Abstract
Small heat shock proteins are chaperones with variable mechanisms of action. The function of cardiac family member Hspb7 is unknown, despite being identified through GWAS as a potential cardiomyopathy risk gene. We discovered that zebrafish hspb7 mutants display mild focal cardiac fibrosis and sarcomeric abnormalities. Significant mortality was observed in adult hspb7 mutants subjected to exercise stress, demonstrating a genetic and environmental interaction that determines disease outcome. We identified large sarcomeric proteins FilaminC and Titin as Hspb7 binding partners in cardiac cells. Damaged FilaminC undergoes autophagic processing to maintain sarcomeric homeostasis. Loss of Hspb7 in zebrafish or human cardiomyocytes stimulated autophagic pathways and expression of the sister gene encoding Hspb5. Inhibiting autophagy caused FilaminC aggregation in HSPB7 mutant human cardiomyocytes and developmental cardiomyopathy in hspb7 mutant zebrafish embryos. These studies highlight the importance of damage-processing networks in cardiomyocytes, and a previously unrecognized role in this context for Hspb7.
Keywords: zebrafish, hESCs, heart development, cardiomyopathy, FilaminC
Introduction
Prevention, recognition and management of protein damage are essential cellular processes for physiological function at the level of the individual cell and the whole organ. This is clearly demonstrated by diseases that stem from pathologic aggregation of mutant or damaged proteins, or disturbance of these homeostatic regulatory pathways (Boncoraglio et al., 2012; Knowles et al., 2014; Wang and Robbins, 2006). Cytoprotective measures are particularly important in terminally differentiated, non-dividing cells that are more vulnerable to the toxic accumulation of misfolded or dysfunctional proteins (as they are unable to dilute them through cell division). Cell types that fall into this category include neurons and muscle cells, including cardiomyocytes. Myocytes must have particularly robust systems for turning over damaged proteins, since they contain large sarcomeric proteins undergoing constant strain and stress, for example in working skeletal muscle, or beating cardiac tissue. The importance of proteostasis is amplified in the non-dividing mammalian cardiomyocyte.
The small heat shock proteins (sHSPs) are a family of 15–30 kDa putative chaperones that are expressed throughout embryonic development and in adult organs. They have varied patterns of expression and heterogeneous mechanisms of action. While poorly understood compared to their higher molecular weight HSP counterparts, sHSPs have recently been shown to be important for maintaining cell fitness in a range of both physiological and pathophysiological conditions. These include a group of disorders associated with altered protein aggregation and cytoskeletal abnormalities including cataract formation (Litt et al., 1998), myopathies (Cappola et al., 2010; Stark et al., 2010; Vicart et al., 1998) and cancer (Zoubeidi and Gleave, 2012). The sHSPs share a conserved α-crystallin domain but exhibit large variation in expression patterns and mechanisms of action (Kappe et al., 2003; Ke et al., 2011; Morrow and Tanguay, 2012; Vos et al., 2010). They are putative chaperone proteins based on the α-crystallin domain, but the natural target substrates are generally unknown. Recently, three separate genome-wide association studies implicated polymorphisms in small heat shock protein, beta 7 (HSPB7) as potential contributors to idiopathic cardiomyopathies (Cappola et al., 2010; Stark et al., 2010; Villard et al., 2011).
HSPB7 is one of the least characterized sHSPs beyond recognition that it is highly expressed in the developing and adult heart (Elicker and Hutson, 2007; Krief et al., 1999). The structural and functional diversity seen within the sHSP family preclude prediction of HSPB7 function based on other sHSP family members. Recent studies characterizing the action of sHSPs reported that several family members prevent polyQ protein aggregation, including most potently HSPB7 (Vos et al., 2010). The efficacy of HSPB7 in preventing aggregation was striking, however, these were in vitro assays, and given that HSPB7 is expressed in striated muscle rather than in the brain, polyQ proteins are unlikely to be an endogenous target of HSPB7. It was also reported that HSPB7 co-localizes with tachypacing-induced F-actin stress fibers, and is protective against their formation in transformed cultured murine atrial cardiac (HL-1) cells (Ke et al., 2011). A recent publication demonstrated that the in vitro cytoprotective anti-polyQ action of HSPB7 occurs before the formation of aggregates, suggesting that HSPB7 either attenuates protein damage or facilitates early processing of damaged proteins (Eenjes et al., 2016).
However, the physiological target(s) and molecular mechanism(s) of action for HSPB7 in the heart remain obscure. We reported that hspb7 lies downstream of the key cardiac transcription factor Gata4 in the developing zebrafish heart, and that hspb7 depletion leads to defects in cardiomorphogenesis (Rosenfeld et al., 2013). Gata4 has been shown to be cardioprotective (Kitta et al., 2003) and it seemed feasible that Hspb7 could be a mechanistic component in this context. Therefore, we generated hspb7 homozygous mutant zebrafish and characterized heart phenotypes, including an adult cardiac pathology that leads to a predisposition to exercise-induced death. We discovered evidence of upregulation and strict dependency on autophagy in mutant hearts and discovered FilaminC (FLNC) as a novel binding partner of HSPB7. We propose a model in which HSPB7 normally facilitates early post-damage processing of large sarcomeric cytoskeletal proteins, and loss of HSPB7 function leads to increased damaged protein load, protein aggregation, and risk for cardiomyopathy.
Materials and Methods
Zebrafish Husbandry
All zebrafish strains were maintained at 28.5C in our zebrafish facility, and all procedures carried out as approved by the WCMC IACUC. Embryos were staged as described (Westerfield, 1993). The tg(myl7:gfp) strain was originally obtained from H.J. Tsai (Taiwan).
Generation of HSPB7 mutant zebrafish lines
Sequences of TALEN and CRISPRs used in this study are provided in Table 1. TALENs were designed using the Cornell TAL Effector Nucleotide Targeter 2.0 website (Doyle et al., 2012) and plasmids encoding specific TALENs were generated with the TALEN Golden Gate 2.0 kit (Addgene), as previously described (Cermak et al., 2011). The mRNA encoding TALENs was synthesized with the mMessage mMachine kit (Thermo Fisher) and 100–400 pg was injected into zebrafish embryos at the 1–2 cell stage. Guide RNAs (gRNAs) for CRISPR-Cas9 mediated mutagenesis were designed with the CHOPCHOP website (Montague et al., 2014) and ordered as double stranded oligo cassettes with a T7 promoter from IDT. The gRNA was generated with the MEGAshortscript kit (Thermo Fisher) and 250 pg was injected with 500 pg recombinant Cas9 protein (PNA BIO) into embryos at the 1–2 cell stage. Carriers of mutations were identified through PCR across the target locus followed by RFLP analysis or re-naturation of PCR products followed by digestion with T7 endonuclease I (NEB). PCR products were cloned into a TOPO vector (Invitrogen) and sequenced to identify mutant alleles.
Table 1.
sgRNA sequences and TALEN targeted sequences in fish and human HSPB7 genes.
| Gene | Species | gRNA Targeted Sequence (PAM) | |
|---|---|---|---|
| Hspb5b | Zebrafish | 5′-ACGTGATCTCCTCATTGTAC(TCC)-3′ | |
| HSPB7 | Human | 5′-(CCC)ACTCGGAGCCCCTGGCCTTC-3′ | |
| Gene | Species | Targeted Sequence | RVD sequences |
| Hspb7 | Zebrafish | 5′-CCTCCTCATCTTCATCCTCT-3′ | HD HD NG HD HD NG HD NI NG HD NG NG HD NI NG HD HD NG HD NG |
| 5′-TACATGGAGAAGAGCCGAGG-3′ | HD HD NG HD NN NN HD NG HD NG NG HD NG HD HD NI NG NN NG NI | ||
Zebrafish stress test
Equal numbers of hspb7 mutant zebrafish and wildtype sibling controls were placed in a swim tunnel (Loligo Systems, Denmark) and subjected to water flow of 50 cms−1 for 30 minutes, once a day. Zebrafish mortality was tracked by physical daily count, and fish locations tracked with an iPhone camera. Zebrafish location during swim challenge was tracked with MTrackJ for ImageJ (Meijering et al., 2012).
Anatomical analysis of adult zebrafish
Adult male zebrafish were submitted for histopathological processing and blinded analysis by the pathology core at Memorial Sloan Kettering Cancer Center. The fish were fixed in Bouin’s solution for 24 hours, cut parasagitally, processed via automated tissue processor, embedded in paraffin, and two 4 micron sections were taken 100 microns apart. Sections were then stained with hematoxylin and eosin and Masson’s trichrome using standard methods. Adult zebrafish hearts were dissected as described (Arnaout et al., 2014) and fixed in 2.5% glutaraldehyde, 4% paraformaldehyde, (both from Electron Microscopy Biosciences), and 0.02% picric acid (Thermo). Samples were submitted to the Microscopy and Image Analysis Core at Weill Cornell Medicine. Following post-fixation processing as described (de Bruijn, 1973), samples were contrasted with lead citrate and viewed on a JEM 1400 electron microscope (JEOL, USA, Inc. Peabody, MA) operated at 100kV. Digital images were captured on a Veleta 2K × 2K CCD camera (EM-SIS, Germany). Sectioned heart samples were imaged and analyzed cumulatively from 3 independent 18 month wildtype or hspb7 mutant fish. The total area analyzed was 3405 μm2 and 3738 μm2 for the wildtype and the mutant hearts, respectively.
Measuring zebrafish heart size
Hearts were dissected from adult zebrafish as described (Ding et al., 2011). Following dissection, adult zebrafish hearts were placed on a microscope slide and imaged on a dissection microscope (Nikon) with top-lighting. Hearts were rotated to present maximal ventricular area and images were captured with Spot Imaging Software. Ventricular area was calculated with FIJI software (www.fiji.sc/). Prior to dissection, anesthetized zebrafish were partially dried with paper towels and weighed. Zebrafish mass was used as a normalizing factor for heart size index (VA/BW) since it is a greater variable than body length in adult fish.
Small molecule treatment of embryonic zebrafish
Wildtype, hspb7 mutant embryos, or wildtype embryos injected with a previously validated (Rosenfeld et al., 2013) hspb7 morpholino (CCTGTTCTGTCTGATGAAAAACATA, Gene Tools) were manually dechorionated with forceps at 24 hpf and exposed to increasing concentrations of bafilomycin A1, chloroquine diphosphate or 3-methyladenine (all from Sigma) in a solution of E3 medium (5.0mM NaCl, 0.17mM KCl, 0.33mM CaCl, 0.33mM MgSO4) with 1% DMSO (Sigma). 1 nl (0.9mM) of morpholino was injected at the one cell stage. Small molecule compounds were refreshed every 24 hours.
Quantitative RT-PCR (qPCR) analysis of gene expression in zebrafish
Zebrafish embryos or adult tissues were homogenized in Trizol (Invitrogen) and RNA was extracted according to manufacturer’s instructions. The qPCR analysis was performed on a LightCycler 480 II (Roche) using LightCycler 480 Sybr Green master mix (Roche). Primer sequences are provided in Table 2. Relative gene expression was determined as described (Holtzinger et al., 2010).
Table 2.
Sequences for human and zebrafish primers used to genotype and for RT-qPCR experiments.
| Gene | Species | Genotyping Forward | Genotyping Reverse |
|---|---|---|---|
| Hspb7 | Zebrafish | TGAGAAAGCTGCTGAGTGGA | ATCCAAAGTCGTCAGCGAAG |
| Hspb7 | Zebrafish | CTAATGAGCGCGAGCAATTC | GCCTCATGCAGCTGACATATT |
| Hspb5b | Zebrafish | TGCAGTGAAAGAGGCAAATG | ACGCGTGACTCCTCAACTTT |
| HSPB7 | Human | CAGGGACAGTCGGCCTTAT | GTGGCCACATCTGTCTTGGT |
| Gene | Species | qPCR Forward | qPCR Reverse |
| HSPB5 | Human | CTTTGACCAGTTCTTCGGAG | CCTCAATCACATCTCCCAAC |
| HSPB7 | Human | CAGGCAACATCAAGACCCTA | CTTGTGAGCGAAGGTGTTCA |
| HSPB8 | Human | CAGAGGAGTTGATGGTGAAGAC | GTTGAGTAAGGAGGGACCTG |
| 18S | Zebrafish | TCGCTAGTTGGCATCGTTTATG | CGGAGGTTCGAAGACGATCA |
| Beta-Actin | Zebrafish | GACAACGGCTCCGGTATG | CATGCCAACCATCACTCC |
| Hspb11 | Zebrafish | AGAGCTCGCCGTTAAACAAG | AATAGGATCCCTTCCCATCG |
| Hspb12 | Zebrafish | TAACCTGGAGACGGACATCAG | TCCGGGGATGTAGTACTTCTGG |
| Hspb1 | Zebrafish | ATGAACACGGCTTCATTTCC | GCGGAGCTTCAACAGTCAGG |
| Hspb5a | Zebrafish | CCCAGGCTTCTTCCCTTATC | GTGCTTCACATCCAGGTTGA |
| Hspb5b | Zebrafish | CCTACTGACGGCCAAATGTT | GGCATCAGCAGCAGACAATA |
| Hspb7 | Zebrafish | GATCTTTCATGTGTCCGAAGG | GATTGTTTGAGGTGGTGACG |
| Hspb8 | Zebrafish | ACCGACCAAAGTGAGAGGTG | GCATTTACGGAGCTCTGAGG |
| KLF4 | Human | CGAACCCACACAGGTGAGAA | TACGGTAGTGCCTGGTCAGTTC |
| NANOG | Human | TCCTGAACCTCAGCTACAAACA | GGTAGGTGCTGAGGCCTTCT |
| OCT4 | Human | TGGGCTCGAGAAGGATGTG | GCATAGTCGCTGCTTGATCG |
| SOX2 | Human | GCACATGAAGGAGCACCCGGATTA | CGGGCAGCGTGTACTTATCCTTCTT |
| HPRT | Human | ACCAGTCAACAGGGGACATAA | CTTCGTGGGGTCCTTTTCACC |
| GATA4 | Human | AAAGAGGGGATCCAAACCAG | TTGCTGGAGTTGCTGGAAG |
| CTNT | Human | GGAGGAGTCCAAACCAAAGCC | TCAAAGTCCACTCTCTCTCCATC |
Cell culture and generation of HSPB7 mutant lines
HL-1 cells (a kind gift from Dr. W. Claycomb, LSU) were maintained in Claycomb medium (Sigma) according to manufacturer’s instructions. Human embryonic stem cells (hESCs) containing inducible Cas9 (Gonzalez et al., 2014), generously provided by Dr. Danwei Huangfu, were maintained as described (Zhu et al., 2014), or in mTESR or TESR-E8 medium (both from STEMCELL technologies) on Matrigel (VWR). The hESC clonal lines carrying mutations in HSPB7 were generated as described (Zhu et al., 2014). The gRNAs (Table 1) were designed with the CHOPCHOP website (Montague et al., 2014). Mutant alleles were identified by PCR and sequencing.
Immunoprecipitation of HSPB7 and Western Blotting
HL-1 cells were lysed on ice in a modified RIPA buffer (20mM Tris pH7.5, 150mM NaCl, 1% NP-40) with cOmplete mini protease inhibitor (Roche). Debris was cleared from lysed cells by centrifugation (21,000g, 10 minutes, 4°C) and protein concentration was calculated with a QuickStart Bradford Protein Assay kit (BioRad). Lysates were precleared by incubation with 1:10 dilution of Protein G Dynabeads (Invitrogen, 30 minutes, 4°C) and then incubated overnight with 1:100 anti-HSPB7 (ab150390, Abcam) or control rabbit IgG whole molecule (Jackson). Complexes were precipitated with 1:10 Protein G Dynabeads (120 minutes, 4°C) and eluted into Western buffer (65% modified RIPA, 25% NuPAGE Sample Buffer, 10% NuPAGE Reducing Agent (both Invitrogen).
Immunofluorescence techniques
Immunostaining on cultured cells was performed after fixing cells with 4% paraformaldehyde (VWR) and permeabilizing with 0.1% Triton X-100 (MP Biomedicals) at RT or with 50 μg/ml digitonin (Sigma) in PBS at 37°C. Digitonin permeabilization was followed by quenching with 40mM NH4Cl (Sigma). Cells were blocked with 2% IgG-free bovine serum albumin (Jackson), 5% lamb serum (Invitrogen) and incubated overnight at 4°C with the appropriate primary antibodies: anti-cardiac troponin T (13–11, 1:500, Thermo Fischer); anti-HSPB7 (ab150390, 1:250, Abcam); anti-LC3B (ab51520, 1:250, Abcam); anti-FLNC (sc48496, 1:100, Santa Cruz).
Cells were counterstained with DAPI (1:10000, Invitrogen) and imaged on an Axio Observer.Z1, LSM510 or LSM880 confocal microscope (all from Zeiss). The LSM880 was used in the Microscopy and Image Analysis Core at Weill Cornell Medicine.
Filter Trap Assay
The filter trap assay was performed as described (Vos et al., 2010). In brief, bafilomycin A-treated cells were counted and a fixed number were lysed in 200μl FTA buffer (10mM Tris-HCl, pH8.0, 150mM NaCl) containing 2% SDS. Samples were boiled for 10 minutes and loaded onto a pre-washed (FTA, 0.1% SDS) 0.2μm cellulose acetate filter (GE Healthcare) with supporting filter paper (Bio-Rad) in a Bio-Dot 96 well filtration system (Bio-Rad). Suction was applied to filter the samples. Following loading, membranes were processed as described above in Western Blotting.
Statistical Analysis
Data are presented as mean ± SEM derived from at least three independent biological replicates. Statistical analysis was performed using GraphPad Prism 6 software. P values generated by unpaired two-tailed t-test, unless indicated otherwise in the Figure Legends. The significance is indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. n.s. indicates not significant.
Results
Generation of mutant hspb7 lines
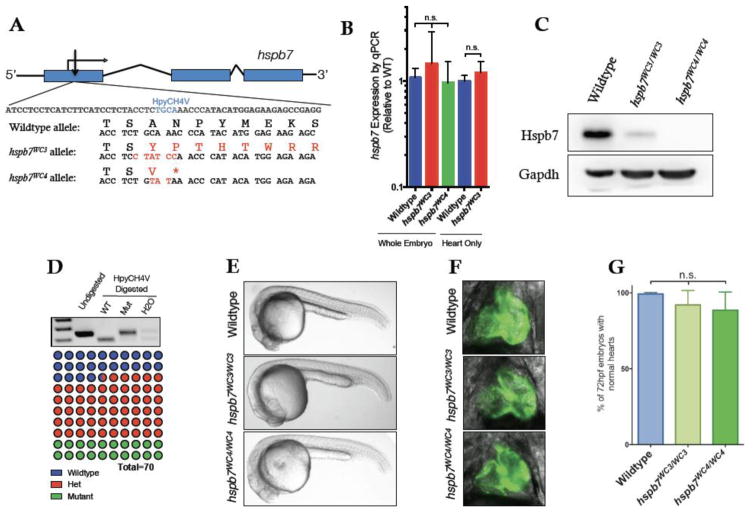
To examine the role of Hspb7 in the zebrafish heart, we generated mutant hspb7 alleles using Transcription Activator-Like Effector Nuclease (TALEN) technology. In the absence of information regarding critical functional domains of Hspb7, we designed TALENs to target mutations near the 5′ end of the gene, reasoning that an indel leading to an early frameshift was likely to cause a deleterious mutation (Fig. 1A). By exploiting the presence of a HpyCH4V restriction site in the wildtype allele, the generation and transmission of mutant alleles was followed by RFLP analysis of PCR products that included the predicted TALEN cut site.
Fig. 1. Generation of mutant hspb7 zebrafish lines.
(A) Structure and partial sequence of the hspb7 gene, showing TALEN targeting proximal to the start codon and resultant mutant alleles WC3 and WC4. (B) qPCR data showing that hspb7WC3 and hspb7WC4 transcripts are expressed at levels comparable to wildtype hspb7 in 5 dpf zebrafish embryos. (C) Western blot of protein lysates from adult zebrafish hearts showing loss of Hspb7 protein in animals homozygous for either of the hspb7 mutant alleles. (D) RFLP analysis showing loss of the HpyCH4V cut site in PCR product from hspb7WC3/WC3 embryos. Quantification of embryonic genotypes demonstrates that hspb7WC3/WC3 alleles are transmitted at Mendelian ratios. (E) Representative brightfield images of wildtype or homozygous hspb7 mutant 24 hpf embryos. (F) Representative 72 hpf hearts from wildtype or hspb7 mutant lines crossed onto a tg(myl7:gfp) background. (G) Quantification of normal hearts at 72 hpf in wildtype or homozygous hspb7 mutant animals (n=200).
We recovered two mutant alleles from separate founder fish that were predicted to encode proteins missing a large proportion of the normal Hspb7 protein (Fig. 1A). Both alleles generate a frameshift immediately after a polyserine stretch near the N-terminus of Hspb7. The first allele, hspb7WC3, is expected to result in the synthesis of an almost entirely novel predicted protein approximately half the mass of wildtype Hspb7. The second frameshift allele causes an early stop codon, truncating the predicted protein to only 27 amino acids, and was designated hspb7WC4. It is a strong prediction that both hspb7WC3 and hspb7WC4 represent null alleles of hspb7 and that animals harboring these alleles will be deficient for any normal role of Hspb7 in the zebrafish. Both mutations lie in the middle of exon 1 and are not predicted to interfere with splicing of hspb7 mRNA. We tested if the premature stop codons predispose mutant hspb7 mRNAs to nonsense-mediated decay, but qPCR analysis indicated that hspb7WC3 and hspb7WC4 transcripts are expressed at levels similar to those from the wildtype hspb7 allele (Fig. 1B). However, as mutant animals could survive to adulthood, we confirmed through western blotting experiments that Hspb7 protein was near or completely absent in these mutant fish (Fig. 1C).
Zebrafish embryos tolerate hspb7 mutant alleles
Embryos homozygous for the mutant hspb7 alleles were recovered from crosses of heterozygous fish in the expected Mendelian ratios (Fig. 1D). Homozygous hspb7WC3 or hspb7WC4 mutants showed no gross morphological defects (Fig. 1E). In order to better visualize the hearts of hspb7 mutant fish, the hspb7WC3 allele was crossed onto a tg(myl7:gfp) background and cardiac development monitored across the first week of life. In the hspb7 mutants, developing hearts were seen to undergo timely jogging and looping and to have normal size and morphology (Fig. 1F, G). Fish homozygous for either of the two mutant hspb7 alleles were viable to adulthood and fertile.
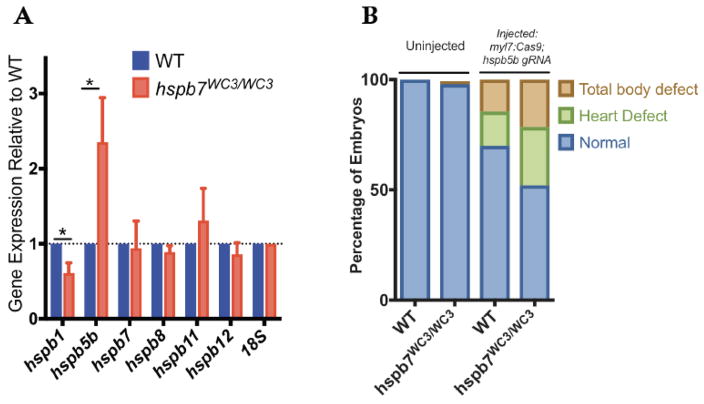
These findings are in contrast with those from previous studies, in which we (Rosenfeld et al., 2013) and others (Lahvic et al., 2013) used morpholinos to knock down Hspb7 during zebrafish embryogenesis and observed embryonic lethality with randomization of left-right asymmetry, cardiac malformation and a change in the size of ventricular cardiomyocytes. It is possible that the morphant phenotypes were caused by off-targeting effects, given recent reports that morphant and mutant phenotypes can be discordant (Kok et al., 2015). Alternatively, consistent with other reports (Rossi et al., 2015), it is possible that the mutants (but not morphants) compensate for loss of Hspb7. Indeed, we found by qPCR experiments that the hspb7 mutants show relatively increased expression levels for hspb5b, a sister sHSP gene that could conceivably compensate for loss of Hspb7 (Fig. 2A). Injection of a sgRNA predicted (and validated) to target hspb5b with Cas9 protein leads to enhanced rates of embryonic abnormalities in the hspb7 mutant background, compared to injection in wildtype embryos (Fig. 2B; p=0.029). This suggests a compensatory role for Hspb5b, which allows the embryos to tolerate hspb7 mutations, and facilitates our ability to evaluate Hspb7 function in adult fish.
Fig. 2. Apparent compensation for Hspb7 by Hspb5b.
(A) qPCR quantification of transcript levels of cardiac-expressed hspb family members in 5 dpf zebrafish embryonic heart tissue demonstrates that hspb5b expression is upregulated in hspb7WC3/WC3 mutant hearts. Changes in gene expression are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method. (B) Injection of cardiac-targeted hspb5b CRISPR sgRNA leads to increased cardiac and body abnormalities at 48 hpf in hspb7WC3/WC3 embryos compared to wildtype controls. Here n for wildtype = 64 larvae, and for hspb7 mutants is 65 larvae. The defects in mutant compared to wildtype is statistically significant according to Fisher’s exact test (p=0.029).
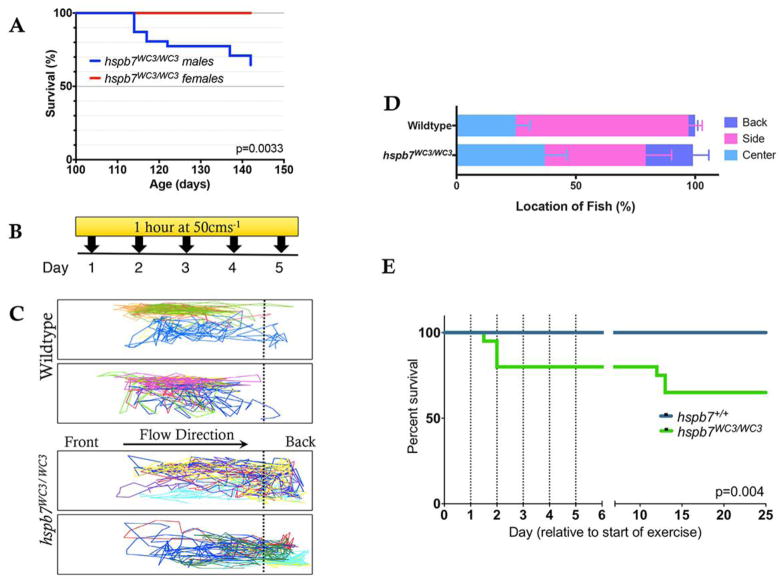
Adult hspb7WC3/WC3 fish have increased susceptibility to exercise-induced stress
When we attempted to breed homozygous null animals, we noticed a statistically significant mortality rate in male fish (Fig. 3A). We hypothesized that death indicated decreased capacity to cope with cardiac stress in hspb7WC3/WC3 animals. To investigate the phenomenon in a controlled manner, batches of male and female adult zebrafish were subjected to an enforced swimming exercise paradigm. Genotyped groups of either hspb7WC3/WC3 fish or wildtype controls were paced at 50 cms−1 for 30 minutes, every day for 5 days (Fig. 3B). This represents a high intensity anaerobic exercise regime, with a water flow approaching the critical swimming speed (Ucrit) of zebrafish (Palstra et al., 2010). We observed that hspb7 mutant animals spent a greater amount of time compared to wildtype controls ‘resting’ at the back of the tank (23.37% of swim time for HSPB7WC3/WC3 versus 2.78% of swim time for wildtype animals, Fig. 3C, D). Strikingly, around 40% of hspb7WC3/WC3 animals died during the first few days of enforced exercise (Fig. 3E), recapitulating our observation of increased mortality following mating. In the swimming stress test, 66% of animals that died were female, indicating that the bias we observed in post-mating mortality does not appear to be a gender-specific compromise of cardiac health.
Fig. 3. hspb7 mutant zebrafish are subject to exercise-induced mortality.
(A) Kaplan-Meier curve showing survival of hspb7 mutant male and female animals following commencement of breeding at 105 days post fertilization. N = 20 animals (10 male, 10 female). (B) High-intensity exercise paradigm for zebrafish. (C) Representative tracking of fish location in the tank during the final minute of exercise shows that hspb7WC3/WC3 fish are more likely to remain at the back of the tank. (D) Quantification of fish location in the tank for each second of the final minute of exercise (n = 10). (E) Kaplan-Meier curve showing increase in mortality in hspb7WC3/WC3 fish following commencement of exercise regime. Dotted lines indicate timing of exercise sessions. For both wildtype and mutant fish n = 20. Significance was calculated using the Graphpad logrank or Mantel-Haenszel test.
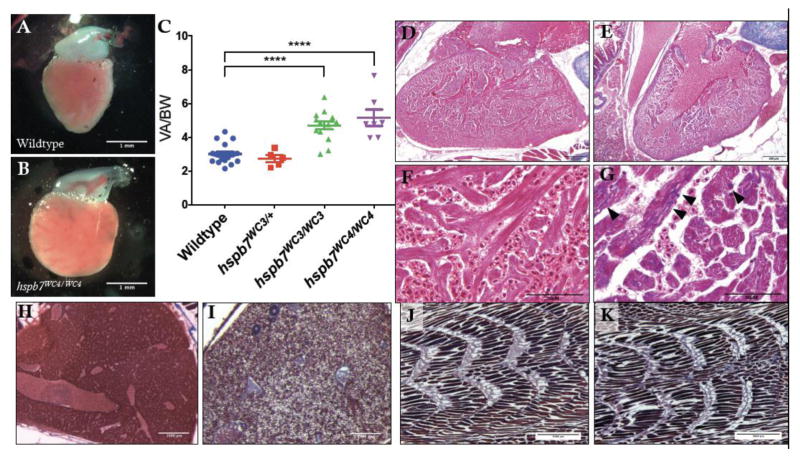
Hspb7WC3/WC3 mutant adults exhibit mild heart pathology
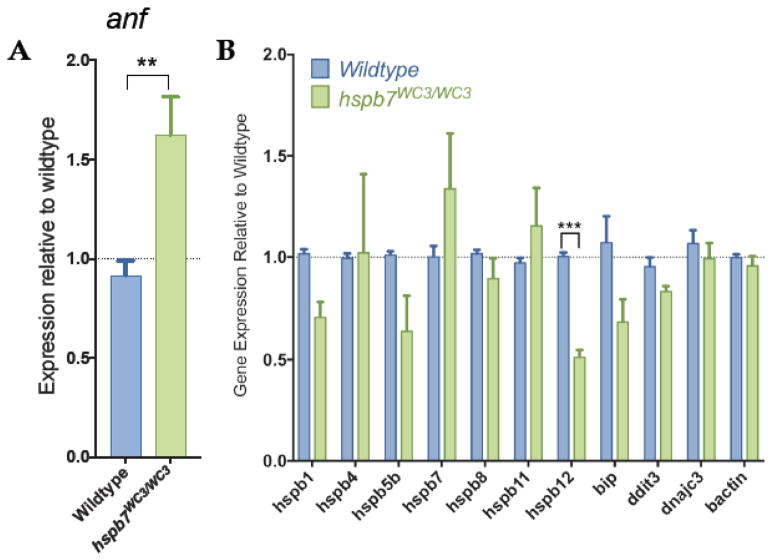
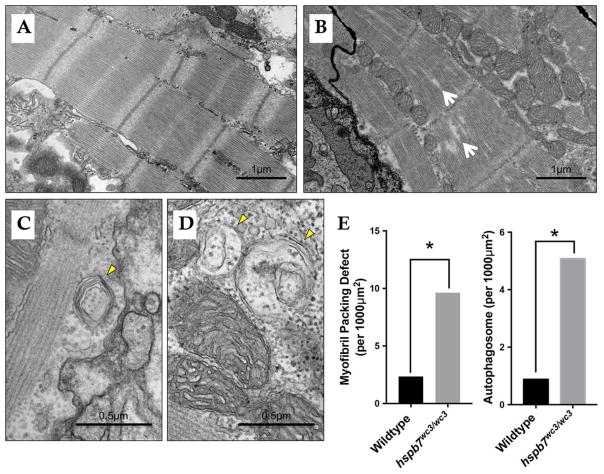
We hypothesized that exercise stress may exacerbate a cardiac dysfunction that is not obvious in unstressed mutants. To explore this idea, adult hspb7WC3/WC3 mutant zebrafish were processed for general histopathological analysis. We discovered that hearts of hspb7 mutants exhibited cardiomegaly, assessed by the ratio of ventricular area to body weight (VA/BW) (Fig. 4A–C). Such increases in the VA/BW ratio have been previously observed in zebrafish models of heart dysfunction23. Inspection of the gross anatomy revealed that mutant fish exhibited hearts that appeared relatively rounded when compared with wildtype controls. Heart pathology was evaluated comparing trichrome stained cardiac sections from mutant or wildtype animals. This revealed mild multifocal cardiac fibrosis in 100% of hspb7WC3/WC3 hearts, which was not seen in control wildtype hearts (Fig. 4D–G). The fibrosis based on Trichrome staining was reproducible and diffuse throughout the myocardium. From our analysis of tissue sections it was not localized to specific regions or structures. We attempted to quantify collagen deposition using the AFOG assay but did not find convincing signals in either wildtype or mutant hearts. We also attempted to measure cell death rates but found few TUNEL+ cells without significant differences in the wildtype or mutant hearts. Unexpectedly, 4/5 mutant fish were found to exhibit diffuse, marked hepatocellular vacuolation (Fig. 4H, I). This phenotype might be indicative of hepatic injury downstream from cardiac dysfunction (Giallourakis et al., 2002; Louie et al., 2015). We note that skeletal muscle did not exhibit fibrotic lesions (Fig. 4J, K). In addition, we observed a reactivation of expression of a marker of cardiac hypertrophy, atrial natriuretic factor (anf), in hspb7 mutant hearts (Fig. 5A). However, we did not observe alteration in the expression of general markers of proteotoxic stress, including the α-crystallins (Fig. 5B). For higher resolution observation of abnormalities in the myocardium of hspb7 mutants, hearts were dissected from adult mutant or wildtype zebrafish and analyzed by transmission electron microscopy (TEM), which revealed sarcomeric abnormalities in homozygous mutant hearts (Fig. 6). Specifically, we noted significantly enhanced numbers of autophagosomes, and myofiber packing defects in the mutant hearts. This indicates that in the absence of Hspb7, there is an underlying cardiopathology, which might predispose mutant animals exposed by exercise-induced stress to cardiac failure.
Fig. 4. Evidence of cardiomyopathy in hspb7WC3/WC3 adult hearts.
Representative images of dissected ventricles and outflow tracts from (A) wildtype and (B) hspb7WC3/WC3 adults. Shown are hearts from 10 month old animals with the atria removed for clear viewing. (C) Quantification of ventricular area to body weight index (VA/BW) showed heart enlargement in animals homozygous for either of the hspb7 mutations. Each point represents one animal. Data are combined for 10 and 18 month zebrafish. Changes in heart size are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method (p<0.0001). (D) Trichrome staining of 3 month wildtype and (E) hspb7 mutant hearts. Focal fibrotic lesions (arrowheads) were seen in all (5/5) hspb7 mutant but never in wildtype samples. Shown are representative images; F and G are higher magnification images for wildtype and mutant hearts, respectively. Arrowheads indicate local fibrotic lesions. (H–K) Representative Trichrome-stained images of adult wildtype (H, J) and mutant (I, K) hspb7WC3/WC3 zebrafish liver (H, I) and skeletal muscle (J, K).
Fig. 5. Analysis of gene expression in hspb7 mutant adult zebrafish hearts.
(A) qPCR analysis of 12 month hspb7WC3/WC3 hearts shows reactivation of anf expression when compared with wildtype controls. (B) qPCR analysis of 12 month hspb7WC3/WC3 hearts shows no upregulation of other markers of general proteotoxic stress. Changes in gene expression are significant according to unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method.
Fig. 6. Evidence of abnormalities in hspb7WC3/WC3 adult hearts by TEM.
Shown are representative transmission electron microscope images of 18 month (A) wildtype and (B) hspb7WC3/WC3 mutant hearts. Wildtype hearts exhibit repeated, regular sarcomeric striations with organized mitochondria. The hspb7 mutant heart tissue shows defects representing breakdown of sarcomeric organization (myofibril packing defects, white arrows). (C and D) Shown are two representative images of hspb7 mutant hearts that also exhibit increased number of autophagosomes (yellow arrowheads). (E) Graphs show quantification of relative occurrence rate (averaged number of defects over the total imaged area) of myofibril packing defects and autophagosomes in the wildtype and hspb7 mutant hearts (n=3 independent hearts for each). The difference in the rate of each defect was determined using an Exact Poisson test in R.
HSPB7 binds to large cytoskeletal proteins FLNC and TITIN
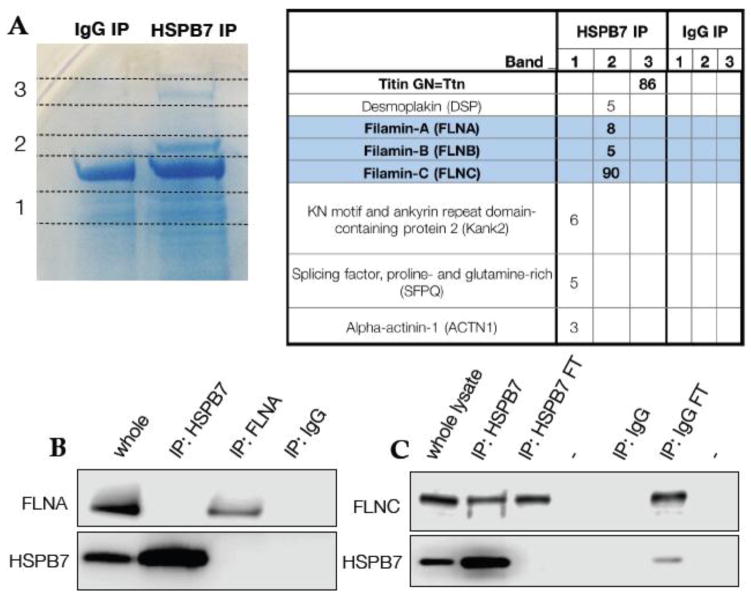
Although the cellular function of HSPB7 is unclear, it has been shown through in vitro assays to prevent aggregation of certain proteins (Ke et al., 2011; Vos et al., 2010), contingent on the presence of intact cellular autophagic machinery (Vos et al., 2010). Seeking clues to its function, we sought to identify proteins that HSPB7 can bind in cardiomyocytes. For this purpose, HSPB7 was immunoprecipitated from lysates of the HL-1 murine atrial cardiac cell line. Protein bands that precipitated specifically with anti-HSPB7 and not with an IgG control were subjected to in-gel trypsinolysis and identified by unbiased tandem mass-spectrometry (Fig. 7A). We identified the large cytoskeletal proteins FilaminC (FLNC) and TITIN as HSPB7 binding partners. FLNC is the muscle-specific isoform of Filamin, with roles in myogenesis (Dalkilic et al., 2006) and sarcomeric maintenance (Fujita et al., 2012; Furst et al., 2013; Leber et al., 2016). HSPB7 was reported previously to bind the ubiquitous isoform FilaminA (Krief et al., 1999). However, we note that coverage for peptides from FLNC was extensive (Fig. 7A), while the few peptides mapped by mass spectrometry as derived from FilaminA or FilaminB are fully conserved in FLNC. Furthermore, we were able to confirm through western blotting experiments that HSPB7 co-immunoprecipitated with FLNC, but not the A isoform (Fig. 7B, C). Given that HSPB7 is expressed exclusively in striated muscle and at a higher level in the heart, FLNC is an excellent candidate for a functional chaperone target, and HSPB7 may aid in either protecting FLNC and other large sarcomeric proteins from damage, or facilitating the processing of these proteins after damage has occurred.
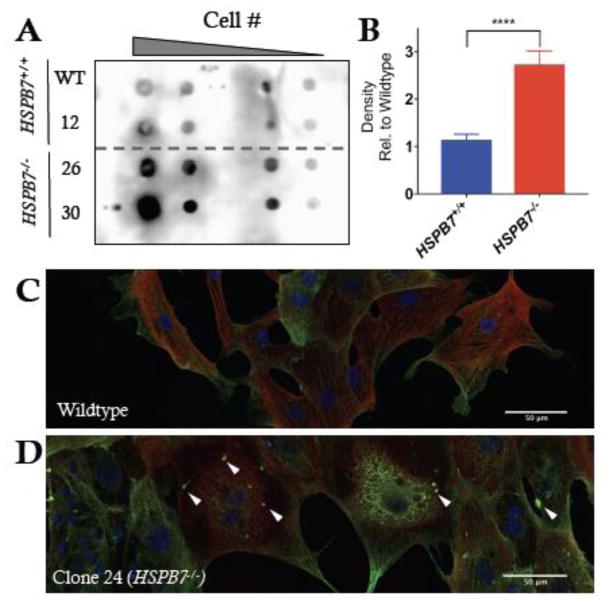
Fig. 7. HSPB7 binds specifically to FLNC.
(A) Coomassie-stained gel with bands specific to immunoprecipitation of HSPB7 identified and labeled. Unbiased mass spectrometer-based peptide identification shows Filamin-C and Titin as HSPB7 interaction partners. (B) Immunoprecipitation of HSPB7 or Filamin-A from HL-1 cell lysates demonstrates that they do not interact. (C) Filamin-C is co-immunoprecipitated with HSPB7 from HL-1 cells.
Generation of human cardiomyocytes lacking HSPB7
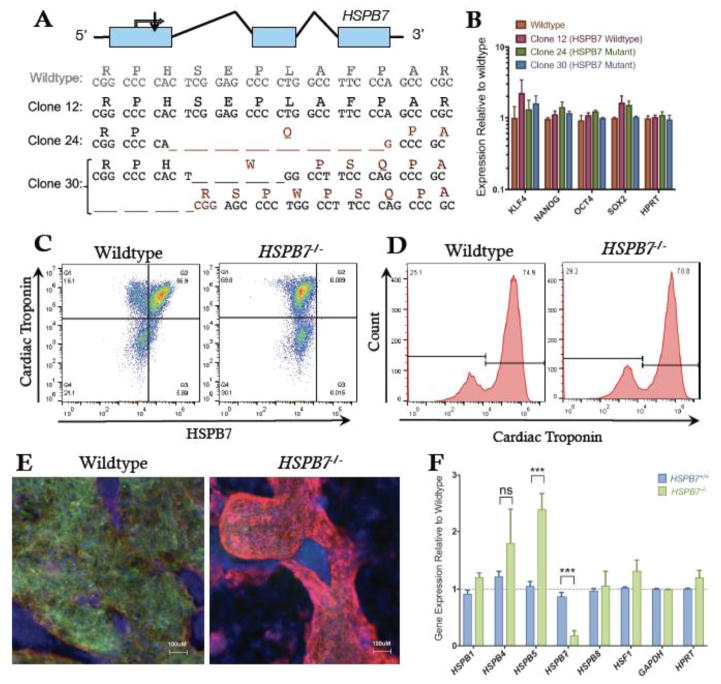
In order to test if HSPB7 function is conserved across vertebrate species, and to facilitate biochemical investigation into the molecular role of HSPB7 in human cardiomyocytes, we used the inducible CRISPR/Cas9 system (Zhu et al., 2014) to target mutations to the HSPB7 locus in human embryonic stem cells (hESCs). We recovered clonal lines in which HPSB7 was predicted to be homozygous null (Fig. 8A). We also identified lines that had gone unsuccessfully through the targeting process and used these as control “wildtype” lines. HSPB7 is not expressed in ESCs, and both wildtype and HSPB7−/− clones maintained normal ESC morphology, expressing comparable levels of pluripotency genes, including SOX2, NANOG and KLF4 (Fig. 8B). We used a directed differentiation protocol, modified from (Lian et al., 2012) to generate beating cardiomyocytes from the hESC lines. Flow cytometry (Figs. 8C, D) and immunofluorescence (Fig. 8E) demonstrated successful generation of cardiomyocytes with efficiency comparable to HSPB7 wildtype lines. These studies also confirmed that HSPB7 protein is absent from the mutant cells (Figs. 8C, E). Interestingly, just as hspb5b was upregulated in the hearts of hspb7WC3/WC zebrafish embryos, we observed a similar upregulation of HSPB5 in HSPB7−/− hESC-derived cardiomyocytes (Fig. 8F). HSPB7 mutant cardiomyocytes were competent to differentiate to cardiac fate and to form beating sheets. However, if HSPB7 functions as a sarcomeric cytoskeletal chaperone protein, we considered that the differentiated mutant cells might display protein aggregation phenotypes.
Fig. 8. Generation of HSPB7 mutant hESCs.
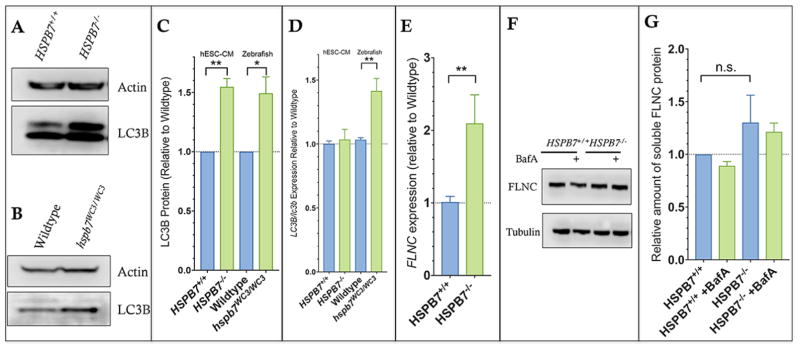
(A) Structure and partial sequence of the human HSPB7 gene, showing CRISPR targeting immediately downstream of the start codon and resultant HSPB7 homozygous and compound heterozygous mutant clones that were recovered. (B) qPCR quantification shows expression levels of pluripotency markers are normal in HSPB7 mutant hESC clones. HSPB7 mutant lines are competent to generate cardiomyocytes, and do not express HSPB7 protein as shown by flow cytometry (C, D) and immunofluorescence (E), both performed on day 10 of differentiation. (F) qPCR quantification of cardiac HSPB family members in wildtype and HSPB7 mutant hESC-derived cardiomyocytes. Changes in gene expression are significant according to unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method.
FLNC aggregates in HSPB7 mutant cardiomyocytes
To compare levels of aggregate formation, the filter trap assay was employed (Vos et al., 2010). Cardiomyocytes derived from HSPB7 mutant hESC lines showed an increase in FLNC aggregates when compared with cardiomyocytes derived from wildtype control lines (Fig. 9A, B). However, the accumulation of FLNC aggregates was dependent on treatment with the autophagy inhibitor bafilomycin A1 (BafA), as the filter assay failed to generate consistent signals when using lysates from untreated wildtype or mutant cells. We also observed an increase in numbers of BafA-dependent puncta when staining for FLNC by immunofluorescence (Fig. 9C, D). These findings demonstrate abnormalities in human cardiomyocytes lacking HSPB7. However, the lack of overt pathology suggests that in human cardiomyocytes, as in zebrafish, the cells can functionally tolerate detrimental effects caused by the absence of HSPB7, perhaps by upregulating pathways that remove aggregated proteins.
Fig. 9. Alteration in FLNC aggregation in HSPB7 mutant CMs.
(A) Filter trap assay probing for Filamin-C in lysates from day 10 hESC-derived cardiomyocytes treated with bafilomycin A shows increase in FLNC-containing aggregates in HPSB7−/− cardiomyocytes compared to wildtype. Shown is a representative image from multiple reproducible experiments. (B) Quantification of filter trap assays. Changes are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method (p<0.0001). (C) Immunofluorescence for cardiac troponin (red) and FLNC (green) in hESC-CMs derived from HSPB7 wildtype or (D) HSPB7 mutant lines following treatment with bafilomycin A. White arrowheads mark representative abnormal puncta.
Autophagy is enhanced in HSPB7 mutant cardiomyocytes
Some members of the sHSP family are known to be involved in protein degradation pathways, including specific types of chaperone-assisted autophagy (Arndt et al., 2010). HSPB7 has been linked to an autophagic process, because the ability of HSPB7 to prevent aggregation of poly-glutamate proteins was attenuated in the absence of the key autophagic protein ATG5 (Vos et al., 2010). Damaged FLNC has recently been shown to be degraded through the process of Chaperone Assisted Selective Autophagy (CASA), in which a CASA protein complex facilitates transport of the damaged protein to the autophagosome (Arndt et al., 2010). Given these links, we hypothesized that to compensate for loss of HSPB7, mutant cardiomyocytes might enhance autophagic processes to facilitate removal of damaged proteins. This was tested by measuring autophagy levels through visualization of the autophagy marker LC3B. We stained differentiated wildtype and HSPB7 mutant hESC-derived cardiomyocytes for LC3B (Fig. 10A, B) in the presence of the inhibitor of autophagy, BafA, and quantified the number and size of LC3B positive puncta as described (Dagda et al., 2008). We observed an increase in the number and size of puncta in HSPB7 mutant cardiomyocytes when compared with control lines (Fig. 10C), consistent with enhanced autophagy in the absence of HSPB7. To better understand alterations to autophagic flux in HSPB7 null hESC-CMs, we evaluated changes to LC3B-II in the presence or absence of BafA, which prevents the fusion of autophagosomes with lysosomes, inhibiting the last step of autophagy. In the absence of inhibitor, levels of LC3B-II in HSPB7 null cells were comparable to levels in wildtype lines (Fig. 10D, E). In addition, BafA increased the LC3B-II protein level in both wildtype and HSPB7 mutant hESC-CMs (Fig. 10D, E), indicating the presence of autophagic flux in these lines. However, the increase in LC3B-II protein level in HSPB7 mutant cells was significantly higher in HSPB7 mutant lines than in wildtype controls, indicating an increased flux through the autophagic pathway in cardiomyocytes lacking HSPB7.
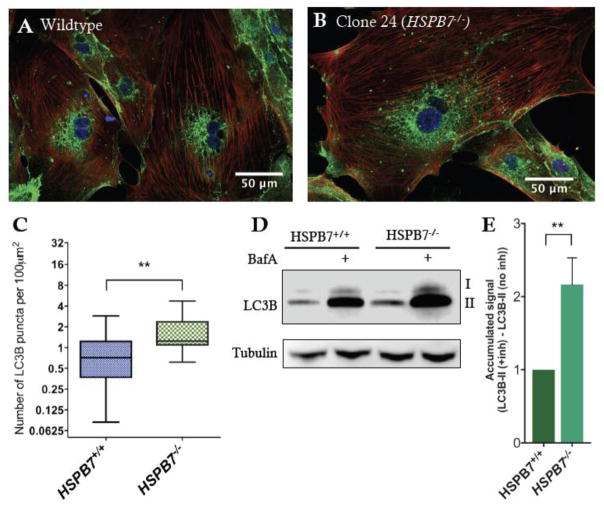
Fig. 10. Upregulation of autophagic processes in HSPB7 null cardiomyocytes.
Shown are representative images of immunofluorescence for LC3B (green) in (A) wildtype or (B) HSPB7 mutant hESC-derived cardiomyocytes. Samples are co-stained with anti-cardiac troponin T (red) and counterstained with DAPI (blue). (C) Quantification of LC3B puncta in hESC-CMs showing an increase in average number of puncta in HSPB7 mutant cardiomyocytes. (D) Western blot for LC3B in the presence or absence of autophagy inhibitor bafilomycin A (BafA). (E) Quantification of LC3BII western blot by densitometry reveals an increase in the accumulation of LC3BII in HSPB7−/− hESC-CMs during bafilomycin A treatment when compared with HSPB7 wildtype controls. For C and E, changes are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method.
Overall LC3B-II levels were significantly increased in mutant cells over wildtype according to western blotting blotting experiments in both HSPB7 mutant hESC-derived cardiomyocytes and adult hspb7WC3/WC3 zebrafish hearts (Fig. 11A–C). qPCR analysis indicated a slight increase in LC3B expression in adult zebrafish hearts which was not seen in hESC-CMs (Fig. 11D), although this may reflect the relative maturity and stress on the two systems. Consistent with a role for CASA in FLNC turnover in HSPB7 null hESC-CMs, we observed upregulation of FLNC mRNA by qPCR (Fig. 11E). We also probed by western blotting experiments whether inhibition of autophagy altered the levels of soluble FLNC, but did not find differences in protein levels (Fig. 11F, G). This observation indicates that the bulk of soluble FLNC may not be processed by autophagy in these cells. However, given the increase in aggregated FLNC in HSPB7 mutant lines, we hypothesize that increased autophagy facilitates processing of insoluble FLNC.
Fig. 11. Increase in LC3B protein in HSPB7 null cardiomyocytes while FLNC protein levels are normal in the absence of HSPB7.
Western blots to quantify LC3B in lysates from hESC-derived cardiomyocytes (A) or adult zebrafish heart (B) in the presence of bafilomycin A (BafA). (C) Quantification by densitometry indicates an increase in LC3B-II levels in cells or hearts that lack HSPB7. (D) qPCR analysis of lc3b expression in adult zebrafish hearts indicates a consistent upregulation that is not seen in HSPB7 in hESC-derived cardiomyocytes. (E) qPCR analysis of flnc expression shows an upregulation in HSPB7−/− hESC-CMs. (F) Western blot for FLNC showing normal levels in HSPB7 null hESC-CMs regardless of autophagy inhibition with BafA. (G) Quantification of western blots with densitometry demonstrates no alteration in levels of soluble FLNC in the absence of HSPB7. For C–E, changes are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method).
Homozygous hspb7 mutant zebrafish embryos exhibit increased sensitivity to inhibition of autophagy
The increase in autophagy measured in HSPB7 mutant human cardiomyocytes indicates a requirement for activation of this pathway to maintain normal cardiac function. This could also be one of the mechanisms by which zebrafish embryos compensate for the loss of Hspb7 during cardiogenesis. In this case, hspb7 mutants are predicted to be hypersensitive to the loss of autophagy. To test this prediction, we exposed wildtype or hspb7 mutant zebrafish embryos to BafA. BafA treatment led to a dose-dependent increase in the incidence of cardiac malformations in both hspb7WC3 and hspb7WC4 homozygous mutants, but not in wildtype controls (Fig. 12A–D). We observed predominantly atrial ballooning with accompanying decrease in ventricular size with a smaller number of embryos exhibiting abnormalities in cardiac looping. Exposure to the proteasome inhibitor MG-132 did not lead to similar phenotypes in hspb7 mutant embryos (Fig. 12E). In a dose-response study, exposure to increasing levels of MG-132 did generate cardiac abnormalities, but mutants were no more sensitive than wildtype embryos. This indicates that it is specifically autophagy, and not general protein degradation processes, that is acting to compensate for loss of Hspb7. We also exposed control or hspb7 mutant embryos to chloroquine diphosphate (CQ), an autophagy inhibitor with a different mechanism of action. Similar to BafA, CQ led to increases in cardiac defects in hspb7 mutant embryos but not in wildtype controls (Fig. 12F). Since both BafA and CQ inhibit primarily the lysosome, we also tested 3-Methyladenine (3-MA) as a more specific blocker of autophagosome formation. Embyros injected with a low dose of morpholino were significantly more sensitive to 3-MA for generation of cardiac abnormalities (Fig. 12G). Overall, these results confirm that normal cardiogenesis in hspb7 mutant embryos is reliant on autophagy.
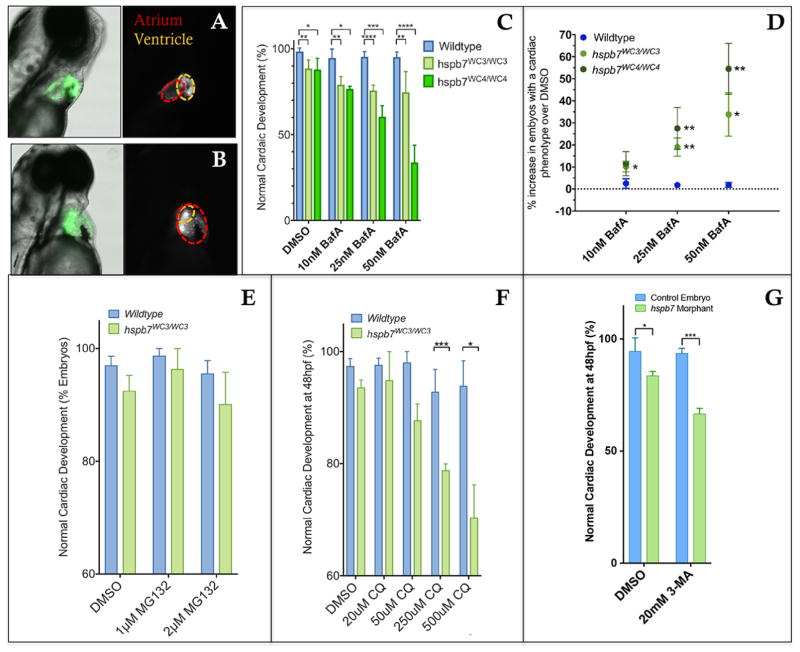
Fig. 12. Inhibition of autophagy leads to cardiac malformation in the absence of Hspb7.
Shown are representative images of 72 hpf, bafilomycin A (BafA)-treated wildtype (A) or hspb7WC3/WC3 mutant (B) embryos with a tg(myl7:gfp) background showing a cardiac malformation with expansion of the atrium and an undersized ventricle in the hspb7WC3/WC3 fish. (C) Quantification of the percentage of embryos with cardiac abnormalities at 48 hpf, following 24 hr treatment with increasing concentrations of Bafilomycin A. n=40 for each treatment group. (D) hspb7 mutant embryos consistently exhibit significant increases in cardiac abnormalities following treatment with Bafilomycin A. (E) Quantification of cardiac abnormalities in embryos treated with proteasome inhibitor MG132 does not lead to increases in abnormalities in cardiac development in hspb7 mutants. (F) Autophagy inhibitor chloroquine diphosphate (CQ) leads to increases in abnormalities in cardiac development in hspb7 mutants at a greater rate than is seen in wildtype embryos. Abnormalities included small ventricles, unlooped hearts and atrial ballooning. (G) Treating larvae with the autophagy inhibitor 3-Methyladenine (3-MA) leads to increases in abnormalities in cardiac development in hspb7 morphants at a rate greater than is seen in wildtype control embryos. Abnormalities included small ventricles, unlooped hearts and atrial ballooning. For C, D, F, and G, changes in cardiac abnormalities are significant according to an unpaired t-test, corrected for multiple comparisons with the Holm-Sidak method.
Discussion
We found that global loss of hspb7 is tolerated during zebrafish embryogenesis but leads to a mild cardiac pathology in adults, including focal fibrotic loci, and sensitizes animals to stress. High-intensity exercise, which causes an acute increase in cardiac workload, leads to exercise-induced mortality in around 40% of adults. Strikingly, we observe a significant cardiomegaly in hspb7 mutant hearts, indicating a pathology of similar magnitude to that seen in a zebrafish model of doxorubicin-induced cardiomyopathy (Ding et al., 2011). In combination with the observation of hepatic vacuolation, this is suggestive of a heart failure phenotype. The hspb7 zebrafish mutant therefore provides a new model for the complex pathogenesis seen in human heart disease, in which environmental stresses and individual variation impact disease severity and outcome.
We discovered that FLNC is a binding partner of HSPB7 and that FLNC aggregates form in human ESC-derived cardiomyocytes in the absence of HSPB7. FLNC is a large sarcomeric protein that interacts with both Z-disc components and sarcolemma-associated proteins, leading to speculation that it may act as both an assembly point for myofibrils in early myogenesis (Dalkilic et al., 2006; van der Ven et al., 2000) and as a mediator of signal transduction from the sarcolemma to the sarcomere (Furst et al., 2013; Leber et al., 2016), facilitating myofibril maintenance. Loss of FLNC is associated with defects in myogenesis (Dalkilic et al., 2006), and human mutations are associated with a range of skeletal and cardiac myopathies (Brodehl et al., 2016; Furst et al., 2013; Valdes-Mas et al., 2014; Vorgerd et al., 2005), many of which are associated with the formation of pathological aggregates containing FLNC. We observed aggregates in cardiomyocytes lacking HSPB7, indicating that loss of HSPB7 may affect the stability or processing of FLNC. Our data indicate that HSPB7 also binds to at least one other large sarcomeric protein, TITIN. Further studies are needed to determine if HSPB7 is a dedicated chaperone for these two, or if this role is generalized to many large sarcomeric proteins. Our results are consistent with a recent report that HSPB7 can bind FLNC in murine skeletal muscle (Juo et al., 2016). Knockout of HSPB7 in skeletal muscle caused lethal progressive myopathy, found to be most severe in the diaphragm (Juo et al., 2016). We did not observe hallmarks of progressive myopathy in the skeletal muscle of hspb7 mutant zebrafish, but it is interesting to note that the diaphragm is the one muscle that, like the heart, must move continuously to maintain life. Zebrafish lack a diaphragm, while Hspb7 is perhaps better compensated in the skeletal muscle of zebrafish compared with mouse. Therefore, the fish model is particularly useful to explore the specific role of HSPB7 in cardiac biology.
We hypothesized that HSPB7 may bind to proteins that need to be processed by autophagy, since this function has been demonstrated for family member HSPB8 (Arndt et al., 2010), a distinct small heat shock protein that is expressed more generally in all striated muscle. We found that autophagic processes are enhanced in HSPB7-depleted cardiomyocytes and that impairing autophagy during embryogenesis causes lethal cardiomyopathy in developing hspb7 mutant embryos. Recent studies have suggested that damaged FLNC is among the proteins degraded by autophagy, a catabolic cellular process for the breakdown and recycling of extraneous or defective proteins and complexes (Arndt et al., 2010). The role of autophagy in the heart is complex. Key autophagic proteins are required for normal cardiac development (Lee et al., 2014), and autophagic turnover is known to occur in the healthy heart (Levine and Klionsky, 2004). Increases in autophagy are seen in situations of cardiac stress, presumably in response to an increase in damaged and dysfunctional proteins downstream of mechanical or oxidative stressors. However, either too little or too much autophagy is pathological under conditions of cardiac stress (Maejima et al., 2015; Nishida et al., 2009; Portbury et al., 2011), indicating that the process must be carefully balanced. A model consistent with our data is that the absence of HSPB7 leads to an increase in damage or unfolding of FLNC, resulting in compensatory activation of autophagic pathways to prevent aggregate formation. Since fibrosis was mild, and cell death not evident, we speculate that abnormalities in the adult heart may be impacted by accelerated autophagy or protein aggregation. This model is in agreement with recent findings that HSPB7 decreases aggregate load through preventing aggregate formation rather than facilitating aggregate removal (Eenjes et al., 2016). Intriguingly, we documented an upregulation of hspb5b in the hearts of hspb7WC3/WC embryos, and observed a similar upregulation of HSPB5 in HSPB7−/− hESC-derived human cardiomyocytes. The Drosophila homolog of CryAB has been shown to act as a chaperone for Cheerio, the Drosophila filamin protein (Wojtowicz et al., 2015). Upregulation of HSPB5 may be at least partially compensating for loss of HSPB7 in our mutant models. A recent study found that knockout of Hspb7 in the mouse is embryonic lethal, associated with elongated actin thin filaments and mislocalization of Tmod1 (Wu et al., 2017). It will be interesting to investigate if this phenotype is also compensated by Hspb5 in the fish mutants.
A recent study also described a potential role for FLNC in the fast repair of myofibrillar microdamage (Leber et al., 2016), indicating a potential role as a signaling hub for the efficient reassembly of sarcomeric structures. Given this finding, a decrease in the availability of functional FLNC caused by mutation of hspb7 could have effects on the ability of the myocyte to repair damage at multiple steps. This multifaceted importance could explain the increase in fibrosis observed in hspb7 mutant zebrafish hearts. We expect that HSPB7 depletion amplifies the effects of high-intensity exercise to a pathological level, leaving cardiomyocytes with a burden of unstable sarcomeric lesions that results in death in a subset of mutant animals.
Our study provides novel insight into how HSPB7 regulates proteostatic mechanisms that function in a cardiomyocyte, and a new model for human stress-dependent cardiomyopathy. Future studies need to investigate the relationship between HSPB7 and the delivery of FLNC to autophagic organelles. Given the complex role of autophagy in the sarcomere, direct therapeutic manipulation is likely to be inherently risky. However, by manipulating specific components like HSPB7 that have a regulatory role, redirection or optimization of autophagic processes might be achieved that will be clinically beneficial for patients with disorders of aggregation.
Highlights.
Targeted mutations in the hspb7 gene were generated in zebrafish and human ESCs
Zebrafish embryos tolerate loss of hspb7 but adult mutants die from exercise stress
FilaminC and Titin are HSPB7 binding partners in cardiac cells
Loss of Hspb7 stimulates autophagic pathways and expression of Hspb5
Inhibiting autophagy causes FilaminC aggregation in mutant human cardiac cells
Inhibiting autophagy causes developmental cardiomyopathy in hspb7 mutant embryos
Acknowledgments
The authors are grateful for expert pathology analysis provided by the Pathology Core at Memorial Sloan Kettering, the Microscopy and Image Analysis Core at Weill Cornell Medicine, and independently by Dr. Domenick Falcone at WCM. Dr. Danwei Huangfu (MSKCC) generously provided the parental iCRISPR hESC line, and Dr. Richard White and his laboratory (MSKCC) provided access and direction in using the swim tunnel. Dr. W. Claycomb generously provided the HL-1 cardiac cell line. Thanks also to Dr. Steven Gross (WCM) for access to mass spectrometry equipment. This work was supported by grants to TE from the NIH (HL111400, HL135778) and the New York State Department of Health (NYSTEM C029156).
Footnotes
Author Contributions
EJM and TE conceived the study and wrote the manuscript. EJM and YFL performed experiments. All authors analyzed data and all authors approved the final manuscript.
Competing Interests
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaout R, Reischauer S, Stainier DY. Recovery of adult zebrafish hearts for high-throughput applications. J Vis Exp. 2014 doi: 10.3791/52248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P, Saint R, Fleischmann BK, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Boncoraglio A, Minoia M, Carra S. The family of mammalian small heat shock proteins (HSPBs): implications in protein deposit diseases and motor neuropathies. Int J Biochem Cell Biol. 2012;44:1657–1669. doi: 10.1016/j.biocel.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Brodehl A, Ferrier RA, Hamilton SJ, Greenway SC, Brundler MA, Yu W, Gibson WT, McKinnon ML, McGillivray B, Alvarez N, et al. Mutations in FLNC are Associated with Familial Restrictive Cardiomyopathy. Hum Mutat. 2016;37:269–279. doi: 10.1002/humu.22942. [DOI] [PubMed] [Google Scholar]
- Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, et al. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–6534. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn WC. Clycogen, its chemistry and morphologic appearance in the electron microscope. I. A modified Os04 fixative which selectively contrasts glycogen. J Ultrastrutural Res. 1973;42:63–73. doi: 10.1016/s0022-5320(73)80004-8. [DOI] [PubMed] [Google Scholar]
- Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, Sivasubbu S, Lin X, Ekker S, Xu X. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109:658–669. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eenjes E, Dragich JM, Kampinga HH, Yamamoto A. Distinguishing aggregate formation and aggregate clearance using cell-based assays. J Cell Sci. 2016;129:1260–1270. doi: 10.1242/jcs.179978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elicker KS, Hutson LD. Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene. 2007;403:60–69. doi: 10.1016/j.gene.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Mitsuhashi H, Isogai S, Nakata T, Kawakami A, Nonaka I, Noguchi S, Hayashi YK, Nishino I, Kudo A. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol. 2012;361:79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Furst DO, Goldfarb LG, Kley RA, Vorgerd M, Olive M, van der Ven PF. Filamin C-related myopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:33–46. doi: 10.1007/s00401-012-1054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giallourakis CC, Rosenberg PM, Friedman LS. The liver in heart failure. Clin Liver Dis. 2002;6:947–967. viii–ix. doi: 10.1016/s1089-3261(02)00056-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzinger A, Rosenfeld GE, Evans T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev Biol. 2010;337:63–73. doi: 10.1016/j.ydbio.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo LY, Liao WC, Shih YL, Yang BY, Liu AB, Yan YT. HSPB7 interacts with dimerized FLNC and its absence results in progressive myopathy in skeletal muscles. J Cell Sci. 2016;129:1661–1670. doi: 10.1242/jcs.179887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1–10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke L, Meijering RA, Hoogstra-Berends F, Mackovicova K, Vos MJ, Van Gelder IC, Henning RH, Kampinga HH, Brundel BJ. HSPB1, HSPB6, HSPB7 and HSPB8 protect against RhoA GTPase-induced remodeling in tachypaced atrial myocytes. PLoS One. 2011;6:e20395. doi: 10.1371/journal.pone.0020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitta K, Day RM, Kim Y, Torregroza I, Evans T, Suzuki YJ. Hepatocyte growth factor induces GATA-4 phosphorylation and cell survival in cardiac muscle cells. J Biol Chem. 2003;278:4705–4712. doi: 10.1074/jbc.M211616200. [DOI] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Faivre JF, Robert P, Le Douarin B, Brument-Larignon N, Lefrere I, Bouzyk MM, Anderson KM, Greller LD, Tobin FL, et al. Identification and characterization of cvHsp. A novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J Biol Chem. 1999;274:36592–36600. doi: 10.1074/jbc.274.51.36592. [DOI] [PubMed] [Google Scholar]
- Lahvic JL, Ji Y, Marin P, Zuflacht JP, Springel MW, Wosen JE, Davis L, Hutson LD, Amack JD, Marvin MJ. Small heat shock proteins are necessary for heart migration and laterality determination in zebrafish. Dev Biol. 2013;384:166–180. doi: 10.1016/j.ydbio.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber Y, Ruparelia AA, Kirfel G, van der Ven PF, Hoffmann B, Merkel R, Bryson-Richardson RJ, Furst DO. Filamin C is a highly dynamic protein associated with fast repair of myofibrillar microdamage. Hum Mol Genet. 2016;25:2776–2788. doi: 10.1093/hmg/ddw135. [DOI] [PubMed] [Google Scholar]
- Lee E, Koo Y, Ng A, Wei Y, Luby-Phelps K, Juraszek A, Xavier RJ, Cleaver O, Levine B, Amatruda JF. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 2014;10:572–587. doi: 10.4161/auto.27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- Louie CY, Pham MX, Daugherty TJ, Kambham N, Higgins JP. The liver in heart failure: a biopsy and explant series of the histopathologic and laboratory findings with a particular focus on pre-cardiac transplant evaluation. Mod Pathol. 2015;28:932–943. doi: 10.1038/modpathol.2015.40. [DOI] [PubMed] [Google Scholar]
- Maejima Y, Chen Y, Isobe M, Gustafsson AB, Kitsis RN, Sadoshima J. Recent progress in research on molecular mechanisms of autophagy in the heart. Am J Physiol Heart Circ Physiol. 2015;308:H259–268. doi: 10.1152/ajpheart.00711.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM. Small heat shock protein expression and functions during development. Int J Biochem Cell Biol. 2012;44:1613–1621. doi: 10.1016/j.biocel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Nishida K, Kyoi S, Yamaguchi O, Sadoshima J, Otsu K. The role of autophagy in the heart. Cell Death Differ. 2009;16:31–38. doi: 10.1038/cdd.2008.163. [DOI] [PubMed] [Google Scholar]
- Palstra AP, Tudorache C, Rovira M, Brittijn SA, Burgerhout E, van den Thillart GE, Spaink HP, Planas JV. Establishing zebrafish as a novel exercise model: swimming economy, swimming-enhanced growth and muscle growth marker gene expression. PLoS One. 2010;5:e14483. doi: 10.1371/journal.pone.0014483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portbury AL, Willis MS, Patterson C. Tearin’ up my heart: proteolysis in the cardiac sarcomere. J Biol Chem. 2011;286:9929–9934. doi: 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld GE, Mercer EJ, Mason CE, Evans T. Small heat shock proteins Hspb7 and Hspb12 regulate early steps of cardiac morphogenesis. Dev Biol. 2013;381:389–400. doi: 10.1016/j.ydbio.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Stark K, Esslinger UB, Reinhard W, Petrov G, Winkler T, Komajda M, Isnard R, Charron P, Villard E, Cambien F, et al. Genetic association study identifies HSPB7 as a risk gene for idiopathic dilated cardiomyopathy. PLoS Genet. 2010;6:e1001167. doi: 10.1371/journal.pgen.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Mas R, Gutierrez-Fernandez A, Gomez J, Coto E, Astudillo A, Puente DA, Reguero JR, Alvarez V, Moris C, Leon D, et al. Mutations in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat Commun. 2014;5:5326. doi: 10.1038/ncomms6326. [DOI] [PubMed] [Google Scholar]
- van der Ven PF, Obermann WM, Lemke B, Gautel M, Weber K, Furst DO. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PF, Bruchertseifer V, Lowe T, Kley RA, Schroder R, Lochmuller H, Himmel M, Koehler K, Furst DO, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MJ, Zijlstra MP, Kanon B, van Waarde-Verhagen MA, Brunt ER, Oosterveld-Hut HM, Carra S, Sibon OC, Kampinga HH. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet. 2010;19:4677–4693. doi: 10.1093/hmg/ddq398. [DOI] [PubMed] [Google Scholar]
- Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book; a laboratory guide for the use of zebrafish (Brachydanio rerio) University of Oregon; 1993. [Google Scholar]
- Wojtowicz I, Jablonska J, Zmojdzian M, Taghli-Lamallem O, Renaud Y, Junion G, Daczewska M, Huelsmann S, Jagla K, Jagla T. Drosophila small heat shock protein CryAB ensures structural integrity of developing muscles, and proper muscle and heart performance. Development. 2015;142:994–1005. doi: 10.1242/dev.115352. [DOI] [PubMed] [Google Scholar]
- Wu T, Mu Y, Bogomolovas J, Fang X, Veevers J, Nowak RB, Pappas CT, Gregorio CC, Evans SM, Fowler VM, et al. HSPB7 is indispensable for heart development by modulating actin filament assembly. Proc Natl Acad Sci U S A. 2017;114:11956–11961. doi: 10.1073/pnas.1713763114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gonzalez F, Huangfu D. The iCRISPR platform for rapid genome editing in human pluripotent stem cells. Methods Enzymol. 2014;546:215–250. doi: 10.1016/B978-0-12-801185-0.00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubeidi A, Gleave M. Small heat shock proteins in cancer therapy and prognosis. Int J Biochem Cell Biol. 2012;44:1646–1656. doi: 10.1016/j.biocel.2012.04.010. [DOI] [PubMed] [Google Scholar]