Abstract
microRNAs (miRNAs) have wide-ranging effects on large-scale gene regulation. As such, they play a vital role in dictating normal development, and their aberrant expression has been implicated in cancer. There has been a large body of research on the role of miRNAs in medulloblastoma, the most common malignant brain tumor of childhood. The identification of the 4 molecular subgroups with distinct biological, genetic, and transcriptional features has revolutionized the field of medulloblastoma research over the past 5 years. Despite this, the growing body of research on miRNAs in medulloblastoma has largely focused on the clinical entity of a single disease rather than the molecular subgroups. This review begins by highlighting the role of miRNAs in development and progresses to explore their myriad of implications in cancer. Medulloblastoma is characterized by increased proliferation, inhibition of apoptosis, and maintenance of stemness programs—features that are inadvertently regulated by altered expression patterns in miRNAs. This review aims to contextualize the large body of work on miRNAs within the framework of medulloblastoma subgroups. The goal of this review is to stimulate new areas of research, including potential therapeutics, within a rapidly growing field.
Keywords: medulloblastoma, miRNA, miRNA therapeutics, pediatric oncology, subgroups
microRNAs (miRNAs) are short 18–25 nucleotide noncoding RNAs that regulate gene expression via posttranscriptional processes.1 The discovery and identification of miRNAs—as a new mechanism of gene regulation—transformed our understanding of the regulation process. Through complementary sequences to their respective target mRNA, miRNAs have been shown to regulate thousands of potential target genes. This complex gene regulatory network is now accepted as a critical process in normal development. miRNAs have wide-ranging effects on large-scale gene regulation, and as such, their aberrant expression has been associated with a variety of different pathological states, including cancer. In this review we highlight the major findings implicating miRNAs in the pathogenesis of medulloblastoma, the most common malignant pediatric brain tumor.
The Biogenesis of miRNAs and Their Role in Normal CNS Development
The biogenesis of miRNAs follows an intricate process of transcription and posttranscriptional modification. The canonical pathway of miRNA biogenesis initiates in the nucleus, where RNA polymerase II transcribes the nascent primary miRNA (pri-miRNA). These transcripts are similar to protein-coding transcripts, which are usually long nucleotide sequences with 5ʹ caps and poly-adenylated 3ʹ tails. Due to the specific stem-loop secondary structures of pri-miRNA, they are recognized and processed by the nuclear ribonuclease Drosha and its partner DiGeorge syndrome critical region 8 (DGCR8), resulting in a shorter precursor miRNA called pre-miRNA. Following this, the pre-miRNA is transported to the cytoplasm via exportin-5 and undergoes further processing by the ribonuclease Dicer1.2 The miRNA commonly undergoes strand separation in which a mature miRNA is incorporated into the RNA-induced silencing complex (RISC) and the passenger miRNA is typically degraded. The mature miRNA and RISC is then able to regulate mRNA translation through the binding of complementary sequences which are generally located in the 3ʹ untranslated regions of the target mRNA.1
Over the past decade much has been revealed about miRNA biology; however, there are still large gaps in our understanding of the full spectrum of miRNA function. Typically, target mRNA is silenced by miRNA through a process called mRNA cleavage. A unique miRNA has the potential to target hundreds if not thousands of mRNA and thus regulate global gene expression. The regulation of miRNAs is not completely understood, but it is now widely accepted that miRNA transcription is facilitated through RNA polymerase II associated transcription factors.3 Furthermore, promoters of protein-coding genes can regulate miRNA expression located within their respective introns, and RNA-binding proteins can also modulate the stability of miRNAs. One prominent example of this is regulation of LIN28, which suppresses the biogenesis of let-7 family miRNAs by inhibiting pri-miRNA cleavage. Epigenetic mechanisms of miRNA transcriptional silencing have also been demonstrated and represent another level of regulation.4
miRNA-directed gene regulation is an essential pathway in virtually all mammalian cell types examined to date.5 Due to the ubiquitous and promiscuous nature of miRNAs in regulating many mRNA targets, miRNA expression must be controlled in both space and time. It is estimated that more than 60% of documented miRNAs are detected in the adult brain, and many of these change their expression over the course of embryonic brain development and maturation.6 Specific species of miRNAs are expressed in different compartments of the neural axis and it has been postulated that miRNA pathways play a dominant role in neuronal fate determination and synaptic plasticity.7
The critical role of miRNAs in embryonic brain development has largely come from mouse knockout studies. The amelioration of miRNA biogenesis via conditional knockout of Dicer1 has been used to study the collective contribution of miRNAs in specific neuronal tissue. Ablation of Dicer1 expression in various neuronal cell populations, including neural stem cells, results in the rapid depletion of the targeted cell populations.8 Further, Dgcr8+/− mutant mice, which lack a component of the complex that is essential for miRNA production, display significant behavioral and cognitive defects.9
One of the most studied regulatory loops involved in neural stem cell differentiation is the repressor element 1-silencing transcription factor (REST) complex (also known as neuron-restrictive silencer factor, or NRSF)/small C-terminal domain phosphatase 1 (SCP1) pathway. This complex normally silences neuronal genes in nonneuronal cells by suppressing miR-9 and miR-124. During neuronal differentiation a negative feedback loop occurs whereby miR-124 suppresses the activity of SCP1 and thereby inhibits the REST complex, allowing for neuronal differentiation.10 miR-124 also appears to promote the transition of neuronal precursors to more mature neurons through the inhibition of SRY (sex determining region Y)-box 9 (SOX9).11 Differentiation from stem cell–fate to a cell-fate lineage involves the coordinated activation of neuronal genes while activating cell cycle exit. Studies have shown that deletion of miR-9 in the hindbrain results in an increase in cell proliferation due to indirect downregulation of the cell cycle inhibitor p27.12 Another potent regulator of neuronal genes, nuclear receptor TLX, has been demonstrated to be a target of both let-7 and miR-9.13,14 An additional mechanism at play involves an interesting switch in alternative splicing patterns whereby miR-124, through inhibition of polypyrimidine tract binding protein 1, represses neuron-specific splicing patterns.15
General Mechanisms of miRNA Disruption in Cancer
Since the discovery of miRNAs, their aberrant expression has been linked to various pathological states, including cancer. In fact, more than 50% of miRNAs are shown to be located in cancer-associated genomic regions.16 Expression profiling and subsequent characterization of cancer-associated miRNAs provided further proof of the relevance of miRNAs to the study of cancer.
It has been observed that miRNA expression predominantly increases over the course of differentiation. As such, since cancer is often a disease characterized by global dedifferentiation or failure of differentiation, it was hypothesized that miRNA expression would be globally downregulated. The first evidence of a causal role of miRNAs at the root of cancer was discovered in chronic lymphoblastic leukemia, where miR-15 and miR-16, frequently deleted in these tumors, were shown to be negative regulators of the anti-apoptotic gene B-cell lymphoma 2 (BCL2).17,18 These structural variations involving miRNAs are a common mechanism for miRNA disruption in the context of cancer. Other major mechanisms of miRNA deregulation involve the silencing of promoters through DNA methylation, activation of major oncogenic transcription factors, and aberrations in the miRNA biogenesis pathway. Global miRNA repression is often brought about by transcriptional repression via oncogenic transcription factors. One prominent example of this regulation is through MYC, which has been shown to repress a number of miRNAs, including let-7 and miR-34 family members, which converge to drive proliferation.19 The let-7 miRNA family have been shown to act as tumor suppressive miRNAs and are thus frequently downregulated. On the opposite spectrum, various oncogenic miRNAs have also been described—an example is the miR-17‒92 cluster, which have been shown to promote cell proliferation, suppress apoptosis, and induce angiogenesis.20 Copy number variations and cytosine-phosphate-guanine methylation have been shown to focus in on various cancer-related miRNAs.21 However, the precise mechanisms underlying miRNA deregulation in tumorigenesis is not well understood.
Medulloblastoma
The most common malignant pediatric brain tumor is medulloblastoma.22 Arising in the cerebellum, these undifferentiated tumors compose a heterogeneous group with significant mortality.23–26 Recent genetic and molecular profiling of disease has significantly increased our understanding of medulloblastoma pathogenesis and uncovered at least 4 molecular subgroups with distinct biology and clinical profiles.27–30 These subgroups are termed WNT, sonic hedgehog (SHH) (based on their associated signaling pathway activity), and the lesser known Group 3 and Group 4.31–33 miRNAs have been identified to be central to the pathogenesis of a wide variety of cancers.34,35 Several mechanisms lead to the dysregulation of miRNAs in medulloblastoma; these are summarized below.
Oncogenic miRNAs
Genome-wide miRNA profiling and candidate gene approaches have allowed the identification of oncogenic miRNAs in medulloblastoma. One of the earliest miRNA integrative genetic studies, using a cohort of 34 primary medulloblastoma samples, revealed the overexpression of miR-18a, -19a, -20a, -21, -25, and -106b.36 Many of these miRNAs belong to a cluster of miRNAs frequently involved in cancer—known as the miR-17‒92 cluster. Using single nucleotide polymorphism arrays, Northcott et al identified the recurrent amplification of the miR-17‒92 proto-oncogene polycistronic cluster in up to 6% of medulloblastomas. Interestingly, the amplification appears to be predominantly involved in SHH tumors.37 Further evidence from mouse studies demonstrated the codependency between SHH signaling and miR-17–92 using the Ptch+/− mouse model.38 Knockout of this miRNA cluster led to a reduction in cerebellar size and folia number, indicative of its critical function in cerebellar development as well as tumorigenesis. Interestingly, miR-17–92 knockdown completely abrogated the formation of medulloblastoma in the Ptch+/− model.39 The use of an 8-mer locked nucleic acid (LNA)-modified antimiR led to prolonged survival in mice injected with intracranial xenografts, highlighting the potential of LNA antimiR in therapy.40
The most devastating predictor of mortality among medulloblastoma patients is the presence of metastasis41; as such, the role of miRNA in metastases has been hotly investigated. In non-SHH medulloblastomas, studies have shown that another cluster, miR-183–96–182, appeared to be highly expressed.42 Functional studies showed that this cluster of miRNAs is involved in cell migration.43 Overexpression of this cluster led to an increase in leptomeningeal spread in a xenograft model. In an independent report, Weeraratne et al demonstrated that the miR-183–96–182 cluster is enriched in MYC-amplified medulloblastomas and appears to target the Akt/phosphatidylinositol-3 kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway.44 An additional miRNA, miR-21, was shown to be upregulated in a cohort of 29 primary samples and patient-derived cell lines; in vitro studies revealed that the metastases suppressor programmed cell death protein 4 (PDCD4) is a direct downstream target of miR-21.45 Knockdown of miR-21 resulted in an increase of negative modulators of metastasis such as E-cadherin and tissue inhibitor of metalloproteinase 2. Other candidate miRNAs overexpressed in medulloblastoma include miR-367, which targets octamer-binding transcription factor 4 (OCT4) and appears to be involved in tumor proliferation and invasion, and miR-106b, which targets the well-known tumor suppressor phosphatase and tensin homolog (PTEN).46,47 Further studies will be required to elucidate the exact mechanisms of these oncogenic miRNAs and develop a comprehensive understanding of their interactions.
Tumor Suppressor miRNAs
Stemness
Stem cells are characterized by their potential to differentiate into more mature cell types and their self-renewal capacity, which allows them to go through several cell division cycles without exhausting either their proliferative capacity or their differentiation potential. These characteristics are very advantageous in tumor homeostasis and, for this reason, signaling pathways involved in maintenance of stemness are frequently overactivated in medulloblastoma as well as other tumors. In 2004, Singh et al demonstrated that a subpopulation of cells with stem cell–like properties in the tumor bulk was responsible for tumor homeostasis and propagation.48,49 miRNAs have been intimately linked to neuronal stem cell differentiation. Various stem cell compartments have been proposed for medulloblastoma, including CD133+ neural stem cells.49 Global miRNA profiling has revealed a distinct profile between CD133+ and CD133− populations,50 and pathway analysis looking at the most differentially expressed miRNA targets identified critical genes involved in neuronal signaling and cancer metastasis.50
The Notch signaling pathway is essential for the maintenance of the neural stem cell pool during CNS development,51 repressing the expression of proneuronal differentiation genes.51 The Notch signaling pathway is frequently overactive in medulloblastoma, and overexpression of hairy and enhancer of split-1 (HES1), a major downstream effector, is considered a poor prognostic factor in this type of tumor.52,53 It is not surprising to find that in medulloblastoma, miRNAs that target Notch signaling pathway effectors are frequently underrepresented. For example, miR-34a and miR-1280 target Notch ligands delta-like canonical Notch ligand 1 (DLL1) and jagged 2 (JAG2), respectively—furthermore, miR-34a has been shown to target NOTCH1 in medulloblastoma.54,55 Similarly, miR-199b-5p and miR-9 target HES1, a major Notch downstream effector, contributing to the silencing of the Notch signaling pathway at the onset of neuronal differentiation.52,53,56 The restoration of the expression of these miRNAs in medulloblastoma cell lines reduced proliferation and clonogenicity and induced differentiation in vitro. In addition, overexpression of miR-199b-5p in medulloblastoma cells reduced the subpopulation of cancer stem cells, resulting in reduced tumorigenicity in xenograft experiments.52
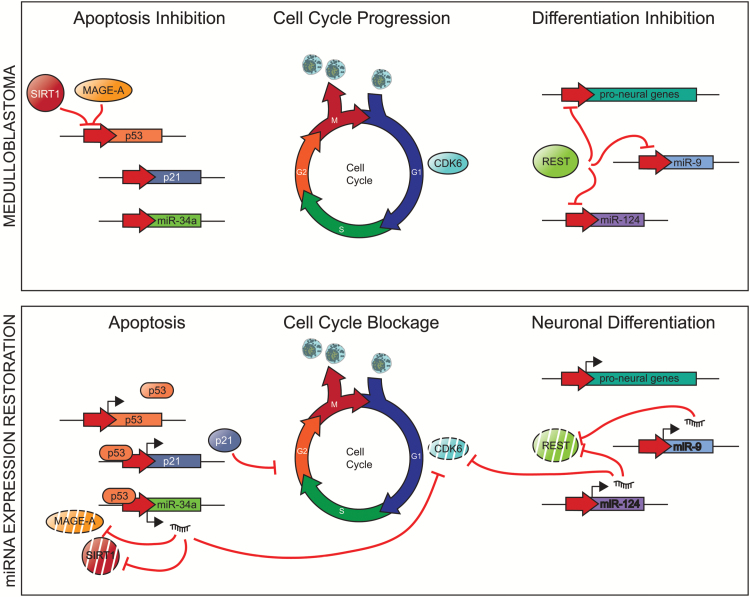
miR-124a and miR-9 are expressed at low levels in neural stem cells, and their levels increase through differentiation, reaching a maximum in the adult brain.57 These miRNAs have a crucial role in the onset of neurogenesis by targeting transcription factors important for the neural precursor function, such as SOX9, Forkhead box G1 (FOXG1), and myeloid ecotropic viral integration site 1 homolog (MEIS1); as such, they promote differentiation.11,58 Mice deficient in miR-9 show severe defects in the thickness and organization of the developing cortex.58 The expression of miR-9 and miR-124a is inhibited by the REST complex, which represses the expression of proneural genes in neural stem cells.59 REST is overexpressed in some medulloblastomas and is correlated with poorer survival. The oncogenic activity of REST has been demonstrated in murine models of medulloblastoma in collaboration with MYC overexpression.60 Some studies have predicted that the 3ʹ untranslated regions of some components of the REST complex present target sequences for miR-9 and miR-124a,61 suggesting the existence of a negative feedback loop between REST and miRNA that would explain the necessity of suppressing miR-9 and miR-124a in medulloblastoma (Figure 1).
Fig. 1.
Dichotomous roles of miR-34a, miR-9, and miR-124 in normal neuronal development and medulloblastoma. In normal neurons, activation of differentiation programs induces miRNAs to repress cell cycle progression. In medulloblastoma, repression of several miRNAs, including miR-34a, miR-9, and miR-124, leads to unregulated cell proliferation. Inhibition of p53 leads to silencing of p21 and miR-34a, which allows cell cycle progression. Expression of REST inhibits proneuronal differentiation miRNAs miR-9 and miR-124.
Other studies show that miR-128a, a miRNA with decreased expression in medulloblastoma, regulates polycomb complex protein BMI-1 (BMI1), a member of the polycomb repressor complex 1.62 Although in this particular study, the authors propose a role for BMI-1 in maintaining levels of reactive oxygen species, BMI1 is known to be involved in inhibiting the expression of proneural genes, and it is essential for neural stem cell maintenance and cerebellar development.63,64 In addition, BMI1 is a proto-oncogene upregulated in medulloblastoma, and its oncogenic activity is due to the inhibition of its downstream target INK4a/ARF, which encodes for the cell cycle inhibitors p16 and p19.49,63
Proliferative Signaling
One of the hallmarks of cancer is the imbalance between proliferation and cell death that leads to an uncontrolled growth of tumor cells. Tumor cells have selected several mechanisms that maintain activated signaling pathways involved in proliferation. Constitutive overactivation of the SHH signaling pathway is characteristic of the aptly named SHH subgroup of medulloblastomas; most of which have germline or somatic mutations of genes in the SHH signaling pathway.65 Ferreti et al observed that miR-324-5p, miR-125b, and miR-326 are significantly downregulated in SHH tumors.66 This set of miRNAs inhibits the expression of Smoothened (SMO) (miR-324-5p, miR-125b, and miR-326) and glioma-associated oncogene (GLI1) (miR-324-5p), both activators of the SHH signaling pathway, and their downregulation contributes to the maintenance of the SHH cascade characteristic of this medulloblastoma subgroup.66
miR-218 is downregulated in medulloblastoma, and the restoration of miR-218 levels has tumor suppressive effects in medulloblastoma cell lines by targeting the rapamycin-insensitive companion of mTOR (RICTOR),67 one of the components of mTOR complex 2 (MTORC2), which phosphorylates and activates Akt, the main effector of the PI3K signaling pathway.68 miR-495 specifically targets growth factor independent 1 (GFI1) transcription factor,69 which is an oncogene overexpressed in medulloblastoma.70 Downregulation of miR-495 is considered a poor prognostic factor, and its level in medulloblastoma inversely correlates with the levels of GFI169; there are currently no data available on the ectopic expression of miR-495 in medulloblastoma cell lines. miR-9 and miR-125a are underexpressed in medulloblastoma, and their expression inversely correlates with tropomyosin receptor kinase C (t-TrkC), which is directly targeted by both miRNAs.36 Neurotrophin receptors regulate proliferation, survival, and differentiation of neural progenitor cells during development, and deregulation of neurotrophin signaling is common in cancer.71 The restoration of miR-9 and miR-125a expression in vitro reduces t-TrkC levels and slows the proliferation of medulloblastoma cell lines.36
Cell Cycle
Recent studies have demonstrated that several miRNAs target different key regulators necessary for cell cycle progression. These miRNAs are frequently downregulated in medulloblastoma. Independent studies have demonstrated that miR-218 and miR-124a, downregulated in medulloblastoma, directly regulate expression of cyclin dependent kinase 6 (CDK6),67,72,73 a key in the progression of G1 phase. CDK6 is recurrently amplified in a subset of Group 4 medulloblastoma tumors, and this overexpression correlates with poor prognosis.65 However, CDK6 amplification is not a sufficient explanation in all CDK6 overexpressing tumors, as it is possible that miR-218 and miR-124a downregulations contribute to CDK6 overexpression. Restoration of miR-218 and miR-124a expression in medulloblastoma cell lines decreases CDK6 levels and, consequently, reduces proliferation.67,72,73 At the same time, miR-31, which is downregulated in medulloblastoma, targets minichromosome maintenance complex component 2 (MCM2), a subunit of the MCM complex necessary for DNA replication initiation during S phase.74 Although not the most ideal preclinical model, forced expression of miR-31 in the Daoy medulloblastoma cell line decreases MCM2 levels, reducing clonogenicity in vitro and tumor growth in xenotransplant assays.74
Cell Cycle Inhibition and Apoptosis
The tumor suppressor gene p53 is a master regulator of cell proliferation. Due to its potent tumor suppressor activity, cancers have evolved different mechanisms to inactivate p53, examples of which can be seen in medulloblastoma. In the recent updated World Health Organization classification for CNS tumors, TP53-mutant SHH medulloblastoma has a characteristically poor prognosis.75 Furthermore, p53 inactivating mutation is frequent at recurrence76 and is one of the reasons why relapsing clones are resistant to chemotherapeutic agents. Expression of miR-34a is activated by p53, which is downregulated in several cancers, including medulloblastoma, and its expression levels correlate, as expected, with p53 levels. Some medulloblastoma cell lines, with low levels of miR-34a, show resistance to chemotherapeutic agents.77,78 The restoration of miR-34a levels decreases proliferation, induces senescence, and triggers apoptosis. This highlights miR-34a as a key regulator in the activation of apoptosis downstream of p53. Some studies have identified new miR-34a targets involved in the epigenetic repression of the p53 promoter, such as sirtuin 1 (SIRT1) and melanoma antigen A (MAGE-A).77–79 Both genes are upregulated in medulloblastoma and could constitute another mechanism for p53–miR-34a apoptosis inactivation. The restoration of the expression of miR-34a represses the expression of SIRT1, and MAGE-A increases p53 levels and hence triggers apoptosis (Figure 1).
Role of miRNAs in Medulloblastoma Metastasis
Given that patients with metastatic medulloblastoma experience the highest degree of mortality, efforts are under way to characterize the molecular signatures of the metastatic compartment, including miRNAs. Work by Garzia et al previously demonstrated a significant decrease in expression of miR-199b-5p in metastatic samples.52 Furthermore, there was a trend towards a better overall survival in the group of high-expression patients. Expression of this miR appears to impair cancer stem cells through the regulation of the Notch pathway via the transcription factor HES1. Using miRNA expression profiling, miR-182 was identified as being overexpressed in non-SHH medulloblastoma. Using xenografts of medulloblastoma cell lines, Bai et al demonstrated that the overexpression of miR-182 contributes to leptomeningeal metastatic dissemination. However, the mechanism and targets of miR-182 in the context of driving metastatic dissemination are currently unknown.42 Another candidate miRNA with a potential role in metastasis is miR-124, which has been shown to be downregulated in medulloblastoma. One of the key targets of miR-124 is SOX9, which is elevated in WNT and SHH medulloblastoma.80 Recent work by Rahmanto et al has shown the importance of SOX9 in driving medulloblastoma metastasis.81 Therapeutic delivery of miR-124 may be a good candidate for the next generation of preclinical trials aimed at controlling metastasis.
miRNA Therapeutics in Medulloblastoma
Various strategies to modify the activity of miRNAs exist, the most broadly used strategy in miRNA therapeutics being the administration of synthetic antisense oligonucleotides, which takes advantage of the property of nucleic acids to interact with each other through sequence complementarity. These therapeutic oligonucleotides can be defined as antimiRs or miRNA mimics depending on their mechanism of action. AntimiRs are single stranded antisense oligonucleotides that bind to a specific miRNA, inhibiting its function, whereas miRNA mimics are double stranded oligonucleotides that, once processed by the miRNA maturation machinery in the cell, mimic the function of a lost/downregulated miRNA.82
Despite the potential of miRNA therapeutics, some technical difficulties are delaying their translation to the clinic. Firstly, administered oligonucleotides have a short half-life in the body, as a consequence of degradation by nucleases both in the serum and in the lysosomes; the degradation makes it difficult for them to reach therapeutic levels. Secondly, given the number of miRNA targets, off-target effects are paramount, and as such, it is necessary to ensure that antimiRs and miRNA mimics are delivered preferentially to the tumor cells.
To address the first issue, several chemical modifications have been introduced into the backbone of the oligonucleotides (eg, LNA, among others); these increase resistance to nucleases and subsequently the miRNAs’ systemic half-life (recently reviewed in Rupaimoole et al82). However, these modifications can affect the binding affinity of miRNA mimics and antimiRs to their specific targets, and hence their function. Despite the stability improvements, chemical modifications do not address the delivery specificity issue.
The encapsulation of oligonucleotides in nanoparticles has drastically changed miRNA therapeutics by protecting, without the necessity of chemical modifications, and delivering oligonucleotides to the target tissue. A wide spectrum of nanoparticles are currently being used in preclinical and clinical trials for miRNA therapeutics, but all seek to maximize delivery of the oligonucleotides to the tumor and, at the same time, reduce toxicity and biodegradability. Nanoparticles can be classified into 2 main groups; cationic polymers (eg, polyethylenimine particles), which deliver the oligonucleotides via endocytosis, or lipid-based nanoparticles (eg, liposomes), which fuse to the plasma membrane of target cells releasing the oligonucleotides directly into the cytosol. The choice of approach depends on the capacity to transport and deliver oligonucleotides, the potential toxicity, and the biodegradability.
Several strategies have achieved targeted delivery to tumor cells by engineering nanoparticles (recently reviewed by Rupaimoole et al82). The most common strategy for tumor delivery takes into account the expression of tumor-specific cell surface markers. One example is the conjugation of nanoparticles with anti–prostate-specific membrane antigen antibody that allows the delivery of antimiR-21 and antimiR-17 specifically to prostate cancer cells, with consequent tumor regression and no accumulation of miRNA in healthy tissues.83 Another approach to achieve tumor-specific delivery exploits the physicochemical characteristics of the tumor microenvironment. This strategy was used in clinical trials for the delivery of miR-34 mimics (MRX-34, Mirna Therapeutics) to different types of cancers (eg, multiple myeloma, renal cell carcinoma, primary liver cancer) (ClinicalTrials.gov NCT01829971). miR-34 mimics were encapsulated in liposomes and, as a consequence of the lower pH, became protonated in the tumor microenvironment and were retained by electrostatic interaction with the tumor cell surface.84 Despite the promising initial response, the trial was terminated due to severe immunological reactions in some patients. It is therefore critical to discern if this was caused by off-target effects of the miR-34 mimic or by an immune reaction to the nanoparticles used as a carrier.
Although there are no current clinical trials for miRNA therapeutics involving medulloblastoma patients, thanks to recent functional studies, we have prioritized a growing list of miRNAs with therapeutic potential (Table 1). Some in vivo preclinical studies show a remarkable antitumor effect when normal miRNA levels are restored in medulloblastoma cells (Table 2). The intravenous administration of LNA antimiRs that target miRNAs from the miRNA-17‒92 cluster—recurrently overexpressed in SHH medulloblastoma—drastically reduces tumor volume in murine models of SHH medulloblastoma.40
Table 1.
Summary of oncogenic and tumor suppressor miRNAs involved in medulloblastoma
| mirR | Chr | Alterations | Targets | Cellular Function | Subgroup | Reference | |
|---|---|---|---|---|---|---|---|
| Oncogenic miRNA (overexpressed) | miR-30b, miR-30d | 8q24.22- q24.23 | Amplification | N/A | 92 | ||
| miR-17–92 | 13q31.3 | Amplification | N-Myc target. Interaction with SHH signaling | SHH | 37 , 38 , 40 | ||
| miR-21 | 17q23.1 | Upregulation | PDCD4 | Metastasis | N/A | 45 | |
| miR-183~96~182 | 7q32 | Upregulation | PI3K-Akt/ mTOR | Cell migration. Apoptosis | Non-SHH/ MYC-amplified | 42 , 44 | |
| miR-367 | 4q25 | Upregulation | RYR3, ITGAV and RAB23 | Proliferation. Invasion | 46 | ||
| miR-106b | 7q22.1 | Upregulation | PTEN | Proliferation. Migration. Invasion | Nonspecific | 47 | |
| Tumor supressor miRNA (downregulated) | miR-let-7 | different locations | HMGB1 | Chemoresistance | 36 , 66 , 91 | ||
| miR-9 | 1q22 | Hypermethylation | HES1, t-TrkC, REST | Anti-proliferation. Differentiation. Pro- apoptosis | All subgroups | 36 , 56 | |
| miR-31 | 9p21.3 | Deletion | MCM2 | Anti-proliferation. Reduces clonogenicity | 36 , 74 | ||
| miR-34a | 1p36.22 | DLL1, MAGE-A, MYCN, SIRT1 | Anti-proliferation. Pro-apoptosis. Reduces tumorigenicity in vivo. Senescence. Sensitivity to chemotherapeutic agents | 54 , 77–79 | |||
| miR-124 | 8p23.1 | Unknown | CDK6, REST, SCL16A1 | Anti-proliferation. Differentiation. Pro- apoptosis. Reduces tumorigenicity in vivo | 66 , 72 , 73 | ||
| mir-125a | 19q13.41 | t-TrkC | Anti-proliferation | 36 | |||
| mir-125b | 11q24.1 | SMO | SHH | 66 | |||
| miR-128a | 2q21.3 | BMI1 | Anti-proliferation. Reduces clonogenicity. Senescence | 36 , 62 | |||
| miR-135a | 3p21.2 | Arhgef6 | Reduces tumorigenicity | SHH | 36 , 66 , 94 | ||
| miR-148a | 7p15.2 | Hypermethylation | NRP1, ROCK1, DNMT1 | Anti-proliferation. Reduces clonogenicity. Reduces invasion. Reduces tumorigenicity | 36 , 93 , 96 , 99 | ||
| miR-199b-5p | 9q34.11 | Hypermethylation | HES1, CD15 | Anti-proliferation. Reduces clonogenicity. Reduces in vivo tumorigenicity. Reduces cancer stem cells | 34 , 35 | ||
| miR-206 | 6p12.2 | OTX2 | Anti-proliferation. Reduces clonogenicity | All subgroups | 93 , 95 , 96 | ||
| miR-218 | 4p15.31 | CDK6, SH3GL1, RICTOR | Anti-proliferation. Reduces clonogenicity. Reduces cell migration and Invasion. Promotes differentiation | 67 , 97 | |||
| miR-219 | 6q21.3 | OTX2, CD164 | Anti-proliferation. Reduces invasion and migration | 36 , 50 , 98 | |||
| miR-324-5p | 17p13.1 | 17p loss | SMO, GLI1 | Reduces clonogenicity | SHH | 66 | |
| miR-326 | 11q13.4 | SMO | Reduces clonogenicity | SHH | 66 | ||
| miR-383 | 8p22 | PRDX3 | Anti-proliferation. Pro-apoptosis | 36 , 100 | |||
| miR-495 | 14q32.31 | GFI1 | 69 |
Table 2.
Preclinical miRNA studies in medulloblastoma
| miRNA | Mechanism | Cell Line | Administration | Carrier Format | Tumor Site | Effect | Reference |
|---|---|---|---|---|---|---|---|
| miR-124 | overexpression | D425 | In vitro | Lentivirus | Flank/intracranial | Growth inhibition | 73 |
| miR-192 | miRNA mimics | D283 | In vivo, intranasal | Nanoparticle | Intracranial | Metastases Inhibition | 85 |
| miR-34a | Overexpression | Daoy | In vitro | Adenovirus | Flank/intracranial | Growth inhibition | 54 |
| miR-199b-5p | Overexpression | Daoy | In vitro | Adenovirus | Flank/intracranial | Growth inhibition | 52 |
| miR-31 | Overexpression | Daoy | In vitro | Transfection | Flank | Growth inhibition | 74 |
| miR17‒92 | Anti-miRNA | Ptch1+/−, Trp53 −/− medulloblastoma cells | In vivo, intra venously | LNA | Flank/intracranial | Growth inhibition | 40 |
The presence of metastatic disease is an indicator of poor prognosis in medulloblastoma patients and, as a consequence of tumor evolution, miRNA expression patterns may be different in primary and metastatic compartments. The downregulation of miR-192 in medulloblastoma samples has been directly correlated with leptomeningeal dissemination. Xenotransplant mouse models of medulloblastoma show a significant reduction in metastatic burden, with the subsequent increase in survival, when mice were treated with miR-192 mimics conjugated with polyethylenimine nanoparticles.85
Other attractive targets for miRNA therapeutics are re-expressions of miR-34 and miR-124, both recurrently downregulated in medulloblastoma.54,72,77–79,86,87 Re-expression of these miRNAs led to a remarkable antitumor effect in vivo using grafted genetically modified medulloblastoma cell lines. This finding still needs to be tested in preclinical trials using miRNA mimics. In the case of miR-124, there are preclinical studies for the administration of miR-124 nanoparticles in Parkinson’s disease and prostate cancer models, and similar approaches could be utilized in medulloblastoma preclinical models.88,89
Moreover, nanoparticles have been used for the co-delivery of miRNA mimics or antimiRs and chemotherapeutic agents to the tumor site; they have shown significant reduction of tumor size compared with chemotherapy alone.90 miRNAs such as miR-7f1 and miR-34a—recurrently downregulated in medulloblastoma—are known to induce sensitization to chemotherapeutic agents when restored,78,91 thus they are promising candidates for future trials that test the benefits of these combination therapies in medulloblastoma. Given the genetic heterogeneity of medulloblastoma and the clonal divergence at metastases, subgroup-specific combination therapy will likely be required to truly control the disease.
Conclusion
Recent genomic studies have shown that medulloblastoma recurrently targets key genes that regulate proliferation, apoptosis, stemness, and differentiation. These observations suggest that medulloblastomas arise from neural stem/progenitor cells that failed to differentiate, thereby maintaining a proliferative and stem cell–like state that favors tumor initiation. In the last few years, the study of miRNA expression profiling in medulloblastoma, and in cancer in general, has received special focus. This roots in the fact that a single miRNA can regulate the expression of hundreds of different genes, and hence an aberrant miRNA expression pattern has a profound impact on tumor initiation and/or progression.
It is common to find that certain miRNAs are frequently downregulated, whereas others are always upregulated in cancer. This suggests that these miRNAs could have tumor suppressor or pro-oncogenic activity, respectively. Thanks to the efforts of the scientific community in identifying and validating some of the targets for these miRNAs in the context of medulloblastoma, we are starting to understand how aberrant miRNA expression plays a critical role in medulloblastoma. Future work will need to concentrate on further characterizing the miRNA profile in a subgroup-specific manner. Identifying specific miRNA targets using targeted approaches can be hugely inefficient given the myriad of targets miRNAs can influence. Genome-wide technologies, such as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), will be needed to illustrate the full tapestry of miRNA regulation in a context-specific manner.
Unfortunately, there is no magical mirror that will reveal the most malignant miRNA in medulloblastoma. Studies to date have demonstrated several promising miRNAs for therapeutic translation. For example, miR-124 and miR-34a, through their roles in stemness and cell cycle, respectively, are promising priority candidates for restoration therapy. Future miRNA studies will need to take subgroup affiliation into consideration given the significant differences in clinical and biological behavior between subgroups. AntimiRs designed against miR-182, which is overexpressed in subsets of medulloblastoma, may be candidates to specifically target metastasis, although their mechanism needs to be elucidated to better understand the impact on normal tissue as well as cancer. This underlies an important principle, which is safety. A recent clinical trial targeting miR-34 was prematurely terminated due to immunologic concerns. As such, further research into more efficient and safer delivery methods is desperately needed. Ultimately, no miRNA behaves alone. Given the genetic divergence of medulloblastoma subclones and the wide-reaching networks of genes that miRNAs regulate, the likely treatment will need to modulate not one, but multiple miRNA interactomes. Unraveling these interactomes will ultimately answer the question, Which combinations of miRs are the most malignant of them all?
Funding
No external funding was used to directly support this review.
Conflict of interest statement. None for all contributing authors.
Acknowledgments
Research in the Taylor laboratory is financially supported by a Program Project Grant from the Terry Fox Research Institute, grants from the CureSearch Foundation and the National Institutes of Health (R01CA148699 and R01CA159859), and a Brain Tumour Foundation of Canada Impact Grant from the Canadian Cancer Society and Brain Canada with the financial assistance of Health Canada (703202). M.D.T. is also supported by the Garron Family Chair in Childhood Cancer Research. X.W. is supported by a CIHR Vanier Canada Graduate Scholarship, McLaughlin Centre for Molecular Medicine MD/PhD Scholarship, and the Ruggles MD/PhD Innovation Award.
References
- 1. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. [DOI] [PubMed] [Google Scholar]
- 2. Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252–263. [DOI] [PubMed] [Google Scholar]
- 5. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. [DOI] [PubMed] [Google Scholar]
- 6. Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54(12):1103–1112. [DOI] [PubMed] [Google Scholar]
- 7. Bak M, Silahtaroglu A, Møller M et al. . MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fénelon K, Mukai J, Xu B et al. . Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108(11):4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15(9):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20(1):19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao C, Sun G, Li S et al. . MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107(5):1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. [DOI] [PubMed] [Google Scholar]
- 17. Calin GA, Dumitru CD, Shimizu M et al. . Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cimmino A, Calin GA, Fabbri M et al. . miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gulino R, Forte S, Parenti R, Memeo L, Gulisano M. MicroRNA and pediatric tumors: future perspectives. Acta Histochem. 2015;117(4–5):339–354. [DOI] [PubMed] [Google Scholar]
- 22. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shih DJ, Northcott PA, Remke M et al. . Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol. 2014;32(9):886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramaswamy V, Remke M, Shih D et al. . Duration of the pre-diagnostic interval in medulloblastoma is subgroup dependent. Pediatr Blood Cancer. 2014;61(7):1190–1194. [DOI] [PubMed] [Google Scholar]
- 25. Ramaswamy V, Remke M, Bouffet E et al. . Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14(12):1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12(7):871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Northcott PA, Shih DJH, Peacock J et al. . Subgroup-specific structural variation across 1000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Northcott PA, Nakahara Y, Wu X et al. . Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taylor MD, Liu L, Raffel C et al. . Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. [DOI] [PubMed] [Google Scholar]
- 30. Wang X, Ramaswamy V, Remke M et al. . Intertumoral and intratumoral heterogeneity as a barrier for effective treatment of medulloblastoma. Neurosurgery. 2013;60(Suppl 1):57–63. [DOI] [PubMed] [Google Scholar]
- 31. Morrissy AS, Garzia L, Shih DJ et al. . Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529(7586):351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Northcott PA, Shih DJ, Remke M et al. . Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123(4):615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramaswamy V, Northcott PA, Taylor MD. FISH and chips: the recipe for improved prognostication and outcomes for children with medulloblastoma. Cancer Genet. 2011;204(11):577–588. [DOI] [PubMed] [Google Scholar]
- 34. Garzia L, Andolfo I, Cusanelli E et al. . MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4(3):e4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andolfo I, Liguori L, De Antonellis P et al. . The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14(5):596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferretti E, De Smaele E, Po A et al. . MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124(3):568–577. [DOI] [PubMed] [Google Scholar]
- 37. Northcott PA, Fernandez-L A, Hagan JP et al. . The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69(8):3249–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uziel T, Karginov FV, Xie S et al. . The miR-17–92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106(8):2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zindy F, Kawauchi D, Lee Y et al. . Role of the miR-17, 92 cluster family in cerebellar and medulloblastoma development. Biol Open. 2014;3(7):597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murphy BL, Obad S, Bihannic L et al. . Silencing of the miR-17–92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013;73(23):7068–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Dubuc AM, Ramaswamy V et al. . Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol. 2015;129(3):449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bai AH, Milde T, Remke M et al. . MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol. 2012;123(4):529–538. [DOI] [PubMed] [Google Scholar]
- 43. Zhang Z, Li S, Cheng SY. The miR-183∼96∼182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J Biomed Res. 2013;27(6):486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weeraratne SD, Amani V, Teider N et al. . Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123(4):539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grunder E, D’Ambrosio R, Fiaschetti G et al. . MicroRNA-21 suppression impedes medulloblastoma cell migration. Eur J Cancer. 2011;47(16):2479–2490. [DOI] [PubMed] [Google Scholar]
- 46. Kaid C, Silva PBG, Cortez BA, Rodini CO, Semedo-Kuriki P, Okamoto OK. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015;106(9):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li KK, Xia T, Ma FM et al. . miR-106b is overexpressed in medulloblastomas and interacts directly with PTEN. Neuropathol Appl Neurobiol. 2015;41(2):145–164. [DOI] [PubMed] [Google Scholar]
- 48. Singh SK, Hawkins C, Clarke ID et al. . Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 49. Wang X, Venugopal C, Manoranjan B et al. . Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene. 2012;31(2):187–199. [DOI] [PubMed] [Google Scholar]
- 50. Genovesi LA, Carter KW, Gottardo NG, Giles KM, Dallas PB. Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PLoS One. 2011;6(9):e23935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449(7160):351–355. [DOI] [PubMed] [Google Scholar]
- 52. Garzia L, Andolfo I, Cusanelli E et al. . MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4(3):e4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Andolfo I, Liguori L, De Antonellis P et al. . The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14(5):596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Antonellis P, Medaglia C, Cusanelli E et al. . MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6(9):e24584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang F, Remke M, Bhat K et al. . A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget. 2015;6(5):2709–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fiaschetti G, Abela L, Nonoguchi N et al. . Epigenetic silencing of miRNA-9 is associated with HES1 oncogenic activity and poor prognosis of medulloblastoma. Br J Cancer. 2014;110(3):636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meza-Sosa KF, Pedraza-Alva G, Pérez-Martínez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci. 2014;8:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103(7):2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Su X, Gopalakrishnan V, Stearns D et al. . Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26(5):1666–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7(9):R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5(6):e10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leung C, Lingbeek M, Shakhova O et al. . Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428(6980):337–341. [DOI] [PubMed] [Google Scholar]
- 64. Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113(2):175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Northcott PA, Jones DT, Kool M et al. . Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12(12):818–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ferretti E, De Smaele E, Miele E et al. . Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Venkataraman S, Birks DK, Balakrishnan I et al. . MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem. 2013;288(3):1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ogita S, Lorusso P. Targeting phosphatidylinositol 3 kinase (PI3K)-Akt beyond rapalogs. Target Oncol. 2011;6(2):103–117. [DOI] [PubMed] [Google Scholar]
- 69. Wang C, Yun Z, Zhao T, Liu X, Ma X. MiR-495 is a predictive biomarker that downregulates GFI1 expression in medulloblastoma. Cell Physiol Biochem. 2015;36(4):1430–1439. [DOI] [PubMed] [Google Scholar]
- 70. Northcott PA, Lee C, Zichner T et al. . Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chopin V, Lagadec C, Toillon RA, Le Bourhis X. Neurotrophin signaling in cancer stem cells. Cell Mol Life Sci. 2016;73(9):1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90(1):1–7. [DOI] [PubMed] [Google Scholar]
- 73. Silber J, Hashizume R, Felix T et al. . Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jin Y, Xiong A, Zhang Z et al. . MicroRNA-31 suppresses medulloblastoma cell growth by inhibiting DNA replication through minichromosome maintenance 2. Oncotarget. 2014;5(13):4821–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Louis DN, Perry A, Reifenberger G et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 76. Morrissy AS, Garzia L, Shih DJ et al. . Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529(7586):351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fan YN, Meley D, Pizer B, Sée V. Mir-34a mimics are potential therapeutic agents for p53-mutated and chemo-resistant brain tumour cells. PLoS One. 2014;9(9):e108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weeraratne SD, Amani V, Neiss A et al. . miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13(2):165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thor T, Künkele A, Pajtler KW et al. . MiR-34a deficiency accelerates medulloblastoma formation in vivo. Int J Cancer. 2015;136(10):2293–2303. [DOI] [PubMed] [Google Scholar]
- 80. Swartling FJ, Savov V, Persson AI et al. . Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21(5):601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Suryo Rahmanto A, Savov V, Brunner A et al. . FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J. 2016;35(20):2192–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. [DOI] [PubMed] [Google Scholar]
- 83. Binzel DW, Shu Y, Li H et al. . Specific delivery of MiRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol Ther. 2016;24(7):1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bader AG. miR-34—a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang SY, Choi SA, Lee JY et al. . miR-192 suppresses leptomeningeal dissemination of medulloblastoma by modulating cell proliferation and anchoring through the regulation of DHFR, integrins, and CD47. Oncotarget. 2015;6(41):43712–43730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li KK, Pang JC, Ching AK et al. . miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol. 2009;40(9):1234–1243. [DOI] [PubMed] [Google Scholar]
- 87. Ferretti E, De Smaele E, Miele E et al. . Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Saraiva C, Ferreira L, Bernardino L. Traceable microRNA-124 loaded nanoparticles as a new promising therapeutic tool for Parkinson’s disease. Neurogenesis (Austin). 2016;3(1):e1256855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shi X, Ma A, Xue L et al. . miR-124 and androgen receptor signaling inhibitors repress prostate cancer growth by downregulating androgen receptor splice variants, EZH2, and Src. Cancer Res. 2015;75(24):5309–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Qian X, Long L, Shi Z et al. . Star-branched amphiphilic PLA-b-PDMAEMA copolymers for co-delivery of miR-21 inhibitor and doxorubicin to treat glioma. Biomaterials. 2014;35(7):2322–2335. [DOI] [PubMed] [Google Scholar]
- 91. Pannuru P, Dontula R, Khan AA et al. . miR-let-7f-1 regulates SPARC mediated cisplatin resistance in medulloblastoma cells. Cell Signal. 2014;26(10):2193–2201. [DOI] [PubMed] [Google Scholar]
- 92. Lu Y, Ryan SL, Elliott DJ et al. . Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS One. 2009;4(7):e6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gokhale A, Kunder R, Goel A et al. . Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J Cancer Res Ther. 2010;6(4):521–529. [DOI] [PubMed] [Google Scholar]
- 94. Hemmesi K, Squadrito ML, Mestdagh P et al. . miR-135a inhibits cancer stem cell-driven medulloblastoma development by directly repressing Arhgef6 expression. Stem Cells. 2015;33(5):1377–1389. [DOI] [PubMed] [Google Scholar]
- 95. Kunder R, Jalali R, Sridhar E et al. . Real-time PCR assay based on the differential expression of microRNAs and protein-coding genes for molecular classification of formalin-fixed paraffin embedded medulloblastomas. Neuro Oncol. 2013;15(12):1644–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Panwalkar P, Moiyadi A, Goel A et al. . MiR-206, a cerebellum enriched miRNA is downregulated in all medulloblastoma subgroups and its overexpression is necessary for growth inhibition of medulloblastoma cells. J Mol Neurosci. 2015;56(3):673–680. [DOI] [PubMed] [Google Scholar]
- 97. Shi J, Yang L, Wang T et al. . miR-218 is downregulated and directly targets SH3GL1 in childhood medulloblastoma. Mol Med Rep. 2013;8(4):1111–1117. [DOI] [PubMed] [Google Scholar]
- 98. Shi JA, Lu DL, Huang X, Tan W. miR-219 inhibits the proliferation, migration and invasion of medulloblastoma cells by targeting CD164. Int J Mol Med. 2014;34(1):237–243. [DOI] [PubMed] [Google Scholar]
- 99. Yogi K, Sridhar E, Goel N et al. . MiR-148a, a microRNA upregulated in the WNT subgroup tumors, inhibits invasion and tumorigenic potential of medulloblastoma cells by targeting Neuropilin 1. Oncoscience. 2015;2(4):334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li KK, Pang JC, Lau KM et al. . MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol. 2013;23(4):413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]