ABSTRACT
The assembly of functional neuronal circuits requires growth cones to extend in defined directions and recognize the correct synaptic partners. Homophilic adhesion between vertebrate Sidekick proteins promotes synapse formation between retinal neurons involved in visual motion detection. We show here that Drosophila Sidekick accumulates in specific synaptic layers of the developing motion detection circuit and is necessary for normal optomotor behavior. Sidekick is required in photoreceptors, but not in their target lamina neurons, to promote the alignment of lamina neurons into columns and subsequent sorting of photoreceptor axons into synaptic modules based on their precise spatial orientation. Sidekick is also localized to the dendrites of the direction-selective T4 and T5 cells, and is expressed in some of their presynaptic partners. In contrast to its vertebrate homologs, Sidekick is not essential for T4 and T5 to direct their dendrites to the appropriate layers or to receive synaptic contacts. These results illustrate a conserved requirement for Sidekick proteins in establishing visual motion detection circuits that is achieved through distinct cellular mechanisms in Drosophila and vertebrates.
KEY WORDS: Sidekick, Photoreceptor, Lamina, Optomotor behavior, Visual motion detection
Summary: Drosophila Sidekick has a conserved function in visual motion detection, but in contrast to vertebrates, it acts in a single cell type rather than mediating recognition of synaptic partners.
INTRODUCTION
Establishing synaptic circuits requires neurons to identify and interact with the correct partners within a highly complex cellular environment. These interactions depend on the combinatorial actions of a wide variety of cell-surface adhesion molecules. Proteins of the cadherin, neurexin, leucine-rich repeat and immunoglobulin (Ig) superfamilies can engage in homophilic or heterophilic interactions, leading either to adhesion or repulsion of the expressing cells (de Wit and Ghosh, 2016). It is not yet clear how such molecular interactions enable the assembly of complex neural circuits with high specificity.
The Drosophila visual system has been used as a model system for many studies of the development and function of neural circuits. Each ommatidium in the compound eye contains eight photoreceptors: R1-R6 express rhodopsin Rh1 and detect visual motion, while R7 and R8 each express one of four different rhodopsins and mediate color vision (Wernet et al., 2014). Motion detection is subdivided into ‘ON’ and ‘OFF’ pathways that are specialized to detect moving bright edges and moving dark edges, respectively (Borst, 2014). Lamina neurons L1 and L2 receive direct synaptic input from R1-R6 (Meinertzhagen and O'Neil, 1991); L1 provides input to the ‘ON’ pathway, while L2 is the major link to the ‘OFF’ pathway (Clark et al., 2011; Joesch et al., 2010; Meier et al., 2014; Silies et al., 2013). Information from the lamina is conveyed to the medulla, where the ‘ON’ and ‘OFF’ signals are transmitted by distinct populations of neurons that converge on the dendrites of T4 (‘ON’) and T5 (‘OFF’) lobula plate neurons in specific layers of the medulla and lobula, respectively (Behnia et al., 2014; Fisher et al., 2015; Serbe et al., 2016; Shinomiya et al., 2014; Strother et al., 2014, 2017; Takemura et al., 2013, 2017). Four subtypes of T4 and T5 neurons are each sensitive to one of the four cardinal directions of motion and provide input to lobula plate tangential neurons that are segregated in four corresponding layers (Maisak et al., 2013).
In the third larval instar, Hedgehog and Spitz signals from the R1-R6 photoreceptor axons induce the final division and neuronal differentiation of their target lamina neurons (Huang and Kunes, 1996; Huang et al., 1998). Interactions between the Ig family members Hibris (Hbs) on postmitotic lamina neurons and Roughest (Rst) on photoreceptor axons then induce the lamina neurons to line up in columns along the retinal axon bundles (Sugie et al., 2010). The R1-R6 axons from a single ommatidium subsequently diverge during pupal development to project to six different synaptic modules known as lamina cartridges, enabling each cartridge to collect information from six photoreceptors located in different neighboring ommatidia that view the same point in visual space (Agi et al., 2014). This sorting process requires the early polarization of the ‘front’ of each photoreceptor growth cone towards its target, while the ‘heels’ of these growth cones form a stable scaffold (Schwabe et al., 2013; Langen et al., 2015). Correct polarization and extension are thought to require a comparison of the relative levels of the cadherin Flamingo (Fmi) on neighboring growth cones, as well as adhesive interactions with other growth cones and target lamina neurons that are mediated by N-cadherin (Ncad) (Prakash et al., 2005; Chen and Clandinin, 2008; Clandinin and Zipursky, 2000; Lee et al., 2003; Schwabe et al., 2013). Photoreceptor axons later develop synaptic terminals that connect to their target lamina neurons through tetrad synapses, at which a single presynaptic active zone contacts four postsynaptic cells. Two of these are always the lamina neurons L1 and L2, while the others may be L3 in combination with amacrine or glial cells (Meinertzhagen and O'Neil, 1991).
The vertebrate retina is comparable in complexity and organization to the combined Drosophila retina, lamina and medulla, and the two visual systems share many design principles (Sanes and Zipursky, 2010). In the chick retina, synapses between different types of bipolar, amacrine and ganglion cells form in distinct sublaminae of the inner plexiform layer (Robles and Baier, 2012) that are specified by homophilic adhesion molecules of the Sidekick (Sdk), Down's syndrome cell adhesion molecule (Dscam) and Contactin families. Each of these proteins is localized to a specific layer and expressed by cells that synapse on each other in that layer (Yamagata and Sanes, 2008, 2012; Yamagata et al., 2002). In the mouse retina, sidekick cell-adhesion molecules 1 and 2 (Sdk1 and Sdk2) label largely non-overlapping subsets of cells, but these are not clearly separated by layer. Sdk2 is required in both VG3 amacrine cells and W3B retinal ganglion cells to form a strong synaptic connection between these neurons, which sense object motion relative to the surroundings (Krishnaswamy et al., 2015).
The Drosophila genome encodes a single Sdk protein. Null sdk alleles have previously been shown to produce small changes in the numbers of photoreceptors and non-neuronal cells in the eye (Nguyen et al., 1997). Here, we show that Sdk is a homophilic adhesion molecule that is expressed in several cells of the visual motion detection circuit and is localized to specific synaptic layers. Consistent with its location, mutants lacking Sdk show defects in optomotor behavior. We find that sdk is required in photoreceptors, but not in their target lamina neurons, for the correct organization of lamina columns and cartridges, indicating that, unlike its vertebrate counterparts, Sdk does not mediate adhesion between pre- and postsynaptic cells. Although Sdk is present on the dendrites of the direction-selective T4 and T5 cells, it is not required for these neurites to arborize in the appropriate layer or to receive synaptic input. These results suggest that Sdk proteins in Drosophila and vertebrates have a common functional role in establishing visual motion detection circuits, but act through distinct cellular mechanisms.
RESULTS
Drosophila Sdk is a homophilic adhesion molecule expressed in the visual system
In the chick and mouse retina, Sdk proteins control layer-specific neuronal projections and synaptic partner selection (Krishnaswamy et al., 2015; Yamagata and Sanes, 2008). The Drosophila genome encodes only a single Sdk homolog (Nguyen et al., 1997). The Sdk protein has a large extracellular domain containing six immunoglobulin (Ig) and thirteen fibronectin type III (FNIII) domains, and a short intracellular domain ending in a PDZ-binding motif (Fig. 1A). Vertebrate Sdk1 and Sdk2 proteins have been shown to act as homophilic adhesion molecules that interact through their four N-terminal Ig domains (Goodman et al., 2016; Yamagata et al., 2002). Cultured S2 cells expressing Drosophila Sdk formed aggregates that did not include control cells expressing mCherry (Fig. 1B-D), indicating that Sdk can also mediate homophilic adhesion. Using in situ hybridization to examine the pattern of sdk expression, we found that sdk mRNA was present throughout the eye-antennal disc in third-instar larvae, and its expression in the central nervous system was highly enriched in the optic lobes (Fig. 1E,F). An antibody raised against the cytoplasmic domain of Sdk revealed increased protein accumulation at the apical surface of cells in the region of the eye disc posterior to the morphogenetic furrow, where photoreceptor differentiation occurs (Fig. 1G,H).
Fig. 1.
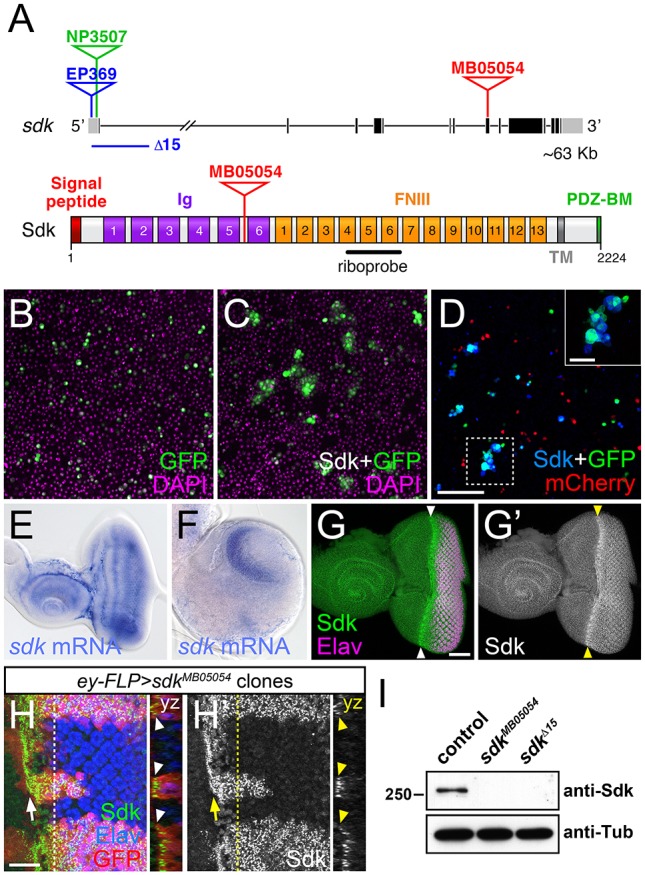
Sdk is a homophilic adhesion molecule expressed in the visual system. (A) The sdk gene (top) with coding exons in black and non-coding exons in gray, showing the positions of the MB05054 Minos insertion, the EP369 P element insertion and derived Δ15 deletion, and the NP3507 GAL4 insertion. The Sdk protein (bottom) has six Ig domains, 13 FNIII domains, a transmembrane domain (TM) and a PDZ-binding motif (PDZ-BM). (B-D) Aggregates formed by S2 cells transfected with Actin5c (Act)-GAL4 and UAS-GFP (green) (B); Act-GAL4, UAS-HA-Sdk and UAS-GFP (C); and a mixture of cells transfected with Act-GAL4, UAS-HA-Sdk and UAS-GFP, and with Act-GAL4 and UAS-mCherry (red) (D). DAPI is in magenta (B,C) and anti-HA in blue (D). Inset provides an enlargement of the boxed region. Sdk-expressing cells form aggregates that lack cells not transfected with Sdk. (E,F) In situ hybridization with a sdk probe on a third-instar eye-antennal imaginal disc (E) and brain (F). sdk is expressed throughout the eye disc and enriched in the optic lobes of the brain. Posterior is towards the right in E-H′. (G) Anti-Sdk (G′, green in G) and anti-Elav (magenta) labeling of an eye-antennal disc, showing enrichment of Sdk protein posterior to the morphogenetic furrow (arrowheads). (H) Anti-Sdk (H′, green in H) and anti-Elav (blue) labeling of an eye disc containing sdkMB05054 mutant clones marked by the absence of GFP (red). Sdk labeling is absent from the clones. An arrow indicates the morphogenetic furrow. yz sections show Sdk accumulation at the apical surface of the disc (arrowheads). (I) Western blot of extracts from w1118 (control), sdkMB05054 and sdkΔ15 embryos, probed with anti-Sdk and anti-Tubulin (Tub). Sdk protein is not detected in either mutant. Scale bars: 100 µm in D; 30 µm in G and inset in D; 10 µm in H,H′.
To investigate whether Sdk plays a role in establishing the visual circuits in Drosophila, we identified mutations disrupting its function. We used both a Minos transposable element insertion in a coding exon, Mi{ET1}sdkMB05054 (Bellen et al., 2011), and a deletion of the 5′ end of the gene made by imprecise excision of P{EP}sdkEP369, a P element inserted in the 5′UTR (Fig. 1A). No Sdk protein was detected in embryos homozygous for either allele (Fig. 1I) or in sdk mutant clones in the eye disc (Fig. 1H), simultaneously confirming that the mutations are null and the antibody specific. As previously reported for other alleles that are no longer available (Nguyen et al., 1997), homozygous sdk mutant flies were viable with slightly rough eyes resulting from mild defects in cone and pigment cell patterning.
Sdk is required in photoreceptors to establish the map for visual motion detection
Antibody labeling revealed that Sdk protein was present in the axons and concentrated in the growth cones of developing R1-R6 photoreceptors, which synapse with lamina neurons to mediate visual motion detection (Fig. 2A,B; Fig. S2D,E), but was not present on the growth cones of R7 and R8, which target medulla neurons and confer color vision (see Fig. 5A,B; Fig. S3F,G) (Hadjieconomou et al., 2011b). We did observe Sdk on glial cells ensheathing the axons of R7 and R8; labeling in this region was removed by knocking down sdk in glia (Fig. S3G) but not photoreceptors (Fig. 2D). In sdk mutants, a few R1-R6 axons penetrated the lamina to terminate in the medulla, but most targeted the correct optic neuropil (Fig. S1).
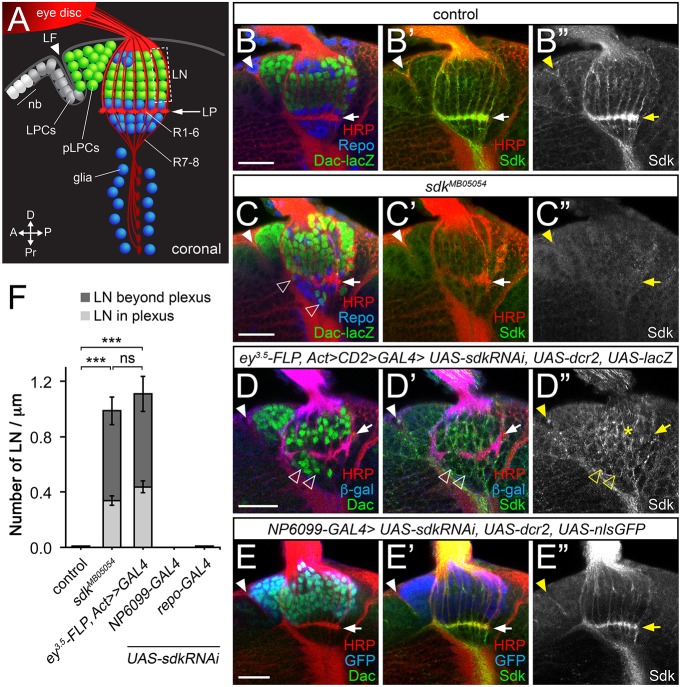
Fig. 2.
Lamina neuron placement requires sdk in photoreceptors. (A) Diagram of a coronal section through the third-instar larval brain, showing neuroblasts (nb) and lamina precursor cells (LPCs), which become postmitotic (pLPCs) behind the lamina furrow (LF; arrowhead) and differentiate into lamina neurons (LN) aligned into columns along the axons of R1-R6. Glia are shown in blue. (B-E″) Confocal images of the same view of larval brains for: (B-B″) control (sdkMB05054/+); (C-C″) sdkMB05054; (D-D″) sdk knockdown in the eye with ey3.5-FLP, Act>CD2>GAL4 [Sdk still accumulates in LNs (asterisk)]; and (E-E″) sdk knockdown in the lamina with NP6099-GAL4. A-E″ are labeled with anti-Sdk (B″,C″,D″,E″, green in B′,C′,D′,E′), anti-HRP to mark photoreceptor axons (red in B,B′,C,C′,D,D′,E,E′), anti-β-galactosidase (β-gal) reflecting dac-lacZ (green in B,C) or anti-Dac (green in D,E) to mark lamina neurons, anti-Repo to mark glia (blue in B,C), or anti-β-gal (blue in D,D′) or anti-GFP (blue in E,E′) to mark the domain of RNAi expression. Lamina neurons are misplaced in sdk mutants and when sdk is knocked down in the eye (empty arrowheads, C,D), but not when it is knocked down in lamina neurons or in glia with repo-GAL4. Filled arrowheads mark the lamina furrow. (F) The number of LNs per µm in or beyond the lamina plexus (LP; arrows in A-E′) in the indicated genotypes. Data are mean±s.e.m. n=15 (sdkΔ7, a precise excision of sdkMB05054 used as a control, and sdkMB05054), n=10 (ey3.5-FLP, Act>CD2>GAL4; sdk RNAi; UAS-dcr2), n=18 (NP6099>sdk RNAi; UAS-dcr2) and n=14 (repo>sdk RNAi; UAS-dcr2). ***P<0.0001 by one-way ANOVA with Tukey's post-hoc test; ns, not significant. Scale bars: 20 µm.
Fig. 5.
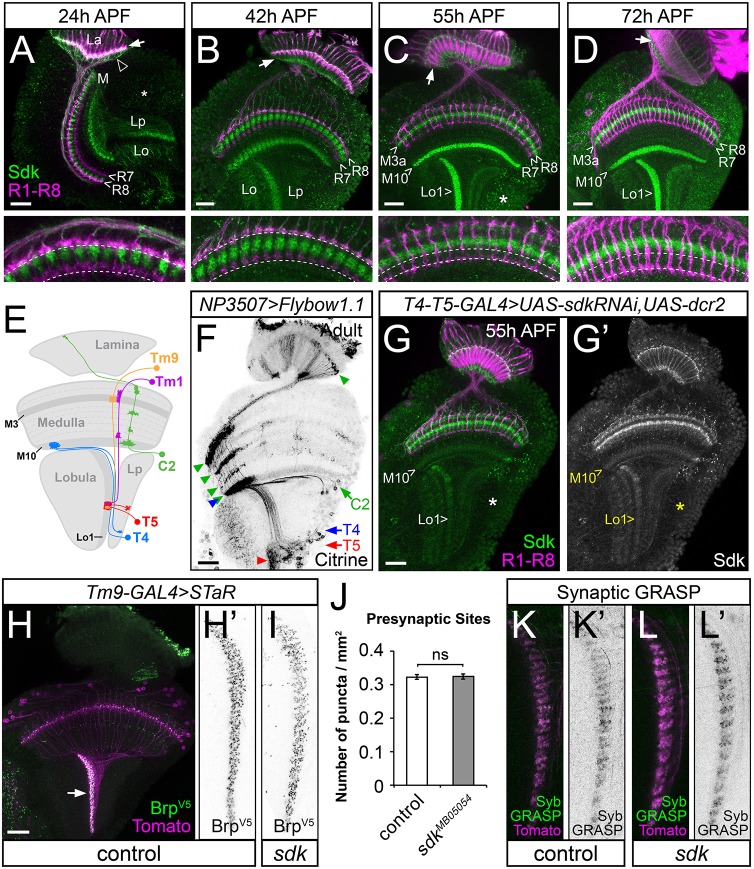
Sdk is localized to the dendrites of T4 and T5, but is not required for them to receive synaptic input. (A-D) Brains labeled with anti-Chp to mark photoreceptor axons (magenta) and anti-Sdk (green). (A) 24 h APF, (B) 42 h APF, (C) 55 h APF and (D) 72 h APF. Lower panels show enlargements of the distal medulla, with dotted lines marking the terminals of R7 and R8. During pupal stages, Sdk is localized to the lamina (La, arrow), to synaptic layers M3a and M10 in the medulla (M), and to Lo1 in the lobula (Lo), but is absent from the R7 and R8 growth cones. Arrowhead in A indicates glial cell bodies. (E) Diagram showing the projection patterns of identified Sdk-expressing neurons in the motion-detection pathways. (F) Adult brain showing subsets of neurons that express NP3507-GAL4, labeled with Citrine using the Flybow system. T4, T5 and C2 cell bodies (arrows) and neurites (arrowheads) are indicated. (G,G′) 55 h APF brain in which sdk RNAi is expressed in T4 and T5 with GMR42F06-GAL4, labeled with anti-Chp (magenta) and anti-Sdk (G′, green). Sdk is lost from the cell bodies in the lobula plate cortex (asterisk, compare with asterisk in C) and from the M10 and Lo1 layers, which contain the dendrites of T4 and T5, respectively. (H-I,K-L′) Adult brains, with the Lo1 layer (arrow in H) enlarged in H′,I,K-L′. (H-I) Endogenous Brp is tagged with V5 in Tm9 to label concentrations of presynaptic active zones (H′,I, green in H). Myr-tdTomato (magenta) labels the Tm9 neurons. (H,H′) sdkΔ7; (I) sdkMB05054. Brp-V5 puncta/µm2 in the Lo1 layer, counted in J (mean±s.e.m.), show no significant difference (P>0.5, t-test) between sdkΔ7 controls and mutants. n=94 sections from eight optic lobes (sdkΔ7), 110 sections from 10 optic lobes (sdkMB05054). (K-L′) GFP reconstituted from presynaptic Syb-GFP1-10 expressed in Tm9 and postsynaptic CD4-GFP11 expressed in T5 (K′,L′, green in K,L). Tm9 terminals are labeled with myr-tdTomato (magenta). (K,K′) sdkΔ7; (L,L′) sdkMB05054. The GRASP signal is taken to indicate that functional synapses are formed between Tm9 and T5 in sdk mutants. Scale bars: 20 µm.
As R1-R6 axons grow into the lamina, signals from these axons induce lamina neuron precursors to complete their final division, differentiate as neurons and align themselves into columns along the photoreceptor axons (Fig. 2A,B; Fig. S2A-C) (Huang and Kunes, 1996; Huang et al., 1998; Sugie et al., 2010). We found that this alignment was defective in sdk mutant larvae. Some lamina neurons failed to join columns and instead were located beneath the lamina plexus formed by the R1-R6 growth cones, which appeared discontinuous in some regions (Fig. 2C,F; Figs S2F and S3B). However, most lamina neurons were still present in sdk mutant adults (Fig. S3H-Q), indicating that sdk is not required for the survival of these cells. To test whether homophilic Sdk adhesion mediates interactions between photoreceptors and lamina neurons, we used RNAi to remove sdk function from different cell populations. Knocking down sdk specifically in the eye reproduced the phenotype of sdk mutants; similar numbers of lamina neurons were located in or beneath the lamina plexus (Fig. 2D,F; Fig. S3C) even though Sdk was still present on lamina neurons (Fig. 2D; Fig. S2H). In contrast, knocking down sdk in lamina neurons or in glia did not affect lamina neuron assembly into columns (Fig. 2E,F; Fig. S3D-G). The requirement for Sdk only in photoreceptors may reflect a requirement for Sdk-mediated adhesion between R1-R6 growth cones to enable the lamina plexus to form a barrier that lamina neurons cannot penetrate.
Beginning around 30 h of pupal development, the six photoreceptor axons from a single column diverge and sort into six different lamina cartridges, such that each cartridge collects visual information from photoreceptors that view the same point in space (Agi et al., 2014). This process, which together with the arrangement of photoreceptor cell visual axes is known as neural superposition, was also abnormal in sdk mutants compared with controls that had a precise excision of the Minos element. Many cartridges received too many or too few photoreceptor axons, and cartridges, which are usually wrapped by glial cells, were frequently fused together (Fig. 3B,E,G; Fig. S4A). In spite of the altered numbers of terminals within each cartridge in sdk mutants, the fine structure of tetrad synapses formed by R1-R6 onto lamina neurons L1, L2 and other cells was normal in all synapses studied in detail (Fig. S4C,D). Again, knocking down sdk exclusively in the eye had the same effect as complete removal of sdk function, but sorting was unaffected by knocking down sdk in lamina neurons (Fig. 3C,F,G; Fig. S4B). When we generated clones of sdk mutant photoreceptors, sorting defects were restricted to regions containing mutant axons, further supporting a requirement for sdk in photoreceptors (Fig. 3H). The axon-sorting phenotype is unlikely to be a secondary consequence of defective photoreceptor differentiation, because sdk mutants had very few extra R1-R6 photoreceptors (Fig. S4E-G) (Nguyen et al., 1997). Again, the requirement for Sdk in photoreceptors but not lamina neurons rules out the possibility that homophilic adhesion between Sdk molecules connects photoreceptors with their synaptic target cells.
Fig. 3.
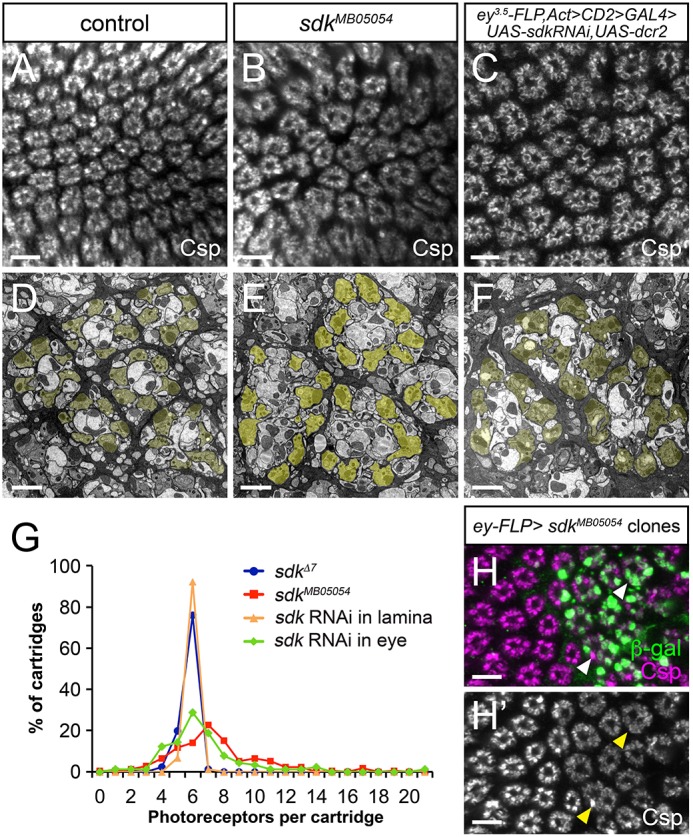
sdk is required in photoreceptors for sorting to the correct cartridges. (A-F) Adult laminas labeled with anti-Cysteine string protein (Csp) to mark the terminals of R1-R6 (A-C) or transmission electron micrographs of adult lamina cartridges with R1-R6 terminals pseudo-colored in yellow (D-F). (A,D) sdkΔ7 control; (B,E) sdkMB05054; (C,F) sdk RNAi in the eye. Many cartridges contain more or fewer photoreceptor terminals than the usual wild-type number (6), indicating mis-sorting. (G) The distribution of the numbers of R1-R6 terminals per cartridge in sdkΔ7, sdkMB05054 and in flies with sdk RNAi expressed in the eye (ey3.5-FLP, Act>CD2>GAL4) or lamina (27G05-FLP, Act>CD2>GAL4); distributions for sdk mutant and RNAi in the eye differ significantly from controls (P<0.0005, t-test). n=171 cartridges from three retinas (sdkΔ7, sdkMB05054); n=90 cartridges from three retinas (sdk RNAi). (H) Adult lamina with sdkMB05054 clones generated with ey-FLP and marked with anti-β-gal (green), labeled with anti-Csp (H′, magenta). Cartridge organization is abnormal in regions where photoreceptors are mutant (arrowheads). Scale bars: 5 µm in A-C,H,H′; 2 µm in D-F.
In the early pupal lamina, Sdk was localized to five spots in a horseshoe shape in each cartridge (Fig. 4A,B). Based on their geometry, these are likely to represent the five contact points between the stable ‘heels’ of the six photoreceptor growth cones, from which the ‘fronts’ extend away in stereotyped directions to reach their target cartridges (Langen et al., 2015). R4 axons can be specifically labeled using the E(spl)mδ-GAL4 driver to express a membrane marker (Chen and Clandinin, 2008). In wild-type laminas, they extend in a consistent direction from their ommatidium of origin (Fig. 4B), whereas in sdk mutants, their distance and direction of extension both varied (Fig. 4C). Two of the Sdk puncta were located on each side of the R4 growth cone, and were specifically depleted by 50% (on average) by expressing sdk RNAi in R4, confirming that they result from sdk expression in photoreceptors (Fig. 4B,D,E). Knocking down Sdk in this way did not disrupt R4 projections (Fig. 4D). This could be due to incomplete removal of Sdk from R4, or it might reflect either a non-autonomous function of sdk in photoreceptor axon sorting, or a requirement for sdk in a specific subset of photoreceptors that does not include R4. Based on the location of Sdk, we favor the model that it mediates adhesion between photoreceptor growth cone heels, providing a stable foundation from which the fronts can extend in a polarized manner to sort into the appropriate cartridges (Fig. 4F).
Fig. 4.
Sdk is present at contacts between photoreceptor axons and promotes growth cone polarization. (A-D) Laminas of sdkΔ7 control (A-B′), sdkMB05054 mutant (C,C′) and E(spl)mδ-GAL4; UAS-sdk RNAi (D,D′) pupae labeled with anti-Sdk (green) and anti-Chaoptin (Chp) to mark photoreceptor axons (blue in A, magenta in A′) and myr-tdTomato expressed in lamina neurons with GH146-GAL4 (red in A) or in R4 with E(spl)mδ-GAL4 (B′,C′,D′, magenta in B,C,D). (A) 30 h after puparium formation (APF). (B-D′) 38-40 h APF. Sdk is localized to five puncta (arrowheads) in a horseshoe shape, two of which are closely apposed to the heel of the R4 growth cone and depleted by sdk RNAi expression in R4 (open arrowheads in D,D′; enlarged in insets). The front of the R4 growth cone is indicated by an arrow in the inset in B′. The orientation of R4 growth cones is variable in sdk mutants but is not affected by knocking down sdk in R4. (E) Quantification of the fluorescence intensity of Sdk labeling at the R4:R5 contact relative to R2:R3 in control and E(spl)mδ-GAL4; UAS-sdk RNAi 30 h pupal laminas. n=67 ommatidia from three laminas, control; n=62 ommatidia from three laminas, sdk RNAi; ***P<0.0001, t-test. (F) Model depicting Sdk stabilizing the contacts between photoreceptor heels to form a scaffold from which the fronts can extend in fixed directions. (G,H) Optomotor responses were tested in a single-fly assay, in which panoramic visual stimuli elicited rotational responses from tethered flies. Graphs show responses to rotating square-wave contrast gratings, with wavelengths of 30°, 60° and 90°. (G) sdkMB05054 compared with sdkΔ7; (H) sdk RNAi expressed in the eye with ey3.5FLP, Act>CD2>GAL4 compared with controls (ey3.5FLP, Act>CD2>GAL4 crossed to attP; UAS-dcr2 or Act>CD2>GAL4 crossed to UAS-sdkRNAi; UAS-dcr2). Loss of sdk in the whole animal or just in the eye produced significantly reduced turning responses compared with controls at a wavelength of 30°, but not at 60° or 90°. Points represent mean responses over flies±s.e.m. n=10 (sdkΔ7); n=13 (sdkMB05054); n=21 (ey3.5FLP, Act>CD2>GAL4 X UAS-sdkRNAi; UAS-dcr2); n=22 (ey3.5FLP, Act>CD2>GAL4 X attP; UAS-dcr2); n=28 (Act>CD2>GAL4 X UAS-sdkRNAi; UAS-dcr2). *P<0.01, **P<0.001 by a rank sum test, Bonferroni-corrected for nine comparisons between the experimental and control genotypes. Scale bars: 5 µm in A-D′,F; 2 µm in insets in A,A′,B,B′,D,D′.
The connections of R1-R6 axons to L1 and L2 are the first synapses in the ‘ON’ and ‘OFF’ motion detection circuits, respectively (Heisenberg, 1971). To test whether the axon-sorting defects observed in sdk mutants had functional consequences for motion detection, we analyzed optomotor behavior in sdk mutant flies. Tethered flies placed on a trackball will attempt to move in the direction of a rotating striped pattern (Fig. 4G). Unlike precise excision controls, sdk mutant flies failed to follow the motion of square wave contrast patterns with short wavelengths over multiple temporal frequencies (Fig. 4G). However, these flies were not blind or unable to move, because longer spatial wavelength patterns elicited strong responses comparable with the control genotype (Fig. 4G). Similar optomotor defects were measured in flies in which sdk was specifically knocked down in the eye (Fig. 4H), suggesting that altered wiring of the retina to lamina connection is responsible for the failure to detect visual motion.
Sdk localizes to synaptic layers in the motion detection circuit
In addition to labeling R1-R6 growth cones, we found that during pupal development Sdk protein was localized to three specific layers within the optic lobes (Fig. 5A-D). Its pattern in medulla layer M10 and lobula layer Lo1 coincided with the location of the dendrites of T4 and T5, two lobula plate neurons that integrate information in the ‘ON’ and ‘OFF’ motion detection pathways, respectively (Fischbach and Dittrich, 1989; Maisak et al., 2013). Using the Flybow technique for single-cell labeling (Hadjieconomou et al., 2011a), we showed that a sdk-GAL4 line is expressed in cells with the arborization pattern characteristic of T4 and T5, as well as the centrifugal feedback neuron C2 (Fischbach and Dittrich, 1989; Meinertzhagen and O'Neil, 1991) (Fig. 5E,F). Sdk labeling was lost from M10 and Lo1 when we expressed sdk RNAi specifically in T4 and T5 (Fig. 5G), confirming that the labeling in these layers reflects Sdk on T4 and T5 dendrites. The strong reduction of immunolabeling when sdk was knocked down in T4 and T5 indicates that Sdk on the axon terminals of neurons that connect with T4 and T5 in those layers accounts for only a small fraction of the labeling. Weaker Sdk labeling was observed on the cell bodies and axons of T4 and T5, and appeared to be present in all four of the lobula plate layers that are innervated by the four subtypes (Fig. S5N,O).
The Sdk expression in medulla layer M3a did not colocalize with the growth cones of any of the five types of lamina output neurons or with Lamina wide-field (Lawf) 1 neurons (Fig. S5A-E,H,I). It is therefore likely to represent expression on the dendrites of a subpopulation of medulla neurons. We identified one such Sdk-expressing neuron as Tm9. The cell bodies of Tm9 cells showed punctate Sdk labeling, their dendrites overlapped the Sdk expression in layer M3a, and their axon terminals contained and were surrounded by Sdk labeling (Fig. S5J-M). Tm9 subserves motion detection and synapses on T5 (Behnia et al., 2014; Fisher et al., 2015; Shinomiya et al., 2014) (Fig. 5E; Fig. S5N,O). However, not all medulla neurons in the motion detection pathway extend dendrites in layer M3a; for example, the dendrites of Mi1, which synapses on T4 (Behnia et al., 2014; Takemura et al., 2013), did not overlap Sdk expression in the distal medulla (Fig. S5F,G).
In contrast to the role of Sdks in targeting neuronal processes to specific layers of the chick retina (Yamagata et al., 2002), T4 and T5 still arborized in the appropriate layers in sdk mutants (Fig. S6A,B), and single-cell labeling revealed no obvious defects in the size or arrangement of the T5 dendritic trees (Fig. S6E-I). As Tm9 and other medulla neurons are predicted to provide cholinergic input to T5 (Shinomiya et al., 2014), we visualized their postsynaptic sites by expressing a tagged acetylcholine receptor subunit, Dα7 (Mosca and Luo, 2014), in T4 and T5. Its localization did not appear to be altered in sdk mutants, although we could not resolve individual synapses with this marker (Fig. S6C,D). Using the synaptic tagging with recombination (STaR) method (Chen et al., 2014) to label the presynaptic active zone protein Bruchpilot (Brp) specifically in Tm9 cells, we observed similar numbers of Brp puncta in the T5 layer in sdk mutants and controls (Fig. 5H-J). Finally, we used a split GFP with one half fused to Synaptobrevin (Syb) and the other to the transmembrane domain of CD4, which specifically detects active synaptic contacts between cells (Macpherson et al., 2015), to show that such contacts between Tm9 and T5 are still present in sdk mutants (Fig. 5K,L). These results strongly suggest that sdk is not essential for synapses to form between Tm9 and T5. In addition, we were unable to detect consistent optomotor behavior defects in flies in which sdk was knocked down specifically in T4 and T5, using either an early driver expressed in the precursors of these cells (IPC-GAL4) (Oliva et al., 2014) or a later onset driver expressed in the differentiated neurons (42F06-GAL4) (Maisak et al., 2013) (Fig. S6J,K). Many aspects of T4 and T5 development thus proceed normally in sdk mutants, although we cannot rule out the possibility that there are subtle defects in their pattern of synaptic connections.
DISCUSSION
We show here that Drosophila Sdk is required to establish a functional pathway for visual motion detection. Defective optomotor behavior in sdk mutant flies appears to result from defects in the organization of connections between R1-R6 photoreceptors and their target lamina neurons. Sdk acts in photoreceptors both to organize lamina neurons into columns and to enable photoreceptor axons to sort to the correct lamina cartridges. Sdk is also expressed in T4 and T5 neurons, the output cells of the ‘ON’ and ‘OFF’ motion detection circuits; however, we could not detect any morphological or functional defects in these cells in the absence of sdk.
In the chicken retina, it was proposed that the homophilic adhesion molecules Sdk1 and Sdk2 each drive neurons to arborize and to form synapses with other neurons that express the same Sdk in a specific sublamina of the inner plexiform layer (Yamagata et al., 2002). Drosophila Sdk is also a homophilic adhesion molecule. However, because it is only required in photoreceptors, Sdk-Sdk binding cannot directly mediate the interaction of lamina neurons with photoreceptor axons. The simplest explanation for the presence of lamina neurons beneath the R1-R6 growth cone layer in sdk mutants or when sdk function is removed from the eye is that Sdk-mediated adhesion between photoreceptor growth cones normally forms a barrier to the movement of lamina neurons. An alternative possibility is that Sdk on photoreceptors might interact with a heterophilic binding partner on lamina neurons. The existence of a splice form of mouse Sdk1 that lacks the first two Ig domains (Kaufman et al., 2004), which are necessary for homophilic adhesion (Goodman et al., 2016; Hayashi et al., 2005), suggests the possibility of functions other than homophilic binding. The only cell-adhesion molecules known to promote interactions between lamina neurons and photoreceptor axons are Rst and Hbs, Ig superfamily members that are homologous to vertebrate Neph and Nephrin proteins, respectively (Fischbach et al., 2009; Sugie et al., 2010). Loss of Sdk does not affect the ability of lamina neurons to associate with photoreceptor axons to the same extent as loss of Rst or Hbs (Sugie et al., 2010), but it is possible that Sdk might modulate the interaction between them. Such an effect might explain why human Sdk1 upregulation contributes to the pathology of kidney diseases such as HIV-induced nephropathy and focal segmental glomerulosclerosis, as interacting Neph and Nephrin molecules form the slit diaphragm in the kidney (Kaufman et al., 2007, 2010).
Homophilic adhesion is also a likely explanation for the requirement for Sdk in photoreceptor axon sorting. Sorting of the axons from a single ommatidium to six different lamina cartridges requires R1-R6 growth cones to polarize such that their mobile ‘fronts’ move away from their stable ‘heels’ at characteristic angles (Schwabe et al., 2013; Langen et al., 2015). The pattern of Sdk labeling in five puncta, two of which arise from and are adjacent to R4, is consistent with a location at the contact points between the six heels, which are arrayed in a similar crescent (Langen et al., 2015). Two cadherins, Fmi and Ncad, have previously been shown to affect photoreceptor growth cone extension and sorting in the lamina. Although they have partially redundant roles, Ncad functions largely autonomously to mediate interactions with target cells and with photoreceptor growth cones within and across ommatidia, whereas a comparison of Fmi levels between neighboring growth cones appears to promote normal polarization (Prakash et al., 2005; Chen and Clandinin, 2008; Clandinin and Zipursky, 2000; Lee et al., 2003; Schwabe et al., 2013). We suggest that Sdk mediates adhesion between growth cone heels, enabling them to form a stable scaffold that may facilitate the comparison of Fmi levels or provide mechanical resistance necessary for polarized extension.
In contrast to knockdown studies in the chick retina, loss of Drosophila sdk does not alter the layer-specific arborization patterns of the Sdk-expressing neurons we have examined. Sdk is localized to the dendrites of T4 and T5, and is present in at least one of their presynaptic partners. Nonetheless, T4 and T5 still extend their dendrites in the correct layers and form arbors of the normal size and shape in the absence of sdk. In the mouse retina, Sdk2 controls the strength of synaptic connections between Sdk2-expressing neurons that detect motion of the central visual field relative to the surround (Krishnaswamy et al., 2015). We find that the Sdk-expressing neuron Tm9 does not require sdk to form synapses with T5, as shown both by punctate localization of the active zone protein Brp at its axon terminals and by activity-dependent synaptic GRASP. It is possible that Sdk is required for synapse formation by other neurons that are presynaptic to T4 or T5, or that it recruits specific postsynaptic proteins to these synapses. Nevertheless, removing sdk function from T4 and T5 does not result in significant defects in optomotor behavior, arguing against an essential role for sdk in these cells.
It has been suggested that the fly and vertebrate visual systems share common design principles reflecting deep evolutionary homology (Sanes and Zipursky, 2010; Shubin et al., 2009). The structural organization of the vertebrate retina resembles the Drosophila retina, lamina and medulla, and the computational mechanisms underlying motion detection also have many common features despite anatomical and molecular differences in the circuits (Clark and Demb, 2016). Our developmental and behavioral studies demonstrate that Sdk shares a physiological function in visual motion detection with its vertebrate counterparts, although their cellular mechanisms of action appear to differ. The functions of other molecules have also diverged between Drosophila and vertebrates; for example, the extensive alternative splicing of Drosophila Dscam1 allows it to mediate self-avoidance during growth and synaptogenesis (Millard et al., 2010; Miura et al., 2013; Schmucker et al., 2000), contrary to the adhesive function of chick Dscams (Yamagata and Sanes, 2008). Self-avoidance in the mouse instead depends on clustered protocadherins, which achieve diversity through promoter duplication (Lefebvre et al., 2012; Mountoufaris et al., 2017). It has been proposed that the formation of visual motion detection circuits in vertebrate and invertebrate visual systems is controlled by a genetic network derived from a common ancestor (Erclik et al., 2009; Gehring, 2004; Harada et al., 2007). Our data suggest that evolutionary changes may include the repurposing of common elements of such a network to mediate distinct functions.
MATERIALS AND METHODS
Genetics
Specific genotypes used for each figure are provided in Table S1. Pupal development hours were calculated from the white prepupal stage (0 h APF) at 25°C. Negatively labeled sdk mutant clones in eye imaginal discs and MARCM clones in adult laminas were made using eyeless (ey)-FLP. The Minos element Mi{ET1}sdkMB05054 was excised from sdk by crossing males with the insertion and the hs-MiT transposase gene to XXY females, heat shocking at 37°C for 1 h daily until the pupal stage, and screening male offspring for the loss of GFP fluorescence in the eyes (Metaxakis et al., 2005). sdkΔ7 was found by sequence analysis to be a precise excision, with an in-frame insertion of 6 bp. The sdkΔ15 allele is a deletion of 1240 bp at the 5′ end of sdk generated by imprecise excision of the P{EP}sdkEP369 element. The UAS-sdk RNAi flies used were P{KK105116}VIE-260B (Vienna Drosophila Resource Center, VDRC), in combination with UAS-dcr2. GMR42F06-GAL4 (Maisak et al., 2013) was used to drive gene expression in T4 and T5, and IPC-GAL4 (Oliva et al., 2014) was used to drive expression in the precursors of these cells. NP6099-GAL4 (Sugie et al., 2010) or 27G05-FLP, Act>CD2>GAL4 (Pecot et al., 2014) was used to drive expression in lamina precursor cells, and GH146-GAL4 to label lamina neurons in the pupa (Schwabe et al., 2014). GMR24C08-GAL4 was used to drive expression in Tm9 (Chen et al., 2014). GAL4 drivers for individual lamina neuron types are described elsewhere (Tuthill et al., 2013). The MultiColor FlpOut system (Nern et al., 2015) was used to label the dendrites of individual T5 neurons in sdk mutants and precise excision controls. Adult flies were heat shocked at 37°C for 1 h to induce recombination.
Histology
Cryosectioning was performed as previously described (Astigarraga et al., 2010). Whole brains were processed as follows prior to their incubation with primary antibodies. Third instar larval brains were fixed in 4% formaldehyde in PBS for 30 min at room temperature, washed in PBT (PBS+0.5% Triton X-100) and blocked in PBT+3% donkey serum. Pupal and adult brains and laminas were fixed in 2% paraformaldehyde/100 mM lysine/0.1 M phosphate buffer (pH 7.4) for 1 h at room temperature, washed in PBT and blocked in PBT+10% donkey serum.
The primary antibodies used were: mouse anti-Chaoptin (Chp) [1:50; Developmental Studies Hybridoma Bank (DSHB) 24B10]; mouse anti-Dachshund (Dac) (1:200; DSHB mAbdac1-1); rat anti-Elav (1:100; DSHB7E8A10); mouse anti-Repo (1:5; DSHB 8D12); mouse anti-Csp (1:10; DSHB 6D6); chicken anti-GFP (1:500; Aves Labs GFP-1010); rabbit anti-β-galactosidase (1:2500; MPBio 555976); rabbit anti-DsRed (1:500; Clontech 632393); mouse anti-V5 (1:400; Invitrogen R960-25); rabbit anti-FLAG (1:200; Sigma F3165); rat anti-HA 3F10 (1:50; Roche 11867423001); mouse anti-HA 6E2 (1:200; Cell Signaling 2367); and Alexa 488, TRITC or Alexa 647-coupled goat anti-HRP, to detect neuronal membranes (1:200; Jackson ImmunoResearch 123-545-021, 1230-025-021, 123-605-021). Secondary antibodies (Jackson ImmunoResearch and Life Technologies) were coupled to the fluorochromes DyLight 405, Cy3, Alexa Fluor 488 or Alexa Fluor 647. DAPI was used at 300 nM. Cryosections were mounted in Fluoromount-G (Southern Biotech) and whole-mount brains in SlowFade Gold antifade reagent (Life Technologies). Confocal pictures were collected using Leica SP5 and Zeiss LSM510 confocal microscopes.
To make the anti-Sdk antibody, a cDNA fragment encoding the intracellular region of Sdk (amino acids 2025 to 2224) was cloned into the pGEX-4T-1 vector. This plasmid was transformed into BL21 E. coli competent cells for translation upon IPTG induction. Bacteria were sonicated and GST-Sdk fusion protein was purified by incubating the supernatant with glutathione-sepharose beads. GST-Sdk was eluted from the beads by incubating with 10 mM glutathione, 50 mM Tris-HCl (pH 8), 100 mM NaCl and dialyzed against PBS overnight at 4°C. Guinea pig antiserum was produced by Covance and affinity purified. Briefly, the GST-Sdk fusion protein was run on an SDS-PAGE gel and transferred to a nitrocellulose membrane, and the antiserum was incubated with the region of the membrane containing the GST-Sdk band for 3 h at 4°C. The membrane was washed with PBS and bound antibody was recovered by incubating the membrane with 200 mM glycine, 1 mM EGTA (pH 2-2.5) for 15 min. The antibody solution was then neutralized to a final pH of 7-8 by adding an equal volume of 100 mM Tris (pH 8.8). This solution was dialyzed against PBS, mixed 50:50 with glycerol and stored at −20°C.
The riboprobe for in situ hybridization was made by cloning a sdk cDNA fragment (bp 3070-3768) into the pGEM-T vector (Promega), followed by in vitro transcription with digoxigenin-labeled UTP, using the DIG RNA Labeling Kit by Roche. Larval discs and brains were fixed in 8% formaldehyde for 1 h on ice, rinsed in PBST (PBS+0.1% Tween-20) and prehybridized in HB4 solution (50% formamide, 5×SSC, 50 µg/ml heparin, 0.1% Tween-20, 5 mg/ml torula yeast RNA extract) for 1 h at 65°C, prior to overnight incubation with HB4+1% riboprobe in the same conditions. Tissue was then rinsed with 50:50 formamide/ 2×SSC /0.1% Tween-20 at 65°C, followed by washes with PBST at room temperature, and anti-digoxigenin antibody incubation for 2 h. Washes with PBST and with alkaline phosphatase (AP) buffer [100 mM NaCl, 50 mM MgCl2, 100 mM Tris (pH 9.5), 0.1% Tween 20] followed. Development was carried out in AP buffer with 0.35% each of NBT and BCIP. The reaction was stopped with PBST.
Cell culture and aggregation assays
S2 cells (originally obtained from Ruth Lehmann and used regularly in our lab) were grown in Schneider's Drosophila Medium (GIBCO Invitrogen) with 10% heat-inactivated fetal bovine serum and 50 units/ml penicillin-50 g/ml streptomycin (GIBCO Invitrogen) at 25°C. To make the UAS-sdk-HA construct, a SpeI-NotI fragment containing three copies of the HA tag was generated by PCR and cloned after the 5′ signal peptide sequence (amino acid 60) of sdk in the LD39520 clone (Drosophila Genomics Resource Center). A BglII-KpnI fragment containing the 5′ end of sdk, including the HA tags was then cloned into pUAST(attB), followed by a KpnI-XbaI fragment, an internal KpnI-KpnI fragment and an XbaI-NheI PCR fragment from the 3′ end of sdk that was ligated into the XbaI site. For aggregation assays, S2 cells were pelleted 48 h after transient transfection using Effectene Transfection Reagent (Qiagen) and washed in fresh medium. Cells (2.5-3 ml at a concentration of 4-5×106 cells/ml) were rocked at 50 rpm for at least 3 h. Plates were then analyzed for the presence of cell aggregates. Pictures were collected using a Zeiss LSM510 confocal microscope.
Transmission EM of the lamina
Male flies had their proboscis excised and their heads were bisected and fixed for 2 h on ice in freshly mixed fixative containing 2.5% glutaraldehyde and 2.5% formaldehyde (as paraformaldehyde) in 0.1 M Na cacodylate buffer (pH 7.3). After primary fixation specimens were washed in 0.1 M Na cacodylate buffer (pH 7.3) for 3×10 min, then postfixed in 2% osmium tetroxide in 0.1 M Na cacodylate for 60 min at 4°C and finally rinsed 3×10 min in distilled water. Dehydration was carried out in an ethanol series 50%, 70%, 80%, 90%, 95% and twice in each of 100% and propylene oxide, for ∼5-10 min each. Infiltration was processed by mixing propylene oxide and Epon (Embed 812: EMS, Hatfield, PA) 1:1 and leaving overnight, then changing to fresh resin the next day for at least 4 h and embedding in fresh Epon in an embedding mold. Polymerization was completed in a 60°C oven for less than 48 h. Sample blocks were trimmed and ultrathin 50-60 nm sections were cut on an Ultratome and collected on Pioloform-coated single-slot EM grids. Grids with sections were post contrasted in 2% uranyl acetate for 10 min and in Reynold's lead citrate for 5 min (Meinertzhagen and O'Neil, 1991). Sections were viewed at 11,500× using an FEI Tecnai 12 electron microscope operated at 80 kV and images were captured using a Gatan 832 Orius SC1000 CCD camera with Gatan DigitalMicrograph software.
Quantifications
To quantify misplaced lamina neurons, Dac-positive cells in or beneath the lamina plexus in third instar larval brains were counted from the posterior view (Figs S2B and 3A-E), at an average tissue thickness of around 30 µm at matched confocal depth. The index provided in Fig. 2F was obtained by dividing the number of cells found in or beyond the lamina plexus by the tissue thickness analyzed (cells/µm).
Quantification of the active zones labeled by synaptic tagging with recombination (STaR) (Chen et al., 2014) was carried out using ImageJ. Confocal sections analyzed were at least 1.5 µm apart to avoid counting the same active zone twice (Chen et al., 2014). A region of interest was drawn around the labeled area, a smooth filter was applied, and puncta (active zones) were counted within that region and divided by the area of the region in µm2.
The number of photoreceptors per cartridge (Fig. 3G) was counted using the electron micrographs. Three flies of each genotype were analyzed, and the photoreceptor profiles counted from 30-60 cartridges in each fly. For quantification of R1-R6 photoreceptors in the adult eye, Rh1-GFP was crossed to sdk alleles and retinas were imaged under water immersion with confocal microscopy (Pichaud and Desplan, 2001). Quantification of Sdk labeling intensity at the contact between R4 and R5 growth cones was measured using ImageJ relative to the contact between R2 and R3 growth cones, after subtracting background.
An explicit power analysis was not used to compute sample sizes in advance. Sample sizes sufficient to detect strongly significant differences between sdk mutants and controls in the assays reported were used for other comparisons, making it unlikely that an effect of similar magnitude would have escaped detection. Biological replicates are defined as different individuals of the same genotype. No outliers have been excluded from the data shown.
Western blotting
To extract proteins from embryos, 0-16 h embryos were dechorionated in bleach, washed and homogenized in 50 mM Tris-HCl (pH 8), 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 5 mM NaF, 5 mM EDTA, Complete Protease Inhibitor Cocktail (Roche). The embryo extracts were quantified by the method of Bradford (Bio-Rad), mixed with Laemmli buffer [4% SDS, 20% glycerol, 120 mM Tris-Cl (pH 6.8), 0.02% Bromophenol Blue] and run on an SDS-PAGE gel. Western blotting was carried out as previously described (Miura et al., 2006), but blocking with BSA for 1 h and incubating with primary antibodies overnight: guinea pig anti-Sdk was used at 1:2000 and mouse anti-β-tubulin (Covance MMS-410P) at 1:40,000.
Optomotor behavior assays
Briefly, flies were cold-anesthetized and tethered in place above a 6 mm diameter polystyrene ball floating on an air cushion (Fig. 4G). An optical mouse measured the rotation of the ball in response to fly-walking behaviors. The fly turned in response to visual stimuli, which were presented on three panoramic screens surrounding the fly and subtending 270° horizontally and 110° vertically (Salazar-Gatzimas et al., 2016). The luminance for all experiments averaged 100 cd/m2 and stimuli were presented in green, centered on a wavelength of ∼525 nm. All experiments were conducted at 34-36°C, which permits thermogenetic experiments and generates robust behavioral responses (Clark et al., 2011). Flies were shown square wave gratings at 0.25 contrast on the screens (i.e. peak intensity was 25% greater than the mean luminance over space) and their rotational responses were measured. Wavelengths for the stimuli were 30°, 60° and 90°, and rotational velocities were chosen so that stimuli had temporal contrast frequencies of 0.5 Hz, 2 Hz and 8 Hz. Positive measured velocities correspond to turning in the direction of visual motion. Moving stimuli were presented in a pseudorandom order both clockwise and counterclockwise; each stimulus was presented for 4 s with a 4 s mean-luminance spatially uniform (i.e. gray) interleave. The response of each fly was measured as the mean turning velocity of the fly over the entire 4 s stimulus presentation, also averaged over presentations of each stimulus type. The mean response of each fly was treated as an independent measurement for statistical purposes. P values were computed using a non-parametric rank sum test, Bonferroni corrected for the nine comparisons made for the nine stimuli. Experimental responses were considered significant if this test gave P<0.05 relative to all relevant control distributions.
Supplementary Material
Acknowledgements
We thank Claude Desplan, Todd Laverty, Chi-Hon Lee, Tim Mosca, Aljoscha Nern, Larry Zipursky, the Bloomington Drosophila Stock Center, the Vienna Drosophila Resource Center, the Kyoto Stock Center, the Drosophila Genomics Resource Center and the Developmental Studies Hybridoma Bank for fly stocks and reagents. We are grateful to Michael Cammer, Maximilien Courgeon, Yan Deng, Hyung Don Ryoo, Justine Oyallon and Tom Clandinin for technical advice. The manuscript was improved by the critical comments of Inés Carrera, Cheuk Hei Ho, Carolyn Morrison and Annabelle Suisse.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.A., J.D., D.A.C., I.A.M., J.E.T.; Methodology: S.A., D.T., M.S.C., O.M.; Formal analysis: M.S.C., O.M., D.A.C., I.A.M., J.E.T.; Investigation: S.A., J.D., D.T., M.S.C., O.M.; Resources: J.E.T.; Writing - original draft: S.A., J.E.T.; Writing - review & editing: S.A., J.D., D.A.C., I.A.M., J.E.T.; Supervision: D.A.C., I.A.M., J.E.T.; Project administration: J.E.T.; Funding acquisition: D.A.C., I.A.M., J.E.T.
Funding
Funding was provided by National Institutes of Health grants GM089799 and EY025540 to J.E.T., and EY026555 to D.A.C.; and by Natural Sciences and Engineering Research Council of Canada grant DIS000065 to I.A.M. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.158246.supplemental
References
- Agi E., Langen M., Altschuler S. J., Wu L. F., Zimmermann T. and Hiesinger P. R. (2014). The evolution and development of neural superposition. J. Neurogenet. 28, 216-232. 10.3109/01677063.2014.922557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S., Hofmeyer K., Farajian R. and Treisman J. E. (2010). Three Drosophila liprins interact to control synapse formation. J. Neurosci. 30, 15358-15368. 10.1523/JNEUROSCI.1862-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R., Clark D. A., Carter A. G., Clandinin T. R. and Desplan C. (2014). Processing properties of ON and OFF pathways for Drosophila motion detection. Nature 512, 427-430. 10.1038/nature13427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K. L. et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, 731-743. 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A. (2014). Neural circuits for motion vision in the fly. Cold Spring Harb. Symp. Quant. Biol. 79, 131-139. 10.1101/sqb.2014.79.024695 [DOI] [PubMed] [Google Scholar]
- Chen P.-L. and Clandinin T. R. (2008). The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 58, 26-33. 10.1016/j.neuron.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Akin O., Nern A., Tsui C. Y., Pecot M. Y. and Zipursky S. L. (2014). Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280-293. 10.1016/j.neuron.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T. R. and Zipursky S. L. (2000). Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron 28, 427-436. 10.1016/S0896-6273(00)00122-7 [DOI] [PubMed] [Google Scholar]
- Clark D. A. and Demb J. B. (2016). Parallel computations in insect and mammalian visual motion processing. Curr. Biol. 26, R1062-R1072. 10.1016/j.cub.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. A., Bursztyn L., Horowitz M. A., Schnitzer M. J. and Clandinin T. R. (2011). Defining the computational structure of the motion detector in Drosophila. Neuron 70, 1165-1177. 10.1016/j.neuron.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J. and Ghosh A. (2016). Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 22-35. [DOI] [PubMed] [Google Scholar]
- Erclik T., Hartenstein V., McInnes R. R. and Lipshitz H. D. (2009). Eye evolution at high resolution: the neuron as a unit of homology. Dev. Biol. 332, 70-79. 10.1016/j.ydbio.2009.05.565 [DOI] [PubMed] [Google Scholar]
- Fischbach K. F. and Dittrich A. P. M. (1989). The optic lobe of Drosophila melanogaster, I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441-475. 10.1007/BF00218858 [DOI] [Google Scholar]
- Fischbach K. F., Linneweber G. A., Andlauer T. F., Hertenstein A., Bonengel B. and Chaudhary K. (2009). The irre cell recognition module (IRM) proteins. J. Neurogenet. 23, 48-67. 10.1080/01677060802471668 [DOI] [PubMed] [Google Scholar]
- Fisher Y. E., Leong J. C., Sporar K., Ketkar M. D., Gohl D. M., Clandinin T. R. and Silies M. (2015). A class of visual neurons with wide-field properties is required for local motion detection. Curr. Biol. 25, 3178-3189. 10.1016/j.cub.2015.11.018 [DOI] [PubMed] [Google Scholar]
- Gehring W. J. (2004). Historical perspective on the development and evolution of eyes and photoreceptors. Int. J. Dev. Biol. 48, 707-717. 10.1387/ijdb.041900wg [DOI] [PubMed] [Google Scholar]
- Goodman K. M., Yamagata M., Jin X., Mannepalli S., Katsamba P. S., Ahlsen G., Sergeeva A. P., Honig B., Sanes J. R. and Shapiro L. (2016). Molecular basis of sidekick-mediated cell-cell adhesion and specificity. Elife 5, e19058 10.7554/eLife.19058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D., Rotkopf S., Alexandre C., Bell D. M., Dickson B. J. and Salecker I. (2011a). Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods 8, 260-266. 10.1038/nmeth.1567 [DOI] [PubMed] [Google Scholar]
- Hadjieconomou D., Timofeev K. and Salecker I. (2011b). A step-by-step guide to visual circuit assembly in Drosophila. Curr. Opin. Neurobiol. 21, 76-84. 10.1016/j.conb.2010.07.012 [DOI] [PubMed] [Google Scholar]
- Harada T., Harada C. and Parada L. F. (2007). Molecular regulation of visual system development: more than meets the eye. Genes Dev. 21, 367-378. 10.1101/gad.1504307 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Kaufman L., Ross M. D. and Klotman P. E. (2005). Definition of the critical domains required for homophilic targeting of mouse sidekick molecules. FASEB J. 19, 614-616. 10.1096/fj.04-2947fje [DOI] [PubMed] [Google Scholar]
- Heisenberg M. (1971). Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J. Exp. Biol. 55, 85-100. [DOI] [PubMed] [Google Scholar]
- Huang Z. and Kunes S. (1996). Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell 86, 411-422. 10.1016/S0092-8674(00)80114-2 [DOI] [PubMed] [Google Scholar]
- Huang Z., Shilo B. Z. and Kunes S. (1998). A retinal axon fascicle uses Spitz, an EGF receptor ligand, to construct a synaptic cartridge in the brain of Drosophila. Cell 95, 693-703. 10.1016/S0092-8674(00)81639-6 [DOI] [PubMed] [Google Scholar]
- Joesch M., Schnell B., Raghu S. V., Reiff D. F. and Borst A. (2010). ON and OFF pathways in Drosophila motion vision. Nature 468, 300-304. 10.1038/nature09545 [DOI] [PubMed] [Google Scholar]
- Kaufman L., Hayashi K., Ross M. J., Ross M. D. and Klotman P. E. (2004). Sidekick-1 is upregulated in glomeruli in HIV-associated nephropathy. J. Am. Soc. Nephrol. 15, 1721-1730. 10.1097/01.ASN.0000128975.28958.C2 [DOI] [PubMed] [Google Scholar]
- Kaufman L., Yang G., Hayashi K., Ashby J. R., Huang L., Ross M. J., Klotman M. E. and Klotman P. E. (2007). The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in HIV-associated nephropathy. FASEB J. 21, 1367-1375. 10.1096/fj.06-7191com [DOI] [PubMed] [Google Scholar]
- Kaufman L., Potla U., Coleman S., Dikiy S., Hata Y., Kurihara H., He J. C., D'Agati V. D. and Klotman P. E. (2010). Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J. Biol. Chem. 285, 25677-25685. 10.1074/jbc.M110.133959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A., Yamagata M., Duan X., Hong Y. K. and Sanes J. R. (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466-470. 10.1038/nature14682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M., Agi E., Altschuler D. J., Wu L. F., Altschuler S. J. and Hiesinger P. R. (2015). The developmental rules of neural superposition in Drosophila. Cell 162, 120-133. 10.1016/j.cell.2015.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Clandinin T. R., Lee C. H., Chen P. L., Meinertzhagen I. A. and Zipursky S. L. (2003). The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat. Neurosci. 6, 557-563. 10.1038/nn1063 [DOI] [PubMed] [Google Scholar]
- Lefebvre J. L., Kostadinov D., Chen W. V., Maniatis T. and Sanes J. R. (2012). Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517-521. 10.1038/nature11305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson L. J., Zaharieva E. E., Kearney P. J., Alpert M. H., Lin T. Y., Turan Z., Lee C. H. and Gallio M. (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 10024 10.1038/ncomms10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisak M. S., Haag J., Ammer G., Serbe E., Meier M., Leonhardt A., Schilling T., Bahl A., Rubin G. M., Nern A. et al. (2013). A directional tuning map of Drosophila elementary motion detectors. Nature 500, 212-216. 10.1038/nature12320 [DOI] [PubMed] [Google Scholar]
- Meier M., Serbe E., Maisak M. S., Haag J., Dickson B. J. and Borst A. (2014). Neural circuit components of the Drosophila OFF motion vision pathway. Curr. Biol. 24, 385-392. 10.1016/j.cub.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Meinertzhagen I. A. and O'Neil S. D. (1991). Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305, 232-263. 10.1002/cne.903050206 [DOI] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A. and Savakis C. (2005). Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171, 571-581. 10.1534/genetics.105.041848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard S. S., Lu Z., Zipursky S. L. and Meinertzhagen I. A. (2010). Drosophila Dscam proteins regulate postsynaptic specificity at multiple-contact synapses. Neuron 67, 761-768. 10.1016/j.neuron.2010.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura G. I., Buglino J., Alvarado D., Lemmon M. A., Resh M. D. and Treisman J. E. (2006). Palmitoylation of the EGFR ligand Spitz by Rasp increases Spitz activity by restricting its diffusion. Dev. Cell 10, 167-176. 10.1016/j.devcel.2005.11.017 [DOI] [PubMed] [Google Scholar]
- Miura S. K., Martins A., Zhang K. X., Graveley B. R. and Zipursky S. L. (2013). Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell 155, 1166-1177. 10.1016/j.cell.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca T. J. and Luo L. (2014). Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. Elife 3, e03726 10.7554/eLife.03726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountoufaris G., Chen W. V., Hirabayashi Y., O'Keeffe S., Chevee M., Nwakeze C. L., Polleux F. and Maniatis T. (2017). Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356, 411-414. 10.1126/science.aai8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D. and Rubin G. M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967-E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. N., Liu Y., Litsky M. L. and Reinke R. (1997). The sidekick gene, a member of the immunoglobulin superfamily, is required for pattern formation in the Drosophila eye. Development 124, 3303-3312. [DOI] [PubMed] [Google Scholar]
- Oliva C., Choi C. M., Nicolai L. J., Mora N., De Geest N. and Hassan B. A. (2014). Proper connectivity of Drosophila motion detector neurons requires Atonal function in progenitor cells. Neural Dev. 9, 4 10.1186/1749-8104-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot M. Y., Chen Y., Akin O., Chen Z., Tsui C. Y. and Zipursky S. L. (2014). Sequential. axon-derived signals couple target survival and layer specificity in the Drosophila visual system. Neuron 82, 320-333. 10.1016/j.neuron.2014.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F. and Desplan C. (2001). A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development 128, 815-826. [DOI] [PubMed] [Google Scholar]
- Prakash S., Caldwell J. C., Eberl D. F. and Clandinin T. R. (2005). Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat. Neurosci. 8, 443-450. 10.1038/nn1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E. and Baier H. (2012). Assembly of synaptic laminae by axon guidance molecules. Curr. Opin. Neurobiol. 22, 799-804. 10.1016/j.conb.2012.04.014 [DOI] [PubMed] [Google Scholar]
- Salazar-Gatzimas E., Chen J., Creamer M. S., Mano O., Mandel H. B., Matulis C. A., Pottackal J. and Clark D. A. (2016). Direct measurement of correlation responses in Drosophila elementary motion detectors reveals fast timescale tuning. Neuron 92, 227-239. 10.1016/j.neuron.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J. R. and Zipursky S. L. (2010). Design principles of insect and vertebrate visual systems. Neuron 66, 15-36. 10.1016/j.neuron.2010.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D., Clemens J. C., Shu H., Worby C. A., Xiao J., Muda M., Dixon J. E. and Zipursky S. L. (2000). Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671-684. 10.1016/S0092-8674(00)80878-8 [DOI] [PubMed] [Google Scholar]
- Schwabe T., Neuert H. and Clandinin T. R. (2013). A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 154, 351-364. 10.1016/j.cell.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T., Borycz J. A., Meinertzhagen I. A. and Clandinin T. R. (2014). Differential. adhesion determines the organization of synaptic fascicles in the Drosophila visual system. Curr. Biol. 24, 1304-1313. 10.1016/j.cub.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbe E., Meier M., Leonhardt A. and Borst A. (2016). Comprehensive characterization. of the major presynaptic elements to the Drosophila OFF motion detector. Neuron 89, 829-841. 10.1016/j.neuron.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Shinomiya K., Karuppudurai T., Lin T. Y., Lu Z., Lee C. H. and Meinertzhagen I. A. (2014). Candidate neural substrates for off-edge motion detection in Drosophila. Curr. Biol. 24, 1062-1070. 10.1016/j.cub.2014.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N., Tabin C. and Carroll S. (2009). Deep homology and the origins of evolutionary novelty. Nature 457, 818-823. 10.1038/nature07891 [DOI] [PubMed] [Google Scholar]
- Silies M., Gohl D. M., Fisher Y. E., Freifeld L., Clark D. A. and Clandinin T. R. (2013). Modular use of peripheral input channels tunes motion-detecting circuitry. Neuron 79, 111-127. 10.1016/j.neuron.2013.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother J. A., Nern A. and Reiser M. B. (2014). Direct observation of ON and OFF pathways in the Drosophila visual system. Curr. Biol. 24, 976-983. 10.1016/j.cub.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Strother J. A., Wu S. T., Wong A. M., Nern A., Rogers E. M., Le J. Q., Rubin G. M. and Reiser M. B. (2017). The emergence of directional selectivity in the visual motion pathway of Drosophila. Neuron 94, 168-182 e110. 10.1016/j.neuron.2017.03.010 [DOI] [PubMed] [Google Scholar]
- Sugie A., Umetsu D., Yasugi T., Fischbach K. F. and Tabata T. (2010). Recognition of pre- and postsynaptic neurons via nephrin/NEPH1 homologs is a basis for the formation of the Drosophila retinotopic map. Development 137, 3303-3313. 10.1242/dev.047332 [DOI] [PubMed] [Google Scholar]
- Takemura S. Y., Bharioke A., Lu Z., Nern A., Vitaladevuni S., Rivlin P. K., Katz W. T., Olbris D. J., Plaza S. M., Winston P. et al. (2013). A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175-181. 10.1038/nature12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura S. Y., Nern A., Chklovskii D. B., Scheffer L. K., Rubin G. M. and Meinertzhagen I. A. (2017). The comprehensive connectome of a neural substrate for ‘ON’ motion detection in Drosophila. Elife 6, e24394 10.7554/eLife.24394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill J. C., Nern A., Holtz S. L., Rubin G. M. and Reiser M. B. (2013). Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79, 128-140. 10.1016/j.neuron.2013.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet M. F., Huberman A. D. and Desplan C. (2014). So many pieces, one puzzle: cell type specification and visual circuitry in flies and mice. Genes Dev. 28, 2565-2584. 10.1101/gad.248245.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M. and Sanes J. R. (2008). Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451, 465-469. 10.1038/nature06469 [DOI] [PubMed] [Google Scholar]
- Yamagata M. and Sanes J. R. (2012). Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J. Neurosci. 32, 14402-14414. 10.1523/JNEUROSCI.3193-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M., Weiner J. A. and Sanes J. R. (2002). Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110, 649-660. 10.1016/S0092-8674(02)00910-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.