Abstract
Profilin 1 (Pfn1) is an important regulator of the actin cytoskeleton and plays a vital role in many actin-based cellular processes. Therefore, identification of a small-molecule intervention strategy targeted against the Pfn1–actin interaction could have broad utility in cytoskeletal research and further our understanding of the role of Pfn1 in actin-mediated biological processes. Based on an already resolved Pfn1–actin complex crystal structure, we performed structure-based virtual screening of small-molecule libraries to seek inhibitors of the Pfn1–actin interaction. We identified compounds that match the pharmacophore of the key actin residues of Pfn1–actin interaction and therefore have the potential to act as competitive inhibitors of this interaction. Subsequent biochemical assays identified two candidate compounds with nearly identical structures that can mitigate the effect of Pfn1 on actin polymerization in vitro. As a further proof-of-concept test for cellular effects of these compounds, we performed proximity ligation assays in endothelial cells (ECs) to demonstrate compound-induced inhibition of Pfn1–actin interaction. Consistent with the important role of Pfn1 in regulating actin polymerization and various fundamental actin-based cellular activities (migration and proliferation), treatment of these compounds reduced the overall level of cellular filamentous (F) actin, slowed EC migration and proliferation, and inhibited the angiogenic ability of ECs both in vitro and ex vivo. In summary, this study provides the first proof of principle of small-molecule–mediated interference with the Pfn1–actin interaction. Our findings may have potential general utility for perturbing actin-mediated cellular activities and biological processes.
Keywords: actin, angiogenesis, cell migration, endothelial cell, profilin, molecular dynamics, pharmacophore, virtual screening
Introduction
Dynamic remodeling of the actin cytoskeleton is an integral aspect of many fundamental cellular activities (for example, cell migration and cell proliferation) and therefore crucial for the regulation of biological processes that rely on actin-based events, whether physiological or pathological. Actin cytoskeletal remodeling is regulated by orchestrated actions of a wide range of actin-binding proteins. One of the key players in the regulation of actin polymerization is profilin 1 (Pfn1),3 a ubiquitously expressed member of the Pfn family of actin monomer–binding proteins. Pfn1 inhibits nucleation and elongation of actin filaments at the pointed end but promotes barbed end elongation of filamentous actin through collaboration with various polyproline domain–containing promoters of actin nucleation and elongating factors. Pfn1 also stimulates actin polymerization through its ability to promote ADP-to-ATP exchange on monomeric actin (reviewed in Ref. 1). There is compelling evidence in the literature that Pfn1 plays an important role in cell migration. With the exception of certain cancer cells, in most physiological contexts, either loss of Pfn1 expression or disruption of the actin-binding ability of Pfn1 is generally associated with defects in membrane protrusion (the initiating event of cell migration that is primarily driven by actin polymerization) and slower cell migration (2–7). Similarly, gene silencing and knockout studies demonstrated defects in cytokinesis (an aspect of cell division that is dependent on the action of the actin cytoskeleton) and reduced cell proliferation upon Pfn1 depletion (3, 8). Collectively, these findings demonstrate the important role of Pfn1 in various actin-dependent cellular functions, further begging the question whether some of these cellular functions are susceptible to chemical inhibition of Pfn1 function. However, currently no inhibitor of Pfn1 function exists. In this study, we demonstrate for the first time that the Pfn1–actin interaction is susceptible to small-molecule inhibition. Guided by structure-based virtual screening of small-molecule libraries, we identified two compounds with similar chemical structures that are capable of reversing the effect of Pfn1 on actin polymerization in vitro. As a further proof-of-concept test for the cellular effects of these compounds, our experiments in endothelial cells (ECs) demonstrated that treatment of these compounds led to attenuation of cellular Pfn1–actin interaction, slower migration and proliferation, and prominent suppression of the angiogenic activity of ECs.
Results
Pharmacophore-based virtual screening of small molecules that can mimic and competitively inhibit the Pfn1–actin interaction in vitro
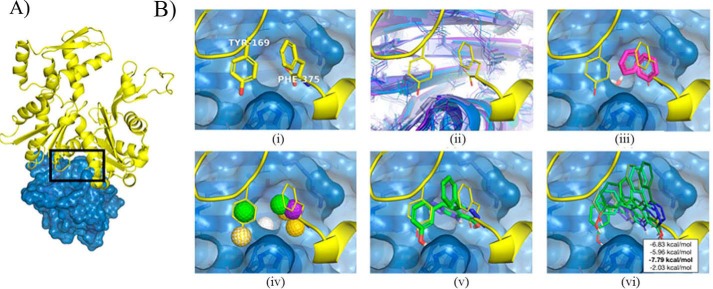
Because the Pfn1–actin complex structure has been resolved (9) (Fig. 1A), we used structure-based virtual screening of small-molecule libraries to identify compounds that can mimic the Pfn1–actin interaction and therefore have the potential to act as competitive inhibitors of this interaction. Structure-based virtual screening has been successfully applied to protein–protein interactions (e.g. p53–MDM2 (10), BH3–Bcl-xL (11), and tumor necrosis factor α (12)). Pfn1–actin has a classical “large and flat” interface, but a few residues are identified as potential hot spots by PocketQuery (13), and their interactions can potentially be exploited for molecular design. The most critical actin residue of the Pfn1–actin structure, as determined by an analysis of PDB code 2BTF, is Y169 of actin, which forms a hydrogen bond with the His-119 residue of Pfn1. Consistent with the computational analysis, the H119E mutant of Pfn1, which eliminates this bond, loses affinity to actin (14, 15). To expand beyond the limited interactions of Tyr-169, we considered the small pocket on Pfn1 adjacent to Tyr-169 in which the C terminus of actin (Phe-375) buries (Fig. 1B, i). As the Phe-375 pocket in the crystal structure was too small to accommodate a ligand, we simulated the Pfn1 monomer with molecular dynamics and identified structures where the pocket was enlarged (Fig. 1B, ii). Potential interactions with residues in this pocket were identified by docking small fragments (benzene and water) to structures extracted from the simulation (Fig. 1B, iii).
Figure 1.
Structure-based workflow for identifying inhibitors of Pfn1–actin interaction. A, the Pfn1-actin interface (PDB code 2BTF) with the targeted interface region highlighted. B, I, interface residues critical to binding were computationally identified. ii, the flexibility of the Pfn1 interface was explored with molecular dynamics. iii, fragments were docked to molecular dynamics snapshots. iv, pharmacophore features were identified from interface residues and docked fragments. v, a pharmacophore search identified compounds matching subsets of these features. vi, energy minimization refined and scored the ligand structures with respect to molecular dynamics snapshots. A consensus score across the receptor and ligand structures was used to select compounds for testing.
The native interactions of actin and the putative interactions identified through fragment docking were combined to construct pharmacophore queries. A pharmacophore represents the spatial arrangement of the essential features of an intermolecular interaction, such as hydrophobic groups and hydrogen bond donors. We generated ten pharmacophore queries, where every query contained a hydrophobic core targeting the Phe-375 pocket and three additional hydrogen bond features selected from five possible interactions. The pharmacophore features used are shown in Fig. 1B, iv, and primarily target the His-119, Arg-74, Asp-86, Leu-87, and Arg-88 residues of Pfn1. Pharmer (16) was used to search the ZINC database (17) for compounds that matched each of the queries. Matching compounds are aligned to the query pharmacophore, as shown in Fig. 1B, v, and need to be optimized with respect to atomistic interactions with the Pfn1 receptor to be effectively ranked. The steric energy of hit compounds was minimized to identify a locally optimal conformation of the ligand and a predicted binding affinity (Fig. 1B, vi). Ligands were minimized and consensus-scored against an ensemble of Pfn1 structures generated using molecular dynamics to account for receptor flexibility. The top 20 scoring hits after filtering for structural diversity were initially acquired for screening (Table S1 for a complete list and structures of the initial batch of 20 compounds; annotated as C1 through C20).
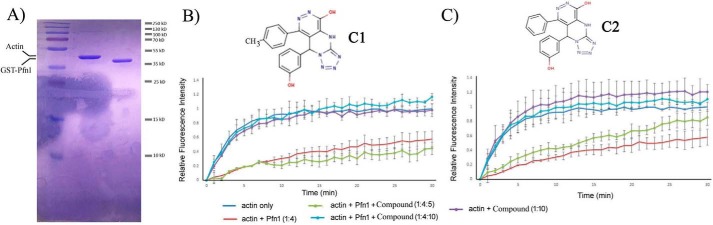
For biochemical screening of the compounds, we performed a pyrene-based actin polymerization assay. This type of assay was used previously to screen a library of chemical compounds to identify inhibitors against the formin family of actin-binding proteins (18). In this assay, Pfn1 reduces actin polymerization by its monomer-sequestering action. Essentially, we searched for compounds that can reverse the inhibitory effect of Pfn1 on actin polymerization but are unable to impact actin polymerization in the absence of Pfn1 (we bacterially expressed and purified GST-tagged Pfn1 as our source of Pfn1 (Fig. 2A)). After screening the compounds, we ascertained that the two highest-scoring small molecules from our virtual screen (identified as compound 1 (C1, 8-(3-hydroxyphenyl)-10-(4-methylphenyl)-2,4,5,6,7,11,12-heptaazatricyclo[7.4.0.03,7]trideca-1(13),3,5,9,11-pentaen-13-ol) and compound 2 (C2, 8-(3-hydroxyphenyl)-10-phenyl-2,4,5,6,7,11,12-heptaazatricyclo[7.4.0.03, 7]trideca-1(13),3,5,9,11-pentaen-13-ol)) showed promise, as demonstrated by recovery of actin polymerization after addition of the compound in a dose-dependent and Pfn1-dependent manner (Fig. 2, B and C). These two compounds, used as racemic mixtures, completely abrogated the inhibitory effect of Pfn1 on actin polymerization at 2.5-fold molar excess compared with Pfn1. In the absence of Pfn1, neither of these two compounds had any significant effect on actin polymerization on their own, further implying that their action on actin polymerization in the Pfn1–actin setting was through inhibition of Pfn1. Mass spectrometry and NMR analyses confirmed the purity and structure of these compounds, respectively (Figs. S1 and S2). Note that compounds C1 and C2 are nearly identical, differing by a single methyl group.
Figure 2.
Effects of C1 and C2 on actin polymerization with or without Pfn1 in vitro. A, Coomassie staining of an SDS-PAGE showing the purity of actin and GST–Pfn1 used in the pyrene–actin assay. B and C, pyrene–actin polymerization assay curves for the indicated experimental conditions (B, C1; C, C2) recorded for 30 min after addition of the polymerization buffer. Each time point represents the mean ± S.D. values of the fluorescence intensity of polymerized pyrene–actin relative to the maximum fluorescence intensity for the actin alone condition (data are summarized from n = 3 experiments). The insets show the chemical structures of the two compounds. The numbers in parentheses indicate relative concentrations of actin, GST–Pfn1, and the compounds. The actual concentrations of actin and Pfn1 were 10 μm and 40 μm, respectively. C1 or C2 was added either at a 50 μm (Pfn1:compound = 1:1.25) or 100 μm (Pfn1:compound = 1:2.5) concentration.
For a limited structure–activity relationship (SAR) analysis, we performed the pyrene–actin assay with 22 additional compounds (annotated as C21 through C42; see Table 2) that had an identical scaffold to C1 and C2. None of these analogous compounds exhibited comparable activity. The SAR suggests that the hydroxyl of the phenol group that mimics the Tyr-169 of actin (Fig. 1B, v) is essential: moving it to the para position (C32) or replacing it with an ether (C24) abrogated activity, as did other transformations of this phenol (C23–C42). Unfortunately, the commercially available compounds with this scaffold did not support the development of an SAR of other aspects of C1 and C2, other than revealing that the addition of a charged nitro group to the phenyl ring removes activity (C22). Considering the positional importance of the hydroxyl of the phenol group that mimics the Tyr-169 residue of actin, it is also not clear why compounds C7 and C8 (which also contained that feature) failed to show any activity. These compounds differ from C1 and C2 through decorations of the solvent-exposed phenyl group (Fig. 1B, v). This suggests that there may be intra- or intermolecular interactions missing from our model, and further refinement of the model remains an avenue of future research.
C1 and C2 inhibit the angiogenic ability of vascular ECs
We showed previously that loss of function of Pfn1 (either by depletion or disruption of the actin-binding function by point mutation) can lead to actin cytoskeletal changes (marked by prominent reduction of actin stress fibers and overall cellular F-actin content), slower migration and proliferation, and prominent reduction in the angiogenic ability of vascular ECs (2, 3). Along this line, another study showed that Pfn1–actin interaction in ECs is stimulated in response to pro-angiogenic growth factor signaling through a specific tyrosine (Tyr-129) phosphorylation-dependent mechanism in vitro and in vivo. Blockade of Tyr-129 phosphorylation (which reduces Pfn1–actin interaction to its basal level) reduces VEGF-induced motility and angiogenesis of EC in vitro and angiogenesis associated with tissue repair and tumors in vivo (19). Pfn1 phosphorylation also promotes angiogenic growth factor secretion in the tumor microenvironment through HIF1 (hypoxia-inducible factor 1) stabilization in a von Hippel–Lindau–dependent mechanism, contributing to tumor progression (20). Therefore, as a proof-of-concept test for the cellular effects of these compounds, we performed a series of experiments in ECs to determine whether some of the above phenotypes can be recapitulated in the settings of chemical inhibition of the Pfn1–actin interaction by compound treatment, as described in the following sections.
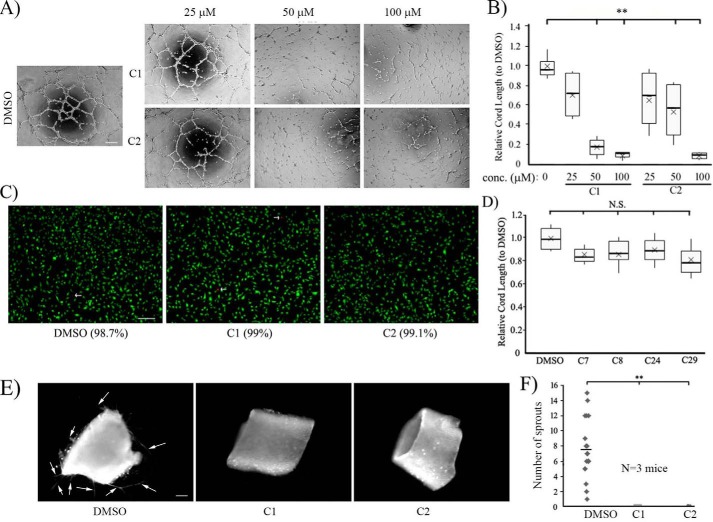
First, to determine the effects of C1 and C2 on angiogenesis, we performed Matrigel cord formation assays (a commonly used in vitro morphogenetic assay to assess the angiogenic potential of ECs) with HmVEC-1 (a widely used immortalized human dermal microvascular cell line, referred to as HmVECs hereafter) at different concentrations (25, 50, and 100 μm) of each of the two compounds. As a control, cells were treated with the vehicle control DMSO. Although there was a general trend of dose-dependent decrease in the cord-forming ability of HmVECs (as measured by the total cord length) in response to either of the compounds, the difference in the cord formation readout between the control and the compound-treated groups was statistically significant only in the 50–100 μm dose range, with the most dramatic reduction (by ∼90%) in angiogenesis seen in response to 100 μm compound treatment (Fig. 3, A and B). Depending on the compound, the average reduction in cord formation ranged from 50–80% for 50 μm compound treatment. To ensure that C1 and C2 were not cytotoxic at this high dose, we performed live/dead staining of HmVECs, which showed nearly 100% cell viability even after overnight treatment of the compounds at 100 μm concentration (Fig. 3C), suggesting a lack of any obvious cytotoxic effects of these compounds in ECs. Note that we also performed cord formation assays with four additional compounds (C7, C8, C24, and C29), given that these compounds had the same general structural scaffold as C1 and C2 but did not show any activity in the actin polymerization assay. Correlated with their lack of biochemical activity in the actin polymerization assay, these compounds also did not have any anti-angiogenic effect in ECs, even when added to the culture at a 100 μm concentration (Fig. 3D). The general correlation between biochemical and biological activities (or lack of activities) of the compounds suggest that the core structural scaffold of compounds C1 and C2 alone per se does not confer the anti-angiogenic action, further underscoring the importance of specific functional groups attached to this scaffold for the biological action of the two compounds. Although widely used, the cord formation assay fails to recapitulate the sprouting behavior of ECs from preexisting blood vessels and therefore does not represent the complexities of multicellular interactions that occur during angiogenesis in vivo. Therefore, we also assessed EC sprouting from mouse aortic rings explanted in ECM in response to C1 and C2. Consistent with our cord morphogenesis assay results, ex vivo endothelial sprouting (identified by lectin staining) of aortic rings was almost completely impaired upon treatment with either C1 or C2 (Fig. 3, E and F; representative phase-contrast images of the sprouts in control versus C2-treated groups are shown in Fig. S3). Collectively, these data demonstrate the anti-angiogenic effects of these compounds.
Figure 3.
Effects of a small-molecule inhibitor of Pfn1–actin interaction on angiogenesis in vitro and ex vivo. A and B, representative phase-contrast images of cord formation by HmVECs on Matrigel after treatment with either DMSO or the indicated concentrations of C1 and C2 (images were acquired with a ×4 objective; scale bar = 200 μm). The associated box and whisker plot in B summarizes the mean values of the cord length for the different experimental conditions relative to the mean value scored for the DMSO control (n = 3 experiments with 2 technical replicates/condition in each experiment; **, p < 0.01). In the box and whisker plots, X represent the mean, the middle lines of the box indicate the median, the top of the box indicates the 75th percentile, the bottom of the box measures the 25th percentile, and the two whiskers indicate the 10th and 90th percentiles, respectively. C, live/dead staining of HmVECs following overnight treatment with either DMSO or C1 or C2 at 100 μm concentration (live and dead (arrows) cells are labeled green and red, respectively; scale bar = 200 μm). The numbers in parentheses indicate the percentage of live cells for different experimental conditions summarized from two experiments. D, a box and whisker plot summarizing the mean values of the cord length for the indicated compounds with a similar structural scaffold as C1 and C2 (as designated in Table S2) relative to the mean value scored for the DMSO control (n = 3 experiments). N.S., not significant. E and F, representative fluorescence images (E) of mouse aortic segments embedded in Matrigel and stained with rhodamine-conjugated lectin for the indicated experimental conditions (arrows show the lectin-positive endothelial sprouts; scale bar = 200 μm). F shows the quantification of sprouts pooled from 20 rings/condition isolated from a total of three mice from three independent experiments (the dashed line shows the mean). **, p < 0.01.
Evidence of inhibition of cellular Pfn1–actin interaction by C2
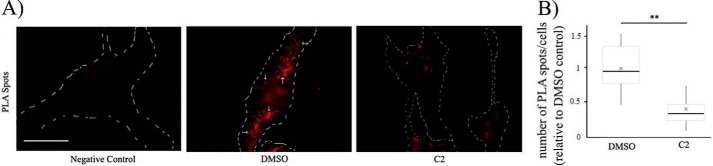
As a proof-of-principle test to verify that our compounds can actually inhibit the Pfn1–actin interaction in ECs, we performed a proximity ligation assay (PLA) to detect endogenous Pfn1–actin interaction with or without overnight treatment of ECs with C2. We found a dramatic 70% reduction in the average number of PLA spots (denotes Pfn1–actin interaction) per cell after treatment with C2 (Fig. 4). These data indicate decreased proximity of Pfn1 and actin, suggestive of reduced interaction of these proteins in ECs resulting from C2 treatment. Note that we also attempted to biochemically verify the ability of these compounds to inhibit the interaction of actin with Pfn1 by either the co-immunoprecipitation method in both HmVECs and HEK-293 cells or a polyproline (PLP) pulldown assay (a method commonly adopted for Pfn1 purification because of the affinity of Pfn1 for PLP sequences) with HEK-293 cell lysate. Unfortunately, these biochemical assays did not reveal any discernible effect of the compounds on Pfn1–actin interaction (data not shown). Possible reasons for the apparent discrepancy in the results between the PLA and biochemical experiments are elaborated under “Discussion.”
Figure 4.
Effect of C2 on endogenous Pfn1–actin interaction in ECs. A, representative fluorescence images of Pfn1–actin PLA spots (arrows) in HmVECs following overnight treatment of either DMSO (control) and C2. An image of the negative control PLA stain (i.e. involving one antibody) only is shown alongside for comparison. Dashed lines indicate the boundaries of the cells (scale bar = 20 μm). B, a dot plot displays the number of PLA spots in C2-treated cells relative to the mean value scored for the DMSO control (the dashed lines represent the mean values (n = 40 cells/group pooled from 2 independent experiments; **, p < 0.01).
C1 and C2 affect the actin cytoskeleton, migration, and proliferation of ECs
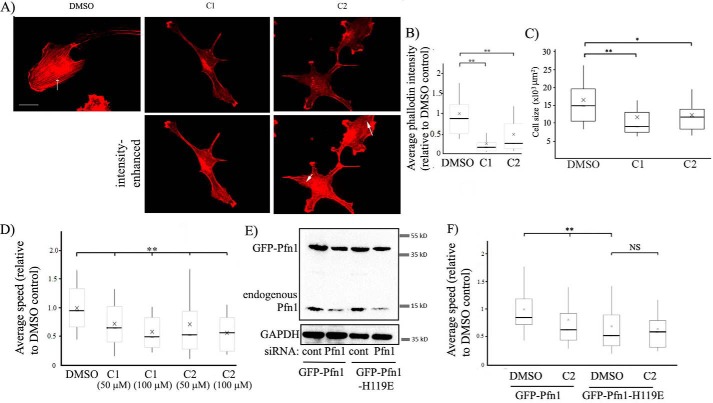
To further test the effect of these compounds on the actin cytoskeleton of ECs, we performed rhodamine–phalloidin staining of HmVECs following overnight treatment with either of the two compounds (at 50 μm concentration) or DMSO (vehicle control). We observed a noticeable decrease in actin stress fibers and overall F-actin content (as measured by the average intensity of rhodamine fluorescence per cell) in HmVECs in response to the treatment with either C1 or C2 (Figs. 5, A and B), suggesting an inhibitory effect of these two compounds on actin polymerization in cells. Morphometric analyses revealed that the average spread area of ECs was significantly lower in the compound-treated groups than in the control group (Fig. 5C), an observation that is consistent with the inhibitory effect of the compounds on actin polymerization (expected to reduce the efficacy of membrane protrusion and, therefore, cell spreading).
Figure 5.
Effects of a small-molecule inhibitor of Pfn1–actin interaction on the actin cytoskeleton and random 2D motility of ECs. A–C, representative fluorescence images (A) of rhodamine-phalloidin–stained HmVECs (acquired with a ×60 objective) following overnight treatment with either DMSO or C1 or C2 at 50 μm concentration (the arrow shows actin stress fibers; scale bar = 20 μm). The top panels show images at identical brightness/contrast settings. The images in the bottom panel were brightness-enhanced to reveal stress fibers (arrows) in compound-treated cells, albeit in much lower abundance than control cells. The box and whisker plot in B summarizes the mean values of the integrated fluorescence intensity of rhodamine–phalloidin relative to the average value of the DMSO control group. The mean values of the cell area for the indicated treatments are summarized in a box and whisker plot in C (n = 50–60 cells/group pooled from 3 experiments; **, p < 0.01; *, p < 0.05). In the box and whisker plots, X represent the mean, the middle lines of the box indicate the median, the top of the box indicates the 75th percentile, the bottom of the box measures the 25th percentile, and the two whiskers indicate the 10th and 90th percentiles, respectively. D, a box and whisker plot depicting the relative speed of randomly migrating HmVECs after treatment with DMSO or the indicated concentrations of compounds C1 or C2 (n = 100–120 cells/group pooled from 3 independent experiments; **, p < 0.01). E, Pfn1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, loading control (cont)) immunoblots of total extracts of HmVECs stably expressing either GFP–Pfn1 or GFP–Pfn1–H119E and following transfection with the indicated siRNAs. F, a box and whisker plot summarizing the effect of C2 on the relative speed of WT Pfn1 versus H119E Pfn1 expressers of HmVECs treated with control or Pfn1 siRNA (n = 60–70 cells/group pooled from 3 independent experiments; **, p < 0.01; NS, not significant).
The interaction of Pfn1 with actin is an important requirement for efficient cell migration (a crucial aspect of angiogenesis). We showed previously that interfering with the actin interaction of Pfn1 by H119E mutation, which reduces the affinity of Pfn1 for actin by ∼25-fold (14), reduces the speed of randomly migrating HmVECs by ∼40% (2). To determine whether the small molecule inhibitors of the Pfn1–actin interaction have a similar effect on EC migration, we analyzed 2D random migration of HmVECs without or with the compounds at two different concentrations (50 and 100 μm). At both concentrations, C1 and C2 reduced the average speed of HmVECs by ∼30% (Fig. 5D). Next we analyzed the effect of C2 on random motility of HmVEC sublines engineered to stably express either the WT form or the actin binding–deficient H119E mutant of Pfn1 as GFP fusion proteins (the expression of endogenous Pfn1 was suppressed by siRNA treatment; see Fig. 5E). The knockdown efficiency of endogenous Pfn1 in WT and H119E-Pfn1 expressers were roughly equal to 80% ± 10% (summarized from three experiments). The expression levels of WT Pfn1 and H119E Pfn1 relative to endogenous Pfn1 (based on the level in control siRNA-transfected cells) were equal to1.05 ± 0.12 and 0.9 ± 0.16, respectively (summarized from three experiments). As expected, the basal speed (in the absence of the compound) of H119E-Pfn1 expressers was ∼30% lower than that of the WT Pfn1 expressers. C2 reduced the speed of WT Pfn1 by 20% (at a slightly lower efficiency than the parental cell line, which may be due to different affinities of C2 for endogenous versus GFP-tagged Pfn1) but did not reduce the average speed of H119E Pfn1 expressers any further (Fig. 5F). Although these data do not necessarily rule out the possibility of off-target effects of the compound, dependence of a functional actin-binding capability of Pfn1 for the anti-migratory effect of the compound is consistent with a scenario in which the anti-migratory effect of any of the compounds is at least partly mediated through inhibition of the Pfn1–actin interaction.
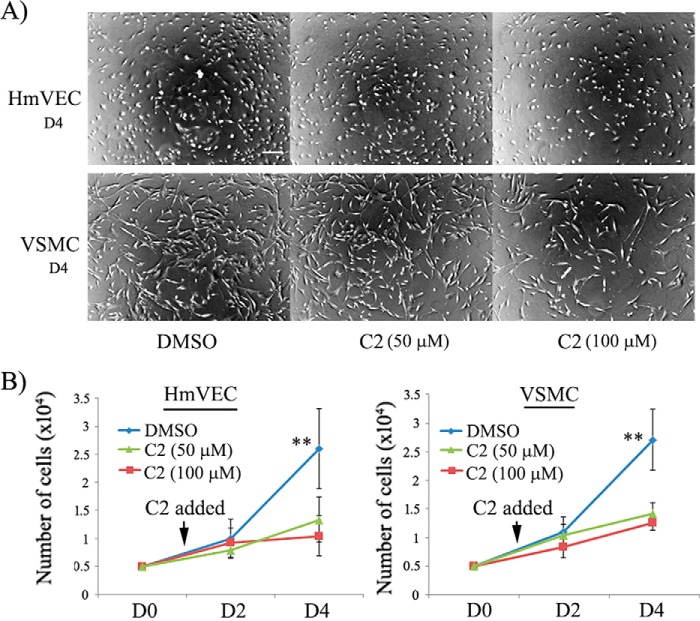
Given that EC proliferation is also an important aspect of angiogenesis and that Pfn1 plays an important role cell proliferation (3, 8), we next examined the effect of C2 on HmVEC proliferation in a 2D culture setting. Our experiments revealed that the average cell counts in response to 3 days of incubation with the two test doses (50 and 100 μm) of C2 were ∼2- and 2.5-fold lower, respectively, than the same in the control group setting (Fig. 6, A and B), indicating an anti-proliferative effect of C2 on ECs.
Figure 6.
Effects of a small-molecule inhibitor of Pfn1–actin interaction on proliferation of ECs and VSMCs in 2D culture. A, representative bright-field images (acquired with a ×4 magnification objective) of HmVECs and VSMCs in 2D culture 3 days after treatment (representing day 4 (D4) of the experiment) with either DMSO or C2 at the indicated concentrations show dose-dependent inhibition of culture growth by C2 (scale bar = 200 μm). B, line graphs summarizing the quantification (mean and standard deviation) of the proliferation assay of HmVECs and VSMCs (cells were plated on day 0 (D0); the mean and standard deviation values are based on cell count data on day 2 and day 4 from three independent experiments with two technical replicates per condition in each experiment. **, p < 0.01 for day 4.
The anti-proliferative action of C2 is not restricted to ECs
Finally, given that Pfn1 is a ubiquitously expressed protein, we wanted to further explore whether C2 has a similar anti-proliferative effect on other types of cells. Therefore, we performed similar proliferation experiments with human vascular smooth muscle cells (VSMCs; we chose this cell type as it is relevant to the vascular system), which also showed robust suppression of VSMC proliferation in 2D culture in response to C2 treatment (Fig. 6, A and B). Although we have not extended these studies to a broad range of cell types, these preliminary findings with VSMCs still suggest that the action of our compound is not specific to ECs only.
Discussion
Pfn is a vital regulator of actin dynamics and cell motility, and, therefore, interfering with the function of Pfn1 could be a useful strategy to perturb physiological and pathological processes that rely on actin-based cell motility. Because no small-molecule inhibitors for Pfn1–actin interaction existed to date, we sought to identify putative inhibitors of Pfn1–actin interaction. This study demonstrates for the first time that the Pfn1–actin interaction is susceptible to small-molecule–mediated inhibition. We provide evidence of anti-migratory, anti-proliferative, and anti-angiogenic actions of the class of compounds identified here. Because the biological effects of these compounds were observed only at a somewhat high concentration range (50–100 μm), these compounds should be only regarded as first-generation inhibitors of Pfn1–actin interaction at this stage. Further characterization and expansion of their chemotype will be necessary to develop a more effective inhibitor class of Pfn1–actin interaction. The structural model used to identify C1 and C2 will serve as a useful guide for the development of such analogs. Although additional experimental characterization, such as X-ray crystallography or NMR, is needed to validate the model, the preliminary SAR (Table S2) and our H119E Pfn1 EC migration results are consistent with the proposed binding mode, where the compound mimics the interaction of Tyr-169 of actin with His-119 of Pfn1.
Despite supportive data for the inhibitory effect of the compound on Pfn1–actin interaction (as suggested by the PLA and pyrene–actin assay results), we were unable to biochemically validate the compound-induced changes in Pfn1–actin interaction by either co-immunoprecipitation or PLP pulldown method. The negative results of biochemical experiments could be due to several plausible reasons. First and foremost, these compounds are likely to be weak inhibitors of the Pfn1–actin interaction, and therefore any subtle change in the interaction would be difficult to detect in a gross biochemical assay. Moreover, use of detergent in the biochemical assays may preclude the binding of the compound to Pfn1 or dissociate weak interactions of Pfn1–compound during the extraction and the pulldown phase. Second, our compound may be only effective in blocking the formation of a direct Pfn1–actin complex that is not associated with other proteins (possibly because of conformational changes induced by other interacting proteins causing steric hindrance). Similarly, indirect protein complexes containing Pfn1 and actin involving other intermediate proteins (e.g. actin nucleation and elongation-promoting factors) may not be targeted by the compounds. Biochemical assays will likely capture different types of complexes containing Pfn1 and actin, whether through direct or indirect interactions. Another potential issue in the PLP pulldown assay is that, in addition to pulling down actin via Pfn1, this technique could, in principle, capture actin through other SH3 domain–containing proteins (that also have strong affinity for PLP) as well. The theoretical maximum distance between two interacting proteins of interest is roughly 40 nm to be able to generate PLA signals. Given the small size of both actin (average diameter, ∼5.5 nm) and Pfn1 (even smaller than actin), although the PLA assay should not be technically limited to detection of only the direct Pfn1–actin complex, we do not know whether Pfn1–actin interaction is detected with equal efficiency by PLA when this interaction is direct versus indirect (part of either small or large protein complexes). Because of the extreme sensitivity of the PLA signal to the distance between the antibodies concerned, it is highly possible that the PLA assay could be more efficient for detecting the direct Pfn1–actin complex (either exclusive or as part of a small complex involving other binding partners) or, at best, small indirect complexes than those involving indirect interactions as a part of large protein complexes. In essence, the PLA assay may detect only a fraction of all cellular Pfn1–actin interactions (that is captured by biochemical assays) and be biased toward interactions that are more likely to be affected by the compounds.
There could be several reasons why at least a 50 μm concentration of our first-generation inhibitor was required for biological effects. First, the literature has reported that the cellular concentration of Pfn1 is generally in the low tens of micromolar range (e.g. human EC, most relevant to this study, 10.6 μm (21); mammary epithelial cells and breast cancer cells, in the range of 13–30 μm (22); chicken hamster ovary cells, ∼10 μm (23)). The compounds identified here may be low-affinity inhibitors of Pfn1–actin interaction, and at least, according to the pyrene–actin assay, a 2.5-fold molar excess of the compound over Pfn1 appeared to be necessary to completely reverse the effect of Pfn1 on actin polymerization. Second, we do not have a clear understanding of the relative stoichiometry of Pfn1 in its free versus bound (either exclusively with actin or PLP ligands or both) states and whether the compound is effective in blocking the Pfn1–actin interaction when Pfn1 is simultaneously bound to other ligands. Third, the actual intracellular concentration of the compounds may be low when they do not efficiently cross through the membrane lipid bilayer. Fourth, although the results of our motility studies in WT versus H119E Pfn1 expressers of EC are consistent with a cellular mechanism of action of the compounds involving Pfn1, we cannot absolutely rule out the possibility of targeting of the compound to some other cellular ligands besides Pfn1, thereby necessitating high concentrations of the compounds for biological effects. However, based on the dependence of a functional actin-binding capability of Pfn1 for the anti-migratory effect of C2 and a general correlation between biochemical and cord angiogenesis data of the compounds, it is highly unlikely that phenotypes elicited by our first-generation compounds are solely attributed to possible off-target effects.
The inhibitory effect of the compounds on angiogenesis (in both cord and aortic ring assays) was much more pronounced than it was on 2D random motility of ECs on tissue culture substrate. One likely explanation could be that EC migration on a compliant ECM substrate (e.g. Matrigel used in the cord angiogenesis assay) and spreading may be more sensitive to a loss of actin binding function of Pfn1 than 2D cell migration on a rigid tissue culture substrate. We and others have shown that loss of function of Pfn1 also leads to reduced cell adhesion (4), a defect that may be more consequential in impacting the motile phenotype of cells in 3D ECM substrate (thus affecting cord morphogenesis) than on 2D rigid tissue culture substrate. Pfn1 also competes with the Arp2/3 complex (a major actin nucleator) for actin binding. Therefore, reduced Pfn1–actin interaction in response to compound treatment can enhance Arp2/3-mediated membrane protrusion, thereby partly mitigating the defect in migration, as demonstrated previously in the case of fibroblasts (25). We also showed that, in addition to affecting migration, our compound had a major suppressive effect on EC proliferation (a key factor for sprouting angiogenesis), and this might also explain why our compound exhibited such a dramatic impact on sprouting angiogenesis. In the future, it will be also interesting to study whether the compounds identified here can lead to regression of already formed vessels.
Finally, the compounds identified here could have potential utility in cell biology research as novel chemical tools for perturbing actin cytoskeletal dynamics and actin-dependent biological processes. We also need to carefully consider the potential implications of our findings related to the anti-angiogenic effects of the compounds. Although angiogenesis is important for growth, development, and tissue repair, it can exacerbate a wide range of human pathologies, including diabetic retinopathy, cancer, atherosclerosis, and arthritis, if uncontrolled. Anti-angiogenic therapies to date have been primarily targeted against the action of VEGF (a potent pro-angiogenic growth factor) signaling. However, the therapeutic benefits of the anti-VEGF strategy have been stymied in many instances because of the involvement of alternative pro-angiogenic signaling pathways and acquired resistance to anti-VEGF treatment (26). Therefore, it is conceivable that targeting molecules that are fundamentally important for angiogenesis-related activities of vascular EC may be an alternative strategy. As an example, attenuation of glycolysis by pharmacological blockade of certain metabolic enzymes has recently shown significant promise in diminishing pathological angiogenesis) (27–29). At least at the conceptual level, there are certain advantages of a Pfn1-centric angiogenesis-modulating strategy. First, Pfn1–actin interaction is fundamentally important for dynamic actin remodeling and EC migration and also stimulated in ECs in response to pro-angiogenic growth factor (2, 19). Second, Pfn1 is not absolutely essential for actin polymerization (this likely explains why differentiated cells generally tolerate even the genetic disruption of Pfn1–actin interaction), but it is utilized by multiple actin assembly pathways to enhance the efficiency of actin polymerization and therefore occupies a critical node of convergent pathways. This nodal positioning may reduce resistance to Pfn1–actin interaction inhibitors that could otherwise occur from alternative compensatory pathways. However, a major disadvantage of targeting Pfn1 is that it is a broadly expressed protein and, presumably, that there is a requirement for Pfn1–actin interaction in all somatic cell types. Clearly, our data showing that the anti-proliferative action of our compound is not restricted to ECs only support this notion. Therefore, there is always a general concern of potential widespread in vivo effects associated with targeting the interaction of Pfn1 with actin. Unless these issues are experimentally addressed through in vivo studies in the future, it is still premature to justify an inhibitor of Pfn1–actin interaction as an anti-angiogenic agent for any therapeutic purpose.
Materials and methods
Pharmacophore query construction
Pharmacophore interaction features were generated for the Pfn1–actin binding pocket by docking fragments (benzene and water) and manually selecting deeply buried interactions. Altogether, we identified two hydrophobic, one aromatic, and five hydrogen bond interaction features in the vicinity of the Tyr-169 residue of actin. As including all these features resulted in an overly specific query without any matches, we generated all 10 possible pharmacophore queries containing any three of the five possible hydrogen bond features. Each query feature was assigned a 1-Å radius, and directionality was not considered.
Molecular dynamics
To account for receptor flexibility, the Pfn1 component of the Pfn1–actin 2BTF complex was simulated by itself using Amber14 and the ff14SB force field (30) with TIP3P explicit water in a truncated octahedral box with a 12-Å buffer, periodic boundary conditions, and particle mesh Ewald electrostatics with a 10-Å cutoff. After two rounds of minimization and a 100-ps equilibration run, the protein was simulated for 139 ns. A greedy algorithm was then used to extract 10 diverse snapshots as measured by backbone root mean square deviation. These snapshots were then aligned to the 2BTF structure used to define the pharmacophore queries.
Consensus scoring and ranking
As the hits identified through pharmacophore search are already aligned with respect to the receptor according to the desired interaction features, there is no need to perform full molecular docking. However, to refine the compound structures and generate a ranking, we performed local energy minimization using smina (31) with the AutoDock Vina scoring function (32). We minimized the pharmacophore-aligned poses from searching the ZINC database (210,802 poses total) with respect to each of the 10 receptor structures generated through molecular dynamics simulation. Poses that moved by more than 2 Å root mean square deviation during minimization were filtered out to retain the desired pharmacophore interactions, and only the top-scoring pose for each compound was retained. The top 100 scoring poses for each of the 10 receptor structures were then merged, and the top 300 unique compounds of this combined set (which included all 39 compounds that were top 100-ranked for at least five of the 10 receptors) were evaluated for acquisition. These compounds were clustered using OpenBabel FP2 fingerprints (33), and, at most, two compounds per cluster were selected for screening, subject to commercial availability, up to a total of 20 compounds.
Cell culture and transfection
HmVECs (ATCC, CRL-3243) were cultured in MCDB-131 (Life Technologies) growth medium with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 10 ng/ml epidermal growth factor, 1 μg/ml hydrocortisone, and 10 mm l-glutamine. Human VSMCs (ATCC, PCS-100-012) were cultured in human smooth muscle cell medium (Cell Applications) with supplement provided by the company, 100 units/ml penicillin, and 100 μg/ml streptomycin. For silencing of genes, SMARTpool Pfn1 siRNA (GE Dharmacon, M-012003-01-0005) was used at 100 nm using transfection reagent 1 (Dharmacon) following the instructions of the manufacturer. HmVECs were treated with either C1 or C2 overnight prior to being assayed.
Angiogenesis assays
The in vitro cord formation assay was performed as described previously (3). Briefly, HmVECs were plated on polymerized Matrigel and allowed to adhere prior to treatment with either test compounds or vehicle control. After 16 h, cord formation was assessed by phase-contrast microscopy. Total cord length was quantified using the Angiogenesis plugin of ImageJ. The ex vivo aortic ring assay was performed using thoracic aortic segments from 9- to 11-week-old C57Bl/6 mice as described recently (24). At the end of the experiments, aortic rings were fixed with 3% (v/v) formalin for 30 min and stained with rhodamine-conjugated lectin (0.05 mg/ml) for 2 h at room temperature and washed before imaging at ×4 magnification.
Cell migration
HmVECs were sparsely plated in a 24-well plate coated with type I collagen (Millipore) and treated with DMSO, C1, or C2 overnight in the cell culture plating medium described earlier. Time-lapse images of randomly migrating HmVEC-1 cells were collected using a ×10 objective (N.A. = 0.4) on an Olympus IX-71 inverted microscope for 2 h at 1-min time intervals using MetaMorph (Universal Imaging). The centroid of the cell nucleus was tracked frame by frame using ImageJ, and the average speed of movement for each cell (= total distance (as measured by the centroid movement)/total time) was calculated.
Protein extraction and immunoblotting
Cell lysates were prepared using a modified radioimmune precipitation assay buffer (25 mm Tris-HCL (pH 7.5), 150 mm NaCl, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol, 1 mm EDTA, 50 mm NaF, 1 mm sodium pervanadate, and protease inhibitors supplemented with 6× sample buffer diluted to 1× with a final SDS concentration in the lysis buffer of 2%). Conditions for the various antibodies were as follows: monoclonal Pfn1 (Abcam, ab124904, 1:3000) and monoclonal glyceraldehyde-3-phosphate dehydrogenase (Sigma, G9545-25UL, 1:3000). 10 μg of total protein lysate was loaded per lane for SDS-PAGE analyses.
PLA
The PLA was performed using the Duolink kit with anti-mouse plus and anti-rabbit minus probes (Sigma, DUO9210). Cells cultured on coverslips were washed with Dulbecco's PBS, fixed with 3.7% formaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 5 min, and then blocked with 10% goat serum for 1 h at room temperature. In situ proximity ligation was performed using two primary antibodies of different species targeting either Pfn1 (Abcam, 1:200, ab124904, rabbit) or actin (BD Biosciences, 1:100, 612656, mouse), both for 1 h at room temperature in 10% goat serum, and a pair of oligonucleotide-labeled secondary antibodies targeting each primary antibody from the Duolink kit according to the protocol of the manufacturer. PLA probe hybridization, ligation, amplification, and detection were performed according to the protocol of the manufacturer. To quantify PLA spots, images (acquired at an emission wavelength of 624 nm with a ×60 oil immersion objective (N.A. = 1.4) on an Olympus IX-71 inverted microscope) were first background-subtracted using the mean fluorescence intensity of cells from the negative control group. A mask was then created from the images of interest, and the Analyze Particles option was used to count the number of spots in each cell from a field of view.
Actin polymerization assay
Bacteria transformed with GST–Pfn1 were grown to optical density 0.6–0.9, induced with 0.1 mm isopropyl β-d-thiogalactopyranoside (Sigma) for 3 h and extracted with lysis buffer (25 mm Tris (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, and 5% glycerol). Following clarification by centrifugation at 18,000 × g for 30 min, cell lysates were mixed with reconstituted glutathione–agarose beads (Thermo Fisher) overnight. After washing the resin with a wash buffer (50 mm Tris and 150 mm NaCl (pH 8.0)), GST–Pfn1 was eluted using an elution buffer (50 mm Tris, 150 mm NaCl (pH 8.0), and 10 mm reduced glutathione). The eluted GST–Pfn1 was then dialyzed in the following buffer prior to use in the polymerization assay: 100 mm Tris (pH 7.4), 100 mm NaCl, 0.2 mm ATP, 5% sucrose, and 1% dextran. Actin polymerization with or without GST–Pfn1 in the presence of DMSO or compound was performed using a fluorescence actin polymerization kit (Cytoskeleton, BK003) according to the protocol of the manufacturer, downscaled to a 96-well format with a 10:1 ratio (9 μm unlabeled and 1 μm pyrene actin) between unlabeled and pyrene actin. The pyrene fluorescence signal was read using a fluorimeter with an excitation wavelength of 360 nm and an emission wavelength of 420 nm.
Phalloidin staining
Cells cultured on coverslips were washed with DPBS, fixed with 3.7% formaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 5 min, and then blocked with 10% goat serum for 1 h at room temperature. Cells were then incubated with ∼33 nm rhodamine–phalloidin (Life Technologies) for 1 h at room temperature. Stained cells were washed twice with PBS containing 0.02% Tween 20, twice with PBS, and once with distilled water before being mounted on a slide and imaged with a ×60 oil immersion objective (N.A. = 1.4) on an Olympus IX-71 inverted microscope. The same constant threshold was set between all fields with parameters that prevented overexposure; cell size and the average fluorescence intensity were scored on a cell-by-cell basis by manual tracing using ImageJ.
Live/dead staining
HmVECs were stained using a live/dead kit (Life Technologies) following the protocol of the manufacturer.
Cell proliferation assay
Five-thousand cells (HmVECs or VSMCs) were plated in the wells of a 24-well plate in duplicate on day 0 and cultured overnight before being subjected to either compound or vehicle treatment on day 1. Cells were trypsinized and counted on days 2 and 4. The medium was replenished on day 3 with appropriate treatment.
Statistics
Statistical tests were performed with either a one-way analysis of variance followed by Tukey's post hoc test or Student's t test when appropriate. p < 0.05 was considered to be statistically significant.
Author contributions
D. G. was involved in the design and execution of experiments and preparation of the manuscript. T. L. and L. M. were involved in the execution of the experiments and made intellectual contributions to the study. P. W. made intellectual contributions to the study and was involved in preparation of the manuscript. D. K. was involved in molecular modeling and preparation of the manuscript. P. R. was involved in the overall planning and execution of the study and preparation of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants 2R01CA108607 (to P. R.) and R01GM108340 (to D. K.) and a pilot grant (to D. K. and P. R.) from the University of Pittsburgh Clinical and Translational Institute (supported by the National Institutes of Health Grant UL1TR001857). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figures S1–S3 and Tables S1 and S2.
- Pfn
- profilin
- EC
- endothelial cell
- GST
- glutathione S-transferase
- C1
- compound 1
- SAR
- structure–activity relationship
- VEGF
- vascular endothelial growth factor
- PLA
- proximity ligation assay
- PLP
- polyproline
- VSMC
- vascular smooth muscle cell
- ECM
- extracellular matrix
- N.A.
- numerical aperture.
References
- 1. Ding Z., Bae Y. H., and Roy P. (2012) Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adh. Migr. 6, 442–449 10.4161/cam.21832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ding Z., Gau D., Deasy B., Wells A., and Roy P. (2009) Both actin and polyproline interactions of profilin-1 are required for migration, invasion and capillary morphogenesis of vascular endothelial cells. Exp. Cell Res. 315, 2963–2973 10.1016/j.yexcr.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding Z., Lambrechts A., Parepally M., and Roy P. (2006) Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 119, 4127–4137 [DOI] [PubMed] [Google Scholar]
- 4. Kullmann J. A., Neumeyer A., Gurniak C. B., Friauf E., Witke W., and Rust M. B. (2011) Profilin1 is required for glial cell adhesion and radial migration of cerebellar granule neurons. EMBO Rep. 13, 75–82 10.1038/embor.2011.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lambrechts A., Jonckheere V., Peleman C., Polet D., De Vos W., Vandekerckhove J., and Ampe C. (2006) Profilin-I-ligand interactions influence various aspects of neuronal differentiation. J. Cell Sci. 119, 1570–1578 10.1242/jcs.02884 [DOI] [PubMed] [Google Scholar]
- 6. Haugwitz M., Noegel A. A., Karakesisoglou J., and Schleicher M. (1994) Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell 79, 303–314 10.1016/0092-8674(94)90199-6 [DOI] [PubMed] [Google Scholar]
- 7. Schweinhuber S. K., Meßerschmidt T., Hänsch R., Korte M., and Rothkegel M. (2015) Profilin isoforms modulate astrocytic morphology and the motility of astrocytic processes. PLoS ONE 10, e0117244 10.1371/journal.pone.0117244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Böttcher R. T., Wiesner S., Braun A., Wimmer R., Berna A., Elad N., Medalia O., Pfeifer A., Aszódi A., Costell M., and Fässler R. (2009) Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 28, 1157–1169 10.1038/emboj.2009.58 10.1038/emboj.2009.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., and Lindberg U. (1993) The structure of crystalline profilin-β-actin. Nature 365, 810–816 10.1038/365810a0 [DOI] [PubMed] [Google Scholar]
- 10. Lawrence H. R., Li Z., Yip M. L., Sung S. S., Lawrence N. J., McLaughlin M. L., McManus G. J., Zaworotko M. J., Sebti S. M., Chen J., and Guida W. C. (2009) Identification of a disruptor of the MDM2-p53 protein-protein interaction facilitated by high-throughput in silico docking. Bioorg. Med. Chem. Lett. 19, 3756–3759 10.1016/j.bmcl.2009.04.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mukherjee P., Desai P., Zhou Y. D., and Avery M. (2010) Targeting the BH3 domain mediated protein-protein interaction of Bcl-xL through virtual screening. J. Chem. Inf. Model. 50, 906–923 10.1021/ci1000373 [DOI] [PubMed] [Google Scholar]
- 12. Chan D. S., Lee H. M., Yang F., Che C. M., Wong C. C., Abagyan R., Leung C. H., and Ma D. L. (2010) Structure-based discovery of natural-product-like TNF-α inhibitors. Angew. Chem. Int. Ed. Engl. 49, 2860–2864 10.1002/anie.200907360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koes D. R., and Camacho C. J. (2012) PocketQuery: protein-protein interaction inhibitor starting points from protein-protein interaction structure. Nucleic Acids Res. 40, W387–W392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ezezika O. C., Younger N. S., Lu J., Kaiser D. A., Corbin Z. A., Nolen B. J., Kovar D. R., and Pollard T. D. (2009) Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: insights from crystal structures of fission yeast profilin. J. Biol. Chem. 284, 2088–2097 10.1074/jbc.M807073200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zou L., Jaramillo M., Whaley D., Wells A., Panchapakesa V., Das T., and Roy P. (2007) Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br. J. Cancer 97, 1361–1371 10.1038/sj.bjc.6604038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koes D. R., and Camacho C. J. (2011) Pharmer: efficient and exact pharmacophore search. J. Chem. Inf. Model. 51, 1307–1314 10.1021/ci200097m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., and Coleman R. G. (2012) ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 52, 1757–1768 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gauvin T. J., Fukui J., Peterson J. R., and Higgs H. N. (2009) Isoform-selective chemical inhibition of mDia-mediated actin assembly. Biochemistry 48, 9327–9329 10.1021/bi901354z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan Y., Arif A., Gong Y., Jia J., Eswarappa S. M., Willard B., Horowitz A., Graham L. M., Penn M. S., and Fox P. L. (2012) Stimulus-dependent phosphorylation of profilin-1 in angiogenesis. Nat. Cell Biol. 14, 1046–1056 10.1038/ncb2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan Y., Potdar A. A., Gong Y., Eswarappa S. M., Donnola S., Lathia J. D., Hambardzumyan D., Rich J. N., Fox, and P. L. (2014) Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat. Cell Biol. 16, 445–456 10.1038/ncb2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moldovan N. I., Milliken E. E., Irani K., Chen J., Sohn R. H., Finkel T., and Goldschmidt-Clermont P. J. (1997) Regulation of endothelial cell adhesion by profilin. Curr. Biol. 7, 24–30 10.1016/S0960-9822(06)00024-8 [DOI] [PubMed] [Google Scholar]
- 22. Mouneimne G., Hansen S. D., Selfors L. M., Petrak L., Hickey M. M., Gallegos L. L., Simpson K. J., Lim J., Gertler F. B., Hartwig J. H., Mullins R. D., and Brugge J. S. (2012) Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell 22, 615–630 10.1016/j.ccr.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finkel T., Theriot J. A., Dise K. R., Tomaselli G. F., and Goldschmidt-Clermont P. J. (1994) Dynamic actin structures stabilized by profilin. Proc. Natl. Acad. Sci. U.S.A. 91, 1510–1514 10.1073/pnas.91.4.1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gau D., Veon W., Capasso T. L., Bottcher R., Shroff S., Roman B. L., Roy P. (2017) Pharmacological intervention of MKL/SRF signaling by CCG-1423 impedes endothelial cell migration and angiogenesis. Angiogenesis 20, 663–672 10.1007/s10456-017-9560-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rotty J. D., Wu C., Haynes E. M., Suarez C., Winkelman J. D., Johnson H. E., Haugh J. M., Kovar D. R., and Bear J. E. (2015) Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54–67 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergers G., and Hanahan D. (2008) and D. Hanahan, Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 10.1038/nrc2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schoors S., De Bock K., Cantelmo A. R., Georgiadou M., Ghesquière B., Cauwenberghs S., Kuchnio A., Wong B. W., Quaegebeur A., Goveia J., Bifari F., Wang X., Blanco R., Tembuyser B., Cornelissen I., et al. (2014) Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab. 19, 37–48 10.1016/j.cmet.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 28. Goveia J., Stapor P., and Carmeliet P. (2014) Principles of targeting endothelial cell metabolism to treat angiogenesis and endothelial cell dysfunction in disease. EMBO Mol. Med. 6, 1105–1120 10.15252/emmm.201404156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carmeliet P., and Jain R. K. (2000) Angiogenesis in cancer and other diseases. Nature 407, 249–257 [DOI] [PubMed] [Google Scholar]
- 30. Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., and Simmerling C. (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koes D. R., Baumgartner M. P., and Camacho C. J. (2013) Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 53, 1893–1904 10.1021/ci300604z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., and Hutchison G. R. (2011) Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.