Abstract
The bacterial strain TR3.2T was isolated from aerobic bioreactor-treated soil from a polycyclic aromatic hydrocarbon (PAH)-contaminated site in Salisbury, NC, USA. Strain TR3.2T was identified as a member of ‘Pyrene Group 2’ or ‘PG2’, a previously uncultivated cluster of organisms associated with the degradation of high-molecular-weight PAHs by stable-isotope probing. Based on its 16S rRNA gene sequence, the strain was classified as a member of the class Gammaproteobacteria but possessed only 90.5 % gene identity to its closest described relative, Methylococcus capsulatus strain Bath. Strain TR3.2T grew on the PAHs pyrene, phenanthrene, anthracene, benz[a]anthracene and fluorene, as well as the azaarene carbazole, and could additionally metabolize a limited number of organic acids. Optimal growth occurred aerobically under mesophilic temperature, neutral pH and low salinity conditions. Strain TR3.2T was catalase and oxidase positive. Predominant fatty acids were C17 : 0 cyclo and C16 : 0. Genomic G+C content of the single chromosome was 67.79 mol% as determined by complete genome sequencing. Due to the high sequence divergence from any cultivated species and its unique physiological properties compared to its closest relatives, strain TR3.2T is proposed as a representative of a novel order, family, genus and species within the class Gammaproteobacteria, for which the name Immundisolibacter cernigliae gen. nov., sp. nov. is proposed. The associated order and family are therefore proposed as Immundisolibacteralesord. nov. and Immundisolibacteraceaefam. nov. The type strain of the species is TR3.2T (=ATCC TSD-58T=DSM 103040T).
Keywords: Gammaproteobacteria, polycyclic aromatic hydrocarbons, Pyrene Group 2
A prior stable-isotope probing (SIP) experiment of polycyclic aromatic hydrocarbon (PAH)-contaminated, aerobic bioreactor-treated soil using the four-ring PAH pyrene identified several clusters of 16S rRNA genes derived from uncultivated Proteobacteria [1]. One group, designated ‘Pyrene Group 2’ or ‘PG2’, had no cultivated relatives and could only be classified to the class Gammaproteobacteria. Subsequent SIP experiments using pyrene, benz[a]anthracene or fluoranthene [2, 3] further implicated bacteria belonging to PG2 in the degradation of high-molecular-weight PAHs as well as the low-molecular-weight PAH phenanthrene [4] and potentially the low-molecular-weight PAH anthracene [5]. The 16S rRNA gene sequences comprising PG2 from these SIP projects possessed a relatively high (≥96 %) intragroup 16S rRNA gene identity. Other globally distributed environmental 16S rRNA gene sequences with high similarity to those designated as PG2 have been detected in hydrocarbon-impacted environments contaminated with oils [6–8], creosote [9], pyridine [10] and aliphatic compounds [11]. Other 16S rRNA gene sequences from PG2 have been recovered from natural asphalts [12] as well as pristine sediment [13] and water samples [14–16]. The most closely related isolate to organisms within PG2 was Methylococcus capsulatus strain Bath (~90 % 16S rRNA gene similarity) [17], a methanotroph isolated from soil [18].
A bacterial strain whose 16S rRNA gene sequence placed it phylogenetically within the cluster of SIP-derived PG2 sequences, designated TR3.2T, was successfully isolated from an aerobic bioreactor-treated, PAH-contaminated soil from the site of a former manufactured gas plant in Salisbury, NC, USA [19]. Strain TR3.2T was isolated through serial dilutions of bioreactor-treated soil onto a modified version of the aerobic cultivation medium for Sulfuritalea hydrogenivorans [20] using a layer of oversprayed pyrene as a carbon source; this medium was also used to obtain the PAH-degrading bacterium ‘Rugosibacter aromaticivorans’ Ca6 [21]. Prior isolation attempts may have been hindered by the lack of required trace minerals or vitamins in the medium in addition to the slow growth of the organism. Subsequent tests of strain TR3.2T grown in the liquid isolation medium excluding various components indicated that the following medium components were required for maximum growth: ‘reactor buffer’ (5 mM Na−K phosphate buffer, 5 mM NH4NO3, pH 7.0), 1 mM MgSO4, 1 mM CaCl2, 1 ml l−1 of a trace element solution [containing (per litre) 12.5 ml HCl (25 %), 2.1 g FeSO4.7H2O, 30 mg H3BO3, 100 mg MnCl2.4H2O, 190 mg CoCl2.6H2O, 24 mg NiCl2.6H2O, 2 mg CuCl2.2H2O, 144 mg ZnSO4.7H2O, 36 mg Na2MoO4.2H2O] and vitamin B12 solution (1 ml l−1; containing 5 mg cyanocobalamin in 100 ml distilled water, filter sterilized and added after autoclaving), to a final pH of 7.0. Carbon was added before autoclaving as either 0.02 % (w/v) pyrene dissolved in acetone to the flask, allowing the solvent to evaporate prior to adding other components, or as 0.2 % (w/v) sodium pyruvate. No growth at all occurred in the absence of MgSO4, CaCl2 or trace element solutions, and reduced growth rates and cell density compared to the complete growth medium were observed when vitamin B12 was omitted from the medium. All subsequent tests used these five components (referred to as ‘sRB2 medium’) with sodium pyruvate as a carbon source unless otherwise noted. For solid medium (‘sRB2-agar’), 1.5 % agar (Acros Organics) was added to liquid medium prior to autoclaving.
The optimal growth conditions of strain TR3.2T in sRB2 medium with pyruvate as a carbon source were determined for a range of temperatures, pH and salinities. Optimal growth was defined as the maximum growth rate during the exponential growth phase as measured by turbidity at OD600 with a HACH DR3000 spectrophotometer (Loveland). Optimal temperature for growth was determined in triplicate 5 ml cultures in liquid sRB2 medium at pH 7.0 and 225 r.p.m. for temperatures of 26, 28, 30, 32, 34, 35, 36 and 37 °C. Additional temperatures were evaluated on solid sRB2 plates at 20 and 4 °C. The effects of pH were tested in triplicate 5 ml tubes of sRB2 medium buffered to pH values of 5.0, 5.5, 6.0, 6.5 (buffered with MES, 50 mM), 7.0, 7.5, 8.0 (HEPES, 50 mM) and 8.5, 9.0 (Tris/HCl, 50 mM), which were incubated at 225 r.p.m. and 30 °C. Salinity effects were determined using triplicate 5 ml cultures of sRB2, pH 7.0, incubated at 30 °C and 225 r.p.m., amended with either 0, 0.25, 0.5, 1, 2, 3, 4 or 5 % (w/v) NaCl. The optimal temperature for growth was from 28 to 30 °C, with obvious growth observed between 20 and 36 °C. Strain TR3.2T grew between pH 6.5 and 8.0 with an optimum at 7.0. No growth was observed at salt concentrations >1 %, and strain TR3.2T had the highest growth rate with no amended NaCl in the medium. Under optimal conditions, inoculated tubes of strain TR3.2T grew to turbidity in liquid culture with constant shaking at 225 r.p.m. in approximately 6 days, with a doubling time of 25 h. Colonies on sRB2 plates with pyruvate as a carbon source developed more slowly over a period of 2 weeks when incubated at 30 °C.
The ability of TR3.2T to grow under anaerobic conditions was assessed using the GasPak 100 system containing a GasPak EZ Anaerobe Container System sachet (BD Biosciences). Plates containing sRB2-agar and pyruvate were inoculated and sealed in the system according to manufacturer’s directions. The system achieves an atmosphere of <1 % oxygen and ≥13 % carbon dioxide within 2.5 h. Anaerobic conditions after 48 h were confirmed by visual observation of an indicator strip soaked in 1 mM resazurin solution. Plates were incubated at 30 °C with concurrent aerobic controls. TR3.2T did not display growth in anaerobic conditions after 4 weeks. Unless otherwise noted, all subsequent tests of TR3.2T occurred at 30 °C, pH 7.0 and 0 % salinity under aerobic conditions. Liquid cultures were additionally incubated with constant shaking at 225 r.p.m.
The cellular morphology of strain TR3.2T was investigated using scanning electron microscopy. For the preparation of samples, cells were grown in liquid sRB2 with pyruvate as a carbon source. Specimens were observed and images taken using a Zeiss Supra 25 FESEM operating at 5 kV, 5 mm working distance and 10 µm aperture (Carl Zeiss SMT). Average sizes were determined from digital micrographs of single, well-distinguished cells. Strain TR3.2T cells were coccobacilli with an average size of 0.56±0.08 µm by 0.35±0.02 µm (n=100 cells). Cells appeared slightly wrinkled, generally as singles or doubles, with no obvious extracellular features (Fig. 1). Gram staining performed using the standard reaction and light microscopy indicated strain TR3.2T was Gram-type negative. Cellular motility was tested using sRB2-agar stab-tubes with agar added at 0.3 % (w/v) and pyruvate as the carbon source. No motility was observed in the stab-tubes, and no flagella were observed in micrographs. Log-phase cells grown in sRB2 stained with the Remel Flagella Stain (Thermo Scientific) and examined under oil-immersion light microscopy did not appear to have flagella. However, analysis of the genome of strain TR3.2T indicated a complete set of genes required for assembly of a flagellum [22]. Expression of flagella by strain TR3.2T therefore may be possible under some conditions, but was not observed under those tested here. Strain TR3.2T colonies on sRB2 plates with pyruvate were small (0.50–0.75 mm in diameter), circular, with a convex elevation and were yellow-orange in colour.
Fig. 1.
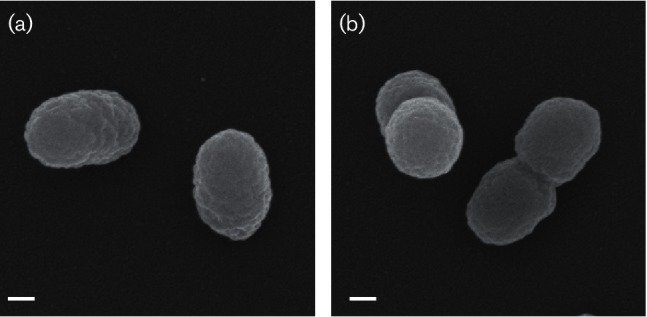
Scanning electron micrographs of strain TR3.2T grown on pyruvate either as (a) single or (b) double cells. Bars, 100 nm.
Metabolism of a variety of carbon substrates was tested using the Biolog GN2 Microplate. For each plate, cells were grown in liquid sRB2 medium amended with pyruvate and washed three times in PBS (pH 7.5) before being resuspended in Biolog GN/GP Inoculating Fluid as per the manufacturer’s instructions. The microplates were incubated overnight at 30 °C and scored through visual examination in comparison to the no-carbon control. In triplicate tests, the only substrates that strain TR3.2T actively metabolized were α-ketobutyric acid, methyl pyruvate, mono-methyl-succinate and α-ketovaleric acid. The other 91 substrates, including a variety of sugars, other organic acids, nucleosides and amino acids, were not metabolized.
Utilization of nitrogen sources was tested by substituting either 5 mM KNO3 or NH4Cl for NH4NO3 in sRB2 liquid medium. Strain TR3.2T displayed growth on pyruvate as a carbon source with both nitrogen sources tested. Nitrate reduction was evaluated using the culture grown in sRB2 medium substituted with 5 mM KNO3. No nitrate reduction to nitrite beyond what was required for assimilation and growth was observed. Starch hydrolysis and cellulase activity were evaluated by supplementing sRB2 plates with either starch (10 g l−1) or cellulose (1 g l−1) (adapted from Kasana et al. [23]). Protease activity was tested by aseptically adding a filter-sterilized skimmed milk solution (10 % skimmed milk powder dissolved in distilled water) to sRB2-agar after autoclaving. Plates were incubated at 30 °C for 4 weeks and monitored for zones of clearing; for starch and cellulose plates, Gram’s iodine was also added at the end of the incubation to help detect zones of clearing. Gelatin hydrolysis was assessed using a nutrient gelatin stab method wherein powdered gelatin (120 g l−1) was added to sRB2, gently heated to dissolve and aliquoted into 5 ml tubes. Gelatin tubes containing TR3.2T were incubated at 30 °C and monitored for liquefaction of the medium. Strain TR3.2T was negative for starch hydrolysis, cellulase, skimmed milk protease and gelatinase activities.
Strain TR3.2T was tested for catalase activity by adding 3 % (v/v) hydrogen peroxide solution to cells freshly scraped from the surface of a sRB2 plate. Oxidase activity was determined by adding a few drops of freshly prepared 1 % N′,N′,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (Acros Organics) to cells scraped from the same plate onto filter paper. Strain TR3.2T was catalase and oxidase positive.
Lipase activity was assessed by adding tributyrin (98 %; Acros Organics) to sRB2-agar (1 %, v/v). Urease activity was determined using a medium prepared with the sRB2 phosphate buffer with urea as a nitrogen source (20 g l−1) and phenol red (10 mg l−1) in addition to the standard concentrations of MgSO4, CaCl2, trace element solution and vitamin B12 solutions with pyruvate as a growth substrate. Strain TR3.2T was positive for lipase activity and negative for urease activity.
Indole production was tested by supplementing sRB2 medium with tryptone (10 g l−1). After growth to turbidity, a few drops of Kovac’s reagent were added to each tube. The lack of a colour change indicated no production of indole by strain TR3.2T. Production of indigo from exogenous indole was assessed by adding a crystal of indole to the lids of inverted Petri plates containing sRB2-pyruvate medium 24 h after inoculation. Indigo production from indole is indicative of activity for some ring-hydroxylating dioxygenases involved in the initial step in the aerobic degradation of PAHs [24, 25]. A purple colour developed by colonies on the plate after exposure to indole confirmed the likely presence of an active ring-hydroxylating dioxygenase.
Growth of strain TR3.2T on select PAHs as sole sources of carbon and energy was tested in sRB2 medium amended with individual PAHs (final concentration 0.2 g l−1). PAHs were added to tubes in either acetone or dichloromethane and the solvent allowed to evaporate prior to adding the remaining medium components. After autoclaving, the tubes were briefly placed in an ultrasonic water bath to disperse PAH crystals before washed TR3.2T cells grown in sRB2-pyruvate were inoculated in triplicate tubes for each PAH and incubated at 30 °C at 225 r.p.m. for up to 42 days. As spectrophotometric measurements of turbidity were not possible due to the presence of PAH crystals in the medium, growth of TR3.2T on PAHs was defined by both protein accumulation and disappearance of the parent compound. Protein concentrations were calculated from aliquots of culture approximately every 48 h using a Pierce BCA Protein Assay according to the manufacturer’s instructions (ThermoFisher Scientific). Disappearance of each PAH was determined using a liquid–liquid extraction with an equal volume of n-hexanes and quantification using a HPLC system with fluorescence detection as previously described [26] and compared to similarly extracted uninoculated tubes with PAH added. For all substrates except naphthalene, this method was able to quantify PAH concentration; naphthalene presumably volatilized from the tubes during either the incubation or extraction. Removal of pyrene (33 % of the initial mass), phenanthrene (47 %), anthracene (45 %), benz[a]anthracene (21 %) and fluorene (36 %) by TR3.2T was correlated with protein accumulation, indicating growth on those substrates. Incubation with chrysene resulted in PAH removal without protein accumulation, suggesting transformation but not growth on this substrate. TR3.2T was negative for growth on or transformation of benzo[a]pyrene, fluoranthene and naphthalene. Positive growth on the azaarene carbazole by strain TR3.2T was determined by visual turbidity and protein accumulation over the course of the incubation; the HPLC assay for quantifying PAHs was not suitable for this compound.
Mineralization of the partially 14C-labelled PAHs phenanthrene, fluoranthene, chrysene, benz[a]anthracene and benzo[a]pyrene was performed as previously described [27]. Strain TR3.2T cells grown in sRB2-pyrene and washed three times with reactor buffer were used to inoculate triplicate flasks. The extent of mineralization was measured after 24 h. Mineralization was assessed as percentage of added PAH mineralized and compared among biotic and acidified replicates. Statistically significant mineralization (P≤0.01) occurred for phenanthrene (56 % mineralization), benz[a]anthracene (29 %), benzo[a]pyrene (41 %) and chrysene (52 %), but not fluoranthene.
Growth on BTEX compounds was investigated by creating an atmosphere in separate, tightly sealed metal containers of either benzene (99 %), toluene (99.8 %), ethylbenzene (99.8 %) or xylenes [o-xylene (98.5 %), m-xylene (99 %), p-xylene (99 %), in equal volumes] by adding 0.5 ml of the chemical(s) to a piece of filter paper in a glass beaker. Inoculated sRB2-agar plates without amended carbon were incubated in sealed containers at room temperature for 1 month and compared to concurrent plates incubated outside the closed systems with and without additional carbon sources. Strain TR3.2T did not display growth on any of the tested BTEX compounds under these conditions.
Cellular fatty acid profiling was performed using the Sherlock Microbial Identification System by Microbial ID on cells freshly grown on sRB2 plates with pyruvate [28]. The dominant fatty acids in TR3.2T were C17 : 0 cyclo (32.1 %), C16 : 0 (29.6 %) and summed feature 3 (C16 : 1ω7c or C16 : 1ω6c) (12.0 %). TR3.2T also contained C19 : 0 cyclo ω8c (8.3 %), C12 : 0 3-OH (6.4 %), C12 : 0 (2.0 %), summed feature 8 (C18 : 1ω7c or C18 : 1ω6c) (1.9 %), summed feature 5 (C18 : 0 anteiso or C18 : 2ω6,9c) (1.6 %), C14 : 0 (1.4 %) and C10 : 0 (1.4 %). Trace amounts (<1 %) of C10 : 0 3-OH, C14 : 0 3-OH, C17 : 1ω7c, C16 : 0 3-OH, C18 : 0, C18 : 0ω7c 11-methyl, C19 : 0, C18 : 1 2-OH and C20 : 2ω6,9c were present. Analysis of respiratory quinones and polar lipids was carried out by the Identification Service, DSMZ, Braunschweig, Germany [29–31]. Major polar lipids in TR3.2T were diphosphatidylglycerol, phosphatidylmethylethanolamine, phosphatidylethanolamine, phosphatidylglycerol and phospholipids (Fig. S1, available in the online Supplementary Material). Also present in minor amounts were lipid, phosphoglycolipid and unidentified pigments. Respiratory quinones present were Q7 (9 %), Q8 (90 %) and Q9 (1 %).
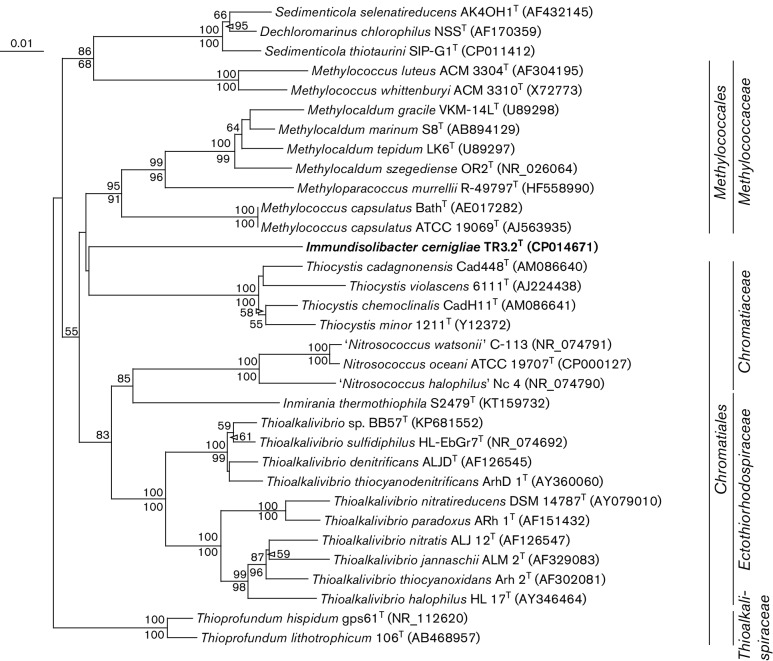
The genome of strain TR3.2T was acquired and annotated as described previously [22]. The genome was a singular, circular chromosome of 3 243 537 bp with a G+C content of 67.79 mol%. No extrachromosomal elements were detected. Annotation predicted 3114 total genes with 3054 protein coding genes, a single rRNA gene operon and 45 tRNA-encoding genes [22]. The complete 16S rRNA gene of strain TR3.2T was compared to publicly available sequences using blastn of the non-redundant GenBank database in separate searches including and excluding environmental sequences. Some of the closest described relatives to TR3.2T based on 16S rRNA gene similarity (Fig. 2) were M. capsulatus strain Bath (90.5 % similarity) [17], M. capsulatus strain ATCC 19069T (strain Texas; 90.2 %), ‘Thioalkalivibrio sulfidophilus’ strain HL-EbGr7 (90.1 %) [32] and Inmirania thermothiophila strain S2479T (90.3 %) [33]. An estimate of DNA–DNA hybridization using the complete genome of strain TR3.2T compared to those of M. capsulatus Bath and ‘Thioalkalivibrio sulfidophilus’ HL-EbGr7 with the genome-to-genome distance calculator [34] predicted a DNA–DNA hybridization value of 18.5 % to both organisms using the recommended formula. Based on the complete 16S rRNA gene, the naive Bayesian classifier of the Ribosomal Database Project [35] indicated that the highest taxonomic level that TR3.2T could reliably be grouped in with confidence (confidence threshold >80 %; training set 16, released June 2016) was the class Gammaproteobacteria. Within the Gammaproteobacteria, strain TR3.2T was equally dissimilar to described bacteria within the orders Methylococcales and Chromatiales. Bootstrapping with multiple algorithms did not reliably cluster strain TR3.2T with members of either order, so its phylogenetic position within the Gammaproteobacteria is uncertain.
Fig. 2.
16S rRNA gene neighbour-joining phylogenetic tree of strain TR3.2T with 16S rRNA gene sequences of the most similar cultivated and described strains. Sequences were aligned and the neighbour-joining tree created using clustalx 2.1 [46] and without considering positions with gaps. The maximum-parsimony tree was determined using the paup [47] implementation within Geneious 9.1.3 [48] and the same alignment file generated for the neighbour-joining tree. Bootstrap values for neighbour-joining and maximum-parsimony algorithms are presented as a percentage of 1000 iterations above and below nodes, respectively. GenBank accession numbers are presented in parentheses. The order and family of cultivated strains are indicated on the right side. Pseudomonas putida strain IAM 1236 (accession no. D84020) was used as an outgroup (not shown). Bar, 0.01 substitutions per nucleotide position.
Despite the lack of phylogenetic similarity to any characterized bacteria, large numbers of 16S rRNA gene sequences with high similarity (>97 %) to strain TR3.2T have been recovered from a variety of environmental sources (Fig. S2). Many of those studies, including those involving SIP of PAH-contaminated soils within our research group, investigated the diversity of communities in which the sample medium was impacted by petroleum or hydrocarbon contamination [7–9, 11, 12, 36]. The prevalence of environmental sequences closely related to strain TR3.2T in similarly contaminated but geographically separated samples suggests a potential widespread association of PG2-type organisms with the degradation of those compounds.
In addition to phylogenetic dissimilarity to the most closely related and characterized bacteria, strain TR3.2T was also physiologically differentiated from those organisms. Members of the order Methylococcales, which includes the genera Methylococcus, Methylocaldum and Methyloparacoccus, are obligate users of C1 compounds for carbon and energy [37, 38]. This is in stark contrast to the complex aromatics and organic acids that strain TR3.2T can use as growth substrates. Additionally, no obvious homologues of particulate methane monooxygenase genes, common among methanotrophic members of the Methylococcales, appear in the annotated TR3.2T genome. Strain TR3.2T is also physiologically distinct from genera within the order Chromatiales. Members of the genus Thioalkalivibrio are obligate chemolithoautotrophs and grow optimally at high pH [39]. While isolated under mesophilic and low salinity conditions, Thiocystis organisms grow optimally in anoxic or microoxic conditions either photolithotrophically or chemoautotrophically [40]. Inmirania thermothiophila is a facultatively anaerobic autotroph that grows optimally at high temperatures and salinity [33]. Members of the genus Nitrosococcus are marine, ammonia-oxidizing chemolithotrophs [41, 42]. While some Sedimenticola strains are capable of growth on aromatic compounds, they are also strict anaerobes that use selenium or nitrogen oxyanions as electron acceptors [43]. Thioprofundum species are thermophilic, piezophilic chemolithoautotrophs isolated from deep-sea hydrothermal vents [44, 45].
On the basis of distinct physiological, phylogenetic and environmental habitat differences to cultivated and characterized bacterial relatives, the novel genus and species Immundisolibacter cernigliae gen. nov., sp. nov. is proposed for this organism, with TR3.2T as the type strain. Due to the physiological and phylogenetic dissimilarity to organisms within described orders of the class Gammaproteobacteria, we also propose a novel order and family of which strain TR3.2T is currently the sole member, and would thus be designated Immundisolibacterales ord. nov. and Immundisolibacteraceae fam. nov., respectively.
Description of Immundisolibacter gen. nov.
Immundisolibacter (Im.mun.di.so.li.bac′ter. L. adj. immundus unclean, impure; L. n. solum soil; N.L. n. bacter rod; N.L. masc. n. Immundisolibacter a rod from unclean soil).
Cells are Gram-negative, non-motile and grow aerobically. Catalase and oxidase positive. Heterotrophic growth occurs on a limited number of organic acids. Predominant fatty acids are C17 : 0 cyclo, C16 : 0 and summed feature 3 (C16 : 1ω7c or C16 : 1ω6c). The major polar lipids are diphosphatidylglycerol, phosphatidylmethylethanolamine, phosphatidylethanolamine, phosphatidylglycerol and phospholipids. The major respiratory quinone is Q8. Phylogenetically, the genus is a member of the class Gammaproteobacteria. The type species is Immundisolibacter cernigliae.
Description of Immundisolibacter cernigliae sp. nov.
Immundisolibacter cernigliae (cer.nig′li.ae. N.L. gen. n. cernigliae named after the prominent researcher of PAH-degrading bacteria Carl Cerniglia).
Cells are coccobacilli approximately 0.56×0.35 µm. Colonies on solid medium are small, convex and circular with a yellow-orange colour. Growth occurs aerobically between 20 and 36 °C (optimum between 28 and 30 °C), pH 6.5–8.0 (optimum 7.0) and at salinity ≤1 % (optimum 0 %). Growth was not observed under anaerobic conditions. Cells can metabolize α-ketobutyric acid, methyl pyruvate, mono-methyl-succinate and α-ketovaleric acid and can grow on polycyclic aromatic compounds as sole sources of carbon, and both nitrate and ammonia as nitrogen sources. Cells are negative for gelatin hydrolysis, cellulase, skimmed milk protease, starch hydrolysis, dissimilatory nitrate reduction and urease, and positive for lipase activity. In addition to those listed in the genus description, fatty acids present in lesser amounts are C19 : 0 cyclo ω8c and C12 : 0 3-OH. In addition to the major respiratory quinone, Q7 and Q9 are also present. Polar lipids are consistent with the genus.
The type strain, TR3.2T (=ATCC TSD-58T=DSM 103040T), was isolated from PAH-contaminated soil from Salisbury, NC, USA. Genomic G+C content is 67.79 mol%.
Description of Immundisolibacteraceae fam. nov.
Immundisolibacteraceae (Im.mun.di.so.li.bac.ter.a.ce′ae. N.L. masc. n. Immundisolibacter type genus of the family; N.L. suff. -aceae ending denoting a family; N.L. fem. pl. n. Immundisolibacteraceae the family of Immundisolibacter).
Description is the same as for the genus Immundisolibacter. The type genus is Immundisolibacter.
Description of Immundisolibacterales ord. nov.
Immundisolibacterales (Im.mun.di.so.li.bac.ter.a′les. N.L. masc. n. Immundisolibacter the type genus of the order; N.L. suff. -ales ending denoting an order; N.L. fem. pl. n. Immundisolibacterales the order of Immundisolibacter).
Order of the class Gammaproteobacteria. Segregation of this genus into a new order is justified by its phylogenetic distance and physiological differences from organisms in the most closely related extant orders. Organisms within the order are Gram-type negative with aerobic growth under mesophilic temperature, neutral pH and low salinity conditions. The type genus is Immundisolibacter.
Funding information
This work was supported by the U.S. National Institute of Environmental Health Sciences (NIEHS) as part of the Superfund Research Program (5 P42ES005948).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
References
- 1.Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol. 2006;8:1736–1745. doi: 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- 2.Jones MD, Crandell DW, Singleton DR, Aitken MD. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ Microbiol. 2011;13:2623–2632. doi: 10.1111/j.1462-2920.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones MD, Singleton DR, Carstensen DP, Powell SN, Swanson JS, et al. Effect of incubation conditions on the enrichment of pyrene-degrading bacteria identified by stable-isotope probing in an aged, PAH-contaminated soil. Microb Ecol. 2008;56:341–349. doi: 10.1007/s00248-007-9352-9. [DOI] [PubMed] [Google Scholar]
- 4.Singleton DR, Hunt M, Powell SN, Frontera-Suau R, Aitken MD. Stable-isotope probing with multiple growth substrates to determine substrate specificity of uncultivated bacteria. J Microbiol Methods. 2007;69:180–187. doi: 10.1016/j.mimet.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 5.Dunlevy SR, Singleton DR, Aitken MD. Biostimulation reveals functional redundancy of anthracene-degrading bacteria in polycyclic aromatic hydrocarbon-contaminated soil. Environ Eng Sci. 2013;30:697–705. doi: 10.1089/ees.2013.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Midgley DJ, Ross JP, Oytam Y, Abell GC, et al. Microbial biodiversity in a Malaysian oil field and a systematic comparison with oil reservoirs worldwide. Arch Microbiol. 2012;194:513–523. doi: 10.1007/s00203-012-0788-z. [DOI] [PubMed] [Google Scholar]
- 7.Liu R, Zhang Y, Ding R, Li D, Gao Y, et al. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J Biosci Bioeng. 2009;108:400–407. doi: 10.1016/j.jbiosc.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Païssé S, Goñi-Urriza M, Coulon F, Duran R. How a bacterial community originating from a contaminated coastal sediment responds to an oil input. Microb Ecol. 2010;60:394–405. doi: 10.1007/s00248-010-9721-7. [DOI] [PubMed] [Google Scholar]
- 9.Tauler M, Vila J, Nieto JM, Grifoll M. Key high molecular weight PAH-degrading bacteria in a soil consortium enriched using a sand-in-liquid microcosm system. Appl Microbiol Biotechnol. 2016;100:3321–3336. doi: 10.1007/s00253-015-7195-8. [DOI] [PubMed] [Google Scholar]
- 10.Bai Y, Sun Q, Sun R, Wen D, Tang X. Bioaugmentation and adsorption treatment of coking wastewater containing pyridine and quinoline using zeolite-biological aerated filters. Environ Sci Technol. 2011;45:1940–1948. doi: 10.1021/es103150v. [DOI] [PubMed] [Google Scholar]
- 11.Militon C, Boucher D, Vachelard C, Perchet G, Barra V, et al. Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol Ecol. 2010;74:669–681. doi: 10.1111/j.1574-6941.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim JS, Crowley DE. Microbial diversity in natural asphalts of the Rancho La Brea tar pits. Appl Environ Microbiol. 2007;73:4579–4591. doi: 10.1128/AEM.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handley KM, Verberkmoes NC, Steefel CI, Williams KH, Sharon I, et al. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 2013;7:800–816. doi: 10.1038/ismej.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gihring TM, Moser DP, Lin L-H, Davidson M, Onstott TC, et al. The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J. 2006;23:415–430. doi: 10.1080/01490450600875696. [DOI] [Google Scholar]
- 15.Liu R, Yu Z, Zhang H, Yang M, Shi B, et al. Diversity of bacteria and mycobacteria in biofilms of two urban drinking water distribution systems. Can J Microbiol. 2012;58:261–270. doi: 10.1139/w11-129. [DOI] [PubMed] [Google Scholar]
- 16.Xia N, Xia X, Zhu B, Zheng S, Zhuang J. Bacterial diversity and community structure in the sediment of the middle and lower reaches of the Yellow River, the largest turbid river in the world. Aquat Microb Ecol. 2013;71:43–55. doi: 10.3354/ame01664. [DOI] [Google Scholar]
- 17.Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittenbury R, Phillips KC, Wilkinson JF. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 19.Singleton DR, Richardson SD, Aitken MD. Pyrosequence analysis of bacterial communities in aerobic bioreactors treating polycyclic aromatic hydrocarbon-contaminated soil. Biodegradation. 2011;22:1061–1073. doi: 10.1007/s10532-011-9463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima H, Fukui M. Sulfuritalea hydrogenivorans gen. nov., sp. nov., a facultative autotroph isolated from a freshwater lake. Int J Syst Evol Microbiol. 2011;61:1651–1655. doi: 10.1099/ijs.0.024968-0. [DOI] [PubMed] [Google Scholar]
- 21.Corteselli EM, Aitken MD, Singleton DR. Rugosibacter aromaticivorans gen. nov., sp. nov., a bacterium within the family Rhodocyclaceae, isolated from contaminated soil, capable of degrading aromatic compounds. Int J Syst Evol Microbiol. 2017;67:311–318. doi: 10.1099/ijsem.0.001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singleton DR, Dickey AN, Scholl EH, Wright FA, Aitken MD. Complete genome sequence of a bacterium representing a deep uncultivated lineage within the Gammaproteobacteria associated with the degradation of polycyclic aromatic hydrocarbons. Genome Announc. 2016;4:e01086-16. doi: 10.1128/genomeA.01086-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram's iodine. Curr Microbiol. 2008;57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- 24.Kanaly RA, Harayama S. Advances in the field of high-molecular-weight polycyclic aromatic hydrocarbon biodegradation by bacteria. Microb Biotechnol. 2010;3:136–164. doi: 10.1111/j.1751-7915.2009.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell MA. Cloning and expression in Escherichia coli of the naphthalene degradation genes from plasmid NAH7. J Bacteriol. 1983;153:822–829. doi: 10.1128/jb.153.2.822-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson SD, Lebron BL, Miller CT, Aitken MD. Recovery of phenanthrene-degrading bacteria after simulated in situ persulfate oxidation in contaminated soil. Environ Sci Technol. 2011;45:719–725. doi: 10.1021/es102420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton DR, Ramirez LG, Aitken MD. Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl Environ Microbiol. 2009;75:2613–2620. doi: 10.1128/AEM.01955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasser M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. 2001. www.microbialid.com/PDF/TechNote_101.pdf MIDI Technical Note 101.
- 29.Tindall BJ. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol. 1990;13:128–130. doi: 10.1016/S0723-2020(11)80158-X. [DOI] [Google Scholar]
- 30.Tindall BJ. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett. 1990;66:199–202. doi: 10.1111/j.1574-6968.1990.tb03996.x. [DOI] [Google Scholar]
- 31.Tindall BJ, Sikorski J, Smibert RM, Kreig NR. Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, editors. Methods for General and Molecular Microbiology. Washington, DC: ASM Press; 2007. pp. 330–393. et al. (editors) [Google Scholar]
- 32.Muyzer G, Sorokin DY, Mavromatis K, Lapidus A, Clum A, et al. Complete genome sequence of "Thioalkalivibrio sulfidophilus" HL-EbGr7. Stand Genomic Sci. 2011;4:23–35. doi: 10.4056/sigs.1483693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slobodkina GB, Baslerov RV, Novikov AA, Viryasov MB, Bonch-Osmolovskaya EA, et al. Inmirania thermothiophila gen. nov., sp. nov., a thermophilic, facultatively autotrophic, sulfur-oxidizing gammaproteobacterium isolated from a shallow-sea hydrothermal vent. Int J Syst Evol Microbiol. 2016;66:701–706. doi: 10.1099/ijsem.0.000773. [DOI] [PubMed] [Google Scholar]
- 34.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, et al. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ Pollut. 2012;162:345–353. doi: 10.1016/j.envpol.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 37.Bowman JP. Order VII. Methylococcales ord. nov. In: Brenner DJ, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Volume 2: The Proteobacteria Part B: The Gammaproteobacteria. New York: Springer; 2005. (editors) [Google Scholar]
- 38.Hoefman S, Van der Ha D, Iguchi H, Yurimoto H, Sakai Y, et al. Methyloparacoccus murrellii gen. nov., sp. nov., a methanotroph isolated from pond water. Int J Syst Evol Microbiol. 2014;64:2100–2107. doi: 10.1099/ijs.0.057760-0. [DOI] [PubMed] [Google Scholar]
- 39.Sorokin DY, Lysenko AM, Mityushina LL, Tourova TP, Jones BE, et al. Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov., and Thioalkalivibrio denitrificancs sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. Int J Syst Evol Microbiol. 2001;51:565–580. doi: 10.1099/00207713-51-2-565. [DOI] [PubMed] [Google Scholar]
- 40.Imhoff JF. Order I. Chromatiales ord. nov. In: Brenner DJ, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Volume 2: The Proteobacteria Part B: The Gammaproteobacteria. New York: Springer; 2005. (editors) [Google Scholar]
- 41.Campbell MA, Chain PS, Dang H, El Sheikh AF, Norton JM, et al. Nitrosococcus watsonii sp. nov., a new species of marine obligate ammonia-oxidizing bacteria that is not omnipresent in the world's oceans: calls to validate the names 'Nitrosococcus halophilus' and 'Nitrosomonas mobilis'. FEMS Microbiol Ecol. 2011;76:39–48. doi: 10.1111/j.1574-6941.2010.01027.x. [DOI] [PubMed] [Google Scholar]
- 42.Klotz MG, Arp DJ, Chain PS, El-Sheikh AF, Hauser LJ, et al. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl Environ Microbiol. 2006;72:6299–6315. doi: 10.1128/AEM.00463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narasingarao P, Häggblom MM. Sedimenticola selenatireducens, gen. nov., sp. nov., an anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst Appl Microbiol. 2006;29:382–388. doi: 10.1016/j.syapm.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Mori K, Suzuki K, Urabe T, Sugihara M, Tanaka K, et al. Thioprofundum hispidum sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing gammaproteobacterium isolated from the hydrothermal field on Suiyo Seamount, and proposal of Thioalkalispiraceae fam. nov. in the order Chromatiales. Int J Syst Evol Microbiol. 2011;61:2412–2418. doi: 10.1099/ijs.0.026963-0. [DOI] [PubMed] [Google Scholar]
- 45.Takai K, Miyazaki M, Hirayama H, Nakagawa S, Querellou J, et al. Isolation and physiological characterization of two novel, piezophilic, thermophilic chemolithoautotrophs from a deep-sea hydrothermal vent chimney. Environ Microbiol. 2009;11:1983–1997. doi: 10.1111/j.1462-2920.2009.01921.x. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings MP, Hancock JM, Zvelebil MJ. Dictionary of Bioinformatics and Computational Biology. Hoboken, NJ: John Wiley & Sons, Ltd; 2004. PAUP* (phylogenetic analysis using parsimony (and other methods)) [Google Scholar]
- 48.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.