Abstract
Background
Mouse models of glioblastoma (GBM), the most aggressive primary brain tumor, are critical for understanding GBM pathology and can contribute to the preclinical evaluation of therapeutic agents. Platelet-derived growth factor (PDGF) signaling has been implicated in the development and pathogenesis of GBM, specifically the proneural subtype. Although multiple mouse models of PDGF-driven glioma have been described, they require transgenic mice engineered to activate PDGF signaling and/or impair tumor suppressor genes and typically represent lower-grade glioma.
Methods
We designed recombinant lentiviruses expressing both PDGFB and a short hairpin RNA targeting Cdkn2a to induce gliomagenesis following stereotactic injection into the dentate gyrus of adult immunocompetent mice. We engineered these viruses to coexpress CreERT2 with PDGFB, allowing for deletion of floxed genes specifically in transduced cells, and designed another version of this recombinant lentivirus in which enhanced green fluorescent protein was coexpressed with PDGFB and CreERT2 to visualize transduced cells.
Results
The dentate gyrus of injected mice showed hypercellularity one week post-injection and subsequently developed bona fide tumors with the pathologic hallmarks of GBM leading to a median survival of 77 days post-injection. Transcriptomic analysis of these tumors revealed a proneural gene expression signature.
Conclusion
Informed by the genetic alterations observed in human GBM, we engineered a novel mouse model of proneural GBM. While reflecting many of the advantages of transgenic mice, this model allows for the facile in vivo testing of gene function in tumor cells and makes possible the rapid production of large numbers of immunocompetent tumor-bearing mice for preclinical testing of therapeutics.
Keywords: GBM, glioma, mouse models, PDGF, proneural glioblastoma
Importance of the study
The development of immunocompetent mouse models that accurately reflect the molecular and histological characteristics of GBM is critical to the understanding of tumorigenesis and for preclinical testing of therapeutics. We have developed a novel mouse model of PDGF-driven GBM mediated by the stereotactic injection of lentiviruses engineered to enhance PDGF signaling while disrupting Cdkn2a function. These viruses were also engineered to contain an inducible Cre recombinase, which allows for tumor cell specific testing of gene function in vivo using mice harboring floxed alleles. The injection of these lentiviruses led to the development of brain tumors with the pathologic hallmarks of GBM and a transcriptome profile corresponding to the proneural subtype of GBM. Further, the penetrance of tumors in this model is very high and the course of tumorigenesis highly predictable, thereby enhancing its usefulness for preclinical testing of therapeutics.
GBM is the most aggressive primary brain tumor and patients with this tumor have a median survival of 12–15 months despite multimodal therapy including chemotherapy, radiation therapy, and surgery.1,2 Although histologically similar, 4 transcriptomic subgroups of GBM have been revealed by extensive molecular profiling: mesenchymal, classical, neural, and proneural.3,4 The proneural subtype, which is particularly resistant to current therapy, is characterized by genomic alterations, including gain-of-function mutations in IDH1 and PDGFRA, and loss-of-function mutations in CDKN2A/B and TP53.3–6
Increased PDGF signaling, which can mimic the consequences of gain-of-function PDGFRA alterations, has been shown to drive gliomagenesis in mouse models, although these models oftentimes develop low-grade glioma.7–9 In these mouse models of brain tumors, the deletion of tumor suppressor genes Cdkn2a, Trp53, or Pten using transgenic mice has been shown to lead to more aggressive and higher histological grade tumors.9–12 The need for multiple genetic alterations to create mouse models of GBM impedes the development of models to examine specific multi-gene oncogenic pathways. Such experimentation requires transgenic mice to be crossed with a large number of mice carrying nonlethal gene deletions and would not likely lead to models in which tumor-specific deletion of therapeutic target candidate genes would occur in established tumors. Even if this was feasible, the use of such models for preclinical evaluation of new therapeutics would be compromised by the variable latency and penetrance that characterizes most transgenic tumor models.
The use of recombinant lentiviruses based on replication incompetent HIV-1 to drive gliomagenesis has been evaluated and used to model gliomas that mimic the mesenchymal subtype.13–15 In these experiments, enhanced HRAS signaling and suppressed Trp53 in a multitude of CNS cell types were used to initiate tumorigenesis.14 We sought to extend this strategy and develop a recombinant lentivirus to mediate a model of proneural GBM. We prepared lentiviruses designed to express human PDGFB and inhibit Cdkn2a expression. We found that these lentiviruses were able to drive tumorigenesis in mice injected in the dentate gyrus, a site in which neural stem cells (NSCs) and progenitor cells are enriched.16 Histological evaluation of these tumors showed that they closely resemble human GBM. We also included in this recombinant virus an inducible CreERT2, which is coexpressed along with PDGFB providing an opportunity to identify potential therapeutic targets when using transgenic mice with floxed alleles of the target being evaluated. Further, we engineered another version of this vector to coexpress enhanced green fluorescent protein (eGFP) in addition to PDGFB and CreERT2, allowing for identification of infected cells and thereby facilitating evaluation of important pathologies such as early stages of tumorigenesis and tumor cell heterogeneity. Transcriptomic profiling of tumors arising in our model suggested that they are closely related to human proneural GBM. Overall, we report the development of a single lentiviral vector mediated mouse model of proneural GBM that is driven by enhanced PDGF signaling and silenced Cdkn2a. This model can be used for preclinical testing of therapeutics and in vivo tumor-specific interrogation of gene function. Used in this manner, this model could be a key reagent to facilitate the development of subtype-specific precision therapy for GBM.
Materials and Methods
Cell Culture
We grew 293FT (ThermoFisher Scientific), primary mouse embryo fibroblast (MEF; isolated at e12.5 from C57BL/6J mice [Jax Stock No: 000664]), and U251 (originally obtained from American Type Culture Collection) cells on tissue culture dishes in growth media consisting of Dulbecco’s modified Eagle’s medium with L-glutamine (Corning) supplemented with 1% penicillin/streptomycin (Corning) and 10% fetal bovine serum (Corning).
Transfections
293FT cells were transfected using LipoD293 (Signagen) as indicated in the product guidelines.
Western Blot Analyses
Western blot analyses were performed as previously described.17 The antibodies used and the working dilutions are shown in Supplementary Table S1.
RNA Preparation and Quantitative Real-Time PCR
For microarray analysis, RNA was purified using the Trizol method (ThermoFisher). For quantitative real-time PCR, RNA was extracted using the RNeasy kit as per product guidelines (Qiagen). Data were quantified relative to short hairpin (sh)Control infected MEF cells and normalized to 18S cycle threshold values using the 2−ΔΔCt method. Primers are provided in Supplementary Table S2.
Microarray Analysis
Mouse RNA preparations were evaluated for purity, quantitated, labeled using the Illumina Epicenter TargetAmp Nano Kit according to manufacturer’s guidelines, and probed using the Illumina mouse ref8 array chip. These data were normalized by variance stabilizing transformation followed by robust spline normalization using the lumi package on R.18 Genes with multiple probe sets were processed so that the probe set with the maximum average hybridization signal was taken as the gene expression level for that gene. The complete normalized dataset is available in Supplementary Table S3 and on Gene Expression Omnibus (accession number GSE99361).
Lentivirus Preparation
Lentiviruses were prepared as previously described19 using a 3-plasmid system with the transfer plasmids constructed in this manuscript (shCdkn2a-PDGFB-T2A-CreERT2 and shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2) in addition to pCMV-Delta8.9 and pVSVG. Concentrated lentiviruses were obtained by precipitating viral containing supernatants with PEG6000 as previously described.20 The viral pellets were resuspended in phosphate-buffered saline (PBS) without calcium or magnesium and stored at −80°C until used.
Lentivirus Titration
Lentiviruses were titered using U251 cells. Fifty thousand cells were plated in each well of a 12-well plate (22.1 mm diameter) in growth media containing 10 μg/mL polybrene. One microliter of undiluted lentivirus was added to the first well, followed by 11 ten-fold serial dilutions of the media from the first well into sequential adjacent wells of the plate. The plates were spun down at 3500 g for 45 minutes and placed in a humidified 37°C incubator supplemented with 5% CO2 for 4 days. For titration of lentiviruses encoding shCdkn2a-PDGFB-T2A-CreERT2, the cells were fixed with 10% formalin (acetate buffered, FisherScientific) and stained as previously described21 using an estrogen receptor alpha antibody (Supplementary Table S1), which recognizes CreERT2, followed by an Alexa-555 fluorescent secondary antibody (Supplementary Table S1). Fluorescent colonies in the rhodamine channel across several dilutions were then counted under a 20X dry objective on an Olympus IX73 inverted fluorescence microscope. For titration of lentiviruses encoding shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2, fluorescent colonies in the GFP channel were counted across several dilutions. All titration experiments were done in duplicates.
Construction of Lentiviral Plasmid
The pTOMO lentiviral transfer plasmid (Addgene #26291, a gift from Inder Verma13) was used as the backbone to construct the shCdkn2a-PDGFB-T2A-CreERT2 and the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 plasmids. Briefly, the pTOMO plasmid was digested with XbaI and SalI. The human PDGFB cDNA was PCR amplified with an XbaI site on the 5ʹ end and a BamhI site on its 3ʹ end from a vector used to create a previous human PDGFB-driven mouse model.7 CreERT2 cDNA was PCR amplified from MSCV CreERT2 puro plasmid (Addgene #22776, a gift from Tyler Jacks22) with an SpeI site on the 5ʹ end and a SalI site on the 3ʹ end. These amplicons were then digested with BamHI and SpeI and ligated to a synthesized T2A sequence (IDT oligos designed and annealed) with a BamHI site on its 5ʹ end and a SpeI site on its 3ʹ end in a 3-fragment ligation reaction. The ligated product was then digested with XbaI and SalI and ligated into the XbaI/SalI digested pTOMO. This recombinant plasmid harbored MfeI and NotI sequences located between HIV-1 ψ and the Rev response element. We then ligated the U6promoter-shCdkn2a-cPPT/CTS, cloned from TRCN0000257162 (pLK0.1 vector, selected from 5 shRNAs tested, data not shown) (Sigma) with an MfeI site on the 5ʹ end and a NotI site on the 3ʹ end, to an MfeI/NotI digested PDGF-T2a-CreERT2 plasmid to obtain shCdkn2a-PDGFB-T2A-CreERT2. The shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 was obtained by cloning an eGFP-E2A cDNA (from pEGFPN1, PCR product ligated to a synthesized E2A sequence, IDT) with an NheI site (compatible sticky ends with SpeI) on the 5ʹ end and an SpeI site on the 3ʹ end. This eGFP-E2A was cloned into an SpeI digested shCdkn2a-PDGFB-T2A-CreERT2 yielding shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2. The full plasmid maps are available in Supplementary Figure S1 and the full plasmid sequences are available in Supplementary Tables S4 and S5. All plasmids were verified using Sanger sequencing.
shRNA
shRNA sequences used in the study are available in Supplementary Table S6.
Mouse Colony and Injection Protocol
All mouse experimentation was performed per an Institutional Animal Care and Use Committee–approved protocol.
F1 progeny mice obtained from a cross between C57BL/6J and 129S1 were crossed again with C57BL/6J to generate F2 progeny, which were inbred and used for experimentation. These mixed background mice were used for all stereotactic injections. The B6.Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze/J (Ai9, Jax Stock No: 007909) mouse was used to verify Cre activity in vivo.23 Mice were injected intraperitoneally (i.p.) with either corn oil or tamoxifen following the Jackson Laboratory protocol (https://www.jax.org/research-and-faculty/tools/cre-repository/tamoxifen, accessed August 8, 2017). One hundred microliters of corn oil with or without tamoxifen (75 mg/kg dose) was injected i.p. for 5 consecutive days. Tissues were collected one week after the last i.p. injection.
Stereotactic injections were done as previously described.20 Eight- to 10-week-old mice were anesthetized using 4% isoflurane on a stereotactic frame (Stoelting) and injected with 4–5 µL (total of 0.75 × 106 plaque-forming units, rate of 0.5 µL/min) in the dentate gyrus using the following coordinates relative to bregma: x = −1.1, y = −1.9, and z = −2.5/−2.4/−2.3, with x representing left(−)/right(+), y: anterior(+)/posterior(−), and z: depth from surface of the skull.
Brain Tissue Processing and Immunohistochemistry Staining
When mice showed symptoms of terminal illness such as lethargy or seizures, they were anesthetized by i.p. injection of avertin (2,2,2 tribromoethanol) (25 mg/kg) in PBS (500 µL total). Mice were then perfused with PBS for 5 minutes through an intracardiac puncture into the left ventricle using a peristaltic pump. This was followed by perfusion with 10% formalin in acetate buffer (FisherScientific) for 5 minutes. Brains were then placed in formalin overnight before being processed and embedded in paraffin. Eight-micrometer sections of these paraffin blocks were cut on a microtome and deparaffinized by 2 sequential xylene incubations followed by rehydration through a decreasing gradient of ethanol. Rehydrated sections were then stained using a Leica Bond Max automated staining instrument (Leica Microsystems). Antibodies used, optimal dilutions, and antigen retrieval protocols are described in Supplementary Table S1. Antigen retrieval was done for 20 minutes using the solutions indicated in Supplementary Table S1. Parallel immunohistochemistry (IHC) experiments in which the primary antibody was omitted were negative (data not shown).
Confocal Imaging
Brain sections from the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 model were analyzed using direct fluorescence on a Zeiss LSM510-META confocal microscope. Briefly 40-µm sections were obtained from embedded brains (2.5% agarose in PBS with 4% sucrose) and cut on a vibratome (Leica VT 1200S). The sections were then placed in a 48-well plate, screened for GFP fluorescence using a fluorescent microscope, and mounted using Vectashield antifade mounting medium (Vector Laboratories) on a microscope slide. The ZEN imaging suite was then used to construct maximal projection images from these sections.
Statistics
R version 3.3.2 was used for statistical analysis. Two-tailed Student’s t-test was used to evaluate differences between 2 groups. The correlation method to subtype mouse tumors used was previously described.9,19 For subtyping according to the glioma subtypes described by Alcantara Llaguno et al,24 we first performed median normalization of transcriptomic data from shCdkn2a-PDGFB-T2A-CreERT2 GBM to data from GSE57038, which includes tumors that developed in Nestin-Cre-ERT2 (type 1) and NG2-CreERT2 (type 2) transgenic mice. We then computed the Spearman correlation score using the same method we used for The Cancer Genome Atlas (TCGA) subtyping with one exception: instead of the clanc840 signature reported by Verhaak et al,25 we used the 310 gene signature reported by Alcantara Llaguno et al. We validated this method by subtyping the tumors reported in the Alcantara Llaguno dataset (GSE57038).24
Gene Set Enrichment Analysis
GSEA was performed as described on the Broad Institute website (http://software.broadinstitute.org/gsea/index.jsp, accessed August 8, 2017). We used the Azare_Neoplastic_Transformation_by_Stat3_Up, Biocarta_Nfkb_Pathway, and KEGG_Wnt_Signaling_Pathway gene sets from the MSigDB collection (http://software.broadinstitute.org/gsea/msigdb/genesets.jsp, accessed August 8, 2017) to probe the signal transducer and activator of transcription 3 (STAT3), nuclear factor-kappaB, and Wnt signaling pathways, respectively. The transcriptome data from our model were median normalized to the data from TCGA prior to GSEA analyses.
Results
Development and Characterization of a Gliomagenic Recombinant Lentiviral Vector Encoding shCdkn2a-PDGFB-T2A-CreERT2
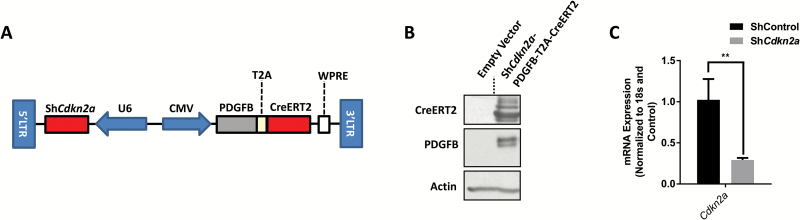
To develop a recombinant virus that could model the development of proneural GBM, we designed a lentiviral vector that expresses human PDGFB driven by a cytomegalovirus (CMV)-derived promoter and an shRNA targeting Cdkn2a driven by a U6 promoter. We also engineered this vector to coexpress CreERT2 along with PDGFB using a T2A cleaving peptide signal (Fig. 1A, Supplementary Table S4). CreERT2 was added to this vector to facilitate future studies to test the importance of specific genes of interest using mice harboring floxed alleles of such genes. The 293FT cells transfected with the recombinant plasmid DNA shown in Supplementary Table S4 and depicted in Fig. 1A expressed both PDGFB and CreERT2 (Fig. 1B). The shCdkn2a used in the recombinant vector suppressed the expression of Cdkn2a in primary MEFs (Fig. 1C).
Fig. 1.
Development and characterization of a gliomagenic recombinant lentiviral vector encoding shCdkn2a-PDGFB-T2A-CreERT2. (A) Vector design and (B) expression of PDGFB and CreERT2 from the vector in 293FT cells. (C) Quantitative real-time PCR analysis of Cdkn2a expression in primary mouse fibroblasts infected with the shCdkn2a virus compared with a control shRNA (2 multiplicity of infection). Mean (C) ± 1 SD of 3 independent experiments are shown. **P < 0.01.
Stereotactic Injection of Lentiviruses Encoding shCdkn2a-PDGFB-T2A-CreERT2 into the Dentate Gyrus Gives Rise to GBM
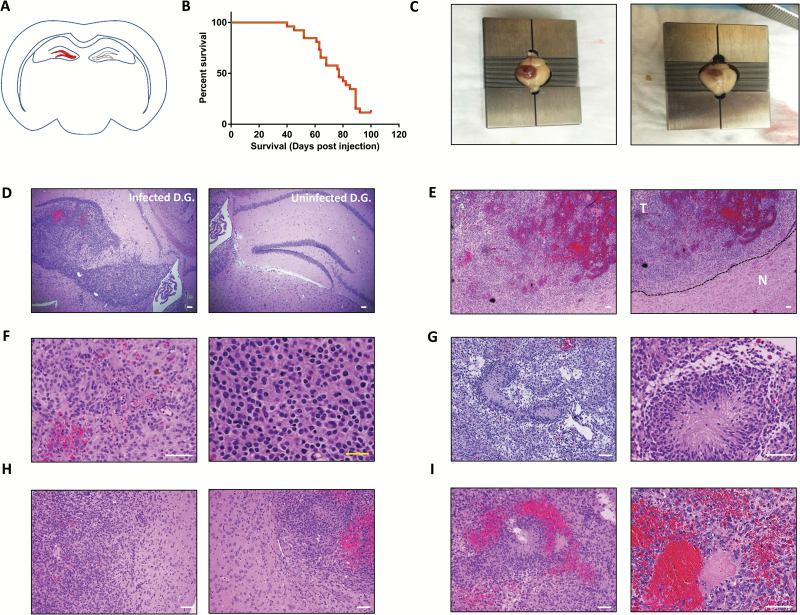
We prepared concentrated lentiviruses (depicted in Fig. 1A) and stereotactically injected the dentate gyrus of immunocompetent adult mice with 750000 transducing units as indicated in the Materials and Methods section (Fig. 2A). Mice injected with lentiviruses encoding shCdkn2a-PDGFB-T2A-CreERT2 had a median survival of 77 days after injection (n = 26 in experiment, 23 animals developed tumors, penetrance of 88.5%; Fig. 2B), variably developed seizures, and became lethargic. Gross examination of brains from these mice invariably (n = 15 independent tumors analyzed) indicated hemorrhagic, neoplastic lesions at the injection site (Fig. 2C). Histological examination of these neoplastic lesions revealed malignant lesions originating in the dentate gyrus (Fig. 2D). These tumors exhibited hypercellularity (Fig. 2E, F), pseudopalisading necrosis (Fig. 2G), invasion of normal stroma (Fig. 2H), and hemorrhage (Fig. 2I), which are pathologic hallmarks of GBM.1,2,26 We describe the frequency of occurrence of these pathologic hallmarks in Supplementary Table S7.
Fig. 2.
Stereotactic injection of lentiviruses encoding shCdkn2a-PDGFB-T2A-CreERT2 into the dentate gyrus gives rise to GBM. (A) A schematic diagram of the injection site (dentate gyrus marked in red) on a coronal section depiction of an adult mouse brain. (B) Kaplan–Meier survival curve showing overall survival of mice injected with shCdkn2a-PDGFB-T2A-CreERT2 lentiviruses. (C) Gross appearance of injected brains during terminal moribund status of mice. Hematoxylin and eosin (H&E) stained tumor sections from mice injected with the shCdkn2a-PDGFB-T2A-CreERT2 lentivirus after (D) 45 days from initial injection and (E) during terminal moribund status. The right panel in (E) displays the sharp contrast in cellularity between tumor and normal tissue (indicated in the image by T and N, respectively, separated by dashed line). H&E stained tumor sections from PDGF-driven tumors showing (F) increased cellularity, (G) pseudopalisading necrosis, (H) invasion of normal stroma, and (I) hemorrhage. (C) and (E–I) contain 2 panels showing 2 representative examples of what we observed in our study. White scale bar: 100 µm. Yellow scale bar: 10 µm.
Immunohistochemical Staining of PDGF-Driven GBM Tumor Sections with Antibodies Against GBM Markers
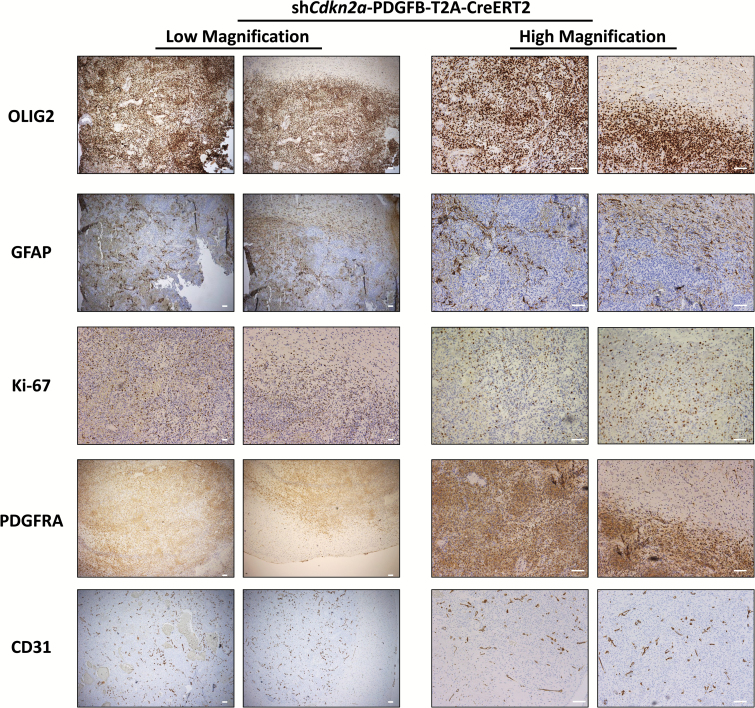
To examine the expression of glioma markers in our model, we used IHC to evaluate sections of tumor tissue (n = 8 independent tumors analyzed) with antibodies that recognize oligodendrocyte transcription factor (OLIG2), glial fibrillary acidic protein (GFAP), Ki-67, and cluster of differentiation (CD)31 (platelet endothelial cell adhesion molecule 1). We also examined the expression of PDGFRA, the major receptor tyrosine kinase that mediates PDGF signaling in glioma.27,28 OLIG2 and PDGFRA were expressed uniformly in tumor cells, while GFAP was detectable in patches. Ki-67 expression was detected in all the tumors analyzed, although not all tumor cells expressed this proliferation marker, which is consistent with staining patterns observed in human GBM.29 CD31 staining confirmed the presence of blood vessels, consistent with the extensive vascularization found in GBM26 (Fig. 3). We also examined the expression of p16INK4A (cyclin-dependent kinase inhibitor 2A [CDKN2A]) in tumors that developed in our model and determined that these tumors do not have detectable p16INK4A protein expression (Supplementary Figure S2).
Fig. 3.
Immunohistochemical staining of shCdkn2a-PDGFB-T2A-CreERT2 GBM tumor sections with antibodies against GBM markers. Stains for OLIG2, GFAP, Ki-67, PDGFRA, and CD31 on tumor sections obtained from mice injected with the gliomagenic lentivirus under low (left panels, 4X magnification) and high (10X) magnification (right panels). Sections were counterstained with hematoxylin. We show 2 representative images for each of the GBM markers under both 4X and 10X magnifications. Scale bar: 100 µm.
Fluorescent Tracking of PDGF-Driven GBM Reveals Early Stages of Tumorigenesis
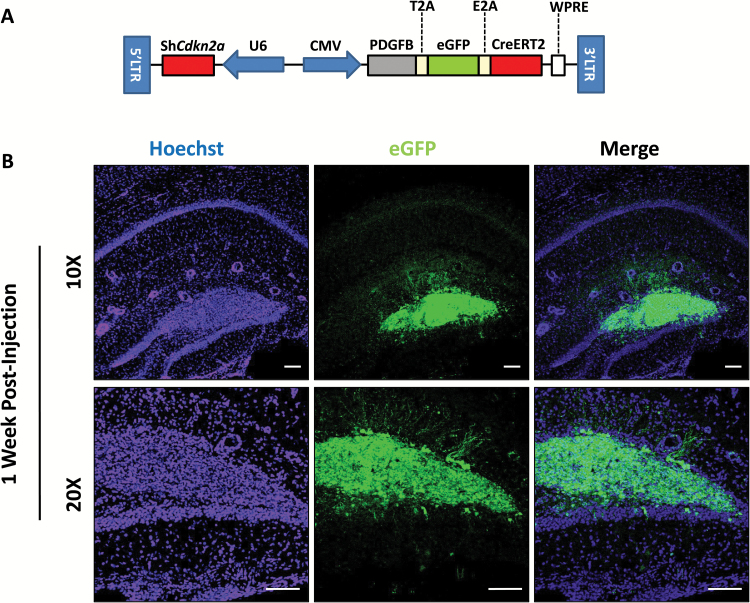
To examine early tumorigenesis in this model, we added eGFP to the same coding locus as PDGFB and CreERT2, leading to expression of eGFP in transduced cells (Fig. 4A). We collected mouse brains one week after injection of the dentate gyrus with the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 lentivirus and examined sections for eGFP fluorescence using confocal microscopy. We observed hypercellularity within the dentate gyrus one week post-injection (Fig. 4B), before significant invasion of adjacent tissue, necrosis, or neo-angiogenesis could be detected. These eGFP-positive cells, however, may not yet be fully transformed tumor cells, though we know that tumors arising in this model are found earliest at this location.
Fig. 4.
Fluorescent tracking of shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 transduced cells reveals early stages of tumorigenesis. (A) Vector design of shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 which expresses eGFP in the same coding locus as PDGFB and CreERT2. Infected cells in the dentate gyrus of an adult mouse (B) 1 week post lentiviral injection under 10X and 20X magnification. GFP (from the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 lentivirus infected cells) and Hoechst (nuclei) channels are shown. Scale bar: 100 µm.
We also used this model to verify the activity of CreERT2. We stereotactically injected the eGFP-expressing virus into a Cre reporter mouse (see the Materials and Methods section), in which a floxed stop cassette was placed between a CAG promoter and a tdTomato red fluorescent protein coding gene and thereby allowed detection of Cre-mediated expression of tdTomato.23 Cre reporter mice injected with the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 lentiviruses and treated with tamoxifen showed tdTomato fluorescence in GFP-positive cells, while mice injected with the same lentiviruses that were treated with corn oil had only a few cells that expressed tdTomato expression (Supplementary Figure S3).
GFP Staining of shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 GBM Tumor Sections Shows that Tumor Cells Are Predominantly Infected with the Lentiviral Vector
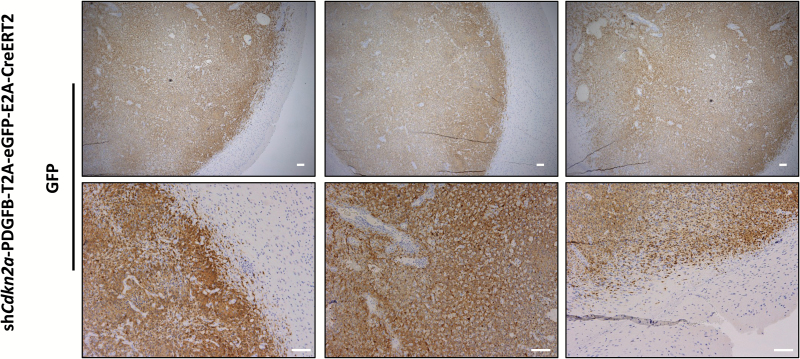
Previous findings in other models of PDGF-driven glioma indicated that expression of PDGFB recruits and transforms stromal cells that do not express PDGFB.12,30 To determine if this occurs in our model, we examined GFP expression in tumor sections from mice injected with the shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 lentivirus (n = 5 independent tumors analyzed). We found that most malignant cells in these tumors express GFP, suggesting strongly that these tumor cells also express the PDGFB transgene (Fig. 5). While stromal cells may be attracted to these tumors and stromal cells present at the site of injection might be infected with inoculated virus, our findings make it unlikely that significant numbers of uninfected cells contribute to the bulk cellular content of these malignancies, a sharp contrast to what occurs in previous PDGF-driven glioma models.12,30
Fig. 5.
GFP staining of shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 GBM tumor sections shows that tumor cells are predominantly infected with the lentiviral vector. Low and high magnification (top and bottom panels, respectively) of shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 GBM tumor sections stained with a GFP antibody. Sections were counterstained with hematoxylin. Scale bar: 100 µm.
Transcriptomic Profiling Classifies shCdkn2a-PDGFB-T2A-CreERT2 Induced Tumors as Proneural GBM
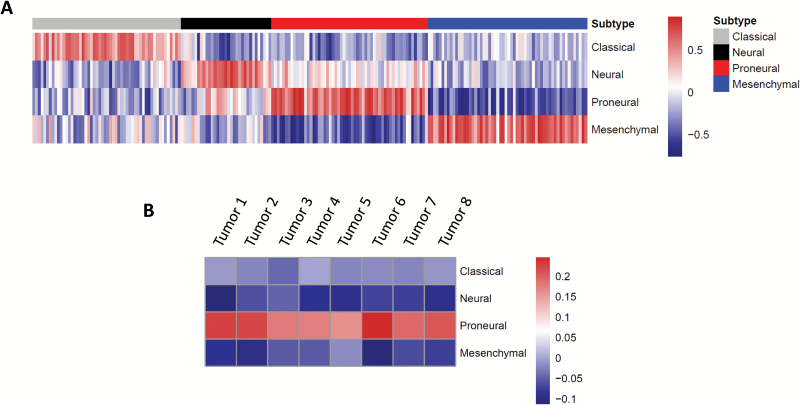
We performed transcriptomic profiling of tumors that arose following lentiviral injection of shCdkn2a-PDGFB-T2A-CreERT2 lentiviruses to determine the molecular subtype to which they belong. We used a previously described correlation method9,19 to subtype murine glioma and revalidated it (Fig. 6A) using transcriptome profiles of primary human GBM from TCGA.31 Our revalidation showed that tumor samples had the highest correlation score to the subtype they are assigned to by TCGA4 with 89% accuracy (Fig. 6A, data not shown). When we performed the same subtyping strategy on 8 independent tumors from shCdkn2a-PDGFB-T2A-CreERT2 injected mice, we observed the highest correlation score with the proneural subtype and negative correlations corresponding to all other subtypes (Fig. 6B). This indicates that lentiviruses encoding shCdkn2a-PDGFB-T2A-CreERT2 induced tumors that correspond to human proneural GBM. Further, we performed GSEA comparing the transcriptomes of tumors arising in our mouse model with human mesenchymal GBM tumors from TCGA. We found that the gene set corresponding to the Wnt pathway was enriched in proneural tumors arising in our mouse model, a finding predicted by the characteristic activation of this pathway in human proneural GBM, while the gene set corresponding to STAT3 signaling was enriched in human mesenchymal GBM, consistent with what has been found in human GBM data (Supplementary Table S8, Supplementary Figure S4).25 Additionally, we performed transcriptomic subtyping using a classification system derived from glioma driven by mutated Pten, Trp53, and Nf1 in different cellular lineages of the central nervous system.24 We found that the majority of tumors arising in our model resemble tumors that arise from nestin-positive neural progenitor cells (Supplementary Figure S5), a finding consistent with the presumed origin of these tumors in neural stem cells of the hippocampus.
Fig. 6.
Transcriptomic profiling classifies shCdkn2a-PDGFB-T2A-CreERT2 induced tumors as proneural GBM. (A) Heatmap showing the subtype signature classification by TCGA (classification bar on top) and validation correlation scores of primary human GBM tumors using Spearman correlation. (B) Heatmap of 8 independent tumor samples from shCdkn2a-PDGFB-T2A-CreERT2 GBM shows an enrichment of a proneural signature using the same method as (A).
Discussion
We report the development and characterization of a single lentiviral vector that can mediate with high efficiency a mouse model of proneural GBM without the need for transgenic mice. The faithful representation of histological and molecular markers in this model to human proneural GBM, in addition to the immunocompetent setting in which these tumors were evaluated, makes this model uniquely efficient in the pursuit of important research questions. This model allows tumor cell specific deletion of any floxed allele, efficient preclinical testing of therapeutic molecules, and the evaluation of immune cell function and immune interventions within immunocompetent animals in GBM pathology, specifically in the proneural subtype of GBM.
We engineered a lentivirus to express PDGFB and an shCdkn2a (Fig. 1A). When this virus infected cells of the dentate gyrus, mice developed GBM. Histological examination (Fig. 2) and IHC evaluation of genes known to be expressed in glioma32–34 (Fig. 3) revealed these tumors to have characteristics of GBM. Further, transcriptomic characterization using a validated correlation subtyping method9,19 (Fig. 6A) indicated that these GBM closely mimic human proneural GBM (Fig. 6B).
The tumors that developed in our shCdkn2a-PDGFB-T2A-CreERT2 model invade normal brain stroma (Fig. 2H). This observation was further validated using our shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 model, in which we were able to evaluate GFP expression and showed that GFP-positive cells were invading the normal brain stroma (Fig. 5 lower left and lower right panels).
Importantly, 2 other models of PDGF-driven glioma showed that a substantial amount of normal cells are recruited to tumor sites and undergo transformation in the tumor microenvironment allowing non-PDGF-expressing cells to contribute to the tumor bulk.12,35 These recruited, transformed cells appear to be dependent on pathways other than PDGF signaling, specifically epidermal growth factor receptor and insulin-like growth factor receptor tyrosine kinase signaling.12 In our shCdkn2a-PDGFB-T2A-eGFP-E2A-CreERT2 model, tumors were mostly composed of transduced cells, as evident by GFP positivity (Fig. 5). We have not pursued experimentation to evaluate why such differences in cellular heterogeneity between our model and the previous models may exist. We postulate that these differences might reflect a different cell of origin or the tumor grade for the tumors examined.24 To that extent, we transduced cells of the dentate gyrus, which include NSCs, progenitor cells of many fates, and differentiated cells,36 using lentiviruses which are capable of infecting proliferating and nonproliferating cells. In contrast, the previously reported models use retrovirus-based systems, which transduce proliferating cells.8,9 Importantly, other lentiviral models of GBM driven by constitutive HRAS signaling and suppressed Trp53 formed tumors that were also mostly composed of transduced cells.13 Our data suggest that the tumors arising in our proneural GBM model represent an expansion of transduced cells. Future studies are warranted to attempt the identification of any GFP-negative malignant cells in tumors arising in our model to better understand the contribution of stromal cells and their transformation in these tumors.
One of the advantages of using the models we developed is that any mouse strain and any transgenic mouse harboring floxed alleles of a gene of interest can be infected easily. Our addition of inducible CreERT2 should allow for tumor-specific deletion of any floxed gene (Supplementary Figure S1). Such experimentation has been done in a mouse model of lentiviral-mediated mesenchymal GBM driven by constitutive HRAS signaling and depleted Trp53. In this model, an inducible Cre expressed from the gliomagenic lentivirus induced the tumor-specific deletion of floxed inhibitor of DNA binding (ID) genes, ID1–2, and evaluated the importance of these genes in maintaining mesenchymal GBM cells.15 This provides a proof of concept that the model we describe here can be used to evaluate the function of floxed genes that might play an important role in the development and maintenance of proneural GBM.
Recent reports have indicated that GBM tumors typically include cells that belong to more than one molecular subtype,37 which may explain tumor subtype switching in patients with recurrent GBM.38 This very important finding calls into attention the relevance of single subtype specific mouse models. We believe that designing faithful mouse models, such as the one we describe here, can provide a setting in which a comprehensive understanding of each subtype can be pursued. Future studies can lead to the development of tumors engineered to reflect multiple molecular subtypes and allow for study of the complex cellular interactions that may underlie molecularly heterogeneous GBM tumors.39
While the tumors developed in this model arise in the stem cell and progenitor cell rich niche of the dentate gyrus, our experimentation does not identify specifically the cell of origin of tumors arising in our model. Future studies can be designed to show evidence for this important line of experimentation, especially considering evidence that the cell of origin can greatly affect the characteristics of the resulting tumor.24,40
This report describes a lentiviral-mediated mouse model of proneural GBM. The versatility of using lentiviruses and the presence of CreERT2 in this model allow for rapid in vivo, tumor-specific evaluation of gene function in any transgenic mouse. These properties, in addition to the short course of tumorigenesis in this model, allow for rapid preclinical testing of therapeutics for proneural GBM, which could help accelerate the development of GBM subtype–specific therapy.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was supported by grants from the Jordan and Kyra Memorial Foundation (M.A.I.), the Andy Fund (M.A.I.), and the Theodora B. Betz Foundation (M.A.I.).
Conflict of interest statement. The authors disclose no potential conflicts of interest.
Supplementary Material
Acknowledgments
We thank all Israel and Luikart laboratory members for helpful discussions. We thank Ms. Tabatha Richardson at Dartmouth for her editorial support. Resources used for this study were the Norris Cotton Cancer Center (NCCC) Genomics Shared Resource, NCCC i.p.IM-Microscopy Shared Resource (Kenneth Orndoff), NCCC Molecular Biology Shared Resource, NCCC Pathology Shared Resource, and the Center for Comparative Medicine and Research (CCMR) at Dartmouth.
References
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brennan CW, Verhaak RG, McKenna A et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. [DOI] [PubMed] [Google Scholar]
- 6. Dunn GP, Rinne ML, Wykosky J et al. . Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hitoshi Y, Harris BT, Liu H, Popko B, Israel MA. Spinal glioma: platelet-derived growth factor B-mediated oncogenesis in the spinal cord. Cancer Res. 2008;68(20):8507–8515. [DOI] [PubMed] [Google Scholar]
- 8. Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15(15):1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lei L, Sonabend AM, Guarnieri P et al. . Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6(5):e20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindberg N, Jiang Y, Xie Y et al. . Oncogenic signaling is dominant to cell of origin and dictates astrocytic or oligodendroglial tumor development from oligodendrocyte precursor cells. J Neurosci. 2014;34(44):14644–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tchougounova E, Kastemar M, Bråsäter D, Holland EC, Westermark B, Uhrbom L. Loss of Arf causes tumor progression of PDGFB-induced oligodendroglioma. Oncogene. 2007;26(43):6289–6296. [DOI] [PubMed] [Google Scholar]
- 12. Fomchenko EI, Dougherty JD, Helmy KY et al. . Recruited cells can become transformed and overtake PDGF-induced murine gliomas in vivo during tumor progression. PLoS One. 2011;6(7):e20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marumoto T, Tashiro A, Friedmann-Morvinski D et al. . Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15(1):110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedmann-Morvinski D, Bushong EA, Ke E et al. . Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338(6110):1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niola F, Zhao X, Singh D et al. . Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest. 2013;123(1):405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gage FH. Neurogenesis in the adult brain. J Neurosci. 2002;22(3):612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Z, Rahme GJ, Chatterjee PD, Havrda MC, Israel MA. ID2 promotes survival of glioblastoma cells during metabolic stress by regulating mitochondrial function. Cell Death Dis. 2017;8(2):e2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. [DOI] [PubMed] [Google Scholar]
- 19. Rahme GJ, Zhang Z, Young AL et al. . PDGF engages an E2F-USP1 signaling pathway to support ID2-mediated survival of proneural glioma cells. Cancer Res. 2016;76(10):2964–2976. [DOI] [PubMed] [Google Scholar]
- 20. Fricano-Kugler CJ, Williams MR, Salinaro JR, Li M, Luikart B. Designing, packaging, and delivery of high titer CRISPR retro and lentiviruses via stereotaxic injection. J Vis Exp. 2016;(111):doi: 10.3791/53783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahme GJ, Israel MA. Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene. 2015;34(1):53–62. [DOI] [PubMed] [Google Scholar]
- 22. Kumar MS, Pester RE, Chen CY et al. . Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madisen L, Zwingman TA, Sunkin SM et al. . A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alcantara Llaguno SR, Wang Z, Sun D et al. . Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell. 2015;28(4):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 27. Nazarenko I, Hede SM, He X et al. . PDGF and PDGF receptors in glioma. Ups J Med Sci. 2012;117(2):99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pietras K, Sjöblom T, Rubin K, Heldin CH, Ostman A. PDGF receptors as cancer drug targets. Cancer Cell. 2003;3(5):439–443. [DOI] [PubMed] [Google Scholar]
- 29. Kayaselçuk F, Zorludemir S, Gümürdühü D, Zeren H, Erman T. PCNA and Ki-67 in central nervous system tumors: correlation with the histological type and grade. J Neurooncol. 2002;57(2):115–121. [DOI] [PubMed] [Google Scholar]
- 30. Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu QR, Park JK, Noll E et al. . Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98(19):10851–10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacque CM, Vinner C, Kujas M, Raoul M, Racadot J, Baumann NA. Determination of glial fibrillary acidic protein (GFAP) in human brain tumors. J Neurol Sci. 1978;35(1):147–155. [DOI] [PubMed] [Google Scholar]
- 34. Raghavan R, Steart PV, Weller RO. Cell proliferation patterns in the diagnosis of astrocytomas, anaplastic astrocytomas and glioblastoma multiforme: a Ki-67 study. Neuropathol Appl Neurobiol. 1990;16(2):123–133. [DOI] [PubMed] [Google Scholar]
- 35. Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scharfman HE. The enigmatic mossy cell of the dentate gyrus. Nat Rev Neurosci. 2016;17(9):562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patel AP, Tirosh I, Trombetta JJ et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J, Cazzato E, Ladewig E et al. . Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snuderl M, Fazlollahi L, Le LP et al. . Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 40. Alcantara Llaguno SR, Parada LF. Cell of origin of glioma: biological and clinical implications. Br J Cancer. 2016;115(12):1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.