ABSTRACT
NKX2-5 is the most commonly mutated gene associated with human congenital heart defects (CHDs), with a predilection for cardiac pole abnormalities. This homeodomain transcription factor is a central regulator of cardiac development and is expressed in both the first and second heart fields (FHF and SHF). We have previously revealed essential functions of nkx2.5 and nkx2.7, two Nkx2-5 homologs expressed in zebrafish cardiomyocytes, in maintaining ventricular identity. However, the differential roles of these genes in the specific subpopulations of the anterior (aSHF) and posterior (pSHF) SHFs have yet to be fully defined. Here, we show that Nkx genes regulate aSHF and pSHF progenitors through independent mechanisms. We demonstrate that Nkx genes restrict proliferation of aSHF progenitors in the outflow tract, delimit the number of pSHF progenitors at the venous pole and pattern the sinoatrial node acting through Isl1 repression. Moreover, optical mapping highlights the requirement for Nkx gene dose in establishing electrophysiological chamber identity and in integrating the physiological connectivity of FHF and SHF cardiomyocytes. Ultimately, our results may shed light on the discrete errors responsible for NKX2-5-dependent human CHDs of the cardiac outflow and inflow tracts.
KEY WORDS: nkx2.5, nkx2.7, Arterial pole, Venous pole, Second heart field, Zebrafish
Summary: Nkx genes play key functions in secondary heart field cardiomyocyte differentiation in zebrafish and shed light on the mechanisms underlying heart malformations and conduction defects in individuals carrying NKX2-5 mutations.
INTRODUCTION
Cardiomyocytes (CMs) are derived from at least two distinct cardiac progenitor populations located in the splanchnic mesoderm. First heart field (FHF) cells differentiate early in the lateral plate mesoderm to give rise to the linear heart tube (Buckingham et al., 2005; Vincent and Buckingham, 2010). Subsequently, second heart field (SHF) cells migrate to the pharyngeal mesoderm, proliferate and differentiate into CMs at the arterial and venous poles. The anterior SHF (aSHF) subpopulation gives rise to the right ventricle and outflow tract (OFT), whereas the posterior SHF (pSHF) contributes primarily to the atria and inflow tract (IFT) (Buckingham et al., 2005; Cai et al., 2003; Dominguez et al., 2012; Dyer and Kirby, 2009; Galli et al., 2008; Rana et al., 2014; Vincent and Buckingham, 2010; Zaffran et al., 2004). Errors in specification and differentiation of the SHF cells under these precise spatial and temporal constraints can lead to a variety of congenital heart defects (CHDs), specifically conotruncal (OFT) and atrial (IFT) abnormalities. These malformations of the cardiac poles are found in 10% and 30%, respectively, of individuals with CHDs and result in significant neonatal morbidity and mortality (Gelb and Chung, 2014; Hoffman and Kaplan, 2002; Hoffman et al., 2004; Loffredo, 2000; Nemer, 2008; Payne et al., 1995; Pierpont et al., 2007; Supino et al., 2006). Despite this clinical impact, the mechanisms underlying the development of the arterial and venous poles of the heart are incompletely understood.
NKX2-5, a homeodomain transcription factor expressed in cells of the FHF and SHF (Stanley et al., 2002), and a regulator of cardiac development (Bruneau, 2008), is associated with a myriad of human CHDs. Moreover, individuals carrying NKX2-5 mutations demonstrate a propensity to develop malformations involving the poles of the heart, such as Tetralogy of Fallot, double outlet right ventricle and atrial septal defects (Abou Hassan et al., 2015; Benson et al., 1999; Chung and Rajakumar, 2016; Elliott et al., 2003; Goldmuntz et al., 2001; McElhinney et al., 2003; Nakajima, 2010; Schott et al., 1998; Srivastava and Olson, 2000). Investigation of the genetic basis of CHDs in model systems has yielded insights into functions of Nkx2-5 in progenitor specification in Drosophila, Xenopus and mouse (Azpiazu and Frasch, 1993; Bodmer, 1993; Grow and Krieg, 1998; Prall et al., 2007), and in cardiac morphogenesis in mouse and zebrafish (Barth et al., 2010; Lyons et al., 1995; Prall et al., 2007; Tanaka et al., 1999; Targoff et al., 2013, 2008; Tu et al., 2009). Furthermore, Nkx2-5 mutants, whether inside or outside the DNA-binding domains (Benson et al., 1999; Gutierrez-Roelens et al., 2002; Ikeda et al., 2002), regulate wild-type targets and also ‘off-targets’, destabilizing the cardiac transcriptional hierarchy in this context (Bouveret et al., 2015). Although previous work has revealed specific roles for Nkx2-5 in the SHF (Cambier et al., 2014; Clark et al., 2013; Dorn et al., 2014; George et al., 2015; Guner-Ataman et al., 2013; Prall et al., 2007; Watanabe et al., 2012; Witzel et al., 2012; Zhang et al., 2014), the distinct cellular and molecular mechanisms operating in the aSHF and pSHF subpopulations remain largely unknown.
Recent studies have revealed essential functions of Nkx2-5 transcriptional regulation in OFT development. In mouse, Nkx2-5 has been shown to drive arterial pole formation via a Bmp2/Smad1 negative-feedback loop (Prall et al., 2007) and through a direct repressive effect on a 1.7 kb Fgf10 enhancer element (Watanabe et al., 2012). In zebrafish, nkx2.5 plays a role in aSHF proliferation (Guner-Ataman et al., 2013) and stabilization of Nkx2.5 by Ajuba protein regulates progenitor specification in this population (Witzel et al., 2012). Moreover, the expression patterns of ltbp3 and mef2cb, aSHF markers in zebrafish, overlap with the nkx2.5-expressing region near the arterial pole (Lazic and Scott, 2011; Zhou et al., 2011). Although these data document the importance of Nkx genes in arterial pole development, it is unclear how Nkx2.5 controls differentiation and expansion of the aSHF progenitor pool to sculpt the precise dimensions of the OFT.
At the sinus venosus (SV), extensive studies have elucidated the function of Nkx2-5 in cardiac conduction system (CCS) development (Briggs et al., 2008; Jay et al., 2004; Moskowitz et al., 2007; Nakashima et al., 2009; Pashmforoush et al., 2004; Ye et al., 2015). Yet the mechanisms by which Nkx genes direct pSHF progenitors to pattern the venous pole have not been fully defined. It is evident from murine models that pSHF cells accumulate in the atrial chambers (Cai et al., 2003; Galli et al., 2008; Lescroart et al., 2012) and pSHF progenitors direct the molecular and cellular events contributing to atrioventricular (AV) septation (Goddeeris et al., 2008; Hoffmann et al., 2009, 2014; Mommersteeg et al., 2006; Snarr et al., 2007a,b; Xie et al., 2012). Interestingly, Nkx2-5 is important in establishing a balance between sinoatrial node (SAN) and atrial myocardial identities (Espinoza-Lewis et al., 2011; Mommersteeg et al., 2007; Nakashima et al., 2014). Another homeodomain transcription factor, Shox2, is also essential for SAN development, in part through repression of Nkx2-5 (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). This Shox2-Nkx2-5 antagonistic mechanism inhibits pacemaker properties that define a ‘default’ primitive cellular phenotype (Ye et al., 2015). Furthermore, studies have shown that Shox2 directly regulates Isl1 (Hoffmann et al., 2013) and Nkx2-5 inhibits Isl1 (Dorn et al., 2014; Prall et al., 2007; Watanabe et al., 2012), a key cardiac transcription factor expressed in the pSHF (Cai et al., 2003; Snarr et al., 2007a; Sun et al., 2007; van den Berg et al., 2009). Variation in ISL1 confers genetic susceptibility for CHDs (Stevens et al., 2010), thus highlighting a crucial role in human disease. Moreover, data have illuminated a pivotal role for Isl1 in SAN development and function (Liang et al., 2015; Tessadori et al., 2012; Vedantham et al., 2015), and these findings offer explanations for CCS abnormalities in individuals harboring NKX2-5 mutations. Given embryonic stem cell work suggesting that direct Isl1 repression by Nkx2-5 is necessary to establish the ventricular lineage (Dorn et al., 2014) and our findings revealing essential roles of Nkx genes in ventricular identity maintenance (George et al., 2015; Targoff et al., 2013), we sought to uncover novel Nkx-Isl1 interactions that are required to pattern the IFT.
Our previous studies demonstrate that the Nkx2-5 homologs expressed in zebrafish CMs, nkx2.5 and nkx2.7 (Lee et al., 1996), act redundantly in cardiac morphogenesis (Targoff et al., 2008; Tu et al., 2009) and are crucial in maintaining ventricular identity by repressing atrial fate (Targoff et al., 2013). Furthermore, temporally controlled expression of nkx2.5 is essential to ensure chamber-specific identity maintenance in both the FHF and SHF during explicit developmental time windows (George et al., 2015). Despite these insights, little is known about the mechanisms by which Nkx genes control the specification and differentiation of SHF progenitors at the cardiac poles. In this study, we find that Nkx genes positively regulate aSHF progenitors at the arterial pole and restrict pSHF-derived CM differentiation at the venous pole. At the arterial pole, Nkx genes are required to promote the aSHF progenitors, within the proliferating pharyngeal region, to differentiate and assemble the OFT. In nkx2.5−/−;nkx2.7−/− embryos, unrestricted expression patterns of SAN genes, bmp4, tbx2b, hcn4 and shox2, highlight severe abnormalities in venous pole development. Furthermore, our findings reveal that Nkx genes normally restrict isl1 expression to the venous pole during heart tube formation, whereas overexpression of nkx2.5 is sufficient to inhibit isl1. Notably, electrophysiological chamber identity is dependent on Nkx gene dose with elimination of all heterogeneity of intercellular coupling in the nkx2.5−/−;nkx2.7−/− embryos. These findings support the innovative concept that CM differentiation is highly graded, with functional partitioning of the heart tube established and maintained through networks of overlapping regulators rather than through binary fate choices. This gradation is consistent with the pleiotropy of phenotypes observed in monogenic forms of CHD and the known redundancy and resilience of cardiac transcriptional networks. Taken together, our results suggest key functions of Nkx genes in SHF CM differentiation in mediating differential roles in anterior and posterior populations. Ultimately, our studies have the potential to shed light on the mechanisms underlying conotruncal and venous pole malformations, and conduction system defects in individuals carrying NKX2-5 mutations.
RESULTS
OFT size is restricted in nkx mutants
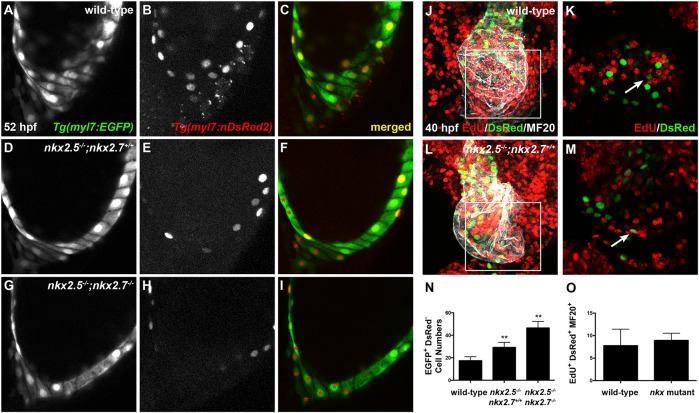
Although our previous studies revealed synergistic functions of nkx2.5 and nkx2.7 in OFT development (George et al., 2015; Targoff et al., 2013), the discrete cellular and temporal mechanisms operating downstream of Nkx genes in the aSHF remain unclear. Previous studies have identified SHF progenitors in the anterior lateral plate mesoderm (ALPM) that express nkx2.5 and contribute to the OFT (Guner-Ataman et al., 2013; Paffett-Lugassy et al., 2017). Thus, we examined the roles of nkx2.5 and nkx2.7 in regulating the spatiotemporal contribution of the aSHF to arterial pole formation. Employing a developmental timing assay to assess the differences in protein maturation time in embryos expressing Tg(myl7:EGFP) and Tg(-5.1myl7:nDsRed2) (de Pater et al., 2009; Huang et al., 2003; Mably et al., 2003) (Fig. 1A-I), we visualized the accretion of the aSHF-derived CMs in the OFT of wild-type and nkx2.5−/−; nkx2.7+/+, nkx2.5−/−; nkx2.7+/− and nkx2.5−/−; nkx2.7−/− (hereafter referred to as nkx mutant) embryos. Progressive loss of Nkx gene function highlights substantial reduction in the volume of OFT myocardium (Fig. 1D-I). Moreover, we detected a statistically significant reduction in aSHF-derived CMs, or EGFP+ DsRed− cells, when comparing wild-type (24 cells±3 cells; n=12) with nkx2.5−/−;nkx2.7−/− (16 cells±4 cells; n=3) embryos (P<0.005). Taken together, these findings underscore that nkx2.5 and nkx2.7 positively regulate the contribution of aSHF progenitors at the zebrafish arterial pole.
Fig. 1.
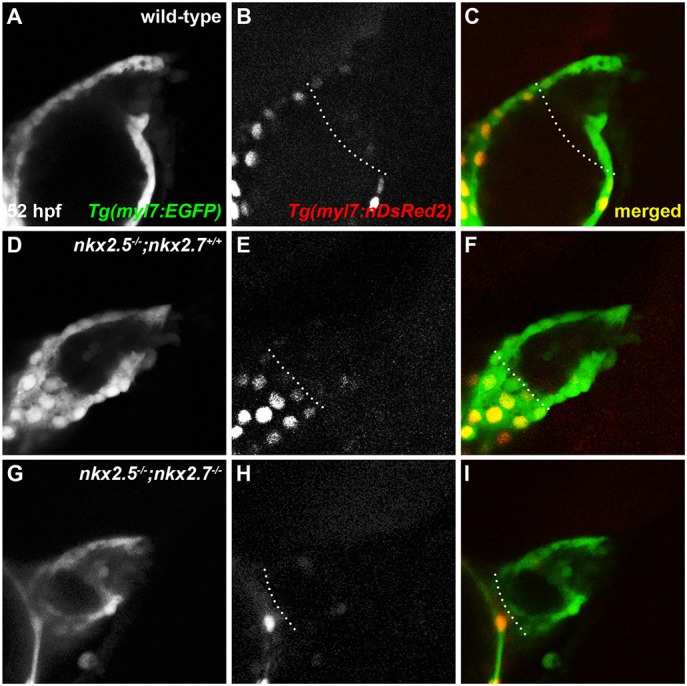
OFT size and CM numbers are reduced in nkx mutants. (A-I) Lateral view, anterior towards the right, at 52 hpf. Representative reconstructions of four z-stacks from a developmental timing assay show diminished cell numbers in the OFT of wild-type (A-C), nkx2.5−/−;nkx2.7+/+ (D-F) and nkx2.5−/−;nkx2.7−/− (G-I) embryos carrying Tg(myl7:EGFP) and Tg(-5.1myl7:nDsRed2).
Nkx genes are dispensable for OFT cardiomyocyte proliferation
Prior work has elucidated an important role for SHF CM proliferation in zebrafish arterial pole development (Guner-Ataman et al., 2013; Lazic and Scott, 2011; Nevis et al., 2013; Schindler et al., 2014; Zeng and Yelon, 2014; Zhou et al., 2011). Given the decreased number of aSHF-derived CMs in the nkx mutant OFT, we tested whether diminished proliferation is responsible for this observation. We pulsed embryos carrying Tg(-5.1myl7:nDsRed2) at 24 hpf, when aSHF-derived CMs proliferate, with the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) (Guner-Ataman et al., 2013; Schindler et al., 2014; Zeng and Yelon, 2014). At 48 hpf, we counted EdU+ DsRed+ MF20+ cells at the arterial pole to quantify proliferating nuclei in the myocardium (Fig. S1). We detected no statistically significant differences in the proliferation indices between wild-type (1.0%±1.3%; n=15) and nkx mutant (1.5%±1.2%; n=8) embryos, indicating that a reduced rate of OFT proliferation is not responsible for the decrease in CMs accrued at the arterial pole.
Nkx genes delimit the OFT progenitor pool
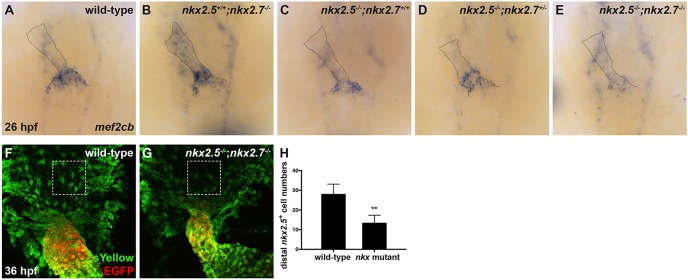
We then postulated that a shrunken arterial pole size in Nkx mutant embryos could represent an earlier influence of Nkx genes on the reservoir of undifferentiated aSHF progenitors. The SHF-derived population at the OFT can be divided into two cell types: differentiated CMs that strongly express myl7, vmhc and nkx2.5, and a progenitor pool that faintly expresses these same markers and strongly expresses mef2cb and ltbp3 (Lazic and Scott, 2011; Zeng and Yelon, 2014; Zhou et al., 2011). Indeed, our findings reveal diminished mef2cb expression in the arterial pole of wild-type compared with nkx mutant embryos (Fig. 2A-E). Specifically, mef2cb expression is slightly decreased in nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7+/− embryos (Fig. 2C,D), and further diminished in nkx2.5−/−;nkx2.7−/− embryos (Fig. 2E) when compared with wild-type and nkx2.5+/+;nkx2.7−/− embryos (Fig. 2A,B). These results corroborate our previously published studies illustrating a progressive decline in ltbp3 expression with successive reduction of Nkx gene dose (Targoff et al., 2013). Altogether, our findings suggest that Nkx gene function is required to establish the proper size of the aSHF progenitor pool. However, in assessing this population through gene expression profiles of SHF markers, we may fail to identify all cells destined to become aSHF-derived CMs.
Fig. 2.
Nkx genes delimit the OFT progenitor pool. (A-E) Dorsal view, anterior towards the top, at 26 hpf. In situ hybridization depicts diminished expression of mef2cb, which labels aSHF-derived CM progenitors, in representative nkx2.5−/−;nkx2.7+/+ (n=6) (C), nkx2.5−/−;nkx2.7+/− (n=3) (D) and nkx2.5−/−;nkx2.7−/− (n=5) (E) embryos compared with wild-type (n=15) (A) and nkx2.5+/+;nkx2.7−/− (n=5) (B) embryos. (F,G) Ventral view of the arterial pole region at 36 hpf. Confocal projections of immunohistochemistry for ZsYellow (green) and EGFP (red) in embryos carrying Tg(nkx2.5:ZsYellow) and Tg(myl7:EGFP) illustrate decreased contribution of nkx2.5+ aSHF-derived CM progenitors (ZsYellow+ EGFP−) to the arterial pole in nkx2.5−/−;nkx2.7−/− (G) compared with wild-type (F) embryos (boxed areas). (H) Quantification of ZsYellow+ EGFP− cells at the arterial pole in wild-type (n=9) and nkx mutant (n=8) embryos reveals a statistically significant decrease in this population following the loss of Nkx gene function. Student's t-test was used to determine statistical significance. Mean and s.e.m. of each data set are shown (**P<0.0001).
Given the importance of the temporal regulation of aSHF precursor accretion to the arterial pole (Cambier et al., 2014; Jahangiri et al., 2016; Zeng and Yelon, 2014), we sought to quantify aSHF progenitors by tracking these late-differentiating cells during OFT recruitment. Recent studies have confirmed the contribution of distal Tg(nkx2.5:ZsYellow)-expressing progenitor cells to the OFT myocardium using Tg(nkx2.5:kaede) (Guner-Ataman et al., 2013; Zeng and Yelon, 2014). Employing Tg(nkx2.5:ZsYellow) and Tg(myl7:EGFP), we examined aSHF progenitor cells in the pharyngeal region distal to the arterial pole where they proliferate prior to differentiation and addition to heart (Hutson et al., 2010; Tirosh-Finkel et al., 2010; van den Berg et al., 2009; Zeng and Yelon, 2014). We observed dispersion and lower levels of expression of Tg(nkx2.5:ZsYellow) in the distal nkx2.5+ OFT progenitors in nkx mutant compared with wild-type embryos at 36 hpf (Fig. 2F,G). We quantified this alteration in expression patterns by counting ZsYellow+ EGFP− cells distal to the arterial pole and between the branchial arches (Fig. 2H). Our results demonstrate a dramatic decrease in the number of distal nkx2.5+ progenitors contributing to the OFT in nkx mutant embryos. Together, these data suggest that loss of Nkx gene function limits the number of undifferentiated aSHF cells prior to their deployment to the arterial pole.
Nkx genes regulate OFT progenitor proliferation timing and differentiation
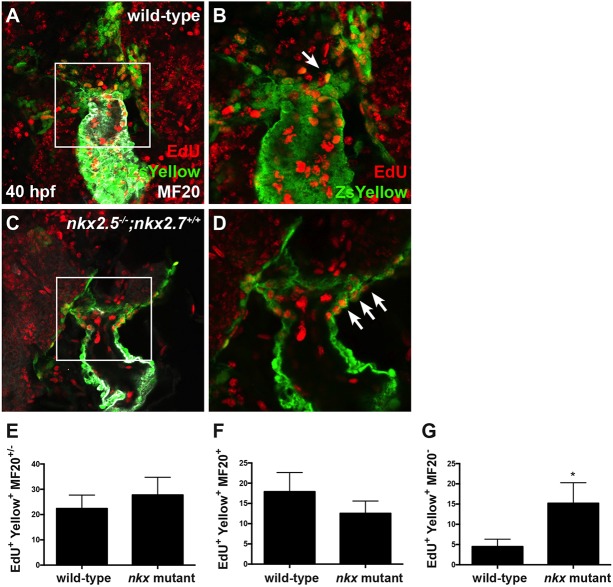
From our findings demonstrating that Nkx genes are required for OFT formation by early regulation of the aSHF progenitor pool size, as opposed to later influence on SHF-derived CM production, we postulated that the decreased number of OFT progenitors in Nkx mutants could be due to diminished progenitor proliferation. To investigate this hypothesis, we performed an EdU incorporation assay in embryos carrying Tg(nkx2.5:ZsYellow) at 18 hpf to identify dividing progenitor cells prior to their migration to the arterial pole. At 40 hpf, we performed MF20 immunostaining to delineate proliferative ZsYellow+ cells that had been incorporated into the OFT in wild-type (Fig. 3A,B) and nkx mutant (Fig. 3C,D) embryos. Surprisingly, we found no significant difference in the proliferation index of the total OFT progenitor population when comparing wild-type with nkx mutant embryos (Fig. 3E). We next tested whether there might be differences in OFT progenitor proliferation indices between wild-type and nkx mutant embryos in the proximal early-differentiating (ZsYellow+ MF20+) and distal late-differentiating (ZsYellow+ MF20−) populations. The proximal OFT progenitor population is recruited to the arterial pole while the distal OFT progenitor population remains in a proliferative state in the pharyngeal region (van den Berg et al., 2009). We found that our results from the EdU incorporation assay are comparable between wild-type and nkx mutant embryos in the proximal precursor population contributing to the arterial pole (Fig. 3F). However, there is a statistically significant increase in the proliferation index of those OFT progenitors that reside in the distal pharyngeal region when comparing wild-type and nkx mutant embryos (Fig. 3B compared with D; Fig. 3G). In summary, our data suggest that Nkx genes regulate OFT size by restricting the proliferation of aSHF progenitors in the distal pharyngeal region and promoting differentiation as they migrate to form the arterial pole.
Fig. 3.
Nkx genes regulate OFT progenitor proliferation timing and differentiation. (A-D) Ventral view of the arterial pole region at 40 hpf. Confocal projections of immunohistochemistry for EdU (red), ZsYellow (green) and MF20 (gray) in embryos carrying Tg(nkx2.5:ZsYellow) following EdU incubation at 18 hpf in wild-type (A,B) and nkx2.5−/−;nkx2.7+/+ (C,D) embryos. White arrows indicate EdU+ ZsYellow+ cells. The boxed areas in A,C are shown at higher magnification in B,D. (E-G) Proliferation indices showing a statistically significant increase in undifferentiated, ZsYellow+ MF20− progenitors in nkx mutant (n=6) compared with wild-type (n=6) embryos (G); the proliferation index is comparable in the proximal, ZsYellow+ MF20+ CM population contributing to the arterial pole (F) as well as in the total OFT, ZsYellow+ MF20+/− CM population (E). Student's t-test was used to determine statistical significance. The mean and s.e.m. of each data set are shown (*P<0.005).
isl1 functions downstream of Nkx genes at the venous pole
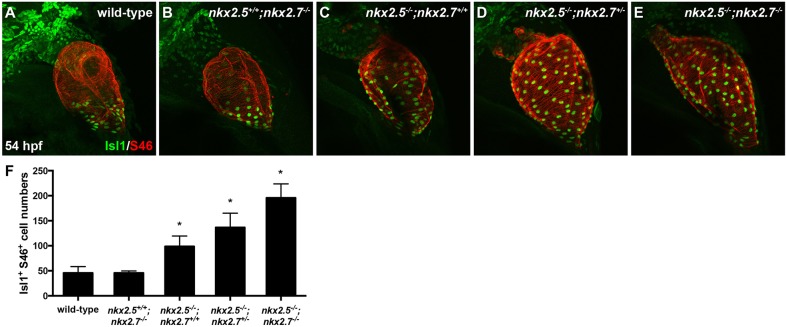
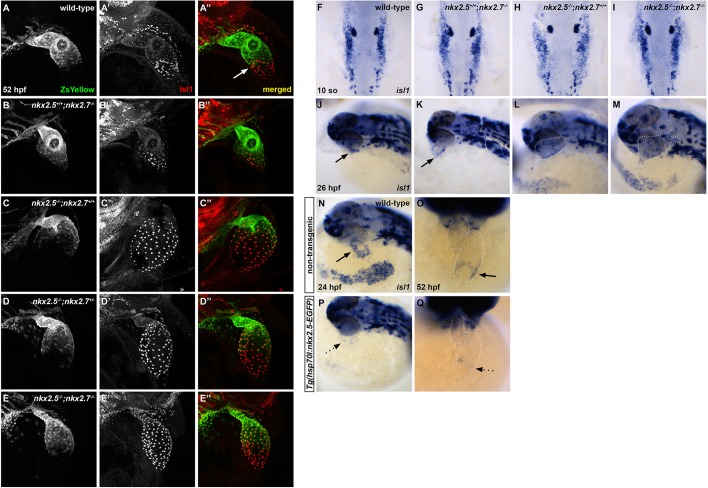
In light of our evidence suggesting that Nkx genes positively regulate the aSHF OFT progenitor pool, we examined possible roles in pSHF development given the relatively high incidence of atrial septal defects in individuals carrying NKX2-5 mutations. In the absence of clearly defined regulators of the IFT in zebrafish, we investigated Isl1, a LIM homeodomain transcription factor, as a potential candidate downstream of Nkx genes at the venous pole. Extensive studies elucidated that isl1 is essential for normal cardiac development across species (Brade et al., 2007; Cai et al., 2003; de Pater et al., 2009; Pandur et al., 2013; Witzel et al., 2012), delineates the SHF at both arterial and venous poles (Cai et al., 2003; Hami et al., 2011; Snarr et al., 2007a; Stoyek et al., 2015; Witzel et al., 2012), and is required for proper pacemaker development and function (Liang et al., 2015; Tessadori et al., 2012; Vedantham et al., 2015). In zebrafish, using an antibody that recognizes both Isl1 and Isl2 proteins (D'Amico et al., 2007; Hutchinson and Eisen, 2006; Witzel et al., 2012), Isl1/2+ cells are found near the OFT and IFT (de Pater et al., 2009; Hami et al., 2011; Witzel et al., 2012), and co-expression of Nkx2.5 in this population is evident during somitogenesis (Witzel et al., 2012). However, Isl2b appears to play a role in arterial pole development (Witzel et al., 2017) and Isl1 in venous pole development with a loss of late-differentiating CMs in the IFT in isl1−/− embryos (de Pater et al., 2009). Thus, we postulated that Nkx genes inhibit isl1 expression at the SV. In support of our model, we found that Isl1 immunostaining is localized specifically to the IFT of wild-type embryos (Fig. 4A), while Isl1+ CMs extend throughout the atrium (S46+ region) in nkx2.5−/−;nkx2.7+/+ embryos (Fig. 4C) and reach the OFT in nkx2.5−/−;nkx2.7−/− embryos (Fig. 4E). We quantified Isl1+ nuclei and detected a progressive increase in the number of cells expressing this transcription factor as Nkx gene dose is diminished (Fig. 4F). Notably, the number of Isl1+ CMs in the nkx2.5−/−;nkx2.7−/− heart is approximately equivalent to the total number of CMs in wild-type embryos (Targoff et al., 2013). Next, we investigated Nkx2.5 and Isl1 expression patterns employing immunohistochemistry in embryos carrying Tg(nkx2.5:ZsYellow) at 52 hpf. This transgene faithfully recapitulates the endogenous nkx2.5 expression observed by in situ hybridization (data not shown). We find that Nkx2.5 and Isl1 are virtually mutually exclusive at the venous pole in wild-type embryos (Fig. 5A-A″). Nkx2.5 is expressed in a decreasing gradient from the OFT to the inflow of the atrium, whereas Isl1 expression is confined to SV with only a few cells co-expressing Nkx2.5 and Isl1 (Fig. 5A″). Alternatively, in nkx mutants, Isl1 expression is derepressed in all atrial CMs that normally express Nkx2.5 (Fig. 5B-E″). Taken together, our studies strongly support the in vivo suppression of Isl1+ cell fate by Nkx genes in the SV during cardiac development.
Fig. 4.
Nkx genes inhibit Isl1 expression at the sinus venosus. (A-E) Lateral view, anterior towards the top, at 54 hpf in wild-type (A), nkx2.5+/+;nkx2.7−/− (B), nkx2.5−/−;nkx2.7+/+ (C), nkx2.5−/−;nkx2.7+/− (D) and nkx2.5−/−;nkx2.7−/− (E) embryos. Representative confocal images of immunofluorescence for Isl1 (green) and S46 (red) demonstrate that Isl1 is restricted to the venous pole of wild-type (A) and nkx2.5+/+;nkx2.7−/− (B) embryos. However, Isl1 expression expands throughout the atrium in nkx mutants (C-E). (F) Quantification of Isl1+ S46+ CMs is depicted showing a gradual increase in cell number with a progressive loss of nkx alleles. Mean and s.e.m. of each data set are shown (*P<0.005) in wild-type (n=16), nkx2.5+/+;nkx2.7−/− (n=2), nkx2.5−/−;nkx2.7+/+ (n=6), nkx2.5−/−;nkx2.7+/− (n=10) and nkx2.5−/−;nkx2.7−/− (n=4) embryos. Student's t-test was used to determine statistical significance.
Fig. 5.
Nkx2.5 is not required for isl1+ progenitor specification, but is sufficient to repress isl1 at the venous pole. (A-E″) Representative confocal images, lateral view, anterior towards the top, at 52 hpf. Immunostaining for Isl1 (red) in embryos carrying Tg(nkx2.5:ZsYellow) demonstrates that Isl1 is restricted to the venous pole and Nkx2.5 expression is absent in this region in wild-type (white arrow) (A-A″) and nkx2.5+/+;nkx2.7−/− (B-B″) embryos. However, Isl1 expression is derepressed in the atrium of nkx2.5−/−;nkx2.7+/+ (C-C″) and nkx2.5−/−;nkx2.7+/− (D-D″) embryos, and expands towards the outflow tract in nkx2.5−/−;nkx2.7−/− (E-E″) embryos. (F-I) Dorsal view, anterior towards the top, at 10 somites. In situ hybridization shows no variation in ALPM isl1 expression between wild-type (F) and nkx mutant (G-I) embryos. (J-M) Lateral view, anterior towards the left, at 26 hpf. In situ hybridization demonstrates that isl1 expression is restricted to a ring at the venous pole of the heart tube in wild-type (J) and nkx2.5+/+;nkx2.7−/− (K) embryos. However, isl1 expression expands to the atrium in nkx2.5−/−;nkx2.7+/+ embryos (L) and throughout the entire heart tube in nkx2.5−/−;nkx2.7−/− embryos (M). Black arrows indicate the ring of isl1 expression near the IFT (J,K). (N-Q) Lateral view, anterior towards the left, at 24 hpf (N,P) and ventral view, anterior towards the top, at 52 hpf (O,Q) in non-transgenic (N,O) and Tg(hsp70l:nkx2.5-EGFP) (P,Q) embryos. In situ hybridization demonstrates that isl1 expression is restricted to a ring at the venous pole (black arrows) at 24 hpf (n=9) (N) and 52 hpf (n=7) (O), and that overexpression of nkx2.5 inhibits isl1 expression specifically in this region (dotted arrows) at 24 hpf (n=9/11) (P) and 52 hpf (n=8/9) (Q).
We then explored the timing of the Nkx-Isl1 interaction in further detail. We evaluated isl1 expression during early stages of cardiac differentiation in the ALPM. Our data illustrate that specification of isl1+ progenitors occurs normally at 10 somites and 15 somites (Fig. 5F-I and data not shown), but isl1 expression is enhanced at 26 hpf in nkx mutant compared with wild-type embryos (Fig. 5J-M). These findings indicate that Nkx genes are first required to inhibit isl1 at the heart tube stage. Using a novel transgene generated in our lab for ubiquitous nkx2.5 overexpression, Tg(hsp70l:nkx2.5-EGFP) (George et al., 2015), we tested the sufficiency of nkx2.5 to inhibit isl1 expression (Fig. 5N-Q). Following heat-shock activation of this inducible transgene at 21 somites, isl1 expression was repressed specifically in the inflow region at both 24 hpf and 52 hpf (Fig. 5P,Q). Altogether, these results demonstrate that Nkx2.5 is not required for isl1+ progenitor specification, but activation of Nkx2.5 transcription prior to heart tube elongation is sufficient to repress isl1 at the venous pole during chamber emergence.
Nkx genes regulate differentiation of pSHF progenitors
Although recent work has shown the contribution of late-differentiating CMs to the venous pole in zebrafish (de Pater et al., 2009; Mosimann et al., 2015), little is known about the transcriptional regulatory networks guiding accretion and differentiation of this pSHF progenitor population. Given the ability of nkx2.5 to restrict Isl1 to the IFT (Fig. 5N-Q), we hypothesize that Nkx genes are essential in patterning the pSHF-derived CMs in this region. To test this hypothesis, we first performed a developmental timing assay to delineate the kinetics of pSHF progenitor accumulation using embryos expressing Tg(myl7:EGFP) and Tg(-5.1myl7:nDsRed2) (as in Fig. 1A-I). Intriguingly, our data reveal a statistically significant increase in pSHF-derived CMs, or EGFP+ DsRed− CMs, at the venous pole in nkx2.5−/−;nkx2.7+/+ embryos along with further enhancement in nkx2.5−/−;nkx2.7−/− embryos when compared with wild-type embryos (Fig. 6A-I,N). However, the source of the increased late-differentiating CMs at the IFT remains unknown. This finding may reflect either increased proliferation of pSHF progenitors or enhanced accrual of pSHF-derived CMs.
Fig. 6.
Nkx genes regulate cardiomyocyte accretion to the atrium. (A-I) Lateral images, anterior towards the top, at 52 hpf. Representative reconstructions of 10 z-stacks from a developmental timing assay highlight the venous pole of wild-type (A-C), nkx2.5−/−;nkx2.7+/+ (D-F) and nkx2.5−/−;nkx2.7−/− (G-I) embryos carrying Tg(myl7:EGFP) (green) and Tg(-5.1myl7:nDsRed2) (red). Asterisks indicate EGFP+ DsRed− cells. (J-M) Ventral view, anterior towards the top, at 40 hpf of wild-type (J,K) and nkx2.5−/−;nkx2.7+/+ (L,M) embryos carrying Tg(-5.1myl7:nDsRed2). Confocal projections (J,L) of immunohistochemistry for EdU (red), DsRed (green) and MF20 (gray) following EdU incubation at 18 hpf and four z-stack reconstructions (K,M) were used to count EdU+ DsRed+ MF20+ CMs in the atrium. The boxed areas in J,L are shown at higher magnification in K,M, respectively. White arrows indicate EdU+ DsRed+ CMs. (N,O) Quantification of EGFP+ DsRed− cells at the venous pole for wild-type (n=12), nkx2.5−/−;nkx2.7+/+ (n=5) and nkx2.5−/−;nkx2.7−/− (n=4) embryos, and proliferation indices in wild-type (n=5) and nkx mutant (n=3) embryos. Student's t-test was used to determine statistical significance. Mean and s.e.m. of each data set are shown (**P<0.0001). There is no statistically significant difference between proliferation indices in the DsRed+ MF20+ populations of wild-type and nkx mutant embryos.
Consistent with our data illustrating normal CM production at the arterial pole following the loss of Nkx gene function (Fig. S1), we observed no difference in proliferation indices at the venous pole following EdU incorporation at 24 hpf between nkx mutant and wild-type embryos (data not shown). Thus, we hypothesized that Nkx genes are required for pSHF progenitor proliferation prior to accretion to the IFT. We performed an EdU incorporation assay at 18 hpf in embryos carrying Tg(-5.1myl7:nDsRed2) to detect dividing CMs at the venous pole. Quantification of the proliferation indices for EdU+ DsRed+ MF20+ cells in the IFT region at this time point also revealed no statistically significant difference between wild-type and nkx mutant embryos (Fig. 6J-M,O). These data support the conclusion that the IFT expansion in nkx mutant embryos is most likely due to enhanced pSHF-derived CM differentiation.
Nkx genes pattern specialized nodal tissue in the sinus venosus via isl1
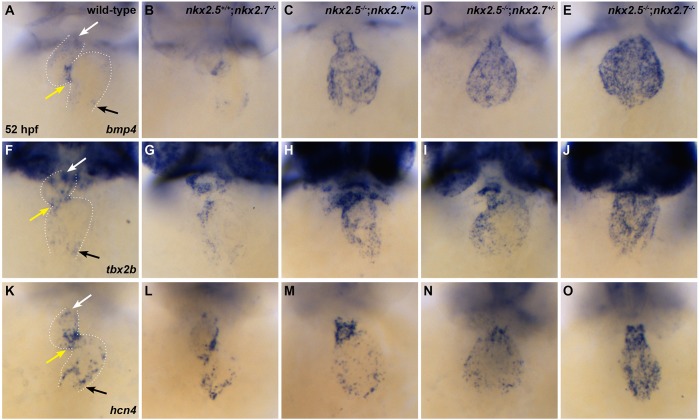
Although recent studies have elucidated the existence of specialized conduction tissue in the zebrafish heart (Arrenberg et al., 2010; Chi et al., 2008; Milan et al., 2006) and have identified isl1+ as molecular marker of this population (Tessadori et al., 2012), the pathways controlling the development of the sinoatrial node (SAN) are still under investigation. Therefore, from evidence of conduction system abnormalities in individuals with NKX2-5 mutations (Benson et al., 1999; Elliott et al., 2003; Gutierrez-Roelens et al., 2002; Schott et al., 1998) and from our findings revealing that Nkx genes regulate isl1 expression (Figs 4 and 5), we posited that Nkx genes are fundamental in patterning the pSHF-derived CMs that contribute to the specialized nodal tissue in the SV. To address this issue, we explored the expression of a number of pacemaker cell genes: bmp4, tbx2b, hcn4 and shox2 (Blaschke et al., 2007; Espinoza-Lewis et al., 2009; Tessadori et al., 2012). In wild-type embryos, bmp4, tbx2b and hcn4 expression is restricted to the OFT, atrioventricular canal (AVC) and IFT at 52 hpf (Fig. 7A,F,K). Intriguingly, all three genes are expressed throughout the myocardium in nkx mutant embryos (Fig. 7C-E,H-J,M-O). We observe a similar expression pattern for shox2 when comparing wild-type and nkx mutant embryos (Fig. S2). Taken together, our findings illustrate that Nkx genes regulate regionalization of SAN markers to the IFT.
Fig. 7.
Nkx genes regulate SAN markers. (A-O) Ventral view, anterior towards the top, at 52 hpf. Dotted white lines illustrate the cardiac silhouettes. In situ hybridization for bmp4 (A-E), tbx2b (F-J) and hcn4 (K-O). Expression of bmp4 is restricted to the OFT (white arrow), AVC (yellow arrow) and IFT (black arrow) in wild-type (n=7/7) (A) and nkx2.5+/+;nkx2.7−/− (n=4/4) (B) embryos. However, its expression is expanded throughout the cardiac chambers in nkx2.5−/−;nkx2.7+/+ (n=3/4) (C), nkx2.5−/−;nkx2.7+/− (n=3/3) (D) and nkx2.5−/−; nkx2.7−/− (n=3/3) (E) embryos. Similar findings are observed in the wild-type (n=6/7) (F), nkx2.5+/+;nkx2.7−/− (n=3/4) (G), nkx2.5−/−;nkx2.7+/+ (n=3/3) (H), nkx2.5−/−;nkx2.7+/− (n=2/2) (I) and nkx2.5−/−;nkx2.7−/− (n=2/2) (J) embryos using the tbx2b probe, and in wild-type (n=8/8) (K), nkx2.5+/+;nkx2.7−/− (n=5/6) (L), nkx2.5−/−;nkx2.7+/+ (n=9/11) (M), nkx2.5−/−;nkx2.7+/− (n=3/3) (N) and nkx2.5−/−;nkx2.7−/− (n=3/3) (O) embryos using the hcn4 probe.
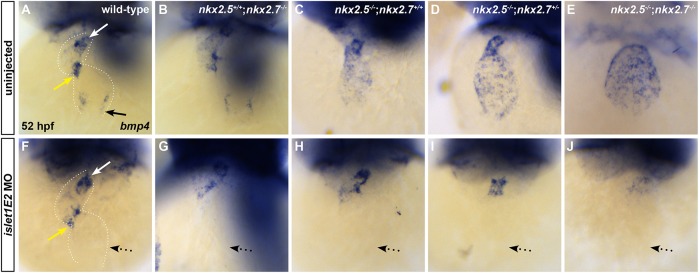
Next, we postulated that Isl1 acting downstream of Nkx genes is responsible for patterning the SAN and establishing the pacemaker activity of the zebrafish heart. Using a previously validated morpholino (MO) to knockdown Isl1 function (Hutchinson and Eisen, 2006), we observed that bmp4 expression is absent in wild-type and nkx2.5+/+;nkx2.7−/− embryos specifically at the IFT following islet1E2 MO injection, whereas normal AVC and OFT expression of bmp4 persists (compare Fig. 8A,B with F,G). These findings are consistent with evidence of selective inhibition of bmp4 expression in the IFT of the isl1sa29 mutant (de Pater et al., 2009). Interestingly, inhibition of isl1 splicing in nkx mutant embryos reveals specific downregulation of ectopic atrial and IFT bmp4 expression (compare Fig. 8H-J with C-E) in the context of otherwise normal development. Together, our results demonstrate that the misregulation of bmp4 expression in nkx mutant embryos is mediated by isl1 overexpression, ultimately placing isl1 downstream of Nkx genes in defining the pSHF-derived specialized conduction cells of the IFT.
Fig. 8.
isl1 acts downstream of Nkx genes at the venous pole. (A-J) Ventral view, anterior towards the top, at 52 hpf. Dotted white lines illustrate the cardiac silhouettes. In situ hybridization for bmp4 (A-J) in wild-type (A,F), nkx2.5+/+;nkx2.7−/− (B,G), nkx2.5−/−;nkx2.7+/+ (C,H), nkx2.5−/−;nkx2.7+/− (D,I) and nkx2.5−/−;nkx2.7−/− (E,J) embryos. Inhibition of isl1 expression using islet1E2 MO in wild-type (n=4/4) (compare A with F) and nkx2.5+/+;nkx2.7−/− (n=4/4) (compare B and G) embryos leads to downregulation of bmp4 expression specifically at the venous pole of the heart (black arrow) without affecting the AVC (yellow arrow) and OFT (white arrow) expression. In nkx2.5−/−;nkx2.7+/+ (n=2/2), nkx2.5−/−;nkx2.7+/− (n=5/6) and nkx2.5−/−;nkx2.7−/− (n=3/3) embryos, isl1 inhibition rescues the misregulation of bmp4 expression in the atrium (dotted arrows), yet AVC and OFT expression patterns remain unchanged (compare C-E with H-J).
Nkx genes regulate electrophysiological patterning
Finally, we explored the effects of the derepression of SAN genes on the electrophysiological phenotype in nkx mutant hearts. The establishment of electrical gradients enables the transition from peristaltic to sequential contraction in the developing heart and this patterning is regulated by differential contributions of FHF and SHF progenitor populations (Mosimann et al., 2015). We hypothesized that the loss of Nkx gene function could lead to a disruption in regional patterns of intercellular coupling given both the absence of ventricular identity (Targoff et al., 2013, 2008) and the expanded SAN gene expression domain in nkx mutant compared with wild-type embryos (Fig. 7).
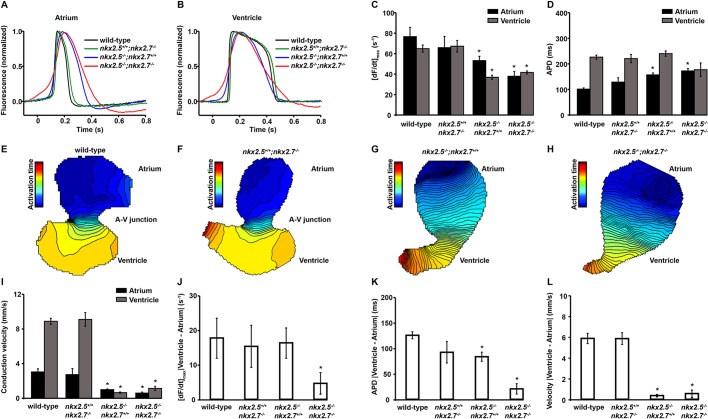
To explore these issues, we measured action potential characteristics and conduction properties in isolated wild-type and nkx mutant hearts using high-resolution optical mapping (Fig. 9, Fig. S3). We did not detect any electrophysiological phenotypes in embryos carrying single-allele deficiencies in one or both nkx genes (nkx2.5+/−;nkx2.7+/+, nkx2.5+/+;nkx2.7+/− and nkx2.5+/−;nkx2.7+/−). When comparing wild-type and nkx2.5+/+;nkx2.7−/− embryos, there is no significant difference in the detailed action potential morphology (Fig. 9A,B), maximum upstroke velocity, [dF/dt]max (Fig. 9C), or atrial and ventricular action potential durations (Fig. 9D). Furthermore, there are no observed differences between the atrial and ventricular conduction velocities in wild-type and nkx2.5+/+;nkx2.7−/− embryos (Fig. 9E,F,I). In contrast, disruption of nkx2.5 gene function affects multiple components of the action potential with significant slowing of the upstroke phase in both chambers (Fig. 9A-C), marked diminution of the maximum slope of the action potential upstroke (Fig. 9C), as well as a substantial prolongation of atrial action potential duration and accentuation of phase 4 depolarization in atrial CMs (Fig. 9A,D). Moreover, homozygous loss of nkx2.5 function leads to severe slowing in action potential propagation, as demonstrated by closer spacing of the isochrones and significantly lower mean conduction velocities measured across both cardiac chambers (Fig. 9G-I). Interestingly, although there is failure of the normal specification of physiological AV delay in the hearts of nkx2.5−/−;nkx2.7+/+ embryos (Fig. 9K,L), there remain significant differences in action potential characteristics between the ventricle and atrium (Fig. 9J,K and Fig. S3A-C,E-G). Although the hearts from nkx2.5+/+;nkx2.7−/− embryos did not exhibit any detectable electrophysiological phenotype (Fig. 9F), combined loss of function of both nkx homologs in nkx2.5−/−;nkx2.7−/− embryos leads to complete loss of electrophysiological chamber identity and further slowing of the action potential upstroke phase in the atrium (Fig. 9A,C,J and Fig. S3D). The loss of action potential duration differences between chambers is also evident in nkx2.5−/−;nkx2.7−/− embryos (Fig. 9D,K and Fig. S3H), including the emergence of a pacemaker phenotype in ventricular cells (Fig. 9B) and slowing of the conduction velocity (Fig. 9H,I,L). Interestingly, there are subtle, but significant, paradoxical effects on absolute atrial and ventricular conduction velocities in nkx2.5−/−;nkx2.7−/− when compared with nkx2.5−/−;nkx2.7+/+ embryos. In the atrium, the nkx2.5−/−;nkx2.7−/− embryos exhibit even slower conduction than the nkx2.5−/−;nkx2.7+/+ embryos. However, in the ventricle, there is an increase in the conduction velocity in the nkx2.5−/−;nkx2.7−/− embryos, although conduction remains markedly slowed and chamber identity is lost from an electrophysiological standpoint. Although these electrophysiological attributes of the nkx mutant embryos reflect severe defects in intercellular coupling, the individual CM properties exhibit a phenotype that is more advanced developmentally than that observed in the primitive heart tube. Altogether, these data highlight the independent regulation of discrete CM parameters throughout early cardiac development and chamber identity maintenance.
Fig. 9.
Nkx genes regulate electrophysiological patterning. (A,B) Representative atrial (A) and ventricular (B) action potentials showing the time courses of cellular depolarization and repolarization in wild-type, nkx2.5+/+;nkx2.7−/−, nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7−/− hearts. (C) Mean maximum slope of the atrial and ventricular action potentials measured as the maximum of the time derivative of the action potential upstroke phase, [dF/dt]max, in wild-type, nkx2.5+/+;nkx2.7−/−, nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7−/− hearts. (D) Action potential duration (APD) in wild-type, nkx2.5+/+;nkx2.7−/−, nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7−/− hearts. (E-H) Representative isochronal maps illustrating the positions of the depolarizing wave front in 5 ms time intervals for wild-type (E), nkx2.5+/+;nkx2.7−/− (F), nkx2.5−/−;nkx2.7+/+ (G) and nkx2.5−/−;nkx2.7−/− (H) hearts. (I) Mean atrial and ventricular action potential propagation velocities in wild-type, nkx2.5+/+;nkx2.7−/−, nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7−/− hearts. (J-L) Absolute differences in the maximum slope of the upstroke phase, [dF/dt]max (J), APD (K) and the conduction velocity (L) between the ventricle and the atrium in wild-type, nkx2.5+/+;nkx2.7−/−, nkx2.5−/−;nkx2.7+/+ and nkx2.5−/−;nkx2.7−/− hearts. Student's t-test was used to determine statistical significance. The mean and s.e.m. of each data set are shown (*P<0.05) for wild-type (n=5), nkx2.5+/+;nkx2.7−/− (n=5), nkx2.5−/−;nkx2.7+/+ (n=6) and nkx2.5−/−;nkx2.7−/− (n=3) hearts.
DISCUSSION
Together, these findings offer new insights into the roles of Nkx genes in SHF development, highlighting the differential regulation at the arterial and venous poles. We show that Nkx genes promote the establishment of the SHF CM progenitor pool and differentiation of this population at the OFT. Our studies also reveal that Nkx genes are essential for the proper patterning of the venous pole by regulating the maintenance of working myocardial properties through repression of isl1. Following the loss of Nkx gene function, the entire myocardium adopts an Isl1+ identity associated with activation of the SAN genes bmp4, tbx2b, hcn4 and shox2. Misregulation of these SAN genes is associated with electrophysiological evidence of expansion of the pacemaker phenotype across both chambers, with cellular characteristics suggesting diminished sodium channel availability at the membranes, features of calcium-dependent action potentials, and diastolic phase 4 potentials. Together, our findings implicate Nkx genes in combinatorial roles for anterior-posterior patterning of chamber identity, regional integration of FHF and SHF territories, and maturation of discrete cassettes of physiological CM differentiation within the heart tube. Moreover, these cellular abnormalities induced by the loss of Nkx gene function in the two-chambered zebrafish heart offer a potential window into the morphological and conduction system defects in individuals with CHD that harbor NKX2-5 mutations.
Through illustration of the differential functions of Nkx factors in the aSHF and pSHF, our studies complement and extend work in other model organisms. Previous studies have revealed a role for Nkx2-5 in limiting cardiac progenitor specification and favoring SHF proliferation (Prall et al., 2007). Yet in zebrafish, early specification of SHF progenitors appears normal given the expression patterns of nkx2.5, nkx2.7, hand2 (Targoff et al. 2013) and isl1 (Fig. 5F-I) in nkx mutant embryos. However, nkx2.5 has been shown to regulate SHF progenitor proliferation at the proximal OFT via ltbp3 (Guner-Ataman et al., 2013). Through detailed dissection of the proliferative potential of the proximal and distal OFT progenitors, our results further elucidate the role that Nkx genes play in limiting the aSHF-derived CM population by regulating the timing of progenitor differentiation at the arterial pole. In view of studies illuminating a common progenitor of heart and head muscle derivatives in other models (Harel et al., 2012; Lescroart et al., 2015, 2010; Nagelberg et al., 2015; Wang et al., 2013), we postulate that aSHF progenitors could also adopt a head muscle fate in the absence of Nkx gene function. Future experiments would be required to test this hypothesis, which may offer additional insights into the diminished late-differentiating cells at the OFT of nkx mutant embryos. Taken together, although other mechanisms may play a role, our findings highlight a novel function of Nkx genes in establishing an aSHF progenitor pool necessary to assemble an arterial pole of the proper size and morphology.
Our data also illustrates that, in the absence of nkx2.5, the atrial myocardium adopts a venous pole identity characterized by misexpression of isl1 and SAN genes associated with failure of specification of the cardiac conduction system (CCS) tissue at the AV boundary. Further graded expansion of this abnormal CM differentiation, including manifestation of increased cellular automaticity and pacemaker-like behaviors throughout the entire ventricle, is observed following the combinatorial loss of nkx2.5 and nkx2.7. Knockdown of isl1 is sufficient to rescue the effects of nkx mutations on SAN gene expression, suggesting that Isl1 repression by Nkx genes patterns the venous pole of the heart and delineates the specialized CCS tissue. However, the molecular mechanisms by which Nkx genes control IFT development through refinement of isl1 expression have yet to be determined. Specifically, the role of bmp4 in establishing electrophysiological CM characteristics requires further investigation. Yet our results offer a potential explanation for the electrophysiological malfunction associated with the loss of fast-conducting properties and defects in specification of the ventricular conduction system lineage in Nkx2-5-deficient models (Moskowitz et al., 2007; Pashmforoush et al., 2004). Furthermore, we propose that the pivotal role for nkx genes in establishing a venous pole phenotype and SAN identity could explain progressive AV conduction defects into adulthood that are exhibited in individuals with NKX2-5 mutations (Jay et al., 2003).
We have previously shown that ventricular CMs transdifferentiate into atrial CMs, demonstrating plasticity of the differentiated myocardium in nkx mutant embryos (Targoff et al., 2013). It is possible that flexibility of the differentiated CMs results in sarcomeric rearrangement that allows for fate transformation in the nkx2.5−/−;nkx2.7−/− embryos. In support of this concept, a recent report demonstrates that ablation of ventricular CMs in the zebrafish embryos leads to atrial-to-ventricular transdifferentiation with associated sarcomeric restructuring (Zhang et al., 2013). Our electrophysiological studies further substantiate this hypothesis through their demonstration that graded loss of nkx alleles regulates multiple elements of the functional identity of the atrial and ventricular myocardium. Although atrial and ventricular action potential features and conduction properties are distinct in wild-type, nkx2.5+/+;nkx2.7−/− and nkx2.5−/−;nkx2.7+/+ hearts, each of the individual null alleles exhibits discrete effects on resting membrane potentials, CM activation, action potential morphology and conduction velocity. Moreover, the electrophysiological differences between atrial and ventricular CMs are completely abolished in nkx2.5−/−;nkx2.7−/− embryos. Despite these findings, there are discordances in the effects of individual genes on electrophysiology in the atrium and ventricle that implicate additional regulatory partners in the specification of the complex regional electrical function necessary for a vertebrate heart.
Taken together, these studies on the functions of Nkx genes in the aSHF and pSHF highlight important concepts in our understanding of CM differentiation. Changes in the expression of individual transcription factors, signaling molecules and ion channels reveal discrete physiological modules and can remodel nuanced functional characteristics in CMs. Given this malleability, the notion of a stable differentiated state does not fully reflect the resolution of modern cellular biology, which has revealed phenotypes that are a result of combinations of discrete and overlapping protein repertoires. These data parallel in vitro studies in iPS-derived differentiated cell types. Despite a physiological resemblance to mature CMs, these populations demonstrate remarkable cellular heterogeneity in detailed electrophysiological phenotypes and transcriptional profiles that more closely resemble one of the parent fibroblast than of the CM being studied. Our data suggest that individual proteins may be sufficient and necessary to induce and maintain plastic states where the regenerative potential in the heart might be realized. Thus, it would be valuable to address the fundamental nature of cardiomyocyte differentiation at the level of individual physiologic functions in order to obtain a better understanding of the different potential for cardiac regeneration between zebrafish and higher vertebrates.
MATERIALS AND METHODS
Zebrafish mutations and transgenes
We used zebrafish carrying the following previously described mutations and transgenes: nkx2.5vu179 (Targoff et al., 2013), nkx2.7vu413 (Targoff et al., 2013), Tg(-5.1myl7:nDsRed2)f2 (Mably et al., 2003), Tg(myl7:EGFP)twu277 (Huang et al., 2003), Tg(nkx2.5:ZsYellow)fb7 (Zhou et al., 2011) and Tg(hsp70l:nkx2.5-EGFP) (Targoff et al., 2013). All zebrafish work followed Institutional Animal Care and Use Committee-approved protocols.
Injection
Embryos were injected at the one-cell stage with 6 ng of the validated morpholino (MO) islet1E2 (5′-TTAATCTGCGTTACCTGATGTAGTC-3′) (Hutchinson and Eisen, 2006). Uninjected sibling embryos were processed as controls.
Heat shock conditions
Embryos from outcrosses of fish carrying Tg(hsp70l:nkx2.5-EGFP) were maintained at 28.5°C and exposed to heat shock at 37°C at desired stages (21 somites to 24 somites). To implement heat shock, 50 embryos were placed in 2.5 ml of embryo medium in a 35 mm Petri dish on top of a covered heat block for 1 h. Three hours after the initiation of heat shock, transgenic embryos were identified by visualization of ubiquitous EGFP expression. Non-transgenic sibling embryos exposed to heat shock served as controls.
Depigmentation of embryos
When necessary, embryos were depigmented after overnight fixation in 4% paraformaldehyde at 4°C. They were washed twice in PBT (PBS-0.1%Tween) then incubated in 3% H2O2, 0.5% KOH and 0.1% Tween for 8 min at 26 h post fertilization (hpf) and 18 min at 52 hpf. Subsequently, the embryos were washed twice again in PBT and stored in methanol at −20°C.
In situ hybridization
We performed whole-mount in situ hybridization as previously described (Yelon et al., 1999) for mef2cb (ZDB-GENE-040901-7), isl1 (ZDB-GENE-980526-112), bmp4 (ZDB-GENE-980528-2059), tbx2b (ZDB-GENE-990726-27), hcn4 (ZDB-GENE-050420-360) and shox2 (ZDB-GENE-040426-1457).
Immunofluorescence
Whole-mount double immunofluorescence was conducted using a protocol described previously (Alexander et al., 1998) for the following primary antibodies: anti-Islet1/2 (1:50; 39.4D5, Developmental Studies Hybridoma Bank), anti-atrial myosin heavy chain (1:20; S46, Developmental Studies Hybridoma Bank), anti-RCFP (1:50; Clontech, 632475) and anti-GFP (1:100; Life Technologies, A11122). When employing the following primary antibodies, anti-Eln2 (1:500; Miao et al., 2007) and anti-sarcomeric myosin heavy chain (1:20; MF20, Developmental Studies Hybridoma Bank), another published protocol was used (Zhou et al., 2011). The following secondary antibodies were employed: goat anti-mouse IgG2b Alexa Fluor 488 or 568, goat anti-mouse IgG1 Alexa Fluor 488 or 568, goat anti-mouse IgG2a Alexa Fluor 488, goat anti-rabbit IgG Alexa Fluor 488 or 568, and goat anti-mouse IgG Cy5 (all at 1:200; Invitrogen).
Proliferation assay
Cardiomyocyte proliferation in embryos expressing Tg(-5.1myl7:nDsRed2)f2 or Tg(nkx2.5:ZsYellow)fb7 was assessed using the Click-iT EdU Imaging Kit (Life Technologies) with reported protocols (Cheesman et al., 2011; Mahler et al., 2010; Schindler et al., 2014). Following EdU incubation for 30 min on ice, embryos were chased for 24 h. Embryos were then fixed in 2% formaldehyde, washed once in PBS and washed twice in PBS-0.2% saponin. Subsequently, immunostaining was performed as described above followed by the Click-iT staining reaction. EdU+ ZsYellow+ MF20+/− and EdU+ DsRed+ MF20− cells were counted using ImageJ software, going through all z-stacks. We calculated the proliferation indices as the percentage of cells that incorporated EdU. Student's t-test (homoscedastic, two-tailed distribution) determined statistical significance between the means of cell number data sets.
Imaging
Images were captured with a Zeiss M2Bio microscope and a Zeiss AxioCam digital camera, prior to processing with Zeiss AxioVision and Adobe Creative Suite software. Confocal imaging was performed with a Nikon A1R MP confocal microscope and z-stacks were analyzed with ImageJ.
Developmental timing assay and cell counting
For the developmental timing assay, embryos carrying Tg(-5.1myl7:nDsRed2)f2 and Tg(myl7:EGFP)twu277 were fixed at 52 hpf for 1 h in 1% formaldehyde, washed once in PBS, washed three times in PBS-0.2% saponin, deyolked and flat-mounted. Newly differentiated CMs were identified as green, but not red cells (EGFP+ DsRed− CMs) and were counted on z-stacks using ImageJ software. To count Isl1+ cells, we performed immunostaining for Isl1 and S46 (as described above) and quantified individual CMs that were positive for Isl1 using ImageJ software on reconstructed z-stacks. Student's t-test (homoscedastic, two-tailed distribution) determined statistical significance between the means of cell number data sets.
Genotyping
PCR genotyping was performed on genomic DNA extracted from individual embryos following in situ hybridization, immunofluorescence or live imaging. Detection of nkx2.5vu179 was performed using primers 5′-CAAACTCACCTCCACACAGG-3′ and 5′-TTACCATCCCGAACCAAAAC-3′ to generate a 144 bp fragment. Digestion of the mutant PCR product with Hinf1 generates 30 bp and 114 bp fragments. Analysis of nkx2.5vu179 in embryos carrying Tg(hsp70l:nkx2.5-EGFP) was performed as previously described (George et al., 2015). Detection of nkx2.7vu413 was executed using primers 5′-TTGTTGCAAATGGACACGTT-3′ and 5′-GGAGTGTCTCGCCGTTTTTA-3′ to generate a 158 bp fragment. Digestion of the mutant PCR product with MseI creates 28 bp and 130 bp fragments.
Optical mapping
Embryonic zebrafish hearts were isolated at 72 hpf and stained with the transmembrane potential-sensitive dye, di-8-ANEPPS (Invitrogen), to measure action potentials using high-speed optical mapping as previously described (Mosimann et al., 2015; Panáková et al., 2010). Fluorescence intensities were recorded with a high-speed CCD camera (CardioCCD-SMQ, Redshirt Imaging) at 2000 frames per second. Activation times were determined as the times at which action potentials reached 50% of their amplitude during the depolarization phase at every location. This criterion for defining electrical activation was found in computer simulations to correlate with the time at which the maximum depolarizing Na+ current occurs during propagation (Fast and Kleber, 1995). Action potentials were temporally (800 Hz low-pass cutoff) and spatially (3×3 pixel average) filtered to enhance signal-to-noise ratios. To quantify the slope of the action potential upstroke phase, the maximum time derivative of the normalized action potential, [dF/dt]max, was calculated. Action potential duration (APD) was defined as the absolute time difference between the decay phase and the upstroke phase of the local action potential at 50% depolarization. By fitting a second-order polynomial hypersurface to the three-dimensional image plane-activation time relationship, conduction velocity vectors were subsequently derived using an established algorithm (Bayly et al., 1998) with custom scripts. Conduction velocity vector fields were estimated from the local action potentials (Panáková et al., 2010). All electrophysiological measurements were averaged across the relevant anatomic segments for each heart.
Supplementary Material
Acknowledgements
We are grateful to Joshua Barber for expert zebrafish care and members of the Targoff Laboratory for their constructive input. We appreciate the critical reading of the manuscript by Lilianna Solnica-Krezel and Joshua S. Waxman. We also thank C. Geoffrey Burns and Caroline E. Burns for sharing the Tg(nkx2.5:ZsYellow) line.
Footnotes
Competing interests
S.C. is currently a consultant for Pairnomix, but the work presented in this paper has no connection with this company.
Author contributions
Conceptualization: S.C., K.L.T.; Methodology: S.C., C.A.M., K.L.T.; Validation: S.C., C.d.S.-T., A.A.W., C.A.M., K.L.T.; Formal analysis: S.C., C.d.S.-T., V.G., A.A.W., S.K., C.A.M., K.L.T.; Investigation: S.C., C.d.S.-T., V.G., A.A.W., S.K., C.A.M., K.L.T.; Resources: S.C., C.A.M., K.L.T.; Data curation: S.C., C.d.S.-T., V.G., A.A.W., S.K.; Writing - original draft: S.C., A.A.W., C.A.M., K.L.T.; Writing - review & editing: C.A.M., K.L.T.; Visualization: K.L.T.; Supervision: C.A.M., K.L.T.; Project administration: K.L.T.; Funding acquisition: C.A.M., K.L.T.
Funding
This work was supported by grants from the National Institutes of Health (K12HD043389, K08HL088002 and R01HL131438 to K.L.T., and R24OD017870 to C.A.M.] and the Burroughs Wellcome Fund [IRSA to C.A.M.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.161497.supplemental
References
- Abou Hassan O. K., Fahed A. C., Batrawi M., Arabi M., Refaat M. M., DePalma S. R., Seidman J. G., Seidman C. E., Bitar F. F. and Nemer G. M. (2015). NKX2-5 mutations in an inbred consanguineous population: genetic and phenotypic diversity. Sci. Rep. 5, 8848 10.1038/srep08848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J., Stainier D. Y. R. and Yelon D. (1998). Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev. Genet. 22, 288-299. [DOI] [PubMed] [Google Scholar]
- Arrenberg A. B., Stainier D. Y. R., Baier H. and Huisken J. (2010). Optogenetic control of cardiac function. Science 330, 971-974. 10.1126/science.1195929 [DOI] [PubMed] [Google Scholar]
- Azpiazu N. and Frasch M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340. 10.1101/gad.7.7b.1325 [DOI] [PubMed] [Google Scholar]
- Barth J. L., Clark C. D., Fresco V. M., Knoll E. P., Lee B., Argraves W. S. and Lee K.-H. (2010). Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Dev. Dyn. 239, 2024-2033. 10.1002/dvdy.22341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly P. V., KenKnight B. H., Rogers J. M., Hillsley R. E., Ideker R. E. and Smith W. M. (1998). Estimation of conduction velocity vector fields from epicardial mapping data. IEEE Trans. Biomed. Eng. 45, 563-571. 10.1109/10.668746 [DOI] [PubMed] [Google Scholar]
- Benson D. W., Silberbach G. M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S., Smalls O., Johnson M. C., Watson M. S., Seidman J. G. et al. (1999). Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Invest. 104, 1567-1573. 10.1172/JCI8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke R. J., Hahurij N. D., Kuijper S., Just S., Wisse L. J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S. E. et al. (2007). Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830-1838. 10.1161/CIRCULATIONAHA.106.637819 [DOI] [PubMed] [Google Scholar]
- Bodmer R. (1993). The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719-729. [DOI] [PubMed] [Google Scholar]
- Bouveret R., Waardenberg A. J., Schonrock N., Ramialison M., Doan T., de Jong D., Bondue A., Kaur G., Mohamed S., Fonoudi H. et al. (2015). NKX2-5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. Elife 4, e06942 10.7554/eLife.06942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T., Gessert S., Kühl M. and Pandur P. (2007). The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev. Biol. 311, 297-310. 10.1016/j.ydbio.2007.08.004 [DOI] [PubMed] [Google Scholar]
- Briggs L. E., Takeda M., Cuadra A. E., Wakimoto H., Marks M. H., Walker A. J., Seki T., Oh S. P., Lu J. T., Sumners C. et al. (2008). Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ. Res. 103, 580-590. 10.1161/CIRCRESAHA.108.171835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau B. G. (2008). The developmental genetics of congenital heart disease. Nature 451, 943-948. 10.1038/nature06801 [DOI] [PubMed] [Google Scholar]
- Buckingham M., Meilhac S. and Zaffran S. (2005). Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826-837. 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- Cai C.-L., Liang X., Shi Y., Chu P.-H., Pfaff S. L., Chen J. and Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877-889. 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier L., Plate M., Sucov H. M. and Pashmforoush M. (2014). Nkx2-5 regulates cardiac growth through modulation of Wnt signaling by R-spondin3. Development 141, 2959-2971. 10.1242/dev.103416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman S. E., Neal J. T., Mittge E., Seredick B. M. and Guillemin K. (2011). Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci. USA 108 Suppl. 1, 4570-4577. 10.1073/pnas.1000072107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N. C., Shaw R. M., Jungblut B., Huisken J., Ferrer T., Arnaout R., Scott I., Beis D., Xiao T., Baier H. et al. (2008). Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6, e109 10.1371/journal.pbio.0060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I.-M. and Rajakumar G. (2016). Genetics of congenital heart defects: the NKX2-5 gene, a key player. Genes 7, 6 10.3390/genes7020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C. D., Zhang B., Lee B., Evans S. I., Lassar A. B. and Lee K.-H. (2013). Evolutionary conservation of Nkx2.5 autoregulation in the second heart field. Dev. Biol. 374, 198-209. 10.1016/j.ydbio.2012.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico L., Scott I. C., Jungblut B. and Stainier D. Y. R. (2007). A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr. Biol. 17, 252-259. 10.1016/j.cub.2006.12.023 [DOI] [PubMed] [Google Scholar]
- de Pater E., Clijsters L., Marques S. R., Lin Y.-F., Garavito-Aguilar Z. V., Yelon D. and Bakkers J. (2009). Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136, 1633-1641. 10.1242/dev.030924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J. N., Meilhac S. M., Bland Y. S., Buckingham M. E. and Brown N. A. (2012). Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ. Res. 111, 1323-1335. 10.1161/CIRCRESAHA.112.271247 [DOI] [PubMed] [Google Scholar]
- Dorn T., Goedel A., Lam J. T., Haas J., Tian Q., Herrmann F., Bundschu K., Dobreva G., Schiemann M., Dirschinger R. et al. (2014). Direct Nkx2-5 transcriptional repression of Isl1 controls cardiomyocyte subtype identity. Stem Cells 33, 1113-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer L. A. and Kirby M. L. (2009). The role of secondary heart field in cardiac development. Dev. Biol. 336, 137-144. 10.1016/j.ydbio.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D. A., Kirk E. P., Yeoh T., Chandar S., McKenzie F., Taylor P., Grossfeld P., Fatkin D., Jones O., Hayes P. et al. (2003). Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J. Am. Coll. Cardiol. 41, 2072-2076. 10.1016/S0735-1097(03)00420-0 [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J. F., Wang D., Yang J. et al. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376-385. 10.1016/j.ydbio.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R. A., Liu H., Sun C., Chen C., Jiao K. and Chen Y. P. (2011). Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 356, 359-369. 10.1016/j.ydbio.2011.05.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast V. G. and Kleber A. G. (1995). Cardiac tissue geometry as a determinant of unidirectional conduction block: assessment of microscopic excitation spread by optical mapping in patterned cell cultures and in a computer model. Cardiovasc. Res. 29, 697-707. 10.1016/S0008-6363(96)88643-3 [DOI] [PubMed] [Google Scholar]
- Galli D., Dominguez J. N., Zaffran S., Munk A., Brown N. A. and Buckingham M. E. (2008). Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 135, 1157-1167. 10.1242/dev.014563 [DOI] [PubMed] [Google Scholar]
- Gelb B. D. and Chung W. K. (2014). Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb. Perspect. Med. 4, a013953 10.1101/cshperspect.a013953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George V., Colombo S. and Targoff K. L. (2015). An early requirement for nkx2.5 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol. 400, 10-22. 10.1016/j.ydbio.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddeeris M. M., Rho S., Petiet A., Davenport C. L., Johnson G. A., Meyers E. N. and Klingensmith J. (2008). Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development 135, 1887-1895. 10.1242/dev.016147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmuntz E., Geiger E. and Benson D. W. (2001). NKX2.5 mutations in patients with tetralogy of fallot. Circulation 104, 2565-2568. 10.1161/hc4601.098427 [DOI] [PubMed] [Google Scholar]
- Grow M. W. and Krieg P. A. (1998). Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev. Biol. 204, 187-196. 10.1006/dbio.1998.9080 [DOI] [PubMed] [Google Scholar]
- Guner-Ataman B., Paffett-Lugassy N., Adams M. S., Nevis K. R., Jahangiri L., Obregon P., Kikuchi K., Poss K. D., Burns C. E. and Burns C. G. (2013). Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 140, 1353-1363. 10.1242/dev.088351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Roelens I., Sluysmans T., Gewillig M., Devriendt K. and Vikkula M. (2002). Progressive AV-block and anomalous venous return among cardiac anomalies associated with two novel missense mutations in the CSX/NKX2-5 gene. Hum. Mutat. 20, 75-76. 10.1002/humu.9041 [DOI] [PubMed] [Google Scholar]
- Hami D., Grimes A. C., Tsai H.-J. and Kirby M. L. (2011). Zebrafish cardiac development requires a conserved secondary heart field. Development 138, 2389-2398. 10.1242/dev.061473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I., Maezawa Y., Avraham R., Rinon A., Ma H.-Y., Cross J. W., Leviatan N., Hegesh J., Roy A., Jacob-Hirsch J. et al. (2012). Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc. Natl. Acad. Sci. USA 109, 18839-18844. 10.1073/pnas.1208690109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J. I. E. and Kaplan S. (2002). The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890-1900. 10.1016/S0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- Hoffman J. I. E., Kaplan S. and Liberthson R. R. (2004). Prevalence of congenital heart disease. Am. Heart J. 147, 425-439. 10.1016/j.ahj.2003.05.003 [DOI] [PubMed] [Google Scholar]
- Hoffmann A. D., Peterson M. A., Friedland-Little J. M., Anderson S. A. and Moskowitz I. P. (2009). sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 136, 1761-1770. 10.1242/dev.034157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S., Berger I. M., Glaser A., Bacon C., Li L., Gretz N., Steinbeisser H., Rottbauer W., Just S. and Rappold G. (2013). Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res. Cardiol. 108, 339 10.1007/s00395-013-0339-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. D., Yang X. H., Burnicka-Turek O., Bosman J. D., Ren X., Steimle J. D., Vokes S. A., McMahon A. P., Kalinichenko V. V. and Moskowitz I. P. (2014). Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 10, e1004604 10.1371/journal.pgen.1004604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-J., Tu C.-T., Hsiao C.-D., Hsieh F.-J. and Tsai H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Hutchinson S. A. and Eisen J. S. (2006). Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development 133, 2137-2147. 10.1242/dev.02355 [DOI] [PubMed] [Google Scholar]
- Hutson M. R., Zeng X. L., Kim A. J., Antoon E., Harward S. and Kirby M. L. (2010). Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development 137, 3001-3011. 10.1242/dev.051565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Hiroi Y., Hosoda T., Utsunomiya T., Matsuo S., Ito T., Inoue J., Sumiyoshi T., Takano H., Nagai R. et al. (2002). Novel point mutation in the cardiac transcription factor CSX/NKX2.5 associated with congenital heart disease. Circ. J. 66, 561-563. 10.1253/circj.66.561 [DOI] [PubMed] [Google Scholar]
- Jahangiri L., Sharpe M., Novikov N., Gonzalez-Rosa J. M., Borikova A., Nevis K., Paffett-Lugassy N., Zhao L., Adams M., Guner-Ataman B. et al. (2016). The AP-1 transcription factor component Fosl2 potentiates the rate of myocardial differentiation from the zebrafish second heart field. Development 143, 113-122. 10.1242/dev.126136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay P. Y., Berul C. I., Tanaka M., Ishii M., Kurachi Y. and Izumo S. (2003). Cardiac conduction and arrhythmia: insights from Nkx2.5 mutations in mouse and humans. Novartis Found. Symp. 250, 227-238; discussion 238-241, 276-229 10.1002/0470868066.ch14 [DOI] [PubMed] [Google Scholar]
- Jay P. Y., Harris B. S., Maguire C. T., Buerger A., Wakimoto H., Tanaka M., Kupershmidt S., Roden D. M., Schultheiss T. M., O'Brien T. X. et al. (2004). Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Invest. 113, 1130-1137. 10.1172/JCI19846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic S. and Scott I. C. (2011). Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 354, 123-133. 10.1016/j.ydbio.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Lee K.-H., Xu Q. and Breitbart R. E. (1996). A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev. Biol. 180, 722-731. 10.1006/dbio.1996.0341 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Kelly R. G., Le Garrec J.-F., Nicolas J.-F., Meilhac S. M. and Buckingham M. (2010). Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development 137, 3269-3279. 10.1242/dev.050674 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Mohun T., Meilhac S. M., Bennett M. and Buckingham M. (2012). Lineage tree for the venous pole of the heart: clonal analysis clarifies controversial genealogy based on genetic tracing. Circ. Res. 111, 1313-1322. 10.1161/CIRCRESAHA.112.271064 [DOI] [PubMed] [Google Scholar]
- Lescroart F., Hamou W., Francou A., Théveniau-Ruissy M., Kelly R. G. and Buckingham M. (2015). Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc. Natl. Acad. Sci. USA 112, 1446-1451. 10.1073/pnas.1424538112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zhang Q., Cattaneo P., Zhuang S., Gong X., Spann N. J., Jiang C., Cao X., Zhao X., Zhang X. et al. (2015). Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256-3268. 10.1172/JCI68257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo C. A. (2000). Epidemiology of cardiovascular malformations: prevalence and risk factors. Am. J. Med. Genet. 97, 319-325. [DOI] [PubMed] [Google Scholar]
- Lyons I., Parsons L. M., Hartley L., Li R., Andrews J. E., Robb L. and Harvey R. P. (1995). Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 9, 1654-1666. 10.1101/gad.9.13.1654 [DOI] [PubMed] [Google Scholar]
- Mably J. D., Mohideen M. A., Burns C. G., Chen J.-N., Fishman M. C. and Mohideen M.-A. P. K. (2003). heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 13, 2138-2147. 10.1016/j.cub.2003.11.055 [DOI] [PubMed] [Google Scholar]
- Mahler J., Filippi A. and Driever W. (2010). DeltaA/DeltaD regulate multiple and temporally distinct phases of notch signaling during dopaminergic neurogenesis in zebrafish. J. Neurosci. 30, 16621-16635. 10.1523/JNEUROSCI.4769-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney D. B., Geiger E., Blinder J., Benson D. W. and Goldmuntz E. (2003). NKX2.5 mutations in patients with congenital heart disease. J. Am. Coll. Cardiol. 42, 1650-1655. 10.1016/j.jacc.2003.05.004 [DOI] [PubMed] [Google Scholar]
- Miao M., Bruce A. E. E., Bhanji T., Davis E. C. and Keeley F. W. (2007). Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 26, 115-124. 10.1016/j.matbio.2006.09.011 [DOI] [PubMed] [Google Scholar]
- Milan D. J., Giokas A. C., Serluca F. C., Peterson R. T. and MacRae C. A. (2006). Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133, 1125-1132. 10.1242/dev.02279 [DOI] [PubMed] [Google Scholar]
- Mommersteeg M. T. M., Soufan A. T., de Lange F. J., van den Hoff M. J., Anderson R. H., Christoffels V. M. and Moorman A. F. (2006). Two distinct pools of mesenchyme contribute to the development of the atrial septum. Circ. Res. 99, 351-353. 10.1161/01.RES.0000238360.33284.a0 [DOI] [PubMed] [Google Scholar]
- Mommersteeg M. T. M., Brown N. A., Prall O. W. J., de Gier-de Vries C., Harvey R. P., Moorman A. F. M. and Christoffels V. M. (2007). Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ. Res. 101, 902-909. 10.1161/CIRCRESAHA.107.161182 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Panáková D., Werdich A. A., Musso G., Burger A., Lawson K. L., Carr L. A., Nevis K. R., Sabeh M. K., Zhou Y. et al. (2015). Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 6, 8146 10.1038/ncomms9146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz I. P. G., Kim J. B., Moore M. L., Wolf C. M., Peterson M. A., Shendure J., Nobrega M. A., Yokota Y., Berul C., Izumo S. et al. (2007). A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell 129, 1365-1376. 10.1016/j.cell.2007.04.036 [DOI] [PubMed] [Google Scholar]
- Nagelberg D., Wang J., Su R., Torres-Vázquez J., Targoff K. L., Poss K. D. and Knaut H. (2015). Origin, specification, and plasticity of the great vessels of the heart. Curr. Biol. 25, 2099-2110. 10.1016/j.cub.2015.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y. (2010). Second lineage of heart forming region provides new understanding of conotruncal heart defects. Congenit. Anom. 50, 8-14. 10.1111/j.1741-4520.2009.00267.x [DOI] [PubMed] [Google Scholar]
- Nakashima Y., Ono K., Yoshida Y., Kojima Y., Kita T., Tanaka M. and Kimura T. (2009). The search for Nkx2-5-regulated genes using purified embryonic stem cell-derived cardiomyocytes with Nkx2-5 gene targeting. Biochem. Biophys. Res. Commun. 390, 821-826. 10.1016/j.bbrc.2009.10.056 [DOI] [PubMed] [Google Scholar]
- Nakashima Y., Yanez D. A., Touma M., Nakano H., Jaroszewicz A., Jordan M. C., Pellegrini M., Roos K. P. and Nakano A. (2014). Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system. Circ. Res. 114, 1103-1113. 10.1161/CIRCRESAHA.114.303219 [DOI] [PubMed] [Google Scholar]
- Nemer M. (2008). Genetic insights into normal and abnormal heart development. Cardiovasc. Pathol. 17, 48-54. 10.1016/j.carpath.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Nevis K., Obregon P., Walsh C., Guner-Ataman B., Burns C. G. and Burns C. E. (2013). Tbx1 is required for second heart field proliferation in zebrafish. Dev. Dyn. 242, 550-559. 10.1002/dvdy.23928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy N., Novikov N., Jeffrey S., Abrial M., Guner-Ataman B., Sakthivel S., Burns C. E. and Burns C. G. (2017). Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development 144, 4616-4624. 10.1242/dev.153411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panáková D., Werdich A. A. and Macrae C. A. (2010). Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature 466, 874-878. 10.1038/nature09249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P., Sirbu I. O., Kühl S. J., Philipp M. and Kühl M. (2013). Islet1-expressing cardiac progenitor cells: a comparison across species. Dev. Genes Evol. 223, 117-129. 10.1007/s00427-012-0400-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashmforoush M., Lu J. T., Chen H., Amand T. S., Kondo R., Pradervand S., Evans S. M., Clark B., Feramisco J. R., Giles W. et al. (2004). Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373-386. 10.1016/S0092-8674(04)00405-2 [DOI] [PubMed] [Google Scholar]
- Payne R. M., Johnson M. C., Grant J. W. and Strauss A. W. (1995). Toward a molecular understanding of congenital heart disease. Circulation 91, 494-504. 10.1161/01.CIR.91.2.494 [DOI] [PubMed] [Google Scholar]
- Pierpont M. E., Basson C. T., Benson D. W. Jr, Gelb B. D., Giglia T. M., Goldmuntz E., McGee G., Sable C. A., Srivastava D., Webb C. L. et al. (2007). Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115, 3015-3038. 10.1161/CIRCULATIONAHA.106.183056 [DOI] [PubMed] [Google Scholar]
- Prall O. W. J., Menon M. K., Solloway M. J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J. J., Robertson B. R., Chaulet H. et al. (2007). An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947-959. 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M. S., Theveniau-Ruissy M., De Bono C., Mesbah K., Francou A., Rammah M., Dominguez J. N., Roux M., Laforest B., Anderson R. H. et al. (2014). Tbx1 coordinates addition of posterior second heart field progenitor cells to the arterial and venous poles of the heart. Circ. Res. 115, 790-799. [DOI] [PubMed] [Google Scholar]
- Schindler Y. L., Garske K. M., Wang J., Firulli B. A., Firulli A. B., Poss K. D. and Yelon D. (2014). Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141, 3112-3122. 10.1242/dev.106336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J. J., Benson D. W., Basson C. T., Pease W., Silberbach G. M., Moak J. P., Maron B. J., Seidman C. E. and Seidman J. G. (1998). Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281, 108-111. 10.1126/science.281.5373.108 [DOI] [PubMed] [Google Scholar]
- Snarr B. S., O'Neal J. L., Chintalapudi M. R., Wirrig E. E., Phelps A. L., Kubalak S. W. and Wessels A. (2007a). Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ. Res. 101, 971-974. 10.1161/CIRCRESAHA.107.162206 [DOI] [PubMed] [Google Scholar]
- Snarr B. S., Wirrig E. E., Phelps A. L., Trusk T. C. and Wessels A. (2007b). A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev. Dyn. 236, 1287-1294. 10.1002/dvdy.21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. and Olson E. N. (2000). A genetic blueprint for cardiac development. Nature 407, 221-226. 10.1038/35025190 [DOI] [PubMed] [Google Scholar]
- Stanley E. G., Biben C., Elefanty A., Barnett L., Koentgen F., Robb L. and Harvey R. P. (2002). Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 46, 431-439. [PubMed] [Google Scholar]
- Stevens K. N., Hakonarson H., Kim C. E., Doevendans P. A., Koeleman B. P. C., Mital S., Raue J., Glessner J. T., Coles J. G., Moreno V. et al. (2010). Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS ONE 5, e10855 10.1371/journal.pone.0010855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyek M. R., Croll R. P. and Smith F. M. (2015). Intrinsic and extrinsic innervation of the heart in zebrafish (Danio rerio). J. Comp. Neurol. 523, 1683-1700. 10.1002/cne.23764 [DOI] [PubMed] [Google Scholar]
- Sun Y., Liang X., Najafi N., Cass M., Lin L., Cai C.-L., Chen J. and Evans S. M. (2007). Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 304, 286-296. 10.1016/j.ydbio.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supino P. G., Borer J. S., Preibisz J. and Bornstein A. (2006). The epidemiology of valvular heart disease: a growing public health problem. Heart Fail. Clin. 2, 379-393. 10.1016/j.hfc.2006.09.010 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Wechsler S. B., Lee I. W., Yamasaki N., Lawitts J. A. and Izumo S. (1999). Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development 126, 1439-1450. [DOI] [PubMed] [Google Scholar]
- Targoff K. L., Schell T. and Yelon D. (2008). Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev. Biol. 322, 314-321. 10.1016/j.ydbio.2008.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targoff K. L., Colombo S., George V., Schell T., Kim S.-H., Solnica-Krezel L. and Yelon D. (2013). Nkx genes are essential for maintenance of ventricular identity. Development 140, 4203-4213. 10.1242/dev.095562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F., van Weerd J. H., Burkhard S. B., Verkerk A. O., de Pater E., Boukens B. J., Vink A., Christoffels V. M. and Bakkers J. (2012). Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE 7, e47644 10.1371/journal.pone.0047644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh-Finkel L., Zeisel A., Brodt-Ivenshitz M., Shamai A., Yao Z., Seger R., Domany E. and Tzahor E. (2010). BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development 137, 2989-3000. 10.1242/dev.051649 [DOI] [PubMed] [Google Scholar]
- Tu C.-T., Yang T.-C. and Tsai H.-J. (2009). Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebrafish embryos. PLoS ONE 4, e4249 10.1371/journal.pone.0004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg G., Abu-Issa R., de Boer B. A., Hutson M. R., de Boer P. A. J., Soufan A. T., Ruijter J. M., Kirby M. L., van den Hoff M. J. B. and Moorman A. F. M. (2009). A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 104, 179-188. 10.1161/CIRCRESAHA.108.185843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedantham V., Galang G., Evangelista M., Deo R. C. and Srivastava D. (2015). RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 116, 797-803. 10.1161/CIRCRESAHA.116.305913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. D. and Buckingham M. E. (2010). How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1-41. 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- Wang W., Razy-Krajka F., Siu E., Ketcham A. and Christiaen L. (2013). NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 11, e1001725 10.1371/journal.pbio.1001725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Zaffran S., Kuroiwa A., Higuchi H., Ogura T., Harvey R. P., Kelly R. G. and Buckingham M. (2012). Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc. Natl. Acad. Sci. USA 109, 18273-18280. 10.1073/pnas.1215360109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel H. R., Jungblut B., Choe C. P., Crump J. G., Braun T. and Dobreva G. (2012). The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev. Cell 23, 58-70. 10.1016/j.devcel.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel H. R., Cheedipudi S., Gao R., Stainier D. Y. R. and Dobreva G. D. (2017). Isl2b regulates anterior second heart field development in zebrafish. Sci. Rep. 7, 41043 10.1038/srep41043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Hoffmann A. D., Burnicka-Turek O., Friedland-Little J. M., Zhang K. and Moskowitz I. P. (2012). Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev. Cell 23, 280-291. 10.1016/j.devcel.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W., Wang J., Song Y., Yu D., Sun C., Liu C., Chen F., Zhang Y., Wang F., Harvey R. P. et al. (2015). A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521-2532. 10.1242/dev.120220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D., Horne S. A. and Stainier D. Y. R. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23-37. 10.1006/dbio.1999.9406 [DOI] [PubMed] [Google Scholar]
- Zaffran S., Kelly R. G., Meilhac S. M., Buckingham M. E. and Brown N. A. (2004). Right ventricular myocardium derives from the anterior heart field. Circ. Res. 95, 261-268. 10.1161/01.RES.0000136815.73623.BE [DOI] [PubMed] [Google Scholar]
- Zeng X.-X. I. and Yelon D. (2014). Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 7, 951-960. 10.1016/j.celrep.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Han P., Yang H., Ouyang K., Lee D., Lin Y.-F., Ocorr K., Kang G., Chen J., Stainier D. Y. R. et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497-501. 10.1038/nature12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nomura-Kitabayashi A., Sultana N., Cai W., Cai X., Moon A. M. and Cai C.-L. (2014). Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 390, 68-79. 10.1016/j.ydbio.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Cashman T. J., Nevis K. R., Obregon P., Carney S. A., Liu Y., Gu A., Mosimann C., Sondalle S., Peterson R. E. et al. (2011). Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645-648. 10.1038/nature10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.