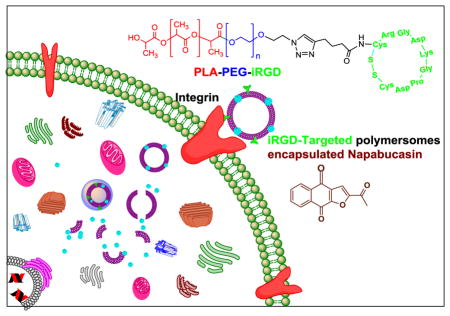
Abstract
Often cancer relapses after an initial response to chemotherapy because of the tumor’s heterogeneity and the presence of progenitor stem cells, which can renew. To overcome drug resistance, metastasis, and relapse in cancer, a promising approach is the inhibition of cancer stemness. In this study, the expression of the neuropilin-1 receptor in both pancreatic and prostate cancer stem cells was identified and targeted with a stimuli-responsive, polymeric nanocarrier to deliver a stemness inhibitor (napabucasin) to cancer stem cells. Reduction-sensitive amphiphilic block copolymers PEG1900-S-S-PLA6000 and the N3-PEG1900-PLA6000 were synthesized. The tumor penetrating iRGD peptide-hexynoic acid conjugate was linked to the N3-PEG1900-PLA6000 polymer via a Cu2+ catalyzed “Click” reaction. Subsequently, this peptide-polymer conjugate was incorporated into polymersomes for tumor targeting and tissue penetration. We prepared polymersomes containing 85% PEG1900-S-S-PLA6000, 10% iRGD-polymer conjugate, and 5% DPPE-lissamine rhodamine dye. The iRGD targeted polymersomes encapsulating the cancer stemness inhibitor napabucasin were internalized in both prostate and pancreatic cancer stem cells. The napabucasin encapsulated polymersomes significantly (p < 0.05) reduced the viability of both prostate and pancreatic cancer stem cells and decreased the stemness protein expression notch-1 and nanog compared to the control and vesicles without any drug. The napabucasin encapsulated polymersome formulations have the potential to lead to a new direction in prostate and pancreatic cancer therapy by penetrating deeply into the tumors, releasing the encapsulated stemness inhibitor, and killing cancer stem cells.
Keywords: Prostate cancer stem cells, pancreatic cancer stem cells, neuropilin-1, iRGD peptide, napabucasin, polymersomes, cancer stemness inhibitor
Graphical abstract
1. Introduction
Cancer stem cells which contribute to tumor heterogenicity are a subpopulations of cells with tumor-initiating capability.[1] These cells have the stem-like properties of normal stem cells, such as self-renewal [2] and multi-lineage differentiation.[3] It is hypothesized that cancer stem cells might be the origin of solid tumors of prostate, colon, breast, and lung.[4] Cancer stem cells express both embryonic stem cells markers (Nanog, Oct4, and Sox-2) and progenitor cell markers (CD33, CD44, and Nestin).[5] Pancreatic cancer stem cells isolated from pancreatic tumors were defined by the CD133+[6] and the CD44+ CD24+ EPCAM+ cells.[7] Prostate cancer stem cells isolated from prostate tumors displayed CD44, CD 133, SSEA3/4, and Oct4 markers (http://www.celprogen.com). Cancer stem cells are mostly responsible for the metastasis, recurrence, and drug resistance in cancer.[8]
Napabucasin (BBI608) is a cancer stemness inhibitor that interferes with gene transcription through the STAT3-dependent pathway. In vitro and in vivo studies demonstrated the ability of napabucasin to inhibit cell proliferation and induce cell death.[1, 9, 10] Napabucasin is currently in Phase III clinical trials for the treatment of gastric, colon, and pancreatic cancers (www.clinicaltrials.gov).
Stimuli-responsive nanoparticles respond to an altered physical or chemical microenvironment in the tumor and release their encapsulated contents only when the abnormalities are encountered.[11] In this study, we have used reduction sensitive polymersomes to deliver napabucasin to prostate and pancreatic cancer stem cells. Polymersomes are bilayer vesicles prepared from amphiphilic block copolymers. These block copolymers spontaneously form a vesicular structure in an aqueous solution, with an aqueous core, enabling the encapsulation of hydrophobic drugs in the bilayer and hydrophilic drugs in the core.[11] Polymersomes offer several advantages as drug carriers over liposomes. Due to the higher molecular weights, polymersomes are more stable compared to the liposomes. The use of polyethylene glycol (PEG) as the hydrophilic block renders the vesicles long circulating and biocompatible. The size of the vesicles and the thickness of the bilayer can be controlled by the molecular weights of the hydrophobic and the hydrophilic blocks of the amphiphilic copolymers. [12]
To target and penetrate the tumor, we have used the reported cell penetrating cyclic iRGD peptide. This peptide interacts with the integrin and neuropilin receptors on the cancer cell surface.[13] The C-terminal of the peptide that stimulates the neuropilin-1 receptor and vascular permeability is known as the C-end rule (CendR).[13] The neuropilin-1 receptor is expressed in cancer and vascular endothelial cells and contributes to tumor progression and angiogenesis.[14] Nanoparticles, antibodies, and drugs conjugated to the iRGD peptide were observed to accumulate in the tumors both in vitro and in animal studies.[15]
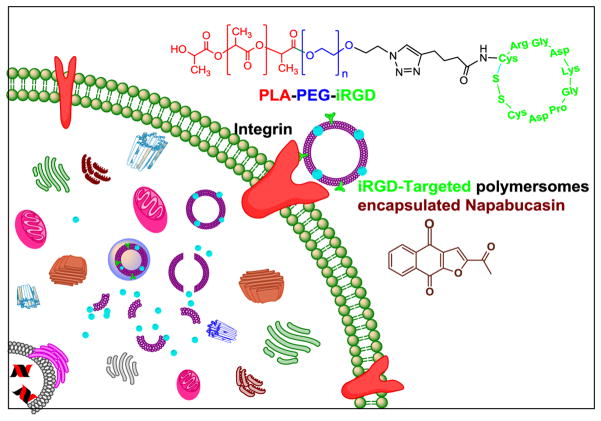
Our goal is to develop a unique, polymeric drug delivery system to target cancer stem cells. In this study, the expression of neuropilin-1 receptor on the surface of both pancreatic and prostate cancer stem cells was identified; thus the cyclic iRGD peptide was synthesized and conjugated with a polymer. We prepared the iRGD targeted, reduction sensitive polymersomes encapsulating the cancer stemness inhibitor napabucasin. We hypothesized that iRGD peptide decorated polymersomes would penetrate a solid tumor and internalize in cancer stem cells. Subsequently, the high reducing agent concentration in the cytosol will reduce the disulfide linker of the amphiphilic polymer, disturb the bilayer structure of the vesicles, and release the encapsulated napabucasin (Figure 1).[11] We observed that iRGD-targeted polymersomes encapsulating napabucasin significantly (p < 0.05) decreased the viability of prostate and pancreatic cancer stem cell microtumors compared to the controls. The cancer stemness marker proteins also decreased after treatment with iRGD-targeted polymersomes encapsulating napabucasin.
Fig 1.
Illustration of the internalization of iRGD peptide-targeted, napabucasin encapsulated polymersomes in the cancer stem cells.
2. Materials and methods
2.1. Synthesis and characterization of Hex-iRGD Peptide
The iRGD peptide was synthesized by using a microwave assisted, solid phase peptide synthesizer (Liberty Blue, CEM Corporation) using commercially available CLEAR-amide resin (Peptides International), and synthesized the peptide was synthesized at a 0.2 mM scale. The peptide sequence was hexynoic acid-Cyclo (Arg-Gly-Asp-Lys-Gly-Pro-Asp-Asp-Cys). After the solid phase synthesis, the resin was washed with 15 mL acetone, centrifuged for 10 minutes at 2000 RPM and kept in a desiccator overnight. Subsequently, the resin was stirred with thallium trifluoroacetate (0.1 mmol) in dimethylformamide at room temperature for 3 hours to cyclize the peptide on the resin. The resin was filtered and washed with a mixture of DMF and dichloromethane (3X), and the residue was collected. Next, the peptide was cleaved from the resin with trifluoroacetic acid (19 mL), triisopropylsilane (0.5 mL), and distilled water (0.5 mL) at room temperature for 2 hours. The resin was filtered, and ice-cold diethyl ether was added to the filtrate. Subsequently, the precipitate was filtered, and dried in a vacuum desiccator. The peptide was characterized using mass spectrometry and circular dichroism spectroscopy.
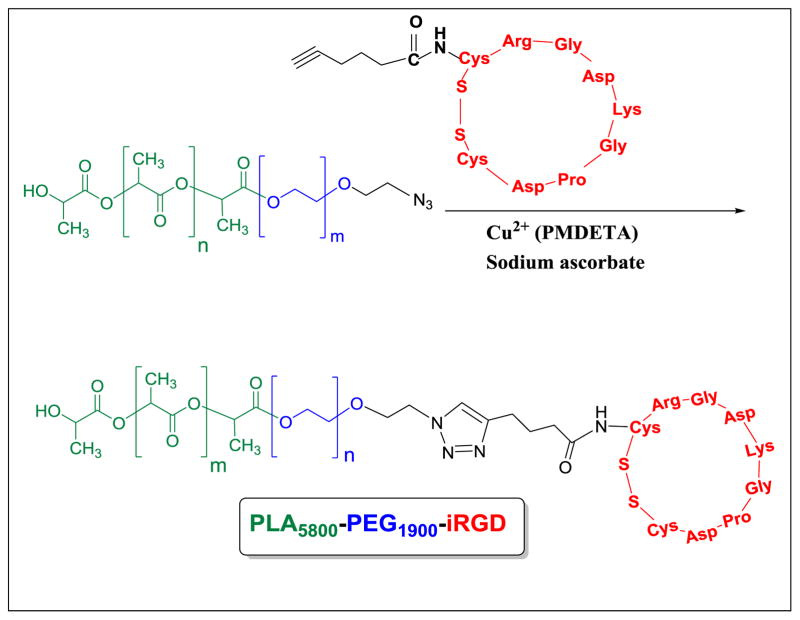
2.2. Synthesis of iRGD-hex-N3-PEG1900-PLA6000 polymer conjugate
We synthesized N 3-PEG1900-PLA6000 polymers following a reported protocol from our laboratory.[16] To conjugate the iRGD-hexynoic acid to N3-PEG1900-PLA6000 polymer, we used the Click chemistry ([2+3]-cycloaddition).[17] Briefly, the N3-PEG1900-PLA6000 polymer (20 mg) and iRGD-hexynoic acid (2 mg) were dissolved in 5 mL of deionized water. The reaction was catalyzed by 400 μL copper (II) sulfate complex (0.053 M) and 400 μL of sodium ascorbate (27 mg/mL). The copper (II) complex was papered by mixing the copper (II) sulfate (0.53 mmol) and pentamethyl diethylenetriamine (2 mmol). The solution was stirred at room temperature for 3 hours, and 4 mL of water was added to produce the final concentration of 0.053M. The reaction was stirred at room temperature for 24 hours. The compound was dialyzed against water (molecular weight cutoff: 1,000) for 48 hours. The product was characterized by FT-IR and circular dichroism spectroscopy (Supporting Information).
2.3. Preparation of iRGD-targeted and non-targeted polymersomes encapsulating napabucasin
The iRGD-targeted polymersomes encapsulating napabucasin were prepared from PEG1900-S-S-PLA6000 (85%), iRGD-PEG1900-PLA6000 (10%), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl ammonium salt (LR, 5%). The non-targeted vesicles encapsulating napabucasin contained PEG1900-S-S-PLA6000 (95%) and LR (5%). The polymers were dissolved in THF (9 mg/mL for the disulfide polymer, 2 mg/mL for the iRGD-PEG1900-PLA6000, 3 mg/mL for napabucasin and 0.01 mg/mL LR in chloroform). The chloroform solution (1.1 mL) of LR lipid was evaporated to form a thin film. The polymers and napabucasin (200 μL, 2.5 mg/mL) were added to the thin layer film. The final mixture was added dropwise to an aqueous solution (10 mM HEPES buffer, pH 7.4) and stirred for 1 hour. To remove the organic solvents, air was passed through the mixture for 45 minutes. Subsequently, the polymersomes were sonicated in a bath for 60 minutes (Symphony 117 V, 60 Hz, Power 9). Then polymersomes (1 mg/mL) were filtrated through a Sephadex™ G-100 size exclusion column to remove the unencapsulated drug. Polymersomes drug loading efficacies (DLE) were determined by UV–Vis spectroscopy. After passing through the size-exclusion column, the absorption of the polymersomes was recorded at 235 nm.
2.4. Preparation of control polymersomes
Control polymersomes were prepared by using PEG1900-S-S-PLA6000 (95%) and LR (5%). The thin film of LR was prepared by evaporating chloroform solution of the dye. Then, a solution of the PEG1900-S-S-PLA6000 polymer in THF (9 mg/mL, 160 μL) was added slowly to the thin film, and the mixture was added dropwise to a stirred aqueous buffer solution (10 mM HEPES buffer, pH 7.4). The resultant polymersomes were stirred for 45 minutes at room temperature, and air was passed for 45 minutes through the mixture to remove the organic solvent. The polymersomes were sonicated for 60 minutes (Symphony 117 V, 60 Hz) and passed through a Sephadex G100 (GE Healthcare) size exclusion column to collect lissamine rhodamine B dye incorporated vesicles. These control polymersomes were used for cell viability assays.
2.5. Polymersomes’ size analysis
The iRGD-targeted and the non-targeted polymersomes encapsulating napabucasin, and the control polymersomes were characterized by dynamic light scattering at 90° using a Zeta Sizer Nano ZS 90 (Malvern Instrument). The polymersomes were equilibrated for 2 minutes; five measurements were recorded with 10 repeats each.
2.6. Atomic Force Microscopic (AFM) imaging
The samples were prepared by incubating 10 μL (1 mg/mL) of each polymersome solution on silicon substrates for 10 minutes in a closed container to prevent evaporation at room temperature. Then, the samples were rinsed with de-ionized water (Millipore) and dried under purified air flow. A commercial atomic force microscope (NT-MDT NTEGRA AFM) was used to perform the imaging measurements under ambient conditions in semi-contact mode with a resonant frequency of 190 kHz AFM probes (Budget sensors).
2.7. Release studies
Napabucasin release from polymersomes was monitored in the presence of different concentrations of glutathione (GSH) as a function of time. The polymersomes encapsulating napabucasin (500 μL of 1 mg/mL solution) was placed in Spectra/Por Float-A-Lyzer G2 Dialysis Tubes, MWCO 500–1000 Da. Subsequently, glutathione was added to make the final concentration of 2 μM, 50 μM, 1 mM, and 5 mM. The absorbance of fluid outside of dialysis tubes was measured at 235 nm using a UV-VIS spectrophotometer (Spectramax M5, Molecular Devices) every 5 minutes. The percent release was calculated from the calibration curve. After the release study, the polymersomes were imaged using atomic force microscopy.
2.8. Cell culture
Human prostate and pancreatic cancer stem cells were purchased from Celprogen. Prostate cancer stem cells were maintained in human prostate cancer stem cell complete growth media with serum and antibiotics and pancreatic cancer stem cell were maintained in human pancreatic cancer stem cell complete growth media with serum and antibiotics from Celprogen. Human prostate and pancreatic cancer stem cell extracellular matrix-coated plates and flasks (Celprogen) were used in all experiments. The cell culture flasks were maintained in an incubator at 37 °C in a 5% CO2 atmosphere. Cell passages between 3–6 were used in all the experiments.
2.9. Uptake studies employing monolayer and three-dimensional spheroids
Prostate and pancreatic cancer stem cells (5×103) were seeded in 12-well cancer stem cell extracellular matrix-coated plates for prostate and pancreatic cancers 24 hours before the experiments. Once the culture was 80–90% confluent, the iRGD-targeted (20 μL) and the control polymersomes (20 μL) were incubated with either prostate or pancreatic cancer stem cells for 3 hours. Then, the cell culture media and treatment were removed, and the cells were washed three times with PBS to remove the non-internalized vesicles. The cell nuclei were stained with HOESCHT 33342 dye (1:1000 dilution) and imaged using the 20X objective of a Lucia fluorescence microscope.
For the uptake experiments in the three-dimensional (3D) spheroids, the 35-well 3D petri dishes (Microtissues) were used to prepare prostate and pancreatic cancer stem cells spheroids. Agarose solution (2% w/v) was prepared and autoclaved. Prostate and pancreatic cancer stem cells suspensions (1×104cells in 70 μL media) were then added to each 3D scaffold. Prostate and pancreatic cancer stem cell spheroids were allowed to grow for 2–3 days. Then iRGD-targeted (20 μL) and control polymersomes (20 μL) were incubated with either prostate or pancreatic cancer stem cell spheroids for 7 hours. After incubation cell culture media and polymersomes were removed, and the cells were washed three times with PBS to remove the non-internalized vesicles. Cell nuclei were stained with HOESCHT 33342 dye (1:1000 dilution). Subsequently, the spheroids were cut into 15 μm sections and placed on slides. Images of spheroids were acquired with a Zeiss AxioObserver Z1 microscope equipped with LSM700 laser scanning module (Zeiss, Thornwood, NY), at 40X magnification with 40x/1.3 Plan-Apochromat lens using a 590 nm solid-state laser for Lissamine rhodamine B and a 405 nm lasers for DAPI excitation. Following the acquisition of 3D microscopy, images were imported to Imaris 8.3 (Bitplane) software where 50 microns thick computer-generated representation of the spheroids were created. (Supporting Information).
2.10. Viability of prostate and pancreatic cancer stem cells in monolayer cultures
Prostate and pancreatic cancer stem cells were seeded at a density of 103/well in a 96-well human prostate and pancreatic cancer stem cell extracellular matrix-coated plates. Once they reached 90% confluency, the plate was divided into five groups: control, control polymersomes, polymersomes encapsulating napabucasin, iRGD-targeted polymersomes encapsulating napabucasin, and free drug (napabucasin). In the control group, we added only the media. Cells treated with napabucasin received 1 μM and 4 μM of free and an equivalent amount of encapsulated drug in iRGD-targeted and non-targeted polymersomes. The cells were treated for 48 hours at 37 °C, in a 5% CO2 atmosphere. Subsequently, the cells were washed with sterile PBS twice and replaced with 200 μL fresh media. Then 20 μL Alamar Blue was added to all the wells and fluorescence was recorded at excitation 565 nm and emission at 590 nm after 4 hours. The data presented are normalized to the control.
2.11. Viability of prostate and pancreatic cancer stem cells in spheroids
Prostate and pancreatic cancer stem cell spheroids were prepared by using 35-well 3D petri dishes (Microtissues). Once the microtumors formed (after 3–5 days), the plates were divided into five groups: control, control polymersomes, non-targeted polymersomes encapsulating napabucasin, iRGD-targeted polymersomes encapsulating napabucasin, and free drug (napabucasin). We treated the spheroids for 48 hours at 37 °C, in a 5% CO2 atmosphere. After 48 hours, the solutions were removed, and the microtumors were washed twice with PBS. The spheroids were incubated with 100 μL of recombinant trypsin (TryPLE, Life Technologies) for 10 minutes. Then spheroids were removed and subjected to the Alamar Blue assay. The data presented are normalized to the control.
2.12. Western blotting
The prostate cancer stem cells were treated with different formulations of polymersomes (buffer encapsulated, napabucasin encapsulated, iRGD targeted with napabucasin encapsulation, and free napabucasin) for 5 hours. Subsequently, the cells were collected and lysed in RIPA buffer with complete protease inhibitor cocktail (Roche), and 0.1% sodium dodecyl sulfate (SDS). Protein concentration in each sample was verified by Bio-Rad DC protein assay. Protein samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in 5% non-fat milk in TRIS buffer containing 1% Tween 20 (1X) for 30 minutes and then incubated with the primary antibody (1:1000 dilution in 5% non-fat milk) overnight and subsequently with appropriate secondary antibody (1:2000). ECL reagents (Pierce, Rockford, IL, USA) were used to developed signals and exposed to X-ray films. The anti-notch-1, Nanog, and GAPDH polyclonal antibody were purchased from Cell Signaling Technology and Neuropilin-1 monoclonal antibody from Santa Cruz Biotechnology.
2.13. Cell apoptosis assay by flow cytometry
After treatment of both prostate and pancreatic cancer stem cells with different formulations of polymersomes for 48 hours, the cells were collected and washed with PBS and diluted in annexin-binding buffer to 1 × 106 cells in 0.5 mL. Then cells were stained with Annexin V-FITC and PI (5 μL) for 10 minutes in the dark at room temperature. After incubation, samples were analyzed by flow cytometry using an Accuri C6 flow cytometer. Apoptosis was determined by data analysis using FlowJo software (FlowJo, LLC).
2.14. Statistical analysis
Graphpad Prism 7 software was used to perform statistical analysis. All the results presented are representative of at least four independent experiments. Error bars denote the mean ± SEM. Statistical analysis: Student’s t-test was used to find the significance between two groups, where significance *p < 0.05.
3. Results and discussion
3.1. iRGD peptide characterization
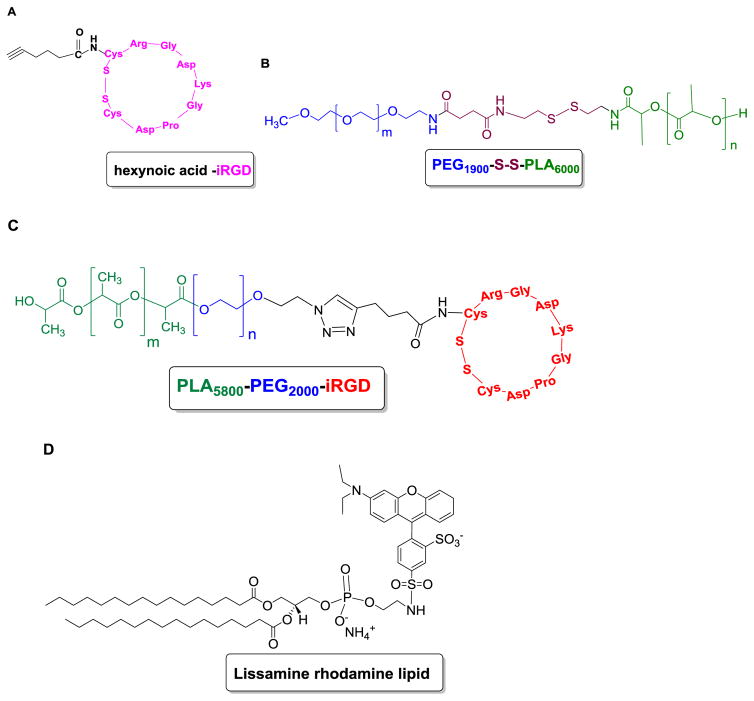
To prepare iRGD targeted polymersomes encapsulating napabucasin, we synthesized the cyclic iRGD peptide conjugated to hexynoic acid (Figure 2A) using a solid phase microwave assisted peptide synthesizer. The synthesized hexynoic acid conjugated cyclic iRGD was characterized by mass spectrometry (Supporting Information).
Fig 2. Structure of synthesized polymers and the fluorescent lipid.
(A) Structure of the synthesized cyclic iRGD peptide conjugated to hexynoic acid, (B) PEG1900-S-S-PLA6000, (C) PLA6000-PEG1900-iRGD polymer, and (D) 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl ammonium salt (commercially available).
3.2. Polymer synthesis and polymersome formation
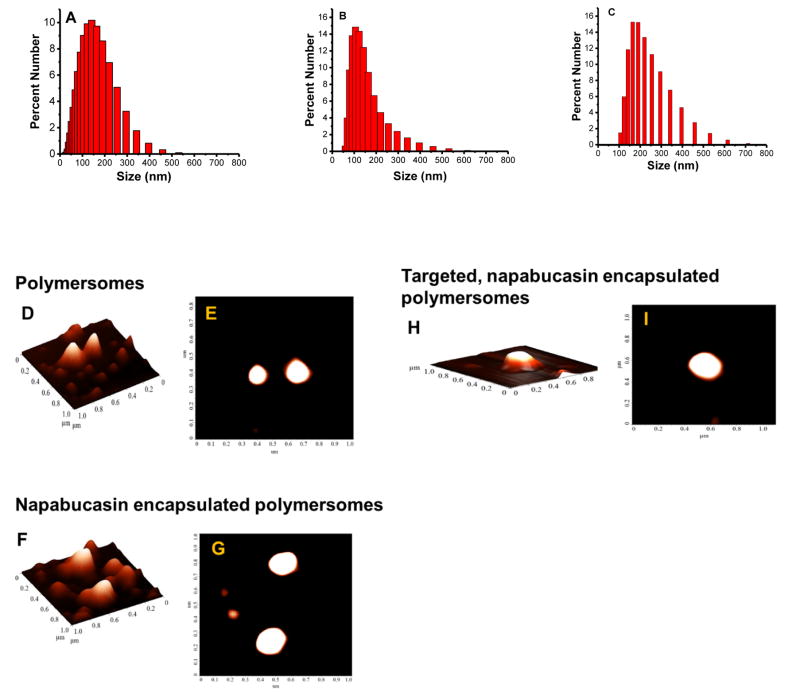
The redox-sensitive polymer PEG1900-S-S-PLA6000 (Figure 2B) was synthesized as previously reported.[11] We employed the Click reaction [2+3 cycloaddition] to conjugate the cyclic iRGD peptide to the N3-PEG1900-PLA6000 polymer. The resultant iRGD-PEG1900-PLA6000 (Figure 2C) was characterized by FT-IR and circular dichroism spectroscopy (Supporting Information). We prepared the targeted polymersomes from the redox-sensitive polymer PEG1900-S-S-PLA6000 (85%), iRGD-PEG1900-PLA6000 (10%), and DPPE–lissamine rhodamine lipid (LR, 5%) using the solvent exchange method.[11] The hydrophobic PLA block of the amphiphilic polymers forms the bilayer structure and the hydrophilic PEG block interacts with the aqueous phase inside and outside of the vesicles. [18] The LR lipid dye was used to visualize the polymersomes. We anticipated that the hydroprobic dipalmitoyl chains of the amphiphilic dye would partition into the hydrophobic polymer bilayer of the vesicles, exposing the hydrophilic lissamine rhodamine moiety to the aqueous phase. [19] The hydrodynamic diameters of the polymersomes were determined by dynamic light scattering (Figure 3A – C) and the spherical structures confirmed by atomic force microscopy (Figure 3D – I). The stemness gene transcription inhibitor napabucasin [9, 10] was encapsulated in the polymersomes with an efficiency of (67 ± 6%). We observed that the iRGD-targeted and the non-targeted polymersomes encapsulating napabucasin had hydrodynamic diameters of 220 ± 10 nm (polydispersity index, PDI 0.2 ± 0.02) and 220 ± 5 nm (PDI 0.2 ± 0.01) respectively. The average hydrodynamic diameter for the control polymersomes was 137 ± 10 nm with a PDI of 0.2 ± 0.02. We observed the targeted and non-targeted polymersomes encapsulating napabucasin were larger than the control polymersomes. It is hypothesized that the incorporation napabucasin in the hydrophobic bilayer of the vesicles increases the size. The cyclic iRGD–PEG1900–PLA6000 polymer in polymersomes composition targets the neuropilin-1 receptor. The Cend R motif on iRGD peptide is hypothesized to enable them to penetration into microtumors (through the neuropilin-1 receptor) and internalization in the cells. The disulfide bond in the redox-sensitive polymer will subsequently be reduced in the cytosol, releasing the encapsulated cancer stemness inhibitor napabucasin.
Fig 3. The hydrodynamic diameters of the polymersomes by DLS (A–C) and AFM (D–I).
(A) Polymersomes, (B) polymersomes encapsulating napabucasin, and (C) peptide-targeted polymersomes encapsulating the stemness inhibitor napabucasin, as determined by dynamic light scattering. Atomic force microscopy (AFM) images of a different formulation of polymersomes (Panels D – I). The top (Panels E, G, and I) and the side views (Panels D, F, and H) for each of the formulations (control, napabucasin encapsulated, and iRGD peptide targeted, napabucasin encapsulated) are shown.
Release of napabucasin from the polymersomes and structural characterization
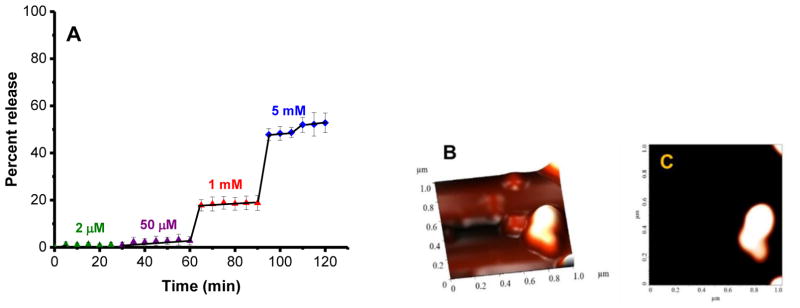
The reduction triggered release of napabucasin from the polymersomes was investigated with various concentrations of glutathione (GSH) as a function of time (Figure 4A). GSH is the most abundant tripeptide (γ-glutamyl-cysteinyl-glycine) in the cells. It has a major role in protecting cells from reactive oxygen species (ROS), oxidant-induced toxicity, toxins, mutagens, and drugs.[20] The polymersomes released 57% of the encapsulated napabucasin in the presence of 1–5 mM GSH (mimicking the cytosol[11]) within 2 hours (Figure 4A, red and blue traces). However, less than 2% release of napabucasin was observed with 50 μM GSH (mimicking the extracellular environment of cancer cells[11], Figure 4A, purple trace). The release of napabucasin from polymersomes was not observed in the presence of 2 μM GSH (mimicking the reducing agent concentration in the blood[11], green trace, Figure 4A). Atomic force microscopy after the release of napabucasin revealed that the morphology and structure of polymersomes were distorted (Figure 4B, C).
Fig 4.
(A) Reduction-mediated release pro le of encapsulated napabucasin from the polymersomes. The drug-encapsulated vesicles were treated with 2 μM (green), 50 μM (purple), 1 mM (red), and 5 mM (blue) concentrations of GSH. The lines connecting the data points are also shown. (B, C) Structural characterization of the polymersomes after release study employing atomic force microscopy.
3.3. Cellular internalization of polymersomes
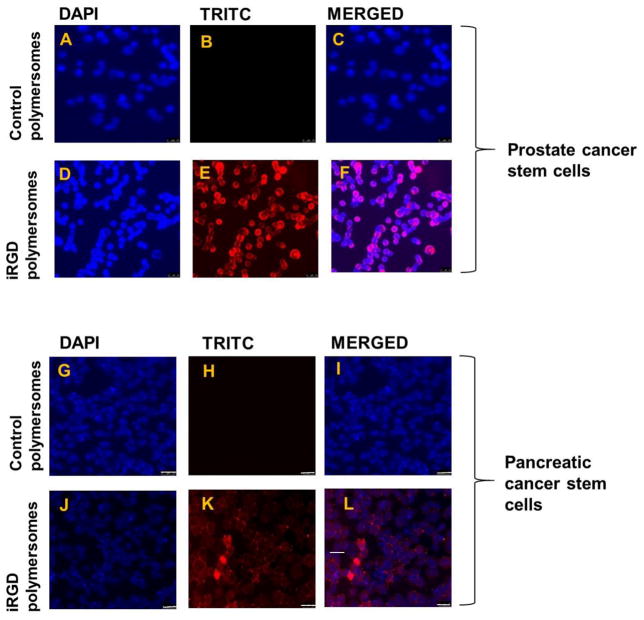
To investigate whether the iRGD peptide has any advantage for internalization in the cancer stem cells, we prepared the iRGD targeted and non-targeted control polymersomes incorporating 5% of the DPPE–lissamine rhodamine lipid (structure shown in Figure 2). DPPE–lissamine rhodamine lipid dye were used for visualizing the polymersomes employing fluorescence microscopy. We incubated the prostate and pancreatic cancer stem cells (monolayer cultures) with iRGD targeted and control polymersomes for 3 hours, the cell nuclei were stained with the HOESCHT 33342 dye and imaged employing a fluorescence microscope. Red fluorescence (from the lissamine rhodamine dye) was observed inside both prostate (Figure 5D–F) and pancreatic cancer stem cells (Figure 5J–L), indicating cellular internalization of the iRGD-decorated polymersomes. However, the control polymersomes without the iRGD peptide did not internalize in the cells (Figure 5A–C, and G–I).
Fig 5. Cellular uptake studies in monolayer and microtumor cultures of prostate and pancreatic cancer stem cells.
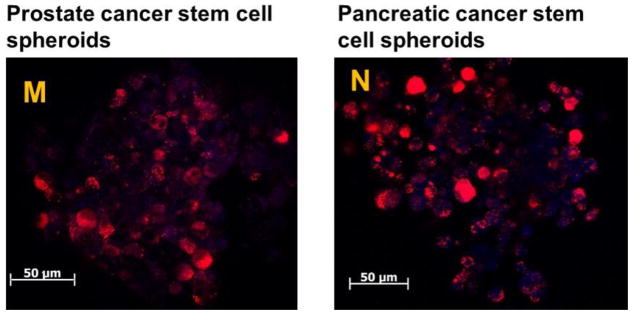
Panels (A–F): prostate cancer stem cells (scale bar: 25 μm), and Panels (G–L): pancreatic cancer stem cells (scale bar: 25μm). (M, N) Cellular uptake studies in a microtumor slice 200 μm from the surface of prostate cancer stem cells (M) and pancreatic cancer stem cell spheroids (N). The co-localization of the red fluorescence from the lissamine rhodamine dye and the blue from HOECHST 33342 indicated that peptide-targeted polymersomes were internalized in the cells. The blue fluorescence from the HOECHST 33342 dye indicates the cell nuclei.
To investigate the depth of the penetration of iRGD targeted polymersomes, three-dimensional spheroids of prostate and pancreatic cancer stem cells were prepared using the 35-well 3D petri dishes (Microtissues). The microtumors fully formed after 3–4 days. The microtumors were incubated with the iRGD targeted polymersomes for 8 hours. Subsequently, the microtumors were washed with sterile PBS, and the nuclei were stained with HOESCHT 33342. Finally, microtumors were frozen and sliced with a microtome into 15 μm sections. Red fluorescence from the polymersome-incorporated LR dye was observed in both prostate and pancreatic cancer stem cells colocalized with the blue nuclear stain in a slice 200 μm from the surface. These images show iRGD targeted polymersomes penetrating at least 200 μm into microtumors of prostate and pancreatic cancer stem cells (Figure 5M, N).
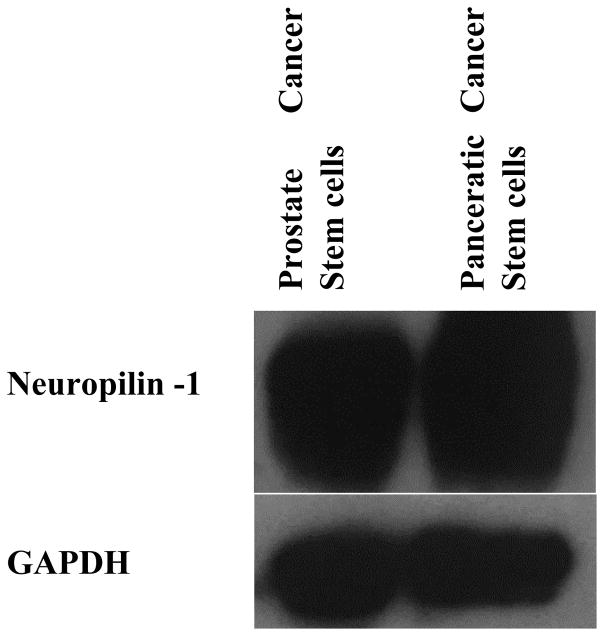
3.4. Neuropilin-1 expression on the cancer stem cells
Neuropilin-1 (NR-1) is a 130–140 kDa type I transmembrane protein [21] that interacts with an isoform of vascular endothelial growth factor (VEGF). It is an endothelial cell-specific mitogen and acts as a coreceptor for class 3 semaphorins. The neuropilin receptor has roles in cell survival, migration, and invasion [22] Expression of NR-1 in the epithelial cells of uterus, endometrium, lung, and kidney have been reported.[21] Glinka at el showed that NR-1 is expressed in breast cancer stem-like cells, activates the NF-KB, and aids in the tumor spheroids formation.[23] Since there was no prior report, the expression of the neuropilin-1 receptor on prostate and pancreatic cancer stem cells were investigated by employing western blotting. Prostate and pancreatic cancer stem cells where shown to express the neuropilin-1 receptor (Figure 6). Hence, it is likely that that the cellular internalization of the iRGD-targeted polymersomes (Figure 5) is mediated by the neuropilin-1 receptor on the surface of the cancer stem cells.
Fig 6.
Expression of neuropilin-1 in prostate and pancreatic cancer stem cells as determined by Western Blotting.
3.5. Viability of prostate and pancreatic cancer stem cells in monolayer cultures
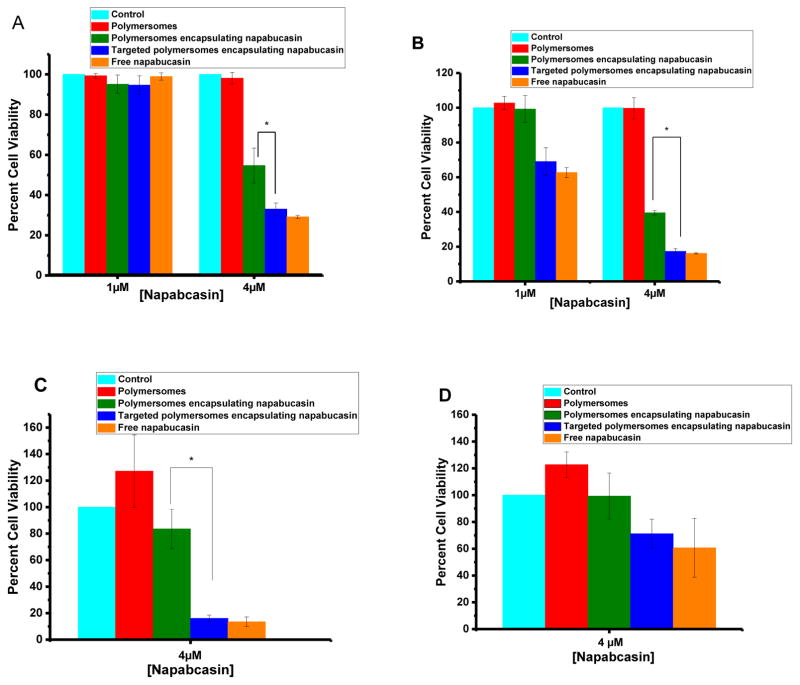
After validating the expression of neuropilin-1 on the cancer stem cells, the effectiveness of the peptide-targeted, drug-encapsulated polymersomes was determined in both prostate and pancreatic cancer stem cells. The monolayer cultures of both prostate and pancreatic cancer stem cells were treated with the polymersomes, polymersomes encapsulating napabucasin, peptide-targeted napabucasin-encapsulating polymersomes, and the free drug (napabucasin) for 48 hours. The cell viability was determined by the Alamar Blue assay (Figure 7A, B). The result shows that the targeted polymersomes encapsulating napabucasin significantly (p<0.5) reduced cell viability in both prostate and pancreatic cancer stem cells (30% and 19%, respectively) compared to the control polymersomes and vesicles encapsulating napabucasin. We also observed that increasing the concentration of encapsulated napabucasin from 1μM to 4 μM led to a pronounced decrease in the cell viability (Figures 7A and B).
Fig 7. The viability of prostate and pancreatic cancer stem cells in monolayer and spheroid cultures.
(A) Monolayer cultures of prostate cancer stem cells, (B) monolayer cultures of pancreatic cancer stem cells, (C) microtumors of prostate cancer stem cells, and (D) microtumors of pancreatic cancer stem cells in cultures. Cell viability with media only (control, cyan bar), polymersomes (red bar), non-targeted polymersomes encapsulating napabucasin (green bar), peptide-targeted polymersomes encapsulating napabucasin (blue bar), and free napabucasin (orange bar, N = 4).
3.5. Viability of prostate and pancreatic cancer stem cells in microtumors
Microtumors of prostate and pancreatic cancer stem cells were prepared in 35-well 3D petri dishes. The formed microtumors were randomly assigned to five groups: control, polymersomes, non-targeted polymersomes encapsulating napabucasin, peptide-targeted polymersomes encapsulating napabucasin, and free napabucasin. The cultured microtumors were treated for 48 hours, and subsequently, Alamar Blue assay was performed to determine the cell viability. Targeted-polymersomes encapsulating napabucasin were observed to reduce the viability of prostate cancer stem cells to 19% and pancreatic cancer stem cell to 65% (Figure 7C and D). The significantly reduced cell viability compared to the control groups indicated that the peptide-targeted polymersomes could penetrate the spheroids through the neuropilin-1 receptor and internalize in the cells. The interaction between CendR motif and NR-1 contributed to the cellular internalization of the targeted polymersomes.[24] Furthermore, the cytotoxicity increased when the vesicles were targeted with the cyclic iRGD peptide, which facilitates the internalization of polymersomes into the cells. However, the increased effectiveness of the targeted polymersomes for the prostate cancer stem cells compared to the pancreatic cancer stem cells remains unexplained.
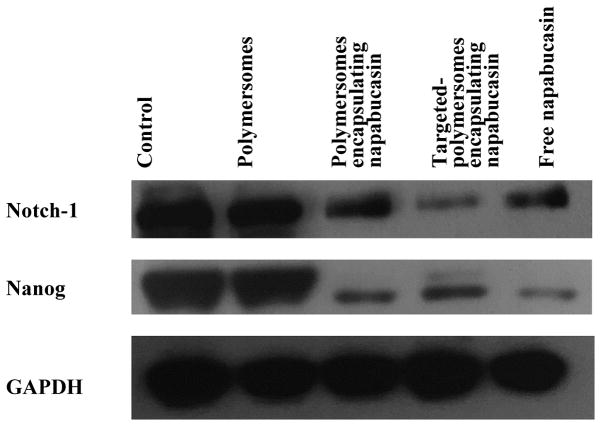
3.6. Cancer stemness protein expression
The stemness protein expression of prostate cancer stem cells was investigated by western blot. Proteins were extracted from the prostate cancer stem cells after treatment with a different formulation of polymersomes. The expression of cancer stemness markers such as Notch-1 and Nanog were significantly decreased after treatment with polymersomes encapsulating napabucasin, targeted polymersomes encapsulating napabucasin, and free napabucasin compared to the control groups (Figure 8 and Supplementary Data). Zhang and Li at el also reported the decreased expression of nanog after treatment with napabucasin.[1, 9]
Fig 8.
Effect of cancer stemness inhibitor (napabucasin) on the expression of two stemness marker proteins, Notch and Nanog.
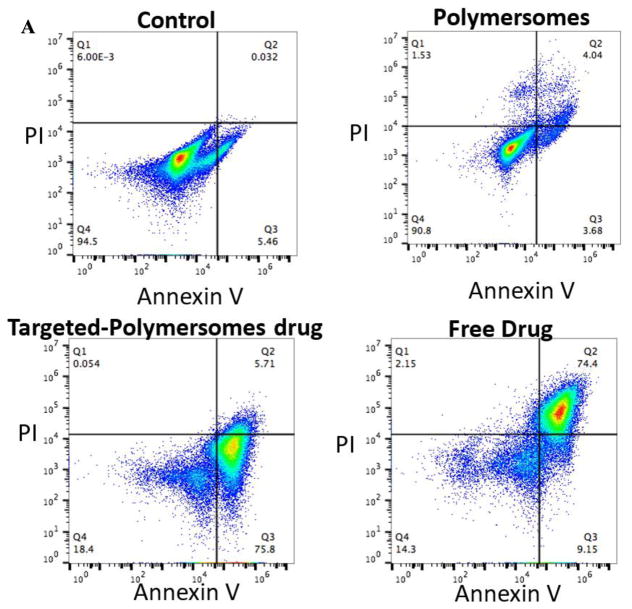
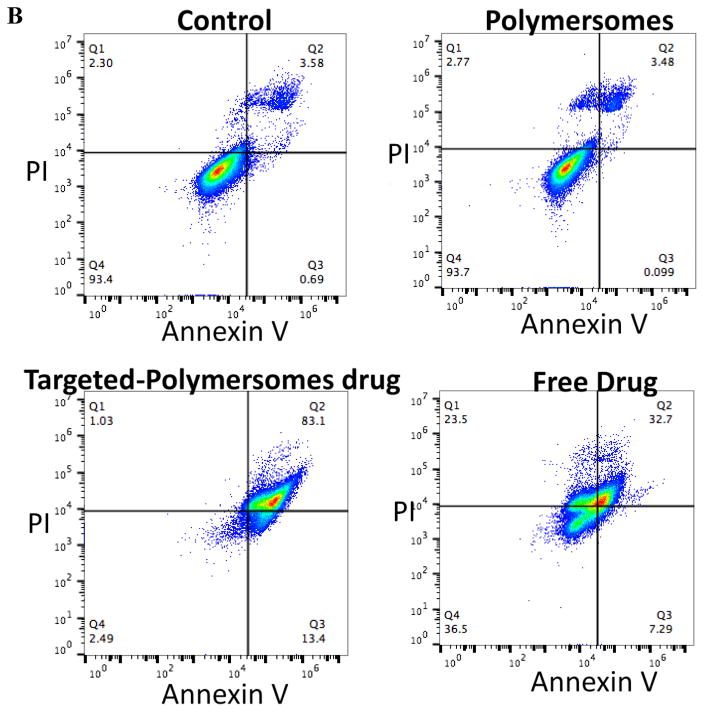
3.7. Cell apoptosis assay by flow cytometry
The apoptotic effects of the polymersomes on the prostate and pancreatic cancer stem cells were examined by staining of the cells with Annexin V-FITC and propidium iodide (PI) followed by flow cytometry analysis. Annexin V is a Ca2+ dependent phospholipid binding protein[25] that has a high affinity for phosphatidylserine. Phosphatidylserine is present in the inner leaflet of healthy cell membranes but is rapidly translocated to the outer leaflet during the early stages of apoptosis[26] and therefore serves as a marker for those cells that have committed to apoptotic cell death. In contrast, PI crosses the membranes of dead or dying cells, intercalates with DNA, emits red fluorescence[25, 26], and serves as a non-specific marker of cell death regardless of mechanism. After treatment with the targeted-polymersomes encapsulating napabucasin, 76% of the prostate cancer stem cells showed early apoptosis; whereas, the polymersomes themselves were not toxic (Figure 9A). Treatment of pancreatic cancer stem cells with targeted polymersomes encapsulating napabucasin led to 83% of cells staining positive for PI, and 13% exhibiting signs of early apoptosis (Figure 9B). We observed that RGD-targeted polymersomes encapsulating encapsulated napabucasin leads to significant stem cell death with signs of apoptosis in both prostate and pancreatic cancer stem cells.
Fig 9.
Flow cytometry analysis of the effect of napabucasin on prostate (A) and pancreatic cancer stem cells (B) with Annexin V and PI staining.
CONCLUSION
We have demonstrated that the europilin-1 is receptor over expressed in both pancreatic and prostate cancer stem cells. The iRGD peptide-decorated, reduction-sensitive polymersomes encapsulating the cancer stemness inhibitor napabucasin were developed to provide effective targeting to the neuropilin-1 receptors. Significant growth inhibition and decreased cancer stemness protein expressions were observed in prostate and pancreatic cancer stem cells in the presence of napabucasin-encapsulated, targeted polymersomes. The iRGD targeted polymersomes penetrated the microtumors of prostate and pancreatic cancer stem cells to at least 200 μm. The result of this study will provide new directions in targeting cancer stem cells for overcoming recurrence in prostate and pancreatic cancers. Further pharmacokinetic and immunogenicity studies of the targeted polymersomes encapsulating napabucasin would overcome the chemotherapeutic side effects of cancer treatment.
Supplementary Material
Scheme 1.
[2+3]-Cycloaddition reaction of N3-PEG1900-PLA6000 polymer and iRGD-hexynoic acid peptide.
Highlights.
Expression of neuropilin-1 receptor is demonstrated in cancer stem cells.
Peptide-decorated reduction-sensitive polymersomes target neuropilin-1 receptors.
The vesicles penetrate at least 200 μm into micro tumors of the cancer stem cells.
Drug released from the polymersomes decrease viability of the cancer stem cells.
Acknowledgments
Funding Sources
This research was supported by NIH grant 1R15CA206067 to JCW, NSF grant DMR 1306154, and NIH grant 1 R01GM 114080 to SM. SM also acknowledges support from the Grand Challenge Initiative and the Office of the Vice President for Research and Creative Activity, North Dakota State University.
We acknowledge the Core Biology Facility supported through the COBRE Center for Protease Research NDSU (1P30GM103332). The neuropilin-1 monoclonal antibody was a gift from Santa Cruz Biotechnology.
Footnotes
Author Contributions
FK and SM planned the experiments. FK conducted the cellular experiments, analyzed the data, and wrote the manuscript. SM supervised the experiments, data analysis, and edited the manuscript. JF and YC conducted the atomic force microscopy imaging. PB imaged the sections of three-dimensional spheroids. JCW designed and analyzed the flow cytometry experiments. All authors have approved the final version of the manuscript.
Declaration of interest
We do not have any financial or personal interest in the research reported.
Supplementary data related to this research is available in separate PDF file.
CD spectra of hexynoic acid conjugated iRGD peptide before and after reaction [2+3]-cycloaddition with N3-PEG-PLA, FT-IR characterization of iRGD peptide after and after reaction with N3-PEG1900-PLA6000, MALDI mass spectrum for hexynoic acid conjugated iRGD peptide, band Intensity of neuropilin-1 receptor in prostate and pancreatic cancer stem cells, band intensity of Nanog expression in prostate cancer stem cells after treatment, band intensity of Nanog expression in prostate cancer stem cell after treatment (PDF file). Internalization of iRGD-targeted polymersomes in prostate and pancreatic cancer stem cell.(MP3 files)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klieser E, Swierczynski S, Mayr C, Schmidt J, Neureiter D, Kiesslich T, Illig R. Role of histone deacetylases in pancreas: Implications for pathogenesis and therapy. World journal of gastrointestinal oncology. 2015;7(12):473–83. doi: 10.4251/wjgo.v7.i12.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shackleton M. Seminars in cancer biology. Elsevier; 2010. Normal stem cells and cancer stem cells: similar and different; pp. 85–92. [DOI] [PubMed] [Google Scholar]
- 3.Lee HE, Kim JH, Kim YJ, Choi S, Kim S, Kang E, Chung I, Kim I, Kim E, Choi Y. An increase in cancer stem cell population after primary systemic therapy is a poor prognostic factor in breast cancer. British journal of cancer. 2011;104(11):1730. doi: 10.1038/bjc.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermann PC, Bhaskar S, Cioffi M, Heeschen C. Seminars in cancer biology. Elsevier; 2010. Cancer stem cells in solid tumors; pp. 77–84. [DOI] [PubMed] [Google Scholar]
- 5.Gu G, Yuan J, Wills M, Kasper S. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer research. 2007;67(10):4807–4815. doi: 10.1158/0008-5472.CAN-06-4608. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer research. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 7.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell stem cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen L, Melo FdSe, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. The lancet oncology. 2012;13(2):e83–e89. doi: 10.1016/S1470-2045(11)70257-1. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Jin Z, Zhou H, Ou X, Xu Y, Li H, Liu C, Li B. Suppression of prostate cancer progression by cancer cell stemness inhibitor napabucasin. Cancer medicine. 2016;5(6):1251–1258. doi: 10.1002/cam4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubbard JM, Grothey A. Napabucasin: An Update on the First-in-Class Cancer Stemness Inhibitor. Drugs. 2017;77(10):1091–1103. doi: 10.1007/s40265-017-0759-4. [DOI] [PubMed] [Google Scholar]
- 11.Karandish F, Haldar MK, You S, Brooks AE, Brooks BD, Guo B, Choi Y, Mallik S. Prostate-Specific Membrane Antigen Targeted Polymersomes for Delivering Mocetinostat and Docetaxel to Prostate Cancer Cell Spheroids. ACS omega. 2016;1(5):952–962. doi: 10.1021/acsomega.6b00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anajafi T, Mallik S. Polymersome-based drug-delivery strategies for cancer therapeutics. Therapeutic delivery. 2015;6(4):521–34. doi: 10.4155/tde.14.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadonosono T, Yamano A, Goto T, Tsubaki T, Niibori M, Kuchimaru T, Kizaka-Kondoh S. Cell penetrating peptides improve tumor delivery of cargos through neuropilin-1-dependent extravasation. Journal of Controlled Release. 2015;201:14–21. doi: 10.1016/j.jconrel.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Miao H-Q, Lee P, Lin H, Soker S, Klagsbrun M. Neuropilin-1 expression by tumor cells promotes tumor angiogenesis and progression. The FASEB journal. 2000;14(15):2532–2539. doi: 10.1096/fj.00-0250com. [DOI] [PubMed] [Google Scholar]
- 15.Ruoslahti E. Tumor penetrating peptides for improved drug delivery. Advanced drug delivery reviews. 2017;110:3–12. doi: 10.1016/j.addr.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anajafi T, Yu J, Sedigh A, Haldar MK, Muhonen WW, Oberlander S, Wasness H, Froberg J, Molla MS, Katti KS, Choi Y, Shabb JB, Srivastava DK, Mallik S. Nuclear Localizing Peptide-Conjugated. Redox-Sensitive Polymersomes for Delivering Curcumin and Doxorubicin to Pancreatic Cancer Microtumors. Molecular pharmaceutics. 2017;14(6):1916–1928. doi: 10.1021/acs.molpharmaceut.7b00014. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Zhao Y. Controlled Release from Cleavable Polymerized Liposomes upon Redox and pH Stimulation. Bioconjugate Chemistry. 2011;22(4):523–528. doi: 10.1021/bc1003197. [DOI] [PubMed] [Google Scholar]
- 18.Mohammadi M, Ramezani M, Abnous K, Alibolandi M. Biocompatible polymersomes-based cancer theranostics: Towards multifunctional nanomedicine. International journal of pharmaceutics. 2017;519(1–2):287–303. doi: 10.1016/j.ijpharm.2017.01.037. [DOI] [PubMed] [Google Scholar]
- 19.Anajafi T, Scott MD, You S, Yang X, Choi Y, Qian SY, Mallik S. Acridine Orange Conjugated Polymersomes for Simultaneous Nuclear Delivery of Gemcitabine and Doxorubicin to Pancreatic Cancer Cells. Bioconjug Chem. 2016;27(3):762–71. doi: 10.1021/acs.bioconjchem.5b00694. [DOI] [PubMed] [Google Scholar]
- 20.Schnelldorfer T, Gansauge S, Gansauge F, Schlosser S, Beger HG, Nussler AK. Glutathione depletion causes cell growth inhibition and enhanced apoptosis in pancreatic cancer cells. Cancer. 2000;89(7):1440–1447. [PubMed] [Google Scholar]
- 21.Bielenberg DR, Pettaway CA, Takashima S, Klagsbrun M. Neuropilins in neoplasms: Expression, regulation, and function. Experimental Cell Research. 2006;312(5):584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Fuh G, Garcia KC, de Vos AM. The interaction of neuropilin-1 with vascular endothelial growth factor and its receptor flt-1. Journal of Biological Chemistry. 2000;275(35):26690–26695. doi: 10.1074/jbc.M003955200. [DOI] [PubMed] [Google Scholar]
- 23.Glinka Y, Mohammed N, Subramaniam V, Jothy S, Prud’homme GJ. Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-κB activation and tumor sphere formation. Biochemical and Biophysical Research Communications. 2012;425(4):775–780. doi: 10.1016/j.bbrc.2012.07.151. [DOI] [PubMed] [Google Scholar]
- 24.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer cell. 2009;16(6):510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermes I, Haanen C, Reutelingsperger C. Flow cytometry of apoptotic cell death. Journal of immunological methods. 2000;243(1):167–190. doi: 10.1016/s0022-1759(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 26.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.