 1995) (Figure 3). For instance, the thyroid hormone, T3, has been reported to promote the binding of monomeric TRβ to TRE DNA (Ribeiro, Kushner, Apriletti, West, & Baxter, 1992). In these studies, T3 is also observed to increase the gel mobility of these TR(+T3) monomer-DNA complexes suggesting a ligand-induced conformational change in the TR(+T3) molecular complex. Furthermore, T3 appears to subtly change the specificity of TR to different TREs. In nuclear receptors, the DBD and LBD have been shown to interact directly, and structural changes in the LBD can influence DNA binding (Chandra, et al., 2008; Putcha & Fernandez, 2009). Through biochemical studies Rastinejad et al. have shown that a single point mutation within the PPARγ LBD can reduce the DNA-binding affinity of the PPARγ:RXRα heterodimer (Chandra, et al., 2008). Likewise, point mutations within the androgen receptor (AR) LBD – associated with androgen insensitivity syndromes – decrease the DNA-binding affinity of the AR homodimer, while leaving intact the ligand-binding affinity (Helsen, et al., 2012). Additionally, distinct ligand-dependent recognition patterns of promoter DNA by CAR have been identified (Cherian, Lin, Wu, & Chen, 2015; Hosoda, et al., 2015; Li, et al., 2015; H. Wang, et al., 2003).
Figure 3. Ligand-to-DNA Allostery.
Different conformational changes in the ligand-binding pocket induced by distinct ligands (green shapes) can lead to the binding of discrete DNA HRE sequences by the distal DBD through interdomain LBD↔DBD allosteric pathways (blue arrow).
Ligand identity can also affect nuclear receptor target gene expression and DNA binding in vivo, further suggesting that ligand molecules directly influence the DNA binding-site specificity of nuclear receptors, and not just the binding-affinity for DNA. For instance, a recently identified human CAR inverse agonist (CAR inhibitor not PXR activator 1, CINPA1) is observed to induce the dissociation of CAR from the promoter when used alone (Cherian, et al., 2015). It is speculated that this LBD-targeted inverse agonist also functions through allostery to induce conformational changes within the DBD that decreases the CAR-CARE binding affinity. In other studies, when the two ERα ligands E2 and 4-hydroxytamoxifen, 4-OHT are used in concert with different EREs there are notable differences in the sensitivity of each ER(+ligand)-ERE molecular complex to digestion by the protease chymotrypsin (Klinge, Jernigan, Smith, Tyulmenkov, & Kulakosky, 2001). Thus, different ERE-ligand combinations appear to induce distinct conformations in ERα. It is also reported in the study above that transcriptional activity correlates both, with distinct ligand-ERE combinations and with the ER-ERE binding affinity (Klinge, et al., 2001). Additionally and as noted above, multiple genome scale studies have shown that the TZD-ligand activation of PPARγ can elicit overlapping but discrete patterns of promoter binding and target gene expression (Camp, et al., 2000; Sears, et al., 2007). Similarly, different GR ligands such as the arylpyrazole compounds prednisolone and dexamethasone can induce different gene expression patterns and lead to distinct GR ChIP-seq-defined GRE-binding patterns (J. C. Wang, et al., 2006). Also, comparative studies by Wang et al. on AR show how different ligands (bound to the LBD) can result in the switching of DNA ARE motifs by the receptor DBD (Z. Chen, et al., 2014).
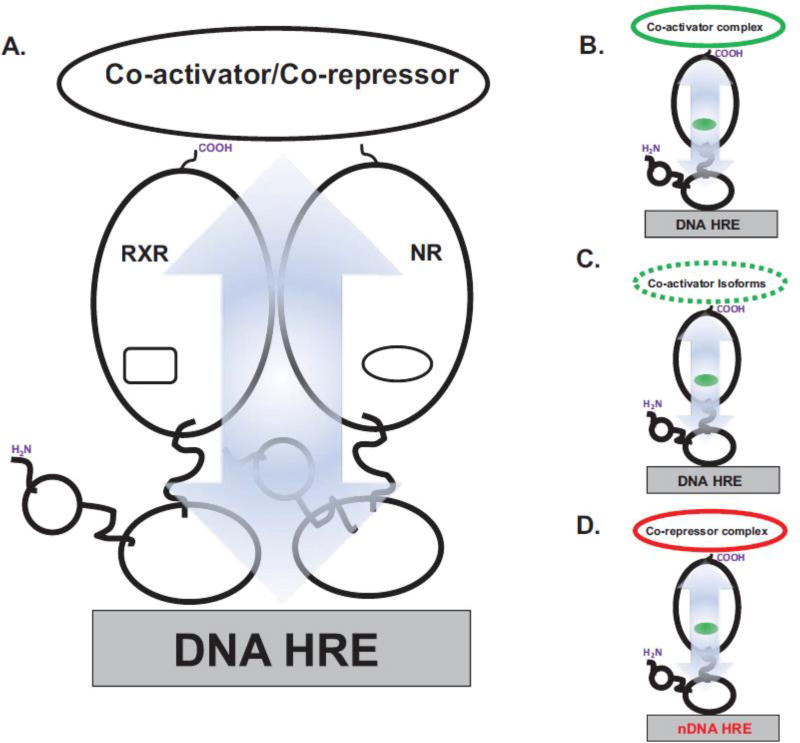
8. Linking co-activator/co-repressor and DNA through allostery
Conversely, DNA HREs can also function as allosteric regulators of co-activator and co-repressor recruitment by nuclear receptors (Figure 4A & 3B) (Gronemeyer & Bourguet, 2009; Putcha & Fernandez, 2009). Biophysical studies using isothermal titration calorimetry (ITC) show significant differences in both binding affinity, KD, and the thermodynamic binding parameters for SRC-derived LXXLL peptides by monomeric TR and TR:RXR heterodimers in the presence and absence of DNA TREs (Putcha & Fernandez, 2009). In separate studies, different DNA ERE sequences conferred distinct binding preferences for LXXLL-containing peptides to both ERα and ERβ (Hall, McDonnell, & Korach, 2002; Wood, Likhite, Loven, & Nardulli, 2001). Also, while the LXXLL peptides derived from SRC3/ACTR are recruited equally well by ERβ to four different EREs, analogous peptides from the co-activators SRC1 and GRIP1 show distinct preferences for different EREs (Figure 4C). Studies on GR have shown that by knocking down expression levels of the GR co-activators Brahma and the co-activator-associated arginine methyltransferase 1 (CARM1) co-activators the specific sequence of the DNA GRE is able to regulate GR transcriptional activity by specifically altering the molecular composition of the transcriptional co-activator complex (Meijsing, et al., 2009).
Figure 4. DNA-to-Co-activator/Co-repressor Allostery.
A. Allosteric pathways (blue arrow) link the DNA HRE-binding and co-activator/co-repressor-binding sites. B. Binding of DNA HRE can impact the binding-affinity of co-activators and conversely, the presence of co-activator protein can change the binding of the DNA HRE. C. DNA HREs can also control the specificity for distinct co-activator isoforms (broken line). D. Negative DNA HREs (nDNA HRE) overturn the ligand-agonist paradigm. Thus, nDNA HREs can promote the recruitment of transcriptional silencing co-repressor molecules to nuclear receptors bound to their cognate agonists.
Indeed, nuclear receptor structure is strongly affected by the presence and even sequence of the DNA response element, and this provides a mechanism for co-activator-DNA allosteric communication. The source of these differential interactions has been shown to derive from conformational changes within the DBD as specifically observed in structural analyses on GR bound to multiple GREs (Lefstin & Yamamoto, 1998; Meijsing, et al., 2009; Watson, et al., 2013). These studies by Yamamoto et al. show that the conformation of a ‘lever arm’ region in the DBD which is known to regulate the transcriptional activity of GR, is affected by the GRE DNA sequence. NMR studies further reveal that the specificity of interactions with GRE bases affects the conformation of distal regions of the GR DBD (Watson, et al., 2013). It is likely that analogous conformational changes within the DBD are propagated to the DBD-LBD interface and may explain the DNA-dependent interactions between the DBD and LBD within TR (Putcha & Fernandez, 2009). Likewise, the propagation of such DNA-induced conformational changes within the vitamin D receptor (RXR:VDR) heterodimer are manifested as fluctuating structural dynamics of the co-activator binding surfaces in the DNA-bound RXR:VDR which are initiated by distinct sequences of the DNA response element (Zhang, et al., 2011).
Remarkably, the base composition of the DNA HRE can also reverse the canonical role of ligands (Figure 4D). As noted above, GR can also mediate gene repression by recruiting transcriptional repressors to specific negative GREs that differ in sequence from canonical, activating GREs (Surjit, et al., 2011). Similar studies on the TR agonist-ligand triiodothyronine (T3) response element (TRE) within the promoter of the secretory phospholipase A2 group IIa (PLA2g2a) gene suggest that this promoter sequence functions as a negative regulator of T3 agonist-bound TRβ (Sharma, et al., 2013). Furthermore, when associated with the PLA2g2a promoter, the TRβ(+T3) complex actively recruits co-repressors to inhibit PLA2g2a expression.
9. Linking the N-terminal A/B domain and DNA through allostery
The N-terminal domain of nuclear receptors is the least understood (Figure 1A). This N-terminal A/B region is diverse in size, sequence and is conformationally malleable suggesting that this domain plays a role in conferring cell type and/or promoter specificity (Hill, Roemer, Churchill, & Edwards, 2012; Lavery & McEwan, 2005; Uversky, Oldfield, & Dunker, 2005; Warnmark, Treuter, Wright, & Gustafsson, 2003). Often, nuclear receptors differ most significantly in the amino-acid composition of the N-terminal A/B domain, implying that this region may play a significant role in mediating different effects of these receptors (Baniahmad, et al., 1993; Evans & Mangelsdorf, 2014; Hadzic, et al., 1995; McEwan & Gustafsson, 1997; Tian, Mahajan, Wong, Habeos, & Samuels, 2006; Tomura, Lazar, Phyillaier, & Nikodem, 1995). The absence of 3-dimensional data on the atomic resolution structure of any nuclear receptor A/B domain has necessitated a broad reliance on the use of spectroscopy and other biophysical tools to infer conformational changes within this domain. Multiple lines of evidence suggest that the nuclear receptor A/B domains are flexible and can adopt distinct conformations through allostery initiated by DNA:DBD interactions (D. L. Bain, Franden, McManaman, Takimoto, & Horwitz, 2000; David L. Bain, Franden, McManaman, Takimoto, & Horwitz, 2001; Baskakov, et al., 1999; Brodie & McEwan, 2005; Connaghan-Jones, Heneghan, Miura, & Bain, 2007; Fernandez, Gahlot, Rodriguez, & Amburn, 2017; Kumar, Lee, Bolen, & Thompson, 2001; Kumar, et al., 2013; Lavery & McEwan, 2005; McEwan, Lavery, Fischer, & Watt, 2007; Reid, Kelly, Watt, Price, & McEwan, 2002; Simons, Edwards, & Kumar, 2014). A common observation is that the A/B domains in all nuclear receptors studied to date, the DNA-initiated allostery elicits a conformational change in the N-terminal A/B domain (Figure 5). This structural flexibility has been proposed to enable the A/B domains to achieve multiple inter-molecular interactions have also been observed for AR (Lavery & McEwan, 2008), GR (Ford, McEwan, Wright, & Gustafsson, 1997; Khan, et al., 2012), PR (Hill, et al., 2012) and TR (Fondell, Brunel, Hisatake, & Roeder, 1996; Fondell, Roy, & Roeder, 1993). Furthermore, the A/B domain observed to fine-tune DNA recognition is finely tuned by the domains flanking the DBD (C domain) (Fernandez, et al., 2017). Thus even subtle changes within these flanking domains (A/B or E/F domains) such as mutations (Helsen, et al., 2012) and molecular interactions with cellular factors (Putcha & Fernandez, 2009) or small-molecule ligands (Chandra, et al., 2008) can affect DNA binding.
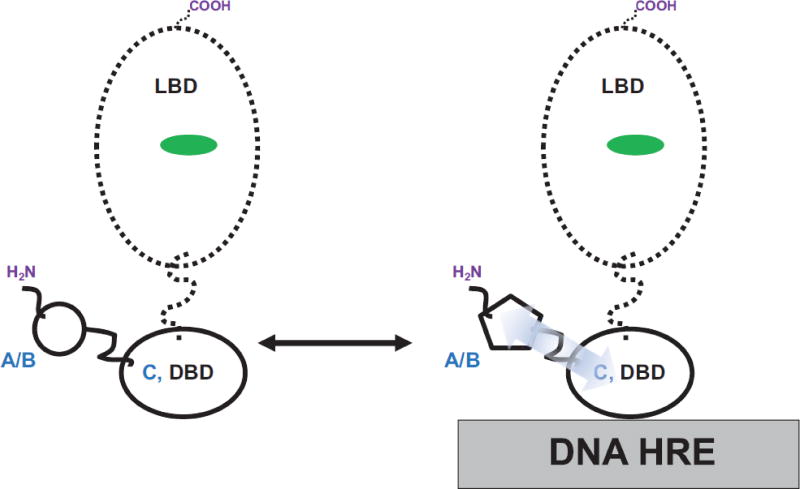
Figure 5. A/B domain-to-DNA Allostery.
Biophysical studies with isolated A/B+DBD and DBD-only domain constructs indicate that the A/B domain and DBD also communicate through allosteric pathways (blue arrow). The A/B domain is shown to influence DNA HRE binding to the DBD and in turn, the DNA HRE induces observable conformational changes within the A/B domain (note the different shapes of the A/B domain sketches: left (DNA-free) to right (DNA-bound)).
10. Conclusions and Outlook
Although significant advances have been made in our understanding of how these transcription factors respond to specific stimuli and regulate gene expression, critical details remain uncharacterized, such as, (1) how these nuclear receptors with overlapping DNA-binding specificity control specific gene transcription, and (2) how the finely-tuned gene-specific recruitment of distinct coregulatory molecules is achieved. Detailed structural analyses on multiple nuclear receptors that include the PPAR:RXR heterodimer (Chandra, et al., 2008), AR (Helsen, et al., 2012) and the GR homodimers (Meijsing, et al., 2009; Watson, et al., 2013) have already identified a few residues that transmit allosteric conformational changes. Already, the allosteric ‘BF-3’ surface site on the AR LBD has been targeted by transcriptional-activity modulating small molecules (Estebanez-Perpina, et al., 2007). However, at the genomic level, the role of ligand-to-DNA, DNA-to-co-activator and ligand-to-ligand allostery remains less characterized. Evidence strongly suggests that DNA HRE sequences and the ligand conformation have considerable long-range effects on nuclear receptor structure. Allosteric communication between co-activator-DNA-binding sites provides a link between target genes and distinct co-activators; however, studies have been restricted to only a few DNA sequences which have limited our understanding of the role of allostery on a genomic scale. Yet, its impact and prevalence in nuclear receptor signaling is of potentially substantial importance in nuclear receptor biology and pharmacology. Such studies will establish a functional connection between the nuclear receptor ligand, co-activators and DNA-binding sites. The genome-scale analysis of allosteric effects on gene transcription, combined with the proposed structural analysis of nuclear receptor-ligand interactions, will provide a unique opportunity for future drug-design studies and will explore how novel ligands will reshape nuclear receptor structure, HRE-DNA recognition and gene transcription. Also, these studies may inherently be collaborative to be performed in an iterative feed-back process where data from one study will inform towards designing more efficacious nuclear receptor modulators through rational ligand design. Comprehensive studies to predict residues that propagate intermolecular interactions across the heterodimer resulting in finely tuned activity states, novel high-throughput computational methods will need to be developed.
Acknowledgments
EJF is a recipient of funding from the NIH (GM123469)
Abbreviations
- 9c
9-cis retinoic acid
- AF1 (or 2)
activation function 1(or 2)
- AR
androgen receptor
- CAR
constitutive androstane receptor
- ChIP
chromatin immunoprecipitation
- DBD
DNA binding domain
- ER
estrogen receptor
- ERE
estrogen response elements
- GR
glucocorticoid receptor
- HRE
hormone response element
- LBD
ligand binding domain
- LBP
ligand binding pocket
- LXR
liver X receptor
- NCoR
nuclear receptor corepressor
- NR
nuclear receptor
- PPARɣ
Peroxisome proliferator-activated receptor ɣ
- RXR
retinoid X receptor
- SMRT
silencing mediator of retinoid and thyroid-hormone receptors
- SRC
steroid receptor coactivator
- T3
triiodothyronine
- TR
thyroid receptor
- TRE
thyroid receptor response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The author declares that there are no conflicts of interest.
Reference List
- Alexander SPH, Cidlowski JA, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Southan C, Davies JA, Collaborators, C The Concise Guide to PHARMACOLOGY 2015/16: Nuclear hormone receptors. British Journal of Pharmacology. 2015;172:5956–5978. doi: 10.1111/bph.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold KA, Eichelbaum M, Burk O. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept. 2004;2:1. doi: 10.1186/1478-1336-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Baulieu E, Beato M, Becker-Andre M, Burbach PH, Camerino G, Chambon P, Cooney A, Dejean A, Dreyer C, Evans RM, Gannon F, Giguere V, Gronemeyer H, Gustafson JA, Laudet V, Lazar MA, Mangelsdorf DJ, Milbrandt J, Milgrom E, Moore DD, O'Malley B, Parker M, Parker K, Perlmann T, Pfahl M, Rosenfeld MG, Samuels H, Schutz G, Sladek FM, Stunnenberg HG, Spedding M, Thummel C, Tsai MJ, Umesono K, Vennstrom B, Wahli W, Weinberger C, Willson TM, Yamamoto K, Comm NRN. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. J Biol Chem. 2000;275:7313–7320. doi: 10.1074/jbc.275.10.7313. [DOI] [PubMed] [Google Scholar]
- Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal Region of Human Progesterone B-receptors: BIOPHYSICAL AND BIOCHEMICAL COMPARISON TO A-RECEPTORS. Journal of Biological Chemistry. 2001;276:23825–23831. doi: 10.1074/jbc.M102611200. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O'Malley BW. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci U S A. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskakov IV, Kumar R, Srinivasan G, Ji YS, Bolen DW, Thompson EB. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem. 1999;274:10693–10696. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge GR. High-dose bicalutamide monotherapy for the treatment of prostate cancer. Urology. 1996;47:44–47. doi: 10.1016/s0090-4295(96)80008-2. discussion 48–53. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical Profiling of Nuclear Receptor Expression Reveals a Hierarchical Transcriptional Network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie J, McEwan IJ. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. J Mol Endocrinol. 2005;34:603–615. doi: 10.1677/jme.1.01723. [DOI] [PubMed] [Google Scholar]
- Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, Lundasen T, Hughes T, Kojetin DJ. Nuclear Receptors and Their Selective Pharmacologic Modulators. Pharmacological Reviews. 2013;65:710–778. doi: 10.1124/pr.112.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp HS, Li O, Wise SC, Hong YH, Frankowski CL, Shen X, Vanbogelen R, Leff T. Differential activation of peroxisome proliferator-activated receptor-gamma by troglitazone and rosiglitazone. Diabetes. 2000;49:539–547. doi: 10.2337/diabetes.49.4.539. [DOI] [PubMed] [Google Scholar]
- Cato ACB, Henderson D, Ponta H. The Hormone Response Element of the Mouse Mammary-Tumor Virus-DNA Mediates the Progestin and Androgen Induction of Transcription in the Proviral Long Terminal Repeat Region. Embo Journal. 1987;6:363–368. doi: 10.1002/j.1460-2075.1987.tb04763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lan X, Thomas-Ahner JM, Wu D, Liu X, Ye Z, Wang L, Sunkel B, Grenade C, Chen J, Zynger DL, Yan PS, Huang J, Nephew KP, Huang THM, Lin S, Clinton SK, Li W, Jin VX, Wang Q. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. The EMBO Journal. 2014 doi: 10.15252/embj.201490306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian MT, Lin W, Wu J, Chen T. CINPA1 is an inhibitor of constitutive androstane receptor that does not activate pregnane X receptor. Mol Pharmacol. 2015;87:878–889. doi: 10.1124/mol.115.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Lee S, Yeom SY, Kim GH, Lee JW, Kim SW. Characterization of activating signal cointegrator-2 as a novel transcriptional coactivator of the xenobiotic nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1711–1719. doi: 10.1210/me.2005-0066. [DOI] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AK, Wilder JH, Grayson AW, Johnson QR, Lindsay RJ, Nellas RB, Fernandez EJ, Shen T. The Promiscuity of Allosteric Regulation of Nuclear Receptors by Retinoid X Receptor. J Phys Chem B. 2016 doi: 10.1021/acs.jpcb.6b02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL. Thermodynamic analysis of progesterone receptor-promoter interactions reveals a molecular model for isoform-specific function. Proc Natl Acad Sci U S A. 2007;104:2187–2192. doi: 10.1073/pnas.0608848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, Lindzey J, Grandien K, Gustafsson J-Å, Korach KS. Tissue Distribution and Quantitative Analysis of Estrogen Receptor-α (ERα) and Estrogen Receptor-β (ERβ) Messenger Ribonucleic Acid in the Wild-Type and ERα-Knockout Mouse. Endocrinology. 1997;138:4613–4621. doi: 10.1210/endo.138.11.5496. [DOI] [PubMed] [Google Scholar]
- Cullingford TE, Bhakoo K, Peuchen S, Dolphin CT, Patel R, Clark JB. Distribution of mRNAs Encoding the Peroxisome Proliferator-Activated Receptor α, β, and γ and the Retinoid X Receptor α, β, and γ in Rat Central Nervous System. Journal of Neurochemistry. 1998;70:1366–1375. doi: 10.1046/j.1471-4159.1998.70041366.x. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O'Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieken ES, Miesfeld RL. Transcriptional transactivation functions localized to the glucocorticoid receptor N terminus are necessary for steroid induction of lymphocyte apoptosis. Mol Cell Biol. 1992;12:589–597. doi: 10.1128/mcb.12.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem. 2006;281:26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, Forman BM. A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol. 2002;22:5270–5280. doi: 10.1128/MCB.22.15.5270-5280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez EJ, Gahlot V, Rodriguez C, Amburn J. DNA-induced unfolding of the thyroid hormone receptor alpha A/B domain through allostery. FEBS Open Bio. 2017;7:854–864. doi: 10.1002/2211-5463.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Brunel F, Hisatake K, Roeder RG. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Roy AL, Roeder RG. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- Ford J, McEwan IJ, Wright AP, Gustafsson JA. Involvement of the transcription factor IID protein complex in gene activation by the N-terminal transactivation domain of the glucocorticoid receptor in vitro. Mol Endocrinol. 1997;11:1467–1475. doi: 10.1210/mend.11.10.9995. [DOI] [PubMed] [Google Scholar]
- Forman BM, Samuels HH. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol Endocrinol. 1990;4:1293–1301. doi: 10.1210/mend-4-9-1293. [DOI] [PubMed] [Google Scholar]
- Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gronemeyer H, Bourguet W. Allosteric effects govern nuclear receptor action: DNA appears as a player. Sci Signal. 2009;2:e34. doi: 10.1126/scisignal.273pe34. [DOI] [PubMed] [Google Scholar]
- Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka BM, Samuels HH. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor alpha is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- Helsen C, Dubois V, Verfaillie A, Young J, Trekels M, Vancraenenbroeck R, De Maeyer M, Claessens F. Evidence for DNA-binding domain--ligand-binding domain communications in the androgen receptor. Mol Cell Biol. 2012;32:3033–3043. doi: 10.1128/MCB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KK, Roemer SC, Churchill ME, Edwards DP. Structural and functional analysis of domains of the progesterone receptor. Mol Cell Endocrinol. 2012;348:418–429. doi: 10.1016/j.mce.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser VJ, Thompson EB. Structural dynamics, intrinsic disorder, and allostery in nuclear receptors as transcription factors. J Biol Chem. 2011;286:39675–39682. doi: 10.1074/jbc.R111.278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hosoda K, Kanno Y, Sato M, Inajima J, Inouye Y, Yanai K. Identification of CAR/RXRα Hetero-dimer Binding Sites in the Human Genome by a Modified Yeast One-Hybrid Assay. Advances in Biological Chemistry. 2015;5:83. [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol. 2004;18:2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- Johnson AB, O'Malley BW. Steroid receptor coactivators 1, 2, and 3: critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol Cell Endocrinol. 2012;348:430–439. doi: 10.1016/j.mce.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson QR, Lindsay RJ, Nellas RB, Fernandez EJ, Shen T. Mapping allostery through computational glycine scanning and correlation analysis of residue-residue contacts. Biochemistry. 2015;54:1534–1541. doi: 10.1021/bi501152d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends in Pharmacological Sciences. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Khan SH, Awasthi S, Guo C, Goswami D, Ling J, Griffin PR, Simons SS, Jr, Kumar R. Binding of the N-terminal region of coactivator TIF2 to the intrinsically disordered AF1 domain of the glucocorticoid receptor is accompanied by conformational reorganizations. J Biol Chem. 2012;287:44546–44560. doi: 10.1074/jbc.M112.411330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. Estrogen response element sequence impacts the conformation and transcriptional activity of estrogen receptor alpha. Mol Cell Endocrinol. 2001;174:151–166. doi: 10.1016/s0303-7207(01)00382-3. [DOI] [PubMed] [Google Scholar]
- Klock G, Strahle U, Schutz G. Oestrogen and glucocorticoid responsive elements are closely related but distinct. Nature. 1987;329:734–736. doi: 10.1038/329734a0. [DOI] [PubMed] [Google Scholar]
- Kojetin DJ, Matta-Camacho E, Hughes TS, Srinivasan S, Nwachukwu JC, Cavett V, Nowak J, Chalmers MJ, Marciano DP, Kamenecka TM, Shulman AI, Rance M, Griffin PR, Bruning JB, Nettles KW. Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat Commun. 2015;6 doi: 10.1038/ncomms9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Lee JC, Bolen DW, Thompson EB. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem. 2001;276:18146–18152. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- Kumar R, Moure CM, Khan SH, Callaway C, Grimm SL, Goswami D, Griffin PR, Edwards DP. Regulation of the structurally dynamic N-terminal domain of progesterone receptor by protein-induced folding. J Biol Chem. 2013;288:30285–30299. doi: 10.1074/jbc.M113.491787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther. 2004;311:811–821. doi: 10.1124/jpet.104.069310. [DOI] [PubMed] [Google Scholar]
- Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery DN, McEwan IJ. Functional characterization of the native NH2-terminal transactivation domain of the human androgen receptor: binding kinetics for interactions with TFIIF and SRC-1a. Biochemistry. 2008;47:3352–3359. doi: 10.1021/bi702220p. [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Li D, Mackowiak B, Brayman TG, Mitchell M, Zhang L, Huang SM, Wang H. Genome-wide analysis of human constitutive androstane receptor (CAR) transcriptome in wild-type and CAR-knockout HepaRG cells. Biochem Pharmacol. 2015 doi: 10.1016/j.bcp.2015.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madretsma GS, Dijk AP, Tak CJ, Wilson JH, Zijlstra FJ. Inhibition of the production of mediators of inflammation by corticosteroids is a glucocorticoid receptor-mediated process. Mediators Inflamm. 1996;5:100–103. doi: 10.1155/S0962935196000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- McEwan IJ, Gustafsson J. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci U S A. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal. 2007;5:e001. doi: 10.1621/nrs.05001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. SnapShot: Nuclear Receptors I. Cell. 142:822–822. e821. doi: 10.1016/j.cell.2010.08.026. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. SnapShot: Nuclear Receptors II. Cell. 142:986–986. e981. doi: 10.1016/j.cell.2010.08.041. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Elbi C, Luecke HF, Hager GL, Yamamoto KR. The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release. Mol Cell Biol. 2007;27:2442–2451. doi: 10.1128/MCB.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CNA, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W. International Union of Pharmacology. LXI. Peroxisome Proliferator-Activated Receptors. Pharmacological Reviews. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- Min G, Kemper JK, Kemper B. Glucocorticoid receptor-interacting protein 1 mediates ligand-independent nuclear translocation and activation of constitutive androstane receptor in vivo. J Biol Chem. 2002;277:26356–26363. doi: 10.1074/jbc.M200051200. [DOI] [PubMed] [Google Scholar]
- Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- Moore DD, Kato S, Xie W, Mangelsdorf DJ, Schmidt DR, Xiao R, Kliewer SA. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor beta, liver X receptor alpha, liver X receptor beta, and vitamin D receptor. Pharmacol Rev. 2006;58:742–759. doi: 10.1124/pr.58.4.6. [DOI] [PubMed] [Google Scholar]
- Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. Chem Med Chem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- Moutinho M, Landreth GE. Therapeutic potential of nuclear receptor agonists in Alzheimer's disease. J Lipid Res. 2017 doi: 10.1194/jlr.R075556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3) J Biol Chem. 2009;284:34785–34792. doi: 10.1074/jbc.M109.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. The Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW, Schrader WT, Mani S, Smith C, Weigel NL, Conneely OM, Clark JH. An alternative ligand-independent pathway for activation of steroid receptors. Recent Prog Horm Res. 1995;50:333–347. doi: 10.1016/b978-0-12-571150-0.50020-2. [DOI] [PubMed] [Google Scholar]
- Olefsky JM. Nuclear receptor minireview series. J Biol Chem. 2001;276:36863–36864. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- Osz J, Brélivet Y, Peluso-Iltis C, Cura V, Eiler S, Ruff M, Bourguet W, Rochel N, Moras D. Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proceedings of the National Academy of Sciences. 2012;109:E588–E594. doi: 10.1073/pnas.1118192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlin MR, Brunzelle JS, Fernandez EJ. Agonist Ligands Mediate the Transcriptional Response of Nuclear Receptor Heterodimers through Distinct Stoichiometric Assemblies with Coactivators. J Biol Chem. 2014;289:24771–24778. doi: 10.1074/jbc.M114.575423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Kumar A. Role of Nuclear Receptor on Regulation of BDNF and Neuroinflammation in Hippocampus of β-Amyloid Animal Model of Alzheimer’s Disease. Neurotoxicity Research. 2014;25:335–347. doi: 10.1007/s12640-013-9437-9. [DOI] [PubMed] [Google Scholar]
- Putcha BD, Fernandez EJ. Direct interdomain interactions can mediate allosterism in the thyroid receptor. J Biol Chem. 2009;284:22517–22524. doi: 10.1074/jbc.M109.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcha BD, Wright E, Brunzelle JS, Fernandez EJ. Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers. Proc Natl Acad Sci U S A. 2012;109:6084–6087. doi: 10.1073/pnas.1119852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Huang P, Chandra V, Khorasanizadeh S. Understanding nuclear receptor form and function using structural biology. J Mol Endocrinol. 2013;51:T1–T21. doi: 10.1530/JME-13-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002;277:20079–20086. doi: 10.1074/jbc.M201003200. [DOI] [PubMed] [Google Scholar]
- Ribeiro RC, Kushner PJ, Apriletti JW, West BL, Baxter JD. Thyroid hormone alters in vitro DNA binding of monomers and dimers of thyroid hormone receptors. Mol Endocrinol. 1992;6:1142–1152. doi: 10.1210/mend.6.7.1508227. [DOI] [PubMed] [Google Scholar]
- Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, Svergun DI, Moras D. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- Safe S, Jin U-H, Hedrick E, Reeder A, Lee S-O. Minireview: Role Of Orphan Nuclear Receptors in Cancer and Potential as Drug Targets. Molecular Endocrinology. 2014;28:157–172. doi: 10.1210/me.2013-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sande S, Privalsky ML. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- Schmuth M, Jiang YJ, Dubrac S, Elias PM, Feingold KR. Thematic review series: skin lipids. Peroxisome proliferator-activated receptors and liver X receptors in epidermal biology. J Lipid Res. 2008;49:499–509. doi: 10.1194/jlr.R800001-JLR200. [DOI] [PubMed] [Google Scholar]
- Schulman IG, Li C, Schwabe JW, Evans RM. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- Sears DD, Hsiao A, Ofrecio JM, Chapman J, He W, Olefsky JM. Selective modulation of promoter recruitment and transcriptional activity of PPARgamma. Biochem Biophys Res Commun. 2007;364:515–521. doi: 10.1016/j.bbrc.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol W, Mahon MJ, Lee YK, Moore DD. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- Shan L, Vincent J, Brunzelle JS, Dussault I, Lin M, Ianculescu I, Sherman MA, Forman BM, Fernandez EJ. Structure of the murine constitutive androstane receptor complexed to androstenol: a molecular basis for inverse agonism. Mol Cell. 2004;16:907–917. doi: 10.1016/j.molcel.2004.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Thakran S, Deng X, Elam MB, Park EA. Nuclear corepressors mediate the repression of phospholipase A2 group IIa gene transcription by thyroid hormone. J Biol Chem. 2013;288:16321–16333. doi: 10.1074/jbc.M112.445569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Sakai N, Kanaya E, Jingami H. Activation of orphan nuclear constitutive androstane receptor requires subnuclear targeting by peroxisome proliferator-activated receptor gamma coactivator-1 alpha. A possible link between xenobiotic response and nutritional state. J Biol Chem. 2003;278:11344–11350. doi: 10.1074/jbc.M212859200. [DOI] [PubMed] [Google Scholar]
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- Simons SS, Jr, Edwards DP, Kumar R. Minireview: dynamic structures of nuclear hormone receptors: new promises and challenges. Mol Endocrinol. 2014;28:173–182. doi: 10.1210/me.2013-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Rosenfeld JM, Xu L, Evans RM, Xie W. A nuclear receptor-mediated xenobiotic response and its implication in drug metabolism and host protection. Curr Drug Metab. 2003;4:59–72. doi: 10.2174/1389200033336739. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The Nuclear Xenobiotic Receptor CAR; Structural Determinants of Constitutive Activation and Heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Tang Q, Chen Y, Meyer C, Geistlinger T, Lupien M, Wang Q, Liu T, Zhang Y, Brown M, Liu XS. A Comprehensive View of Nuclear Receptor Cancer Cistromes. Cancer Research. 2011;71:6940. doi: 10.1158/0008-5472.CAN-11-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Mahajan MA, Wong CT, Habeos I, Samuels HH. The N-Terminal A/B domain of the thyroid hormone receptor-beta2 isoform influences ligand-dependent recruitment of coactivators to the ligand-binding domain. Mol Endocrinol. 2006;20:2036–2051. doi: 10.1210/me.2005-0437. [DOI] [PubMed] [Google Scholar]
- Tomura H, Lazar J, Phyillaier M, Nikodem VM. The N-terminal region (A/B) of rat thyroid hormone receptors alpha 1, beta 1, but not beta 2 contains a strong thyroid hormone-dependent transactivation function. Proc Natl Acad Sci U S A. 1995;92:5600–5604. doi: 10.1073/pnas.92.12.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Umesono K, Giguere V, Glass CK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- Wang JC, Shah N, Pantoja C, Meijsing SH, Ho JD, Scanlan TS, Yamamoto KR. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward HW. Anti-oestrogen therapy for breast cancer: a trial of tamoxifen at two dose levels. Br Med J. 1973;1:13–14. doi: 10.1136/bmj.1.5844.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- Watson LC, Kuchenbecker KM, Schiller BJ, Gross JD, Pufall MA, Yamamoto KR. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol. 2013;20:876–883. doi: 10.1038/nsmb.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherman RV, Fletterick RJ, Scanlan TS. Nuclear-receptor ligands and ligand-binding domains. Annu Rev Biochem. 1999;68:559–581. doi: 10.1146/annurev.biochem.68.1.559. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Welboren W-J, Sweep FCGJ, Span PN, Stunnenberg HG. Genomic actions of estrogen receptor α: what are the targets and how are they regulated? Endocrine-Related Cancer. 2009;16:1073–1089. doi: 10.1677/ERC-09-0086. [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. The EMBO Journal. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werman A, Hollenberg A, Solanes G, Bjørbæk C, Vidal-Puig AJ, Flier JS. Ligand-independent Activation Domain in the N Terminus of Peroxisome Proliferator-activated Receptor γ (PPARγ): DIFFERENTIAL ACTIVITY OF PPARγ1 AND-2 ISOFORMS AND INFLUENCE OF INSULIN. Journal of Biological Chemistry. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- Wright E, Busby SA, Wisecarver S, Vincent J, Griffin PR, Fernandez EJ. Helix 11 dynamics is critical for constitutive androstane receptor activity. Structure. 2011;19:37–44. doi: 10.1016/j.str.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Vincent J, Fernandez EJ. Thermodynamic characterization of the interaction between CAR-RXR and SRC-1 peptide by isothermal titration calorimetry. Biochemistry. 2007;46:862–870. doi: 10.1021/bi061627i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A Structural Basis for Constitutive Activity in the Human CAR/RXRalpha Heterodimer. Mol Cell. 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci U S A. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wang Z, Feng Q, Pintilie GD, Foulds CE, Lanz RB, Ludtke SJ, Schmid MF, Chiu W, O'Malley BW. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol Cell. 2015;57:1047–1058. doi: 10.1016/j.molcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]