Abstract
Background:
Salt Overly Sensitive (SOS) pathway is a well-known pathway in arabidopsis, essential for maintenance of ion homeostasis and thus conferring salt stress tolerance. In arabidopsis, the Ca2+ activated SOS3 interacts with SOS2 which further activates SOS1, a Na+/H+ antiporter, responsible for removing toxic sodium ions from the cells. In the present study, we have shown that these three components of SOS pathway, BjSOS1, BjSOS2 and BjSOS3 genes exhibit differential expression pattern in response to salinity and ABA stress in contrasting cultivars of Brassica. It is also noticed that constitutive expression of all the three SOS genes is higher in the tolerant cultivar B. juncea as compared to the sensitive B. nigra. In silico interaction of BjSOS2 and BjSOS3 has been reported recently and here we demonstrate in vivo interaction of these two proteins in onion epidermal peel cells. Further, overexpression of BjSOS3 in corresponding arabidopsis mutant ΔAtsos3 was able to rescue the mutant phenotype and exhibit higher tolerance towards salinity stress at the seedling stage.
Conclusion:
Taken together, these findings demonstrate that the B. juncea SOS3 (BjSOS3) protein is a functional ortholog of its arabidopsis counterpart and thus show a strong functional conservation of SOS pathway responsible for salt stress signalling between arabidopsis and Brassica species.
Keywords: Arabidopsis, Brassica, Contrasting genotypes, Protein interaction, Salinity stress, SOS1, SOS2, SOS3, Transcript abundance
1. INTRODUCTION
Among the various abiotic stresses, plant growth and development is significantly affected by soil salinity [1]. High salt concentration affects the plant, mainly by osmotic and ionic stress [2]. Plants develop various defense mechanisms to maintain appropriate ion homeostasis by actively excluding the ions from the cell or partitioning them into the vacuole [3, 4]. Among these ions, sodium ion (Na+) is the primary source of toxicity in cells, which causes plant growth inhibition. Higher Na+ yields low concentrations of K+ in the cell (because of its ability to compete with K+), thereby affecting the activity of many essential enzymes [5]. Plant cells maintain ion homeostasis by maintaining appropriate K+/Na+ ratio in the cytoplasm, to provide optimal conditions for biochemical and metabolic activities [6, 7]. SOS (Salt Overly Sensitive) pathway, which is responsible for the maintenance of Na+ and K+ concentration in a cell, comprises of three proteins namely, SOS1, SOS2 and SOS3 [6, 8]. It has been reported earlier that the activation of Na+/H+ antiporter AtSOS1 by salt stress is controlled via phosphorylation by AtSOS3 and AtSOS2 proteins [9]. The carboxy-terminal regulatory domain of AtSOS2 interacts with AtSOS3 through the FISL motif [10, 11]. SOS3 is a calcium binding protein and belongs to EF-hand type family of proteins in arabidopsis. This unique family of proteins show similarity with animal β-subunit of calcineurin and animal neuronal calcium sensors [12, 13]. Co-expression of SOS2 and SOS3 protein along with SOS1 has shown higher tolerance to salt stress in sodium transport deficient yeast mutant than expression of independent SOS2 or SOS3 proteins, thereby indicating the role of SOS2-SOS3 complex in the activation of SOS1 [11]. SOS2-SOS3 interaction in SOS pathway has also been shown by sos3/sos2 double-mutant analysis in arabidopsis [14]. It has also been observed that sos1, sos2 and sos3 mutants are hypersensitive to Na+ and Li+ ions, thus confirming the importance of these proteins in salinity tolerance in plant [8]. Further, transgenic arabidopsis plants overexpressing different SOS proteins, individually or in combinations, have shown increased tolerance to salinity stress [15, 16].
Brassica is an important oil seed crop, which diverged from arabidopsis around 14.5-20.4 million years ago from a common ancestor [17]. In the family Brassicaceae, comparative genetic mapping has revealed co-linear chromosome segments [18, 19] with linkage arrangements been reported between arabidopsis and B. oleracea [20]. Rana et al. [21] have reported that the Brassica genome has duplicated or possibly triplicated with the corresponding homologous segments of arabidopsis. Sizes of genomes of various Brassica species are significantly higher than arabidopsis (125Mb) and range from 529-696 Mb for the diploids and 1068-1284 Mb for polyploids [22]. It is also presumed that few novel gene interactions might have evolved potentially by sub-functionalization and/or neo-functionalization of paralogs [23]. Recently, we have reported the transcriptome-based genome assembly of B. juncea var. CS52 - a salt tolerant variety [24]. One of the key findings of this study is that B. juncea has higher transcript abundance of various stress related genes from different metabolic pathway and it is able to tolerate salinity stress by efficient ROS scavenging machinery. However, the detailed investigations about how this homeostasis is maintained, is yet to be discovered at molecular level.
In an earlier study, we have shown that SOS pathway-related genes show strong correlation with salinity tolerance in Brassica species [25, 26]. We have also shown the direct effect of Ca2+ chelator, EGTA, on stress inducibility of BjSOS3, thus establishing its calcium sensing nature. Recently, in silico interactions of BjSOS2 and BjSOS3 of Brassica juncea have also been shown [27].
In the present study, based on BiFC analysis, we have shown in vivo interaction of BjSOS2 with BjSOS3 by co-expressing these proteins in onion peel epidermal cells. We have also studied qRT-PCR-based transcript abundance of BjSOS1, BjSOS2 and BjSOS3 in contrasting genotypes of Brassica, under salinity stress and ABA treatment. Most importantly, we have shown that BjSOS3 allele isolated from B. juncea could functionally complement the ΔAtsos3 mutant, thereby showing the structural and functional conservation of SOS pathway across the plant genera.
2. MATERIALS AND METHODs
2.1. Plant Growth
Seeds of B. juncea var. CS52 and B. nigra obtained from ICAR-CSSRI, Karnal, India were thoroughly washed with de-ionized water and germinated in hydroponic system, filled with half strength Hoagland’s medium. The hydroponic setup was kept in the plant growth chamber for 48 h in dark and then exposed to light and kept under control condition 25±2°C, 12 h light and dark cycles. Brassica seedlings were grown for nine days in hydroponics with continuous air bubbling and renewal of nutrient media after every two days.
2.2. Stress Treatment, RNA Isolation and cDNA Synthesis
For salinity and ABA stress treatment, hydroponically grown nine day old seedlings of Brassica spp. were transferred to ½ Hoagland media containing either 200 mM NaCl or 100 µM ABA respectively. Shoots of Brassica seedlings (200 mg), in triplicates were harvested at 0 h, 8 h and 24 h of treatment and total RNA was extracted using Tri reagent (Life Technologies, Rockville, USA) as described by Kumar et al. [26]. Total RNA was analysed for its integration and purity, before proceeding for cDNA synthesis. Using cDNA synthesis kit (Fermentas Life Sciences, Burlington, Canada), cDNA was synthesised from 2 µg of total RNA from each sample, as described by the manufacturer.
2.3. qRT-PCR-based Expression Analysis
Primers for BjSOS1, BjSOS2 and BjSOS3 genes were designed with Primer Express 3.0 software (Applied Biosystems, California, USA) using default parameters. To ensure high specificity, we have selected the 3’UTR region of these genes for the purpose of primer designing. BLASTn using the KOME and NCBI databases confirmed the uniqueness of each primer pair to amplify selective genes. The PCR mixture contained 5 μl of cDNA (10 times diluted), 10μl of 2× SYBR Green PCR Master Mix (Applied Biosystems, California, USA) and 4 nM of each gene-specific primer in a final volume of 20μl. The real-time PCR was performed employing StepOneTM Real-Time PCR System (Applied Biosystems, California, USA). All the PCR reactions were performed under the following conditions, 10 min at 95°C and 40 cycles of 15 s at 95°C, 1 min at 58°C in 96-well optical reaction plates. The specificity of amplification was tested by dissociation curve analysis and agarose gel electrophoresis. To minimize any error, two biological samples with three technical replicates each, were taken for the expression analysis of each gene. The expression of each gene in different RNA samples was normalized with the expression of internal control (actin gene from Brassica; BjAct). Fold change in transcript abundance for each gene in different samples was calculated relative to its expression in B. nigra control seedlings using 2-ddCt method [24]. For statistical significance, student’s t-test was performed using STATISTICA Data Miner software (Version 7, StatSoft, Oklahoma, USA). Results are represented as mean +/- Standard Error (SE).
2.4. Interaction Study of BjSOS2 and BjSOS3 by Bimolecular Fluorescence Complementation Assay (BiFC)
For in vivo interaction study of BjSOS2 and BjSOS3, we have employed pSAT series of vectors pair (pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1) wherein two putative interacting proteins are tagged with n-terminal and c-terminal enhanced YFP fragment, respectively (Fig. S1 (3.1MB, pdf) ). Both full length BjSOS3 and BjSOS2 were cloned in pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1 pSAT vector respectively at EcoRI and BamHI sites. For revalidation of in vivo interaction data, we did the vector swapping and cloned BjSOS3 and BjSOS2 full length gene in reverse vectors i.e. pSAT-1A-ceYFP-N1 and pSAT-1A-neYFP-N1 respectively. Thereafter, three combinations of constructs – vector control (pSAT-1A-neYFP-N1 + pSAT-1A-ceYFP-N1), BjSOS3 + BjSOS2 (pSAT-1A-neYFP-N1BjSOS3 and pSAT-1A-ceYFP-N1BjSOS2) and BjSOS2 + BjSOS3 (pSAT-1A-neYFP-N1BjSOS2 and pSAT-1A-ceYFP-N1BjSOS3) were coated on 1 mm gold particles and bombarded on cultured epidermal cell of onion on Murashige and Skoog (MS) agar plates by using PDS-1000 particle delivery system (Bio-Rad, USA). The bombardment parameters were set as distance 85 mm from the macrocarrier to samples; pressure 1100 psi and vacuum 28 inch Hg. After bombardment, onion epidermis on MS agar plate was incubated at 25±2°C in dark for 16 h. YFP fluorescent signals for all the three co-transformed combinations were examined with a confocal laser scanning microscope (Olympus fluoviewTMFV1000, Japan) in the 514 nm-excitation wavelength at Advanced Instrument Research Facility (AIRF), JNU, New Delhi, India.
2.5. Cloning of BjSOS3 in Plant Transformation Vector and Raising of Transgenic Arabidopsis
To raise arabidopsis plants overexpressing BjSOS3, the full length BjSOS3 gene was cloned at NcoI and SpeI sites of pCAMBIA1304 under the control of 35SCaMV promoter. Positive clone was confirmed by colony PCR as well as restriction digestion. For complementation assay, construct carrying the 35SCaMV-BjSOS3 was transformed into Agrobacterium strain GV3101 and transformation of arabidopsis mutant line ΔAtsos3 (CS3869 procured from ABRC) was carried out (Fig. S2 (3.1MB, pdf) ). Putative transgenic plants (T1) harboring BjSOS3 were screened on Murashige and Skoog (MS) agar medium containing 30 mg l-1 hygromycin. To reconfirm the stable insertion of the gene in the genome of plant, tissue PCR was performed in these lines using gene specific primers. Seeds form homozygous plants (which showed positive for tissue PCR of transgene) were further selected (T2) and analyzed for transgene expression. Seeds from homozygous T2 plants were harvested and T3 seedlings were used for complementation assay.
2.6. Complementation Assay for Salinity and ABA Response
For complementation assay, T3 arabidopsis seeds were sterilized and vernalized for four days at 4ºC. Seeds were plated on MS agar media and kept in plant tissue culture room for 2 days at 22±2 °C for germination. After germination, four days old seedlings were transferred to MS media or MS media supplemented with NaCl (100 or 200 mM) and root bending assay was performed as described by Zhu et al. [8]. After 5 days, seedling growth and survival percentage were compared between BjSOS3-complemented lines with the ΔAtsos3 mutant and wild type plants (WT). For ABA response, four days old seedlings were transferred to MS media or MS media supplemented with 25 µM ABA and photographed after 7 days. Three independent experiments were performed and results are represented as mean +/- Standard Error (n = 15).
2.7. Plant Growth Analysis and Measurement of Na+ and K+ Content
For root growth and total biomass accumulation analysis under salt stress, 4 days old seedlings were transferred on MS plate containing 0 or 100 mM NaCl, and after 8 days, root growth and total biomass observations were compared. For estimation of sodium (Na+) and potassium (K+), seedlings were harvested from the plates and briefly rinsed with distilled water, and dried at 65°C for 24 h. The seedlings were weighed and digested with HNO3, and K+ and Na+ concentrations were determined with flame photometer as described by Xu et al. [28]. Three independent experiments were performed for each analysis and data are shown as mean +/- SE.
2.8. Statistical Analysis
For assessing the statistical significance of data, student’s t-test was performed using STATISTICA Data Miner software (Version 7, StatSoft, Oklahoma, USA).
3. RESULTS
3.1. BjSOS1, BjSOS2 and BjSOS3 Transcripts are Induced in Response to Salinity and ABA Stress in Contrasting Genotypes of Brassica
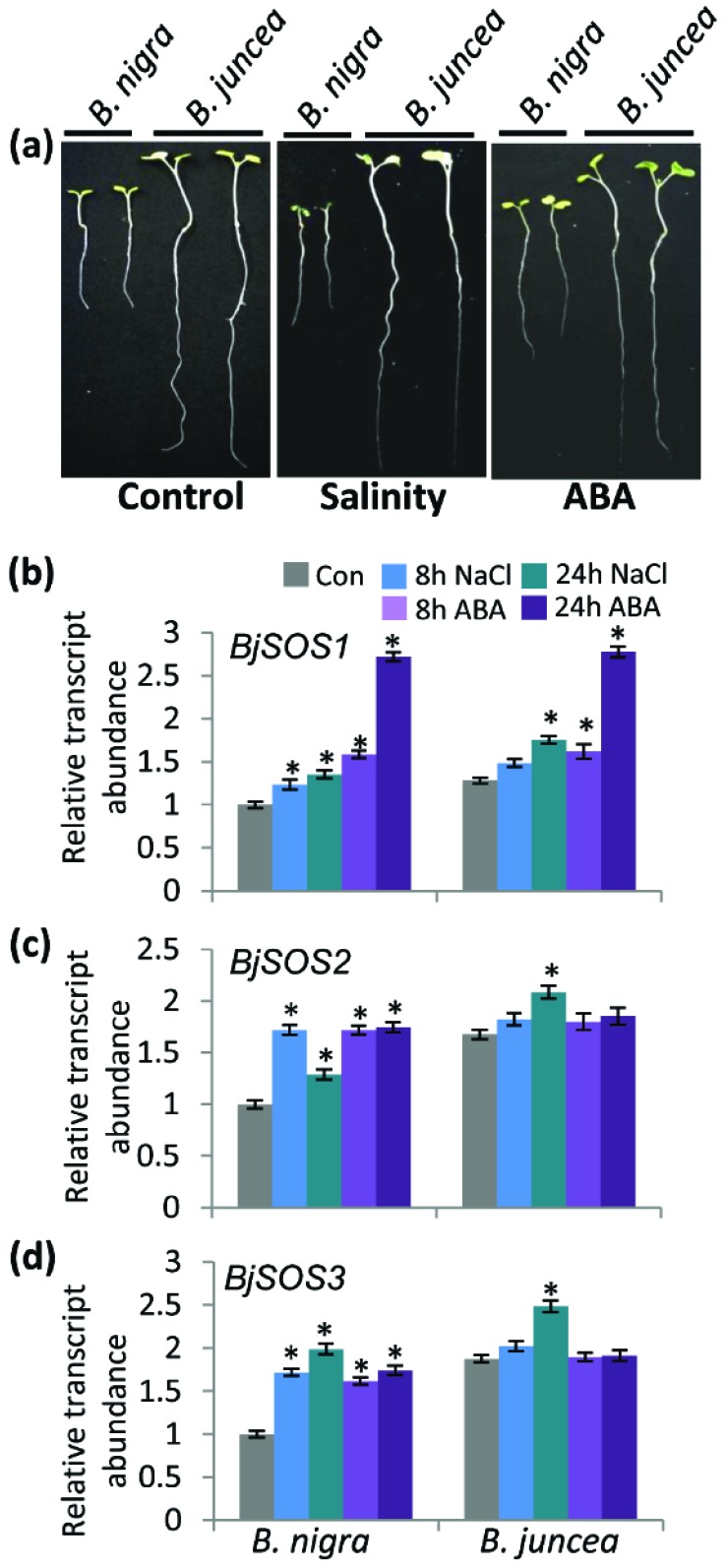
In order to analyze the constitutive and salinity induced transcript abundance for SOS components, Brassica seedlings were subjected to 200 mM NaCl or 100 µM ABA for early (8 h) or late (24 h) duration. Phenotypically, no significant differences were observed after 8 h (early) of salinity or ABA stress in contrasting Brassica genotypes. However, at late (24 h) duration of salinity stress, leaf rolling and seedling drooping was observed in B. nigra (salt sensitive) genotype (Fig. 1a). Similarly, at late (24 h) duration of ABA stress, B. juncea seedlings appeared more healthy and robust than B. nigra seedlings (Fig. 1a). Further, transcript levels for BjSOS1, BjSOS2 and BjSOS3 were analyzed in shoot tissues by qRT-PCR. It was observed that, constitutive expression of BjSOS1, BjSOS2 and BjSOS3 was higher in salt tolerant cultivar B. juncea as compared to salt sensitive cultivar B. nigra. BjSOS1 expression in B. nigra was found to be induced in response to salinity stress. Under ABA stress, BjSOS1 expression was induced upto 3 folds higher as compared to the control in B. nigra. In B. juncea, constitutive expression of BjSOS1 was relatively higher than B. nigra. Upon NaCl and ABA treatment, the expression pattern was similar to B. nigra (Fig. 1b). Constitutive expression of BjSOS2 in B. nigra was relatively lower than B. juncea, but got induced upon salinity and ABA treatment. However, no significant induction in BjSOS2 expression was observed under ABA treatment in the latter (Fig. 1c). Transcript abundance for BjSOS3 was found to be induced by ~2 folds in response to both salinity and ABA again, in B. nigra. (Fig. 1d). In response to salinity stress, B. juncea also showed almost 2.5 fold induction. In summation, contrasting genotypes of Brassica spp. showed differential regulation of BjSOS genes in response to NaCl and ABA stress, with the tolerant genotype maintaining higher transcript levels under both late and early duration of various stress applications, as compared to the sensitive genotype.
Fig. (1).
Growth of seedlings of contrasting Brassica genotypes in response to high salinity and ABA, and transcript abundance for various SOS pathway members. Nine days old hydroponically grown Brassica seedlings were subjected to salinity (200 mM NaCl) or ABA (100 µM). qRT-PCR was performed using the cDNA prepared from total RNA isolated from shoots of the seedlings subjected to salinity stress (200 mM NaCl); or ABA (100 µM) for 8 h and 24 h time period. (a). Representative photograph of seedlings of Brassica genotypes after 24 h of salinity (200 mM NaCl) or ABA (100 µM). Relative expression of (b) BjSOS1, (c) BjSOS2 and (d) BjSOS3. Bar graphs were plotted between stress duration (X-axis) and log 2-ddCt value in number (Y-axis). Gene expression data was normalised with the plant reference gene ‘actin’ as an internal control. Relative expression of genes was plotted against the expression of B. nigra control. The values represented are the mean of two biological and three technical replicates, standard error is shown above the bar. For statistical significance, student t-test was performed and asterisk above the graph means significant differences from their respective control (Con) at P ≤ 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
3.2. BjSOS3 Interacts with BjSOS2 In Vivo
For investigating the predicted protein-protein interactions, bimolecular fluorescence complementation (BiFC) assay was performed. For this purpose, we have employed pSAT series of vectors (pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1) wherein two putative interacting proteins are tagged with n-terminal or c-terminal enhanced YFP fragment, respectively. Both full length BjSOS3 and BjSOS2 were cloned at EcoRI and BamHI sites of pSAT vector pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1 respectively.
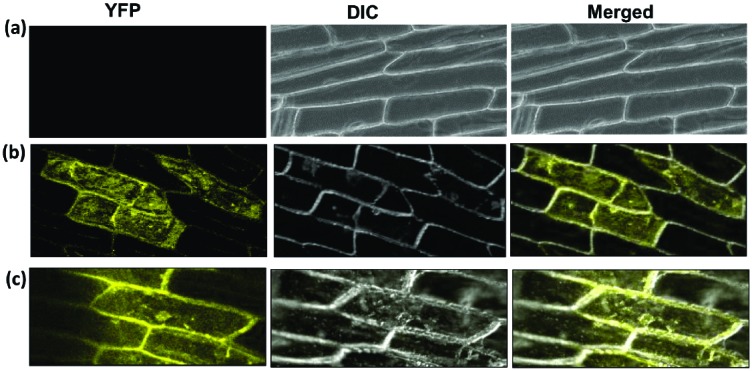
Cloning of BjSOS3 and BjSOS2 in both pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1 vectors was confirmed through colony PCR (Figs. S1a (3.1MB, pdf) and S1b (3.1MB, pdf) ) as well as restriction digestion (Figs. S1c (3.1MB, pdf) and S1d (3.1MB, pdf) ) to yield a positive band of 633 bp and 1333 bp, respectively. Thereafter, vector pair pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1, pSAT-1A-neYFP-N1BjSOS3 and pSAT-1A-ceYFP-N1BjSOS2, pSAT-1A-neYFP-N1BjSOS2 and pSAT-1A-ceYFP-N1BjSOS3 were used for bombardment in onion peel using the particle gun. Onion peel cells transformed with empty pSAT vectors did not show any fluorescence under confocal microscope (Fig. 2a). However, both the vector pairs harbouring BjSOS3 and BjSOS2, in fusion with either N or C-terminal YFP showed strong fluorescence throughout the cytoplasm indicating a positive interaction between these proteins in vivo (Figs. 2b and 2c). These results confirm the physical interaction between BjSOS2 and BjSOS3 proteins.
Fig. (2).
In vivo interaction of full length BjSOS2 and BjSOS3 proteins. (a). Onion peel co-transformed with empty vectores, pSAT-1A-neYFP-N1 and pSAT-1A-ceYFP-N1. (b). Onion peel co-transformed with constructs, pSAT-1A-neYFP-N1BjSOS3 and pSAT-1A-ceYFP-N1BjSOS2. (c). Onion peels transformed with reverse constructs, pSAT-1A-neYFP-N1BjSOS2 and pSAT-1A-ceYFP-N1BjSOS3. YFP, Yellow fluorescent protein; DIC, Differential interference contrast.
3.3. BjSOS3 Could Rescue the ΔAtsos3 Mutant of Arabidopsis Under Salinity Stress
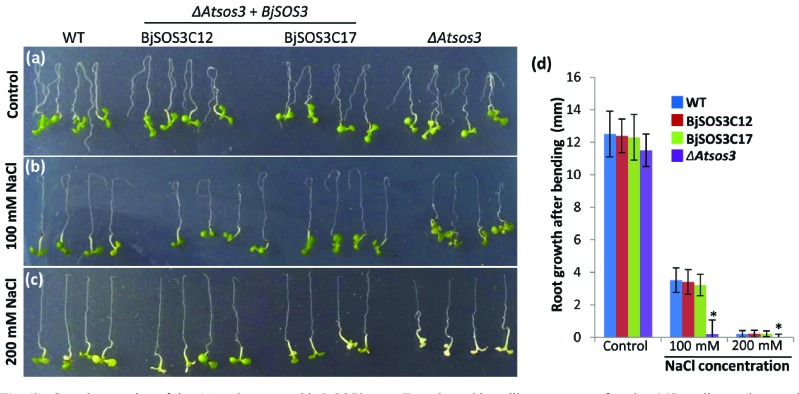
Arabidopsis seeds with knock out AtSOS3 gene (Mutant stock CS3869) were procured from ABRC and used for the complementation assays. Full length BjSOS3 gene cloned in plant expression vector, pCAMBIA1304 at NcoI and SpeI sites was used for transformation of the ΔAtsos3 mutant (Fig. S2 (3.1MB, pdf) ). Root bending assays were performed in T3 homozygous seedlings and phenotypes of BjSOS3-complemented lines with the ΔAtsos3 mutant and WT arabidopsis seedlings were compared. Morphological observation showed that WT, BjSOS3-complemented lines as well as the ΔAtsos3 mutant seedlings grew well and appeared healthy on control Murashige-Skoog (MS) medium without NaCl supplement (Fig. 3a). In contrast, on the media containing 100 mM NaCl, the growth of the ΔAtsos3 mutant was substantially compromised while the complemented lines grew very well (Fig. 3b). On plates with MS agar media containing 200 mM NaCl, the growth of the ΔAtsos3 mutant was drastically compromised and chlorophyll bleaching was observed. However, seedlings of the ΔAtsos3 mutant complemented with BjSOS3, showed higher tolerance to salinity and survived on MS agar media supplemented with 200 mM NaCl (Fig. 3c). It was also found that on the plates containing MS agar media supplemented with 0 or 100 mM of NaCl, all the seedlings of WT, the ΔAtsos3 mutant as well as the ΔAtsos3 mutant complemented with BjSOS3 could survive for 5 days. However, in the presence of higher salinity (200 mM NaCl), the ΔAtsos3 lines showed a significant reduction in survival, where only 6% seedlings could survive in contrast to 80% survival for the WT seedlings. Interestingly, seedlings of the ΔAtsos3 mutant complemented with BjSOS3 showed a response similar to that of the WT where 80% survival rate was recorded under similar stress conditions.
Fig. (3).
Complementation of the ΔAtsos3 mutant with BjSOS3 gene. Four days old seedlings were transferred to MS media supplemented with (a) 0, (b) 100 or (c) 200 mM of NaCl. Plates were placed upside down to allow seedlings to grow in inverted position for 5 days and pictures were taken. (d). Measurement of root growth after bending of the ΔAtsos3 mutant complemented with BjSOS3, the ΔAtsos3 mutant and WT seedlings shown above. Error bars represent the standard deviation (n = 15). For statistical significance, student’s t-test was performed and asterisk above the graph means significant differences from WT at P ≤ 0.05. WT, wild type; BjSOS3C12 and BjSOS3C17 are the two representative of the ΔAtsos3 mutant complemented with BjSOS3; ΔAtsos3, arabidopsis sos3 mutant.
The assay was further extended to the analysis of root bending, which is quick and fast way to determine the relative tolerance in seedlings of arabidopsis towards salinity stress. On plates containing the normal MS media, all the genotypes showed significant growth in the primary root after the plates were kept in inverted position, thereby indicating healthy status of these seedlings. In contrast, on saline growth media (100 mM NaCl), the roots of the ΔAtsos3 mutant could not grow beyond 1 mm in inverted position, while the WT seedlings showed a growth up to 4 mm in the primary root after the plates were kept in inverted position (Fig. 3d). This observation clearly shows that 100 mM NaCl is detrimental for the growth of WT arabidopsis seedlings, but it is more so for the ΔAtsos3 mutant seedlings. Interestingly, the ΔAtsos3 mutant lines complemented with BjSOS3 gene showed a response similar to that of the WT seedlings, where the primary root could grow up to 4mm in length (Fig. 3d). Under severe salinity stress, i.e. on media containing 200 mM NaCl, none of the seedlings could grow their roots after the plates were kept in the inverted position indicating thereby that these conditions are not physiological for arabidopsis seedlings. Thus, these results conclusively prove the requirement of SOS3 protein for survival under salinity stress conditions. These results also showed that AtSOS3 and BjSOS3 are functionally conserved.
3.4. The ΔAtsos3 Mutant Complemented with BjSOS3 Show Mutant Phenotype Reversion by Maintaining Ion Homeostasis Under Salinity Stress
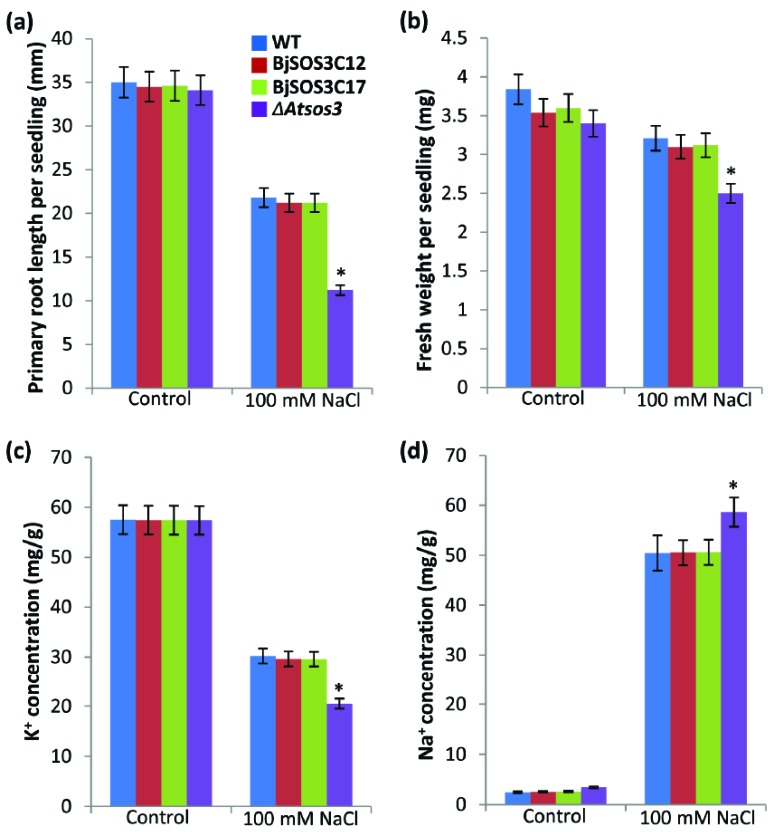
For root growth and total biomass accumulation analysis under salt stress, we transferred 4 days old seedlings of WT, the ΔAtsos3 mutant and the ΔAtsos3 mutant complemented with BjSOS3 on MS plate containing 0 or 100 mM NaCl, and observed their growth parameters for 8 days. Under control conditions, wild type plants, the ΔAtsos3 mutants and BjSOS3 complemented lines displayed relatively uniform growth phenotype. However, when treated with 100 mM NaCl, growth of the ΔAtsos3 mutant was drastically compromised, as compared to BjSOS3 complemented lines. Thus, expression of BjSOS3 in the ΔAtsos3 mutant plants substantially alleviates the growth retardation imposed by moderate salt stress, hence rescuing the mutant phenotype. Analysis of growth parameters such as root length and fresh weight of seedlings showed that all the rescued mutants generally displayed a near-wild-type level of salt tolerance, although with differential extents between different lines (Fig. 4a). Total fresh weight of seedlings of ΔAtsos3 transformed with BjSOS3 was found to be higher than the ΔAtsos3, but comparable to WT (Fig. 4b). This data shows that the Brassica SOS3 protein can substitute for the corresponding arabidopsis counterpart.
Fig. (4).
Growth and ion accumulation in seedlings of the ΔAtsos3 mutant complemented with BjSOS3 under salinity. Four days old seedlings were transferred to MS agar plates supplemented with 0 or 100 mM NaCl and allowed to grow for 8 days before the observation were taken. (a). Primary root length. (b). Fresh weight of each seedling. (c). K+ content in seedlings. (d). Na+ content in seedlings. For primary root length per seedling and fresh weight per seedling, results are the average from three independent replicates. For K+ and Na+ estimation, three independent experiments were performed and results are mean +/- SE. Error bars represent the standard deviation (n = 15). For statistical significance, student’s t-test was performed and asterisk above the graph means significant differences from WT at P ≤ 0.05. WT, wild type; BjSOS3C12 and BjSOS3C17 are the two representative of the ΔAtsos3 mutant complemented with BjSOS3; ΔAtsos3, arabidopsis sos3 mutant.
Maintenance of K+/Na+ homeostasis is an important requirement for salt tolerance. (Fig. 4c) shows that the K+ content in the ΔAtsos3 plants complemented with BjSOS3 was equal to the mutant and WT under control conditions. However, under salinity stress, the seedlings of ΔAtsos3 transformed with BjSOS3 accumulated the same level of K+ as the WT plants, but more K+ than the ΔAtsos3 plants.
Since the outcome of the functional SOS pathway operative in plants is the efficient ion homeostasis, we decided to check the levels of Na+ in these plants, in response to salinity stress. These results indicated that the ΔAtsos3 accumulated high levels of Na+ under salinity stress. On the other hand, Na+ contents in the ΔAtsos3 plants complemented with BjSOS3 were not different from those of the wild type plants (Fig. 4d), thereby proving the functional conservation of SOS3 across the species.
3.5. The ΔAtsos3 Mutant Complemented with BjSOS3 Show Enhanced Tolerance to ABA
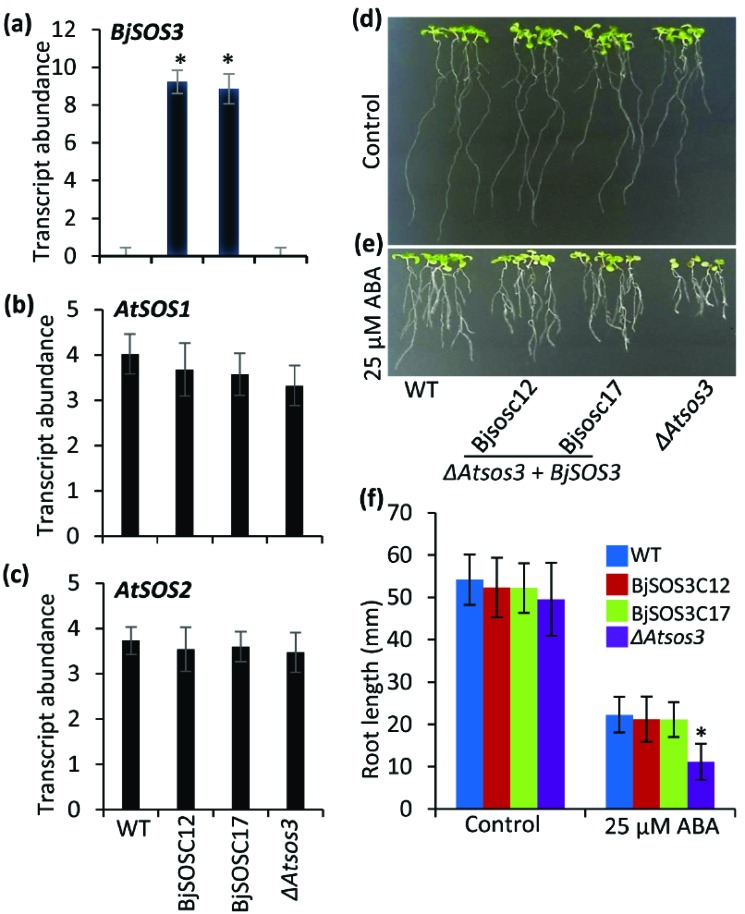
To see the role of BjSOS3 in ABA response, BjSOS3 complemented arabidopsis mutants were transferred to MS agar plate supplemented with 25 µM ABA and phenotype of these seedlings was observed. BjSOS3 transcript accumulation in complemented arabidopsis lines BjSOS3C12 and BjSOS3C17 at seedling stage showed constitutive expression of BjSOS3 gene under control conditions (Fig. 5a). WT and the ΔAtsos3 mutant arabidopsis does not show any transcript corresponding to BjSOS3 under similar growth condition. Expression of arabidopsis native AtSOS1 and AtSOS2 in
Fig. (5).
Transcript abundance of SOS3 members in BjSOS3 complemented transgenic arabidopsis and their ABA stress response. Transgenic arabidopsis seedlings were grown on MS agar plate and samples were harvested for expression analysis. qRT-PCR was performed using the cDNA prepared from total RNA isolated from whole seedlings. Relative expression of (a) BjSOS3, (b) AtSOS1 (c) AtSOS2. Bar graphs were plotted between stress duration (X-axis) and log 2-dCt value in number (Y-axis). Gene expression data was normalised with the plant reference gene ‘actin’ as an internal control. The values represented are the mean of two biological and three technical replicates, standard error is shown above the bar. For statistical significance, student t-test was performed and asterisk above the graph means significant differences from WT at P ≤ 0.05. Four days old seedlings were transferred to MS media supplemented with (d) 0 or (e) 25 µM of ABA. After 7 days of growth, pictures were taken and (f) root length was measured. Error bars represent the standard deviation (n = 15). For statistical significance, student’s t-test was performed and asterisk above the graph means significant differences from WT at P ≤ 0.05.
WT, BjSOS3 complemented lines and the ΔAtsos3 mutant was also observed using qRT-PCR (Figs. 5b and 5c). No significant difference was observed in the ΔAtsos3 mutant, complemented mutant and WT arabidopsis seedlings as far as the transcript abundance of AtSOS1 and AtSOS2 is concerned.
Four days old seedlings of ΔAtsos3 mutant complemented with BjSOS3 were transferred to MS agar media and observed for the next 7 days. These seedlings showed no phenotypic difference as compared to the ΔAtsos3 mutant and WT seedlings (Fig. 5d). Analysis of root growth indicated no significant difference between complemented and mutant arabidopsis seedlings. However, leaf bleaching and root growth inhibition was observed in the ΔAtsos3 mutant grown on MS media supplemented with 25 µM of ABA (Fig. 5e). Under ABA stress, BjSOS3 complemented ΔAtsos3 mutant seedling regain root growth, as compared to the WT or the ΔAtsos3 mutant seedlings (Fig. 5f).
DISCUSSION and conclusion
Salinity is known to be a major impediment to achieve potential yield of a crop plant. Additionally, soil salinization of cultivable agricultural land at alarming rate is of great concerns for fulfilling the food requirement of expanding world population. Genetic diversity in the plant kingdom provides the treasure for selecting salinity tolerant genotypes [29]. It is more useful to search the tolerant plants within same genotype as compared to finding in another genotype [25]. Additionally, study of diploidy and polyploidy within the genera is another convenient strategy for screening tolerant genotypes [30, 31]. In most of the cases, polyploid plants are reported to possess higher salinity tolerance than their respective diploids [25, 32, 33]. SOS (Salt Overly Sensitive) pathway comprising of three major proteins namely, SOS1, SOS2 and SOS3, is an important known mechanism involved in maintenance of Na+ and K+ homeostasis in plants [6, 8]. In terms of gene sequences, this pathway has been shown to be highly conserved, in terms of the gene
sequences, among various plant species [26]. However, it remains to be seen if the members of the family are functionally conserved among plant species. In the present work, we have made an attempt to dissect out such functional conservation between the two members of Brassicaceae family i.e. Arabidopsis and Brassica.
Gene expression analysis has revealed that constitutive transcript abundance of BjSOS1, BjSOS2 and BjSOS3 were higher in B. juncea (salt tolerant) as compared to B. nigra (salt sensitive) cultivar (Fig. 1). Under salinity stress, these three SOS genes BjSOS1, BjSOS2 and BjSOS3 members showed differential regulation in contrasting genotypes of Brassica species. Expression dynamics presented in this paper are consistent with the previous study in Brassica seedlings under salinity stress [26, 27]. However, these results are based on qRT-PCR and hence more confirmatory. Under ABA stress, expression of SOS genes increased with increase in stress duration in both the contrasting genotypes. This analysis showed that, these SOS genes are also induced by ABA. Several reports suggest the co-expression of many stress responsive genes under both salinity and ABA [34]. Shi et al. [15] have shown that one of the salt stress responsive SOS members SOS5 showed increased expression under 100 µm ABA treatment in arabidopsis.
In SOS pathway, SOS2 interacts with SOS3 and subsequently SOS2-SOS3 complex phosphorylates the Na+/H+ antiporter SOS1 [9] to stimulate its Na+/H+ exchange activity at the plasma membrane [11, 35]. Using in silico tools, Kushwaha et al. [26] have shown that BjSOS2 interacts with BjSOS3. We did the interaction study by co-expressing BjSOS2 and BjSOS3 in onion peel epidermal cells and found that they are interacting with each other in vivo (Fig. 2). In a similar study, it was observed that PtSOS2 and PtSOS3 of Populus showed in vivo interaction in yeast and plant cell [36]. In-depth study of SOS pathway establish the function of SOS3, a calcium sensor with four EF-hand domains and a N-myristoylation signal peptide, senses the salinity induced increase of intracellular calcium [12, 37, 38], and then activate SOS2, a Ser/Thr protein kinase [10, 14, 39, 40]. Further, we have complemented the ΔAtsos3 mutant in arabidopsis with BjSOS3 and performed root-bending assay. Root-bending assay have shown that BjSOS3-complemented lines as well as the ΔAtsos3 mutant seedlings grew relatively well and appeared healthy on MS agar media. However, on MS agar plate supplemented with 100 or 200 mM NaCl, BjSOS3 complemented lines showed higher survival and more growth in root after bending as compared to the ΔAtsos3 mutant (Fig. 3). Root-bending assay is a powerful method to measure sensitivity of plants for salinity in arabidopsis sos mutant [7, 8]. Shi et al. [9] complemented the sos5 arabidopsis mutant with SOS5 gene isolated from WT and observed similar functional regain and increased tolerance to salinity in root-bending experiments.
We have further performed salinity stress tolerance assay on MS agar plate supplemented with 100 mM of salinity, which showed higher plant vigor and K+ content in BjSOS3 complemented arabidopsis plants as compared to the ΔAtsos3 mutant. Complemented lines accumulate relatively very less Na+ as compared to the ΔAtsos3 mutant (Fig. 4). These results clearly showed that BjSOS3 of Brassica juncea var. CS52 is capable of functionally complementing the ΔAtsos3 in arabidopsis.
Our findings are in corroboration with the findings of many other researchers showing lesser survival rate and reduced growth with lesser K+ content and higher Na+ accumulation in the ΔAtsos3 mutant in Arabidopsis [8, 9, 41]. SOS3 is the first protein in SOS pathway and we investigated the expression of other downstream SOS members i.e. SOS1 and SOS2 in the ΔAtsos3 mutant under control conditions. A relatively lower constitutive transcript abundance of SOS1 was observed in the ΔAtsos3. However, almost same level of SOS1 expression in WT and BjSOS3 complemented line was observed under control conditions (Fig. 5). Our data is corroborated with similar findings showing very low constitutive expression of SOS1 in shoot and root of sos3-1 mutant under control condition [9]. However, induction in SOS1 expression was reported under salinity stress in root tissue but expression was unchanged in the shoots. SOS2 expression in the ΔAtsos3 mutant was equal in magnitude in WT and complemented transgenic lines. In support of our expression analysis, complementation study of SOS2 in arabidopsis sos3-1 mutant have revealed the low expression levels for SOS2 in arabidopsis sos3-1 mutant and higher accumulation in complemented arabidopsis seedlings under control conditions [40].
Regaining of original phenotype in the ΔAtsos3 mutant after complementation with BjSOS3 under ABA stress showed the functional capability of Brassica SOS3 protein with the SOS machinery in arabidopsis. In Aeluropus lagopoids, salinity and ABA individually or in combination or with signaling molecule Ca2+ leads to higher transcript accumulation of SOS pathway genes which supports our stress tolerance assay in arabidopsis seedling [42]. Regaining of near original phenotypes in BjSOS3 complemented arabidopsis may also be linked to the divergence of arabidopsis and Brassica from a common ancestor [17]. Supporting the divergence, comparative genetic mapping has also affirmed co-linear chromosome segments [18, 19] in the family Brassicaceae and linkage arrangements between arabidopsis and Brassica oleracea [20]. In Popular, PtSOS3 complements the ΔAtsos3 and restore the SOS pathway in arabidopsis [36]. Based on these studies, it can be concluded that SOS pathway components are functionally conserved in Brassica species and arabidopsis. We hope that this study will open up new avenues for understanding the salinity tolerance mechanisms operative in diverse plant species, as mediated via the SOS pathway.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
We acknowledge the financial support received from Council of Scientific and Industrial Research and Department of Science and Technology, Ministry of Science and Technology, New Delhi (India). Financial support received from UGC-PURSE scheme to AP and Department of Science and Technology to KKN is acknowledged. We also acknowledge AIRF-JNU for confocal microscope facility.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Pareek A., Sopory S.K., Bohnert H. Govindjee. Abiotic Stress Adaptation in Plants: physiological, molecular and genomic foundation. New York: Springer; 2010. [Google Scholar]
- 2.Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 3.Apse M.P., Aharon G.S., Snedden W.S., Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 4.Parida A., Das A.B., Mittra B. Effects of salt on growth, ion accumulation, photosynthesis and leaf anatomy of the mangrove, Bruguiera parviflora. Trees (Berl.) 2004;108:167–174. [Google Scholar]
- 5.Xiong L., Schumaker K.S., Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl.):s165–s183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J.K., Liu J., Xiong L. Genetic analysis of salt tolerance in Arabidopsis evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H., Ishitani M., Kim C., Zhu J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfter U., Ishitani M., Zhu J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintero F.J., Ohta M., Shi H., Zhu J.K., Pardo J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishitani M., Liu J., Halfter U., Kim C.S., Shi W., Zhu J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S. Schumaker. K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-Like Calcium Binding Protein8 By SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 2009;21:1607–1619. doi: 10.1105/tpc.109.066217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y., Halfter U., Ishitani M., Zhu J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H., Lee B.H., Wu S.J., Zhu J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q., Chen Z.Z., Zhou X.F., Yin H.B., Li X., Xin X.F., Hong X.H., Zhu J.K., Gong Z. Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant. 2009;2:22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers J.E., Abbey C., Anderson S., Chang C., Draye X., Hoppe A.H., Jessup R., Lemke C., Lennington J., Li Z., Lin Y.R., Liu S.C., Luo L., Marler B.S., Ming R., Mitchell S.E., Qiang D., Reischmann K., Schulze S.R., Skinner D.N., Wang Y.W., Kresovich S. Schertz. K.F.; Paterson, A.H. A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics. 2003;165(1):367–386. doi: 10.1093/genetics/165.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paterson A.H., Bowers J.E., Burow M.D., Draye X., Elsik C.G., Jiang C.X., Katsar C.S., Lan T.H., Lin Y.R., Ming R., Wright R.J. Comparative genomics of plant chromosomes. Plant Cell. 2000;12(9):1523–1540. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt R., Acarkan A., Boivin K. Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 2001;39:253–262. [Google Scholar]
- 20.Lukens L., Zou F., Lydiate D., Parkin I., Osborn T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics. 2003;164:359–372. doi: 10.1093/genetics/164.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rana D., van den Boogaart T., O’Neill C.M., Hynes L., Bent E., Macpherson L., Park J.Y., Lim Y.P., Bancroft I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004;40:725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnston J.S., Pepper A.E., Hall A.E., Chen Z.J., Hodnett G., Drabek J., Lopez R., Price H.J. Evolution of genome size of Brassicacae. Ann. Bot. 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth C., Rastogi S., Arvestad L., Dittmar K., Light S., Ekman D., Liberles D.A. Evolution after gene duplication: models, mechanisms, sequences, systems and organisms. J. Exp. Zool. 2007;306B:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R., Mishra M., Gupta B., Parsania C., Singla-Pareek S.L., Pareek A. De novo assembly and characterization of stress transcriptome in a salinity-tolerant variety CS52 of Brassica juncea. PLoS One. 2015;10(5):e0126783. doi: 10.1371/journal.pone.0126783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar G., Purty R.S., Sharma M.P., Singla-Pareek S.L., Pareek A. Physiological response among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J. Plant Physiol. 2009;166:507–520. doi: 10.1016/j.jplph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Kumar G., Purty R.S., Singla-Pareek S.L., Pareek A. Maintenance of stress related transcripts in tolerant cultivar at a level higher than sensitive one appears to be a conserved salinity response among plants. Plant Signal. Behav. 2009;4(5):431–434. doi: 10.4161/psb.4.5.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kushwaha H.R., Kumar G., Verma P.K., Singla-Pareek S.L., Pareek A. Analysis of a salinity induced BjSOS3 protein from Brassica indicate it to be structurally and functionally related to its ortholog from Arabidopsis. Plant Physiol. Biochem. 2011;49(9):996–1004. doi: 10.1016/j.plaphy.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Kumari S., Panjabi Nee Sabharwal V., Kushwaha H.R., Sopory S.K., Singla-Pareek S.L., Pareek A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genomics. 2009;9:109–123. doi: 10.1007/s10142-008-0088-5. [DOI] [PubMed] [Google Scholar]
- 30.Ashraf M. Relationships between growth and gas exchange characteristics in some salt-tolerant amphidiploid Brassica species in relation to their diploid parents. Environ. Exp. Bot. 2001;45:155–163. doi: 10.1016/s0098-8472(00)00090-3. [DOI] [PubMed] [Google Scholar]
- 31.Munns R., James R.A. Screening methods for salt tolerance: a case study with tetraploid wheat. Plant Soil. 2003;253:201–218. [Google Scholar]
- 32.Chao D.Y., Dilkes B., Luo H., Douglas A., Yakubova E., Lahner B., Salt D.E. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science. 2013;341(6146):658–659. doi: 10.1126/science.1240561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C., Zhao L., Zhang H., Yang Z., Wang H., Wen S., Zhang C., Rustgi S., von Wettstein D., Liu B. Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. USA. 2014;111(32):11882–11887. doi: 10.1073/pnas.1412839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi S., Seki M., Ishida J., Satou M., Sakurai T., Narusaka M., Kamiya A., Nakajima M., Enju A., Akiyama K., Yamaguchi-Shinozaki K., Shinozaki K. Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. 2004. [DOI] [PubMed]
- 35.Qiu Q.S., Barkla B.J., Vera-Estrella R., Zhu J.K., Schumaker K.S. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 2003;132:1041–1052. doi: 10.1104/pp.102.010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang R.J., Liu H., Bao Y., Lv Q.D., Yang L., Zhang H.X. The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 2010;74(4-5):367–380. doi: 10.1007/s11103-010-9680-x. [DOI] [PubMed] [Google Scholar]
- 37.Liu J., Zhu J.K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Barrena M.J., Fujii H., Angulo I., Martínez-Ripoll M., Zhu J.K., Albert A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell. 2007;26:427–435. doi: 10.1016/j.molcel.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., Ishitani M., Halfter U., Kim C.S., Zhu J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA. 2000;97:3703–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y., Qiu Q.S., Quintero F.J., Pardo J.M., Ohta M., Zhang C., Schumaker K.S., Zhu J.K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement of its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis. Plant Cell. 2004;16:435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J., Zhang W., Guo Y. Arabidopsis SOS3 plays an important role in salt tolerance by mediating calcium-dependent microfilament reorganization. Plant Cell Rep. 2013;32(1):139–148. doi: 10.1007/s00299-012-1348-3. [DOI] [PubMed] [Google Scholar]
- 42.Jannesar M., Razavi K., Saboora A. Effects of salinity on expression of the salt overly sensitive genes in Aeluropus lagopoides. Aust. J. Crop Sci. 2014;1:1–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.